-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
Cationic antimicrobial peptides (cAMPs) are small proteins naturally produced by the immune system to limit bacterial growth mainly through pore formation in the membrane. It has recently been suggested that sub-inhibitory concentrations of cAMPs promote bacterial mutagenesis, similarly to antibiotics. However, we previously reported that cAMPs do not increase mutation rate and do not activate bacterial stress responses. Here we resolve this contradiction. We report that free iron in the culture medium increases mutagenesis in the presence of cAMPs. We show that sub-inhibitory concentrations of cAMPs facilitate entry of free iron into bacterial cells, where it interacts with hydrogen peroxide, thereby resulting in production of DNA-damaging reactive oxygen species and increased mutagenesis. Moreover, these results may have clinically-relevant implications: while very little free iron is normally present in healthy individuals, this is not the case in patients suffering from cystic fibrosis, where elevated bacterial mutagenesis promotes antibiotic resistance and contributes to persistence and severity of infection. Thus, an intervention aimed at reduction of free iron in the lungs could reduce the cAMPs-facilitation of iron-mediated mutagenesis; hence antibiotic resistance and pathoadaptation.
Published in the journal: . PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005546
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005546Summary
Cationic antimicrobial peptides (cAMPs) are small proteins naturally produced by the immune system to limit bacterial growth mainly through pore formation in the membrane. It has recently been suggested that sub-inhibitory concentrations of cAMPs promote bacterial mutagenesis, similarly to antibiotics. However, we previously reported that cAMPs do not increase mutation rate and do not activate bacterial stress responses. Here we resolve this contradiction. We report that free iron in the culture medium increases mutagenesis in the presence of cAMPs. We show that sub-inhibitory concentrations of cAMPs facilitate entry of free iron into bacterial cells, where it interacts with hydrogen peroxide, thereby resulting in production of DNA-damaging reactive oxygen species and increased mutagenesis. Moreover, these results may have clinically-relevant implications: while very little free iron is normally present in healthy individuals, this is not the case in patients suffering from cystic fibrosis, where elevated bacterial mutagenesis promotes antibiotic resistance and contributes to persistence and severity of infection. Thus, an intervention aimed at reduction of free iron in the lungs could reduce the cAMPs-facilitation of iron-mediated mutagenesis; hence antibiotic resistance and pathoadaptation.
Introduction
Pseudomonas aeruginosa is an important opportunistic pathogen involved in chronic respiratory and hospital-acquired infections [1]. In cystic fibrosis (CF) patients, one of the most common genetic diseases in humans, this bacterium causes chronic lung infections that result in significant morbidity and mortality [2]. P. aeruginosa infections are difficult to treat due the inherent resistance to many drug classes, its ability to acquire resistance, via mutations, to all relevant treatments and its high and increasing rates of resistance locally [3].
Mutagenesis plays a crucial role in adaptation of this pathogen for persistence and antibiotic resistance acquisition in CF [4], being found a high proportion of hypermutable bacteria among P. aeruginosa isolates [5]. Recently, Limoli et al [6] reported an increased mutant frequency after treatment of P. aeruginosa with the human cationic peptide LL-37. Based on this finding they proposed that cationic peptides elevated bacterial mutation rates. In a recent study, we reported mutation rates of E. coli in the presence of cationic antimicrobial peptides (cAMPs) and antibiotics. LL-37 was present in the panel of AMPs that we tested and we did not find any increase in mutation rates. We also used transcription reporter assays and qRT-PCR and showed that none of the AMPs elicited the main mutagenic stress pathways of bacteria SOS or rpoS [7].
Here, we aim to resolve this apparent contradiction based on the observation that the different studies used different media. Limoli et al. [6] used M63 for P. aeruginosa and LB for E. coli, while we used non-cation adjusted Mueller-Hinton Broth (MHB), commonly used for cAMP susceptibility testing. The most striking difference in culture media between the two studies is that both, M63 [8] and LB are iron-rich [9,10], while MHB is not [11]. Fe2+ catalyses hydroxyl radical formation by reacting with hydrogen peroxide both intra - and extra-cellularly, the Fenton reaction [12]. Without free iron hydrogen peroxide reactivity is low at physiological pH and iron metabolism is strictly controlled to avoid DNA and other damage caused by oxygen radicals [13,14]. In most natural situations iron is in short supply, but in cystic fibrosis [15,16] due to accult bleeding of highly vascularised lung tissue and haemoptysis particularly during acute exacerbations, and damage of the respiratory tract epithelium where Fe2+ is present intermittently. Although ferrous iron is prone to oxidation, the anaerobic growth of at to high density of bacteria [17] may contribute to stabilise this metal in the reduced state.
Many cationic antimicrobial peptides change membrane permeability properties [18], and this led us to hypothesize that sub-inhibitory concentrations of LL-37 increase uncontrolled iron transport from the extracellular space to the cytoplasm without the intervention of the bacterial iron trafficking system. An intracellular surplus of iron should then result in DNA damage caused by the Fenton reaction.
Here, we (i) estimate the mutation rate of P. aeruginosa in the presence of LL-37, iron or both; (ii) we then investigate the hypothesis that ferrous iron (Fe+2) is causal to an increase in mutation rate and that LL-37 and colistin facilitate this process; and finally (iii) we investigate the mutational spectra to find out if promoted mutations match with any of the molecular signatures of Fenton reactions.
Results and Discussion
Mutation rate is increased in the presence of ferrous Fe2+ and LL-37
First we determined the mutation rate of P. aeruginosa using a fluctuation assay, in the presence of either LL-37 (32 μg/ml), Fe2+ (40μM), or both (Fig 1). Mutation rate was only increased when LL-37 and Fe2+ were added simultaneously, while addition of LL-37 or Fe2+ separately did not produce the same effect. To investigate the effect of different concentrations of LL-37 and Fe2+, we estimated mutant frequencies. None of the LL-37 concentrations tested, ranging from 4 to 32 μg/ml, yielded any detectable changes in mutant frequency (S1 Fig). Fe2+ alone, at different concentrations, also did not increase the mutant frequencies in comparison with the control group (S2 Fig).
Fig. 1. The joint action of LL-37 and ferrous iron induces an increase in the mutation rate of P. aeruginosa. 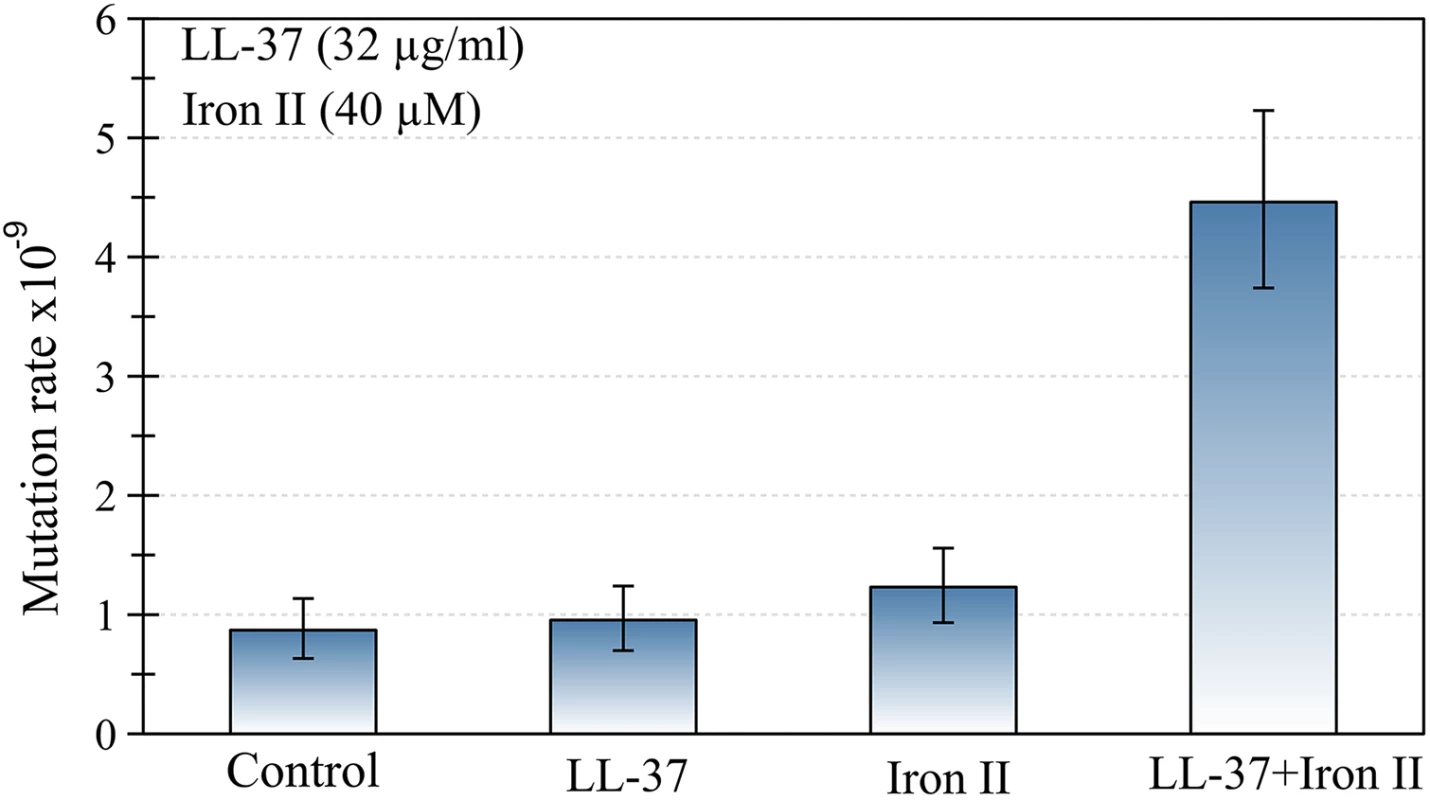
No changes occur when LL-37 or iron are added to the culture separately. Error bars represent 95% confidence intervals for mutation rates. Subsequently, we assayed the MIC50 (32 μg/ml) of LL-37 with several iron concentrations and measured the impact on rifampicin mutant frequency of P. aeruginosa strain PAO1. We found that added concentrations of 10, 20, 40 and 80 μM of Fe2+, increased the mutant frequency between three to five times (S3 Fig).
Limoli et al. [6] observed increased mutant frequencies in both, P. aeruginosa and E. coli, proposing that LL-37 induces mutagenesis in these bacteria. Taken at face value, this contrasts with our work, where we did not find such effect in E. coli [7]. P. aeruginosa in the Limoli et al. study however, was cultured in an iron-rich medium, M63 [8], which contains 0.5 mg of iron sulphate per litre (Fe2+). To confirm the results in a different bacterial model, the experiment was repeated with E. coli using LB as a culture medium. However, LB is also iron-rich (~16 μM of iron) [9,10]. In these experiments, just before the treatment with LL-37 in a saline solution, bacteria were washed. It has, however, been shown that Gram-negative bacteria can actively accumulate iron in the periplasmic space [19]. This led us to hypothesise that sub-lethal concentrations of cationic antimicrobial peptides can facilitate iron transport into the cells.
LL-37 facilitates mutagenesis but iron is causal
To confirm that iron is causing the observed increase in mutagenesis, we first repeated the assay in the presence of an iron chelator, 2–2′ bipyridyl. 2–2′ bipyridyl completely abolished the increase in mutant frequency in the tested concentration of LL-37+Fe2+ combination. The sole action of the chelator plus Fe2+ did not increase the mutant frequency (Fig 2).
Fig. 2. The mutagenesis of LL-37+Fe2+ combination is suppressed by the addition of an iron chelator. 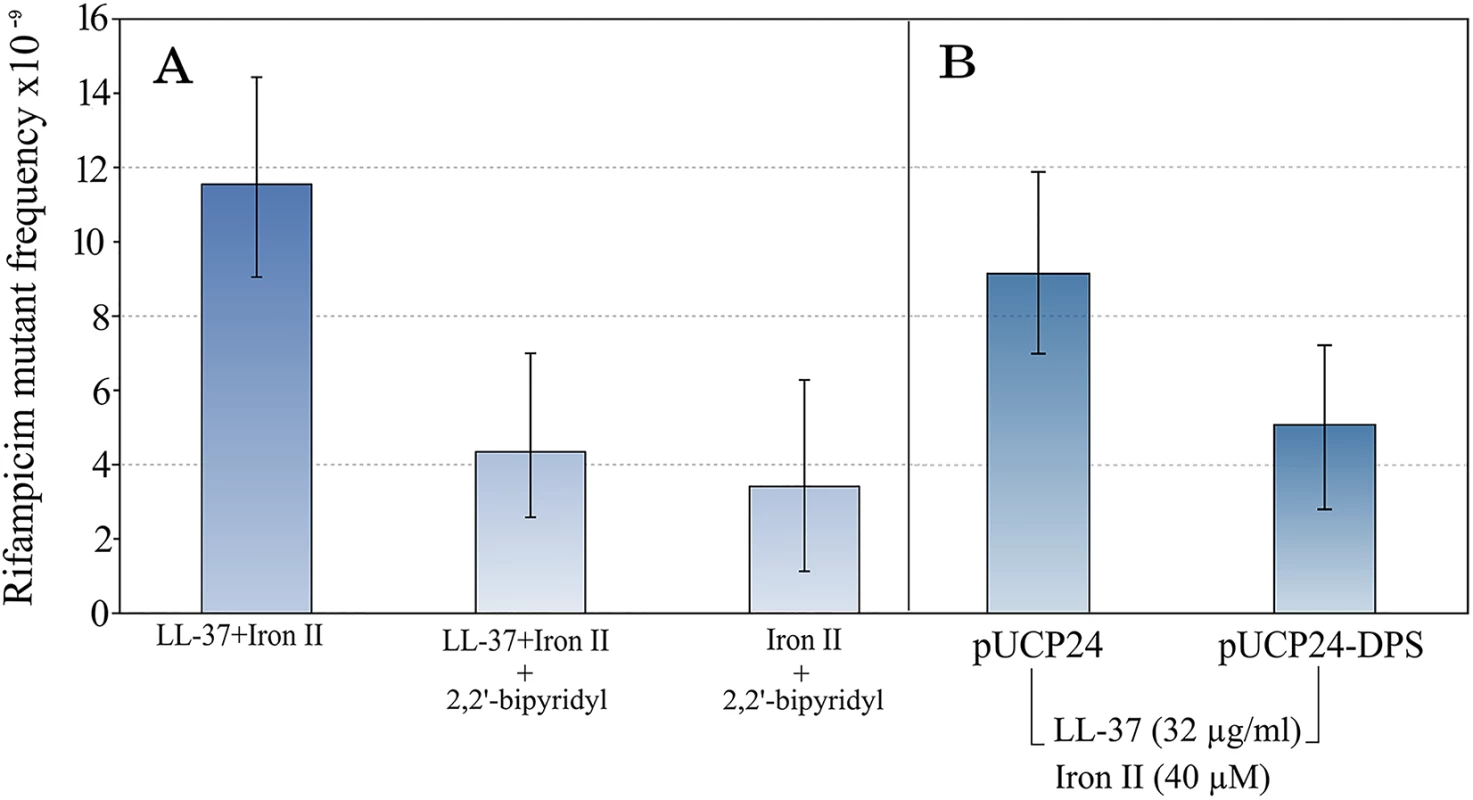
This supports the notion that iron is causal in increasing mutagenesis (A). In the same line, over-expression of Dps, a natural iron chelator in bacteria, also decreases the mutant frequency to rifampicin (B). Error bars represent 95% confidence intervals for mutant frequencies. Bacteria have a number of mechanisms to keep free iron as low as possible inside the cell. One of these is Dps, a natural iron chelating protein. To confirm the results obtained with 2–2′ bipyridyl, we used a Dps over-expressing strain by cloning the dps gene into a multi-copy plasmid under a constitutive promoter. The mutagenesis was not completely reverted in comparison with non-treated controls, but there was a 1.8 fold-reduction (Fig 2).
The iron uptake assay showed that there was a significant difference (Mann-Whitney U test, P = 0.00015) in total iron content in bacterial cells between LL-37+ Fe2+ (17.50 ± 5.10 nmoles/ml, mean ± standard deviation) and iron alone (5.65 ± 4.13 nmoles/ml, mean ± standard deviation) treatments after 30 minutes of incubation, indicating that LL-37 indeed promotes a non-physiological free iron entrance.
We also tested a further AMP: colistin, which is of bacterial origin, and is one of the most effective agents against P. aeruginosa in CF infections [20]. We treated cultures of P. aeruginosa PAO1 with MIC50 of colistin (1.8 μg/ml) in high and low concentrations of iron in the media. The effects of colistin were very similar to the effects we found for LL-37 (Fig 3). Although the mechanism of action of colistin is not fully understood [21], our results suggest pore forming mechanism or permeability changes in the cell as likely mechanisms. It has been demonstrated that colistin promotes the uptake of hydrophilic antibiotics, explaining their synergism with them [22].
Fig. 3. Mutagenesis induced by colistin and Fe2+ combination. 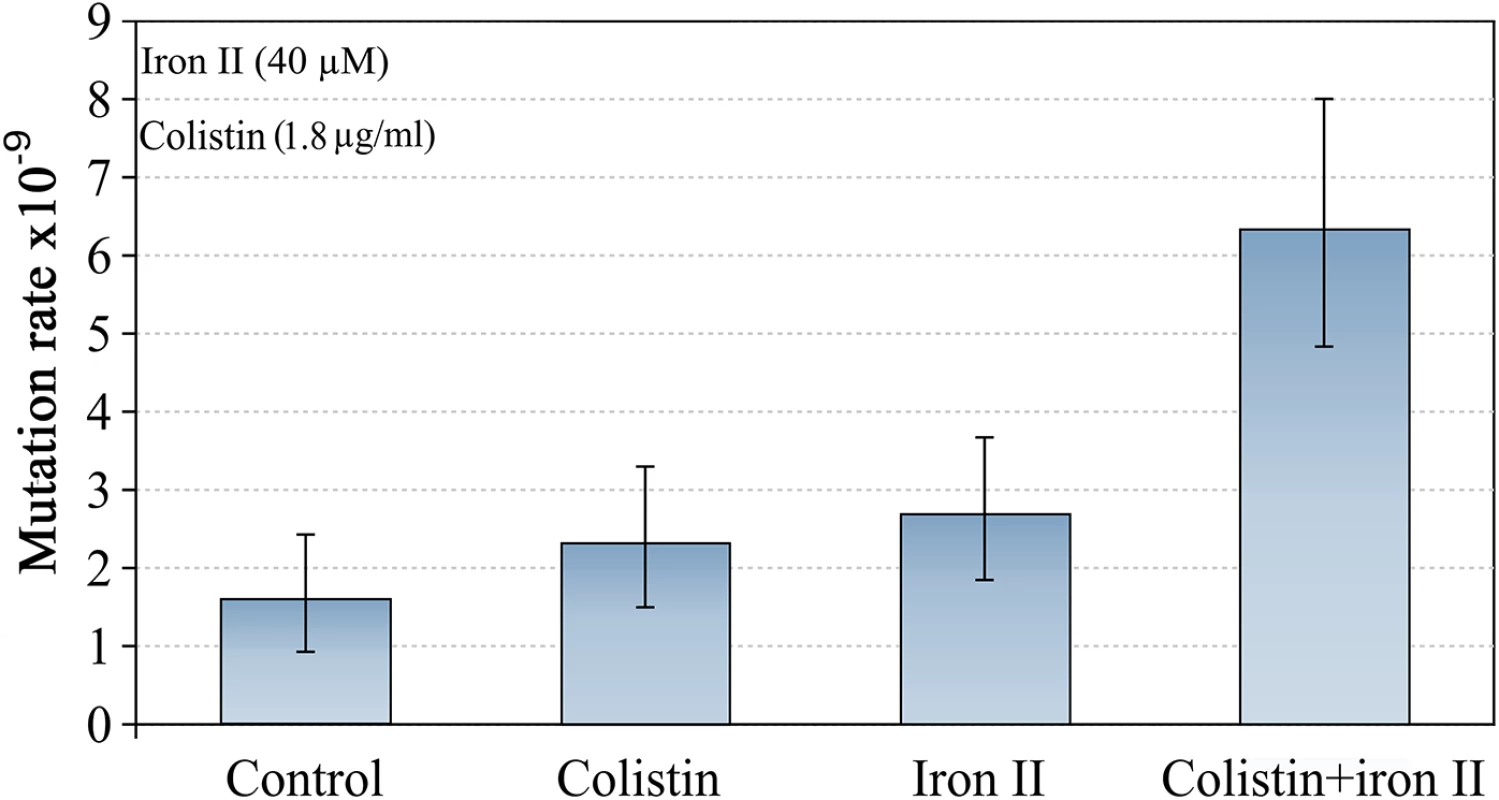
The mutation rate is only increased if both, iron and colistin, are present. Error bars represent 95% confidence intervals for mutation rates. Previous work has suggested that iron is involved in Pseudomonas aeruginosa pathoadaptation and antibiotic resistance acquisition [15]. All experiments were carried out with the more soluble Fe2+ because ferrous iron is abundant in the CF lung (~39 μM on average for severely ill patients) and significantly correlates with disease severity [23]. Moreover, iron activates the two-component signal transduction system BqsRS in P. aeruginosa, which is transcriptionally active in CF sputum, and promotes tolerance to cationic stressors [24]. This increases the tolerance to both host peptides such as LL-37 and colistin, which is administered in CF therapy.
The new mechanism proposed here has almost certainly important implications. Under specific pathologies such as cystic fibrosis or other respiratory chronic diseases, iron that is usually in short supply, is abundant. Disturbances in iron metabolism have been shown to promote evolution of antibiotic resistance in E. coli [25]. Given that cationic antimicrobial peptides usually induce membrane permeability in all susceptible microbes and Fenton chemistry is universal, the mechanism we propose here is likely applicable to other AMPs. For example, beta-defensin-2 was shown to double the mucoid conversion rate, a mutagenic process, of P. aeruginosa [6]. Yet free iron is rarely available in most physiological situations but for a few pathologies such as cystic fibrosis
Additionally, it has been reported that some cationic antimicrobial peptides interact with bacterial membrane proteins and delocalise them [26]. It is conceivable then that LL-37 may interact and interfere with iron transport systems, which in turn may contribute to iron homeostasis disruption and enhance mutagenesis. This possibility requires additional investigation and will be the goal for future studies.
In the context of cystic fibrosis, the increase in salt concentration may lead to the reduced activity or complete inactivation of other antimicrobial peptides, as observed for human beta-defensin-1 [27], while other components could eventually contribute to mutagenesis in the same way that LL-37 does. Moreover, the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems of P. aeruginosa may play an important role in antimicrobial peptide tolerance. This resistance is reproduced in vitro when magnesium concentrations are low [28]. Our experimental conditions, where divalent cations are depleted or in low concentrations, seem comparable. In the light of our results this suggest that bacteria under certain conditions that elicit the expression of these two component systems alter the lipidA structure resulting in increased resistance to colistin [29]. The same systems up-regulate ferrous iron uptake which is mediated by feoAB operon [28,30]. This phenomenon could potentially contribute to saturate intracellular iron storage systems and to generate an excess of iron that eventually can participate in Fenton reaction operating by the mechanisms that we propose here.
How would the mechanism we propose here enhance the overall mutation rate of bacterial populations? In the context of cystic fibrosis, there is a high proportion of hypermutable bacteria due to the inactivation of their DNA miss-match repair (MMR) genes [31]. We may expect a synergistic effect of both types of mutagenesis as we proposed in the past for the mutagenic effect of cystic fibrosis lung environment and the intrinsic mutagenesis of P. aeruginosa [4]. It can be speculated that iron-mutagenesis can facilitate the rise of mutator bacteria by enhancing the inactivation of MMR genes. This could then weaken genetic constraints that impede the evolution of bacteria to resist antibiotics by multiple pathways as previously described [32].
Mutational spectra of iron-induced mutagenesis
The results above strongly suggest that sub-lethal concentrations of cationic antimicrobial peptides facilitate the access of free iron to the cytoplasmic compartment of P. aeruginosa. Given that the Fenton reaction results in DNA damage [33] and reactive oxygen species (ROS) damages to the DNA display specific molecular fingerprints, we investigated if there was evidence of the Fenton reaction effects on the mutational spectrum.
It would be very difficult to investigate the mutational spectrum in rpoB (sub-unit of RNA polymerase) that confers resistance to rifampicin in P. aeruginosa PAO1. This gene is essential and detectable mutations are mostly point mutations, which would constrain the analysis to a few types of changes. We therefore decided to use another strain, PA14, where mutations that confer resistance to fosfomycin are well characterized [34–36]. Resistance mutations to fosfomycin in P. aeruginosa PA14 depend on a single non-essential locus encoding the glycerol-3-phosphate transporter GlpT, making it a much better marker for studying the entire repertoire of mutations compared to rpoB [36,37] [38].
The treatment of P. aeruginosa P14, which shows a similar sensitivity to LL-37 as the PAO1 strain used above, with the mutagenic combination of LL-37+Fe2+ showed almost a four fold-increase in mutant frequency (S4 Fig).
To assess how addition of free iron and antimicrobial peptides affect the mutation spectrum of the glpT gene of PA14 strain we exposed bacteria to either free iron, antimicrobial peptide LL-37, or both. All of the resulting resistant mutants contained non-synonymous substitutions or deletions in glpT that potentially affect the stability of GlpT transporter and likely disrupt the function of the protein (Figs 4, S5 and S6 and S1 Table). LL-37+Fe2+-treated bacteria displayed striking differences in mutational spectra when compared to the other treatments (Fig 4 and S1 Table). We used Monte Carlo hypergeometric test implemented in iMARS, a mutation analysis reporting software [39], to assess the overall differences between mutational spectra. The probability of the mutational spectra to be the same stood at 0.554706 (P-value confidence limits 0.531080–0.578) when the iron treatment was compared to LL-37, but was below 0.0000001 when either were compared with LL-37+Fe2+ treatment. Moreover, we found a mutation hotspot (R93 to W change, 10/20 clones) in the LL-37+Fe2+ treatment group that was significantly (P = 0.0004, two-tailed Fisher’s exact-test) different from the two other treatments (S2, S3 and S4 Tables). This mutation hotspot is a single nucleotide transition from C to T, which is one of the most frequent types of mutations caused by ROS [40]. We found that twelve out of twenty clones from the LL-37+Fe2+ condition had C to T transitions, whereas none were present in either iron or LL-37 - treated bacteria (two-tailed Fisher's exact-test (P< 0.0001) (S7 Fig). It is striking that although several C to T mutations can potentially lead to glpT inactivation are in principle possible, that the majority are concentrated in a single point. Such mutation hotspots can be driven by a specific topology on the chromosome [41], or a particular sequence prone to double strand breaks resulting in mutations after repair [42]. In general, observable mutations are the results of mutation-repair balance and not all mutations are repaired with the same efficiency. A good example in P. aeruginosa is the mutational inactivation of the anti-sigma factor gene, mucA, with the mutated allele mucA22 most prevalent (25 to 40%). This inactivation seems to be spectrum dependent [43].
Fig. 4. Mutation hotspots in glpT. 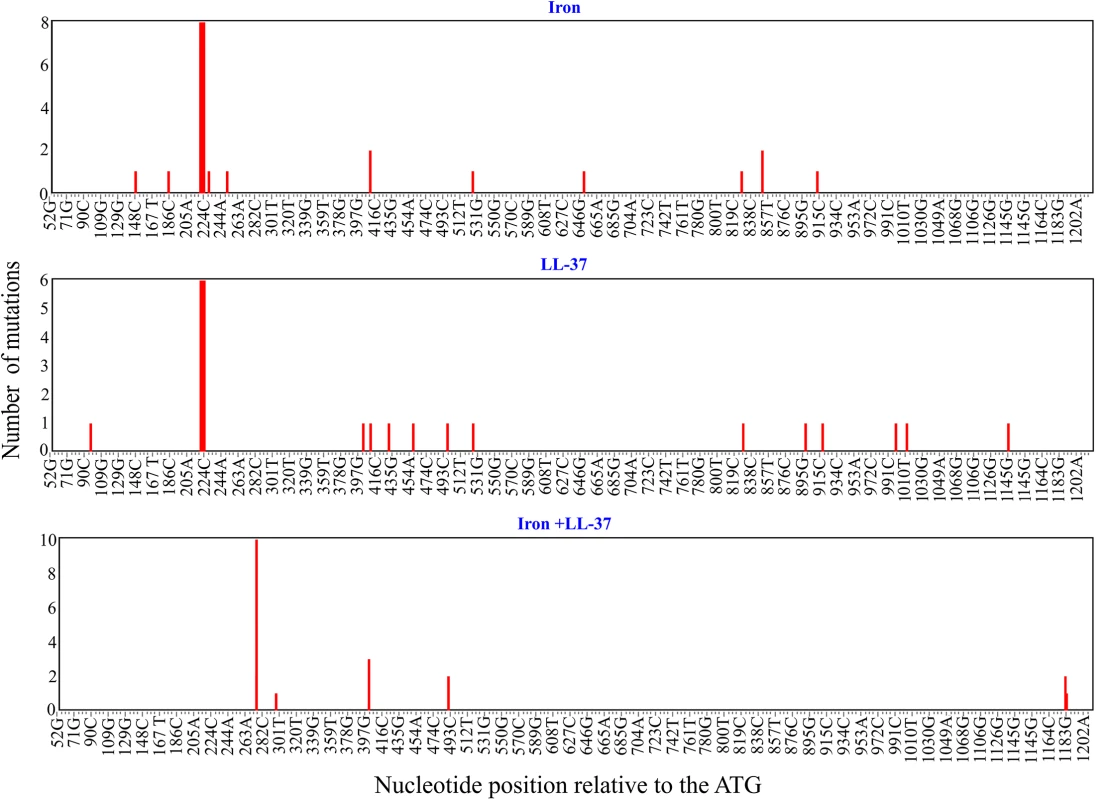
Distribution of the mutations in the 1160 bp-long fragment of glpT in P. aeruginosa PA14 fosfomycin-resistant clones treated with iron, LL-37, or a combination of both. The number of mutations (nucleotide substitutions, indels) is plotted against respective nucleotide positions within the gene fragment. Note the overlapping mutations at positions 220–225, 409 and 524 bp in iron and LL-37 treatments and absence of common mutations in LL-37+Fe2+ treatment. The fig was generated using the mutational spectrum analysis software iMARS [39]. Interestingly, C to T transitions were one of the most frequent types of single nucleotides changes in the genomes of Salmonella typhimurium evolved to increasing concentrations of LL-37 in modified LB medium short of sodium chloride and anions [44], which is fully consistent with our results.
A number of studies [45–48], which caused some controversy [49–51], suggested that hydroxyl radicals can be generated as a consequence of antibiotic treatments and this aggressive by-product may take part in the killing mechanism of bactericidal drugs or promote mutagenesis. Most of these studies were carried out in the iron-rich LB medium and whether processes as described here for the interaction between an AMP and iron apply to antibiotics remains to be explored.
Conclusions
Our results support the notion that under certain pathological situations, sub-inhibitory concentrations of cAMPs facilitate uncontrolled uptake of free iron by bacterial cells, which results in increased mutagenesis by Fenton reaction (Fig 5). According to our results, this could be a general mechanism underlying mutagenesis by joint action of antimicrobial peptides and free iron in specific situations where iron is not limited.
Fig. 5. A model for LL-37-mediated, iron–induced mutagenesis in Pseudomonas aeruginosa. 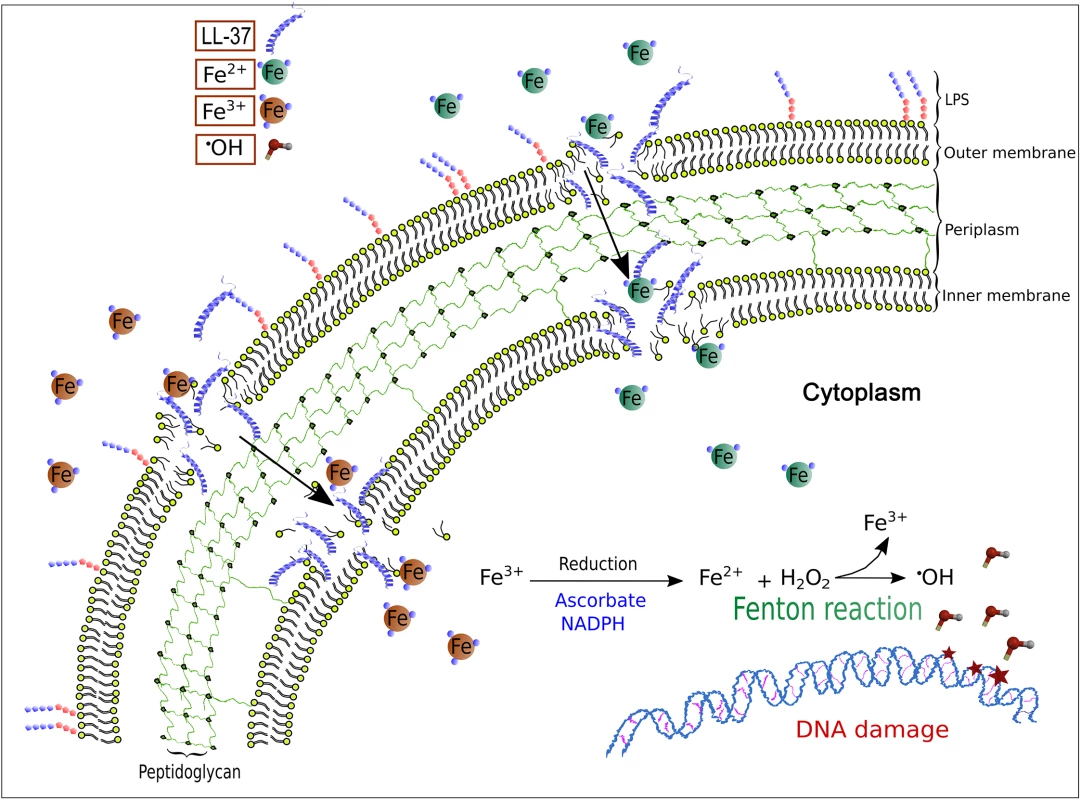
The interactions of LL-37 with the membrane at sub-inhibitory concentrations lead to transient permeability changes in the membranes that promote iron movement in favour of the electrochemical gradient. Uncontrolled uptake of ferrous ions stimulates Fenton reaction that leads to hydroxyl radical formation and results in DNA damage and mutagenesis. Free iron levels are kept as low as possible due to its toxic effect for all living beings but especially in bacteria that lack proper cell compartments. In fact, iron withdrawal is part of the natural innate immune response in inflammation that makes free iron even scarcer. During inflammation and infection a “hypoferremic response” (anaemia of inflammation) is observed [52,53]. Many chelating proteins such as transferrin and ferritin exhibit antibacterial activity simply by making iron availability incompatible with bacterial proliferation [54].
Despite showing that many cationic antimicrobial peptides are unable to increase mutation rates [7], the particular situation of iron-induced mutagenesis can be of great interest for certain types of infections where iron homoeostasis is compromised. In cystic fibrosis, bleeding happens frequently. Considering that P. aeruginosa is one of the most common pathogens that acquire all antibiotic resistance by mutations, the mechanism proposed here is likely very relevant for pathoadaptation.
Finally, our work has potential implications for the development of future treatment of chronic respiratory infections by Pseudomonas aeruginosa. For example, modulation of iron-chelating agent in CF therapy could potentially slow down the pathoadaptation and development of resistance in CF, and diminish lung damaging by ROS.
Materials and Methods
Bacteria and growth conditions
The P. aeruginosa PAO1 wild-type strain was kindly provided by M. A. Jacobs. The P. aeruginosa PA14 was kindly provided by Nicole T. Liberati and Frederick Ausubel. All bacterial strains were cultured in Mueller-Hinton Broth, non-cation adjusted (Sigma), with total iron content 0.22 ± 0.02 μM, following recommendations of CLSI for cationic antimicrobial peptide susceptibility testing. The MHB pH was adjusted to 6 in all cases with acetic acid to enhance solubility of both, LL-37 and iron compound. All experiments were performed at 37°C, under agitation in liquid culture. For genetic manipulation, Escherichia coli DH5α strain was used and routinely cultured in Lysogeny Broth (LB medium), supplemented with antibiotic when appropriate.
Minimal inhibitory concentration (MIC)
MICs were determined according to CLSI recommendations by a microdilution method with some modifications for antimicrobial peptides. The MIC was defined as the antimicrobial concentration that inhibited growth after 24 hours of incubation in liquid MHB medium at 37°C. Polypropylene non-binding multi-well plates (Th. Geyer, Germany) were used for all experiments.
Determination of MIC50
The MIC50s for all antimicrobials were determined by inoculating strains grown to mid-log phase into the wells of a 96-microwell plate. Approximately 102 cells from overnight cultures of PAO1 and PA14 strains were inoculated into 50 ml tubes containing 10 ml of MBH and incubated at 37°C with strong agitation until the mid-log phase of growth (approximately 108 cfu/ml). Then, 100 μl of 2 ×108 cells from these cultures were inoculated in each well of polypropylene non-binding 96-multiwell plates containing 100 μl of fresh Mueller-Hinton medium with growing concentration of serially diluted LL-37. The plates were incubated at 37°C during four hours with continuous agitation in a plate reader (Synergy HT, BioTeK). Four replicates per concentration were prepared and the experiments were repeated twice. MIC50s at 4 hours were defined as the concentrations at which 50% of growth reduction in comparison to the control at OD600 were observed.
Estimation of mutant frequencies and mutation rates
For spontaneous-mutation rate measurements of PAO1 strain, 1/100 dilutions of overnight cultures were inoculated into four tubes per group, each containing two ml of MHB medium. The cultures were incubated at 37°C with strong agitation to reach ~108 cfu/ml. At this point, appropriate concentration of LL-37, colistin, iron sulphate (FeSO4) or combinations of antimicrobial peptides and iron, were added to the cultures. The tubes were allowed to continue their normal growth overnight until saturation. In the experiments with colistin, the bacterial suspensions were washed twice with saline solution 0.9% NaCl before plating. The cultures were appropriately diluted and plated on MHB agar plates with or without rifampicin (100 μg/ml). The mutant frequency was estimated by the number of colonies growing on rifampicin divided by the number of total cfu/ml. To confirm the results, relevant concentrations were also assayed with ten replicates to see the influence of the treatment on the population mutation rates (the number of mutations per cell per generation). Mutation rates were calculated by maximum verisimilitude method and data were processed using the on-line web-tool Falcor (http://www.mitochondria.org/protocols/FALCOR.html) as recommended [55,56]. Falcor software was used to estimate the mutant frequency too. The mutation rates for the strain PA14 under the selected treatments were determined in the same way as described for PAO1, but fosfomycin (128 μg/ml) was used instead of rifampicin.
Sequencing of fosfomycin resistant mutants (Fos-R) and sequence analysis
To assess the effects of iron and antimicrobial peptides and their combination on the mutation spectrum of P. aeruginosa strain PA14, the glpT gene of twenty randomly selected Fos-R clones from independent cultures for each treatment group (MHB supplemented either with 40 μM Fe2+, LL-37 (32 μg/ml) or a combination of LL-37+Fe2+ at same concentrations for both), was amplified by colony PCR using glpT-P14-F1 (5-AGCGGAGCTCGCGATGTTC-3) and glpT-P14-R1 (5-TCAGCCGGCTTGCTGCGG-3) primers [36] and Kapa2G Fast ReadyMix PCR with dye kit (KAPA Biosystems, Boston, US). Cycling conditions were as follows: 95°C 7’/(95°C 15”/60°C 15”/72°C 40”)x 35/72°C 7’/4°C hold. The PCR products were purified and sequenced at Macrogen Europe using the forward and reverse primers described above. Sequences were assembled using SeqTrace software. Assembled sequences were imported into CLC Sequence Viewer 6 and aligned using default settings. Low quality flanking sequences were removed and the alignment was trimmed to the 1160 bp fragment (the 52–1212 bp region relative to the A in the start ATG codon of the 1347bp-long glpT ORF). A tridimensional homology model of GlpT was generated using Cn3D software by performing a BLASTP search using PA14 strain GlpT protein sequence as a query and mapping the resulting alignment against the experimentally determined Escherichia coli K-12 GlpT protein structure. To evaluate the potential effects of amino acid substitution on protein stability, the online tool I-Mutant (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) was used. TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used for prediction of transmembrane helices in glpT protein sequence. Mutational spectrum differences were analysed using the software iMARS [39].
Influence of 2–2′ bipyridyl on LL-37+Fe2+ mutagenesis
The effect of 2–2′ bipyridyl, an iron chelating agent, on LL-37+Fe2+ mutagenesis was determined by measuring its influence on mutant frequency on a selected concentration of LL-37+Fe2+ combination (32 μg/ml and 40 μM of Fe2+respectively), where mutagenesis was observed. The experiment consisted of adding a titrating concentration of 2–2′ bipyridyl (114 μM) to chelate 95% of the added iron of treated cultures, in order to make the treatment compatible with bacterial growth. Cultures with the described LL-37+Fe2+ combination with no addition of 2–2′ bipyridyl were used as a control. The mutant frequencies of both groups were determined as described elsewhere in this section. LL-37, iron and 2–2′ bipyridyl were simultaneously added to the exponentially growing cultures.
Cloning of dps gene and mutagenesis experiment
DNA fragment containing the PAO1 dps gen from genomic DNAs was amplified by PCR using the oligonucleotides PA-DPS-F1 (5′-ATGGAAATCAATATCGGAATCG-3′) and PA-DPS-R1 (5′-CTACTCAAATCAAGCGGTTGGC-3′) as forward and reverse primers, respectively. The fragment contains the ATG codon and 50 nucleotides downstream of the stop codon. The PCR product was directly cloned into the SmaI-digested and T-tailed pUCP24 plasmid vector (replicative in both P. aeruginosa and E. coli), which harbours Gentamicin resistance markers [57]. E. coli DH5α was used following standard protocols for genetic manipulations. The resulting plasmids, termed pUCP24-DPS, were introduced by electroporation into PAO1 wild type strain. The cloning vector was also transformed into the same strain as control. An experiment similar to the one designed for 2–2′ bipyridyl was carried out. A mutagenic combination of LL-37+Fe2+ was assayed in the strains carrying pUCP24-DPS plasmid or the empty vector pUCP24 and mutant frequencies were determined for both groups.
Quantification of iron concentrations
Total iron quantification was carried our as previously described with minor modifications [58,59]. Cultures of P. aeruginosa PAO1 were grown to an OD600 of approximately 0.5 at 37°C with agitation in a volume of two ml in MHB. The cultures were centrifuged at 4000 g during ten minutes at 20°C. The pellets were re-suspended in fresh MHB and three different groups were prepared. The treatments consisted of LL-37 (32 μg/ml) (I), iron sulphate to a final concentration of 40 μM (II), a combination iron sulphate and LL-37 (III), both of them at the same concentrations of their respective group I and II, and a control group (IV) to which the proportional amounts of LL-37 and iron sulphate solvent were added (sterile dH20 and dH20, pH = 5 respectively). The cultures were incubated for up to 30 minutes and harvested by centrifugation as before but at 4°C. The cell pellets were washed twice with ice-cold phosphate-buffered saline (PBS) and re-suspended in 1 ml of TE buffer containing 5 mg/ml of egg lysozyme (Sigma) and incubated during 10 minutes at room temperature. To quantify total iron, the lysate (one ml) was mixed with one ml of HCl 10 mM and 1 ml of iron-releasing reagent containing HCl 1.4 M + 4.5% (weight/volume) aqueous solution of KMnO4; 1/1 and incubated at 60°C for two hours. After cooling, 0.06 ml iron-detection reagent (6.5 mM ferrozine, 6.5 mM neocuproine, 2.5 M ammonium acetate, 1 M ascorbic acid in water) was added and the sample absorbance was read at 550 nm in a plate reader Synergy HT (Biotek). The iron concentrations were determined based on a standard curve obtained with increasing concentrations of ferric chloride and normalized to protein concentration of the lysates. Each group consisted of five cultures. We determined the content of total iron in our cultures media MHB and LB, using the same procedure describe above, starting by the addition of 1 ml of HCl 10 mM and 1 ml of the iron-releasing reagent. Under the suspicion that MHB had lower iron content, the samples of this medium were prepared ten-fold concentrated.
Statistical analyses
An unpaired Student's t test or Mann-Whitney U test was used where appropriate for statistical analysis, according to the nature of the data (parametric or nonparametric adjustment). Two-tailed Fisher’s exact test or Monte Carlo hypergeometric test were used to calculate statistical significance of differences in mutation frequencies at each codon site of the alignment between treatments in the mutational spectrum analysis. P values less than 0.05 were considered statistically significant. All tests were performed with statistic software R except for mutational spectrum analysis where iMARS [39] was used instead.
Supporting Information
Zdroje
1. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000/09/13 ed. 2000;406 : 959–964. doi: 10.1038/35023079 10984043
2. Hassett DJ, Korfhagen TR, Irvin RT, Schurr MJ, Sauer K, Lau GW, et al. Pseudomonas aeruginosa biofilm infections in cystic fibrosis: insights into pathogenic processes and treatment strategies. Expert Opin Ther Targets. 2010/01/09 ed. 2010;14 : 117–130. doi: 10.1517/14728220903454988 20055712
3. Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34 : 634–40. doi: 10.1086/338782 11823954
4. Rodríguez-Rojas A, Oliver A, Blázquez J. Intrinsic and environmental mutagenesis drive diversification and persistence of Pseudomonas aeruginosa in chronic lung infections. J Infect Dis. 2012;205 : 121–7. doi: 10.1093/infdis/jir690 22080096
5. Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science (80-). 2000/05/20 ed. 2000;288 : 1251–1254. 8507 [pii]
6. Limoli DH, Rockel AB, Host KM, Jha A, Kopp BT, Hollis T, et al. Cationic antimicrobial peptides promote microbial mutagenesis and pathoadaptation in chronic infections. Ausubel FM. PLoS Pathog. Public Library of Science; 2014;10: e1004083. doi: 10.1371/journal.ppat.1004083 24763694
7. Rodríguez-Rojas A, Makarova O, Rolff J. Antimicrobials, stress and mutagenesis. PLoS Pathog. 2014;10: e1004445. doi: 10.1371/journal.ppat.1004445 25299705
8. Miller JH. Experiments in molecular genetics [Internet]. Cold Spring Harbor Laboratory; 1972. Available: http://books.google.co.uk/books/about/Experiments_in_molecular_genetics.html?id=PtVpAAAAMAAJ&pgis=1
9. Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, et al. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181 : 1415–28. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=93529&tool=pmcentrez&rendertype=abstract 10049371
10. Yang Y, Harris DP, Luo F, Xiong W, Joachimiak M, Wu L, et al. Snapshot of iron response in Shewanella oneidensis by gene network reconstruction. BMC Genomics. 2009;10 : 131. doi: 10.1186/1471-2164-10-131 19321007
11. Girardello R, Bispo PJM, Yamanaka TM, Gales AC. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J Clin Microbiol. 2012;50 : 2414–8. doi: 10.1128/JCM.06686-11 22553247
12. Imlay J, Chin S, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science (80-). 1988;240 : 640–642. doi: 10.1126/science.2834821
13. Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27 : 215–37. Available: http://www.ncbi.nlm.nih.gov/pubmed/12829269 12829269
14. Yamamoto Y, Fukui K, Koujin N, Ohya H, Kimura K, Kamio Y. Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus mutans. J Bacteriol. 2004;186 : 5997–6002. doi: 10.1128/JB.186.18.5997–6002.2004 15342568
15. Reid DW, Carroll V, O’May C, Champion A, Kirov SM. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur Respir J. 2007;30 : 286–92. doi: 10.1183/09031936.00154006 17504792
16. Reid DW, Carroll V, O’May C, Champion A, Kirov SM. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur Respir J. 2007/05/17 ed. 2007;30 : 286–292. 09031936.00154006 [pii] doi: 10.1183/09031936.00154006 17504792
17. Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002/02/06 ed. 2002;109 : 317–325. doi: 10.1172/JCI13870 11827991
18. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415 : 389–95. doi: 10.1038/415389a 11807545
19. Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. Annual Reviews 4139 El Camino Way, P.O. Box 10139, Palo Alto, CA 94303–0139, USA; 2000;54 : 881–941. doi: 10.1146/annurev.micro.54.1.881 11018148
20. Nasnas R, Saliba G, Hallak P. [The revival of colistin: an old antibiotic for the 21st century]. Pathol Biol. 2008/01/08 ed. 2009;57 : 229–235. S0369-8114(07)00223-4 [pii] doi: 10.1016/j.patbio.2007.09.013
21. Biswas S, Brunel J-M, Dubus J-C, Reynaud-Gaubert M, Rolain J-M. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10 : 917–34. doi: 10.1586/eri.12.78 23030331
22. Colistin: An Update on the Antibiotic of the 21st Century [Internet]. [cited 27 Aug 2015]. Available: http://www.medscape.com/viewarticle/772588_6
23. Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot A, et al. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. MBio. 2013;4: e00557–13–. doi: 10.1128/mBio.00557-13 23963183
24. Kreamer NN, Costa F, Newman DK. The ferrous iron-responsive BqsRS two-component system activates genes that promote cationic stress tolerance. MBio. 2015;6: e02549. doi: 10.1128/mBio.02549-14 25714721
25. Méhi O, Bogos B, Csörgő B, Pál F, Nyerges A, Papp B, et al. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol Biol Evol. 2014;31 : 2793–804. doi: 10.1093/molbev/msu223 25063442
26. Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci U S A. 2014;111: E1409–18. doi: 10.1073/pnas.1319900111 24706874
27. Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88 : 553–60. Available: http://www.ncbi.nlm.nih.gov/pubmed/9038346 9038346
28. McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, et al. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol. 2006/05/19 ed. 2006;188 : 3995–4006. 188/11/3995 [pii] doi: 10.1128/JB.00053-06 16707691
29. Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 2004;186 : 575–9. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=305751&tool=pmcentrez&rendertype=abstract 14702327
30. Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci U S A. 2002;99 : 16243–8. doi: 10.1073/pnas.242338299 12446835
31. Oliver A. High Frequency of Hypermutable Pseudomonas aeruginosa in Cystic Fibrosis Lung Infection. Science (80-). 2000;288 : 1251–1253. doi: 10.1126/science.288.5469.1251
32. Couce A, Rodríguez-Rojas A, Blázquez J. Bypass of genetic constraints during mutator evolution to antibiotic resistance. Proc Biol Sci. 2015;282 : 20142698. doi: 10.1098/rspb.2014.2698 25716795
33. Imlay J a. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77 : 755–76. doi: 10.1146/annurev.biochem.77.061606.161055 18173371
34. Rodriguez-Rojas A, Couce A, Blazquez J. Frequency of spontaneous resistance to fosfomycin combined with different antibiotics in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010/08/18 ed. 2010;54 : 4948–4949. AAC.00415-10 [pii] doi: 10.1128/AAC.00415-10 20713658
35. Rodríguez-Rojas A, Maciá MD, Couce A, Gómez C, Castañeda-García A, Oliver A, et al. Assessing the Emergence of Resistance: the Absence of Biological Cost in vivo May Compromise Fosfomycin Treatments for P. aeruginosa Infections. PLoS One. 2010;(In press). doi: 10.1371/journal.pone.0010193 20419114
36. Castaneda-Garcia A, Rodriguez-Rojas A, Guelfo JR, Blazquez J. The Glycerol-3-Phosphate Permease GlpT Is the Only Fosfomycin Transporter in Pseudomonas aeruginosa. J Bacteriol. 2009/09/08 ed. 2009;191 : 6968–6974. JB.00748-09 [pii] doi: 10.1128/JB.00748-09 19734311
37. Rodriguez-Rojas A, Blazquez J. The Pseudomonas aeruginosa pfpI gene plays an antimutator role and provides general stress protection. J Bacteriol. 2008/11/26 ed. 2009;191 : 844–850. JB.01081-08 [pii] doi: 10.1128/JB.01081-08 19028889
38. Rodríguez-Rojas A, Maciá MD, Couce A, Gómez C, Castañeda-García A, Oliver A, et al. Assessing the emergence of resistance: the absence of biological cost in vivo may compromise fosfomycin treatments for P. aeruginosa infections. PLoS One. 2010;5: e10193. doi: 10.1371/journal.pone.0010193 20419114
39. Morgan C, Lewis PD. iMARS—Mutation analysis reporting software: An analysis of spontaneous cII mutation spectra. Mutat Res—Genet Toxicol Environ Mutagen. 2006;603 : 15–26. doi: 10.1016/j.mrgentox.2005.09.010
40. McBride TJ, Preston BD, Loeb LA. Mutagenic spectrum resulting from DNA damage by oxygen radicals. Biochemistry. 1991;30 : 207–13. Available: http://www.ncbi.nlm.nih.gov/pubmed/1703014 1703014
41. Juurik T, Ilves H, Teras R, Ilmjärv T, Tavita K, Ukkivi K, et al. Mutation frequency and spectrum of mutations vary at different chromosomal positions of Pseudomonas putida. PLoS One. 2012;7: e48511. doi: 10.1371/journal.pone.0048511 23119042
42. Shee C, Gibson JL, Rosenberg SM. Two mechanisms produce mutation hotspots at DNA breaks in Escherichia coli. Cell Rep. 2012;2 : 714–21. doi: 10.1016/j.celrep.2012.08.033 23041320
43. Moyano AJ, Luján AM, Argaraña CE, Smania AM. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol. 2007;64 : 547–59. doi: 10.1111/j.1365-2958.2007.05675.x 17493134
44. Lofton H, Pränting M, Thulin E, Andersson DI. Mechanisms and fitness costs of resistance to antimicrobial peptides LL-37, CNY100HL and wheat germ histones. PLoS One. 2013;8: e68875. doi: 10.1371/journal.pone.0068875 23894360
45. Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336 : 315–9. doi: 10.1126/science.1219192 22517853
46. Kohanski M a, DePristo M a, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. Elsevier Ltd; 2010;37 : 311–20. doi: 10.1016/j.molcel.2010.01.003 20159551
47. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007/09/07 ed. 2007;130 : 797–810. S0092-8674(07)00899-9 [pii] doi: 10.1016/j.cell.2007.06.049 17803904
48. Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009/08/04 ed. 2009;12 : 482–489. S1369-5274(09)00090-3 [pii] doi: 10.1016/j.mib.2009.06.018 19647477
49. Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339 : 1213–6. doi: 10.1126/science.1232688 23471410
50. Liu Y, mlay J a Cell Death from Antibiotics Without the Involvement of Reactive Oxygen Species. Science (80-). 2013;339 : 1210–1213. doi: 10.1126/science.1232751
51. Imlay JA. Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin Microbiol. 2015;24C: 124–131. doi: 10.1016/j.mib.2015.01.004
52. Ward CG, Bullen JJ, Rogers HJ. Iron and infection: new developments and their implications. J Trauma. 1996/08/01 ed. 1996;41 : 356–364. Available: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8760553 8760553
53. Wessling-Resnick M. Iron homeostasis and the inflammatory response. Annu Rev Nutr. 2010;30 : 105–22. doi: 10.1146/annurev.nutr.012809.104804 20420524
54. Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6: e1000949. doi: 10.1371/journal.ppat.1000949 20711357
55. Couce A, Blazquez J. Estimating mutation rates in low-replication experiments. Mutat Res. 2011/07/09 ed. 2011;In Press. S0027-5107(11)00147-3 [pii] doi: 10.1016/j.mrfmmm.2011.06.005 21736881
56. Hall BM, Ma C - X, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25 : 1564–5. doi: 10.1093/bioinformatics/btp253 19369502
57. West SE, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994/10/11 ed. 1994;148 : 81–86. 0378-1119(94)90237-2 [pii] 7926843
58. Vilchèze C, Hartman T, Weinrick B, Jacobs WR. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat Commun. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2013;4 : 1881. doi: 10.1038/ncomms2898 23695675
59. Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem. 2004;331 : 370–5. doi: 10.1016/j.ab.2004.03.049 15265744
Štítky
Genetika Reprodukční medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



