-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
SOX2 is a master regulator of both pluripotent embryonic stem cells (ESCs) and multipotent neural progenitor cells (NPCs); however, we currently lack a detailed understanding of how SOX2 controls these distinct stem cell populations. Here we show by genome-wide analysis that, while SOX2 bound to a distinct set of gene promoters in ESCs and NPCs, the majority of regions coincided with unique distal enhancer elements, important cis-acting regulators of tissue-specific gene expression programs. Notably, SOX2 bound the same consensus DNA motif in both cell types, suggesting that additional factors contribute to target specificity. We found that, similar to its association with OCT4 (Pou5f1) in ESCs, the related POU family member BRN2 (Pou3f2) co-occupied a large set of putative distal enhancers with SOX2 in NPCs. Forced expression of BRN2 in ESCs led to functional recruitment of SOX2 to a subset of NPC-specific targets and to precocious differentiation toward a neural-like state. Further analysis of the bound sequences revealed differences in the distances of SOX and POU peaks in the two cell types and identified motifs for additional transcription factors. Together, these data suggest that SOX2 controls a larger network of genes than previously anticipated through binding of distal enhancers and that transitions in POU partner factors may control tissue-specific transcriptional programs. Our findings have important implications for understanding lineage specification and somatic cell reprogramming, where SOX2, OCT4, and BRN2 have been shown to be key factors.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003288
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003288Summary
SOX2 is a master regulator of both pluripotent embryonic stem cells (ESCs) and multipotent neural progenitor cells (NPCs); however, we currently lack a detailed understanding of how SOX2 controls these distinct stem cell populations. Here we show by genome-wide analysis that, while SOX2 bound to a distinct set of gene promoters in ESCs and NPCs, the majority of regions coincided with unique distal enhancer elements, important cis-acting regulators of tissue-specific gene expression programs. Notably, SOX2 bound the same consensus DNA motif in both cell types, suggesting that additional factors contribute to target specificity. We found that, similar to its association with OCT4 (Pou5f1) in ESCs, the related POU family member BRN2 (Pou3f2) co-occupied a large set of putative distal enhancers with SOX2 in NPCs. Forced expression of BRN2 in ESCs led to functional recruitment of SOX2 to a subset of NPC-specific targets and to precocious differentiation toward a neural-like state. Further analysis of the bound sequences revealed differences in the distances of SOX and POU peaks in the two cell types and identified motifs for additional transcription factors. Together, these data suggest that SOX2 controls a larger network of genes than previously anticipated through binding of distal enhancers and that transitions in POU partner factors may control tissue-specific transcriptional programs. Our findings have important implications for understanding lineage specification and somatic cell reprogramming, where SOX2, OCT4, and BRN2 have been shown to be key factors.
Introduction
Transcription factors bind DNA in a sequence-specific manner and regulate gene expression patterns in response to developmental cues. Thus, transcription factors often direct a hierarchy of events controlling cellular identity [1], [2]. The HMG box containing transcription factor SOX2 is essential for the development of the epiblast in the early mammalian embryo [3] and for the maintenance of embryonic stem cells (ESCs) in vitro [4]. SOX2 is also necessary for the function and maintenance of neural progenitor cells (NPCs) in the nervous system [5], [6]. Further, SOX2 functions in other adult stem cell and progenitor populations in the gastrointestinal and respiratory tract, as well as in the developing lens, inner ear, taste buds, and testes [7]–[12]. Thus, SOX2 is a critical regulator of distinct stem cell states, but how it can serve this multifunctional role is not fully understood.
In ESCs, SOX2 is a component of the core transcriptional regulatory circuitry that controls pluripotency. Together with OCT4 (Pou5f1) and NANOG, SOX2 binds to the proximal promoters of large cohort of genes with known roles in pluripotency (including Oct4, Sox2, and Nanog) as well as those that function later in development [13]–[16]. These data suggest that SOX2 regulates ESC state by actively promoting pluripotency and by marking the regulatory regions of developmental genes for future activation. Consistent with this, SOX2 can act as a pioneer factor at a subset of genes in ESCs, and can be sequentially replaced by other SOX family members during differentiation, leading to activation of genes [17], [18]. SOX2 is also a critical factor in somatic cell reprogramming, whereby adult cells are converted into a pluripotent ESC-like state by the exogenous expression of a small set of transcription factors [19]–[21], with SOX2 being at the top of a gene expression hierarchy during the late phase of reprogramming [22].
In the central nervous system (CNS), Sox2 is required for proper NPC function during embryonic development and for maintenance of NPCs postnatally [23]–[25]. Specifically, loss of Sox2 in the developing CNS leads to multiple brain defects, including precocious progenitor differentiation and a reduced proliferating cell population in the brain, resulting in perinatal lethality [5], [6], [26], [27]. In contrast, forced expression of Sox2 blocks terminal differentiation of NPCs [26]–[29]. While a critical role for Sox2 in distinct stem cell populations has been firmly established both in vivo and in vitro, the molecular mechanisms by which SOX2 regulates cell type-specific gene expression programs are not clear.
Analysis of genome-wide binding profiles indicates that SOX2 occupies the promoters of thousands of genes [17], [30], however, a direct comparison of SOX2 targets in ESCs and NPCs has not been reported. Emerging evidence indicates that transcription factors drive tissue specific gene expression programs through interactions with distal enhancer elements [31]–[33]. Recent studies have shown that histone modification patterns, specifically monomethylation of lysine 4 of histone H3 (H3K4me1) and acetylation of lysine 27 on histone H3 (H3K27ac), mark distal enhancers [34]–[37]. Using this set of histone marks, we previously identified thousands of enhancer elements in ESCs and NPCs [34]. Thus far, the binding of SOX2 at enhancers has only been clearly demonstrated at a few genes in both ESCs and NPCs. For example, SOX2 occupies the proximal and distal enhancers upstream of the Oct4 promoter in ESCs whereas binding at an intronic enhancer (Nes30) in the Nestin gene was observed in NPCs [14], [38]–[40]. Thus, knowledge of SOX2-bound enhancers in these two cell types will contribute significant new insights into understanding control of cell state.
SOX family members weakly bind DNA and cannot robustly activate transcription alone, suggesting roles for additional partner factors in target selection [41]. Consistent with this, cooperation between SOX and POU transcription factor families has been highly conserved across metazoans where these factors are important regulators of developmental programs [42]. For example, SOX2 cooperates with the Class V POU family member OCT4 in ESCs to maintain pluripotency [13]–[16], however transcription factors that function with SOX2 genome-wide in NPCs are largely unknown. Thus, the identification of factors that bind to genomic sites with SOX2 will also be key to understanding how this master regulator can control distinct phenotypic outcomes.
Here, we defined the genome-wide binding patterns of SOX2 in ESCs and NPCs and show that SOX2 occupied a largely distinct set of genomic regions within promoters and distal enhancer elements in the two cell types. Similar to its cooperation with OCT4 (Pou5f1) in ESCs, we identified the Class III POU transcription factor BRN2 (Pou3f2) as a candidate SOX2 partner factor that co-bound a large fraction of distal enhancers with SOX2 in NPCs. Consistent with a functional role, forced expression of BRN2 in differentiating ESCs led to recruitment of SOX2 to a subset of NPC distal enhancers. This recruitment was associated with changes in chromatin structure, activation of neighboring genes, and ultimately precocious differentiation toward a neural-like state. Further analysis of bound sequences showed differences in the arrangement of a SOX-POU binding in ESCs and NPCs and revealed enrichment for additional transcription factor motifs. Together, these data reveal new insights into how SOX2 can function in a context-dependent manner to specify distinct stem cell states. Our work also has important implications for understanding development as well as the process of somatic cell reprogramming.
Results
SOX2 occupies distinct genomic regions in ESCs and NPCs
SOX2 is a master regulator of pluripotent ESCs and multipotent NPCs, yet how the same transcription factor can specify distinct stem cell states remains an open question. We reasoned that detailed analysis of genomic binding patterns in the two cell types might reveal how SOX2 can regulate diverse gene expression programs. To this end, we differentiated ESCs toward NPCs using established protocols [43], and interrogated SOX2 binding sites by chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq). Analysis of SOX2 binding in genetically identical ESC and NPC lines identified 13,717 and 16,685 enriched regions, respectively (Table S1). Our results were highly consistent with prior work in ESCs [16], however we observed a lower correlation compared to published data sets in neural progenitor cells (Figure S1A and Discussion). We found that >95% of bound regions are unique to each cell type (only 1,274 of the total regions are common to both datasets) (Figure 1A and Figure S1A, S1B). Thus, we identified a union set of 29,128 enriched regions at high confidence and found that SOX2 occupied a largely non-overlapping set of genomic sites in ESCs and NPCs.
Fig. 1. SOX2 binds promoters with cell-type-specific functions in ESCs and NPCs. 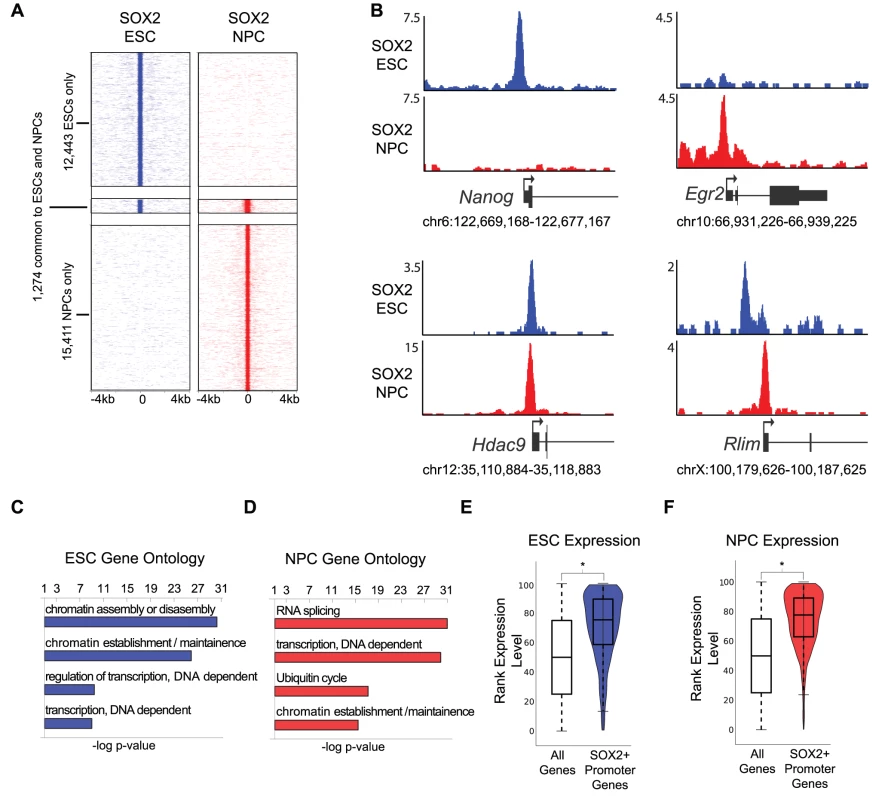
(A) Heat maps of SOX2 enrichment in ESCs and NPCs centered on peaks of enrichment and extended 4 kb in each direction. (B) Gene plots showing SOX2 density at Nanog, Erg2, Hdac9, and Rlim in ESCs (blue) and in NPCs (red). y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). (C, D) GOstat gene ontology analysis of genes linked to SOX2 bound TSSs. x-axis corresponds to negative log base ten p-value of enrichment of genes in target list compared to a whole genome background set. (E, F) Box and Violin plots representing expression data from Affymetrix arrays of genes linked to SOX2 target TSSs. y-axis corresponds to percentile expression rank, * denotes p-value<0.01, Student's T-test, two tailed. SOX2 is thought to bind to regulatory regions of genes with roles in stem cell maintenance and neural differentiation [13]–[16], however, a direct comparison of genome-wide binding in ESCs and NPCs has not been reported. Thus, we first mapped binding sites within 1 kb of a transcription start site (TSS) and found that SOX2 occupied 893 and 3,821 sites within promoters in ESCs and NPCs, respectively (Table S2). While ∼one-third (36%) of bound TSSs in ESCs were common to NPCs, SOX2 largely occupied distinct sites within promoters in the two cell types (Figure S1C). For example, SOX2 occupied the Nanog promoter only in ESCs, while the Egr2 (Krox20) promoter was bound only in NPCs, and a site within the Hdac9 promoter was occupied in both cell types (Figure 1B). Nanog and Egr2 are critical regulators of the ESC state and neural development, respectively, and Hdac9 is a broadly expressed chromatin regulator with a known role in brain development [44]–[48]. Furthermore, we also found examples where SOX2 occupied different sites in ESCs and NPCs but within the promoters of the same gene, such as the Rlim promoter, which encodes a regulator of both X-inactivation and later neural patterning [49], [50] (Figure 1B). Consistent with this, while roughly one-third of TSS-associated regions overlapped in ESCs and NPCs, 58% of the genes bound by SOX2 in ESCs were also NPC targets (Figure 1B, Figure S1C and S1D). These data suggest that SOX2 can utilize different binding sites to regulate genes in a context-dependent manner.
On a global level, SOX2 bound to a set of genes that code for chromatin and transcriptional regulators in both ESCs and NPCs in accordance with previous data [13]–[16] (Figure 1C, 1D and Table S3). While many of these targets were common to both cell types, a large group of chromatin and transcriptional regulators (490) were occupied uniquely in NPCs. Moreover, SOX2 bound more promoter regions in NPCs compared to ESCs and also occupied genes with diverse functions such as RNA splicing, regulation of the ubiquitin cycle, and translation (Figure 1D). While RNA splicing is a general cellular function, alternative splicing is known to play a key role in brain development [51]. For example, in NPCs, SOX2 occupied the promoters of the alternative splicing factors PTB and nPTB, which constitute a molecular switch regulating neuronal commitment [52]. We also found that SOX2 occupied genes displayed higher expression compared to all genes (Figure 1E, 1F and Table S4) suggesting that SOX2 has a positive regulatory role at promoters in each cell type.
SOX2 binds to distal enhancer elements in ESCs and NPCs
While SOX2 occupied proximal promoter regions in the two cell types, the vast majority of bound sites (>93% and >77% in ESCs and NPCs, respectively) mapped greater than 1 kb from annotated TSSs (Figure 2A). Distal enhancers are important non-coding DNA elements that control tissue specific gene expression patterns at variable distances from the promoters they regulate through binding of transcriptional and chromatin regulators [31]–[33]. We previously identified thousands of putative enhancers in ESCs and NPCs by genome-wide analysis of H3K4me1 and H3K27Ac occupancy, two histone marks known to mark distal enhancer elements [34]. SOX2 bound ∼17% (4,947) and ∼24% (6,842) of these putative enhancers in ESCs and NPCs, respectively (Figure 2B and Table S2). Currently, distal enhancers are presumed to regulate the nearest gene [34], [37], and after assigning each enhancer to the nearest upstream or downstream gene, we found that the SOX2-bound enhancers corresponded to 3,372 and 3,990 genes in ESCs and NPCs, respectively (Table S2). While these sites were largely distinct in the two cell types (Figure S2A), ∼44% of genes associated with SOX2 enhancers in ESCs also had a bound enhancer assigned to the same gene in NPCs (Figure S2B) and included many factors with specific roles in neural specification. Notably, analysis of bound enhancers revealed thousands of additional genes that may be regulated by SOX2 in both cell types which would not have been identified by analysis of only TSSs (Figure S2C, S2D). These data are consistent with the idea that, while enhancer utilization is highly cell type-specific, individual genes can be regulated by different enhancers [31], [53].
Fig. 2. SOX2 binds distinct enhancer regions in ESCs and NPCs. 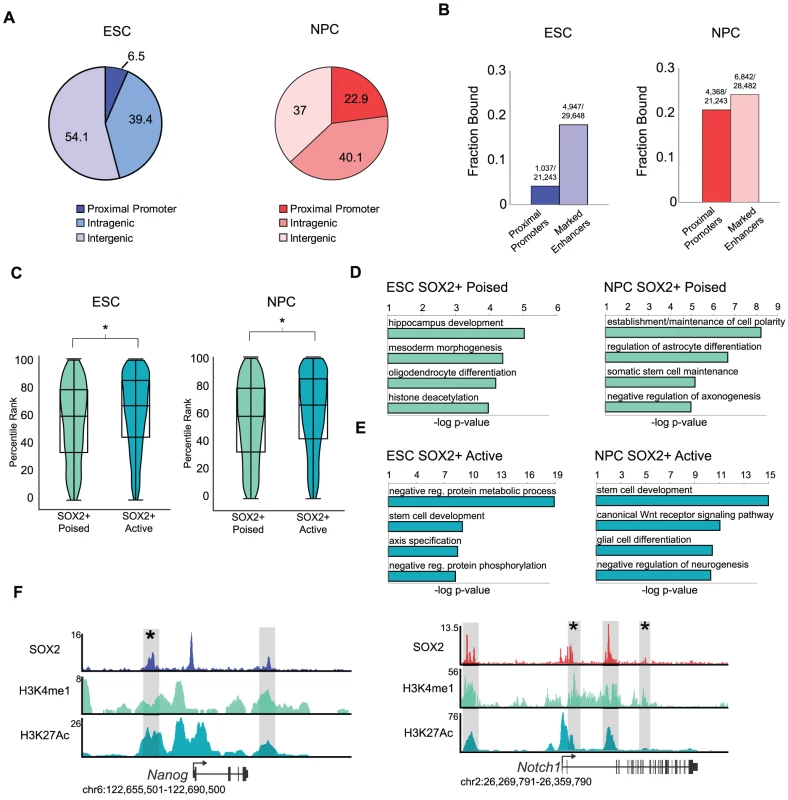
(A) SOX2 peaks mapping to annotated transcriptional start sites, intragenic regions, and intergenic regions. Numbers on pie chart indicate fraction of bound regions that fall into each category. (B) Fraction of total start sites or total marked enhancers associated with SOX2 enriched regions. Numbers above bars reflect absolute numbers of bound regions and genomic features. (C) Box and Violin plots representing expression data from Affymetrix arrays of genes linked to SOX2-bound poised and active enhancers. y-axis corresponds to percentile expression rank, * denotes p-value<0.01, Student's T-test, two tailed. (D, E) Analysis of GO biological processes enriched in SOX2 bound poised and active enhancer datasets in ESCs and NPCs. x-axis reflects negative log base 10 of binomial raw p-value for enrichment versus a whole genome background. (F) Gene plots showing SOX2, H3K4me1, and H3K27Ac density at Nanog in ESCs and Notch1 in NPCs. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). Gray boxes indicate putative enhancers occupied by SOX2. * denotes known enhancer region. The pattern of H3K4me1 and H3K27Ac occupancy can distinguish a given enhancer as active (H3K4me1+/−; H3K27Ac+) or poised (H3K4me1+; H3K27Ac−), states which correlate with high expression of a neighboring gene or the potential of that gene to be expressed later during development, respectively [34], [37], [54], [55]. Thus, globally genes nearest active enhancers are expressed at a higher level than those linked to poised elements. By comparison of SOX2-bound regions with the set of active and poised enhancers in our previous study [34], we found that SOX2 occupied 2,100 and 4,037 poised enhancers and 2,847 and 2,805 active enhancers in ESCs and NPCs, respectively (Table S2). Consistent with the idea that enhancers regulate transcriptional output, expression of genes closest to SOX2-bound active enhancers is significantly higher than genes associated with SOX2-bound poised enhancers (Figure 2C).
To gain deeper biological insights, we used the GREAT algorithm to perform Gene Ontology (GO) analysis to determine the function of genes associated with SOX2-bound enhancers. SOX2-bound poised enhancers in ESCs were nearest genes that function in commitment to the neural lineage and morphogenesis and included Jag1, Neurog3, and Nkx2-2, whereas those associated with poised enhancers in NPCs included genes with roles in terminal differentiation into neurons and glia such as Atoh1, Lhx8, Id2 and Id4 (Figure 2D, Tables S5 and S6). Notably, SOX2 bound to active enhancers nearest genes with functions in stem cell development in both cell types. Enriched categories in ESCs also revealed genes that function in early development and axis specification whereas genes linked to active enhancers in NPCs have roles in WNT signaling and neurogenesis (Figure 2E). For example, SOX2 occupied a known enhancer in the 5′ region of the Nanog locus in ESCs [56] and bound to intronic enhancers in Notch1 in NPCs [57], [58], known regulators of pluripotency and neurogenesis, respectively (Figure 2F). Thus, we identified thousands of stage-specific enhancers including many previously known enhancers in both cell types.
Despite the low overlap of SOX2-bound enhancer regions in ESCs and NPCs, genes linked to SOX2-bound poised enhancers in ESCs had functions in neural development, similar to genes linked to SOX2-bound enhancers in NPCs. Thus, we hypothesized that SOX2 might be regulating a subset of targets in both cell types by occupying distinct enhancer elements. Indeed, direct comparison of these genes revealed that ∼50% of genes (821 of 1,654) associated with SOX2-bound poised enhancers in ESCs also had a bound enhancer associated with that gene in NPCs (Figure S2E), despite the regions of SOX2 binding being largely cell-type specific. Importantly, genes where enhancers remained poised showed no significant difference in expression whereas those genes that gained active enhancers were expressed at higher levels (Figure S2F). These data are consistent with the idea that, while enhancer utilization is highly cell type-specific, individual genes can be regulated by different enhancers [31], [53]. Along those lines, using the GREAT algorithm to query the MGI gene expression database, we determined that 2,253 of the 4,037 SOX2-bound poised enhancers in NPCs were linked to genes expressed in the postnatal mouse nervous system (binomial p-value = 1.91e-35) (Table S6). Together, our data support the idea that poised enhancers can predict future developmental potential and suggest that SOX2 regulates a larger network of genes than previously anticipated by binding to distal enhancer elements.
BRN2 co-occupies distal enhancers with SOX2 in NPCs
SOX2 binds DNA weakly and it is insufficient to strongly activate transcription without cooperation with additional factors [59]. Consistent with this idea, we identified the canonical SOX2 motif, 5′-CTTTGTT-3′ [60]–[63] as highly enriched in ESCs and NPCs despite the difference in binding patterns (Figure 3A). Thus, we sought to identify additional factors that may function with SOX2 in ESCs and NPCs. SOX2 partners with the Class V POU-domain containing transcription factor OCT4 in ESCs to regulate a large cohort of genes important for pluripotency [13]–[16] however, partner factors in NPCs have not been clearly defined.
Fig. 3. BRN2 co-occupies distal enhancers with SOX2 in NPCs. 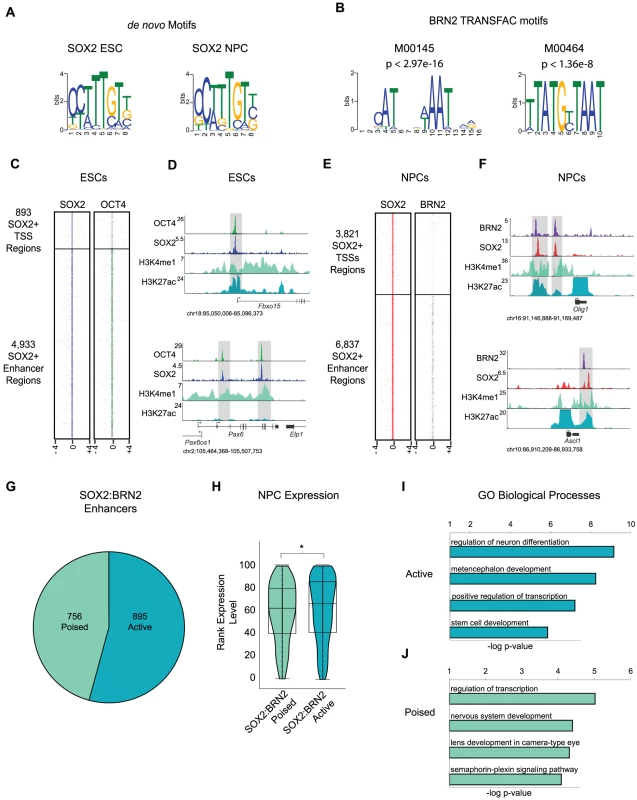
(A) de novo MEME motif analysis of SOX2 bound regions in ESCs and NPCs revealed canonical SOX2 motif. (B) TRANSFAC BRN2 motifs enriched in SOX2 target regions in NPCs. p-values represent significance of enrichment based on Mann-Whitney Wilcoxon ranked sum test with Benjamini-Hochberg multiple hypothesis testing correction. (C) Heat maps of SOX2 and OCT4 at SOX2 bound promoters and enhancers centered on peaks of SOX2 enrichment and extended 4 kb in each direction. (D) Gene plots showing SOX2, OCT4, H3K4me1, and H3K27Ac density at the Fbxo15 promoter and at poised enhancers of Pax6 in ESCs. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). Gray boxes indicate regions co-occupied by SOX2 and OCT4. (E) Heatmaps of SOX2 and BRN2 enrichment at SOX2-bound promoters and enhancers in NPCs centered on peaks of SOX2 enrichment and extended 4 kb in each direction. (F) Gene plots showing SOX2, BRN2, H3K4me1, and H3K27Ac density at Olig1 and Ascl1 loci in NPCs. y-axis corresponds to reads per million. Due to the high enrichment of H3K27Ac at active promoters, y-axis was cut off to show full dynamic range of enhancer-associated H3K27Ac density. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). Gray boxes indicate regions of SOX2-BRN2 co-occupancy. * indicates known enhancer. (G) Breakdown of number of SOX2-BRN2 target enhancers that are H3K4me1+, H3K27Ac− (poised) or H3K4me1+/−, H3K27Ac+ (active). (H) Box and Violin plots representing expression data from Affymetrix arrays of genes linked to poised and active SOX2-BRN2 target enhancers in NPCs. y-axis corresponds to percentile expression rank, * denotes p-value<0.01, Student's T-test, two tailed. (I, J) GREAT analysis of genes linked to poised and active SOX2-BRN2 target enhancers in NPCs. Interactions between SOX and POU factors are conserved in all metazoans and play key roles in embryonic development [42], thus, we hypothesized that SOX2 may also function with POU factors in NPCs. To test this, we interrogated a 100 bp window surrounding peaks of SOX2 enrichment in NPCs and determined enrichment for all known vertebrate transcription factor-binding motifs in the TRANSFAC database. Notably, we identified several enriched motifs, including two highly similar motifs recognized by the Class III POU factor BRN2 (Pou3f2) (Figure 3B and Table S7). BRN2 was of particular interest for several reasons. First, our transcriptome analysis showed that Brn2 is highly expressed in NPCs, but not in ESCs (Table S7). Moreover, Brn2 and Sox2 are both expressed in neurogenic regions of the brain and SOX2 and BRN2 are known to co-occupy a small number of loci in this tissue [38], [64], [65]. Like Sox2, Brn2 loss-of-function causes pleiotropic defects and NPC impairment [66]–[69]. Furthermore, Sox2, Brn2, and the forkhead transcription factor Foxg1 are sufficient to reprogram fibroblasts toward a multipotent NPC-like state [70]. These data suggest that transitions in POU partner factors of SOX2 may control cell identity in distinct stem cell populations.
Although neurogenesis and maintenance of cell identity in the brain require BRN2, its target genes in NPCs were not known. To address this, we performed ChIP-Seq and identified 6,574 BRN2 occupied regions in NPCs (Table S1). Similar to SOX2-bound regions, more BRN2-bound regions mapped to previously identified distal enhancers [34] than to promoter regions (Figure S3A). Motif analysis revealed enrichment for a canonical Octamer (OCT) motif (5′-ATGCATAT -3′) [71], [72] within BRN2 bound sites validating the high quality of our data set (Figure S3B).
We next examined the overlap between SOX2 and the two POU factors (BRN2 and our previously published OCT4-ESC dataset [16], Table S1) in ESCs and NPCs. Regions occupied by OCT4 and BRN2 showed little overlap (Figure S3C), indicating that these factors occupied cell-type-specific targets. Our data confirmed that SOX2 and OCT4 co-occupied many genomic sites in ESCs [13]–[16] (Figure 3C and Figure S3D-S3G). For example, SOX2 and OCT4 co-bound the promoter of Fbxo15 and to two putative enhancers of Pax6 that have been previously identified based on evolutionary sequence conservation and histone modification patterns [73] (Figure 3D). Notably, whereas BRN2 was absent from most SOX2-bound promoters in NPCs (Figure 3E), BRN2 occupied a subset of distal enhancers and bound many of these sites with SOX2, including known SOX2-BRN2 targets such as enhancers of Sox2 and Nestin [38], [65] (Figure S3H-S3K). For example, SOX2 and BRN2 co-occupied putative 3′ enhancer regions of Olig1 [74], and a known regulatory region 3′ of the Ascl1 (Mash1) locus [75] (Figure 3F). Together, these data suggest that SOX2 functions with BRN2 at a subset of distal enhancers to regulate target genes in NPCs.
Whereas SOX2-OCT4 bound enhancers associated with genes that have roles in pluripotency and lineage commitment, SOX2-BRN2 enhancers neighbored genes that function in NPC identity. Overall, SOX2 and BRN2 occupied 756 poised and 895 active enhancers in NPCs (Figure 3G). SOX2-BRN2 bound active enhancers correlated with genes that were expressed at higher levels than those associated with poised enhancers (Figure 3H). Further analysis revealed genes linked to active enhancers included transcription factors that play roles in neural development such as Notch1, Rfx4 and Sox2 itself (Figure 3I and Table S8). Interestingly, genes linked to the co-bound poised enhancers in NPCs included regulators of later stages of neuronal developmental such as the pro-neural transcription factor Atoh1 [76], [77] and Dab1, a critical regulator of neuroblast migration [78] (Figure 3J and Table S8). Notably, ∼24% of genes associated with SOX2-OCT4 poised enhancers in ESCs overlapped with genes associated with SOX2-BRN2 bound enhancers in NPCs that included known regulators of neural development such as Atoh1 and Ncam1, despite differences in the bound regions. Thus, SOX2-POU partnerships may control neural development by differentially targeting specific subsets of enhancers in pluripotent ESCs and multipotent NPCs, in order to establish the development potential of this tissue from very early stages of embryogenesis.
BRN2 expression in ESCs promotes neural differentiation
The significant overlap between BRN2 and SOX2 in NPCs predicts that BRN2 is also an important driver of neural commitment. To test this idea, we generated ESC lines that harbored a drug-inducible Brn2 transgene (TetO-Brn2) and assayed the potential of these cells to differentiate toward the neural lineage (Figure S4A–S4C). Upon Brn2 induction, ESCs showed distinct morphological changes from round cells that grew in colonies to polarized, Nestin-positive cells at day 1 of differentiation compared to control cells (Figure 4A and Figure S4D). Consistent with these changes, neural lineage genes such as Nestin and Sox1 showed higher expression in ESCs upon Brn2 expression (Figure 4B). Notably, Brn2 induction led to changes in gene expression and cell fate in the absence of additional growth factors whereas control cells did not show significant differences under these conditions. Thus, forced expression of Brn2 can promote differentiation of ESCs toward a neural-like fate.
Fig. 4. Brn2 biases ES cells towards neural differentiation. 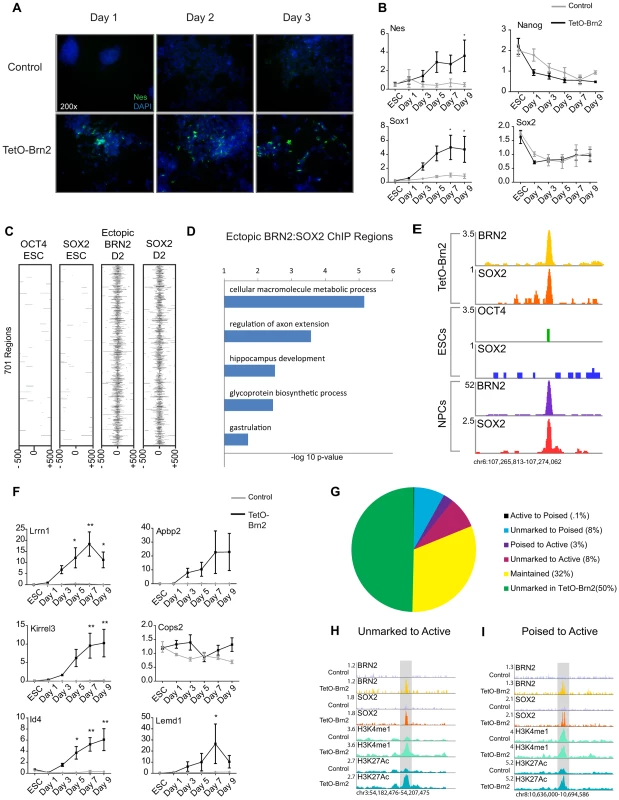
(A) Staining with DAPI (blue) and immunocytochemistry of NESTIN (green) in ESCs induced to differentiate in adherent cultures with or without ectopic Brn2. (B) qRT-PCR of the indicated genes in ESCs with (black lines) and without (gray lines) ectopic Brn2 expression through differentiation. y-axis represents relative expression normalized to Gapdh in 3 biological replicates, measured in triplicate. ESC time point is ESCs without doxycycline, and d1–d9 time points represent time in differentiation medium. * denotes p-value<0.05, ** denotes p-value<0.01 ANOVA with Bonferroni correction (C) Heatmap of OCT4 and SOX2 enrichment in ESCs and ectopic BRN2 and SOX2 in TetO-Brn2 cells of 701 genomic regions occupied by only ectopic BRN2 and SOX2. (D) GREAT GO biological processes enriched in 701 regions in (C). x-axis reflects negative log base 10 of raw binomial p-value for enrichment versus a whole genome background. (E) Gene plots depicting peaks of enrichment in indicated datasets at a locus distal to Lrrn1. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). (F) qRT-PCR of genes associated with SOX2-BRN2 binding in TetO-Brn2 cells and NPCs, in ESCs with (black lines) and without (gray lines) ectopic Brn2 expression through differentiation. y-axis represents relative expression normalized to Gapdh in 3 biological replicates, measured in triplicate. ESC time point is ESCs without doxycycline, and d1–d9 time points represent time in differentiation medium. * denotes p-value<0.05, ** denotes p-value<0.01 ANOVA with Bonferroni correction. (G) Pie chart reflecting overlap between SOX2-BRN2 regions and enhancer chromatin marks in TetO-Brn2 cells. Percentages in legend reflect fraction of 1,533 SOX2-BRN2 regions in each category. (H) Example region that was occupied by ectopic BRN2 in TetO-Brn2 cells, leading to recruitment of endogenous SOX2 and the deposition of H3K4me1 and H3K27Ac. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). Gray box indicates region of SOX2-BRN2 co-occupancy in TetO-Brn2 cells which was not occupied by SOX2 in control cells. (I) Example poised enhancer occupied by ectopic BRN2 in induced cells, leading to recruitment of endogenous SOX2, and deposition of H3K27Ac. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). Gray box indicates region of SOX2-BRN2 co-occupancy in TetO-Brn2 cells which was not occupied by SOX2 in control cells. Our data suggested that POU factor expression may be a key determinant of cell-type-specific SOX2 target selection, so we hypothesized that ectopic BRN2 might be sufficient to recruit endogenous SOX2 to genomic regions de novo. To test this, we collected TetO-Brn2 cells two days after induction (Figure S4D) and performed ChIP-Seq. We identified 12,362 and 8,401 regions occupied by SOX2 and BRN2 in these cells, respectively (Table S1). Similar to SOX2 and BRN2 in NPCs, these factors occupied more distal sites than promoters (Figure S4E). Strikingly, ∼18% (1,034 regions) occupied by BRN2 in the induced ESCs were also bound by BRN2 in NPCs, indicating that ectopic BRN2 retained some of its NPC target specificity. These regions were distal to loci encoding neurodevelopmental regulators such as Ephrin Receptors (Epha3, Epha4, Epha5, Epha7) and transcription factors such as Id2 and Id4 (Table S9).
Importantly, we defined 1,533 regions co-occupied by BRN2 and SOX2 in these cells. Comparison of these regions to SOX2 and OCT4 targets in ESCs and SOX2 in control cells at day 2 (Table S1) revealed 701 (46%) of these sites were bound uniquely by SOX2-BRN2 in the induced cells. These data suggested that BRN2 was necessary for SOX2 binding at these sites (Figure 4C and Figure S4F, S4G). Analysis of enriched GO categories showed that genes closest to these novel targets had roles in the development and function of the nervous system (Figure 4D and Table S9). Notably, 21% of these novel sites (144 regions) were also bound by SOX2 and/or BRN2 in NPCs, including enhancers linked to genes with demonstrated roles in neural development such as Lrrn1 and Abpa2 (X11l) [79]–[81] (Figure 4E). Expression analysis by qRT-PCR of a subset of these TetO-Brn2/NPC, SOX2-BRN2 genes, including Lrrn1, Abpa2, Kirrel3, Cops2, Id4, and Lemd1, revealed that some were significantly induced in TetO-Brn2 cells compared to controls (Figure 4F). Thus, ectopic BRN2 was sufficient to recruit SOX2 to NPC-specific sites and to induce the expression of nearby genes, indicating that POU-factor partners are sufficient to functionally recruit SOX2 to a subset of cell-type-specific target loci.
Given that SOX2-BRN2 binding in NPCs correlated with cell-type-specific distal enhancers, we hypothesize that SOX2-BRN2 might play a role in regulating the state of these elements. Thus, we next examined the distribution of the enhancer chromatin marks, H3K4me1 and H3K27Ac, in TetO-Brn2 and control cells at day 2 in order to determine whether the ectopic binding of these factors could alter local chromatin structure (Table S1). We found that 777 of the 1,533 co-bound sites (∼51%) were coincident with H3K4me1 and/or H3K27Ac regions in TetO-Brn2 cells and 488 of these regions (∼32%) displayed similar patterns in both induced and control cells (Figure 4G). Interestingly, 165 of the co-occupied regions (∼11%) gained H3K27Ac upon Brn2 induction, and were closest to genes involved in neural development such as Atoh1, NeuroD1, and Tcf7l (Tcf3). This included 125 regions (∼8%) that were unmarked (i.e. lacked H3K4me1 or H3K27Ac) in control cells (Figure 4H) and 40 regions (∼3%) that transitioned from a poised state to active (Figure 4I). Thus, ectopic BRN2-SOX2 binding was sufficient to activate both poised and unmarked enhancers, supporting a role for these factors in controlling global gene expression networks by regulating the activity of cis-regulatory elements. Collectively, these data support a role for distinct POU factors in SOX2 binding site selection and gene regulation, and suggests a model by which BRN2 functions with SOX2 to mediate developmental transitions in the neural lineage.
Binding configurations of SOX2 and POU factors at genomic targets in ESCs and NPCs
Given that most SOX and POU family members bind highly similar motifs, we hypothesized that distinct motif configurations may explain, in part, the diverse binding patterns in ESCs and NPCs. For example, SOX2 and OCT4 bind DNA in distinct conformations depending on the arrangement of binding sites 82–86 and these configurations have consequences on factor binding and transcriptional outcome [38], [82], [86], [87]. We found that SOX2 frequently occupied sites within 25-bp of OCT4 (∼25%), and that relatively few sites were greater than 100–200 bp from OCT4 (∼8%) (Figure 5A). In contrast, while a significant fraction of regions showed SOX2 and BRN2 bound within 25-bp in NPCs (∼12%), a larger fraction (∼33%) occurred at distances of 100–200 bp. For example, an intragenic region of the Wwc1 locus was bound by SOX2 and BRN2 in NPCs and peaks of enrichment were 100 bp apart (Figure 5B), while in ESCs neither SOX2 nor OCT4 recognized this element. These data indicate that while SOX2 and POU factors occupied similar motifs in ESCs and NPCs, these factors bound to different arrangements of these motifs in a cell type-specific manner.
Fig. 5. Motif configuration affects binding by SOX2 and cell-type-specific POUs. 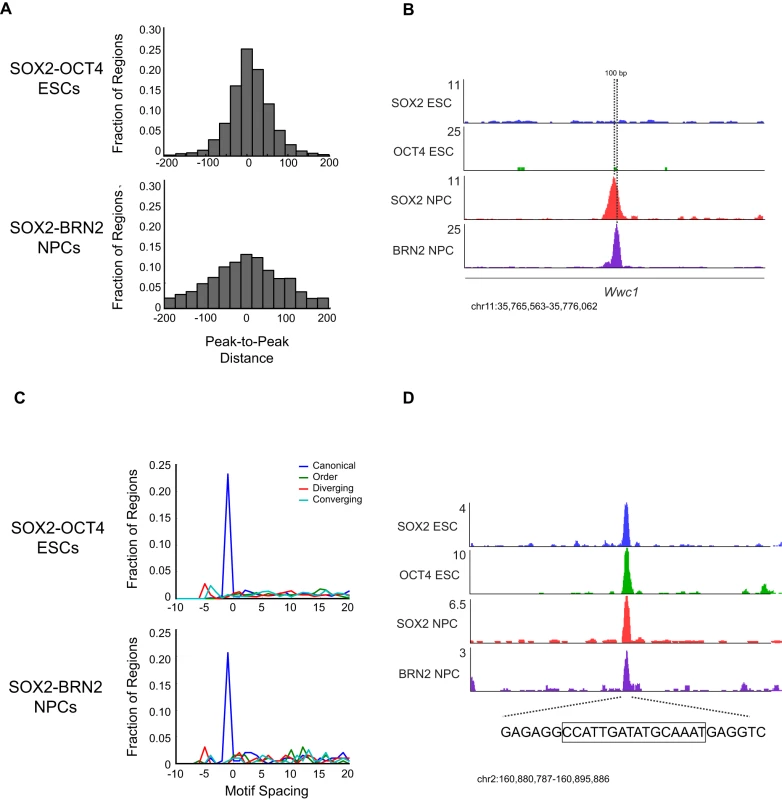
(A) Frequency distribution of distances in 25 base pair bins between peaks of OCT4 (top) and BRN2 (bottom) from SOX2 bound peaks. (B) Gene plots at the Wwc1 locus. Direction of transcription (5′-3′) is left to right. Hashed line represents position of peaks of SOX2 and BRN2 enrichment separated by 100 bp. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). (C) Distribution of orientation of and distance in base pairs between of SOX and POU motifs within SOX2-OCT4 (top), SOX2-BRN2 (bottom) bound regions. Negative values on x-axis reflect instances where SOX and OCT TRANSFAC motifs overlap. y-axis reflects fraction of occurrences of indicated spacing and orientation of all bound regions which contain a SOX and OCT motif. (D) Gene plots 3′ of the Chd6 locus, which contains a SOX-OCT motif in the canonical orientation with a −1 bp spacer. Hashed line represents sequence under the peaks of enrichment, and boxed sequence represents the canonical SOX-OCT motif with a −1 bp spacer at this locus. y-axis corresponds to reads per million. Genomic positions reflect NCBI Mouse Genome Build 36 (mm8). Many known SOX2-OCT4 target sites comprise a composite SOX-Octamer (OCT) motif, consisting of a 5′-SOX motif followed by a 3′ OCT site [15], [88], [89]. Therefore, we further analyzed the configuration of the SOX2 and OCT motifs by directly inspecting the sequence within the co-occupied regions. SOX-OCT composite motifs can exist in several configurations that were previously termed “canonical”, “order”, “diverging”, and “converging” [86] (Figure 5C). Interestingly, these configurations were shown to determine which combinations of SOX and POU factors could co-occupy a given site. Surprisingly, we observed that the canonical orientation with a 1 bp overlap between the native TRANSFAC motifs was the most highly represented configuration in both SOX2-OCT4 co-bound regions in ESCs (∼23% of motif pairs) and SOX2 and BRN2 co-bound regions in NPCs (∼21% of motif pairs) (Figure 5C). For example, at a locus on chromosome 2 distal to Chd6, SOX2-OCT4 occupied a canonical motif with a 1 bp overlap in ESCs, and SOX2-BRN2 occupied the same site in NPCs (Figure 5D). Thus, SOX2-OCT4 and SOX2-BRN2 prefer the same composite SOX-OCT motif at genomic targets in ESCs and NPCs.
Distinct SOX-POU sites harbor recognition motifs for other transcription factor families
Combinatorial interactions among transcription factors are important for driving specific transcriptional responses [90]–[93]. In ESCs, SOX2 and OCT4 are known to co-occupy genomic sites with a cohort of other transcription factors, including NANOG, SALL4, and TCF7L1 [13]–[16], [94]–[96]. Thus, we sought to identify additional transcription factors that may interact with SOX2 and BRN2 in NPCs. To this end, we analyzed SOX2-BRN2 bound regions for enrichment of known transcription factor motifs (Table S10). To discover factors that may function specifically with SOX2 and BRN2 in NPCs, we contrasted these motifs with those that were enriched in SOX2-OCT4 co-bound regions. Notably, the enriched motifs in SOX2-BRN2 regions corresponded to transcription factors that were highly expressed in NPCs relative to ESCs (Monte Carlo analysis, p-value = 0.03, Materials and Methods) (Figure 6A). For example, NF-I motifs were highly enriched in SOX2-BRN2 regions in NPCs and family members such as NF-Ia, NF-Ib, and NF-Ix were expressed at significantly higher level in NPCs than ESCs (Table S10). NF-I factors have known roles in central nervous system formation and in NPC function [97]. Motifs associated with the RFX family were also enriched in SOX2-BRN2 regions (Table S10). RFX family members play essential roles in early nervous system patterning [98], [99]. While Rfx3, Rfx4, and Rfx7 were expressed at significantly higher levels in NPCs, Rfx2 expression was higher in ESCs (Table S10). Interestingly, a recent proteomic study identified RFX3 and NF-IB as putative SOX2 interaction partners in NPCs [30]. Thus, our analysis has identified additional transcription factors that may regulate specialized gene networks with SOX2 and POU factors in ESCs and NPCs.
Fig. 6. Additional transcription factor motifs are enriched in SOX2-POU-bound regions. 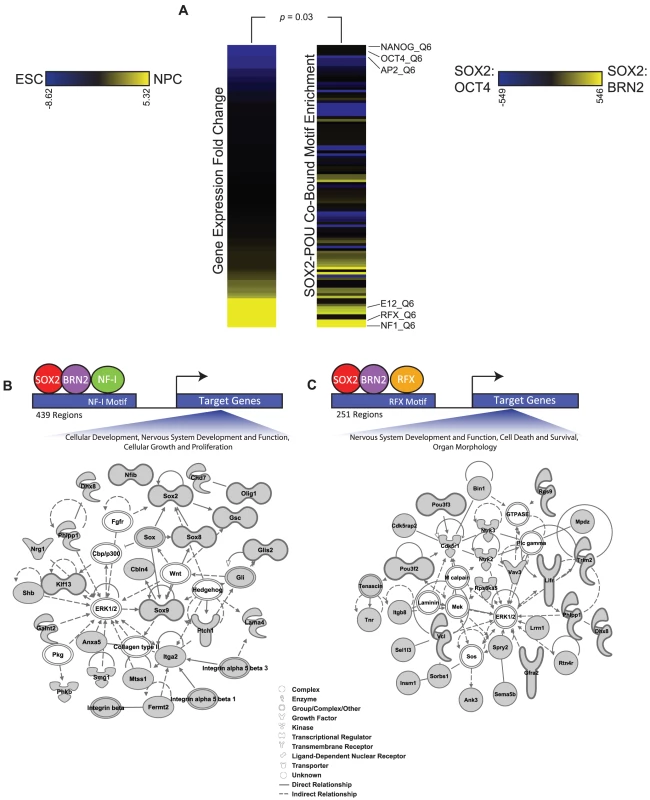
(A) Heatmaps display the relationship between expression changes in transcription factors and the enrichment of their motifs in SOX2-BRN2 bound regions compared to SOX2-OCT4-bound regions. The full set of TRANSFAC motifs were ranked by statistical significance of enrichment in SOX2-BRN2-bound regions and in SOX2-OCT4-bound regions using the Mann-Whitney Wilcoxon test. The heat map on the right displays the change in rank of 108 TRANSFAC motifs between the two datasets. Only motifs that were ranked in the top 200 in either dataset are shown. The heat map on the left shows the fold change in gene expression between ESCs and NPCs of a transcription factor that recognizes the corresponding motif on the right. Scale bars: Left, fold gene expression change of transcription factors between ESCs (blue) and NPCs (yellow); Right, change in rank of TRANSFAC motifs between SOX2-OCT4 bound regions (blue) and SOX2-BRN2 bound regions (yellow). p-value reflects correlation of motif enrichment and gene expression of transcription factors which can recognize the motifs by a Monte-Carlo analysis. (B, C) Ingenuity Pathway Analysis to visualize the functional interconnection among genes associated with SOX2-BRN2 regions that also contain either an NF-I or RFX motif. We identified 439 SOX2-BRN2-NF-I-motif and 251 SOX2-BRN2-RFX-motif regions in NPCs (see Materials and Methods). Further analysis showed that SOX2-BRN2 regions containing NF-I or RFX motifs were largely exclusive (only 34 common regions) suggesting that SOX2-BRN2 sites could be further classified by interactions with specific sets of transcription factors. Consistent with this observation, SOX2-BRN2 regions containing an NF-I motif were linked to genes with functions in nervous system development and cell growth, including Sox2 and NF-Ib themselves as well as Olig1 and Integrin genes (Figure 6B). In contrast, SOX2-BRN2-RFX-motif regions were linked to a largely distinct set of regulators of neural development including regulators of neuronal apoptosis such as Ntrk2 (TrkB) [100], Ntrk3 (TrkC) [101], and Cdk5r1 (p35) [102], [103], an important process regulating the development of the CNS (Figure 6C). Interestingly, conditional ablation of Sox2 in NPCs is associated with increased apoptosis in the developing brain [6]. Thus, RFX and NF-I family members represent additional candidate partner factors in NPCs that may further contribute to specific regulation at SOX2-BRN2 target genes. Collectively, our work reveals a detailed picture of how SOX2 coordinates gene expression programs during lineage commitment and provides novel insights into the key principles that underpin regulation of diverse stem cell states.
Discussion
The HMG-box transcription factor SOX2 has critical roles in the function of multiple stem cell types including pluripotent embryonic stem cells (ESCs) and multipotent neural progenitor cells (NPCs). How this master regulator can control diverse transcriptional programs has remained an important and unresolved question in the field. While SOX2 occupied many promoters in both cell types, the major class of genomic elements occupied by SOX2 in ESCs and NPCs were distal enhancers (Figure 1 and Figure 2). While our data displayed high concordance among replicates and with published data sets in ESCs, SOX2 binding in NPCs was less correlated with prior data [17], [30] (Figure S1). This is likely due to the different protocols used to derive and culture NPCs. NPCs with similar developmental potential but distinct molecular profiles exist throughout development [24], and these populations respond differently to external signaling cues present in culture media [104]–[106]. Thus, it is perhaps not surprising that SOX2 binding is more variable in NPCs relative to ESCs.
We derived NPCs directly from genetically identical ESCs allowing us to directly analyze SOX2 binding as these cells transition between states. Several criteria support the high quality of our data. First, we identified many known SOX2 binding sites including promoters and enhancers in both ESCs and NPCs. Second, while many binding sites were distinct, we identified a canonical SOX2 motif as highly enriched in both cell types. Third, SOX2 overlapped significantly with POU partner factors in ESCs and NPCs consistent with the expectation that these transcription factor families function together to regulate developmental progression. In addition, we identified a SOX-OCT composite motif as enriched in these co-bound sites.
SOX2 occupied largely exclusive sites in ESCs and NPCs, despite using the same DNA motif to recognize these genomic targets. Moreover, SOX2 occupied distinct regions in the same promoter and distinct enhancers associated with the same gene. These data indicated that additional factors dictated SOX2 binding site specificity. While SOX2 co-occupied many binding sites with OCT4 in ESCs, partner factors in NPCs have not been well defined. We found the recognition motif for the Class III POU factor BRN2 was enriched in SOX2 bound regions in NPCs. The evolutionary conservation of the SOX-POU interaction, the co-expression of Sox2 and Brn2 in neurogenic regions of the brain, and the neurodevelopmental defects associated with Brn2 loss-of-function suggested that SOX2 and BRN2 together regulate a subset of genes important for neural fate. Consistent with this, we defined a large group of enhancer elements co-bound by SOX2 and BRN2 in NPCs. We identified known functional enhancers bound by SOX2 and BRN2 in NPCs, such as the Nes30 enhancer of the Nestin locus [38], [40] and the 3′ enhancer of the Sox2 locus, SRR2 [5], and extended this list to include hundreds of additional neural-specific enhancers.
Consistent with a positive role in regulating neural cell state, forced expression of Brn2 led to up-regulation of neural markers and to differentiation toward the neural lineage. Our work is in agreement with several studies that have implicated Brn2 as an early marker of neural commitment [40], [107], [108]. Notably, ectopic BRN2 was sufficient to recruit SOX2 to hundreds of novel sites in differentiating ESCs that corresponded to a subset of enhancers also bound in NPCs. The recruitment of SOX2 by BRN2 to specific loci was sufficient to induce expression of nearby genes and to alter chromatin state in some cases. These data are in agreement with the notion that SOX proteins require partner factors to tightly bind to genomic targets and modulate transcriptional outcomes [59]. Interestingly, ectopic expression of OCT4 alone in NPCs was sufficient to reprogram cells into induced pluripotent stem cells, presumably by partnering with endogenous SOX2 [109], consistent with the idea that POU factors can recruit SOX2 to specific targets. Furthermore, ectopic expression of Sox2, Brn2, and the forkhead factor Foxg1 can transdifferentiate fibroblasts to NPC-like cells [70]. Taken together, these data may facilitate efforts to define the minimal set of genes needed to promote the transition from undifferentiated cells to the neural lineage. Thus, our results implicate BRN2 as a SOX2 partner factor and suggest that together these factors are important for neural specification and NPC function.
While the motifs occupied by these factors were highly similar, the arrangement of SOX and OCT motifs in SOX2-POU target sites displayed differences in ESCs and NPCs. Regulation of SOX-POU target genes appears to depend not only on the presence of a SOX and an OCT motif in close proximity to each other, but also on other DNA sequence determinants, including the spacing and orientation of these motifs with respect to each other [82], [84], [87], [110]–[113]. However, these observations related to only a few genes and had not been extended genome-wide. While we found that SOX2-OCT4 and SOX2-BRN2 preferred similar composite motifs when they were bound in close proximity to each other, examination of co-bound regions found that peaks of SOX2 and BRN2 in NPCs were often spaced farther apart than peaks of SOX2 and OCT4 in ESCs. Thus, allosteric interactions between transactivation domains of SOX and POU factors may be key in stabilizing ternary complexes and in setting the stage for additional interactions that determine binding specificity and transcriptional output at target genes [84]–[87], [114], [115].
Combinatorial interactions among transcription factors allow cells to respond to environmental and developmental cues in a tissue-specific manner. A classical example involves the regulation of interferon-β expression through cooperative binding of transcription factors and chromatin proteins to an enhancer, collectively known as the interferon-β enhancesome [116]. In ESCs, SOX2 and OCT4 are known to physically interact with other transcription factors at many loci, including enhancers [14], [94], [117]–[120], suggesting that SOX2-POU factors may also nucleate specific enhancesomes. We identified a set of candidate factors that may interact with SOX2-BRN2 that included RFX and NF-I family members. NF-I factors are expressed in NPCs in vivo and their loss in development leads to defects in central nervous system formation and specifically NPC dysfunction [97], [121]–[124]. RFX family members also play essential roles in proper brain development [99], [125]. For example, RFX4 regulates Sonic Hedgehog (SHH) signaling in the developing nervous system and loss of function resulted in pleiotropic brain defects linked to SHH signaling [99], [125]. Defects associated with conditional ablation of Sox2 in the brain were also shown to be partially mediated by aberrant SHH signaling [6]. Additional studies revealed that SOX2 co-localized with the ATP-dependent histone remodeler CHD7 in NPCs [30]. Thus, interactions with chromatin modifiers or other epigenetic regulators may also be critical for binding site selection and establishment of NPC-specific gene expression programs in response to particular signals.
Recent data showed that SOX2 functions as a pioneer factor in ESCs by marking a subset of genes for activation by other SOX family members, namely SOX4 in the B-cell lineage, SOX3 in NPCs and SOX11 in immature neurons [17], [18]. Interestingly, the POU factor Brn5, like Sox11, is expressed in differentiated cell types in the CNS and thought to play a role in regulating cell state [126]–[130], thus elucidation of BRN5 targets in these cells may reveal another layer of SOX-POU regulation of neurogenesis. Taken together, these data suggest that transitions in SOX-POU partners regulate the earliest stages of development through terminal differentiation. Ultimately, characterization of combinatorial interactions among transcription factors and chromatin regulators at distal enhancers will be central to understanding the complex mechanisms that control cell state throughout development.
Materials and Methods
Data deposition
ChIP-Seq and Affymetrix microarray data are deposited on GEO database under the accession numbers GSE38850 and GSE35496.
Cell growth and culture conditions
C57/BL6-129JAE (V6.5) mouse embryonic stem cells were cultured in as described [34]. Neural progenitors were derived via in vitro differentiation from V6.5 ESCs as described [43] and cultured on 15 µg/ml polyornithine and 1 µg/ml laminin in N3 medium, supplemented with 5 ng/ml bFGF, 20 ng/ml EGF, and 1 µg/ml laminin. In the presence of growth factors the vast majority of these cells can be labeled homogenously with antibodies against NESTIN, SOX2, and PAX6. Upon growth factor withdrawal, the cells differentiate into TUJ1-positive neurons.
Chromatin immunoprecipitation (ChIP) and library preparation
ChIP in NPCs was performed as described previously [131]. Briefly, approximately 5×108 cells were cross-linked and chromatin fractions were sheared by sonication. ChIP-enriched and input DNA were purified and genomic libraries were prepared using the ChIP-Seq Sample Prep Kit (Illumina 1003473) according to the manufacturers protocol (Illumina 11257047) for selecting library fragments between 200 and 350 bp. Samples were run using the GA2X genome sequencer (SCS v2.6, pipeline 1.5).
For ChIP in ESCs, and in TetO-Brn2 cells and control cells were cross-linked and harvested as above. Approximately 5×107 formaldhyde-crosslinked cells were lysed and as above on an IP-Star (Diagenode). Chromatin was sonicated on the Bioruptor (Diagenode) to an average size of 0.2–1 kb. ChIP was performed on chromatin from approximately 5 million cells with 3 µg of antibody (above) using the IP-Star Automated System (Diagenode) and 2.5% of chromatin was used for each whole cell extract (WCE). Following reversal of crosslinks, sample and WCE DNA was purified. ChIP and WCE DNA was dissolved in water and barcoded genomic libraries were prepared using the TruSeq DNA Sample Prep Kit (Illumina) and multiplexed on the HiSeq 2000 (Illumina).
ChIP antibodies
Antibodies used in ChIP experiments are as follows: SOX2 (R and D Systems AF2018 goat polyclonal); BRN2 (Santa Cruz Biotechnology sc-6029 goat polyclonal); H3 (rabbit polyclonal Abcam ab1791) H3K4me1 (rabbit polyclonal Abcam ab8895); H3K27Ac (rabbit polyclonal Abcam ab4729).
ChIP–seq analysis pipeline
Images acquired from the Illumina/Solexa sequencer were processed using the bundled Solexa image extraction pipeline. Sequences were aligned using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) using murine genome NCBI Build 36 (UCSC mm8) as the reference genome with default settings for mismatch tolerance and non-unique mapping events. Mapped reads were analyzed as described [16]. Briefly, sequence reads were extended 200 bp for transcription factors and 400 bp for histone modifications and allocated in 25 bp bins. Statistically significant enriched bins were identified using a Poissonian background model, with a p-value threshold of 10−8 to minimize false positives. We then used an empirical background model (whole cell extracts (WCE) for transcription factors or pan-histone histone H3 ChIP-Seq (H3) for chromatin marks) that requires bins to be enriched relative to background to eliminate non-random enrichment. Replicate datasets were combined and analyzed in one batch. Previously published datasets for enhancer associated histone marks were analyzed as described [34], [132]. SOX2, BRN2, and OCT4 enriched regions within 1 kb of a TSS were assigned to the associated gene, while bound enhancers were identified as regions that overlap H4K4me1 and/or H3K27Ac regions that are >1 kb from a TSS [34] and were assigned to the nearest gene using the GREAT algorithm for gene ontology studies and using the Galaxy web tool for all other analyses.
ChIP–enrichment plots
ChIP–seq plots for individual genes were generated using the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway).. wig files were generated from ChIP-Seq reads and density was normalized to reads-per-million. Published datasets were used to correlate SOX2 bound regions to histone modification patterns for enhancer analysis [34], [132].
Comparison of ChIP–seq datasets
We used a 1-bp minimum cutoff for the overlap between regions to define common genomic targets, as described throughout the manuscript to define co-bound SOX2-POU sites and sites occupied by SOX2 or POU factors across cell types. Correlation of ChIP-Seq datasets in figure S1 was performed using a similarity metric based on a correlation coefficient [133]. This analysis generates a correlation coefficient between zero and one reflecting the similarity of genomic regions occupied in two datasets.
RNA isolation and microarray analysis
RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's protocol and DNAse treated using the DNA-Free RNA kit (Zymo Research R1028). Samples were then prepared for Affymetrix GeneChip Expression Array analysis. 5 µg total RNA was used to prepare biotinylated cRNA according to the manufacturer's protocol (Affymetrix One Cycle cDNA Synthesis Kit). Samples were prepared for hybridization, hybridized to arrays, and washed according the Affymetrix hybridization manual using the Affymetrix GeneChip Hybridization, Wash and Stain Kit. GeneChip arrays (Mouse 430) were hybridized in a GeneChip Hybridization Oven at 45°C for 16 hours at 60 RPM. Arrays were scanned on a GeneChip Scanner 3000 and images were extracted and analyzed using GeneChip Operating Software v1.4.
To define expression levels of genes linked to bound promoters and enhancers, and to define fold change of expression levels of transcription factors linked to enriched TRANSFAC motifs between ESCs and NPCs, biological replicates were analyzed using the Affymetrix GCOS program and the mean intensity for each probe across three arrays was calculated. Maximum probe mean values for each gene were taken as gene expression levels. Box and Violin plots were constructed depicting median values as the center line, and bottom and top of the box representing the 25th and 75th percentiles, respectively. Whiskers depict+1.5*IQR (interquartile range) for top, −1.5*IQR for the bottom. To define differentially expressed genes, array data was RMA normalized using updated annotation from the BrainCDF the site and remapped from Ensembl Gene ID to Gene Name using Biomart table. For finding differentially expressed (DE) genes, the biological replicates were subjected to moderated welch test (MWT). Genes were called differentially expressed if the MWT FDR<0.05 and the fold change of the mean of the replicates was more than 1.5 fold up or down.
Gene ontology
Gene ontology analysis was performed using GOSTAT (http://gostat.wehi.edu.au/cgi-bin/goStat.pl) for genes linked to SOX2 bound promoters or the GREAT algorithm [134] (http://great.stanford.edu/) for regions associated with SOX2 bound enhancers. GOSTAT was performed using the mgi (mouse) GO annotation database for promoter-associated regions. Since GREAT analysis requires inputs in the mm9 genome build, lift-over of mm8 called regions was performed using the Galaxy web tool prior to input into GREAT (main.g2.bx.psu.edu/). In general, terminal “GO Biological Process” terms were presented in figures to maximize the specificity of the information presented. In some cases terminal terms contained few genes and were thus misleading, so more informative parent terms encompassing less specific but more relevant descriptions of biological processes are presented.
De novo motif enrichment analysis
MEME (meme.sdsc.edu/) [135] was used to find DNA sequences enriched in SOX2-, OCT4-, and BRN2-bound regions in ESCs and NPCs. Plus/minus 75 base pairs surrounding a subset of the highest peaks of enrichment for each factor (minimum peak height 100 for SOX2, 148 for OCT4, or 225 for BRN2) were input into MEME and motif logos were generated from obtained position weight matrices.
Generation of inducible Brn2 ESC lines
Brn2 inducible ESCs were generated using the “flp-in” system described previously [136]. Briefly, a single copy of a tetracycline inducible mouse Brn2 cDNA were flipped into the Col1a1 locus of KH2 ESCs harboring an M2-rtTA gene in the Rosa26 locus.
ESC differentiation
Inducible Brn2 and control (KH2) ESCs were passaged off feeders and cultured in ESC medium with 2 µg/ml Dox. Twenty-four hours after passage, cells were culture in N2B27 (without Vitamin A) media without LIF or serum for the duration of the experiment [137]. Gene expression for differentiation markers was assayed by quantitative Real-Time PCR at 24-hour intervals. For immunostaining, cells were fixed with 4% paraformaldehyde in PBS and stained with anti-Nestin (Developmental Hybridoma Bank) and DAPI.
Quantitative real-time PCR
Trizol-isolated RNA from three biologically independent samples was purified, DNAse treated (DNA free RNA Kit, Zymo Research) and reverse transcribed using a First Strand Synthesis Kit (Invtirogen). cDNA was analyzed in triplicate for each biological sample by quantitative PCR using an ABI Prism 7000 (Applied Biosystems) with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). All primers used in this study are listed in Table S11. Data were extracted from the linear range, and the standard curve method was used to obtain relative expression values. Technical replicates were averaged and then biological replicates were averaged. Statistical significance was determined using Graphpad Prism to perform an ANOVA with Bonferroni Correction for multiple testing.
Definition of SOX2-BRN2-bound H3K4me1 and H3K27Ac Regions in TetO-Brn2 cells
Regions of SOX2-BRN2 co-occupancy in TetO-Brn2 cells were defined as above. To define regions of differential chromatin state between TetO-Brn2 and control cells, we first compared H3K4me1 enrichment in these cells to define regions common to both cell types or unique to one or the other. Common regions were merged and treated as one enhancer if detected in both cell types. A similar analysis was performed for H3K27Ac enrichment. SOX2-BRN2 regions were then compared to regions of H3K4me1 and H3K27Ac in order to define SOX2-BRN2 binding events that resulted in changes in chromatin state between TetO-Brn2 cells and controls.
Identification of DNA binding motifs within SOX2-bound regions
100 base pair windows around the max peak of SOX2-bound regions (in ESCs and NPCs) regions were analyzed for the presence of overrepresented DNA binding motifs. Similarly, 150 base pair windows around the midpoints between the max peaks of SOX2 and OCT4 or BRN2 (in ESCs or NPCs, respectively) in co-bound regions were analyzed for the presence of overrepresented DNA binding motifs. We used a hypothesis-based approach to identify known protein-DNA recognition elements enriched in each dataset. The set of hypotheses are derived from all vertebrate position-specific scoring matrices (PSSMs) from TRANSFAC [138] filtered for sufficient information content (IC>8 total bits). As many of these motifs are redundant, we clustered them based on pairwise distance by KL-divergence of the PSSMs using Affinity Propagation. The TAMO programming environment [139] was used to store the PSSMs and score sequences. We used two approaches to identify overrepresented motifs. All motifs discussed in the paper were found by both methods except for M00145 in SOX2 bound sites in NPCs which was only found by the first approach described below.
In the first approach, we determined whether motifs were overrepresented in a foreground set of all bound regions (SOX2-bound or SOX2-POU co-bound, depending on the analysis) compared to a background set of randomly generated sequences which matched the GC content of the foreground using the Mann-Whitney-Wilcoxon (MWW) ranked sum test. For each independent motif test, sequences were ranked by the maximum motif score in each sequence (across all k-mers in the sequence for a motif of width k). This ranked list was used to compute the U statistic for the foreground set from which we computed a p-value and applied a Benjamini-Hochberg multiple hypothesis correction. Because many motifs in the databases are very similar to each other, we present the motif within each cluster with the most significant p-value.
In the second approach, we determined whether motifs were overrepresented in a foreground set of 1,000 randomly selected bound regions compared to a background set of randomly generated sequences matching the GC content of the foreground using THEME [140]. A β value of 0.7 and 5-fold cross-validation (CV) were used as THEME parameters. Statistical significance of the CV-error was calculated using randomization of 25 trials and multiple hypothesis corrected using the Benjamini-Hochberg procedure. As in the MWW tables, we present the motif within each cluster with the most significant p-value.
Genome-wide distances between SOX2 and cell-type-specific POU factors
Distances between SOX2 bound sites and cofactor bound sites in ESCs and NPCs were calculated as follows. Overlapping regions of SOX2 and POU factors were defined as regions with at least 1-bp of overlap. Peaks from these overlapping regions were then used to define distances between the bound factors. In particular, we calculated distances between SOX2-BRN2 site pairs (NPCs), and SOX2-OCT4 site pairs (ESCs). Site pairs were defined by matching each SOX2 bound site to the closest cofactor bound site within 200 bases. Distance was calculated as the cofactor chromosomal coordinate subtracted from the SOX2 chromosomal coordinate.
Analysis of spacing between motif matches
Spacing between SOX and OCT sites was determined using a motif-based approach to determine specific spatial arrangement of the motifs in SOX2-OCT4 (ESCs) and SOX2-BRN2 (NPCs) co-bound regions. Max motif scores were calculated as described above and normalized as in Equation 1. Motif matches to SOX were defined as normalized scores greater than 0.85 to a general SOX TRANSFAC matrix, M01308. Similarly, OCT family motif matches were defined as normalized scores greater than 0.85 to a general OCT TRANSFAC matrix, M00342. For each sequence i and motif j, a motif score sij:
was calculated. Spacing was defined as the number of base positions between the OCT4 and SOX2 motif matches relative to the SOX2 motif match. OCT-SOX motif pairs were associated with the previously defined “canonical”, “order”, “diverging”, and “converging” orientations [86].Gene expression/motif enrichment heat maps
The Mann-Whitney Z-score test result was used to rank all vertebrate TRANSFAC motifs in order of enrichment for SOX2 and OCT4 bound regions in ESCs, and SOX2 and BRN2 bound regions in NPCs. The change in rank (ΔRank) from ESC SOX2-OCT4 bound regions to SOX2-BRN2 bound regions was determined for each motif. Motifs were filtered to include only motifs with a rank less than or equal to 200 in the two ranked lists. Gene expression fold change was determined for each transcription factor associated with at least one TRANSFAC motif. After assigning a pseudocount of 1 to the normalized Affymetrix gene expression values for each transcription factor at the ESC and NPC stages, the log (base 2) of the fold change was calculated. Motifs were mapped to associated transcription factors according to the vertebrate all profile accessible on ExPlain 3.0 containing 656 motifs. The TF with the fold change that best agreed with the ΔRank of the associated motif was chosen as the ‘representative’ factor for the motif (for instance, if ΔRank was negative, the associated transcription factor that had the most negative log-transformed fold change was chosen.) Scaled motif ΔRank values and the associated log-transformed gene expression fold change values were sorted in order of log-transformed gene expression fold change, and viewed in a heatmap (Spotfire, TIBCO). Only motifs that had associated transcription factor expression values were considered.
Agreement between ΔRank of motif and expression fold-change of associated factor
A spearman correlation coefficient was calculated between the ΔRank values of the motifs and the expression-fold change of their associated transcription factors. This required each motif to be associated with a single transcription factor. In the case where multiple transcription factors are known to bind a single motif, the TF was selected as described in the previous section. The significance of the spearman correlation coefficient was assessed by a Monte-Carlo algorithm. The input to the Monte-Carlo algorithm was a table in which each row of the first column was a motif and each row of the second column was the set of transcription factors known to bind the motif. The column containing the motifs was randomly permuted 100,000 times (thereby randomizing the associations of transcription factors to motifs), and the process of selecting a single transcription factor to be associated with each motif was repeated. After associating each motif with a single random transcription factor, the spearman correlation between the ΔRank of the motifs and the log-fold-change in expression of the transcription factors was computed. The fraction of randomized tables that produced a higher spearman correlation than the original table was reported as the p-value. Only motifs for which the rank in either the SOX2-OCT4 list or the SOX2-BRN2 list was in the top 200 were used. The motifs also had to have at least one associated transcription factor for which gene expression data was available.
Target gene networks
Genomic intervals corresponding to enriched transcription factor binding motifs in SOX2-BRN2 bound regions were determined. The single nearest gene to a given region was determined using the GREAT algorithm. Genes associated with a motif having a motif similarity score of equal to or greater than 0.85 (439 NF-I motif regions and 251 RFX motif regions) were used to generate a non-redundant target gene list. This gene list was then used as the input for Ingenuity Pathway Analysis. Ingenuity recognized 431 NF-I associated genes and 249 RFX associated genes. Overlap of genomic regions containing motif sequences was performed using Galaxy.
Supporting Information
Zdroje
1. YoungRA (2011) Control of the Embryonic Stem Cell State. Cell 144 : 940–954.
2. HolmbergJ, PerlmannT (2012) Maintaining Differentiated Cellular Identity. Nature Reviews Genetics 13 : 429–439.
3. AvilionAA, NicolisSK, PevnyLH, PerezL, VivianN, et al. (2003) Multipotent Cell Lineages in Early Mouse Development Depend on Sox2 Function. Genes & Development 17 : 126.
4. MasuiS, NakatakeY, ToyookaY, ShimosatoD, YagiR, et al. (2007) Pluripotency Governed by Sox2 Via Regulation of Oct3/4 Expression in Mouse Embryonic Stem Cells. Nature Cell Biology 9 : 625–635.
5. MiyagiS, MasuiS, NiwaH, SaitoT, ShimazakiT, et al. (2008) Consequence of the Loss of Sox2 in the Developing Brain of the Mouse. FEBS Letters 582 : 2811–2815.
6. FavaroR, ValottaM, FerriALM, LatorreE, MarianiJ, et al. (2009) Hippocampal Development and Neural Stem Cell Maintenance Require Sox2-Dependent Regulation of Shh. Nature Neuroscience 12 : 1248–1256.
7. KamachiY, SockanathanS, LiuQ, BreitmanM, Lovell-BadgeR, et al. (1995) Involvement of Sox Proteins in Lens-Specific Activation of Crystallin Genes. The EMBO Journal 14 : 3510.
8. KiernanAE, PellingAL, LeungKKH, TangASP, BellDM, et al. (2005) Sox2 Is Required for Sensory Organ Development in the Mammalian Inner Ear. Nature 434 : 1031–1035.
9. OkuboT, PevnyLH, HoganBLM (2006) Sox2 Is Required for Development of Taste Bud Sensory Cells. Genes & Development 20 : 2654.
10. GontanC, de MunckA, VermeijM, GrosveldF, TibboelD, et al. (2008) Sox2 Is Important for Two Crucial Processes in Lung Development: Branching Morphogenesis and Epithelial Cell Differentiation. Developmental Biology 317 : 296–309.
11. DomyanET, FerrettiE, ThrockmortonK, MishinaY, NicolisSK, et al. (2011) Signaling through Bmp Receptors Promotes Respiratory Identity in the Foregut Via Repression of Sox2. Development 138 : 971–981.
12. ArnoldK, SarkarA, YramMA, PoloJM, BronsonR, et al. (2011) Sox2+ Adult Stem and Progenitor Cells Are Important for Tissue Regeneration and Survival of Mice. Cell Stem Cell 9 : 317–329.
13. BoyerLA, LeeTI, ColeMF, JohnstoneSE, LevineSS, et al. (2005) Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 122 : 947–956.
14. ChenX, XuH, YuanP, FangF, HussM, et al. (2008) Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell 133 : 1106–1117.
15. LohYH, WuQ, ChewJL, VegaVB, ZhangW, et al. (2006) The Oct4 and Nanog Transcription Network Regulates Pluripotency in Mouse Embryonic Stem Cells. Nature Genetics 38 : 431–440.
16. MarsonA, LevineSS, ColeMF, FramptonGM, BrambrinkT, et al. (2008) Connecting Microrna Genes to the Core Transcriptional Regulatory Circuitry of Embryonic Stem Cells. Cell 134 : 521–533.
17. BergslandM, RamsköldD, ZaouterC, KlumS, SandbergR, et al. (2011) Sequentially Acting Sox Transcription Factors in Neural Lineage Development. Genes & Development 25 : 2453–2464.
18. LiberD, DomaschenzR, HolmqvistPH, MazzarellaL, GeorgiouA, et al. (2010) Epigenetic Priming of a Pre-B Cell-Specific Enhancer through Binding of Sox2 and Foxd3 at the ESC Stage. Cell Stem Cell 7 : 114–126.
19. TakahashiK, TanabeK, OhnukiM, NaritaM, IchisakaT, et al. (2007) Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131 : 861–872.
20. TakahashiK, YamanakaS (2006) Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126 : 663–676.
21. WernigM, MeissnerA, ForemanR, BrambrinkT, KuM, et al. (2007) In Vitro Reprogramming of Fibroblasts into a Pluripotent Es-Cell-Like State. Nature 448 : 318–324.
22. BuganimY, FaddahDA, ChengAW, ItskovichE, MarkoulakiS, et al. (2012) Single-Cell Expression Analyses During Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell 150 : 1209–1222.
23. TempleS, Alvarez-BuyllaA (1999) Stem Cells in the Adult Mammalian Central Nervous System. Current Opinion in Neurobiology 9 : 135–141.
24. TempleS (2001) The Development of Neural Stem Cells. Nature 414 : 112–117.
25. SuhH, ConsiglioA, RayJ, SawaiT, D'AmourKA, et al. (2007) In Vivo Fate Analysis Reveals the Multipotent and Self-Renewal Capacities of Sox2+ Neural Stem Cells in the Adult Hippocampus. Cell Stem Cell 1 : 515–528.
26. BylundM, AnderssonE, NovitchBG, MuhrJ (2003) Vertebrate Neurogenesis Is Counteracted by Sox1–3 Activity. Nature Neuroscience 6 : 1162–1168.
27. GrahamV, KhudyakovJ, EllisP, PevnyL (2003) Sox2 Functions to Maintain Neural Progenitor Identity. Neuron 39 : 749–765.
28. Bani-YaghoubM, TremblayRG, LeiJX, ZhangD, ZurakowskiB, et al. (2006) Role of Sox2 in the Development of the Mouse Neocortex. Developmental Biology 295 : 52–66.
29. LeN, NagarajanR, WangJYT, ArakiT, SchmidtRE, et al. (2005) Analysis of Congenital Hypomyelinating Egr2lo/Lo Nerves Identifies Sox2 as an Inhibitor of Schwann Cell Differentiation and Myelination. Proc Natl Acad Sci USA 102 : 2596.
30. EngelenE, AkinciU, BryneJC, HouJ, GontanC, et al. (2011) Sox2 Cooperates with Chd7 to Regulate Genes That Are Mutated in Human Syndromes. Nature Genetics 43 : 607–611.
31. BueckerC, WysockaJ (2012) Enhancers as Information Integration Hubs in Development: Lessons from Genomics. Trends in Genetics 6 : 276–284.
32. BulgerM, GroudineM (2011) Functional and Mechanistic Diversity of Distal Transcription Enhancers. Cell 144 : 327–339.
33. OngCT, CorcesVG (2011) Enhancer Function: New Insights into the Regulation of Tissue-Specific Gene Expression. Nature Reviews Genetics 12 : 283–293.
34. CreyghtonMP, ChengAW, WelsteadGG, KooistraT, CareyBW, et al. (2010) Histone H3k27ac Separates Active from Poised Enhancers and Predicts Developmental State. Proc Natl Acad Sci USA 107 : 21931.
35. HeintzmanND, HonGC, HawkinsRD, KheradpourP, StarkA, et al. (2009) Histone Modifications at Human Enhancers Reflect Global Cell-Type-Specific Gene Expression. Nature 459 : 108–112.
36. HeintzmanND, StuartRK, HonG, FuY, ChingCW, et al. (2007) Distinct and Predictive Chromatin Signatures of Transcriptional Promoters and Enhancers in the Human Genome. Nature Genetics 39 : 311–318.
37. Rada-IglesiasA, BajpaiR, SwigutT, BrugmannSA, FlynnRA, et al. (2010) A Unique Chromatin Signature Uncovers Early Developmental Enhancers in Humans. Nature 470 : 279–283.
38. TanakaS, KamachiY, TanouchiA, HamadaH, JingN, et al. (2004) Interplay of Sox and Pou Factors in Regulation of the Nestin Gene in Neural Primordial Cells. Molecular and Cellular Biology 24 : 8834.
39. ChewJL, LohYH, ZhangW, ChenX, TamWL, et al. (2005) Reciprocal Transcriptional Regulation of Pou5f1 and Sox2 Via the Oct4/Sox2 Complex in Embryonic Stem Cells. Molecular and Cellular Biology 25 : 6031–6046.
40. JinZ, LiuL, BianW, ChenY, XuG, et al. (2009) Different Transcription Factors Regulate Nestin Gene Expression During P19 Cell Neural Differentiation and Central Nervous System Development. Journal of Biological Chemistry 284 : 8160–8173.
41. KondohH, KamachiY (2010) Sox-Partner Code for Cell Specification: Regulatory Target Selection and Underlying Molecular Mechanisms. The International Journal of Biochemistry and Cell Biology 42 : 391–399.
42. DaileyL, BasilicoC (2001) Coevolution of Hmg Domains and Homeodomains and the Generation of Transcriptional Regulation by Sox/Pou Complexes. Journal of Cellular Physiology 186 : 315–328.
43. OkabeS, Forsberg-NilssonK, SpiroAC, SegalM, McKayRDG (1996) Development of Neuronal Precursor Cells and Functional Postmitotic Neurons from Embryonic Stem Cells in Vitro. Mechanisms of Development 59 : 89–102.
44. MitsuiK, TokuzawaY, ItohH, SegawaK, MurakamiM, et al. (2003) The Homeoprotein Nanog Is Required for Maintenance of Pluripotency in Mouse Epiblast and Es Cells. Cell 113 : 631–642.
45. ChambersI, ColbyD, RobertsonM, NicholsJ, LeeS, et al. (2003) Functional Expression Cloning of Nanog, a Pluripotency Sustaining Factor in Embryonic Stem Cells. Cell 113 : 643–655.
46. Schneider-MaunouryS, TopilkoP, SeitandouT, LeviG, Cohen-TannoudjiM, et al. (1993) Disruption of Krox-20 Results in Alteration of Rhombomeres 3 and 5 in the Developing Hindbrain. Cell 75 : 1199–1214.
47. ShamMH, VesqueC, NonchevS, MarshallH, FrainM, et al. (1993) The Zinc Finger Gene Krox20 Regulates Hoxb2 (Hox2.8) During Hindbrain Segmentation. Cell 72 : 183–196.
48. SugoN, OshiroH, TakemuraM, KobayashiT, KohnoY, et al. (2010) Nucleocytoplasmic Translocation of Hdac9 Regulates Gene Expression and Dendritic Growth in Developing Cortical Neurons. European Journal of Neuroscience 31 : 1521–1532.
49. ShinJ, BossenzM, ChungY, MaH, ByronM, et al. (2010) Maternal Rnf12/Rlim Is Required for Imprinted X-Chromosome Inactivation in Mice. Nature 467 : 977–981.
50. OstendorffHP, TursunB, CornilsK, SchluterA, DrungA, et al. (2006) Dynamic Expression of Lim Cofactors in the Developing Mouse Neural Tube. Developmental Dynamics 235 : 786–791.
51. GrabowskiP (2011) Alternative Splicing Takes Shape During Neuronal Development. Current Opinion in Genetics & Development 4 : 388–94.
52. BoutzPL, StoilovP, LiQ, LinCH, ChawlaG, et al. (2007) A Post-Transcriptional Regulatory Switch in Polypyrimidine Tract-Binding Proteins Reprograms Alternative Splicing in Developing Neurons. Genes & Development 21 : 1636.
53. WamstadJA, AlexanderJM, TrutyRM, ShrikumarA, LiF, et al. (2012) Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell 151 : 206–220.
54. ErnstJ, KheradpourP, MikkelsenTS, ShoreshN, WardLD, et al. (2011) Mapping and Analysis of Chromatin State Dynamics in Nine Human Cell Types. Nature 473 : 43–49.
55. ZentnerGE, TesarPJ, ScacheriPC (2011) Epigenetic Signatures Distinguish Multiple Classes of Enhancers with Distinct Cellular Functions. Genome Research 21 : 1273–1283.
56. JiangJ, ChanYS, LohYH, CaiJ, TongGQ, et al. (2008) A Core Klf Circuitry Regulates Self-Renewal of Embryonic Stem Cells. Nature Cell Biology 10 : 353–360.
57. TzatzalosE, SmithSM, DohST, HaoH, LiY, et al. (2012) A Cis-Element in the Notch1 Locus Is Involved in the Regulation of Gene Expression in Interneuron Progenitors. Developmental Biology 372 (2)
217–28.
58. TaranovaOV, MagnessST, FaganBM, WuY, SurzenkoN, et al. (2006) Sox2 Is a Dose-Dependent Regulator of Retinal Neural Progenitor Competence. Genes & Development 20 : 1187–1202.
59. KondohH, KamachiY (2010) Sox-Partner Code for Cell Specification: Regulatory Target Selection and Underlying Molecular Mechanisms. The International Journal of Biochemistry & Cell Biology 42 : 391–399.
60. YuanH, CorbiN, BasilicoC, DaileyL (1995) Developmental-Specific Activity of the Fgf-4 Enhancer Requires the Synergistic Action of Sox2 and Oct-3. Genes & Development 9 : 2635.
61. CollignonJ, SockanathanS, HackerA, Cohen-TannoudjiM, NorrisD, et al. (1996) A Comparison of the Properties of Sox-3 with Sry and Two Related Genes, Sox-1 and Sox-2. Development 122 : 509.
62. KamachiY, CheahKSE, KondohH (1999) Mechanism of Regulatory Target Selection by the Sox High-Mobility-Group Domain Proteins as Revealed by Comparison of Sox1/2/3 and Sox9. Molecular and Cellular Biology 19 : 107.
63. Salmon-DivonM, DvingeH, TammojaK, BertoneP (2010) Peakanalyzer: Genome-Wide Annotation of Chromatin Binding and Modification Loci. BMC Bioinformatics 11 : 415.
64. CatenaR, TiveronC, RonchiA, PortaS, FerriA, et al. (2004) Conserved Pou Binding DNA Sites in the Sox2 Upstream Enhancer Regulate Gene Expression in Embryonic and Neural Stem Cells. Journal of Biological Chemistry 279 : 41846–41857.
65. MiyagiS, NishimotoM, SaitoT, NinomiyaM, SawamotoK, et al. (2006) The Sox2 Regulatory Region 2 Functions as a Neural Stem Cell-Specific Enhancer in the Telencephalon. Journal of Biological Chemistry 281 : 13374–13381.
66. HeX, TreacyMN, SimmonsDM, IngrahamHA, SwansonLW, et al. (1989) Expression of a Large Family of Pou-Domain Regulatory Genes in Mammalian Brain Development. Nature 340 : 35–41.
67. HaraY, RovescalliAC, KimY, NirenbergM (1992) Structure and Evolution of Four Pou Domain Genes Expressed in Mouse Brain. Proc Natl Acad Sci USA 89 : 3280.
68. McEvillyRJ, de DiazMO, SchonemannMD, HooshmandF, RosenfeldMG (2002) Transcriptional Regulation of Cortical Neuron Migration by Pou Domain Factors. Science 295 : 1528.
69. SugitaniY, NakaiS, MinowaO, NishiM, JishageK, et al. (2002) Brn-1 and Brn-2 Share Crucial Roles in the Production and Positioning of Mouse Neocortical Neurons. Genes & Development 16 : 1760.
70. LujanE, ChandaS, AhleniusH, SüdhofTC, WernigM (2012) Direct Conversion of Mouse Fibroblasts to Self-Renewing, Tripotent Neural Precursor Cells. Proc Natl Acad Sci USA 109 : 2527–2532.
71. StaudtLM, SinghH, SenR, WirthT, SharpPA, et al. (1986) A Lymphoid-Specific Protein Binding to the Octamer Motif of Immunoglobulin Genes. Nature 323 : 640–643.
72. PhillipsK, LuisiB (2000) The Virtuoso of Versatility: Pou Proteins That Flex to Fit1. Journal of Molecular Biology 302 : 1023–1039.
73. KleinjanDA, SeawrightA, ChildsAJ, van HeyningenV (2004) Conserved Elements in Pax6 Intron 7 Involved in (Auto)Regulation and Alternative Transcription. Developmental Biology 265 : 462–477.
74. FriedliM, BardeI, ArcangeliM, VerpS, QuazzolaA, et al. (2010) A Systematic Enhancer Screen Using Lentivector Transgenesis Identifies Conserved and Non-Conserved Functional Elements at the Olig1 and Olig2 Locus. PLoS ONE 5: e15741 doi:10.1371/journal.pone.0015741.
75. Verma-KurvariS, SavageT, SmithD, JohnsonJE (1998) Multiple Elements Regulate Mash1 Expression in the Developing Cns. Developmental Biology 197 : 106–116.
76. RoseMF, AhmadKA, ThallerC, ZoghbiHY (2009) Excitatory Neurons of the Proprioceptive, Interoceptive, and Arousal Hindbrain Networks Share a Developmental Requirement for Math1. Proc Natl Acad Sci USA 106 : 22462–22467.
77. LaiHC, KlischTJ, RobertsR, ZoghbiHY, JohnsonJE (2011) In Vivo Neuronal Subtype-Specific Targets of Atoh1 (Math1) in Dorsal Spinal Cord. The Journal of Neuroscience 31 : 10859–10871.
78. HondaT, KobayashiK, MikoshibaK, NakajimaK (2011) Regulation of Cortical Neuron Migration by the Reelin Signaling Pathway. Neurochemical Research 36 : 1270–1279.
79. AndreaeLC, PeukertD, LumsdenA, GilthorpeJD (2007) Analysis of Lrrn1 Expression and Its Relationship to Neuromeric Boundaries During Chick Neural Development. Neural Development 2 : 22.
80. TaguchiA, WanakaA, MoriT, MatsumotoK, ImaiY, et al. (1996) Molecular Cloning of Novel Leucine-Rich Repeat Proteins and Their Expression in the Developing Mouse Nervous System. Brain Res Mol Brain Res 35 : 31–40.
81. SanoY, Syuzo-TakabatakeA, NakayaT, SaitoY, TomitaS, et al. (2006) Enhanced Amyloidogenic Metabolism of the Amyloid Beta-Protein Precursor in the X11l-Deficient Mouse Brain. Journal of Biological Chemistry 281 : 37853–37860.
82. BotquinV, HessH, FuhrmannG, AnastassiadisC, GrossMK, et al. (1998) New Pou Dimer Configuration Mediates Antagonistic Control of an Osteopontin Preimplantation Enhancer by Oct-4 and Sox-2. Genes & Development 12 : 2073–2090.
83. NishimotoM, FukushimaA, OkudaA, MuramatsuM (1999) The Gene for the Embryonic Stem Cell Coactivator Utf1 Carries a Regulatory Element Which Selectively Interacts with a Complex Composed of Oct-3/4 and Sox-2. Molecular and Cellular Biology 19 : 5453–5465.
84. AmbrosettiDC, SchölerHR, DaileyL, BasilicoC (2000) Modulation of the Activity of Multiple Transcriptional Activation Domains by the DNA Binding Domains Mediates the Synergistic Action of Sox2 and Oct-3 on the Fibroblast Growth Factor-4 enhancer. Journal of Biological Chemistry 275 : 23387.
85. ReményiA, LinsK, NissenLJ, ReinboldR, SchölerHR, et al. (2003) Crystal Structure of a Pou/Hmg/DNA Ternary Complex Suggests Differential Assembly of Oct4 and Sox2 on Two Enhancers. Genes & Development 17 : 2048–2059.
86. JauchR, AksoyI, HutchinsAP, NgCKL, TianXF, et al. (2011) Conversion of Sox17 into a Pluripotency Reprogramming Factor by Reengineering Its Association with Oct4 on DNA. Stem Cells 29 : 940–951.
87. AmbrosettiDC, BasilicoC, DaileyL (1997) Synergistic Activation of the Fibroblast Growth Factor 4 Enhancer by Sox2 and Oct-3 Depends on Protein-Protein Interactions Facilitated by a Specific Spatial Arrangement of Factor Binding Sites. Molecular and Cellular Biology 17 : 6321.
88. FerrarisL, StewartAP, KangJ, DeSimoneAM, GemberlingM, et al. (2011) Combinatorial Binding of Transcription Factors in the Pluripotency Control Regions of the Genome. Genome Research 21 : 1055–1064.
89. NgCKL, LiNX, CheeS, PrabhakarS, KolatkarPR, et al. (2012) Deciphering the Sox-Oct Partner Code by Quantitative Cooperativity Measurements. Nucleic Acids Research
90. RemenyiA, ScholerHR, WilmannsM (2004) Combinatorial Control of Gene Expression. Nature Structural and Molecular Biology 11 : 812–815.
91. Rochette-EglyC, GermainP (2009) Dynamic and Combinatorial Control of Gene Expression by Nuclear Retinoic Acid Receptors (RARs). Nuclear Receptor Signalling 7: e005.
92. SundrudMS, NolanMA (2010) Synergistic and Combinatorial Control of T Cell Activation and Differentiation by Transcription Factors. Curr Opin Immunol 22 : 286–292.
93. MichelD (2010) How Transcription Factors Can Adjust the Gene Expression Floodgates. Progress in Biophysics and Molecular Biology 102 : 16–37.
94. KimJ, ChuJ, ShenX, WangJ, OrkinSH (2008) An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. Cell 132 : 1049–1061.
95. YangJ, ChaiL, FowlesTC, AlipioZ, XuD, et al. (2008) Genome-Wide Analysis Reveals Sall4 to Be a Major Regulator of Pluripotency in Murine-Embryonic Stem Cells. Proc Natl Acad Sci USA 105 : 19756–19761.
96. ColeMF, JohnstoneSE, NewmanJJ, KageyMH, YoungRA (2008) Tcf3 Is an Integral Component of the Core Regulatory Circuitry of Embryonic Stem Cells. Genes & Development 22 : 746.
97. das NevesL, DuchalaCS, Tolentino-SilvaF, HaxhiuMA, ColmenaresC, et al. (1999) Disruption of the Murine Nuclear Factor I-a Gene (Nfia) Results in Perinatal Lethality, Hydrocephalus, and Agenesis of the Corpus Callosum. Proc Natl Acad Sci USA 96 : 11946–11951.
98. BaasD, MeinielA, BenadibaC, BonnafeE, MeinielO, et al. (2006) A Deficiency in Rfx3 Causes Hydrocephalus Associated with Abnormal Differentiation of Ependymal Cells. Europena Journal Neuroscience 24 : 1020–1030.
99. AshiqueAM, ChoeY, KarlenM, MaySR, PhamluongK, et al. (2009) The Rfx4 Transcription Factor Modulates Shh Signaling by Regional Control of Ciliogenesis. Science Signalling 2: ra70.
100. AlcantaraS, FrisenJ, del RioJA, SorianoE, BarbacidM, et al. (1997) Trkb Signaling Is Required for Postnatal Survival of Cns Neurons and Protects Hippocampal and Motor Neurons from Axotomy-Induced Cell Death. The Journal of Neuroscience 17 : 3623–3633.
101. PostigoA, CalellaAM, FritzschB, KnipperM, KatzD, et al. (2002) Distinct Requirements for Trkb and Trkc Signaling in Target Innervation by Sensory Neurons. Genes & Development 16 : 633–645.
102. O'HareMJ, KushwahaN, ZhangY, AleyasinH, CallaghanSM, et al. (2005) Differential Roles of Nuclear and Cytoplasmic Cyclin-Dependent Kinase 5 in Apoptotic and Excitotoxic Neuronal Death. The Journal of Neuroscience 25 : 8954–8966.
103. RochetJC (2007) Novel Therapeutic Strategies for the Treatment of Protein-Misfolding Diseases. Expert Rev Mol Med 9 : 1–34.
104. GrittiA, ParatiE, CovaL, FrolichsthalP, GalliR, et al. (1996) Multipotential Stem Cells from the Adult Mouse Brain Proliferate and Self-Renew in Response to Basic Fibroblast Growth Factor. The Journal of Neuroscience 16 : 1091–1100.
105. TropepeV, SibiliaM, CirunaBG, RossantJ, WagnerEF, et al. (1999) Distinct Neural Stem Cells Proliferate in Response to Egf and Fgf in the Developing Mouse Telencephalon. Developmental Biology 208 : 166–188.
106. QianX, ShenQ, GoderieSK, HeW, CapelaA, et al. (2000) A Programmed Sequence of Neuron and Glial Cell Production from Isolated Murine Cortical Stem Cells. Neuron 28 : 69–80.
107. YasuharaN, ShibazakiN, TanakaS, NagaiM, KamikawaY, et al. (2006) Triggering Neural Differentiation of ES Cells by Subtype Switching of Importin-A. Nature Cell Biology 9 : 72–79.
108. Iwafuchi-DoiM, YoshidaY, OnichtchoukD, LeichsenringM, DrieverW, et al. (2010) The Pou5f1/Pou3f-Dependent but SoxB-Independent Regulation of Conserved Enhancer N2 Initiates Sox2 Expression During Epiblast to Neural Plate Stages in Vertebrates. Developmental Biology 352 (2)
354–66.
109. KimJB, SebastianoV, WuG, Araúzo-BravoMJ, SasseP, et al. (2009) Oct4-Induced Pluripotency in Adult Neural Stem Cells. Cell 136 : 411–419.
110. KuhlbrodtK, HerbarthB, SockE, EnderichJ, Hermans-BorgmeyerI, et al. (1998) Cooperative Function of Pou Proteins and Sox Proteins in Glial Cells. Journal of Biological Chemistry 273 : 16050.
111. GhislainJ, CharnayP (2006) Control of Myelination in Schwann Cells: A Krox20 Cis-Regulatory Element Integrates Oct6, Brn2 and Sox10 Activities. EMBO Rep 7 : 52–58.
112. NakatakeY, FukuiN, IwamatsuY, MasuiS, TakahashiK, et al. (2006) Klf4 Cooperates with Oct3/4 and Sox2 to Activate the Lefty1 Core Promoter in Embryonic Stem Cells. Molecular and Cellular Biology 26 : 7772–7782.
113. ReiprichS, KrieschJ, SchreinerS, WegnerM (2010) Activation of Krox20 Gene Expression by Sox10 in Myelinating Schwann Cells. Journal of Neurochemistry 112 : 744–754.
114. NowlingTK, JohnsonLR, WiebeMS, RizzinoA (2000) Identification of the Transactivation Domain of the Transcription Factor Sox-2 and an Associated Co-Activator. Journal of Biological Chemistry 275 : 3810–3818.
115. WilliamsDCJr, CaiM, CloreGM (2004) Molecular Basis for Synergistic Transcriptional Activation by Oct1 and Sox2 Revealed from the Solution Structure of the 42-Kda Oct1· Sox2· Hoxb1-DNA Ternary Transcription Factor Complex. Journal of Biological Chemistry 279 : 1449–1457.
116. Maniatis T, Falvo J, Kim T, Kim T, Lin C, et al.. Structure and Function of the Interferon-B Enhanceosome; 1998. Cold Spring Harbor Laboratory Press. pp. 609–620.
117. WangJ, RaoS, ChuJ, ShenX, LevasseurDN, et al. (2006) A Protein Interaction Network for Pluripotency of Embryonic Stem Cells. Nature 444 : 364–368.
118. LiangJ, WanM, ZhangY, GuP, XinH, et al. (2008) Nanog and Oct4 Associate with Unique Transcriptional Repression Complexes in Embryonic Stem Cells. Nature Cell Biology 10 : 731–739.
119. Van Den BergDLC, SnoekT, MullinNP, YatesA, BezstarostiK, et al. (2010) An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell 6 : 369–381.
120. ChambersI, TomlinsonSR (2009) The Transcriptional Foundation of Pluripotency. Development 136 : 2311–2322.
121. Steele-PerkinsG, PlachezC, ButzKG, YangG, BachurskiCJ, et al. (2005) The Transcription Factor Gene Nfib Is Essential for Both Lung Maturation and Brain Development. Molecular and Cellular Biology 25 : 685–698.
122. PiperM, BarryG, HawkinsJ, MasonS, LindwallC, et al. (2010) NfIA Controls Telencephalic Progenitor Cell Differentiation through Repression of the Notch Effector Hes1. The Journal of Neuroscience 30 : 9127–9139.
123. CampbellCE, PiperM, PlachezC, YehYT, BaizerJS, et al. (2008) The Transcription Factor NfIX Is Essential for Normal Brain Development. BMC Developmental Biology 8 : 52.
124. PlachezC, LindwallC, SunnN, PiperM, MoldrichRX, et al. (2008) Nuclear Factor I Gene Expression in the Developing Forebrain. Journal of Computational Neurology 508 : 385–401.
125. BlackshearPJ, GravesJP, StumpoDJ, CobosI, RubensteinJLR, et al. (2003) Graded Phenotypic Response to Partial and Complete Deficiency of a Brain-Specific Transcript Variant of the Winged Helix Transcription Factor Rfx4. Development 130 : 4539–4552.
126. OkamotoK, WakamiyaM, NojiS, KoyamaE, TaniguchiS, et al. (1993) A Novel Class of Murine Pou Gene Predominantly Expressed in Central Nervous System. Journal of Biological Chemistry 268 : 7449–7457.
127. AndersenB, SchonemannM, PearseRV, JenneK, SugarmanJ, et al. (1993) Brn-5 Is a Divergent Pou Domain Factor Highly Expressed in Layer Iv of the Neocortex. Journal of Biological Chemistry 268 : 23390.
128. CuiH, BulleitRF (1997) Expression of the Pou Transcription Factor Brn-5 Inhibits Proliferation of Ng108-15 Cells. Biochemical and Biophysical Research Communications 236 : 693–696.
129. CuiH, BulleitRF (1998) Expression of the POU Transcription Factor Brn‐5 Is an Early Event in the Terminal Differentiation of CNS Neurons. Journal of Neuroscience Research 52 : 625–632.
130. WuR, JurekM, SundarababuS, WeinsteinDE (2001) The Pou Gene Brn-5 Is Induced by Neuregulin and Is Restricted to Myelinating Schwann Cells. Molecular and Cellular Neuroscience 17 : 683–695.
131. LeeTI, JohnstoneSE, YoungRA (2006) Chromatin Immunoprecipitation and Microarray-Based Analysis of Protein Location. Nature Protocols 1 : 729–748.
132. MikkelsenTS, KuM, JaffeDB, IssacB, LiebermanE, et al. (2007) Genome-Wide Maps of Chromatin State in Pluripotent and Lineage-Committed Cells. Nature 448 : 553–560.
133. BilodeauS, KageyMH, FramptonGM, RahlPB, YoungRA (2009) Setdb1 Contributes to Repression of Genes Encoding Developmental Regulators and Maintenance of ES Cell State. Genes & Development 23 : 2484.
134. McLeanCY, BristorD, HillerM, ClarkeSL, SchaarBT, et al. (2010) Great Improves Functional Interpretation of Cis-Regulatory Regions. Nature Biotechnology 28 : 495–501.
135. BaileyTL, ElkanC (1994) Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc Int Conf Intell Syst Mol Biol 2 : 28–36.
136. BeardC, HochedlingerK, PlathK, WutzA, JaenischR (2006) Efficient Method to Generate Single-Copy Transgenic Mice by Site-Specific Integration in Embryonic Stem Cells. Genesis 44 : 23–28.
137. YingQL, StavridisM, GriffithsD, LiM, SmithA (2003) Conversion of Embryonic Stem Cells into Neuroectodermal Precursors in Adherent Monoculture. Nature Biotechnology 21 : 183–186.
138. WingenderE, DietzeP, KarasH, KnuppelR (1996) Transfac: A Database on Transcription Factors and Their DNA Binding Sites. Nucleic Acids Research 24 : 238–241.
139. GordonDB, NekludovaL, McCallumS, FraenkelE (2005) Tamo: A Flexible, Object-Oriented Framework for Analyzing Transcriptional Regulation Using DNA-Sequence Motifs. Bioinformatics 21 : 3164–3165.
140. MacisaacKD, GordonDB, NekludovaL, OdomDT, SchreiberJ, et al. (2006) A Hypothesis-Based Approach for Identifying the Binding Specificity of Regulatory Proteins from Chromatin Immunoprecipitation Data. Bioinformatics 22 : 423–429.
141. HochedlingerK, YamadaY, BeardC, JaenischR (2005) Ectopic Expression of Oct-4 Blocks Progenitor-Cell Differentiation and Causes Dysplasia in Epithelial Tissues. Cell 121 : 465–477.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




