-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
The formation of leaf vein patterns has fascinated biologists for centuries. Transport of the plant signal auxin has long been implicated in vein patterning, but molecular details have remained unclear. Varied evidence suggests a central role for the plasma-membrane (PM)-localized PIN-FORMED1 (PIN1) intercellular auxin transporter of Arabidopsis thaliana in auxin-transport-dependent vein patterning. However, in contrast to the severe vein-pattern defects induced by auxin transport inhibitors, pin1 mutant leaves have only mild vein-pattern defects. These defects have been interpreted as evidence of redundancy between PIN1 and the other four PM-localized PIN proteins in vein patterning, redundancy that underlies many developmental processes. By contrast, we show here that vein patterning in the Arabidopsis leaf is controlled by two distinct and convergent auxin-transport pathways: intercellular auxin transport mediated by PM-localized PIN1 and intracellular auxin transport mediated by the evolutionarily older, endoplasmic-reticulum-localized PIN6, PIN8, and PIN5. PIN6 and PIN8 are expressed, as PIN1 and PIN5, at sites of vein formation. pin6 synthetically enhances pin1 vein-pattern defects, and pin8 quantitatively enhances pin1pin6 vein-pattern defects. Function of PIN6 is necessary, redundantly with that of PIN8, and sufficient to control auxin response levels, PIN1 expression, and vein network formation; and the vein pattern defects induced by ectopic PIN6 expression are mimicked by ectopic PIN8 expression. Finally, vein patterning functions of PIN6 and PIN8 are antagonized by PIN5 function. Our data define a new level of control of vein patterning, one with repercussions on other patterning processes in the plant, and suggest a mechanism to select cell files specialized for vascular function that predates evolution of PM-localized PIN proteins.
Published in the journal: . PLoS Genet 9(2): e32767. doi:10.1371/journal.pgen.1003294
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003294Summary
The formation of leaf vein patterns has fascinated biologists for centuries. Transport of the plant signal auxin has long been implicated in vein patterning, but molecular details have remained unclear. Varied evidence suggests a central role for the plasma-membrane (PM)-localized PIN-FORMED1 (PIN1) intercellular auxin transporter of Arabidopsis thaliana in auxin-transport-dependent vein patterning. However, in contrast to the severe vein-pattern defects induced by auxin transport inhibitors, pin1 mutant leaves have only mild vein-pattern defects. These defects have been interpreted as evidence of redundancy between PIN1 and the other four PM-localized PIN proteins in vein patterning, redundancy that underlies many developmental processes. By contrast, we show here that vein patterning in the Arabidopsis leaf is controlled by two distinct and convergent auxin-transport pathways: intercellular auxin transport mediated by PM-localized PIN1 and intracellular auxin transport mediated by the evolutionarily older, endoplasmic-reticulum-localized PIN6, PIN8, and PIN5. PIN6 and PIN8 are expressed, as PIN1 and PIN5, at sites of vein formation. pin6 synthetically enhances pin1 vein-pattern defects, and pin8 quantitatively enhances pin1pin6 vein-pattern defects. Function of PIN6 is necessary, redundantly with that of PIN8, and sufficient to control auxin response levels, PIN1 expression, and vein network formation; and the vein pattern defects induced by ectopic PIN6 expression are mimicked by ectopic PIN8 expression. Finally, vein patterning functions of PIN6 and PIN8 are antagonized by PIN5 function. Our data define a new level of control of vein patterning, one with repercussions on other patterning processes in the plant, and suggest a mechanism to select cell files specialized for vascular function that predates evolution of PM-localized PIN proteins.
Introduction
Branched structures pervade all levels of organization in living organisms, from molecules to organelles, cells, multicellular organs and entire organisms, and the principles that guide the formation of these branched structures have long been object of interest of biologists and mathematicians. However, few branched structures have historically captured more widespread attention than the vein networks of plant leaves. From a developmental standpoint, such attention seems justified as files of vein precursor cells are selected from within a population of seemingly identical cells [1], [2]. Furthermore, in most species the product of this patterning process is both reproducible and variable: reproducible as vein networks supply all areas of the leaf; variable as the exact sites of vein formation are unpredictable [3]. These observations argue against rigid specification of leaf vein patterns and suggest a self-organizing control mechanism that reconciles the plasticity of vein formation with the stringent requirement for organ vascularization [4].
Though the identity of the molecules involved is largely unknown, varied evidence supports a decisive role for the polar, cell-to-cell transport of the plant signal auxin in leaf vein patterning: auxin application induces formation of new veins oriented towards pre-existing veins [3]; the inductive and orienting effect of applied auxin on vein formation is suppressed by auxin transport inhibitors [5]; and auxin transport inhibitors induce characteristic defects in vein patterns [6], [7]. During leaf development, expression of the plasma-membrane (PM)-localized PIN-FORMED1 (PIN1) auxin transporter of Arabidopsis thaliana [8], [9] is initiated in broad tissue domains and becomes gradually restricted to single files of vascular precursor cells [10], [11]. Narrowing of PIN1 expression domains is associated with polarization of PIN1 subcellular localization to the presumed auxin-efflux side of PIN1-expressing cells. Auxin levels define the initial size of PIN1 expression domains, and both domain narrowing and PIN1 polarization are obstructed by auxin transport inhibitors. These observations agree with conceptual formulation [12], and mathematical modeling (reviewed in [14]–[18]) of progressive restriction of nondirectional auxin transport across tissues to polar transport in single files of vascular precursor cells by positive feedback between auxin flow through a cell and the cell's auxin conductivity. However, in contrast to the severe vein-pattern defects induced by auxin transport inhibitors [6], [7], vein pattern defects in pin1 mutant leaves are mild [6], [19], [20]. PIN1 is member of a family comprising four other PM-localized PIN proteins and three, evolutionary older, endoplasmic reticulum (ER)-localized PIN proteins [21]–[24]. Thus, the mild vein-pattern defects of pin1 have been attributed to redundancy among PM-localized PIN proteins (e.g., [6], [10], [19], [25]), redundancy that has been shown to underlie, to different extents, many other developmental processes ([20], [26], [27] and references therein).
Here we show that the vein network of the Arabidopsis leaf is patterned by two distinct and convergent auxin transport pathways: intercellular auxin transport mediated by PM-localized PIN1 and intracellular auxin transport mediated by ER-localized PIN6, PIN8 and PIN5. Our results suggest an ancestral auxin-transport-dependent mechanism to specify cell files to vascular function that predates evolution of PM-localized PIN proteins.
Results
Vein patterning functions of Arabidopsis PIN genes
WT Arabidopsis grown under normal conditions forms separate leaves, whose vein patterns are defined by reproducible features [28], [29] (Figure 1A and 1B): a narrow, central midvein that runs the length of the leaf; lateral veins that branch from the midvein and join distal veins to form closed loops; minor veins that branch from midvein and loops and either end freely or join other veins; and minor veins and loops that curve near the leaf margin, lending a scalloped outline to the vein network. By contrast, WT leaves developed in the presence of auxin transport inhibitors often show separation defects (‘fused leaves’), and vein patterns of auxin-transport-inhibited leaves deviate from the norm in at least four respects [6], [7]. First, the midvein bifurcates near the leaf tip. Second, the vein network consists of more lateral veins. Third, lateral veins do not join the midvein at the centre of the leaf but run parallel to one another to form a wider midvein. Fourth, lateral veins end in a marginal vein that closely parallels the leaf margin, lending a smooth outline to the vein network. Mutation of PIN1 (AT1G73590) approximates these defects, but vein patterns are only mildly affected in pin1 [6], [19] (Figure 1). As PIN1 is one of eight PIN genes [21]–[24] and other gene families have been implicated in auxin transport (e.g., [30]–[32]), the mild vein-pattern defects of pin1 might reflect contribution of other auxin transporters to vein patterning; here we tested whether other PIN genes contribute to vein patterning.
Fig. 1. Vein patterning functions of Arabidopsis PIN genes. 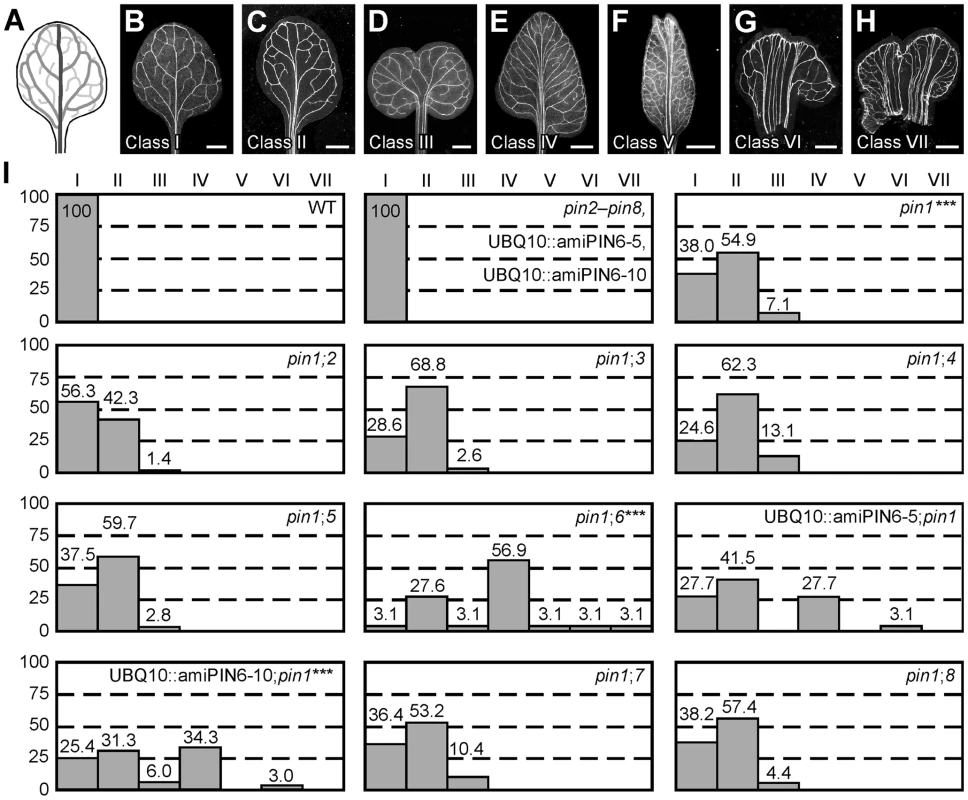
(A,B) Vein pattern of WT mature first leaf. In (A), dark grey, midvein; grey, loops; light grey, minor veins. (B–H) Dark-field illumination of cleared mature first leaves illustrating phenotype classes: unbranched, narrow midvein and scalloped vein-network outline (B); bifurcated midvein and scalloped vein-network outline (C); fused leaves with scalloped vein-network outline (D); conspicuous marginal vein (E); fused leaves with conspicuous marginal vein (F); wide midvein (G); fused leaves with wide midvein (H). (I) Percentages of leaves in phenotype classes. Difference between pin1 and WT, between pin1;6 and pin1, and between UBQ10::amiPIN6;pin1 and pin1 was significant at P<0.001 (***) by Kruskal-Wallis and Mann-Whitney test with Bonferroni correction. Sample population sizes: WT, 65; pin2, 68; pin3, 68; pin4, 68; pin5, 68; pin6, 68; UBQ10::amiPIN6-5, 65; UBQ10::amiPIN6-10, 65; pin7, 68; pin8, 68; pin1, 71; pin1;2, 71; pin1;3, 77; pin1;4, 69; pin1;5, 72; pin1;6, 65; UBQ10::amiPIN6-5;pin1, 65; UBQ10::amiPIN6-10;pin1, 67; pin1;7, 77; pin1;8, 68. Bars: (B–F) 1.5 mm; (G,H) 0.75 mm. We first explored this possibility by analyzing vein patterns of single mutants in PIN2 (AT5G57090), PIN3 (AT1G70940), PIN4 (AT2G01420), PIN5 (AT5G16530), PIN6 (AT1G77110), PIN7 (AT1G23080) and PIN8 (AT5G15100). pin2–pin8 (Table S1) had WT vein patterns (Figure 1), limiting nonredundant vein-patterning functions to PIN1. Thus, to uncover potential vein-patterning functions of PIN2–PIN8, we next analyzed vein patterns of double mutants between pin1 and pin2–pin8. Only mutation of the auxin-transporter-encoding PIN6 [9] had a significant effect on pin1 phenotype spectrum, with ∼65% of pin1pin6 (pin1;6) leaves belonging to new, stronger classes (Figure 1). pin6 had a similar effect on pin1 phenotype spectrum in seedlings (Figure S1): ∼95% penetrance of cotyledon defects in pin1;6 vs. ∼35% in pin1, and appearance in pin1;6 of a cup-shaped cotyledon phenotype resembling that of embryos developed in the presence of auxin transport inhibitors [33], [34]. In both leaves and seedlings, the pin1;6 phenotype spectrum was mimicked by expressing an artificial microRNA targeting PIN6 (UBQ10::amiPIN6) in the pin1 background (Figure 1 and Figure S1). We conclude that PIN6 is the PIN gene that most contributes to auxin-transport-dependent vein patterning in the absence of PIN1 function.
PIN6 expression in leaf development
Double-mutant analysis suggests functions for PIN6 in vein patterning (Figure 1; see Discussion). We thus asked whether PIN6 was expressed during vein formation. To address this question, we imaged expression of a functional (Table S1) translational fusion of PIN6 to GFP driven by the PIN6 promoter (PIN6::PIN6:GFP). During leaf development, PIN6::PIN6:GFP expression was initiated in broad subepidermal domains that narrowed to sites of vein formation (Figure 2A–2H), suggesting PIN6 expression during vein formation. Expression of PIN6::PIN6:GFP was recapitulated by expression of a PIN6::YFPnuc transcriptional fusion (PIN6 promoter driving expression of a nuclear YFP) (Figure S2A–S2E), which overlapped with expression of PIN1::PIN1:CFP [35] in leaf subepidermal tissues (Figure 2I). In pin1, PIN6::PIN6:GFP expression remained confined to subepidermal tissues, but expression was weaker (Figure 2J–2L).
Fig. 2. PIN6 expression in leaf development. 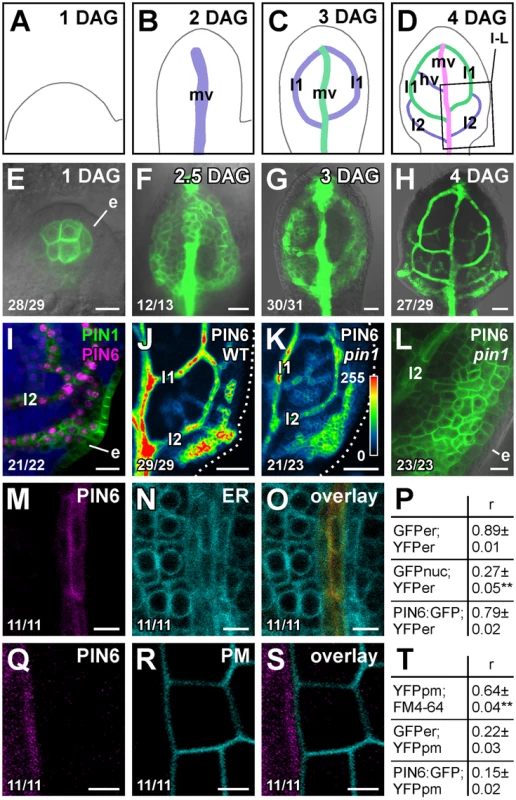
(A–O,Q–S) Top right: leaf age in days after germination (DAG), marker and genotype. Bottom left: reproducibility index. First leaves. (A–D) Midvein, loops and minor veins differentiate progressively later in the same region of the developing leaf, and loops and minor veins differentiate in a tip-to-base sequence during leaf development [6], [7], [57]; purple, green and magenta: successive stages of vein differentiation. Box in (D) illustrates position of close-ups in (I–L). (E–O,Q–S) Confocal laser scanning microscopy with (E–H,L) or without (I–K,M–O,Q–S) transmitted light. (E–H) PIN6::PIN6:GFP expression. (I) Expression of PIN6::YFPnuc and PIN1:PIN1:CFP at 4 DAG; blue: chlorophyll. (J–L) PIN6::PIN6:GFP expression in WT (J) and pin1 (K,L) at 4 DAG. LUT (in K) visualizes expression levels. Dotted line: leaf outline. (M–O,Q–S) Expression of PIN6::PIN6:GFP (M,Q), 35S::YFPer (N), 35S::LTI6B:YFP (R) and respective overlays displayed with a dual-channel LUT [66] (O,S): prevalence of cyan over colocalized magenta signal is shown in green, opposite in red, and colocalized cyan and magenta signals of equal intensity in yellow. (P,T) Quantification of colocalized GFP and YFP signals (as mean ± SE of Manders' coefficient ‘r’) in populations (n = 11) of positive controls: J1721::GFPer;35S::YFPer (P), 35S::YFPpm;FM4-64 (T); negative controls: ATHB8::GFPnuc;35S::YFPer (P), J1721::GFPer; 35S::YFPpm (T); and samples: PIN6::PIN6:GFP;35S::YFPer (P), PIN6::PIN6:GFP; 35S::YFPpm (T). (P) Difference between negative control and positive control, and between negative control and sample was significant at P<0.01 (**) by one-way ANOVA and Tukey's test. (T) Difference between positive control and negative control, and between positive control and sample was significant at P<0.01 (**) by one-way ANOVA and Tukey's test. e, epidermis; hv, minor vein; l1, first loop; l2, second loop; mv, midvein. Bars: (E,I,L) 10 µm; (F,G) 20 µm; (H,J,K) 50 µm; (M–O) 5 µm; (Q–S) 2.5 µm. A PIN6 translational fusion localizes to the ER in tobacco suspension cells [36]. To determine PIN6 subcellular localization in Arabidopsis leaves, we quantified degree of colocalization between expression of PIN6::PIN6:GFP and expression of ER, or PM, markers. Expression of PIN6::PIN6:GFP correlated with expression of ER, but not PM, markers (Figure 2M–2T and Figure S3), suggesting ER-localization of PIN6 in vein development.
Genetic interaction between PIN1, PIN5, PIN6, and PIN8 in vein patterning
PIN6 belongs to a subfamily of proteins that includes the ER-localized PIN5 and PIN8 auxin transporters [36]–[39]. Thus, we asked whether frequencies of vein pattern phenotypes in pin1;6 were affected by additional mutation of PIN5 or PIN8. While pin8 shifted the distribution of pin1;6 phenotypes towards stronger classes, pin5 partially normalized pin1;6;8 phenotype spectrum (Figure 3A). pin5 and pin8 had similar effects in seedlings (Figure S1): (1) complete penetrance of cotyledon defects in pin1;6;8 vs. ∼95% in pin1;6; (2) ∼95% penetrance of the cup-shaped cotyledon phenotype in pin1;6;8 vs. ∼60% in pin1;6; and (3) partial normalization of cotyledon defects of pin1;6;8 by pin5.
Fig. 3. Genetic interaction between PIN1, PIN5, PIN6, and PIN8 in vein patterning. 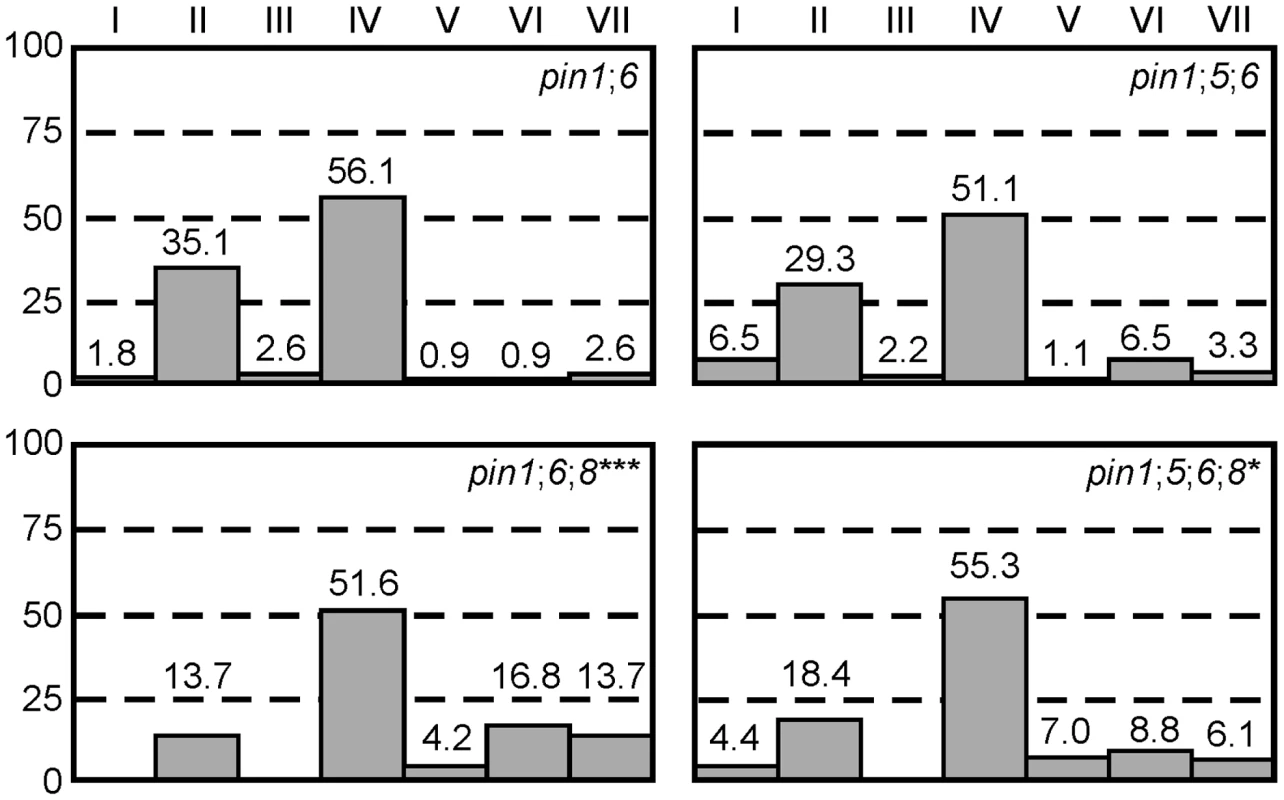
Percentages of leaves in phenotype classes (defined in Figure 1). Difference between pin1;6;8 and pin1;6, and between pin1;5;6;8 and pin1;6;8 was significant at P<0.05 (*) or P<0.001 (***) by Kruskal-Wallis and Mann-Whitney test with Bonferroni correction. Sample population sizes: pin1;6, 114; pin1;5;6, 92; pin1;6;8, 95; pin1;5;6;8, 114. PIN8 expression in leaf development
Triple-mutant analysis suggests functions for PIN8 in vein patterning (Figure 3; see Discussion). We thus asked whether PIN8 was expressed, as PIN1 [10], [11], PIN5 [36] and PIN6 (Figure 2 and Figure S2A–S2E), during vein development. To address this question, we imaged expression of a functional (Table S1) translational fusion of PIN8 to GFP driven by the PIN8 promoter (PIN8::PIN8:GFP). Expression of PIN8::PIN8:GFP was restricted to narrow sites of vein formation in both WT and pin1;6 (Figure 4A–4E), suggesting PIN8 expression in vein development, a conclusion further supported by expression of a PIN8::YFPnuc transcriptional fusion (PIN8 promoter driving expression of a nuclear YFP) (Figure 4H–4K).
Fig. 4. PIN8 expression in leaf development. 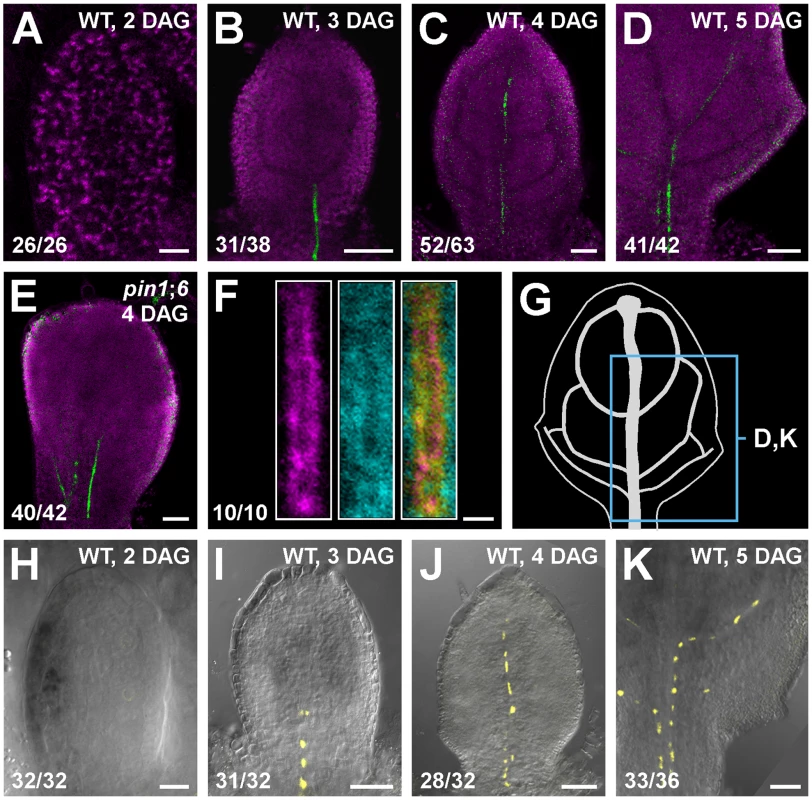
(A–F,H–K) Top right: genotype, leaf age in days after germination (DAG). Bottom left: reproducibility index. Confocal laser scanning microscopy without (A–F) or with (H–K) transmitted light; first leaves. (A–E) Green: PIN8::PIN8:GFP expression; magenta: chlorophyll. (F) Expression at 3.25 DAG of PIN8::PIN8:GFP (left), staining by ER-Tracker Red (centre) and their overlay displayed with a dual-channel LUT (defined in Figure 2) (right). (G) 5-DAG first leaf illustrating positions of close-ups in (D) and (K). (H–K) PIN6::YFPnuc expression. Bars: (A,H) 10 µm; (B–E,I–K) 50 µm; (F) 2 µm. In pollen, PIN8 translational fusions localize to the ER [38], [39]. To determine PIN8 subcellular localization in leaves, we quantified degree of colocalization between expression of PIN8::PIN8:GFP and staining by the glibenclamide-derived ER-Tracker Red dye, which selectively stains the ER (e.g., [40]–[42]). Expression of PIN8::PIN8:GFP correlated with staining by ER-Tracker Red (Figure 4F) (mean Manders' coefficient ‘r’ ± SE: 0.82±0.03; n = 10) and overlapped with staining by the dapoxyl-derived ER-Tracker Blue-White DPX dye [43] (Figure S2G), suggesting ER-localization of PIN8 in vein development.
Necessary functions of PIN5, PIN6, and PIN8 in vein network formation
Single mutation of PIN5, PIN6 or PIN8 had no effect on vein patterns (Figure 1). Thus, to test whether a function in vein patterning could be assigned to ER-localized PIN proteins in the presence of PIN1-mediated intercellular auxin-transport, we analyzed vein patterns of combinations of pin5, pin6 and pin8. Double and triple mutants had WT vein patterns (sample population sizes: WT, 26; pin5;6, 22; pin5;8, 24; pin6;8, 39; pin5;6;8, 30), but pin6;8 had a more complex vein network, a defect that was normalized by pin5 (Figure 5A–5D).
Fig. 5. Necessity of PIN5, PIN6, and PIN8 for vein network formation. 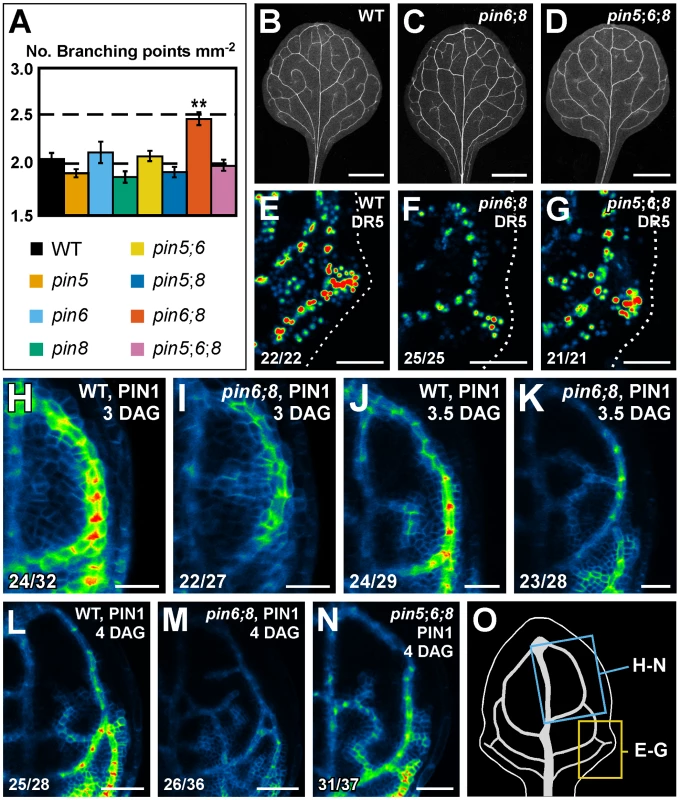
(A) Vein network complexity as mean ± SE number of vein branching points per first-leaf area unit in mm2 [28]. Difference between pin6;8 and all other genotypes was significant at P<0.01 (**) by one-way ANOVA and Tukey's test. Sample population sizes: WT, 30; pin5, 30; pin6, 30; pin8, 27; pin5;6, 28; pin5;8, 28; pin6;8, 28; pin5;6;8, 28. (B–N) Top right: genotype, markers and leaf age in days after germination (DAG). Bottom left: reproducibility index. (B–D) Dark-field illumination of cleared mature first leaves. (E–N) Confocal laser scanning microscopy; first leaves. LUT (in Figure 2K) visualizes expression levels. (E–G) DR5rev::YFPnuc expression, 4 DAG. Dotted line: leaf outline. (H–N) PIN1::PIN1:YFP expression. (O) 4-DAG first leaf illustrating positions of close-ups in (E–G) and (H–N). Bars: (B–D) 1.5 mm; (E–G,L–N) 50 µm; (H–K) 25 µm. We next asked whether pin6;8 defects in vein network formation were associated with changes in auxin response levels. To address this question, we imaged expression of the DR5rev::YFPnuc auxin response reporter [44]. In subepidermal tissues of WT leaves, DR5rev::YFPnuc was strongly expressed at sites of vein formation (Figure 5E); in subepidermal tissues of pin6;8 leaves, DR5rev::YFPnuc expression was weaker (Figure 5F), a defect that was normalized by pin5 (Figure 5G).
We then asked whether pin6;8 defects in auxin response levels and vein network formation were associated with changes in auxin-responsive PIN1 expression [10], [44]–[46]. To address this question, we imaged expression of PIN1::PIN1:YFP [47] in the leaf area enclosed by the first loop. In both WT and pin6;8, PIN1::PIN1:YFP expression was initiated in broad domains that narrowed to sites of vein formation (Figure 5H–5M). However, in pin6;8 PIN1::PIN1:YFP expression was weaker (Figure 5H–5M), vein-associated domains of PIN1::PIN1:YFP expression became visible at earlier time-points (Figure 5H and 5I), and at each time point we observed more vein-associated domains of PIN1::PIN1:YFP expression (Figure 5H–5M). Defects in PIN1::PIN1:YFP expression levels of pin6;8 were normalized by pin5 (Figure 5N).
In summary, pin6;8 had defects in expression of DR5 and PIN1 and in formation of vein networks, and such defects were normalized by pin5.
Sufficient functions of PIN5, PIN6, and PIN8 in vein network formation
We next asked whether PIN6 function was sufficient to control expression of DR5 and PIN1 and formation of vein networks. To address this question, we expressed the PIN6 cDNA by the promoter of RIBOSOMAL PROTEIN S5A (AT3G11940) (RPS5A::PIN6) and by the promoter of MONOPTEROS (AT1G19850) (MP::PIN6), each highly active in developing leaves [48] (Figure S2F). Expression of the auxin-responsive markers DR5rev::YFPnuc, ATHB8::CFPnuc [49] and PIN1::PIN1:YFP was stronger in leaves of RPS5A::PIN6 and MP::PIN6 (Figure 6A–6G). Furthermore, in both these backgrounds lateral domains of PIN1::PIN1:YFP expression often failed to join distal veins (Figure 6E–6G), defects that correlated with the simpler, open vein networks of the mature leaves (Figure 6I–6M).
Fig. 6. Sufficiency of PIN5, PIN6, and PIN8 for vein network formation. 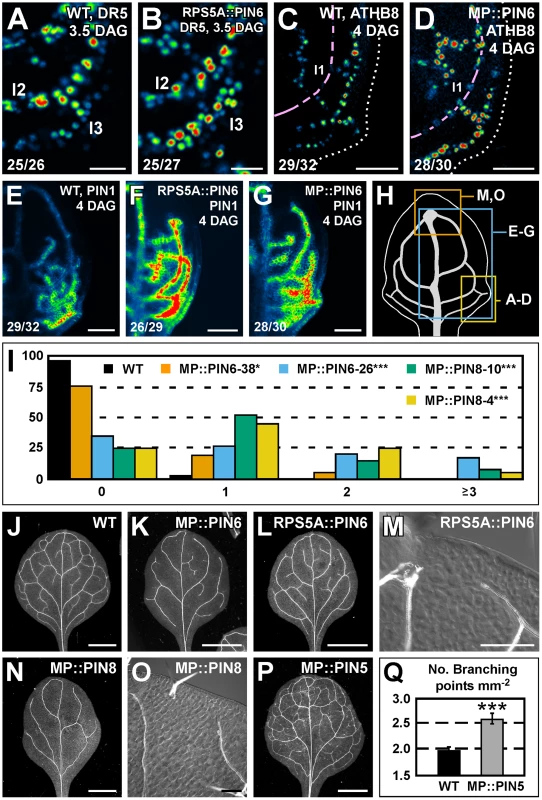
(A–G,J–P) Top right: genotype, markers and leaf age in days after germination (DAG). Bottom left: reproducibility index. (A–G) Confocal laser scanning microscopy; first leaves. LUT (in Figure 2K) visualizes expression levels. (J–P) Dark-field (J–L,N,P) or differential-interference-contrast (M,O) illumination of cleared mature first leaves. (A,B) DR5rev::YFPnuc expression. (C,D) ATHB8::CFPnuc expression. Magenta line connects nuclei in the first loop (l1). Dotted line: leaf outline. (E–G) PIN1::PIN1:YFP expression. (H) 4-DAG first leaf illustrating positions of close-ups in (A–D), (E–G) and (M,O). (I) Percentage of first leaves with 0, 1, 2 or ≥3 open loops. Difference between MP::PIN6 and WT, and between MP::PIN8 and WT was significant at P<0.05 (*) or P<0.001 (***) by Kruskal-Wallis and Mann-Whitney test with Bonferroni correction. Sample population sizes: WT, 43; MP::PIN6-38, 40; MP::PIN6-26, 42; MP::PIN8-10, 40; MP::PIN8-4, 40. (Q) Vein network complexity as mean ± SE number of vein branching points per first-leaf area unit in mm2 [28]. Difference between MP::PIN5 and WT was significant at P<0.001 (***) by unpaired, two-tailed t-test. Sample population sizes: WT, 28; MP::PIN5, 28. l1, first loop; l2, second loop; l3, third loop. Bars: (A,B) 25 µm; (C–G) 50 µm; (J–L,N,P) 1.5 mm; (M,O) 0.2 mm. We then asked whether ectopic expression of PIN8 or PIN5 had any effect on vein network formation. To address this question, we analyzed vein patterns of plants expressing the cDNA of PIN8 or PIN5 by the MP promoter (MP::PIN8 or MP::PIN5, respectively). We found that MP::PIN8 leaves had simpler, open vein networks and that MP::PIN5 leaves had a more complex vein network (Figure 6I and 6N–6Q).
In summary, ectopic PIN6 expression was sufficient to control expression of DR5 and PIN1. Furthermore, ectopic expression of PIN6 or PIN8, on the one hand, and of PIN5, on the other, had opposite effects on vein network formation.
Simulation of pin1;6 defects by reduction of auxin levels in pin1
Weaker expression of auxin responsive markers in pin6;8 leaves and stronger expression of auxin responsive markers in leaves of RPS5A::PIN6 and MP::PIN6 suggest that PIN6-mediated auxin transport increases intracellular levels of biologically active auxin (see Discussion). If this were the case, pin1;6 phenotype spectrum might be mimicked by expressing in pin1 the bacterial gene iaaL, which decreases levels of free auxin by its conjugation to lysine [50], [51]. Expression of PIN6::iaaL (PIN6 promoter driving expression of iaaL) in pin1 mimicked the phenotype spectrum of pin1;6 leaves (Figure 7) and cotyledons (Figure S1), a finding that is consistent with the hypothesis that PIN6-mediated intracellular auxin transport increases auxin levels.
Fig. 7. Control of PIN1-dependent vein patterning by intracellular auxin levels. 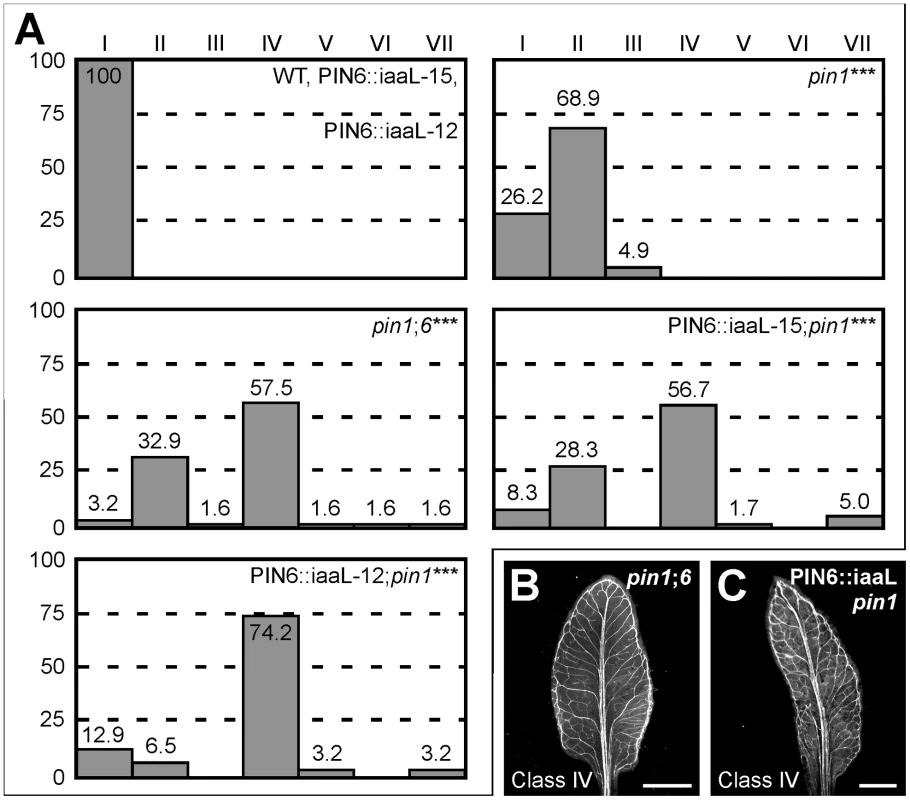
(A) Percentages of leaves in phenotype classes (defined in Figure 1). Difference between pin1 and WT, between pin1;6 and pin1, and between PIN6::iaaL;pin1 and pin1 was significant at P<0.001 (***) by Kruskal-Wallis and Mann-Whitney test with Bonferroni correction. Sample population sizes: WT, 50; PIN6::iaaL-15, 50; PIN6::iaaL-12, 50; pin1, 61; pin1;6, 61; PIN6::iaaL-15;pin1, 60; PIN6::iaaL-12;pin1, 62. (B,C) Dark-field illumination of cleared mature first leaves. Top right: genotype. Bottom left: phenotype class. Bars: (B,C) 2 mm. Discussion
The mechanisms that control the patterning of vein networks of plant leaves have long fascinated biologists and mathematicians. Varied evidence has increasingly been accumulating that supports an inductive and orienting role for the transport of the plant signal auxin in leaf vein patterning [3, 5–7, 10–13], but molecular details have remained unclear. Here we show that the vein network of the Arabidopsis leaf is patterned by two distinct and convergent auxin-transport pathways: an intercellular pathway mediated at the plasma membrane (PM) by the PIN1 auxin transporter and an intracellular pathway mediated at the endoplasmic reticulum (ER) by the PIN6, PIN8 and PIN5 auxin transporters. While a role for other families of auxin transporters (e.g., [30]–[32]) in vein patterning is by no means precluded, our results suggest a new, unsuspected level of control of vein patterning by PIN-mediated auxin transport, one with repercussions on patterning of other plant features.
The localization of PIN1 to the PM [8] and of PIN6 to the ER together with the appearance in pin1;6 of vein pattern defects that exceed the sum of the single-mutant defects suggest that PIN1 and PIN6 act in distinct auxin-transport pathways whose functions converge on vein patterning. By contrast, the overlapping subcellular localizations of PIN6 and PIN8 [36]–[39] together with the purely quantitative enhancement of pin1;6 vein pattern defects by pin8 suggest redundant function of PIN6 and PIN8 in PIN1-dependent vein patterning. Further, the overlapping subcellular localization of PIN8 [36]–[39] and PIN5 [36], [37], [39] together with the partial normalization of pin1;6;8 vein pattern defects by pin5 suggest antagonistic functions of PIN8 and PIN5 in PIN1/PIN6-dependent vein patterning. The interaction between the intercellular auxin-transport pathway controlled by PIN1 and the intracellular auxin-transport pathway controlled by PIN6, PIN8 and PIN5 is relevant for at least two other patterning events, separation of cotyledons and of leaves, and might have implications for other developmental processes. The redundancy between PIN6 and PIN8 and the antagonism between PIN8 and PIN5 are, however, independent of PIN1-mediated intercellular auxin transport: in the presence of PIN1 function, PIN6 and PIN8 redundantly control vein network formation, and such redundant functions are antagonized by PIN5. Redundant functions of PIN6 and PIN8 and antagonistic functions of PIN5, as inferred from loss-of-function data, are also consistent with gain-of-function evidence: ectopic PIN8 expression induces vein pattern defects similar to those induced by ectopic PIN6 expression and opposite to those of pin6;8, suggesting that PIN8 can supply vein patterning functions similar to those of PIN6; and ectopic PIN5 expression induces vein pattern defects opposite to those induced by ectopic expression of PIN6 or PIN8 and similar to those of pin6;8, suggesting that PIN5 can supply vein patterning functions opposite to those of PIN6 and PIN8. The internally consistent relationship between the effects of loss of function of PIN6, PIN8 and PIN5 and the effects of gain of function of these genes is not limited to vein patterning but extends to auxin response: auxin response levels are decreased by simultaneous mutation of PIN6 and PIN8 and by ectopic expression of PIN5 [36] and increased by ectopic expression of PIN6 or PIN8 [39] and by mutation of PIN5 [36]. Thus, the genetic interaction between PIN6, PIN8 and PIN5 might reflect general properties of the mechanism with which these proteins function, a conclusion that is also supported by the antagonistic functions of PIN8 and PIN5 in a process unrelated to vein patterning such as pollen development [39]. The most parsimonious explanation for the antagonistic interaction between PIN6/PIN8 and PIN5, which transports auxin from the cytoplasm to the ER lumen [36], is that PIN6 and PIN8 transport auxin from the ER lumen to the cytoplasm or to the nucleus, whose envelope is continuous with the ER membrane [52], [53] (Figure 8C). This scenario is also supported by the observation that reducing auxin levels in pin1 leads to pin1;6 characteristic defects, and is consistent with higher levels of auxin measured in PIN8 overexpressors [38], [39] and pin5 mutants [36]. Alternatively, or in addition, PIN6, PIN8 and PIN5 could all transport in the same direction but have different affinities for auxins or auxin conjugates with different, even opposing, developmental functions (reviewed in [54]) (Figure 8D).
Fig. 8. Summary and interpretation. 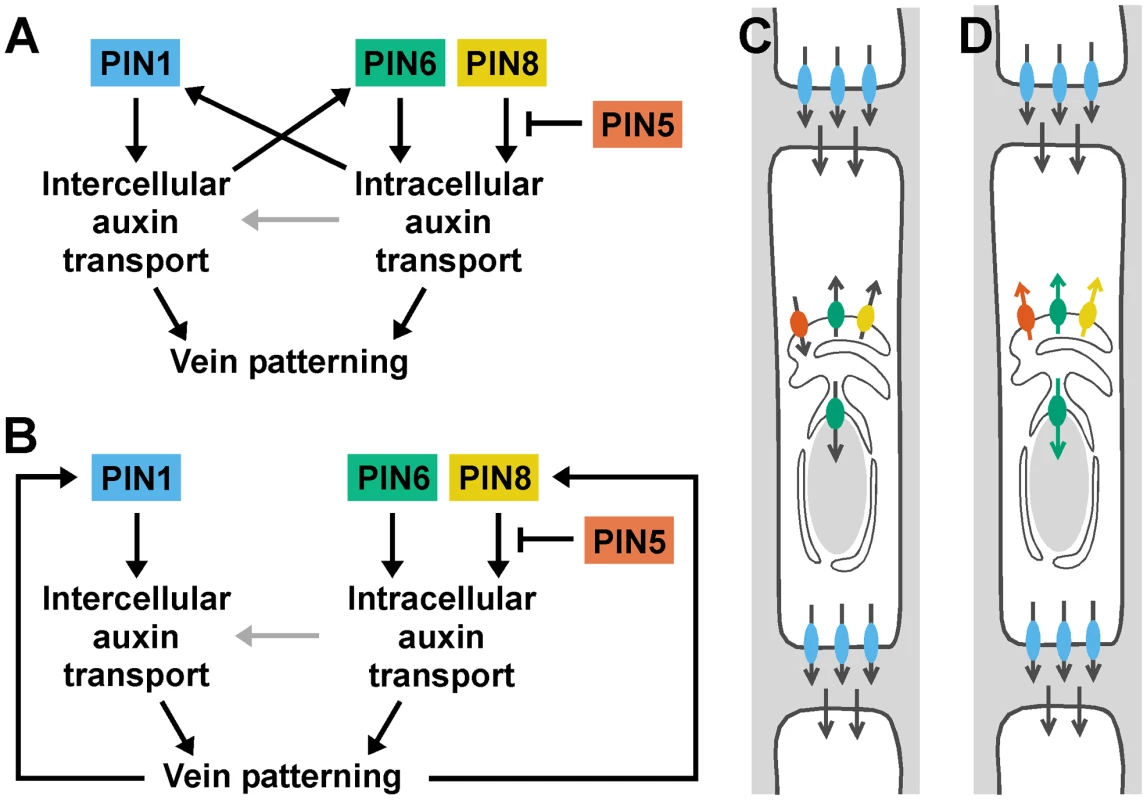
(A,B) Two potential, non-mutually-exclusive regulatory circuits for PIN1 and PIN5/PIN6/PIN8 in vein patterning. Arrows indicate positive regulation; blunt-ended lines indicate negative regulation. Distinct functions of PIN1-mediated intercellular auxin transport and of PIN5/PIN6/PIN8-mediated intracellular auxin transport converge on vein patterning. Additionally, PIN1-mediated intercellular auxin transport could control vein patterning through regulation of PIN6 expression, and PIN5/PIN6/PIN8-mediated intracellular auxin transport could control vein patterning through regulation of PIN1 expression (A). Alternatively, vein patterning could feed back on expression of PIN1 and PIN6 (B). PIN1-independent functions of PIN5/PIN6/PIN8-mediated intracellular auxin transport in vein patterning could include intercellular auxin transport by a mechanism nonhomologous to that used by PM-localized PIN proteins [63] (grey arrows). (C,D) Localization of PIN1 (blue) at the basal plasma membrane and of PIN5 (orange), PIN6 (green) and PIN8 (yellow) at the endoplasmic reticulum/nuclear envelope (ER/NE) of vascular cells. For simplicity, PIN5, PIN6 and PIN8 are shown to be expressed in the same cell, only PIN6 is shown to be also localized at the NE, and localization of AUX/LAX [30], ABCB/PGP/MDR [32] and PILS [31] auxin transporters is not shown. Arrows indicate presumed directions of auxin transport. Antagonism between pin5 and pin6;8 in vein patterning could reflect opposite directions in which PIN5 and PIN6/PIN8 transport auxin (C), different abilities of PIN5 and PIN6/PIN8 to transport different auxins or auxin conjugates (orange, green and yellow arrows) (D; for simplicity, the mirror-image scenario, i.e. transport to the ER/NE lumen, is not shown), or varied combinations of the two. See text for additional details. Our data suggest that auxin transported by PIN6 and PIN8 increases levels of auxin-responsive PIN1 expression during vein formation (Figure 8A). In pin6;8, low auxin levels at sites of vein formation could accelerate formation of narrow vein-associated domains of PIN1 expression, thus resulting in high-complexity vein networks [13], [55]–[58]. Conversely, in leaves ectopically expressing PIN6, high auxin levels at sites of vein formation could hinder or prevent formation and connection of vein-associated PIN1-expression domains, thus resulting in simple, open vein networks [13], [56], [57], [59]–[61]. Alternatively, PIN6/PIN8-mediated auxin transport could control vein network formation exclusively through PIN1-independent pathways; defects in auxin response or PIN1 expression would then be consequence, rather than cause, of defective vein-network formation (Figure 8B). In any case, the synthetic enhancement of pin1 defects by pin6 and pin6;8, as opposed to the epistasis of pin6;8 to pin1, suggests vein patterning functions of PIN6/PIN8-mediated intracellular auxin transport beyond regulation of PIN1 expression or of intercellular auxin transport mediated by PM-localized PIN proteins (Figure 8A and 8B). We can probably exclude that such functions of PIN6 and PIN8 require physical interaction with PIN1 because PIN1 and PIN6/PIN8 localize to different cellular compartments and because PIN1/pin1;pin6/pin6 lacks defects in cotyledons and vein patterns (n>50), defects that would be expected for interacting proteins (reviewed in [62]). Because the localization of PIN6 and PIN8 is maintained in pin1 backgrounds, it seems unlikely that these two proteins have auxin transport functions that are redundant and homologous with those of PIN1. Nevertheless, PIN6/PIN8/PIN5-mediated intracellular auxin-transport could contribute to intercellular auxin transport by a mechanism nonhomologous to that used by PM-localized PIN proteins [63] (Figure 8A and 8B), a function of ER-localized PIN proteins that would be particularly relevant in primitive land plants like mosses, which seem to lack PM-localized PIN proteins [36] but do select cell files for vascular function [64].
Materials and Methods
Plants
Origin and nature of lines, genotyping strategies and oligonucleotide sequences are in Tables S1, S2, S3, respectively. Seeds were sterilized and germinated, and plants were grown and transformed as in [65].
Imaging
For plasma-membrane stainings, seedlings were incubated in 10 µg ml−1 FM 4-64 (Invitrogen) for ∼4 min under vacuum before mounting. For endoplasmic-reticulum stainings, dissected leaves were incubated in 10 µM ER-Tracker Red (Invitrogen/Life Technologies) or ER-Tracker Blue-White DPX (Invitrogen/Life Technologies) for ∼30 min under vacuum before mounting. Leaf primordia were mounted in water under 0.17-mm-thick coverslips (Fischer Scientific) and imaged with the 20×/0.8 Plan-Apochromat or 40×/1.2 W C-Apochromat objective of an Axio Imager.M1/LSM 510 META confocal microscope (Carl Zeiss). 512×512-pixel frames were scanned unidirectionally at 8-bit depth with 1.6-µsec pixel dwell time and 8-fold averaging. Scanning zoom was adjusted to set pixel size to half the spacing of the features to be resolved, with minimum pixel size equal to half the objective lateral resolving power. Emission was collected from ∼5-µm-thick (single-fluorophore imaging) or ∼1-µm-thick (multi-fluorophore imaging) optical slices. Amplifier gain was set at 1; detector gain at ∼50–60%. Laser transmission and offset value were adjusted to match signal to detector's input range. Marker-line-specific imaging parameters are in Tables S4 and S5. For multi-fluorophore imaging, sequential excitation and collection of emission were performed in line-by-line channel-switching mode, which did not result in signal crossover under our imaging conditions. Signal levels in images acquired at identical settings were visualized as in [65]. Signal colocalization was visualized as in [66] and quantified as in [67]. Mature leaves were imaged as in [49]. Images were analyzed and processed with ImageJ (National Institutes of Health), and figures were generated with Canvas (ACD Systems International Inc.).
Supporting Information
Zdroje
1. FosterAS (1952) Foliar Venation in Angiosperms from an Ontogenetic Standpoint. American Journal of Botany 39 : 752–766.
2. PrayTR (1955) Foliar venation in Angiosperms. II. Histogenesis of the venation of Liriodendron. American Journal of Botany 42 : 18–27.
3. SachsT (1989) The development of vascular networks during leaf development. Current Topics in Plant Biochemistry and Physiology 8 : 168–183.
4. BerlethT, MattssonJ, HardtkeCS (2000) Vascular continuity and auxin signals. Trends in Plant Science 5 : 387–393.
5. GersaniM (1987) The Induction of Differentiation of Organized Vessels in a Storage Organ. Annals of Botany 59 : 31–34.
6. MattssonJ, SungZR, BerlethT (1999) Responses of plant vascular systems to auxin transport inhibition. Development 126 : 2979–2991.
7. SieburthLE (1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiology 121 : 1179–1190.
8. GalweilerL, GuanC, MullerA, WismanE, MendgenK, et al. (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 : 2226–2230.
9. PetrasekJ, MravecJ, BouchardR, BlakesleeJJ, AbasM, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 : 914–918.
10. ScarpellaE, MarcosD, FrimlJ, BerlethT (2006) Control of leaf vascular patterning by polar auxin transport. Genes & Development 20 : 1015–1027.
11. WenzelCL, SchuetzM, YuQ, MattssonJ (2007) Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. The Plant Journal 49 : 387–398.
12. SachsT (1991) Cell Polarity and Tissue Patterning in Plants. Development Supplement 1 : 83–93.
13. SachsT (1981) The control of the patterned differentiation of vascular tissues. Advances in Botanical Research 9 : 151–262.
14. GarnettP, SteinacherA, StepneyS, ClaytonR, LeyserO (2010) Computer simulation: the imaginary friend of auxin transport biology. Bioessays 32 : 828–835.
15. KrupinskiP, JonssonH (2010) Modeling auxin-regulated development. Cold Spring Harbor Perspectives in Biology 2: a001560 doi:001510.001101/cshperspect.a001560.
16. SantosF, TealeW, FleckC, VolpersM, RupertiB, et al. (2010) Modelling polar auxin transport in developmental patterning. Plant Biology 12 Supplement 1 : 3–14.
17. SmithRS, BayerEM (2009) Auxin transport-feedback models of patterning in plants. Plant, Cell & Environment 32 : 1258–1271.
18. WabnikK, GovaertsW, FrimlJ, Kleine-VehnJ (2011) Feedback models for polarized auxin transport: an emerging trend. Molecular Biosystems 7 : 2352–2359.
19. BilsboroughGD, RunionsA, BarkoulasM, JenkinsHW, HassonA, et al. (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences, USA 108 : 3424–3429.
20. GuenotB, BayerE, KierzkowskiD, SmithRS, MandelT, et al. (2012) PIN1-Independent Leaf Initiation in Arabidopsis. Plant Physiology 159 : 1501–1510.
21. KrecekP, SkupaP, LibusJ, NaramotoS, TejosR, et al. (2009) The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biology 10 : 249 doi:10.1186/gb-2009-10-12-249.
22. ZazimalovaE, MurphyAS, YangH, HoyerovaK, HosekP (2010) Auxin transporters–why so many? Cold Spring Harbor Perspectives in Biology 2: a001552 doi:10.1101/cshperspect.a001552.
23. PaponovIA, TealeWD, TrebarM, BlilouI, PalmeK (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends in Plant Science 10 : 170–177.
24. Viaene T, Delwiche CF, Rensing SA, Friml J (2012) Origin and evolution of PIN auxin transporters in the green lineage. Trends in Plant Science In press.
25. ClayNK, NelsonT (2005) Arabidopsis thickvein mutation affects vein thickness and organ vascularization, and resides in a provascular cell-specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiology 138 : 767–777.
26. SassiM, LuY, ZhangY, WangJ, DhonuksheP, et al. (2012) COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1 - and PIN2-dependent auxin transport in Arabidopsis. Development 139 : 3402–3412.
27. FischerU, IkedaY, LjungK, SerralboO, SinghM, et al. (2006) Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Current Biology 16 : 2143–2149.
28. CandelaH, Martinez-LabordaA, MicolJL (1999) Venation pattern formation in Arabidopsis thaliana vegetative leaves. Developmental Biology 205 : 205–216.
29. NelsonT, DenglerN (1997) Leaf Vascular Pattern Formation. Plant Cell 9 : 1121–1135.
30. PeretB, SwarupK, FergusonA, SethM, YangY, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24 : 2874–2885.
31. BarbezE, KubesM, RolcikJ, BeziatC, PencikA, et al. (2012) A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485 : 119–122.
32. GeislerM, MurphyAS (2006) The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Letters 580 : 1094–1102.
33. LiuC, XuZ, ChuaNH (1993) Auxin Polar Transport Is Essential for the Establishment of Bilateral Symmetry during Early Plant Embryogenesis. Plant Cell 5 : 621–630.
34. HadfiK, SpethV, NeuhausG (1998) Auxin-induced developmental patterns in Brassica juncea embryos. Development 125 : 879–887.
35. GordonSP, HeislerMG, ReddyGV, OhnoC, DasP, et al. (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134 : 3539–3548.
36. MravecJ, SkupaP, BaillyA, HoyerovaK, KrecekP, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459 : 1136–1140.
37. GangulyA, LeeSH, ChoM, LeeOR, YooH, et al. (2010) Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiology 153 : 1046–1061.
38. BoscoCD, DovzhenkoA, LiuX, WoernerN, RenschT, et al. (2012) The endoplasmic reticulum localized PIN8 is a pollen specific auxin carrier involved in intracellular auxin homeostasis. The Plant Journal 71 : 860–870.
39. DingZ, WangB, MorenoI, DuplakovaN, SimonS, et al. (2012) ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nature Communications 3 : 941 doi:10.1038/ncomms1941.
40. YamasakiS, Sakata-SogawaK, HasegawaA, SuzukiT, KabuK, et al. (2007) Zinc is a novel intracellular second messenger. Journal of Cell Biology 177 : 637–645.
41. DonAS, Martinez-LamencaC, WebbWR, ProiaRL, RobertsE, et al. (2007) Essential requirement for sphingosine kinase 2 in a sphingolipid apoptosis pathway activated by FTY720 analogues. Journal of Biological Chemistry 282 : 15833–15842.
42. TanY, DourdinN, WuC, De VeyraT, ElceJS, et al. (2006) Ubiquitous calpains promote caspase-12 and JNK activation during endoplasmic reticulum stress-induced apoptosis. Journal of Biological Chemistry 281 : 16016–16024.
43. ColeL, DaviesD, HydeGJ, AshfordAE (2000) ER-Tracker dye and BODIPY-brefeldin A differentiate the endoplasmic reticulum and golgi bodies from the tubular-vacuole system in living hyphae of Pisolithus tinctorius. Journal of Microscopy 197 : 239–249.
44. HeislerMG, OhnoC, DasP, SieberP, ReddyGV, et al. (2005) Patterns of Auxin Transport and Gene Expression during Primordium Development Revealed by Live Imaging of the Arabidopsis Inflorescence Meristem. Current Biology 15 : 1899–1911.
45. VietenA, VannesteS, WisniewskaJ, BenkovaE, BenjaminsR, et al. (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 : 4521–4531.
46. NemhauserJL, HongF, ChoryJ (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 : 467–475.
47. XuJ, HofhuisH, HeidstraR, SauerM, FrimlJ, et al. (2006) A molecular framework for plant regeneration. Science 311 : 385–388.
48. WeijersD, Franke-van DijkM, VenckenRJ, QuintA, HooykaasP, et al. (2001) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128 : 4289–4299.
49. DonnerTJ, SherrI, ScarpellaE (2009) Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136 : 3235–3246.
50. RomanoCP, HeinMB, KleeHJ (1991) Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes & Development 5 : 438–446.
51. JensenPJ, HangarterRP, EstelleM (1998) Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiology 116 : 455–462.
52. GraumannK, EvansDE (2011) Nuclear envelope dynamics during plant cell division suggest common mechanisms between kingdoms. Biochemistry Journal 435 : 661–667.
53. OdaY, FukudaH (2011) Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. The Plant Journal 66 : 629–641.
54. Ludwig-MullerJ (2012) Auxin conjugates: their role for plant development and in the evolution of land plants. Journal of Experimental Botany 62 : 1757–1773.
55. Rolland-LaganAG, PrusinkiewiczP (2005) Reviewing models of auxin canalization in the context of leaf vein pattern formation in Arabidopsis. The Plant Journal 44 : 854–865.
56. WabnikK, Kleine-VehnJ, BallaJ, SauerM, NaramotoS, et al. (2010) Emergence of tissue polarization from synergy of intracellular and extracellular auxin signaling. Molecular Systems Biology 6 : 447 doi:10.1038/msb.2010.103.
57. ScarpellaE, FrancisP, BerlethT (2004) Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131 : 3445–3455.
58. McKownAD, DenglerNG (2009) Shifts in leaf vein density through accelerated vein formation in C(4) Flaveria (Asteraceae). Annals of Botany 104 : 1085–1098.
59. PrusinkiewiczP, CrawfordS, SmithRS, LjungK, BennettT, et al. (2009) Control of bud activation by an auxin transport switch. Proceedings of the National Academy of Sciences, USA 106 : 17431–17436.
60. BallaJ, KalousekP, ReinohlV, FrimlJ, ProchazkaS (2011) Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. The Plant Journal 65 : 571–577.
61. SachsT (1970) A control of bud growth by vascular tissue differentiation. Israel Journal of Botany 19 : 484–498.
62. HawleyRS, GillilandWD (2006) Sometimes the result is not the answer: the truths and the lies that come from using the complementation test. Genetics 174 : 5–15.
63. WabnikK, Kleine-VehnJ, GovaertsW, FrimlJ (2011) Prototype cell-to-cell auxin transport mechanism by intracellular auxin compartmentalization. Trends in Plant Science 16 : 468–475.
64. LigroneR, DuckettJG, RenzagliaKS (2000) Conducting tissues and phyletic relationships of bryophytes. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 355 : 795–813.
65. SawchukMG, DonnerTJ, HeadP, ScarpellaE (2008) Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiology 148 : 1908–1924.
66. DemandolxD, DavoustJ (1997) Multicolour analysis and local image correlation in confocal microscopy. Journal of Microscopy 185 : 21–36.
67. MandersEMM, VerbeekFJ, AtenJA (1993) Measurement of Colocalization of Objects in Dual-Color Confocal Images. Journal of Microscopy 169 : 375–382.
Štítky
Genetika Reprodukční medicína
Článek MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease MiceČlánek Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 2- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- A Meta-Analysis of Thyroid-Related Traits Reveals Novel Loci and Gender-Specific Differences in the Regulation of Thyroid Function
- Genetic Landscape of Open Chromatin in Yeast
- Deleterious Alleles in the Human Genome Are on Average Younger Than Neutral Alleles of the Same Frequency
- Age-Dependent Transition from Cell-Level to Population-Level Control in Murine Intestinal Homeostasis Revealed by Coalescence Analysis
- Next-Generation Sequencing Identifies the Danforth's Short Tail Mouse Mutation as a Retrotransposon Insertion Affecting Expression
- ImmunoChip Study Implicates Antigen Presentation to T Cells in Narcolepsy
- Massive Mitochondrial Gene Transfer in a Parasitic Flowering Plant Clade
- Comment on “Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome”
- The Prefoldin Bud27 Mediates the Assembly of the Eukaryotic RNA Polymerases in an Rpb5-Dependent Manner
- Genetic Determinants of Trabecular and Cortical Volumetric Bone Mineral Densities and Bone Microstructure
- Encodes a Novel and -Genus-Specific Regulator of Photoperiodic Flowering in Rice
- Only One Isoform of CTP Synthase Forms the Cytoophidium
- Mechanisms Involved in the Functional Divergence of Duplicated GroEL Chaperonins in DK1622
- A Genome-Wide RNAi Screen in Identifies the Nicotinic Acetylcholine Receptor Subunit ACR-7 as an Antipsychotic Drug Target
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Ancient DNA Reveals Prehistoric Gene-Flow from Siberia in the Complex Human Population History of North East Europe
- Inflammation-Mediated Genetic and Epigenetic Alterations Drive Cancer Development in the Neighboring Epithelium upon Stromal Abrogation of TGF-β Signaling
- MicroRNA-3148 Modulates Allelic Expression of Toll-Like Receptor 7 Variant Associated with Systemic Lupus Erythematosus
- RNAi–Based Functional Profiling of Loci from Blood Lipid Genome-Wide Association Studies Identifies Genes with Cholesterol-Regulatory Function
- CELF Family RNA–Binding Protein UNC-75 Regulates Two Sets of Mutually Exclusive Exons of the Gene in Neuron-Specific Manners in
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- The Ubiquitin Ligase Subunit Acts in Target Tissue to Restrict Tracheal Terminal Cell Branching and Hypoxic-Induced Gene Expression
- Mitotic Evolution of Shows a Stable Core Genome but Recombination in Antigen Families
- Tysnd1 Deficiency in Mice Interferes with the Peroxisomal Localization of PTS2 Enzymes, Causing Lipid Metabolic Abnormalities and Male Infertility
- A Regulatory Pathway, Ecdysone-Transcription Factor Relish-Cathepsin L, Is Involved in Insect Fat Body Dissociation
- PcG-Mediated Higher-Order Chromatin Structures Modulate Replication Programs at the BX-C
- MSH3 Polymorphisms and Protein Levels Affect CAG Repeat Instability in Huntington's Disease Mice
- JNK-Interacting Protein 3 Mediates the Retrograde Transport of Activated c-Jun N-Terminal Kinase and Lysosomes
- Discovery of a Splicing Regulator Required for Cell Cycle Progression
- Rearrangements of 2.5 Kilobases of Noncoding DNA from the Locus Define Predictive Rules of Genomic -Regulatory Logic
- Admixture Mapping in Lupus Identifies Multiple Functional Variants within IFIH1 Associated with Apoptosis, Inflammation, and Autoantibody Production
- Roles of the Developmental Regulator Homothorax in Limiting Longevity in
- miR-199a-5p Is Upregulated during Fibrogenic Response to Tissue Injury and Mediates TGFbeta-Induced Lung Fibroblast Activation by Targeting Caveolin-1
- A Kinome-Wide RNAi Screen in Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
- SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State
- Retrotransposon Activates Ectopic Expression: A Short Tail
- Confounding by Repetitive Elements and CpG Islands Does Not Explain the Association between Hypomethylation and Genomic Instability
- Cell Reprogramming Requires Silencing of a Core Subset of Polycomb Targets
- Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
- Essential Developmental, Genomic Stability, and Tumour Suppressor Functions of the Mouse Orthologue of
- Conditional Inactivation of the DNA Damage Response Gene in Mouse Testis Reveals Separable Roles for Components of the RAD9-RAD1-HUS1 Complex in Meiotic Chromosome Maintenance
- Genome-Wide Analysis Points to Roles for Extracellular Matrix Remodeling, the Visual Cycle, and Neuronal Development in Myopia
- Patterning of Leaf Vein Networks by Convergent Auxin Transport Pathways
- An Evolutionary Perspective on Epistasis and the Missing Heritability
- A Retrotransposon Insertion in the 5′ Regulatory Domain of Ptf1a Results in Ectopic Gene Expression and Multiple Congenital Defects in Danforth's Short Tail Mouse
- The Mub1/Ubr2 Ubiquitin Ligase Complex Regulates the Conserved Dsn1 Kinetochore Protein
- Mutations Can Cause Enamel-Renal Syndrome (ERS)
- Yemanuclein and HIRA Cooperate for Assembly of H3.3-Containing Nucleosomes in the Male Pronucleus
- Hepatocyte Growth Factor, a Determinant of Airspace Homeostasis in the Murine Lung
- ISWI and CHD Chromatin Remodelers Bind Promoters but Act in Gene Bodies
- COM-1 Promotes Homologous Recombination during Meiosis by Antagonizing Ku-Mediated Non-Homologous End Joining
- Control of Multicellular Development by the Physically Interacting Deneddylases DEN1/DenA and COP9 Signalosome
- Antagonism Versus Cooperativity with TALE Cofactors at the Base of the Functional Diversification of Hox Protein Function
- Dynamic Association of NUP98 with the Human Genome
- Ectopic Expression of Induces Spinal Defects, Urogenital Defects, and Anorectal Malformations in Mice
- Regulation of Contributes to the Lineage Potential of Neurogenin3+ Endocrine Precursor Cells in the Pancreas
- Gene-Based Testing of Interactions in Association Studies of Quantitative Traits
- The Amidation Step of Diphthamide Biosynthesis in Yeast Requires , a Gene Identified through Mining the - Interaction Network
- Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci
- Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture
- DNA Methylation Mediated Control of Gene Expression Is Critical for Development of Crown Gall Tumors
- Identification of the SlmA Active Site Responsible for Blocking Bacterial Cytokinetic Ring Assembly over the Chromosome
- Expression of a Novel P22 ORFan Gene Reveals the Phage Carrier State in Typhimurium
- Altered Cohesin Gene Dosage Affects Mammalian Meiotic Chromosome Structure and Behavior
- Quantitative Analysis of Histone Modifications: Formaldehyde Is a Source of Pathological N-Formyllysine That Is Refractory to Histone Deacetylases
- Duplicate Abalone Egg Coat Proteins Bind Sperm Lysin Similarly, but Evolve Oppositely, Consistent with Molecular Mimicry at Fertilization
- Lessons from on the Strengths and Weaknesses of Structured Association Mapping
- DNA–Methylome Analysis of Mouse Intestinal Adenoma Identifies a Tumour-Specific Signature That Is Partly Conserved in Human Colon Cancer
- Transposon Variants and Their Effects on Gene Expression in
- Polygenic Modeling with Bayesian Sparse Linear Mixed Models
- Single Transmembrane Peptide DinQ Modulates Membrane-Dependent Activities
- The JNK Signaling Pathway Activates Expression of Stress Response Genes by Derepressing the Fos/HDAC Repressor Complex
- The Interaction of CtIP and Nbs1 Connects CDK and ATM to Regulate HR–Mediated Double-Strand Break Repair
- Regulation of Metamorphosis by Xenobiotic Response Regulators
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Complex Inheritance of Melanoma and Pigmentation of Coat and Skin in Grey Horses
- Coordination of Chromatid Separation and Spindle Elongation by Antagonistic Activities of Mitotic and S-Phase CDKs
- Autophagy Induction Is a Tor- and Tp53-Independent Cell Survival Response in a Zebrafish Model of Disrupted Ribosome Biogenesis
- Assembly of the Auditory Circuitry by a Genetic Network in the Mouse Brainstem
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



