-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
Colicins are considered model proteins for studying bacterial toxins. These narrow spectrum antibiotics can kill by a variety of mechanisms, e.g. by forming pores in the membranes of susceptible cells or by degrading their nucleic acids. Colicin genes are plasmid-encoded and repressed by the master regulator of the DNA damage response, LexA. Induction of several pore-forming colicin genes is also repressed by IscR, which ensures that colicin genes are switched on as a last resort in DNA damaged cells, when nutrients are depleted. Here we show that nuclease colicin genes are not controlled by IscR but that the AsnC protein, in concert with LexA, is directly responsible for uncoupling the immediate expression of the DNase colicin E8 from the main induction of the SOS response. AsnC wraps the DNA of the colicin E8 promoter into a complex nucleoprotein assembly and the architecture of this complex is altered by the presence of the amino acid L-asparagine. Thus, repression by metabolite-responsive and DNA-damage responsive regulators operates at the regulatory regions of different colicins. Hence, the response to several environmental signals have been integrated to ensure that, following DNA damage, colicin synthesis is tightly repressed and induced only in terminally damaged cells.
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005354
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005354Summary
Colicins are considered model proteins for studying bacterial toxins. These narrow spectrum antibiotics can kill by a variety of mechanisms, e.g. by forming pores in the membranes of susceptible cells or by degrading their nucleic acids. Colicin genes are plasmid-encoded and repressed by the master regulator of the DNA damage response, LexA. Induction of several pore-forming colicin genes is also repressed by IscR, which ensures that colicin genes are switched on as a last resort in DNA damaged cells, when nutrients are depleted. Here we show that nuclease colicin genes are not controlled by IscR but that the AsnC protein, in concert with LexA, is directly responsible for uncoupling the immediate expression of the DNase colicin E8 from the main induction of the SOS response. AsnC wraps the DNA of the colicin E8 promoter into a complex nucleoprotein assembly and the architecture of this complex is altered by the presence of the amino acid L-asparagine. Thus, repression by metabolite-responsive and DNA-damage responsive regulators operates at the regulatory regions of different colicins. Hence, the response to several environmental signals have been integrated to ensure that, following DNA damage, colicin synthesis is tightly repressed and induced only in terminally damaged cells.
Introduction
Colicins are high-molecular-weight toxic proteins that are produced by and specifically target Escherichia coli and its close relatives [1]. These narrow-spectrum antibiotics kill by either targeting the DNA, RNA or cell membranes of susceptible cells. Cytoplasmic colicins are released upon the synthesis of a lysis protein, the expression of which is independent of intracellular colicin accumulation [2]. This causes the stochastic lysis of producing cells and is suggested to assist surviving sister cells by killing potential competing sensitive cells [3]. Colicin-mediated competition has been suggested to have functions in modulating population dynamics and maintaining diversity of microbial communities [4–7]. Nutrient limitation and DNA damage seem to be the major signals that control colicin production, enabling interference competition among strains [8].
Colicins are plasmid-encoded and are expressed from strong promoters whose activity is tightly repressed by the LexA transcription factor, the master regulator for the SOS DNA damage repair response in bacteria [1,9]. Most of the SOS genes involved in DNA repair and cell division arrest are expressed immediately after DNA damage, but induction of colicin genes is delayed. This presumably provides cells time to repair DNA in order to preserve the integrity of their genome, before the induction of colicin production [10]. In previous work, we established that the global transcriptional repressor IscR delays the induction of the pore-forming colicin K gene (cka) [11]. We showed that IscR participates in a double-locking mechanism, in concert with LexA, by stabilizing the LexA SOS repressor at the promoter and this links colicin expression to the nutritional status of the cell. Thus, the IscR protein uncouples the induction of colicin expression from the temporal induction of the SOS response that deals with repairable DNA damage. This mechanism also operates at other promoters, which control the expression of bactericidal pore-forming colicins [11], however, it is not known if a similar fail-safe double-lock system has also evolved for the nuclease colicin genes.
Here we report that IscR does not modulate the expression of nuclease colicin genes. Hence, we studied the regulation of the DNA degrading colicin E8 gene (cea8) in more depth and identified the AsnC transcription factor as directly responsible for the delay in cea8 expression. AsnC is a member of Lrp/AsnC family of transcriptional regulators that modulate cellular metabolism in both archaea and bacteria [12,13]. In E. coli, AsnC is required to activate the expression of the L-asparagine synthetase A gene (asnA) and this stimulation is abolished in the presence of the amino acid L-asparagine [14]. In addition to this, expression of asnC is negatively autoregulated by AsnC and also repressed by the nitrogen assimilation control (Nac) protein, under nitrogen-limiting conditions [15], however, this regulation is not modulated by the presence of L-asparagine [14]. Functional E. coli AsnC is an octamer, whose structure was resolved by X-ray crystallography [13]. We show that AsnC binds to the cea8 regulatory region at multiple sites, likely wrapping the DNA into a nucleoprotein assembly and that its binding is affected by L-asparagine. In the LexA-AsnC-cea8 complex, two LexA dimers are flanked by multiple AsnC octamers, and the presence of L-asparagine influences AsnC modulated promoter region geometry. Data presented here shows that double locking by LexA and AsnC operates at the cea8 promoter region to delay induction of the colicin E8 gene, thereby linking cea8 expression to DNA damage and L-asparagine availability. Thus, AsnC provides colicinogenic cells with time for DNA damage repair and limits colicin E8 induction to terminally damaged cells.
Results and Discussion
IscR does not regulate the induction of the DNA or RNA degrading colicins
To study the induction of various nuclease colicins after DNA damage, we assayed the activity of colicin promoters in E. coli K-12 strain BW25113. In this experiment, different DNA fragments, carrying colicin promoters, were cloned into the lac expression vector, pRW50, to give colicin promoter::lac fusions. After inducing the DNA damage response with a sub-inhibitory concentration of nalidixic acid, we observed that the promoters of the DNA degrading colicins E2, E7 and E8, of colicins E5 and D, targeting tRNA, and of the rRNA cleaving colicin E6, are only induced after a prolonged delay (Fig 1A). Note that there is little expression from nuclease colicin promoters in the absence of DNA damage (S1 Fig).
Fig. 1. IscR does not modulate the temporal induction of nuclease colicins. 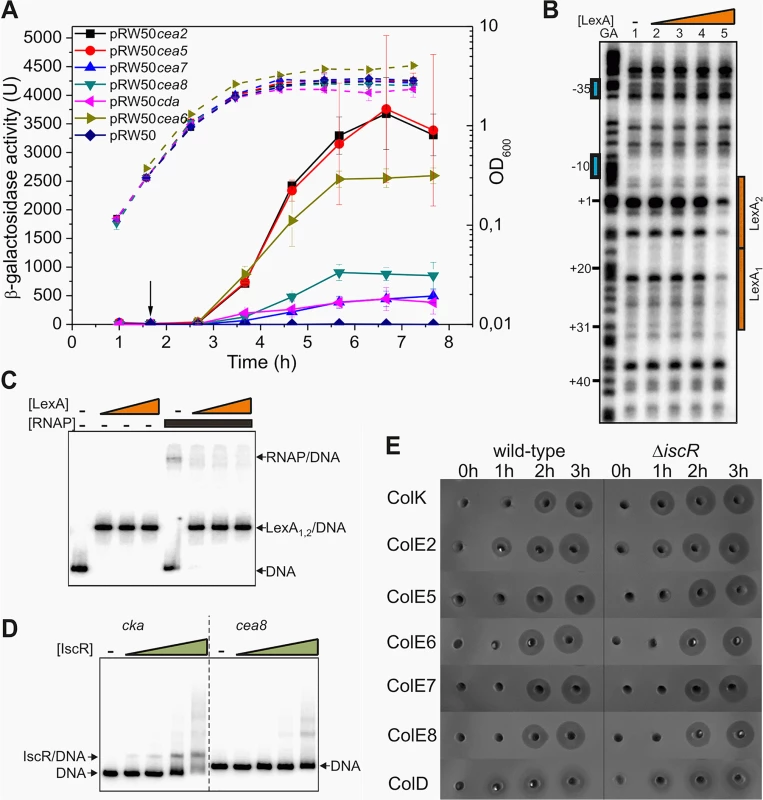
A) Measured β-galactosidase activities of strain BW25113, carrying the lac expression vector pRW50 containing various nuclease colicin promoter fragments. Each value represents the mean ± SD of at least three independent measurements, the arrow indicates the time of addition of a sub-lethal concentration of nalidixic acid (37 μM) and the dashed lines represent optical density (OD600). B) The binding of purified LexA protein to a P32 end-labelled cea8 fragment was investigated using DNase I footprint analysis. The location of the two LexA binding sites, LexA1 and LexA2 is indicated by orange boxes and the -10 and -35 promoter elements by blue boxes. The final concentration of LexA protein used in reactions was 12.5, 25, 50 and 100 nM (lanes 2–5). C) The binding of purified LexA and RNA polymerase (RNAP) to a P32 end-labelled cea8 fragment was investigated using EMSA analysis. DNA fragments were first incubated for 15 min with various concentrations of LexA (100, 200 or 400 nM) followed by the addition of 100 nM RNAP with further incubation for 15 min at 37°C. The location of free DNA, the LexA/DNA and RNAP/DNA complexes is indicated. D) The binding of purified IscR protein to P32 end-labelled colicin K (cka) or cea8 promoter fragments was assayed using EMSA analysis. The concentration of IscR used was 0.4, 0.8, 1.6 and 3.2 μM. The location of free DNA and the IscR/DNA complex is marked. E) Assays of colicin production in BW25113 and its ΔiscR derivative cells, carrying various colicin-encoding plasmids. Equal amounts of cells were collected at hourly time points after the addition of nalidixic acid (0 h) and crude cell extracts were placed into wells in agar plates overlaid with soft agar, harbouring the E. coli K-12 indicator strain DH5α pBR322. Experiments were performed in duplicate and representative growth curves are shown in S2 Fig. We previously established that the delayed expression of several pore-forming colicins, is due to co-repression by the global transcriptional repressors LexA and IscR [11]. At the colicin K promoter, the LexA repressor was shown to bind to the tandem operators just downstream of the -10 promoter element and prevented RNA polymerase binding. The IscR protein was suggested to increase stability of LexA at these targets. To determine if a similar mechanism controls the expression of the nuclease colicins, we investigated the regulation of the DNA degrading colicin E8 by electrophoretic mobility-shift assays (EMSA) and DNase I footprinting. Our results reveal that at the promoter region of cea8, the LexA repressor binds to two overlapping targets and blocks the access of RNA polymerase to the promoter (Fig 1B and 1C). To investigate the possible binding of IscR to the cea8 promoter, an EMSA assay was again used. Results in Fig 1D show that IscR binds specifically to the cka promoter region but not to that of cea8. In addition, we tested whether the IscR protein is directly responsible for the delayed production of colicin E8 and other nuclease colicins, by comparing the colicin production in our wild-type and ΔiscR strains. Results, illustrated in Fig 1E, show that IscR has a negligible effect on the synthesis of many nuclease colicins. This contrasts with the pore-forming colicin K, where the ΔiscR allele causes a 100 increase after the first hour of induction (S3 Fig) [11]. This indicates that IscR does not regulate any of the nuclease colicin genes and prompted us to search for other transcription factors, involved in controlling the timing of their expression.
AsnC delays cea8 expression
To investigate the delay in cea8 induction in SOS-induced cells we used a pull-down assay [11], using a cleared cell extract from mid-logarithmic grown, SOS induced, E. coli cells, and a biotinylated 179 bp cea8 promoter fragment as a bait. Eluted proteins were separated by SDS-polyacrylamide gel electrophoresis and nine bands were analysed by mass spectroscopy (Fig 2A). We identified 30 transcription regulators and nucleoid associated factors that had associated with the bait (S1 Table). To screen for their ability to regulate cea8 expression after DNA damage induction with nalidixic acid, we measured cea8::lac activity in deletion mutants from the Keio collection [16] and we selected strains in which a 3-fold increase in cea8 promoter activity, in comparison to the wild-type strain, was observed (S2 Table). Thus, we focused on the AsnC, StpA, OmpR, YbjK, YihW, YegW and MngR proteins and measured cea8 promoter activity following SOS induction using pRW50 cea8::lac fusion in the corresponding deletion mutant strains throughout the bacterial growth curve. Results presented in Fig 2B show that disruption of asnC resulted in the biggest effects on cea8 promoter induction after DNA damage. An intermediate increase in promoter activity was observed in the strain deficient for stpA, whilst the other deletions had a minimal effect, with our data confirming that IscR does not regulate colicin E8 expression. The StpA protein, a paralogue of the nucleoid-associated protein H-NS, forms a rigid filament along DNA, and can cause DNA bridging [17]. Furthermore, StpA can act as an RNA chaperone [18] and a transcriptional repressor [19,20], thus, it may be involved in colicin gene expression. However, here we focused on AsnC, and assayed its binding to cea8 promoter region and its effect on colicin E8 synthesis. To do this, we introduced the ΔasnC allele into a strain that harbours a cea8-encoding plasmid. After treatment of cells with a subinhibitory concentration of nalidixic acid that induced DNA damage, cell growth and colicin production was compared in the wild-type and the ΔasnC mutant. Our results show that AsnC enhances viability of the strain harbouring the colicin E8-encoding plasmid (Fig 2C). Bioassays were also used to follow colicin levels in crude cell extracts prepared from cells before and after SOS induction. The results show that, in the ΔasnC strain, colicin E8 was produced an hour earlier in comparison to the delayed synthesis in the wild-type strain (Fig 2D). This suggests that AsnC directly represses cea8 promoter activity and, in concert with LexA, ensures regulated and delayed expression of the cea8 gene.
Fig. 2. AsnC delays cea8 expression. 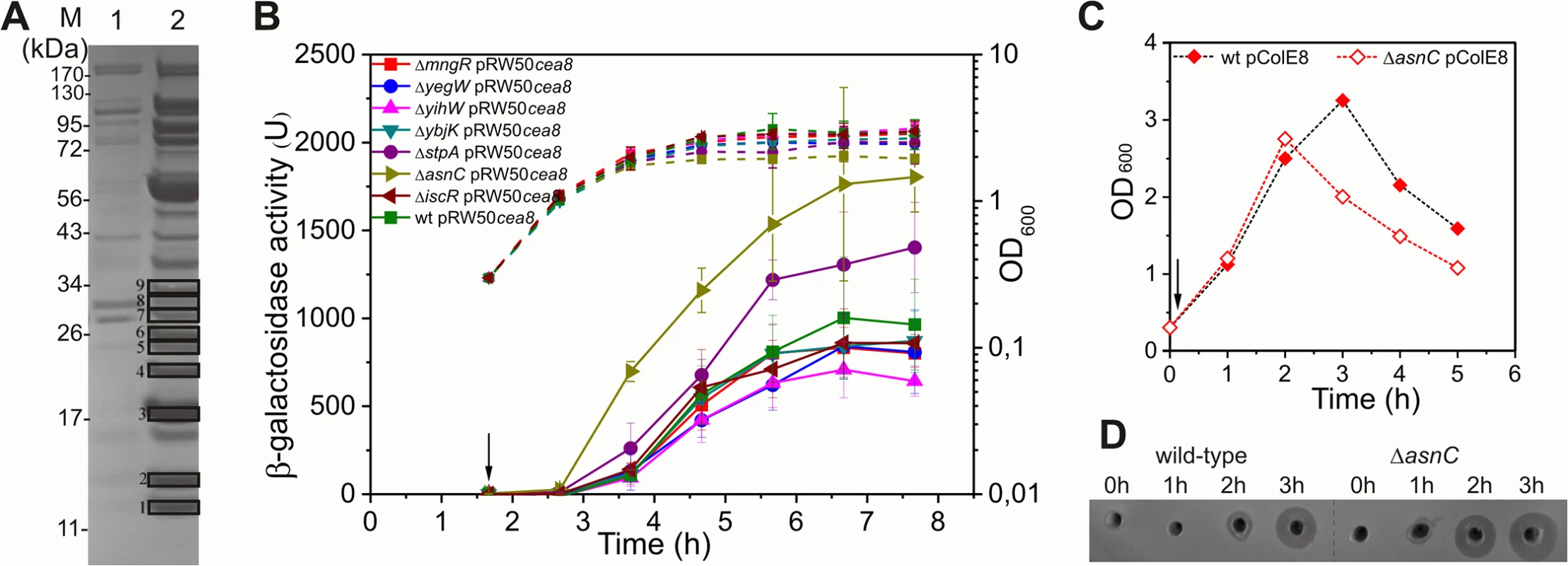
A) A Coomassie blue stained SDS-PAGE protein gel, which shows the protein profile of the eluates obtained from a control (lane 1) and cea8 affinity chromatography (lane 2) experiment. Proteins 1–9, which are denoted by boxes, were trypsin digested and analysed by mass spectrometry. Candidate proteins identified by affinity chromatography are listed in S2 Table. B) The cea8 promoter region was cloned into the lac expression vector pRW50 and transformed into E. coli strain BW25113 (wt) and various mutant derivatives. The arrow indicates the time of addition of nalidixic acid (37 μM) and the dashed line represents optical density (OD600). β-galactosidase activity was determined at hourly intervals. Each value represents the mean ± SD of at least three independent measurements. C) Representative growth curves of BW25113 (wt) and ΔasnC cells, carrying plasmid pColE8, which naturally expresses the DNA degrading E8 colicin. The arrow indicates the time of addition of nalidixic acid. D) Assays of colicin production in BW25113 and ΔasnC cells, carrying the pColE8. Equal amounts of cells were collected at hourly time points after the time of addition of nalidixic acid (0 h), indicated by an arrow in panel C, and crude cell extracts were applied into wells in agar plates overlaid with soft agar, harbouring the colicin sensitive strain DH5α pBR322. All experiments were performed in duplicate. L-asparagine affects interaction of AsnC with cea8 promoter region
The AsnC protein is a member of the Lrp/AsnC family of regulators, which often assemble to form wheel-like octamers and whose DNA binding activity can be modified by small molecules, such as amino acids [13]. AsnC regulates the expression of its own gene, asnC, and the asnA gene, encoding for a synthetase that catalyses the ammonia-dependent conversion of aspartate to asparagine [14]. To investigate the binding of AsnC to the cea8 promoter (Fig 3A), we over-expressed and purified the AsnC protein and performed in vitro experiments in the presence or absence of the amino acid L-asparagine. EMSA experiments show that several AsnC molecules can interact with cea8 (Fig 3) and that in the presence of L-asparagine a number of distinct complexes can be observed (Fig 3B). In the absence of L-asparagine, at higher AsnC concentrations, DNA remained in the wells of the gel, indicating that high molecular weight nucleoprotein complexes had formed.
Fig. 3. Binding of AsnC protein to cea8 is altered by L-asparagine. 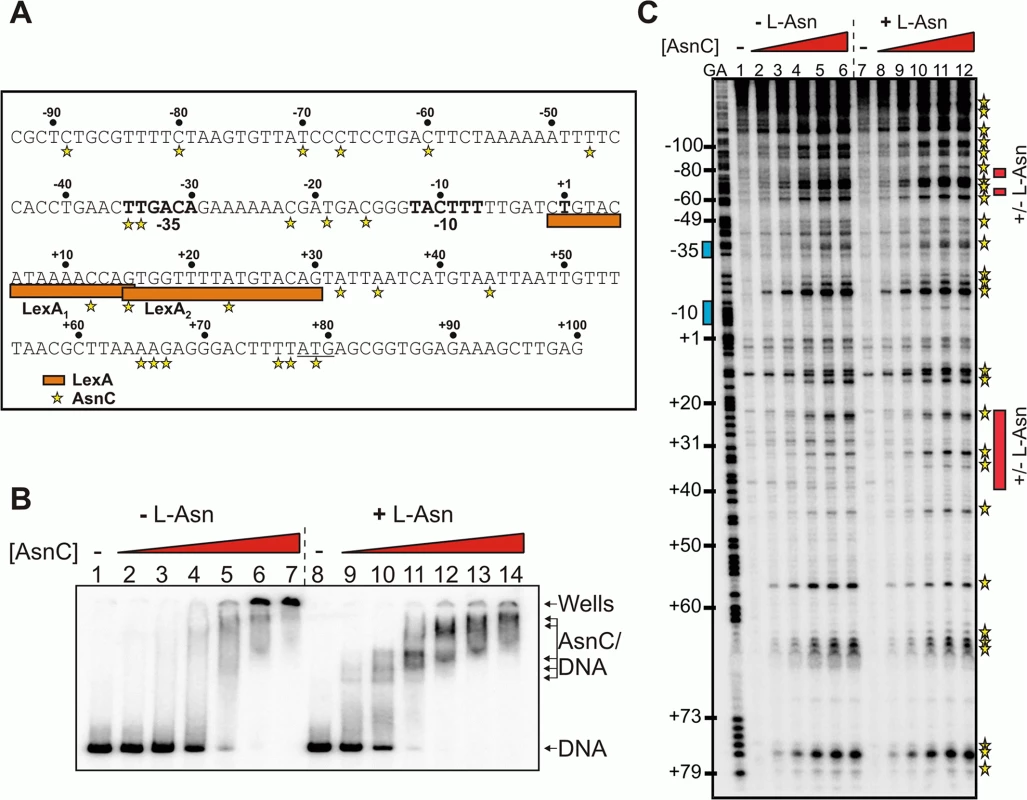
A) Organisation of the colicin E8 promoter region. The figure shows the DNA sequence of the cea8 promoter from position -93 to +100. The -35 and -10 core promoter elements and the predicted start of transcription (+1) are shown in bold type and the start of translation (ATG) is underlined. The location of the two LexA binding sites (LexA1 and LexA2) is shown by orange boxes. The AsnC-induced hypersensitive sites observed by DNase I footprinting are starred. B) The binding of purified AsnC protein to a P32 end-labelled cea8 fragment in the presence and absence of L-asparagine (± L-Asn) was investigated using EMSA analysis. The concentration of AsnC in lanes 2–7 and 9–14 was 0.5, 1.05, 2.1, 4.2, 8.4 and 12.6 μM, respectively. The location of free DNA, the position of the wells and the various AsnC/DNA complexes is indicated. C) An in vitro DNase I footprint experiment analysing the binding of purified AsnC to the cea8 promoter. The concentration of AsnC in lanes 2–6 and 8–12 was 0.5, 1.0, 2.1, 4.2, and 6.3 μM, respectively. The AsnC-induced hypersensitive sites are starred (also see panel A) and the location of the different protection patterns observed due to L-asparagine is indicated by red boxes. DNase I footprinting was also used to study the location of AsnC binding to the cea8 promoter sequence, again in the presence or absence of L-asparagine. Results in Fig 3C show that AsnC interacts along the entire length of the 179 bp cea8 promoter region. Inspection of the cea8 region, interacting with AsnC, revealed 27 DNase I hypersensitive sites, which is indicative of local bending and distortion of the DNA helix. This results in a widening of the minor groove and makes the DNA more susceptible to DNase I attack, leading to the production of hypersensitive bands [21]. In several locations the presence of L - asparagine altered the binding of AsnC to the cea8 promoter (see red boxes in Fig 3C). The position of the red boxes in Fig 3C was determined by comparing the AsnC footprint gels in the presence or absence of L-asparagine. Our in vitro analysis indicates that AsnC binds to the cea8 promoter region at multiple sites, likely wrapping the DNA into a complex nucleoprotein assembly, and that the architecture of this complex is altered by the presence of L-asparagine (Fig 3).
LexA and AsnC can bind to the cea8 promoter region simultaneously
Since our data show that AsnC binds at multiple locations and alters the architecture of colicin E8 regulatory region, as well as binding within LexA target sites (Fig 3), we tested if AsnC and LexA can simultaneously bind to the cea8 promoter region. To investigate this, we performed EMSA analysis on the cea8 promoter fragment. Using purified LexA and AsnC, in the presence of L-asparagine, we observed a large nucleoprotein complex composed of at least two AsnC functional oligomers, presumably octamers [13], and two LexA dimers interacting at cea8 (Fig 4A). Note, that LexA was used at a concentration of 400 nM at which LexA repressor occupies both LexA binding sites within cea8 (Fig 1B). To determine whether occupancy of the DNA by AsnC affects the binding of LexA at the cea8 promoter region, we performed DNase I footprint analysis and compared signatures of LexA and AsnC in the presence or absence of L-asparagine. In both conditions, LexA repressors bound to tandem targets just downstream of the -10 promoter element (Fig 4B). As observed for AsnC binding in Fig 3C, the addition of L-asparagine also modulated the binding of AsnC in the LexA-AsnC nucleoprotein complex (Fig 4B and 4C). In the AsnC-LexA-cea8 complex, specific hypersensitive sites were apparent (determined by stars in Fig 4B), suggesting that the binding of both proteins subtly alters the structure or trajectory of the DNA around -10 element (Fig 4D–4F). Thus, we conclude that concurrent binding of LexA and AsnC to the cea8 regulatory region ensures delayed induction of the DNase E8 synthesis after DNA damage.
Fig. 4. LexA and AsnC can bind to cea8 promoter region simultaneously. 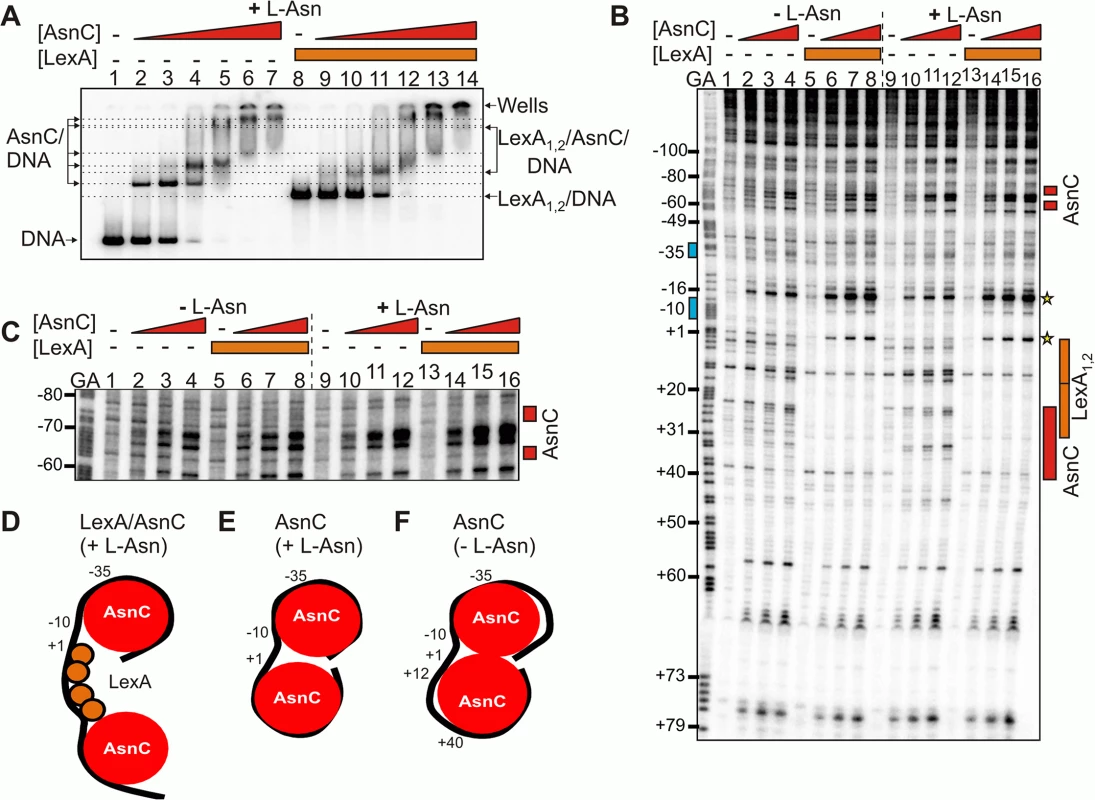
A) The panel shows an EMSA analysing the binding of purified LexA and AsnC protein to a P32 end-labelled cea8 fragment in the presence of L-asparagine (+ L-Asn). The concentration of AsnC in lanes 2–7 and 9–14 was 0.5, 1.05, 2.1, 4.2, 8.4 and 12.6 μM, respectively. LexA protein was present in reactions at concentration of 400 nM. The location of free DNA, the position of the wells and the various protein/DNA complexes is marked. B) The binding of purified LexA and AsnC proteins to the cea8 promoter fragment was studied by DNase I footprinting in the presence and absence of L-asparagine (± L-Asn). AsnC was included at concentrations of 1.0, 2.1 and 4.2 μM and LexA at a concentration of 400 nM. The prominent hypersensitive bands, corresponding to positions +1 and -16, that are produced by the concurrent binding of AsnC and LexA, are starred. Red boxes indicate the position of AsnC interactions, which are affected by L-Asn. The location of the two LexA binding sites, LexA1 and LexA2, is indicated by orange boxes and the -10 and -35 promoter elements by blue boxes. C) The panel shows an extended run of the footprint analysis from panel B, focusing on positions -80 to -60 upstream of the transcription start site. Labels are identical to those described above. Panels D-F) show models of the different nucleoprotein complexes formed at the cea8 promoter by the binding of LexA and AsnC. The cea8 promoter is sensitive to L-asparagine
Our data suggest that tight repression of DNase colicin E8 might be affected by the availability of amino acid L-asparagine and this signal is relayed via AsnC. To test this hypothesis we measured cea8 promoter activity in SOS-induced wild-type cells grown in the M9 minimal medium containing either 10 mM NH4Cl or 20 mM L-asparagine as the sole source of nitrogen. Fig 5 shows that in the L-asparagine containing medium, the expression from cea8 remains low, whilst it increases in medium containing higher levels of NH4Cl. This is in agreement with our in vitro data (Figs 3 and 4) and suggests that L-asparagine is needed to stabilize a specific AsnC assembly at the cea8 promoter region. Hence, we suggest that depletion of L-asparagine is the signal for AsnC de-repression at the cea8 promoter.
Fig. 5. The cae8 promoter is sensitive to the amino acid L-asparagine. 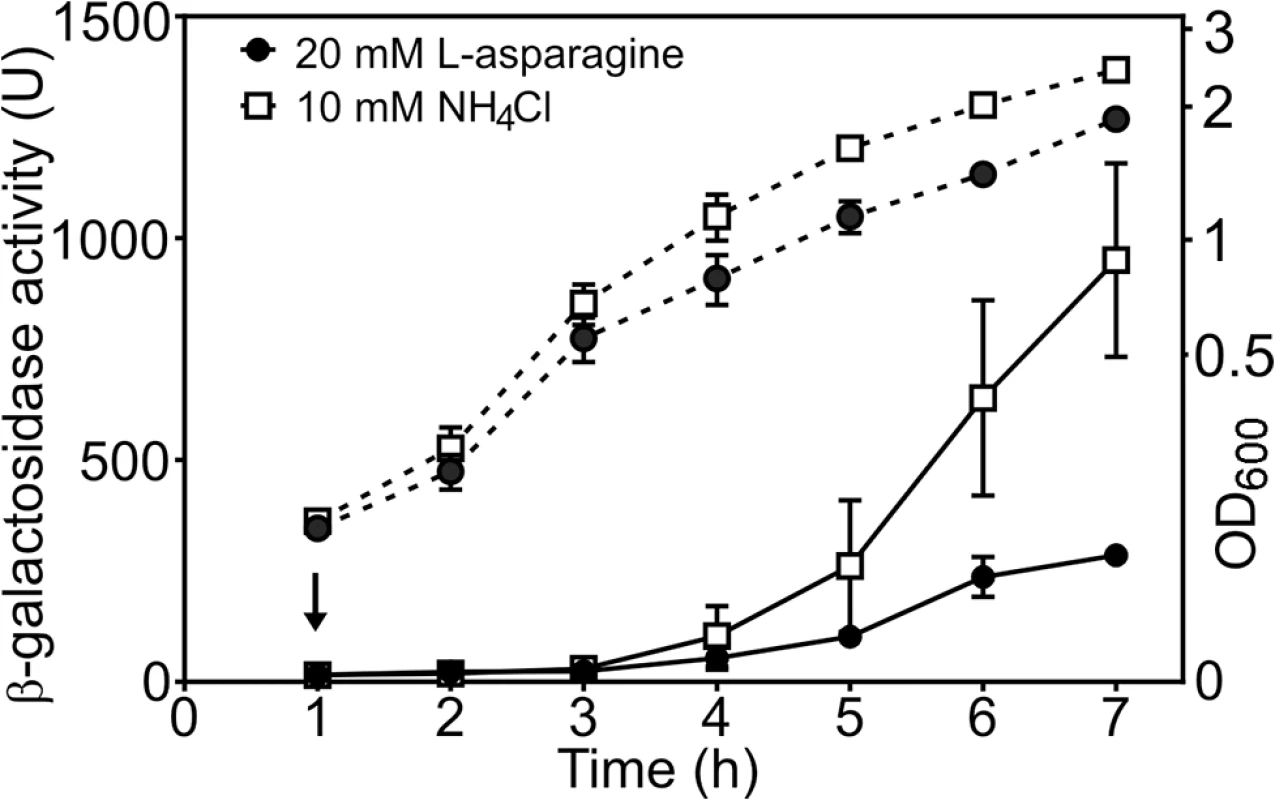
The figure shows measured β-galactosidase activities from wild-type BW25113 cells, carrying the cea8 promoter region subcloned into pRW50. Cells were grown in M9 minimal medium containing 0.5 mM NH4Cl until an OD600 of ~0.2, when the culture was split in to two and grown further in the presence of either 10 mM NH4Cl or 20 mM L-asparagine. The arrow indicates the time of addition of nalidixic acid and the dashed lines represent OD600. Each value is the average of duplicate experiments and the standard deviation is shown. AsnC regulates the expression of different nuclease colicins
To determine whether AsnC modulates the expression of other colicins, we assayed DNase colicin E2 (cea2) and rRNase colicin E6 (cea6) promoter activities following SOS induction with nalidixic acid using cea6::lacZ and cea2::lacZ promoter fusions in the wild-type, ΔiscR and ΔasnC strains. Results illustrated in Fig 6A and 6B show that the disruption of asnC resulted in elevated cea2 and cea6 promoter activity immediately after DNA damage induction, when compared to the wild-type and the ΔiscR strains. This indicates that AsnC, rather than IscR, is a key transcriptional repressor of the cea2 and cea6 promoters. In addition, we transferred the colicinogenic plasmids for these DNase and RNase colicins into the ΔasnC and wild-type strains. Following the induction of DNA damage, cell growth (Fig 6C) and colicin production (Fig 6D) was monitored in both strains. In the absence of asnC, cells failed to reach as high optical density as in the wild-type strain, suggesting that elevated colicin expression and cell lysis had taken place (Fig 6C). Cell extracts were prepared from cultures before and after DNA damage induction and colicin levels compared by a colicin production bioassay (Fig 6D). After SOS induction, nuclease colicin synthesis was induced earlier for colicins E2, E5 and E6 in the ΔasnC strain, in comparison to the wild-type strain, indicating that AsnC directly modulates the expression of a number of other colicin genes. Alignment of the cea8 promoter region sequence with corresponding sequences from colicin E2, E5 and E6 indicated that these promoters are very similar (S4 Fig) and, thus, similar co-ordinated regulation is perhaps to be expected.
Fig. 6. AsnC modulates the temporal induction of different DNA- and RNA-degrading colicins. 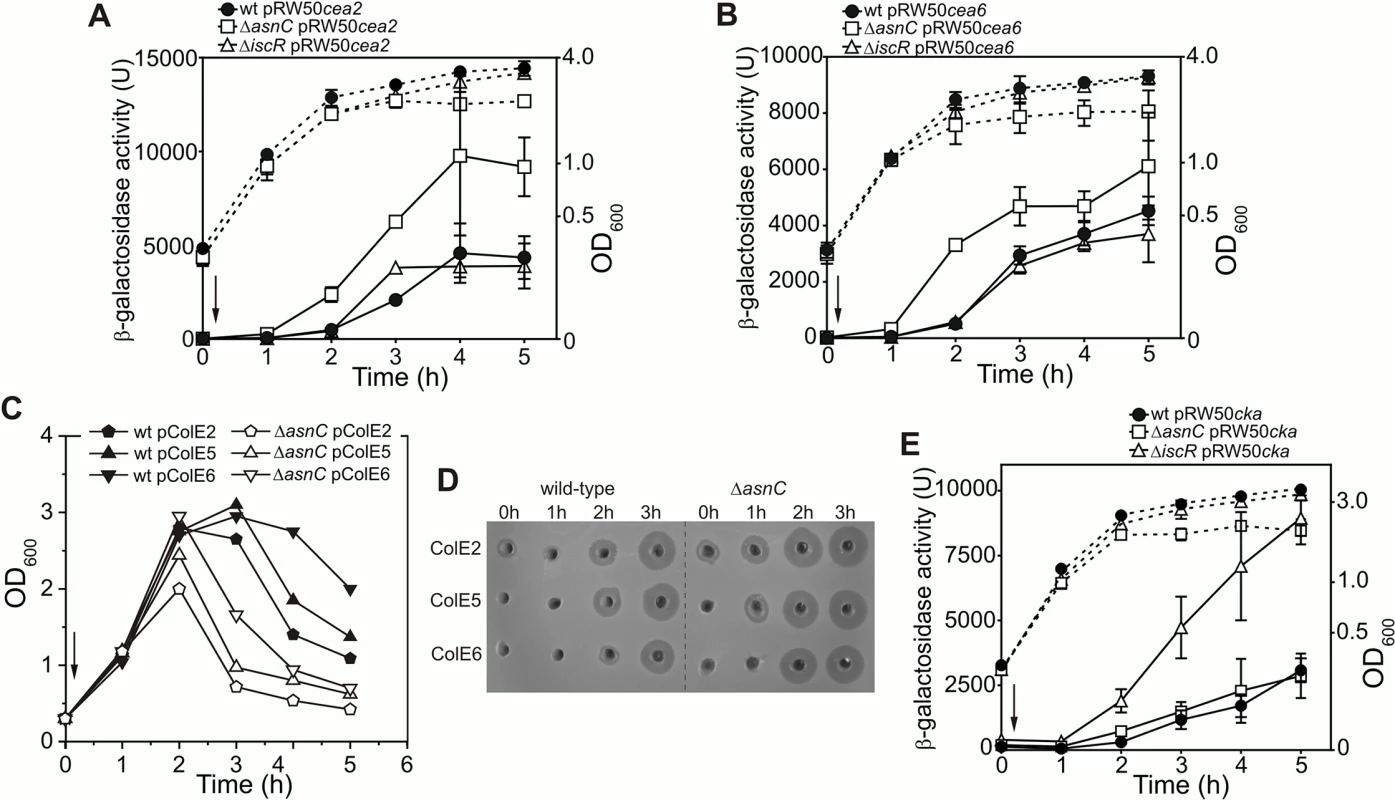
Expression of the (A) cea2::lacZ and the (B) cea6::lacZ fusions in strain BW25113 (wt) and the ΔiscR and ΔasnC mutants. For panels A and B each value represents the mean ± SD of at least two independent measurements, the arrow indicates the time of the addition of a sub-lethal concentration of nalidixic acid (37 μM) and the dashed lines represent OD600. C) Growth curves of BW25113 (wt) and ΔasnC cells harbouring naturally occurring plasmids encoding either the DNA degrading colicin E2, the tRNA cleaving colicin E5 or the rRNA cleaving colicin E6. The arrow indicates the time of addition of nalidixic acid. Experiments were performed in duplicate and representative growth curves are shown. D) Assays of colicin production in BW25113 and ΔasnC cells, carrying various colicin-encoding plasmids. Equal amounts of cells were collected at hourly time points after the addition of nalidixic acid (0 h) and crude cell extracts were applied into wells in agar plates, overlaid with soft agar harbouring the colicin sensitive strain DH5α pBR322. The experiments were performed in duplicate. E) AsnC protein has little effect on the expression of the pore-forming colicin K. The activity of the cka promoter was measured in strain BW25113 (wt) and its ΔiscR or ΔasnC mutant derivatives. Each value represents the mean ± SD of four independent measurements, the arrow indicates the time of addition of a sub-lethal concentration of nalidixic acid (37 μM) and the dashed lines represent the OD600. It is clear from S4 Fig that regions of the cka promoter are similar to that of cea8, particularly around the two LexA SOS boxes. As this region was bound differentially by AsnC at cea8 in the presence of L-asparagine, we examined whether purified AsnC could bind in vitro to a radiolabelled cka promoter fragment using EMSA. Results in S5A Fig indicated that AsnC does bind to cka and its DNA binding was modulated by L-asparagine. As this raises the possibility that AsnC could regulate colicin K production in vivo, we measured cka promoter activity after SOS-induction in wild-type, ΔasnC and ΔiscR cells, carrying a cka::lacZ fusion cloned into pRW50. Results in Fig 6E indicate that AsnC had little effect on cka expression, but confirmed that IscR is a major repressor of the cka promoter. In addition, colicin K expression was also examined in our wild-type, ΔasnC and ΔiscR strains, whilst carrying a colicin K-encoding plasmid (S5B and S5C Fig). These experiments again showed that IscR is the major regulator of colicin K expression and that AsnC has little effect on the expression of this pore-forming colicin.
Concluding remarks
E. coli harbours many promoters that are regulated by multiple transcription factors, each of which ensures that different intra - or extracellular signals are integrated into gene expression [22]. At the DNase colicin E8 regulatory region we identified a large nucleoprotein complex composed of two LexA repressors flanked by at least two AsnC octamers, that likely wrap DNA in a nucleosome-like structure to firmly prevent transcription initiation (Fig 4D–4F).
The AsnC protein belongs to the Lrp/AsnC family of transcriptional regulators that are widely distributed among prokaryotes and affect cellular metabolism, often in response to exogenous amino acids [12,13]. In contrast to other members of the family, which are global regulators and affect a variety of bacterial functions [23], the AsnC protein was thought to be a gene specific regulator, controlling only two genes in E. coli (asnA and asnC) [14]. Here we report a novel role for AsnC, in which the promoters of the nuclease colicins have “recruited” this protein, enabling regulation in response to L-asparagine levels.
Our data show that an amino acid effector modulates AsnC interaction at the colicin E8 promoter, which influences regulation of cea8 expression. We predict that, as for the Neisseria meningitidis AsnC ortholog, [24], L-asparagine binding modulates the stability of a certain protein oligomeric state and also the mode of binding in the E. coli AsnC-cea8-LexA nucleoprotein complex. Furthermore, as expression of asnC is negatively autoregulated and dependent on the Nac protein under nitrogen-limiting conditions [15], nutrient conditions, specifically nitrogen levels, and nitrogen metabolism might coordinate the cea8 expression through altering the amount of AsnC within the cell. Note that nutrients were recently reported to modulate the release of the DNase colicins by modulating the translation efficiency of the colicin E2 lysis gene transcript [25]. Therefore, AsnC appears to couple metabolic signals to the induction of colicin operon components, in order to synchronise accumulation with the release of the colicin.
In our previous work, we showed that the global transcriptional factor, IscR, in response to the nutritional status of the cell, and, co-dependently with LexA, delays induction of the pore-forming colicin genes after SOS induction [11]. This was a surprising finding as E. coli IscR had been thought to be primarily involved with controlling housekeeping iron sulphur cluster biogenesis, anaerobic respiration enzymes and biofilm formation [26,27]. Here our data strongly suggests that temporal induction of DNA and RNA targeting colicins is IscR independent, and show that the key regulator is the AsnC repressor. At the cea8 promoter, AsnC repression seems to reflect L-asparagine levels and presumably serves as an indicator of general amino acid abundance and availability. In contrast, AsnC does not affect the expression of pore-forming colicin K gene expression. Thus, our data imply that the promoters of the nuclease and pore-forming colicins have adopted different transcription regulators to co-ordinately regulate transcription in conjunction with the LexA repressor and distinct metabolic inputs are integrated at these promoters, which both affect the timing and level of colicin induction.
It is clear that colicinogenic plasmids seem to have evolved to exploit transcriptional factors that are of the host origin. We suggest that ubiquitous regulators, present in most E. coli strains were “picked” in order that the colicinogenic plasmids can be swapped between strains [1], with the colicin promoters being silenced in the same manner. In addition, colicin production and subsequent lysis protein driven colicin release causing death of the producing bacteria, may enable eradication of strains that lack or synthesize a non-functional regulator and cannot efficiently respond or adapt to different environmental signals.
Material and Methods
Bacterial strains and plasmids
The bacterial strains, plasmids, promoter fragments and oligodeoxynucleotide primers used in the present study are listed in S3 Table. The E. coli Keio collection wild-type strain, BW25113, and its derivatives were used throughout the study [16]. To verify the Keio collection deletion strains, a transposon-specific primer Keio1Kn [16] and the gene specific primer (named as gene pre) was used (S3 Table) in 30 cycles of PCR reactions (30 sec 94°C, 30 sec 55°C, 60 sec 72°C). PCR products were analysed on 1.2% agarose gels and stained with ethidium bromide. The colicin D (cda), E2 (cea2), E5 (cea5), E6 (cea6), E7 (cea7) and E8 (cea8) promoter fragments were amplified by PCR from natural colicin encoding plasmids using primers colX_beta_F and colX_beta_R (X denotes the relevant colicin), which introduce flanking EcoRI and HindIII sites (S3 Table). For testing colicin promoter activities, each promoter fragment was cloned into the lac expression vector, pRW50. Plasmid constructs were named as pRW50cxay (x and y denotes each colicin). As a source of DNA fragments for in vitro analysis, EcoRI-HindIII colicin E8 and colicin K promoter fragments were cloned into pSR. To assay colicin synthesis and ensure plasmid selection, the transposon Tn3 (ApR) was inserted into the naturally occurring colicinogenic plasmids of the Pugsley colicin collection [28], harbouring operons for either colicin D, E2, E5, E6, E7 or E8. Strain CL127 carrying Tn3 on the conjugative plasmid pHly152-T8 was used as a donor strain. To generate plasmid pAsnC, for the overexpression of the N-terminal His6 AsnC fusion protein, primers asnC_u and asnC_d were used to PCR amplify the asnC open reading frame and introduce flanking BamHI and MluI restriction sites. Purified PCR product was subsequently cloned into expression vector pET8c (Novagen) to generate pAsnC.
Proteins
E. coli RNA polymerase holoenzyme harbouring σ70 (RNAP) was obtained from Epicentre Technologies (Madison). The His6-LexA protein was overexpressed and purified as described in [29] and stored in 20 mM Tris (pH 7.3), 200 mM NaCl at -80°C. The His6-IscR protein was overexpressed, purified and its concentration determined as described in [11]. To induce the synthesis of AsnC protein, an overnight culture of E. coli BL21 (DE3)pLysE strain grown on an agar plate, containing ampicilin (100 μg ml-1) and chloramphenicol (25 μg ml-1), harbouring pAsnC was grown to an optical density at 600 nm (OD600) of 0.6 when 0.8 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the culture. After 4 h of growth the cells were harvested and the N-terminally His6-tagged AsnC was affinity purified by Ni-chelate chromatography (Quiagen) and stored at 4°C in 50 mM NaH2PO4 (pH 8), 300 mM NaCl, 250 mM imidazole. The concentrations of the LexA and AsnC proteins were determined using a NanoDrop 1000 (Thermo Scientific) using the extinction coefficients at 280 nm of 6990 M-1 cm-1 and of 10555 M-1 cm-1, respectively.
β-Galactosidase assays
The low-copy number lac expression vector, pRW50 [30], was used to measure the activity of the colicin promoters. Plasmids harbouring colicin promoter fragments (S3 Table) were transformed into the relevant strains. Cells were grown aerobically (180 r.p.m.) at 37°C in Lysogeny Broth (LB) supplemented with tetracycline (12.5 μg ml-1). To induce the SOS response, a sub-inhibitory concentration [31], 37 μM, of nalidixic acid (Sigma-Aldrich) was added to the culture when the OD600 reached 0.3. Culture samples were assayed for β-galactosidase activity according to the Miller method [32]. The presented values are the averages of at least three independent experiments and are shown with standard deviations. To measure the L-asparagine effect on cea8 promoter activity, the relevant wild type strain carrying pRW50cea8 was grown to an OD600 ~0.2 in M9 medium [33], containing a low concentration of NH4Cl (0.5 mM) to sustain growth. The bacterial culture was then split and supplemented with either 10 mM NH4Cl or 20 mM L-asparagine. After the addition of 37 μM nalidixic acid, as indicated, samples were taken and analysed as described above.
Affinity purification of proteins interacting with the colicin E8 promoter region
A biotinylated 179 bp colicin E8 promoter fragment from position -169 to position +10 from the translation start site (TSS) was generated by PCR using primers Pull_FE8 and Pull_RE8 with pColE8-Tn3 as a template. The DNA fragment was purified by GeneJET PCR purification kit (Thermo Scientific). Immobilisation of the biotinylated DNA (50 μg) to 5 mg of M-280 streptavidin Dynabeads (Invitrogen) was carried out in 15 minutes at room temperature as described [11]. An overnight culture of the E. coli BW25113, harbouring the pRW50cea8, was diluted 1 : 200 into 0.5 l LB broth supplemented with tetracycline (12.5 μg ml-1) and induced with nalidixic acid (37 μM) once the OD600 had reached 0.3. After 45 min, cells were harvested and cell extract prepared as described [31]. Cleared lysates (~20 ml) were mixed with streptavidin beads with or without cross-linked biotinylated cea8 promoter fragment in 50 ml centrifuge tubes (Costar) and incubated for 10 min with gentle mixing on ice. Dynabeads were collected using a magnet and washed four times in 20 mM Hepes-Na (pH 7.4), 100 mM NaCl, 0.1% (v/v) Tween 20. Proteins were eluted with 500 μl of buffer (20 mM Hepes-Na, 800 mM NaCl, 0.1% (v/v) Tween 20) and concentrated by TCA precipitation. Proteins were resolved on a 12% SDS-PAGE gel (Invitrogen), and visualized by Coomassie blue staining. To identify proteins, nine 1 mm gel slices were excised and analysed by the Functional Genomics, Proteomics and Metabolomics Facility at the University of Birmingham using a Thermo-Finnigan LTW Orbitrap mass spectrometer. Candidate proteins that exhibited DNA binding properties were analysed further.
Bacteriocin production assays
Colicin synthesis was monitored in the wild-type E. coli BW25113 strain or its ΔasnC derivative JW3721 [16], harbouring one of the colicinogenic plasmids, and was grown aerobically at 37°C in LB broth supplemented with ampicillin. Nalidixic acid (Sigma-Aldrich) was added to the culture at a final concentration of 37 μM, when the OD600 reached 0.3. Samples were taken before induction and at 1, 2 and 3 h after. Cells were diluted to obtain 1 ml samples with an OD600 of 0.3 and the crude cell extracts were prepared by sonication and the cell debris cleared by centrifugation for 1 min at 17000 x g. 100 μl of each extract was injected into wells in an agar plate containing tetracycline (12.5 μg ml-1) overlaid with the lawn of the indicator strain (DH5α harbouring pBR322) as described in [11]. As an alternative approach for colicin determination in the crude cell extracts, 5 μl of a ten-fold or five-fold dilution series of extracts were applied to an agar plate overlaid with the indicator strain as above. Indicator strains were grown at 37°C and the plates photographed using a G:Box (Syngene).
In vitro experiments
EMSA analysis, using purified LexA, IscR, AsnC and RNAP, with the cea8 and cka promoter regions, was performed as described in [34]. DNA fragments were excised from pSRcea8 or pSRcka using EcoRI and HindIII restriction enzymes and purified promotror fragments were labelled at the HindIII end with [γ-32P]-ATP using polynucleotide kinase (NEB). Approximately 0.5 ng of DNA fragment was incubated with varying amounts of purified proteins, as indicated. The reaction buffer contained 20 mM Hepes (pH 8), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM DTT, 5% (v/v) glycerol and 0.5 mg/ml BSA and the final reaction volume was 10 μl. Where AsnC was used, the EMSA buffer contained 5 mM L-asparagine (L-asn), where indicated. Samples were incubated at 37°C for 15 min before electrophoresis. For competitive EMSA experiments, DNA fragments were first incubated with various concentrations of LexA (for 15 min at 37°C) followed by the addition of RNAP and incubated for another 15 min at 37°C. Herring sperm DNA was included at a concentration of 6.5 μg ml-1 for these experiments. After incubation, all samples were immediately run on a 5% polyacrylamide gel at 12 V cm-1 in 0.25 x TBE, running under tension, and were visualised using a Bio-Rad Molecular Imager FX and Quantity One Software (Bio-Rad).
DNase I footprinting of AsnC and LexA at the cea8 promoter region was performed as described [35], using purified proteins in the presence or absence of 5 mM L-asparagine and a purified EcoRI-HindIII cea8 fragment that had been 32P-end labelled at the HindIII site using polynucleotide kinase and [γ-32P]ATP.
Supporting Information
Zdroje
1. Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71 : 158–229. 17347522
2. Mader A, von Bronk B, Ewald B, Kesel S, Schnetz K, Frey E, et al. Amount of Colicin Release in Escherichia coli Is Regulated by Lysis Gene Expression of the Colicin E2 Operon. PLoS One. 2015;10: e0119124. doi: 10.1371/journal.pone.0119124 25751274
3. Lloubes R, Bernadac A, Houot L, Pommier S. Non classical secretion systems. Res Microbiol. 2013;164 : 655–663. doi: 10.1016/j.resmic.2013.03.015 23542424
4. Walker D, Rolfe M, Thompson A, Moore GR, James R, Hinton JCD, et al. Transcriptional profiling of colicin-induced cell death of Escherichia coli MG1655 identifies potential mechanisms by which bacteriocins promote bacterial diversity. J Bacteriol. 2004;186 : 866–869. 14729715
5. Czaran TL, Hoekstra RF, Pagie L. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci U S A. 2003;99 : 786–790.
6. Majeed H, Gillor O, Kerr B, Riley MA. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J. 2011;5 : 71–81. doi: 10.1038/ismej.2010.90 20664553
7. Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428 : 412–414. 15042087
8. Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11 : 285–293. doi: 10.1038/nrmicro2977 23456045
9. Butala M, Žgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;66 : 82–93. doi: 10.1007/s00018-008-8378-6 18726173
10. Salles B, Weisemann JM, Weinstock GM. Temporal control of colicin E1 induction. J Bacteriol. 1987;169 : 5028–5034. 3117771
11. Butala M, Sonjak S, Kamenšek S, Hodošček M, Browning DF, Žgur-Bertok D, et al. Double locking of an Escherichia coli promoter by two repressors prevents premature colicin expression and cell lysis. Mol Microbiol. 2012;86 : 129–139. doi: 10.1111/j.1365-2958.2012.08179.x 22812562
12. Perez-Rueda E, Collado-Vides J. Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J Mol Evol. 2001;53 : 172–179. 11523004
13. Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, et al. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34 : 1439–1449. 16528101
14. Kolling R, Lother H. AsnC: an autogenously regulated activator of asparagine synthetase A transcription in Escherichia coli. J Bacteriol. 1985;164 : 310–315. 2864330
15. Poggio S, Domeinzain C, Osorio A, Camarena L. The nitrogen assimilation control (Nac) protein represses asnC and asnA transcription in Escherichia coli. FEMS Microbiol Lett. 2002;206 : 151–156. 11814655
16. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 : 2006 0008.
17. Lim CJ, Whang YR, Kenney LJ, Yan J. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 2012;40 : 3316–3328. doi: 10.1093/nar/gkr1247 22187157
18. Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35 : 1257–1269. 17267410
19. Shiraishi K, Ogata Y, Hanada K, Kano Y, Ikeda H. Roles of the DNA binding proteins H-NS and StpA in homologous recombination and repair of bleomycin-induced damage in Escherichia coli. Genes Genet Syst. 2007;82 : 433–439. 17991999
20. Wolf T, Janzen W, Blum C, Schnetz K. Differential dependence of StpA on H-NS in autoregulation of stpA and in regulation of bgl. J Bacteriol. 2006;188 : 6728–6738. 16980475
21. Craig ML, Suh WC, Record MT Jr. HO. and DNase I probing of E sigma 70 RNA polymerase—lambda PR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34 : 15624–15632. 7495790
22. Lee DJ, Minchin SD, Busby SJ. Activating transcription in bacteria. Annu Rev Microbiol. 2012;66 : 125–152. doi: 10.1146/annurev-micro-092611-150012 22726217
23. Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Mol Microbiol. 2003;48 : 287–294. 12675791
24. Ren J, Sainsbury S, Combs SE, Capper RG, Jordan PW, Berrow NS, et al. The structure and transcriptional analysis of a global regulator from Neisseria meningitidis. J Biol Chem. 2007;282 : 14655–14664. 17374605
25. Hol FJ, Voges MJ, Dekker C, Keymer JE. Nutrient-responsive regulation determines biodiversity in a colicin-mediated bacterial community. BMC Biol. 2014;12 : 68. doi: 10.1186/s12915-014-0068-2 25159553
26. Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, et al. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A. 2001;98 : 14895–14900. 11742080
27. Wu Y, Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol. 2009;191 : 1248–1257. doi: 10.1128/JB.01086-08 19074392
28. Pugsley AP. The ins and outs of colicins. Part I: Production, and translocation across membranes. Microbiol Sci. 1984;1 : 168–175. 6444117
29. Butala M, Klose D, Hodnik V, Rems A, Podlesek Z, Klare JP, et al. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res. 2011;39 : 6546–6557. doi: 10.1093/nar/gkr265 21576225
30. Lodge J, Fear J, Busby S, Gunasekaran P, Kamini NR. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett. 1992;74 : 271–276. 1526459
31. Butala M, Busby SJ, Lee DJ. DNA sampling: a method for probing protein binding at specific loci on bacterial chromosomes. Nucleic Acids Res. 2009;37: e37. doi: 10.1093/nar/gkp043 19181705
32. Miller JH. Experiments in molecular genetics. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1972.
33. Sambrook J, Fritsch EF, Maniatis M.Molecular cloning: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1987.
34. Browning DF, Cole JA, Busby SJ. Regulation by nucleoid-associated proteins at the Escherichia coli nir operon promoter. J Bacteriol. 2008;190 : 7258–7267. doi: 10.1128/JB.01015-08 18757534
35. Squire DJ, Xu M, Cole JA, Busby SJ, Browning DF. Competition between NarL-dependent activation and Fis-dependent repression controls expression from the Escherichia coli yeaR and ogt promoters. Biochem J. 2009;420 : 249–257. doi: 10.1042/BJ20090183 19245365
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



