-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
Carbon assimilation in Bacteria is governed by a mechanism known as carbon catabolite repression (CCR). In contrast to several other bacterial clades CCR in Pseudomonas species appears to be primarily regulated at the post-transcriptional level. In this study, we have identified the RNA chaperone Hfq as the principle post-transcriptional regulator of CCR in P. aeruginosa (PAO1). Hfq is shown to act as a translational regulator and to prevent ribosome loading through binding to A-rich sequences within the ribosome binding site of mRNAs, which encode enzymes involved in carbon utilization. It has been previously shown that the synthesis of the RNA CrcZ is augmented in the presence of non-preferred carbon sources. Here, we show that the CrcZ RNA binds to and sequesters Hfq, which in turn abrogates Hfq-mediated translational repression of mRNAs, the encoded functions of which are required for the breakdown of non-preferred carbon sources. This novel mechanistic twist on Hfq function not only highlights the central role of RNA based regulation in CCR of PAO1 but also broadens the view of Hfq-mediated post-transcriptional mechanisms.
Published in the journal: . PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004440
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004440Summary
Carbon assimilation in Bacteria is governed by a mechanism known as carbon catabolite repression (CCR). In contrast to several other bacterial clades CCR in Pseudomonas species appears to be primarily regulated at the post-transcriptional level. In this study, we have identified the RNA chaperone Hfq as the principle post-transcriptional regulator of CCR in P. aeruginosa (PAO1). Hfq is shown to act as a translational regulator and to prevent ribosome loading through binding to A-rich sequences within the ribosome binding site of mRNAs, which encode enzymes involved in carbon utilization. It has been previously shown that the synthesis of the RNA CrcZ is augmented in the presence of non-preferred carbon sources. Here, we show that the CrcZ RNA binds to and sequesters Hfq, which in turn abrogates Hfq-mediated translational repression of mRNAs, the encoded functions of which are required for the breakdown of non-preferred carbon sources. This novel mechanistic twist on Hfq function not only highlights the central role of RNA based regulation in CCR of PAO1 but also broadens the view of Hfq-mediated post-transcriptional mechanisms.
Introduction
The opportunistic human pathogen Pseudomonas aeruginosa causes acute as well as chronic infections in immunocompromised individuals. Moreover, airway epithelia of patients suffering from cystic fibrosis are frequently colonized by the pathogen [1]. P. aeruginosa is a metabolically versatile organism with the ability to utilize numerous carbon sources, which allows the bacterium to thrive in different environments such as soil, marine habitats as well as on/in different organisms [2].
In Bacteria, the uptake and utilization of carbon compounds is controlled in a hierarchical manner by a mechanism known as carbon catabolite repression (CCR). Generally speaking, CCR prevents the utilization of less preferred carbon sources until the preferred one is consumed. In Escherichia coli CCR prevents the expression of catabolic genes, the transcription of which requires the transcriptional activator CRP (cyclic AMP receptor protein) in conjunction with cAMP, whereas in Bacillus subtilis CCR is mediated by the transcriptional repressor CcpA (catabolite control protein A). In both organisms CCR is regulated by a signal transduction pathway inherent to the phosphoenolpyruvate-carbohydrate phosphotransferase system [3].
In most studied Pseudomonas spp. the presence of organic acids (for example succinate) results in CCR, which leads to repression of catabolic genes required for the consumption of other carbon sources. During CCR catabolic genes were deemed to be down-regulated by the translational repressor Crc (catabolite repression control protein) [4]. It has been suggested that Crc binds to CA-rich motifs within or adjacent to ribosome binding sites (RBS) of multiple target mRNAs, and thereby prevents their translation [5]–[7]. Upon relief of CCR, the regulatory RNAs, CrcZ in PAO1 [7], CrcZ/CrcY in P. putida [8] and CrcZ/CrcX in P. syringae [9] were proposed to bind to and to counteract Crc by trapping the protein. This hypothesis was in line with the observation that the CrcZ levels increase in the presence of poor carbon sources, and that they are reduced in the presence of a preferred carbon source [7]. However, our recent structural and biochemical studies challenged the role of Crc as a direct translational repressor of genes governed by CCR in PAO1. Recombinant Crc purified to homogeneity did neither bind to amiE mRNA, encoding aliphatic amidase, nor to CrcZ RNA [10], [11]. Rather, the previously reported RNA binding activity of His-tagged Crc purified by nickel affinity chromatography [6], [7] was attributed to a contamination of the Crc-His preparations with the RNA chaperone Hfq [10], [11].
In Enterobacteriaceae Hfq is pivotal for riboregulation [12], [13], which results on the one hand from binding to and protection of sRNAs from nucleolytic decay [14], and on the other hand from accelerating base-pairing between sRNAs and their target mRNAs [15]–[17]. E. coli Hfq hexamers have dedicated RNA binding sites, preferably binding uridine-rich stretches of sRNAs around the central pore of the proximal surface [18], [19] and A-rich sequences on the distal surface [20]. In addition, the lateral surface of the hexamer can as well contribute to sRNA binding [21]. The dedicated sRNA and mRNA binding surfaces on either site of the Hfq-hexamer may serve to transiently increase the local concentration of two RNA substrates. Moreover, the inherent capacity of Hfq to induce conformational changes in RNAs together with the observed structural flexibility of RNA ligands bound to Hfq could stochastically facilitate base-pairing [22], [23].
Although many sRNA candidates have been identified in PAO1 [24]–[27], the function of only a few has been revealed. The sRNAs PhrS [28] and PrrF [29] have been shown and inferred, respectively, to act by base-pairing with target mRNAs, whereas the protein binding RNAs RsmY and RsmZ are known to antagonize the function of the translational regulator RsmA [30]. PAO1 Hfq was shown to stabilize the protein binding RNA RsmY [31], [32] and to affect expression of some sRNAs including PhrS [25]. In PAO1, Hfq acts as a pleiotropic regulator, impacting on growth, virulence, motility, and quorum sensing [31], [33]. A transcriptome analysis of a PAO1hfq - strain revealed that ∼15% of all genes were de-regulated. These included a number of genes encoding proteins involved in carbon compound catabolism, which were up-regulated in the absence of Hfq [31].
Here, we studied the impact of Hfq on CCR in PAO1. In vivo and in vitro studies revealed that Hfq acts as a translational repressor of several catabolic genes. Moreover, we present evidence that the regulatory RNA CrcZ binds to and sequesters Hfq, which in turn results in translation of Hfq-regulated mRNAs. Hence, this study revealed a novel mechanistic twist on post-transcriptional regulation by Hfq and highlights its role in regulating the central metabolism in P. aeruginosa.
Results
Hfq represses catabolic genes at the post-transcriptional level
Several observations suggested a link between Hfq and CCR in PAO1. A comparative transcriptome analysis of a PAO1 wt and a PAO1hfq- strain disclosed transcripts encoding functions related to carbon compound and amino-acid catabolism that were up-regulated in the absence of Hfq ([31]; Table S1). Many of these transcripts comprise A-rich stretches (Table S1) within or adjacent to the RBS, which could serve as recognition motifs for the distal poly-A binding site of Hfq [20], [34]. Mutations within these A-rich stretches in certain mRNAs abrogated CCR [7], [35].
Crc was deemed to act as a translational regulator in PAO1 CCR [5]–[7]. However, Crc was recently shown to be deficient in binding to CrcZ and to the CCR regulated amiE mRNA [10]. In fact, these studies identified Hfq as a contaminant RNA binding activity in Crc preparations [10], [11]. This observation prompted us to test (i) whether Hfq serves as the principle post-transcriptional regulator of CCR in PAO1, and (ii) whether the regulatory RNA CrcZ, displaying several A-rich motifs [7], might abrogate Hfq-mediated regulation by sequestering Hfq.
We first revisited post-transcriptional regulation of amiE mRNA, encoding an aliphatic amidase. Strain PAO1(pTCamiE) and strain PAO1(pME9655) harboring a plasmid borne transcriptional amiE-lacZ and a translational amiE::lacZ fusion, respectively, were grown in BSM medium either in the presence of succinate/acetamide (CCR) or mannitol/acetamide (no CCR). Acetamide was added to induce transcription of the chimeric amiE genes, i.e. to mimic CCR. As shown in Figure S1A (left panel), the β-galactosidase activities conferred by the transcriptional fusion was comparable in either medium, i.e. in the presence and absence of CCR. However, when compared with growth in the presence of mannitol/acetamide (no CCR), amiE::lacZ translation was repressed in strain PAO1(pME9655) (Figure S1A; right panel) during cultivation in the presence of succinate/acetamide, i.e. when CCR was in place.
We next tested whether amiE mRNA and CrcZ RNA associate with Hfq upon induction of amiE transcription during CCR. Strain PAO1hfq - harboring plasmid pMMBhfqFlag (encodes flag-tagged Hfq) and the control plasmid pMMB67HE, respectively, was grown to an OD600 of 1.5 in BSM medium supplemented with succinate. In addition, acetamide was added to induce transcription of the amiE gene, i.e. to mimic CCR. Then, cell lysates were prepared and Hfq-associated RNAs were co-immunoprecipitated (CoIP) with Hfq-specific antibodies. As revealed by RT-PCR both, amiE and CrcZ, were found in complex with Hfq (Figure S2). In contrast RsmZ, which does not bind to Hfq [7], could not be detected among the RNAs that co-immunoprecipitated with Hfq (Figure S2). Taken together, these initial studies validated amiE as a model mRNA to scrutinize the hypothesized role of Hfq in CCR.
To obtain first hints whether Hfq is involved in post-transcriptional regulation of amiE during CCR, the strains PAO1, PAO1hfq-, PAO1Δcrc and PAO1hfq-Δcrc were transformed with plasmid pTCamiE harboring the transcriptional amiE-lacZ fusion and with plasmid pME9655 harboring the translational amiE::lacZ fusion, respectively. The strains were grown in BSM medium in the presence of succinate to establish CCR, and in the presence of acetamide to induce amiE-lacZ/amiE::lacZ transcription. The β-galactosidase activity conferred by the transcriptional amiE-lacZ fusion was comparable in the presence and absence of Hfq and/or Crc in either strain (Figure S1B). In contrast, the β-galactosidase activity conferred by the translational amiE::lacZ fusion differed in strains PAO1, PAO1hfq-, PAO1Δcrc and PAO1hfq-Δcrc when grown in BSM medium containing succinate and acetamide (CCR). In contrast to PAO1, amiE::lacZ translation was greatly increased in the PAO1hfq - strain and in the PAO1hfq-Δcrc double mutant, respectively (Figure 1A). The absence of Crc resulted as well in marked amiE::lacZ translation, albeit at a lower level when compared with the hfq- mutant or the hfq-Δcrc double mutant, suggesting that Hfq exerts a more pronounced negative effect on amiE translation during CCR than Crc. To verify these experiments, we tested whether Hfq likewise affects translation of the estA and the phzM genes, which are also known to be regulated by CCR [7], [35], [36]. The impact of Hfq was again monitored using translational lacZ reporter gene fusions. The results obtained mirrored those obtained with the amiE::lacZ reporter gene. Their translation was repressed during growth of PAO1 in BSM medium containing succinate, whereas in the absence of Hfq, Crc or both, the synthesis of the encoded fusion proteins increased (Figures 1B and C). In contrast, the expression of the heterologous lacZ gene (variation control), was comparable in strains PAO1, PAO1hfq-, PAO1Δcrc and PAO1hfq-Δcrc (Figure S1C). Taken together, these initial studies supported our hypothesis that Hfq is involved in post-transcriptional regulation of CCR regulated genes.
Fig. 1. Repression of catabolic genes by Hfq. 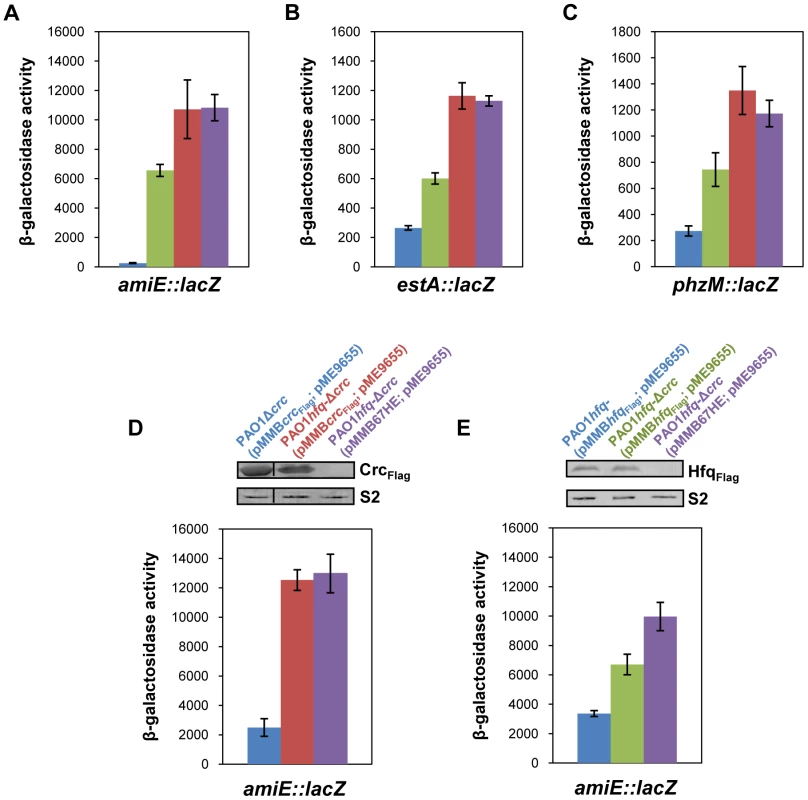
(A–C) The strains were grown to an OD600 of 2.0 in BSM medium supplemented with (A) 40 mM succinate and 40 mM acetamide (to establish CCR and to induce amiE::lacZ transcription) or (B, C) only with 40 mM succinate (CCR). Then, the cells were harvested and the β-galactosidase activities were determined. The bars depict β-galactosidase values conferred by the translational amiE::lacZ fusion encoded by plasmid pME9655 (A), the estA::lacZ fusion encoded by plasmid pTLestA (B) and the phzM::lacZ fusion encoded by plasmid pME10011 (C) in strains PAO1 (blue bar), PAO1Δcrc (green bar), PAO1hfq- (red bar) and PAO1hfq-Δcrc (purple bar), respectively. The error bars represent standard deviations from three independent experiments. (D, E) The strains were grown in BSM medium supplemented with succinate and acetamide. (D) At an OD600 of 1.0 IPTG (1 mM final concentration) was added to strains PAO1Δcrc(pMMBcrcFlag; pME9655) (blue bar), PAO1hfq-Δcrc(pMMBcrcFlag; pME9655) (red bar) and PAO1hfq-Δcrc(pMMB67HE; pME9655) (purple bar), respectively. The bars represent the β-galactosidase values conferred by the plasmid pME9655 encoded translational amiE::lacZ fusion 3 h after induction of the plasmid borne crcFlag gene. The error bars represent standard deviations from three independent experiments. The corresponding CrcFlag levels were determined by western-blot analysis using anti-Flag antibodies in strains PAO1Δcrc(pMMBcrcFlag; pME9655), PAO1hfq-Δcrc(pMMBcrcFlag; pME9655) and PAO1hfq-Δcrc(pMMB67HE; pME9655). Immunodetection of ribosomal protein S2 (loading control) was performed as described in Materials and Methods. (E) At an OD600 of 1.0 IPTG (1 mM final concentration) was added to strains PAO1hfq-(pMMBhfqFlag; pME9655) (blue bar), PAO1hfq-Δcrc(pMMBhfqFlag; pME9655) (green bar) and PAO1hfq-Δcrc(pMMB67HE; pME9655) (purple bar), respectively. The bars represent the β-galactosidase values conferred by the plasmid pME9655 encoded translational amiE::lacZ fusion 3 h after hfqFlag induction. The error bars represent standard deviations from three independent experiments. The corresponding HfqFlag levels were determined by western-blot analysis using anti-Flag antibodies in strains PAO1hfq-(pMMBhfqFlag; pME9655), PAO1hfq-Δcrc(pMMBhfqFlag; pME9655) and PAO1hfq-Δcrc(pMMB67HE; pME9655) (top panel). Immunodetection of ribosomal protein S2 (loading control) was performed as described in Materials and Methods. Hfq acts as the principle post-transcriptional regulator and Crc has an auxiliary function in CCR
As shown in Figure 1A–C, both, Crc and Hfq, were required for full repression of all three CCR regulated genes. Although translation occurred in the absence of Crc, a pronounced increase in translation only required the absence of Hfq, i.e. the observed de-repression was comparable in the hfq- strain and in the hfq-Δcrc double mutant. We interpreted this as showing that Hfq acts as the principal translational repressor, whereas Crc seemed to act as an auxiliary factor, somehow amplifying the negative regulation exerted by Hfq. To further test this hypothesis, amiE::lacZ translation was monitored in the PAO1hfq-Δcrc double mutant complemented with a plasmid borne crcFlag and hfqFlag gene, respectively. In consideration that crc could impinge on hfq expression and vice versa, the plasmid pMMBcrcFlag and pMMBhfqFlag borne crcFlag and hfqFlag genes, respectively, were equipped with the same expression signals, i.e. their expression was controlled by the Ptac promoter and identical translation initiation signals. The different strains used in this experiment were grown in BSM medium supplemented with succinate and acetamide (CCR). At an OD600 of 1.0, IPTG was added to induce ectopic expression of the crcFlag and hfqFlag genes, respectively. Three hours thereafter, amiE::lacZ translation was monitored by determination of the β-galactosidase activities and the Crc-Flag and Hfq-Flag levels were determined by quantitative western-blot analysis.
As shown in Figure 1D (blue bar), under these conditions amiE::lacZ translation was repressed in strain PAO1Δcrc(pMMBcrcFlag; pME9655). In contrast, translation of the amiE::lacZ fusion gene was de-repressed in the absence of Hfq in the double mutant PAO1hfq-Δcrc(pMMBcrcFlag; pME9655) (Figure 1D; red bar). Moreover, when compared with the control strain PAO1hfq-Δcrc(pMMB67HE; pME9655) (Figure 1D, purple bar), ectopic expression of crcFlag in strain PAO1hfq-Δcrc(pMMBcrcFlag; pME9655) did not affect amiE::lacZ translation in the absence of Hfq.
Next, amiE::lacZ translation was monitored in strains PAO1hfq-(pMMBhfqFlag, pME9655) and PAO1hfq-Δcrc(pMMBhfqFlag; pME9655) after growth in BSM succinate/acetamide medium (CCR) and 3 h after ectopic expression of the hfqFlag gene. The absence of Crc in the double mutant strain PAO1hfq-Δcrc(pMMBhfqFlag; pME9655) (Figure 1D; green bar) resulted in an increased de-repression of amiE::lacZ translation when compared with strain PAO1hfq-(pMMBhfqFlag;, pME9655) (Figure 1D; blue bar) despite comparable levels of HfqFlag in both strains. Taken together, these experiments showed that Crc only impacts on amiE::lacZ translation in the presence of Hfq. Hence, they corroborate the idea that Crc does not act per se as a translational regulator but functions as an ancillary factor in Hfq-mediated repression of target genes.
Hfq binds to the translation initiation region of amiE mRNA and prevents in vitro ribosome binding and translation
Next, we tested whether Hfq directly represses translation of amiE mRNA by binding to the translation initiation region (TIR). First, a filter binding assay was performed with purified PAO1 Hfq and an amiE mRNA fragment encompassing nucleotides (nt) from position −134 to +20 with regard to the A (+1) of the start codon. This experiment revealed that Hfq binds to amiE−134–+20 with a Kd of ∼67.0±1.4 nM (Figure S3). Next, the Hfq binding site(s) were mapped on amiE RNA. Enzymatic probing was performed with riboendonucleases T1 (G-specific cleavage) and A (C/U-specific cleavage) in the absence and presence of Hfq. As shown in Figure 2, Hfq protected the segment of amiE RNA extending from G−28 to U−5. This region includes the Shine and Dalgarno sequence (SD) sequence of amiE mRNA. Thus, Hfq binding to this region would readily explain the observed translational repression of amiE::lacZ mRNA (Figure 1A, D, E).
Fig. 2. Hfq binding to the RBS of amiE mRNA revealed by enzymatic probing. 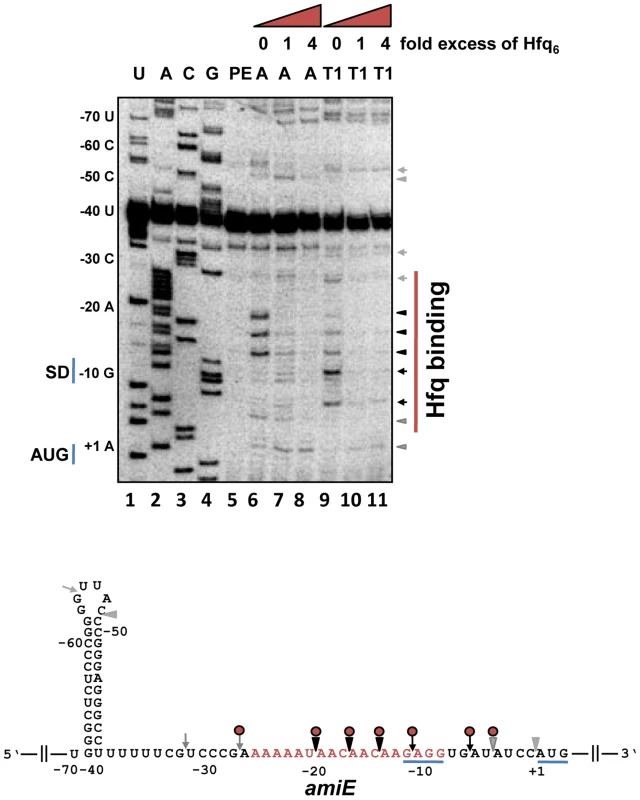
Lanes 1–4: sequencing reactions. Lane 5, primer extension (PE). Lanes 6–8, RNase A (A) cleavage in the absence and in the presence of increasing amounts of Hfq-hexamer (Hfq6). Lanes 9–11, RNase T1 (T1) cleavage in the absence and in the presence of increasing amounts of Hfq6. The nucleotides indicated on the left of the autoradiograph and in the RNA sequence (below) are numbered with regard to the A (+1) of the amiE start codon. Arrowheads and arrows denote RNase A and RNase T1 cleavage, respectively. Black and grey symbols indicate strong and weak cleavage, respectively. Red circles indicate protection from RNase cleavage by Hfq. The region protected by Hfq is indicated by a red bar on the right of the autoradiograph. Only the relevant part of the autoradiograph is shown. The A-rich stretch from nt position −26 to −8, which can be potentially accommodated in the six distal binding pockets of Hfq, are indicated in red in the amiE RNA sequence. The Shine and Dalgarno sequence and the start codon of amiE are underlined. To further test whether Hfq acts as a translational repressor of amiE mRNA, the PURExpress system was employed. The in vitro translation system was programmed with amiEFlag mRNA, encoding a Flag-tagged amidase. As shown in Figure S4, translation of amiEFlag mRNA was already impeded at a 1∶1 molar ratio of Hfq to mRNA. As the in vitro system is reconstituted from purified components of the translation machinery of E. coli, no additional PAO1 component was apparently required for repression.
To further demonstrate that Hfq directly interferes with ribosome binding, a toeprinting assay was performed with amiE−134–+76 mRNA in the presence and absence of Hfq. Briefly, in the presence of tRNAfMet, 30S ribosomes form a stable ternary complex at the RBS of mRNAs, which can be visualized by inhibition of cDNA synthesis primed downstream of the start codon [37]. A toeprint signal usually occurs at position +15 to +17 with regard to the A (+1) of the start codon. As shown in Figure 3, lane 7, a toeprint signal at the amiE RBS was observed in the absence of Hfq, whereas the addition of Hfq to amiE−134–+76 mRNA inhibited ternary complex formation (Figure 3, lanes 8 and 9). In contrast, the presence of Hfq alone did not result in a stop signal (Figure 3, lane 6), which is in accordance with earlier observations that translational repressors do not always provide a roadblock for cDNA synthesis by reverse transcriptase under these conditions [38]. Taken together, these in vitro studies strongly supported the idea that Hfq acts as a translational repressor that prevents ribosome loading on amiE mRNA.
Fig. 3. Hfq inhibits translation initiation complex formation on amiE mRNA. 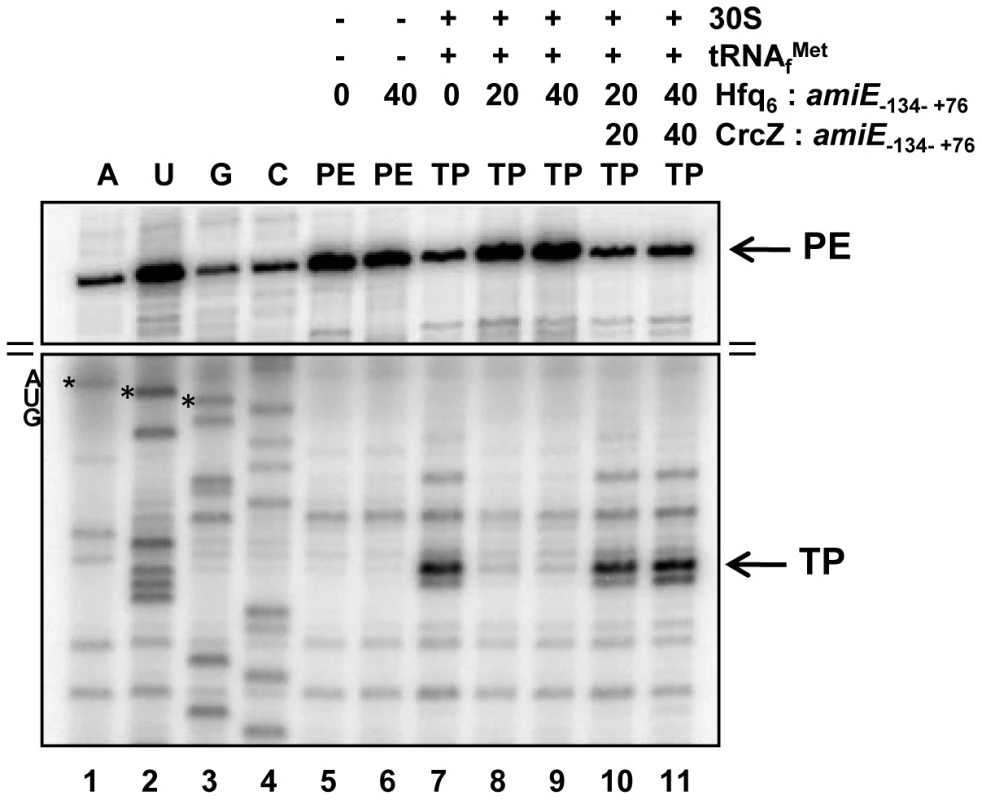
Lanes 1–4, sequencing reactions. Lane 5, primer extension (PE) in the absence of 30S subunits, tRNAfMet, and Hfq. Lane 6, primer extension in the presence of Hfq. Lane 7, toeprinting with 30S subunits and tRNAfMet in the absence of Hfq. Lanes 8 and 9, toeprinting with 30S subunits and tRNAfMet in the presence of Hfq. Lanes 10 and 11, toeprinting with 30S subunits and tRNAfMet in the presence of CrcZ and Hfq. CrcZ and/or Hfq were added concomitantly with amiE mRNA, 30S subunits and tRNAfMet. The molar ratios of Hfq-hexamer (Hfq6) and CrcZ RNA to amiE mRNA are indicated on top. Hfq and CrcZ were added in equimolar ratios. The arrows denote the toeprint signal (TP) at position +17 with regard to the A (+1) of the start codon and the primer extension signals (PE). Only the relevant parts of the autoradiogram are shown. The position of the AUG start codon is indicated. The distal site of Hfq is required for regulation of amiE mRNA
Link et al. [20] reported a crystal structure of E. coli Hfq in complex with poly(A15), wherein the poly(A) tract is bound to the distal face using tripartite binding motifs. They consist of an adenosine specific site (A - site), a purine nucleotide selectivity site (R-site) and a sequence-non-discriminating E-site. The amino-acids involved in building the A-R-N motifs are fully conserved in the Hfq protein of PAO1 [34]. As the hexamer Hfq could accommodate the entire A-rich stretch from nt −26 to −8 of amiE mRNA (Figure 2) in the six binding pockets, we next tested whether the distal binding site of Hfq is required and sufficient for translational repression of amiE mRNA. We therefore engineered the PAO1 hfq variants hfqY25DFlag and hfqK56AFlag as the corresponding E. coli mutant proteins were shown to be deficient in binding to polyA - and polyU-tracts [18], respectively. In contrast to the PAO1 hfqFlag gene and the PAO1 hfqK56AFlag allele, ectopic expression of the hfqY25DFlag allele did not result in repression of amiE::lacZ translation, albeit all three proteins, HfqFlag, HfqY25DFlag and HfqK56AFlag, were present at comparable levels (Figure 4A). Basically the same results were obtained when translation of the estA::lacZ (Figure 4B) and phzM::lacZ (Figure 4C) fusion genes was monitored in the presence of HfqFlag, HfqY25DFlag and HfqK56AFlag, respectively. Hence, translational repression of these reporter genes apparently required an intact distal polyA binding site of Hfq.
Fig. 4. The distal poly-A binding site of Hfq is required for amiE repression. 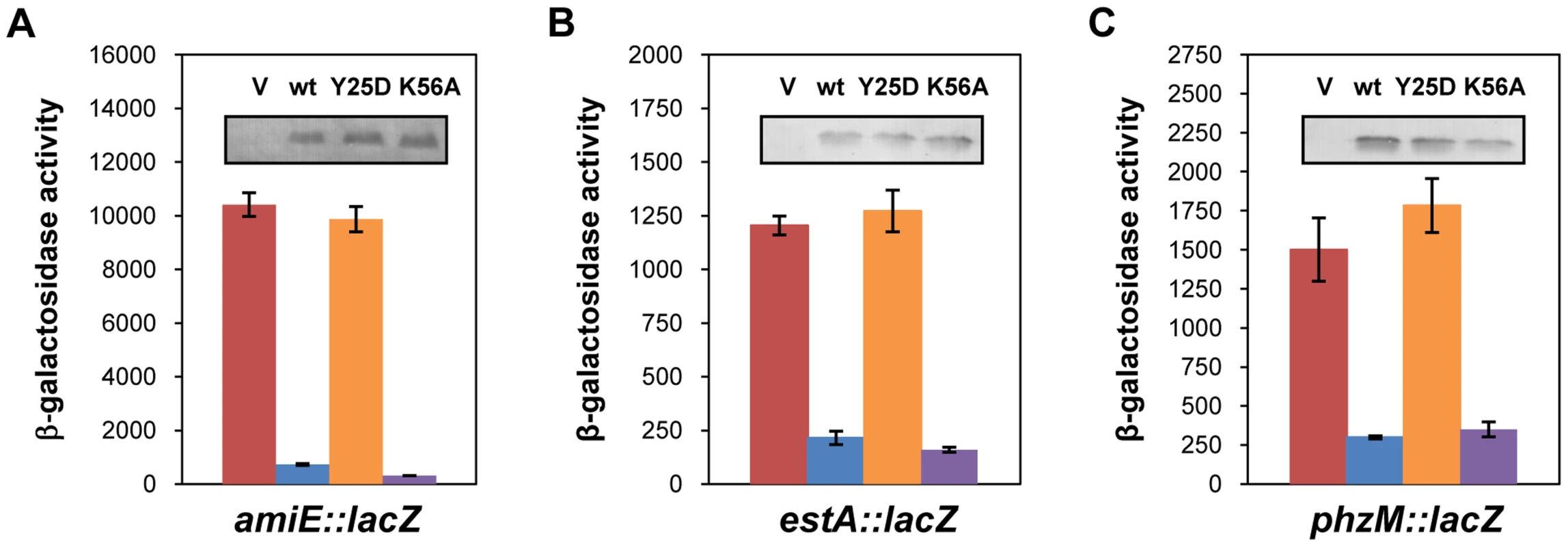
The cultures were grown to an OD600 of 2.0 in BSM medium supplemented with (A) 40 mM succinate and 40 mM acetamide or (B, C) only with 40 mM succinate. The β-galactosidase activity conferred by the translational amiE::lacZ fusion encoded by plasmid pME9655 (A), the estA::lacZ fusion encoded by plasmid pTLestA (B) and the phzM::lacZ fusion encoded by plasmid pME10011 (C) was determined in strain PAO1hfq-(pME9655) harboring the control plasmid pME4510 (red bar), plasmid pME4510hfqFlag (blue bar), plasmid pME4510hfqY25DFlag (orange bar) and pME4510hfqK56AFlag (purple bar), respectively. Inset: the levels of HfqFlag, HfqY25DFlag and HfqK56AFlag were determined by western-blot analysis using anti-Flag antibodies. The error bars represent standard deviations from three independent experiments. To verify these in vivo data, the PAO1 Hfq variants HfqY25D and HfqK56A were purified, and binding to amiE−134–+20 mRNA was assessed using electrophoretic mobility shift assays (EMSA). As shown in Figure S5A, while PAO1 Hfq and HfqK56A bound to the mRNA fragment, the HfqY25D protein failed to bind. With increasing concentrations of either Hfq or HfqK56A two shifted bands were observed (Figure S5A), suggesting that Hfq binds at two sites of the amiE−134–+20 fragment. This observation can be explained by the Hfq binding site mapped between nucleotides G−28 to U−5 (Figure 2) and by a probable second binding site, comprising an A-rich region (nucleotides −92 to −71), which was inferred from an RNomics approach after CoIP with Hfq-specific antibodies [25]. Taken the in vivo and in vitro studies together, these experiments strongly suggested that Hfq binds with its distal face to the RBS of amiE, and by inference most likely also to estA and phzM mRNA.
To further corroborate the idea that Hfq directly represses amiE translation the experiment was also performed in the heterologous E. coli hfq- strain JW4130. The strain was transformed with the control plasmid pME4510 and derivatives thereof harboring the PAO1 hfqFlag gene, the PAO1 hfqY25DFlag allele and the PAO1 hfqK56AFlag allele, respectively. In addition, these strains were transformed with plasmid pME9658, wherein the amiL terminator preceding the amiE gene was deleted. As shown in Figure S5B, the experimental results paralleled that performed in PAO1. The translation of the amiE::lacZ gene was repressed in the presence of HfqFlag and HfqK56AFlag, whereas the reporter gene was translated in the presence of HfqY25DFlag. Thus, the presence of Hfq was apparently necessary and sufficient for repression of amiE translation in E. coli.
The regulatory RNA CrcZ titrates Hfq in vitro
The regulatory RNA CrcZ is present at lower levels during CCR, when compared to conditions when CCR is not in place, e.g. in BSM medium containing mannitol as the sole carbon source [7]. The 407 nt long CrcZ RNA contains six A-rich stretches (Figure S6A) to which Hfq can potentially bind with its distal surface, i.e. with the same binding surface as it binds to the RBS of amiE (Figure S5A). Binding of Hfq to the first 151 nt of CrcZ, containing 3 A-rich stretches (Figure S6A) was confirmed using EMSA assays (Figure S6B). Three shifted bands could be discerned (Figure S6B, lane 2), which would be consistent with one Hfq-hexamer binding to either A-rich stretch. As anticipated, the PAO1 HfqY25D mutant protein was defective in binding to CrcZ151, whereas the HfqK56A variant bound to the CrcZ fragment like native Hfq. Thus, CrcZ binds to the distal face of Hfq like amiE mRNA, and therefore has the potential to titrate Hfq, which in turn would explain the observed increase in translation of amiE::lacZ mRNA in BSM mannitol medium, i.e. in the absence of CCR (see Figure S1A).
In the next set of experiments we therefore asked whether CrcZ can abrogate Hfq-mediated translational repression in vitro. First, a mobility shift assay was performed with radioactively labeled amiE−134–+20 RNA in the presence of unlabelled specific CrcZ151 and non-specific RsmZ competitor RNAs, respectively. As shown in Figure 5A, CrcZ151 competed with amiE−134–+20 for binding to Hfq (Figure 5A, lanes 5 and 6), whereas the addition of RsmZ RNA, which does not bind to Hfq [7], did not result in a downshift of labeled amiE−134–+20 RNA (Figure 5A, lane 7). When compared with the addition of unlabelled amiE−134–+20 RNA, the addition of an equimolar concentration of CrcZ151 resulted already in a significant downshift, i.e. loss of Hfq binding to amiE−134–+20. Hence, CrcZ151 acted as a better competitor for Hfq than amiE−134–+20, which is anticipated as the used CrcZ151 has three Hfq binding sites, whereas the amiE fragment has only two.
Fig. 5. CrcZ competes with amiE for Hfq. 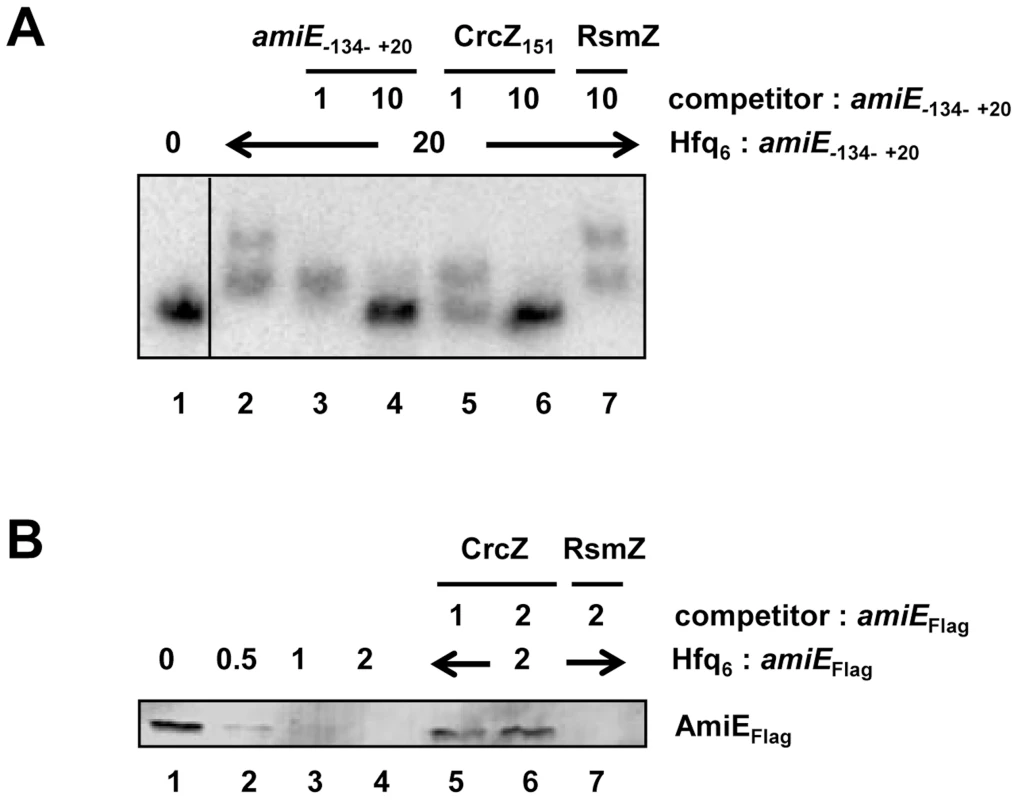
(A) Electrophoretic mobility shift assay with 10 nM radioactively labeled amiE−134–+20 mRNA in the presence of Hfq protein and unlabelled specific or non-specific competitor RNAs. Lane 1, no protein was added to labeled amiE−134–+20 mRNA. Lane 2–7, Hfq-hexamer was added in 20-fold molar excess over labeled amiE−134–+20 RNA. Lanes 3 and 4, 5 and 6, and 7, unlabeled amiE−134–+20 RNA, unlabelled CrcZ151 RNA and unlabelled RsmZ RNA were added, respectively. The molar ratios of the competitor RNAs to amiE−134–+20 mRNA are shown on top. (B) Repression of amiEFlag mRNA translation is relieved in the presence of CrcZ. Lane 1, in vitro translation of amiEFlag mRNA. Lanes 2–4, inhibition of amiEFlag mRNA translation in the presence of increasing amounts of Hfq. The molar ratios of Hfq-hexamer to amiEFlag mRNA are denoted on top. Lanes 5–7, Hfq was added in 2-fold molar excess over amiEFlag mRNA. CrcZ was concomitantly added in equimolar concentration (lane 5) or in two-fold molar excess (lane 6) over Hfq. Lane 7, RsmZ was added in 2-fold molar excess over amiEFlag mRNA. Next, amiEFlag mRNA was translated in vitro in the presence of Hfq as well as in the presence of Hfq and CrcZ mRNA. As shown before (Figure S4), Hfq inhibited translation of amiEFlag, (Figure 5B, lanes 2–4). In contrast, the mRNA was translated in the presence of CrcZ (Figure 5B, lanes 5 and 6), whereas the Hfq-mediated translational repression was not relieved in the presence of the non-specific competitor RNA RsmZ (Figure 5B, lane 7).
Similarly, the addition of CrcZ RNA to the Hfq-amiE complex, under conditions that inhibited translation initiation complex formation, resulted in reappearance of a toeprint signal (Figure 3, lanes 10 and 11). Thus, CrcZ counteracted the function of Hfq. Taken together, these in vitro experiments indicated that CrcZ can titrate Hfq, and that it can prevent it from binding to the amiE RBS.
CrcZ-mediated regulation of catabolic genes in vivo depends on Hfq
In our model titration of Hfq by CrcZ would result from increased CrcZ levels [7] in the presence of non-preferred carbon sources, which in turn would lead to de-repression of Hfq-regulated genes. Hence, it would be anticipated that ectopic over-expression of CrcZ would cause de-repression of Hfq-regulated genes even during CCR, i.e. under conditions when the CrcZ levels are low [7]. With this line of reasoning, we next asked whether ectopic expression of crcZ during CCR results in increased expression of the estA gene, which was assessed by monitoring the esterase activity. To avoid any interference of Crc, the experiments were performed in a PAO1Δcrc strain. The control strain PAO1Δcrc(pMMB67HE) and the crcZ over-expressing strain PAO1Δcrc(pMMBcrcZ) were grown in BSM medium supplemented with 40 mM succinate to an OD600 of 1. Then, crcZ expression was induced and samples were taken 60 minutes thereafter. Esterase was produced even without induction of crcZ, which can be reconciled with the absence of Crc (see Figure 1B) and by endogenous background expression of crcZ in strain PAO1Δcrc(pMMB67HE) (Figure 6A, upper panel). Nevertheless, ectopic over-expression of crcZ (Figure 6A, upper panel) resulted in elevated activities of esterase (Figure 6A), reflecting de-repression of estA translation.
Fig. 6. CrcZ relieves repression by Hfq in vivo. 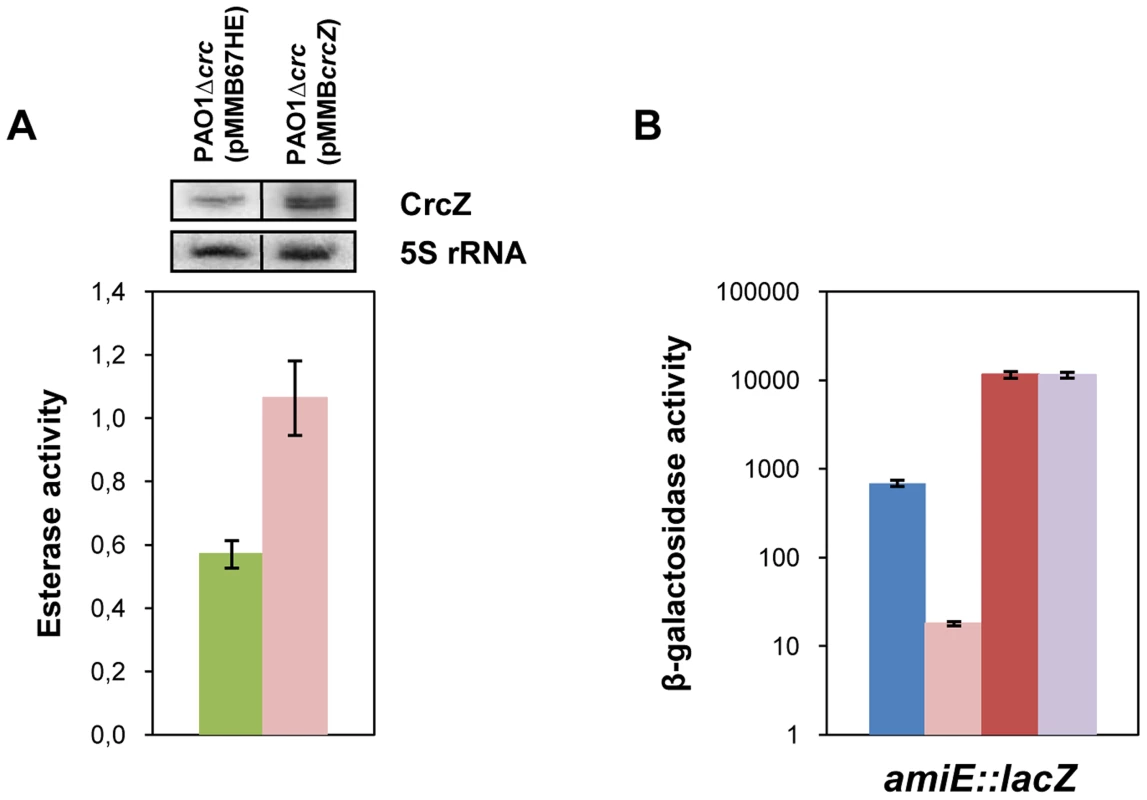
(A) The esterase activity was measured as change in A410 min−1 per OD600 in strain PAO1Δcrc harboring either the control plasmid pMMB67HE (green bar) or the crcZ encoding plasmid pMMBcrcZ (pink bar). The strains were grown to an OD600 of 1 in BSM medium supplemented with 40 mM succinate. Then, crcZ expression was induced by addition of IPTG and samples were withdrawn 60 minutes thereafter. The error bars represent standard deviations from three independent experiments. The CrcZ levels (top panel) were determined by Northern-blot analysis. 5S rRNA served as a loading control. (B) The strains were grown to an OD600 of 2.0 in BSM medium supplemented with 40 mM succinate and 40 mM acetamide. Then, the cells were harvested and the β-galactosidase activities were determined. The bars depict the β-galactosidase values conferred by the translational amiE::lacZ fusion encoded by plasmid pME9655 in strain PAO1 (blue bar), PAO1ΔcrcZ (pink bar), PAO1hfq- (red bar) and PAO1hfq-ΔcrcZ (purple bar). The error bars represent standard deviations from three independent experiments. Similarly, over-expression of crcZ in PAO1 resulted in de-repression of amiE::lacZ translation during CCR in BSM succinate/acetamide medium (Figure S7A), which is in agreement with our model wherein CrcZ titrates Hfq. In contrast to strain PAO1(pMMBcrcZ;pME9655) (Figure S7A), ectopic expression of crcZ did not affect translation of amiE::lacZ in the hfq- strain PAO1hfq-(pMMBcrcZ;pME9655) (Figure S7B), clearly showing that CrcZ-mediated regulation requires Hfq.
The model would further specify that a deletion of the crcZ gene should increase repression of amiE::lacZ during CCR. As shown in Figure 6B, when compared with strain PAO1(pME9655), amiE::lacZ translation was even further repressed in strain PAO1ΔcrcZ (pME9655) when grown in BSM medium supplemented with succinate and acetamide (CCR).
Moreover, amiE::lacZ translation during CCR was indistinguishable in the absence of Hfq in strain PAO1hfq-(pME9655) and in the double mutant PAO1hfq-ΔcrcZ(pME9655). Taken together, these experiments showed that the hfq deletion is epistatic to crcZ, in other words that CrcZ exerts regulation on amiE::lacZ only in the presence of Hfq. Hence, these in vivo experiments lend further support to the hypothesis that CrcZ titrates Hfq, and thereby abrogates Hfq-mediated translational repression.
Discussion
Target genes of Hfq in PAO1
In this study, we provided evidence that Hfq represses three CCR regulated genes, amiE, estA and phzM. Given that Hfq blocked translation initiation of amiE mRNA by binding to A-rich stretches encompassing the RBS, we revisited catabolic and transport genes that were found to be up-regulated in a comparative transcriptome analysis of a PAO1 and a PAOhfq - mutant [31]. Out of 126 putative target mRNAs, 72 transcripts contained A-rich stretches in the TIR (Table S1). Out of the latter, 28 transcripts contain several single non-consecutive A-R-N motifs, whereas 44 transcripts contained consecutive (A-R-N)n repeats with n≥3 either within or in close proximity to the RBS (Table S1), indicating that Hfq could interfere with ribosome binding. Although it remains to be tested for either candidate transcript it is tempting to speculate that Hfq is involved in translational regulation of several genes controlled by CCR. In addition, it is conceivable that several CCR controlled genes, which do not contain A-rich regions in the TIR [39], are indirectly controlled by Hfq.
Hfq in CCR of Pseudomonas aeruginosa: regulating a few at a time
Based on our in vivo and in vitro studies with the amiE model mRNA, we suggest that Hfq acts as a translational repressor of transcripts subjected to CCR in PAO1 (Figure 7). As there appear to be several Hfq regulated genes (Table S1) and our preliminary results show that Hfq levels are more or less constant during growth in different carbon sources (E. Sonnleitner, unpublished), the question arises how Hfq can regulate numerous mRNAs. Many if not all catabolic genes including the corresponding transporter genes are primarily regulated at the transcriptional level [4], [40], and their transcription usually requires the presence of the respective catabolite. Therefore, it is rather unlikely that many of the putative target genes (Table S1) are concomitantly induced at the same time. Thus, in the presence of a preferred carbon source, e.g. succinate, only a few other catabolites may induce concomitant transcription of the corresponding catabolic genes. Moreover, translational repression most likely leads to degradation of mRNAs [41], [42] encoding catabolic enzymes other than those required for the breakdown of succinate, and thus to recycling of Hfq. Therefore, CCR control may not require vast amounts of Hfq. This hypothesis is supported by the experiment shown in Figure S8. When compared with the absence of CCR (presence of acetamide only) or with the PAO1hfq- strain, amiE mRNA was faster degraded in PAO1 during CCR in the presence of succinate and acetamide. In addition, we have estimated the number of Hfq hexamers/cell in BSM - succinate medium with 2160+/−56 per PAO1 cell (Figure S9), which might suffice to silence “a few catabolic transcripts” that are induced during CCR (Figure 7).
Fig. 7. Simplified model for CCR in Pseudomonas aeruginosa orchestrated by Hfq and CrcZ. 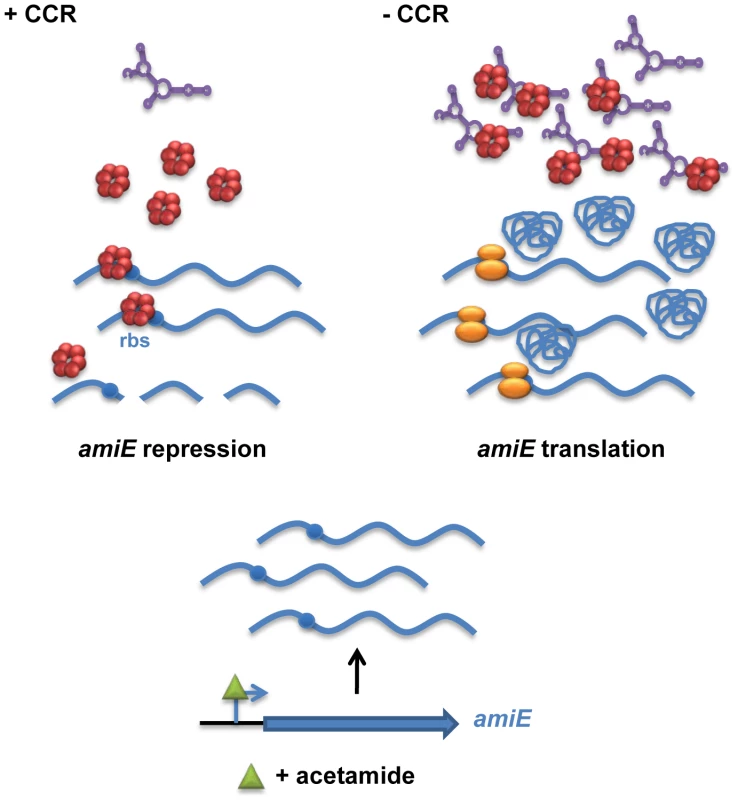
During CCR (left), for example during growth in the presence of succinate, the regulatory RNA CrcZ (in purple) is poorly transcribed. Upon catabolite-dependent induction of catabolic genes, for example induction of amiE transcription by acetamide, Hfq (red hexamer) represses translation of the corresponding transcripts, which in turn results in their degradation. When CCR is alleviated (right) the abundance of CrcZ increases. CrcZ binds to and titrates Hfq, which permits translation of catabolic genes, for example amiE. In the presence of non-preferred carbon sources, the two component system CbrAB is activated; phosphorylated CbrB binds to the RpoN-dependent promoter of crcZ and stimulates CrcZ synthesis [7], [43]. Our data (Figures 5 and 6) suggest that CrcZ then binds to and titrates Hfq by virtue of its A-rich binding motifs. This in turn would allow translation of transcripts that are induced by available catabolites, and thus synthesis of the cognate degradative enzymes (for example aliphatic amidase) (Figure 7). For the following reasons we favor the hypothesis that no other sRNAs are required for regulation of the catabolic genes examined in this study. Most E. coli sRNAs bind to the proximal site of Hfq, and binding is usually abrogated in the HfqK56A mutant protein [13]. However, in contrast to the HfqY25D variant, the HfqK56A mutant protein was still capable to repress the fusion genes governed by CCR in PAO1 (Figure 4) and in E. coli (Figure S5B). It seems therefore reasonable to suggest that regulatory sRNAs other than CrcZ are not involved in Hfq-mediated regulation of these catabolic genes.
A novel concept for Hfq function: from mediating riboregulation to being regulated
Although binding of E. coli Hfq to A-rich stretches in mRNA has been demonstrated in several model systems (see below), Hfq seems to work predominantly in conjunction with sRNAs [44]–[46]. Hfq-mRNA binding may recruit the sRNA to the target mRNA and stimulate sRNA-mRNA pairing. In this scenario, the sRNA competes with initiating 30S ribosomes, whereas Hfq has a rather indirect function.
Some deviations from the canonical model of Hfq assisted and sRNA-mediated regulation of target mRNAs have been reported. In E. coli evidence has been provided that Hfq acts as an autogenous repressor on its own mRNA. As for PAO1 amiE mRNA (Figure 2), E. coli Hfq was shown to bind to an A-rich sequence encompassing the SD sequence of hfq mRNA [47]. Another example for translational repression by Hfq entails regulation of the E. coli sdhC mRNA by the sRNA Spot 42. Desnoyers and Massé [48] reported that Spot 42 binds upstream of the RBS and recruits Hfq, which then directly represses translation. Moreover, E. coli Hfq was recently shown to act as a repressor of cirA mRNA translation in the absence of a sRNA [49]. Interestingly, the translational block exerted by Hfq was shown to be abrogated by RyhB RNA pairing to cirA mRNA [49].
Similarly, the model shown in Figure 7 entails direct repression of target mRNAs by PAO1 Hfq. However, in contrast to Spot 42 [48] and RyhB [49] RNA, which recruit Hfq to and abrogate Hfq-mediated translational repression of the target mRNA, respectively, CrcZ RNA acts as a decoy for Hfq. The CrcZ RNA contains six (A-R-N)n repeats (n≥4) (Figure S6A) that can potentially be exploited for binding to the distal face of Hfq. Thus, in the absence of CCR Hfq is most likely sequestered by CrcZ, and therefore not available to act as a translational repressor on target mRNAs. In this way the Hfq/CrcZ regulatory system is reminiscent to the Pseudomonas RsmA/RsmY/RsmZ system and the CsrA/CsrB system of several Gram-negative Bacteria [30], [50], [51]. The ability of Hfq to bind to RsmY [31] in fact poses the question whether the regulatory systems are interlinked. This could provide an explanation why different carbon sources can affect virulence traits such as biofilm formation and antibiotic resistance [39], [52].
Moreover, given its constellation with Hfq, it seems possible that CrcZ on the one hand can interfere with any mRNA-dependent role of Hfq. On the other hand, the role of Hfq in sRNA-mediated riboregulation remains ill defined in PAO1. Although Hfq appears to be required for the PAO1 sRNAs PrrF1/PrrF2 to regulate target mRNAs [53], it is unclear whether Hfq exerts this function by stabilizing the sRNAs and/or by stimulating sRNA-mRNA pairing. As CrcZ binds to the distal site of Hfq it is conceivable that Hfq might even still be able to bind sRNAs such as PrrF at the proximal site, and thus to protect them from degradation. In any case it will be interesting to study whether sequestration of Hfq by CrcZ indirectly affects other Hfq-mediated processes in P. aeruginosa.
The elusive function of Crc in CCR of Pseudomonas
We have recently shown that the Crc protein is devoid of RNA binding activity [10], and that the previously observed RNA binding activity of Crc could be attributed to Hfq [11]. However, Crc contributes to Hfq-mediated repression during CCR (Figure 1). In addition, Crc was shown to impact on biofilm formation [54], [55], virulence and antibiotic susceptibility [52], traits which are also affected by Hfq [31], [33]. This raises the question how Crc impacts on Hfq function. Crc appears not to interfere with hfq expression. The translation of a hfq::lacZ reporter gene, whose transcription is driven by the authentic hfq promoter, was indistinguishable in PAO1 and in the isogenic PAO1Δcrc strain (Figure S10A). In addition, the levels of Hfq, as determined by quantitative western-blot analysis, were not significantly altered in PAO1Δcrc when compared with the wild-type strain (Figure S10B). Conversely, Hfq seems not to affect the cellular concentration of Crc (Figure S10B). Thus, Crc seems not to impact on the Hfq levels, and vice versa Hfq seems not to affect that of Crc.
RelA was recently shown to enhance multimerization of E. coli Hfq, and thereby to stimulate binding to sRNAs [56]. We therefore considered the possibility that Crc might interact with Hfq to increase the specificity of Hfq for A-rich sequences. However, as revealed by EMSA assays, the presence of Crc did not increase the affinity of PAO1 Hfq for amiE−134–+20 RNA (E. Sonnleitner, unpublished), making it less likely that Crc acts similar to RelA. Nevertheless, we are currently exploring the possibility whether Crc is associated with Hfq in vivo.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The strains and plasmids used in this study are listed in Table S2. Unless indicated otherwise, the cultures were grown at 37°C in BSM minimal medium supplemented with 40 mM succinate. The strains PAO1hfq-Δcrc and PAO1hfq-ΔcrcZ were constructed by homologous recombination. Briefly, plasmid pME9672 and plasmid pME9673, respectively, were mobilized into strain PAO1hfq - with the aid of E. coli strain HB101(pRK2013), and then chromosomally integrated through selection for tetracycline resistance. Excision of the vector by a second crossover event was achieved by enrichment for tetracycline-sensitive cells [57]. If required E. coli and PAO1 were grown in the presence of 100 µg ml−1 ampicillin, 25 µg ml−1 tetracycline or 25 µg ml−1 kanamycin and 50 µg ml−1 gentamicin, 100 µg ml−1 tetracycline or 250 µg ml−1 carbenicillin, respectively. Details on the construction of plasmids used in this study are provided in Text S1.
β-Galactosidase assays
The β-galactosidase activities were determined as described by Miller [58]. The cells were permeabilized with 5% toluene. The β-galactosidase units in the different experiments were derived from three independent experiments. The error bars in the different Figures represent standard deviations.
Western-blot analyses
The protein levels of Hfq and Crc fused to C-terminal Flag-tags (DYKDDDDK) were determined in the respective strains and under the growth conditions as specified in the legends to the Figures. 1 ml aliquots of the respective cultures were withdrawn; the cells were harvested by centrifugation, resuspended and boiled in protein sample buffer. Equal amounts of total protein were separated on 12% SDS-polyacrylamide gels and then electro-blotted to a nitrocellulose membrane. The blots were blocked with 5% dry milk in TBS buffer, and then probed with rabbit anti-DYKDDDDK polyclonal antibody (Roth). The antibody-antigen complexes were visualized with alkaline-phosphatase conjugated secondary antibodies (Sigma) using the chromogenic substrates nitro blue tetrazolium chloride (NBT) and 5-Bromo-4-chloro-3-indolyl phosphate (BCIP).
Protein purification
Hfq protein, HfqY25D and HfqK56A protein were produced in the hfq deficient E. coli strain AM111F′ harboring plasmid pHfqPae, pHfqPaeY25D or pHfqPaeK56A. Protein purifications were performed as described in detail by Beich-Frandsen et al. [59].
In vitro transcription
For in vitro transcription of amiE (1172 nt), CrcZ (426 nt) and RsmZ (141 nt) RNAs the AmpliScribe T7-Flash Transcription Kit (Epicentre Biotechnologies) was used according to the manufacturer's instructions. First, PCR fragments were generated with the primer pairs (see Table S3) A5/A75 (amiE), E6/C6 (crcZ) and W26/X26 (rsmZ), whereby the forward primers contained T7 promoter sequences. Primer A75 encoded in addition a Flag-tag sequence. For the filter binding and gel mobility shift assays truncated versions of amiE−134–+20 (first 154 nt) and crcZ151 (first 151 nt) were used. The corresponding PCR fragments were amplified with the primers A5/C1 (amiE−134–+20) and E6/E2 (crcZ151). For the toeprint assay the amiE−134–+76 fragment was in vitro transcribed using oligonucleotides A5 and Q99 (Table S3).
Enzymatic probing of RNA
Five pmol of in vitro transcribed full length amiE RNA was incubated in RT-buffer (50 mM Tris pH 8.3, 60 mM NaCl, 6 mM Mg-acetate, 10 mM DTT) at 37°C for 30 min in a 10 µl reaction with 0, 5 and 20 pmol of purified Hfq protein. Then 2 U RNase T1 (1 µl), 10 pg RNase A (1 µl) or 1 µl of RNase free H2O was added and incubated for additional 10 min followed by phenol/chloroform extraction and precipitation. 200 fmol RNase treated or untreated RNAs were further used for the primer extension reaction with AMV reverse transcriptase (Promega), which was primed with the 5′-[32P]-labeled C78 oligonucleotide to test for protection by Hfq of the proximal part of amiE mRNA. For sequencing amiE RNA and ddNTPs (Fermentas) were used in primer extension reaction(s).
Toeprint analysis
The amiE−134–+76 RNA used for toeprinting was obtained as described above. The [32P]-5′-end labeled oligonucleotide Q99 was annealed to amiE mRNA (+57 to +76 with regard to the A (+1) of the start codon) and used to prime cDNA synthesis by MMuLV reverse transcriptase (Thermo Scientific). The toeprinting assay was carried out with purified E. coli 30S ribosomal subunits and E. coli initiator-tRNA (tRNAfMet) as described by Hartz et al. [37]. The mRNA (0.05 pmol) was pre-incubated at 37°C for 10 min with or without 4 pmol 30S subunits and 16 pmol tRNAfMet before reverse transcriptase was added. To test whether Hfq interferes with translation initiation, 0.05 pmol amiE−134–+76 mRNA was pre-incubated at 37°C for 10 min with 4 pmol 30S subunits, 16 pmol tRNAfMet and Hfq-hexamer (1 or 2 pmol, respectively) before the reverse transcriptase reaction was performed. To test whether CrcZ can abrogate the Hfq mediated repression of amiE translation initiation, 0.05 pmol amiE−134–+76 mRNA was pre-incubated at 37°C for 10 min with 4 pmol 30S subunits, 16 pmol tRNAfMet and equimolar amounts of Hfq-hexamer and CrcZ (1 or 2 pmol, respectively) before reverse transcriptase was added.
Electro mobility shift assays
The amiE−134–+20 and crcZ151 RNAs (see above) were dephosphorylated with FastAP thermo sensitive alkaline phosphatase (Thermo Scientific) and subsequently 5′-end labeled using [γ-32P]-ATP (Hartmann Analytic) and polynucleotide kinase (Thermo Scientific). The labeled RNAs were gel-purified and dissolved in diethylpyrocarbonate-treated water. Labeled RNA (10 nM) was incubated with increasing amounts of purified Hfq, the HfqY25D or HfqK56A mutant proteins in 10 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 60 mM NaCl, 10 mM NaH2PO4, 10 mM DTT, and 25 ng tRNA in a total volume of 10 µl. Unlabeled RNA was used as competitor as stated in the legend to Figure 5A. The reaction mixtures were incubated at 37°C for 30 min to allow protein–RNA complex formation. The samples were mixed with 4 µl loading dye (25% glycerol, 0.2 mg/l xylencyanol and bromphenol blue) immediately before loading and separated on 4% polyacrylamide gels using Tris-borate buffer. The radioactively labeled bands were visualized with a PhosphorImager (Molecular Dynamics) and quantified with ImageQuant software 5.2.
In vitro translation
In vitro translation was performed with the PURExpress in vitro protein synthesis kit (New England BioLabs). 5 pmol in vitro transcribed amiEFlag mRNA was used in a 12.5 µl reaction. Increasing amounts of purified Hfq protein (as specified in legends of Figures 5B and S4) were added. For competition CrcZ (5 and 10 pmol) or RsmZ (10 pmol) RNA were added. After 1 h of incubation at 37°C, 5 µl of the reaction was mixed with 5 µl protein loading buffer and a western-blot was performed using anti-Flag antibodies as described above.
Esterase assay
Esterase activity was assayed as described previously [60]. Briefly, the cells were harvested by centrifugation and washed in 100 mM potassium phosphate buffer pH 7.2. The substrate (25 µl p-nitrophenyl-caproate dissolved in 5 ml ethanol) was added to 100 ml potassium phosphate buffer (100 nM; pH 7.2) containing MgSO4 to a final concentration of 10 mM. 1 ml of the test solution and 50 µl of cells were used to determine esterase activity by monitoring the change in absorbance at 410 nM min−1, which was normalized to the optical density (OD600) of the culture.
Northern-blot analyses
Total RNA of the respective strains as specified in the legends to the Figures was purified using hot phenol. The steady state levels of CrcZ and 5S rRNA (loading control) was determined by Northern-blotting using 4 µg of total RNA. The RNA samples were denatured for 5 min at 65°C in loading buffer containing 50% formamide, separated on a 8% polyacrylamide/8 M urea gel, and then transferred to a nylon membrane by electroblotting. The RNAs were cross-linked to the membrane by exposure to UV light. The membranes were hybridized with gene-specific 32P-end-labelled oligonucleotides (CrcZ: K3; 5S rRNA: I26; Table S3). The hybridization signals were visualized using a PhosphorImager (Molecular Dynamics).
Supporting Information
Zdroje
1. FolkessonA, JelsbakL, YangL, JohansenHK, CiofuO, et al. (2012) Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10 : 841–851.
2. CogganKA, WolfgangMC (2012) Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 14 : 47–70.
3. GörkeB, StülkeJ (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6 : 613–624.
4. RojoF (2010) Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34 : 658–684.
5. MorenoR, Ruiz-ManzanoA, YusteL, RojoF (2007) The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Mol Microbiol 64 : 665–675.
6. MorenoR, MarziS, RombyP, RojoF (2009) The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res 37 : 7678–7690.
7. SonnleitnerE, AbdouL, HaasD (2009) Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci 106 : 21866–21871.
8. MorenoR, FonsecaP, RojoF (2012) Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol 83 : 24–40.
9. FiliatraultMJ, StodghillPV, WilsonJ, ButcherBG, ChenH, et al. (2013) CrcZ and CrcX regulate carbon source utilization in Pseudomonas syringae pathovar tomato strain DC3000. RNA Biol 10 : 245–255.
10. MilojevicT, GrishkovskayaI, SonnleitnerE, Djinovic-CarugoK, BläsiU (2013) The Pseudomonas aeruginosa catabolite repression control protein Crc is devoid of RNA binding activity: false positive results caused by Hfq impurities. PLoS One 8: e64609.
11. MilojevicT, SonnleitnerE, RomeoA, Djinović-CarugoK, BläsiU (2013) False positive RNA binding activities after Ni-affinity purification from Escherichia coli. RNA Biol 10 : 1066–1069.
12. StorzG, VogelJ, WassarmanKM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43 : 880–891.
13. VogelJ, LuisiBF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9 : 578–589.
14. MollI, AfonyushkinT, VytvytskaO, KaberdinVR, BläsiU (2003) Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9 : 1308–1314.
15. VečerekB, RajkowitschL, SonnleitnerE, SchroederR, BläsiU (2008) The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res 36 : 133–143.
16. HopkinsJF, PanjaS, WoodsonSA (2011) Rapid binding and release of Hfq from ternary complexes during RNA annealing. Nucleic Acids Res 39 : 5193–5202.
17. PanjaS, SchuDJ, WoodsonSA (2013) Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res 41 : 7536–7546.
18. MikuleckyPJ, KawMK, BresciaCC, TakachJC, SledjeskiDD, et al. (2004) Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol 11 : 1206–1214.
19. WangW, WangL, ZouY, ZhangJ, GongQ, et al. (2011) Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev 25 : 2106–2117.
20. LinkTM, Valentin-HansenP, BrennanRG (2009) Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci 106 : 19292–19297.
21. SauerE, SchmidtS, WeichenriederO (2012) Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc Natl Acad Sci 109 : 9396–9401.
22. RibeiroEAJr, Beich-FrandsenM, KonarevPV, ShangW, VečerekB, et al. (2012) Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res 40 : 8072–8084.
23. VincentHA, HendersonCA, StoneCM, CaryPD, GowersDM, et al. (2012) The low-resolution solution structure of Vibrio cholerae Hfq in complex with Qrr1 sRNA. Nucleic Acids Res 40 : 8698–8710.
24. LivnyJ, BrencicA, LoryS, WaldorMK (2006) Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res 34 : 3484–3493.
25. SonnleitnerE, Sorger-DomeniggT, MadejMJ, FindeissS, HackermüllerJ, et al. (2008) Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology 154 : 3175–3187.
26. FerraraS, BrugnoliM, De BonisA, RighettiF, DelvillaniF, et al. (2012) Comparative profiling of Pseudomonas aeruginosa strains reveals differential expression of novel unique and conserved small RNAs. PLoS One 7: e36553.
27. Gómez-LozanoM, MarvigRL, MolinS, LongKS (2012) Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol 14 : 2006–2016.
28. SonnleitnerE, GonzalezN, Sorger-DomeniggT, HeebS, RichterAS, et al. (2011) The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol 80 : 868–885.
29. WildermanPJ, SowaNA, FitzGeraldDJ, FitzGeraldPC, GottesmanS, et al. (2004) Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci 101 : 9792–9797.
30. LapougeK, SchubertM, AllainFH, HaasD (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67 : 241–253.
31. SonnleitnerE, SchusterM, Sorger-DomeniggT, GreenbergEP, BläsiU (2006) Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol Microbiol 59 : 1542–1558.
32. Sorger-DomeniggT, SonnleitnerE, KaberdinVR, BläsiU (2007) Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun 352 : 769–773.
33. SonnleitnerE, HagensS, RosenauF, WilhelmS, HabelA, et al. (2003) Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog 35 : 217–228.
34. MurinaV, LekontsevaN, NikulinA (2013) Hfq binds ribonucleotides in three different RNA-binding sites. Acta Crystallogr D Biol Crystallogr 69 : 1504–1513.
35. HuangJ, SonnleitnerE, RenB, XuY, HaasD (2012) Catabolite repression control of pyocyanin biosynthesis at an intersection of primary and secondary metabolism in Pseudomonas aeruginosa. Appl Environ Microbiol 78 : 5016–5020.
36. SonnleitnerE, ValentiniM, WennerN, HaicharFZ, HaasD, et al. (2012) Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS One 7: e44637.
37. HartzD, McPheetersDS, TrautR, GoldL (1988) Extension inhibition analysis of translation initiation complexes. Methods Enzymol 164 : 419–425.
38. WinterRB, MorrisseyL, GaussP, GoldL, HsuT, et al. (1987) Bacteriophage T4 regA protein binds to mRNAs and prevents translation initiation. Proc Natl Acad Sci 84 : 7822–7826.
39. LinaresJF, MorenoR, FajardoA, Martínez-SolanoL, EscalanteR, et al. (2010) The global regulator Crc modulates metabolism, susceptibility to antibiotics and virulence in Pseudomonas aeruginosa. Environ Microbiol 12 : 3196–3212.
40. Drew R, Haq M (2004) Lessons from the ami operon. In: Ramos JL, editor. Pseudomonas: Virulence and Gene Regulation. New York: Kluwer Academic/Plenum. pp. 425–449.
41. DeanaA, BelascoJG (2005) Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev 19 : 2526–2533.
42. KaberdinVR, BläsiU (2006) Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev 30 : 967–979.
43. AbdouL, ChouHT, HaasD, LuCD (2011) Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa. J Bacteriol 193 : 2784–2792.
44. GeissmannTA, TouatiD (2004) Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J 23 : 396–405.
45. UdekwuKI, DarfeuilleF, VogelJ, ReimegårdJ, HolmqvistE, et al. (2005) Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 19 : 2355–2366.
46. SoperTJ, DoxzenK, WoodsonSA (2011) Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA 17 : 1544–1550.
47. VečerekB, MollI, BläsiU (2005) Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA 11 : 976–984.
48. DesnoyersG, MasséE (2012) Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 26 : 726–739.
49. SalvailH, CaronMP, BélangerJ, MasséE (2013) Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J 32 : 2764–2778.
50. TimmermansJ, Van MelderenL (2010) Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 67 : 2897–2908.
51. HerovenAK, BöhmeK, DerschP (2012) The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol 9 : 379–391.
52. YeungAT, BainsM, HancockRE (2011) The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol 193 : 918–931.
53. OglesbyAG, FarrowJM3rd, LeeJH, TomarasAP, GreenbergEP, et al. (2008) The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem 283 : 15558–15567.
54. O'TooleGA, GibbsKA, HagerPW, PhibbsPVJr, KolterR (2000) The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J Bacteriol 182 : 425–431.
55. ZhangL, ChiangWC, GaoQ, GivskovM, Tolker-NielsenT, et al. (2012) The catabolite repression control protein Crc plays a role in the development of antimicrobial-tolerant subpopulations in Pseudomonas aeruginosa biofilms. Microbiology 158 : 3014–3019.
56. ArgamanL, Elgrably-WeissM, HershkoT, VogelJ, AltuviaS (2012) RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc Natl Acad Sci 109 : 4621–4626.
57. YeRW, HaasD, KaJO, KrishnapillaiV, ZimmermannA, et al. (1995) Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J Bacteriol 177 : 3606–3609.
58. Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbour: Cold Spring Harbor Press.
59. Beich-FrandsenM, VecerekB, KonarevPV, SjöblomB, KloiberK, et al. (2011) Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res 39 : 4900–4915.
60. WilhelmS, GdyniaA, TielenP, RosenauF, JaegerKE (2007) The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J Bacteriol 189 : 6695–6703.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 6- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



