-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Integrative and conjugative elements (ICEs) are a relatively newly recognized class of mobile elements in bacteria, which integrate at one or more positions in a host chromosome, can be excised, circularized, and transfer by conjugation to a new recipient cell. Genome sequencing indicated that ICEs often carry genes with potential adaptive functions for the host. Various ICE-types have been described and ICEclc is a useful model for a wide class of elements found in Beta - and Gammaproteobacteria. Because ICEs normally remain “silent” in the host chromosome and often lack selectable markers, their lifestyle is difficult to study. One of the characteristics of ICEclc is that transfer is initiated in only 3-5% of donor cells in a population during stationary phase. Here, we describe an operon of three regulatory genes, two of which control the transfer initiation of ICEclc. Our findings suggest that the low transfer rate results from the repression of an activator and that this is essential to minimize the deleterious effect of hyper-activation of transfer initiation. While the individual regulatory genes are quite common on ICEs, they rarely occur in this configuration.
Published in the journal: . PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004441
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004441Summary
Integrative and conjugative elements (ICEs) are a relatively newly recognized class of mobile elements in bacteria, which integrate at one or more positions in a host chromosome, can be excised, circularized, and transfer by conjugation to a new recipient cell. Genome sequencing indicated that ICEs often carry genes with potential adaptive functions for the host. Various ICE-types have been described and ICEclc is a useful model for a wide class of elements found in Beta - and Gammaproteobacteria. Because ICEs normally remain “silent” in the host chromosome and often lack selectable markers, their lifestyle is difficult to study. One of the characteristics of ICEclc is that transfer is initiated in only 3-5% of donor cells in a population during stationary phase. Here, we describe an operon of three regulatory genes, two of which control the transfer initiation of ICEclc. Our findings suggest that the low transfer rate results from the repression of an activator and that this is essential to minimize the deleterious effect of hyper-activation of transfer initiation. While the individual regulatory genes are quite common on ICEs, they rarely occur in this configuration.
Introduction
Comparisons between ever-increasing numbers of sequenced genomes reveal the large extent to which prokaryotic genomes have undergone horizontal gene transfer (HGT) [1]-[5]. HGT has traditionally been viewed as the consequence of natural transformation, or of the action of mobile elements such as conjugative plasmids and phages [6], [7]. During the last decade, however, other types of mobile genetic elements such as integrative and conjugative elements (ICEs) have been recognized, which are widespread and thus may significantly contribute to HGT [8]-[13]. In contrast to phages and plasmids, however, we still know little about the life styles of the diverse ICE types, their modes of self-transfer and regulatory pathways controlling self-transfer. Like temperate phages, ICEs mostly exist in an integrated form at one or more specific sites in the host's chromosome (often in genes for tRNA), and are vertically transmitted to daughter cells by chromosome replication and segregation [8], [10], [14]. In order to transfer horizontally, ICEs excise themselves by site-specific recombination (attL and attR, Figure 1). This produces a circular double-stranded DNA molecule, which can transfer by conjugation to a new recipient cell, where it can reintegrate [14]. Autonomous plasmid-like replication of the excised form may occur [15]-[17], but is not required for the transfer itself.
Fig. 1. Schematic overview of ICEclc and the location of the genes relevant to this study. 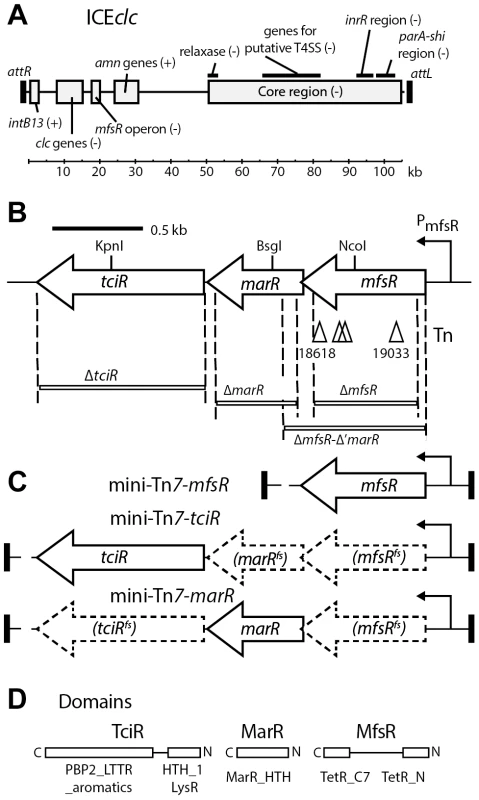
(A) ICEclc integrated form with the two flanking 18-bp repeats (as black rectangles, attR and attL). Previously determined gene regions are placed at their approximate location. Important functional regions are depicted as grey boxes accompanied by legends. + or -, indicate the orientation of the coding strand for the respective gene(s) (the + strand being the one of intB13). clc genes, chlorocatechol degradation; amn genes, 2-aminophenol degradation. kb, kilo-basepairs. (B) Detail of the mfsR operon. Arrows, predicted open reading frames (the right-to-left orientation indicates the minus strand). Triangles, positions of the Tn5-mediated kanamycin gene insertions (nucleotide positions indicated below, according to the AJ617740.2 numbering). Regions deleted in this study are displayed as white bars with the names of the mutations noted below. The hooked arrow indicates repression by the mfsR gene product on the PmfsR promoter. (C) Detail of the fragments inserted by mini-Tn7 delivery for complementation of mfsR and tciR deletions. (D) BlastP-predicted domains for each of the three regulatory genes in the mfsR operon. C and N, carboxy and amino terminus, respectively. The regulatory mechanisms that control the switch from integrated to excised state vary widely among different ICE types insofar as this has been studied. In several ICEs, this switch is the consequence of a cascade of a variety of regulatory factors, such as PhrI/RapI and ImmR/ImmA in ICEBs1 [18], SetR/SetCD in ICESXT [19], [20], KorSA/Pra in pSAM2 [21]-[23] or QseM/TraR in ICEMlSymR7 [24], [25]. Most wild-type ICEs transfer at low frequencies (i.e., less than 1 per 103 donors), suggesting that the regulatory cascades keep extremely tight control and allow only a small subset of cells in a population to follow the path of ICE excision and transfer, but the need for such tight control is a priori unclear. This bistability is most pronounced and well-studied for a model ICE named ICEclc in Pseudomonas [26], [27], which is evolutionary very distinct from the afore-mentioned ICEs [8], [28]. ICEclc is originally found in two copies in Pseudomonas knackmussii B13 and is member of a family of ICEclc-like elements widely distributed among proteobacterial species [29]. ICEclc is integrated at the 3′-end of tRNAGly genes but can excise itself by the action of the IntB13 integrase encoded on the element (Figure 1A). Expression of intB13 in the integrated form is under control of the promoter Pint, which by single cell reporter gene analysis was shown to become active only in 3-5% of a bacterial population during stationary phase [26]. Direct single cell visualization further confirmed that only cells which express reporter gene fused to Pint above a threshold are capable of transferring ICEclc to new recipients, a bistable state which we recently named “transfer competence” (tc) [30]. Irrespective of the success of ICEclc transfer, tc cells can only divide a few times once they re-enter exponential phase before they arrest growth. We recently showed that this is due to the expression of the ICEclc genes shi and parA [30]. Expression of intB13 is dependent on a variety of factors, most notably a gene named inrR (Figure 1A), which itself is also bistably expressed [26]. Both inrR and intB13 expression are dependent on the abundance of the stationary phase sigma factor RpoS, with cells having highest RpoS levels being more likely to activate Pint and PinR [29]. RpoS and InrR are important for activating ICEclc excision and transfer, but are not sufficient. Therefore, we hypothesized that additional factors are necessary for the tc state to develop [29].
In this study, we report a locus of three consecutive regulatory genes on ICEclc, which is essential for controlling its transfer. The locus was uncovered by random transposon mutagenesis, and further studied by creation of deletion mutants and complementation. The effect of mutations was studied at the level of ICEclc expression through microarray hybridizations, RT-PCR and reporter gene-based single cell fluorescence microscopy, and further in ICEclc transfer assays. Fitness of mutants compared to wild-type was examined in growth assays and individual cell fates were followed by microscopy. Bioinformatics was used to analyze the configuration of the ICEclc regulatory locus within this ICE family, and to possibly reconstruct the steps that may have led to selection of the specific regulatory control mechanism of ICEclc. The results of our study help to explain why a careful balance has to be maintained between ICE transfer frequency and fitness loss.
Results
Discovery of an ICEclc transfer control locus by transposon mutagenesis
In order to discover ICEclc-located factors involved in its self-transfer, a library of P. knackmussii B13 mutants was generated by using random Tn5 mutagenesis [31]. Next, we recovered ICEclc elements with Km-insertions by conjugating the pool of B13 mutants en masse to Pseudomonas putida UWC1 and selecting for Km-resistant P. putida (Figure S1). We hypothesized that mutant ICEclc with insertions in genes implicated in self-transfer could still be transferred to UWC1, when the second copy of ICEclc in the same B13 donor cell is intact and complements transfer of the mutant copy. A total of 1920 Km-resistant P. putida transconjugants was recovered and subsequently conjugated each individually with a second P. putida recipient, resistant to nalidixic acid (Figure S1). For those conjugations in which no Km - and nalidixic acid-resistant transconjugant growth was detected, the corresponding P. putida donor was recovered and the location of the KmR-gene insertion on ICEclc was mapped. A total of 18 clones was recovered, which had insertions in an ICEclc open reading frame numbered orf18502, that we renamed mfsR (Figure 1B). Surprisingly, apart from one donor with an insertion in intB13, no other mutants with impaired ICEclc transfer were found in this screening. The KmR-gene had been inserted in four different positions in mfsR, at ICEclc nucleotide positions 19033, 18758, 18730 and 18618 (Figure 1B, accession number: AJ617440.2). This suggests that transposon insertions in strain B13 were sufficiently frequent to cover all genes, but that the selection procedure was biased for the recovery of the mfsR insertion, which may have been due to the function of mfsR as regulator in ICEclc transfer (see below). Alternatively, it is possible that insertions in ICEclc genes needed for transfer might not be efficiently complemented by the second ICEclc copy and would thus be underrepresented in the P. putida library. Frequencies of ICEclc transfer of the strains P. putida UWC1-2961 (KmR-gene insertion at 19033) and UWC1-2962 (insertion at 18618) in a filter-based conjugation assay were 103-fold and 102-fold lower than of a P. putida with one integrated wild-type ICEclc copy, respectively (Figure 2A).
Fig. 2. ICEclc transfer frequencies from P. putida UWC1 donors with different ICEclc genotypes. 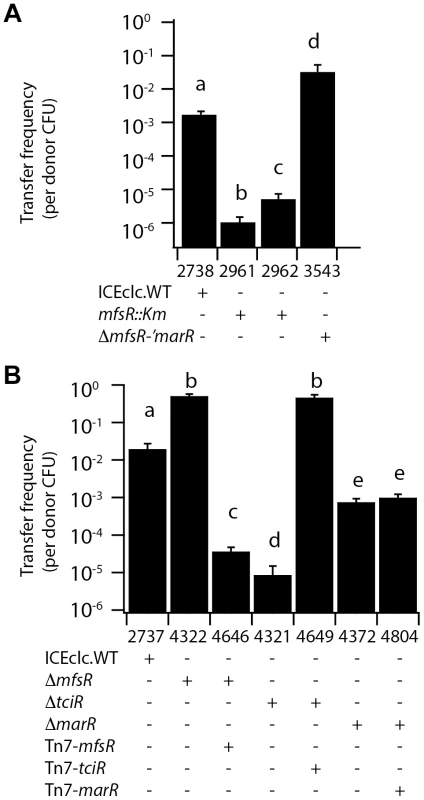
(A) and (B), Independently carried out transfer experiments using the indicated strain sets. Bars show mean transfer frequencies as transconjugant colony forming units (CFU/ml, growing on 3CBA, Km- or Gm-resistant) per donor CFU/ml from biological triplicates, and the corresponding standard deviations. Letters above bars indicate statistically significantly different groups per panel in an Anova with post hoc Tukey-Kramer test (P<0.001), with the same letter pointing to the absence of statistically significant differences. mfsR is part of an operon formed by three consecutive transcriptional regulators
Closer inspection indicated mfsR to be the first open reading frame in a series of three consecutive transcriptional regulators, previously designated as orf18502, orf17984 and orf17162 (Figure 1B). mfsR encodes a TetR-like regulator harboring helix-turn-helix motifs TetR_N and TetR_C_7 (pfam0040 and pfam14246, respectively, see Figure 1D). The orf17984 gene overlaps with the end of the mfsR open reading frame by 4 bp and encodes a putative regulator of the MarR family (smart00347 HTH_MARR motif). The last gene of this cluster starts 24 bp downstream of the stop codon of orf17984 and is predicted to code for a LysR-type transcriptional regulator, harboring an N-terminal HTH_1 motif (pfam00126) and a C-terminal substrate-binding domain (PBP2_LTTR_aromatics_like; cd08414). The gene orf17162 was renamed tciR (transfer competence inducer regulator) in anticipation of the results described further below. Reverse transcription of P. putida UWC1 (ICEclc) RNA isolated from exponential phase-grown cells, followed by specific PCR amplification confirmed that the three genes are transcribed on the same mRNA, which ends downstream of tciR (Figure S2). This implies that mfsR-marR-tciR form a single polycistronic unit.
Effects of tciR, marR, and mfsR deletions on ICEclc transfer
In order to more precisely investigate the role of the three regulators on ICEclc transfer, their open reading frames were each individually and partially deleted in separate strains, namely P. putida UWC1 (ICEclc-ΔmfsR, strain 4322), UWC1 (ICEclc-ΔmarR, strain 4372), UWC1 (ICEclc-ΔmfsR-Δ'marR, strain 3453) and P. putida UWC1 (ICEclc-ΔtciR, strain 4321) (Figure 1B, Table 1). ICEclc transfer frequencies in plate-mating assays with a gentamicin-resistant P. putida UWC1 as recipient were 2·103-fold lower for UWC1 donors with ICEclc having an internal deletion in tciR compared to intact ICEclc (Figure 2B). Complementation of the ICEclc-ΔtciR mutation with a single copy mini-Tn7 transposed fragment containing the tciR gene under the PmfsR-promoter (strain 4649, Figure 1C) restored transfer, even to much higher levels than wild-type ICEclc (Figure 2B).
Tab. 1. Strains used in this study and their specifications. 
ICEclc transfer frequencies were 27-fold lower for UWC1 donors with ICEclc having an internal deletion in marR compared to intact ICEclc (Figure 2B). Complementation of the ICEclc-ΔmarR mutation with a similar single copy mini-Tn7-marR insertion did not change transfer rates (strain 4804, Figure 2B). This suggests that the effect of the marR deletion on ICEclc transfer is rather due to polar disturbance of the expression of the downstream-located tciR.
In contrast, ICEclc elements with mfsR deletions [i.e., P. putida UWC1 (ICEclc-ΔmfsR) and UWC1 (ICEclc-ΔmfsR-Δ'marR)] transferred with 25 - and 15-fold higher frequencies than wild type ICEclc, respectively (Figure 2A, B). Complementation of the ICEclc-ΔmfsR mutation with a single copy mini-transposed mfsR gene under control of its own promoter reduced ICEclc-ΔmfsR transfer frequencies by 104-fold, also here much stronger than predicted from wild-type ICEclc itself (Figure 2B). These results suggested that tciR is the actual regulator of ICEclc transfer, and further that mfsR is regulating expression of the mfsR-marR-tciR operon. Since MfsR is expected to be a repressor, its deletion would lead to higher expression of the downstream genes marR and tciR, which results in increased ICEclc transfer. The effect of the transposon insertions in mfsR (i.e., lower ICEclc transfer rates, Figure 2A) seems therefore due to a polar effect on marR-tciR expression.
tciR encodes a global activator of the genes in the ICEclc core region
Next, we examined the effect of regulatory gene deletions on gene expression of ICEclc as a whole, using semi-tiling microarray analyses (Figure 3). When P. putida UWC1 (ICEclc) wild-type cells are growing exponentially on 3-chlorobenzoate (3CBA), expression from the genes in the ICEclc core region (roughly the second half of ICEclc) plus the integrase intB13 is silent, whereas they are highly transcribed when cells are in stationary phase (Figure 3A). Among others, the core region encodes genes implicated in ICEclc conjugative transfer [32], [33]. P. putida with mutant ICEclc lacking either marR or tciR strongly diminished expression in the core region and of the integrase gene in stationary phase when compared to wild type (Figure 3C, Figure S3). Lower core and integrase gene expression explains the lower ICEclc transfer rates from these mutants (Figure 2B). In contrast, mfsR deletion resulted in much higher expression from the ICEclc core genes in exponentially growing cells (Figure 3B), and even slightly higher expression in stationary phase than in wild-type ICEclc (Figure S4), which explains the 10 - to 100-fold ICEclc higher transfer rates (Figure 2B). Expression of the mfsR-marR-tciR cluster itself was the same in the tciR and marR deletion mutants, and no different to the wild-type (Figure 4A, C, E). In contrast, expression of the mfsR-marR-tciR cluster was higher in the mfsR deletion mutants than in wild-type, both in exponential and stationary phase cells (Figure 4B, D). Since gene expression from ICEclc is similar in mutants lacking mfsR alone or mfsR plus the first 117 bp of marR (Figure S4), we conclude that it is the LysR-type regulator encoded by tciR, which is the main activator for ICEclc core gene expression.
Fig. 3. Differential expression of the ICEclc gene region from micro-array data in selected mutant ICEclc versus wild type in P. putida UWC1. 
(A) Differential expression of the ICEclc region between stationary and exponential phase cells of wild type P. putida UWC1 (ICEclc). (B) Differential expression of the ICEclc region between the mfsR deletion mutant and wild-type, in exponentially growing cells. (C) Comparison of the tciR deletion mutant and wild-type, in stationary phase cells. (D) Comparison of the mfsR transposon insertion mutant versus wild-type, in stationary phase cells. Dots indicate the 2log-fold change of hybridization signal per microarray probe in the comparison, plotted at their distance along the ICEclc sequence (X-axis; in kb). Regions of interest on ICEclc are redrawn as grey boxes at the bottom of each section (+ or - indicate the DNA strand on which the region is encoded). Separate displays indicate expression differences on the plus- (open symbols) or the minus-strand (closed symbols). Grey bars in the background indicate the two-fold cut-off level. For a complete set of microarray results, see Figures S3 and S4. Fig. 4. Detailed view on the differential expression of the mfsR operon in P. putida ICEclc wild-type or mutants. 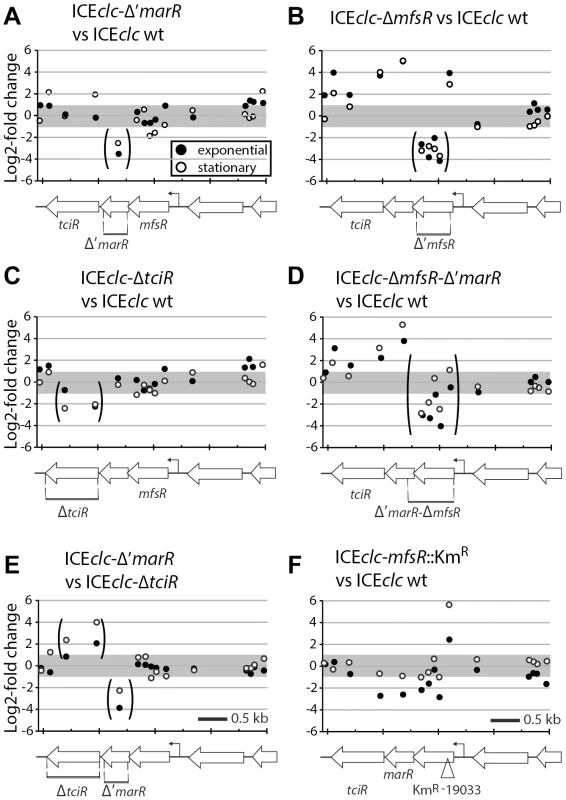
(A) MarR deletion mutant versus wild-type. (B) mfsR deletion mutant versus wild type. (C) tciR deletion mutant versus wild-type. (D) mfsR-'marR deletion mutant versus wild-type. (E) marR versus tciR deletions. (F) mfsR-transposon insertion mutant versus wild-type. Panels show 2log-fold change of expression level per microarray probe in this region of ICEclc for exponential (dark dots) and stationary phase cells (white dots). Genetic map of the region drawn at the bottom of each section for clarity. Arrows represent genes, deleted regions are indicated by stippled bars and corresponding probes are within brackets. Microarray analysis also helped to understand the behaviour of the mfsR Km-insertion mutant (Figure 3D). As for the tciR deletion mutant, expression of the ICEclc core region and of the integrase was dramatically lower than wild-type in stationary phase cells (Figure 3D, Figure S4). On the other hand, both mfsR deletion and mfsR Km-insertion mutants showed increased expression of a group of genes on ICEclc coding for a putative efflux system (Figure 3B, D). Detailed inspection of mfsR-marR-tciR expression in the Km-insertion mutant revealed that the first 160 bp of mfsR, upstream of the KmR-gene insertion were higher expressed than in wild-type cells (Figure 4F). In contrast, the downstream genes marR and tciR were lower expressed compared to wild-type and to the mfsR deletion mutant (Figure 4B, D, F). This confirmed, therefore, that insertion of the KmR-gene had caused a polar effect on expression of marR and tciR, which explains the strongly diminished expression of the ICEclc core genes in stationary phase in the mfsR Km-insertion mutant, and decreased ICEclc transfer.
Inserting the presumed mfsR promoter region upstream of a promoterless mcherry gene in single copy on the chromosome of P. putida UWC1 without ICEclc produced strong and homogenous mCherry expression among all cells (Figure 5A, strain 3482). Inserting into this strain a single copy mfsR gene expressed from its own promoter abolished mCherry expression (Figure 5A, strain 4302). Expression of mCherry from PmfsR in P. putida UWC1 (ICEclc) was very low, whereas disruption of mfsR on ICEclc again resulted in high mCherry expression (Figure 5A, strain 3606). Complementation of this strain by a single copy mfsR gene under its own promoter caused repression of mCherry expression (Figure 5A). All these data are consistent with the hypothesis that MfsR is repressing expression of itself and the downstream located marR and tciR genes.
Fig. 5. Effect of mutations in the mfsR region on the expression of the PmfsR-, Pint- and PinR-promoters of ICEclc in P. putida UWC1. 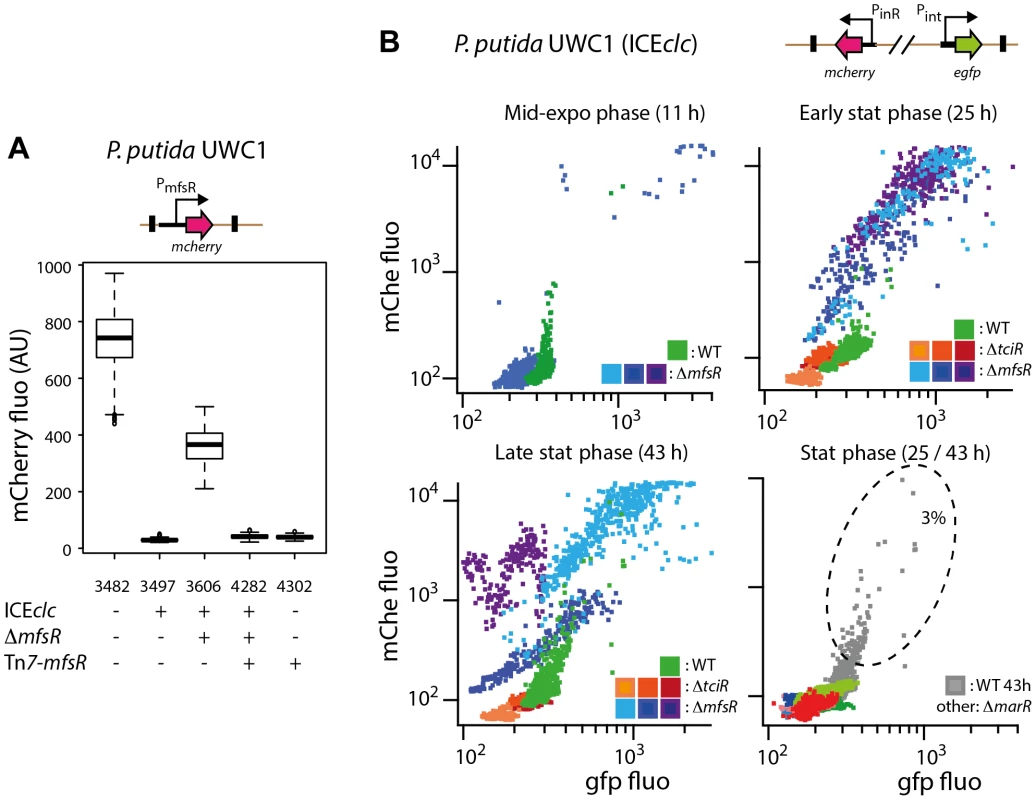
(A) mCherry expression from the mfsR promoter added in single copy to the chromosome of the indicated P. putida UWC1 strains (relevant genotypes and strain numbers specified below the graph). mCherry expression measured on individual cells (n = 1000) by epifluorescence microscopy in late exponential phase of cultures grown on 10 mM succinate and expressed as box plots (AU, arbitrary units at 20 ms exposure time). (B) Scatter plot of GFP and mcherry fluorescence in single cells of P. putida UWC1 (ICEclc) wild-type, ΔmfsR, ΔtciR or ΔmarR deletions, equipped with a single copy mini-transposon containing the Pint-egfp and PinR-mcherry fusions. Panels show expression of both markers at different growth phases, as indicated, with colors representing genotypes with independent mini-Tn5 insertions. Note as example the subpopulation of wild-type cells (dotted ellips) expressing both reporters, compared to the majority of cells in the mfsR deletion mutant but a complete absence of such subpopulation in the tciR and marR deletion mutants. mfsR deletion leads to an increase in the number of cells activating ICEclc
We then tested whether changed ICEclc transfer rates and core gene expression were in fact due to changes in the proportion of cells activating ICEclc. Hereto, a double promoter-reporter construct, carrying Pint-gfp and PinR-echerry was inserted in single copy on the chromosomes of P. putida UWC1 (ICEclc), P. putida UWC1 (ICEclc-ΔmfsR), P. putida UWC1 (ICEclc-Δ'marR) and P. putida UWC1 (ICEclc-ΔtciR) (Table 1). Pint and PinR are the respective promoters for the integrase gene intB13 (integrated form) and the integrase activator gene inrR. Previous studies showed that both promoters are active only in a small subpopulation of cells, which are representative for transfer competent cells [26], [29], [30]. Consistent with previous data, the subpopulation of P. putida UWC1 (ICEclc) wild-type cells expressing Pint and PinR in stationary phase suspended cultures represented a few percent (Figure 5B). In contrast, deletion in mfsR resulted in 80-100% of cells expressing Pint - and PinR-promoters (Figure 5B). Expression of both promoters in P. putida UWC1 (ICEclc-ΔmfsR) occurred in early stationary phase whereas in wild-type cells their expression is maximal in late stationary phase (Figure 5B). Conversely, P. putida UWC1 (ICEclc-ΔtciR) and P. putida UWC1 (ICEclc-ΔmarR) did not produce any detectable Pint - or PinR-expressing cells, neither in exponential nor in stationary phase (Figure 5B). Considering a detection limit by microscopy of ∼1 fluorescent cell among 1000-10,000 non fluorescent cells, the absence of detectable Pint - or PinR-expressing cells in those mutants would be in accordance with absence of ICEclc core gene activation on microarrays (Figure 3D) and lower transfer frequencies (Figure 2B).
Mutants with mfsR deletion in ICEclc face a strong fitness cost
Given that P. putida UWC1 carrying ICEclc-ΔmfsR transferred at a much higher rate than wild-type ICEclc, and also expressed both Pint - and PinR - promoters in almost all cells, we wondered why such mutants did not become selected spontaneously. Both P. putida UWC1 (ICEclc) wild-type and (ICEclc-ΔmfsR) displayed statistically indistinguishable generation times during exponential growth on minimal medium with either 3CBA or succinate as carbon source (Table 2), although P. putida UWC1 (ICEclc-ΔmfsR) went through a longer lag phase (Figure S5A). In contrast, the proportion of colony forming units (CFU) in samples taken from stationary phase cultures on 3CBA or succinate and plated on 3CBA solid medium was dramatically reduced for P. putida UWC1 (ICEclc-ΔmfsR) (Figure 6). Whereas the number of colonies formed on 3CBA plates was the same as that on succinate for P. putida UWC1 (ICEclc), only 0.3-5.6% appeared on 3CBA plates for UWC1 (ICEclc-ΔmfsR) (Table 2). Also in absolute terms, the number of CFU/ml for UWC1 (ICEclc-ΔmfsR) cells taken from stationary phase cultures both on 3CBA and succinate was lower than that for UWC1 (ICEclc) (Figure 6). Moreover, 8 of 10 tested colonies of UWC1 (ICEclc-ΔmfsR) grown on MM plates with succinate did no longer amplify the clcA gene of ICEclc (not shown), the remaining two still being able to grow on 3CBA. Furthermore, half or more of microcolonies formed from UWC1 (ICEclc-ΔmfsR) with a single copy Pint-egfp insertion showed incidence of malformations and cell lysis, similar to what was reported previously for nutrient-reactivated tc cells [30] (Figure 6B), but cells in the other microcolonies divided with generation times even slightly faster (1.49±0.15 h) than those in microcolonies of P. putida UWC1 (ICEclc, 1.79±0.09 h). This indicates that the mfsR deletion in ICEclc imposes a strong fitness cost on P. putida UWC1. Survival of UWC1 was restored to wild-type level when the ICEclc-ΔmfsR was complemented by the mini-Tn7 inserted mfsR gene (Figure S6). In contrast, neither P. putida UWC1 (ICEclc) with marR or tciR deletion, nor the mini-Tn7 complemented strains of P. putida UWC1 (ICEclc-ΔtciR) and (ICEclc-ΔmarR) were impaired in survival compared to UWC1 (ICEclc) (Figure S6).
Fig. 6. Fitness loss of P. putida UWC1 (ICEclc) caused by the mfsR deletion. 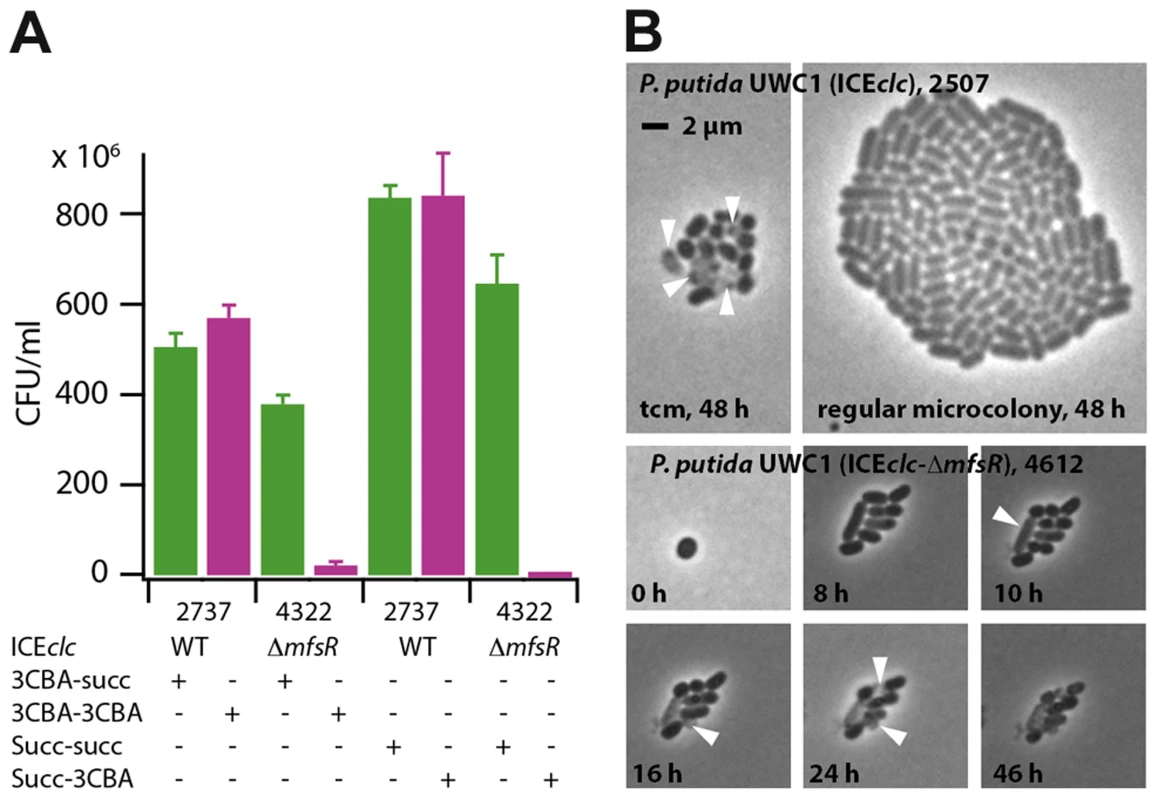
(A) Survival of P. putida UWC1 (ICEclc, 2737) and P. putida UWC1 (ICEclc-ΔmfsR, 4322) pregrown in suspended culture to stationary phase on 3-chlorobenzoate (3CBA) or succinate (succ), and plated from there on 3CBA or succinate agar. (E.g., 3CBA-3CBA, suspended culture on 3CBA, plated on 3CBA agar). Survival expressed as colony forming units (CFU) on the agar plate per ml of stationary phase culture. Data bars indicate the average from independent biological triplicates. Error bars indicate the calculated standard deviation from the average. (B) Phase-contrast micrographs at 1000-fold magnification of microcolonies of P. putida UWC1 (ICEclc) and P. putida (ICEclc-ΔmfsR Pint-egfp, 4612) growing on agarose surface supplemented with 0.1 mM 3CBA. Shown are a regular stationary phase microcolony of P. putida with wild-type ICEclc and a transfer competent microcolony (tcm), occurring at 1-3% frequency as reported previously [30]. For comparison, massive lysis (white arrows) and cellular malformations formed in many microcolonies of P. putida UWC1 with the mfsR deletion. Tab. 2. Effects of the mfsR deletion on the growth characteristics of P. putida UWC1 carrying ICEclc. 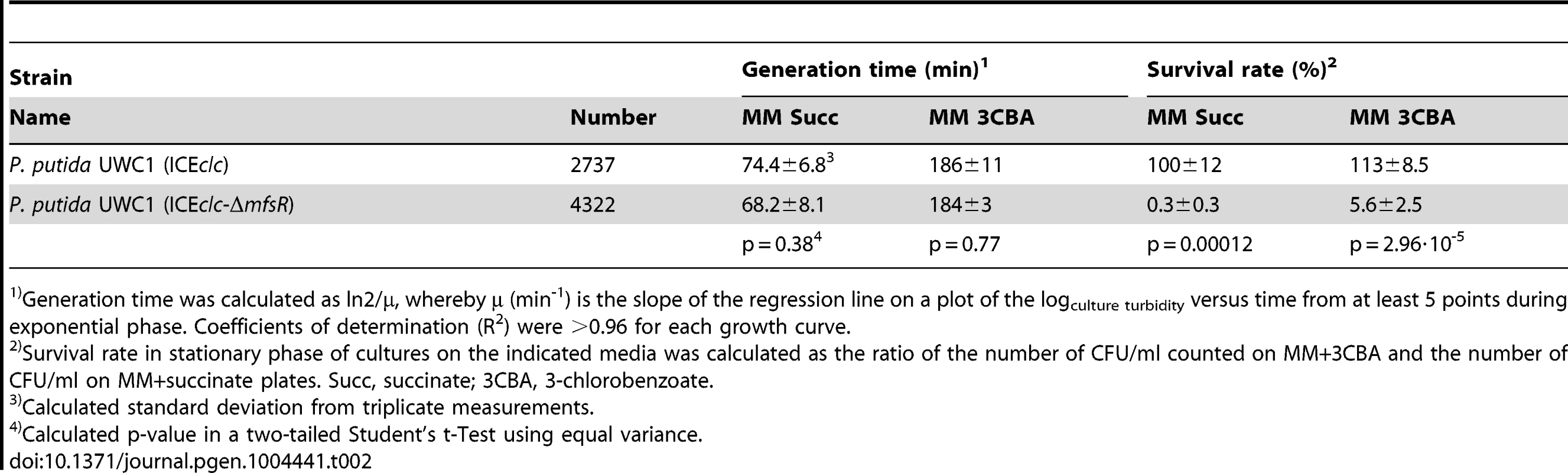
Generation time was calculated as ln2/μ, whereby µ (min-1) is the slope of the regression line on a plot of the logculture turbidity versus time from at least 5 points during exponential phase. Coefficients of determination (R2) were >0.96 for each growth curve. tciR is a very widespread ortholog among ICE closely related to ICEclc
Using bioinformatic queries, we retrieved orthologs to tciR and mfsR from sequenced bacterial genomes, and examined manually whether they occur in chromosomal regions qualifying as ICE (e.g., presence of an integrase gene nearby, see Materials and Methods). Interestingly, orthologs to the individual components of the mfsR operon are widespread but rarely occur in the same configuration (Figure 7). So far, the ICEclc mfsR-marR-tciR configuration is only found in the ICEclc variant of Burkholderia xenovorans LB400, whereas the Tn4371-element of Acidovorax sp. strain JS42 and Aeromonas hydropohila SSU (accession number AGWR01000022.1) both carry an mfsR homolog and a MarR-type regulator immediately downstream, but no tciR equivalent nearby. There is a tciR ortholog in Acidovorax sp. strain JS42, but not on the same chromosomal region as mfsR (Figure 7).
Fig. 7. Conservation of tciR analogues in putative ICEclc-like regions in a variety of other bacterial genomes. 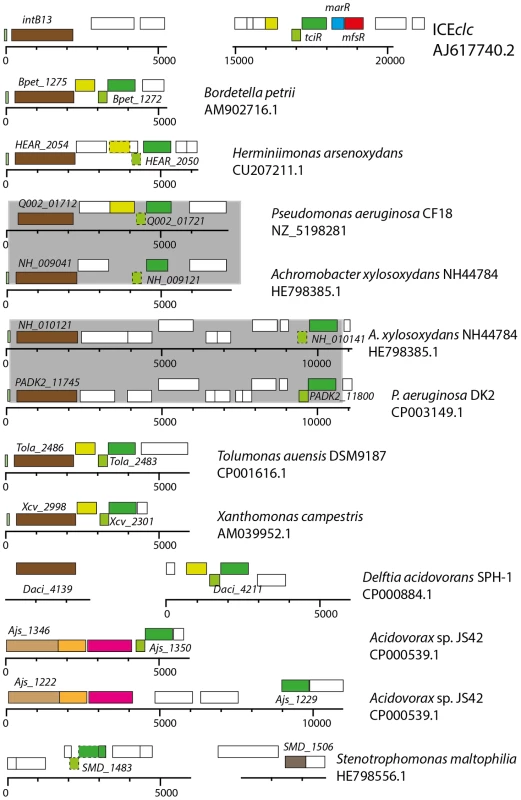
Illustration represents tciR analogues (identified on the basis of a BLASTN E-value lower than 1·10-15), and surrounding relevant gene regions in the indicated bacterial genomes (species name, accession numbers) compared to ICEclc. Genes are indicated as in the respective genome accession. Rectangles show annotated genes and their orientation (top, orientation towards the left; bottom, gene orientation towards to right); common colors indicate similar predicted functions. Stippled rectangles indicate common gene regions inferred from Artemis comparison, but not present in the respective annotation. ICE were inferred from (i) more than 75% nucleotide identities across the complete core region of ICEclc, and within a 1-100 kb window from the tciR position, and (ii) the presence of an integrase gene (in brown) within a 5-20 kb window from the tciR-analogue. Note how some genomes carry multiple different ICE from the same family (e.g., Achromobacter xylosoxidans, Acidovorax sp. strain JS42), and further how pair-wise identical ICE regions (shaded in grey) occur between different genomes. Finally note how the tciR-analogues often co-occur with a xer-type regulatory gene on the other strand (light green), and a further lysR gene member (yellow), but in none of the cases shown here with an mfsR counterpart (in red). On the other hand, tciR seems much more widespread among ICEs, as homologs can be found in ICEclc-like elements GI1 and GI6 of Bordetella petrii DSM12804, in PAGI-2 of P. aeruginosa strain C, in diverse ICEs of X. campestris pv. vesicatoria str. 85-106, suspected ICEs in Herminiimonas arsenicoxydans, Cupriavidus metallidurans CH34, and Tolumonas auensis DSM 9187, among several dozens of others (Figure 7). Given that TciR is such a common regulator found in ICEs of the ICEclc family, it might be similarly implicated in their transfer control. This regulation is likely different in detail from ICEclc, given the absence of an mfsR and marR.
Discussion
ICEclc has two distinctive modes of existence: the integrated form, which is transmitted vertically, and the circular form, which can be horizontally transferred. Previous work in our laboratory has shown that the transition between these two states occurs in only a few percent of cells in a population under stationary phase conditions [26], [30], [34]. We have recently suggested to name cells in which the molecular decision occurs to activate the ICEclc horizontal transfer mode transfer competent (tc) cells [30]. Single cell time-lapse experiments indicated that ICEclc transfer - at least insofar as detectable by microscopy, only occurs from tc cells, which can be distinguished through simultaneous expression of fluorescent proteins from single copy transcriptional fusions to the Pint and PinR promoters of ICEclc [30]. Activation of those two promoters is the likely outcome of a multi-step regulatory cascade that orchestrates expression of some fifty genes [32], but the key factors that determine the onset of this cascade and control the extent of bistability are still obscure. Previous work provided evidence for the role of the stationary phase sigma factor RpoS in activation of ICEclc promoters, and we could show that tc cells on average have higher levels of RpoS [34]. In the present study, we report the discovery of a cluster of three regulatory genes, two of which globally control ICEclc activation and transfer, and additionally maintain a cap on fitness loss induced by the ICE (Figure 8).
Fig. 8. Model for regulation of ICEclc transfer competence. 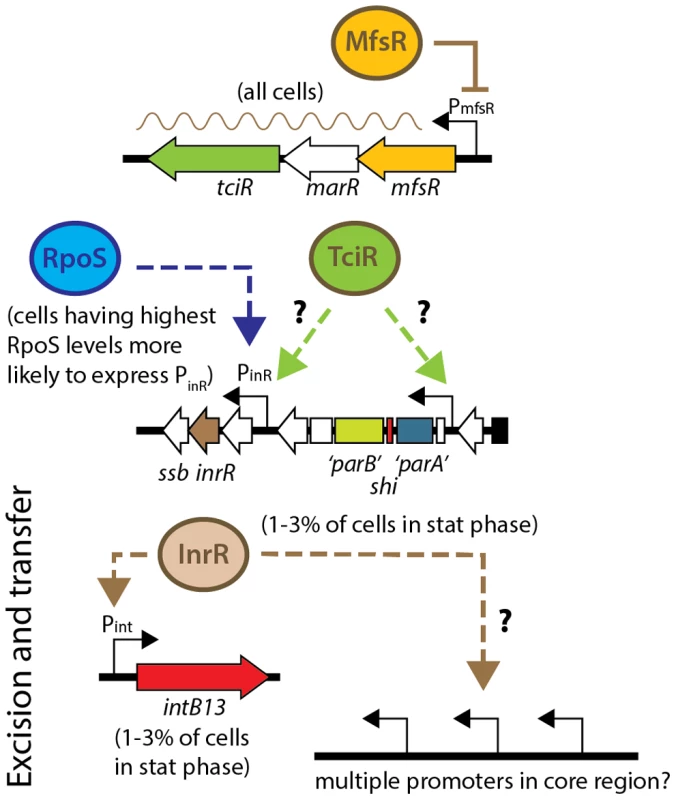
MfsR autoregulates expression of itself and of the TciR activator, without which ICEclc transfer decreases by 2·103-fold. TciR may activate specifically one or more promoters on ICEclc, such as the RpoS-dependent PinR-promoter [29], or a promoter upstream of the parA-like gene [32]. Expression of the PinR-promoter occurs preferentially in cells having highest RpoS levels, and only 1-3% of cells in a population in stationary phase visibly express reporter gene from PinR [29]. InrR transmits bistable activation through an unknown process to the intB13 promoter [26], and possibly simultaneously to other promoters for the genes for the conjugative system in the ICEclc core region [32]. The exact mechanism of arisal of bistability is unclear as yet. For gene locations on ICEclc, see Figure 1A. The three regulatory genes occur in a unique configuration of a TetR-type repressor (encoded by mfsR), followed by a MarR-type (orf17984) and a LysR-type regulator (tciR). Transcript and microarray analysis of the locus in wild-type and mutant ICEclc, plus analysis of reporter gene expression from the mfsR promoter in a variety of host backgrounds, showed that the three genes are expressed as a polycistronic unit and are under autoregulatory control by MfsR (Figure 5A).
Precise gene deletions and complementations indicated that tciR is likely the main global regulator of ICEclc transfer activation. Deletion of tciR caused a 2·103-fold lower frequency of ICEclc transfer compared to wild type (Figure 2B), silenced expression of the ICEclc core region (Figure 3B) and reduced the proportion of cells expressing Pint and PinR in stationary phase (Figure 5B). Complementation of the tciR deletion on ICEclc with a single copy mini-Tn7 inserted mfsRfs-marRfs-tciR fragment (Figure 1C) fused to the mfsR promoter region restored the expected phenotype (Figure 2B). TciR may act either directly as a regulator on a variety of individual ICEclc core promoters, or as a “master” regulator in a hierarchical activation cascade (Figure 8).
The role of marR is less clear and as yet unsolved. Deletion in marR resulted in essentially the same ICEclc transcriptome profile as deletion in tciR (Figure 3D). It also resulted in a lower transfer frequency than wild-type but not as low as the deletion in tciR (Figure 2B), and produced no detectable reporter gene expression from Pint or PinR (Figure 5). In contrast, complementation of the marR deletion on ICEclc by a single copy marR gene through mini-Tn7 delivery (Figure 1C) did not restore ICEclc transfer (Figure 2B). Furthermore, mutants with deletions in mfsR or mfsR plus the first 116 bp of marR (Figure 1B), behaved quite similar in transfer frequency (Figure 2) and showed similar ICEclc transcriptomes (Figure S4). The marR gene therefore seems to have no clear role in ICEclc core gene expression.
The most surprising effect of deletions in mfsR was a complete deregulation of the ICEclc core gene expression. This became obvious from frequencies of ICEclc transfer being 10-100 fold higher than wild type, approximating 1 transfer per donor cell (Figure 2B). The deregulation was also obvious in microarray data showing the ICEclc core region in the mfsR deletion mutants being already transcribed in exponential phase (Figure 3B, Figure S3). Finally, 80-100% of individual cells in stationary phase expressed the reporter genes from Pint and PinR in the mfsR deletion strain compared to 3-5% in wild-type (Figure 5B). This can be explained by the fact that deletion of mfsR would abolish autorepression, which would lead to constant high expression of marR and tciR. This overinitiates ICEclc core gene expression, leads to more cells entering the tc state and to higher transfer rates. The balance of mfsR control appears to be extremely delicate, since even complementation with a single gene copy under control of the original promoter results in a stronger effect than the wild-type, both for tciR and mfsR (Figure 2B). The delicate balance became also obvious from the polar effects of insertion of the Km-resistance gene within mfsR, leading to decreased transcription of marR and tciR, diminished core gene expression (Figure 3D) and reduced transfer rates (Figure 2A).
The finding that deletion and complementation of mfsR or tciR drastically changes the proportion of cells activating ICEclc, could imply that the bistability seen in wild-type situation (i.e., 3-5% of cells in stationary phase becoming transfer competent) is a result of feedback at this locus. Cells activating ICEclc in the wild-type situation could arise as a consequence of “sloppy” control by MfsR, incidentally causing a few cells to escape its control and transcribing marR and tciR. We think this is an unlikely scenario, because mCherry expression from the mfsR promoter is homogenous among cells (Figure 5A). Alternatively, there might be a chemical ligand that specifically binds to MfsR in a small subset of cells, upon which its repression is relieved in those cells. The resulting TciR would then be the necessary activator to trigger ICEclc core expression in cells with on average highest RpoS levels [29] (Figure 8). On the other hand, even though mfsR may be the first level of control, bistability may also originate at later checkpoints in the regulatory cascade, which depend on the presence of sufficient TciR.
Quasi-global appearance of transfer competence across all cells in the mfsR deletion mutant resulted in massive fitness loss (Figure 6), which became evident at two levels. First of all, time-lapse observations indicated lysis and aberrant cell growth in more than 50% of microcolonies (Figure 6B). This lysis and growth arrest are similar to what we previously described as being a side consequence of becoming transfer competent in wild-type cells [30], and is caused by the parA-shi gene products on ICEclc [30]. Secondly, there was a strong loss of the capacity to grow on 3CBA among cells sampled from stationary phase cultures of the mfsR mutant compared to wild-type (Figure 6A, Figure S6), indicative for loss of ICEclc and counterselection against maintaining ICEclc-ΔmfsR. However, those cells that maintained ICEclc-ΔmfsR could still grow on 3CBA and showed indistinguishable exponential growth rate (Table 2). This paradox can be understood when modeling the number of tc cells in batch culture populations for ICEclc wild-type (probability of tc arisal, Ptc, in stationary phase of 0.025) and for the ICEclc-ΔmfsR mutant (Ptc = 0.5). This model (Figure S5B) shows that whereas a large proportion of tc cells appear in ICEclc-ΔmfsR mutant cultures in stationary phase, these can only divide 2-3 times upon reinoculation into fresh medium before lysing. This causes an apparent prolongation of a lag phase visible as stagnant culture turbidity, but does not influence the overall predicted population exponential growth rate in batch culture (Figure S5B).
As expected from the postulated role of TciR, its complementation in trans also leads to increased ICEclc transfer, but interestingly, only the mfsR deletion caused strongly decreased cell survival (Figure S6). We therefore hypothesize that ICEclc activation may follow two separate processes: transfer and tc cell growth arrest [30], that may both be initiated at the mfsR locus. Deleting mfsR would then deregulate both processes, whereas expressing tciR in trans would only increase activation through the transfer branch (Figure 8).
The configuration of the mfsR-marR-tciR operon of ICEclc is unique, but tciR alone is a very common part of ICE similar to ICEclc (Figure 7). We therefore speculate that mfsR-marR are a more recent acquisition in ICEclc, which drastically changed the expression of the tciR gene. Unfortunately, expression of tciR analogs in other ICEs has not been studied and very little has been reported on the transferability of ICEs related to ICEclc. The exceptions being GI3 of B. petrii that transfers at extremely low frequencies (∼10-7) [35], and the P. aeruginosa PAGI-2 element for which transfer has not been detected at all [16]. In comparison, wild-type ICEclc transfers at rates of 10-2 to 10-3 per donor (Figure 2), suggesting that it was perhaps the acquisition of the mfsR regulatory control that led to expression of transfer activity in a larger proportion of cells in the population. As we show here, the downside of increasing the proportion of ICEclc tc cells is an increase of the proportion of cells displaying growth arrest through the shi-parA pathway [30]. Likely, the MfsR autoregulation evolved to a stage of permitting efficient transfer but avoiding too much fitness loss to the population. Even though the mechanistic details are different for ICEclc, double control layers are more common for various ICEs and typically involve a variety of regulators acting on each other and/or in response to specific chemical ligands [18], [19], [23]-[25]. It will be highly interesting to further study the mechanistic details of the control systems that maintain very low ICE transfer rates, and to understand whether and how such control can evolve to allow hyperefficient transfer.
Materials and Methods
Strains and culture conditions
Table 1 lists the strains used in this study. Escherichia coli DH5α (Gibco Life Technologies, Gaithersburg, Md.), E. coli DH5α λpir, E. coli BW20767/pRL27 were cultured at 37°C on Luria-Bertani (LB) medium [36]. Pseudomonas species were cultured at 30°C on LB or 21C minimal medium (MM) [37] complemented with one of the following carbon sources: 0.5, 5, or 10 mM 3-chlorobenzoate (3CBA), 15 mM succinate or 10 mM fructose. Antibiotics were supplemented to the growth medium to select for maintenance of genetic constructions at the following concentrations: kanamycin (Km) 25 µg/ml, chloramphenicol (Cm) 20 µg/ml, rifampicin (Rif) 50 µg/ml, nalidixic acid (Nal) 50 µg/ml, gentamicin (Gm) 20 µg/ml, and ampicillin (Ap) 100 µg/ml.
Strain constructions and DNA techniques
DNA purification, PCR, restriction enzyme digestions, DNA ligations and electro-transformations were performed according to standard procedures [36]. Deletions in ICEclc genes were created by double recombination techniques as described elsewhere [29], [38]. Nucleotide positions are given according to AJ617740 (ICEclc). Primers used for strain constructions are listed in table S1.
For complementation of P. putida UWC1 (ICEclc-ΔmfsR, strain 4322) we first amplified the mfsR gene plus the 429 bp upstream region containing the mfsR promoter using PCR. This fragment was cloned into pGEM-T-easy and verified for correctness by DNA sequencing. The fragment containing the correct mfsR region was then recovered by restriction enzyme digestion with PstI and BamHI, and ligated into the mini-Tn7 vector pUC-miniTn7-Gm [39]. After transformation and verification in E. coli, the mini-Tn7 construct was introduced into P. putida UWC1 (ICEclc-ΔmfsR) by using the pUX-BF13 helper plasmid [40]. Clones resistant to Gm were selected and verified by PCR for correct insertion of the mfsR DNA in the attTn7 locus. To complement P. putida UWC1 (ICEclc-ΔtciR, strain 4321) and (ICEclc-ΔmarR, strain 4372) we amplified the complete mfsR-marR-tciR locus including the 429-bp upstream region. This fragment was cloned into pGEM-T-easy and again verified for correctness by DNA sequencing. The fragment was recovered by digestion with BamHI and StuI, and ligated with the mini-Tn7 vector. A frameshift was then introduced in the mfsR coding region by digestion at the unique NcoI-site, filling in using Klenow and religation. This will cause premature ending of the mfsR gene product (mfsRfs). A second frameshift was subsequently introduced to inactivate the marR gene product, using the unique BsgI-site (marRfs). After transformation and verification in E. coli, the construct was introduced in P. putida UWC1 (ICEclc-ΔtciR) as outlined above. This procedure was repeated to create a fragment with frameshifts in mfsR and in tciR (using the unique KpnI site), but maintaining an intact marR. This construct was introduced into P. putida UWC1 (ICEclc-ΔmarR). Gm-resistant clones were verified by PCR for the correct insertion at the attTn7-site, and for the presence of ICEclc.
A 656-bp region upstream of mfsR was amplified by PCR and fused to a promoterless mcherry gene. This fragment was introduced in single copy on the chromosome of P. putida UWC1, P. putida UWC1 (ICEclc) or P. putida UWC1 (ICEclc-ΔmfsR) using mini-Tn5 delivery. Three independent Km-resistant colonies were verified by PCR for the correct insertion and stored individually. P. putida UWC1 mini-Tn5-PmfsR-mcherry and P. putida UWC1 (ICEclc-ΔmfsR) mini-Tn5-PmfsR-mcherry were then further used as recipient to introduce the mini-Tn7-mfsR construct.
Random mutagenesis and screening
Random mini-transposon insertions in P. knackmussii B13 were generated by mobilization of the pRL27 suicide plasmid from E. coli BW20767 in a biparental mating. Hereto both strains were each cultured overnight in 3 ml LB, pelleted down, resuspended in 50 µl sterile saline solution (0.9% NaCl), mixed in a 1∶1 (v/v) ratio and incubated on the surface of an LB agar plate for 24 hours at 30°C. The mixture was then resuspended with 1 ml saline solution, which was inoculated in 100 ml MM with 0.5 mM 3CBA plus Km to select for the mini-transposon insertion and Cm to counterselect against E. coli, and incubated at 30°C for 16 h with orbital shaking (180 rpm). An aliquot of 3 ml of this pool of enriched KmR B13 mutants was used en masse as donor in a subsequent mating procedure. Hereto, cells from the 3 ml suspension were pelleted by centrifugation, washed with 3 ml sterile saline and mixed with 3 ml of suspension of P. putida UWC1 recipient, that had been grown for 16 h on LB, was pelleted by centrifugation and resuspended in sterile saline. The mating mixture was again centrifuged, the cell pellet was resuspended in 50 µl sterile saline solution and spotted on the surface of a MM agar plate containing 0.5 mM 3CBA. The mixture was incubated for 72 hours at 30°C, after which the cells were washed from the plate with 1 ml sterile saline, which was further serially diluted and plated on MM agar plates with 5 mM 3CBA plus Km and Rif to select for transconjugants carrying mutant ICEclc. Individual colonies were purified, recultured in organized 96-well format and stored at -80°C after addition of and mixing with glycerol to 15% (v/v). Libraries were replicated and regrown in 100 µl LB plus Rif for 16 h in 96-well microtiter plates, mixed with 100 µl P. putida UWC1 NalR recipient suspension, and incubated at 30°C for 48 h. Then 50 µl of each well was reinoculated into 170 µl of MM containing 5 mM 3CBA plus Km, Rif and Nal, and growth was measured by continuous OD-measurements in a multiplate reader (FluoStar Omega, BMG labtech). Absence of growth was taken as indication for absence of ICEclc transfer, in which case the donor culture was recovered for mapping of the transposon insertion.
Insertion mappings
DIG-labeled primers 070934 or 070935, annealing to one of the ends of the KmR insert but facing outward, were used (separately) in single-primer PCR with DNA from mutant UWC1 donors as templates. The reactions produced oligonucleotide probes with the 5′-DIG label, the sequence of the end of the KmR gene and the adjacent sequence of the ICEclc insert position. Such products were used for rough localization of the insertion position by hybridizing to macroblot membranes (Eurogentec, UK), whose set of 55-mer oligonucleotides covers most of the ICEclc genes. Hybridization and detection of the DIG-marker were carried out according to the manufacturer's instructions (Roche Diagnostics GmbH, Mannheim, Germany). Once the insertion was roughly mapped on ICEclc, PCR-based sequencing was used to determine exact position of the KmR-gene insertion.
ICEclc transfer assays
The frequency of ICEclc transfer was determined in experimental conditions described previously [33]. P. putida UWC1 ICEclc wild-type or mutant derivatives were used as donors, whereas P. putida UWCGC (constitutively fluorescent, GmR) or P. putida UWC1 KmR were used as the recipient (Table 1). Briefly, donors and recipient were each cultivated on 5 mM 3CBA MM and 10 mM fructose MM, respectively, and combined on 0.5 mM 3CBA agar plates as a single concentrated pellet. After 48 hours incubation at 30°C, mating mixes were resuspended, diluted and plated on 5 mM 3CBA MM agar (counting of donor CFU) or 5 mM 3CBA Gm or Km agar (counting of transconjugant CFU). Transconjugants were checked by PCR and frequencies were expressed as the number of transconjugant CFU per donor CFU. Donor survival was used for the data shown in Figure S6.
ICEclc transcriptome analysis by microarrays
The ICEclc transcriptomes of P. putida UWC1 (ICEclc), P. putida UWC1 (ICEclc), P. putida UWC1 (ICEclc-KmR19033), P. putida UWC1 (ICEclc-Δ'marR), P. putida UWC1 (ICEclc-Δ'tciR), P. putida UWC1 (ICEclc-ΔmfsR-Δ'marR), P. putida UWC1 (ICEclc-ΔmfsR) were investigated by microarray analysis, as described previously [32]. Total RNA was extracted from cells grown on 10 mM 3CBA MM, and harvested at mid exponential phase (OD600 = 0.6) and 48 h after entrance in stationary phase. Reverse transcription using cyanine-dCTP among the dNTPs produced labeled cDNA that was further purified and hybridized on 8×15 K microarray slides (Agilent, Santa Clara, CA, USA). Slides were washed and scanned according to manufacturer's instructions (Agilent). Data were recovered and analyzed using GeneSpring GX. Microarray data can be accessed from the GEO database (accession number: GSE51391).
Time-lapse microscopy
P. putida UWC1 strains were precultured for 16 h at 30°C in LB medium, after which 100 µl were transferred to 20 ml fresh MM 4 mM 3CBA medium in presence of the appropriate antibiotics. This culture was incubated for 96 hours at 30°C and 200 rpm shaking, after which the cells were 100-fold diluted in MM without C-source and inoculated on agarose surfaces (gel patches) for time-lapse microscopy [41]. Medium for gel patches consisted of 1% agarose dissolved by heating into MM with 0.1 mM 3CBA. Gel patches were created by pipetting 130 µl of the agarose-MM-3CBA solution kept at 55 °C on the surface of a circular cover glass (42 mm ø and 0.17-mm thick), placed in an autoclaved perfusion chamber (POC, H. Saur, Reutlingen, Germany), separated with a 0.5 mm thick silicon spacer ring and covering them with a second cover glass. After solidification of the agarose, the upper cover slip was removed and 6 µl of the diluted cell suspension was placed onto the agarose gel patch. As soon as the drops were dried on the surface, the patches were turned upside down and placed bacteria-facing-down on a new round cover glass [41]. A second silicon spacer ring was added to allow air circulation within the closed chamber and the glass sandwich was fixed into the metal cast POC chamber with a metal ring. Up to four patches could be placed simultaneously within a single glass sandwich in a POC chamber.
Microcolony development was followed directly on a Nikon Inverted Microscope Eclipse Ti-E, equipped with a Perfect Focus System (PFS), pE-100 CoolLED and a Plan Apo λ 100×1.45 Oil objective (Nikon), installed in a controlled temperature room (22°C). Ten random regions of every patch were imaged automatically during 48 hours with intervals of 1 h, in Phase Contrast mode (10 ms exposure), eGFP (500 ms) and eCherry (500 ms). Images were recorded using Micro-Manager 1.4 (http://www.micro-manager.org/) and fluorescence values were extracted using MetaMorph (Series 7.5, MDS, Analytical Technologies).
P. putida UWC1 ICEclc and mutant fitness tests
Triplicates of strains UWC1 (ICEclc) and UWC1 (ICEclc-ΔmfsR) were grown for 16 h in LB medium at 30°C. Both strains were then 500-fold diluted (starting OD600 0.001) in MM with 5 mM 3CBA or 10 mM succinate. Upon reaching early stationary phase, strains were again diluted into fresh MM (starting OD 0.001) with the same carbon source, and growth was followed by frequent culture turbidity measurements (OD600). 24 h after reaching stationary phase, each replicate culture was serially diluted in MM and plated onto MM agar plates with 5 mM 3CBA or with 10 mM succinate. The number of CFU/ml was scored and the ratio was calculated between the number of CFU/ml on MM agar with 5 mM 3CBA and the number of CFU/ml on MM with 10 mM succinate.
Ten randomly chosen colonies of UWC1 (ICEclc-ΔmfsR) cultivated in MM with succinate and grown on MM-succinate agar plates were retested for growth on MM agar with 5 mM 3CBA. The presence of ICEclc was determined by colony PCR on the same colonies by amplifying the clcA gene, which is carried by ICEclc and the gene product of which is essential for 3CBA metabolism.
Bioinformatic screening for ICE related to ICEclc
Homologues to tciR of ICEclc were detected by BLASTN to the nr/nt database at E-value <1·10-15. The corresponding whole or draft genome sequences were retrieved and compaired by aligning to ICEclc (Accession number AJ617740.2) using Megablast. Detected regions were manually recovered and searched for the tciR homologue and an intB13 homologue within a 1-100 kb window. If annotated, the presence of a gene for tRNA-Gly nearby the intB13 homologue was scored. Regions covering all criteria (i.e., homology to ICEclc core region, tciR homologue and presence of integrase gene) were retained as containing putative ICE. Selected regions were further individually pair-wise compared by using the Artemis Comparison Tool within the WebACT service [42].
Supporting Information
Zdroje
1. KooninEV, WolfYI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36 : 6688–6719.
2. KloesgesT, PopaO, MartinW, DaganT (2011) Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol 28 : 1057–1074.
3. RankinDJ, RochaEP, BrownSP (2011) What traits are carried on mobile genetic elements, and why? Heredity 106 : 1–10.
4. BeikoRG, HarlowTJ, RaganMA (2005) Highways of gene sharing in prokaryotes. Proc Natl Acad Sci U S A 102 : 14332–14337.
5. GogartenJP, TownsendJP (2005) Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol 3 : 679–682.
6. FrostLS, LeplaeR, SummersAO, ToussaintA (2005) Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3 : 722–732.
7. ThomasCM, NielsenKM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3 : 711–721.
8. JuhasM, van der MeerJR, GaillardM, HardingRM, HoodDW, et al. (2009) Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33 : 376–393.
9. GuglielminiJ, QuintaisL, Garcillan-BarciaMP, de la CruzF, RochaEPC (2011) The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7: e1002222.
10. WozniakRA, WaldorMK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8 : 552–563.
11. BurrusV, WaldorMK (2004) Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol 155 : 376–386.
12. BurrusV, PavlovicG, DecarisB, GuédonG (2002) Conjugative transposons: the tip of the iceberg. Mol Microbiol 46 : 601–610.
13. DobrindtU, HochhutB, HentschelU, HackerJ (2004) Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2 : 414–424.
14. Bellanger X, Payot S, Leblond-Bourget N, Guedon G (2013) Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 10.1111/1574-6976.12058 [epub ahead of print].
15. KlockgetherJ, RevaO, LarbigK, TümmlerB (2004) Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 186 : 518–534.
16. KlockgetherJ, WürdemannD, RevaO, WiehlmannL, TümmlerB (2007) Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J Bacteriol 189 : 2443–2459.
17. LeeCA, BabicA, GrossmanAD (2010) Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 75 : 268–279.
18. AuchtungJM, LeeCA, MonsonRE, LehmanAP, GrossmanAD (2005) Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102 : 12554–12559.
19. BeaberJW, WaldorMK (2004) Identification of operators and promoters that control SXT conjugative transfer. J Bacteriol 186 : 5945–5949.
20. BurrusV, WaldorMK (2003) Control of SXT integration and excision. J Bacteriol 185 : 5045–5054.
21. SezonovG, HagègeJ, FriedmannA, GuérineauM (1995) Characterization of pra, a gene for replication control in pSAM2, the integrating element of Streptomyces ambofaciens. Mol Microbiol 17 : 533–544.
22. SezonovG, DuchêneA-M, FriedmannA, GuérineauM, PernodetJ-L (1998) Replicase, excisionase, and integrase genes of the Streptomyces element pSAM2 constitute an operon positively regulated by the pra gene. J Bacteriol 180 : 3056–3061.
23. SezonovG, PossozC, FriedmannA, PernodetJ-L, GuérineauM (2000) KorSA from the Streptomyces integrative element pSAM2 is a central transcriptional repressor: target genes and binding sites. J Bacteriol 182 : 1243–1250.
24. RamsayJP, SullivanJT, JambariN, OrtoriCA, HeebS, et al. (2009) A LuxRI-family regulatory system controls excision and transfer of the Mesorhizobium loti strain R7A symbiosis island by activating expression of two conserved hypothetical genes. Mol Microbiol 73 : 1141–1155.
25. RamsayJP, MajorAS, KomarovskyVM, SullivanJT, DyRL, et al. (2013) A widely conserved molecular switch controls quorum sensing and symbiosis island transfer in Mesorhizobium loti through expression of a novel antiactivator. Mol Microbiol 87 : 1–13.
26. MinoiaM, GaillardM, ReinhardF, StojanovM, SentchiloV, et al. (2008) Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc Natl Acad Sci U S A 105 : 20792–20797.
27. GaillardM, VallaeysT, VorhölterFJ, MinoiaM, WerlenC, et al. (2006) The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J Bacteriol 188 : 1999–2013.
28. Miyazaki R, Minoia M, Pradervand N, Sentchilo V, Sulser S, et al.. (2011) The clc element and related genomic islands in Proteobacteria. In: Roberts AP, Mullany P, editors. Bacterial integrative mobile genetic elements: Landes Bioscience.
29. MiyazakiR, MinoiaM, PradervandN, SulserS, ReinhardF, et al. (2012) Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet 8: e1002818.
30. ReinhardF, MiyazakiR, PradervandN, van der MeerJR (2013) Cell differentiation to “mating bodies” induced by an integrating and conjugative element in free-living bacteria. Curr Biol 23 : 255–259.
31. LarsenRA, WilsonMM, GussAM, MetcalfWW (2002) Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178 : 193–201.
32. GaillardM, PradervandN, MinoiaM, SentchiloV, JohnsonDR, et al. (2010) Transcriptome analysis of the mobile genome ICEclc in Pseudomonas knackmussii B13. BMC Microbiol 10 : 153.
33. MiyazakiR, van der MeerJR (2011) A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol Microbiol 79 : 743–758.
34. SentchiloVS, RavatnR, WerlenC, ZehnderAJB, van der MeerJR (2003) Unusual integrase gene expression on the clc genomic island of Pseudomonas sp. strain B13. J Bacteriol 185 : 4530–4538.
35. LechnerM, SchmittK, BauerS, HotD, HubansC, et al. (2009) Genomic island excisions in Bordetella petrii. BMC Microbiol 9 : 141.
36. Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
37. Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, et al., editors (1981) Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology.
38. Martinez-GarciaE, de LorenzoV (2011) Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol 13 : 2702–2716.
39. ChoiKH, GaynorJB, WhiteKG, LopezC, BosioCM, et al. (2005) A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2 : 443–448.
40. KochB, JensenLE, NybroeO (2001) A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J Microbiol Methods 45 : 187–195.
41. Reinhard F, van der Meer JR (2010) Microcolony growth assays In: Timmis KN, de Lorenzo V, McGenity T, van der Meer JR, editors. Handbook of Hydrocarbon and Lipid Microbiology: Springer Verlag. pp. 3562-3570.
42. AbbottJC, AanensenDM, BentleySD (2007) WebACT: an online genome comparison suite. Methods Mol Biol 395 : 57–74.
43. DornE, HellwigM, ReinekeW, KnackmussH-J (1974) Isolation and characterization of a 3-chlorobenzoate degrading Pseudomonad. Arch Microbiol 99 : 61–70.
44. McClureNC, WeightmanAJ, FryJC (1989) Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol 55 : 2627–2634.
45. SentchiloV, CzechowskaK, PradervandN, MinoiaM, MiyazakiR, et al. (2009) Intracellular excision and reintegration dynamics of the ICEclc genomic island of Pseudomonas knackmussii sp. strain B13. Mol Microbiol 72 : 1293–1306.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 6- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



