-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Transport of Magnesium by a Bacterial Nramp-Related Gene
Magnesium ions are essential for life, and, correspondingly, all organisms must encode for proteins to transport them. Three classes of bacterial proteins (CorA, MgtE and MgtA/B) have previously been identified for transport of the ion. This current study introduces a new route of magnesium import, which, moreover, is unexpectedly provided by proteins distantly related to Natural resistance-associated macrophage proteins (Nramp). Nramp metal transporters are widespread in the three domains of life; however, most are assumed to function as transporters of transition metals such as manganese or iron. None of the previously characterized Nramps have been shown to transport magnesium. In this study, we demonstrate that certain bacterial proteins, distantly related to Nramp homologues, exhibit transport of magnesium. We also find that these new magnesium transporters are genetically controlled by a magnesium-sensing regulatory element. Importantly, we find numerous additional examples of similar genes sharing this regulatory arrangement, suggesting that these genes may be a frequent occurrence in bacteria, and may represent a class of magnesium transporters. Therefore, our aggregate data discover a new and perhaps broadly important path of magnesium import in bacteria.
Published in the journal: . PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004429
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004429Summary
Magnesium ions are essential for life, and, correspondingly, all organisms must encode for proteins to transport them. Three classes of bacterial proteins (CorA, MgtE and MgtA/B) have previously been identified for transport of the ion. This current study introduces a new route of magnesium import, which, moreover, is unexpectedly provided by proteins distantly related to Natural resistance-associated macrophage proteins (Nramp). Nramp metal transporters are widespread in the three domains of life; however, most are assumed to function as transporters of transition metals such as manganese or iron. None of the previously characterized Nramps have been shown to transport magnesium. In this study, we demonstrate that certain bacterial proteins, distantly related to Nramp homologues, exhibit transport of magnesium. We also find that these new magnesium transporters are genetically controlled by a magnesium-sensing regulatory element. Importantly, we find numerous additional examples of similar genes sharing this regulatory arrangement, suggesting that these genes may be a frequent occurrence in bacteria, and may represent a class of magnesium transporters. Therefore, our aggregate data discover a new and perhaps broadly important path of magnesium import in bacteria.
Introduction
Metal ions are essential and serve many cellular purposes, including functioning as cofactors for numerous metalloenzymes. The latter are responsible for a diverse array of biochemical reactions and, together, comprise one third of all cellular proteins [1]–[3]. Conversely, all metals elicit toxic effects when they accrue to excess. Therefore, specific mechanisms are required for maintaining intracellular pools. In many instances, metal-sensing regulatory proteins (metalloregulatory proteins) control expression of transport proteins, or of metal-sequestering cellular factors [4]–[6].
One broadly important class of metal transporters is that of Nramps (natural resistance-associated macrophage proteins). The Nramp1 family has been discovered in mammals to transport metals out of the macrophage phagosome. Mutational disruption of the gene results in increased susceptibility to infection by intracellular pathogens [7]–[9]. This suggests that deprivation of essential metals is a strategy used by hosts for compromising the phagosome as a niche for bacterial growth and replication. Nramp1 transports manganese, and possibly iron, while a second Nramp (Nramp2) transports primarily iron. In general, these Nramp genes are members of a large gene family, with numerous representatives in all three domains of life. For example, the sequence identity between bacterial and mammalian Nramps is high, often in excess of 35% [10]. Interestingly, just as mammalian Nramps are involved in microbial resistance, bacterial Nramps may be simultaneously required during infection by intracellular pathogens [11]–[13], although the importance of these proteins during infection remains a subject of debate [14], [15]. Essentially, bacterial and mammalian Nramps may compete for the same metals within the phagosome at the interface of host-pathogen interactions [16], [17]. Also, in addition to their important roles during microbial pathogenesis, genes encoding Nramps are required by many bacterial genomes as fundamental transporters of divalent ions.
Nramps share common structural features, including 10–12 transmembrane domains and conserved residues interspersed throughout. High-resolution structural data from X-ray crystallography is still lacking though, restricting knowledge of the structural basis of metal selectivity and transport. It is generally presumed that Nramp family members are employed for transport of manganese or iron, although some family members have been shown to exhibit broad transport activity for other divalent ions [18]–[21]. However, magnesium has not been shown to serve as a substrate for Nramps that have been characterized. Indeed, the initial characterization of a bacterial proton-dependent manganese transporter (MntH), which is a member of the Nramp family of proteins, demonstrated that it could import iron or manganese even in the presence of 5 mM magnesium, suggesting that magnesium was unlikely to serve as an MntH substrate [22].
Magnesium is the most abundant divalent metal in living cells and is required for numerous cellular activities, including serving as cofactor for enzymatic reactions and maintaining the structures of membranes and ribosomes [23]–[25]. Cytoplasmic levels of most transition metals are maintained at relatively low concentrations through action of high affinity metalloproteins [6], [26]–[28]. In contrast, intracellular free magnesium is maintained at a higher level (0.5–2.0 mM), which requires specific magnesium transport proteins [24], [29]–[33].
Three families of magnesium transporters have been discovered in bacteria: CorA, MgtE, and MgtA/MgtB P-type ATPase proteins [33]–[38]. While many metalloregulatory proteins have been identified as sensors of transition metals, less is known regarding control of magnesium homeostasis. One mechanism is through cytoplasmic gating domains of CorA and MgtE, which help couple intracellular magnesium demand with transport activity. In addition, a few genetic regulatory mechanisms have been discovered for magnesium homeostasis. Best studied in this regard is a two component regulatory system in Salmonella enterica that completes phosphoryl transfer from a sensor kinase (PhoQ) to response regulator (PhoP) in response to fluctuations in extracellular magnesium [39]. PhoP activates genes for magnesium homeostasis, such as mgtA, as well as genes important for growth and replication within a host cell. Interestingly, the mgtA transcript is also subject to a second, post-initiation layer of magnesium-responsive genetic regulation [40]. Changes in magnesium alter the secondary structure within the mgtA 5′ leader RNA; stabilization of one particular configuration is coupled with control of transcription elongation, which has the effect of limiting mgtA transcription to conditions of low magnesium [40].
Signal-responsive RNA elements, akin to S. enterica mgtA, which coordinate chemical cues with regulation of downstream gene expression, are referred to as riboswitches. A second, and mechanistically distinct, magnesium riboswitch, sometimes called the ‘M-box’, has also been discovered in bacteria. Originally discovered upstream of the Bacillus subtilis mgtE gene this riboswitch is also broadly conserved in numerous distantly-related bacteria [41]–[43]. It is almost always located upstream of one of the three known classes of magnesium transporters: CorA, MgtA, and MgtE. Riboswitches are generally composed of two portions: a signal-responsive aptamer and a downstream region that couples conformational changes of the aptamer with control of transcription, translation, or mRNA stability [44]–[48]. The structure of the magnesium-bound M-box aptamer domain has been resolved by X-ray crystallography and its mechanism for sensing magnesium has been investigated by various biochemical and biophysical experiments [49], [50]. Together, the aggregate data on this riboswitch suggest strongly that it serves as a metalloregulatory RNA for control of magnesium transport genes [42].
Given the close regulatory relationship between the M-box riboswitch and magnesium transporter genes, we were surprised to discover a subset of riboswitches situated upstream of Nramp-related genes. This observation established an intriguing conundrum. While M-box riboswitches respond to magnesium fluctuations in vivo, no Nramp or Nramp-related proteins have been found to transport this divalent ion. Therefore, one of these general assumptions must be incorrect. Either these particular riboswitches have been adapted to sense a divalent ion other than magnesium, or, alternatively, these particular Nramp-related homologs exhibit an unexpected role in magnesium homeostasis. Our combined data support the latter. We find that a Clostridium acetobutylicum ATCC 824 magnesium riboswitch controls expression of an Nramp-related gene and, moreover, that this particular transporter is surprisingly proficient in magnesium transport. These data, therefore, identify this subset of solute carrier proteins as a fourth class of magnesium transporters, designated herein as NrmT (Nramp-related magnesium transporter).
Results
Identification of magnesium riboswitches upstream of Nramp-related genes
M-box magnesium riboswitches [41], [42] are widespread in bacteria, and are almost always positioned upstream of putative magnesium transport genes (i.e., corA, mgtE, or mgtA). The riboswitch is presumed to control expression of the transport protein in a magnesium-responsive manner, as it does for Bacillus subtilis mgtE [41]. Most magnesium riboswitches affect gene expression by controlling formation of an intrinsic transcription terminator (Fig. 1A). A three-dimensional structural model of the aptamer (ligand-binding) domain revealed the presence of between 6 and 9 functionally important divalent ion binding sites [41], [49], [50]. However, this structural model alone cannot rule out the intriguing hypothesis that there might still exist aptamer variants that sense divalent ions other than magnesium. Motivated by this hypothesis, we searched using Infernal [51] for instances where M-box riboswitches were located upstream of genes for transport of metals other than magnesium. This search uncovered multiple instances where it appeared that putative Nramp family genes were located immediately downstream of M-box riboswitch candidates, mostly in Clostridia and Deltaproteobacteria. Since Nramp transporters are generally assumed to mediate transport of manganese and/or iron, and have never been show to transport magnesium, we chose to examine more closely a few representative examples of M box-regulated Nramp-related genes. For this, we chose two separate loci within the Clostridium acetobutylicum ATCC 824 genome (Ca_c0685 and Ca_c3329) (Fig. 1B).
Fig. 1. Identification of M-box RNAs located upstream of bacterial Nramp-related genes. 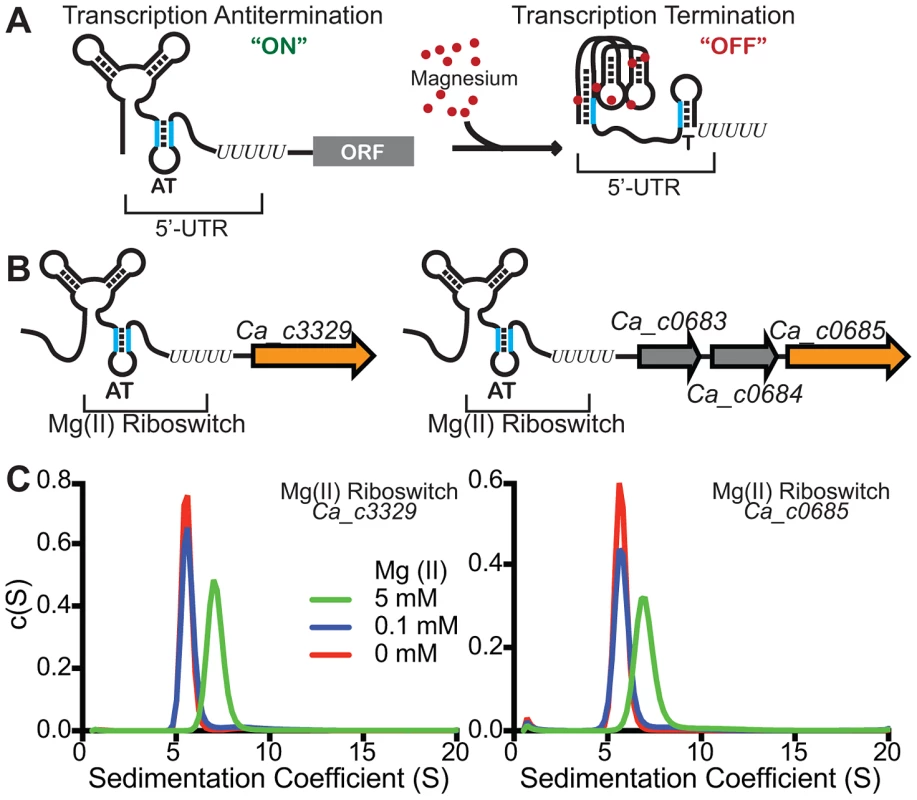
(A) The magnesium riboswitch consists of two portions – a divalent-sensing aptamer and downstream sequences which couple the conformational status of the aptamer with formation of an intrinsic transcription termination site. An increase in intracellular magnesium triggers a compacted conformation of the aptamer domain [42] and sequestration of an oligonucleotide tract that would otherwise disrupt terminator formation (“T”). Therefore, increased magnesium promotes transcription termination, repressing downstream gene expression. (B) We searched for instances of putative magnesium riboswitches located upstream of Nramp-related genes and identified many such occurrences. Two were identified for Clostridium acetobutylicum Nramp-related sequences, Ca_c0685 and Ca_c3329, and are shown schematically herein. (C) To determine whether the Ca_c0685 and Ca_c3329 RNA elements were likely to function as magnesium riboswitches, the respective aptamer domains were incubated with varying magnesium and analyzed by analytical ultracentrifugation. Prior studies of the magnesium riboswitch revealed a striking compaction of the aptamer domain in response to magnesium [41]–[42], [49]. The sedimentation velocity measurements of the Ca_c0685 and Ca_c3329 riboswitches revealed an identical compaction with magnesium, suggesting they are likely to function similar to the previously characterized riboswitch. Magnesium-specific regulation by the Ca_c0685 riboswitch
As a biochemical test of magnesium riboswitch function, the aptamer portions of the putative Ca_c0685 and Ca_c3329 riboswitches were transcribed in vitro and subjected to analytical ultracentrifugation measurements (Fig. 1C). Previous data using this technique demonstrated a large, and characteristic, change in hydrodynamic radius for B. subtilis magnesium riboswitches in response to binding of magnesium [41], [42], [49]. For example, the aptamer domain of the B. subtilis magnesium riboswitch exhibited a Svedberg coefficient of approximately 5.6 in low (100 µM) magnesium, and approximately 6.9 under conditions of elevated (5 mM) magnesium, indicating significant divalent ion-induced compaction. Sedimentation velocity measurements of the Ca_c0685 and Ca_c3329 riboswitch aptamer domains revealed strikingly similar changes in sedimentation coefficients upon addition of magnesium (e.g., 5.8 and 7.0 in low and high magnesium, respectively, for Ca_c0685), consistent with significant metal-induced compaction of the Clostridial RNAs. Therefore, the Ca_c0685 and Ca_c3329 riboswitches appear by this biochemical test to resemble previously characterized magnesium riboswitches. For an in vivo test of riboswitch function, the Ca_c0685 and Ca_c3329 riboswitches were sub-cloned downstream of a constitutive promoter (PrpsD), but upstream of a yellow fluorescent reporter gene (yfp), for examination of their regulatory activity in vivo. The reporter fusions were integrated single copy into the Bacillus subtilis genome and yfp abundance was measured by S1 mapping (Fig. 2). Total RNA was extracted from cells cultured in rich media, after treatment for one hour by 2 mM EDTA or 2 mM magnesium chloride. A PrpsD-yfp control revealed almost no change in yfp upon treatment. In contrast, the Ca_c0685 riboswitch-yfp fusion exhibited an increase in yfp upon EDTA treatment, and a reduction in yfp in response to 2 mM magnesium, consistent with regulatory control by a magnesium riboswitch. To explore this further, cells were cultured to mid-logarithmic growth phase and exposed to 2 mM EDTA, followed by resuspension in medium containing a range of magnesium concentrations (Fig. 3A). Under these conditions, the yfp transcript was increasingly reduced by the Ca_c0685 riboswitch as extracellular magnesium was increased. However, only minor repression was observed with Ca_c3329.
Fig. 2. Control of gene expression by a C. acetobutylicum M-box RNA. 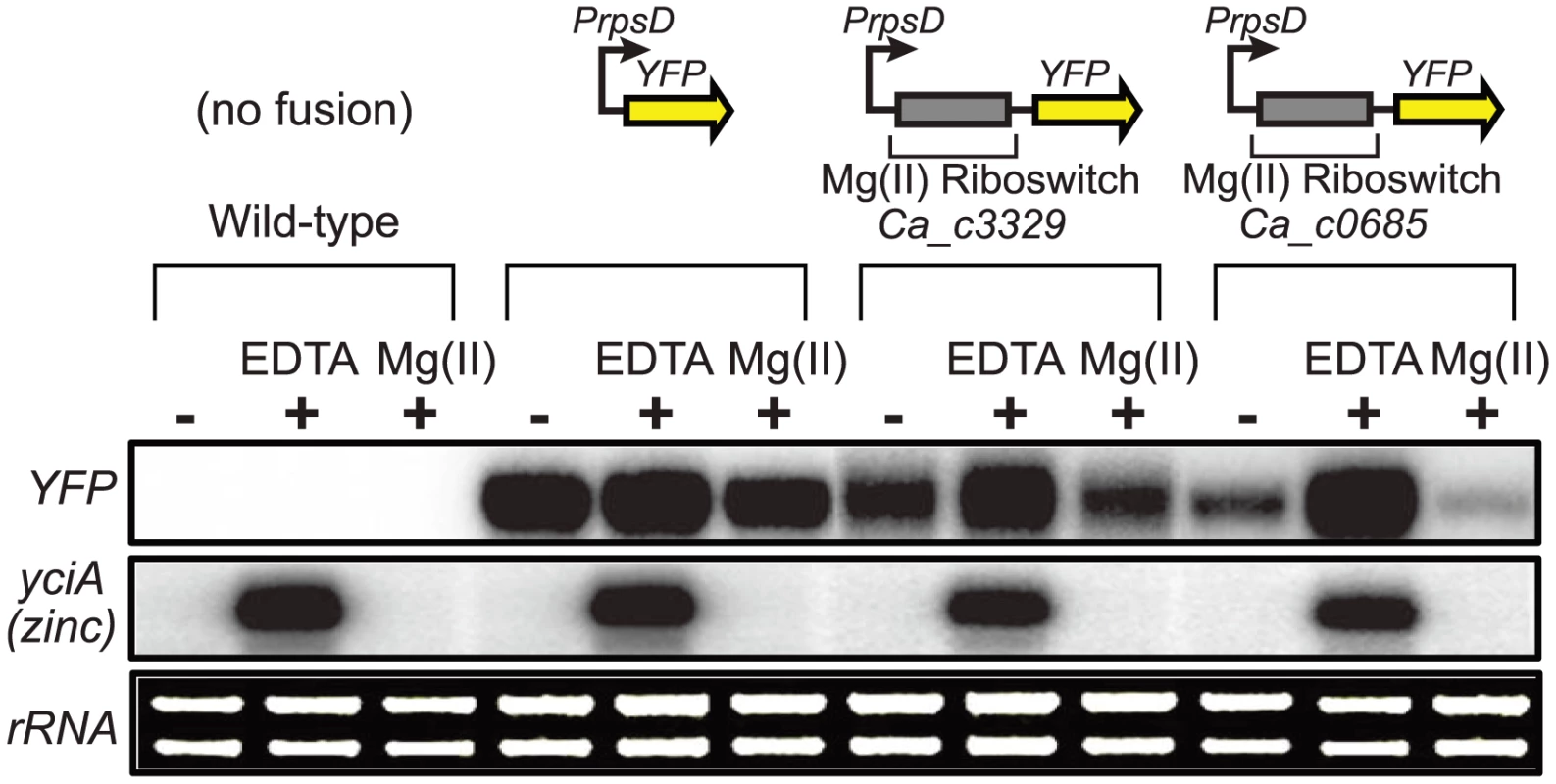
The Ca_c0685 and Ca_c3329 riboswitches were fused downstream of a constitutive promoter (PrpsD) and upstream of the yellow fluorescent reporter gene (yfp) to determine whether they could control heterologous gene expression in a divalent cation-dependent manner. Control strains either lacking the yfp reporter construct or containing a constitutive PrpsD-yfp fusion were included in this study. Cells were cultured to mid-logarithmic growth phase in 2xYT rich medium supplemented with 50 µM MgCl2 (-), then incubated with 2 mM chelating agent, EDTA, or incubated in the presence of excess magnesium (2 mM) for one hour. Total RNA was assessed by staining of rRNA bands. Abundance of the yfp gene and of a zinc-responsive control transcript were monitored by S1 mapping. Radiolabeled DNA probes (Table S2) were used for S1 mapping of the yfp transcript. Fig. 3. Metal specificity of C. acetobutylicum riboswitch-yfp reporter fusions. 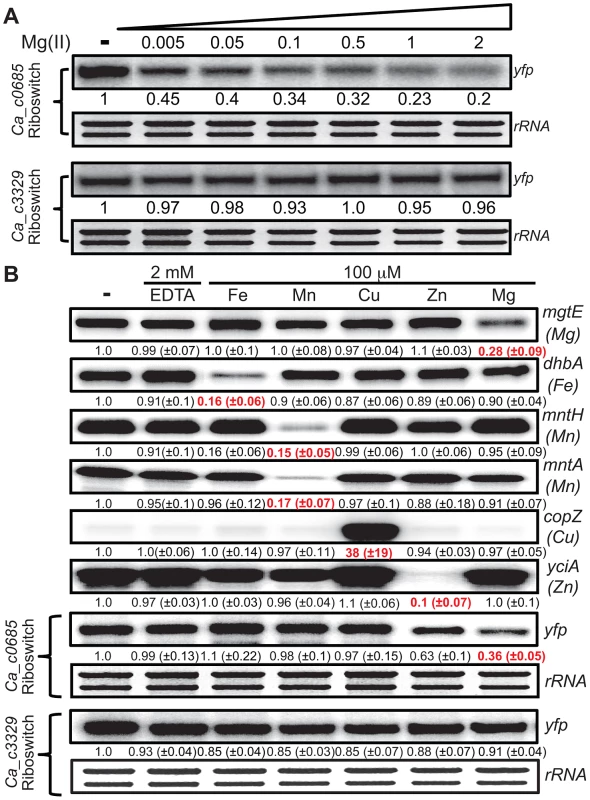
(A) The strains expressing either the Ca_c0685 riboswitch-yfp or Ca_c3329 riboswitch-yfp reporter fusions were cultured in glucose minimal medium supplemented with 50 µM magnesium until reaching an OD600 of ∼0.5–0.7, at which point 2 mM EDTA was added and cells were incubated for 1 hour. These cells were harvested by centrifugation and the pellet was washed three times and resuspended with an equal volume of chelated glucose minimum medium (chelated with Chelex-100). Either EDTA (2 mM final concentration) or varying magnesium concentrations were added and the cells were incubated for another 1 hour before harvesting. Total RNA was assessed by staining of rRNA bands. Abundance of the yfp gene was monitored by S1 mapping. (B) To assess the specificity of the C. acetobutylicum riboswitch-yfp reporter fusion, we cultured cells expressing either the Ca_c0685 riboswitch-yfp or Ca_c3329 riboswitch-yfp reporter fusions in glucose minimal medium supplemented with 50 µM magnesium and appropriate antibiotics until reaching an OD600 of ∼0.5–0.7, at which point 2 mM EDTA was added for 1 hour. These cells were harvested by centrifugation and the pellet was washed three times and resuspended with an equal volume of chelated glucose minimum medium (chelated with Chelex-100). Either EDTA (2 mM final concentration) or 100 µM various metals were added and the cells were incubated for another 1 hour before harvesting. Radiolabeled DNA probes (Table S2) were used for S1 mapping of the yfp transcript, and for several control transcripts that are known to respond to other metals (e.g., mgtE (magnesium), dhbA (iron), mntH (manganese), mntA (manganese), copZ (copper), yciA (zinc)). These data demonstrate that the Ca_c0685 specifically controls gene expression in response to magnesium, although addition of zinc also resulted in a moderate reduction in gene expression. Shown is a representative gel with quantification derived from experimental triplicates. In order to examine metal specificity in vivo for regulation by the Ca_c0685 riboswitch, cells were cultured to mid-logarithmic growth phase, exposed to 2 mM EDTA, and resuspended in medium containing excess iron, manganese, copper, zinc or magnesium (Fig. 3B). Expression of the Ca_c0685-yfp reporter was evaluated alongside control measurements of B. subtilis metal transport genes. This analysis confirmed a three-fold reduction of yfp in response to magnesium by the Ca_c0685 riboswitch. Excess zinc also moderately reduced the yfp transcript (∼37%); however, excessive levels of iron, manganese, or copper had no effect on yfp, although the presence of these metals did affect transcripts known to be under their regulatory influence. These data together revealed that the Ca_c0685 riboswitch is a magnesium-sensing regulatory element. In contrast, it remains unclear from these experiments why the Ca_c3329 riboswitch appears to be unresponsive within the B. subtilis host organism.
Expression of Ca_c0685 and Ca_c3329 does not complement a deficiency in manganese transport
Prior experimental evidence has primarily demonstrated that many bacterial Nramp homologues are for transport of manganese or iron [22], [52]–[56]. Although the Ca_c00685 and Ca_c3329 proteins are only distantly related to Nramp family proteins, the genes encoding Ca_c0685 and Ca_c3329 were heterologously expressed in B. subtilis under IPTG-inducible control (Fig. S1). To investigate a potential role in manganese transport, markerless deletion mutants of B. subtilis manganese transport genes, mntH and mntABCD, were introduced into these strains. The ΔmntH/ΔmntABCD double mutant (bCAW2105) exhibited a growth defect in defined medium, which could be rescued with addition of >10 µM manganese, or by ectopic expression of B. subtilis MntH (Fig. 4A; Fig. S1), consistent with prior findings [50]. In contrast, neither heterologous expression of Ca_c0685 or Ca_c3329 was able to rescue the manganese transport deficiency. Therefore, these C. acetobutylicum genes are unlikely to encode for manganese transport.
Fig. 4. Heterologous expression of Ca_c0685 and Ca_c3329. 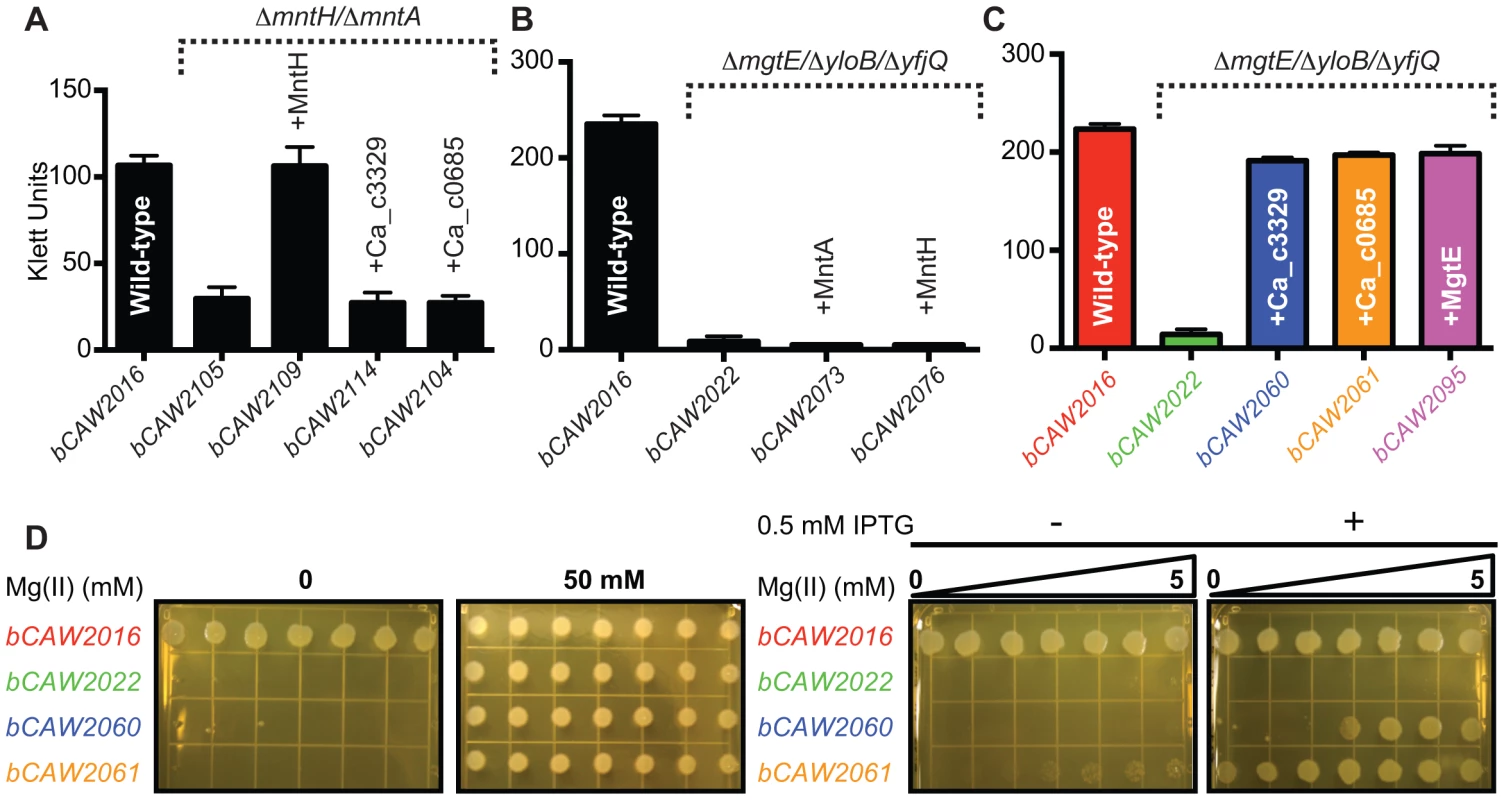
The ca_c0685 and ca_c3329 genes were subcloned under IPTG-inducible control and integrated single-copy into the B. subtilis amyE gene. These expression cassettes were also integrated into various strains containing deletions of different divalent cation transporters. In all instances, expression of ca_c0685 and ca_c3329 was monitored by S1 mapping (Figure S1, S2, S3). To investigate the effect of gene expression in these various strains, cells were cultured alongside control strains. Shown herein are bar graphs plots of stationary phase growth for these respective strains. (A) Growth after entry into stationary phase is shown for B. subtilis control strains, including a wild-type and a manganese transport-deficient strain, and transport-deficient strains complemented with IPTG-inducible control of B. subtilis MntH, Ca_c0685 or Ca_c3329. These strains were cultured in minimal medium with no added manganese in the presence of 0.5 mM IPTG. (B) Heterologous expression of B. subtilis MntH and MntABCD do not rescue a magnesium-transport deficient phenotype. Growth measurements immediately after entry into stationary phase are shown for B. subtilis control strains, including wild-type, a magnesium transport-deficient strain, and transport-deficient strains complemented with either IPTG-inducible MntABCD or MntH. Full growth curves are shown in Figure S2. These strains were cultured in rich medium in the presence of 0.5 mM IPTG. (C) Heterologous expression of Ca_c0685 and Ca_c3329 in a magnesium transport-deficient strain. Growth measurements immediately after entry into stationary phase (full growth curves are included in Figure S3) are shown for B. subtilis strains, including wild-type, a magnesium transport-deficient control strain, and transport-deficient strains complemented with inducible Ca_c0685, Ca_c3329, or the magnesium transporter MgtE. The strains were cultured in rich medium in the presence of 0.5 mM IPTG and 2.5 mM magnesium. Expression of Ca_c0685 and Ca_c3329 both fully rescued growth in this medium. (D) In addition to the liquid culture growth experiments, 3 µl of each of these strains (∼1×104/µl) was spotted onto solid medium containing a gradient of magnesium that ranged from 0 to 5 mM magnesium, respectively. These plates were incubated for 10 hours at 37°C before they were photographed. Complementation of magnesium transport activity by Ca_c0685 and Ca_c3329
B. subtilis contains examples of all three magnesium transporter families [33]–[39], including ykoK (mgtE homolog), yloB (mgtA homolog), and two corA homologues (yfjQ, yqxL). A triple mutant (bCAW2022) containing markerless deletions of mgtE, yloB, and yfjQ exhibited a strong defect in magnesium transport activity (Fig. 4B; Fig. S2), and requires ∼50 mM extracellular magnesium to restore growth in rich medium [57]. As a preliminary check of the specificity of this magnesium transport-deficient phenotype, the known B. subtilis manganese transporters, mntH and mntABCD, were ectopically integrated into the genome under inducible control (creating bCAW2073 and bCAW2076). Expression of these manganese transporters was unable to complement the severe magnesium deficiency exhibited by bCAW2022. This supports the hypothesis that the bCAW2022 strain exhibits a specific defect in magnesium transport activity.
To examine the impact of Ca_c0685 and Ca_c3329 expression on magnesium transport, they were integrated single-copy into the bCAW2022 genome under inducible control and growth was assessed under conditions of magnesium limitation. Expression of Ca_c0685 fully restored growth to resemble that of wild-type cells (Fig. 4C; Fig. S3). Moreover, when this strain was inoculated onto solid medium that contained a gradient of magnesium from sub-micromolar to 5.0 mM, growth was observed on all portions of the plate, in contrast to bCAW2022 (Fig. 4D; Fig. S3). This suggests that Ca_c0685 rescued growth even under conditions of sub-micromolar magnesium. Also, Ca_c3329 was able to rescue the magnesium transport-deficient phenotype for bCAW2022; however, it rescued growth only when magnesium was included at >2 mM. Therefore, these data demonstrated that both Ca_c0685 and Ca_c3329 are capable of magnesium transport activity, with Ca_c0685 potentially showing higher affinity for the magnesium ion.
Magnesium repression of Ca_c0685 and Ca_c3329 in Clostridium acetobutylicum
Our observation that Ca_c3329 acts as a magnesium transporter seems at first glance to be inconsistent with our prior result showing that the PrpsD-Ca_c3329-yfp fusion was unresponsive to magnesium (or any other divalent ions tested). However, heterologous expression of riboswitch-reporter fusions is not always successful. This is, in certain instances, likely to be due to differences in the molecular environment, such as changes in RNase preferences or in RNA recognition by transcription elongation factors. Therefore, it is possible that the Ca_c3329 riboswitch is nonfunctional when expressed in B. subtilis but is still functional in the C. acetobutylicum host for regulation of a transporter. However, it is also possible that the Ca_c3329 riboswitch is nonfunctional in both organisms. As a test of these possibilities, C. acetobutylicum was cultured to OD600 of ∼0.8, and then treated with 2 mM EDTA for one hour, at which point the cells were harvested and resuspended in magnesium-free medium. These cells were then aliquoted into media containing varying magnesium concentration and incubated for 2 additional hours before extraction of total RNA. S1 mapping of Ca_c0685 and Ca_c3329 revealed that both genes were subjected to repression of transcription as magnesium was increased (Fig. 5). This indicates that both Ca_c0685 and Ca_c3329 are likely to be repressed by magnesium within the context of their host organism, and that the Ca_c3329 riboswitch is likely to be functionally responsive to magnesium in C. acetobutylicum.
Fig. 5. Magnesium repression of Ca_c0685 and Ca_c3329 in C. acetobutylicum. 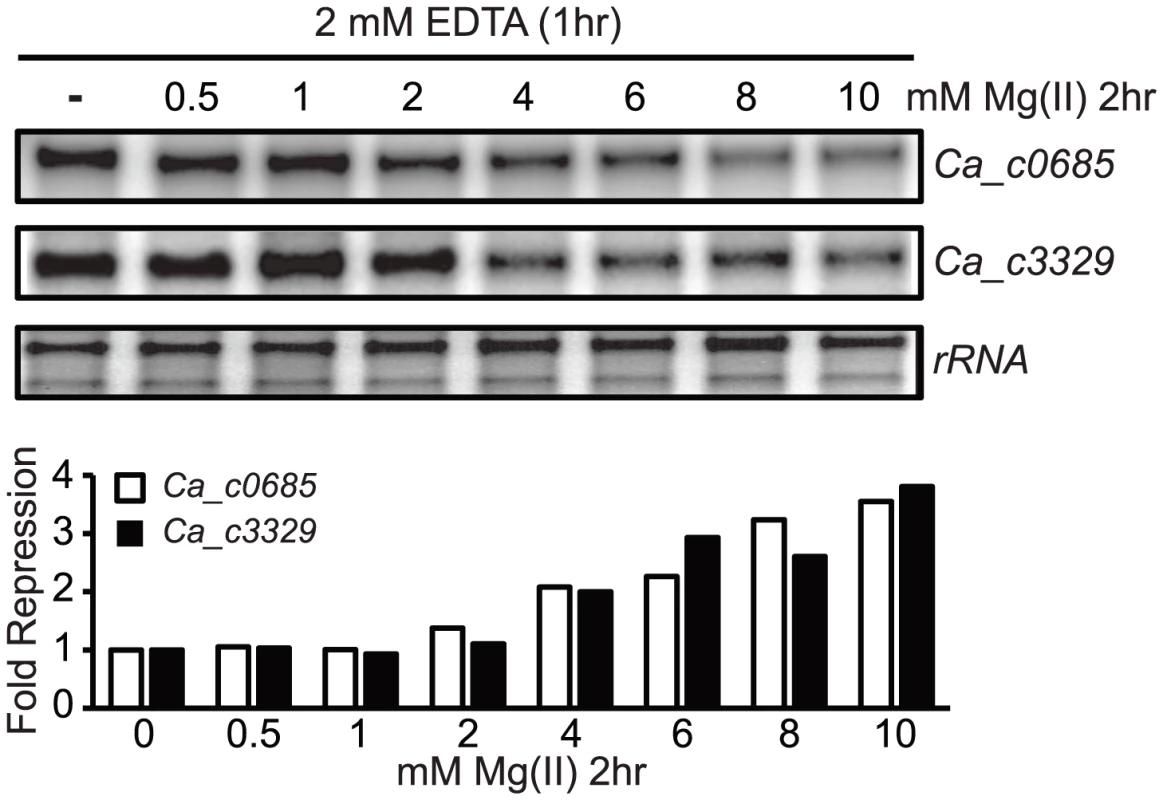
C. acetobutylicum cells were cultured in growth medium to an OD600 of ∼0.8. They were then treated with 2 mM EDTA for one hour, followed by centrifugation and resuspension in magnesium-free growth medium. The cells were then aliquoted into growth medium containing a range of magnesium concentrations and incubated for 2 hours under standard conditions. Abundance of the Ca_c3329 and Ca_c0685 transcripts was then measured by S1 mapping, as shown by the representative experiment herein. Phylogenetic analysis of Nramp-related transporters associated with magnesium riboswitches
Nramp family transporters are widespread among bacteria and eukaryotes [58] and are hypothesized to have emerged early in evolution. We performed a phylogenetic analysis of putative Nramp-related genes located immediately downstream of M-box RNAs. In addition, we collected representatives from three previously identified groups of bacterial MntH-like proteins (groups A, B and C, according to a previous classification, [58]), Nramp homologs from human, Arabidopsis and yeast, and several examples of an Nramp outgroup that was identified previously [59]–[61]. Members of the branched-chain amino acid transporter family, a part of the APC superfamily containing similar LeuT folds, were used as an outgroup, as in a previous characterization of Nramp phylogeny [60], [62]. We constructed the multiple sequence alignment (Fig. S4) and the maximum likelihood phylogenetic tree (Fig. 6) for the 47 selected representatives. All M-box-regulated homologs clustered into a single branch on the phylogenetic tree adjacent to the Nramp outgroup genes, whereas other bacterial Nramp transporters, including known manganese/iron transporters, were distributed in their respective groups. Interestingly, all Nramp-related transporters that appeared to be regulated by the magnesium riboswitch clustered together, suggesting a relationship between magnesium and members of this branch. This analysis indicates that these riboswitch-associated Nramp-related transporters form a distinct clade that is derived from a more distant common ancestor than those of the Nramp family, but that shares a more recent common ancestor with the Nramp outgroup. Given the phylogenetic relatedness of these proteins, we renamed them NrmT, for Nramp-related magnesium transporter.
Fig. 6. Bayesian phylogenetic tree of 45 Nramp family transporters, Nramp outgroup proteins, riboswitch-associated outgroup members, and branched-chain amino acid transporters. 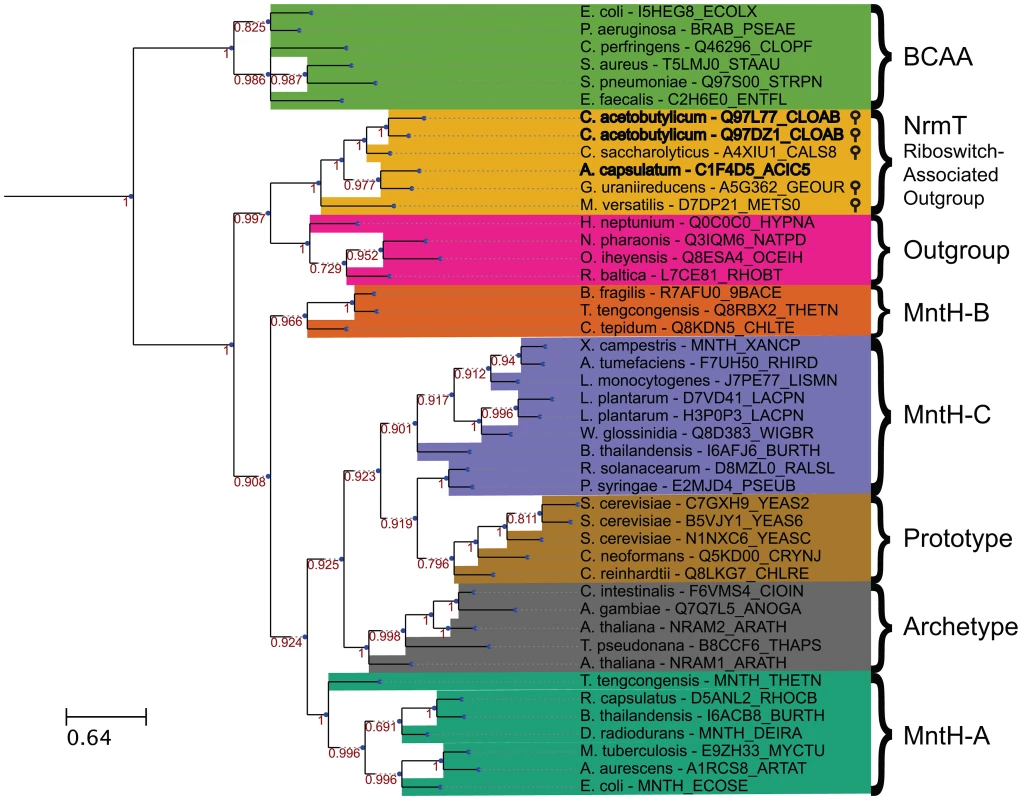
The sequences were aligned using MAFFT v7 using the L-INS-I algorithm [75]. This tree is the consensus of four replicate trees constructed using MrBayes 3.2 [76], [77]. Each replicate tree was constructed using four total chains for Metropolis coupling, and 1,000,000 generations with 25% relative burn-in. Priors used were the defaults for amino-acid models with an equal mixture of amino-acid substitution models. Transporter genes preceded by putative magnesium riboswitches are denoted by stem-loops. Branch support values are indicated in red by each internal node. Branch lengths represent expected number of substitutions per position. Similar trees were obtained with different approaches including maximum likelihood (MetaPIGA), minimum evolution, and maximum parsimony (MEGA6), not shown [78], [79]. To search for unique features of NrmT group, we examined the genomic context of representative genes (Fig. S5). Surprisingly, this revealed that the local genomic context of most group members includes a common, additional gene. This latter gene appears by sequence homology to encode for a protein that is specifically homologous to the N-terminal cytoplasmic domain of the MgtE transporter. This protein appears to be encoded by a single gene, except in C. acetobutylicum where the riboswitch-regulated operon Ca_c0685 includes a homolog that is split into two smaller genes (Fig. 1B).
Together, these observations appear to suggest a possible relationship between magnesium homeostasis and the Nramp-like outgroup (NrmT) identified herein. As a preliminary test of this possibility, another member of this grouping, but that lacked a magnesium riboswitch, was arbitrarily chosen for heterologous expression in B. subtilis. Specifically, we identified an nrmT gene from Acidobacterium capsulatum, located downstream of the small gene exhibiting homology to the MgtE cytoplasmic domain. Heterologous expression of the A. capsulatum Nramp-related gene (Fig. S6) in bCAW2022 (i.e., the magnesium transporter-deficient strain) rescued growth, but only in the presence of low millimolar magnesium. The partial MgtE gene was then integrated at a separate locus of the genome and co-expressed with the A. capsulatum putative transporter; expression of both proteins did not further improve rescue of the magnesium transport defect. However, expression of this A. capsulatum transporter gene was fully capable of rescuing the manganese transport defect exhibited by bCAW2105 (ΔmntH/ΔmntABCD) (Fig. S6). These data suggest that the A. capsulatum NrmT protein is likely to function as a manganese transporter, although what role the partial MgtE gene may play in metal transport remains unknown. Therefore, only a subset of the NrmT proteins, sometimes associated with magnesium riboswitches, is likely to exhibit high affinity magnesium transport. These data also illustrate the potential value of using the magnesium riboswitch as an identifying feature of magnesium specificity in associated transporters.
Discussion
The three major classes of bacterial magnesium transporters (CorA, MgtE, MgtA) were discovered using complementation strategies similar to that described herein [35], [63]–[65]. While other, minor routes of magnesium import may be possible, organisms from all three domains of life are generally expected to encode at least a subset of these three protein families, as magnesium acquisition is essential. In this study, we employed a similar complementation approach to suggest that C. acetobutylicum proteins, unrelated to the known classes of transporters, are capable of magnesium transport. As these proteins are distantly related to the Nramp family of proteins, which have not been found to transport magnesium, this was an unexpected discovery that suggests either the substrate range for Nramp transporters must be expanded to include this divalent ion, or, more likely, that a new class of Nramp-related divalent metal transporters has been introduced. Therefore, these observations together suggest that a subset of Nramp-related transport proteins constitutes a fourth class of dedicated magnesium transporters in bacteria, designated herein as NrmT.
Our data also revealed that a riboswitch upstream of Ca_c0685 is proficient within the confines of a heterologous host in coupling intracellular magnesium fluctuation with control of downstream gene expression. This observation strengthens the overall body of evidence showing that magnesium is the central signal perceived by the M-box riboswitch. It also suggests that identification of these riboswitches can, in certain instances, be used to help predict which Nramp-related homologues are likely to function as dedicated magnesium transporters, rather than transporters of other divalent cations such as manganese. This is also bolstered by our observations that Ca_c3329 was both repressed by magnesium in C. acetobutylicum and provided magnesium transport activity in B. subtilis, albeit at higher concentrations of the ion. Therefore, in total, we speculate that the C. acetobutylicum magnesium riboswitches control expression of two magnesium-transporting Nramp-related genes, which may be functionally specialized for different ranges of extracellular magnesium. There are two MntH homologues that are also encoded by this organism (Ca_p0063 and Ca_c0628, representing a MntH-A and MntH-B protein, respectively) [55], which are not associated with magnesium riboswitches and that we speculate are likely to provide transport of manganese or iron.
MntH homologues are widespread members of the Nramp family of proteins. In addition to two groups of eukaryotic Nramps, phylogenetic analyses have identified three groups of bacterial MntH proteins, designated as group A, B, and C. The group A proteins were characterized as proton motive force-dependent transporters of divalent ions, typically manganese or iron [20], [22], [53], [59], [66]. Also, group A MntH genes have been shown to be expressed by intracellular bacteria during host infection, and to be regulated by external availability of metal ions by the manganese-responsive MntR repressors [22], [53]. Group C MntH share a closer sequence relationship with eukaryotic Nramp homologues, while group B exhibits an origin closer to the root of the Nramp tree. Group B MntH derive mostly from strict anaerobes, such as Chlorobium, proto-photosynthetic bacteria and Clostridium species, suggesting it may have appeared before onset of aerobiosis [55], [58]. In contrast, the C. acetobutylicum transporters characterized herein do not belong in any of these Nramp subgroups (Fig. 6). Instead, they display only a moderate relationship to previously established Nramp groups. Their closest relationship to established Nramp groupings is with group B members; however, there is still considerable distance exhibited between them. Instead, phylogenetic analysis indicates that the magnesium riboswitch-regulated proteins are more closely related to a phylogenetic outgroup of the Nramp family than to the family itself. This outgroup was previously identified [61] as an evolutionary intermediate between the Nramp family and a superfamily characterized by the LeuT 3D fold [62]. The taxonomic distribution of this particular outgroup is broad and is not restricted to species where magnesium riboswitches are associated. When analyzed with the phylogenetic data, a structural modeling analysis previously suggested that the Nramp phylogenetic outgroup segregates between the large cluster of transporters known to share the LeuT 3D fold and the Nramp family [61], [67]. Based on this work [61], [67] the Nramp outgroup may be viewed as an evolutionary link between cation-driven transporters that act on non-metal substrates and those transporting divalent metals. In this study, we find that the riboswitch-regulated proteins are related to this previously described outgroup, but may also form their own subclass (Fig. 6).
It is not yet obvious what sequence features might differentiate the magnesium-transporting proteins from Nramp-related proteins transporting other divalent ions. However, prior multiple sequence alignments suggested a few amino acids that are conserved and that may be involved in metal coordination. For example, several individual intramembrane sites that distinguish Nramps from the outgroup have been implicated in cation transport [61]. Some of these residues are maintained while others are replaced in the proteins associated with magnesium riboswitches (Fig. S4). Therefore, subsequent site-directed analyses of these and other residues may eventually provide important insight into the metal selectivity and transport cycling properties exhibited by this newly discovered magnesium-transporter.
Further study is also required for determining the functional role(s) of the ancillary gene, which encodes for a small protein homologous to the N-terminal cytoplasmic domain of the MgtE transporter. This ORF was not strictly required for magnesium transport activity of Ca_c0685 and Ca_c3329, as it was not included in our genetic complementation assays; however, it may play an additional role. The cytosolic domain of MgtE has been suggested by prior biochemical and structural data to contain magnesium sites that regulate transport activity. From this, we speculate that the MgtE-like fragment is likely to provide cytoplasmic feedback regulation of transport for the NrmT proteins, or, alternatively, may affect metal chaperone activity. A role for the MgtE-like fragment in metal homeostasis is also supported by the observation that many of the MgtE-like fragments are regulated by magnesium riboswitches, co-transcribed with the NrmT outgroup members. However, our complementation analysis of the A. capsulatum outgroup protein, which lacks a magnesium riboswitch, revealed that it is likely to primarily transport manganese, rather than magnesium, despite the presence of an adjacent MgtE-like fragment. Therefore, the MgtE-like fragment that is associated with the A. capsulatum protein would be expected to serve a function other than magnesium regulation, perhaps instead responding to manganese ions. Subsequent biochemical analysis of transport activity will be required to test these predictions, and to reveal the function of the small ORF.
Nramps are believed to be important in most organisms for transport of manganese or iron. We demonstrate herein that an outgroup of Nramp-related proteins are likely to function as dedicated magnesium transporters. Therefore, when considering the potential routes of magnesium transport activity for a target organism, this family of proteins must be considered as potential suspects, along with previously identified magnesium transport classes. Future studies will be required to compare these newly discovered magnesium transporters with CorA, MgtE and MgtA/B proteins, and to determine whether they are also important to infection by bacterial pathogens.
Materials and Methods
Strains and culture conditions
All B. subtilis strains used in this study were isogenic with common laboratory strains listed in Table S1. Depending on the experiment, they were cultured in liquid rich medium [2xYT; (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl)], solid rich medium [Tryptone Blood Agar Base (TBAB)], and glucose minimal medium [20 g/L (NH4)2SO4, 183 g/L K2HPO4*3H2O, 60 g/L KH2PO4, 10 g/L sodium citrate, 0.5% glucose, 0.5 mM CaCl2, 5 µM MnCl2, and 2 g/L MgSO4*7H2O when appropriate] at 37°C. When appropriate, antibiotics were included at: 100 µg/mL spectinomycin, 5 µg/mL chloramphenicol and 1 µg/mL erythromycin plus 25 µg/mL lincomycin. To chelate divalent cations from media, 5 g/100 mL of Chelex-100 resin (Bio-Rad) was added to dissolved medium and equilibrated with stirring for 2 hours, followed by removal of resin by filtration. DNA was transformed into B. subtilis using a modified version of a previously published protocol [68].
Construction of riboswitch-yfp reporter strains
For construction of reporter fusions between C. acetobutylicum riboswitches and yellow fluorescent protein gene (yfp), the putative magnesium riboswitches were amplified by PCR and subcloned into pDG1662 (Table S2). The constitutive promoter from B. subtilis rpsD was subcloned upstream while the yfp gene was placed downstream of the putative riboswitches, respectively. These plasmids were transformed into B. subtilis PY79 for integration into amyE.
Construction of strains for complementation experiments
The wild-type strain for all complementation experiments was derived from B. subtilis 168 by integration of empty pHyperspank vector at the amyE locus [69]. Manganese and magnesium transporter knockout strains were created by in-frame markerless deletion [57], [70]. Complementation strains were created by sub-cloning of the transporter genes into pHyperspank under the control of an IPTG-inducible promoter and double homologous recombination of the resulting construct into the amyE locus of the appropriate transporter knockout strain. Correct strains were verified by diagnostic PCR and Sanger sequencing. All strains used in this experiment are listed in Table S1.
Growth measurements
B. subtilis strains were cultured on TBAB plates, supplemented when necessary with MgCl2 concentrations indicated in the text, and the appropriate antibiotics. These cells were used to inoculate 5 mL of either 2xYT or glucose minimal media (GMM) [43], that was cultured overnight while shaking at 37°C. An aliquot was then diluted 1∶100 in 25 mL 2xYT or GMM supplemented as necessary with antibiotics. These cells were incubated shaking at 37°C until reaching an OD600 of ∼0.5, whereupon they were pelleted, washed twice with 10 mL 2xYT or GMM, and resuspended to an OD600 of ∼0.05 in 25 mL, including the indicated amounts of MgCl2 and/or 0.5 mM IPTG. Klett readings were recorded at regular time intervals using 250 mL flasks. Stationary phase OD600 measurements were taken at the second time point after the intersection of exponential and stationary growth phases.
Gradient plate assays
For preparation of agar plates containing a gradient of magnesium, we utilized a procedure described previously [71], [72]. Briefly, a slanted 2% agar medium base containing the maximum desired concentration of magnesium was prepared in a standard petri dish, upon which a magnesium-free, 0.8% top agar medium was poured, thus allowing a gradient of magnesium to be established by diffusion from the slanted bottom layer. Approximately 3 µl culture (at ∼1×104 cells/µl) were spotted onto magnesium gradient plates (from 0 mM magnesium to either 2.5 or 5.0 mM magnesium), with or without 0.5 mM IPTG and incubated for exactly 10 hours at 37°C at which point the plates were imaged by photography. In a related experiment, a serially diluted culture (from 6.25×103 to 100 cells) was spotted onto glucose minimal medium plates with and without 10 µM manganese chloride, and with and without 0.5 IPTG, and incubated for exactly 10 hours at 37°C at which point they were photographed.
S1 mapping analysis
Total RNA was harvested from cells that were grown to mid-logarithmic phase in 2xYT or in glucose minimal medium [41]. When appropriate, the glucose minimal medium was first subjected to chelation of divalent metals by incubation with various amounts of EDTA, as described in the manuscript. Total RNA was extracted by hot phenol after fixation of cell pellet with RNAprotect reagent (Qiagen), according to the manufacturer instructions and as described previously [28], [73]. The quality and quantity of RNA was measured by absorbance spectroscopy and confirmed by resolution on 1.3% formaldehyde-agarose gels. Gene-specific oligonucleotide probes (Table S2) for Ca_c3329, Ca_c0685, mntA, mntH, yfp, mgtE, dhbA, copZ, and yciA transcripts were used for PCR amplification using Clostridium acetobutylicum and B. subtilis genomic DNA as template. Each specific DNA probe was radiolabeled with [γ-32P] ATP and T4 polynucleotide kinase and 30,000–40,000 cpm of labeled probe was used in each reaction. 100 µg of total RNA was pelleted and lyophilized; this pellet was then carefully resuspended in 20 µl hybridization buffer [40 mM PIPES (pH 6.4), 400 mM NaCl, 1 mM EDTA, 80% (v/v) formamide]. Individual samples were incubated at 80°C for 25 min and slow cooled to 42°C. 300 µl of S1 nuclease mix (∼100 units in S1 nuclease buffer [280 mM NaCl, 30 mM NaOAc (pH 4.4), 4.5 mM ZnOAc]) was added and incubated at 37°C for 45 min. The reaction was terminated by addition of 75 µl of S1 nuclease termination solution (2.5 M NH4OAc, 0.05 M EDTA). The DNA-RNA hybrid was precipitated by adding 400 µl of isopropanolol and the pellet was washed with 70% (v/v) ethanol, vacuum dried, and resuspended in 10 µl alkaline loading dye. The protected DNA fragments were then resolved by 6% (wt/vol) polyacrylamide gels containing 7 M urea. The dried gels were exposed to a phosphor imaging screen (FLA-2000; Fuji) and bands were quantified using Multi Gauge V3.0 or ImageJ.
Growth of Clostridium acetobutylicum
C. acetobutylicum 824 was cultured in 400 mL Clostridial growth medium (CGM) [74] until it reached an OD600 of ∼0.8. EDTA (pH 8.0) was added to a final concentration of 2 mM. After one-hour incubation, the cells were harvested by centrifugation and resuspended in magnesium-free CGM. 40 mL of this cell suspension was then aliquoted into separate containers containing designated concentrations of magnesium. After a two-hour incubation, cells were harvested by centrifugation and stored at −80°C until lysis and RNA extraction.
Supporting Information
Zdroje
1. AndreiniC, BertiniI, CavallaroG, HollidayGL, ThorntonJM (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem 13 : 1205–1218 doi:10.1007/s00775-008-0404-5
2. WaldronKJ, RutherfordJC, FordD, RobinsonNJ (2009) Metalloproteins and metal sensing. Nature 460 : 823–830 doi:10.1038/nature08300
3. GuerraAJ, GiedrocDP (2012) Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch Biochem Biophys 519 : 210–222 doi:10.1016/j.abb.2011.11.021
4. MooreCM, HelmannJD (2005) Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol 8 : 188–195 doi:10.1016/j.mib.2005.02.007
5. PennellaMA, GiedrocDP (2005) Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals 18 : 413–428 doi:10.1007/s10534-005-3716-8
6. GiedrocDP, ArunkumarAI (2007) Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans 63 : 3107–3120 doi:10.1039/b706769k
7. SkameneE, GrosP, ForgetA, KongshavnPAL, St CharlesC, et al. (1982) Genetic regulation of resistance to intracellular pathogens. Nature 297 : 506–509 doi:10.1038/297506a0
8. HuJ, BumsteadN, BarrowP, SebastianiG, OlienL, et al. (1997) Resistance to salmonellosis in the chicken is linked to NRAMP1 and TNC. Genome Res 7 : 693–704 doi:10.1101/gr.7.7.693
9. LangT, PrinaE, SibthorpeD, BlackwellJM (1997) Nramp1 transfection transfers Ity/Lsh/Bcg-related pleiotropic effects on macrophage activation: influence on antigen processing and presentation. Infect Immun 65 : 380–386.
10. PinnerE, GruenheidS, RaymondM, GrosP (1997) Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a member of the mammalian natural resistance-associated macrophage protein family. J Biol Chem 272 : 28933–28938 doi:10.1074/jbc.272.46.28933
11. SupekF, SupekovaL, NelsonH, NelsonN (1997) Function of metal-ion homeostasis in the cell division cycle, mitochondrial protein processing, sensitivity to mycobacterial infection and brain function. J Exp Biol 200 : 321–330.
12. ZwillingBS, KuhnDE, WikoffL, BrownD, LafuseW (1999) Role of iron in Nramp1-mediated inhibition of mycobacterial growth. Infect Immun 67 : 1386–1392.
13. GomesMS, AppelbergR (1998) Evidence for a link between iron metabolism and Nramp1 gene function in innate resistance against Mycobacterium avium. Immunology 95 : 165–168.
14. BoechatN, BordatY, RauzierJ, HanceAJ, GicquelB, et al. (2002) Disruption of the Gene Homologous to Mammalian Nramp1 in Mycobacterium tuberculosis Does Not Affect Virulence in Mice. 70 : 4124–4131 doi:10.1128/IAI.70.8.4124
15. DomenechP, PymAS, CellierM, BarryCE, ColeST (2002) Inactivation of the Mycobacterium tuberculosis Nramp orthologue (mntH) does not affect virulence in a mouse model of tuberculosis. FEMS Microbiol Lett 207 : 81–86.
16. GovoniG, GrosP (1998) Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm Res 47 : 277–284 doi:10.1007/s000110050330
17. NelsonN (1999) Metal ion transporters and homeostasis. EMBO J 18 : 4361–4371.
18. ForbesJR, GrosP (2003) Iron, manganese, and cobalt transport by Nramp1 (Slc11a1) and Nramp2 (Slc11a2) expressed at the plasma membrane. Blood 102 : 1884–1892 doi:10.1182/blood-2003-02-0425
19. ForbesJR, GrosP (2001) Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol 9 : 397–403 doi:10.1016/S0966-842X(01)02098-4
20. KehresDG, ZaharikML, FinlayBB, MaguireME (2000) The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol 36 : 1085–1100 doi:[]mmi1922 [pii]
21. GunshinH, MackenzieB, Berger UV, GunshinY, RomeroMF, et al. (1997) Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388 : 482–488 doi:10.1038/41343
22. MakuiH, RoigE, ColeST, HelmannJD, GrosP, et al. (2000) Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol 35 : 1065–1078.
23. RomaniAM, ScarpaA (2000) Regulation of cellular magnesium. Front Biosci 5: D720–D734 doi:10.2741/Romani
24. MoomawAS, MaguireME (2008) The unique nature of mg2+ channels. Physiol Bethesda Md 23 : 275–285.
25. ReinhartRA (1988) Magnesium metabolism. A review with special reference to the relationship between intracellular content and serum levels. Arch Intern Med 148 : 2415–2420 doi:10.1001/archinte.1988.00380110065013
26. ChenK, YuldashevaS, Penner-HahnJE, O'HalloranTV (2003) An atypical linear Cu(I)-S2 center constitutes the high-affinity metal-sensing site in the CueR metalloregulatory protein. J Am Chem Soc 125 : 12088–12089 doi:10.1021/ja036070y
27. ChiversPT, SauerRT (2000) Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem 275 : 19735–19741 doi:10.1074/jbc.M002232200
28. ShinJ-H, JungHJ, AnYJ, ChoY-B, ChaS-S, et al. (2011) Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci U S A 108 : 5045–5050.
29. GrubbsRD, MaguireME (1987) Magnesium as a regulatory cation: criteria and evaluation. Magnesium 6 : 113–127.
30. WilliamsRJ (1970) Cation distributions and the energy status of cells. J Bioenerg 1 : 215–225.
31. FlatmanPW (1991) Mechanisms of magnesium transport. Annu Rev Physiol 53 : 259–271 doi:10.1146/annurev.ph.53.030191.001355
32. FroschauerEM, KolisekM, DieterichF, SchweigelM, SchweyenRJ (2004) Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett 237 : 49–55.
33. MaguireME (2006) The structure of CorA: a Mg(2+)-selective channel. Curr Opin Struct Biol 16 : 432–438 doi:10.1016/j.sbi.2006.06.006
34. GardnerRC (2003) Genes for magnesium transport. Curr Opin Plant Biol 6 : 263–267 doi:10.1016/S1369-5266(03)00032-3
35. HmielSP, SnavelyMD, FlorerJB, MaguireME, MillerCG (1989) Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J Bacteriol 171 : 4742–4751.
36. KehresDG, MaguireME (2002) Structure, properties and regulation of magnesium transport proteins. Biometals 15 : 261–270.
37. SmithRL, MaguireME (1995) Distribution of the CorA Mg2+ transport system in gram-negative bacteria. J Bacteriol 177 : 1638–1640.
38. SmithRL, MaguireME (1998) Microbial magnesium transport: unusual transporters searching for identity. Mol Microbiol 28 : 217–226.
39. PerezJC, ShinD, ZwirI, LatifiT, HadleyTJ, et al. (2009) Evolution of a bacterial regulon controlling virulence and Mg(2+) homeostasis. PLoS Genet 5: e1000428.
40. CromieMJ, ShiY, LatifiT, GroismanEA (2006) An RNA sensor for intracellular Mg(2+). Cell 125 : 71–84 doi:10.1016/j.cell.2006.01.043
41. DannCE, WakemanCA, SielingCL, BakerSC, IrnovI, et al. (2007) Structure and mechanism of a metal-sensing regulatory RNA. Cell 130 : 878–892 doi:10.1016/j.cell.2007.06.051
42. RameshA, WinklerWC (2010) Magnesium-sensing riboswitches in bacteria. RNA Biol 7 : 77–83 doi:10.4161/rna.7.1.10490
43. WakemanCA, WinklerWC, DannCE (2007) Structural features of metabolite-sensing riboswitches. Trends Biochem Sci 32 : 415–424.
44. NudlerE, MironovA, NudlerE, MironovAS (2004) The riboswitch control of bacterial metabolism. Trends Biochem Sci 29 : 11–17 doi:10.1016/j.tibs.2003.11.004
45. SchwalbeH, BuckJ, FürtigB, NoeskeJ, WöhnertJ (2007) Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl 46 : 1212–1219 doi:10.1002/anie.200604163
46. MontangeRK, BateyRT (2008) Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys 37 : 117–133 doi:10.1146/annurev.biophys.37.032807.130000
47. DambachMD, WinklerWC (2009) Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol 12 : 161–169.
48. RothA, BreakerRR (2009) The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78 : 305–334 doi:10.1146/annurev.biochem.78.070507.135656
49. RameshA, WakemanCA, WinklerWC (2011) Insights into metalloregulation by M-box riboswitch RNAs via structural analysis of manganese-bound complexes. J Mol Biol 407 : 556–570.
50. WakemanCA, RameshA, WinklerWC (2009) Multiple metal-binding cores are required for metalloregulation by M-box riboswitch RNAs. J Mol Biol 392 : 723–735.
51. BarrickJE (2009) Predicting riboswitch regulation on a genomic scale. Methods Mol Biol 540 : 1–13 doi:_10.1007/978-1-59745-558-9_1
52. JakubovicsNS, JenkinsonHF (2001) Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147 : 1709–1718.
53. QueQ, HelmannJD (2000) Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35 : 1454–1468.
54. KehresDG, JanakiramanA, SlauchJM, MaguireME (2002) Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+). J Bacteriol 184 : 3151–3158.
55. RicherE, CourvilleP, BergevinI, CellierMFM (2003) Horizontal gene transfer of “prototype” Nramp in bacteria. J Mol Evol 57 : 363–376 doi:10.1007/s00239-003-2472-z
56. HohleTH, O'BrianMR (2009) The mntH gene encodes the major Mn(2+) transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol Microbiol 72 : 399–409.
57. WakemanCA, GoodsonJR, ZachariaVM, WinklerWC (2014) An Assessment of the Requirements for Magnesium Transporters in Bacillus subtilis. J Bacteriol 196(6): 1206–14 doi:10.1128/JB.01238-13
58. CellierMF, BergevinI, BoyerE, RicherE (2001) Polyphyletic origins of bacterial Nramp transporters. Trends Genet 17 : 365–370 doi:10.1016/S0168-9525(01)02364-2
59. ChaloupkaR, CourvilleP, VeyrierF, KnudsenB, Tompkins Ta, et al. (2005) Identification of functional amino acids in the Nramp family by a combination of evolutionary analysis and biophysical studies of metal and proton cotransport in vivo. Biochemistry 44 : 726–733 doi:10.1021/bi048014v
60. Cellier M, Gros P (2004) The Nramp Family. Georgetown: Eurekah.com.
61. CourvilleP, UrbankovaE, RensingC, ChaloupkaR, QuickM, et al. (2008) Solute carrier 11 cation symport requires distinct residues in transmembrane helices 1 and 6. J Biol Chem 283 : 9651–9658 doi:10.1074/jbc.M709906200
62. CellierMFM (2012) Nutritional immunity: homology modeling of Nramp metal import. Adv Exp Med Biol 946 : 335–351 doi:_10.1007/978-1-4614-0106-3_19
63. TownsendDE, EsenwineAJ, GeorgeJ, BrossD, MaguireME, et al. (1995) Cloning of the mgtE Mg2+ transporter from Providencia stuartii and the distribution of mgtE in gram-negative and gram-positive bacteria. J Bacteriol 177 : 5350–5354.
64. SmithRL, BanksJL, SnavelyMD, MaguireME (1993) Sequence and topology of the CorA magnesium transport systems of Salmonella typhimurium and Escherichia coli. Identification of a new class of transport protein. J Biol Chem 268 : 14071–14080.
65. TaoT, SnavelyMD, FarrSG, MaguireME (1995) Magnesium transport in Salmonella typhimurium: mgtA encodes a P-type ATPase and is regulated by Mg2+ in a manner similar to that of the mgtB P-type ATPase. J Bacteriol 177 : 2654–2662.
66. AgranoffD, MonahanIM, ManganJA, ButcherPD, KrishnaS (1999) Mycobacterium tuberculosis expresses a novel pH-dependent divalent cation transporter belonging to the Nramp family. J Exp Med 190 : 717–724.
67. CzachorowskiM, Lam-Yuk-TseungS, CellierM, GrosP (2009) Transmembrane topology of the mammalian Slc11a2 iron transporter. Biochemistry 48 : 8422–8434 doi:10.1021/bi900606y
68. AnagnostopoulosC, SpizizenJ (1961) Requirements for transformation in Bacillus subtilis. J Bacteriol 81 : 741–746.
69. Van OoijC, LosickR (2003) Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol 185 : 1391–1398.
70. ArnaudM, ChastanetA, DébarbouilléM (2004) New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70 : 6887–6891 doi:10.1128/AEM.70.11.6887-6891.2004
71. ZhangH, DavisonW, MillerS, TychW (1995) In situ high resolution measurements of fluxes of Ni, Cu, Fe, and Mn and concentrations of Zn and Cd in porewaters by DGT. Geochim Cosmochim Acta 59 : 4181–4192 doi:10.1016/0016-7037(95)00293-9
72. TobinMB, PeeryRB, SkatrudPL (1997) Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200 : 11–23 doi:10.1016/S0378-1119(97)00281-3
73. DeikusG, BabitzkeP, BechhoferDH (2004) Recycling of a regulatory protein by degradation of the RNA to which it binds. Proc Natl Acad Sci U S A 101 : 2747–2751.
74. RoosJW, McLaughlinJK, PapoutsakisET (1985) The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of Clostridium acetobutylicum. Biotechnol Bioeng 27 : 681–694 doi:10.1002/bit.260270518
75. KatohK, StandleyDM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30 : 772–780 doi:10.1093/molbev/mst010
76. RonquistF, HuelsenbeckJP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 : 1572–1574 doi:10.1093/bioinformatics/btg180
77. AltekarG, DwarkadasS, HuelsenbeckJP, RonquistF (2004) Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20 : 407–415 doi:10.1093/bioinformatics/btg427
78. HelaersR, MilinkovitchMC (2010) MetaPIGA v2.0: maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics 11 : 379 doi:10.1186/1471-2105-11-379
79. TamuraK, StecherG, PetersonD, FilipskiA, KumarS (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30 : 2725–2729 doi:10.1093/molbev/mst197
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 6- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



