-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
Pathogenic microbes often evolve complex traits to adapt to their respective hosts, and this evolution is ongoing: for example, microorganisms are developing resistance to antimicrobial compounds in the clinical setting. The ability of the common human pathogenic fungus, Candida albicans, to switch from yeast to hyphal (filamentous) growth is considered a central virulence attribute. For example, hyphal formation allows C. albicans to escape from macrophages following phagocytosis. A well-investigated signaling network integrates different environmental cues to induce and maintain hyphal growth. In fact, deletion of two central transcription factors in this network results in a mutant that is both nonfilamentous and avirulent. We used experimental evolution to study the adaptation capability of this mutant by continuous co-incubation within macrophages. We found that this selection regime led to a relatively rapid re-connection of signaling between environmental cues and the hyphal growth program. Indeed, the evolved mutant regained the ability to filament and its virulence in vivo. This bypass of central transcription factors was based on a single nucleotide exchange in a gene encoding a component of the general transcription regulation machinery. Our results show that even a complex regulatory network, such as the transcriptional network which governs hyphal growth, can be remodeled via microevolution.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004824
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004824Summary
Pathogenic microbes often evolve complex traits to adapt to their respective hosts, and this evolution is ongoing: for example, microorganisms are developing resistance to antimicrobial compounds in the clinical setting. The ability of the common human pathogenic fungus, Candida albicans, to switch from yeast to hyphal (filamentous) growth is considered a central virulence attribute. For example, hyphal formation allows C. albicans to escape from macrophages following phagocytosis. A well-investigated signaling network integrates different environmental cues to induce and maintain hyphal growth. In fact, deletion of two central transcription factors in this network results in a mutant that is both nonfilamentous and avirulent. We used experimental evolution to study the adaptation capability of this mutant by continuous co-incubation within macrophages. We found that this selection regime led to a relatively rapid re-connection of signaling between environmental cues and the hyphal growth program. Indeed, the evolved mutant regained the ability to filament and its virulence in vivo. This bypass of central transcription factors was based on a single nucleotide exchange in a gene encoding a component of the general transcription regulation machinery. Our results show that even a complex regulatory network, such as the transcriptional network which governs hyphal growth, can be remodeled via microevolution.
Introduction
The incidence of invasive fungal infections has steadily increased within the past decades, largely because of a growing population of susceptible individuals, reflecting the progress of modern medicine in prolonging life even with severe underlying diseases and the increasing rate of immuno-deficient patients. One of the most frequently isolated fungi is Candida albicans, an ubiquitous and normally harmless commensal of the alimentary tract and mucocutaneous membranes. As an opportunistic pathogen, it can cause superficial infections like oropharyngeal candidiasis, especially in HIV patients, as well as life-threatening systemic infections with mortality rates up to 40%, even with current antifungal treatment options [1].
The transition from the commensal to a pathogenic state depends on the microbiota, the host response, and C. albicans activities, such as adhesion, secretion of hydrolases, metabolic adaptation, biofilm formation and, importantly, morphological plasticity, which includes the yeast-to-filament transition [2]–[7]. To survive and thrive in the many different niches inside the host, C. albicans must be able to adapt to changing environments and different stresses. In the short term, this occurs primarily by changes in gene expression and translation, and via post-translational modifications, but ultimately microevolutionary processes will play an important role. As a prominent example, White et al. [8] have shown that microevolution is the driving force behind the emergence of antifungal drug resistance. They demonstrated the de novo appearance of fluconazole resistance in evolving C. albicans strains in vivo [8]. Furthermore, clinical isolates generally exhibit large genetic variations, and microevolution can be observed both in vitro and in vivo [9], [10], indicating that this process plays an important role in host-pathogen interactions. Therefore, microevolution provides a source of variation for the adaptive response of C. albicans to challenging (host) environments.
Different mechanisms account for the generation of new genotypic variants, including point mutations, amplification or deletion of chromosomal segments, chromosomal translocation or inversion, and whole chromosome aneuploidy. These genetic variations can affect expression of single genes or the structure of their encoded proteins as well as whole transcriptional networks via a mechanism known as transcriptional rewiring. In this process, the interaction between promoter regions and their corresponding regulators can be switched to different pairings, which in turn cause new connections to be formed between a signal and a transcriptional response [11], [12]. Whereas many studies have explored the underlying mechanisms of drug resistance, the role that microevolution plays in host-pathogen interactions has rarely been investigated: Forche et al. [13] found that a C. albicans strain, passaged through a mouse host, responded by undergoing chromosome-level genetic variations, which were sufficient to generate new variants of C. albicans.
The yeast-to-hyphae transition of C. albicans is central for pathogenicity [14], [15]. Filamentation plays a pivotal role for adhesion to, invasion into and damage of epithelial and endothelial cells [2], [16], [17]. Upon internalization by macrophages, C. albicans induces host cell death by triggering pyroptosis, a form of programmed cell death [18], [19]. However, later in the infection process the yeast-to-hyphae transition contributes to escape from the phagosome [19], [20]. Morphology also plays a key role in host recognition [21]. Given the importance of morphology of C. albicans for pathogenicity, it is not surprising that the yeast-to-filament transition is induced by a wide range of environmental factors and conditions like high pH, host body temperature, CO2, starvation and presence of serum, all of which act via several signaling pathways. Among them, the cAMP-dependent protein kinase A (cAMP-PKA) and the mitogen-activated protein kinase (MAPK) pathways, which target the transcription factors Efg1 and Cph1, respectively, play a central role in hyphal formation [22], [23]. This is demonstrated by a cph1Δ/efg1Δ double mutant, which is unable to form hyphae under almost all hyphae-inducing conditions in vitro (except agar embedded conditions) and which is probably the most commonly used mutant of C. albicans in a wide range of experiments [14], [15], [22], [24].
Due to the central role of the yeast-to-filament transition for C. albicans virulence, we used the cph1Δ/efg1Δ double mutant as a model for evolutionary adaptation. To this end, we performed a series of co-culture passages of this mutant with macrophages. We expected that the hostile environment of the phagosome imposes a high selective pressure on the fungus favoring either intracellular adaptation or return to filamentation in order to escape. We performed phenotypic, transcriptomic and genomic analyses of the pre - and post-passaged strains to elucidate the degree of genetic plasticity of C. albicans when facing host stresses. We show that adaptation to macrophages leads to distinct phenotypic differences between the pre - and post-passaged strains with regained filamentation in the latter. As the causative mutation, we identified a heterozygous, non-synonymous single nucleotide exchange in the gene SSN3, which encodes the cyclin-dependent kinase of a regulatory module of the Mediator complex. Our results demonstrate that the regulation of the morphological switch in C. albicans can be subject to microevolution.
Results
Experimental microevolution causes a reversion of the nonfilamentous phenotype of the cph1Δ/efg1Δ mutant strain
To determine the ability of C. albicans to adapt to stresses inside phagocytes and to test the adaptability of the hyphal regulatory network, we first screened for mutants which are unable to escape from macrophages via filamentation response. We tested multiple C. albicans deletion strains with known defects in hyphal formation: strains lacking RAS1, RIM101, DFG16, TEC1, HGC1, EED1, or UME6 and the avirulent double deletion mutant lacking CPH1 and EFG1 [22]. Of these, only the cph1Δ/efg1Δ double mutant was completely unable to escape from macrophages even after 24 hours, while all other mutants still formed filaments inside the host cell and pierced the phagocyte membrane to some extent (S1A Figure). Microscopy with FITC-labeled cph1Δ/efg1Δ cells revealed that these cells were viable and still able to replicate in the yeast form after ingestion by macrophages (S1B Figure).
Therefore, we chose the cph1Δ/efg1Δ strain for the following microevolution experiment. Cells of the murine macrophage cell line J774A.1 were infected with the cph1Δ/efg1Δ double mutant at a macrophage-to-fungal ratio of 2∶1 and co-incubated. Every 24 hours, non-phagocytosed cells were removed and macrophages were lysed to harvest the phagocytosed cells. A defined fraction of this population was then transferred to a fresh macrophage population.
After 19 passages, a significant morphological alteration became visible, as several phagocytosed cells started to form filaments. These filamenting cells became fixed in the population after additional 23 rounds of co-incubation. This morphologically distinct variant, evolutionary derived from the cph1Δ/efg1Δ mutant, was termed Evo. The absence of CPH1 and EFG1 in the Evo strain was verified by Southern blot analysis (S1C Figure). To exclude temporary or epigenetic effects, the Evo strain was repassaged daily in liquid rich (YPD) medium without any selection pressure by host cells for 14 passages. The phenotype remained stable and no reversal was detected.
To test whether the regained ability to form filaments was restricted to macrophage interactions or observed under additional hypha-inducing conditions, we analyzed the morphology of the Evo strain in the absence of host cells. In the cell culture medium DMEM with 10% serum at 37°C and 5% CO2, clear filament formation of the Evo strain, but not the cph1Δ/efg1Δ strain, was observed (Fig. 1). Filamentous growth is associated with highly polarized ergosterol inclusion in membranes, which can be visualized by filipin staining [25]. As shown in Fig. 1, Evo cells grown in the presence of serum exhibited intense filipin staining at the filament tips, equal to the wild type cells. Consistent with the defect in polarized growth, the cph1Δ/efg1Δ strain showed a more uniform filipin staining. Staining with calcofluor white for morphology analyses showed the expected true hyphae for the wild type and elongated yeasts for the cph1Δ/efg1Δ strain (Fig. 1). Interestingly, the Evo strain showed heterogeneous cell morphologies, i.e. a mixture of pseudohyphae with constrictions at the septa and true hyphae with parallel-sided walls (Fig. 1). The percentage of the different morphological forms was quantified using the morphological index (MI) [26] of individual cells after 4 and 12 hours of growth in serum (S2A Figure). The MI for cph1Δ/efg1Δ was <2.5 at both time points, indicating yeast morphology. In contrast, most cells of the Evo strain grew as pseudohyphae (MI 2.5–3.4) after 4 hours, while after 12 hours true hyphae were evident (MI>3.4) in approx. 50% of the population. Both morphologies will be referred to as filaments.
Fig. 1. Co-incubation with macrophages led to regained filamentation in the cph1Δ/efg1Δ strain. 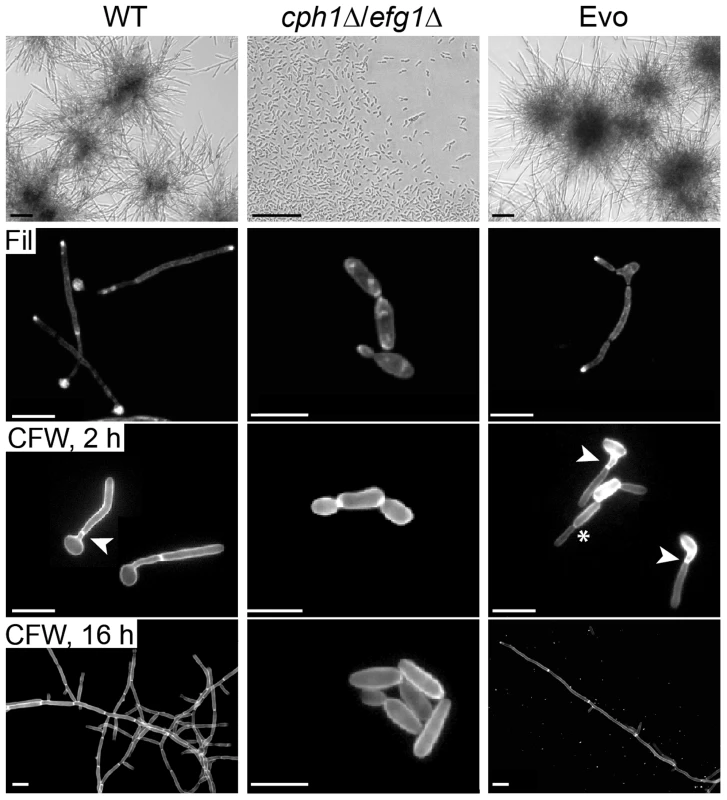
Morphology of wild type (WT), cph1Δ/efg1Δ and Evo strain after 18 h of incubation in DMEM+10% FBS at 37°C and 5% CO2 demonstrate the re-appearance of filamentation in the Evo strain (scale bar: 100 µm, upper panel). All strains were grown for 4 h for filipin (Fil) staining, and for 2 h or 16 h for calcofluor white (CFW) staining on cover slips, and analyzed by fluorescence microscopy (scale bar: 10 µm, lower panels). Arrow heads highlight septa (true hyphae), while asterisks indicate constrictions (pseudohyphae). We then tested different classical hyphae-induction media for C. albicans to assess the extent of phenotype reversal to wild type morphology. In response to serum-containing YPD medium with 5% CO2, the Evo strain initially formed filaments but switched back to yeast growth much earlier than the wild type (S2B Figure). Filamentation (mainly pseudohyphae) also occurred in response to the amino sugar N-acetyl-D-glucosamine as sole carbon source and 5% CO2 (S2B Figure). Finally, cells of the Evo strain were incubated in serum-containing water at 37°C in atmospheric air. Again, stable filamentation was induced, demonstrating that high CO2 is not absolutely necessary for filamentation of the Evo strain (S2B Figure). In embedded media at 23°C (S2C Figure.), deletion of EFG1 causes a hyperfilamentous phenotype [24]. Accordingly, the cph1Δ/efg1Δ strain was hyperfilamentous under these conditions. Interestingly, while cells of the Evo strain displayed an even more pronounced hyperfilamentous phenotype, it did not undergo filamentation on solid medium at 37°C, as seen in the cph1Δ/efg1Δ strain (S2D Figure).
In conclusion, our microevolution experiment led to the regained ability of filamentous growth in the cph1Δ/efg1Δ mutant in response to a diverse range of hyphae-inducing conditions, indicating that microevolutionary events had enabled this strain to bypass the dependency on Cph1 and Efg1 for filamentation in these media.
The Evo strain regained virulence potential
Filamentous growth is an important contributing factor for the escape from macrophages. We therefore determined the amount of Evo cells that escaped from macrophages by piercing through their membranes after 4 h, 6 h and 8 h of co-incubation (Fig. 2A). Both Evo and wild type, but not the cph1Δ/efg1Δ double mutant, were able to escape from macrophages. However, the piercing rate of the Evo strain was significantly lower than for the wild type at all time points. After 8 h of co-incubation nearly all wild type cells had escaped from the macrophages, but only about 25% of Evo cells. The delay in filamentation and the presence of pseudohyphae in the Evo strain may explain these differences. Next, we assessed the fungus' ability to invade oral epithelial cells. Invasion requires previous adhesion, and the cph1Δ/efg1Δ strain was almost entirely unable to adhere to epithelial cells (Fig. 2B). Adhesion of the Evo strain was still reduced compared to the wild type, but significantly higher than for the double mutant (Fig. 2B). This is reflected by the invasion capacity of the Evo strain, which was significantly lower than the wild type strain, but substantially higher than the cph1Δ/efg1Δ strain. Finally, we also investigated the potential of the Evo strain to damage macrophages and epithelial cells by measuring the release of lactate dehydrogenase (LDH). After 32 hours of co-incubation, the Evo strain had damaged macrophages to the same extent as the wild type strain, and epithelial cells to a significantly higher degree than the cph1Δ/efg1Δ strain (Fig. 2C).
Fig. 2. Characterization of Evo strain interaction with host cells and virulence potential. 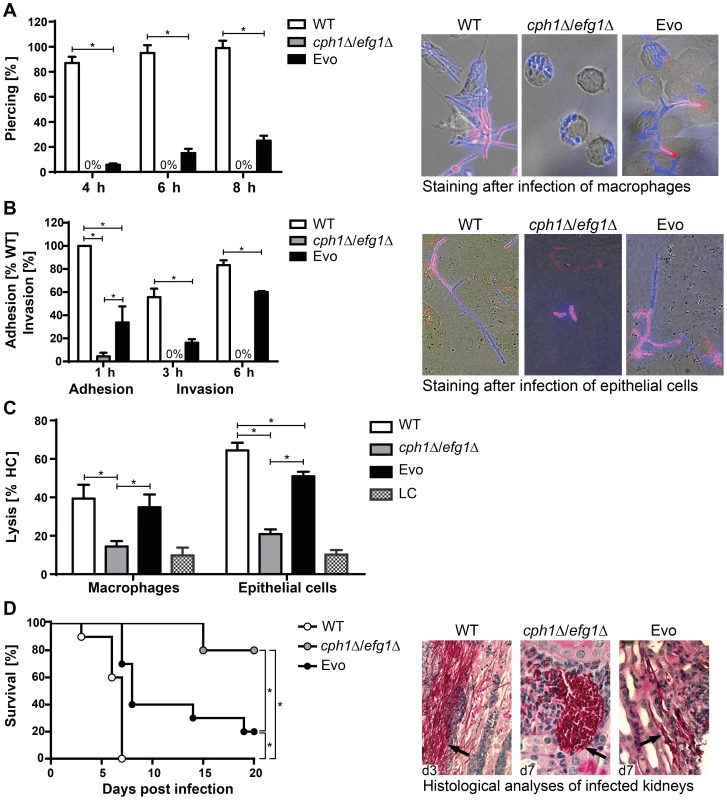
(A) Escape of C. albicans cells by piercing of macrophages (J774A.1) after different timepoints (left). Micrographs of strains after 6 h of co-incubation with J774A.1 cells (right). Intracellular C. albicans appears blue (CFW), extracellular section of the cells red (Concanavalin A, ConA). Cells of the cph1Δ/efg1Δ strain cannot escape from macrophages, while Evo cells regained this property during the evolution experiment. (B) Adhesion to and invasion of oral epithelial cells (TR-146). Adhesion values are given as percentage of adherent WT cells (left). Micrographs show filamentation of C. albicans WT and Evo strains after 6 h of incubation with TR-146 cells (right). The regained ability to filament enabled the Evo strain to invade epithelial cells. Staining was performed as described in (A). (C) Damage to macrophages and epithelial monolayers, determined by lactate dehydrogenase (LDH) assay after 32 h of co-incubation (LC = low control, medium only). WT and Evo strain, but not the cph1Δ/efg1Δ strain caused clear damage to both cell types. For piercing, adhesion, invasion and cell damage assay results are given as mean+SD of three independent experiments (*p<0.05). (D) Survival of BALB/c mice challenged intravenously (left; n = 10/strain). Nearly all mice infected with the Evo strain succumbed to the infection, while almost all animals infected with cph1Δ/efg1Δ strain survived (*p<0.05). PAS-hematoxylin-stained kidney sections from different days (d) post challenge (right) show fungal cells (arrows) either in the filamentous form (WT and Evo strain) or yeast form (cph1Δ/efg1Δ strain). The Evo strain had thus regained abilities putatively relevant for systemic infections. Hence, the virulence of the Evo strain was tested in a murine model of hematogenously disseminated candidiasis. Survival was monitored over a period of 21 days. As predicted, mice infected with the Evo strain showed an intermediate and significantly different survival rate compared to mice infected with the wild type and cph1Δ/efg1Δ strains (Fig. 2D). Histological examination of kidneys from infected animals revealed that the Evo strain retained its filamentous morphology in vivo, even though filaments formed by the Evo strain were shorter than by the wild type, and invasion into deeper layers of the kidney tissue was less pronounced (Fig. 2D).
In summary, the evolved changes in response to macrophages enabled the Evo strain not only to form filaments in vitro, but also in contact with host cells, which correlated with a higher virulence potential both in vitro and in vivo.
The Evo strain expresses hyphal-associated genes and responds to farnesol
Hyphal-associated virulence of C. albicans is not only due to filamentation per se, but also to the expression of hyphae-associated genes. In order to monitor the expression of typical hyphae-associated genes in the Evo strain, we measured the mRNA levels of HWP1, ECE1 and ALS3, all encoding hyphal cell surface proteins, and of EED1, a gene that is associated with hyphal cell elongation [27]. An upregulation of all four genes in the Evo strain was confirmed by qRT-PCR after 1 hour of incubation in DMEM+10% FBS at 37°C and 5% CO2 (Fig. 3A). HWP1 expression was similar in the Evo and WT strain, whereas ECE1 and ALS3 were higher expressed in the WT strain, and EED1 was more strongly upregulated in the Evo strain. Furthermore, we observed Als3 exposure on the surface of wild type and Evo cells by immunofluorescence, but not on the cph1Δ/efg1Δ strain (Fig. 3B). This regained cell-surface exposure of the Als3 adhesin [28] is in accordance with the increased adhesion potential of the Evo strain.
Fig. 3. Analysis of hyphae-associated gene expression, Als3 surface expression and response to farnesol. 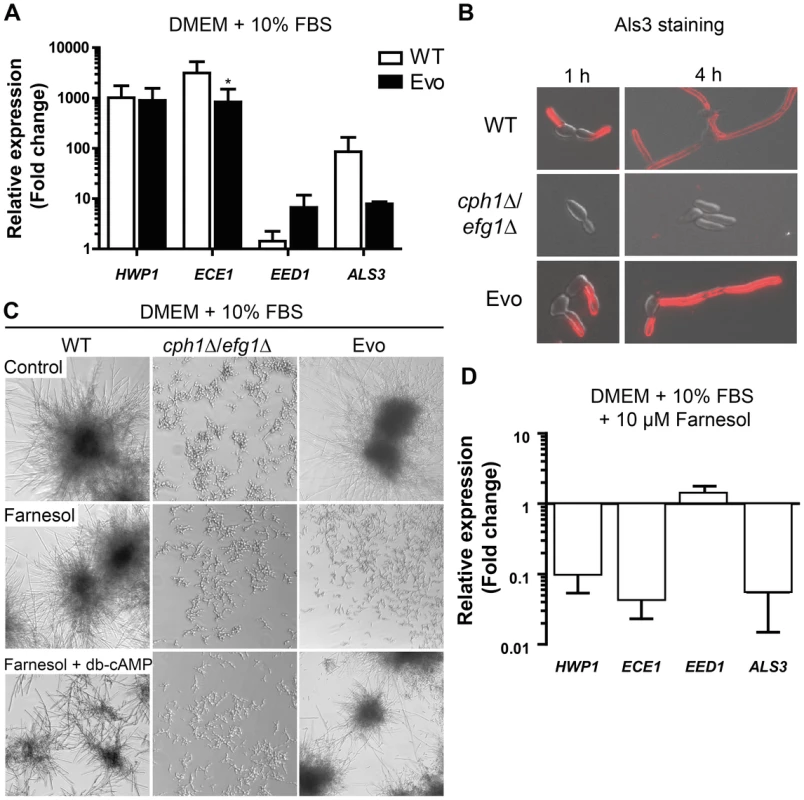
(A) The Evo strain expresses hyphae-associated genes after growth for 1 h at 37°C at 5% CO2 on a plastic surface similar to WT. Relative gene expression of filament-inducing conditions was compared to yeast promoting conditions (YPD, 30°C) for three independent experiments. Expression was normalized against three housekeeping genes (ACT1, EFB1 and PMA1) and data are shown as mean+SD of three biological experiments (*p<0.05). (B) Immunofluorescence micrographs of cells immuno-stained for Als3 after growth in DMEM+10% FBS at 37°C and 5% CO2 on cover slips. Wild type (WT) and Evo cells are Als3-positive, while cph1Δ/efg1Δ cells show no signal (representative samples). (C) Morphogenetic response to farnesol treatment alone or in combination with exogenous dibutyryl-cyclic AMP (db-cAMP). All strains were exposed to either methanol (control), 1 µM farnesol or 1 µM farnesol+10 mM db-cAMP and incubated at 37°C and 5% CO2 for 18 h (representative pictures from three independent experiments are shown). Note that in Evo cells inhibition of filamentation by farnesol treatment was completely abrogated when db-cAMP was added. (D) Repression of hyphae-associated gene expression in the Evo strain by 10 µM farnesol. Expression was normalized against three housekeeping genes (ACT1, EFB1 and PMA1). The fold change in expression relative to filament-inducing conditions alone is shown as mean+SD of three biological experiments. We were next interested if the filamentation program can be blocked by the quorum-sensing molecule farnesol. Very low concentrations (1 µM) of farnesol in the medium resulted in a complete repression of filament formation in the Evo strain, whereas wild type cells still formed hyphae (Fig. 3C). Consistently, farnesol treatment led to a dramatic repression of filament-associated gene expression (Fig. 3D). By addition of exogenous dibutyryl-cyclic AMP (db-cAMP) to the farnesol-containing medium, filamentation was rescued in the Evo strain (Fig. 3C). These data suggest a critical role for cAMP signaling in the filamentation process of the Evo strain.
The Evo strain shows wild type levels of filamentation-associated transcription factor gene expression
The yeast-to-filament regulatory network comprises many different transcription factors (TFs). The filament-associated biofilm formation is controlled by a network formed by Bcr1, Tec1, Brg1, Rob1, Ndt80 and Efg1 [29]. Efg1 positively regulates all other TF genes in this network except ROB1. We measured the transcription of these central TF genes at 30 min and 60 min after filament induction. As shown in Fig. 4A, we found an at least 1.5-fold upregulation of ROB1 and TEC1 after 30 min, and of BCR1 and BRG1 at both timepoints in the Evo strain. The wild type, however, showed only an increased expression of TEC1 at both timepoints and of BRG1 after 30 min. In contrast, most of these TF genes were down - or scarcely upregulated in the cph1Δ/efg1Δ strain (Fig. 4A).
Fig. 4. Expression of transcription factors under filament-inducing conditions. 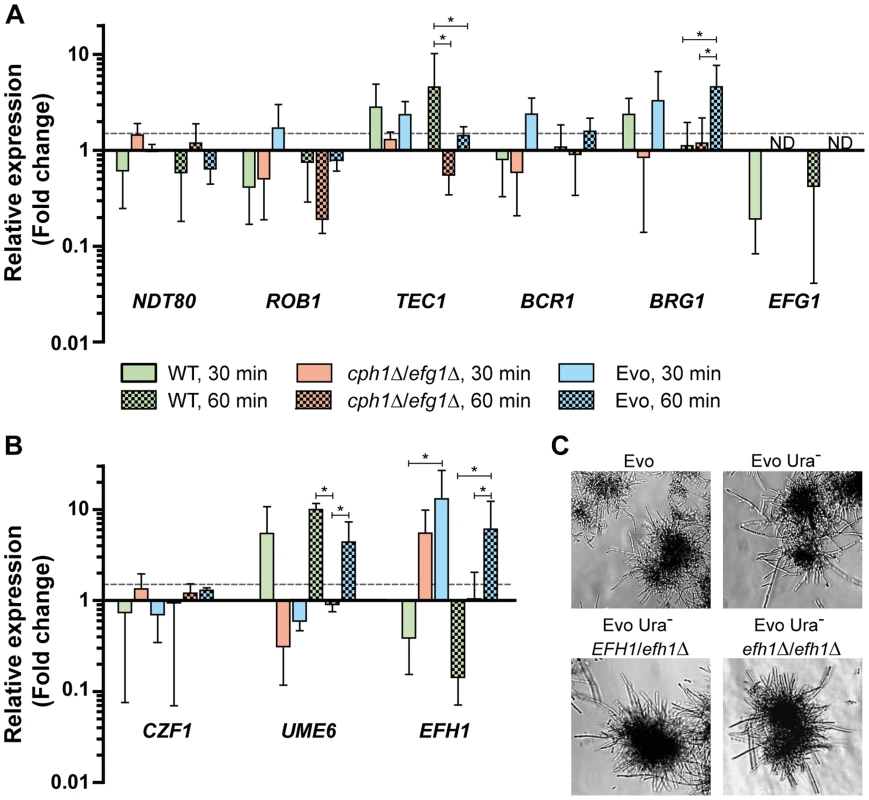
(A+B) Relative expression of nine central transcription factor genes in the analyzed strains after growth in DMEM+10% FBS at 37°C and 5% CO2 on a plastic surface. Fold change between filament-inducing and yeast promoting conditions (YPD, 30°C) is shown, normalized to three housekeeping genes (ACT1, EFB1 and PMA1). Means+SD of n = 3 (dotted line indicates threshold at 1.5; *p<0.05). (C) Deletion of EFH1 in the Evo strain did not affect hyphal growth. Cells were incubated for 18 h at 37°C and 5% CO2 in DMEM+10% FBS (representative pictures). Formation of wild type filaments is also regulated in part by CZF1 under certain conditions [30]. An increased expression of CZF1, however, is not the cause for filamentation in the Evo strain. The CZF1 mRNA levels under serum induction did not greatly differ from the mRNA level in the cph1Δ/efg1Δ strain (Fig. 4B). In addition, the mRNA level of UME6, a key TF gene necessary for the maintenance of filamentation [31], was upregulated in wild type cells at both time points but not in the cph1Δ/efg1Δ strain (Fig. 4B). Interestingly, UME6 expression was more than 4-fold upregulated in the Evo strain after 60 min growth in serum-containing medium.
C. albicans possesses an EFG1 homolog, EFH1, and overexpression of this gene is known to induce pseudohyphal growth. In addition, like EFG1, EFH1 is involved in the regulation of expression of filament-associated genes [24]. We found that EFH1 showed the strongest upregulation (7.4-fold) among the tested TF genes in the Evo strain. However, deletion of EFH1 did not abolish filamentation of an Evo strain derivative (Fig. 4C). Hence, the filamentation phenotype of the Evo strain was not linked to this TF.
In summary, the Evo strain has regained most of the transcriptional hallmarks of filament production, including the upregulation of the central transcription factor genes TEC1, BRG1 and UME6. The few discrepancies to the wild type may partially explain the remaining differences in morphology. However, the late-phase upregulation of UME6 indicates that the filament maintenance of the Evo strain is similar to the wild type at the transcriptional level. Furthermore, the function of Efg1 was not replaced by Efh1 in the Evo strain.
The cell wall defects of the cph1Δ/efg1Δ mutant are reverted in the Evo strain
Our data indicated that the Evo strain regained the potential to produce hyphae, showed upregulation of transcription factor genes involved in filamentous growth and other hyphal associated genes, and regained a high virulence potential. The reduced virulence of the cph1Δ/efg1Δ strain is likely predominantly caused by the filamentation defects, however, Efg1 has also an important role in cell wall architecture [32] and the cell wall is essential for adhesion and invasive growth and thus for pathogenicity [33]. We therefore tested the Evo strain for cell wall defects by treatment with cell wall perturbing agents, i.e. congo red (CR), calcofluor white (CFW) and sodium dodecyl sulfate (SDS). As shown in Fig. 5A, the cph1Δ/efg1Δ strain was hypersensitive to all tested agents. In contrast, the Evo strain was as resistant as the wild type to CR and CFW, agents that disturb glucan and chitin architecture, respectively. The same phenotypic reversal was observed for the cell membrane disturbing agent SDS, suggesting a loose structure of the cell wall only in the cph1Δ/efg1Δ strain.
Fig. 5. Microevolution led to decreased sensitivity of the Evo strain to different cell wall perturbing agents. 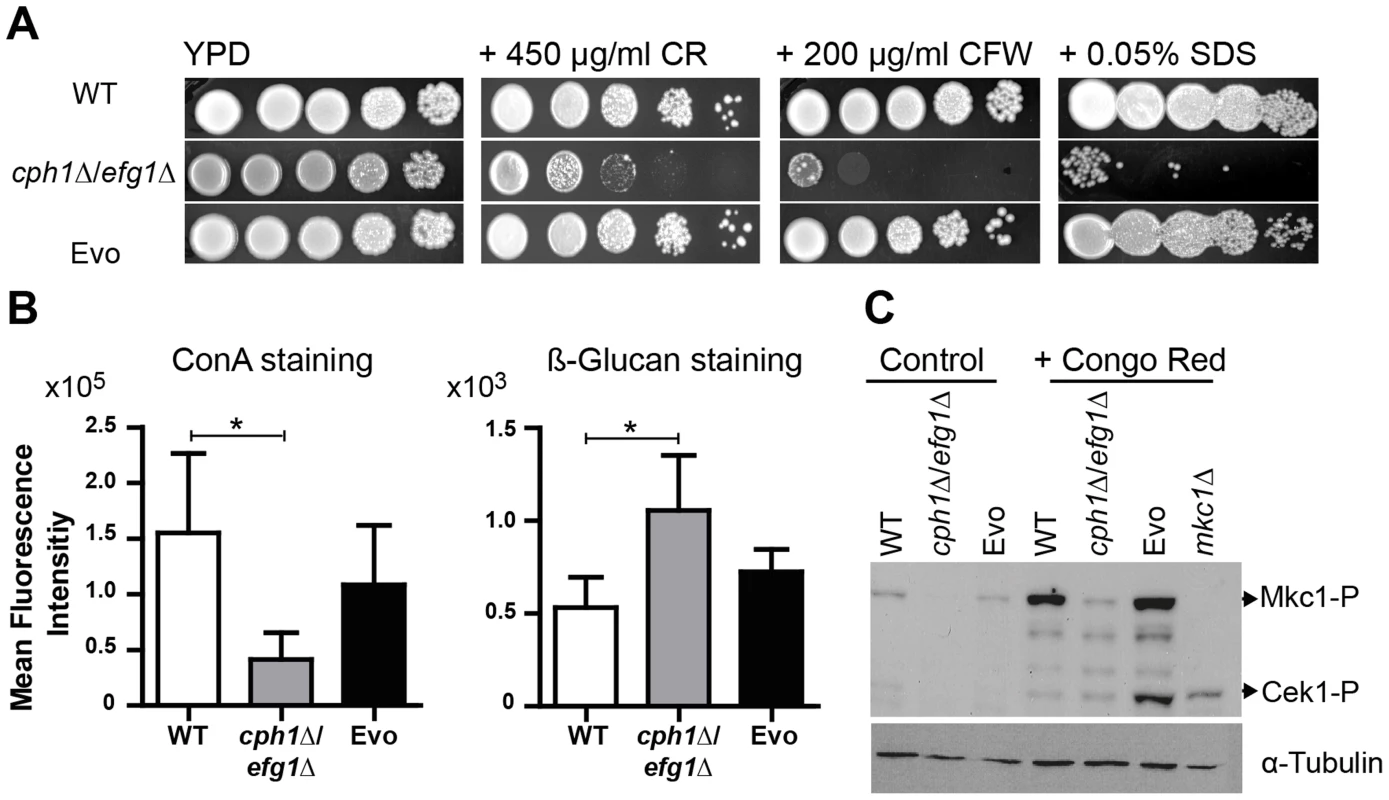
(A) Resistance of analyzed strains against different cell wall stressors. The cph1Δ/efg1Δ strain was sensitive to all stresses while the Evo strain regained WT resistance (representative pictures of three experiments are shown). (B) Flow cytometry analysis of mannan and β-glucan exposure on the surface of live cells. Differences in fluorescence intensity between cph1Δ/efg1Δ strain and Evo strain point to an altered cell wall composition. Mean fluorescence intensity+SD of n = 3 (*p<0.05). (C) Western blot analysis to identify phosphorylated Mkc1 and Cek1 in C. albicans strains grown under non-stress conditions (control) or conditions of cell wall stress (450 µg/ml congo red) for 4 hours. Cell wall stress triggered phosphorylation of Mkc1 and Cek1 in the Evo strain, but not in the cph1Δ/efg1Δ strain. Tubulin served as loading control. These results indicate that the altered cell wall composition of the cph1Δ/efg1Δ strain was at least partially restored in the Evo strain. We therefore stained exposed mannan and β-1,3-glucan with fluorescently labeled concanavalin A (ConA) and anti-β-1,3-glucan antibody, respectively (Fig. 5B). Quantification by FACS analysis displayed significantly reduced mannan and increased β-1,3-glucan signals on the surface of the cph1Δ/efg1Δ strain compared to the wild type strain. The Evo strain showed an intermediate mannan and wild type-like glucan exposure.
The two MAP kinases, Cek1 and Mkc1, become activated in wild type C. albicans upon treatment with cell wall disturbing agents [34]–[36]. After treatment with CR, both Cek1 and Mkc1 were phosphorylated in the Evo strain but not in the cph1Δ/efg1Δ strain (Fig. 5C). Unusually, only phosphorylated Mkc1 could be detected in the wild type strain, which may be due to changes in the CR treatment protocol compared to previous experiments performed by another group [34]. However, these results show that the Evo strain regained the ability to phosphorylate Mkc1 and Cek1 in response to cell wall stress.
Global transcriptional analysis of the evolved strain by RNA-Seq
To gain insight into the regulatory program of filamentation in the absence of CPH1 and EFG1, we performed gene expression analysis by RNA sequencing under hyphae - and non-hyphae-inducing conditions. Both, the cph1Δ/efg1Δ and the Evo strain, were analyzed with a sequencing depth sufficient to cover the genome 75–350×. Expression (RPKM≥1) was detected for 5,854 of the C. albicans open reading frames (94%), as well as for 561 nTARs (novel transcriptionally active regions, [37]), 67 small nuclear RNAs and for 24 tRNAs (see Materials and Methods for details and S2 Table for a complete list of all detected transcripts). Differential expression of selected genes was subsequently validated by qRT-PCR using biological replicates (S3A Figure).
After the transfer to filament-inducing conditions, 379 transcripts were significantly upregulated (≥2-fold, p<0.01) and 279 downregulated in the Evo strain. In the cph1Δ/efg1Δ strain, 255 transcripts were up - and 252 downregulated under the same condition. Within the group of upregulated transcripts, 209 genes were induced in both strains, while 46 transcripts were specifically induced in the cph1Δ/efg1Δ strain and 170 transcripts specifically in the Evo strain. 186 of the downregulated transcripts were repressed in both strains, whereas 66 and 93 transcripts were specifically repressed in the cph1Δ/efg1Δ and Evo strains, respectively (S3B Figure).
We investigated the expression of individual marker genes for filamentation [38] more closely (S3C Figure). As expected, all eight genes of the core filamentation response (ALS3, ECE1, DCK1, HGT2, HWP1, IHD1, RBT1 and orf19.2457) were significantly upregulated in the Evo strain under filament-inducing conditions. Four of these genes (ECE1, HWP1, IHD1 and RBT1) and further filament-associated genes, like ALS1, BRG1 and HGC1 were also upregulated in the non-filamenting cph1Δ/efg1Δ strain. Expression of filament-associated genes independent of any morphological transition has previously been described in the cph1Δ/efg1Δ mutant [38]–[40]. However, these genes were expressed at a significantly higher level in the Evo strain compared to the cph1Δ/efg1Δ strain under filament-inducing condition (S2 DS5 Table).
Overall, genes most highly expressed (≥5-fold) in the Evo strain under filament-inducing condition are mainly hyphal-associated genes (HWP1, ECE1, ALS3, RBT1, FRG2, ALS1 and IHD1). Furthermore, the expression of YWP1, encoding a yeast-form cell wall protein, is downregulated in the Evo strain, while its expression did not change in the cph1Δ/efg1Δ strain. These results suggest that genes associated with C. albicans hyphae formation are also associated with filamentation of the Evo strain.
Upregulation (>1.5-fold, p<0.01; S2 DS1 and DS4 Table) of DCK1, LMO1 and CEK1, which are required for filamentation under embedded conditions and for cell wall integrity [41], was found solely in the Evo strain. This provides a possible explanation for the hyper-filamentous phenotype under embedded conditions as well as the increased resistance to cell wall perturbants compared to the double mutant (Figs. 1+5).
To determine whether changes in the regulation of effector genes are reflected by an upregulation of specific TF genes, we also analyzed the expression levels of TF genes in the cph1Δ/efg1Δ and Evo strains under filament-inducing conditions in more depth (S2 DS7 Table). A significantly higher expression of 21 TF genes was shared by both strains, and only five TF genes were specifically upregulated in the cph1Δ/efg1Δ strain as compared to the levels in the Evo strain. Interestingly, 17 TF genes had significantly higher expression specifically in the Evo strain and not in cph1Δ/efg1Δ, including three genes known to be important hyphal morphogenesis regulators: UME6 (in agreement with previous qRT-PCR results), RIM101 and HAC1. Eight of the higher expressed TF genes in the Evo strain have unknown biological functions.
In the cph1Δ/efg1Δ strain, but not in the Evo strain, CPH2, TEC1 and ACE2, which encode TFs involved in hyphal growth, were significantly downregulated under filament-inducing conditions. Finally, a significantly lower expression was observed for NRG1 in the Evo strain, which codes for a repressor of hyphal development [42]. Hence, we scanned for Nrg1 binding sites (A/C)(A/C/G)C3T [43] in the putative promoter regions of genes specifically upregulated twofold in the Evo strain and detected the sequence motifs in 70% of these promoter regions (S2 DS8 Table). With this, the Nrg1 binding motif is statistically overrepresented in promoters of upregulated genes (p<0.01) when compared to promoters of all other genes. The downregulation of NRG1 in the Evo strain may therefore facilitate expression of filament-associated genes and hence filament formation.
Further analyses indicated a significant upregulation of genes encoding for secreted aspartyl proteases (SAP5, SAP6, SAP10). In addition, significant differences in expression of genes associated with cell wall biosynthesis (CHK1, KRE6, GLC3, MP65, ALG11 and MNT2), alkalinisation (ACH1) as well as of genes involved in glucose and galactose interconversion and uptake (GAL10, GAL1, HGT2, HGT4, HGT12 and GSY1) were observed.
In summary, our transcriptional analysis indicated that serial passage through macrophages led to substantial alterations of the global transcriptional profile. The programs and pattern we found differed clearly from the cph1Δ/efg1Δ mutant, and resembled more the well-known programs of the wild type strain. This is concomitant with and likely correlated with the regained ability of the Evo strain to induce filaments and to induce damage to host cells in vitro and in vivo.
Comparative whole genome re-sequencing identifies mutations potentially linked to Cph1/Efg1-independent filamentation
We went on to determine the genetic basis for the observed phenotypical differences. No obvious large-scale structural variations were detectable between the karyotypes of wild type, the cph1Δ/efg1Δ and Evo strains using pulsed field gel electrophoresis (PFGE; S4A Figure). To detect possible loss of heterozygosity (LOH) events [44], we analyzed four SNP-restriction fragment length polymorphism (RFLP) markers per chromosome [45]. No differences were detected between double mutant and Evo strain (S3 DS1 Table). Taken together, these data show that no gross chromosomal rearrangements have occurred in the Evo strain.
We re-sequenced the genomes of the Evo and the cph1Δ/efg1Δ strains to identify single nucleotide polymorphisms (SNP) that may have arisen during the microevolution experiment. Sequencing depth for cph1Δ/efg1Δ and Evo were 99× and 108× in average, respectively, with 98.8% of the C. albicans SC5314 reference genome covered in both cases. Comparison of both sequences revealed a chromosome 7 trisomy in the cph1Δ/efg1Δ strain, an aneuploidy that appears to have been lost during the evolution experiment (S4B Figure). This is also reflected by a 1.5× higher mean transcription level of genes on chromosome 7 in the cph1Δ/efg1Δ strain (S4C Figure). In addition, an amplification of URA3 on chromosome 3 was observed. URA3 was originally used as a marker to delete CPH1 and EFG1 in the cph1Δ/efg1Δ strain, and is now present in three copies in this mutant. The Evo strain contained 7–8 copies (S4B Figure). A qPCR analysis on isolated gDNA supported these findings (S4D Figure). PFGE and subsequent hybridization with a URA3 specific probe further revealed that all copies were located on the same chromosome (S4E Figure). To exclude any possible contribution of multiple URA3 gene copies to the filamentous phenotype, the Evo strain was cured from URA3 with 5-fluoroorotic acid treatment [46]. This Evo Ura− strain was still able to filament, showing that URA3 copy number is not responsible for the filamentous phenotype (S4F Figure). Additionally, after re-introduction of a single URA3 using the standard CIp10 plasmid at the RPS10 locus [47], these strains exhibited the same adhesion, invasion and macrophage damage properties as their multi-URA3 counterparts (S4G Figure). This indicates that the excessive URA3 copies do not have an influence on classical virulence properties of C. albicans.
We observed a high number of SNPs in the cph1Δ/efg1Δ strain: altogether, 70,197 heterozygous and 3,156 homozygous SNPs were identified in cph1Δ/efg1Δ relative to the C. albicans SC5314 consensus reference genome (Assembly 21, [48]). Similarly, 72,315 heterozygous and 3,294 homozygous SNPs were identified in the Evo strain. These figures are consistent with those achieved when reads obtained by sequencing the genome of C. albicans SC5314 are aligned on the reference genome and reflect the high level of heterozygosity in C. albicans as well as putative sequencing errors and ambiguous positions in the reference genome (homozygous SNPs). After combining these sets and filtering, only 329 putative SNPs were found to distinguish the cph1Δ/efg1Δ and Evo strains. Notably, polymorphisms at 209 of these positions are observed in the genomes of 19 clinical isolates, distributed over several C. albicans phylogenetic groups (CdE, unpublished data). This suggests that they were most likely not responsible for the restoration of filamentation. Of the 120 remaining positions, 83 were in non-coding regions, 22 resulted in synonymous changes and 15 resulted in non-synonymous changes (S3 DS2-4 Table). Finally, the RNA-Seq dataset was used as an additional source to detect SNPs specifically in expressed genes (see Materials and Methods & S3 DS6&7 Table,): A total of 65 putative transcribed SNPs, both heterozygous and homozygous, were found in the Evo strain, of which 21 were located in non-coding regions. Inside ORFs, 26 caused a synonymous and 13 a non-synonymous nucleotide exchange. Of all 39 SNPs detected in coding regions, 24 were located in genes of the ALS gene family (ALS2 and ALS4), although these are likely false positives, as genes of the ALS family possess a very high sequence similarity and tandem repeat regions complicating read-mapping and SNP resolution [49]. Comparison of SNPs detected by RNA-Seq and Whole-Genome Sequencing revealed three SNPs shared by both detection methods. One SNP was located in a non-coding region between two uncharacterized genes (orf19.351 and orf19.352), while the other two were located inside ORFs. A SNP in ATP18 (orf19.2066.1) resulted in a synonymous amino acid exchange, while the second SNP in SSN3 (orf19.794) resulted in a heterozygous, non-synonymous Arg/Arg to Arg/Gln amino acid change.
A Mutation in SSN3 is essential for the filamentous phenotype in the Evo strain
As the SNP at nucleotide position 1,055 in the SSN3 ORF (Fig. 6A) was detected in both analyses, we focused our investigation on this specific mutation. Ssn3 has been well characterized in Saccharomyces cerevisiae as an RNA polymerase II holoenzyme-associated cyclin-dependent kinase of the Mediator complex contributing to transcriptional control [50]. It was shown that Ssn3 promotes the degradation of the transcription factor Ste12 by phosphorylation and thereby regulates S. cerevisiae filamentous growth [51]. As depicted in Fig. 6B, the heterozygous Arg352Gln mutation of Ssn3 in the Evo strain is located within the activation segment of the protein kinase catalytic domain. An amino acid sequence comparison of C. albicans Ssn3 to sequences from S. cerevisiae, Cryptococcus neoformans, Mus musculus and Homo sapiens demonstrated this arginine residue to be conserved from fungi to mammals. The activation segment comprises several conserved structural features: the magnesium binding loop, the activation loop and the P+1 loop, in which the mutation occurred. While the activation loop is the site of regulatory phosphorylation in many kinases, the P+1 loop forms a pocket that recognizes the substrate protein [52].
Fig. 6. Single nucleotide polymorphism in SSN3 of the Evo strain and location of the mutated amino acid. 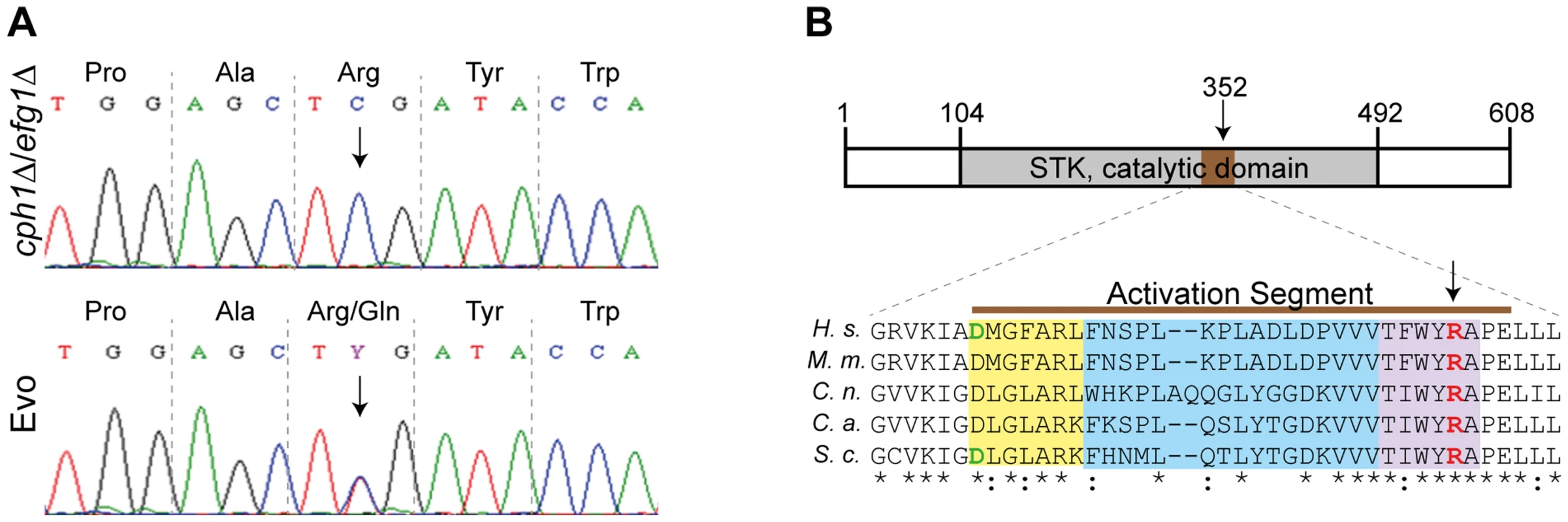
(A) Partial SSN3 sequence for cph1Δ/efg1Δ and Evo strains flanking SNP 1055 (marked with an arrow). Notice the heterozygosity in the Evo strain. (B) Schematic view of the catalytic domain of Ssn3 (STK = serine/threonine kinase) with the position of the activation segment highlighted in brown and the amino acid exchange indicated by an arrow (top). Sequence alignment of the Ssn3 activation segment in different species (H. s. Homo sapiens [NP_001251.1], M. m. Mus musculus [NP_705827.2], C. n. Cryptococcus neoformans [XP_568416.1], C. a. C. albicans [XP_720918.1] and S. c. Saccharomyces cerevisiae [NP_015283.1]). The arrow indicates the amino acid exchange in the Evo strain. The Mg-binding loop is highlighted in yellow, the activation loop in blue and the P+1 loop in purple. Amino acids that are known to abrogate kinase activity when mutated are colored in green [51], [98]. Asterisks underneath the alignment indicate positions with conserved amino acids and colons indicate highly similar residues (bottom). The mutated arginine (red) is part of the highly conserved P+1 substrate recognition loop. To ascertain the impact of the SNP on filamentation induction, we selectively deleted either the mutated or the wild type SSN3 allele in the Evo strain, using the dominant selection marker SAT1 [53]. Sanger sequencing confirmed the exclusive presence of either one allele in the genome (Fig. 7A). Strikingly, when incubated in DMEM with 10% serum at 37°C and 5% CO2 only the strain with the mutated allele still present (Evo ssn3Δ/SSN3m) was able to induce and maintain filamentation. The mutant containing only the wild type allele (Evo SSN3/ssn3mΔ) remained in the elongated yeast form, and thus presented the typical ancestral (cph1Δ/efg1Δ) phenotype (Fig. 7A). In addition, only the Evo ssn3Δ/SSN3m strain could escape from macrophages by forming filaments like the wild type (Fig. 7A). The damage capacity correlated with this ability to produce filaments: While Evo and Evo ssn3Δ/SSN3m strains showed the same levels of phagocyte lysis, the Evo SSN3/ssn3mΔ strain caused significantly less damage during co-incubation with macrophages. In fact, damage was indistinguishable from the original cph1Δ/efg1Δ strain (Fig. 7B). In contrast, the deletion of the mutated allele had no influence on the hyphal development defect on solid medium (S5A Figure) and sensitivity to cell wall disturbing agents (S5B Figure).
Fig. 7. A single nucleotide polymorphism in SSN3 is essential for filamentation. 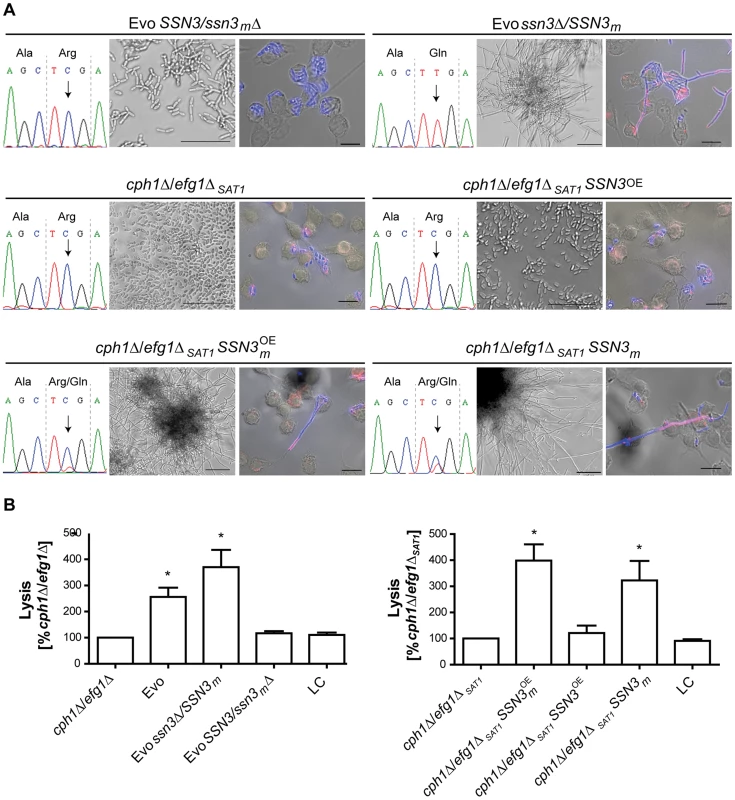
(A) Distinct impact on morphology by: deleting either the mutated (SSN3/ssn3mΔ) or the wild type SSN3 allele (ssn3Δ/SSN3m) in the Evo strain, by overexpressing either the wild type SSN3 allele (cph1Δ/efg1ΔSAT1SSN3OE) or the mutated SSN3 allele (cph1Δ/efg1ΔSAT1SSN3mOE) or by expressing the mutated SSN3 allele from its native locus (cph1Δ/efg1ΔSAT1SSN3m) in a newly generated cph1Δ/efg1ΔSAT1 strain. The partial SSN3 sequences demonstrate the homozygosity or heterozygosity of the SSN3 allele (left). Filamentous growth is visible in the Evo ssn3Δ/SSN3m, cph1Δ/efg1ΔSAT1SSN3mOE and cph1Δ/efg1ΔSAT1SSN3m strains after growth for 18 h at 37°C and 5% CO2 in DMEM+10% FBS and during co-incubation with macrophages, but not with the Evo SSN3/ssn3mΔ strain, cph1Δ/efg1ΔSAT1 and cph1Δ/efg1ΔSAT1SSN3OE strains (scale bars: 18 h, 50 µm and MΦ, 20 µm; representative pictures are shown) (right). (B) Cell damage of macrophages caused by the different strains, as determined by lactate dehydrogenase (LDH) assay after 32 h of co-incubation. Robust host cell damage depends on the presence of the mutated allele SSN3m. Mean and SD of n = 4 (*p<0.05; compared to cph1Δ/efg1Δ and cph1Δ/efg1ΔSAT1 respectively; LC = low control, medium only). To further ascertain that the SSN3 mutation alone is sufficient to allow filamentation in a cph1Δ/efg1Δ background, we created an independent cph1Δ/efg1Δ double mutant using the dominant selection marker SAT1 (see S3 DS1 Table). Importantly, this cph1Δ/efg1ΔSAT1 strain contained neither the URA3 amplification nor the trisomy of chromosome 7 or other genetic alterations of the original cph1Δ/efg1Δ strain. In all our filamentation assays, this mutant behaved identical to the original cph1Δ/efg1Δ strain by not forming any hyphae (Fig. 7A) and hence not escaping from or damaging macrophages (Figs. 7A & 7B). To isolate the effect of the mutated SSN3, we followed several strategies with this new mutant: SSN3 overexpression strains were created of both the wild type and mutated (SSN3m) allele under the control of the strong ADH1 promoter (see S1 Protocol). Strikingly, only the mutated allele allowed hyphae formation under inducing conditions in the cph1Δ/efg1ΔSAT1 strain (Fig. 7A, lower left corner), even in the presence of the two native SSN3 alleles. Similarly, macrophage lysis was increased in the SSN3m overexpressing strain, but not under SSN3 overexpression (Fig. 7B, right panel). Finally, we integrated the mutated SSN3 (together with a SAT1 cassette) into the SSN3 locus of cph1Δ/efg1ΔSAT1, replacing one SSN3 allele and essentially reproducing the heterozygous situation of the Evo strain. Again, this strain behaved virtually identical to the Evo strain, both in forming hyphae (Fig. 7A) and in damaging macrophages (Fig. 7B, right panel).
In summary, these data show that a non-synonymous mutation in SSN3 that arose during our microevolution experiment is alone sufficient for regaining the ability to filament even in the absence of Efg1 and Cph1.
Discussion
Previous experimental studies on the acquisition of antifungal drug resistance and on stress-induced chromosome rearrangements have elegantly demonstrated the adaptive potential of C. albicans [44], [54]. Here, we demonstrate – to our knowledge for the first time – that a complex trait such as the hyphal formation program of C. albicans can be subject to microevolution in the laboratory.
The yeast-to-hyphae transition is of crucial importance for full C. albicans pathogenicity, which is reflected by its complex regulation [14]. Multiple overlapping as well as separate signaling pathways are activated by various environmental signals to regulate hyphae formation. Wild type hyphae are an important contributor to the fungus' ability to escape from engulfing macrophages. In contrast, the cph1Δ/efg1Δ mutant strain cannot escape by filament formation, yet is able to replicate inside macrophages and to block phagosome maturation. Therefore, we expected that the mutant strain would survive in the phagosome, albeit with reduced fitness compared to the wild type. We monitored the phenotypic changes of the cph1Δ/efg1Δ strain co-passaged with macrophages for 42 passages. On a comparatively short evolutionary timescale our experiment resulted in a strain which not only regained the ability to filament, but also re-acquired other important characteristics, like a more wild type-like cell wall structure and increased virulence. We were able to show that a minimal sequence alteration accounts for the striking phenotypic reversal to wild type-like filamentation: a single missense mutation in SSN3. SSN3 encodes a fungal protein kinase, which phosphorylates various regulators in S. cerevisiae. Our data shows that it can become important for bypassing the requirements of Efg1 and Cph1 for filamentation in C. albicans.
The in-depth characterization of the evolved strain revealed that the hyphal morphogenesis program can be induced by certain, but not all conditions which induce filamentation in the wild type strain. The fact that the Evo strain filaments in liquid, but not on solid media indicates an involvement of cAMP signaling and hence argues for a bypass of Efg1 functions rather than Cph1 [55]–[57]. This was further supported by three additional findings. First, the yeast-to-filament switch occurred in response to either serum, GlcNAc or CO2, stimuli all known to trigger the activation of PKA signaling [23], [58]–[60]. Second, filamentation was entirely blocked by the addition of the quorum-sensing molecule farnesol which represses both cAMP-PKA and MAPK signaling pathways [61], [62]. The full restoration of filamentation when cAMP was added supports the involvement of the cAMP-PKA pathway. Third, the repressor of hyphae formation, Nrg1 is normally downregulated by the cAMP-PKA pathway, except in the presence of farnesol [63]. Transcriptome analysis showed NRG1 expression to be downregulated in the Evo strain, but not in the cph1Δ/efg1Δ mutant. As 70% of the upregulated genes in the Evo strain contain an Nrg1 binding site, these data emphasize the likely importance of Nrg1 levels on filamentation of the Evo strain.
Given that the cph1Δ/efg1Δ mutant is strongly reduced in virulence [22], the almost wild type-level virulence in the Evo strain in our murine model was striking. Examination of kidney sections revealed filament formation of the Evo strain in vivo. Compared to wild type filaments, these were shorter and resulted in less pronounced tissue invasion, which is likely associated with the lower overall virulence compared to the wild type.
Three factors are likely to have contributed to the increased virulence of the Evo strain in the absence of Efg1 and Cph1: First, its ability to escape from macrophages like the wild type; second, its adhesion to host cells which was significantly higher than the cph1Δ/efg1Δ strain; and third, the ability to form filaments upon contact with epithelial cells, which is a prerequisite for both active penetration into and induced endocytosis by host cells [64]. Wächtler et al. [17] showed that filamentation alone is insufficient to cause damage of host cells. We therefore compared the damage capacities of the cph1Δ/efg1Δ and the Evo strains. The Evo strain exhibited a significantly increased potential to damage both macrophages and epithelial cells compared to the double mutant. The adaptation to macrophages was accompanied by differences in additional traits, such as resistance to cell wall stresses. In the cph1Δ/efg1Δ strain, the higher sensitivity to cell wall disturbing agents, as well as the modified exposure of cell wall components, likely reflect an altered cell wall organization which was restored in the Evo strain. This is supported by findings from a recent study by Zavrel et al. [32] which showed that deletion of EFG1 alone affects cell wall architecture. In our strains, these modifications of the cell wall seemed to be mediated by the kinases Mkc1 and Cek1. Previous analyses carried out in cek1Δ and mkc1Δ mutants already indicated their direct relationship to cell wall composition and integrity [35], [65], [66].
By analyzing the differences in gene expression acquired during co-culture passaging with macrophages, we found that all genes belonging to the core filamentation network [38] were upregulated in the Evo strain. This suggests that during filamentation the Evo strain transcriptionally utilizes the complete filamentation program. The transcription factors Tec1, Brg1, Ume6, Rim101, Hac1 and Efh1, which are known to be involved in regulation of filamentation [24], [67]–[71], were also upregulated in the Evo strain. Together with Nrg1, they likely orchestrate filament formation in the Evo strain. For UME6, it has been shown that its transcription is repressed by Nrg1-Tup1 and that ectopic Ume6 expression in cph1Δ/efg1Δ can rescue the filamentation defect under certain conditions [69].
For the maintenance of hyphal extension, both UME6 and EED1 are central [27], [31] and both showed an increased expression in the evolved strain. Thus, the mechanisms of hyphal extension seems similar between Evo and wild type cells [27]. Hence, the transcriptional conditions for initiation and maintenance of filamentation, which comprise the release of repression and the upregulation of positive regulators of filamentation, are met in the Evo strain. Furthermore, a considerable number of transcripts specifically up - and downregulated in the Evo strain are both Candida-specific and uncharacterized. It is feasible, therefore, that these uncharacterized transcripts assumed a novel role specifically during filament formation in the Evo strain. This is especially true as the morphological switch is one of the best-investigated characteristics in C. albicans, and genes involved in this process are generally well studied. However, differential regulation of genes not clearly linked to the yeast-to-hyphal switch, including these genes, but also WOR1 and NAT4 (both involved in the white-opaque switching) and SST2 (involved in the mating response pathway), could have been caused by the mutated Ssn3 kinase (see below). Finally, it also should be noted that, even in the absence of filamentation, cph1Δ/efg1Δ was able to upregulate certain genes described as hyphae-associated under the condition tested here (incubation in DMEM+10% FBS at 37°C and 5% CO2 on a plastic surface). This is in disagreement with previous data showing that EFG1 is required for expression of several hyphae-associated genes [23], [72]. It is possible, however, that alternative pathway(s), such as the Rim101 pH response pathway, are involved, as the cells were simultaneously exposed to diverse stimuli for filamentation. However, these genes still showed an increased induction in the Evo strain compared to the cph1Δ/efg1Δ strain, which argues for a further adaptation-induced, filament-associated change in regulation.
It has been demonstrated that acquired drug resistance in C. albicans is often accompanied by aneuploidy and/or isochromosome formation [54], [73] and that several stress conditions can enhance the rates of LOH events likely by mitotic recombination [44]. However, we did not detect any LOH events between the cph1Δ/efg1Δ and the Evo strain. The chromosome 7 trisomy was present initially in the cph1Δ/efg1Δ strain [74] and the Evo strain restored disomy by loss of one copy. The remaining gross genetic difference, an URA3 amplification in the Evo strain can be explained by an insufficient Ura3 expression from the EFG1 locus. An amplification of the gene may have increased fungal fitness during our experiment by ensuring more transcripts and hence more efficient growth. Prior studies suggested that ectopic expression of URA3 influences the phenotypes of a diverse range of mutants [75], [76], and duplication of a hisG-URA3-hisG cassette resulted in restored filamentation of an hwp1Δ mutant [77]. We were able to exclude these Ura3 effects as causes for the Evo strain filamentation, as the acquired filamentation phenotype was maintained after removal of the multiple URA3 copies. Furthermore, after re-introduction of a single copy of URA3, no differences in virulence traits, like adhesion or invasion, were detectable as compared to the multi-copy strains. Overall, these data and the fact that the observed filamentation and other phenotypes persisted even after repassaging in rich (YPD) medium, argued for small-scale genomic alterations, rather than epigenetic changes, acquired by cph1Δ/efg1Δ cells adapting to macrophages.
Comparative genome sequencing (by WGS and RNA-Seq) of cph1Δ/efg1Δ and Evo strains allowed us to pinpoint the microevolutionary changes in the Evo strain at the single nucleotide level. By combining the different approaches, we detected an expressed SNP in SSN3, which resulted in an Arg-to-Gln change at a highly conserved position within the presumable protein kinase domain. This SNP and thus gain of heterozygosity was found to be central for the yeast-to-filament transition of the Evo strain. Deletion of the mutated SSN3 allele prevented the morphological switch in the Evo strain during growth under filament-inducing conditions and interaction with several types of host cells. Other phenotypes specific to the Evo strain were not affected by the deletion of the mutated SSN3 allele, suggesting that they evolved independently from filamentation.
Importantly, introduction of a single mutated allele into an independent efg1Δ/cph1Δ strain fully copied the filamentation and host cell damage phenotype of the Evo strain. This strain contained neither the multiple URA3 copies nor the trisomy of chromosome 7 or any other possible genetic alterations of the original efg1Δ/cph1Δ strain. Hence, the SSN3 mutation alone bypassed the lack of the central transcription factors Cph1 and Efg1 and restored the ability to cause host cell damage in vitro, and likely to induce higher virulence in vivo.
Ssn3 itself (also referred as Srb10 or Cdk8) is part of the CDK (cyclin-dependent kinase) module (SRB10/11) of the Mediator complex, which is a regulator of RNA-polymerase II (RNAP II) activity [78], [79]. This CDK module phosphorylates the largest subunit of RNAP II, and Ssn3 additionally has roles in both transcriptional activation and repression in response to physiological signals, coordinating gene expression. By regulating the stability of the two important regulators, Ste12 (ortholog of Cph1) and Phd1, Ssn3 in S. cerevisiae is involved in the differentiation of yeasts into pseudohyphae under nutrient-limiting conditions [51], [80]. Interestingly, a kinase-deficient Asp290Ala Ssn3 only weakly phosphorylates Ste12 in vitro, and the lack of phosphorylation increases its stability [51]. Moreover, the catalytic activity of Ssn3 contributes to the repression of a subset of Tup1-regulated genes [81]–[83] in S. cerevisiae. Tup1 is recruited to promoters by Nrg1 [84], a factor which was downregulated in the Evo strain.
Although the precise signaling pathway(s) controlling Ssn3 remain to be determined, Chang et al. [85] showed that the activity of Srb9, another subunit of the CDK kinase module, is regulated by the PKA signaling pathway in S. cerevisiae. Based on our data it is tempting to speculate that the activation of the cAMP-PKA pathway results in activation of Ssn3 kinase activity, and the observed filament-specific transcriptional changes may thus depend on either a reduced or absent substrate recognition or on impaired substrate phosphorylation activity due to the Arg352Gln substitution. It is supposable that a loss of the substrate-specific kinase activity increases the stability of positive regulator(s) of filamentation by reducing their phosphorylation. Alternatively (or in addition), the impaired kinase activity could lead to a derepression of genes associated with positive regulation of filamentous growth. Importantly, in this model the kinase-deficient Ssn3 remains part of the Mediator complex, and could fulfill any additional function it may have (e.g. in the structure or recruitment of additional proteins). In both models, a decreased kinase activity would reduce inhibitory effects on filamentation, and hence would increase the sensitivity of the filamentation network to external stimuli. This would likely allow to bypass the need for additional Efg1 signaling. Additional genes outside of the immediate filamentation network may also be affected, as this model implies a pleiotropic effect of the Ssn3 mutation, with several transcription factors as possible clients.
Thus, it seems that not the disrupted cAMP-PKA signaling pathway itself evolved in our microevolution experiment, but instead a regulatory hub for filamentation which the pathway probably targets in addition to Efg1. In this hub, even single or few mutations seem to be able to lead to striking phenotypic alterations, as many filament-associated genes are directly or indirectly targeted. Finally, it is interesting to speculate why only one SSN3 allele was mutated, and we did not observe any LOH event to homozygosity at this locus. It seems possible that one mutated allele alone was sufficient to promote filamentation in macrophages, while the other wild type allele, still capable of full phosphorylation activity, was still required for additional functions of Ssn3. This is somewhat supported by the observation that overexpression of the mutated SSN3 allele in a background with the native SSN3 alleles still in place was sufficient to allow hyphae formation. In our model, the mutated Ssn3 competes with the wild type Ssn3, and overexpression allows the mutated protein to gain entry into a sufficient number of Mediator complexes.
In conclusion, using the nonfilamentous mutant cph1Δ/efg1Δ, we have shown that C. albicans can rescue one of its key virulence traits, the yeast-to-hyphal switch, with a single nucleotide change when put under adequate selection pressure. A mutation in the transcriptional regulator Ssn3 adaptively rewired the transcription network to enable filamentation in response to external cues while bypassing the need for Efg1 and Cph1. This shows an unexpected robustness of the whole filamentation system even to severe disruptions, and a high degree of adaptability. The selection scenario we used - co-incubation with macrophages - clearly reflects a condition C. albicans encounters in the host and thus might be an evolutionary pressure that can shape the infection biology of this fungus. In fact, this hypothesis is supported by another evolution experiment, which analyzed the adaptation of C. glabrata to macrophages. There, the selection pressure resulted in the appearance of a strain with pseudohyphae-like structures and increased virulence again by a single nucleotide mutation [86]. This demonstrates that during interaction with the host or host cells, significant changes in morphology and virulence are possible on a very short evolutionary time-scale.
Materials and Methods
Ethic statement
All animal experiments were in compliance with the German animal protection law and were approved by the responsible Federal State authority (Thüringer Landesamt für Lebensmittelsicherheit und Verbraucherschutz) and ethics committee (beratende Kommission nach § 15 Abs. 1 Tierschutzgesetz; permit no. 03-007/07).
Body surface temperature and body weight were recorded daily and animals were monitored twice a day for disease progression. Mice showing severe signs of illness (isolation from the group, apathy, hypothermia and drastic weight loss) were humanely sacrificed by ketamine/xylazine overdose and exsanguination.
Strains and growth conditions
Candida albicans strains and mutants used in this study are listed in S1 Table. Strains were grown in YPD medium (1% peptone, 1% yeast extract, 2% glucose and optionally 2% agar) or SD medium (2% dextrose, 0.17% yeast nitrogen base, 0.5% ammonium sulfate and optionally 2% agar) at 30°C. Uridine (50 µg/ml) or nourseothricin (NAT; 100 µg/ml) were added as required. If not stated otherwise, stationary phase cells were used in the experiments. Mutants were constructed as described in Protocol S1.
Cell lines
The murine peritoneal macrophage-like cell line J774A.1 (DSMZ) and the human buccal carcinoma epithelial cell line TR-146 (Cancer Research Technology) were grown in Dulbecco's Modified Eagle's Medium (DMEM, PAA) supplemented with 10% FBS (PAA) and routinely cultured until passage 20. Both cell lines were maintained at 37°C under 5% CO2. J774A.1 cells were removed from tissue-culture flasks by gentle scraping, while TR-146 cells were enzymatically harvested by Accutase (PAA) treatment.
Evolution experiment
About 8×106 J774A.1 macrophages were seeded into a 75 cm2 cell culture flask with DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin (PAA). For the evolution experiment, macrophages were initially infected with 4×106 cells of the cph1Δ/efg1Δ strain. After that, 4×106 re-isolated C. albicans cells were transferred to a fresh macrophage culture. After 24 h of co-incubation, infected macrophages were washed (3× with PBS) and lysed with 2 ml lysis buffer (50 mM Tris, 5 mM EDTA, 150 mM NaCl and 0.5% Nonidet P40 [Sigma-Aldrich]). The lysate was transferred to a 2 ml reaction tube and fungal cells were collected by centrifugation. The C. albicans cells were washed two times with DMEM and counted before infection of fresh macrophages.
To verify the absence of EFG1 and CPH1, Southern blot analysis was performed for the Evo strain as described previously [22]. Briefly, genomic DNA (gDNA) was digested with AvaII or KpnI to verify EFG1 or CPH1 deletion, respectively. DIG-labeled probes were generated (Roche) using genomic DNA from the strain SC5314 and primers EFG-A/EFG-B and P33/CPH-B (S1 Table).
Phenotypic characterization
A detailed description of the phenotypic analyses can be found in Protocol S1.
Staining procedures and detection of β-1,3-glucans and mannans
Fungal cells were grown in DMEM+10% FBS on glass coverslips in a 24 well microtiter plate for filipin (Sigma), calcofluor white (CFW) and Als3 immunostaining. Flow cytometry was used to quantify mannan and β-1,3-glucan exposure on the surface of stationary C. albicans cells after staining with concanavalin A and anti - β-1,3-glucan. Piercing and invasion rate were determined by differential staining. All staining procedures are described in Protocol S1. Epifluorescence (Leica DM5500B, Leica DFC360 FX) was used to detect CFW and filipin (DAPI filter), Alexa Fluor 488 (FITC filter) and Alexa Fluor 647 (Cy5 filter). Micrographs were taken with a Leica Digital Camera DFC360 FX or a Zeiss AxiCam ICc3.
Replication and piercing assay
Two times 105 J774A.1 macrophages were seeded onto glass cover slips placed in 24 well microtiter plates and allowed to adhere overnight. Non-adherent macrophages were removed by washing with PBS. To monitor intracellular replication, C. albicans cells were labeled with 100 µg/ml fluorescein isothiocyanate (FITC, Sigma-Aldrich) in carbonate buffer (0.1 M Na2CO3, 0.15 M NaCl, pH 9.0) for 30 min at 37°C and washed 3× with PBS. To quantify piercing rates, cells were washed without prior staining. Two times 105 fungal cells were added to macrophages in DMEM+10% FBS. The plates were incubated for indicated timepoints (see figure legends). Cells were then washed once with PBS and fixed with 4% paraformaldehyde. Intracellular replication was detected by fluorescence microscopy after mounting the samples in ProLong Gold Antifade Reagent with DAPI (Invitrogen). After co-incubation, piercing of macrophages by filaments was quantified by differential staining. The assays were performed in biological triplicates.
Adherence and invasion assay
Two times 105 TR-146 epithelial cells were seeded onto glass cover slips placed in 24 well microtiter plates and cultured for 2–3 days to 95%-100% confluency. Adherence and invasion assays were performed as previously described [17]. Briefly, to determine the adhesion rate, TR-146 monolayers were infected with 1×106 C. albicans cells. After one hour of co-incubation, non-adherent yeast cells were removed by rinsing 3× with PBS. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and adherent C. albicans cells were stained with CFW for fluorescence microscopy. Invasion rates were determined by infecting TR-146 monolayers with 1×105 C. albicans cells. After incubation, cells were fixed and differentially stained for fluorescence microscopy. Both assays were repeated at least three times.
Quantification of damage to host cells
Five times 104 host cells (J774A.1 or TR-146) were seeded in 96 well microtiter plates. J774A.1 macrophages were cultured for 1 day before use, while TR-146 epithelial cells were cultured for 2 days to 95%–100% confluency. Damage of macrophages and epithelial cells was determined by measuring the release of lactate dehydrogenase (LDH) with the Cytotoxicity Detection Kit (Roche Applied Science) following 32 h of co-incubation with 5×104 C. albicans cells according to the manufacturer's protocol. The experiments were performed as previously described [17] and repeated at least three times.
Murine infection model
For survival studies the intravenous challenge model for disseminated C. albicans infection was used. Six to eight weeks old female BALB/c mice (18–20 g) purchased from Charles River were used for the experiments. Mice were challenged intravenously with 5×105 C. albicans cells in 200 µl PBS via the lateral tail vein. All mice surviving to day 20 were humanely sacrificed. For histology, kidneys were collected and fixed with buffered formalin and paraffin-embedded sections were stained with Periodic acid-Schiff (PAS) according to standard protocols.
Western blot analysis
To detect phosphorylated Mkc1 and Cek1 as well as α-tubulin, cells of an overnight culture were adjusted to an OD600 of 0.5 in SD medium (control) or SD medium supplemented with 450 µg/ml congo red, and incubated for 4 hours at 30°C. Cell disruption, protein extraction and western blot analysis using anti-phospho-p44/42 MAP kinase antibody (Cell Signalling Technology) and rat anti-α-tubulin antibody (AbD Serotec), respectively, were performed as previously described [87].
RNA sample preparation and isolation
C. albicans cells from an overnight culture were diluted to OD600 = 0.2 in YPD medium and grown to log-phase for 4 h at 30°C. Cells were collected by centrifugation and a zero time point sample was frozen in liquid nitrogen until RNA extraction. In addition, 1×107 cells were incubated one hour under filament-inducing conditions (DMEM+10% FBS at 37°C and 5% CO2 in a 75 cm2 cell culture flask). For farnesol experiments, 10 µM farnesol was added to the medium just prior to the experiment. After incubation, medium and non-adherent cells were removed and 5 ml ice-cold PBS was added. The cells were collected by scraping and then centrifuged for 5 min at 6,000 g at 4°C. Cell pellets were snap frozen in liquid nitrogen. Total RNA was isolated using the Ribopure-Yeast Kit (Ambion) and treated with Turbo DNase (Ambion). RNA quality was determined in a Bioanalyzer with an RNA 6000 Nano LabChip Kit (Agilent Technologies) according to the manufacturer's protocol. RNA concentration was determined with a Nanodrop ND1000 (Peqlab).
Copy number determination and quantitative gene expression analysis
Copy number and expression levels of selected genes were analyzed with a my-Budget 5× EvaGreen QPCR Mix II (Bio&Sell) in a C1000TM Thermal Cycler (BioRad) using gene-specific primers (S1 Table). For expression analysis, 600 ng of total RNA was reversely transcribed with the SuperScript III First-Strand Synthesis Kit (Invitrogen) according to the manufacturer's instructions. URA3 gene copy number was determined from 100 ng of gDNA with primers URA3-fw and URA3-re (S1 Table). PCR conditions were as followed: 95°C for 15 min, 40 cycles of each 95°C for 15 s, 60°C for 40 s and 72°C for 15 s. A melting profile was generated to confirm PCR product specificity. Relative gene expression levels were determined by the 2ΔΔCt method [88] with ACT1, EFB1 and PMA1 as internal controls. URA3 copy number was calculated with ACT1 internal control and gDNA from SC5314 (containing two copies of URA3) as reference. Three independent experiments were performed.
Pulsed-field gel electrophoresis (PFGE) and SNP-RFLP analysis
PFGE and SNP-RFLP are described in Protocol S1.
RNA sequencing, transcriptional profiling and SNP discovery from RNA-Seq data
In order to use only high quality reads, trimming was performed using Btrim (window size = 15, average quality score = 20) [89]. For differential gene expression analysis high quality trimmed reads were mapped against the sequence Assembly 21 of strain SC5314 [48] using the spliced read mapper TopHat 2.0.6 [90] with the “known transcripts” (-G option) and uniquely mapped reads were counted using HTSeq [86]. Raw counts for each gene were loaded into R and differentially expressed genes were identified using the packages edgeR and DESeq [91], [92] and filtered by adjusted p-values (<0.01) and RPKM value (≥1). Data were deposited at the Gene Expression Omnibus (GSE56174) and can be found in S2 Table. Nrg1 binding sites (A/C)(A/C/G)C3T in putative promoter regions (usually −1000 bp/+50 bp) of all C. albicans genes were determined by SiTaR [93] allowing no mismatch. Fisher's exact test was used to determine if the Nrg1 motif-containing promoters were overrepresented in genes specifically upregulated twofold in the Evo strain, as compared to all remaining genes. For SNP calling quality trimmed reads from all samples of each strain were merged and the protocol of GATK [94] with slight changes was followed (i.e. reads were mapped using BWA algorithm [95], duplicates were removed and realignment around indels and base recalibration was performed). Next, we used bam-readcount (www.github.com/genome/bam-readcount), which determines the nucleotide distribution at each single base. Heterozygous SNPs were defined as positions where 25% or more of the reads showed an alternative nucleotide. Homozygous SNPs were defined as positions where more than 90% of the reads differed from the reference. Minimum nucleotide sequence depth was 20. Clustal Omega [96] was used for multiple sequence alignments.
Whole genome sequencing and sequence analysis
Genomic DNA isolated from the cph1Δ/efg1Δ and Evo strains were processed to prepare libraries for Illumina sequencing, and the TruSeq DNA Sample Prep kit (Illumina) was used according to the manufacturer's recommendations. DNAs were randomly fragmented by sonication to an average fragment length of 500 bp and Illumina adapters were blunt-end ligated to the fragments. The final libraries were amplified by PCR followed by sequencing on an Illumina Genome Analyzer platform (Illumina GAII). 60 nt single-end reads were aligned to the C. albicans strain SC5314 reference genome [48] downloaded on 02/24/2012 using shore 5.0 [97]. Sequencing depth scores were computed for each 1 kb region across the genomes and for ORFs using sequencing depth data for each nucleotide located within the 1 kb region or the ORF. Sequencing depth scores were normalized based on the overall sequencing depth obtained for each genome. Single nucleotide polymorphisms were identified using shore 5.0 [97] at positions covered at least 30 times with a minimum quality of 25. Homozygous SNPs were defined as positions where 90% of the reads meeting these criteria differed from the reference genome. Heterozygous SNPs were defined as positions where 20% or more of the reads showed one allele and 80% or less of the reads showed a second allele.
Statistical analysis
Data were visualized and statistically analyzed using GraphPad Prism version 5.00 (GraphPad Software, USA). Statistical analyses were performed by 1-way ANOVA (mannan and β-1,3-glucan exposure) or 2-way ANOVA (piercing, adhesion, invasion, damage and gene expression) followed by a Bonferroni correction. Differences in survival of mice were evaluated by Log-rank (Mantel-Cox) test.
Supporting Information
Zdroje
1. PfallerMA, DiekemaDJ (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20 : 133–163.
2. KumamotoCA, VincesMD (2005) Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol 7 : 1546–1554.
3. EneIV, BrunkeS, BrownAJ, HubeB (2014) Metabolism in Fungal Pathogenesis. Cold Spring Harb Perspect Med
4. KvaalCA, SrikanthaT, SollDR (1997) Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun 65 : 4468–4475.
5. MayerFL, WilsonD, HubeB (2013) Candida albicans pathogenicity mechanisms. Virulence 4 : 119–128.
6. van de VeerdonkFL, PlantingaTS, HoischenA, SmeekensSP, JoostenLA, et al. (2011) STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med 365 : 54–61.
7. van EnckevortFH, NeteaMG, HermusAR, SweepCG, MeisJF, et al. (1999) Increased susceptibility to systemic candidiasis in interleukin-6 deficient mice. Med Mycol 37 : 419–426.
8. WhiteTC, PfallerMA, RinaldiMG, SmithJ, ReddingSW (1997) Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis 3 Suppl 1: S102–109.
9. Rustchenko-BulgacEP, ShermanF, HicksJB (1990) Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J Bacteriol 172 : 1276–1283.
10. ShinJH, ParkMR, SongJW, ShinDH, JungSI, et al. (2004) Microevolution of Candida albicans strains during catheter-related candidemia. J Clin Microbiol 42 : 4025–4031.
11. TsongAE, TuchBB, LiH, JohnsonAD (2006) Evolution of alternative transcriptional circuits with identical logic. Nature 443 : 415–420.
12. TuchBB, GalgoczyDJ, HerndayAD, LiH, JohnsonAD (2008) The evolution of combinatorial gene regulation in fungi. PLoS Biol 6: e38.
13. ForcheA, MageePT, SelmeckiA, BermanJ, MayG (2009) Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182 : 799–811.
14. SudberyPE (2011) Growth of Candida albicans hyphae. Nat Rev Microbiol 9 : 737–748.
15. JacobsenID, WilsonD, WächtlerB, BrunkeS, NaglikJR, et al. (2012) Candida albicans dimorphism as a therapeutic target. Expert Rev Anti Infect Ther 10 : 85–93.
16. FillerSG, SheppardDC (2006) Fungal invasion of normally non-phagocytic host cells. PLoS Pathog 2: e129.
17. WächtlerB, WilsonD, HaedickeK, DalleF, HubeB (2011) From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One 6: e17046.
18. WellingtonM, KoselnyK, SutterwalaFS, KrysanDJ (2014) Candida albicans triggers NLRP3-mediated pyroptosis in macrophages. Eukaryot Cell 13 : 329–340.
19. UwamahoroN, Verma-GaurJ, ShenHH, QuY, LewisR, et al. (2014) The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. MBio 5: e00003–00014.
20. LorenzMC, BenderJA, FinkGR (2004) Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3 : 1076–1087.
21. GowNA, van de VeerdonkFL, BrownAJ, NeteaMG (2011) Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10 : 112–122.
22. LoHJ, KöhlerJR, DiDomenicoB, LoebenbergD, CacciapuotiA, et al. (1997) Nonfilamentous C. albicans mutants are avirulent. Cell 90 : 939–949.
23. StoldtVR, SonnebornA, LeukerCE, ErnstJF (1997) Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. Embo J 16 : 1982–1991.
24. DoedtT, KrishnamurthyS, BockmühlDP, TebarthB, StempelC, et al. (2004) APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell 15 : 3167–3180.
25. MartinSW, KonopkaJB (2004) Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell 3 : 675–684.
26. Merson-DaviesLA, OddsFC (1989) A morphology index for characterization of cell shape in Candida albicans. J Gen Microbiol 135 : 3143–3152.
27. MartinR, MoranGP, JacobsenID, HeykenA, DomeyJ, et al. (2011) The Candida albicans-specific gene EED1 encodes a key regulator of hyphal extension. PLoS One 6: e18394.
28. ZhaoX, OhSH, ChengG, GreenCB, NuessenJA, et al. (2004) ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150 : 2415–2428.
29. NobileCJ, FoxEP, NettJE, SorrellsTR, MitrovichQM, et al. (2012) A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148 : 126–138.
30. BrownDHJr, GiusaniAD, ChenX, KumamotoCA (1999) Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol 34 : 651–662.
31. BanerjeeM, ThompsonDS, LazzellA, CarlislePL, PierceC, et al. (2008) UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell 19 : 1354–1365.
32. ZavrelM, MajerO, KuchlerK, RuppS (2012) Transcription factor Efg1 shows a haploinsufficiency phenotype in modulating the cell wall architecture and immunogenicity of Candida albicans. Eukaryot Cell 11 : 129–140.
33. GowNA, HubeB (2012) Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol 15 : 406–412.
34. EismanB, Alonso-MongeR, RomanE, AranaD, NombelaC, et al. (2006) The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5 : 347–358.
35. Navarro-GarciaF, Alonso-MongeR, RicoH, PlaJ, SentandreuR, et al. (1998) A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology 144 (Pt 2) 411–424.
36. RomanE, NombelaC, PlaJ (2005) The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol Cell Biol 25 : 10611–10627.
37. BrunoVM, WangZ, MarjaniSL, EuskirchenGM, MartinJ, et al. (2010) Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res 20 : 1451–1458.
38. MartinR, Albrecht-EckardtD, BrunkeS, HubeB, HünnigerK, et al. (2013) A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One 8: e58613.
39. PierceJV, DignardD, WhitewayM, KumamotoCA (2013) Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot Cell 12 : 37–49.
40. SamaranayakeYH, CheungBP, YauJY, YeungSK, SamaranayakeLP (2013) Human serum promotes Candida albicans biofilm growth and virulence gene expression on silicone biomaterial. PLoS One 8: e62902.
41. HopeH, SchmauchC, ArkowitzRA, BassilanaM (2010) The Candida albicans ELMO homologue functions together with Rac1 and Dck1, upstream of the MAP Kinase Cek1, in invasive filamentous growth. Mol Microbiol 76 : 1572–1590.
42. BraunBR, KadoshD, JohnsonAD (2001) NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. Embo J 20 : 4753–4761.
43. MuradAM, LengP, StraffonM, WishartJ, MacaskillS, et al. (2001) NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. Embo J 20 : 4742–4752.
44. ForcheA, AbbeyD, PisithkulT, WeinzierlMA, RingstromT, et al. (2011) Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio 2.
45. ForcheA, SteinbachM, BermanJ (2009) Efficient and rapid identification of Candida albicans allelic status using SNP-RFLP. FEMS Yeast Res 9 : 1061–1069.
46. FonziWA, IrwinMY (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134 : 717–728.
47. MuradAM, LeePR, BroadbentID, BarelleCJ, BrownAJ (2000) CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16 : 325–327.
48. InglisDO, ArnaudMB, BinkleyJ, ShahP, SkrzypekMS, et al. (2012) The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40: D667–674.
49. HoyerLL (2001) The ALS gene family of Candida albicans. Trends Microbiol 9 : 176–180.
50. KuchinS, YeghiayanP, CarlsonM (1995) Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci U S A 92 : 4006–4010.
51. NelsonC, GotoS, LundK, HungW, SadowskiI (2003) Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421 : 187–190.
52. NolenB, TaylorS, GhoshG (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell 15 : 661–675.
53. ReussO, VikA, KolterR, MorschhauserJ (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341 : 119–127.
54. SelmeckiAM, DulmageK, CowenLE, AndersonJB, BermanJ (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5: e1000705.
55. LiuH, KöhlerJ, FinkGR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266 : 1723–1726.
56. BockmühlDP, ErnstJF (2001) A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157 : 1523–1530.
57. LebererE, HarcusD, BroadbentID, ClarkKL, DignardD, et al. (1996) Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 93 : 13217–13222.
58. JungWH, StatevaLI (2003) The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology 149 : 2961–2976.
59. CastillaR, PasseronS, CantoreML (1998) N-acetyl-D-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell Signal 10 : 713–719.
60. KlengelT, LiangWJ, ChaloupkaJ, RuoffC, SchröppelK, et al. (2005) Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 15 : 2021–2026.
61. RomanE, Alonso-MongeR, GongQ, LiD, CalderoneR, et al. (2009) The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans. FEMS Yeast Res 9 : 942–955.
62. Davis-HannaA, PiispanenAE, StatevaLI, HoganDA (2008) Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 67 : 47–62.
63. LuY, SuC, WangA, LiuH (2011) Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol 9: e1001105.
64. ZakikhanyK, NaglikJR, Schmidt-WesthausenA, HollandG, SchallerM, et al. (2007) In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 9 : 2938–2954.
65. Navarro-GarciaF, EismanB, FiuzaSM, NombelaC, PlaJ (2005) The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology 151 : 2737–2749.
66. Galán-DiezM, AranaDM, Serrano-GomezD, KremerL, CasasnovasJM, et al. (2010) Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect Immun 78 : 1426–1436.
67. SchweizerA, RuppS, TaylorBN, RollinghoffM, SchröppelK (2000) The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol 38 : 435–445.
68. ClearyIA, LazzellAL, MonteagudoC, ThomasDP, SavilleSP (2012) BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol Microbiol 85 : 557–573.
69. ZeidlerU, LettnerT, LassnigC, MüllerM, LajkoR, et al. (2009) UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res 9 : 126–142.
70. DavisD, WilsonRB, MitchellAP (2000) RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol 20 : 971–978.
71. WimalasenaTT, EnjalbertB, GuillemetteT, PlumridgeA, BudgeS, et al. (2008) Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol 45 : 1235–1247.
72. SharkeyLL, McNemarMD, Saporito-IrwinSM, SypherdPS, FonziWA (1999) HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol 181 : 5273–5279.
73. SelmeckiA, Gerami-NejadM, PaulsonC, ForcheA, BermanJ (2008) An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol 68 : 624–641.
74. ArbourM, EppE, HoguesH, SellamA, LacroixC, et al. (2009) Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res 9 : 1070–1077.
75. ChengS, NguyenMH, ZhangZ, JiaH, HandfieldM, et al. (2003) Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect Immun 71 : 6101–6103.
76. SundstromP, CutlerJE, StaabJF (2002) Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect Immun 70 : 3281–3283.
77. SharkeyLL, LiaoWL, GhoshAK, FonziWA (2005) Flanking direct repeats of hisG alter URA3 marker expression at the HWP1 locus of Candida albicans. Microbiology 151 : 1061–1071.
78. MyersLC, KornbergRD (2000) Mediator of transcriptional regulation. Annu Rev Biochem 69 : 729–749.
79. LewisBA, ReinbergD (2003) The mediator coactivator complex: functional and physical roles in transcriptional regulation. J Cell Sci 116 : 3667–3675.
80. RaithathaS, SuTC, LourencoP, GotoS, SadowskiI (2012) Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol Cell Biol 32 : 664–674.
81. SchüllerJ, LehmingN (2003) The cyclin in the RNA polymerase holoenzyme is a target for the transcriptional repressor Tup1p in Saccharomyces cerevisiae. J Mol Microbiol Biotechnol 5 : 199–205.
82. GreenSR, JohnsonAD (2004) Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol Biol Cell 15 : 4191–4202.
83. KuchinS, CarlsonM (1998) Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol 18 : 1163–1171.
84. ParkSH, KohSS, ChunJH, HwangHJ, KangHS (1999) Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol Cell Biol 19 : 2044–2050.
85. ChangYW, HowardSC, HermanPK (2004) The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol Cell 15 : 107–116.
86. BrunkeS, SeiderK, FischerD, JacobsenI, KasperL, et al. (2014) Brunke S, Seider K, Fischer D, Jacobsen ID, Kasper L, et al. (2014) One Small Step for a Yeast - Microevolution within Macrophages Renders Candida glabrata Hypervirulent Due to a Single Point Mutation. PLoS Pathog 10 (10) e1004478 doi:10.1371/journal.ppat.1004478
87. MayerFL, WilsonD, JacobsenID, MiramónP, GrosseK, et al. (2012) The novel Candida albicans transporter Dur31 Is a multi-stage pathogenicity factor. PLoS Pathog 8: e1002592.
88. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔCT Method. Methods 25 : 402–408.
89. KongY (2011) Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98 : 152–153.
90. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
91. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140.
92. AndersS, HuberW (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106.
93. FaziusE, ShelestV, ShelestE (2011) SiTaR: a novel tool for transcription factor binding site prediction. Bioinformatics 27 : 2806–2811.
94. Van der AuweraGA, MauricioOC, HatlC, PoplinR, del AngelG, et al. (2013) From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics 11.10.1–11.10.33.
95. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
96. SieversF, WilmA, DineenD, GibsonTJ, KarplusK, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7 : 539.
97. OssowskiS, SchneebergerK, ClarkRM, LanzC, WarthmannN, et al. (2008) Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res 18 : 2024–2033.
98. AkoulitchevS, ChuikovS, ReinbergD (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407 : 102–106.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



