-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A ABC Transporter Regulates Lifespan
The drug transporters are often known for their ability to transport different physiological-related compounds across cell membranes. Although the abnormal up-regulation of some these transporters is believed to be the common cause of the clinic problem called drug resistance, the biological functions of these transporters remain largely unknown. Here we show that a Drosophila homolog of the mammalian drug transporter plays a role in lifespan regulation. Mutations of this gene increase the sensitivity to oxidative stress and reduce lifespan, while overexpression of this gene increases resistance to oxidative stress and extends lifespan. By molecular and genetic analyses, we have linked functions of this gene to a key signaling transduction pathway that has been known to be important in lifespan regulation.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004844
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004844Summary
The drug transporters are often known for their ability to transport different physiological-related compounds across cell membranes. Although the abnormal up-regulation of some these transporters is believed to be the common cause of the clinic problem called drug resistance, the biological functions of these transporters remain largely unknown. Here we show that a Drosophila homolog of the mammalian drug transporter plays a role in lifespan regulation. Mutations of this gene increase the sensitivity to oxidative stress and reduce lifespan, while overexpression of this gene increases resistance to oxidative stress and extends lifespan. By molecular and genetic analyses, we have linked functions of this gene to a key signaling transduction pathway that has been known to be important in lifespan regulation.
Introduction
In Drosophila, one important feature of the aging process appears to be the similarity between the changes in gene expression that occur during aging and oxidative stress response [1], [2], [3]. For instance, the up-regulation of genes encoding for some chaperones and/or detoxification agents in response to oxidative stress has been found to highly correlate with the aging process [1], [2], [3]. Hsp proteins may promote longevity by facilitating the clearance of damaged proteins that accumulate during aging [4]. Another example is the JNK signaling pathway which can be triggered by a variety of insults, including oxidative stress, and has been shown to be a genetic determinant of aging in Drosophila [5]. Mutations in the JNK cascade increase stress sensitivity and lead to shortened lifespan. Conversely, flies with increased JNK activity can sustain oxidative stress and live longer [6]. Although genome-wide surveys [1] are powerful and have linked a set of genes between stress response and aging, the majority of them have not been tested experimentally for lifespan; some genes involved in both processes may still be missing by genome-wide surveys. Here we report that a new gene, namely dMRP4, which has not been reported on the survey list [1], clearly plays a role in both aging process and oxidative stress.
The multidrug resistance-associated protein 4 (MRP4) belongs to the subfamily C (also known as ABCC) of the ATP-binding cassette (ABC) transporter protein family. It has been classified as a detoxification protein that is implicated in transport of structurally diverse endogenous and xenobiotic compounds, including antivirus and anticancer drugs that usually induce oxidative stress in cells and lead to toxicity [7], [8], [9]. MRP4 mRNA and protein are widely expressed in many tissues of mammals including humans [10], suggesting that this transporter may be involved in different physiological processes. However, several recent studies have shown that mammalian MRP4 is not essential for development, since MRP4-knockout mice are viable and do not reveal any abnormalities [11], [12], [13], [14]. Therefore, the biological function of MRP4 remains largely unknown.
MRP-associated drug resistance has represented an important clinical problem in the treatment of cancers. Some cancer cells seem to adopt a survival strategy to protect against chemotherapy-induced oxidative stress by increasing transport of chemotherapeutics out of cells, as a result of induction of MRP, including MRP4 [15], [16], [17], [18], [19]. Indeed, up-regulation of MRP4 expression has been linked to a variety of human cancers [20], [21], [22], [23], [24]. The induction of hepatic MRP4 by oxidative stress has also been observed in mammalian liver injury after chemical treatments and this response appears to be regulated primarily at a transcriptional level [25], [26]. However, oxidative stress-inducing agents do not always induce MRP4 [27], [28], [29], [30], raising the possibility that the induction of MRP4 expression during oxidative stress may be agent-dependent and/or cell type-specific. Furthermore, no study has attempted to address whether MRP4 is required for general oxidative stress resistance at a whole organismal level.
We have previously identified the Drosophila homolog of mammalian MRP4, called dMRP4, during an unbiased screen for genes whose overexpression causes an abnormal response to hypoxia in adult flies [31]. dMRP4 encodes a protein sharing 43% overall amino acid identity and 63% similarity with the human MRP4 [32], [33]. In this study, we have investigated the possible involvement of dMRP4 in resistance to oxidative stress. By genetic manipulation, we present evidence that dMRP4 is associated with changes in lifespan under both oxidative stress and normal conditions, likely through a mechanism that is linked to JNK signaling in Drosophila.
Results
dMRP4 is an oxidative stress-responsive gene and is required for oxidative stress resistance
To test our hypothesis that the expression of dMRP4 may be regulated by oxidative stress in Drosophila, we first analyzed dMRP4 transcriptional activity in response to oxidative stimuli by feeding flies with paraquat, which generates superoxide in mitochondria [34] and has been widely used as an oxidative stress inducer in vivo. The expression of dMRP4 was strongly induced in wild-type flies fed with 10 mM paraquat for 12 hours (Fig. 1A). Similar induction patterns were observed in parallel with two known oxidative stress-responsive genes [3], [6], [35], puc (puckered) and gstD1 (glutathione s transferase D1). To test whether dMRP4 responds to other oxidative stressors, we analyzed its transcriptional changes in flies treated with hydrogen peroxide as well as hyperoxia. Up-regualtion of dMRP4 was clearly observed after hydrogen peroxide or hyperoxia treatment, in parallel with two known up-regulated markers, gstD1 and hsp22, under these conditions [1] (Fig. 1B–C). These results indicate that Drosophila dMRP4 is a bona fide oxidative stress-responsive gene.
Fig. 1. dMRP4 is up-regulated in response to oxidative stress. 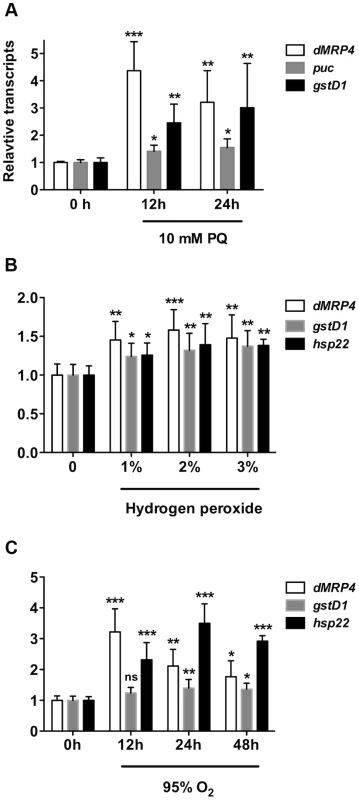
(A) Quantitative RT-PCR analyses of RNA isolated from wild-type flies (w1118) after exposed to paraquat (A), hydrogen peroxide (B,) or hyperoxia (C) for indicated times. Data is showed as means ± S.D. from at least 5 independent experiments. One way ANOVA followed by post hoc t-test: * p<0.05, ** p<0.01, *** p<0.001, ns: No significance (p>0.05). To test whether dMRP4 indeed might play a role in oxidative stress resistance, we generated two mutations by excision of two independent EP elements near the dMRP4 gene (Fig. 2A). Analysis of the dMRP4 expression by RT-PCR indicated that dMRP4 RNA was undetectable in these mutants (Fig. 2B). However, the more sensitive assay with qt-PCR revealed about 8% dMRP4 mRNA retaining in both homozygous mutations (Fig. 2C). Currently it is not clear if this transcript residual was resulted from splice forms of the predicted full length mRNA or from an alternative transcription start site of the remaining dMRP4 transcript after the truncation. Nevertheless, these results indicate that the two dMRP4 alleles represent strong loss-of-function mutations. In addition, flies homozygous for both mutations were viable and fertile, suggesting that dMRP4 may not be an essential gene for development. However, it cannot be ruled out that the remaining residual in these mutations might still retain some vital function during development.
Fig. 2. dMRP4 is required for oxidative stress resistance. 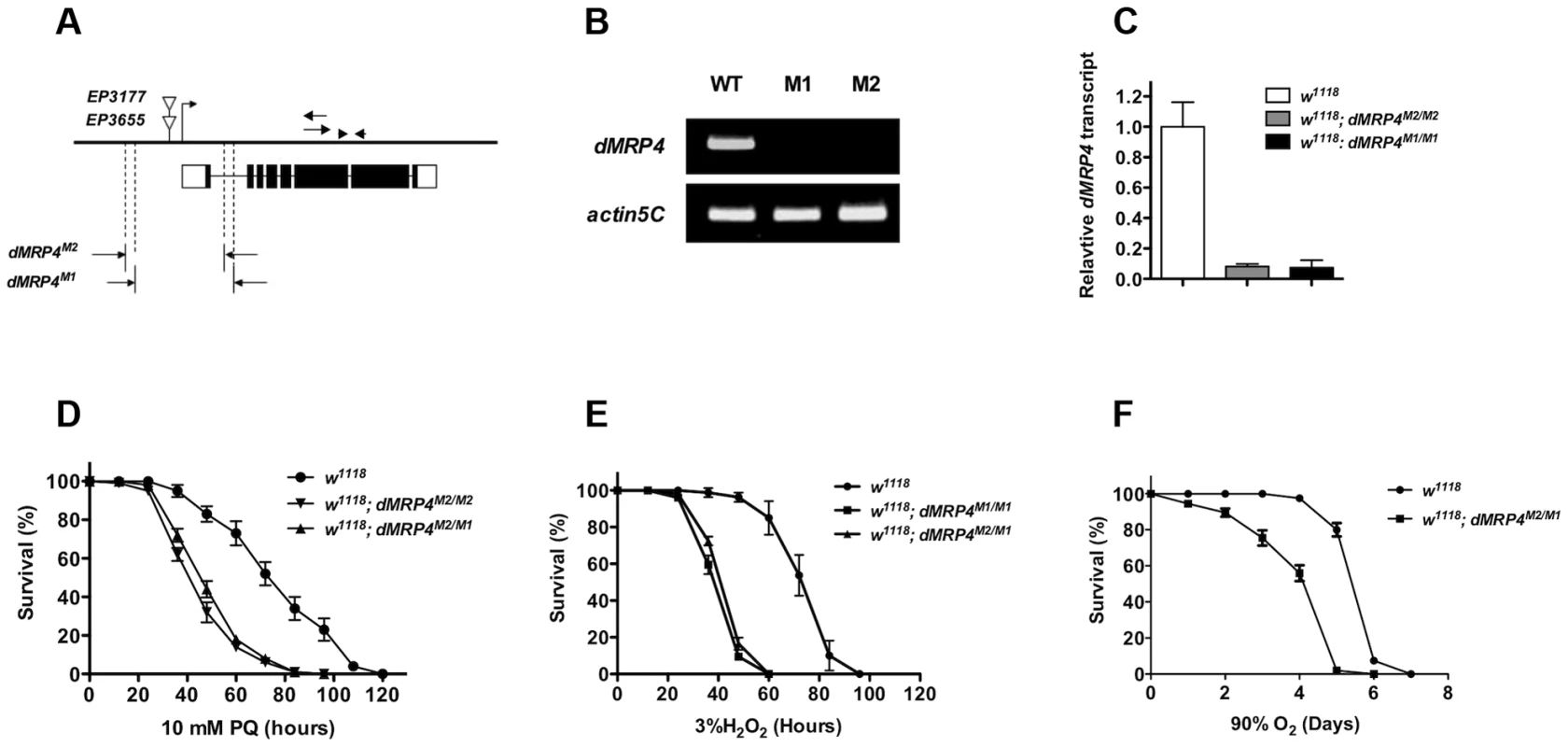
(A) Molecular analysis of dMRP4 mutants. The solid bar represents the genomic region of dMRP4. The bent arrow indicates the transcription start site of dMRP4 gene. The open triangles show the insertion positions of EP3655 and EP3177. Open boxes below the solid bar represent exons of dMRP4 transcript and filled boxes indicate the encoding protein sequences. The span of both deletions was determined by sequencing the corresponding regions with specific primers. The arrows were primers for dMRP4-related semi-quantitative RT-PCR and arrowheads for qt-PCR experiments. The deleted sequences were described in Materials and Methods. (B) Expression of dMRP4 mRNA in two mutant alleles. Semi-quantitative RT-PCR was used to determine the levels of dMRP4 mRNA expression. dMRP4 mRNA was under-detectable in dMRPM1/M1 (M1) or dMRPM2/M2 (M2). Actin5C served as an internal standard. (C) qt-PCR analysis of dMRP4 mRNA in two dMRP4 alleles. (D) Effects of paraquat-induced oxidative stress on dMRP4 mutant flies (n = 180 for each group). (E) Effects of hydrogen peroxide-induced oxidative stress on dMRP4 mutant flies (n = 200 for each group). (F) Effects of hyperoxia-induced oxidative stress on dMRP4 mutant flies (n = 200 for each group). Error bars represent S.E. To address whether induction of dMRP4 is required for defense against oxidative stress, we monitored the survival of adult flies treated with three most commonly used oxidative stressors: paraquat, hydrogen peroxide, or hyperoxia. In each condition the two dMRP4 alleles or their transheterozygous combination displayed similar and reproducible phenotypes: flies lacking dMRP4 reduced profoundly their viability under oxidative stress relative to controls (Fig. 2D–F, Log-rank test, p<0.001). These results demonstrate that wild-type dMRP4 is required for oxidative stress resistance in Drosophila.
dMRP4 is required for JNK-dependent induction of gene expression
Oxidative stress is known to activate a protective program involving induction of a number of stress-responsive genes in cells [3], [6], [35], [36]. JNK signaling is activated in response to oxidative stress and is a major genetic factor in control of oxidative stress tolerance and aging process [3], [6], [35], [36], [37]. Since puc (a phosphatase inhibitor of JNK) is often used as a marker for activation of the JNK pathway [3], [6], [35], [36], [38], we tested whether there were any differential expression changes of JNK signaling by examining puc induction in dMRP4 mutant flies fed with paraquat. Compared to the pattern in wild-type flies, puc expression was completely diminished in dMRP4 mutant flies under oxidative stress (Fig. 3A). To further evaluate whether dMRP4 might play a general role in JNK signaling, induction of other JNK-mediated marker genes, such as gstD1 [6], hsp68 and Jafrac1, was also examined. Although expression of all these marker genes was induced in wild-type flies after paraquat feeding, their induction, with exception for gstD1, was significantly reduced in the dMRP4 mutant flies (Fig. 3C–D), indicating that activation of JNK signaling by oxidative stress requires a wild-type dMRP4 function. Because flies deficient for JNK signaling become more susceptible to stress [6], a phenotype resembling what we have observed with flies deficient for dMRP4, impairment of JNK signaling in dMRP4 mutants may be an important cause for increased lethality when animals face oxidative insults. There was also a possibility that dMRP4 itself may be a component of the JNK pathway.
Fig. 3. dMRP4 is required for JNK-mediated gene expression under oxidative stress. 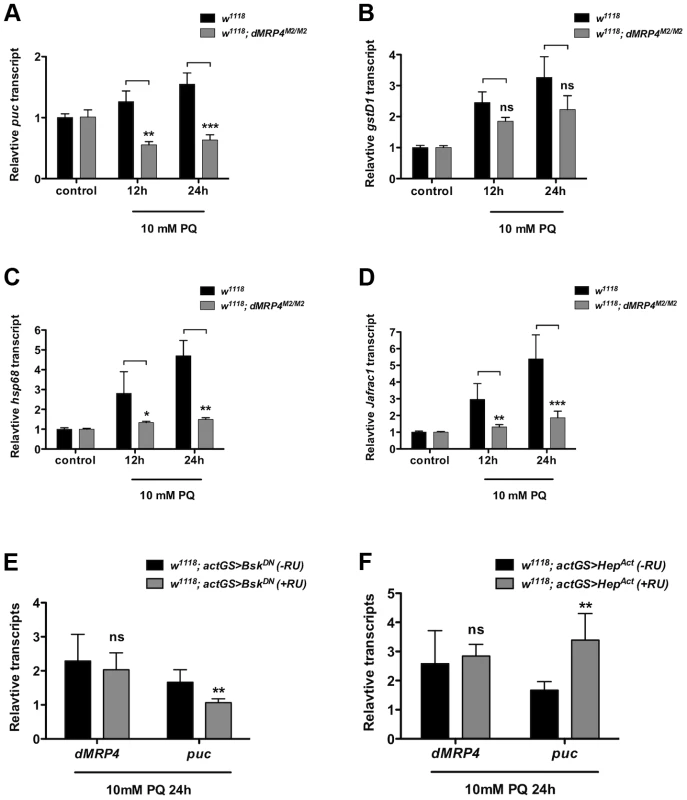
qt-PCR analysis of mRNA isolated from WT flies (w1118) after exposed to 10 mM paraquat (PQ) for indicated time points. (A–D) Relative mRNA levels were compared between w1118 and dMRP4 to the respective controls (no paraquat feeding) where the basal values were set at 1.0. (E) Relative mRNA levels of dMRP4 from flies carrying actGS-BskDN were compared between RU486 feeding (+RU, 150 ug/ml) and non-feeding groups [75]. (F) Relative mRNA levels of dMRP4 from flies carrying actGS-HepAct were compared between RU486 feeding (+RU, 150 ug/ml) and non-feeding groups [75]. puc expression served as a marker for JNK activity in response to paraquat. Data was presented as means ± SD from 3–5 independent experiments. Student's t-test: * p<0.05, ** p<0.01, *** p<0.001. ns: No significance (p>0.05). To test whether dMRP4 might be a component of the JNK pathway, we examined dMRP4 response in flies with reduced activities of JNK signaling by the expression of a dominant negative form of Bsk (BskDN) (Basket, a Drosophila homolog of JNK). BskDN can mimic bsk mutant phenotypes in flies and cells [39]. In this experiment, BskDN expression was induced in adult flies by actin-GeneSwitch-Gal4 (actGS-Gal4), a RU486-mediated system [40] that drives ubiquitous expression in whole fly. In the presence of drug RU486, BskDN expression was activated from the UAS driven transgene. The relative mRNA levels from RU486-fed flies were compared to control flies carrying the same induction system (actGS>dMRP4) without drug feeding. Inhibition of JNK activity by BskDN, as shown by puc expression, did not repress dMRP4 induction in response to paraquat (Fig. 3E), indicating that JNK signaling is not required for dMRP4 induction under this stress. Next we asked whether stimulation of JNK signaling might influence dMRP4 induction. This was achieved by conditionally expressing an activated version of Hep (HepAct) (hemipterus, a Drosophila homolog of JNKK). HepAct has been shown to be a JNK gain-of-function mutant [39]. Constitutive activation of JNK signaling by HepAct did not change dMRP4 expression in paraquat-fed flies relative to controls (Fig. 3F). These results indicate that unlike those direct targets of JNK, dMRP4 induction by paraquat is independent of JNK activity, and therefore dMRP4 is not a direct component, but instead acts in parallel on a signaling that perhaps only regulates expression of some downstream effectors, of the JNK pathway.
Overexpression of dMRP4 in adults confers oxidative resistance
If dMRP4 is essential for oxidative resistance in Drosophila, an increased dMRP4 expression may increase oxidative resistance in wild-type flies. To test this hypothesis, we used the RU486-system to test the role of dMRP4 overexpressing in paraquat resistance. Adult flies carrying tub5GS>dMRP4, after being fed with RU486 for dMRP4 induction (Fig. 4I), significantly improved survival rates following acute treatment with paraquat (30 mM) compared to control flies (Fig. 4A). Importantly, RU486 feeding itself had no effect on survival under the same condition (Fig. 4B). These experiments underline the protective role of dMRP4 from paraquat challenge. It also implies that this protection does not need dMRP4 to be elevated before reaching adulthood.
Fig. 4. Elevated dMRP4 expression increases oxidative resistance. 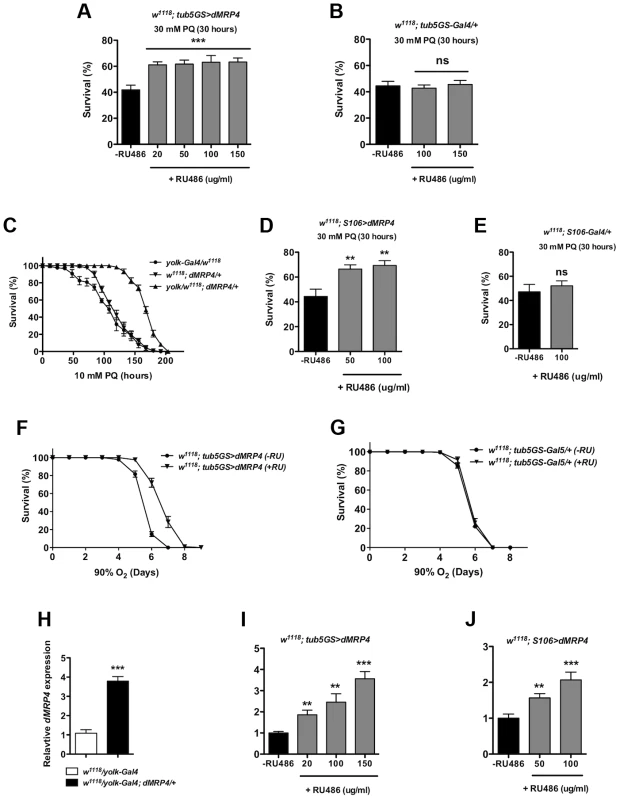
Overexpression of dMRP4 globally (A and F) or tissue-specifically (C–D), significantly promoted adult fly survival of paraquat (PQ)-induced oxidative stress. Since yolk-Gal4 is expressed specifically in the female fat body, female flies were used in the yolk>dMRP4 experiment (C). Male flies were otherwise used in all other experiments. (H–J) qt-PCR analysis of dMRP4 induction by different Gal4 drivers. Concentrations of paraquat and RU486 used in individual experiment were indicated, except for (F–G) where concentration of RU486 used was 150 ug/ml. Student's t-test was used in (E and F) and ANOVA was used in (D) and (I–J). * p<0.05, ** p<0.01, *** p<0.001. ns: No significance (p>0.05). Sample size: (A) tub5GS>dMRP4 [75], n = 160; tub5GS>dMRP4 (+RU), n = 180; (B) tub5GS-Gal4/+ [75], n = 160; tub5GS-Gal4/+ (+RU), n = 160; (C) yolk-Gal4/w1118, n = 160; dMRP4/+, n = 160; yolk-Gal4/w1118; dMRP4/+, n = 160; (D) S106>dMRP4 [75], n = 180; S106>dMRP4 (+RU), n = 180; (E) S106-Gal4/+ [75], n = 160; S106-Gal4/+ (+RU), n = 160. (F) tub5GS>dMRP4 [75], n = 200; tub5GS>dMRP4 (+RU), n = 200; (G) tub5GS-Gal4/+ [75], n = 200; tub5GS-Gal4/+ (+RU), n = 180. Because mammalian MRP4 has been implicated in protecting the liver from oxidative stress [25], [26], we sought to investigate whether it was also the case in Drosophila. Drosophila fat body is an analogous tissue to mammalian liver and white adipose tissue [41], [42]. yolk-Gal4 is expressed specifically in the female fat body [43]. We tested whether overexpression of dMRP4 in the fat body could provide overall protection against oxidative damage to the whole fly. Induction of dMRP4 in female fat body by yolk-Gal4 led 4-fold increase in the dMRP4 transcript (Fig. 4H) and rendered flies much more tolerant to paraquat treatment as compared to controls (yolk-Gal4/+ or dMRP4/+) (Fig. 4C, Log-rank test, p<0.01). Similarly, overexpression of dMRP4 by S106-Gal4, an inducible driver expressed predominantly in adult fat body [40], [44], [45], significantly increased survival of paraquat-fed flies in the presence of RU486 (Fig. 4D). Again, RU486 treatment showed dose-dependent induction of dMRP4 expression (Fig. 4J) but played no role in mortality under the same condition (Fig. 4E). Thus, the Drosophila fat body appears to be an important tissue for dMRP4 to sustain paraquat-induced oxidative stress. Furthermore, the protective role of dMRP4 under paraquat challenge is applicable for both sexes.
The anti-oxidative effect of dMRP4 on lifespan was further tested by exposing flies to hyperoxia. Flies overexpressing dMRP4 by RU484 induction clearly lived longer under 90% oxygen environment compared to controls (Fig. 4F, Log-rank test, p<0.001). We conclude that wild-type dMRP4 function is to promote resistance to oxidative stress in Drosophila.
dMRP4 regulates normal lifespan
Aging shares many features with oxidative stress [1]. The free radical theory has proposed a link between aging and oxidative stress [46], [47]. Recent studies from genetic manipulation of many genes in Drosophila have presented evidence that resistance to oxidative stress genetic often correlate with increased lifespan [6], [48], [49], [50]. Since manipulation of dMRP4 can influence lifespan under oxidative stress, it would be important to examine whether dMRP4 regulates lifespan under non-stress conditions. We observed that mutations in dMRP4 dramatically caused a shortened normal adult lifespan (Fig. 5A, Log-rank test, p<0.0001). In particular, dMRP4M2/M2 flies had a mean lifespan (as measured by 50% survival) of 45 days and a maximum lifespan (as measured by the 90 percent survival) of 60 days. Compared to wild-type controls, dMRP4M2/M2 flies had a major reduction in the mean lifespan of about 47% and a decrease in maximum lifespan of 24% (Fig. 5A). Similar results were observed with dMRP4M1/M1 flies (Fig. 5A). The overall mortality rates of these groups were compared using Partial Slopes Rank-Sum Test [51] over the linear portion of the increase in mortality. Despite an apparent initiation of early mortality before day 30 in survival of dMRP4 mutants, there was no significant difference in slopes between the mutants and wild type (Fig. 5B), indicating that loss of dMRP4 decreased lifespan by lowing the whole mortality trajectory, but not the rate of increase in mortality with age. Thus, although dMRP4 is not required for normal development, it is required for normal lifespan under non-stress conditions.
Fig. 5. dMRP4 affects lifespan. 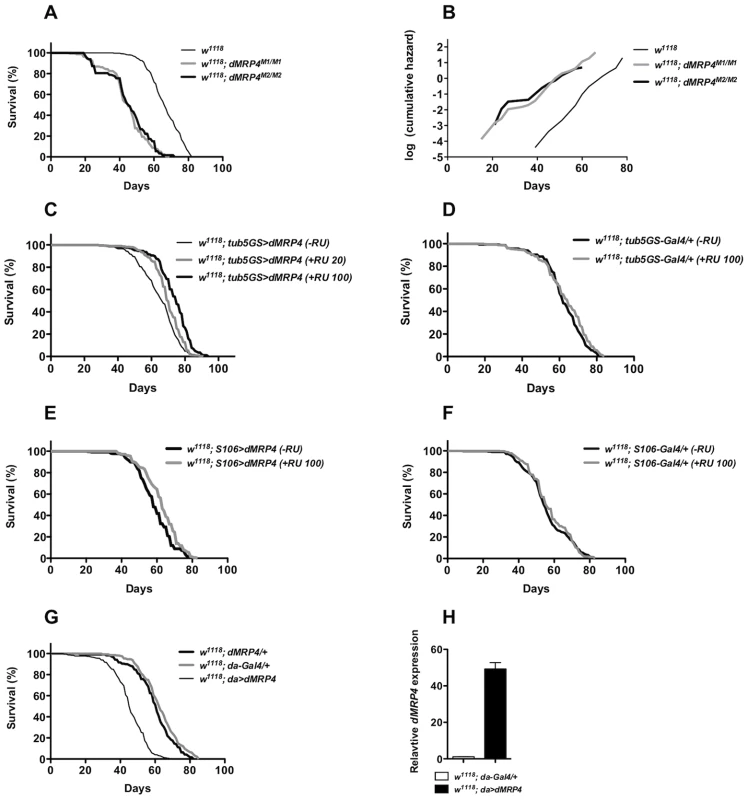
(A) Lifespan of adult flies lacking dMRP4. Survival was presented as mean of at least 300 males with different genotypes. Homozygous dMRP4 mutant flies lived significantly shorter than their sibling controls (Log-rank test, p<0.0001): the mean lifespan (50% mortality) was 45 days for dMRP4M2/M2 and 45 days for dMRP4M2/M1, respectively, compared to 66 days for wild-type control (w1118); the maximum lifespan (90% mortality) was 63 for dMRP4M2/M2 and 60 days for dMRP4M2/M1, respectively, compared to 78 days for wild-type control. (B) Analysis of age-specific mortality. The log cumulative hazard plots were presented for different genotypes of survival data. The ratio of slopes from Partial Slopes Rank-Sum Test: w1118 vs dMRP4M1/M1 = 1.24 (p = 0.2005), w1118 vs dMRP4M2/M2 = 1.32 (p = 0.0947), dMRP4M1/M1 vs dMRP4M2/M2 = 1.07 (p = 0.7182). (C) Lifespan of adult flies with genotype tub5GS>dMRP4 (tubulin5-GS-Gal4/EP3177) between treatments: −RU486 group, mean = 64 days, maximum = 78 days (n = 383), +RU486 group (100 ug/ml), mean = 74 day, maximum = 84 (n = 427), and +RU486 group (20 ug/ml), mean = 70 days, maximum = 82 days (n = 438). The cohort was derived from a combination of two independent cohorts (about 200 flies for each cohort) which were conducted over different time periods, and the individual cohorts were similar to each other. (D) Effects of RU486 treatment on control groups. tub5GS-Gal4/+ (−RU486), mean = 62 days (n = 300) and tub5GS-Gal4/+ (+RU486 100 ug/ml), mean = 62 days (n = 300). (E) Lifespan of adult flies with genotype S106>dMRP4 (S106-Gal4/+; EP3177/+) between treatments: −RU486, mean = 60 days (n = 408), or +RU486, mean = 62 days (n = 460). (F) Effects of RU486 treatment on control groups. S106-Gal4/+ (−RU486), mean = 54 days (n = 300) and S106-Gal4/+ (+RU486 100 ug/ml), mean = 55 days (n = 300). (G) Lifespan of adult flies with genotype da>dMRP4 was compared to the parent controls. The mean lifespan of these flies was 62 days for da-Gal4/+ (n = 340), 61 days for dMRP4/+ (n = 320), and 45 days for da>dMRP4 (n = 380). The maximum lifespan was 77 days for da-Gal4/+, 77 days for dMRP4/+, and 56 days for da>dMRP4. (H) qt-PCR analysis of the dMRP4 expression driven by da-Gal4. Since flies overexpressing dMRP4 were more resistant to oxidative stress, we tested whether overexpressing dMRP4 would be sufficient to extend lifespan. RU486-mediated overexpression was used to minimize the influence of genetic background on lifespan assays. RU486-fed tub5GS>dMRP4 flies lived significantly longer than their siblings without RU486 feeding (Fig. 5C, Log-rank test, p<0.0001). The lifespan extension by tub5GS>dMRP4 expression appeared to be correlated with the dose of RU 486. In one case, the mean lifespan was extended to 16% and the maximum lifespan to 8% (Fig. 5C, RU486 100 ug/ml). In the other case, when flies were fed with 20 ug/ml RU486, this group of flies showed only about 9% of increase in the mean lifespan and 5% of increase in the maximum lifespan, even though their overall lifespan appeared to significantly increase (Fig. 5C, Log-rank test, p<0.0001). Increased lifespan was not due to chronic RU486 treatment because no significant difference in lifespan was seen between treated or untreated tub5GS-Gal4 groups (Fig. 5D, Log-rank test, p = 0.3). We conclude that another dMRP4 function is to promote normal lifespan in Drosophila.
In these experiments the lifespan extension clearly correlated with increased expression of dMRP4, but it remained unclear whether tissue-specific dMRP4 overexpression was sufficient to extend lifespan and whether the overall levels and/or timing of such expression would be critical. Interestingly, S106>dMRP4 flies treated with RU486 did not live longer (Fig. 5E, Log-rank test, p = 0.37) even though the fat body-specific expression of dMRP4 did show resistance to paraquat, suggesting that there might be different requirements between resistance to oxidative stress and lifespan extension. Again, RU486 treatment showed no difference between parallel controls (Fig. 5F, Log-rank test, p = 0.09). Moreover, high levels of ubiquitous dMRP4 expression by da-Gal4 throughout development were not beneficial and instead, there was a negative correlation with lifespan (Fig. 5G, Log-rank test, p<0.0001). These observations suggest that in order for dMRP4 overexpression to be beneficial for lifespan extension, the spatial and temporal such expression with proper levels have to be tightly controlled.
dMRP4 regulates the basal transcription of stress - and longevity - associated genes
In order to learn the molecular mechanism by which dMRP4 regulates lifespan, we selectively studied transcription profiling of several genes whose expression changes have been linked to both aging and stress [1]. Among five hsp (heat shock protein) genes examined, expression of three genes, hsp68, hsp70 and l(2)efl (lethal (2) essential for life, a small hsp gene) was severely down-regulated in dMRP4 mutant flies (Fig. 6A), while they were significantly up-regulated when dMRP4 was overexpressed (Fig. 6B). Overexpression of dMRP4 was also sufficient to increase expression of other two hsp genes, hsp22 and hsp83 (Fig. 6B). Since l(2)efl is a known target of dFOXO (Drosophila forkhead transcription factor) in lifespan regulation [52], it raised the possibility that dMRP4 might regulate expression of other dFOXO-dependent genes. Indeed, expression of the dFOXO target gene thor, which encodes 4E-BP (eIF4E binding protein), was also greatly enhanced when dMRP4 was overexpressed. Since both thor and hsp68 are target genes of JNK signaling [6], [52], we further examined expression patterns of several other JNK targets (Fig. 3A–D). Like hsp68, basal expression of puc and gstD1 was down-regulated in dMRP4 mutant flies and was up-regulated with dMRP4 overexpression (Fig. 6A–B). Furthermore, basal expression of Jafrac1 was increased when dMRP4 was overexpressed, even though its expression was not affected by dMRP4 mutation under normal condition. Thus, in addition to regulating the JNK-dependent gene expression under oxidative stress, dMRP4 also regulates the basal transcription of such genes under normal conditions.
Fig. 6. dMRP4 regulates expression of some stress- and aging-related genes. 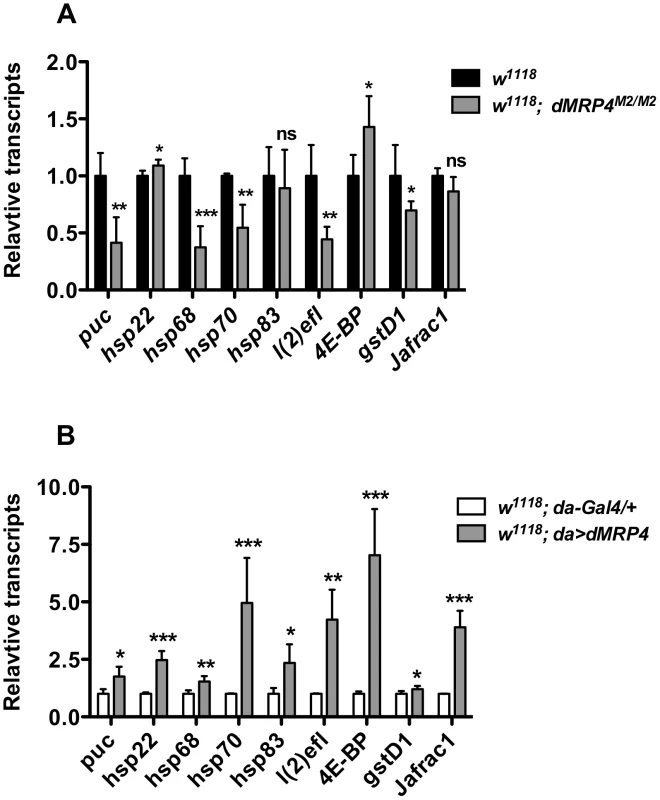
(A) dMRP4 was required for basal expression of several stress- and aging-related genes. The mRNA levels of these genes were compared between w1118 and dMRP4 mutant (dMRP4M2/M2) by qt-PCR analysis. (B) Elevated expression of dMRP4 increased the basal expression of stress- and aging-related genes. The mRNA levels of these genes were compared between control Gal4 (da/+) and dMRP4 overexpression (da>dMRP4) by qt-PCR analysis. Data was presented as means ± SD from 3–5 independent experiments. Student's t-test: * p<0.05, ** p<0.01, *** p<0.001. ns: No significance (p>0.05). Increased expression of hsp22 [53], hsp68 [6], [54], hsp70 [55], l(2)efl [52], Jafrac1 [54], [56], has been reported to increase Drosophila lifespan. We hence suggest that increased expression of these genes by elevated dMRP4 expression may account for, at least in part, the dMRP4-mediated lifespan extension.
dMRP4 regulates the aging process
Increasing age is accompanied with physiological decline. The locomotor decline is one of prominent physiological changes as they grow older. The climbing ability, measured by negative geotaxis, of adult fly reflects a function of age in Drosophila [57], [58]. To determine whether the onset of aging associated with dMRP4, we performed a negative geotaxis test for flies with different ages. Although there was no difference in negative geotaxis behavior between 5-days old dMRP4 mutant and wild-type adults, the age-associated functional decline became visible in dMRP4 mutant flies already at day 10 of adulthood, at a time when no mortality was seen regardless of mutant or wild-type controls (Fig. 7C). By age 40 days, although there was a progressive functional decline in the control group, it was clearly worse in dMRP4 mutant groups (w1118 vs dMRP4M2/M1, Fig. 7C). Thus, the functional decline as they aged was faster in dMRP4 mutants than in controls.
Fig. 7. Extending lifespan of dMRP4 deficiency by a mild increase in JNK signaling under both paraquat resistance and normal condition. 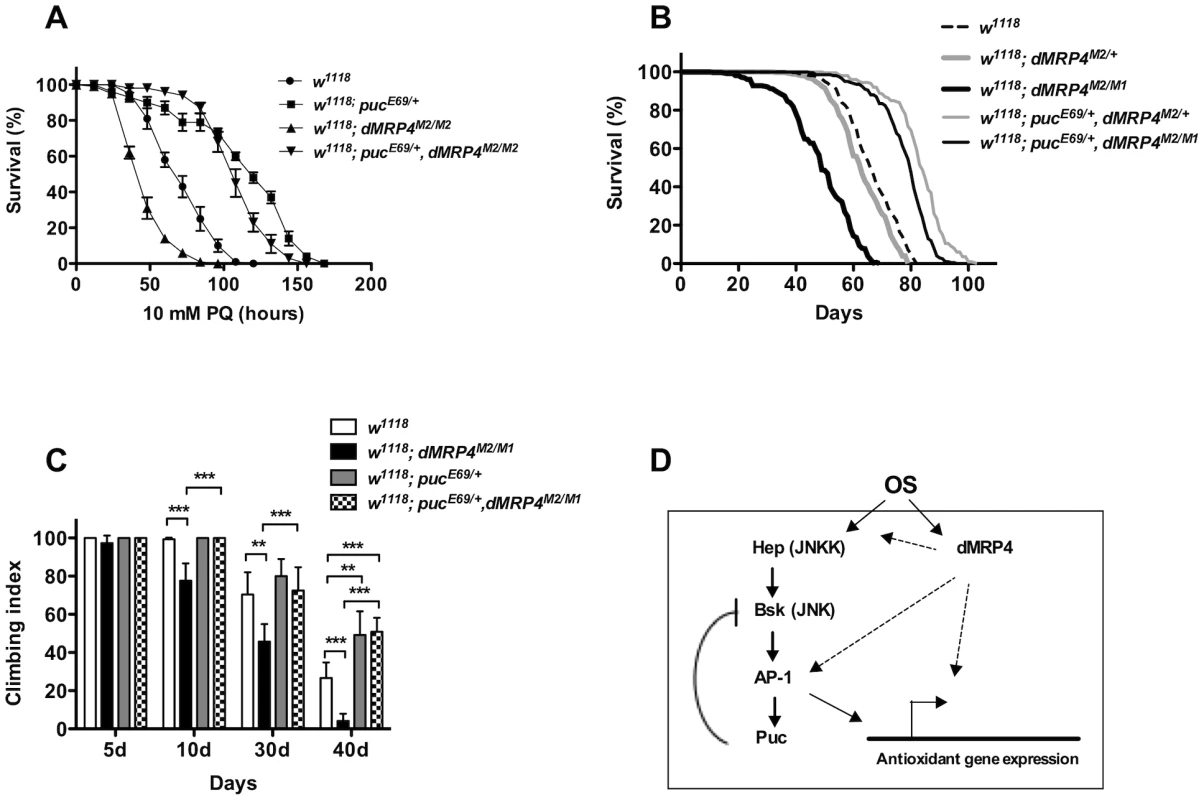
Flies heterozygous for puc (pucE69/+) were significantly resistant to paraquat-induced oxidative stress (A) and had a remarkably longer lifespan under non-stress condition (B) compared to controls. (A) The lifespan of dMRP4 mutant flies (dMRP4M2/M1) under paraquat stress was compared to control (w1118) and puc, dMRP4 double mutant flies (pucE69/+, dMRP4M2/M1). Each group represented 180 flies. (B) The lifespan of dMRP4 mutant flies (dMRP4M2/M1) under normal condition was compared to controls (dMRP4M2/+ and w1118) and puc, dMRP4 double mutant flies (pucE69/+, dMRP4M2/M1). The mean lifespan (50% mortality) was 48 days for dMRP4M2/M1 (n = 360), 62 days for heterozygous control dMRP4M2/+ (n = 360), and 67 days for w1118 (n = 300), 83 days for pucE69/+, dMRP4M2/+ (n = 320), and 79 days for pucE69/+, dMRP4M2/M1 (n = 360). The maximum lifespan (90% mortality) was 62 days for dMRP4M2/M1, 75 days for heterozygous control (dMRP4M2/+), 93 days for flies pucE69/+, dMRP4M2/+), 77 days for w1118, and 88 days for flies (pucE69/+, dMRP4M2/M1). (C) The locomotor defect, indicated by negative geotaxis performance, of dMRP4 mutant flies was completely restored by pucE69/+. Each column was derived from a pool of 80–100 male flies with indicated ages. Student's t-test: ** p<0.01, *** p<0.001. Error bars were S.D. (D) A model for role of dMRP4 in JNK-mediated oxidative resistance and lifespan extension. Paraquat-induced oxidative stress (OS) was sensed through JNK and dMRP4, respectively. The OS signaling may then be converged at levels of the AP-1 transcription factors, the major target of JNK, because dMRP4 is necessary and sufficient for transcription of some JNK-dependent antioxidant genes and because genetic manipulation of puc can fully rescue dMRP4 mutant phenotypes under both oxidative and normal conditions. However, it is also possible that dMRP4 is required for JNK to fully respond to OS at any levels upstream of AP-1. Involvement of dMRP4 in regulation of other JNK-independent antioxidant genes cannot be excluded. The solid arrows represent the work from published studies, and dash arrows indicate the work from this study. Activation of JNK signaling rescues dMRP4 deficiency
Activation of JNK signaling can increase stress resistance and extend lifespan in both Drosophila [6], [52], [59], and C.elegans [60]. Our observations (Fig. 3A–D, Fig. 6A–B) suggest that the deficiency in basal transcription and stress response of JNK signaling may be an important cause for loss of stress tolerance and normal lifespan with dMRP4 mutant flies. If this were the case, increasing JNK signaling might be expected to correct dMRP4 deficiency. We tested this hypothesis by recombination of a pucE69 chromosome into the dMRP4 mutant background. pucE69/+ flies were more resistant to paraquat and lived longer under normal conditions [6] (Fig. 6A and B). When dMRP4 mutant flies also heterozygous for pucE69 were challenged with paraquat, they behaved like pucE69/+ flies alone: they lived significantly longer not only than dMRP4 mutant flies, but also longer than wild-type controls (Fig. 7A, p<0.01). Consistent with a previous report [6], pucE69/+ flies extended normal lifespan (27% mean lifespan and 24% maximum lifespan) of control flies (dMRP4M2/+) under non-stress conditions (Fig. 7B, p<0.0001). More strikingly, the puc, dMRP4 double mutant flies remarkably extended the mutant mean lifespan by 61% (dMRP4M2/M1 vs pucE69/+, dMRP4M2/M1) and maximum lifespan by 42% (Fig. 7B, p<0.0001). These results demonstrate that dMRP4 deficiency in stress resistance and lifespan regulation is correlated with a defect in JNK signaling. These results also place puc genetically in epistatic interaction with dMRP4 in both stress resistance and lifespan regulation.
We tested whether the functional decline with age might also be associated with JNK activity by comparing the climbing ability between wild-type and pucE69/+ flies. Increased JNK signaling did not appear to benefit wild-type flies before 30 days of age, as climbing tests did not reveal a significant difference in locomotor function between wild-type and pucE69/+ flies (Fig. 7C). However, after 40 days of age, increased JNK activity indeed improved climbing ability, and therefore functional aging in wild-type flies (w1118 vs pucE69/+ in the 40 d group, Fig. 7C), suggesting that JNK activity is required for fitness of older flies. We then tested whether the age-associated functional decline of dMRP4 mutants could be caused by impaired JNK signaling as well. The climbing ability of puc, dMRP4 double mutant flies was restored to the level comparable to that of wild-type flies in the first 30 days of age. Therefore, early functional decline of dMRP4 mutants is possibly associated with a decline of JNK signaling (Fig. 7C). Furthermore, by age of 40 days, puc, dMRP4 double mutant flies behaved like pucE69/+ flies, showing better climbing performance even over wild-type flies (Fig. 7C). Thus, the JNK activity can seemingly rescue all defects that are associated with dMRP4 phenotypes. We conclude that dMRP4 plays a critical role in regulation of JNK-mediated oxidative resistance and aging process.
Discussion
The MRP4 subfamily and its homologs have not been reported in any lifespan-related studies including genome-wide surveys. In this study, we have investigated the physiological function of dMRP4 gene in Drosophila. A main finding from our work is that dMRP4 regulates lifespan under both normal conditions and oxidative stress, concomitantly with changes of JNK activity in vivo. Our main finding is based on several observations: First, dMRP4 is required for induction of some JNK-dependent genes in response to paraquat-induced oxidative stress. Second, elevated dMRP4 expression stimulates basal transcription of some JNK-dependent genes downstream of JNK signaling. Third, increased JNK activity in dMRP4 mutant background can rescue dMRP4-related phenotypes identified in this work, supporting our hypothesis that dMRP4 may regulate oxidative resistance and lifespan, at least in part, through JNK signaling.
The finding that dMRP4 has a role in lifespan is particularly intriguing because we are able to show for the first time that a drug transporter like MRP4 is involved in lifespan regulation. Like Drosophila dMRP4, MRP4 KO mice show no visible phenotype [11], [12], [13], [14], and mrp-4 knockdown in C.elegans with RNAi results in no observed phenotype either [61], [62]. These observations together suggest that MRP4 and its homologs across species do not contribute to normal development in the animal world. However, unlike in other species, we found that the Drosophila dMRP4 is required for adult lifespan. Flies deficient for dMRP4 live significantly shorter, under both stressful and normal conditions. Subsequently, our work reveals that dMRP4 acts as a modulator of a network of gene expression since loss - or gain-of dMRP4 function leads to major changes in the transcriptional profiling of a number of genes that may contribute to lifespan regulation. Therefore we suggest that gene expression changes mediated by dMRP4 may represent a molecular mechanism by which dMRP4 regulates lifespan. For instance, hsp genes have been implicated in regulation of both stress resistance and lifespan extension [4], [63], and are among the best-known biomarkers of aging in C.elegans [63], [64], in Drosophila [1], [65], and perhaps even in humans [66]. Given the fact that the expression of hsp reporters in young individual flies has been observed to be partially predictive of remaining lifespan [65], down-regulation of several hsp gene expression (i.e. hsp68, hsp70, l(2)efl) in dMRP4 mutant background could explain the shorter lifespan of these flies, while their up-regulation (i.e. hsp22, hsp68, hsp70, hsp83, l(2)efl) at a young age by dMRP4 overexpression may help protect against oxidative stress and extend lifespan of wild-type flies. This scenario is consistent with previous notions that genes are involved in stress responses generally share similar involvement with aging [1].
In addition to hsp genes, the interaction of dMRP4 with JNK signaling may provide an alternative mechanism to explain dMRP4 functions. Because the JNK pathway is known to be crucial in stress resistance and aging, impairment of JNK signaling in dMRP4 mutant flies, indicated by transcriptional down-regulation of several known JNK-related effecters, could result in dMRP4-associated phenotypes. The acute phenotype is seen particularly when the animal faces stressors such as paraquat-induced oxidative stress, which recapitulates the phenotype shown by mutations in the JNK pathway [6]. The effect of the JNK pathway on lifespan has also been observed during aging under normal conditions. Flies with reduced JNK activity have a shorter lifespan [6], a phenotype similar to that seen in dMRP4 mutant flies. Furthermore, some downstream effectors in the JNK pathway also exhibit phenotypes that are reminiscent of dMRP4. For instance, loss of Jafrac1 function leads to an exaggerated sensitivity to paraquat-induced oxidative stress and a shortened lifespan, while overexpression of Jafrac1 increases oxidative resistance and extends lifespan [56]. Interestingly, expression of Jafrac1 transcription is down-regulated in the dMRP4 mutant in response to oxidative stress (Fig. 2D) and is up-regulated by dMRP4 overexpression (Fig. 6B). How dMRP4 regulates Jafrac1 remains to be investigated. One possible scenario is that dMRP4 executes its functions through interacting with JNK signaling to modulate the expression of downstream effectors such as Jafrac1 especially that the expression of Jafrac1 itself is regulated by JNK signaling [56]. After all, the most compelling evidence for the relationship between dMRP4 and JNK signaling comes from our genetic epistatic assays. When JNK signaling is enhanced in dMRP4 mutant background, all dMRP4-related defects are restored, and puc, dMRP4 double mutant flies now phenocopy pucE69/+ flies, clearly proving that JNK signaling plays a central role in realizing dMRP4 functions. Our work also suggests that promoting lifespan by increasing JNK signaling may be a result of its ability to antagonize oxidation on macromolecules, thereby postponing aging. Compared to JNK signaling, the effect of increased dMRP4 expression on lifespan extension seems less dramatic. Yet this phenotype, together with the results showing that loss - or gain-of JNK function does not alter dMRP4 expression, indicates that dMRP4 functions as a modulator of, but not a component within, JNK signaling. Furthermore, if dMRP4 is one of upstream modulators of JNK/Puc signaling, it is conceivable that its overexpression cannot entirely recapitulate the effect of JNK/Puc activation and consequently, it may not be as effective as a direct manipulation of JNK/Puc signaling with respect to lifespan.
Together our results, we propose a working model to summarize how dMRP4 executes its functions in conjunction with JNK signaling (Fig. 7D). Future work needs to explore how a transmembrane protein such as dMRP4 could integrate its signal into the JNK pathway under both stress and normal conditions.
Although in human and mammalian models of cholestasis, MRP4 has been implicated in providing protection against oxidative stress, the genetic basis for this resistance has not yet been addressed. Therefore, the connection between tissue oxidative stress, survival of the animal, and the physiological function of MRP4, has been lacking. In this work we show that overexpression of dMRP4 in Drosophila fat body, the equivalent tissue of mammalian liver and white adipose tissue, can confer oxidative resistance to the whole animal, suggesting a functional importance of dMRP4 in the fat body in the protection of Drosophila against oxidative stress. Drosophila fat body has recently been reported as a primary site of lipid oxidative damage after paraquat treatment [67]. dFOXO, whose expression is predominately restricted to the fat body, appears to regulate sensitivity of paraquat-induced oxidative damage and age-associated degeneration of behavioral rhythms through this tissue [68]. Furthermore, overexpression of dFOXO in the adult fat body can increase stress resistance and retard aging process [44], [69], supporting the physiological role of fat body in stress defense for the whole organism. Strikingly, we show in this work that expression of two targets of dFOXO, l()efl and thor, are greatly induced when dMRP4 is overexpressed, raising the intriguing possibility that dMRP4 may promote stress resistance and lifespan extension by activation of dFOXO, for instance through JNK signaling [52]. However, unlike the finding that global induction of dMRP4 can promote lifespan, we have not observed a significant lifespan extension when dMRP4 overexpression is restricted in fat body. This observation suggests that the ability of stress resistance may not be an absolute factor associated with longevity in a particular tissue. It is also possible that in order for dMRP4 to benefit for longer life, more tissues with its elevated expression need to be involved. Our studies in fact have not ruled out the roles of dMRP4 in tissues other than the fat body to survival even under oxidative stress.
The main function of MRP4 family is known for their ability to transport a variety of diverse endogenous and xenobiotic compounds. An interesting speculation could be raised as to whether dMRP4 might function simply as a transport in paraquat resistance. In this scenario, flies deficient in dMRP4 might not be able to efficiently exclude paraquat out of cells, thereby leading to substrate-related toxic effects. However, this assumption would hardly explain why flies deficient in dMRP4 lose their resistance to hydrogen peroxide and hyperoxia. In addition, there is no report for paraquat as a potential substrate of any MRP4 members thus far.
The deteriorate influence by da>dMRP4 overexpression is notable because this phenotype has not been seen in overexpression studies of mammalian MRP4. Although use of the whole animal in this study clearly differs from use of cultured cells in mammalian researches, it is more likely that high levels of dMRP4 expression may interfere with normal development, resulting in a pleotropic impact on later assays. An early report did observed that overexpression of two EP lines, which all targeted dMRP4, in larvae caused neuromuscular phenotypes [70].
Given the considerable conservation of pathways between Drosophila and mammals, it will be interesting to test if manipulating MRP4 in mammalian liver cells could confer resistance to the liver, or even to the whole animal subjected to chemotherapy-induced oxidative stress. Finally, our proposed mechanism that interactions between dMRP4 and JNK signaling may shed new light on the clinic problems for long-lived cancer cells with drug resistance due to elevated expression of MRP including MRP4 proteins.
Materials and Methods
Fly strains and genetics
EP3177 and EP3655 were described previously [31]. Other stocks: w1118, w; TM3,Sb,Ser/TM6B,Tb, w; Sco/CyO; MKRS/TM6B,Tb, daughterless (da)-Gal4, S106-Gal4, pucE69, UAS-BskDN and UAS-HepAct strains were obtained from Bloomington stock center. These strains have been backcrossed to w1118 for 8–10 times before experiments. yolk-Gal4 [43] was kindly provided by Norbert Perrimon and was backcrossed into w1118 background for 8 times. Actin-GeneSwitch-Gal4 (actGS-Gal4, [71], [72]) was a gift from Dirk Bohmann. tublin5-GeneSwitch-Gal4 (tub5GS-Gal4, [73]) was a gift from Scott Pletcher. These Gal4 strains have been backcrossed into w1118 background for 6 times before use. Flies were raised on standard Drosophila food (per liter: 17.3 g of yeast, 73.1 g of cornmeal, 10 g of soy flour, 77 ml of light corn syrup, 4.8 ml of propionic acid, and 5.7 g of agar).
To generate dMRP4 mutant flies, two independent EP lines, EP3177 and EP3655 were first backcrossed into w1118 background for 8 times. EP males were crossing to w1118; Δ2-3 Sb/TM3 females that provides with transposase. Males with mosaic color eyes were excised and subsequently balanced with w1118; TM3,Sb,Ser/TM6B,Tb strain. The balanced excisions were then repeatedly backcrossed via the balancer strain for 8 times to establish excision stocks. They were identified by loss of the expression of the mini-white gene. The genomic deletions were determined by sequencing with specific primers spanning the EP insertion region. Two deletions obtained had truncated the 5′-end of putative dMRP4 transcript, which was designated as dMRP4 mutation 1 (w1118; dMRP4M1) and dMRP4 mutation 2 (w1118; dMRP4M2) (Fig. 2A). dMRP4M1 was excised from EP3655, which inserted at 47 bp from the transcription start site of the predicted gene CG14709, resulting a 2.7 kb deletion that removed 1179 bp upstream of dMRP4 transcript and a 1521 bp region including 585 bp of the entire exon 1 encoding the first 25 amino acids of the protein, as well as 936 bp of the intron 1. dMRP4M2 was resulted from an excision of EP3177, which inserted at 88 bp from the transcription start site of the predicted gene CG14709. This led to a 3 kb deletion that has removed 2117 bp upstream of dMRP4 transcript and an 883 bp region spanning the entire exon 1 and part of intron 1.
The pucE69, dMRPM2 recombination strain was generated by recombination of pucE69 and dMRPM2 onto the same 3rd chromosome. Both the balanced pucE69 and dMRPM2 were repeatedly backcrossed via w1118; TM3,Sb,Ser/TM6B,Tb for 8 times before the recombination experiments. The presence of both mutations after meiotic recombination was verified by genetic cross and by PCR with specific primers. Resultant pucE69/+, dMRPM2/M2 double mutants were then continuously backcrossed via w1118; TM3,Sb,Ser/TM6B,Tb for more than 10 times and were kept with the balancer as a parent stock.
To induce dMRP4 overexpression, adult flies carrying different Gal4 drives were crossed to homozygous EP3177 lines. For RU486 induction, a 25 mg/ml RU486 (mifepristone, Sigma) stock solution made in 100% ethanol was diluted with water for desired concentrations. 250 ul of diluted RU486 solution was added onto the surface of standard fly food. This “on food” method has been shown to be simple and effective over other RU486 supply methods [74]. The vials were allowed to dry for 24 hours before use. The same solution without RU486 was added to fly food for control experiments.
Paraquat treatment
In most experiments, three to four day-old males, grouped with 20 flies per vial, were fed on a 3 mm Whatmann paper soaked with 10 mM paraquat (N,N′-dimethyl-4,4′-bipyridinium dichloride, Sigma) in 5% sucrose/PBS. Flies of different genotypes were also fed only with 5% sucrose/PBS as experimental controls. Under this condition all flies can live up for 10 days perfectly. Scores were done every 12 hours for the number of dead flies. Fresh paraquat was added daily. All tests were performed at 25°C. Flies were not starved before adding paraquat in this test to avoid unnecessary stress. Survival comparisons were analyzed by Kaplan–Meier Log-rank Test using Graph Pad Prism4. p<0.05 was considered statistically significant.
In RU486-induced experiments, 20 adult males (2–4 days old) per vial were fed with different concentrations of RU486 for 4–6 days. They were then transferred on a 3 mm Whatmann paper soaked with 30 mM paraquat in 5% sucrose for acute survival test, or with 10 mM paraquat in 5% sucrose for mRNA induction at 24 hours. Control flies were from the same collection and were treated in parallel.
For RNA, all samples were collected at the end of treatments and were immediately frozen in dry ice for RNA preparations.
Hydrogen peroxide treatment
Eight day-old males were fed with different concentrations of hydrogen peroxide (v/v, Sigma) in 5% sucrose/PBS. Control flies were fed with 5% sucrose/PBS only. RNA for qt-PCR was extracted from these flies after 24 hours treatment. For survival tests, ten day-old males with different genotypes were fed with 3% hydrogen peroxide. Fresh hydrogen peroxide was added every day. Scoring and analysis were done essentially as described in paraquat treatment.
Hyperoxia treatment
Eight day-old males, grouped with 20 flies per vial on regular food, were exposed to a steady flow of 95% or 90% oxygen bubbled through water in a sealed chamber. RNA for qt-PCR was extracted from these flies after indicated time points. For survival tests, twelve day-old males with different genotypes were treated with 90% oxygen as above. For RU486 induction, flies from the same breeding were divided into two groups, one group fed on food containing RU486 (150 ug/ml) and the other on normal food through the experiments. Flies were transferred to fresh vials every 2–3 days. Scoring was done every day.
Lifespan
Flies were collected within 24 hours of eclosion and grouped into 20 males per vial. Tests were performed at 25°C. For each experiment, at least 200 flies of each genotype were tested.
For GeneSwitch experiments, males of genotypes w1118; tub5GS-Gal4, w1118; actGS-Gal4, or w1118; S106-Gal4 were crossed to w1118; EP3177 or w1118 females, respectively. Male progeny from these crosses were aged for 3 days after eclosion, and then were divided into 20 flies per vial, with or without indicated concentrations of RU486 in food. Flies were transferred to fresh vials with or without RU486 every other day and dead flies were scored at the time of transfer.
All experiments were conducted at least two times from independent biological breeding. The maximum lifespan was the mean lifespan of last 10% of survival animals in each cohort.
Negative geotaxis
10–20 male flies, ages from 5–40 days at 25°C, were transferred to a clean plastic vial, rested for 3 min, and then measured for bang-induced vertical climbing distance at room temperature (20–21°C). The performance was scored as percentage of flies crossing 7 cm within 10 seconds in a single vial, which was expressed as average of 5 repeated tests for a single vial. 80–100 flies were tested for each genotype at each time point.
RT-PCR
Total RNA was isolated from whole flies using RNeasy Mini Kit (Qiagen, Maryland, USA) according to the manufacturer's instructions. cDNA synthesis was performed with oligo-dT and random primers using SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA). Semiquantitative PCR was performed as described [31]. Real-time PCR was performed in duplicate using SYBR Green on an ABI 7900HT Real-Time PCR system (Applied Biosystems) according to the manufacture's protocol. All samples were analyzed from at least 3 independent of experiments. Data was normalized first to the level of the rp49 mRNA prior to quantifying the relative levels of mRNA between controls and experimentally treated samples. All detailed primers are available upon request.
Statistics
All survival data were analyzed by Kaplan–Meier Log-rank Test for overall survival and by the Student's t-test for mean and maximum lifespan using Graph Pad Prism4. The log mortality was determined by OASIS program [51]. Treated data were then plotted using Graph Pad Prism4. Other comparisons were determined either by Student's t-test or One way ANOVA followed by post hoc t-test. p<0.05 was considered statistically significant.
Zdroje
1. LandisG, ShenJ, TowerJ (2012) Gene expression changes in response to aging compared to heat stress, oxidative stress and ionizing radiation in Drosophila melanogaster. Aging 4 : 768–789.
2. PletcherSD, LibertS, SkorupaD (2005) Flies and their golden apples: the effect of dietary restriction on Drosophila aging and age-dependent gene expression. Ageing research reviews 4 : 451–480.
3. ZouS, MeadowsS, SharpL, JanLY, JanYN (2000) Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 97 : 13726–13731.
4. TowerJ (2011) Heat shock proteins and Drosophila aging. Experimental gerontology 46 : 355–362.
5. BiteauB, KarpacJ, HwangboD, JasperH (2011) Regulation of Drosophila lifespan by JNK signaling. Experimental gerontology 46 : 349–354.
6. WangMC, BohmannD, JasperH (2003) JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Developmental cell 5 : 811–816.
7. BorstP, EversR, KoolM, WijnholdsJ (2000) A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst 92 : 1295–1302.
8. DeanM (2005) The genetics of ATP-binding cassette transporters. Methods Enzymol 400 : 409–429.
9. ToyodaY, HagiyaY, AdachiT, HoshijimaK, KuoMT, et al. (2008) MRP class of human ATP binding cassette (ABC) transporters: historical background and new research directions. Xenobiotica 38 : 833–862.
10. BorstP, de WolfC, van de WeteringK (2007) Multidrug resistance-associated proteins 3, 4, and 5. Pflugers Arch 453 : 661–673.
11. BelinskyMG, GuoP, LeeK, ZhouF, KotovaE, et al. (2007) Multidrug resistance protein 4 protects bone marrow, thymus, spleen, and intestine from nucleotide analogue-induced damage. Cancer Res 67 : 262–268.
12. LeggasM, AdachiM, SchefferGL, SunD, WielingaP, et al. (2004) Mrp4 confers resistance to topotecan and protects the brain from chemotherapy. Mol Cell Biol 24 : 7612–7621.
13. LinZP, ZhuYL, JohnsonDR, RiceKP, NottoliT, et al. (2008) Disruption of cAMP and prostaglandin E2 transport by multidrug resistance protein 4 deficiency alters cAMP-mediated signaling and nociceptive response. Mol Pharmacol 73 : 243–251.
14. MennoneA, SorokaCJ, CaiSY, HarryK, AdachiM, et al. (2006) Mrp4−/ − mice have an impaired cytoprotective response in obstructive cholestasis. Hepatology 43 : 1013–1021.
15. KruhGD, BelinskyMG, GalloJM, LeeK (2007) Physiological and pharmacological functions of Mrp2, Mrp3 and Mrp4 as determined from recent studies on gene-disrupted mice. Cancer Metastasis Rev 26 : 5–14.
16. GomiA, ShinodaS, MasuzawaT, IshikawaT, KuoMT (1997) Transient induction of the MRP/GS-X pump and gamma-glutamylcysteine synthetase by 1-(4-amino-2-methyl-5-pyrimidinyl)methyl-3-(2-chloroethyl)-3 - nitrosourea in human glioma cells. Cancer research 57 : 5292–5299.
17. GottesmanMM (2003) Cancer gene therapy: an awkward adolescence. Cancer gene therapy 10 : 501–508.
18. KuoMT, BaoJ, FuruichiM, YamaneY, GomiA, et al. (1998) Frequent coexpression of MRP/GS-X pump and gamma-glutamylcysteine synthetase mRNA in drug-resistant cells, untreated tumor cells, and normal mouse tissues. Biochemical pharmacology 55 : 605–615.
19. YamaneY, FuruichiM, SongR, VanNT, MulcahyRT, et al. (1998) Expression of multidrug resistance protein/GS-X pump and gamma-glutamylcysteine synthetase genes is regulated by oxidative stress. The Journal of biological chemistry 273 : 31075–31085.
20. CaiC, OmwanchaJ, HsiehCL, ShemshediniL (2007) Androgen induces expression of the multidrug resistance protein gene MRP4 in prostate cancer cells. Prostate Cancer Prostatic Dis 10 : 39–45.
21. GradiloneA, PulcinelliFM, LottiLV, TrifiroE, MartinoS, et al. (2008) Celecoxib upregulates multidrug resistance proteins in colon cancer: lack of synergy with standard chemotherapy. Curr Cancer Drug Targets 8 : 414–420.
22. HoLL, KenchJG, HandelsmanDJ, SchefferGL, StrickerPD, et al. (2008) Androgen regulation of multidrug resistance-associated protein 4 (MRP4/ABCC4) in prostate cancer. Prostate 68 : 1421–1429.
23. NorrisMD, SmithJ, TanabeK, TobinP, FlemmingC, et al. (2005) Expression of multidrug transporter MRP4/ABCC4 is a marker of poor prognosis in neuroblastoma and confers resistance to irinotecan in vitro. Mol Cancer Ther 4 : 547–553.
24. LemosC, KathmannI, GiovannettiE, BelienJA, SchefferGL, et al. (2009) Cellular folate status modulates the expression of BCRP and MRP multidrug transporters in cancer cell lines from different origins. Mol Cancer Ther 8 : 655–664.
25. AleksunesLM, SlittAM, CherringtonNJ, ThibodeauMS, KlaassenCD, et al. (2005) Differential expression of mouse hepatic transporter genes in response to acetaminophen and carbon tetrachloride. Toxicological sciences: an official journal of the Society of Toxicology 83 : 44–52.
26. AleksunesLM, SlittAL, MaherJM, DieterMZ, KnightTR, et al. (2006) Nuclear factor-E2-related factor 2 expression in liver is critical for induction of NAD(P)H:quinone oxidoreductase 1 during cholestasis. Cell stress & chaperones 11 : 356–363.
27. ChenC, KlaassenCD (2004) Rat multidrug resistance protein 4 (Mrp4, Abcc4): molecular cloning, organ distribution, postnatal renal expression, and chemical inducibility. Biochemical and biophysical research communications 317 : 46–53.
28. MaherJM, DieterMZ, AleksunesLM, SlittAL, GuoG, et al. (2007) Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46 : 1597–1610.
29. GuX, ManautouJE (2010) Regulation of hepatic ABCC transporters by xenobiotics and in disease states. Drug metabolism reviews 42 : 482–538.
30. Lin-LeeYC, TatebeS, SavarajN, IshikawaT, Tien KuoM (2001) Differential sensitivities of the MRP gene family and gamma-glutamylcysteine synthetase to prooxidants in human colorectal carcinoma cell lines with different p53 status. Biochemical pharmacology 61 : 555–563.
31. HuangH, HaddadGG (2007) Drosophila dMRP4 regulates responsiveness to O2 deprivation and development under hypoxia. Physiol Genomics 29 : 260–266.
32. TarnayJN, SzeriF, IliasA, AnniloT, SungC, et al. (2004) The dMRP/CG6214 gene of Drosophila is evolutionarily and functionally related to the human multidrug resistance-associated protein family. Insect Mol Biol 13 : 539–548.
33. DeanM, RzhetskyA, AllikmetsR (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11 : 1156–1166.
34. HassanHM, FridovichI (1979) Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. The Journal of biological chemistry 254 : 10846–10852.
35. SykiotisGP, BohmannD (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell 14 : 76–85.
36. FinkelT, HolbrookNJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408 : 239–247.
37. WestonCR, DavisRJ (2007) The JNK signal transduction pathway. Current opinion in cell biology 19 : 142–149.
38. Martin-BlancoE, GampelA, RingJ, VirdeeK, KirovN, et al. (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes & development 12 : 557–570.
39. WeberU, ParicioN, MlodzikM (2000) Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development 127 : 3619–3629.
40. RomanG, EndoK, ZongL, DavisRL (2001) P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 98 : 12602–12607.
41. HotamisligilGS (2006) Inflammation and metabolic disorders. Nature 444 : 860–867.
42. BakerKD, ThummelCS (2007) Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell metabolism 6 : 257–266.
43. GeorgelP, NaitzaS, KapplerC, FerrandonD, ZacharyD, et al. (2001) Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Developmental cell 1 : 503–514.
44. HwangboDS, GershmanB, TuMP, PalmerM, TatarM (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429 : 562–566.
45. GiannakouME, GossM, JungerMA, HafenE, LeeversSJ, et al. (2004) Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305 : 361.
46. HarmanD (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11 : 298–300.
47. SohalRS, MockettRJ, OrrWC (2000) Current issues concerning the role of oxidative stress in aging: a perspective. Results Probl Cell Differ 29 : 45–66.
48. SohalRS, SohalBH, OrrWC (1995) Mitochondrial superoxide and hydrogen peroxide generation, protein oxidative damage, and longevity in different species of flies. Free radical biology & medicine 19 : 499–504.
49. JohnsonFB, SinclairDA, GuarenteL (1999) Molecular biology of aging. Cell 96 : 291–302.
50. WheelerJC, KingV, TowerJ (1999) Sequence requirements for upregulated expression of Drosophila hsp70 transgenes during aging. Neurobiology of aging 20 : 545–553.
51. YangJS, NamHJ, SeoM, HanSK, ChoiY, et al. (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PloS one 6: e23525.
52. WangMC, BohmannD, JasperH (2005) JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121 : 115–125.
53. MorrowG, BattistiniS, ZhangP, TanguayRM (2004) Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. The Journal of biological chemistry 279 : 43382–43385.
54. BiteauB, KarpacJ, SupoyoS, DegennaroM, LehmannR, et al. (2010) Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS genetics 6: e1001159.
55. TatarM, KhazaeliAA, CurtsingerJW (1997) Chaperoning extended life. Nature 390 : 30.
56. LeeKS, Iijima-AndoK, IijimaK, LeeWJ, LeeJH, et al. (2009) JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends life span. The Journal of biological chemistry 284 : 29454–29461.
57. Cook-WiensE, GrotewielMS (2002) Dissociation between functional senescence and oxidative stress resistance in Drosophila. Experimental gerontology 37 : 1347–1357.
58. GarganoJW, MartinI, BhandariP, GrotewielMS (2005) Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Experimental gerontology 40 : 386–395.
59. KarpacJ, Hull-ThompsonJ, FalleurM, JasperH (2009) JNK signaling in insulin-producing cells is required for adaptive responses to stress in Drosophila. Aging cell 8 : 288–295.
60. OhSW, MukhopadhyayA, SvrzikapaN, JiangF, DavisRJ, et al. (2005) JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proceedings of the National Academy of Sciences of the United States of America 102 : 4494–4499.
61. SonnichsenB, KoskiLB, WalshA, MarschallP, NeumannB, et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434 : 462–469.
62. KamathRS, FraserAG, DongY, PoulinG, DurbinR, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 : 231–237.
63. ReaSL, WuD, CypserJR, VaupelJW, JohnsonTE (2005) A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nature genetics 37 : 894–898.
64. WalkerGA, WhiteTM, McCollG, JenkinsNL, BabichS, et al. (2001) Heat shock protein accumulation is upregulated in a long-lived mutant of Caenorhabditis elegans. The journals of gerontology Series A, Biological sciences and medical sciences 56: B281–287.
65. YangJ, TowerJ (2009) Expression of hsp22 and hsp70 transgenes is partially predictive of drosophila survival under normal and stress conditions. The journals of gerontology Series A, Biological sciences and medical sciences 64 : 828–838.
66. TerryDF, WyszynskiDF, NolanVG, AtzmonG, SchoenhofenEA, et al. (2006) Serum heat shock protein 70 level as a biomarker of exceptional longevity. Mechanisms of ageing and development 127 : 862–868.
67. ZhengJ, MutchersonR2nd, HelfandSL (2005) Calorie restriction delays lipid oxidative damage in Drosophila melanogaster. Aging cell 4 : 209–216.
68. ZhengX, YangZ, YueZ, AlvarezJD, SehgalA (2007) FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proceedings of the National Academy of Sciences of the United States of America 104 : 15899–15904.
69. GiannakouME, PartridgeL (2004) The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends in cell biology 14 : 408–412.
70. KrautR, MenonK, ZinnK (2001) A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Current biology: CB 11 : 417–430.
71. FordD, HoeN, LandisGN, TozerK, LuuA, et al. (2007) Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycyline or RU486/Mifepristone. Experimental gerontology 42 : 483–497.
72. RahmanMM, SykiotisGP, NishimuraM, BodmerR, BohmannD (2013) Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging cell 12 : 554–562.
73. KuoTH, FedinaTY, HansenI, DreisewerdK, DierickHA, et al. (2012) Insulin signaling mediates sexual attractiveness in Drosophila. PLoS genetics 8: e1002684.
74. PoirierL, ShaneA, ZhengJ, SeroudeL (2008) Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging cell 7 : 758–770.
75. KlionskyDJ, AbdallaFC, AbeliovichH, AbrahamRT, Acevedo-ArozenaA, et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8 : 445–544.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



