-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
Perturbation of the expression level of IKAROS, a transcription factor critical during hematopoiesis, is associated with malignant transformation in mice and humans. The importance of IKAROS expression levels for the control of target-gene regulation was addressed in hematopoietic progenitor cells. The collaboration between IKAROS and the Nucleosome Remodeling and Deacetylase (NuRD) complex can promote gene activation or repression. IKAROS can also interact with the Positive-Transcription Elongation Factor b (P-TEFb) and the Protein Phosphatase 1 (PP1), an important P-TEFb regulator. Immunoaffinity purification of IKAROS interacting proteins and Fast Protein Liquid Chromatography analysis revealed a dynamic interaction between IKAROS, PP1 and the newly defined NuRD-P-TEFb complex. This complex can be targeted to specific genes in cells expressing high levels of IKAROS to promote productive transcription elongation. Based on our results we suggest that, in addition to P-TEFb, the NuRD complex and PP1 are required to facilitate transcription elongation of IKAROS-target genes and normal differentiation of hematopoietic progenitor cells.
Published in the journal: . PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004827
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004827Summary
Perturbation of the expression level of IKAROS, a transcription factor critical during hematopoiesis, is associated with malignant transformation in mice and humans. The importance of IKAROS expression levels for the control of target-gene regulation was addressed in hematopoietic progenitor cells. The collaboration between IKAROS and the Nucleosome Remodeling and Deacetylase (NuRD) complex can promote gene activation or repression. IKAROS can also interact with the Positive-Transcription Elongation Factor b (P-TEFb) and the Protein Phosphatase 1 (PP1), an important P-TEFb regulator. Immunoaffinity purification of IKAROS interacting proteins and Fast Protein Liquid Chromatography analysis revealed a dynamic interaction between IKAROS, PP1 and the newly defined NuRD-P-TEFb complex. This complex can be targeted to specific genes in cells expressing high levels of IKAROS to promote productive transcription elongation. Based on our results we suggest that, in addition to P-TEFb, the NuRD complex and PP1 are required to facilitate transcription elongation of IKAROS-target genes and normal differentiation of hematopoietic progenitor cells.
Introduction
The tumor suppressor IKAROS is a transcription factor critical for hematopoietic multi-lineage priming, cell fate and lineage determination [1]–[5]. Mice homozygote for the Ikaros null mutation (IkNULL) display severe defects in lymphocyte development and function, and develop leukemias and lymphomas with complete penetrance [6]. These phenotypes reflect the requirement of IKAROS to activate the lymphoid program in hematopoietic progenitor cells (HPCs) [4]. IKAROS is also involved in transcriptional regulation of erythroid - and myeloid-specific genes [7]–[13]. The hematopoietic differentiation is affected not only by the presence or absence of IKAROS, but also by its relative expression level [14]. In particular, during B-cell progenitor differentiation, dynamic change of IKAROS expression level has been identified as a key regulator for the expression of multiple target genes [15], [16].
IKAROS controls chromatin organization mainly through association with the Nucleosome Remodeling and Deacetylase (NuRD) complex [5], [17], [18]. NuRD was initially identified as a repressive complex but it was demonstrated afterwards to promote transcription of specific genes as well [5], [19]–[22]. It remains to be defined how this HDAC-containing complex activates transcription. IKAROS contributes to the assembly and stability of the pre-initiation complex (PIC) at promoters [13], [23]–[26] and interacts directly with CDK9, the catalytic subunit of P-TEFb (Positive-Transcription Elongation Factor b) [27]. CDK9 phosphorylates the C-terminal domain (CTD) of the large subunit of RNA Polymerase II (POL II) on Ser2 as well as the SPT5 subunit of DSIF and the E subunit of NELF. These events are required to release promoter-proximal paused POL II and thus, allow productive transcription elongation. Most nuclear P-TEFb is sequestered in the 7SK snRNP repressive complex. This repressive complex is characterized by the snRNP molecule and the proteins HEXIM (HEXIM1 or 2), LARP7 and MePCE [28]. Of interest here, is the dissociation of the P-TEFb from this repressive complex promoted by the sequential activity of the protein phosphatase 2B (PP2B) that favors conformation changes of the 7SK snRNP and protein phosphates 1α (PP1α), involved in CDK9 dephosphorylation at different residues including Thr186 and Ser175 [29], [30]. Dephosphorylated CDK9/P-TEFb is preferentially recruited to promoters by the general factor BRD4 or specific transcription factors such as HIV TAT [31]–[33]. Then, CDK9/P-TEFb becomes catalytically active and promotes the release of promoter-proximal paused POL II when it is “re-phosphorylated” by the TFIIH associated CDK7 [30]. PP1α is one of the three catalytic subunits (α, β or γ) which, together with a regulatory subunit, forms each PP1 enzyme [34]. Interestingly, IKAROS interacts with PP1 and is dephosphorylated by this phosphatase [35]. Whether the IKAROS-PP1 interaction is important for Cdk9/P-TEFb activation and thus, transcription elongation of IKAROS-target genes is not known.
Here, we sought to define the importance of these protein associations for IKAROS and NuRD to function as transcriptional activators. We demonstrate that IKAROS is an adaptor molecule required for the recruitment of the newly identified NuRD-P-TEFb complex to IKAROS-target genes. IKAROS binding to the promoter region of specific genes is also associated with the local recruitment of the CDK9/P-TEFb activator, PP1α. Interestingly, the Mi2/NuRD occupancy at IKAROS-target genes is enhanced when transcription elongation is proficient, and the release of POL II promoter-proximal pausing is decreased in the absence of Mi2. Our data also reveal that the dynamic interaction of IKAROS with the NuRD-P-TEFb complex depends on IKAROS protein levels. Low-levels of IKAROS suffice for Mi2/NuRD recruitment to promoters, but higher levels of IKAROS are required for NuRD-P-TEFb and PP1α to chromatin, CDK9/P-TEFb activation, and productive transcription elongation. Finally, we demonstrate that IKAROS-PP1α protein interaction is required for normal hematopoietic differentiation of primary HPCs.
Results
IKAROS interacts with the newly characterized NuRD-P-TEFb complex
Several studies have demonstrated physical interaction between IKAROS and NuRD [5], [17], [18], [36] or IKAROS and P-TEFb [23], [27]. To define how IKAROS coordinates chromatin remodeling and transcription elongation activities, we first conducted tandem immunoaffinity purification and mass spectrometry (LC-MS/MS) analysis in Jurkat cells expressing Flag - and HA-tagged IKAROS (Flag-HA-Ik) (Figures 1A, S1A). LC-MS/MS of Jurkat/Flag-HA-Ik eluates indicated that IKAROS associates with PP1α NuRD (Mi2, MTA2, RBBP4 and MBD3) and P-TEFb (CDK9, CYCT1) components (Table 1), whereas the P-TEFb inhibitor HEXIM1 and the negative elongation factor NELF were not amongst the purified proteins. In addition, HEXIM1 did not immunoprecipitate with IKAROS (Figure S1B), thus suggesting that IKAROS interacts with the elongation-competent and catalytically active P-TEFb.
Fig. 1. IKAROS interacts with the newly characterized NuRD-P-TEFb complex. 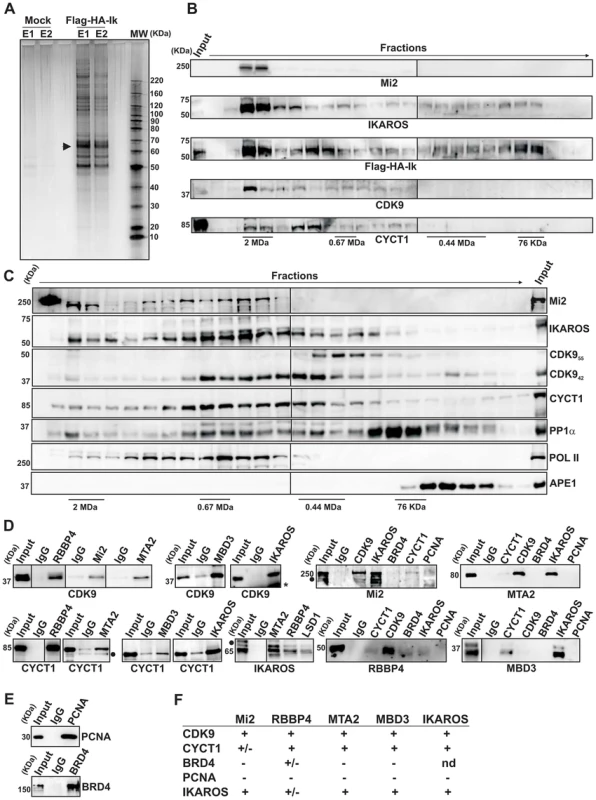
A) Purification of Flag-HA-Ik associated proteins. Nuclear extracts from Jurkat cells carrying the pOZ-N-Flag-HA-IKAROS-IRES-IL2R vector and expressing a double tagged IKAROS (Flag-HA-Ik) or Jurkat cells expressing the pOZ-N-Flag-HA-IRES-IL2R empty vector (Mock) were used for sequential immunoaffinity purification using Flag- followed by HA-conjugated matrix. A fraction of the purified complexes were loaded on SDS-PAGE and silver stained; E1: first HA elution; E2: second HA elution; MW: molecular weights (in KDa); the Flag-HA-Ik protein is indicated by an arrow; B, C) Molecular weight fractionation of the IKAROS-associated complexes. Flag-HA-Ik immunoaffinity purified complexes (panel B) or nuclear extracts (panel C) were fractionated using a Superose 6 10/300GL column; 0.5 ml fractions were collected, TCA-precipitated and loaded on SDS-PAGE for western blot analysis; immunoblots were probed with the antibodies indicated at the bottom or on the right side of the panels; the 37 KDa APE1 nuclear protein was used as control; D) Protein co-immunoprecipitation of Jurkat cell nuclear extracts. Immunoprecipitations were carried out with the antibodies indicated on top of each panel; these antibodies were specific for: IKAROS, the general P-TEFb activator BRD4, NuRD-associated proteins (Mi2, RBBP4, MTA2, MBD3) or P-TEFb components (CDK9, CYCT1). Immunoblots were probed with the antibodies indicated at the bottom of the panels. Input samples represent 2% of nuclear extracts; IgG: isotype-matched immunoglobulins; asterisk: IgG light chains; filled dots: non-specific bands; n≥3; E) The nuclear protein PCNA was used as negative control for the immunoprecipitation procedure; F) Summary of the most relevant protein-protein interactions; +: strong interaction; +/−: weak interaction; −: no interaction; nd: not determined. Tab. 1. Immunoaffinity purification of Flag-HA-IKAROS complexes from Jurkat cells and their identification by LC-MS/MS analysis. 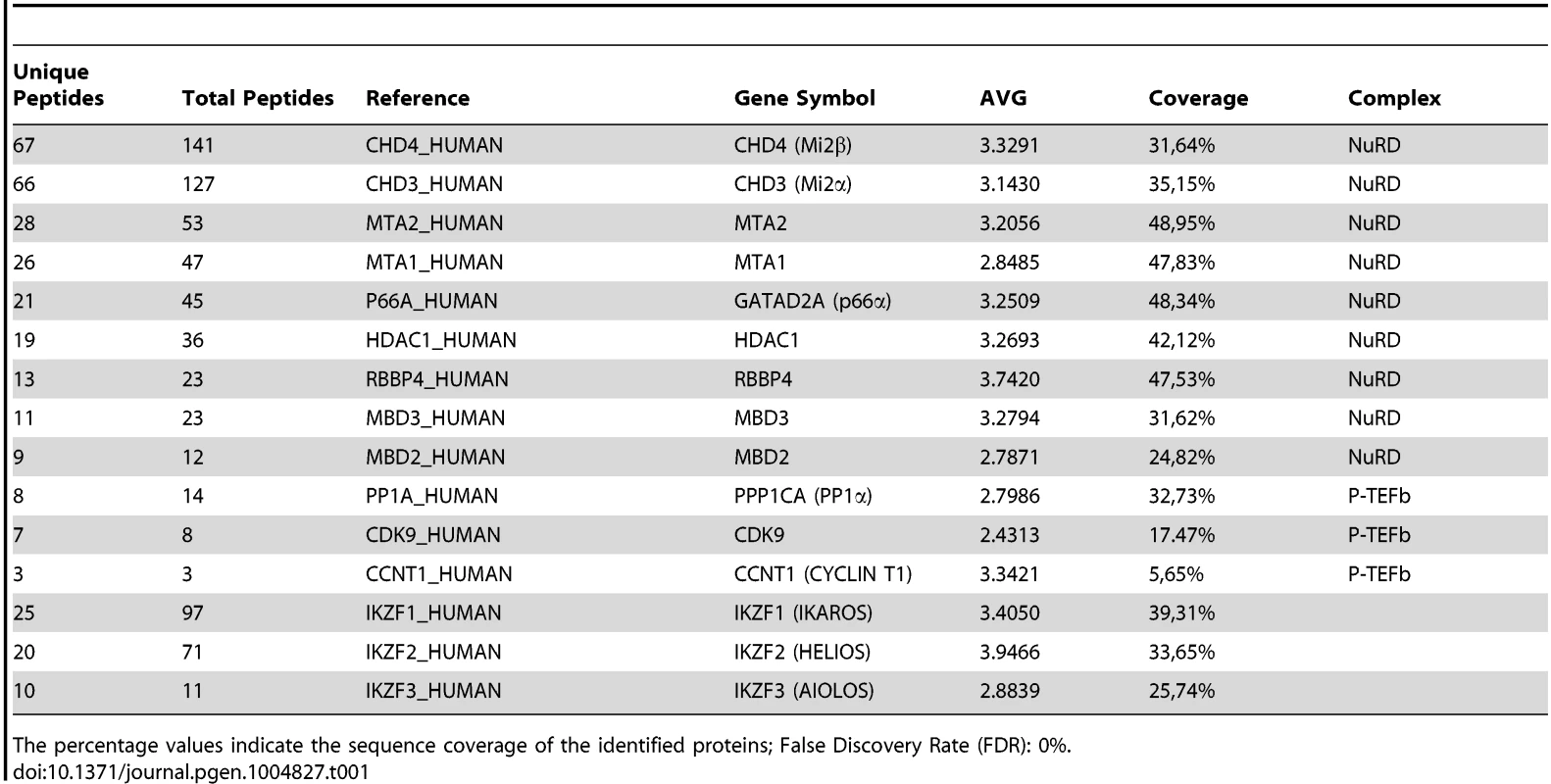
The percentage values indicate the sequence coverage of the identified proteins; False Discovery Rate (FDR): 0%. To determine whether IKAROS interactions with Mi2/NuRD and CDK9/P-TEFb define a single complex or multiple complexes, Flag-HA-Ik immunoaffinity-purified complexes were analyzed by size exclusion chromatography. Flag-HA-Ik co-fractionated with NuRD and P-TEFb components as a major peak of ∼2 MDa (Figure 1B). Additionally, Mi2, CDK9, CYCT1, PP1α and IKAROS co-eluted in nuclear extracts of wild type Jurkat cells (Figure 1C). Since IKAROS mainly eluted at 0.5 MDa (Figure 1C), it is likely that only a minority of the total nuclear IKAROS stably associates with the NuRD-P-TEFb complex. Accordingly, silver staining of IKAROS-purified complexes (Figure 1A) allowed the detection of two weak, but above-background, protein bands migrating at the range of the two known CDK9 isoforms [37] (Figure S1C). That only a small fraction of the nuclear CDK9/P-TEFb complex is highly active and capable of stimulating transcription elongation rate has also been shown in different systems [38], [39].
Next, we investigated whether the interaction of IKAROS with CDK9 or Mi2 could be differently modulated by fluctuation of IKAROS protein levels. Protein co-immunoprecipitation (co-IP) experiments were carried out in Ikaros knock-down Jurkat cells (Figure S1D). Co-IP results indicated that IKAROS-CDK9 interaction is more sensitive to IKAROS levels than the IKAROS-Mi2 interaction because IKAROS-CDK9 interaction is impaired in Ikaros knock-down Jurkat cells whereas IKAROS-Mi2 interaction is comparable in non-target as well as Ikaros knock-down Jurkat cells (Figure S1E). This suggests that in hematopoietic cells a fraction of nuclear IKAROS proteins can be dynamically assembled in a multifunctional protein complex containing both NuRD and P-TEFb components and that formation and/or stability of IKAROS-P-NuRD-P-TEFb interaction is influenced by the expression level of IKAROS.
Protein interactions between NuRD and P-TEFb components were then assessed by protein co-IP of Jurkat nuclear extracts. The NuRD-associated proteins Mi2, RBBP4, MTA2, and MBD3 interacted with the P-TEFb components CDK9 and CYCT1 (Figure 1D, S1F), whereas none of these factors interacted with the negative control nuclear protein PCNA (Figures 1D, E). Among the NuRD-associated proteins, only RBBP4 weakly interacted with the general P-TEFb activator BRD4 [40] (Figures 1D, S1G). CDK9, CYCT1, Mi2, MTA2, RBBP4, and MBD3 also co-immunoprecipitated with IKAROS (Figure 1D). DNase1 did not affect Mi2-CDK9 protein interaction (Figure S1H), indicating that DNA is dispensable. Thus, tandem immunoaffinity purification, size-exclusion chromatography and co-IP experiments demonstrate that IKAROS can assemble with the newly defined NuRD-P-TEFb multifunctional complex (Figure 1F).
The simultaneous recruitment of P-TEFb and NuRD is IKAROS dose-dependent
We examined whether IKAROS could be a licensing factor for NuRD-P-TEFb complex recruitment to chromatin and whether the variation of IKAROS thresholds, which naturally occurs during hematopoiesis [5], [15], [41], could influence NuRD-P-TEFb association with IKAROS and thereby, regulate transcription elongation. To clarify this issue we studied two IKAROS-target genes, c-Kit and Flt3 (Figure S2A), which are cell receptors critical for HPC survival that are downregulated in IkNULL HPCs [42]. We used: (i) bone marrow lineage negative (lin−) HPCs obtained from Ikaros wild type (IkWT), Ikaros heterozygote null (IkHT) or Ikaros homozygote null (IkNULL) mice [6]; and (ii) the mouse G1E2 cells, which are proliferating hematopoietic progenitor-like cells [43]. The expression level of IKAROS in G1E2 cells was sufficient to detect Mi2-CDK9 interaction (Figure 2A).
Fig. 2. The simultaneous recruitment of P-TEFb and NuRD is IKAROS dose-dependent. 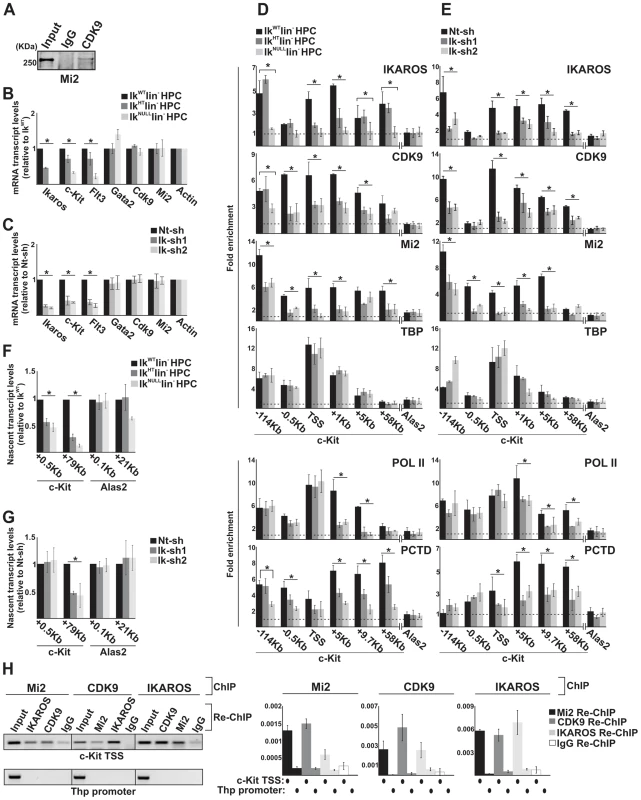
A) Protein co-immunoprecipitation of G1E2 total cell lysates. Immunoprecipitations were carried out with CDK9 antibodies; immunoblots were probed with Mi2 antibodies; Input samples represent 2% of protein extracts; B, C) Gene expression profiles. mRNA samples were retro-transcribed with oligo-dT nucleotides; cDNA was used as template for qPCR with gene-specific primer sets; β-Actin was used as internal control; y axis: relative mRNA transcript enrichment levels; ratios are plotted as the mean ± Standard Deviation (SD) of the measurements; n≥4; D, E) Chromatin Immunoprecipitation (ChIP). ChIP assays were carried out with the antibodies indicated on the top of each panel; POL II: is an antibody against the N-terminal region of the large subunit of POL II and binds POL II in a phosphorylation-independent manner; PCTD: is an antibody against the CTD repeats phosphorylated at Ser2; y-axis: fold enrichments of c-Kit (−114 Kb enhancer; −0.5 Kb; TSS; +1 Kb, +5 Kb, +9.7 Kb, +58 Kb ORF regions) or Alas2 (TSS; used as negative control) regions relative to Thp promoter and input samples are plotted as the mean ± SD of the measurements; a value of 1 (dotted lines) indicates no enrichment. Lin− HPCs: bone marrow-derived lineage negative hematopoietic progenitor cells; IkWT lin−: Ikaros wild type lin− HPCs; IkHT lin−: Ikaros heterozygote null lin− HPCs; IkNULL lin−: Ikaros homozygote null lin− HPCs; Nt-sh: non-target sh-RNA G1E2 clones; Ik-sh: Ikaros-specific sh-RNA (Ik-sh1 and Ik-sh2) G1E2 clones. *: P≤0.05 by Student's t-test; F, G) Gene expression profiles. RNA samples were retro-transcribed with random oligonucleotides to amplify nascent transcripts, which were used as templates for qPCR with intron-specific c-Kit (+0.5 Kb and +79 Kb regions), Alas2 (+0.1 Kb and +21 Kb regions) or Gapdh (used as internal control) primer sets; y axis: relative nascent transcript enrichment levels; ratios are plotted as the mean ± Standard Deviation (SD) of the measurements; n≥4; H) Sequential ChIP (re-ChIP) assays carried out on G1E2 cells. Mi2, CDK9 or IKAROS antibodies were used for the first ChIP; IKAROS, CDK9, Mi2 antibodies or IgG controls were used for the second ChIP; left panel: representative examples of semi-quantitative PCR of re-ChIP sample carried out with primer sets specific for c-Kit TSS or Thp promoter; right panel: quantitative re-ChIP analysis carried out with the percent input method; re-ChIP samples were used as templates for quantitative PCR with primer sets specific for c-Kit TSS or Thp promoter; y-axis: fold enrichments of c-Kit TSS or Thp promoter (used as negative control) regions in re-ChIP samples relative to input samples are plotted as the mean ± SD of the measurements; n = 3. c-Kit and Flt3 expression correlated with the level of Ikaros expression in lin− HPCs (Figure 2B). Similarly, the expression level of c-Kit and Flt3 was significantly decreased in Ikaros knock-down G1E2 cells (Figure 2C). However, the expression level of additional c-Kit regulators, such as Gata2 [44], Cdk9 and Mi2 (see below) was not reduced in IkNULL or Ikaros knock-down G1E2 cells (Figures 2B, C and S2B).
To investigate whether reduced c-Kit expression is a direct effect of the Ikaros loss, we carried out chromatin immunoprecipitation (ChIP) assays. ChIP experiments indicated that IKAROS was recruited to critical regulatory regions of the c-Kit locus [44] e.g., the −114 Kb enhancer, the c-Kit Transcriptional Start Site -TSS-, and +1 Kb, +5 Kb as well as +58 Kb regions of the Open Reading Frame (ORF) in IkWT lin− HPCs and G1E2 cells (Figure 2D, E). The same regions were occupied by CDK9, and Mi2 in IkWT lin− HPCs, whereas the reduced level of IKAROS affected CDK9 and Mi2 recruitment at c-Kit (Figure 2D, E) but not at c-Fos promoter (Figure S2C), which was selected since c-Fos gene is expressed in HPCs but it is not regulated by IKAROS [45]. ChIP performed with TBP or POL II specific antibodies revealed that the PIC is assembled at c-Kit TSS regardless of Ikaros expression level (Figure 2D, E). However, the recruitment of POL II and POL II with phosphorylated Ser2 at the C-terminal domain (PCTD) at c-Kit ORF was reduced in IkHT, IkNULL lin− HPCs as well as the Ikaros knock-down G1E2 cells (Figure 2D, E), suggesting that IKAROS can positively control c-Kit transcription elongation. This assumption was supported by the significant decrease of CDK9 binding to c-Kit TSS and ORF (Figure 2D, E) and the consequent reduction of c-Kit nascent transcript levels towards the 3′ end of the gene (Figure 2F, G) in Ikaros-deficient cells.
The correlation between IKAROS expression level and the release of promoter-proximal paused POL II was further demonstrated by the analysis of c-Kit traveling ratio, which is determined by the relative ratio of POL II density in gene ORF vs. promoter-proximal regions [46], [47]. As indicated in Table 2, c-Kit traveling ratio decreased in IkHT and IkNULL lin− HPCs, when compared to IkWT.
Tab. 2. c-Kit POL II traveling ratio. 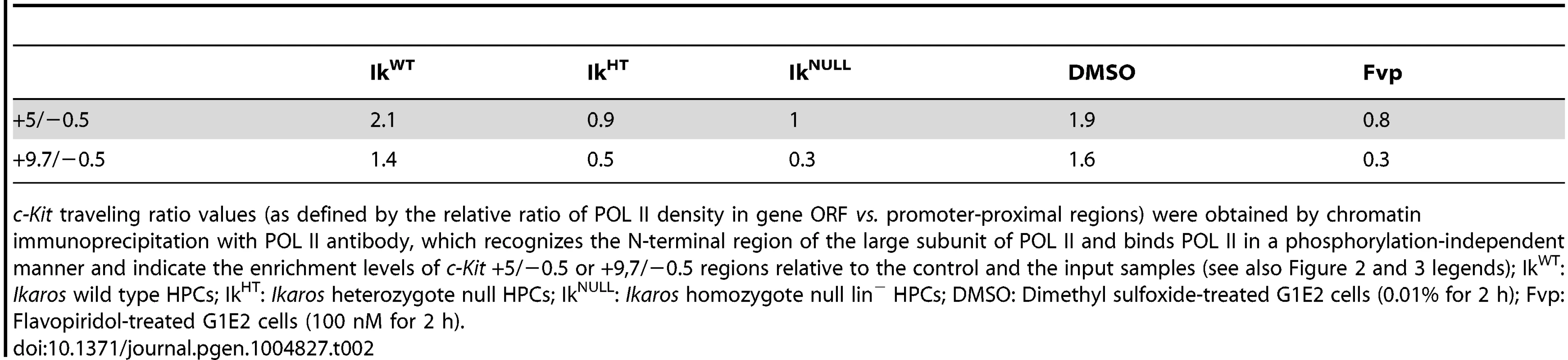
c-Kit traveling ratio values (as defined by the relative ratio of POL II density in gene ORF vs. promoter-proximal regions) were obtained by chromatin immunoprecipitation with POL II antibody, which recognizes the N-terminal region of the large subunit of POL II and binds POL II in a phosphorylation-independent manner and indicate the enrichment levels of c-Kit +5/−0.5 or +9,7/−0.5 regions relative to the control and the input samples (see also Figure 2 and 3 legends); IkWT: Ikaros wild type HPCs; IkHT: Ikaros heterozygote null HPCs; IkNULL: Ikaros homozygote null lin− HPCs; DMSO: Dimethyl sulfoxide-treated G1E2 cells (0.01% for 2 h); Fvp: Flavopiridol-treated G1E2 cells (100 nM for 2 h). To address whether IKAROS, CDK9 and Mi2 simultaneously co-occupy the c-Kit TSS, sequential ChIP (re-ChIP) [48] was carried out. Sequential ChIP analysis provided the evidence that in hematopoietic progenitors, these proteins can associate to the c-Kit TSS together (Figure 2H), hence suggesting that transcription elongation is promoted by the IKAROS-mediated recruitment of the NuRD-P-TEFb complex.
Collectively, these results indicate that in HPCs, IKAROS facilitates productive transcription elongation at c-Kit (Figure 2) and Flt3 (Figure S2D, E; Table S1) genes and that reduced IKAROS levels, which do not affect the PIC assembly, can perturb transcription elongation.
The IKAROS-NuRD-P-TEFb complex assists POL II during transcription elongation
Since IKAROS, CDK9 and Mi2 were detected at c-Kit TSS and ORF in IkWT HPCs (Figure 2D, E), we tested whether these factors play an active role during transcription elongation or associate with chromatin at c-Kit ORF independently of productive transcription elongation. Thus, transcription elongation was inhibited with Flavopiridol, which is a specific inhibitor of CDK9 activity [49]. G1E2 cells were treated with 100 nM Flavopiridol, for a short period of time (2 hours) to avoid indirect effects due to general inhibition of gene expression [50]. As expected, CDK9, POL II and CYCT1 protein levels were not reduced in G1E2-treated cells (Figure 3A), and IKAROS phosphorylation [35] appeared to be unmodified (Figure S3A). However, c-Kit and Flt3 nascent transcript levels were significantly reduced (Figures 3B, S3B). Flavopiridol treatment of G1E2 cells did not affect IKAROS recruitment to the c-Kit (Figure 3C) or Flt3 (Figure S3C) promoter region. Furthermore, decreased detection of POL II within the gene ORF, POL II accumulation at c-Kit TSS, and lack of PCTD association at c-Kit TSS and ORF, confirmed that Flavopiridol did not affect POL II loading, hence PIC assembly (Figures 3C, S3C) but substantially reduced transcription elongation (Table 2). Similar results (reduced c-Kit expression, unchanged IKAROS recruitment and lack of PCTD association to c-Kit promoter) were obtained in Cdk9 knock-down G1E2 cells (Figure S3D–F). Thus, transcription elongation block (obtained either by Flavopiridol treatment or Cdk9 knock-down) does not affect loading of IKAROS, CDK9/P-TEFb and Mi2/NuRD at c-Kit TSS region, but impedes P-TEFb and, surprisingly, IKAROS and NuRD recruitment within the gene ORF.
Fig. 3. The IKAROS-NuRD-P-TEFb complex assists POL II during transcription elongation. 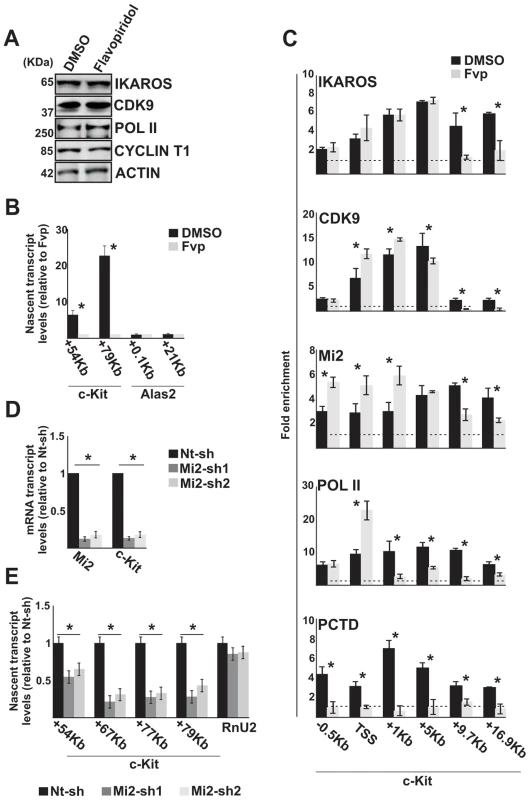
In panels A–C, G1E2 cells were treated for 2 h either with 0.01% DMSO (DMSO) or 100 nM Flavopiridol (Fvp). A) Protein expression analysis. Western blot assays of total cell lysates of DMSO- or Flavopiridol-treated G1E2 cells; ACTIN was used as an internal control; the antibodies used are indicated on the right side of the panels; B, E) Gene expression profiles. RNA samples were retro-transcribed with random oligonucleotides to amplify nascent transcripts, which were used as templates for qPCR with intron-specific c-Kit (+54 Kb, +67 Kb, +77 Kb, +79 Kb regions) or Alas2 (+0.1 Kb, +21 Kb regions) primer sets; Rnu2-1 (a Flavopiridol-insensitive gene) was used as internal control in panel B; Gapdh was used as internal control in panel E; y axis: relative nascent transcript enrichment levels; ratios are plotted as the mean ± Standard Deviation (SD) of the measurements; n≥4; C) ChIP assays were carried out with the antibodies indicated on the top of each panel; POL II: is an antibody against the N-terminal region of the large subunit of POL II and binds POL II in a phosphorylation-independent manner; PCTD: is an antibody against the CTD repeats phosphorylated at Ser2; y-axis: fold enrichments of c-Kit regions (−.5 Kb; TSS, +0.5 Kb, +1 Kb, +5 Kb, +9.7 Kb, +16.9 Kb ORF regions) relative to Thp promoter and input samples are plotted as the mean ± SD of the measurements; a value of 1 (dotted lines) indicates no enrichment; n≥4; D) Gene expression profiles. mRNA samples were retro-transcribed with oligo-dT nucleotides; cDNA was used as template for qPCR with gene-specific primer sets; β-Actin was used as internal control; y axis: relative mRNA enrichment levels; ratios are plotted as the mean ± SD of the measurements; n≥4. *: P≤0.05 by Student's t-test. Since the elongation block by Flavopiridol led to an accumulation of Mi2 at c-Kit TSS and reduced association with the downstream regions (Figure 3C), we assessed whether NuRD could directly influence transcription elongation by knock-down of one of the NuRD components, Mi2, in G1E2 cells. The Mi2 knock-down cells showed a reduction of c-Kit and Flt3 mature as well as nascent transcripts (Figure 3D, E). The fact that Mi2 knock-down had a greater effect on transcripts generated at distal rather than proximal regions, suggests that the NuRD complex can assist and facilitate POL II progression during transcription elongation.
IKAROS contributes to PP1α association with the NuRD-P-TEFb complex
IKAROS is a substrate of PP1 activity [35] and PP1α can act as a positive regulator of CDK9/P-TEFb [29]. IKAROS interacts with the α catalytic subunit of PP1 in G1E2 cells (Figure 4A), in COS-7 cells transfected with IKAROS (Figure S4A) and in Jurkat cells (Table 1). Among the PP1 catalytic subunits, we identified PP1α as the most relevant and abundant interacting partner of IKAROS (Table S2). Thus, we investigated whether, by binding to PP1α and CDK9, IKAROS could deliver PP1α to the large P-TEFb complex, thereby activating CDK9/P-TEFb and favoring transcription elongation. In accordance with this assumption, PP1α recruitment was demonstrated at c-Kit TSS in IkWT and much less efficiently in IkHT or IkNULL lin− HPCs (Figure 4B). PP1α recruitment to c-Kit TSS also decreased in Ikaros knock-down G1E2 clones (Figure 4C), even though PP1α expression was similar to control cells (Figure 4D).
Fig. 4. IKAROS contributes to PP1α association with the NuRD-P-TEFb complex. 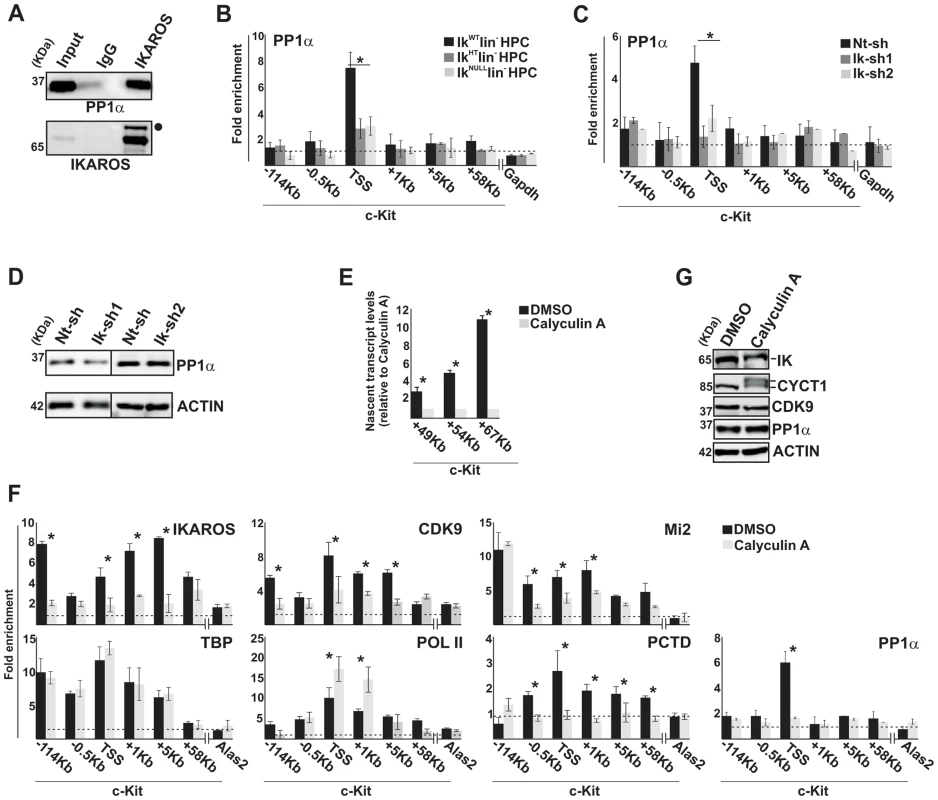
A) Protein co-immunoprecipitation of G1E2 nuclear extracts. Immunoprecipitations were carried out with IKAROS antibodies or isotype-matched immunoglobulins (IgG) and immunoblots were probed with PP1α or IKAROS antibodies; Input samples represent 2% of nuclear extracts; filled dot: non-specific band; B, C, F) ChIP assays were carried out with the antibodies labeled on the top of each panel; POL II: is an antibody against the N-terminal region of the large subunit of POL II and binds POL II in a phosphorylation-independent manner; PCTD: is an antibody against the CTD repeats phosphorylated at Ser2. Lin− HPCs: bone marrow-derived lineage negative hematopoietic progenitor cells; IkWT lin−: Ikaros wild type lin− HPCs; IkHT lin−: Ikaros heterozygote null lin− HPCs; IkNULL lin−: Ikaros homozygote null lin− HPCs; Nt-sh: non-target sh-RNA G1E2 clones; Ik-sh: Ikaros-specific sh-RNA (Ik-sh1 and Ik-sh2) G1E2 clones; DMSO: G1E2 cells treated for 30 min with 0.05% DMSO (diluent control); Calyculin A: G1E2 cells treated for 30 min with100 nM Calyculin A; y-axis: fold enrichments of c-Kit (−114 Kb; −0.5 Kb; TSS; +1 Kb, +5 Kb, +58 Kb ORF regions), Alas2 (TSS) or Gapdh (TSS) regulatory regions relative to Thp promoter and input samples are plotted as the mean ± Standard Deviations (SD); a value of 1 (dotted lines) indicates no enrichment; n≥4; Alas2, Gapdh and Thp TSS regions were used as negative controls; D, G) Protein expression analysis. Western blot assays of total cell lysates were performed with non-target (Nt-sh) or Ikaros-specific (Ik-sh1 and Ik-sh2) sh-RNA G1E2 clones and with DMSO- or Calyculin A-treated G1E2 cells; the antibodies used are indicated on the right side of the panels; E) Gene expression profiles. RNA samples were retro-transcribed with random oligonucleotides to amplify nascent transcripts, which were used as templates for qPCR with intron-specific c-Kit (+49 Kb, +54 Kb, +67 Kb regions) or Gapdh (used as internal control) primer sets; y axis: relative nascent transcript enrichment levels; ratios are plotted as the mean ± SD of the measurements; n≥4. *: P≤0.05 by Student's t-test. The role of the IKAROS-PP1α interaction for CDK9/P-TEFb activation was further delineated by exploring the influence of Calyculin A, a PP1/PP2A inhibitor [34], on c-Kit and Flt3 transcription elongation. Short-term treatment with Calyculin A did not cause toxic effects or cell death (Figure S4B), but induced significant reduction of c-Kit nascent transcripts (Figure 4E), whereas transcription elongation was not reduced upon treatment with inhibitors specific for PP2A (Okadaic Acid) or PP2B (Cyclosporin A) (Figure S4C, D), which are two phosphatases that do not dephosphorylate IKAROS proteins [35]. ChIP assay revealed that IKAROS was not significantly recruited to the c-Kit locus in Calyculin A-treated cells (Figure 4F). In general, CDK9, Mi2 and PCTD occupancy at c-Kit TSS, +1 and +5 regions, and PP1α recruitment at c-Kit TSS were also diminished, whereas POL II accumulated at TSS and +1 region and TBP recruitment did not vary (Figure 4F). Similar outcomes were observed at the Flt3 locus (Figure S4E–H). Calyculin A treatment did not affect CDK9 or PP1α protein levels, but rather led to the appearance of two slow-migrating CYCT1 - and IKAROS-specific bands (Figure 4G). These bands, which are likely corresponding to phosphorylated forms of CYCT1 and IKAROS proteins, were expressed at detectable levels. Thus, it can be concluded that the IKAROS-PP1α interaction: (i) contributes to IKAROS dephosphorylation and efficient binding to chromatin; (ii) allows PP1α association with c-Kit and Flt3 TSS and (iii) promotes CDK9/P-TEFb activation in order to release promoter-proximal paused POL II.
The IKAROS-PP1 interaction is required for normal hematopoietic cell differentiation
The functional role of IKAROS-PP1 interaction during hematopoiesis was further investigated by ex vivo assays in methylcellulose (Figure 5A). IkWT and IkNULL lin− HPCs were transduced with the control pMSCV empty vector (IkWT/GFP or IkNULL/GFP); IkNULL lin− HPCs were also transduced with pMSCV/Ik1 (Ikaros1 isoform, identified as IkNULL/Ik1 HPCs) or pMSCV/Ik1ΔPP1 (a mutant of Ikaros1 carrying the A465/7 mutation, identified as IkNULL/Ik1ΔPP1 HPCs). Ik1ΔPP1 is an IKAROS mutant unable to bind PP1 and therefore, resistant to dephosphorylation [35]. This mutant is reported to possess a shorter half-like than wild type IKAROS and thus, we assessed the stability of Ik1ΔPP1 in IkNULL/Ik1ΔPP1 HPCs by single-cell immunofluorescence (IF) analysis. Since these transgenes possess a Flag-HA tag, IF analysis was carried out with anti-HA antibodies. IF studies indicated that Ik1ΔPP1 accumulates in the nucleus of Ik1ΔPP1-infected lin− HPCs (Figure S5A). As well, in 293T transfected cells, whereby Ik1ΔPP1 was expressed at slightly lower levels than Ik1, Ik1ΔPP1 could interact with several known IKAROS protein partners (Mi2, CDK9, CYCT1 and MTA2) as demonstrated by protein co-IP experiments (Figure S5B).
Fig. 5. The IKAROS-PP1 interaction is required for normal hematopoietic differentiation. 
Ikaros homozygote null (IkNULL) bone marrow-derived lineage negative (lin−) hematopoietic progenitor cells (HPCs) were transduced with pMSCV/Ik (IkNULL/Ik), pMSCV/IkΔPP1 (IkNULL/IkΔPP1) or the empty pMSCV vector (IkNULL/GFP); Ikaros wild type (IkWT) lin− HPCs were transduced with the empty pMSCV vector only (IkWT/GFP); A) Scheme of the experimental procedure; Epo: Erythropoietin; ΔEpo: absence of Erythropoietin; B) Gene expression profiles in lin− transduced HPCs; mRNA samples were retro-transcribed with oligo-dT nucleotides; cDNA was used as templates for qPCR with Ikaros, c-Kit, Flt3 or Gapdh specific primer sets; β-Actin was used as internal control; y axis: relative mRNA enrichment levels; ratios are plotted as the mean ± SD of the measurements; n = 3. *: P≤0.05 by Student's t-test; C) In vitro hematopoietic differentiation of lin− HPCs. Clonogenic assays in methylcellulose of uninfected (IkWT lin− and IkNULL lin−) or transduced (IkWT/GFP; IkNULL/GFP; IkNULL/Ik; IkNULL/IkΔPP1) lin− HPCs; cells were seeded on methylcellulose and colonies were scored at day 14; CFU-GEMM: colony forming unit granulocyte, erythrocyte, macrophage, megakaryocyte; CFU-G/M/GM: collectively identifies granulo-macrophage colonies, BFU-E: burst-forming unit erythrocyte; BFU-E/Mk: immature erythroid colonies with elevated megakaryocytic content; BFU-Mk/E: almost pure megakaryocyte colonies that contain only few clusters of immature erythroid cells; the data shown are representative of three independent experiments; table: absolute number of methylcellulose colonies; CFC: Colony Forming Cell; Ratio: ratio of methylcellulose colonies per number of plated lin− HPCs; D–F) Cell proliferation and gene expression profiles in IkWT/GFP and IkNULL/GFP transduced lin− HPCs upon SCF (Stem Cell Factor) and FL (Flt3 ligand) treatment; IkWT/GFP and IkNULL/GFP transduced lin− HPCs were treated for 3 days with SCF and/or FL at the concentrations indicated at the bottom of the panels; D: cell proliferation and cell viability was determined by Trypan blue staining; y axis: cell number fold increase at day 0 and day 3; E, F: gene expression profile studies were carried out as detailed in panel B using β-Actin as internal control; y axis: mRNA enrichment levels relative to IkNULL-GFP/β-Actin values; the data shown in panels D–F are representative results of three independent experiments. In transduced lin− HPCs, Ikaros1 and Ikaros1ΔPP1 transgene expression levels were about 5 - and 8-fold higher than endogenous Ikaros (Figure 5B), which is comparable to other IKAROS rescue models [3]. As in IkNULL lin− HPCs, c-Kit and Flt3 expression was impaired in IkNULL/GFP lin− HPCs. Importantly, infection of IkNULL lin− HPCs with pMSCV/Ik1 increased c-Kit expression to 74% and Flt3 expression to 46.7% (if their expression levels in IkWT/GFP lin− HPCs is considered as 100%), whereas expression of c-Kit and Flt3 genes was only increased to 33.3% (for c-Kit) and 11.5% (for Flt3) upon pMSCV/Ik1ΔPP1 infection (Figure 5B). Thus, even though Ik1ΔPP1 could slightly increase c-Kit and Flt3 gene expression in lin− transduced HPCs, Ik1 restored transcription elongation of these target genes more efficiently, indicating the functional importance of the IKAROS-PP1 interaction for gene regulation.
Clonogenic assays in methylcellulose revealed that IkNULL lin− HPCs displayed decreased Colony Forming Cell (CFC) activity (1/166 IkNULL lin− HPCs vs. 1/33 IkWT lin− HPCs) as well as reduced colony size, which suggests reduction of cell proliferative potential. IkNULL lin− HPCs produced less mixed CFC (CFU-GEMM) and increased erythroid/megakaryocyte (BFU-E/Mk) colonies (Figure 5C).
Although the expression of the c-KIT receptor at the cell surface of IkNULL HPCs is minimal compared to IkWT HPCs [42], we investigated whether a close-to-normal c-KIT and FLT3 activation could be obtained by the stimulation of IkNULL HPCs with increasing amounts of Stem Cell Factor (SCF or KIT ligand) as well as FLT3 ligand (FL). We observed that IkNULL lin− HPCs were refractory to SCF and FL treatments. This functional limitation was demonstrated by the absence of: (i) induced cell proliferation (Figure 5D); (ii) c-Kit or Flt3 induction (Figure 5E, F); and (iii) hematopoietic commitment and differentiation (Figure S5C) upon SCF and/or FL treatment of IkNULL lin− HPCs. These results along with previously published work [51], suggest that IKAROS regulation of c-Kit and Flt3 expression is critical for CFC activity and differentiation ability of HPCs.
To define the biological contribution of IKAROS-dependent control over transcription elongation, we studied the hematopoietic potential of IkNULL lin− HPCs transduced with Ikaros1 or the elongation-incompetent Ikaros1ΔPP1 mutant. IkWT/GFP lin− HPCs maintained the same hematopoietic potential than non-transduced IkWT lin− HPCs, both in term of colony number as well as colony types. However, IkNULL/GFP lin− HPCs had increased CFU-GEMM and BFU-Mk/E and reduced BFU-E potential compared to non-transduced IkNULL lin− HPCs (Figure 5C; IkNULL vs. IkNULL/GFP). The gain in CFU-GEMM might be caused by an advantageous transduction ability of multipotent progenitor cells, whereas the gain in megakaryocyte vs. erythroid component, as indicated by increased BFU-Mk/E colonies, might be related to the Erythropoietin deprivation during the infection procedure (Figure 5A). Infection of IkNULL lin− HPCs with Ikaros1 significantly augmented CFC activity of transduced HPCs (Figure 5C, table), which correlates with the increased number of CFU-GEMM, BFU-E/Mk and, moreover, BFU-E colonies (Figure 5C, table; IkNULL lin− vs. IkNULL/Ik1 lin− HPCs). Interestingly, CFC activity of IkNULL/Ik1ΔPP1 lin- HPCs was lower than in IkNULL/Ik1 lin− HPCs (Figure 5C, table), but the hematopoietic potential of IkNULL/GFP lin− HPCs and IkNULL/Ik1ΔPP1 lin− HPCs was very similar (Figure 5C, table; IkNULL/GFP lin− vs. IkNULL/Ik1ΔPP1 lin− HPCs). Thus these results suggest that the Ikaros1ΔPP1 mutant is defective in restoring hematopoietic functions and that IKAROS interaction with PP1, which is required for productive transcription elongation, is important for normal hematopoiesis.
Discussion
P-TEFb can be recruited to gene promoters either by the general P-TEFb activator BRD4 [40] or by transcriptional activators that bind to specific consensus DNA sequences [37]. We previously reported that IKAROS acts as a template for the recruitment of P-TEFb to specific genes in hematopoietic cells [23], [27]. The partnership of IKAROS with the NuRD complex, which is critical for chromatin organization is also very-well established [5], [7], [18], [25]. The results presented here show that NuRD and P-TEFb can be associated together in a multifunctional complex, and that IKAROS is required for NURD-P-TEFb complex recruitment to specific genes in HPCs. Additionally, our results suggest that Mi2/NuRD actively participates in relieving POL II promoter-proximal pausing and contributes to the control of transcription elongation of IKAROS-target genes.
IKAROS association with NuRD-P-TEFb complex: A combination of chromatin organization and transcription elongation activities
Size fractionation analysis of nuclear extracts and IKAROS immunoaffinity-purified protein complexes revealed that a modest but significant percentage of nuclear CDK9 and CYCT1 (P-TEFb) is stably associated with IKAROS and NuRD components. This association resembles to the small and highly active portion of P-TEFb that is included in the Super Elongation Complexes (SECs), known to stimulate transcription elongation rate [38], [39]. Interestingly, based on LC-MS/MS analysis, it appears that several subunits of the SECs interact with IKAROS in hematopoietic cells (Table S3). Furthermore, the absence of the transcription elongation inhibitor, NELF, and the P-TEFb inhibitor, HEXIM1, among the IKAROS-NuRD-P-TEFb interacting proteins supports the notion that the newly defined complex favors transcription elongation in hematopoietic cells.
Nucleosomes form barriers for the elongating POL II. The main remodeling complex known to facilitate POL II passage through nucleosomes is the histone chaperone complex FACT [52]. Although we cannot exclude that FACT is implicated together with IKAROS and the NuRD-P-TEFb complex in the control of c-Kit elongation, FACT components were not identified by LC-MS/MS analysis of IKAROS-associated proteins. Instead, our data indicate that POL II release and the rate of transcription elongation of these genes are affected by Mi2 knockdown. In addition, Mi2/NuRD occupancy at c-Kit and Flt3 ORF decreased when transcription elongation was blocked by the CDK9 inhibitor Flavopiridol. Therefore, our results suggest that in HPCs, the NuRD complex can act as a chromatin complex that both destabilizes and restores nucleosomal structure in order to assist and facilitate the passage of POL II during transcription elongation. Accordingly, the NuRD complex can support nucleosome remodeling through the Mi2-associated helicase activity and histone deacetylation through the HDAC-associated activity [53], [54].
Initially regarded as a co-repressing complex because of its association with histone deacetylases and transcriptional repressors, the NuRD complex has also been associated with permissive chromatin and gene activation [5], [19]. More recently, it has been reported that the NuRD-mediated repression of a subset of pluripotency genes in ES cells occurs in a dynamic equilibrium with activation signals in order to fine-tune expression of these genes in response to differentiation stimuli [20]–[22]. Our findings bring a novel perspective to the role of NuRD in transcription regulation since we demonstrate a functional link between chromatin-associated activities that are required for the control of early transcription regulation i.e., NuRD-dependent chromatin remodeling associated with gene priming and PIC assembly [55], and productive transcription elongation whereby Mi2/NuRD contributes to P-TEFb-dependent relief of paused POL II and transcription elongation.
IKAROS recruits the NuRD-P-TEFb complex at chromatin in dosage-dependent manner
During differentiation of B-cell progenitors, the dynamic change of IKAROS expression level has been identified as a key mechanism for multiple target gene expression [15], [16]. Perturbation of IKAROS expression level has deleterious effects in double-positive thymocytes and in pre-B cells whereby reduced IKAROS function is associated with malignant transformation in mice and humans [41], [56], [57]. Furthermore, the IKAROS haploinsufficiency is reported to promote acute lymphoblastic leukemia with a high risk of relapse [58]. Our results suggest that chromatin association of the NuRD-P-TEFb complex to IKAROS target-genes depends on IKAROS expression level, hence providing a reasonable explanation for IKAROS dosage effects observed in hematological malignancies. In IkWT lin− HPCs and G1E2 cells, a fraction of IKAROS associates with P-TEFb and NuRD and this complex is recruited to c-Kit and Flt3 genes. In IkHT lin− HPCs and Ikaros knockdown G1E2 cells, IKAROS, Mi2/NuRD and CDK9/P-TEFb are recruited less efficiently to gene regulatory regions and transcription elongation is affected although transcription initiation occurs normally and POL II is in a promoter-proximal paused configuration. Finally, in IkNULL HPCs, transcription initiation occurs normally but P-TEFb and NuRD recruitment to c-Kit and Flt3 loci is impaired. Then, the enrichment of elongation-competent Ser2-phosphorylated POL II at gene ORF drops to almost background values. As a result, transcription elongation is profoundly reduced.
A large set of hematopoietic lineage-specific genes are characterized by permissive chromatin organization and PIC assembly at their promoters in HPCs [59]–[65]. IKAROS is required for the establishment of lineage-specific transcriptional programs [4], [5], [51]. Based on our results, we posit that when expressed at higher levels in HPCs and lymphoid progenitors, IKAROS induces rapid and dynamic expression of multiple genes poised for expression through the relief of the promoter-proximal paused POL II and transcription elongation. During alternative lineage-specification, IKAROS levels decrease, the IKAROS-NuRD-P-TEFb complex associates less efficiently to chromatin, and transcription elongation of different activated genes targeted by IKAROS declines.
IKAROS-PP1α functional interaction
PP1α is involved in CDK9 dephosphorylation at Thr-186 and Ser-175 [30]. Dephosphorylation of Thr-186 and Ser-175 facilitates P-TEFb dissociation from the 7SK snRNP repressive complex. While the importance of Ser-175 phosphorylation/dephosphorylation is debated, dephosphorylation of Thr-186 is known to promote binding of the CDK9/P-TEF complex at genes TSS [29]. At the TSS, Thr-186 is phosphorylated by the TFIIH-associated CDK7, an event required for the catalytic activity of CDK9 (P-TEFb activation) and thus, the release of promoter-proximal paused POL II [66]. Hyperphosphorylation of IKAROS negatively affects its stability and DNA binding affinity [67], [68]. IKAROS dephosphorylation by PP1 [35] enhances IKAROS binding to DNA and, based on results presented here, it can also favor the transfer of this phosphatase to CDK9, thereby contributing to P-TEFb release from the 7SK snRNP and CDK9 activation [29]. Indeed, immunoaffinity purification and FPLC analyses suggest that PP1α associates with IKAROS, NuRD and P-TEFb in a 2 MDa complex that does not include subunits of the 7SK snRNP repressive complex. Since we found that PP1α is particularly abundant in close proximity to the TSS of the transcriptionally active c-Kit and Flt3 genes, and PP1α recruitment to these TSS is highly influenced by IKAROS concentration, it can be argued that IKAROS contributes to chromatin recruitment of PP1α at the TSS of target genes hence, facilitating CDK9/P-TEFb association through dephosphorylation of CDK9 Thr-186 and possibly, Ser-175 [29], [69]. Furthermore, although CDK9 phosphorylation by CDK7 activates CDK9 and promotes POL II release, it has been demonstrated that imbalanced CDK9 hyperphosphorylation by CDK7 can have the opposite effect [30]. Thus, the relative higher abundance of PP1α at the TSS (when compared to the other regions bound by the IKAROS-NURD-P-TEFb) might be required to prevent excessive phosphorylation of TSS-bound CDK9 by the TFIIH-associated CDK7.
The importance of the IKAROS-PP1α network was demonstrated by results obtained with the PP1α inhibitor Calyculin A and with the Ik1ΔPP1 mutant. First, Calyculin A treatment, which leads to IKAROS hyperphosphorylation and reduced DNA binding [35], resulted in decreased recruitment of PP1α, Mi2 and CDK9 to c-Kit and Flt3 promoters but did not affect TBP and POL II recruitment. Thus, PP1α activity is critical for IKAROS as well as NuRD and P-TEFb recruitment to these promoters and for transcription elongation, while being dispensable for PIC formation. Second, the rescue of IkNULL HPCs attempted with the Ik1ΔPP1 mutant, which interacts with Mi2, MTA2, CYCT1 and CDK9 (Figure S5B) but does not efficiently interact with PP1 [35], demonstrated that without the interaction with PP1, IKAROS cannot exert its normal function during hematopoiesis.
It is worth noting that PP1 can contribute to co-transcriptional pre-mRNA splicing control [70], [71], an event that could also favor transcription elongation of IKAROS target genes. In fact, LC-MS/MS analysis suggested that some proteins implicated in the process of transcription termination can interact with IKAROS in hematopoietic cells (Table S3). Based on our results we suggest that IKAROS, NuRD and P-TEFb are not only active at the TSS but they facilitate transcription elongation and could also influence transcription termination.
Thus, IKAROS control over transcription elongation at genes such as c-Kit and Flt3 is likely to provide a rapid adjustment of their expression levels, and be a critical mechanism affecting HSC/HPCs interaction with the niche, cell fate and stress response [72]–[75].
In conclusion, we have demonstrated that IKAROS is the DNA-binding subunit of the newly characterized NuRD-P-TEFb multifunctional complex, a chromatin-associated complex that contains chromatin remodeling (NuRD) as well as gene transcription elongation (P-TEFb) activities. IKAROS is important for CDK9 and Mi2 interaction and combined recruitment to actively transcribed genes in hematopoietic cells. We demonstrate that low IKAROS expression level does not preclude appropriate promoter organization but impairs productive elongation, whereas higher IKAROS levels are necessary to relieve promoter-proximal paused POL II and efficient transcription elongation of target genes (Figure 6). Our results suggest that NuRD can assist POL II transcription elongation complex throughout the ORF of IKAROS target genes and provides mechanistic cues to explain the central role played by IKAROS as transcriptional activator during hematopoietic lineage decision, differentiation, and interaction with the niche.
Fig. 6. Model of IKAROS dosage effect for formation and recruitment of the NuRD-P-TEFb complex and transcription elongation control of IKAROS-target genes. 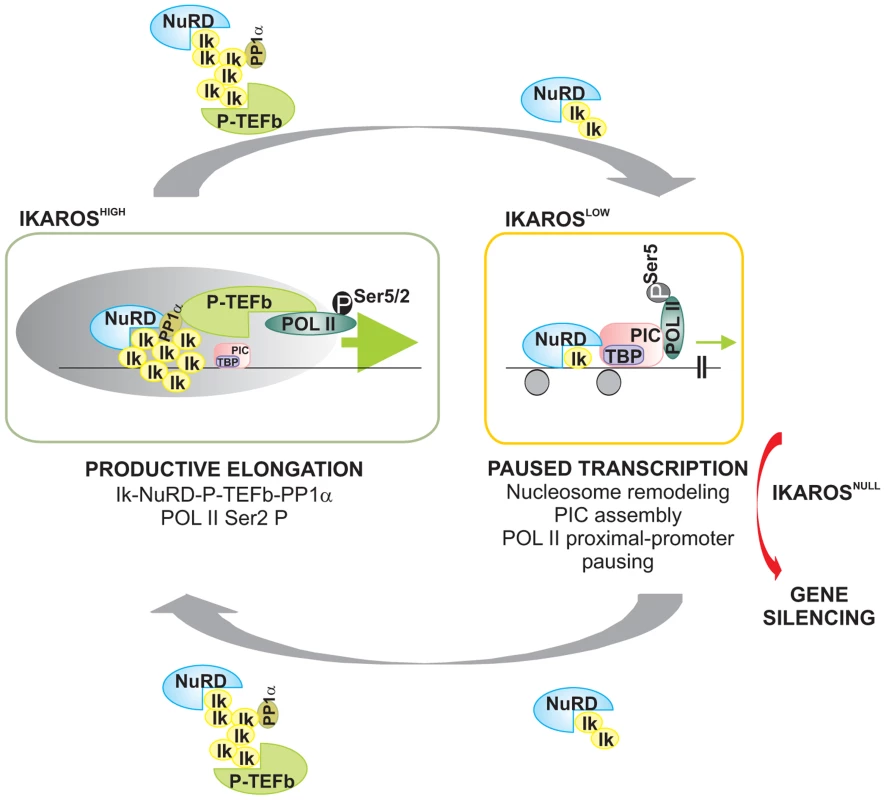
Productive elongation: when expressed at high levels, as in lineage negative hematopoietic progenitor cells, IKAROS (Ik) associates Mi2/NuRD and CDK9/P-TEFb activities into one complex at gene transcription start site (TSS) and throughout the ORF of IKAROS-target genes. IKAROS dependent enrichment of PP1α at TSS provides a control to P-TEFb activation by the dephosphorylation of CDK9. Once activated, CDK9/P-TEFb phosphorylates Ser-2 on POL II CTD as well as NELF and DSIF, two negative regulators of POL II elongation and thereby, releases the promoter-proximal paused POL II. Then, IKAROS and the NuRD-P-TEFb complex assist POL II during elongation. Based on results obtained by LC-MS/MS analysis, the latter might occurs along with SEC subunits. Paused transcription: the gradual decrease of IKAROS protein concentration, such as normally observed at defined stages of hematopoietic cell differentiation, still allows Mi2/NuRD loading at gene regulatory regions as well as PIC formation at gene promoters. However, lack of P-TEFb and PP1α recruitment results in promoter-proximal pausing of POL II. Gene silencing: in absence of IKAROS, as observed in lymphoma and leukemia clones carrying mutated and/or deleted alleles of IKAROS, the IKAROS-target genes are embedded in inactive chromatin conformation (not depicted) and they are characterized by the absence of PIC formation. To simplify the model, some of the proteins or complexes mentioned are not indicated. Materials and Methods
Protein co-IP, ChIP, re-ChIP, qPCR and qRT-PCR assays
These assays were done essentially as previously reported [7], [23], [27], [48].
Fast Protein Liquid Chromatography (FPLC)
Protein complexes were purified from Jurkat nuclear extract or IKAROS immunoaffinity-purified complexes on an AKTA Purifier FPLC System (GE Healthcare) using a Superose 6 10/300GL column (GE Healthcare).
Sequential immunoaffinity purification of IKAROS-associated proteins
Flag-HA sequential immunoaffinity (or Tap-Tag) purification was carried out as described in Nakatani et al. [76] starting from nuclear extracts prepared from pOZ-N-Flag-HA-IRES-ILR2 (mock sample) or pOZ-N-Flag-HA-IKAROS-IRES-ILR2 - (Flag-HA-Ik sample) Jurkat cell clones. LC-MS/MS analysis was performed at the Taplin Mass Spectrometry Facility (Harvard University, Boston).
Mouse transgenic lines and primary cell isolation
Homozygote (IkNULL) and heterozygote (IkHT) Ikaros null mice were genotyped by PCR as described [6]. Animals were sacrificed by cervical dislocation. Animal experiments were conducted in accordance with the Canadian Council on Animal Care (CCAC) guidelines and approved by the Maisonneuve-Rosemont Hospital animal care committee (approval numbers 2011-16 and 2011-17). Lineage negative (lin−) HPCs were purified using the easySep mouse hematopoietic progenitor enrichment kit (StemCell Technology).
Retrovirus infection and clonogenic assays in methylcellulose
pMSCV/Ik1 and pMSCV/Ik1ΔPP1 vectors were generated by cloning the murine Ikaros1 cDNA or the mutant Ikaros1ΔPP1 cDNA, both with a Flag and HA tag at their N-terminal regions, into the MSCV-pgk-EGFP vector (Dr G. Sauvageau, IRIC). Retroviral infection of lin− HPCs was carried out as described [77]. Lin− HPCs were cultured for 3 days in medium containing 50 ng/ml SCF, 10 ng/ml IL3, 10 ng/ml IL6 and 5×10−5 M β-mercaptoethanol, without Erythropoietin. Transduced lin−/GFP+ HPCs were isolated on a FACS Aria III sorter (BD Biosciences) based on green fluorescence. HPCs were seeded in triplicate on complete methylcellulose medium (MethoCult Stemcell Technologies). Colonies were scored at day 14.
Statistical analysis
Unpaired Student's t-test was used to determine the level of statistical significance (P-value).
Additional Materials and Methods information can be found in Text S1.
Supporting Information
Zdroje
1. HeizmannB, KastnerP, ChanS (2013) Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med 210 : 2823–2832.
2. JohnLB, WardAC (2011) The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol 48 : 1272–1278.
3. MalingeS, ThiollierC, ChlonTM, DoreLC, DieboldL, et al. (2013) Ikaros inhibits megakaryopoiesis through functional interaction with GATA-1 and NOTCH signaling. Blood 121 : 2440–51.
4. NgSY, YoshidaT, ZhangJ, GeorgopoulosK (2009) Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30 : 493–507.
5. ZhangJ, JacksonAF, NaitoT, DoseM, SeavittJ, et al. (2012) Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13 : 86–94.
6. WangJH, NichogiannopoulouA, WuL, SunL, SharpeAH, et al. (1996) Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5 : 537–549.
7. BottardiS, RossJ, BourgoinV, Fotouhi-ArdakaniN, Affar elB, et al. (2009) Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes. Mol Cell Biol 29 : 1526–1537.
8. ChoSJ, HuhJE, SongJ, RheeDK, PyoS (2008) Ikaros negatively regulates inducible nitric oxide synthase expression in macrophages: involvement of Ikaros phosphorylation by casein kinase 2. Cell Mol Life Sci 65 : 3290–3303.
9. DijonM, BardinF, MuratiA, BatozM, ChabannonC, et al. (2008) The role of Ikaros in human erythroid differentiation. Blood 111 : 1138–1146.
10. DumortierA, KirstetterP, KastnerP, ChanS (2003) Ikaros regulates neutrophil differentiation. Blood 101 : 2219–2226.
11. PapathanasiouP, AttemaJL, KarsunkyH, HosenN, SontaniY, et al. (2009) Self-renewal of the long-term reconstituting subset of hematopoietic stem cells is regulated by Ikaros. Stem Cells 27 : 3082–3092.
12. RossJ, MavoungouL, BresnickEH, MilotE (2012) GATA-1 utilizes Ikaros and polycomb repressive complex 2 to suppress Hes1 and to promote erythropoiesis. Mol Cell Biol 32 : 3624–3638.
13. YoshidaT, NgSY, GeorgopoulosK (2010) Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol 22 : 154–160.
14. GeorgopoulosK (2002) Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol 2 : 162–174.
15. Ferreiros-VidalI, CarrollT, TaylorB, TerryA, LiangZ, et al. (2013) Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood 121 : 1769–1782.
16. SchwickertTA, TagohH, GultekinS, DakicA, AxelssonE, et al. (2014) Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 15 : 283–293.
17. KimJ, SifS, JonesB, JacksonA, KoipallyJ, et al. (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10 : 345–355.
18. O'NeillDW, SchoetzSS, LopezRA, CastleM, RabinowitzL, et al. (2000) An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol 20 : 7572–7582.
19. MiccioA, WangY, HongW, GregoryGD, WangH, et al. (2010) NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J 29 : 442–456.
20. ReynoldsN, LatosP, Hynes-AllenA, LoosR, LeafordD, et al. (2012) NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell 10 : 583–594.
21. WhyteWA, BilodeauS, OrlandoDA, HokeHA, FramptonGM, et al. (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482 : 221–225.
22. YildirimO, LiR, HungJH, ChenPB, DongX, et al. (2011) Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147 : 1498–1510.
23. BottardiS, ZmiriFA, BourgoinV, RossJ, MavoungouL, et al. (2011) Ikaros interacts with P-TEFb and cooperates with GATA-1 to enhance transcription elongation. Nucleic Acids Res 39 : 3505–3519.
24. HarkerN, NaitoT, CortesM, HostertA, HirschbergS, et al. (2002) The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell 10 : 1403–1415.
25. KoipallyJ, HellerEJ, SeavittJR, GeorgopoulosK (2002) Unconventional potentiation of gene expression by Ikaros. J Biol Chem 277 : 13007–13015.
26. QuirionMR, GregoryGD, UmetsuSE, WinandyS, BrownMA (2009) Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol 182 : 741–745.
27. BottardiS, MavoungouL, BourgoinV, MashtalirN, Affar elB, et al. (2013) Direct Protein Interactions Are Responsible for Ikaros-GATA and Ikaros-Cdk9 Cooperativeness in Hematopoietic Cells. Mol Cell Biol 33 : 3064–3076.
28. ZhouQ, LiT, PriceDH (2012) RNA polymerase II elongation control. Annu Rev Biochem 81 : 119–143.
29. ChenR, LiuM, LiH, XueY, RameyWN, et al. (2008) PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev 22 : 1356–1368.
30. NekhaiS, PetukhovM, BreuerD (2014) Regulation of CDK9 activity by phosphorylation and dephosphorylation. Biomed Res Int 2014 : 964964.
31. D'OrsoI, FrankelAD (2010) HIV-1 Tat: Its Dependence on Host Factors is Crystal Clear. Viruses 2 : 2226–2234.
32. JangMK, MochizukiK, ZhouM, JeongHS, BradyJN, et al. (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19 : 523–534.
33. YangZ, YikJH, ChenR, HeN, JangMK, et al. (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19 : 535–545.
34. CeulemansH, BollenM (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84 : 1–39.
35. PopescuM, GurelZ, RonniT, SongC, HungKY, et al. (2009) Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J Biol Chem 284 : 13869–13880.
36. SridharanR, SmaleST (2007) Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem 282 : 30227–30238.
37. RomanoG (2013) Deregulations in the cyclin-dependent kinase-9-related pathway in cancer: implications for drug discovery and development. ISRN Oncol 2013 : 305371.
38. LuoZ, LinC, ShilatifardA (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13 : 543–547.
39. LinC, SmithER, TakahashiH, LaiKC, Martin-BrownS, et al. (2010) AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37 : 429–437.
40. WuSY, LeeAY, LaiHT, ZhangH, ChiangCM (2013) Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell 49 : 843–857.
41. WinandyS, WuP, GeorgopoulosK (1995) A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83 : 289–299.
42. NichogiannopoulouA, TrevisanM, NebenS, FriedrichC, GeorgopoulosK (1999) Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med 190 : 1201–1214.
43. WeissMJ, YuC, OrkinSH (1997) Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol 17 : 1642–1651.
44. JingH, VakocCR, YingL, MandatS, WangH, et al. (2008) Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29 : 232–242.
45. NovershternN, SubramanianA, LawtonLN, MakRH, HainingWN, et al. (2011) Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144 : 296–309.
46. RahlPB, LinCY, SeilaAC, FlynnRA, McCuineS, et al. (2010) c-Myc regulates transcriptional pause release. Cell 141 : 432–445.
47. ReppasNB, WadeJT, ChurchGM, StruhlK (2006) The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell 24 : 747–757.
48. DengC, LiY, LiangS, CuiK, SalzT, et al. (2013) USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet 9: e1003524.
49. ChaoSH, FujinagaK, MarionJE, TaubeR, SausvilleEA, et al. (2000) Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem 275 : 28345–28348.
50. SambolEB, AmbrosiniG, GehaRC, KennealeyPT, DecarolisP, et al. (2006) Flavopiridol targets c-KIT transcription and induces apoptosis in gastrointestinal stromal tumor cells. Cancer Res 66 : 5858–5866.
51. YoshidaT, NgSY, Zuniga-PfluckerJC, GeorgopoulosK (2006) Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 7 : 382–391.
52. OrphanidesG, LeRoyG, ChangCH, LuseDS, ReinbergD (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92 : 105–116.
53. KimT, XuZ, Clauder-MunsterS, SteinmetzLM, BuratowskiS (2012) Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150 : 1158–1169.
54. LiB, GogolM, CareyM, LeeD, SeidelC, et al. (2007) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316 : 1050–1054.
55. HuG, WadePA (2012) NuRD and pluripotency: a complex balancing act. Cell Stem Cell 10 : 497–503.
56. MullighanCG, GoorhaS, RadtkeI, MillerCB, Coustan-SmithE, et al. (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446 : 758–764.
57. MullighanCG, MillerCB, RadtkeI, PhillipsLA, DaltonJ, et al. (2008) BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453 : 110–114.
58. MullighanCG, SuX, ZhangJ, RadtkeI, PhillipsLA, et al. (2009) Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360 : 470–480.
59. AnguitaE, HughesJ, HeyworthC, BlobelGA, WoodWG, et al. (2004) Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. Embo J 23 : 2841–2852.
60. BottardiS, RossJ, Pierre-CharlesN, BlankV, MilotE (2006) Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. Embo J 25 : 3586–3595.
61. CaloE, WysockaJ (2013) Modification of enhancer chromatin: what, how, and why? Mol Cell 49 : 825–837.
62. JimenezG, GriffithsSD, FordAM, GreavesMF, EnverT (1992) Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci U S A 89 : 10618–10622.
63. PilonAM, AjaySS, KumarSA, SteinerLA, CherukuriPF, et al. (2011) Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood 118: e139–148.
64. WuW, ChengY, KellerCA, ErnstJ, KumarSA, et al. (2011) Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res 21 : 1659–1671.
65. YeM, IwasakiH, LaiosaCV, StadtfeldM, XieH, et al. (2003) Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19 : 689–699.
66. LarochelleS, AmatR, Glover-CutterK, SansoM, ZhangC, et al. (2012) Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 19 : 1108–1115.
67. Gomez-del ArcoP, MakiK, GeorgopoulosK (2004) Phosphorylation controls Ikaros's ability to negatively regulate the G(1)-S transition. Mol Cell Biol 24 : 2797–2807.
68. GurelZ, RonniT, HoS, KucharJ, PayneKJ, et al. (2008) Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J Biol Chem 283 : 8291–8300.
69. AmmosovaT, ObukhovY, KotelkinA, BreuerD, BeullensM, et al. (2011) Protein phosphatase-1 activates CDK9 by dephosphorylating Ser175. PLoS One 6: e18985.
70. MisteliT, SpectorDL (1996) Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell 7 : 1559–1572.
71. ShiY, ReddyB, ManleyJL (2006) PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell 23 : 819–829.
72. KimuraY, DingB, ImaiN, NolanDJ, ButlerJM, et al. (2011) c-Kit-mediated functional positioning of stem cells to their niches is essential for maintenance and regeneration of adult hematopoiesis. PLoS One 6: e26918.
73. MassonK, RonnstrandL (2009) Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell Signal 21 : 1717–1726.
74. PapathanasiouP, PerkinsAC, CobbBS, FerriniR, SridharanR, et al. (2003) Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 19 : 131–144.
75. ShinJY, HuW, NaramuraM, ParkCY (2014) High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J Exp Med
76. NakataniY, OgryzkoV (2003) Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 370 : 430–444.
77. KroonE, KroslJ, ThorsteinsdottirU, BabanS, BuchbergAM, et al. (1998) Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J 17 : 3714–3725.
78. HahmK, ErnstP, LoK, KimGS, TurckC, et al. (1994) The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol 14 : 7111–7123.
79. MolnarA, GeorgopoulosK (1994) The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol 14 : 8292–8303.
80. SunL, LiuA, GeorgopoulosK (1996) Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. Embo J 15 : 5358–5369.
81. CarottaS, DakicA, D'AmicoA, PangSH, GreigKT, et al. (2010) The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32 : 628–641.
Štítky
Genetika Reprodukční medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



