-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach
Integration of the viral DNA into host chromosomes was found in most of the hepatitis B virus (HBV)–related hepatocellular carcinomas (HCCs). Here we devised a massive anchored parallel sequencing (MAPS) method using next-generation sequencing to isolate and sequence HBV integrants. Applying MAPS to 40 pairs of HBV–related HCC tissues (cancer and adjacent tissues), we identified 296 HBV integration events corresponding to 286 unique integration sites (UISs) with precise HBV–Human DNA junctions. HBV integration favored chromosome 17 and preferentially integrated into human transcript units. HBV targeted genes were enriched in GO terms: cAMP metabolic processes, T cell differentiation and activation, TGF beta receptor pathway, ncRNA catabolic process, and dsRNA fragmentation and cellular response to dsRNA. The HBV targeted genes include 7 genes (PTPRJ, CNTN6, IL12B, MYOM1, FNDC3B, LRFN2, FN1) containing IPR003961 (Fibronectin, type III domain), 7 genes (NRG3, MASP2, NELL1, LRP1B, ADAM21, NRXN1, FN1) containing IPR013032 (EGF-like region, conserved site), and three genes (PDE7A, PDE4B, PDE11A) containing IPR002073 (3′, 5′-cyclic-nucleotide phosphodiesterase). Enriched pathways include hsa04512 (ECM-receptor interaction), hsa04510 (Focal adhesion), and hsa04012 (ErbB signaling pathway). Fewer integration events were found in cancers compared to cancer-adjacent tissues, suggesting a clonal expansion model in HCC development. Finally, we identified 8 genes that were recurrent target genes by HBV integration including fibronectin 1 (FN1) and telomerase reverse transcriptase (TERT1), two known recurrent target genes, and additional novel target genes such as SMAD family member 5 (SMAD5), phosphatase and actin regulator 4 (PHACTR4), and RNA binding protein fox-1 homolog (C. elegans) 1 (RBFOX1). Integrating analysis with recently published whole-genome sequencing analysis, we identified 14 additional recurrent HBV target genes, greatly expanding the HBV recurrent target list. This global survey of HBV integration events, together with recently published whole-genome sequencing analyses, furthered our understanding of the HBV–related HCC.
Published in the journal: . PLoS Genet 8(12): e32767. doi:10.1371/journal.pgen.1003065
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003065Summary
Integration of the viral DNA into host chromosomes was found in most of the hepatitis B virus (HBV)–related hepatocellular carcinomas (HCCs). Here we devised a massive anchored parallel sequencing (MAPS) method using next-generation sequencing to isolate and sequence HBV integrants. Applying MAPS to 40 pairs of HBV–related HCC tissues (cancer and adjacent tissues), we identified 296 HBV integration events corresponding to 286 unique integration sites (UISs) with precise HBV–Human DNA junctions. HBV integration favored chromosome 17 and preferentially integrated into human transcript units. HBV targeted genes were enriched in GO terms: cAMP metabolic processes, T cell differentiation and activation, TGF beta receptor pathway, ncRNA catabolic process, and dsRNA fragmentation and cellular response to dsRNA. The HBV targeted genes include 7 genes (PTPRJ, CNTN6, IL12B, MYOM1, FNDC3B, LRFN2, FN1) containing IPR003961 (Fibronectin, type III domain), 7 genes (NRG3, MASP2, NELL1, LRP1B, ADAM21, NRXN1, FN1) containing IPR013032 (EGF-like region, conserved site), and three genes (PDE7A, PDE4B, PDE11A) containing IPR002073 (3′, 5′-cyclic-nucleotide phosphodiesterase). Enriched pathways include hsa04512 (ECM-receptor interaction), hsa04510 (Focal adhesion), and hsa04012 (ErbB signaling pathway). Fewer integration events were found in cancers compared to cancer-adjacent tissues, suggesting a clonal expansion model in HCC development. Finally, we identified 8 genes that were recurrent target genes by HBV integration including fibronectin 1 (FN1) and telomerase reverse transcriptase (TERT1), two known recurrent target genes, and additional novel target genes such as SMAD family member 5 (SMAD5), phosphatase and actin regulator 4 (PHACTR4), and RNA binding protein fox-1 homolog (C. elegans) 1 (RBFOX1). Integrating analysis with recently published whole-genome sequencing analysis, we identified 14 additional recurrent HBV target genes, greatly expanding the HBV recurrent target list. This global survey of HBV integration events, together with recently published whole-genome sequencing analyses, furthered our understanding of the HBV–related HCC.
Introduction
Chronic human hepatitis B virus (HBV) infection leads to a range of liver diseases, from chronic hepatitis and liver cirrhosis to hepatocellular carcinoma (HCC). HBV infects approximately 2 billion people worldwide with 350 million suffering from chronic infection. HBV infection causes 500,000 to 1.2 million deaths annually, 320,000 of which are attributable to HCC [1]. Chronic HBV infection has been found to play a causal role in the development HCC from many epidemiological studies [2]. In addition, integrated HBV DNA sequences and episomal HBV genomes have been found in 85–90% of HBV-related HCCs [3].
The role of HBV integration in the development of HCC is rather complicated and yet not fully elucidated. HBV DNA integration was found to be distributed throughout different chromosomal sites in the host genome [4]. These integrations could induce chromosome changes, genome instability, or changes in the expression of human genes. HBV integration events were reported to be associated with chromosome fragile sites or repetitive sequences, and usually followed by local rearrangement, all of which relate to a higher genomic instability [5]. Furthermore, recent studies using genetic approaches and microarray technologies showed that HBV-related HCCs displayed higher rates of chromosomal alterations than HCCs of other risk factors [6]. HBV insertion was found to target the retinoic acid receptor-beta (RARB) gene [7] or the human cyclin A2 (CCNA2) [8], and generate chimeric oncogenic proteins.
A comprehensive analysis of HBV insertion sites targeting new genes would be desirable and would allow us to understand the mechanism of HCC development and to identify novel therapeutic targets. Towards this comprehensive goal, a moderate high-throughput approach using Alu-PCR approach has identified 68 cellular flanking sequences (seven repetitive or unidentified sequences, 42 cellular genes, and 19 sequences potentially coding for unknown proteins) for HBV integration in the HCC genomes [3]. The targeted genes belong to distinct pathways: calcium signaling related genes, 60s ribosomal protein encoding genes, and platelet derived growth factor and mixed lineage leukemia encoding genes. Recurrent HBV insertions nearby the telomerase reverse transcriptase (TERT) have also been reported [3], [9], [10].
Recently, many novel approaches have been developed to identify unique integration sites (UISs) for retrovirus in high-throughput manner using the next-generation sequencing (NGS) technologies including novel nonrestrictive approaches [11]–[13], which avoided preferential amplification and biased identification of UISs for the previous methods that use specific restriction enzymes.
More recently, complete genomic sequencing has been used to identify HBV integration genome-wide, taking advantage of the advance in next-generation sequencing technologies. Sung et al. conducted WGS at more than 30 fold coverage for 81 HBV positive HCCs and identified 399 HBV integration events using a criteria of more than 2 read pairs with close mapping positions linking an end of hg19 to an end of HBV, which resulted in the identification of an average of 4.9 HBV integration events per individual patient [14]. In another recently publication, Jiang et al. sequenced 4 HCC patients at 80X coverage and identified 255 HBV integration sites, which would generate at about 64 HBV integrations per individual patient [15]. Considering that WGS is still expensive and cost prohibitive for sequencing large number of samples, we have developed, in this study, an alternative method that integrates ligation-mediated PCR with Illumina's paired-end (PE) adapters for amplification and deep sequencing. Here we reported the analysis of 286 unique integration sites (UISs) from 40 pairs of HBV-related HCC and tumor-adjacent tissues, which resulted in the identification of about 7 HBV integrations per individual. We were able to further estimate the abundance of the insertion sites because our method used random fragmentation. Our analysis provided a global profile of HBV DNA integration for HCC. An integrated analysis with recently published whole genome sequencing analyses of HBV-related HCCs [14]–[16] provided us with 14 additional recurrent HBV target genes, thus greatly expanding the HBV recurrent target list.
Results
Development of an NGS–based approach for genome-wide analysis of HBV integration
We developed a massive anchored parallel sequencing (MAPS) approach to isolate and sequence integrants. The method combines thoughtful designs integrating ligation-mediated PCR with Illumina's PE adapters for amplification and deep sequencing. The MAPS approach stems from the intelligent integration of adapters used in Illumina library preparation with adapters used in LM-PCR, developed for HIV integration site analysis [17]. Illumina uses adapters for flow cell surface annealing, amplification and sequencing while LM-PCR uses adapters (or linkers) as primer matching sites for amplification of unknown nearby sequences. Adopting designing principles for linkers in established genome walking studies [18], we designed a functional adapter building upon Illumina's PE adapter sequence. The PE 2 Walking Adapter was partially complementary and ‘Y’ shaped, with a ‘T’ overhanging on the blunt end (Figure 1A).
Fig. 1. An overview of MAPS. 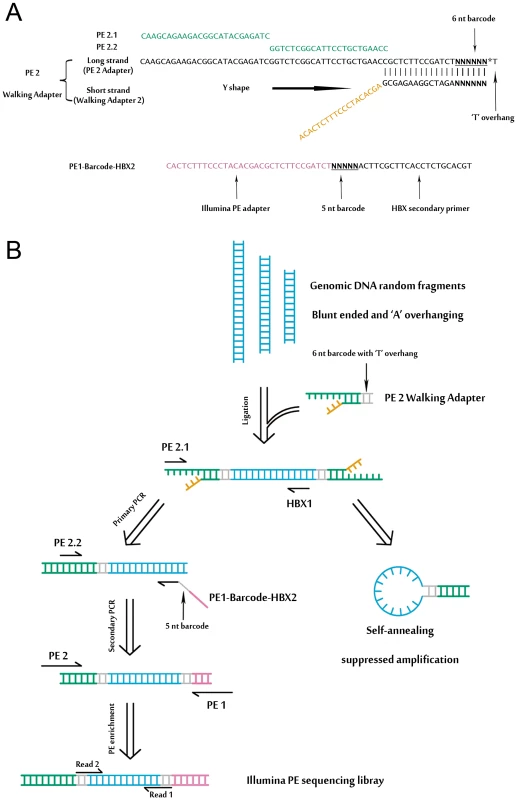
(A) Detailed illustration of the ‘Y’ shaped PE 2 Walking Adapter and the barcoded HBX nested primer. (B) Schematic of the MAPS approach. Adapters and primers are colored in accordance with (A). Detailed primer sequence information is provided in the Table S1, S2 and S3. A schematic outline of the MAPS is illustrated in Figure 1B, and all MAPS primers are listed in Table S1. Genomic DNA was sheared by sonication, resulting in fragment sizes ranging from 1 kb to 100 bp. The ligation of PE 2 Walking Adapter to the fragments generates an intermediate library with two important characteristics: first, one adapter of the single strand template is identical to Illumina's PE PCR Primer 2 (PE 2); second, the adapter sequence is long enough to suppress unspecific amplification due to single-primer PCR effect [19] during the nested PCR. Targeted sequences were further enriched and full length Illumina PE sequencing adapters were introduced by PE PCR Primer 1 and 2 (PE 1 and PE 2). The library fragments between sizes of 500–600 bp were selected for sequencing.
DNA bar coding and Illumina PE sequencing of HBV integrations
To make the MAPS approach more efficient, we introduced barcodes to primers for multiplex analysis (Figure 1, Table S2 and S3). In particular, the 6 nt barcode (with a common ‘T’ overhang) was introduced through the barcoded PE 2 Walking Adapters. The 5 nt barcode on Read 1 was introduced by barcoded primers through the secondary PCR.
We applied our MAPS approach in identifying HBV integrations in 40 pairs (cancer and adjacent tissues) of HBV-positive HCCs. According to the previous screen study [3], viral half of chimeric HBV-human DNA was preferentially truncated at a region between 1500–2000 base pairs on the HBV (numbering from the hypothetical EcoRI site of the HBV subtype adw). Therefore, a nested pair of primers in the gene hepatitis B virus x (HBX) of HBV was designed based on 4 HBV complete genomes (AY800389.1, AY800390.1, AY800391.1, and AY800392.1) isolated from the same local hospital where we collected the HCC tissues. The nested HBX primer (1583–1602), given the preference of viral truncation during integration, was able to generate chimeric HBV-human amplicons suited to the MAPS approach described above. Two negative controls were included, one with the human genomic DNA from an HBV-negative individual, and the other with the above plus purified HBV genomic DNA (i.e. a mixture of HBV DNA with the human genomic DNA, but no integration).
After NGS analysis, sequences from both ends were obtained. The Read 1 sequence starts from the same position after the anchored nested HBX primer; and the Read 2 sequence, which was obtained from the opposite end, differs in the alignment to the human genome due to the DNA random fragmentation from sonication (Figure 2A). We relied on the DNA shear point deduced in Read 2 to identify putative integrations. To be conservative, integration was determined as authentic if there is a Read 2 sequence present at the corresponding viral-cellular junction.
Fig. 2. Interpretation of the results of MAPS. 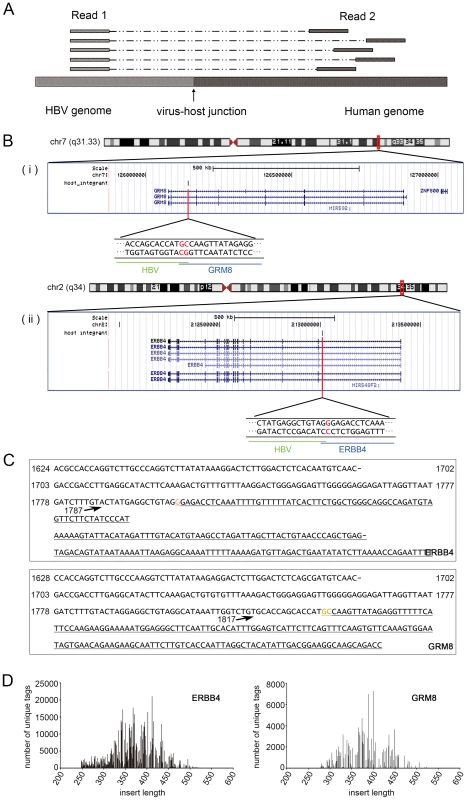
(A). A representative mapping of the paired reads from MAPS at integration sites. Read 1 sequences are anchored, since they extend from the fixed HBX nested primer; while the positions of Read 2 sequences vary due to the random fragmentation. (B). Insertion sites illustration in UCSC genome browser. (i) Insertion in ERBB4. (ii) Insertion in GRM8. (C). Integrant sequences of ERBB4 and GRM8. Breakpoint is indicated by arrow. The numbers are HBV genome coordinates, based on the reference of GQ205441. The human gene sequences are underlined. Orange bases indicate the common base between HBV and Human at the junction. (D). Distribution of integrant lengths in the sequenced library of ERBB4 and GRM8 integrations. Identification of global HBV integrations in the human genome and their distribution features
In the end, we identified 296 HBV DNA integrations (Table S4 and Table 1). 10 of which are paired, identical integrations found in both cancer and their adjacent tissues of the same patient, therefore there were 286 UISs. Among them, 42 sites were found in cancers, while 254 sites were found in adjacent tissues, which is about 6 times more than that found in cancers.
10.1371/journal.pgen.1003065.t001 Table 1Summary of HBV targeted genes identified.
Tissue disease state chr Insertion orientation Nubmer of unique end fragments Number of total reads Description Gene symbol (10 kb annotation)* Adjacent tissue chrX + 25 15550 Septin 6 SEPT6 Adjacent tissue chr16 − 6 4565 RNA binding protein, fox-1 homolog (C. elegans) 1 A2BP1 (RBFOX1) Adjacent tissue chr16 + 3 358 RNA binding protein, fox-1 homolog (C. elegans) 1 A2BP1 (RBFOX1) Adjacent tissue chr2 − 3 122 Abl interactor 2 ABI2 Adjacent tissue chr16 + 4 1449 acyl-CoA synthetase medium-chain family member 2A ACSM2A Adjacent tissue chr12 − 23 2062 Acyl-CoA synthetase short-chain family member 3 ACSS3 Adjacent tissue chr14 − 41 4606 chromosome 14 open reading frame 55 ADAM21 Adjacent tissue chr4 − 4 811 Alcohol dehydrogenase 1B (class I), beta polypeptide ADH1B Adjacent tissue chr10 − 5 1453 Aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4) AKR1C4 Adjacent tissue chr4 − 125 61810 Albumin ALB Cancer tissue chr12 − 9 30554 Asparagine-linked glycosylation 10 homolog B (yeast, alpha-1,2-glucosyltransferase) ALG10B Adjacent tissue chrX − 9 131 Asparagine-linked glycosylation 13 homolog (S. cerevisiae) ALG13 Adjacent tissue chr2 − 31 15568 Anaplastic lymphoma kinase (Ki-1) ALK Adjacent tissue chr2 − 18 11626 Alkaline phosphatase, intestinal ALPI Adjacent tissue chr7 − 52 44022 Amphiphysin AMPH Adjacent tissue chr22 + 4 536 Apolipoprotein L, 1 APOL1 Adjacent tissue chr11 − 16 862 Rho guanine nucleotide exchange factor (GEF) 12 ARHGEF12 Adjacent tissue chr11 − 6 3607 Rho guanine nucleotide exchange factor (GEF) 12 ARHGEF12 Adjacent tissue chr8 − 7 6932 ATPase, H+ transporting, lysosomal 38 kDa, V0 subunit d2 ATP6V0D2 Adjacent tissue chr22 + 18 9882 BCL2-interacting killer (apoptosis-inducing) BIK Adjacent tissue chr7 + 6 658 V-raf murine sarcoma viral oncogene homolog B1 BRAF Adjacent tissue chr12 − 20 615 Chromosome 12 open reading frame 40 C12orf40 Adjacent tissue chr15 + 6 326 DNM1 pseudogene 46 C15orf51 Adjacent tissue chr4 + 37 2363 C1q and tumor necrosis factor related protein 7 C1QTNF7 Adjacent tissue chr9 − 9 250 Chromosome 9 open reading frame 93 C9orf93 Adjacent tissue chr4 + 9 835 coiled-coil domain containing 158 CCDC158 Cancer tissue chr4 − 130 73841 Cyclin A2 CCNA2 Adjacent tissue chr4 − 14 9198 Cyclin A2 CCNA2 Adjacent tissue chr6 − 6 878 Cyclin C CCNC Adjacent tissue chr17 − 11 2684 Cell division cycle 27 homolog (S. cerevisiae) CDC27 Adjacent tissue chr7 − 4 2255 cyclin-dependent kinase 13 CDC2L5 Adjacent tissue chr5 + 18 470 Cadherin 10, type 2 (T2-cadherin) CDH10 Adjacent tissue chr16 + 14 6882 Cadherin 8, type 2 CDH8 Adjacent tissue chr3 + 5 187 Centrosomal protein 63 kDa CEP63 Adjacent tissue chr1 + 3 174 Chitinase 3-like 2 CHI3L2 Adjacent tissue chr12 + 38 693 Citron (rho-interacting, serine/threonine kinase 21) CIT Cancer chr3 − 236 66086 Contactin 6 CNTN6 Adjacent tissue chr21 − 42 2632 Collagen, type XVIII, alpha 1 COL18A1 Adjacent tissue chrX − 45 1849 Collagen, type IV, alpha 6 COL4A6 Adjacent tissue chr21 − 4 322 Collagen, type VI, alpha 2 COL6A2 Adjacent tissue chr2 + 95 91537 Carbamoyl-phosphate synthetase 1, mitochondrial CPS1 Adjacent tissue chr15 − 28 2082 CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 CTDSPL2 Adjacent tissue chr11 + 9 7279 CUGBP, Elav-like family member 1 CUGBP1 Adjacent tissue chr10 − 5 92 CUGBP, Elav-like family member 2 CUGBP2 Cancer tissue chr10 − 298 261343 Cytochrome P450, family 2, subfamily C, polypeptide 8 CYP2C8 Adjacent tissue chr10 + 29 6742 Cytochrome P450, family 2, subfamily C, polypeptide 8 CYP2C8 Adjacent tissue chr11 + 5 168 Discs, large homolog 2, chapsyn-110 (Drosophila) DLG2 Adjacent tissue chr8 − 3 46 Dihydropyrimidinase DPYS Adjacent tissue chr6 − 61 5013 Ectonucleotide pyrophosphatase/phosphodiesterase 1 ENPP1 Adjacent tissue chr1 + 4 60 Epidermal growth factor receptor pathway substrate 15 EPS15 Adjacent tissue chr2 + 301 792607 V-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) ERBB4 Cancer chr16 + 16 6616 Endoplasmic reticulum to nucleus signaling 2 ERN2 Adjacent tissue chr1 + 58 7369 Estrogen-related receptor gamma ESRRG Adjacent tissue chr4 − 15 8951 Ellis van Creveld syndrome EVC Adjacent tissue chr17 + 14 10665 Family with sequence similarity 117, member A FAM117A Adjacent tissue chr3 − 25 1844 family with sequence similarity 162, member A FAM162A Adjacent tissue chr5 + 4 957 F-box and leucine-rich repeat protein 17 FBXL17 Adjacent tissue chr5 + 9 196 F-box and leucine-rich repeat protein 7 FBXL7 Adjacent tissue chr4 − 23 318 KIAA1727 protein FHDC1 Adjacent tissue chr18 + 6 5412 Formin homology 2 domain containing 3 FHOD3 Adjacent tissue chr2 − 169 36268 Fibronectin 1 FN1 Adjacent tissue chr2 − 9 1054 Fibronectin 1 FN1 Adjacent tissue chr2 − 47 631 Fibronectin 1 FN1 Adjacent tissue chr2 − 8 301 Fibronectin 1 FN1 Cancer tissue chr2 − 58 33209 Fibronectin 1 FN1 Adjacent tissue chr17 + 9 30405 Fructosamine-3-kinase-related protein FN3KRP Adjacent tissue chr3 + 89 15775 Fibronectin type III domain containing 3B FNDC3B Adjacent tissue chr4 + 47 30826 Fraser syndrome 1 FRAS1 Adjacent tissue chr4 + 32 1331 Follistatin-like 5 FSTL5 Adjacent tissue chr14 − 3 35 exonuclease 3′-5′ domain containing 2 GALNTL1 Adjacent tissue chr7 − 3 2771 UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 GALNTL5 Cancer tissue chr5 − 30 2690 Glucosamine-6-phosphate deaminase 1 GNPDA1 Adjacent tissue chr13 − 4 1057 Glypican 6 GPC6 Adjacent tissue chr4 − 5 125 Glutamate receptor, ionotropic, delta 2 GRID2 Adjacent tissue chr7 + 156 127769 Glutamate receptor, metabotropic 8 GRM8 Adjacent tissue chr12 + 22 453 Glycogen synthase 2 (liver) GYS2 Adjacent tissue chr11 + 6 22742 Mps one binder kinase activator-like 2 HCCA2 Adjacent tissue chr6 − 6 3624 HCG27 Adjacent tissue chr6 + 6 3221 Histone cluster 1, H2bd HIST1H2BD Adjacent tissue chr19 − 6 256 Histocompatibility (minor) HA-1 HMHA1 Adjacent tissue chr10 + 25 1214 Heparanase 2 HPSE2 Adjacent tissue chr3 + 18 504 Histidine-rich glycoprotein HRG Adjacent tissue chr5 + 5 568 Interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) IL12B Adjacent tissue chr3 + 5 2776 Interleukin 1 receptor accessory protein IL1RAP Adjacent tissue chr10 − 4 2968 Interleukin 2 receptor, alpha IL2RA Adjacent tissue chr6 + 5 431 Interphotoreceptor matrix proteoglycan 1 IMPG1 Adjacent tissue chr19 − 10 3440 Inositol 1,4,5-trisphosphate 3-kinase C ITPKC Adjacent tissue chr17 + 124 1293 K(lysine) acetyltransferase 2A KAT2A Adjacent tissue chr14 + 8 291 KIAA0317 KIAA0317 Adjacent tissue chr3 + 8 636 Kelch-like 6 (Drosophila) KLHL6 Adjacent tissue chr6 − 9 304 Lin-28 homolog B (C. elegans) LIN28B Cancer tissue chr9 − 53 4292 Leucine rich repeat and Ig domain containing 2 LINGO2 Adjacent tissue chr7 + 45 1917 Limb region 1 homolog (mouse) LMBR1 Adjacent tissue chr12 − 4 119 WAS protein family homolog 1 pseudogene LOC100288778 Adjacent tissue chr5 + 7 10522 Hypothetical protein LOC285627 LOC285627 Adjacent tissue chr17 − 5 246 LOC644172 Adjacent tissue chr6 + 62 1101 Leucine rich repeat and fibronectin type III domain containing 2 LRFN2 Adjacent tissue chr2 + 45 703 Low density lipoprotein-related protein 1B (deleted in tumors) LRP1B Adjacent tissue chr15 − 5 582 Leucine-rich repeat kinase 1 LRRK1 Adjacent tissue chr4 + 14 14654 Mastermind-like 3 (Drosophila) MAML3 Adjacent tissue chr1 + 6 2023 Mannan-binding lectin serine peptidase 2 MASP2 Adjacent tissue chr3 − 3 48 Muscleblind-like (Drosophila) MBNL1 Adjacent tissue chr6 − 43 3090 Mediator complex subunit 23 MED23 Adjacent tissue chr19 + 12 9041 Meis homeobox 3 MEIS3 Adjacent tissue chr7 + 5 68 microRNA 182 MIR182 Adjacent tissue chr6 + 4 90 microRNA 206 MIR206 Adjacent tissue chr7 + 11 631 Myeloid/lymphoid or mixed-lineage leukemia 3 MLL3 Cancer tissue chr19 − 80 4468 Myeloid/lymphoid or mixed-lineage leukemia 4 MLL4 Cancer chr2 − 26 840 Melanoregulin MREG Adjacent tissue chr5 + 16 650 Myosin X MYO10 Adjacent tissue chr18 + 31 17222 Myomesin 1, 185 kDa MYOM1 Cancer tissue chr7 + 192 76342 long intergenic non-protein coding RNA 174 NCRNA00174 Adjacent tissue chr11 + 3 129 NEL-like 1 (chicken) NELL1 Adjacent tissue chr6 − 127 123237 Na+/K+ transporting ATPase interacting 2 NKAIN2 Adjacent tissue chr10 − 88 73961 Neuregulin 3 NRG3 Adjacent tissue chr2 − 43 31154 Neurexin 1 NRXN1 Adjacent tissue chr3 − 6 7428 Poly (ADP-ribose) polymerase family, member 9 PARP9 Adjacent tissue chr2 + 4 84 Phosphodiesterase 11A PDE11A Adjacent tissue chr1 − 4 278 Phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4 dunce homolog, Drosophila) PDE4B Adjacent tissue chr8 + 8 10926 Phosphodiesterase 7A PDE7A Adjacent tissue chr6 + 14 442 enoyl-CoA delta isomerase 2 PECI Adjacent tissue chr14 + 4 1864 Placental growth factor, vascular endothelial growth factor-related protein PGF Cancer chr5 − 289 3293 Protein geranylgeranyltransferase type I, beta subunit PGGT1B Adjacent tissue chr1 − 14 216 Phosphatase and actin regulator 4 PHACTR4 Adjacent tissue chr1 − 11 491 Phosphatase and actin regulator 4 PHACTR4 Cancer tissue chr12 − 54 6030 Polyhomeotic homolog 1 (Drosophila) PHC1 Cancer chr3 − 107 7765 Phosphoinositide-3-kinase, catalytic, beta polypeptide PIK3CB Adjacent tissue chr20 − 7 1858 Phospholipase C, beta 1 (phosphoinositide-specific) PLCB1 Adjacent tissue chr1 − 12 5993 Phospholipase D family, member 5 PLD5 Adjacent tissue chr7 − 29 10098 Plexin A4 PLXNA4 Adjacent tissue chr12 + 6 256 Protein phosphatase 1H (PP2C domain containing) PPM1H Adjacent tissue chr8 + 20 700 Protein kinase, DNA-activated, catalytic polypeptide PRKDC Adjacent tissue chr17 + 7 2875 Proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 PSMD12 Adjacent tissue chr11 + 43 9668 Protein tyrosine phosphatase, receptor type, J PTPRJ Adjacent tissue chr17 + 18 714 RAB5C, member RAS oncogene family RAB5C Adjacent tissue chr17 − 6 320 Retinoic acid induced 1 RAI1 Adjacent tissue chr17 − 77 9045 Retinoic acid receptor, alpha RARA Cancer chr10 − 306 96735 Hypothetical protein LOC220980 RASSF4 Adjacent tissue chr10 − 4 127 Retinol binding protein 4, plasma RBP4 Adjacent tissue chr6 + 39 2877 RALBP1 associated Eps domain containing 1 REPS1 Adjacent tissue chrX + 4 1297 Regucalcin (senescence marker protein-30) RGN Adjacent tissue chr17 + 11 4735 Rho GTPase activating protein 44 RICH2 Adjacent tissue chr15 + 3 120 S phase cyclin A-associated protein in the ER SCAPER Adjacent tissue chr6 + 138 23599 Sex comb on midleg-like 4 (Drosophila) SCML4 Adjacent tissue chr2 + 4 1532 SEC14 and spectrin domains 1 SESTD1 Adjacent tissue chr8 + 3 554 Secreted frizzled-related protein 1 SFRP1 Adjacent tissue chr14 + 77 795 surfactant associated 3 SFTA3 Adjacent tissue chr5 + 6 579 Superkiller viralicidic activity 2-like 2 (S. cerevisiae) SKIV2L2 Adjacent tissue chr12 − 27 22758 Solute carrier family 2 (facilitated glucose transporter), member 13 SLC2A13 Adjacent tissue chr3 + 26 14482 Sarcolemma associated protein SLMAP Adjacent tissue chr4 − 6 8441 SMAD family member 1 SMAD1 Cancer chr5 + 570 20953 SMAD family member 5 SMAD5 Adjacent tissue chr5 + 31 310 SMAD family member 5 SMAD5 Cancer chr5 + 30 1124 SMAD family member 5 SMAD5 Adjacent tissue chr2 − 14 2397 Son of sevenless homolog 1 (Drosophila) SOS1 Adjacent tissue chr12 + 16 7109 SRY (sex determining region Y)-box 5 SOX5 Adjacent tissue chr11 + 4 56 Spondin 1, extracellular matrix protein SPON1 Adjacent tissue chr1 + 7 8119 Single stranded DNA binding protein 3 SSBP3 Adjacent tissue chr8 + 11 4638 Suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) ST18 Adjacent tissue chr1 − 17 1196 Syntaxin 6 STX6 Adjacent tissue chr5 + 7 469 Synaptic vesicle glycoprotein 2C SV2C Adjacent tissue chr8 + 42 8798 Transforming, acidic coiled-coil containing protein 1 TACC1 Adjacent tissue chr8 + 16 4216 TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150 kDa TAF2 Cancer tissue chr5 − 382 46659 Telomerase reverse transcriptase TERT Cancer chr5 − 86 2724 Telomerase reverse transcriptase TERT Cancer tissue chr5 − 63 3789 Telomerase reverse transcriptase TERT Cancer chr5 − 230 2853 Telomerase reverse transcriptase TERT Cancer tissue chr5 − 270 7445 Telomerase reverse transcriptase TERT Adjacent tissue chr5 − 5 259 Telomerase reverse transcriptase TERT Cancer tissue chr5 − 392 47491 Telomerase reverse transcriptase TERT Adjacent tissue chr5 − 55 2941 Telomerase reverse transcriptase TERT Adjacent tissue chr12 + 47 3611 Transmembrane protein 117 TMEM117 Adjacent tissue chr12 + 3 105 Transmembrane protein 132B TMEM132B Adjacent tissue chr15 − 11 6194 Transmembrane protein 87A TMEM87A Adjacent tissue chr1 + 7 412 Ubiquitin specific peptidase 33 USP33 Adjacent tissue chr2 + 4 225 Xin actin-binding repeat containing 2 XIRP2 Adjacent tissue chr11 − 3 244 Zinc finger and BTB domain containing 16 ZBTB16 Adjacent tissue chr16 − 81 13492 Zinc finger protein 1 homolog (mouse) ZFP1 Adjacent tissue chr9 + 12 5969 Zinc finger protein 618 ZNF618 Adjacent tissue chr7 − 9 370 Zinc finger protein 804B ZNF804BWe randomly picked 28 putative insertions and used a standardized nested-PCR assay, we confirmed 27 out of 28 putative insertions (96%) (Table S5). Our confirmation rate is similar to what Sung et al. reported for the HBV insertions from whole genome sequencing, which they randomly selected 32 breakpoints at the 6 affected genes for PCR analysis in 22 samples and were able to successfully validate 82% of these integration sites [14]. To further validate those putative sites, we sequenced two amplified integrant PCR products (HBV integration in ERBB4 and GRM8) by the Sanger method. BLAST analysis revealed that both sequences extend from the HBX gene to the identified sites in the human genome (Figure 2B and 2C). We also examined the length of DNA fragments in the sequencing library. The lengths of the integrants (without the adapter and barcode sequences) clustered around 350–450 bp (Figure 2D).
We annotated insertions the nearest RefSeq genes within 10 kb from the HBV-Human junction points (Table S4) and this annotation was used in the other analyses in this manuscript. However, we also provided the annotations to within 150 kb in the Table S4 as previous studies showed that the HBV genome includes enhancer elements capable of activating promoters even 150 kb away, in a position and orientation independent manner [9], [20].
We compared the genes targeted by HBV from our study to three recently published studies. Fujimoto et al. only identified 6 genes (ADAM5P, FRAS1, THRB, HNF1A, FAM18B2, TTLL9) in addition to TERT (their supplementary table 13) from 11 HBV-related HCCs in 23 integration events [16]. Only the TERT gene is in common with our HBV targeted gene list. Sung et al. identified 399 HBV integration events from 81 HBV positive HCCs (from their supplementary table S3). As they did not indicate what distance they allowed from the HBV integration to the start or end site of genes, we re-annotated their list with the same criterion we used in our annotation to within 10 kb of genes, and then performed the comparison. Jiang et al. reported the identification of 255 HBV integration events in three HBV positive HCCs [15]. We also re-annotated Jiang et al.'s list to within 10 kb of the gene for the comparison. Comparing our list (Table 1) to Sung et al.'s list, we found that there are 9 genes in common (Table 2). Comparing our list to Jiang et al.'s list, we found that they are four genes in common. Two genes, FN1 (fibronectin 1) and MLL4 (Myeloid/lymphoid or mixed-lineage leukemia 4) were found in all three studies (Table 2). Except for FN1 and MLL4, there were no additional common genes between the Sung et al.'s list and Jiang et al.'s list.
10.1371/journal.pgen.1003065.t002 Table 2Common HBV targeted genes among the three studies.
Our study Sung et al. Jiang et al. Description ADH1B ADH1B Alcohol dehydrogenase 1B (class I), beta polypeptide CPS1 CPS1 Carbamoyl-phosphate synthetase 1, mitochondrial ESRRG ESRRG Estrogen-related receptor gamma FN1 FN1 FN1 Fibronectin 1 LRFN2 LRFN2 Leucine rich repeat and fibronectin type III domain containing 2 MLL4 MLL4 MLL4 Myeloid/lymphoid or mixed-lineage leukemia 4 MYOM1 MYOM1 Myomesin 1, 185 kDa RAI1 RAI1 Retinoic acid induced 1 TERT TERT Telomerase reverse transcriptase CTDSPL2 CTDSPL2 CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 LRP1B LRP1B Low density lipoprotein-related protein 1B (deleted in tumors)We also checked all the HBV integration events against the miRNA clusters using the genomic coordinates of all miRNAs in the miRBase (www.mirbase.org). We found one integration event at ChrX:108297754 that maps 18 nucleotides upstream of miRNA has-mir-6087. Has-mir-6087 was recently identified in human ES cells [21].
We also mapped the position of each UIS to chromosomes and generated a superimposed diagram illustrating integration of UISs to within cytogenetic bands of the human genome (Figure 3). To analyze whether the HBV integrations had any preferences over transcript units and individual chromosomes, a set of 8015 computer-simulated random integrations was generated as the control. We calculated the frequency of HBV integration within each chromosome, showing that chromosome 17 was favored target for HBV integration (Figure 4). 60.5% of the HBV integrations (178/294) landed in RefSeq genes (Table S4). This is significantly different from the result of the random dataset, of which only 42.68% landed in genes reflecting the feature of the human genomes (Pearson's chi-squared test, P<0.0001).
10.1371/journal.pgen.1003065.g003 Figure 3Distribution of the identified UISs.
Locations of UISs in the human genome. Circles indicate the UISs found only in cancer tissue, triangles represent UISs only found in the adjacent tissue, and rectangles mean UISs found in both. The nearest gene in the vicinity of UIS was annotated, and the filled shapes marks UISs that integrate in the transcript unit.
10.1371/journal.pgen.1003065.g004 Figure 4Number of UISs per chromosome.
A frequency of 1 (represented by the dotted line) stands for the same level of integration as the random data, i.e., this chromosome is neither favored nor disfavored for HBV integration.
Functional annotations of the HBV targeted genes
To understand the function of the genes targeted by HBV, we performed Gene Ontology (GO) analysis using the GO miner program. We found that they are enriched in 3 GO terms related to cAMP metabolic processes and several GO terms related to T-cell differentiation and activation involved in immune response (Table S6). Interestingly, GO terms related to GO:0034661 ncRNA catabolic process, GO:0031050 dsRNA fragmentation, GO:0070918 production of small RNA involved in gene silencing by RNA, and GO:0071359 cellular response to dsRNA were also enriched (Table S6). These suggest that the HBV targeted host genes related to responding, or to defending or eliminating the virus such as the immune response and degradation of RNAs. We also found that GO terms related to TGF beta receptor pathway were enriched (Table S6).
To understand whether HBV targeted genes are enriched with particular protein domains, we used the DAVID (Database for Annotation, Visualization and Integrated Discovery) Functional Annotation tool to analyze the combined list of HBV targeted genes (Table 1). The enriched INTERPRO terms with EASE score, which is a modified Fisher Exact P-Value, <0.05, were shown is Table S7. We found that 7 genes (PTPRJ, CNTN6, IL12B, MYOM1, FNDC3B, LRFN2, FN1) contain the IPR003961 (Fibronectin, type III domain), 7 genes (NRG3, MASP2, NELL1, LRP1B, ADAM21, NRXN1, FN1) with the IPR013032 (EGF-like region), conserved site, and three genes (PDE7A, PDE4B, PDE11A) containing the IPR002073 (3′, 5′-cyclic-nucleotide phosphodiesterase) (Table S7). Mapping the HBV targeted genes to the KEGG pathways, we found that they were enriched in several KEGG pathways including hsa04512 (ECM-receptor interaction), hsa04510 (Focal adhesion), and hsa04012 (ErbB signaling pathway) (Table S8).
Semi-quantification of HBV insertion sites based on tags covering the integration sites
The abundance of the UISs was quantified by the count of different shear points near the junction, a method that was also utilized by Gillet et al. [11]. In addition, we tested the quantitative potential of the UISs abundance by a serial dilution PCR, and found that the number of unique tags covering an integration site correlated with the abundance of the integration determined by serial dilution PCR, both for an individual sample (Figure 5A) and across different samples (Figure 5B).
10.1371/journal.pgen.1003065.g005 Figure 5Quantitative feature of MAPS.
(A) Semi-quantitative PCR results in HCC DNA sample N13. Each panel represents different integration frequencies, judged by the number of unique tags identified by MAPS, as indicated by the number followed by the sample name on the left column labels. The amounts of genomic DNA used for PCR were indicated at the top of the lanes. (B) Semi-quantitative PCR results in multiplexed DNA samples. The left column indicates the HCC DNA sample identifications and the number of unique tags identified. (C) Correlation co-efficiency (R2 = 0.9278) of the numbers of unique tags covering the common insertions retrieved between two technical replicate runs. (D) Rarefaction analysis of integration sites recovered in MAPS.
To examine technical reproducibility of the quantification, we compared duplicate results using same DNA samples but sheared separately. The correlation co-efficiency of the number of unique tags covering the common integration sites, even between two technical replicate experiments with slight modifications of primers used (one with the single 5 nt barcode and the other with the paired barcodes), was very high (R2 = 0.9278; Figure 5C). We also randomized the total reads into 39 subsets and plotted a rarefaction curve showing the number of putative integrations identified with accumulating subsets. The curve plateaus in Figure 5D suggested that further sequencing would not yield many additional integration sites. The reliability of quantification was also reflected by the consistency in the number of unique tags covering the opposite integration junctions from the same integrations (data not shown).
Fewer UISs were identified in HCC comparing to adjacent tissues supporting the theory of clonal expansion in HCC development
As each integration event provides a unique genetic marker for a cell, we can assess the clonal expansion of HBV integrated hepatocytes using UISs. We found that the cancer tissues showed clonal expansion compared to tumor-adjacent tissues, as judged by the number of shear sites near individual integration (Figure 6, Mann–Whitney test, p<0.0001). Furthermore, the number of UISs (median = 1) found in cancer was less than that of the tumor-adjacent tissue (median = 7, Figure 6B, Mann–Whitney test, p<0.0001), consistent with the results from previous studies [3], [22]. Interestingly, there was no difference in the UIS abundance between clones inserted in the vicinity of genes and clones that were not (Mann–Whitney test, p = 0.8576) (Data not shown).
10.1371/journal.pgen.1003065.g006 Figure 6More clonal expansion but fewer insertions were found in cancer than in the adjacent tissue.
(A) We counted the number of unique tags to quantify the relative abundance of each UIS. The bar is the median of the number of unique tags. (B) Number of HBV integrations identified in cancer and adjacent tissue. The bar represents the median of the number of integrations.
Recurrent HBV integration into 8 human host genes including TERT and FN1 genes
We annotated the 286 UISs with RefSeq genes and found that 8 genes were targeted by HBV integration for more than once (Table 3), including ARHGEF12 (Rho guanine nucleotide exchange factor (GEF) 12), CYP2C8 (cytochrome P450, family 2, subfamily C, polypeptide 8), FN1 (fibronectin 1), PHACTR4 (Phosphatase and actin regulator 4), PLXNA4 (Plexin A4), RBFOX1 [RNA binding protein, fox-1 homolog (C. elegans) 1], SMAD5 (SMAD family member 5), and TERT (telomerase reverse transcriptase).
10.1371/journal.pgen.1003065.t003 Table 3Recurrent HBV targeted genes identified by the MAPS approach.
Sample ID Tissue disease state chr insertion orientation number of unique end fragments number of total reads Gene symbol Description transcript direction junction nucleotide position read covering the junction point (Underlined for HBV) N23 Adjacent tissue chr11 − 16 862 ARHGEF12 Rho guanine nucleotide exchange factor (GEF) 12 + 120259620 TTTATTTTGCGACAGAGTTTTGCTCTTGTTTCCCAGGCTGCCTACAGCCTCCTAGTACAAAGACCTTTAACCTCATCTCCTCCCCCAA N22 Adjacent tissue chr11 − 6 3607 ARHGEF12 Rho guanine nucleotide exchange factor (GEF) 12 + 120345441 CTATGACAAAGGGAAGAGGAGAGAACCATTCTCACTCATGCACTGATTAGTTGCATGGTGCTGGTGAACAGACCAATTTATGCCTACA C23 Cancer tissue chr10 − 298 261343 CYP2C8 Cytochrome P450, family 2, subfamily C, polypeptide 8 − 96804432 TCTCACAGTTTCCTGGCCCTGTAAAGTGTCGCTCATAGTACAAAGACCTTTAACCTCATCTCCTACCCCAACTCCTCCCACTCATTAA N102 Adjacent tissue chr10 + 29 6742 CYP2C8 Cytochrome P450, family 2, subfamily C, polypeptide 8 − 96817072 CCATATATCACCCAATTTAAAGTGTACACTTTAGTACATAAATCTTTAACCTAATCTCCTCCCCCAACTCCTCCCAGTCCTTAAACCA N113 Adjacent tissue chr2 − 169 36268 FN1 Fibronectin 1 − 216249015 TTTATAGCAGCATGTAAATCACATCTTCTTGGTGAACAGACCAATTTATGCCTACAGCCTCCTAGTACAAA N19 Adjacent tissue chr2 − 9 1054 FN1 Fibronectin 1 − 216252251 AAGACATTATCCCAGACAAATTACAATGCTGATAATTCCTCGTGAATTGACAAGAAAAAGTTGCATGGTGCTGGTGAACAGACCAATT N118 Adjacent tissue chr2 − 47 631 FN1 Fibronectin 1 − 216254297 GGGCTTGTAATCCCAGCACTTTGGGAGGCCGAGGAGGGTGGAGCACTTGAGTACTGGTGAACAGACCAATT N106 Adjacent tissue chr2 − 8 301 FN1 Fibronectin 1 − 216274425 GGCTGTGATTTCGGTCACAGATTCAGAAGTGGCCACAAGAAGTACAAAGATCATTAACCTAATCTCCTCCCCCAACTCCTACCAGTCT C25 Cancer tissue chr2 − 58 33209 FN1 Fibronectin 1 − 216293206 TTCATATTCAGCTCTGGTGCTGCAAGGTATTACAGCCTCCTAGTACAAAGACCTTTAACCTAATCTCCTCCCCCAACTCCTCCCAGTC N125 Adjacent tissue chr1 − 14 216 PHACTR4 Phosphatase and actin regulator 4 + 28785619 TTTTCATGTAACAGAGGAAGCAGACCAGCCCACTACAGTGTGGCATGGTGCTGGTGAACAGACCAATTTAT N23 Adjacent tissue chr1 − 11 491 PHACTR4 Phosphatase and actin regulator 4 + 28819591 AAAAGCTCCGAGATGGAGGTTCATGAAGAGAGCAATCGAACACACCAATTTATGCCTACAGCCTCCTAGTACAAAGACCTTTAACCTC N9 Adjacent tissue chr7 − 29 10098 PLXNA4 Plexin A4 − 132158848 AAACAAAACAAAACAAAAAAAAACAGGGCAATAAACGGTGCACAGACCAATTTATGCCTACAGCCTCCTAGTACAAAGATCATTAACC C116 Cancer tissue chr7 − 4 521 PLXNA4 upstream within 150 kb of Plexin A4 − 132358062 AAGACTTCTCCTTTGGGGGATTCAAGAGGCAGGTTCTTGATCATTAACCTAGTCTCCTCCCCCAACTCCTC N13 Adjacent tissue chr16 − 6 4565 RBFOX1 RNA binding protein, fox-1 homolog (C. elegans) 1 + 6075152 TATGTCTTAAACATTATGGGAAAATTGTTAGGTGTTTTTGTATGCCTACAGCCTCCTAGTACAAAGATCTTTAATCTAATCTACTCCC N104 Adjacent tissue chr16 + 3 358 RBFOX1 RNA binding protein, fox-1 homolog (C. elegans) 1 + 7464068 TGTAACATGTACAGATTCTTATCTCACTCACCAATTTATGCCTACAGCCTCCTAGTACAAAGACCTTTAACCTAACCTCTTCCCCCAA C109 Cancer chr5 + 570 20953 SMAD5 SMAD family member 5 + 135491847 GAATGTGAACTACCTATAAAGAAGGCTATGTGCAGAGGTGAAGCGAAGTCGAAGAGAT N109 Adjacent tissue chr5 + 31 310 SMAD5 SMAD family member 5 + 135491847 TTGAAGAAGCTGTCATGAATGTGAACTACCTATAAAGAAGGCTACGTGCAGAGGTGAAGCGAAGT C110 Cancer chr5 + 30 1124 SMAD5 SMAD family member 5 − 135491851 TCATGAATGTGAACTACCTATAAAGAAGGCTACGTGCAGAGGTGAAGCGAAGT C18 Cancer tissue chr5 − 382 46659 TERT Telomerase reverse transcriptase − 1295135 ATCGCGGGGGTGGCCGGGGCCAGGGCTTCCCACAAAGTTGCATGGTGCTGGTGAACAGACCAATTTATGCCTACAGCCTCCTAGTACA C100 Cancer chr5 − 86 2724 TERT Telomerase reverse transcriptase − 1295396 GTGTCTGTGCCCGCGAATCCACTGGGAGCCCGGCGAAAAAGGTGCATGGGGCTTGTGGACCGACCAATTTATGCCTACCGCCCACTAG C116 Cancer tissue chr5 − 63 3789 TERT Telomerase reverse transcriptase − 1295441 CGCAGCTGCTCCGGGCGGACCCGGGGGTCTGGTGCCTACAGCCTCCTAGTACAAAGACCTTTAGCCTAATC C105 Cancer chr5 − 230 2853 TERT Telomerase reverse transcriptase − 1295563 GCTGTGGGGTAACCCGAGGGAGGGGCCATGATAGGACAAAGATCATTAACCTAATCTCCTCCCCCGACTCCTCCCAGTCTTTAAACAC C117 Cancer tissue chr5 − 270 7445 TERT Telomerase reverse transcriptase − 1295715 TGAAGGGGAGGACGGAGGCGCGTAGACGCTGGTGGACAGACCAATTTATGCCTACAGCCTCCGAGCACAAA N117 Adjacent tissue chr5 − 5 259 TERT Telomerase reverse transcriptase − 1295715 GAATGCCGGACGTGAAGGGGAGGACGGAGGCGCGTAGACGCTGGTGAACAGACCAATTTATGCCTACAGCC C124 Cancer tissue chr5 − 392 47491 TERT Telomerase reverse transcriptase − 1298846 CCTGCCATGGGGTGAAATTCTTTCTTTTTGAAAATACACAGGCTTTGAAGGATGCCTCAAGTGAGGTCGCT N124 Adjacent tissue chr5 − 55 2941 TERT Telomerase reverse transcriptase − 1298846 TCCCCTGCCATGGGGTGAAATTCTTTCTTTTTGAAAACACACAGTCTTTGAAGTATGCCTCAAGGTCGGTCSung et al. completed whole genome sequencing for 81 HBV-positive HCCs and identified 6 recurrent genes (TERT, MLL4, CCNE1, SENP5, ROCK1 and FN1) [14]. FN1 and TERT were found as recurrent in both our and Sung et al.'s studies. The TERT gene was also reported by Fujimoto as a recurrent targeted gene of HBV [16]. If we consider the matches of our genes to the previous identified HBV targeted genes as recurrent, there are additional 9 recurrent genes (Table 2).
To understand the consequences of HBV integration and the exact genomic features that is disrupted by the HBV integration, we created a BED format file to be used as custom track for UCSC genome browser to display the full list of HBV UIS with the chromosome integration junction (a 100 nucleotides were added to the junction position to create the ChromEnd column in the BED file) and with the total number of sequence reads covering the junction (Table S9). The positions of the recurrent integration sites in relationship to the gene structures were displayed using UCSC genome browser custom track (Figure S1). All 6 unique integrations targeting TERT inserted at the gene's promoter region. In particular, five HBV integrations in TERT located at the upstream the transcription start site, and one located in the 5′ UTR (Figure 7), similar to the HBV integrations target TERT identified in previous studies [9], [10]. Sung et al. identified 18 HBV integrations to intron 2 and 6, and to promoter regions of TERT [14], among which majority (15 of 18) of the integrations were in the promoter regions of TERT, similar to our observation. We also identified 5 unique HBV integrations into the FN1 gene. Four integrations were at the introns (at intron 5, 26, 27, 29) and a fifth integration into the exon 15 (Figure 8), which could either create a chimeric FN1-HBV fusion protein or could generate a truncated FN1 protein missing the exon 16–46. For the rest of HBV targets, the integrations were all at the introns of the genes except for an insertion into the exon of PHACTR4 (Figure S1).
10.1371/journal.pgen.1003065.g007 Figure 7Illustration of the integrations targeted TERT.
The sequences were representative Read 2 tags that cover the HBV integration sites to the TERT gene. HBV DNA was shown in lowercase, and the Human DNA was in uppercase and underlined. The numbered boxes represent nearby exons. The nucleotide position at the human and HBV were also indicated (based on GenBank AY800389.1 and Human hg19 assemble).
10.1371/journal.pgen.1003065.g008 Figure 8Illustration of the integrations in FN1.
The sequences were representative Read 2 tags that cover the junction points of the HBV integration to the FN1 gene. HBV DNA was shown in lowercase, and the Human DNA was in uppercase and underlined. The numbered boxes represent nearby exons. The nucleotide position at the human and HBV were also indicated (based on GenBank AY800389.1 and Human hg19 assemble).
We performed RT-PCR on the available samples. For TERT gene, we compared one pair of samples where the only the cancer tissue harbors the integration, another pair of samples where both the HCC and the adjacent tissues harbor the HBV integration, and 6 pairs of samples where none of the tissues harbor the HBV integration. We found that expression ratios of TERT gene in HCC tissues to the tumor-adjacent tissues were 18.7, 1.0, and 0.33 (median of 6) respectively (Figure S2A). This is consistent with Sung et al.'s data showing HBV integration increase the expression of TERT gene [14]. This may due to the fact that HBV integrations to the TERT gene that we identified were all to the promoter regions.
We also conducted real time RT-PCR of the FN1 genes in three pairs of samples that were available. In two pairs of samples where FN1 integration was found only in the adjacent tissues, one showed no significant change (1.5 fold only), and the other showed reduction in the adjacent tissues harboring FN1 integration (Figure S2B). For the third pair of samples where FN1 integration was found in the cancer tissue only, the expression of FN1 was increased in the cancer tissue (Figure S2B). This might suggest that, for FN1 integration event, the effects on the FN1 might be position dependent or context (tumor vs. adjacent normal) dependent. The expression ratios of FN1 in the cancer to adjacent tissues in samples without HBV integration also varied greatly (Figure S2B, right panel). There was no significant association between the HBV integration in the FN1 gene and the expression changes (overall T-test P = 0.506 comparing samples with or without HBV integration, regardless of the HBV integrations were in the adjacent or cancer tissues) (Figure S2B, right panel). This is consistent with Sung et al.'s observation that there was no statistically significant difference for the FN1 expression between samples with or without HBV integration (P = 0.42), as shown in their figure 4b [14].
We also conducted RT-PCR with the available pairs of samples for other recurrent HBV targeted gene including ARHGEF12, CYP2C8, PHACTR4, PLXNA4, and RBFOX1. For ARHGEF12, the integration was found in the tumor-adjacent tissues in two pairs of available samples, and both integrations increased the expression of ARHGEF12 in the tumor-adjacent samples (7.4 and 1.8 fold respectively, tumor-adjacent to tumor ratios), which is statistically significant different from ARHGEF12 expression in 9 pairs of samples without ARHGEF12 integration (the average expression ratio of normal to tumor tissues was 1.1, T-test P = 0.04) (Figure S2C).
RT-PCR analysis of two HBV integrations into the PHACTR4 gene in the tumor-adjacent tissues revealed that the integration increased the expression 35.8 fold in one case, but did not increase the expression in another case (only 1.44 fold), compared with the matched cancer tissues that did not harbor the integration (data not shown). RT-PCR analysis of HBV integration to the PLXNA4 and CYP2C8 did not reveal significant changes in gene expression (data not shown).
For the RBFOX1 gene, HBV integration events reduced the expression of RBFOX1 in the tumor-adjacent tissues comparing to the cancer tissue in two match pairs of samples. In additional 6 pairs of samples without HBV integration in RBFOX1 that we analyzed, RBFOX1 gene were over expressed in 5 of the 6 pairs, but was greatly under expressed in one of the 6 pairs (maybe an outlier) (Figure S2D). Thus, the overall P value is not significant (P = 0.9).
Considering that a HBV target identified both in our study and the whole genome sequencing analyses of HCCs [14]–[16] is a recurrent event, we conducted an integrative analysis and identified a total of 20 recurrent HBV targeted genes (Table 4). Among them, 15 genes were either first identified in our studies or derived from the integrative analysis of our data with the recently published whole genome sequencing analysis.
10.1371/journal.pgen.1003065.t004 Table 4Recurrent targeted combining the integrative analysis.
Gene Symbol Studies identified Descriptions ARHGEF12 our study Rho guanine nucleotide exchange factor (GEF) 12 CYP2C8 our study Cytochrome P450, family 2, subfamily C, polypeptide 8 PHACTR4 our study Phosphatase and actin regulator 4 PLXNA4 our study Plexin A4 RBFOX1 our study SMAD5 our study SMAD family member 5 ADH1B integrated analysis* Alcohol dehydrogenase 1B (class I), beta polypeptide CPS1 integrated analysis Carbamoyl-phosphate synthetase 1, mitochondrial ESRRG integrated analysis Estrogen-related receptor gamma LRFN2 integrated analysis Leucine rich repeat and fibronectin type III domain containing 2 MYOM1 integrated analysis Myomesin 1, 185 kDa RAI1 integrated analysis Retinoic acid induced 1 CTDSPL2 integrated analysis CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 LRP1B integrated analysis Low density lipoprotein-related protein 1B (deleted in tumors) FN1 our study, Sung et al., Jiang et al. Fibronectin 1 TERT our study, Sung et al. Telomerase reverse transcriptase MLL4 Sung et al., integrated analysis Myeloid/lymphoid or mixed-lineage leukemia 4 CCNE1 Sung et al. Cyclin E1 SENP5 Sung et al. SUMO1/sentrin specific peptidase 5 ROCK1 Sung et al. Rho-associated, coiled-coil containing protein kinase 1 *Integrated analysis of our HBV target list with the recently published whole genome sequencing studies.
Discussion
PCR-based methods including Alu-PCR [23], LM-PCR [24] and RS-PCR [10] were employed for isolating HBV integrants, but with biased amplification and limited sequencing capacity. Our massive anchored parallel sequencing (MAPS) approach avoided restriction bias by random fragmentation and takes full advantage of NGS with dual-functional adapters for comprehensive profile of HBV DNA integrations. Our approach is complementary to or an alternative approach to recent reports of applications of whole genome sequencing in the identification of HBV integration events [14]–[16].
We identified 286 UISs from 40 pairs of HBV-related HCC and tumor-adjacent tissues (Table S4), averaging at about 7 HBV integrations per individual. Recently, Sung et al. conducted whole genome sequencing at more than 30 fold coverage for 81 HBV positive HCCs and identified 399 HBV integration events using a criteria of more than 2 read pairs with close mapping positions linking an end of hg19 to an end of HBV, which resulted in the identification of an average of 4.9 HBV integration events per individual patient [14]. In another recently publication, Jiang et al. sequenced four HCC patients (only three are HBV positives) at 80X coverage and identified 255 HBV integration sites, which would generate at about 85 HBV integrations per HBV positive HCC patient [15]. Comparing the number of HBV integration per individual, our findings that the 7 HBV integrations per individual in our analysis is similar to the 4.9 HBV integrations per individual in Sung et al.'s study [14]. However, this might not be surprising as we sequenced two tissues (HCC and adjacent tissues) per individual while Sung et al. [14] only sequenced one tissue sample per individual. We found that adjacent tissues also harbor HBV integration events. However, the average numbers of HBV integration in our study and in Sung et al.'s analysis (about 7 and 5 respectively) are far off from the average number of HBV integrations (about 85) in Jiang et al.'s study. It is worth noting that, in the same issue of Nature Genetics, Fujimoto et al. found 23 HBV breakpoints using a criteria of 3 or more supporting read-pairs from 11 hepatitis B virus (HBV)-related HCCs with about 40X coverage whole genome sequencing, averaging at only about 2 HBV breakpoints per sample [16]. Further investigation would be necessary to resolve the discrepancy. There might be many potential reasons: first, there might be great HCC heterogeneities. Different subclasses of HCC, not yet defined, could have been used in the studies. Secondly, different HBV strains or sub-genotypes might have different capability of integration. Future analysis of the correlation between HBV subtypes and their integration frequencies would be necessary to address the question. Thirdly, different technologies might have different false positive and false negative HBV integration identification rates. The Complete Genomics' technology was used in Jiang et al.'s study [15] but the Illumina's platform was used in Sung et al.'s study [14]. The false positives might be derived from different methods of library construction and ligation procedures (which might generate chimeric ligation events). The false negative rate might be related to the sequence depth, sequence quality, the mapping parameters for the sequence reads and so on. To control the false positive identification, we made the efforts to use two controls: one negative genomic DNAs (without HBV integration) and another with a mixture of free HBV DNA and human genomic DNAs. The latter would control for chimeric ligation events. Using our analysis pipeline and criteria, we did not find any HBV integration events in either of the controls. Fourthly, the criteria to call an HBV integration event might be another factor affecting the number of HBV events called. Sung et al. and Jiang et al. called an HBV integration events if there are supported by at least two paired-end reads [14] or two chimeric reads [15]. Fujimoto et al. [16] and we used the criteria of at least 3 mapped read pairs.
We found more (about 6 times) HBV integration events in adjacent tissues (254 of 296 events, about 86%) than those in cancer tissues (42 of 296, about 14%). The median number of UISs in cancers was 1, whereas it is much higher at 7 in our tumor-adjacent tissues, consistent with previous reports showing nearly one insertion per HCC nodule [3], [25] and several (less than 10) integrations in chronic hepatitis patients [22], [26]. However, our observation is different from Sung et al.'s study where they found that HBV integration is observed more frequently in the tumors (86.4%) than in adjacent liver tissues (30.7%) [14]. We do not yet know the reason of this difference. We postulate that definition of adjacent tissues might be different in the two studies. In our study, the tumor-adjacent tissues might be closer to the cancerous tissues than the tumor-adjacent tissues in Sung et al.'s study [14]. In other word, one could postulate that the tumor-adjacent tissues closer to the cancer tissues might already contain precursor lesions while the tumor-adjacent tissues further away might be normal liver tissues. Therefore, we have adopted the term ‘tumor-adjacent’ tissue instead of the term normal tissue or the ‘adjacent non-tumor’ tissue in this manuscript.
In addition to the lower number of UISs in cancer tissues described above, the HBV insertional frequencies in cancer tissues were statistically larger than that in cancer-adjacent tissues (p<0.0001), which was also observed by Jiang et al. in their study [15]. Taking together, this might suggest a higher level of clonal expansion of cancer hepatocytes in liver cancer tissues compared to cancer-adjacent tissues, which was also suggested by Jiang et al.'s whole genome sequencing of 4 hepatocellular carcinoma patients [15]. As suggested by a previous study [3], it seems that viral integration occurred during HBV chronic infection before HCC initiation. Hepatocytes with HBV integration underwent certain rounds of expansion during chronic hepatitis; some of the cells participated in the formation of so called focal proliferative lesions and gained growth advantages, resulting in clonal expansions during tumorigenesis.
We also compared our list with the list of common CISs for HCC-associated genes identified in a transposon-based (Sleeping Beauty) insertional mutagenesis screening in mice [27]. There are no common genes between the lists. However, two genes in the mouse CISs belong to the same family members with two genes in our list: zbtb20 in mouse vs. ZBTB16 in human, and slc25a13 in mouse vs. SLC2A13 in human. This prompted us to analyze the two lists to see if they have common pathways. Using the DAVID program (david.abcc.ncifcrf.gov/) to analyze the pathways enriched in both lists, we found that they shared three enriched (P<0.05) pathways, which are the renal cell carcinoma pathway, focal adhesion pathway, and the ErbB signaling pathway (data not shown).
Once, integrations of virus into host genomic DNA was thought to be totally random [28]. More recent studies, however, suggested that integration of various viruses into human genome has distinct preferences [29]–[31]. The profile of HBV insertion sites in this study also demonstrated that HBV integration preferentially landed in transcription units and specific chromosomes. Using a randomly generated in silico integration site dataset, we demonstrated that viral integration by HBV favored transcription units (P<0.0001) and chromosome 17. Preference for HBV integration to chromosomes 11 and 17 has been reported in a previous study [4] and a preference for chromosome 3 has been reported in chronic hepatitis tissues without HCC by Alu-PCR [26].
A possible oncogenic contribution of HBV DNA integration involves the insertion of viral DNA into cellular genomic regulatory regions or coding regions, resulting in modification of gene expression (cis-activation) or production of structurally and functionally aberrant cellular or hybrid proteins. Previously identified targeted genes include the myc family oncogenes [32], [33], CCNA2 [8], [34], [35], the RARB [7], [36], the mevalonate kinase (MK) [37], [38], the carboxypeptidase N (CPN2) [39], sarco/endoplasmic reticulum ralcium ATPase (SERCA) [40] and TERT [9]. Whether any oncogenic proteins such as chimeric HBV-host gene fusion proteins would be generated from the 286 UISs remains to be investigated. We found that 8 target genes were inserted by HBV for more than once (Table 3), including ARHGEF12, CYP2C8, FN1, PHACTR4, RBFOX1, SMAD5 and TERT. Previously, TERT and MLL4 (myeloid/lymphoid or mixed-lineage leukemia 4) were identified as recurrent HBV targeted genes [10], [16], [41]. Here we identified additional 8 genes as recurrent target of HBV integration, expanding the list of human host genes targeted by recurrent HBV integration. Further additional 9 recurrent genes were identified if we consider the matches of our HBV targeted genes to the previously identified HBV targeted genes [14], [15], expanding the novel recurrent target genes to 14 (Table 4, identified under our study or integrated analysis).
Fibronectin 1 is a high-molecular weight (∼440 kDa) glycoprotein of the extracellular matrix and contain binding domains for binding to cell surface integrins and other extracellular matrix components such as collagen, fibrin, actin and heparan sulfate proteoglycans (e.g. syndecans), and it plays a major role in cell adhesion, motility, opsonization, as well as in wound healing and embryonic development [42]. Altered fibronectin expression, degradation, and organization have been implicated in cancer and fibrosis [43]. We identified five HBV integration sites in the FN1 genes: four integrations into the intron 5, 26, 27 and 29, and the fifth into the exon 15 (Figure 8). Whether these HBV integrations could alter FN1 expression or had any causative role in HCC carcinogenesis remains to be determined. HBV integration into FN1 exon 15 could either create a chimeric FN1-HBV fusion protein or could generate a truncated FN1 protein missing the exon 16–46, which would shorten the transcript from 8449 nucleotides (NM 002026) to less than 2565 nucleotide (exon 15 ends at nucleotide 2565). The protein product would shorten from 2355 AA to less than 766 AA, which would reduce the number of fibronectin type-III repeats from 16 in FN1 to about 1.5 in the truncated version. Furthermore, the truncated FN1 would miss the heparin-binding and syndecan-binding domains. Sung et al. recently also reported 5 HBV integrations into the FN1 gene; however, all are to the introns [14].
Using PCR, we were able to confirm the HBV genomic integration into FN1 using the genomic DNAs used for MAPS library preparation and sequencing. However, when we attempted to take a second section of the tissue to prepare RNA and to perform RT-PCR to identify chimeric transcripts that could be potentially generated from the integration, we were not able to identify chimeric HBV-FN1 RNA transcripts. It could be possible that integration prevented transcription or due to different section of the tissues were used as tumors usually show great heterogeneity. Further experimentation would be necessary to figure out the consequence of the HBV integration into the FN1 region.
Considering that 7 genes (PTPRJ, CNTN6, IL12B, MYOM1, FNDC3B, LRFN2, FN1)) contain the IPR003961: Fibronectin, type III domain (Table S6), we speculate that fibronectin type III repeat or like fibronectin type III repeat fold domain might play a role in HCC. Zhang et al. recently showed that up-regulated miR-143 transcribed by nuclear factor kappa B (NFKB) promotes the invasion of HBV-HCC by repressing expression of fibronectin type III domain containing 3B (FNDC3B) [44]. Further investigation is warranted. However, we believe that would be the task for a future paper, as it will take considerable effort and time to investigate the mechanism involved. When Tomlins et al. first identified recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer in a science publication [45], it took the field several years to figure out the mechanism involved [46].
We used quantitative RT-PCR to assess the effects of HBV integration on the targeted gene expression in available matched pairs of tumor-adjacent and tumor samples. We found that HBV integration into the TERT gene increases its expression (Figure S2A). This is consistent with Sung et al.'s data showing HBV integration increase the expression of TERT gene [14], and might also reflect the fact that HBV integrations to the TERT gene that we identified were all in the promoter regions. In addition, we also found that HBV integration to the ARHGEF12 gene in the tumor-adjacent tissues increased their expression (Figure S2C). However, for the rest of recurrent targeted genes, the effects on the expression are more complicated: they either showed no effects or had sample-specific effects. Our observation on the effect of HBV integration to its targeted genes is similar to what Sung et al. observed, where they found that three of the six HBV targeted genes showed changed expression level, but another three of the six targeted genes showed no statistically significant changes in gene expression (Figure 4b in [14]).
To determine the sensitivity of our MAPS approach, we tested to see if we can identify the same HBV integrations in data from replicate runs generated during our optimization of the protocol. The two replicate experiments were performed using the same pool of DNAs of 6 pairs of samples plus two controls, but differ slightly in the modifications of primers used: the first was with the single 5 nt barcode and the second with the paired barcodes. For the first replicate, we identified 54 HBV integration events, and for the second replicate, we identified 68 HBV integration events. 40 HBV integration events are in common (data not shown). We identified more HBV integration events for the second data sets as we had a deeper sequencing coverage (a total of 30,761,630 reads vs. 24,886,345 in the first dataset). A total of 82 HBV integration events were identified combining both replicates. 14 were uniquely identified in the first replicate. Therefore, it is possible that we might miss 14 of 82 (17%) of the HBV integration events if we only sequenced 30 million per 14 samples (i.e. a sequence depth of about 2.2 million reads per sample). In a random sampling analysis (Figure 5D), we showed that indeed the curve for the number of HBV integration events identified per million of additional sequence reads starts to become flat at 10 million reads (for 14 samples, this would correspond to an average at about 700 K per sample), but it continues to rise, although slowly. As the average sequence depth is about 2.2–2.5 million reads per sample in our analysis, we would miss at least 17% of the HBV integration events. It is worth to point out that our sensitivity is not 100%, but our specificity is high as we controlled for the false positive identification using two controls: one negative genomic DNAs (without HBV integration) and another with a mixture of free HBV DNA and human genomic DNAs. The latter would control for chimeric ligation events. Using our analysis pipeline and criteria, we did not find any HBV integration events in either of the controls.
There are 47 HBV integration events that could not be annotated (Table S4). We searched them against the human repeat sequence using the RepeatMasker program (www.repeatmasker.org). Seven of the integrations were found in the repeat regions including LINE, LTR/ERV1 and SINE/Alu repeats (Table S11).
In summary, this study highlights these novel findings: 1) higher number of HBV integration events were found in cancer adjacent tissues than in HCC tissues, suggesting a clonal expansion process during HCC development; 2) fibronectin and its associated genes including fibronectin type III-like fold domain containing genes are frequently targeted by HBV DNA integration, and 3) 14 novel recurrent HBV targeted genes were identified when combined our list with recently published HBV targeted gene lists [14]–[16], greatly expanding the recurrent HBV human genome integration gene list. This study, together with recent global survey of HBV integration events [14]–[16] provides a good foundation for further experimentation and understanding the mechanism of HBV-related HCC.
Materials and Methods
Ethics statement
Informed consent was obtained from each patient and the ethics committee of the First Affiliated Hospital, College of Medicine, and Zhejiang University approved all aspects of the study.
Isolation and identification of HBV DNA integrations
The study population comprised 40 HBV-positive HCCs (Table S10).
Two micrograms of the genomic DNA in a volume of 60 µl of Elution Buffer (EB, Qiagen) were fragmented by Bioruptor (Diagenode Inc.). The operating conditions were the following: ice-water batch refreshed every 5 minutes of sonication, intensity level H, 5 sec power on and 5 sec power off, a total of 25–35 minutes of sonication depends on the results of gel electrophoresis. Sonication resulted in the sizes of DNA fragments ranging from 1000 bp to 100 bp. DNA ends were then end-repaired using 15 units of T4 DNA polymerase (New England Biolabs, NEB), 5 units of DNA polymerase I Klenow fragment (NEB), 50 units of T4 polynucleotide kinase (NEB) and 0.8 mM of dNTP (NEB) in T4 DNA ligase buffer (NEB) at 20°C during 30 min. DNA fragments was then purified using a Qiaquick PCR purification kit (Qiagen) and eluted in 30 µl of EB. Addition of an adenosine at the 3′ ends of the DNA was performed by adding 0.2 mM of dATP (TaKaRa) and 15 units of Klenow Fragment 3′ to 5′ exo - (NEB) in NEB2 buffer (NEB) at 37°C for 30 min. DNA was then purified using a Qiaquick PCR purification kit and eluted in 30 µl of EB. Twenty-six different linkers were constructed, each one with a specific 6 bp tag (Table S2) to mark each DNA sample before amplification.
Two rounds of PCR were performed to amplify target sequences (integrants) using KOD-Plus-Neo DNA polymerase (Toyobo Co., Ltd.). A first round of PCR was performed with the HBX1 primer and the walking primer PE 2.1 for 25 cycles. Target sequences were amplified in 50 µl reaction volume containing 1X PCR Buffer for KOD-Plus-Neo DNA polymerase, 1.5 mM MgSO4, 200 µM each of dATP, dCTP, dGTP and dTTP, 300 nM each of the HBX primer and the walking primer, 500 ng of the walking library and 0.02 units/µl KOD-Plus-Neo DNA polymerase. Amplification condition comprised of a 2 minutes activation step at 94°C followed by 25 cycles of 95°C for 15 seconds, and 68°C for 1 min. A second round of PCR was performed with the PE1-Barcode-HBx2 primer and the walking primer PE 2.2. One micro liter of the first round PCR product was used as the template in a 50 µl reaction. All components were at the same concentrations as the first round PCR, and cycling conditions were the same as well. Reactions of nested PCR were diluted 10 fold in sterile distilled water.
Five microliters of the diluted product were used to enrich target templates while introducing the full-length Illumina PE sequencing adapters. Enriched libraries were then purified by gel electrophoresis separately. For each sample, a gel slice containing the DNA material in around 500 bp was excised with clean scalpels. Libraries were purified with the QIAquick Gel Extraction Kit (Qiagen Inc.), and then the 12 barcoded samples were pooled together with two control libraries at equimolar ratios. These two negative control libraries were used to help evaluate this method. HBV-free genomic DNAs isolated from the glioblastoma cell line U251, either mixed with HBV Genomic DNA or alone, were subjected to the MASP approach described above. Pooled PE sequencing libraries were quantified with Qubit (Invitrogen Inc.). 95×95 or 78×78 cycles of PE sequencing reactions were performed on an Illumina GA II according the standard protocol.
Double barcodes for multiplex analysis
Six or sixteen pairs (cancer and adjacent tissues) of HBV-positive hepatocellular carcinomas (HCCs) were analyzed in each multiplex lane using DNA bar coding, a 5 nt barcode on Read 1 and a 6 nt barcode on Read 2. The barcoded primers were listed in Table S2 and S3.
Illumina image analysis algorithms use the first 4 or 5 cycles of pictured images to identify the positions of individual cluster (11). In MAPS, Read 1 ends sequences extended from specific nest primers (HBX secondary primer) are of low-diversity. If there is no barcode bases added before the primer, the position determining base signals on the image tend to be severely skewed (data not shown), which compromises much of the sequencing capacity. As a result, we used the 5 nt barcode in PE1-Barcode-HBX2 primers to analyze pooled DNA samples.
Multiple parallel nested PCR reactions provide an ideal scenario for migration of amplicons between samples. Barcoded HBX primers crosstalk with integrant templates from other samples, and the crosstalk products were simultaneously sequenced. In this study, however, insertions were assigned to original samples by comparing their frequencies in each sample (data not shown). To secure sequence assignment and suppress PCR contaminations, we additionally introduced the 6 nt barcode to Read 2. Using the paired barcodes, we eliminated potential contaminations and assign insertions to their samples with confidence.
Sequencing data analysis and insertion identification
Images deconvolution and quality values calculation were performed using the Goat module of the Illumina pipeline. Read 1 sequences of Illumina PE sequencing were assigned to individuals according to the 5 nt barcodes using SplitFastQIndex.py from http://bioinf.eva.mpg.de/multiplex/. HBV containing sequences were filtered from non-HBV sequences using SOAP (Short Oligonucleotide Analysis Package) (http://soap.genomics.org.cn/) matching with all HBV genomes references we downloaded from NCBI. The corresponding Read 2 sequences with the correct 6 nt barcode of each sample were picked out using the SplitFastQIndex.py script. The selected Read 2 sequences were then aligned with HBV genomes to filter out HBV derived sequences by SOAP. The remaining sequences were aligned with Human Genome to map potential insertions.
HBV containing sequences from Read 1 were identified by aligning them to all HBV genome references from the GenBank database isolated in China. The corresponding Read 2 was aligned to the HBV genomes to filter out HBV derived sequences. The remaining Read 2 was aligned to the human genome (build hg19) to map their locations for putative insertion site identification.
We calculated the frequency and coverage of the insertion sites. Read 2 sequences were viewed as redundant copies of unique tags, if they mapped the same coordinates of the Human Genome. The coverage is the average copy numbers of all the unique tags. To be conservative, we considered different tags offset by less than 2 nucleotides at the ends as one effective unique tag. We also set the criteria that a putative integration should have at least three effective unique tags. Based on the sequencing data from the negative controls, we selected candidate integration site when there is more than 10 fold of overall coverage, and 2/3 of the unique tags have the coverage over 3 have 10 fold of overall coverage. We viewed those putative integrations as authentic if there is Read 2 sequence covering the integration junction point.
Validation by nested PCR assays
The first round of the PCR was performed with the HBX1 primer and a site-specific first round primer for 25 cycles, with 50 ng of the genomic DNA as templates. The 20 µl reaction contained 1X PCR Buffer for KOD-Plus-Neo DNA polymerase, 1.5 mM MgSO4, 200 µM each of dATP, dCTP, dGTP and dTTP, 200 nM each of HBX primer and site-specific primer, and 0.02 units/µl KOD-Plus-Neo DNA polymerase. Thermal conditions were consisted of a 2 minutes activation step at 94°C followed by 25 cycles of 95°C for 15 seconds, and 68°C for 1 min. 1 µl of the products were diluted by adding 49 µl sterile distilled water to each. 0.5 µl of the dilution was subjected to the second round, 30 cycles of 10 µl reactions with the nest site-specific primer and HBX 2 primer (ACTTCGCTTCACCTCTGCACGT). Concentrations and thermal condition were similar to the first round; except that extension time at 68°C was reduced to 45 sec. Final products were visualized on 2% gels.
Semi-quantitative analysis of UIS abundance
The relative abundance of a given UIS was calculated from the number of amplicons of different length (i.e. different shear site). Semi-quantitative PCR was performed to evaluate the quantification within a single sample and among multiplex samples. Duplicate experiments were compared to examine the consistency of the quantification. To evaluate the sequencing depth for quantitative analysis, we also did a rarefaction analysis of raw sequence data using a random sampling approach.
Generation of random dataset
To determine if there were favored chromosomes for HBV integration in vivo, we generated a comparative random set of 8,015 genomic positions in silico. A set of 10,000 random numbers was generated by Perl script, between 1 and 3,095,677,412, the size of the human genome. These random numbers were then converted into chromosomal positions. We retrieved 35 nt sequences that ended with these positions. We then mapped those sequences to the Human Genome using SOAP, resulting in 8,015 unique matches.
Rarefaction analysis
We randomized the total sequence reads 10 times into 39 size bins to evaluate saturation of the insertion pool by current MAPS strategy. As the total reads was 30,761,630, the largest size of randomized sets was set to 30 million (M). The first 9 smallest sizes of subset range from 0.1 M to 0.9 M, at an interval of 0.1M. The other 30 subsets range from 1 M to 30 M, at an interval of 1M. We determined the numbers of putative integrations in all the randomized subsets according to the criteria above. The results were plotted using Sigma Plot (version 11.0; SPSS Inc.). Regression analysis was also performed to fit the rarefaction curve into the double rectangular hyperbola curve model.
Mapping and functional annotation of UIS
RefSeq genes or its vicinity (150 kb upstream of the transcript start sites and 150 kb downstream of the transcript stop sites) inserted by HBV DNA were determined by home-written Perl scripts. The nearest RefSeq gene was selected for each integration site, and was referred as HBV integration target gens in the text. We used the Idiographica (http://www.ncrna.org/idiographica/) to generate a high-quality ideogram of human chromosomes with HBV integration sites. We performed Gene Ontology (GO) analysis using the GO miner program (http://discover.nci.nih.gov/gominer/index.jsp). Additional Functional annotation of the target genes tagged by integration was performed using DAVID (http://david.abcc.ncifcrf.gov/) Bioinformatics Resources. Categories of KEGG PATHWAY, and INTERPRO were included for further interpretation.
Real-time PCR assay
Total RNAs were extracted using mirVana miRNA Isolation Kit (Ambion, Life Technologies) according to the manufactory's instruction on total RNA extraction. The concentration and quality of RNAs were determined by Nanodrop and gel electrophoresis. RT-PCRs were performed with M-MLV Reverse Transcriptase (Promega) using 2 µg RNAs, and real-time PCRs were performed using SYBR Premix Ex Taq II (TaKaRa) in 20 µl in duplicates.
Statistical analysis
Statistical tests were performed using SAS 9.2. The Shapiro-Wilk test or the Kolmogorov-Smirnov test was used for determining normality of data. The Mann–Whitney test was performed to analyze the difference in UISs abundance and viral integration frequency between tumor and non-tumorous groups. Integration rate in RefSeq genes compared to random was tested using the Pearson's chi-squared test. A p-value <0.05 was considered as significant for all the tests.
Supporting Information
Zdroje
1. ParkinDM (2006) The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118 : 3030–3044.
2. SzmunessW (1978) Hepatocellular carcinoma and the hepatitis B virus: evidence for a causal association. Prog Med Virol 24 : 40–69.
3. MurakamiY, SaigoK, TakashimaH, MinamiM, OkanoueT, et al. (2005) Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 54 : 1162–1168.
4. TokinoT, MatsubaraK (1991) Chromosomal sites for hepatitis B virus integration in human hepatocellular carcinoma. J Virol 65 : 6761–6764.
5. FeitelsonMA, LeeJ (2007) Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett 252 : 157–170.
6. MarchioA, PineauP, MeddebM, TerrisB, TiollaisP, et al. (2000) Distinct chromosomal abnormality pattern in primary liver cancer of non-B, non-C patients. Oncogene 19 : 3733–3738.
7. GarciaM, de TheH, TiollaisP, SamarutJ, DejeanA (1993) A hepatitis B virus pre-S-retinoic acid receptor beta chimera transforms erythrocytic progenitor cells in vitro. Proc Natl Acad Sci U S A 90 : 89–93.
8. WangJ, ZindyF, ChenivesseX, LamasE, HengleinB, et al. (1992) Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene 7 : 1653–1656.
9. HorikawaI, BarrettJC (2001) cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J Natl Cancer Inst 93 : 1171–1173.
10. FerberMJ, MontoyaDP, YuC, AdercaI, McGeeA, et al. (2003) Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 22 : 3813–3820.
11. GilletNA, MalaniN, MelamedA, GormleyN, CarterR, et al. (2011) The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood 117 : 3113–3122.
12. BradyT, RothSL, MalaniN, WangGP, BerryCC, et al. (2011) A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Research 39: e72–e72.
13. KoudijsMJ, KlijnC, van der WeydenL, KoolJ, Ten HoeveJ, et al. (2011) High-throughput semiquantitative analysis of insertional mutations in heterogeneous tumors. Genome Res 21 : 2181–2189.
14. SungWK, ZhengH, LiS, ChenR, LiuX, et al. (2012) Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet doi:10.1038/ng.2295.
15. JiangZ, JhunjhunwalaS, LiuJ, HavertyPM, KennemerMI, et al. (2012) The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res 22 : 593–601.
16. FujimotoA, TotokiY, AbeT, BoroevichKA, HosodaF, et al. (2012) Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet doi:10.1038/ng.2291.
17. WangGP, CiuffiA, LeipzigJ, BerryCC, BushmanFD (2007) HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17 : 1186–1194.
18. JeungJU, ChoSK, ShinJS (2005) A partial-complementary adapter for an improved and simplified ligation-mediated suppression PCR technique. J Biochem Biophys Methods 64 : 110–120.
19. ShaginDA, LukyanovKA, VagnerLL, MatzMV (1999) Regulation of average length of complex PCR product. Nucleic Acids Res 27: e23.
20. ShamayM, AgamiR, ShaulY (2001) HBV integrants of hepatocellular carcinoma cell lines contain an active enhancer. Oncogene 20 : 6811–6819.
21. YooJK, KimJ, ChoiSJ, NohHM, KwonYD, et al. (2012) Discovery and Characterization of Novel MicroRNAs During Endothelial Differentiation of Human Embryonic Stem Cells. Stem Cells Dev 21 : 2049–2057.
22. MasonWS, LiuC, AldrichCE, LitwinS, YehMM (2010) Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J Virol 84 : 8308–8315.
23. MinamiM, PoussinK, BrechotC, PaterliniP (1995) A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics 29 : 403–408.
24. TamoriA, YamanishiY, KawashimaS, KanehisaM, EnomotoM, et al. (2005) Alteration of gene expression in human hepatocellular carcinoma with integrated hepatitis B virus DNA. Clin Cancer Res 11 : 5821–5826.
25. Paterlini-BrechotP, SaigoK, MurakamiY, ChamiM, GozuacikD, et al. (2003) Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 22 : 3911–3916.
26. MinamiM, DaimonY, MoriK, TakashimaH, NakajimaT, et al. (2005) Hepatitis B virus-related insertional mutagenesis in chronic hepatitis B patients as an early drastic genetic change leading to hepatocarcinogenesis. Oncogene 24 : 4340–4348.
27. KengVW, VillanuevaA, ChiangDY, DupuyAJ, RyanBJ, et al. (2009) A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol 27 : 264–274.
28. BrechotC, GozuacikD, MurakamiY, Paterlini-BrechotP (2000) Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol 10 : 211–231.
29. WuX, LiY, CriseB, BurgessSM (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300 : 1749–1751.
30. BushmanF, LewinskiM, CiuffiA, BarrS, LeipzigJ, et al. (2005) Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol 3 : 848–858.
31. MitchellRS, BeitzelBF, SchroderAR, ShinnP, ChenH, et al. (2004) Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2: e234 doi:10.1371/journal.pbio.0020234.
32. HansenLJ, TennantBC, SeegerC, GanemD (1993) Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol 13 : 659–667.
33. FourelG, TrepoC, BougueleretL, HengleinB, PonzettoA, et al. (1990) Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature 347 : 294–298.
34. WangJ, ChenivesseX, HengleinB, BrechotC (1990) Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature 343 : 555–557.
35. BerasainC, PatilD, PeraraE, HuangSM, MoulyH, et al. (1998) Oncogenic activation of a human cyclin A2 targeted to the endoplasmic reticulum upon hepatitis B virus genome insertion. Oncogene 16 : 1277–1288.
36. DejeanA, BougueleretL, GrzeschikKH, TiollaisP (1986) Hepatitis B virus DNA integration in a sequence homologous to v-erb-A and steroid receptor genes in a hepatocellular carcinoma. Nature 322 : 70–72.
37. GraefE, CaselmannWH, WellsJ, KoshyR (1994) Insertional activation of mevalonate kinase by hepatitis B virus DNA in a human hepatoma cell line. Oncogene 9 : 81–87.
38. GraefE, CaselmannWH, HofschneiderPH, KoshyR (1995) Enzymatic properties of overexpressed HBV-mevalonate kinase fusion proteins and mevalonate kinase proteins in the human hepatoma cell line PLC/PRF/5. Virology 208 : 696–703.
39. PineauP, MarchioA, TerrisB, MatteiMG, TuZX, et al. (1996) A t(3;8) chromosomal translocation associated with hepatitis B virus intergration involves the carboxypeptidase N locus. J Virol 70 : 7280–7284.
40. ChamiM, GozuacikD, SaigoK, CapiodT, FalsonP, et al. (2000) Hepatitis B virus-related insertional mutagenesis implicates SERCA1 gene in the control of apoptosis. Oncogene 19 : 2877–2886.
41. SaigoK, YoshidaK, IkedaR, SakamotoY, MurakamiY, et al. (2008) Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum Mutat 29 : 703–708.
42. PankovR, YamadaKM (2002) Fibronectin at a glance. J Cell Sci 115 : 3861–3863.
43. LongcopeC, FeminoA, JohnstonJO (1988) Inhibition of peripheral aromatization in baboons by an enzyme-activated aromatase inhibitor (MDL 18,962). Endocrinology 122 : 2007–2011.
44. ZhangX, LiuS, HuT, HeY, SunS (2009) Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 50 : 490–499.
45. TomlinsSA, RhodesDR, PernerS, DhanasekaranSM, MehraR, et al. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310 : 644–648.
46. TomlinsSA, LaxmanB, VaramballyS, CaoX, YuJ, et al. (2008) Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 10 : 177–188.
Štítky
Genetika Reprodukční medicína
Článek Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction DiseaseČlánek Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination SchemesČlánek Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 12- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- A Mystery Unraveled: Essentiality of RNase III in Is Caused by Resident Prophages
- Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction Disease
- Insertion/Deletion Polymorphisms in the Promoter Are a Risk Factor for Bladder Exstrophy Epispadias Complex
- Mi2β Is Required for γ-Globin Gene Silencing: Temporal Assembly of a GATA-1-FOG-1-Mi2 Repressor Complex in β-YAC Transgenic Mice
- Dissection of a Quantitative Trait Locus for PR Interval Duration Identifies as a Novel Modulator of Cardiac Conduction
- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Construction of a Global Pain Systems Network Highlights Phospholipid Signaling as a Regulator of Heat Nociception
- Genes Contributing to Pain Sensitivity in the Normal Population: An Exome Sequencing Study
- Identification of , a Locus Controlling Dominant Resistance to Autoimmune Orchitis, as Kinesin Family Member 1C
- ATX1-Generated H3K4me3 Is Required for Efficient Elongation of Transcription, Not Initiation, at ATX1-Regulated Genes
- Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome
- Identification of Novel Type 2 Diabetes Candidate Genes Involved in the Crosstalk between the Mitochondrial and the Insulin Signaling Systems
- The Genetic Architecture of Adaptations to High Altitude in Ethiopia
- Population Genomics of the Endosymbiont in
- Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination Schemes
- Testicular Differentiation Occurs in Absence of R-spondin1 and Sox9 in Mouse Sex Reversals
- A Yeast GSK-3 Kinase Mck1 Promotes Cdc6 Degradation to Inhibit DNA Re-Replication
- Genetic Adaptation Associated with Genome-Doubling in Autotetraploid
- The Essential Function of RNase III Is to Silence Foreign Toxin Genes
- Long-Range Regulatory Polymorphisms Affecting a GABA Receptor Constitute a Quantitative Trait Locus (QTL) for Social Behavior in
- A New Isolation with Migration Model along Complete Genomes Infers Very Different Divergence Processes among Closely Related Great Ape Species
- Chromosome Fragile Sites in Harbor Matrix Attachment Regions That May Be Associated with Ancestral Chromosome Rearrangement Events
- Genome-Wide Association Study Implicates Testis-Sperm Specific as a Susceptibility Locus for Impaired Acrosome Reaction in Stallions
- A Mechanism of Gene Amplification Driven by Small DNA Fragments
- Base Damage within Single-Strand DNA Underlies Hypermutability Induced by a Ubiquitous Environmental Agent
- Integrative Analysis of a Cross-Loci Regulation Network Identifies as a Gene Regulating Insulin Secretion from Pancreatic Islets
- Reciprocal Signaling between the Ectoderm and a Mesendodermal Left-Right Organizer Directs Left-Right Determination in the Sea Urchin Embryo
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Genome-Wide Fine-Scale Recombination Rate Variation in
- Deciphering the Transcriptional-Regulatory Network of Flocculation in
- On Lung Function and Interactions Using Genome-Wide Data
- Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function
- The Effective Population Size of Malaria Mosquitoes: Large Impact of Vector Control
- Recessive Mutations in Implicate β-III Spectrin in Both Cognitive and Motor Development
- An Excess of Gene Expression Divergence on the X Chromosome in Embryos: Implications for the Faster-X Hypothesis
- Reduced Life- and Healthspan in Mice Carrying a Mono-Allelic MVA Mutation
- Natural Variation at the MATE Transporter Locus Reveals Cross-Talk between Fe Homeostasis and Zn Tolerance in
- Histone Deacetylase HDA6 Is Functionally Associated with AS1 in Repression of Genes in
- A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling
- A Polycomb Group Protein Is Retained at Specific Sites on Chromatin in Mitosis
- Diapause Formation and Downregulation of Insulin-Like Signaling via DAF-16/FOXO Delays Axonal Degeneration and Neuronal Loss
- Genes That Act Downstream of Sensory Neurons to Influence Longevity, Dauer Formation, and Pathogen Responses in
- A Genome-Wide RNAi Screen Reveals MAP Kinase Phosphatases as Key ERK Pathway Regulators during Embryonic Stem Cell Differentiation
- Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
- Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
- Systems Genetic Analysis of Osteoblast-Lineage Cells
- Population Genomics of Inversion Polymorphisms in
- Spreading of Heterochromatin Is Limited to Specific Families of Maize Retrotransposons
- DNA Topoisomerases Maintain Promoters in a State Competent for Transcriptional Activation in
- A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences during Morphogenesis
- Approaching the Functional Annotation of Fungal Virulence Factors Using Cross-Species Genetic Interaction Profiling
- Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization
- Systematic Identification of Rhythmic Genes Reveals as a New Element in the Circadian Clockwork
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



