-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Long-Range Regulatory Polymorphisms Affecting a GABA Receptor Constitute a Quantitative Trait Locus (QTL) for Social Behavior in
Aggregation is a social behavior that varies between and within species, providing a model to study the genetic basis of behavioral diversity. In the nematode Caenorhabditis elegans, aggregation is regulated by environmental context and by two neuromodulatory pathways, one dependent on the neuropeptide receptor NPR-1 and one dependent on the TGF-β family protein DAF-7. To gain further insight into the genetic regulation of aggregation, we characterize natural variation underlying behavioral differences between two wild-type C. elegans strains, N2 and CB4856. Using quantitative genetic techniques, including a survey of chromosome substitution strains and QTL analysis of recombinant inbred lines, we identify three new QTLs affecting aggregation in addition to the two known N2 mutations in npr-1 and glb-5. Fine-mapping with near-isogenic lines localized one QTL, accounting for 5%–8% of the behavioral variance between N2 and CB4856, 3′ to the transcript of the GABA neurotransmitter receptor gene exp-1. Quantitative complementation tests demonstrated that this QTL affects exp-1, identifying exp-1 and GABA signaling as new regulators of aggregation. exp-1 interacts genetically with the daf-7 TGF-β pathway, which integrates food availability and population density, and exp-1 mutations affect the level of daf-7 expression. Our results add to growing evidence that genetic variation affecting neurotransmitter receptor genes is a source of natural behavioral variation.
Published in the journal: . PLoS Genet 8(12): e32767. doi:10.1371/journal.pgen.1003157
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003157Summary
Aggregation is a social behavior that varies between and within species, providing a model to study the genetic basis of behavioral diversity. In the nematode Caenorhabditis elegans, aggregation is regulated by environmental context and by two neuromodulatory pathways, one dependent on the neuropeptide receptor NPR-1 and one dependent on the TGF-β family protein DAF-7. To gain further insight into the genetic regulation of aggregation, we characterize natural variation underlying behavioral differences between two wild-type C. elegans strains, N2 and CB4856. Using quantitative genetic techniques, including a survey of chromosome substitution strains and QTL analysis of recombinant inbred lines, we identify three new QTLs affecting aggregation in addition to the two known N2 mutations in npr-1 and glb-5. Fine-mapping with near-isogenic lines localized one QTL, accounting for 5%–8% of the behavioral variance between N2 and CB4856, 3′ to the transcript of the GABA neurotransmitter receptor gene exp-1. Quantitative complementation tests demonstrated that this QTL affects exp-1, identifying exp-1 and GABA signaling as new regulators of aggregation. exp-1 interacts genetically with the daf-7 TGF-β pathway, which integrates food availability and population density, and exp-1 mutations affect the level of daf-7 expression. Our results add to growing evidence that genetic variation affecting neurotransmitter receptor genes is a source of natural behavioral variation.
Introduction
Most animal and human behaviors are variable, in part due to genetic variation between individuals. Genetic mapping of strain differences in anxiety, learning, activity levels, and the response to addictive drugs in mice, as well as aggression, locomotor activity, and sensory behaviors in Drosophila, has demonstrated a complex genetic basis for these traits [1]–[3]. Typically, multiple loci contribute to each behavior, the contribution of each locus is small, and the effect of many individual loci depends on the genotype at other loci and on environmental conditions [1]. This genetic complexity poses challenges for the discovery of specific genetic variants that modulate behavior, and consequently only a few genes contributing to natural behavioral diversity have been definitively identified. Defining quantitative behavioral genes at a molecular level has the potential to point to classes of genes that generate behavioral variation and to provide new insights into the neuronal control of behavior.
Social behaviors are central to the survival and reproductive success of humans and animals, and defects in social cognition and interaction are core features of human autism and schizophrenia [4], [5]. Social behaviors are also variable within and between animal species, providing a starting point for genetic analysis. Animals within a species display different social behaviors based on their sex, developmental stage, reproductive status and environmental conditions [6]. In addition, these behaviors are shaped by individual genetic variation that interacts with environmental factors. For example, genetic variation among Drosophila males affects their territorial aggressive behavior, and this genetic variation interacts with an environmental regulator of aggression, population density [7]–[9].
Aggregation between members of a species is a social interaction that can provide direct benefits, such as the conservation of body heat [10], as well as facilitating more complex behaviors such as reproduction, migration, defense, or communal foraging [11]. However, aggregation increases competition for local resources and facilitates disease transmission, so it is not always favorable [11], [12]. Accordingly, many animals aggregate under certain conditions but not others. Aggregation behavior and its regulation by environmental and genetic factors have been studied extensively in the nematode Caenorhabditis elegans. C. elegans aggregates spontaneously on food at high population density [13], a behavior that is enhanced at high oxygen levels [14], when food is depleted [15], or under other stressful conditions. Similar behaviors are observed in other nematode species at high density [16]. The role of aggregation in C. elegans biology is poorly understood, but it might promote mating [17], lower oxygen to a preferred intermediate level [14], or expose young animals to pheromones that drive entry into the stress-resistant dauer larva stage [17].
Different wild-type C. elegans strains vary in their propensity to aggregate. The “solitary” laboratory strain N2 aggregates infrequently on a lawn of bacterial food, whereas wild-caught “social” strains aggregate at much higher rates [13], [18]. In addition to aggregating, wild social animals move quickly on food, have a stronger preference for certain pheromones, and accumulate on the oxygen-poor border of a bacterial lawn; the latter behavior is called bordering [14], [18], [19]. Aggregation and bordering typically occur under the same conditions, in part because oxygen regulates both behaviors [13]–[15], [18]–[23]. The extreme solitary behavior of the N2 strain arose as an adaptation to laboratory conditions [24], and results from a gain-of-function point mutation in the neuropeptide receptor gene npr-1 [18] together with a loss-of-function rearrangement of the sensory globin gene glb-5 [24], [25]. Both NPR-1 and GLB-5 act in a circuit that regulates sensitivity to aggregation-promoting environmental signals. GLB-5 acts in sensory neurons to regulate sensitivity to environmental oxygen [24], [25], whereas NPR-1 acts in an interneuron hub of a circuit that integrates aggregation-promoting sensory cues from oxygen, noxious chemicals, and pheromones [19]. Thus the genetic and environmental regulation of aggregation converge on a shared neuronal circuit to regulate behavior.
A second genetic pathway that regulates aggregation is controlled by the TGF-β homolog DAF-7, which serves a key neuroendocrine role in integrating nutrient availability, overpopulation stress, and physiology in C. elegans [20]. The presence of food induces daf-7 expression in the ASI sensory neurons, whereas population density pheromones suppress daf-7 in ASI [26], [27]. DAF-7 protein is secreted from ASI and activates the TGF-β receptors DAF-1 and DAF-4 in a variety of neurons to regulate dauer larva development, aggregation, fat accumulation, gene expression, and lifespan [20], [21], [26]–[30]. Loss-of-function mutations in daf-7 or its receptors result in aggregation and bordering in the N2 genetic background, and aggregation is enhanced in daf-7; npr-1 double mutants, suggesting that these two pathways act at least partly independently of one another [20], [21].
By characterizing intercrosses between the N2 laboratory strain and the Hawaiian strain CB4856, we observed variability in aggregation and bordering behavior that were not explained by the laboratory-induced npr-1 and glb-5 mutations. This variability suggested the existence of additional quantitative trait loci for these behaviors. Here we use a set of recombinant inbred advanced intercross lines (RIAILs) and chromosome substitution strains to probe the genetic architecture and molecular basis of natural variation in aggregation and bordering behavior between the two C. elegans strains, structuring the analysis to control for the strong effect of npr-1. We show that aggregation and bordering are genetically complex, with multiple contributing quantitative trait loci (QTLs), and refine one QTL to identify a new gene affecting the daf-7 pathway, the GABA receptor EXP-1.
Results
At least five loci that differ between N2 and CB4856 affect social behavior
The Hawaiian CB4856 (HW) C. elegans strain is highly divergent from the N2 laboratory strain at many loci [31], including the neuropeptide receptor gene npr-1. To probe the combined effects of loci other than npr-1 on aggregation and bordering behavior, we examined two near-isogenic lines (NILs) that differed from each parental strain by the genetic substitution of a small region near the npr-1 gene (Figure 1A, 1B; see Methods). As shown previously, a NIL in which the HW allele of npr-1 was introduced into an N2 background resulted in increased levels of aggregation and bordering [18], [24] (Figure 1B). These levels were, however, significantly lower than those of the HW strain under the conditions examined. Conversely, introducing the N2 allele of npr-1 into the HW background did not restore behavior to N2-like levels, instead resulting in a strain with intermediate levels of aggregation and bordering (Figure 1A, 1B). These results indicate that loci in addition to npr-1 affect aggregation and bordering behaviors.
Fig. 1. Identification of QTLs for social behavior. 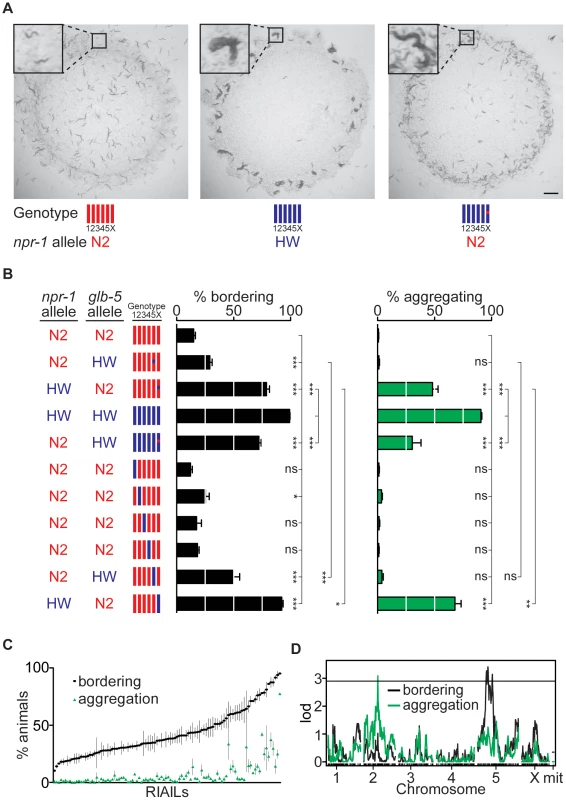
(A) Behavior of N2 animals (left), HW animals (middle), and HW animals with the N2 npr-1 allele (right). Photographs were taken three days after three adult hermaphrodites produced self-progeny on plates seeded with E. coli OP50. Scale bar, 2 mm. Insets show individual solitary N2 animals, a group of aggregating HW animals, and aggregating and non-aggregating animals from the HW strain with the N2 npr-1 allele. For genotypes in all figures, red denotes N2 DNA, blue denotes HW DNA. (B) Bordering and aggregation behaviors of npr-1 near-isogenic lines (NILs) and of chromosome-substitution strains. In this and other figures, bordering and aggregation were measured on 150 adult animals two hours after transferring to E. coli OP50 seeded plates (see Methods); values represent the mean of at least three assays per strain. Error bars, s.e.m. * P<0.05, ** P<0.01, *** P<0.001, by ANOVA with Bonferroni test. ns, not significant. (C) Behaviors of 102 N2-HW recombinant inbred advanced intercross lines (RIAILs) that carry the N2 npr-1 allele. (D) QTL analysis of RIAILs shown in (C). The horizontal line denotes the P<0.05 genome-wide significance threshold. lod, log likelihood ratio. To systematically define genetic differences between N2 and HW, we characterized chromosome substitution strains (CSS) in which each of the six N2 chromosomes was individually replaced by a HW chromosome [32]. Strains bearing HW chromosomes II or V had significantly higher bordering than N2, and the strain bearing the HW X chromosome had high levels of both bordering and aggregation, identifying three chromosomes with loci affecting these behaviors (Figure 1B). The known laboratory-derived mutations in npr-1 and glb-5 are on X and V, respectively. However, the effects of X chromosome substitution were significantly greater than those of the HW npr-1 NIL, and effects of chromosome V substitution were significantly greater than those of a similar NIL bearing the HW allele of glb-5. These results imply the existence of at least one additional QTL on each of chromosomes V and X (Figure 1B). Thus the combination of CSS and NIL analysis indicates that aggregation and bordering are affected by at least five loci that differ between N2 and HW: one or more loci on II, glb-5 and at least one additional locus on V, and npr-1 and at least one additional locus on X.
In a parallel approach, QTL analysis was performed on recombinant inbred advanced intercross lines (RIAILs) derived from crosses between N2 and HW [31]. To set aside the large effect of the npr-1 mutation, we examined only strains with the N2 allele of npr-1. These 102 RIAILs had a continuous quantitative distribution of bordering and aggregation behaviors, implying the existence of multiple QTLs rather than one locus of large effect (Figure 1C). Bordering and aggregation behaviors were strongly but not perfectly correlated in the RIAILs (r = 0.73, 99%C.I. = 0.58–0.83), suggesting that genetic contributions to bordering and aggregation in these strains are similar but perhaps not identical.
QTL analysis of the RIAILs identified a significant QTL on chromosome II (II-QTL) for aggregation and a significant QTL on chromosome V (V-QTL) for bordering (Figure 1D). The II-QTL explains 8.2% (P<0.01) of the aggregation variance and 5.3% (P = 0.019) of the bordering variance in the RIAILs, whereas the V-QTL explains 14% (P<0.01) of the bordering variance and an insignificant fraction of the aggregation variance (see Methods). The chromosome V QTL overlaps glb-5, as well as covering a broader region that may encompass the second bordering QTL inferred from the chromosome V substitution strain (Figure 1B, 1D). The II-QTL does not correspond to a previously characterized locus, and was analyzed further.
A QTL for aggregation and bordering maps to a 6.2 kb interval
The QTL analysis of RIAILs placed the II-QTL near 6.62 Mb (1.5-LOD support interval = 2.48–9.55 Mb). To confirm this map position, we created a NIL, kyIR20, containing the peak of the HW II-QTL (from 4.77 to 6.65 Mb) in an N2 background. The behavior of kyIR20 resembled that of the chromosome substitution strain bearing all of HW chromosome II, with 2.5-fold more bordering than the N2 strain, and a small effect on aggregation (Figure 2A). The effects of the II-QTL on bordering and aggregation cosegregated throughout fine-mapping of the QTL, suggesting that they have a common genetic basis (see below).
Fig. 2. A social behavior II-QTL maps to a 6.2 kb region. 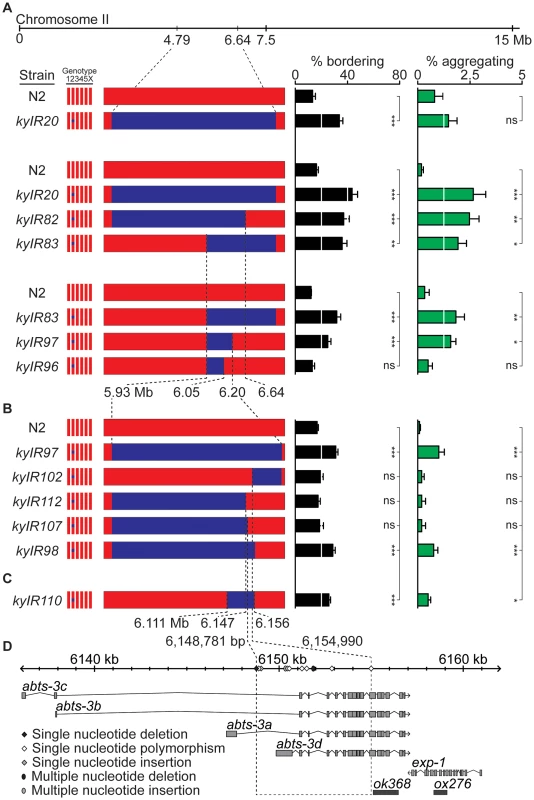
(A) Bordering and aggregation behaviors of recombinants in the II-QTL region introduced as NILs into an N2 background. (B) Behaviors of NILs derived from kyIR97. (C) Behaviors of kyIR110, a near-isogenic line containing 45 kb of HW DNA in an N2 background. (D) Expansion of the 6.2 kb QTL, showing polymorphisms between N2 and HW, location of transcripts (see Methods), and location of deletion alleles used in Figure 3 and Figure 4. Error bars, s.e.m. * P<0.05, ** P<0.01, *** P<0.001 by t-test or ANOVA with Dunnett test. ns, not significant. To identify the specific HW region(s) that affect behavior, we generated recombinants between N2 and the kyIR20 NIL, deriving NILs with less HW DNA than kyIR20. These NILs defined an interval from 6.047 to 6.192 Mb, a region encompassing 56 genes, as a minimal region sufficient to promote bordering and aggregation (Figure 2A). A NIL that behaved significantly different from N2, kyIR97, contained only 270 kb of HW DNA (Figure 2A). To fine-map this II-QTL, 5000 F2 progeny of crosses between the kyIR97 NIL and N2 were screened at the DNA level for recombination events within the QTL interval, and the recombinants were tested for aggregation and bordering behaviors. Five informative recombination events split this region. The behavior of the five recombinants indicated the presence of genetic changes necessary for bordering and aggregation in a 6.2 kb interval (6,148,781–6,154,990) (Figure 2B, 2D). kyIR110, a NIL with less than 45 kb of HW DNA covering the 6.2 kb interval, retained higher bordering and aggregation behaviors than N2 (Figure 2C). These genetic results definitively placed a QTL within the 45 kb associated with kyIR110, and support a location for one quantitative trait nucleotide within the smaller 6.2 kb interval. However, a trend toward smaller behavioral effects as the QTL was refined leaves open the possibility that additional causative polymorphisms between N2 and HW are present in the larger kyIR20 interval.
The 6.2 kb minimal QTL interval fell within a single gene, abts-3, which encodes a predicted anion transporter (Figure 2D). Sequencing this region uncovered 11 polymorphisms between HW and N2 (Figure 2D and Table S1): five noncoding single nucleotide polymorphisms (SNPs), two coding SNPs (abts-3a G615D, abts-3d Q118P), one single nucleotide deletion, one single nucleotide insertion, a three-nucleotide single amino acid insertion (abts-3d 115I116), and a 23-nucleotide deletion in HW. Sequence analysis of 59 additional wild strains indicated that 10 of the 11 sequence polymorphisms were represented in other wild C. elegans populations, whereas one SNP was present only in HW (Table S1 and data not shown). Notably, the N2 sister strain LSJ2, which separated from N2 soon after their isolation from the wild [24], was identical in sequence to N2 at this locus. Thus the N2 sequences are likely to be present in wild populations, and not laboratory-derived. The existence of one private polymorphism in HW is consistent with the fact that HW is one of the most divergent wild C. elegans strains, harboring many polymorphisms not found in any other strain [33].
The GABA receptor exp-1 is a Quantitative Trait Gene affected by the II-QTL
The mapping of the II-QTL defined the location of the relevant sequence change, but not necessarily the affected gene, as noncoding regulatory changes could act at a distance to affect neighboring genes [34]–[38]. To define the gene affected by the II-QTL, we performed quantitative complementation tests between the II-QTL and loss-of-function mutations in genes in the region [1], [39]. In this test, the N2 and HW QTLs were examined as heterozygotes with null alleles of candidate genes, with the expectation that the null allele would fail to complement the QTL with reduced activity.
Initial experiments indicated that the bordering and aggregation behaviors of an N2 II-QTL/HW II-QTL heterozygote resembled the N2 II-QTL homozygote (Figure 3A). The recessive nature of the HW II-QTL suggested that the HW phenotype should be observed in a HW/null heterozygote. Since the 6.2 kb minimal QTL interval was fully contained within abts-3, this gene was considered the most promising candidate. However, a deletion allele of abts-3(ok368) complemented the HW II-QTL, as well as the N2 II-QTL, to give N2-like behavior in heterozygotes (Figure 3B; the location of the mutation is shown in Figure 2D). This result argued against abts-3 being the gene affected by the II-QTL.
Fig. 3. exp-1 is a quantitative trait gene for bordering and aggregation. 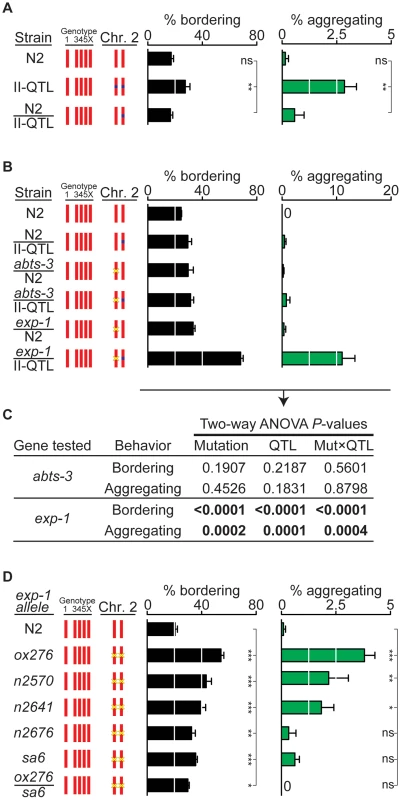
(A) Dominance test between N2 and the II-QTL (n = 7). (B) Quantitative complementation tests between the HW II-QTL, N2, and deletion mutants abts-3(ok368) and exp-1(ox276). Heterozygote F1 progeny from crosses were identified using a fluorescent marker (n = 6 assays for each cross). Yellow ‘x’ denotes a deletion mutation. A transgenic fluorescent marker used to identify F1s in this cross elevated bordering slightly and may also accentuate aggregation (see Figure S4). (C) Analysis of variance of data in panel (B). Two-way ANOVA used Mutation (N2 vs mutant allele) and QTL (N2 vs II-QTL) as variables and was performed for both abts-3(ok368) and exp-1(ox276) mutations. A significant P value for the Mut×QTL interaction indicates failure to complement, a defining feature of quantitative trait genes [1], [39]. (D) Behaviors of exp-1(ox276), a deletion allele, exp-1(n2570), exp-1(n2641), exp-1(n2676), and exp-1(sa6) missense alleles, and exp-1(ox276)/exp-1(sa6) trans-heterozygotes. Yellow ‘x’ denotes a mutation. Error bars, s.e.m. In (A) and (D), * P<0.05, ** P<0.01, *** P<0.001 by ANOVA with Dunnett test. ns, not significant. A second gene close to the II-QTL is exp-1, which encodes a γ-aminobutyric acid (GABA)-gated cation channel [40]; the stop codon of exp-1 is 2.2 kb away from the 6.2 kb QTL (Figure 2D). In a quantitative complementation test, a loss-of-function mutation in exp-1(ox276) failed to complement the HW II-QTL, with the heterozygote showing substantial bordering and aggregation behaviors (Figure 3B). Control experiments demonstrated that the exp-1 mutation was fully complemented by the N2 II-QTL, excluding dominant effects of exp-1 (Figure 3B). Two-way ANOVA provided strong statistical support for an interaction between the II-QTL and exp-1, but not abts-3 (Figure 3C). These results suggest that exp-1 is a quantitative trait gene that affects bordering and aggregation.
To explain these results, we suggest that noncoding variation 3′ of the exp-1 transcript, within the abts-3 gene, modifies aggregation and bordering behavior (at least in part) by affecting the activity of exp-1. The overall abundance of exp-1 mRNA measured by quantitative RT-PCR was similar in N2 and in the HW II-QTL strain (Figure S1), suggesting that the 3′ sequences in the II-QTL may confer specific spatial or temporal patterns of expression, rather than affecting total mRNA levels.
EXP-1 and the neurotransmitter GABA regulate social behavior
As an independent test of the role of exp-1 in social behavior, we examined five exp-1 mutant alleles in an N2 genetic background: a predicted null allele, exp-1(ox276), and four missense alleles, exp-1(n2570), exp-1(n2641), exp-1(n2676), and exp-1(sa6) [40]. All exp-1 mutants had high levels of bordering behavior, and exp-1(ox276), exp-1(n2570), and exp-1(n2641) had significantly increased aggregation (Figure 3D). In a complementation test, exp-1(ox276)/exp-1(sa6) trans-heterozygotes failed to complement for bordering behavior, as expected if they affect the same complementation group (gene) (Figure 3D). These results indicate that normal exp-1 activity suppresses bordering and aggregation in the N2 strain, and suggest that the II-QTL from HW has a reduced level of exp-1 activity.
Previous studies of exp-1 in the enteric nervous system defined a region sufficient for rescue of exp-1 phenotypes related to defecation [40], but this clone did not include the 6.2 kb downstream region defined by the II-QTL. A transgene containing the minimal exp-1 region involved in defecation failed to rescue the bordering and aggregation defects of the exp-1(ox276) deletion mutant (Figure S2, transgene ‘6’), suggesting that it lacked regulatory sequences for exp-1 expression relevant to the bordering phenotype. Among multiple tested transgenes spanning the exp-1 locus and adjacent sequences, only transgenes covering 70 kb encompassing both exp-1 and abts-3 effectively rescued the social behaviors of exp-1(ox276) (Figure S2, transgene ‘4+5’). Similar transgenes reduced bordering in the HW II-QTL strain (Figure S2). These results suggest that exp-1 regulation of aggregation and bordering requires long-range regulation, including substantial sequences 3′ of exp-1 (see Discussion).
exp-1 encodes an unconventional GABA-gated cation channel that depolarizes cells in the presence of GABA [40]; it is one of four genetically characterized C. elegans GABA receptors. All GABAergic signaling in C. elegans is blocked by mutations in unc-25 [41], [42], which encodes the GABA biosynthetic enzyme glutamate decarboxylase, so unc-25 mutants would be expected to have phenotypes related to those of exp-1. Indeed, unc-25(n2324) mutants showed bordering behaviors, albeit weaker than those of the exp-1 deletion mutant (Figure 4A). An exp-1(ox276); unc-25(n2324) double mutant did not have enhanced defects, and in fact resembled the milder unc-25 mutant rather than the stronger exp-1 mutant (Figure 4A). The shared effects of exp-1 and unc-25 are consistent with the hypothesis that GABA regulates bordering behavior by activating exp-1. The milder phenotype of unc-25 and the double mutants suggests that GABA might also activate a second GABA receptor with an opposite effect to exp-1.
Fig. 4. exp-1 acts with GABA and daf-7 TGF-β to regulate bordering and aggregation. 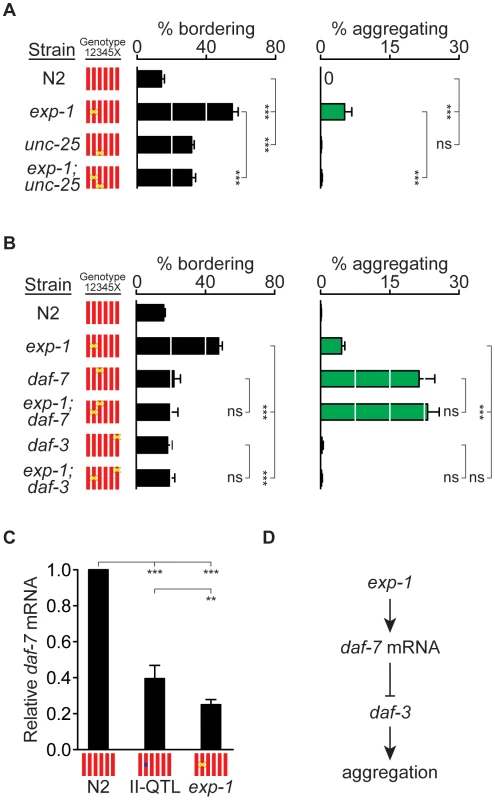
(A) Behaviors of exp-1(ox276), unc-25(n2324), and double mutants. (B) Behaviors of exp-1(ox276), daf-7(e1372), daf-3(e1376), and double mutants. (C) Relative amounts of daf-7 mRNA in N2, HW II-QTL kyIR110 near-isogenic line, and exp-1(ox276), measured by quantitative RT-PCR, (D) Model for exp-1 effects on aggregation. In (A) and (B), error bars, s.e.m. *** P<0.001 by ANOVA with Bonferroni test. ns, not significant. In (C), error bars, 95% C.I. ** P<0.01, *** P<0.001 by t-tests with Bonferroni correction. exp-1 acts in the daf-7 TGF-β pathway
The daf-7 neuroendocrine pathway, which is regulated by food and pheromone levels, inhibits aggregation in the N2 genetic background [20]. In the standardized aggregation assay used here, daf-7(e1372) mutant adults showed high levels of aggregation and low levels of bordering compared to exp-1 mutants (Figure 4B). Adult exp-1(ox276); daf-7(e1372) double mutant animals did not have enhanced behavioral phenotypes compared to single mutants, suggesting that the two genes act in a common pathway (Figure 4B). In agreement with this possibility, a mutation in the downstream daf-3 co-SMAD transcriptional regulator, which suppresses daf-7 aggregation phenotypes [20], also suppressed the bordering and aggregation of exp-1 (Figure 4B).
The relatively low levels of bordering in daf-7 adults were unexpected based on the literature [20], but may be explained by developmentally-regulated differences in behaviors: we observed that L4-stage daf-7 animals bordered and aggregated more extensively than adult animals (Figure S3), and previous studies did not analyze these stages separately. In L4 animals, as in adults, the bordering and aggregation of exp-1(ox276); daf-7(e1372) double mutant animals was no stronger than that of the most severe single mutant, suggesting participation in a common pathway (Figure S3).
Transcription of daf-7 is regulated by environmental conditions: daf-7 mRNA levels increase when food is abundant and decrease in the presence of density pheromones [26], [27]. This transcriptional regulation is partly intrinsic to the ASI sensory neuron that expresses daf-7, and partly determined by intercellular signaling between other sensory neurons and ASI [43]. Using RT-PCR, we asked whether daf-7 transcriptional regulation was altered in exp-1 mutants. exp-1(ox276) mutant animals had a 4-fold decrease in daf-7 mRNA levels compared to controls (Figure 4C). Moreover, daf-7 mRNA levels in the minimal II-QTL strain kyIR110 were intermediate between those of N2 and those of exp-1 mutants, consistent with the possibility that the II-QTL causes a partial reduction of exp-1 activity (Figure 4C). In combination with the genetic epistasis results, these observations suggest that exp-1 and the II-QTL stimulate aggregation and bordering at least partly through effects on daf-7 expression (Figure 4D).
Discussion
Using chromosome substitution strains and RIAILs between N2 and HW C. elegans strains, we found that at least five QTLs affect bordering and aggregation behavior. Two QTLs correspond to the known laboratory-acquired mutations in npr-1 and glb-5, two remain to be identified, and one QTL maps to a region near the abts-3 and exp-1 genes. The minimal 6.2 kb QTL is entirely contained within abts-3, but our results argue that the transcription unit affected by the QTL is the neighboring gene exp-1, which encodes a GABA receptor. exp-1 and abts-3 genes are adjacent and convergently transcribed in all five Caenorhabditis species for which data is available (www.wormbase.org), suggesting that this synteny might be functionally relevant.
Both association studies and linkage studies map causal genetic variants (i.e. functionally relevant sequence polymorphisms), but if the causal variants affect regulatory sequences, they can be located far from the affected gene or genes [1], [38]. The exp-1 transcript ends 2.2 kb from the nearest polymorphism in the II-QTL, suggesting that the QTL affects a regulatory region 3′ to the exp-1 coding region. Most known C. elegans regulatory sites are upstream of or within coding regions, but there are precedents for 3′ transcriptional regulatory elements, including an element located 5.6 kb 3′ of the egl-1 cell death gene [37] and elements located 3′ to the osm-9 and ocr-2 family of sensory ion channel genes [44], [45], and there are also many mammalian precedents for 3′ regulation [46], [47]. In the context of natural variation, a recent study identified a 3′ regulatory region of the Nasonia unpaired-like gene that affects wing width differences between wasp species [48]. 3′ regulatory regions were not represented in over 300 other genetic variants that cause phenotypic variation within and between species; this may represent either a real difference or an ascertainment bias toward 5′ regulatory elements (studies compiled in [49], [50]). Human SNPs that affect gene expression (eQTNs) are three times more abundant 5′ of transcriptional start sites than 3′ of transcriptional end sites, and human enhancers are also more commonly 5′ of the gene they regulate, suggesting that the bias toward 5′ regulatory elements is real but not absolute [51], [52].
Most C. elegans mutations can be rescued by transgenes that cover relatively short regions surrounding the gene of interest. By contrast, the aggregation and bordering behaviors of the II-QTL and the exp-1(ox276) mutant were refractory to rescue with small transgenes that could rescue the enteric nervous system defects of the exp-1 mutation [40]. The only genomic DNA fragments that successfully rescued exp-1(ox276) aggregation and bordering covered 26 kb 5′ and 40 kb 3′ of the exp-1 coding region, including all abts-3 transcripts and several additional transcripts. One possible explanation for this result is that appropriate exp-1 expression requires several long-range cis-regulatory elements, including the 6.2 kb QTL and other distal sequences. A second possibility is that exp-1 is unusually sensitive to general DNA context, resembling genes expressed in the C. elegans germline, which are transcriptionally silenced unless they are embedded in a complex genomic context [53].
exp-1 encodes an unconventional GABA-gated cation channel that depolarizes cells rather than inhibiting them [40]. It was first identified based on its excitatory action on enteric muscles, but exp-1–GFP fusions are also expressed in some neurons (PDA, RID, ADE, SABD) [40]. The relevant site of exp-1 expression for social behavior is unknown, because the minimal transgene that rescues the enteric defect did not rescue social behavior, and therefore may not encompass the full expression pattern of exp-1. A fuller characterization of the 70 kb genomic region that does rescue social behavior may provide insights into sites of exp-1 expression. GABA-deficient animals have mild bordering and aggregation behavior phenotypes, consistent with a likely function of exp-1 as a GABA receptor. GABA is produced by just 26 neurons in C. elegans; identification of the relevant GABAergic neurons may assist in defining the exp-1 circuit.
exp-1 and the daf-7 TGF-β pathway show genetic interactions, suggesting a common role in aggregation. In agreement with this interaction, the level of daf-7 mRNA was reduced in exp-1 null mutants and in animals with the HW exp-1 QTL. exp-1 may be part of the system for detecting environmental stresses that regulates daf-7 expression and neuroendocrine function. The daf-7 pathway and the GABA neurotransmitter system also cooperate to regulate C. elegans dauer development [54], perhaps by using the same transcriptional mechanism defined here.
These results add to evidence that genetic polymorphisms affecting neurotransmitter receptors are sources of natural behavioral variation, and reinforce the importance of studying natural variation as a means toward new biological insights. In humans, very few genetic variants that affect behavioral traits have been mapped. Among these are the ligand-gated ion channels CHRNA3 and CHRNA5, which encode nicotinic acetylcholine receptor subunits implicated in cigarette smoking behavior by genome-wide association studies [55]. Thus ligand-gated ion channels represent sites of behavioral variation both in C. elegans and in humans.
Previous studies in C. elegans and in humans have suggested that G protein-coupled neurotransmitter receptors may be preferred sites of behavioral variation. Among the G protein-coupled receptors implicated in behavioral variation are the C. elegans tyramine receptor tyra-3, which modifies exploratory behavior [56], the C. elegans neuropeptide receptor npr-1, which affects social behavior [18], and human variation in receptors for serotonin, dopamine, and several neuropeptides that has been associated with psychiatric traits [57]–[60]. Both G protein-coupled receptors and nicotinic acetylcholine receptors in the brain are considered modulatory because they are not essential for fast neurotransmission [61], and in both cases the receptors belong to large families of related genes that could provide a reservoir for genetic variation. These properties may allow polymorphisms in modulatory receptors to generate variation in neuronal circuits while sparing the core functions of the nervous system. Our results demonstrate the ability to identify such genetic variants even when they have small quantitative effects on behavior.
Methods
Nematode growth
Strains were grown and maintained under standard conditions at 22–23°C (room temperature) on Nematode Growth Medium (NGM) 2% agar plates [62]. All animals used for behavioral assays were grown on plates seeded with Escherichia coli OP50.
Behavioral assays
Aggregation and bordering behaviors were measured essentially as described [18], with modifications from [19]. Briefly, 2–3 week old 2% agar NGM plates (stored at 4°C) were seeded with 200 µL of a saturated E. coli OP50 bacterial culture in LB 2 days before the assay and left at room temperature. 150 adult animals were picked onto the assay lawn. After two hours at 22–23°C, bordering and aggregation behavior were quantified by eye using a dissecting microscope. An animal was considered to be bordering if its whole body resided within 1 mm of the border of the bacterial lawn. Aggregation behavior was measured as the fraction of animals that were in contact with two or more other animals along at least 50% of their body length; this criterion is highly stringent but unambiguous. Each strain was tested at least five times, except for CB4856 (Figure 1B, four assays), RIAILs (Figure 1C, at least three assays each), and transgenic rescued lines (Figure S3, at least three assays per line, and at least three lines per tested clone). Introgression lines (Figure 2) were tested at least seven times.
Quantitative trait locus analysis
The N2-HW recombinant inbred advanced intercross lines (RIAILs) used in this study represent the terminal generation of a 20-generation pedigree founded by reciprocal crosses between N2 and HW. The lines were constructed through 10 generations of intercrossing followed by 10 generations of selfing [31]. They have been genotyped at 1454 nuclear and one mitochondrial markers and have a 5.3-fold expansion of the F2 genetic map [31]. Each RIAIL was tested at least three times. QTL analysis was performed on the mean bordering and aggregation of N2-HW RIALs by nonparametric interval mapping at 1 cM intervals in R/qtl [63]. Significance levels were estimated from 10,000 permutations of the data.
Percent variance explained by QTLs was based on QTLs defined by the marker with the highest LOD score in the II-QTL (which reached genome-wide significance for aggregation) and the V-QTL (which reached genome-wide significance for bordering). Percent variance explained was calculated for each QTL with ANOVA. While it is not common to measure effect sizes for traits for which a statistically significant QTL is not found, we report the effect sizes of the II-QTL and the V-QTL for both bordering and aggregation behaviors in the text because the two traits have a strong genetic correlation.
Generation of near-isogenic lines
Near-isogenic lines were created by backcrossing a chromosomal region or allele into the desired genetic background as described below. Desired segments were then inbred to homozygosity. Marker positions are based on Wormbase release WS229.
CX11922 (CB4856>N2) kyIR20 II: QX111, a RIAIL containing the HW II-QTL was backcrossed to clr-1 dpy-10 (in an N2 background) for 9 generations, picking non-Clr, non-Dpy males each generation. The introgression breakpoints are, on the left, between 4,783,398 (indel) and 4,800,876 (marker haw25011), and on the right, between 6,627,080 (marker haw25929) and 6,672,356 (marker haw25938).
CX13072 (CB4856>N2) kyIR82 II: CX11922 kyIR20 was crossed to N2 and an F2 recombinant was made homozygous. The introgression breakpoints are, on the left, between 4,783,398 (indel) and 4,800,876 (marker haw25011), and on the right, between 6,215,940 (marker haw25805) and 6,442,763 (indel).
CX13073 (CB4856>N2) kyIR83 II: CX11922 kyIR20 was crossed to N2 and an F2 recombinant was made homozygous. The introgression breakpoints are, on the left, between 5,926,596 (indel) and 5,941,581 (indel), and on the right, between 6,627,080 (marker haw25929) and 6,672,356 (marker haw25938).
CX13602 (CB4856>N2) kyIR97 II: CX13073 kyIR83 was crossed to N2 and an F2 recombinant was made homozygous. The introgression breakpoints are, on the left, between 5,926,596 (indel) and 5,941,581 (indel), and on the right, between 6,195,603 (marker haw25802) and 6,198,696 (marker haw25803).
CX13601 (CB4856>N2) kyIR96 II: CX13073 kyIR83 was crossed to N2 and an F2 recombinant was made homozygous. The introgression breakpoints are, on the left, between 5,926,596 (indel) and 5,941,581 (indel), and on the right, between 6,047,397 (marker haw25707) and 6,064,898 (marker haw25718).
Fine-mapping of the II-QTL for social behavior
N2 animals were crossed to CX13602 kyIR97, F1 hermaphrodites were selfed and 5,000 individual F2 hermaphrodites were dispensed into single wells of 96-well plates with the use of a worm sorter (COPAS Biosort system; Union Biometrica). These F2s were grown on 200 µL of an E. coli OP50 suspension in S-basal buffer with cholesterol, rotating at 230 RPM, at 22°C for 6 days (1–2 generations). The progeny of each F2 were genotyped at markers 5,941,581 (indel) and 6,195,603 (marker haw25802), a 0.41 cM interval, and recombinants between these markers were identified. Animals homozygous for the recombinant chromosome were then tested for social behavior.
CX13854 (CB4856>N2) kyIR102 II: The left breakpoint fell at 6,155,430 (marker haw25773); the breakpoint on the right is between 6,195,603 (marker haw25802) and 6,198,696 (marker haw25803).
CX14013 (CB4856>N2) kyIR112 II: The introgression breakpoints are, on the left, between 5,926,596 (indel) and 5,941,581 (indel), and on the right, between 6,132,624 (marker haw25748) and 6,146,765 (marker haw25767).
CX14008 (CB4856>N2) kyIR107 II: The introgression breakpoints are, on the left, between 5,926,596 (indel) and 5,941,581 (indel), and on the right, between 6,147,008 (marker haw25768) and 6,148,900 (marker haw25769).
CX13845 (CB4856>N2) kyIR98 II: The introgression breakpoints are, on the left, between 5,926,596 (indel) and 5,941,581 (indel), and on the right, between 6,156,160 (marker haw25774) and 6,156,620 (marker haw25775).
CX14011 (CB4856>N2) kyIR110 II: The introgression breakpoints are, on the left, between 6,111,057 (marker haw25735) and 6,117,083 (marker haw25738), and on the right, between 6,156,160 (marker haw25774) and 6,156,620 (marker haw25775).
Identification of the abts-3d isoform
The abts-3d isoform was not present in the standard Gene Models of Wormbase (WS229). The existence of an alternative first exon in abts-3d was inferred from the following evidence: (1) presence of Illumina sequence reads from cDNA derived from polyA+ RNA of larval and adult C. elegans covering the putative exon, including reads that span an inferred 3′ exon junction but absence of 5′ exon junction reads (as shown in Wormbase), (2) presence of a long open reading frame that overlaps the cDNA reads, (3) high degree of conservation of the putative exon with other Caenorhabditis species, relative to other introns in the gene, (4) high evolution rate across Caenorhabditis species of third positions of putative codons in the region relative to first and second positions, (5) presence of a 3-bp indel in the region, an unusual feature in non-coding sequences.
Quantitative RT–PCR
Animals were synchronized in the L1 stage (16 to 20 h post-egg laying) by allowing adults to lay eggs on seeded plates for four hours. The L1 stage was chosen because (1) it is an important time point for daf-7 regulation, (2) neuronal genes are expressed at the highest relative level with respect to total RNA in L1 animals, and (3) it is easy to maintain tight developmental synchrony at this stage. Total RNA from these L1 stage synchronized cultures was isolated with Trizol-chloroform, precipitated with an equal volume of 70% ethanol and cleaned with Zymo Quick-RNA MicroPrep according to the manufacturer's instuructions. 800 ng of RNA and oligo-dT were used for reverse transcription using SuperScript III First-Strand Synthesis (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed with Fast SYBR Green Master Mix (Applied Biosystems) on a 7900HT Real-Time PCR System (Applied Biosystems). cdc-42 was used as the calibrator for relative quantitation [64]. Primers used were:
exp-1_F, ttttggcagatttcaacagc
exp-1_R, ttcatcattttcctccatcaag
daf-7_F, gcaccaactcaggtgtttgtat
daf-7_R, aatccctttggtgcctcttt
cdc-42_F, cggatgttggagagaagttgg
cdc-42_R, ctgttgtggtgggtcgagag
Extrachromosomal transgenes
Transgenes were made by injection of DNA clones into the gonads of young adult hermaphrodites together with a fluorescent coinjection marker [65]. To control for variation among transgenes, at least three independent lines from each injection were characterized.
The following fosmids were injected alone or in combination, as described below in Transgenic strains: WRM066cE09, WRM0620bF02, WRM0610bG09, H35N03, WRM0612dD07. Prior to injection, fosmid structures were confirmed by restriction digest with diagnostic enzymes.
pAB05 is a 7.6 kb genomic exp-1 NsiI/ScaI fragment that contains an in-frame GFP fusion in the intracellular loop between M3 and M4 of EXP-1 [40].
Strains
“Wild-type” strains
Strain – Origin:
AB1 – Adelaide, Australia
CB4853 – Altadena, California, USA
CB4856 (HW) – Hawaii, USA
JU258 – Madeira, Portugal
LSJ2 – Bristol, England
MY1 – Lingen, Germany
MY10 – Roxel, Germany
MY14 – Mecklenbeck, Germany
MY16 – Mecklenbeck, Germany
MY18 – Roxel, Germany
N2 – Bristol, England
QX1216 – San Francisco, California, USA
N2-HW RIAILs for bordering and aggregation QTL analyses
QX1, QX3, QX4, QX5, QX6, QX8, QX9, QX15, QX16, QX17, QX18, QX20, QX25, QX26, QX27, QX29, QX32, QX33, QX38, QX43, QX44, QX47, QX48, QX49, QX51, QX52, QX53, QX54, QX55, QX56, QX57, QX62, QX68, QX70, QX71, QX72, QX73, QX74, QX76, QX78, QX79, QX80, QX81, QX82, QX83, QX84, QX85, QX87, QX90, QX92, QX93, QX94, QX95, QX96, QX97, QX99, QX100, QX102, QX103, QX110, QX112, QX114, QX115, QX120, QX121, QX128, QX129, QX137, QX140, QX147, QX156, QX157, QX161, QX163, QX165, QX171, QX174, QX175, QX176, QX177, QX178, QX181, QX186, QX187, QX189, QX190, QX192, QX193, QX194, QX203, QX206, QX210, QX212, QX216, QX217, QX218, QX220, QX221, QX227, QX228, QX230, QX231.
Near-isogenic lines in an N2 background
CX14180 kyIR122 [X: ∼4.24–∼4.83 Mb, CB4856>N2]
CX11922 kyIR20 [II: ∼4.79–∼6.65 Mb, CB4856>N2]
CX13072 kyIR82 [II: ∼4.79–∼6.30 Mb, CB4856>N2]
CX13073 kyIR83 [II: ∼5.93–∼6.65 Mb, CB4856>N2]
CX13602 kyIR97 [II: ∼5.93–∼6.97 Mb, CB4856>N2]
CX13601 kyIR96 [II: ∼5.93–∼6.06 Mb, CB4856>N2]
CX13854 kyIR102 [II: ∼6.16–∼6.20 Mb, CB4856>N2]
CX14013 kyIR112 [II: ∼5.93–∼6.14 Mb, CB4856>N2]
CX14008 kyIR107 [II: ∼5.93–∼6.15 Mb, CB4856>N2]
CX13845 kyIR98 [II: ∼5.93–∼6.16 Mb, CB4856>N2]
CX14011 kyIR110 [II: ∼6.11–∼6.16 Mb, CB4856>N2]
Chromosome substitution strains
WE5236 [I, CB4856>N2]
WE5237 [II, CB4856>N2]
WE5238 [III, CB4856>N2]
WE5239 [IV, CB4856>N2]
WE5240 [V, CB4856>N2]
WE5241 [X, CB4856>N2]
Transgenic strains
CX13765–CX13767 kyIR97; kyEx4224–kyEx4226 [fosmid WRM066cE09 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX14588–CX14590 kyIR110; kyEx4407, kyEx4408, kyEx4410 [fosmid WRM0620bF02 @10 ng/µL+WRM066cE09 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX14308–CX14310 exp-1(ox276); kyEx4544–kyEx4546 [fosmid WRM0610bG09 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX14305–CX14307 exp-1(ox276); kyEx4444, kyEx4445, kyEx4518 [fosmid H35N03 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX14585–CX14587 exp-1(ox276); kyEx4224–kyEx4226 [fosmid WRM066cE09 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX14298–CX14300 exp-1(ox276); kyEx4426, kyEx4428, kyEx4429 [fosmid WRM0620bF02 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX14141–CX14143, CX14145 exp-1(ox276); kyEx4406–kyEx4408, kyEx4410 [fosmid WRM0620bF02 @10 ng/µL+WRM066cE09 @10 ng/µL+Pelt-2::GFP @4.5 ng/µL]
CX13841–CX13844 exp-1(ox276); kyEx4250–kyEx4253 [pAB05 @25 ng/µL+Pelt-2::mCherry @2 ng/µL]
CX14301–CX14304 exp-1(ox276); kyEx4431–kyEx4434 [fosmid WRM0612dD07 @10 ng/µL+pAB05 @25 ng/µL+Pelt-2::mCherry @2 ng/µL]. A total of 16 transgenic lines were tested.
CX11693 kyIs538 [Pglb-5::p12 hCaspase 3::SL2 GFP, Pelt-2::mCherry]
CX13602 kyIR97 kyIs538 [Pglb-5::p12 hCaspase 3::SL2 GFP, Pelt-2::mCherry]
Mutant strains
CX13840 abts-3(ok368) II, autosomes outcrossed 3 times and X chromosome outcrossed completely to N2
CX13975 exp-1(ox276) II, outcrossed 10 times to N2
CX15377 exp-1(n2570) II, autosomes outcrossed 9 times and X chromosome outcrossed completely to N2
CX15378 exp-1(n2641) II, autosomes outcrossed 4 times and X chromosome outcrossed completely to N2
CX15379 exp-1(n2676) II, autosomes outcrossed 3 times and X chromosome outcrossed completely to N2
CX14520 exp-1(sa6) II, autosomes outcrossed 6 times and X chromosome outcrossed completely to N2
CX14453 unc-25(n2324) III, outcrossed 4 times to N2
CX14455 exp-1(ox276) II; unc-25(n2324) III
CB1372 daf-7(e1372) III
CX14451 exp-1(ox276) II; daf-7(e1372) III
CB1376 daf-3(e1376) X
CX14452 exp-1(ox276) II; daf-3(e1376) X
Supporting Information
Zdroje
1. BendeskyA, BargmannCI (2011) Genetic contributions to behavioural diversity at the gene-environment interface. Nat Rev Genet 12 : 809–820.
2. FlintJ (2003) Analysis of quantitative trait loci that influence animal behavior. J Neurobiol 54 : 46–77.
3. FlintJ, MackayTF (2009) Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res 19 : 723–733.
4. MundyP, SigmanM, UngererJ, ShermanT (1986) Defining the social deficits of autism: the contribution of non-verbal communication measures. J Child Psychol Psychiatry 27 : 657–669.
5. TandonR, NasrallahHA, KeshavanMS (2009) Schizophrenia, “just the facts” 4. Clinical features and conceptualization. Schizophr Res 110 : 1–23.
6. Wilson EO (2000) Sociobiology: the new synthesis. Cambridge: The Belknap Press of Harvard University Press.
7. DierickHA, GreenspanRJ (2006) Molecular analysis of flies selected for aggressive behavior. Nat Genet 38 : 1023–1031.
8. WangL, DankertH, PeronaP, AndersonDJ (2008) A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci U S A 105 : 5657–5663.
9. EdwardsAC, RollmannSM, MorganTJ, MackayTF (2006) Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet 2: e154 doi:10.1371/journal.pgen.0020154..
10. HaigD (2008) Huddling: brown fat, genomic imprinting and the warm inner glow. Curr Biol 18: R172–174.
11. Székely T, Moore Aj, Komdeur J (2010) Social behavior: genes, ecology and evolution: Cambridge University Press.
12. ParrishJK, Edelstein-KeshetL (1999) Complexity, pattern, and evolutionary trade-offs in animal aggregation. Science 284 : 99–101.
13. HodgkinJ, DoniachT (1997) Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics 146 : 149–164.
14. GrayJM, KarowDS, LuH, ChangAJ, ChangJS, et al. (2004) Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430 : 317–322.
15. RogersC, PerssonA, CheungB, de BonoM (2006) Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol 16 : 649–659.
16. Croll NA, Matthews BE (1977) Biology of nematodes. London: Wiley & Sons.
17. SrinivasanJ, KaplanF, AjrediniR, ZachariahC, AlbornHT, et al. (2008) A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454 : 1115–1118.
18. de BonoM, BargmannCI (1998) Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94 : 679–689.
19. MacoskoEZ, PokalaN, FeinbergEH, ChalasaniSH, ButcherRA, et al. (2009) A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458 : 1171–1175.
20. ThomasJH, BirnbyDA, VowelsJJ (1993) Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134 : 1105–1117.
21. de BonoM, TobinDM, DavisMW, AveryL, BargmannCI (2002) Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature 419 : 899–903.
22. CoatesJC, de BonoM (2002) Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419 : 925–929.
23. CheungBH, Arellano-CarbajalF, RybickiI, de BonoM (2004) Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol 14 : 1105–1111.
24. McGrathPT, RockmanMV, ZimmerM, JangH, MacoskoEZ, et al. (2009) Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61 : 692–699.
25. PerssonA, GrossE, LaurentP, BuschKE, BretesH, et al. (2009) Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature 458 : 1030–1033.
26. RenP, LimCS, JohnsenR, AlbertPS, PilgrimD, et al. (1996) Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274 : 1389–1391.
27. SchackwitzWS, InoueT, ThomasJH (1996) Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17 : 719–728.
28. GreerER, PerezCL, Van GilstMR, LeeBH, AshrafiK (2008) Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metab 8 : 118–131.
29. NolanKM, Sarafi-ReinachTR, HorneJG, SafferAM, SenguptaP (2002) The DAF-7 TGF-beta signaling pathway regulates chemosensory receptor gene expression in C. elegans. Genes Dev 16 : 3061–3073.
30. ShawWM, LuoS, LandisJ, AshrafJ, MurphyCT (2007) The C. elegans TGF-beta dauer pathway regulates longevity via insulin signaling. Curr Biol 17 : 1635–1645.
31. RockmanMV, KruglyakL (2009) Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet 5: e1000419 doi:10.1371/journal.pgen.1000419..
32. GlauserDA, ChenWC, AginR, MacinnisBL, HellmanAB, et al. (2011) Heat avoidance is regulated by transient receptor potential (TRP) channels and a neuropeptide signaling pathway in Caenorhabditis elegans. Genetics 188 : 91–103.
33. AndersenEC, GerkeJP, ShapiroJA, CrissmanJR, GhoshR, et al. (2012) Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet 44 : 285–290.
34. LetticeLA, HorikoshiT, HeaneySJ, van BarenMJ, van der LindeHC, et al. (2002) Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A 99 : 7548–7553.
35. EnattahNS, SahiT, SavilahtiE, TerwilligerJD, PeltonenL, et al. (2002) Identification of a variant associated with adult-type hypolactasia. Nat Genet 30 : 233–237.
36. TishkoffSA, ReedFA, RanciaroA, VoightBF, BabbittCC, et al. (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39 : 31–40.
37. ConradtB, HorvitzHR (1999) The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98 : 317–327.
38. MauranoMT, HumbertR, RynesE, ThurmanRE, HaugenE, et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337 : 1190–1195.
39. MackayTF (2001) Quantitative trait loci in Drosophila. Nat Rev Genet 2 : 11–20.
40. BegAA, JorgensenEM (2003) EXP-1 is an excitatory GABA-gated cation channel. Nat Neurosci 6 : 1145–1152.
41. McIntireSL, JorgensenE, HorvitzHR (1993) Genes required for GABA function in Caenorhabditis elegans. Nature 364 : 334–337.
42. JinY, JorgensenE, HartwiegE, HorvitzHR (1999) The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J Neurosci 19 : 539–548.
43. KimK, SatoK, ShibuyaM, ZeigerDM, ButcherRA, et al. (2009) Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326 : 994–998.
44. ColbertHA, SmithTL, BargmannCI (1997) OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci 17 : 8259–8269.
45. JoseAM, BanyIA, ChaseDL, KoelleMR (2007) A specific subset of transient receptor potential vanilloid-type channel subunits in Caenorhabditis elegans endocrine cells function as mixed heteromers to promote neurotransmitter release. Genetics 175 : 93–105.
46. AgarwalS, AvniO, RaoA (2000) Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12 : 643–652.
47. NaganawaS, GinsbergHN, GlickmanRM, GinsburgGS (1997) Intestinal transcription and synthesis of apolipoprotein AI is regulated by five natural polymorphisms upstream of the apolipoprotein CIII gene. J Clin Invest 99 : 1958–1965.
48. LoehlinDW, WerrenJH (2012) Evolution of shape by multiple regulatory changes to a growth gene. Science 335 : 943–947.
49. SternDL, OrgogozoV (2008) The loci of evolution: how predictable is genetic evolution? Evolution 62 : 2155–2177.
50. WittkoppPJ, KalayG (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13 : 59–69.
51. VeyrierasJB, KudaravalliS, KimSY, DermitzakisET, GiladY, et al. (2008) High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4: e1000214 doi:10.1371/journal.pgen.1000214..
52. SanyalA, LajoieBR, JainG, DekkerJ (2012) The long-range interaction landscape of gene promoters. Nature 489 : 109–113.
53. KellyWG, XuS, MontgomeryMK, FireA (1997) Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146 : 227–238.
54. HobertO, TessmarK, RuvkunG (1999) The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126 : 1547–1562.
55. Tobacco and Genetics Consortium (2010) Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42 : 441–447.
56. BendeskyA, TsunozakiM, RockmanMV, KruglyakL, BargmannCI (2011) Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472 : 313–318.
57. VacicV, McCarthyS, MalhotraD, MurrayF, ChouHH, et al. (2011) Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 471 : 499–503.
58. BevilacquaL, DolyS, KaprioJ, YuanQ, TikkanenR, et al. (2010) A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature 468 : 1061–1066.
59. ResslerKJ, MercerKB, BradleyB, JovanovicT, MahanA, et al. (2011) Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 470 : 492–497.
60. WuJ, XiaoH, SunH, ZouL, ZhuLQ (2012) Role of dopamine receptors in ADHD: a systematic meta-analysis. Mol Neurobiol 45 : 605–620.
61. MacDermottAB, RoleLW, SiegelbaumSA (1999) Presynaptic ionotropic receptors and the control of transmitter release. Annu Rev Neurosci 22 : 443–485.
62. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
63. BromanKW, WuH, SenS, ChurchillGA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 : 889–890.
64. HoogewijsD, HouthoofdK, MatthijssensF, VandesompeleJ, VanfleterenJR (2008) Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9 : 9.
65. MelloC, FireA (1995) DNA transformation. Methods Cell Biol 48 : 451–482.
Štítky
Genetika Reprodukční medicína
Článek Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction DiseaseČlánek Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination SchemesČlánek Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
-
Všechny články tohoto čísla
- A Mystery Unraveled: Essentiality of RNase III in Is Caused by Resident Prophages
- Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction Disease
- Insertion/Deletion Polymorphisms in the Promoter Are a Risk Factor for Bladder Exstrophy Epispadias Complex
- Mi2β Is Required for γ-Globin Gene Silencing: Temporal Assembly of a GATA-1-FOG-1-Mi2 Repressor Complex in β-YAC Transgenic Mice
- Dissection of a Quantitative Trait Locus for PR Interval Duration Identifies as a Novel Modulator of Cardiac Conduction
- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Construction of a Global Pain Systems Network Highlights Phospholipid Signaling as a Regulator of Heat Nociception
- Genes Contributing to Pain Sensitivity in the Normal Population: An Exome Sequencing Study
- Identification of , a Locus Controlling Dominant Resistance to Autoimmune Orchitis, as Kinesin Family Member 1C
- ATX1-Generated H3K4me3 Is Required for Efficient Elongation of Transcription, Not Initiation, at ATX1-Regulated Genes
- Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome
- Identification of Novel Type 2 Diabetes Candidate Genes Involved in the Crosstalk between the Mitochondrial and the Insulin Signaling Systems
- The Genetic Architecture of Adaptations to High Altitude in Ethiopia
- Population Genomics of the Endosymbiont in
- Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination Schemes
- Testicular Differentiation Occurs in Absence of R-spondin1 and Sox9 in Mouse Sex Reversals
- A Yeast GSK-3 Kinase Mck1 Promotes Cdc6 Degradation to Inhibit DNA Re-Replication
- Genetic Adaptation Associated with Genome-Doubling in Autotetraploid
- The Essential Function of RNase III Is to Silence Foreign Toxin Genes
- Long-Range Regulatory Polymorphisms Affecting a GABA Receptor Constitute a Quantitative Trait Locus (QTL) for Social Behavior in
- A New Isolation with Migration Model along Complete Genomes Infers Very Different Divergence Processes among Closely Related Great Ape Species
- Chromosome Fragile Sites in Harbor Matrix Attachment Regions That May Be Associated with Ancestral Chromosome Rearrangement Events
- Genome-Wide Association Study Implicates Testis-Sperm Specific as a Susceptibility Locus for Impaired Acrosome Reaction in Stallions
- A Mechanism of Gene Amplification Driven by Small DNA Fragments
- Base Damage within Single-Strand DNA Underlies Hypermutability Induced by a Ubiquitous Environmental Agent
- Integrative Analysis of a Cross-Loci Regulation Network Identifies as a Gene Regulating Insulin Secretion from Pancreatic Islets
- Reciprocal Signaling between the Ectoderm and a Mesendodermal Left-Right Organizer Directs Left-Right Determination in the Sea Urchin Embryo
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Genome-Wide Fine-Scale Recombination Rate Variation in
- Deciphering the Transcriptional-Regulatory Network of Flocculation in
- On Lung Function and Interactions Using Genome-Wide Data
- Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function
- The Effective Population Size of Malaria Mosquitoes: Large Impact of Vector Control
- Recessive Mutations in Implicate β-III Spectrin in Both Cognitive and Motor Development
- An Excess of Gene Expression Divergence on the X Chromosome in Embryos: Implications for the Faster-X Hypothesis
- Reduced Life- and Healthspan in Mice Carrying a Mono-Allelic MVA Mutation
- Natural Variation at the MATE Transporter Locus Reveals Cross-Talk between Fe Homeostasis and Zn Tolerance in
- Histone Deacetylase HDA6 Is Functionally Associated with AS1 in Repression of Genes in
- A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling
- A Polycomb Group Protein Is Retained at Specific Sites on Chromatin in Mitosis
- Diapause Formation and Downregulation of Insulin-Like Signaling via DAF-16/FOXO Delays Axonal Degeneration and Neuronal Loss
- Genes That Act Downstream of Sensory Neurons to Influence Longevity, Dauer Formation, and Pathogen Responses in
- A Genome-Wide RNAi Screen Reveals MAP Kinase Phosphatases as Key ERK Pathway Regulators during Embryonic Stem Cell Differentiation
- Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
- Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
- Systems Genetic Analysis of Osteoblast-Lineage Cells
- Population Genomics of Inversion Polymorphisms in
- Spreading of Heterochromatin Is Limited to Specific Families of Maize Retrotransposons
- DNA Topoisomerases Maintain Promoters in a State Competent for Transcriptional Activation in
- A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences during Morphogenesis
- Approaching the Functional Annotation of Fungal Virulence Factors Using Cross-Species Genetic Interaction Profiling
- Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization
- Systematic Identification of Rhythmic Genes Reveals as a New Element in the Circadian Clockwork
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



