-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Histone Deacetylase HDA6 Is Functionally Associated with AS1 in Repression of Genes in
ASYMMETRIC LEAVES 1 (AS1) is a MYB-type transcription repressor that controls leaf development by regulating KNOX gene expression, but the underlying molecular mechanism is still unclear. In this study, we demonstrated that AS1 can interact with the histone deacetylase HDA6 in vitro and in vivo. The KNOX genes were up-regulated and hyperacetylated in the hda6 mutant, axe1-5, indicating that HDA6 may regulate KNOX expression through histone deacetylation. Compared with the single mutants, the as1-1/axe1-5 and as2-1/axe1-5 double mutants displayed more severe serrated leaf and short petiole phenotypes. In addition, the frequencies of leaf lobes and leaflet-like structures were also increased in as1-1/axe1-5 and as2-1/axe1-5 double mutants, suggesting that HDA6 acts together with AS1 and AS2 in regulating leaf development. Chromatin immunoprecipitation assays revealed that HDA6 and AS1 bound directly to KNAT1, KNAT2, and KNATM chromatin. Taken together, these data indicate that HDA6 is a part of the AS1 repressor complex to regulate the KNOX expression in leaf development.
Published in the journal: . PLoS Genet 8(12): e32767. doi:10.1371/journal.pgen.1003114
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003114Summary
ASYMMETRIC LEAVES 1 (AS1) is a MYB-type transcription repressor that controls leaf development by regulating KNOX gene expression, but the underlying molecular mechanism is still unclear. In this study, we demonstrated that AS1 can interact with the histone deacetylase HDA6 in vitro and in vivo. The KNOX genes were up-regulated and hyperacetylated in the hda6 mutant, axe1-5, indicating that HDA6 may regulate KNOX expression through histone deacetylation. Compared with the single mutants, the as1-1/axe1-5 and as2-1/axe1-5 double mutants displayed more severe serrated leaf and short petiole phenotypes. In addition, the frequencies of leaf lobes and leaflet-like structures were also increased in as1-1/axe1-5 and as2-1/axe1-5 double mutants, suggesting that HDA6 acts together with AS1 and AS2 in regulating leaf development. Chromatin immunoprecipitation assays revealed that HDA6 and AS1 bound directly to KNAT1, KNAT2, and KNATM chromatin. Taken together, these data indicate that HDA6 is a part of the AS1 repressor complex to regulate the KNOX expression in leaf development.
Introduction
The initiation of leaf primordia is established by recruitment of cells from the flanks of the shoot apical meristem (SAM). Meristem activity in the shoot apex is specified in part by the class I KNOTTED-LIKE HOMOBOX (KNOX) genes [1]–[3]. Lateral organs, such as leaves, are initiated on the flank of SAM, and down-regulation of KNOX genes is essential to facilitate this process [1], [4]. Moreover, the silencing of KNOX genes is important in developing organs since the ectopic KNOX expression during organogenesis resulted in patterning defects and over-proliferation of cells [5]–[7]. Thus, the balance between stem cell differentiation and proliferation that is decisive for plant development is attained, in part through the proper regulation of the KNOX expression.
In Arabidopsis, the KNOX family can be further divided into three classes. Class I KNOX genes are similar to KNOTTED1 (KN1) in maize, including BREVIPEDICELLUS (BP)/KNAT1, KNAT2, KNTA6 and SHOOTMERISTEMLESS (STM). These genes are expressed in the SAM and down-regulated in leaf primordia [8]. Class II KNOX genes comprise KNAT3, KNAT4, KNAT5 and KNAT7, which are broadly expressed. Class III only contains KNATM, which is a novel KNOX gene lacking the homeodomain. It was demonstrated that KNATM functions together with KNAT1 and BELL proteins by forming heterodimer [9]. Moreover, ectopic expression of KNATM resulted in the curled down and serrated rosette leaves in wild type plants [9].
KNOX repression is mediated by the orthologous MYB domain proteins ROUGH SHEATH2 (RS2) in maize (Zea mays) and ASYMMETRIC LEAVES1 (AS1) in Arabidopsis thaliana [10]–[13]. In addition, AS1 interacts with the LATERAL ORGAN BOUNDARIES (LOB) domain protein AS2 and directly represses the expression of BP/KNAT1 and KNAT2 [14]–[16]. Previous studies revealed that AS1 and AS2 may recruit a chromatin-remodeling protein Histone Regulatory Homolog 1 (HIRA) to regulate the expression of target genes [17]. Moreover, HIRA has also been shown to interact with a histone deacetylase (HDAC) in animal cells [18].
In this study, we investigated the interaction of AS1 with the histone deacetylase HDA6 and their involvement in leaf development. We demonstrated that HDA6 can interact with AS1 in vivo and in vitro. The hda6 mutant, axe1-5, displayed curling and serrated leaves as well as shorter petioles, suggesting that HDA6 is involved in leaf development. Additionally, HDA6 and AS1 associate directly with the promoters of KNAT1, KNAT2 and KNATM. Taken together, our data suggest that HDA6 is a part of the AS1 repression complex to regulate the expression of KNOX genes.
Results
HDA6 interacts with AS1 in vitro and in vivo
AS1 is a MYB-type transcription repressor that controls leaf patterning by repressing class-1 KNOX gene expression [16]. However, the molecular mechanism how AS1 represses KNOX gene expression is still unclear. In yeast and mammalian cells, many transcription repressors were found to recruit HDACs to regulate their target genes [19]. To further understand the molecular mechanism of AS1-dependent KNOX repression, we analyzed the interaction of AS1 with HDA6, a RPD3-type HDAC in Arabidopsis [20], [21] by using BiFC assays. The coding sequences of HDA6 and AS1 were fused to the N-terminal 174-amino acid portion of yellow fluorescent protein (YFP) in the pEarley-Gate201 vector (pEarleyGate201-YN) or the C-terminal 66-amino acid portion of YFP in the pEarleyGate202 vector (pEarleyGate202-YC) [22]. The Agrobacterium cells containing these constructs were co-transfected into Nicotiana benthamiana leaves. The yellow fluorescence was observed at the nuclear when HDA6-YN and AS1-YC were transient expressed in N. benthamiana leaves, indicating that HDA6 interacted with AS1 in vivo (Figure 1A). In contrast, the yellow fluorescence was not observed in the negative controls (Figure S1).
Fig. 1. HDA6 interacted with AS1. 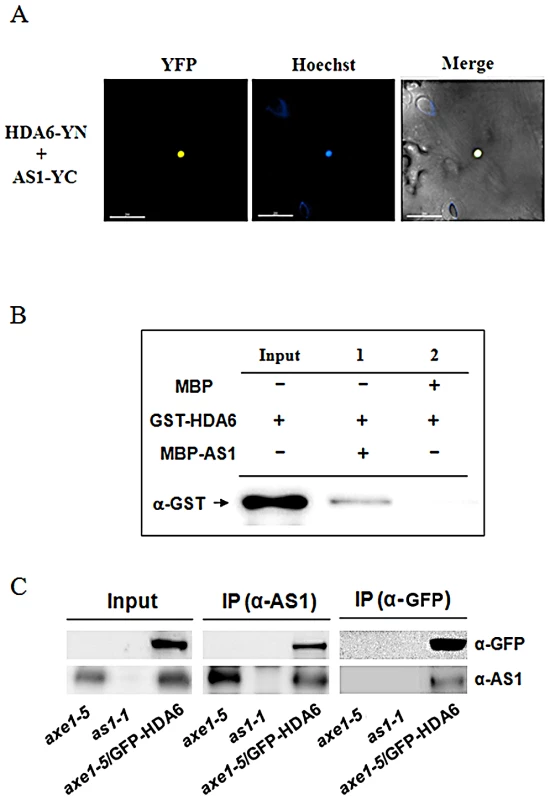
(A) BiFC in N. benthamiana leaves showing interaction between HDA6 and AS1 in living cells. HDA6 and AS1 fused with the N terminus (YN) or C terminus (YC) of YFP were codelivered into tobacco leaves by Agrobacterium GV3101 and visualized using a confocal microscope. The nucleus was stained with Hoechst staining (Blue). Bars = 200 µm. (B) HDA6 interacted with AS1 in a pull-down assay. 2 µg MBP-AS1or MBP was incubated with 2 µg GST-HDA6 and MBP affinity resin, and the bound proteins were then eluted from resin and probed with the anti-GST antibody. (C) In vivo interaction between HDA6 and AS1 in Arabidopsis. Crude extracts of transgenic plants expressing 35S:GFP-HDA6 in axe1-5 background were immunoprecipitated (IP) with AS1 or GFP antibody and analyzed by western blotting. as1-1 and axe1-5 mutant plants were used as the negative control. The interaction between HDA6 and AS1 was further confirmed by in vitro pull down assays. When purified MBP-AS1 recombinant protein was incubated with glutathione S-transferase (GST)-HDA6 protein, HDA6-GST was pulled down by MBP-AS1 (Figure 1B), indicating that HDA6 was directly associated with AS1. Co-immunoprecipitation (CoIP) assays were also used to analyze the interaction between HDA6 and AS1. A stable transgenic plant expressing 35S:GFP-HDA6 in the hda6 mutant (axe1-5) was generated [23]. Overexpressing 35S:GFP-HDA6 in axe1-5 complemented the mutant phenotype, suggesting that the GFP-HDA6 fusion protein is functional. Crude extracts (input) of axe1-5, as1-1 and axe1-5/35S:GFP-HDA6 were immunoprecipitated by the AS1 antibody, then analyzed by western blotting. As shown in Figure 1C, GFP-HDA6 was clearly co-immunoprecipitated by endogenous AS1. Furthermore, AS1 protein was also co-immunoprecipitated by GFP-HDA6 when immunoprecipitated by the GFP antibody (Figure 1C). Taken together, our data strongly indicate that HDA6 interacts with AS1 in vitro and in vivo.
AS1 and AS2 can interact and form homo - and hetero-dimers
Previous studies indicated that AS1 and AS2 can associate together both in yeast cells by yeast two-hybrid assays and in vitro by ELISA experiments using purified His-AS1 and GST-AS2 recombinant proteins [14]. By using BiFC assays, we also found that AS1 and AS2 can interact with each other in N. benthamiana leaves (Figure S2). Furthermore, both AS1 and AS2 can also interact with itself (Figure 2A). These observations indicated that AS1 and AS2 can form both homo - and hetero-dimers. The yellow fluorescence was observed at the nucleus when AS1-YN and AS1-YC, AS2-YN and AS2-YC, or AS1-YN and AS2-YC were transient expressed in N. benthamiana leaves (Figure 2A and Figure S2). Moreover, the in vivo interaction between HDA6 and AS2 was also found by using BiFC (Figure 2B). Collectively, these results together with the finding that HDA6 interacts with AS1 suggested that HDA6, AS1 and AS2 function together in the same protein complex.
Fig. 2. AS1and AS2 formed the homodimer in plants. 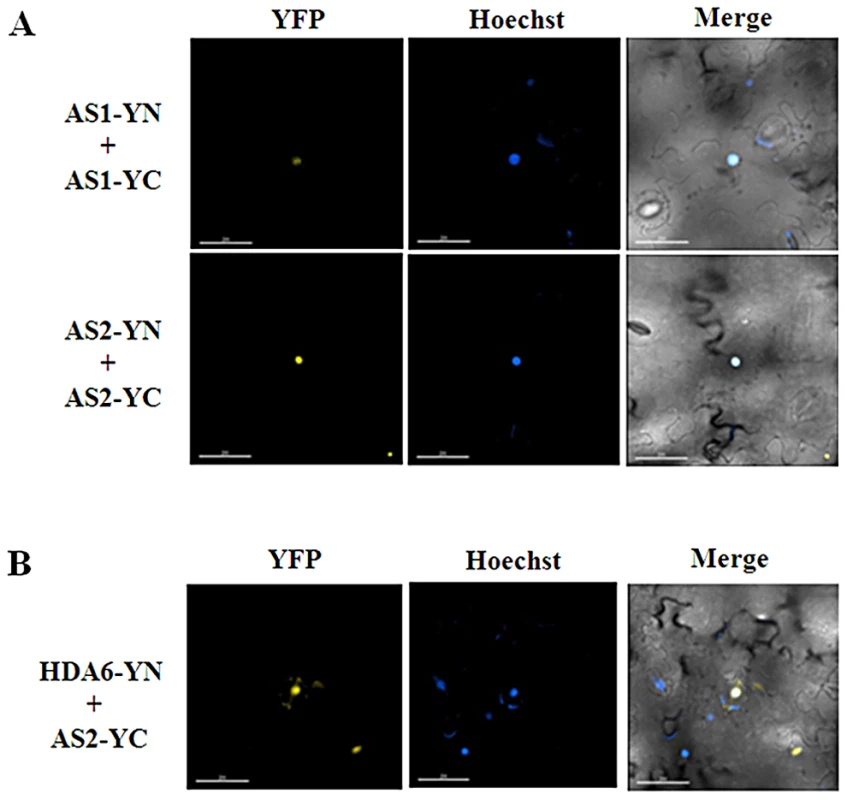
(A) BiFC in N. benthamiana leaves showing the interaction of AS1 and AS2 with itself in living cells. (B) BiFC in N. benthamiana leaves showing the interaction between HDA6 and AS2 in living cells. The nucleus was stained with Hoechst staining (Blue). Bars = 200 µm. We further tested the protein-protein interactions among HDA6, AS1 and AS2 in the protoplasts isolated from the mutants. By using BiFC assays, we found that HDA6 interacted with AS1 in the nucleus of as2-1 mutants (Figure S3). Likewise, the interaction of HDA6 and AS2 was also found in the nucleus of as1-1 mutants. In addition, we also showed that AS1 interacted with AS2 in the nucleus of axe1-5 mutants. Our data indicate that loss of one component of HDA6, AS1 and AS2 does not affect the interaction of two others in Arabidopsis.
hda6 mutants displayed curling and serrated leaves
Previously, we reported that the Arabidopsis HDA6 is required for flowering time control and the hda6 mutant, axe1-5, displayed a delayed flowering phenotype [23]. In addition, axe1-5 mutants also displayed the curling leaves under both long-day (LD) and short-day (SD) conditions (Figure 3A). Similar curling and serrated leaves were also found in another hda6 mutant, sil1 [25] (Figure 3A), and the HDA6-RNAi plants (Figure S4). hda6 mutants displayed the down curling phenotype on both the distal and lateral axis (Figure 3A). These results demonstrated that HDA6 functions not only in controlling adaxial-abaxial axis, but also in proximal-distal axis and in medial-lateral axis.
Fig. 3. Phenotypes of axe1-5, sil1, as1-1, as2-1, as1-1/axe1-5, and as2-1/axe1-5. 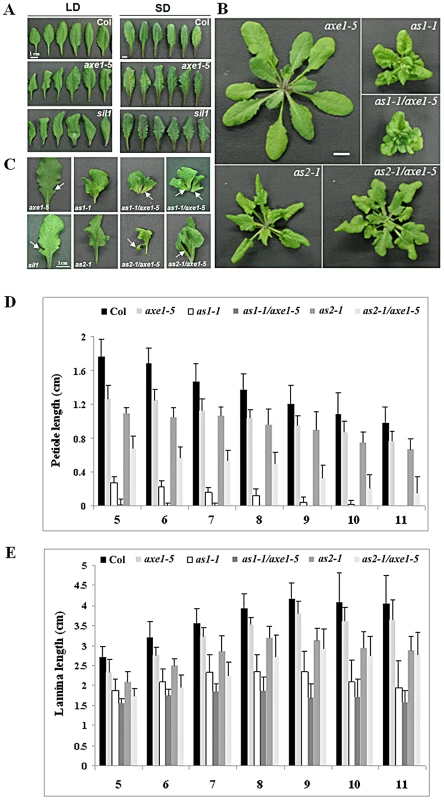
(A) The margin serration and curling leaf phenotype of hda6 mutants. Col, axe1-5 and sil1 plants were grown for 25 days under LD conditions or 75 days under SD conditions. (B) as1-1/axe1-5 and as2-1/axe1-5 double mutants displayed a more severe curling and serration leaf phenotype. (C) The phenotype of leaf lobes and leaflet-like structures in axe1-5, as1-1, as2-1, as1-1/axe1-5 and as2-1/axe1-5. Arrows indicate the leaf lobes in axe1-5 mutants; or leaflet-like structures in as1-1/axe1-5 and as2-1/axe1-5 double mutants. (D) Quantification of the petiole lengths of 5th leaves to 11th leaves. (E) Quantification of the lamina lengths of 5th leaves to 11th leaves. Plants grown under LD conditions for 32 days were used to measure the petiole and lamina lengths. Error bars show SD (n>12). as1-1/axe1-5 and as2-1/axe1-5 double mutants displayed a more severe phenotype compared to the single mutants
The as1 and as2 mutants of Arabidopsis thaliana exhibit pleiotropic phenotypes in leaf development, including the curling and serrated leaves [26]. To examine the genetic interaction between HDA6 and AS1 or AS2, we generated as1-1/axe1-5 and as2-1/axe1-5 double mutants and compared the leaf phenotype of single and double mutants. Under LD conditions, as1-1/axe1-5 and as2-1/axe1-5 double mutant plants showed more severe leaf phenotypes compared with as1-1 and as2-1 single mutant plants (Figure 3B and 3C).
We also measured the lengths of petioles and lamina in wild type and mutant plants. Compared with wild type, the lengths of the petioles were decreased in axe1-5 mutants (Figure 3D). as1-1/axe1-5 and as2-1/axe1-5 double mutants displayed shorter petioles compared with as1-1 and as2-1 single mutant plants (Figure 3B, 3C and Figure 3D). However, the lamina lengths of as1-1/axe1-5 and as2-1/axe1-5 did not show significant changes compared with the single mutants (Figure 3E).
We further measured the frequencies of leaf lobe formation in axe1-5, sil1, as1-1/axe1-5 and as2-1/axe1-5 mutants. The frequencies of leaf lobes were significantly increased in as1-1/axe1-5 and as2-1/axe1-5 double mutants (Table 1). as2 mutants produced leaflet-like structures on the petioles [26]. In as1-1/axe1-5 and as2-1/axe1-5 double mutants, the frequencies of leaflet-like structures were increased (Table 2), and some of the leaf lobes were similar to leaflet-like structures (Figure 3C). These results suggested that HDA6 acts with AS1 and AS2 in regulating leaf development.
Tab. 1. Frequency of leaf lobes. 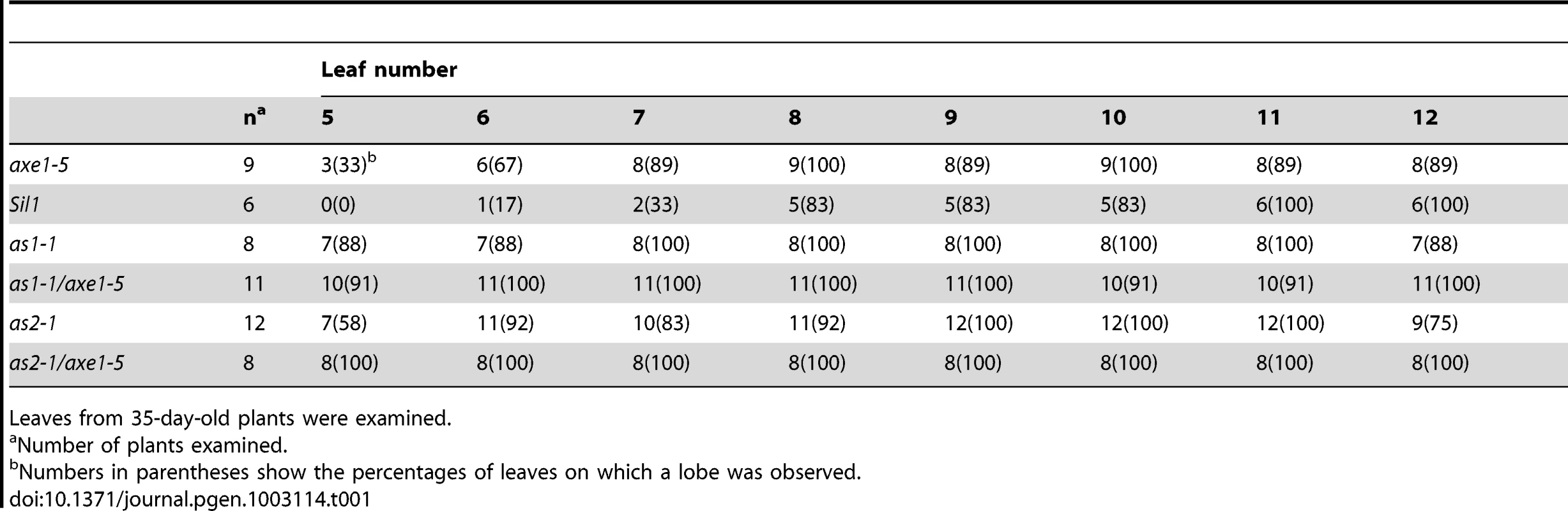
Leaves from 35-day-old plants were examined. Tab. 2. Frequency of leaflet-like structure. 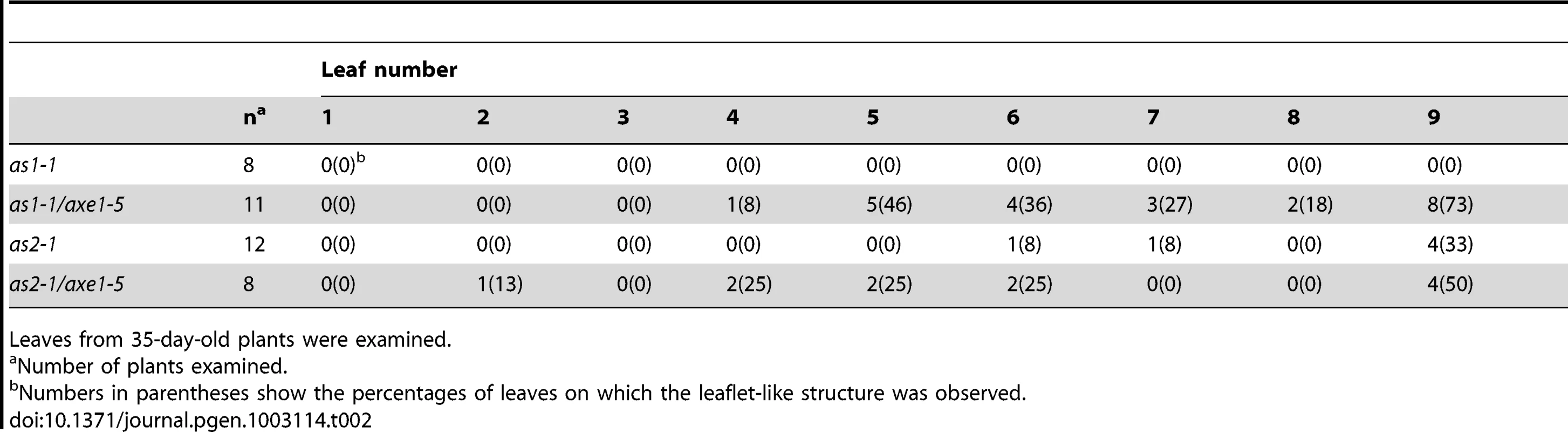
Leaves from 35-day-old plants were examined. Expression of KNOX genes was increased in axe1-5, as1-1, and as2-1 mutant plants
We further analyzed the gene expression by quantitative reverse transcription (qRT)-PCR in mutant plants. Compared with Col wild type, no significant changes were found in the expression of AS1 and AS2 in the axe1-5 (Figure S5). As shown in Figure 4, the expression of KNAT1, KNAT2 and KNATM was increased in axe1-5 compared to Col wild type. Consistent with the previous study [13], the transcript levels of KNAT1 and KNAT2 were elevated in as1-1 and as2-1 mutant plants. In addition, the expression of KNATM was also up-regulated in as1-1 and as2-1 mutant plants. Moreover, the expression of KNAT1, KNAT2 and KNATM was highly increased in as1-1/axe1-5 and as2-1/axe1-5 double mutants compared with their corresponding single mutants. These data indicate that HDA6 may function synergistically with AS1 and AS2 in regulating the expression of KNOX genes.
Fig. 4. Expression of KNOX genes was increased in axe1-5, as1-1, as2-1, as1-1/axe1-5, and as2-1/axe1-5 mutant plants. 
qRT-PCR analysis of gene expression of KNAT1, KNAT2 and KNATM in axe1-5, as1-1, as2-1, as1-1/axe1-5 and as2-1/axe1-5 plants grown under LD conditions for 20 days. Asterisks mark values that are significantly different from the Col wild type (t- test, **P<0.01, *P<0.05). We also analyzed the expression of PHB, PHV, CUC1 and CUC2, which were involved in leaf development through the miRNA regulated pathway [27]–[30]. However, no significant different was found in the expression of PHB, PHV, CUC1 and CUC2 (Figure S5).
Histone H3 acetylation levels of KNOX genes were increased in axe1-5, as1-1/axe1-5, and as2-1/axe1-5 mutant plants
To determine whether the high expression of KNOX genes in the mutants is related to histone hyperacetylation in chromatin, ChIP assays were used to analyze the histone H3 acetylation levels of KNAT1, KNAT2 and KNATM. The relative enrichment of histone H3 acetylation was determined by real-time PCR using primers specific for the proximal promoter (within 500 bp upstream of the transcription starting sites) and transcription start regions of individual genes. As shown in Figure 5, levels of histone H3 acetylation were slight elevated in the proximal promoter and transcription start regions of KNAT1, KNAT2 and KNATM in axe1-5, suggesting that HDA6 may regulate these genes expression by chromatin deacetylation. We further analyzed histone acetylation levels of KNAT1, KNAT2 and KNATM in as1-1, as2-1 and the double mutants. As shown in Figure 5B, hyperacetylation of histone H3 was found in the promoter and first exon of KNAT1, KNAT2 and KNATM in as1-1/axe1-5 and as2-1/axe1-5 double mutants. In contrast, hyperacetylation of histone H3 was not found in as1-1 and as2-1 single mutants. These results suggested that hyperacetylation of histone H3 in KNAT1, KNAT2 and KNATM found in as1-1/axe1-5 and as2-1/axe1-5 double mutants was caused by the hda6 mutation.
Fig. 5. Levels of H3K9K14Ac in KNAT1, KNAT2, and KNATM chromatin in as1-1/axe1-5 and as2-1/axe1-5 double mutants. 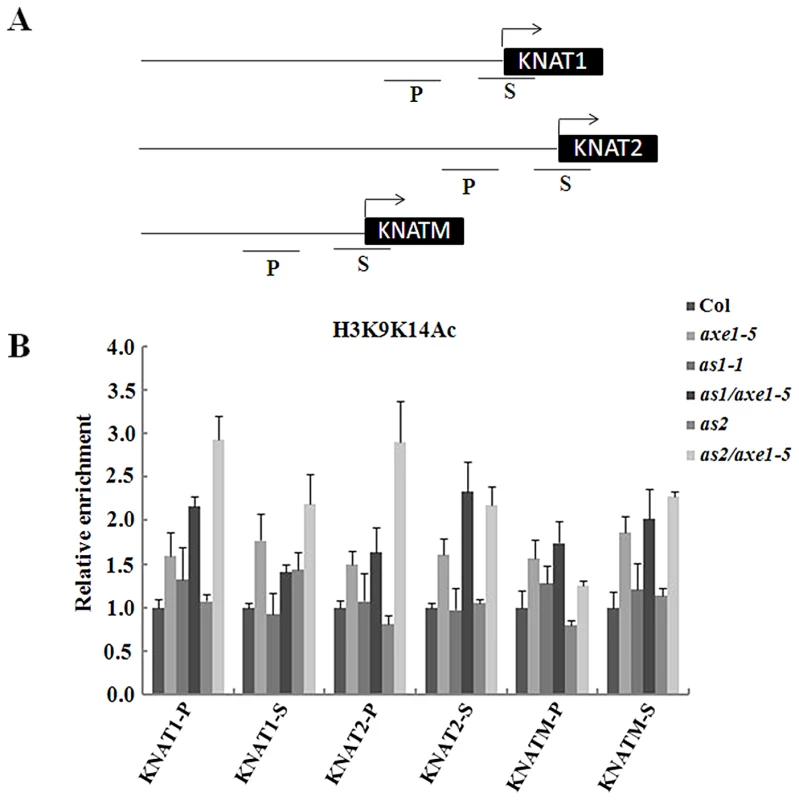
(A) Schematic diagram of KNAT1, KNAT2, KNATM promoter and transcription start regions examined by ChIP. P, promoter region; S, transcription start region. (B) Relative levels of H3K9K14Ac in Col and mutant plants. The amount of DNA after ChIP was quantified and normalized to an internal control (ACTIN2). The values shown are means ± SD. Histone H3K4Me3 is another chromatin mark associated with active genes. We also investigated the histone H3K4Me3 level in axe1-5 mutants. However, no significant changes in the H3K4Me3 of KNAT1, KNAT2 and KNATM were found (Figure S6A). H3K9Me2 was reported as a chromatin marker associated with gene repression. No significant changes in the level of histone H3K9Me2 was found in axe1-5 mutants (Figure S6B).
HDA6 and AS1 bound to KNAT1, KNAT2, and KNATM chromatin
The direct association between AS1 and HDA6 suggested that AS1 may recruit HDA6 to repress the downstream target genes. Previous studies demonstrated that the AS1 repressor complex binds directly to the regulatory motif I (CWGTTD) and motif II (KMKTTGAHW) on the promoters of the KNAT1 and KNAT2 [16]. We also found the conserved motif I and motif II in two promoter regions (KNAMT-X and KNAMT-Y) of KNATM (Figure 6A and Figure S7). To investigate whether AS1 binds directly to KNAT1, KNAT2 and KNATM, ChIP analyses using the AS1 antibody were performed in Col wild type and as1-1 mutants. Consistent with the previous report [16], AS1 can bind to the promoters of KNAT1 and KNAT2 (Figure 6B). In addition, AS1 can also bind directly to KNATM (Figure 6B). In comparison, AS1 cannot bind to the control genes, ACTIN2 and TUB2. To analyze whether the binding of AS1 to KNAT1, KNAT2 and KNATM requires the presence of AS2, we also performed ChIP assays using the as2-1 mutants. We found the loss of binding of AS1 to the KNOX chromatin in the as2-1 mutant (Figure 6C), suggesting that AS2 is required for the binding of AS1 to the KNOX genes.
Fig. 6. AS1 and HDA6 bound to KNAT1, KNAT2, and KNATM promoters. 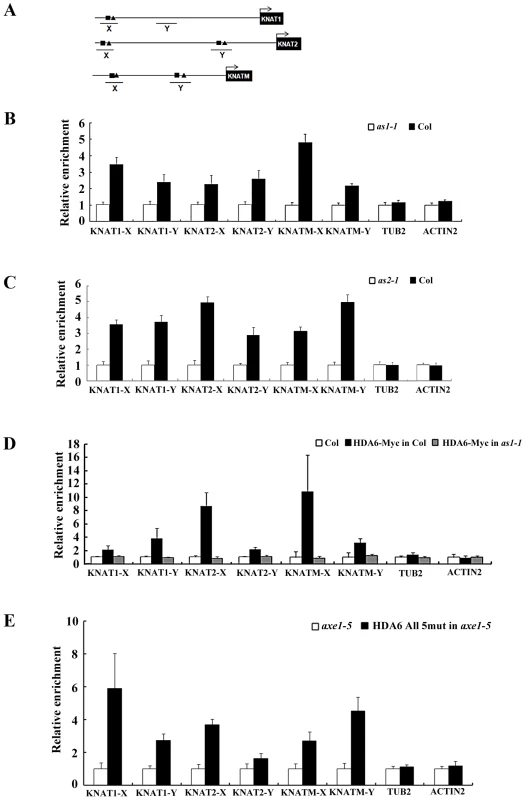
(A) Diagrams of KNAT1, KNAT2, KNATM and the regions examined by ChIP. X and Y indicate regions examined by ChIP. Motif I and II are indicated by square and triangle, respectively. (B, C) Recruitment of AS1 to the KNAT1, KNAT2 and KNATM loci. Relative fold enrichment was calculated by dividing the amount of DNA immunoprecipitated from Col plants by that from the as1-1 or as2-1 plants and compared with input DNA. Plants were grown under LD conditions for 18 days. ACTIN2 and TUB2 were used as the negative controls. The values shown are means ± SD. (D) Recruitment of HDA6 to the KNAT1, KNAT2 and KNATM loci. Relative fold enrichment was calculated by dividing the amount of DNA immunoprecipitated with the anti-Myc antibody from Col plants by that from the HDA6-Myc transgenic plants and compared with input DNA. HDA6-Myc in Col is a transgenic line expressing a Myc-tagged HDA6 in Col background, whereas HDA6-Myc in as1-1 is a transgenic line expressing a Myc-tagged HDA6 in as1-1 mutants. (E) Recruitment of HDA6 to the KNAT1, KNAT2 and KNATM is independent of its catalytic activity. DNA fragments coimmunoprecipitated with the anti-Flag antibody relative to axe1-5 were analyzed by ChIP. HDA6 All 5 mut in axe1-5 is a transgenic line expressing HDA6 bearing the five amino acid mutation of the active site in axe1-5 mutants. Plants were grown under LD conditions for 18 days. ACTIN2 and TUB2 were used as the negative controls. The values shown are means ± SD. To examine whether HDA6 can binds directly to KNAT1, KNAT2 and KNATM., transgenic plants expressing HDA6-Myc were subjected to ChIP analysis using an anti-Myc antibody. As shown in Figure 6D, ChIP analyses revealed that HDA6 can bind to the promoters of KNAT1, KNAT2 and KNATM. We also analyze whether HDA6 recruitment is dependent on AS1. ChiP assays were performed using an anti-Myc antibody in transgenic plants expressing the HDA6-Myc in as1 mutants. As shown in Figure 6D, HDA6 cannot bind to KNAT1, KNAT2 and KNATM in as1 mutants, suggesting that AS1 is required to recruit HDA6.
To analyze whether the HDA6 binding is dependent on its catalytic activity, we performed ChIP assays using an anti-FLAG antibody in transgenic plants (HDA6 all 5 mut in axe1-5) expressing the FLAG-tagged HDA6 bearing the five amino acid mutation of the active site in axe1-5 mutants [31]. As show in Figure 6E, the active site mutant HDA6 can still bind to KNAT1, KNAT2 and KNATM, suggesting that HDA6 recruitment is independent of its catalytic activity. Taken together, our findings suggested that HDA6, AS1 and AS2 act together and directly repress the expression of KNOX genes in Arabidopsis.
Discussion
HDA6 regulates the KNOX expression
The Arabidopsis genome sequence contains 9 KNOX genes, which can be further classified into 3 classes [32]. In leaves, AS1 and AS2 down-regulate class I KNOX genes, but not STM; conversely, STM represses AS1 expression in the SAM [12], [33]. Downregulation of KNOX genes expression is a vital step in leaf initiation, and silencing of these genes needs to be maintained for normal organogenesis [13], [15]. In this study, we demonstrated that hda6 mutants displayed the curling and serrated leaves and shorter petioles. Compared with the single mutants, as1-1/axe1-5 and as2-1/axe1-5 double mutants show more severer phenotypes on curling leaves, petiole lengths, and leaflet-like structures, supporting that HDA6 acts synergistically with AS1 and AS2 in the regulation of leaf development.
KNAT1 and KNAT2 were previously found to be repressed by AS1 and AS2 [14]–[16]. Our results indicated that the transcript levels of KNAT1, KNAT2 and KNATM were altered in axe1-5, as1-1 and as2-1 mutants. Furthermore, the expression of KNAT1, KNAT2 and KNATM was highly increased in as1-1/axe1-5 and as2-1/axe1-5 double mutants compared to their corresponding single mutants. In addition, levels of histone H3 acetylation was elevated in KNAT1, KNAT2 and KNATM loci in axe1-5, as1-1/axe1-5 and as2-1/axe1-5 mutants, suggesting that HDA6 is required for the repression of KNOX genes by chromatin deacetylation. ChIP analyses revealed that HDA6 and AS1 bound directly to the promoters of KNAT1, KNAT2 and KNATM. These data indicate that HDA6 and AS1 function together in controlling KNOX gene expression through histone dacetylation. In addition, AS1 is required to recruit HDA6 in KNOX repression HDA6 cannot bind to KNAT1, KNAT2 and KNATM in as1 mutants, suggesting that AS1 is required to recruit HDA6 in KNOX repression.
Microarray gene expression analyses revealed that a large number of loci are differently expressed in hda6 mutants [23], [34], indicating that HDA6 may play multiple roles in different development processes. Recent studies suggested that the expression of KNOX genes is only one important factor for leaf development [28], [30]. Further analysis is required to determine whether HDA6 is involved in other leaf development pathways.
KNATM is a novel target of the AS1–AS2 complex
AS1 is a Myb domain transcription factor related to RS2 in maize and PHANTASTICA in Antirrhinum [12]. Mutations in AS1 result in abnormal leaves, with marginal outgrowths or lobes [12], [13], [33], [35] AS2 encodes a LOB domain protein containing a leucine-zipper motif [36]–[38]. Mutations in the as2 gene cause a phenotype similar to as1 mutants [13], [33]. Previous studies indicated that AS1 and AS2 can associate together both in vitro and in yeast cells [14]. By using the BiFC assay, we found that AS1 and AS2 can interact and form homo and hetero-dimer in plant cells. These data suggested that AS1 and AS2 function in the same protein complex.
A recent study indicated that AS1 functions as a transcriptional repressor and binds directly to its KNOX targets when in a complex with AS2 [16]. It was found that the AS1–AS2 repressor complex binds directly to the regulatory motif I (CWGTTD) and motif II (KMKTTGAHW) in the promoters of the KNAT1 and KNAT2 [16]. Similar to KNAT1 and KNAT2, we also found the conserved motif I and motif II in the promoter of KNATM. KNATM is a novel Arabidopsis Class III KNOX gene that has a MEINOX domain but lacks the homeodomain [9]. ChIP assayes revealed that AS1 can bind directly to the promoter regions of KNAT1, KNAT2 and KNATM. These data suggested that in addition to KNAT1 and KNAT2, the AS1–AS2 complex is also targeted to KNATM by binding to the conserved motifs I and II. To our knowledge, this is the first study demonstrating that KNATM is regulated by AS1 and AS2.
HDA6 is one of the epigenetic components involved in the AS1–AS2–mediated KNOX repression
Recent studies suggested that the AS1–AS2 complex binds to the KNAT1 and KNAT2 promoters and recruit the chromatin-remodeling protein HIRA to maintain the chromatin in a stable repressive state [15], [16], [39]. In mammalian cells, HIRA was shown to interact with a histone deacetylase [18]. Moreover, it was observed that Arabidopsis seedlings treated with TSA, an inhibitor of HDACs, produced abaxialized filamentous leaves, indicating the involvement of HDACs in leaf morphogenesis [24]. In this study, we provided direct evidence indicating that HDA6 is involved in leaf morphogenesis by interacting with AS1 and AS2 to regulate the KNOX expression. Compared with the single mutants, as1-1/axe1-5 and as2-1/axe1-5 double mutants show more severe phenotypes on curling leaves, petiole lengths, and leaflet-like structures, supporting that HDA6 acts with AS1 and AS2 to regulate leaf development. Taken together, our results demonstrated that histone deacetylation is one of the epigenetic components involved in AS1–AS2 complex-mediated KNOX repression. HDA6 may therefore be part of the AS1–AS2 repression complex to repress the target gene expression. Our data indicate that loss of one component of HDA6, AS1 and AS2 does not affect the interaction of two others in Arabidopsis. Previous studies indicated that the interaction between AS1 and AS2 is required for their binding to the promoters of KNOX genes, because neither AS1 nor AS2 alone was able to bind to the target DNA sequences in vitro [40]. We observed the loss of binding of AS1 to the KNOX chromatin in the as2-1 mutant, suggesting that AS2 is required for the AS1 binding. Furthermore, HDA6 cannot bind to KNOX chromatin in as1-1 mutants, indicating that AS1 is required to recruit HDA6. Taken together, both AS1 and AS2 are required for the recruitment of HDA6 to chromatin in repression of KNOX genes.
A recent work has also shown that the Polycomb Repressive Complexes (PRCs) repress KNOX transcription [40]. It was found that CLF-containing PRC2 regulates KNOX genes by trimethylation of histone H3K27 [41]. Thus, AS1 and AS2 may also recruit other chromatin factors such as PRCs to regulate class I KNOX genes. Taken together, our results suggested that HDA6 is one of the epigenetic components involved in the AS1–AS2 complex-mediated KNOX repression during leaf development in Arabidopsis.
Materials and Methods
Plant materials
Arabidopsis thaliana was grown in 23°C under LD (16 h light/8 h dark) or SD (8 h light/16 h dark) conditions. axe1-5, sil1, as1-1 and as2-1 are in the Col background, whereas the HDA6 RNAi lines CS24038 and CS24039 are in Ws background.
Quantitative RT–PCR analysis
Arabidopsis leaves (0.2 g) were ground with liquid nitrogen in a mortar and pestle and mixed with 1 ml Trizol Reagent (Invitrogen) to isolate total RNA. After treated with DNase (Promega), two microgram of total RNA was used for the first-strand cDNA synthesis. cDNA was synthesized in a volume of 20 µl that contained the Moloney Murine Leukemia Virus Reverse Transcriptase buffer (Promega), 1.5 µM poly(dT) primer, 0.5 mM deoxyribonucleotide triphosphates, 25 units RNasin ribonuclease inhibitor, and 200 units Moloney Murine Leukemia Virus Reverse Transcriptase at 37°C for 1 h.
cDNAs obtained from reverse transcription were used as a template to run real-time PCR. The following components were added to a reaction tube: 9 µL of iQ SYBR Green Supermix solution (Bio-Rad), 1 µL of 5 µM specific primers, and 8 µL of the diluted cDNA template. Thermocycling conditions were 95°C for 3 minutes followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s, with a melting curve detected at 95°C for 1 minute, 55°C for 1 minute, and detected the denature time from 55°C to 95°C. Each sample was quantified at least triplicate and normalized using Ubiquitin 10 as an internal control. The gene-specific primer pairs for quantitative RT-PCR are listed in Table S1.
ChIP assays
ChIP assay was carried out as described [42]. Chromatin extracts were prepared from 10 day old seedlings treated with formaldehyde. The chromatin was sheared to an average length of 500 bp by sonication and immunoprecipitated with specific antibodies including anti-acetylated histone H3K9K14 (Catalogue no. 06-599, Millipore), anti-trimethylated histone H3K4 (Catalogue no. 04-745, Millipore), anti-c-Myc (Catalogue no. M4439, Sigma) and anti-FLAG (Catalogue no. F1804, Sigma). The DNA cross-linked to immunoprecipitated proteins was analyzed by real-time PCR. Relative enrichments of various regions of KNAT1, KNAT2 and KNATM in axe1-5, as1-1 and as1-1/axe1-5 over Col were calculated after normalization to ACTIN2. Each of the immunoprecipitations was replicated three times, and each sample was quantified at least in triplicate. The primers used for real-time PCR analysis in ChIP assays are listed in Table S2.
BiFC assays
To generate the constructs for BiFC, full-length coding sequences of HDA6, AS1 and AS2 were PCR-amplified using Pfu polymerase (Finnzymes). The PCR products were subcloned into the pENTR/SD/D-TOPO or pCR8/GW/TOPO vector and then recombinated into the pEarleyGate201-YN and pEarleyGate202-YC vectors [22]. The resulting constructs were transformed into the Agrobacterium GV3101 and the Agrobacteria containing these constructs were cotransfected into five week old Nicotiana benthamiana leaves. For the protoplast transient expression, HDA6, AS1 and AS2 fused with pEarleyGate201-YN or pEarleyGate201-YC were co-transfected into protoplasts by PEG transfection [43]. Transfected leaves and protoplasts were imaged using TCS SP5 (Leica) Confocal Spectral Microscope Imaging System.
In vitro pull-down assays
Pull-down assays were performed as previously described [44] with some modifications. 2 µg Myelin basic protein (MBP) and MBP-AS1 recombinant proteins were incubated with 30 µl of MBP resin in a total volume of 500 µl of MBP binding buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA) for 2 h at 4°C, and the binding reaction was washed 3 times by the binding buffer, then 2 µg GST-HDA6 recombinant protein was added and incubated for additional 2 h at 4°C. After extensive washing (at least 8 times), the pulled down proteins were eluted by boiling, separated by 10% SDS-PAGE, and detected by western blotting using an anti-GST antibody.
Coimmunoprecipitation assays
Coimmunocipitation assays were performed as previous described [23]. The 20-day-old axe1-5/35S:GFP-HDA6, axe1-5 and as1-1 plants were harvested and ground in liquid nitrogen. Total proteins were extracted in an extraction buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 20% glycerol, and 1% CA-630) containing protease inhibitor cocktail (Roche). Cell debris was pelleted by centrifugation at 14,000 g for 30 min. The supernatant was incubated with anti-AS1 or anti-GFP specific antibody overnight at 4°C by gently rotation, then 50 µl of protein G agarose beads (Millipore) was added. After 3 h of incubation at 4°C by gently rotation, the beads were centrifuged and washed five times with a washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, and 1% CA-630). Proteins were eluted with 40 µl of 2.5× sample buffer and analyzed by western blotting using anti-AS1 and anti-GFP (Santa Cruz Biotechnologies) antibodies.
Supporting Information
Zdroje
1. LongJA, OhnoC, SmithZR, MeyerowitzEM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312 : 1520–1523.
2. VollbrechtE, ReiserL, HakeS (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 : 3161–3172.
3. ScofieldS, MurrayJA (2006) KNOX gene function in plant stem cell niches. Plant Mol Biol 60 : 929–946.
4. JacksonD, VeitB, HakeS (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 : 405–413.
5. SinhaNR, WilliamsRE, HakeS (1993) Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7 : 787–795.
6. ChuckRS, WilliamsJM, GoldbergMA, LubniewskiAJ (1996) Recurrent corneal ulcerations associated with smokeable methamphetamine abuse. Am J Ophthalmol 121 : 571–572.
7. KidnerCA, TimmermansMCP, ByrneME, MartienssenRA (2002) Developmental genetics of the angiosperm leaf. Adv Bot Res 38 : 191–234.
8. ByrneME (2005) Networks in leaf development. Curr Opin Plant Biol 8 : 59–66.
9. MagnaniE, HakeS (2008) KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20 : 875–887.
10. TimmermansMC, HudsonA, BecraftPW, NelsonT (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 : 151–153.
11. TsiantisM, SchneebergerR, GolzJF, FreelingM, LangdaleJA (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284 : 154–156.
12. ByrneME, BarleyR, CurtisM, ArroyoJM, DunhamM, et al. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 : 967–971.
13. OriN, EshedY, ChuckG, BowmanJL, HakeS (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 : 5523–5532.
14. XuL, XuY, DongA, SunY, PiL, et al. (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 : 4097–4107.
15. Phelps-DurrTL, ThomasJ, VahabP, TimmermansMC (2005) Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 : 2886–2898.
16. GuoM, ThomasJ, CollinsG, TimmermansMC (2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20 : 48–58.
17. RingroseL, ParoR (2007) Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134 : 223–232.
18. AhmadA, TakamiY, NakayamaT (2003) WD dipeptide motifs and LXXLL motif of chicken HIRA are necessary for transcription repression and the latter motif is essential for interaction with histone deacetylase-2 in vivo. Biochem Biophys Res Commun 312 : 1266–1272.
19. YangXJ, GregoireS (2005) Class II histone deacetylases: from sequence to function, regulation, and clinical implication. Mol Cell Biol 25 : 2873–2884.
20. AufsatzW, MetteMF, van der WindenJ, MatzkeM, MatzkeAJ (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. Embo J 21 : 6832–6841.
21. MurfettJ, WangXJ, HagenG, GuilfoyleTJ (2001) Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13 : 1047–1061.
22. LuQ, TangX, TianG, WangF, LiuK, et al. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61 : 259–270.
23. YuCW, LiuX, LuoM, ChenC, LinX, et al. (2011) HISTONE DEACETYLASE6 Interacts with FLOWERING LOCUS D and Regulates Flowering in Arabidopsis. Plant Physiol 156 : 173–184.
24. UenoY, IshikawaT, WatanabeK, TerakuraS, IwakawaH, et al. (2007) Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19 : 445–457.
25. LiuX, YuCW, DuanJ, LuoM, WangK, et al. (2012) HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol 158 : 119–129.
26. IkezakiM, KojimaM, SakakibaraH, KojimaS, UenoY, et al. (2010) Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J 61 : 70–82.
27. RhoadesMW, ReinhartBJ, LimLP, BurgeCB, BartelB, et al. (2002) Prediction of plant microRNA targets. Cell 110 : 513–520.
28. EmeryJF, FloydSK, AlvarezJ, EshedY, HawkerNP, et al. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13 : 1768–1774.
29. TangG, ReinhartBJ, BartelDP, ZamorePD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17 : 49–63.
30. NikovicsK, BleinT, PeaucelleA, IshidaT, MorinH, et al. (2006) The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18 : 2929–2945.
31. EarleyKW, PontvianneF, WierzbickiAT, BlevinsT, TuckerS, et al. (2010) Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24 : 1119–1132.
32. HamantO, PautotV (2010) Plant development: a TALE story. C R Biol 333 : 371–381.
33. SemiartiE, UenoY, TsukayaH, IwakawaH, MachidaC, et al. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 : 1771–1783.
34. ToTK, KimJM, MatsuiA, KuriharaY, MorosawaT, et al. (2011) Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 7: e1002055 doi:10.1371/journal.pgen.1002055..
35. TsukayaH, UchimiyaH (1997) Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol Gen Genet 256 : 231–238.
36. LinWC, ShuaiB, SpringerPS (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 : 2241–2252.
37. XuY, SunY, LiangWQ, HuangH (2002) The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. ACTA BOTANICA SINICA-CHINESE EDITION - 44 : 1194–1202.
38. IwakawaH, UenoY, SemiartiE, OnouchiH, KojimaS, et al. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43 : 467–478.
39. LiH, XuL, WangH, YuanZ, CaoX, et al. (2005) The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17 : 2157–2171.
40. XuL, ShenWH (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18 : 1966–1971.
41. SchubertD, PrimavesiL, BishoppA, RobertsG, DoonanJ, et al. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. Embo J 25 : 4638–4649.
42. GendrelAV, LippmanZ, MartienssenR, ColotV (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2 : 213–218.
43. YooSD, ChoYH, SheenJ (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2 : 1565–1572.
44. YangJY, IwasakiM, MachidaC, MachidaY, ZhouX, et al. (2008) betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22 : 2564–2577.
Štítky
Genetika Reprodukční medicína
Článek Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction DiseaseČlánek Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination SchemesČlánek Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- A Mystery Unraveled: Essentiality of RNase III in Is Caused by Resident Prophages
- Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction Disease
- Insertion/Deletion Polymorphisms in the Promoter Are a Risk Factor for Bladder Exstrophy Epispadias Complex
- Mi2β Is Required for γ-Globin Gene Silencing: Temporal Assembly of a GATA-1-FOG-1-Mi2 Repressor Complex in β-YAC Transgenic Mice
- Dissection of a Quantitative Trait Locus for PR Interval Duration Identifies as a Novel Modulator of Cardiac Conduction
- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Construction of a Global Pain Systems Network Highlights Phospholipid Signaling as a Regulator of Heat Nociception
- Genes Contributing to Pain Sensitivity in the Normal Population: An Exome Sequencing Study
- Identification of , a Locus Controlling Dominant Resistance to Autoimmune Orchitis, as Kinesin Family Member 1C
- ATX1-Generated H3K4me3 Is Required for Efficient Elongation of Transcription, Not Initiation, at ATX1-Regulated Genes
- Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome
- Identification of Novel Type 2 Diabetes Candidate Genes Involved in the Crosstalk between the Mitochondrial and the Insulin Signaling Systems
- The Genetic Architecture of Adaptations to High Altitude in Ethiopia
- Population Genomics of the Endosymbiont in
- Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination Schemes
- Testicular Differentiation Occurs in Absence of R-spondin1 and Sox9 in Mouse Sex Reversals
- A Yeast GSK-3 Kinase Mck1 Promotes Cdc6 Degradation to Inhibit DNA Re-Replication
- Genetic Adaptation Associated with Genome-Doubling in Autotetraploid
- The Essential Function of RNase III Is to Silence Foreign Toxin Genes
- Long-Range Regulatory Polymorphisms Affecting a GABA Receptor Constitute a Quantitative Trait Locus (QTL) for Social Behavior in
- A New Isolation with Migration Model along Complete Genomes Infers Very Different Divergence Processes among Closely Related Great Ape Species
- Chromosome Fragile Sites in Harbor Matrix Attachment Regions That May Be Associated with Ancestral Chromosome Rearrangement Events
- Genome-Wide Association Study Implicates Testis-Sperm Specific as a Susceptibility Locus for Impaired Acrosome Reaction in Stallions
- A Mechanism of Gene Amplification Driven by Small DNA Fragments
- Base Damage within Single-Strand DNA Underlies Hypermutability Induced by a Ubiquitous Environmental Agent
- Integrative Analysis of a Cross-Loci Regulation Network Identifies as a Gene Regulating Insulin Secretion from Pancreatic Islets
- Reciprocal Signaling between the Ectoderm and a Mesendodermal Left-Right Organizer Directs Left-Right Determination in the Sea Urchin Embryo
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Genome-Wide Fine-Scale Recombination Rate Variation in
- Deciphering the Transcriptional-Regulatory Network of Flocculation in
- On Lung Function and Interactions Using Genome-Wide Data
- Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function
- The Effective Population Size of Malaria Mosquitoes: Large Impact of Vector Control
- Recessive Mutations in Implicate β-III Spectrin in Both Cognitive and Motor Development
- An Excess of Gene Expression Divergence on the X Chromosome in Embryos: Implications for the Faster-X Hypothesis
- Reduced Life- and Healthspan in Mice Carrying a Mono-Allelic MVA Mutation
- Natural Variation at the MATE Transporter Locus Reveals Cross-Talk between Fe Homeostasis and Zn Tolerance in
- Histone Deacetylase HDA6 Is Functionally Associated with AS1 in Repression of Genes in
- A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling
- A Polycomb Group Protein Is Retained at Specific Sites on Chromatin in Mitosis
- Diapause Formation and Downregulation of Insulin-Like Signaling via DAF-16/FOXO Delays Axonal Degeneration and Neuronal Loss
- Genes That Act Downstream of Sensory Neurons to Influence Longevity, Dauer Formation, and Pathogen Responses in
- A Genome-Wide RNAi Screen Reveals MAP Kinase Phosphatases as Key ERK Pathway Regulators during Embryonic Stem Cell Differentiation
- Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
- Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
- Systems Genetic Analysis of Osteoblast-Lineage Cells
- Population Genomics of Inversion Polymorphisms in
- Spreading of Heterochromatin Is Limited to Specific Families of Maize Retrotransposons
- DNA Topoisomerases Maintain Promoters in a State Competent for Transcriptional Activation in
- A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences during Morphogenesis
- Approaching the Functional Annotation of Fungal Virulence Factors Using Cross-Species Genetic Interaction Profiling
- Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization
- Systematic Identification of Rhythmic Genes Reveals as a New Element in the Circadian Clockwork
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



