-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Transcriptional Rewiring of the Sex Determining Gene Duplicate by Transposable Elements
Control and coordination of eukaryotic gene expression rely on transcriptional and posttranscriptional regulatory networks. Evolutionary innovations and adaptations often require rapid changes of such networks. It has long been hypothesized that transposable elements (TE) might contribute to the rewiring of regulatory interactions. More recently it emerged that TEs might bring in ready-to-use transcription factor binding sites to create alterations to the promoters by which they were captured. A process where the gene regulatory architecture is of remarkable plasticity is sex determination. While the more downstream components of the sex determination cascades are evolutionary conserved, the master regulators can switch between groups of organisms even on the interspecies level or between populations. In the medaka fish (Oryzias latipes) a duplicated copy of dmrt1, designated dmrt1bY or DMY, on the Y chromosome was shown to be the master regulator of male development, similar to Sry in mammals. We found that the dmrt1bY gene has acquired a new feedback downregulation of its expression. Additionally, the autosomal dmrt1a gene is also able to regulate transcription of its duplicated paralog by binding to a unique target Dmrt1 site nested within the dmrt1bY proximal promoter region. We could trace back this novel regulatory element to a highly conserved sequence within a new type of TE that inserted into the upstream region of dmrt1bY shortly after the duplication event. Our data provide functional evidence for a role of TEs in transcriptional network rewiring for sub - and/or neo-functionalization of duplicated genes. In the particular case of dmrt1bY, this contributed to create new hierarchies of sex-determining genes.
Published in the journal: . PLoS Genet 6(2): e32767. doi:10.1371/journal.pgen.1000844
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000844Summary
Control and coordination of eukaryotic gene expression rely on transcriptional and posttranscriptional regulatory networks. Evolutionary innovations and adaptations often require rapid changes of such networks. It has long been hypothesized that transposable elements (TE) might contribute to the rewiring of regulatory interactions. More recently it emerged that TEs might bring in ready-to-use transcription factor binding sites to create alterations to the promoters by which they were captured. A process where the gene regulatory architecture is of remarkable plasticity is sex determination. While the more downstream components of the sex determination cascades are evolutionary conserved, the master regulators can switch between groups of organisms even on the interspecies level or between populations. In the medaka fish (Oryzias latipes) a duplicated copy of dmrt1, designated dmrt1bY or DMY, on the Y chromosome was shown to be the master regulator of male development, similar to Sry in mammals. We found that the dmrt1bY gene has acquired a new feedback downregulation of its expression. Additionally, the autosomal dmrt1a gene is also able to regulate transcription of its duplicated paralog by binding to a unique target Dmrt1 site nested within the dmrt1bY proximal promoter region. We could trace back this novel regulatory element to a highly conserved sequence within a new type of TE that inserted into the upstream region of dmrt1bY shortly after the duplication event. Our data provide functional evidence for a role of TEs in transcriptional network rewiring for sub - and/or neo-functionalization of duplicated genes. In the particular case of dmrt1bY, this contributed to create new hierarchies of sex-determining genes.
Introduction
Control and coordination of eukaryotic gene expression rely on transcriptional and posttranscriptional regulatory networks. From an evolutionary point of view innovations and changes in given functional linkages of regulatory networks have to occur at the DNA level by alteration of the cis-regulatory sequence defining transcription factor binding sites. While such alterations may occur in any cis-regulatory module, they will have fundamentally different effects depending on where in the structure of the network they occur (see [1] for review). After the discovery of the ubiquitousness of repeated sequences, a long standing hypothesis proposed that repeated sequences were likely to be active in the 5′ regions of genes controlling transcription [2] and that they could move and supply evolutionary variations [3].
From an evolutionary perspective, transposable elements (TEs) have recently been attributed an important role in shaping the gene regulation landscape [4],[5],[6]. In spite of and, to some extent, because of their selfish and parasitic nature, the movement and accumulation of TEs have exerted a strong influence on the evolutionary trajectory of their host genome [7]. Many ways have been illustrated through which TEs can directly influence the regulation of nearby gene expression, both at the transcriptional and post-transcriptional levels (for review see [5]). Genome-scale bioinformatic analyses have shown that many promoters and polyadenylation signals of human and mouse genes are derived from primate and rodent–specific TEs respectively [8],[9]. Hence, it is postulated that insertion of TEs harbouring “ready-to-use” cis-regulatory sequences probably contributed to the establishment of lineage-specific patterns of gene expression [10]. In addition to donating cis-elements and creating new regulatory networks, the movement and accumulation of TEs have recently been proposed to participate in the rewiring of pre-established regulatory networks (see [5] for review).
Such rewiring is especially important when rapid adaptation of existing regulatory networks or new networks become necessary. One system where fast changes obviously regularly occurred during evolution is the genetic control of sexual development [11],[12]. It is well documented that different groups of organisms and sometimes even closely related species of different populations of the same species have fundamentally different modes of sex determination. Comparative evolutionary studies of the genetic cascades controlling sex determination in different species revealed that the master genes at the top of the regulatory hierarchy can change dramatically as new species evolve, while the downstream genes at the bottom of the hierarchy remain the same, exerting essentially identical functions from one species to the next (see [12],[13] for review).
The most conserved downstream component characterized to date, a gene with homology to both the Drosophila doublesex and C. elegans mab-3 sex regulatory genes, is the Dmrt1 (Doublesex and Mab-3 Related Transcription factor 1) gene of vertebrates [14]. All three genes encode proteins sharing a common DNA-binding domain and belong to the DM domain gene family, which has been shown to be involved in sex determination and differentiation in organisms as phylogenetically divergent as corals, worms, flies and all vertebrate groups ranging from fish to mammals. Of note, in humans, haploinsufficiency of the genomic region that includes DMRT1 and its paralogs DMRT2 and DMRT3 leads to XY male to female sex reversal [15]. In chicken and other avian species Dmrt1 is located on the Z chromosome, but absent from W, making it an excellent candidate for the male sex-determining gene of birds [16],[17].
In the medaka fish (Oryzias latipes), which has XY-XX sex determination, a duplicated copy of dmrt1, designated dmrt1bY or DMY, on the Y-chromosome was shown to be the master regulator of male development [18],[19], similar to Sry in mammals. Interestingly, also in Xenopus laevis a W-specific duplicate of dmrt1 was shown to participate in primary gonad development [20]. Because medaka dmrt1bY acts, like Sry, as a dominant male determiner [21], it is believed that it is functionally equivalent to the mammalian gene and might share many molecular features [22],[23]. Although many of the early cellular and morphological events downstream of Sry have been characterized, as well as a number of genes involved in these processes (for review see [24]–[26]), little is known about the mode of action and the biological targets of Sry [27]. Interestingly, dmrt1, the ancestor of dmrt1bY, is one of these downstream effectors of Sry. Contrary to the situation with Sry, it is totally unknown how in medaka dmrt1bY expression is regulated. As a prerequisite to elucidate the function of dmrt1bY, information on its expression regulation at the transcriptional level is required. Here, we found a feed back down-regulation of the dmrt1bY promoter. Also dmrt1a, the autosomal ancestor of dmrt1bY, is able to down-regulate transcription of its paralog. Interestingly we found clear evidence that the major cis-regulatory element, pre-existing within a new medaka-specific TE at the time of its insertion, was co-opted in order to confer to Dmrt1bY its specific expression pattern after gene duplication. This is the first experimental evidence supporting a role of TEs for transcriptional network rewiring in sub - and/or neo-functionalization of duplicated genes. Additionally, in the particular case of dmrt1bY, this contributed to create new hierarchies of sex determining genes.
Results
Sequence evolution of the dmrt1bY promoter
To obtain insights into the sequence evolution of the cis-regulatory region of the dmrt1bY gene on the Y-chromosome, we first compared its genomic region and that of its autosomal progenitor, the dmrt1a gene from linkage group 9 (LG9), with those of the available dmrt1 orthologs from other teleosts (stickleback, Tetraodon, Fugu, zebrafish), chicken and human. This phylogenetic footprinting approach should point to the conservation of regulatory elements being putatively essential for vertebrate Dmrt1 gene expression (Figure S1). Furthermore, it could possibly indicate cis-regulatory subfunctionalization between the medaka dmrt1 paralogs as observed for other pairs of duplicated genes with spatio-temporal expression divergence [28],[29]. However, our VISTA plots (Figure S1) did not reveal sequence conservation in the promoter regions of neither dmrt1bY nor dmrt1a with other vertebrates except for stretches corresponding to the teleost MHCL gene, which are pseudogenes in both medaka dmrt1 5′ regions [30]. Conserved non-coding elements were also not found between the other vertebrate sequences suggesting that the regulatory regions of vertebrate Dmrt1 orthologs diverged strongly despite their conserved position in the sex determination cascade. High turn-over of cis-regulatory regions in the face of conserved expression is commonly found for vertebrate genes [31].
However, longer stretches of conservation between promoter regions of dmrt1bY and dmrt1a in medaka were evident (Figure S1). Thus, we compared in more detail the promoter regions of the medaka dmrt1 paralogs upstream of the transcriptional start site to the last exons of their upstream gene, KIAA00172, which is a pseudogene on the Y chromosome but functional on the autosomal LG9 [30] (Figure 1A). This region spans around 9 Kb on the Y chromosome but only around 6 Kb on autosomal LG9.
Fig. 1. Comparative analysis of the dmrt1bY promoter and its transcription factor binding sites. 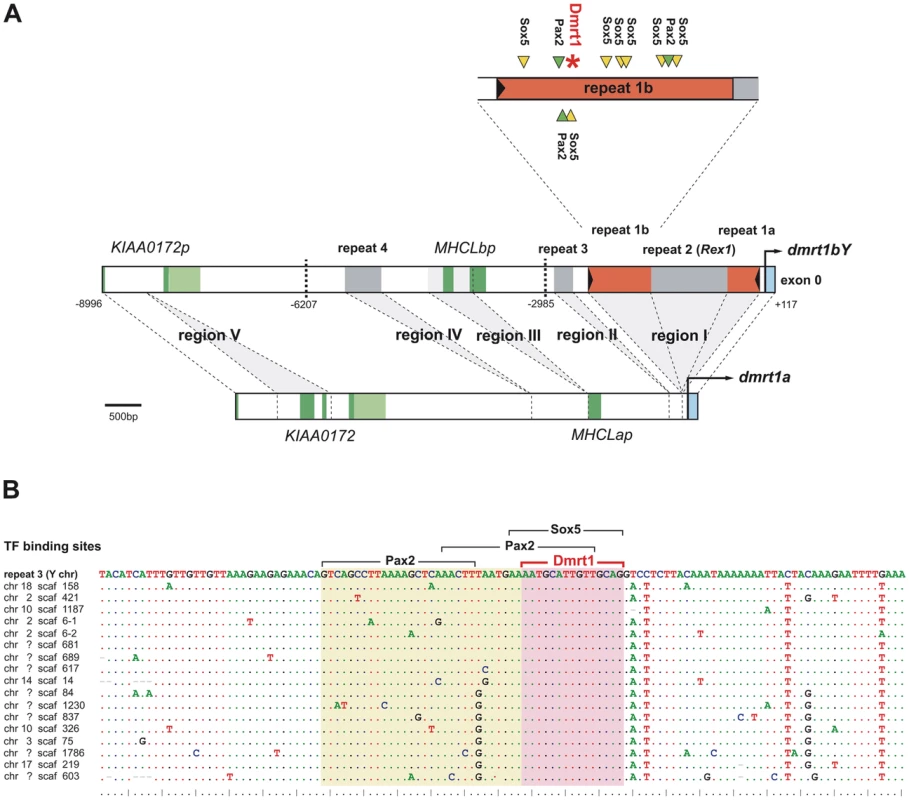
(A) The analyzed promoter region of dmrt1bY in comparison to its dmrt1a paralog. Length differences between the promoters are based on regions I–IV, of which three (I, II, IV) have been added to the dmrt1bY promoter after duplication, while two others have been lost from the dmrt1bY promoter (KIAA0172 region V) or from the dmrt1a promoter (MHCLp region III), respectively. Bold dashed lines indicate cutting sites for transcriptional regulation analyses. Region I contains a putative Izanagi DNA transposons (repeat 1), into which a Rex1 element (repeat 2) was inserted secondarily. The upstream part of split repeat 1 (repeat 1b) in the dmrt1bY promoter contains multiple Sox5 and Pax2 binding sites as well as a Dmrt1 binding site. Dark green indicates coding sequence of genes and pseudogenes, light green indicates their untranslated regions. (B) Alignment of the Y chromosomal repeat 1b with 17 exemplary repeat copies in the medaka genome. Dots indicate conserved sites. The Dmrt1 binding site is perfectly conserved suggesting that it is an integral part of repeat 1 elements. All 28 copies of repeat 1b in medaka with the Dmrt1 binding sites are listed in Table S1. In the upstream sequence of dmrt1 paralogs, five regions contribute to length divergence between autosome and Y chromosome (Table 1, Figure 1A, Figure S1 and Figure S2). Region I located 69 bp upstream of the transcriptional start site of dmrt1bY is over 2 Kb in length and absent from dmrt1a. Similarly, regions II–IV further upstream are only found on the Y chromosome but not on the autosome.
Tab. 1. Characterization of regions contributing to length differences between dmrt1 upstream sequences. 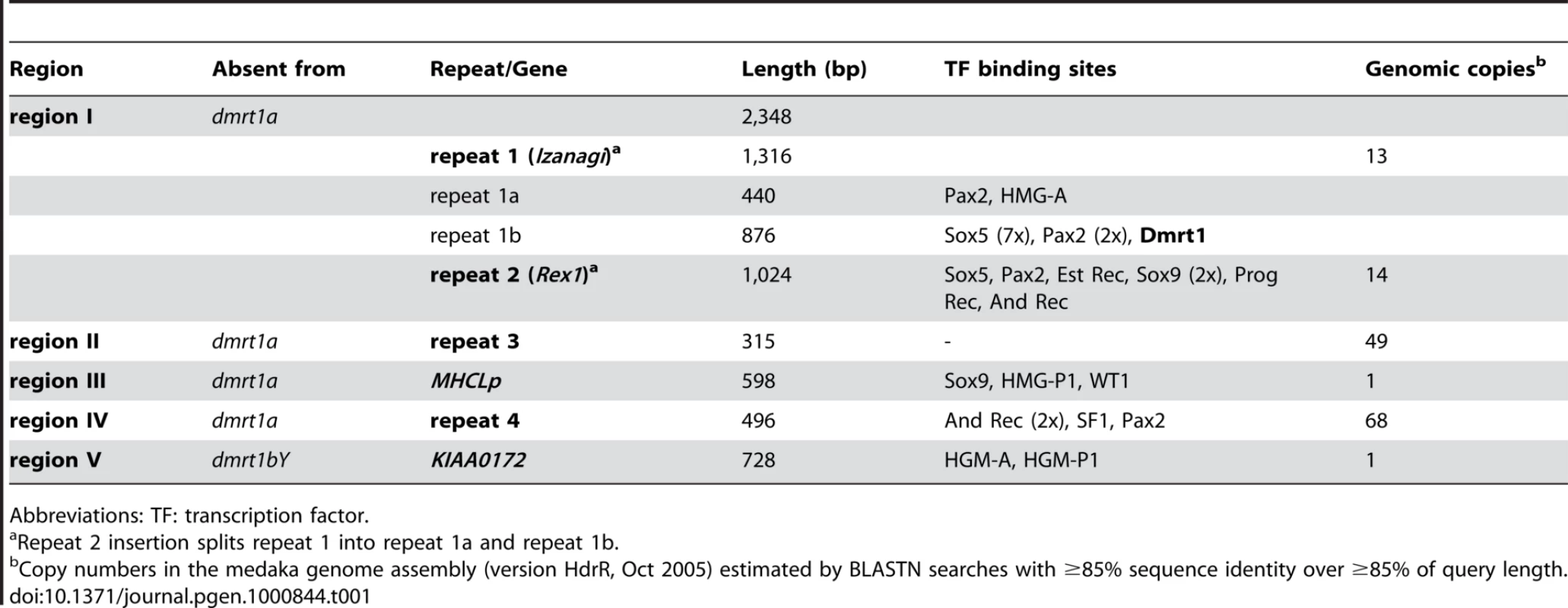
Abbreviations: TF: transcription factor. Region V, in contrast, is missing from the Y chromosome but present on the autosome. This region contains two exons of the KIAA0172 gene and obviously has been lost during the pseudogenization of the Y chromosomal KIAA0172 copy after the duplication of the dmrt1 region [30].
Region III is directly adjacent to the MHCL pseudogenes [30],[32] present in both dmrt1 promoters and a stretch of sequence similarity with other teleost MHCL orthologs is found within region III on the Y chromosome (Figure S1). Furthermore, region III is present only once in the medaka genome and does not constitute a repetitive element according to RepeatMasker analysis. Thus, region III seems to have been lost from the autosomal dmrt1a region after duplication, but has been kept on the Y chromosome.
Next, we further characterized the additional regions I, II and IV for the presence of repetitive sequences. As each of these three regions is present in multiple copies in the medaka genome (Table 1), they constitute repetitive sequences that were rather added to the Y chromosomal region than lost from the autosome. Each repeat element is only present once in the analyzed section of the dmrt1bY region and is often found in poorly assembled regions of the medaka genome emphasizing their repetitive character. BLAST searches in other teleost genomes failed to find the repeat elements identified here in other species suggesting that they are specific to the medaka lineage.
Particularly, region I is subdivided into three parts with a non-LTR retrotransposable Rex1 element of the LINE family (class I transposable elements) as middle segment (“repeat 2”) (Table 1).
The surrounding parts (“repeat 1a”, “repeat 1b”) also represent repetitive sequences with multiple copies in the medaka genome. These two repeat regions are found side by side in other regions of the genome. Thus, they together build a larger repeat element (“repeat 1”) into which repeat 2 was inserted (see below). Repeat 1 has a length of 1316 bp and is characterized by 27 bp terminal inverted repeats (TIRs) (5′-CAATGAGTTATATCACTAGAGGAGACA-3′) assigning it to DNA transposons (class II transposable elements). However, it does not contain a transposase gene or any other open reading frame and thus constitutes a non-autonomous class II element. Only few diagnostic motifs are available to classify such elements [33]. Repeat 1 in the dmrt1bY promoter has a 8 bp target site duplication (5′ - GTGTGGCT-3′) and other copies of this element in the medaka genome have target site duplications of the same length. Repeat 1 is found in multiple copies in the medaka genome (Table 1 and Table S1), which generally have target site duplications. This points to an active state of repeat 1 in the medaka genome.
From the consensus sequence of the multiple repeat 1 elements in the medaka genome, a THAP protein domain composed of three putative exons was deduced (Figure S2 and Figure S3). In the repeat 1 element in the dmrt1bY promoter, the second putative exon of the THAP domain has been disrupted by the insertion of the repeat element (Figure S2 and Figure S3). The THAP domain is a DNA-binding zinc finger motif present in the P element transposases from Drosophila [34]. Furthermore, the terminal motif of repeat 1 is similar to the consensus sequence for the P element superfamily of DNA transposons (5′-YARNG-3′) [7]. Thus, we conclude that we have identified a new, medaka-specific non-autonomous P element element that we term Izanagi (named after an ancient Japanese deity, “the male who invites”; for etymology see Text S1).
Vertebrate mobile DNA transposons of the P element family have been only found so far in zebrafish [35],[36]. However, the THAP domain has been recurrently recruited from domesticated P elements during chordate evolution [37]. In the Izanagi family, the THAP domain is degenerated and, in the case of repeat 1, has been additionally disrupted by the repeat 2 insertion.
Region II (“repeat 3”) and region IV (“repeat 4”) also have multiple copies in the medaka genome (Table 1). They do not contain open reading frames and, like repeat 1, they also lack similarity to known transposable elements. Furthermore, target site duplication or other diagnostic features could not be recognized preventing further classification as putative transposable elements.
Identification of putative transcription factor binding sites within dmrt1bY promoter and transcriptional activity in different cell lines
The sequence of the medaka dmrt1bY promoter region (9.107 Kb) was next analyzed for the presence of putative transcription factor binding sites using the MatInspector program (Figure 1A and Figure S4 and Table 1). Most interestingly, the Izanagi element is characterized by an overrepresentation of putative binding sites for Sox5 (Figure 1 and Figure S4). In this region seven Sox5 binding sites are present while random prediction would expect 15 times less (only 0.46 sites; MatInspector). This, together with the fact that Sox5 expression has been correlated with direct dmrt1 promoter down-regulation in zebrafish [38] suggests that region to be of primary interest for medaka Dmrt1bY transcriptional regulation, but remains to be investigated for the proposed functional role of Sox5.
Additionally, the 9 Kb dmrt1bY promoter region contains several other putative transcription factor binding sites such as Pax2, HMG-box protein 1, HMG-A, Sox9, WT1 and SF1 binding sites that are reasonable candidates for gonadal-specific transcriptional regulation (Figure 1A and Figure S4 and Table 1). Several of them are conserved with the dmrt1a promoter (Figure S4) and might be essentially required for dmrt1 expression.
To evaluate the mechanisms regulating dmrt1bY transcription, a portion of the medaka gene from +117 bp to −8990 bp of the transcriptional start site was cloned upstream of the Gaussia luciferase gene (pBSII-ISceI::9 Kb Dmrt1bY prom::GLuc) and the activity of the promoter was measured in a variety of cell types using transient transfection analysis. Sequential deletions of the 9 Kb promoter were generated from pBSII-ISceI::9 Kb Dmrt1bY prom::GLuc. In all three cell types basal promoter activity was detectable when using the 3 Kb proximal region (Figure S5). In fibroblast cell lines (Xiphophorus A2 and medaka HN2), but not in Sertoli TM4 cells, a dramatic drop in promoter activity was observed when the region from bp −2985 to −6207 was added (Figure S5). Similarly, the same decrease of promoter activity was apparent in Sertoli TM4 cells when the bp −6207 to −8996 region was additionally inserted (Figure S5). This indicates the possible presence of Sertoli cell specific transcriptional repressing sequence(s) within the most distal part of the dmrt1bY promoter. The most proximal part of the promoter always accounted for the basal activity in all the cell lines tested. Interestingly, two adjacent binding sites for Steroidogenic factor 1 (Sf1) are located at positions −5933 and −5524 (Figure S4). Being specifically expressed in Sertoli -and Leydig - cells, it is tempting to assume that the presence of these two distinct SF-1 binding sites nested in this −3 to −6 Kb fragment is accounting for this difference. A similar situation has been shown for the porcine Sry promoter for which SF-1 transactivation occurs at two SF1 binding sites [39].
A unique Dmrt1 binding site is present in medaka dmrt1bY but not in dmrt1 promoter
Using the vertebrate Dmrt1 binding site matrix [40], different dmrt1 promoters -including medaka dmrt1a and dmrt1bY - (up to 9 KB upstream the ORF) were scanned for such target site sequences. A unique and robust Dmrt1 binding site of high prediction probability was found only in the medaka dmrt1bY promoter (CTGCAACAATGCATT; weight: 8.5, pValue: 1.0 e-05, lnPval:−11.492) (Figure 1A and Figures S2, S3, S4) but not in the dmrt1a promoter (lower threshold set to 0). Interestingly, this predicted Dmrt1 binding site is nested within the above newly described Oryzias latipes Izanagi element in the proximal active part of dmrt1bY promoter (Figure 1A and Figure S5). The medaka putative Dmrt1 binding site is present at position −2132 within repeat 1b in close proximity to the Sox5 binding site-rich region (Figure 1A and Figure S4). We first asked about the origin of this Dmrt1 binding site. It might have either evolved de novo from sequence provided by repeat 1b or been an integral part of such repeats and then was inserted into the dmrt1bY promoter after duplication. We therefore blasted the region approximately 300 bp up-and downstream of the Dmrt1 binding site to the medaka genome and aligned the obtained repeat sequences (Figure 1B). In total, we identified 28 elements that are highly similar to repeat 1b and that contain the same Dmrt1 binding site found in the dmrt1bY promoter (Table S1). Furthermore, the predicted Dmrt1 binding site is present in the derived Izanagi consensus sequence. Hence, this putative Dmrt1 binding site is a regular and conserved part of the Izanagi transposon family.
Timing of the Izanagi element insertion into the dmrt1bY promoter
Given that the Dmrt1 binding site donated to the dmrt1bY promoter by the Izanagi element has been important for the evolution of dmrt1bY function within the sex determining cascade, we asked about the timing of the Izanagi insertion in relation to the duplication of the medaka dmrt1 genes. The dmrt1 gene duplication occurred in a common ancestor of medaka (O. latipes), O. curvinotus and O. luzonensis around 10 million years ago [41].
First, we estimated the sequence divergence between repeat 1 from the dmrt1bY promoter and the Izanagi element consensus and mapped it onto a linearized neighbour joining (NJ) tree of dmrt1 genes from the genus Oryzias, which was based on neutral sites only (third codon positions). This analysis showed that the repeat 1 insertion occurred after the split from O. mekongensis but before the divergence of medaka, O. curvinotus and O. luzonensis (Figure S6A). This is exactly the branch on which the dmrt1 duplication occurred. We also estimated the dmrt1 duplication by the same method. There has to be a note of caution with dating the age of the dmrt1 duplication due to the enhanced rate of molecular evolution of dmrt1bY after duplication [41]. Nevertheless, based on sequence divergence data the insertion of repeat 1 is certainly estimated to be younger than the dmrt1 duplication (Figure S6A). Using a different nuclear marker to date the divergence of the Oryzias species, the tyrosinase a gene, a similar result was obtained (Figure S6A). The analogous analysis for the secondary insertion of repeat 2 into repeat 1, in contrast, revealed that this insertion is quite young and must have occurred in Oryzias latipes.
We conclude that our sequence divergence estimates are consistent with an insertion of repeat 1 and thereby of the Dmrt1 binding site shortly after the dmrt1 duplication, supporting its importance for the evolution the Dmrt1bY sex determinator function.
Transcription of dmrt1bY is regulated by its own gene product and by that of its paralog
1-Dmrt1bY that down-regulates activity of its own promoter
Co-transfection analyses were used to examine the predicted interaction between medaka Dmrt1bY and its own promoter (Figure 2A). For this purpose, the proximal 2868 bp dmrt1bY promoter region containing the putative Dmrt1 binding site at position −2132 (see Figure 1 and Figure 2) fused to luciferase was used and co-transfected with different amounts of a plasmid expressing dmrt1bY (Figure 2A). In presence of dmrt1bY expressing plasmids dmrt1bY promoter activity was considerably reduced, up to 74%, in all cell types tested (Mus musculus Sertoli TM4, Xiphophorus xiphidium fibroblast A2 and Oryzias latipes spermatogonial (Sg3) and embryonic stem (MES1) cells) (Figure 2A). To confirm the possible direct interaction with the putative Dmrt1 target binding site, a dmrt1bY mutant promoter with a modified Dmrt1 target site was created (Figure 2B and Materials and Methods). When co-transfected with the dmrt1bY expressing plasmid, the activity of the mutant promoter was clearly increased (up to almost 5 fold) in comparison to the wild-type promoter (Figure 2B) whereas the mutant promoter did not reveal any significant regulation by Dmrt1bY (Figure S7).
Fig. 2. Transient-transfection analysis. 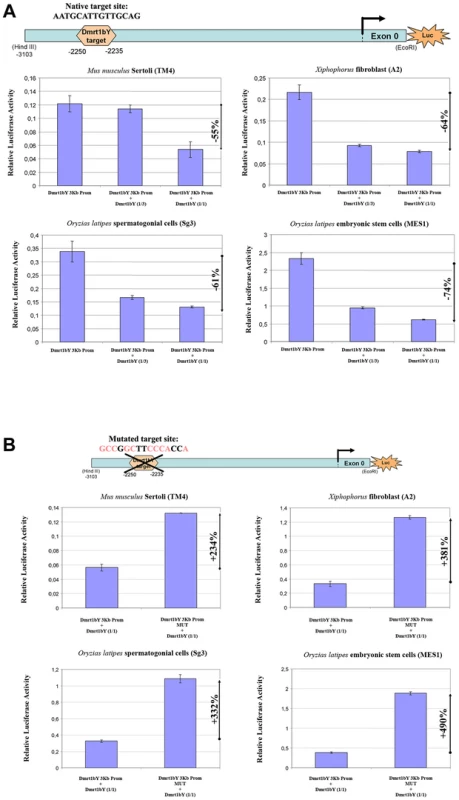
(A) Transient-transfection analysis of proximal dmrt1bY promoter activity co-transfected in different ratios of a dmrt1bY expressing plasmid. The 3 Kb proximal dmrt1bY promoter construct was co-transfected with different amounts of dmrt1bY-expressing plasmid (1∶3 and 1∶1 ratios) in different cell lines. (B) Transient-transfection analysis of mutant proximal dmrt1bY promoter activity. In all the cell lines, when overexpressing Dmrt1bY, the 3 Kb mutant proximal dmrt1bY promoter construct (lacking the Dmrt1 binding site) shows higher activity compared to the “wild-type” construct containing the Dmrt1 target site. The data are presented as the firefly/Gaussia luciferase activity. Transfections were done three times; error bars represent the standard errors of the means. 2-Dmrt1a, the autosomal ancestor of Dmrt1bY, regulates the transcriptional activity of the dmrt1bY promoter
We next addressed the question of a possible cross-regulation of the Dmrt1a protein towards the dmrt1bY promoter. The above-described experiments employing Dmrt1bY were repeated, this time using the autosomal Dmrt1a. When the proximal 2868 bp dmrt1bY promoter region containing the putative Dmrt1 binding site fused to luciferase was co-transfected with a plasmid expressing Dmrt1a, dmrt1bY promoter activity was considerably reduced –up to 92% - (Figure 3A and 3B). This reduction is higher than observed for dmrt1bY (ranging from −55% to 74%). Using the mutant dmrt1bY promoter revealed that removing the Dmrt1 target site was able to restore transcriptional activity (Figure 3C and 3D) in medaka MES1 and Sg3 cell lines.
Fig. 3. Transient-transfection analysis of dmrt1bY promoter activity when co-transfected with different ratios of a Dmrt1a expressing plasmid. 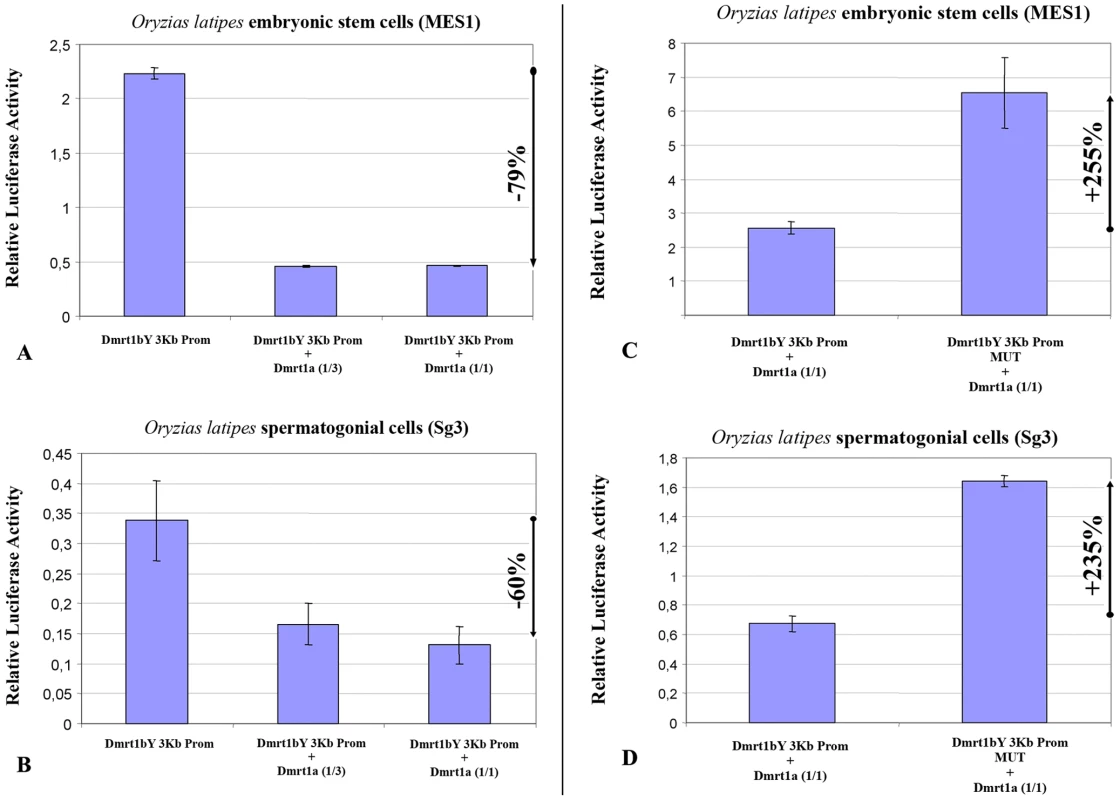
(A,B) The 3 Kb proximal dmrt1bY promoter construct was co-transfected with different amounts of dmrt1a-expressing plasmid (1∶3 and 1∶1 ratios). (C,D) The 3 Kb mutant proximal dmrt1bY promoter construct was co-transfected with different amounts of dmrt1a-expressing plasmid (1∶3 and 1∶1 ratios). The data are presented as the firefly/Gaussia luciferase activity. Transfections were done three times; error bars represent the standard errors of the means. 3-Dmrt1bY and Dmrt1a both bind to the putative Dmrt1 response element within dmrt1bY promoter
Electrophoretic mobility shift assays (EMSAs) were performed to show the direct interaction of Dmrt1a and Dmrt1bY proteins within the target site in the dmrt1bY promoter (Figure 4). DNA binding assays using the dmrt1bY Dmrt1-target sequence and in vitro translated Dmrt1a or Dmrt1bY demonstrated that both proteins are indeed able to bind to the Dmrt1 target sequence (position −2132 bp) (Figure 4). Binding specificity was confirmed using a mutated Dmrt1 binding site as competitor (Figure 4).
Fig. 4. Electrophoretic Mobility Shift Analysis (EMSA) of in vitro translated Dmrt1bY protein interaction with the Dmrt1 binding target derived from its own promoter. 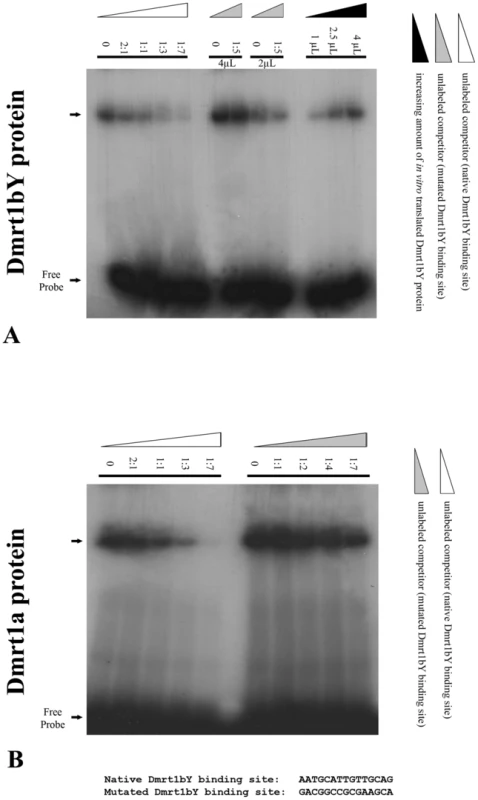
Gel mobility shift using increasing amounts of either in vitro translated Dmrt1bY or Dmrt1a proteins to shift a constant amount of radiolabelled dmrt1 probe. Binding reactions were resolved on a 5% polyacrylamide gel. (A) In vitro translated Dmrt1bY protein was incubated with radiolabelled Dmrt1bY target sequence as a probe, and non-radiolabelled probe was used as a competitor. (B) In vitro translated Dmrt1a protein was incubated with radiolabelled Dmrt1bY target sequence as a probe, and non-radiolabelled control (non-specific) probe was used as a competitor. Analysis of dmrt1bY expression in vivo
Thus far we could show feed back down-regulation of dmrt1bY and regulation by its paralog Dmrt1a in vitro. We next addressed whether this regulation indeed exists in vivo.
In vivo quantification of the 9Kbdmrt1bYpromoter activity in transgenic fish indicates a strong Dmrt1a cross-regulation
To get a first information of the in vivo dmrt1bY transcriptional regulation, a transgenic line where the dmrt1bY promoter drives dmrt1bY::GFP fusion protein expression was established with the goal of quantifying gonadal dmrt1bY promoter activity in either Dmrt1a expressing or non-expressing tissues (Figure 5). These transgenic fish showed a strong correlation of higher gonadal dmrt1bY promoter activity (5.5 times more in average) in absence of dmrt1a expression (ovary) compared to the dmrt1a expressing (testes) background (Figure 5).
Fig. 5. In vivo quantification of the 9KbDmrt1bYpromoter activity in transgenic fish. 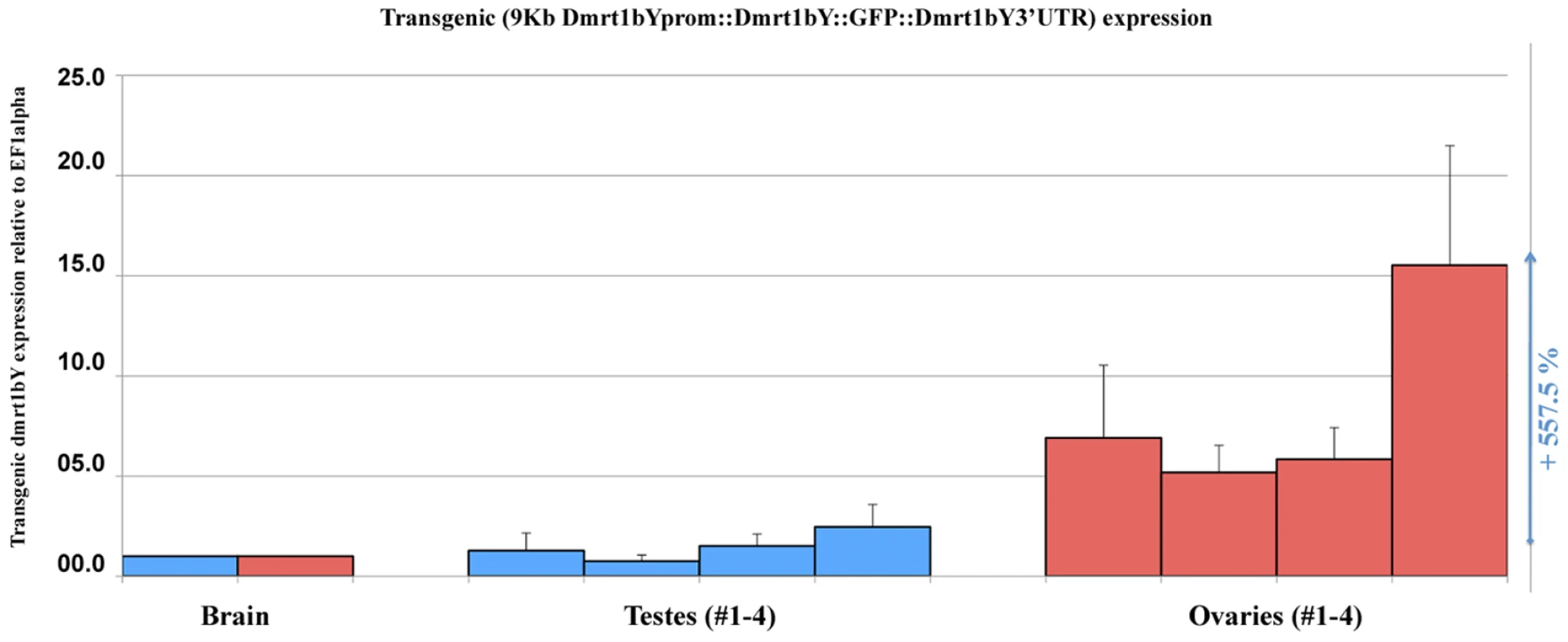
Real-time PCR quantification of the 9KbDmrt1bYpromoter activity in two different gonadal backgrounds either expressing dmrt1a (testes, #1–4) or not (ovaries, #1–4) reveals higher activity (+557.5% expression) in absence of dmrt1a expression. Dmrt1bY expression is relative to elongation factor 1 alpha (ef1 alpha) and normalised to brain background expression (set to 1). In vivo Dmrt1bY binding to the Izanagi Dmrt1 target site
To assess in vivo Dmrt1bY interaction with its own promoter, two additional stable transgenic lines expressing either the full Dmrt1bY protein or a truncated form lacking the DNA binding domain, both fused to GFP, were created. The two lines were used for in vivo Tissue Chromatin ImmunoPrecipitation (Tissue-ChIP) on testis tissue using GFP antibody for immunoprecipitation. An up to 7-fold enrichment compared to the control confirmed the capacity of Dmrt1bY to bind not only to the Dmrt1 promoter-nested Izanagi-target site but also to the Izanagi Dmrt1bY-target site in general (Figure 6).
Fig. 6. Tissue Chromatin immunoprecipitation (Tissue–ChIP) of Dmrt1bY binding to the Izanagi -nested Dmrt1 target site. 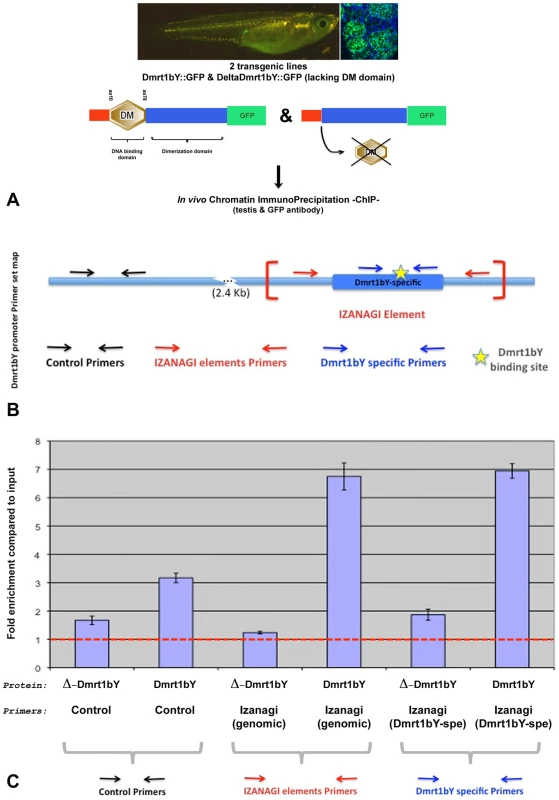
Chromatin immunoprecipitation using both Dmrt1bY::GFP and deltaDmrt1bY::GFP transgenic lines respectively expressing either Dmrt1bY or a control truncated Dmrt1bY (delta DM form lacking the DNA binding domain) fused to GFP revealed specific in vivo Dmrt1bY protein affinity to the Izanagi-nested dmrt1 target site, including the one described within dmrt1bY promoter. (A) Transgenic lines established for in vivo tissue–ChIP. (B) Dmrt1bY promoter primer sets map. (C) Specific enrichment of Izanagi-nested Dmrt1 binding sites subsequent to in vivo Dmrt1bY immunoprecipitation. Expression domains of the two dmrt1 paralogs indicating cross-regulation during male gonad development in vivo
Stable transgenic lines expressing fluorescent reporter protein (GFP and/or mCherry) were established to monitor the expression dynamics of Dmrt1a or Dmrt1bY in vivo. Analysis of early (10 to 30–35 dph) gonadal expression (Figure 7A) in two different (9KbDmrt1bYprom::GFP or 9KbDmrt1bYprom::mCherry) transgenic lines revealed identical fluorescent reporter protein expression in Sertoli cells (Figure 7C and 7D) as well as in interstitial tubule cells (Figure 7B). This exactly matches the protein expression pattern reported earlier from studies using a Dmrt1bY-specific antibody [42]. Later on, fluorescence declined in Sertoli cell (Figure 7E–7G to be compared to Figure 7H–7J). Noteworthy, corroborating the in vitro data, in double transgenic fish (9KbDmrt1bYprom::mCherry and BACdmrt1a::GFP) the decline of dmrt1bY promoter driven fluorescence was paralleled by a rise of dmrt1a promoter expression in Sertoli cells (Figure 7E–7G to be compared to Figure 7I and 7J). In fully mature testes (over 45–50 dph), dmrt1bY promoter expression remained only in few Sertoli cells scattered around the germ cells (Figure 7H) while the dmrt1a promoter was now predominantly expressed (Figure 7I and 7J).
Fig. 7. In vivo visualization of expression of the two medaka Dmrt1 paralogs. 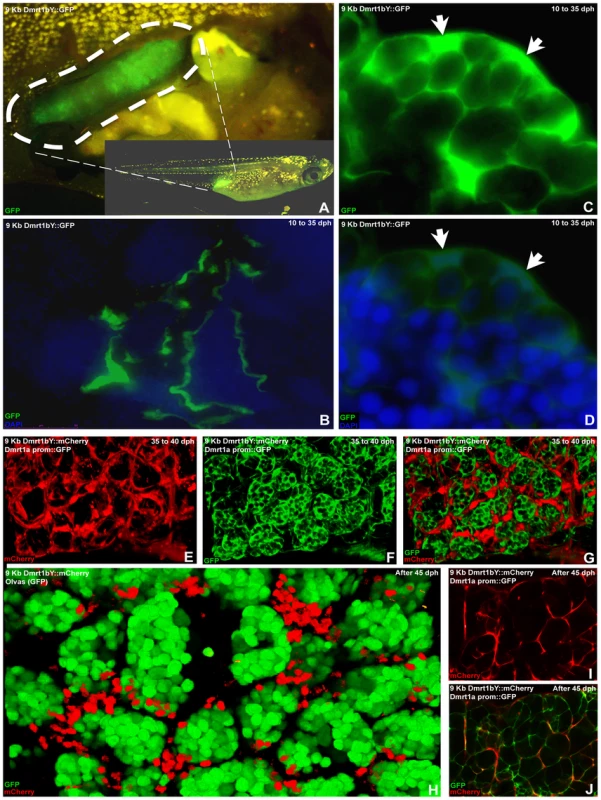
(A) Testis-specific GFP expression in 9KbDmrt1bYprom::GFP transgenic fish. (B) From testis formation (10 dph) up to adulthood, in 9KbDmrt1bYprom::GFP transgenic fish, robust testicular GFP expression is persistently noticeable in the epithelial cells of the intratesticular efferent duct. (C,D) Concomitantly, in agreement with the endogenous dmrt1bY expression, from 10 dph up to 30–35 dph strong specific GFP fluorescence is also detected in Sertoli cells. (E–G,I,J) By 35 dph, Sertoli cell-specific decline in fluorescence (mCherry) could be observed (E,I,H). In double transgenic fish (9KbDmrt1bYprom::mCherry and BACdmrt1a::GFP) this 9KbDmrt1bYprom-driven mCherry decline in expression is balanced by the rise of dmrt1a expression in Sertoli and germ cells (GFP in F,G,J). (H–J) In fully mature testes (after 45–50 dph), 9KbDmrt1bYprom-driven mCherry expression remains in scattered Sertoli cells around the germ cells (GFP in N) while Dmrt1a is now predominantly expressed in Sertoli cells (J). Discussion
Sex determination involves a complex hierarchy of genes. Expression screen analyses have resulted in hundreds of candidate genes that show sex-specific expression pattern. However it has been difficult to place these genes into a network of gene regulation and function. Nevertheless, several genes encoding for transcription factors, with specific temporal and spatial expression patterns during early gonad induction, have been suggested to participate in this process. Among them, from C. elegans to mammals, genetic evidence has suggested that the dmrt1 gene is an important regulator of male development at a downstream position of the regulatory network. In medaka, a duplicated copy of dmrt1 has acquired the upstream position of the sex-determining cascade. The analysis presented here provides evidences that this evolutionary novelty, which is predicted to require a rewiring of the regulatory network is brought about by co-option of “ready-to -use” pre-existing cis-regulatory elements carried by transposing elements. We could show that the master sex determining gene of medaka, dmrt1bY, is able to bind to one of these elements in its own promoter. This binding leads to a significant repression of its own transcription.
During early stages when the primordial gonad is formed, dmrt1bY is exclusively expressed and exerts its sex determining function [18]. The dmrt1a gene, with its proposed specification and maintenance function for the Sertoli cells, is expressed only when the testes are in the process of differentiation. Notably, the master sex determinator gene dmrt1bY, continues to be expressed. In adult testes, where both paralogs have been shown to be expressed, the predominant expression of dmrt1a compared to dmrt1bY (50 fold higher; [43]) argues for a downregulation of dmrt1bY. Although additional post-transcriptional mechanisms accounting for dmrt1bY expression regulation, involving the 3′ UTR [44], have been shown to be essential for spatial expression pattern in the embryo and restricted expression to the gonad in adult fish, the data presented here indicate that a feed back auto-regulation of dmrt1bY promoter activity and trans-regulation by its paralog Dmrt1a is a key mechanism of dmrt1bY transcriptional tuning (Figure 8).
Fig. 8. Model for feedback and cross-regulation of the medaka dmrt1 paralogs. 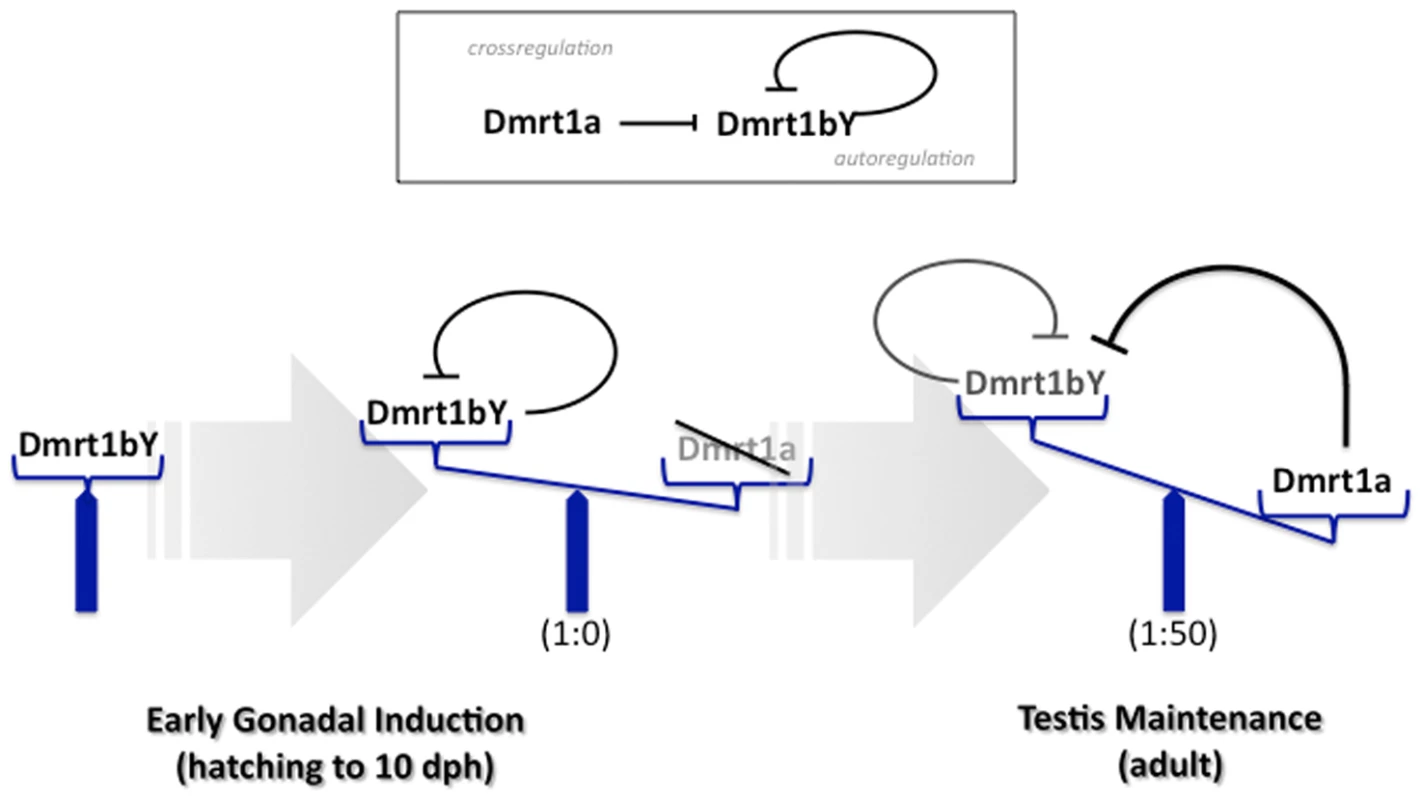
During sex determination stages only dmrt1bY is expressed and dmrt1a is off. Hence, the sex determining function of dmrt1bY is exerted. In adult testes, both paralogs are expressed notably in Sertoli cells, but the autorepression of the dmrt1bY promoter by its own gene product and the cross-regulation by Dmrt1a lead to a predominant expression of dmrt1a compared to dmrt1bY (appox. 50 fold). With respect to the evolutionary history of the two dmrt1 genes in medaka, it is of note that the newly generated paralog dmrt1bY, independently of any functional considerations, is kept back under tight transcriptional regulation of the ancestral dmrt1a gene. Consequently this avoids any kind of expression pattern redundancy in testes after their development is initiated and could then be a reasonable way of preserving both genes from any purification/degeneration processes after duplication, thus favouring a subsequent sub-neo-functionalization.
So far no putative Dmrt1 binding site could be observed within the more than 10 Kb upstream medaka dmrt1a sequence inspected. Similarly such Dmrt1 target sites are absent from the zebrafish, fugu, stickleback, mouse or human 10 kB upstream dmrt1 promoter regions. This together with the apparent loss of Dmrt1 canonical cis-regulatory sequences (such as Gata4) indicates a particular transcriptional context acquired by dmrt1bY during its evolution towards becoming a novel master sex determination gene.
It was previously reported that multiple TEs inserted into the Y-specific region on medaka LG1 [30]. Interestingly, our study revealed that the cis-regulatory element containing the Dmrt1 binding site, pre-existing within the Izanagi element at the time of its insertion, was co-opted in order to confer dmrt1bY its specific expression pattern after gene duplication around 10 million years ago [19],[41]. This fact has interesting evolutionary implications, since TEs are probably the most dynamic part of the genome. Dmrt1 possibly also regulates other genes in the proximity of Izanagi elements via the Dmrt1 binding site (Table S1).
In the context of gene duplication and its correlated process of sub-/neo - functionalization (see [45]–[47] for review), the contribution of TEs to the remodelling of the sex determination cascade (see [12],[48] for review) is of prime interest. The case reported here for the medaka-specific Izanagi element bringing in a novel regulatory element into the dmrt1bY promoter is –at least to our knowledge - the first example showing that TEs not only change/rewire the expression of existing genes but surely lead to the creation of new regulatory hierarchies within recently duplicated genes. The present case is even more interesting since this new TE-derived TFBS confers transcriptional control from the ancestral gene against the duplicate and allows the dmrt1bY gene to take an upstream position in the sex determination cascade without excluding its dmrt1 ancestor from a role in sexual development.
This supports a role of TEs for transcriptional network rewiring in sub - and/or neo - functionalization of duplicated genes in creating new hierarchies of sex determining genes.
Materials and Methods
Bioinformatic analyses
Comparative analysis of vertebrate Dmrt1 genomic regions were performed with mVISTA at http://genome.lbl.gov/vista [49] using the Shuffle-LAGAN alignment program [50]. Medaka dmrt1a (LG9) and dmrt1bY (LG1) region sequences were obtained from [30], all other regions from the Ensembl Genome Browser (http://www.ensembl.org/; release 49, March 2008): stickleback groupXIII, scaf57; Fugu scaf4; Tetraodon chr12, scaf14966; zebrafish chr5, scaf463; chicken chrZ, supercontig194; human chr9, supercontig NT_008413.
Screens for repetitive elements were performed with RepeatMasker (http://www.repeatmasker.org/). Additional copies of repeat elements and their genomic environment in the medaka genome (version HdrR, Oct 2005) were identified with BLASTN with >85% sequence identity over >85% of query length (Table 1). Alignments of repeat elements were obtained with CLUSTALW as implemented in BioEdit [51] followed by manual improvement. 50% threshold frequency was used for inclusion in repeat consensus sequences. The putative THAP domain found in the Izanagi consensus was identified by comparison to the PFAM database (http://pfam.sanger.ac.uk/).
Transcription factor binding sites were determined using MatInspector of the Genomatix portal (http://www.genomatix.de/). Binding sites for Dmrt1 in different genomes were identified using the matrix provided by [40] together with the Regulatory Sequence Analysis Tools portal; RSat (http://rsat.ulb.ac.be/rsat/). MEGA4 [52] was used to estimate sequence divergence between repeat 1 and the Izanagi element consensus (0.034 + − 0.005) as well as between repeat 2 and the Rex1 element consensus (0.010 + − 0.003) using the Kimura-2-parameter model. Linearized neighbor-joining trees of dmrt1 and tyrosinase a gene were obtained as described in ref. [41], with the only exception that they were based on third codon positions only. Other models of sequence evolution gave similar results. Accession numbers are given in Figure S6.
Cloning of the 9.107 Kb 5′ flanking sequence of medaka Dmrt1bY and plasmid constructs
For promoter analysis, a 9107 bp fragment upstream of the Dmrt1bY open reading frame (ORF) was isolated by restriction enzyme digestion (XhoI/EcoRI) from BAC clone Mn0113N21 [30], was cloned into pBSII-ISceI plasmid (pBSII-ISceI::9 Kb Dmrt1bY prom. plasmid). Subsequently, Gaussia luciferase gene from pGLuc-basic (New England Biolabs) plasmid was inserted between EcoRI and NotI sites of pBSII-ISceI::9 Kb Dmrt1bY prom (pBSII-ISceI::9 Kb Dmrt1bY prom::GLuc plasmid). pBSII-ISceI::3 Kb Dmrt1bY prom::GLuc and pBSII-ISceI::6 Kb Dmrt1bY prom::GLuc plasmids were constructed the same way removing 5′ fragments of the 9107 bp Dmrt1bY promoter region using Eco47III and HindIII restriction enzyme digestion respectively and re-ligation.
Mutation of the Dmrt1bY binding site was performed by PCR in the context of pBSII-ISceI::3 Kb Dmrt1bY prom::GLuc plasmid (native form: AATGCATTGTTGCAG; mutated form: GCCGGCTTCCCACCA). All PCR-obtained fragments were sequenced.
To generate plasmids for in vitro transcription, full-length cDNAs encoding medaka dmrt1a or dmrt1bY were subcloned into EcoRI/NotI digested pRN3 plasmid [53].
For establishment of transgenic lines, either GFP or mCherry open reading frames were inserted between EcoRI and NotI sites of pBSII-ISceI::9 Kb Dmrt1bY prom (pBSII-ISceI::9 Kb Dmrt1bY prom::GFP or mCherry plasmids respectively). GFP fusion protein vector (dmrt1bY::GFP and deltadmrt1bY::GFP) were constructed as described in [43].
Cell lines, cell transfection, and Luciferase assay
Mouse TM4 Sertoli cells, Xiphophorus embryonic epithelial A2 cells, and medaka spermatogonial (Sg3) and fibroblast like (HN2) cells were cultured as described [54],[55],[56],[57]. Cells were grown to 80% confluency in 6-well plates and transfected with 5 µg expression vector using FuGene (Roche) or Lipofectamine (Invitrogen) reagents as described by the manufacturers.
Gaussia luciferase activity was quantified using the Luciferase Reporter Assay System from Promega and normalized against co-tranfected firefly luciferase expressing plasmid (ptkLUC+; [58]). When DNA amounts transfected are expressed as a ratio, the total amount of expression vector remained constant (5 µg) by filling in the reaction with empty vector. Experiments for which error bars are shown result from at least three replicates and error bars represent the standard error of the mean.
Electrophoretic Mobility Shift Assays (EMSA)
(Dmrt1bY-Trgt) 5′-AGCTTAATGCATTGTTGCAGAGCT-3′, (Competitor) 5′-AGCTGACGGCCGCGAAGCAAGCT and respective complements were annealed by heating to 90°C for five minutes in 1X T4 PolyNucleotide Kinase (PNK) buffer (70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM dithiothreitol); slow-cooled to 50°C; held at that temperature for 5 minutes and then cooled to room temperature. For radioactive labelling 50 pmol of the duplex 5′ termini were used together with 50 pmol of gamma-[32P]-ATP and 20 units of T4 PNK in 1X adjusted T4 PNK buffer and incubated for 20 minutes at 37°C. Unincorporated nucleotides were removed through a Sephadex G-50 spin column.
For producing Dmrt1a and Dmrt1bY proteins, pRN3::Dmrt1a or pRN3::Dmrt1bY plasmids were linearized using KpnI and then in vitro transcribed using mMessage mMachine kit (Ambion). Finally, Dmrt1a or Dmrt1bY proteins were in vitro translated using Ambion's Retic Lysate Kit from the previously in vitro transcribed capped Dmrt1a or Dmrt1bY RNAs.
DNA binding reaction contained 10 mMTris-HCl (pH 7.9), 100 mM KCl, 10% glycerol, 5 mM MgCl2, 1 µg torula rRNA, 0.075% Triton X-100, 1 mM DTT, 1 µg BSA, 0.5 ng radiolabeled duplex probe and 2 or 4 µL in vitro translation mix in a total volume of 20 µL. 1/10 volume heparin (50 mg/mL) was added just before loading the binding reaction. For control reticulocyte lysate alone together with radiolabeled duplex probe was used and did not result in any shift (data not shown). Binding reactions were performed on ice for ten minutes and complexes were resolved on a 5% native acrylamide (37.5∶1) gel in 0.5 X TBE and then directly subjected to autoradiography.
Expression analyses
Total RNA was extracted from 9KbDmrt1bYprom::Dmrt1bY::GFP::Dmrt1bY3′UTR transgenic fish (Carbio genetic background) using the TRIZOL reagent (Invitrogen) according to the supplier's recommendation. After DNase treatment, reverse transcription was done with 2 micrograms total RNA using RevertAid First Strand Synthesis kit (Fermentas) and random primers. Real-time quantitative PCR was carried out with SYBR Green reagents and amplifications were detected with an i-Cycler (Biorad). All results are averages of at least two independent RT reactions and 2–5 PCR reactions from each RT reaction using each time three set of primer combination (DMTYk: 5′-CCTTCTTCCCCAGCAGCCT-3′/eGFP3 : 5′-AGTCGTGCTGCTTCATGTGGTC-3′; DMTYa2 : 5′-CGACTCCATGAGCAGTGAAA-3′/eGFP3 : 5′-AGTCGTGCTGCTTCATGTGGTC-3′; DMTYa2 : 5′-CGACTCCATGAGCAGTGAAA-3′/eGFP5 : 5′-GAACTTCAGGGTCAGCTTGC-3′. Error bars represent the standard deviation of the mean. Relative expression levels (according to the equation 2–DeltaCT) were calculated after correction of expression of elongation factor 1 alpha (elf1alpha) and brain expression was set to 1 as a reference.
In vivo Chromatin Immunoprecipitation
For in vivo chromatin immunoprecipitation, the EpiQuik Tissue Chromatin Immunoprecipitation kit (Epigentek) was utilized according to the manufacturers instructions, using testis tissue samples either from dmrt1bY::GFP or deltadmrt1bY::GFP transgenic fish (20 testes for each) and GFP antibody (Upstate) for immunoprecipitation. After immunoprecipitation [(Izanagi element Dmrt1bYspeF003) 5′-TCCGGTCTCTCCGGCGTGTGG-3′/(Izanagi element Dmrt1bYspeR00) 5′-TTGTAAGAGGACCTGCAACAATG-3′; (Izanagi element F01) 5′-CTATCTTGGTGAGGTCGACGATGCC-3′/(Izanagi element R01) 5′-AATTTAAATTACATGTCAAAGAGGTC-3′; (Dmrt1bYCtrF04) 5′-GTTCTGACTTTCAGCGTCTCACCTG-3′/(Dmrt1bYCtrR04) 5′-GGTTCTGGTCCAAATCTGTCAGAAG-3′] primer sets were used for enrichment quantification by real-time PCR.
Transgenic fish lines
For the generation of stable transgenic lines the meganuclease protocol [59] was used. Briefly, approximately 15–20 pg of total vector DNA in a volume of 500 pl injection solution containing I-SceI meganuclease was injected into the cytoplasm of one cell stage medaka embryos (Carbio strain). Adult F0 fish were mated to each other and the offspring was tested for the presence of the transgene by PCR from pooled hatchlings. Siblings from positive F1 fish were raised to adulthood and tested by PCR from dorsal fin clips as described [60].
Identically to the transgenic line expressing the Dmrt1bY protein fused to GFP [61] a second line lacking the Dmrt1bY DNA binding domain (DM-domain between aminoacids 10 and 78) was established. These two lines were used for in vivo Chromatin Immunoprecipitation. Similarly, for in vivo Dmrt1bY promoter activity quantification another transgenic line expressing a 9Kbdmrt1bYprom driven dmrt1bY::GFP fusion protein was created. Dmrt1a prom::GFP transgenic medaka was generated following the BAC transgenic method [62]. The BAC clone including dmrt1a genomic region, ola1-171C06 (NCBI accession numbers; DE071574 and DE071575) was obtained from NBRP. The followings were the primers to amplify EGFP fragments for homologous recombination into the BAC clone; Forward: 5′ -tctgacatgagcaaggagaagcagggcaggccggttccggagggcccggcTCAACCGGTCGCCACCATGG-3′ Reverse:5′-ttcagcggagacacgaagccgtggttccggcagcgggagcacttgggcatcGTCGACCAGTTGGTGATTTTG-3′.
Supporting Information
Zdroje
1. Davidson
2006 The regulatory genome. Gene regulatory networks in development and evolution Burlington Elsevier 289
2. BrittenRJ
DavidsonEH
1969 Gene regulation for higher cells: a theory. Science 165 349 357
3. BrittenRJ
DavidsonEH
1971 Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol 46 111 138
4. BejeranoG
LoweCB
AhituvN
KingB
SiepelA
2006 A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441 87 90
5. FeschotteC
2008 Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9 397 405
6. LoweCE
CooperJD
BruskoT
WalkerNM
SmythDJ
2007 Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet 39 1074 1082
7. FeschotteC
PrithamEJ
2007 DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41 331 368
8. Marino-RamirezL
LewisKC
LandsmanD
JordanIK
2005 Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet Genome Res 110 333 341
9. van de LagemaatLN
LandryJR
MagerDL
MedstrandP
2003 Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19 530 536
10. Marino-RamirezL
JordanIK
2006 Transposable element derived DNaseI-hypersensitive sites in the human genome. Biol Direct 1 20
11. CharlesworthD
CharlesworthB
2005 Sex chromosomes: evolution of the weird and wonderful. Curr Biol 15 R129 131
12. HerpinA
SchartlM
2008 Regulatory putsches create new ways of determining sexual development. EMBO Rep
13. GrahamP
PennJK
SchedlP
2003 Masters change, slaves remain. Bioessays 25 1 4
14. RaymondCS
ShamuCE
ShenMM
SeifertKJ
HirschB
1998 Evidence for evolutionary conservation of sex-determining genes. Nature 391 691 695
15. RaymondCS
ParkerED
KettlewellJR
BrownLG
PageDC
1999 A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum Mol Genet 8 989 996
16. NandaI
ShanZ
SchartlM
BurtDW
KoehlerM
1999 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat Genet 21 258 259
17. ShettyS
KirbyP
ZarkowerD
GravesJA
2002 DMRT1 in a ratite bird: evidence for a role in sex determination and discovery of a putative regulatory element. Cytogenet Genome Res 99 245 251
18. MatsudaM
NagahamaY
ShinomiyaA
SatoT
MatsudaC
2002 DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 559 563
19. NandaI
KondoM
HornungU
AsakawaS
WinklerC
2002 A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci U S A 99 11778 11783
20. YoshimotoS
OkadaE
UmemotoH
TamuraK
UnoY
2008 A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A 105 2469 2474
21. MatsudaM
ShinomiyaA
KinoshitaM
SuzukiA
KobayashiT
2007 DMY gene induces male development in genetically female (XX) medaka fish. Proc Natl Acad Sci U S A 104 3865 3870
22. MatsudaM
2005 Sex determination in the teleost medaka, Oryzias latipes. Annu Rev Genet 39 293 307
23. SwainA
2002 Vertebrate sex determination: a new player in the field. Curr Biol 12 R602 603
24. RossAJ
CapelB
2005 Signaling at the crossroads of gonad development. Trends Endocrinol Metab 16 19 25
25. SekidoR
Lovell-BadgeR
2009 Sex determination and SRY: down to a wink and a nudge? Trends Genet 25 19 29
26. WilhelmD
PalmerS
KoopmanP
2007 Sex determination and gonadal development in mammals. Physiol Rev 87 1 28
27. SekidoR
Lovell-BadgeR
2008 Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453 930 934
28. KleinjanDA
BancewiczRM
GautierP
DahmR
SchonthalerHB
2008 Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet 4 e29 doi:10.1371/journal.pgen.0040029
29. WoolfeA
ElgarG
2007 Comparative genomics using Fugu reveals insights into regulatory subfunctionalization. Genome Biol 8 R53
30. KondoM
HornungU
NandaI
ImaiS
SasakiT
2006 Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res 16 815 826
31. ChanTM
LongabaughW
BolouriH
ChenHL
TsengWF
2009 Developmental gene regulatory networks in the zebrafish embryo. Biochim Biophys Acta 1789 279 298
32. BrunnerB
HornungU
ShanZ
NandaI
KondoM
2001 Genomic organization and expression of the doublesex-related gene cluster in vertebrates and detection of putative regulatory regions for DMRT1. Genomics 77 8 17
33. WickerT
SabotF
Hua-VanA
BennetzenJL
CapyP
2007 A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8 973 982
34. RoussigneM
KossidaS
LavigneAC
ClouaireT
EcochardV
2003 The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci 28 66 69
35. HagemannS
HammerSE
2006 The implications of DNA transposons in the evolution of P elements in zebrafish (Danio rerio). Genomics 88 572 579
36. HammerSE
StrehlS
HagemannS
2005 Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol Biol Evol 22 833 844
37. QuesnevilleH
NouaudD
AnxolabehereD
2005 Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol 22 741 746
38. GaoS
ZhangT
ZhouX
ZhaoY
LiQ
2005 Molecular cloning, expression of Sox5 and its down-regulation of Dmrt1 transcription in zebrafish. J Exp Zoolog B Mol Dev Evol 304 476 483
39. PilonN
DaneauI
ParadisV
HamelF
LussierJG
2003 Porcine SRY promoter is a target for steroidogenic factor 1. Biol Reprod 68 1098 1106
40. MurphyMW
ZarkowerD
BardwellVJ
2007 Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol 8 58
41. KondoM
NandaI
HornungU
SchmidM
SchartlM
2004 Evolutionary origin of the medaka Y chromosome. Curr Biol 14 1664 1669
42. KobayashiT
MatsudaM
Kajiura-KobayashiH
SuzukiA
SaitoN
2004 Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka, Oryzias latipes. Dev Dyn 231 518 526
43. HornungU
HerpinA
SchartlM
2007 Expression of the male determining gene dmrt1bY and its autosomal coorthologue dmrt1a in medaka. Sex Dev 1 197 206
44. HerpinA
NakamuraS
WagnerTU
TanakaM
SchartlM
2009 A highly conserved cis-regulatory motif directs differential gonadal synexpression of Dmrt1 transcripts during gonad development. Nucleic Acids Res 37 1510 1520
45. MeyerA
SchartlM
1999 Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 11 699 704
46. MeyerA
Van de PeerY
2005 From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27 937 945
47. PostlethwaitJ
AmoresA
CreskoW
SingerA
YanYL
2004 Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet 20 481 490
48. WrightS
FinneganD
2001 Genome evolution: sex and the transposable element. Curr Biol 11 R296 299
49. FrazerKA
PachterL
PoliakovA
RubinEM
DubchakI
2004 VISTA: computational tools for comparative genomics. Nucleic Acids Res 32 W273 279
50. BrudnoM
MaldeS
PoliakovA
DoCB
CouronneO
2003 Glocal alignment: finding rearrangements during alignment. Bioinformatics 19 Suppl 1 i54 62
51. HallTA
1999 BioEdit: a user -friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp 41 95 98
52. TamuraK
DudleyJ
NeiM
KumarS
2007 MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596 1599
53. LemaireP
GarrettN
GurdonJB
1995 Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell 81 85 94
54. BeverdamA
WilhelmD
KoopmanP
2003 Molecular characterization of three gonad cell lines. Cytogenet Genome Res 101 242 249
55. HongY
LiuT
ZhaoH
XuH
WangW
2004 Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci U S A 101 8011 8016
56. KomuraJ
MitaniH
ShimaA
1988 Fish cell culture: Establishment of two fibroblast-like cell lines (OL-17 and OL-32) from fins of the medaka, Oryzias latipes. In Vitro Cellular & Developmental Biology 24 294 298
57. KuhnC
VielkindU
AndersF
1979 Cell cultures derived from embryos and melanoma of poeciliid fish. In Vitro 15 537 544
58. AltschmiedJ
DuschlJ
1997 Set of optimized luciferase reporter gene plasmids compatible with widely used CAT vectors. Biotechniques 23 436 438
59. ThermesV
GrabherC
RistoratoreF
BourratF
ChoulikaA
2002 I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev 118 91 98
60. AltschmiedJ
HornungU
SchluppI
GadauJ
KolbR
1997 Isolation of DNA suitable for PCR for field and laboratory work. Biotechniques 23 228 229
61. HerpinA
SchindlerD
KraissA
HornungU
WinklerC
2007 Inhibition of primordial germ cell proliferation by the medaka male determining gene Dmrt I bY. BMC Dev Biol 7 99
62. NakamuraS
SaitoD
TanakaM
2008 Generation of transgenic medaka using modified bacterial artificial chromosome. Dev Growth Differ 50 415 419
Štítky
Genetika Reprodukční medicína
Článek Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the GenomeČlánek Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in MiceČlánek Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' DiseaseČlánek Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD TagsČlánek Wing Patterns in the Mist
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Scale of Population Structure in
- Allelic Exchange of Pheromones and Their Receptors Reprograms Sexual Identity in
- Genetic and Functional Dissection of and in Age-Related Macular Degeneration
- A Single Nucleotide Polymorphism within the Acetyl-Coenzyme A Carboxylase Beta Gene Is Associated with Proteinuria in Patients with Type 2 Diabetes
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Cdk2 Is Required for p53-Independent G/M Checkpoint Control
- Uncoupling of Satellite DNA and Centromeric Function in the Genus
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in the Clade
- Use of DNA–Damaging Agents and RNA Pooling to Assess Expression Profiles Associated with and Mutation Status in Familial Breast Cancer Patients
- Cheating by Exploitation of Developmental Prestalk Patterning in
- Replication and Active Demethylation Represent Partially Overlapping Mechanisms for Erasure of H3K4me3 in Budding Yeast
- Cdk1 Targets Srs2 to Complete Synthesis-Dependent Strand Annealing and to Promote Recombinational Repair
- A Genome-Wide Association Study Identifies Susceptibility Variants for Type 2 Diabetes in Han Chinese
- Genome-Wide Identification of Susceptibility Alleles for Viral Infections through a Population Genetics Approach
- Transcriptional Rewiring of the Sex Determining Gene Duplicate by Transposable Elements
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in
- Proteasome Nuclear Activity Affects Chromosome Stability by Controlling the Turnover of Mms22, a Protein Important for DNA Repair
- Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in Mice
- Structure, Function, and Evolution of the spp. Genome
- Human and Non-Human Primate Genomes Share Hotspots of Positive Selection
- A Kinase-Independent Role for the Rad3-Rad26 Complex in Recruitment of Tel1 to Telomeres in Fission Yeast
- Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' Disease
- Molecular Evolution and Functional Characterization of Insulin-Like Peptides
- Molecular Poltergeists: Mitochondrial DNA Copies () in Sequenced Nuclear Genomes
- Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags
- Wing Patterns in the Mist
- DNA Binding of Centromere Protein C (CENPC) Is Stabilized by Single-Stranded RNA
- Genome-Wide Association Study Reveals Multiple Loci Associated with Primary Tooth Development during Infancy
- Mutations in , Encoding an Equilibrative Nucleoside Transporter ENT3, Cause a Familial Histiocytosis Syndrome (Faisalabad Histiocytosis) and Familial Rosai-Dorfman Disease
- Genome-Wide Identification of Binding Sites Defines Distinct Functions for PHA-4/FOXA in Development and Environmental Response
- Ku Regulates the Non-Homologous End Joining Pathway Choice of DNA Double-Strand Break Repair in Human Somatic Cells
- Nucleoporins and Transcription: New Connections, New Questions
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Nucleoporins and Transcription: New Connections, New Questions
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



