-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Molecular Evolution and Functional Characterization of Insulin-Like Peptides
Multicellular animals match costly activities, such as growth and reproduction, to the environment through nutrient-sensing pathways. The insulin/IGF signaling (IIS) pathway plays key roles in growth, metabolism, stress resistance, reproduction, and longevity in diverse organisms including mammals. Invertebrate genomes often contain multiple genes encoding insulin-like ligands, including seven Drosophila insulin-like peptides (DILPs). We investigated the evolution, diversification, redundancy, and functions of the DILPs, combining evolutionary analysis, based on the completed genome sequences of 12 Drosophila species, and functional analysis, based on newly-generated knock-out mutations for all 7 dilp genes in D. melanogaster. Diversification of the 7 DILPs preceded diversification of Drosophila species, with stable gene diversification and family membership, suggesting stabilising selection for gene function. Gene knock-outs demonstrated both synergy and compensation of expression between different DILPs, notably with DILP3 required for normal expression of DILPs 2 and 5 in brain neurosecretory cells and expression of DILP6 in the fat body compensating for loss of brain DILPs. Loss of DILP2 increased lifespan and loss of DILP6 reduced growth, while loss of DILP7 did not affect fertility, contrary to its proposed role as a Drosophila relaxin. Importantly, loss of DILPs produced in the brain greatly extended lifespan but only in the presence of the endosymbiontic bacterium Wolbachia, demonstrating a specific interaction between IIS and Wolbachia in lifespan regulation. Furthermore, loss of brain DILPs blocked the responses of lifespan and fecundity to dietary restriction (DR) and the DR response of these mutants suggests that IIS extends lifespan through mechanisms that both overlap with those of DR and through additional mechanisms that are independent of those at work in DR. Evolutionary conservation has thus been accompanied by synergy, redundancy, and functional differentiation between DILPs, and these features may themselves be of evolutionary advantage.
Published in the journal: . PLoS Genet 6(2): e32767. doi:10.1371/journal.pgen.1000857
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000857Summary
Multicellular animals match costly activities, such as growth and reproduction, to the environment through nutrient-sensing pathways. The insulin/IGF signaling (IIS) pathway plays key roles in growth, metabolism, stress resistance, reproduction, and longevity in diverse organisms including mammals. Invertebrate genomes often contain multiple genes encoding insulin-like ligands, including seven Drosophila insulin-like peptides (DILPs). We investigated the evolution, diversification, redundancy, and functions of the DILPs, combining evolutionary analysis, based on the completed genome sequences of 12 Drosophila species, and functional analysis, based on newly-generated knock-out mutations for all 7 dilp genes in D. melanogaster. Diversification of the 7 DILPs preceded diversification of Drosophila species, with stable gene diversification and family membership, suggesting stabilising selection for gene function. Gene knock-outs demonstrated both synergy and compensation of expression between different DILPs, notably with DILP3 required for normal expression of DILPs 2 and 5 in brain neurosecretory cells and expression of DILP6 in the fat body compensating for loss of brain DILPs. Loss of DILP2 increased lifespan and loss of DILP6 reduced growth, while loss of DILP7 did not affect fertility, contrary to its proposed role as a Drosophila relaxin. Importantly, loss of DILPs produced in the brain greatly extended lifespan but only in the presence of the endosymbiontic bacterium Wolbachia, demonstrating a specific interaction between IIS and Wolbachia in lifespan regulation. Furthermore, loss of brain DILPs blocked the responses of lifespan and fecundity to dietary restriction (DR) and the DR response of these mutants suggests that IIS extends lifespan through mechanisms that both overlap with those of DR and through additional mechanisms that are independent of those at work in DR. Evolutionary conservation has thus been accompanied by synergy, redundancy, and functional differentiation between DILPs, and these features may themselves be of evolutionary advantage.
Introduction
The ability of organisms to respond appropriately to changes in their environment is key to survival and reproductive success. An essential environmental variable for all organisms is their food supply and energetically demanding processes, such as growth, metabolism and reproduction are matched to nutrition by nutrient-sensing pathways, such as the insulin/IGF signalling (IIS) and TOR pathways [1]. An important recent discovery has been that reduced activity of IIS and TOR can slow aging and increase stress resistance and lifespan in the yeast Saccharomyces cerevisiae, the nematode worm Caenorhabditis elegans, the fruit fly Drosophila melanogaster and mice [2]. The mechanisms by which these pathways exert their diverse effects are hence of interest, as are the ways in which these parallel biological roles are achieved in evolutionarily diverse organisms.
The IIS pathway includes both peptide ligands, which can act at a distance, and intracellular components. In mammals, the ligands include insulin, the insulin-like growth factors (IGF) and relaxins. IGFs are mainly involved in growth control during development, whereas insulin secretion from pancreatic β-cells controls carbohydrate and lipid metabolism. Relaxins are produced by the ovary and are involved in reproduction. Insulin-like peptides (ILPs) have also been identified across a broad range of invertebrates, including molluscs, the nematode Caenorhabditis elegans and several insect species [3]. Most invertebrate genomes contain multiple ILPs, including 40 in C. elegans [4] and 7 in Drosophila melanogaster (DILP1-7) [5]. In contrast, while mammals often have up to 4 isoforms of the cellular components of IIS, they are encoded by single genes in Drosophila, including one Drosophila Insulin receptor (DInR), one insulin receptor substrate (chico) and one downstream forkhead box O transcription factor (dFOXO) [1]. The relative simplicity of the cellular IIS pathway, together with the diversification of DILPs, implies that the diverse functions of IIS could be in part mediated by functional diversification of the ligands.
Supporting functional differentiation between ligands, each dilp gene shows a characteristic spatio-temporal expression pattern. For instance, DILP4 is expressed in the embryonic midgut and mesoderm [5], DILP6 predominantly in the larval and adult fat body, with expression strongly up-regulated in the transition from larva to pupa [6]. DILP7 is expressed in specific neurons that innervate the female reproductive tract [7],[8], and inactivation of them results in sterile flies with an “egg-jamming” phenotype, suggesting that DILP7 could be a Drosophila relaxin [7]. DILP1, 2, 3 and 5 are expressed in brain median neurosecretory cells (MNCs) of the larval brain [5],[9],[10], but only DILP2, 3 and 5 could be detected in MNCs of the adult fly [11]. DILP2 is also expressed during development in imaginal discs, and in salivary glands and DILP5 in follicle cells of the female ovary [5].
Targeted ablation of the MNCs during early larval development results in developmental delay, growth defects and elevated carbohydrate levels in the larval hemolymph [10], while later ablation, during the final larval stage, results in lower female fecundity, increased storage of lipids and carbohydrates, elevated resistance to starvation and oxidative stress and increased lifespan [11]. Notably, expression of ILPs in MNC is evolutionarily conserved among insects, suggesting important evolutionarily conserved functions for these cells and their ligands. Furthermore, similar developmental programs are involved in the specification of MNCs in Drosophila and pancreatic beta cells in mammals, suggesting that insulin-producing cells of invertebrates and vertebrates may be derived from a common ancestry [12]. It is not yet clear if the phenotypes of MNC-ablated flies result from loss of one or more of the DILPs, or whether MNCs have functions independent of DILPs. Nor is it known if MNC-expressed DILPs have specific functions or act redundantly.
All seven DILPs possess the ability to promote growth, with DILP2 the most potent, and these ligands therefore probably all act as DInR agonists [9]. Over-expression of DILP2 also suppressed germ line stem cell loss in ageing females, probably through action in the stem cell niche [13]. DILP2 may also modulate lifespan, because its transcript is lowered in various mutant, long-lived flies. Over-expression of dFOXO in the adult fat body [14], over-expression of a dominant negative form of p53 in MNCs [15], increased JNK activity in MNCs [16] and hypomorphic mutants of the Drosophila NPY like protein sNPF [17], have all been reported to both extend lifespan and reduce the level of DILP2 expression. However, direct reduction in the level of DILP2 by in vivo double-stranded RNA interference (RNAi) did not increase lifespan [18], leaving the role of DILP2 unclear.
Dietary restriction (DR), a reduction in food intake without malnutrition, extends lifespan in diverse animals. In both C. elegans and Drosophila extension of lifespan by DR has been suggested to be independent of IIS, because lifespan increases in response to DR in animals lacking the key IIS effector FOXO transcription factor [19]–[21]. However, the finding could indicate instead that, in the absence of a normal increase in FOXO activity during DR, other pathways can act redundantly to increase lifespan. Indeed, other evidence has suggested that reduced IIS and DR may extend lifespan through overlapping mechanisms [19],[22]. DILP3 and DILP5 transcript levels are reduced in starved larvae [9] and DILP5 in DR adult flies [20], potentially indicating a role of DILPs in the fly's response to DR.
Gene families generally expand by gene duplication, and the duplicate copies are retained either if there is a requirement for large amounts of the gene product or if the duplicate copies undergo some sequence divergence and functional differentiation [23]. Recent work has suggested that feedback between partially redundant duplicate genes could itself be an important source of information aiding signal transduction [24]. It is not known if, as well as the functional differentiation implied by the findings described above, there is functional redundancy between the Drosophila ILPs. Nor is it known if there is stability of sequence differentiation and of membership of this gene family over evolutionary time or whether there is gene turnover. In addition, assignment of specific functions to individual dilp genes requires experimental manipulation. Although studies using RNAi have been informative [17],[18], RNAi can be prone to off-target effects [25] and other forms of cellular toxicity, and often results only in hypomorphic phenotypes.
We have used the completed genome sequences of 12 Drosophila species [26] to examine the stability of the dilp gene family, and found that the 7 DILPs have remained present and clearly differentiated from each other in sequence over a period of 40–60 million years. Evolutionary conservation of different regions of the peptides suggests that 6 of the 7 DILPs are cleaved like mammalian insulins, while the seventh may remain uncleaved, like mammalian IGFs. We generated specific null mutants for all seven DILPs, by homologous recombination or P-element mediated excision, and also generated flies that lack two or more DILPs simultaneously. Using these mutants we made a systematic analysis of DILP function in development, metabolism, reproduction, stress resistance, lifespan and response to DR of Drosophila. We show that DILPs can act redundantly, which suggests that redundancy among ILPs may be of evolutionarily advantage. We found both synergy and compensation of expression between DILPs. In particular DILP6 in the fat body compensated for the loss of MNC DILPs, demonstrating that DILPs are part of a complex feedback system between the central nervous system and peripheral tissues such as the fat body, which controls development, metabolism and reproduction. We further show that DILP2 is an important determinant of lifespan, describe a novel role for the fat body derived DILP6 peptide in growth control and demonstrate that dilp7 null mutants have normal fecundity, contrary to the suggestion DILP7 could be a Drosophila relaxin. Finally, we describe a specific interaction between the endosymbiont Wolbachia and IIS in the regulation of lifespan and show that DILPs mediate the responses of lifespan and fecundity to DR in Drosophila.
Results
Phylogenetic analysis of ILPs in the genus Drosophila
Taking advantage of the 12 sequenced Drosophila genomes [26], we investigated the evolution of ILPs across the Drosophila genus. As in D. melanogaster, seven dilp genes were identified in all 12 Drosophila species, with the exception of D. simulans and D. grimshawi (Figure 1). Using the Jones-Thornton-Taylor (JTT) matrix to calculate average amino acid distances, we found that the distances between the seven DILPs within each species is much greater than the distances among the putative orthologues in the different Drosophila species (Figure 1) We did not identify a dilp6 gene in D. simulans, probably because of sequencing gaps in the corresponding genomic region. Thus the seven dilp genes were present before the divergence of the 12 Drosophila species more than 40 million years ago, and none has been turned over during that period. Interestingly, the D. grimshawi genome contains eight dilp genes, as a result of a duplication of dilp2 (Figure 1). D. grimshawi is a member of the endemic Hawaiian Drosophila species, which are characterized by their large body size. The finding that DILP2 is an important regulator of growth in D. melanogaster ([9] and see below) suggests that evolutionary changes in dilp2 gene expression may have contributed to the evolution of body size in the Hawaiian Drosophila species.
Fig. 1. Phylogeny of Drosophila insulin-like peptides. 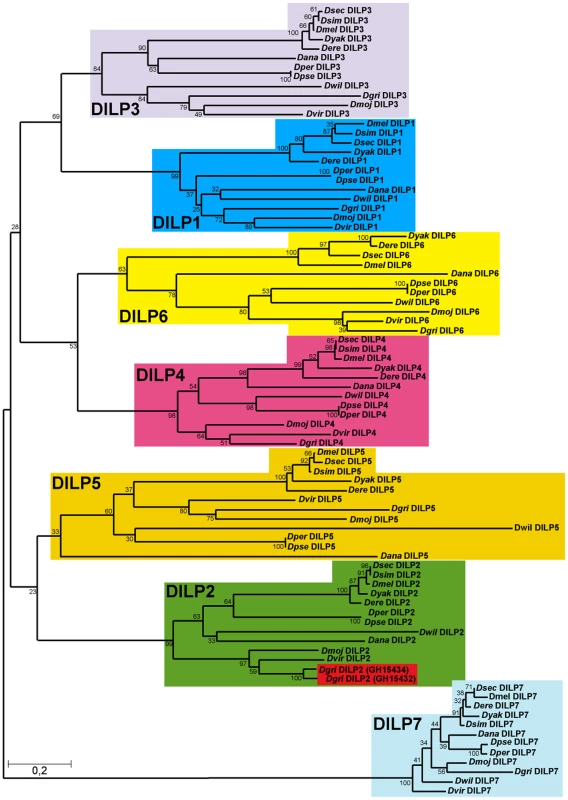
The evolutionary history of the seven DILP families from 12 fully sequenced Drosophila species inferred using the Neighbour-Joining method. Bootstrap replicate support percentages are shown next to the branches. The tree is drawn to scale, with branch lengths proportional to evolutionary distance computed using the JTT matrix-based method. 7 DILPs have remained present and clearly differentiated from each other during the more than 40 million years of evolution of Drosophilidae flies. D. grimshawi contains two DILP2 peptides (shaded in red). In mammals, IIS ligands are synthesized as pre-propeptides, consisting of a signal peptide and contiguous B-C-A peptides. In insulin and relaxin the C-peptide is clipped out by a convertase enzyme targeting basic amino acid cleavage sites, to produce a bioactive peptide consisting of A - and B-chain linked by 2–3 disulfide bridges. In contrast, IGFs contain a shortened C-peptide, which is not removed, resulting in a single chain peptide hormone. We therefore examined evolutionary conservation of different regions of the DILPs across the 12 Drosophila species to determine if the proteins are likely to be cleaved. Amino acid alignments of DILP pre-propeptides from the different Drosophila species shows that the bioactive peptides, i.e. the A and B chains are more conserved than the signal peptides and the C-peptides (Figure S1). The only exception is the shortened C-peptide in DILP6, which shows a similar degree of conservation to the A and B chains. This finding may indicate that although it contains a basic cleavage site the C-peptide is not removed and is part of a single chain bioactive DILP6 peptide, as has been recently suggested for the DILP6-like BIGFLP protein in the silkworm Bombyx mori [6]. Thus, DILP6 may resemble IGF rather than insulin. Despite the low amino acid conservation, functional signal peptides were found to be present in all DILP pre-propeptides using the Signal P prediction software [27], which indicates that all seven DILPs act as secreted peptide hormones in all 12 Drosophila species. Furthermore, cysteine residues involved in disulfide bridge formation as well as basic cleavage sites are highly conserved (Figure S1A), further supporting the view that DILPs, except for DILP6, resemble insulin and consist of heterodimeric peptides of A and B-chain linked by disulfide bridges.
Although the seven DILPs have been stably differentiated and retained in the 12 Drosophila genomes, they show different degrees of amino acid conservation (Figure S1B), suggesting different degrees either of functional constraint or of directional selection. DILP7 is by far the most highly conserved DILP peptide, with an overall amino acid identity of 76% between the pre-propeptides of D. melanogaster and the most distantly related Drosophila species D. grimshawi, increasing to 83% and 94%, respectively, when only the A and B chains are considered (Figure S1B). Indeed, only DILP7 has bona fide orthologues outside the Drosophila family, with 64–66% amino acid identity (A and B chain, 48% overall) with ILP5 from the mosquitoes Anopheles gambiae and Aedes aegypti and 63% and 55% (A and B chain, 48% overall) with ILP7 from the red flour beetle Tribolium castaneum [28],[29]. DILP4 is next most conserved, with only 52% overall amino acid identity and 75% and 70% identity in the A and B chain, respectively (D. melanogaster vs. D. grimshawi). DILPs 1, 2, 3, 5 and 6 evolved faster than DILP4 and DILP7 and at about the same rate as each other (ca. 40% overall amino acid identities, 50–55% identities within the A and B chain, D. melanogaster vs. D. grimshawi, Figure S1B). The higher amino acid sequence conservation of DILPs 7 and 4 suggests that these two DILPs might carry essential functions that are different from the other DILPs and can therefore not be compensated by the other DILPs.
Generation of Drosophila insulin-like peptide mutants
In order to analyse the in vivo function of individual dilp genes, we generated dilp-specific mutants by ends-out homologous recombination [30] for dilp1, 2, 3, 4, 5 and 7 (Figure 2A–2C) and by P-element mediated imprecise excision for dilp6 (Figure 2D). In addition, to address redundancy and synergy among individual dilp genes, we used homologous recombination to generate mutant flies with a combined knock out of two or several DILPs, including 2, 3 (dilp2–3), 2, 3, 5 (dilp2–3,5), 1, 2, 3, 4 (dilp1-4) and 1, 2, 3, 4, 5 (dilp1–4,5).
Fig. 2. Gene locus organization and generation of dilp mutants. 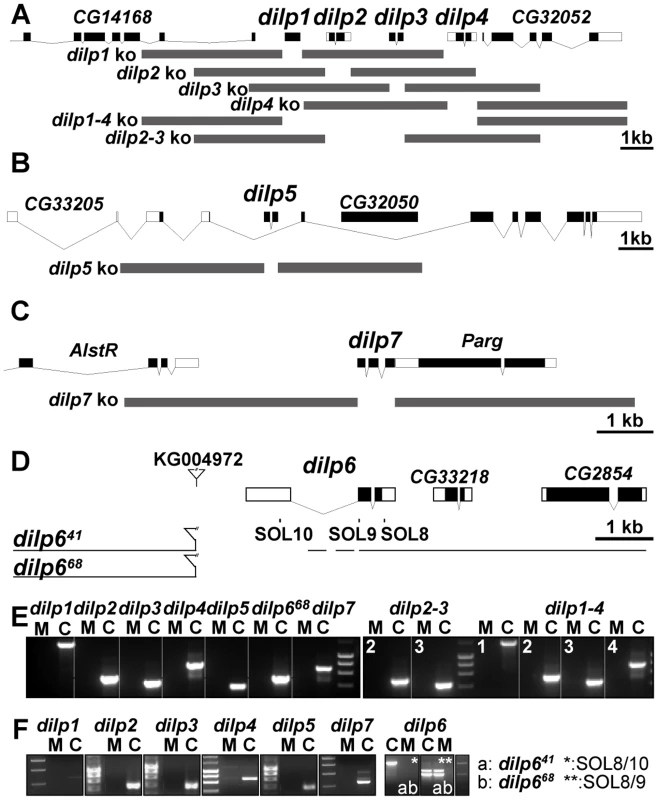
(A–C) Null mutations for dilp1, 2, 3, 4, 5, 7, 2–3, and 1–4 were generated by ends-out homologous recombination and by imprecise P-element excision for dilp6 (D). (A) The dilp1–4 genes cluster at cytological position 67C8 is separated from dilp5 by two intervening genes of unknown function (Flybase: CG32052 and CG33205). (B) dilp5 is located within an intron of the CG33205 gene. (C) dilp7 is located on the X-chromosome at 3E2 immediately downstream of the essential Poly(ADP-ribose) glycohydrolase (Parg) gene. Donor constructs (dilp ko) for ends-out homologous recombination are indicated by grey bars. Gap between grey bars indicates the genomic region replaced by a whitehs marker gene. Coding parts of exons are marked in black, non-coding parts by white boxes. (D) Transposon integration line KG004972 was used to generate dilp6 deletion mutants dilp641 and dilp668. Note: both dilp6 deletion alleles contain remaining P-element sequence, hatched line: region of breakpoint in dilp641. (E) PCR on genomic DNA of dilp mutants with dilp specific primer combinations shows homologous recombination mediated replacement of dilp genes by the whitehs marker gene and deletion of dilp6 in dilp668 mutants (M: mutant, C: control). (F) RT–PCR analysis confirms that dilp mutants are transcript null alleles. dilp641 mutants express ectopic dilp6 transcripts that lack the first exon but contain the full ORF. dilp668 mutants are dilp6 transcript null alleles. Ends out homologous recombination donor constructs were designed to delete the complete coding sequence of the targeted dilp gene, by replacing it with a white marker gene without affecting the genomic sequence of adjacent genes (Figure 2A–2C). Putative homologous recombination events were tested by PCR on genomic DNA with gene-specific primer combinations for the insertion of the white marker gene into the target gene location (Figure 2E). Several independent targeting events were recovered per dilp gene. Long-range PCR analysis on genomic DNA showed that most targeting events were precise homologous recombinations (Figure S2 and data not shown) and only these were used for subsequent experiments. Reverse Transcription (RT) PCR analysis showed that the dilp mutants are transcript-null alleles (Figure 2F). Lack of DILP expression in the mutants was confirmed by immunohistochemistry on adult fly brains (Figure S3) and by Western blot analysis (Figure 3D). The single mutants were specific for individual dilp genes. For instance, dilp2 mutants lacked expression only of DILP2, not of DILP3 or DILP5 (Figure S3D, S3E, S3F).
Fig. 3. Compensatory regulation of gene expression in dilp mutants. 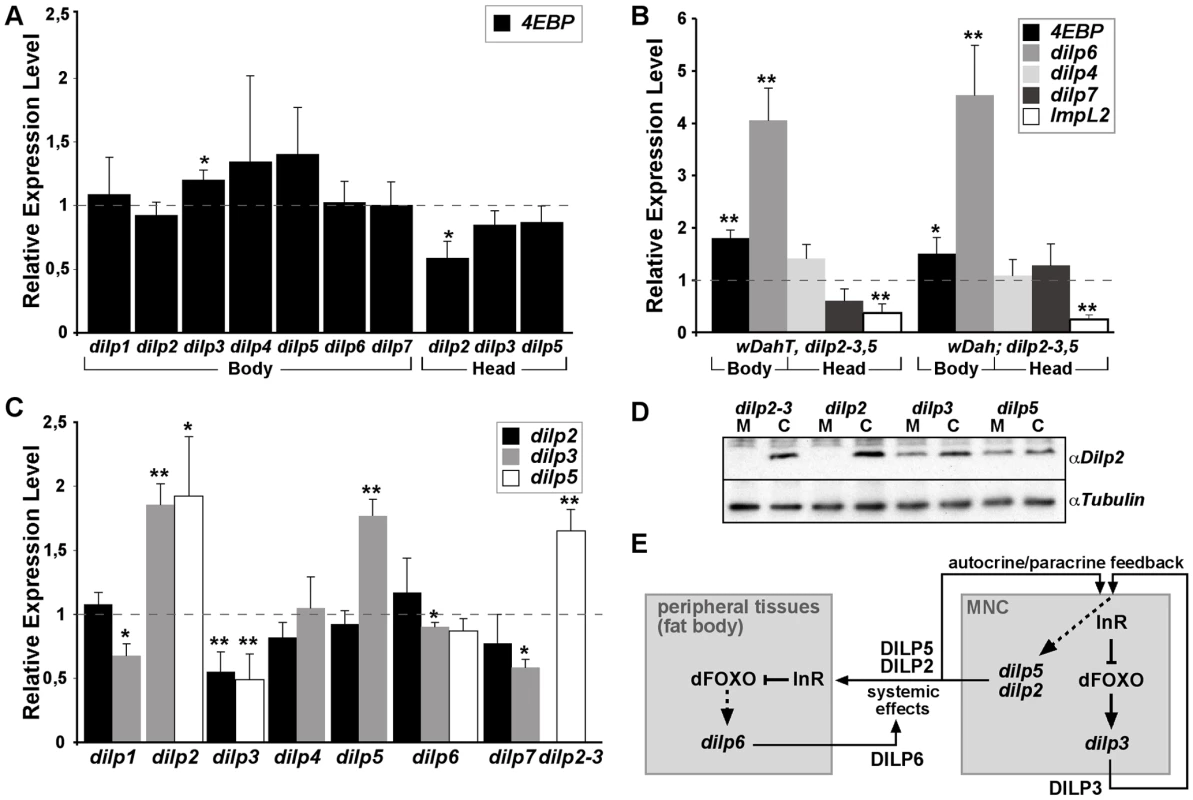
(A) Q-RT-PCR analysis of 4E-BP expression on fly bodies and fly heads of dilp single mutants. Expression of the IIS downstream target gene 4E-BP is not changed in dilp single mutants. (B) Q-RT-PCR analysis demonstrates up-regulation of 4E-BP and dilp6 and down-regulation of ImpL2 in dilp2–3,5 mutants independent of Wolbachia infection status (wDahT: Wolbachia-, wDah: Wolbachia+). (C) Compensatory regulation among MNC-expressed DILPs. Expression levels of DILP2, 3, and 5 were measured by QRT-PCR on RNA extracted from heads of dilp mutants. (D) Western blot analysis of DILP2 expression in total head protein confirms lack of DILP2 expression in dilp2 and dilp2–3 mutants and downregulation of DILP2 levels in dilp3 mutants. M: homozygous mutant, C: w1118 control. An anti-Tubulin antibody was used as loading control. (E) Diagram summarizing the feedback system involved in the control of DILP expression levels. DILP3 is part of a feedback system that acts in an autocrine/paracrine manner to regulate DILP expression in the MNC. DILP expression between MNC in the central nervous system and the peripheral fat body tissue is regulated by a negative feedback system that involves DILP6. See text for further details. Arrows denote activation, blunted lines denote repression. All experiments in (A–D) were done using 8–10 day old females reared on 1x SY-A food. Expression level of mutants were normalised to the corresponding wild type control, which by default was set to 1. * p<0.05, ** p<0.01. dilp6 mutant flies were generated by imprecise P-element excision of KG04972 integrated in the 5′upstream region of the dilp6 gene (Figure 2D). Two different dilp6 alleles were isolated, the small deletion dilp641 covering the dilp6 5′upstream region including the first exon and the large deletion dilp668 covering the complete dilp6 gene as well as at least 4 other genes. RT PCR analysis confirmed dilp668 to be a transcript null allele (Figure 2F). dilp6 expression was still detectable in dilp641 mutants. However, 5′ RACE analysis revealed that these transcripts lacked the first exon of the dilp6 gene and instead contained ectopic sequence including additional ORFs immediately upstream of the dilp6 ORF, which might interfere with the translation of the DILP6 peptide (for details see Text S1). Consistently, whenever tested both dilp6 mutant alleles were phenotypically indistinguishable from each other. In addition, dilp641 mutant flies showed the same developmental growth defects as flies in which dilp6 is knocked down by RNAi [31], suggesting dilp641 to be a strong hypomorph or even a functional null allele.
Compensatory regulation
Reduced expression of DILPs has been associated with changes in IIS pathway activation [15]. We therefore evaluated IIS activity in body tissues of adult dilp mutants by measuring transcript levels of the translational regulator 4E-BP (encoded by Thor), a direct target of dFOXO, which is induced when IIS is repressed and dFOXO is activated (Figure 3A and 3B). As expected, 4E-BP transcript levels were up-regulated in dilp2–3,5 mutants (Figure 3B), consistent with peripheral activation of dFOXO and reduced IIS. However, we did not observe significant up-regulation of 4E-BP transcript levels in dilp single mutants, except for dilp3 mutants, which showed slight up-regulation of 4E-BP levels in bodies but not in heads (Figure 3A). Thus, the knock out of most individual DILPs did not result in systemic down-regulation of IIS that was detectable by measuring 4E-BP transcript levels. A possible explanation for this finding could be that DILPs act redundantly as part of a negative feedback system in which knock-out of one DILP is compensated by the up-regulation of others.
To address the possibility of compensatory regulation we measured dilp transcript levels in dilp mutant flies (Figure 3B and 3C). We did not detect any up-regulation of dilp2 or dilp3 transcript levels in dilp1, 4, 6 or 7 mutants, however, dilp5 was up-regulated in dilp2 and dilp2–3 mutants and dilp3 was up-regulated in dilp2 and dilp5 mutants (Figure 3C), demonstrating compensatory transcriptional regulation among MNC-expressed DILPs. Intriguingly, dilp2 and dilp5 expression was down-regulated in dilp3 mutants (Figure 3C and 3D), suggesting synergy in expression, with dilp3 acting as a positive regulator of dilp2 and dilp5 expression. Remarkably, while expression of dilp4 and dilp7 was not significantly changed in dilp2–3,5 mutants, the fat-body-expressed dilp6 gene was strongly up-regulated (Figure 3B), suggesting the existence of a negative feedback system that acts to coordinate DILP expression between the MNCs in the central nervous system and peripheral tissues like the fat body (Figure 3E). Interestingly, expression of the DILP binding protein ImpL2, a negative regulator of IIS [32], was down-regulated in dilp2–3,5 mutants (Figure 3B), demonstrating that the negative feedback system is not restricted to the regulation of dilp transcription.
Systematic analysis of DILP function
The compensatory regulation among dilp genes may indicate that they act at least in part redundantly. However, each dilp gene is expressed in a tissue - and stage specific manner, suggesting that there is also diversification of their functions. In order to examine the in vivo function of individual dilp genes and to address whether and to what extent they act redundantly and synergistically to execute these functions, we initiated a systematic analysis of the dilp mutants, addressing phenotypes associated with MNC-ablation and reduced IIS, including egg-to-adult survival, development time, organismal growth, stress resistance, energy storage, lifespan and fecundity (summarized in Table 1).
Tab. 1. Systematic analysis of DILP function. 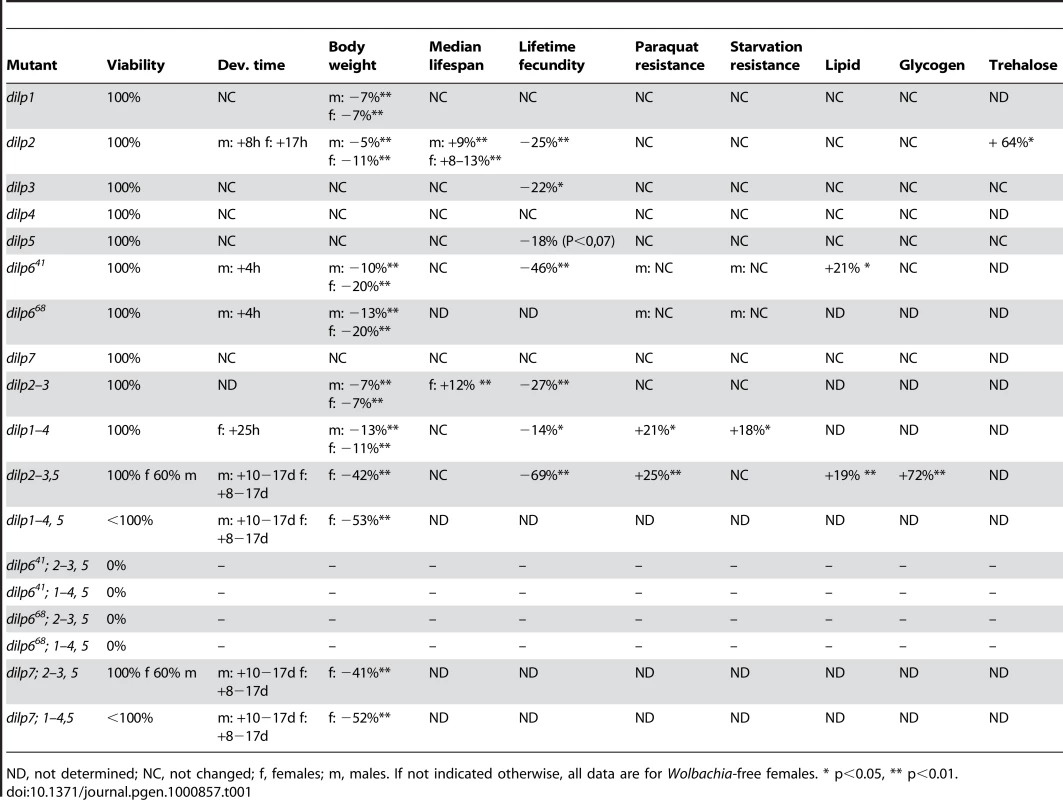
ND, not determined; NC, not changed; f, females; m, males. If not indicated otherwise, all data are for Wolbachia-free females. * p<0.05, ** p<0.01. Egg-to-adult survival
All seven dilp single mutants as well as dilp2–3 and dilp1–4 mutants were homozygous viable (Table 1). In contrast, dilp2–3,5 mutants showed sex-specific lethality; whereas homozygous mutant females showed normal viability, only 50–60% of dilp2–3,5 homozygous males developed into adult flies (Table 1). Survival was not further negatively affected in dilp7;2–3,5 mutant flies, but was reduced in dilp1–4,5 mutants. Intriguingly, animals that lacked all DILPs except for DILP6 (dilp7;1–4,5 mutants) still developed into adult flies. In contrast, combined knock-out of DILPs 2, 3, 5 and 6 caused complete lethality in males and females. This result indicates that DILP6 acts redundantly to MNC-expressed DILPs, consistent with the compensatory up-regulation of DILP6 transcript in dilp2–3,5 mutants (Figure 2B). Lethality in combination with dilp2–3,5 mutants was observed for both dilp6 alleles, further evidence that dilp641 is a dilp6 loss-of function allele.
Development time
dilp2 and dilp6 were the only single mutants that showed a delay in egg-to-adult development (Figure 4A, Table 1). Development time was only slightly further delayed in dilp1–4 mutants compared to dilp2 single mutants. In contrast, dilp2–3,5 mutants had a severe developmental delay (Figure 4A), comparable to flies with ablated MNCs or DInR mutants [10]. The developmental delay was caused by delays in larval or pupal development, because dilp2–3,5 homozygous mutant embryos developed into first instar larvae at the same rate as wild type controls (data not shown). dilp2–3,5 mutants also eclosed over a much longer period; all control flies eclosed within a day while dilp2–3,5 mutant flies continued to hatch over a period of almost ten days (Figure 4A). dilp1–4,5 mutants, dilp7;2–3,5 mutants and dilp7,1–4,5 mutants had similar development times to dilp2–3,5 mutants (Table 1), suggesting that DILPs1, 4 and 7 are not involved in the regulation of developmental timing.
Fig. 4. Development time and body weight of dilp mutants. 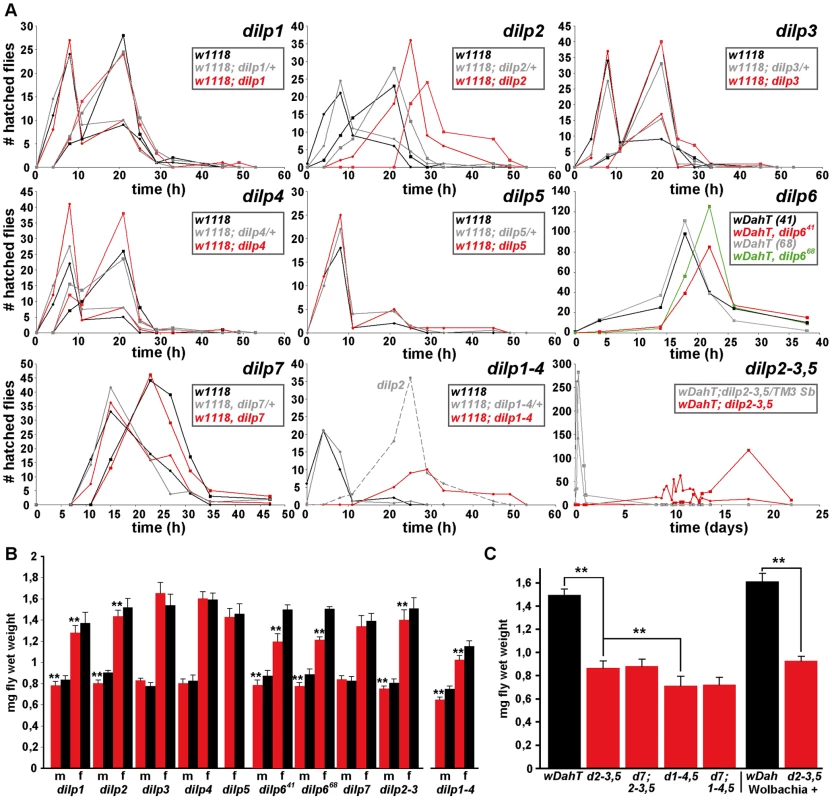
(A) Egg-to-adult development time of dilp mutants. Only the hatching period of the adult flies is shown. Rectangle: males (m), circle: females (f). (Note: number of heterozygous flies was halved to adjust to the number of homozygous and w control flies.) (B) Body weight of dilp mutant (red) males (m) and females (f) compared to controls (black). (n = 20 flies). (C) Body weight of female flies that lack multiple dilp genes. (n = 40 f; dilp1–4,5 n = 20 f; dilp7,1–4,5 n = 18 f). Wolbachia+ flies (wDah) weigh more than Wolbachia- flies (wDahT). Body weight in (B,C): wDahT background, except for dilp1–4 mutants: w1118. ** p<0.01, t-test. Organismal growth
Organismal growth was analysed by measuring the body weight of adult flies (Figure 4B and 4C, Table 1). dilp 3, 4, 5 and 7 single mutants showed normal body weight. dilp1 and dilp2 mutants showed a slight reduction in weight (Figure 4B), consistent with the shorter adult body length seen upon RNAi-mediated knockdown of DILP1 and DILP2 [17]. Although dilp1 mutants weighed less, they developed at the same rate as control flies, demonstrating that growth defects are not necessarily coupled with a delay in development. Intriguingly, dilp6 mutants showed the biggest reduction in body weight of all dilp single mutants (Figure 4B). DILP6 resembles IGFs and is expressed at high levels in the fat body but not in the MNCs [6], suggesting DILP6 to be an IGF-like peptide secreted by the fat body that promotes growth during larval-pupal development.
dilp2–3 mutants weighed as much as dilp2 mutants and only a minor additional decrease in body weight was seen in dilp1–4 mutants (Figure 4B), likely to be the result of the combined lack of DILP1 and DILP2. In contrast, body weight of dilp2–3,5 mutants was severely reduced (Figure 4C), and an even further reduction was observed in dilp1–4, 5 mutants, which were approximately 50% smaller than controls (Figure 4C). Lack of DILP7 did not result in a further decrease in body weight of either dilp2–3,5 or dilp1–4,5 mutants (Figure 4C), suggesting that DILP7 does not contribute to the regulation of organismal growth.
Stress resistance and energy storage
Oxidative stress and starvation resistance of dilp mutants was analysed by monitoring the survival of females on 20 mM paraquat and 1% agar, respectively. None of the dilp single mutants or the dilp2–3 mutants was more resistant to paraquat or starvation treatment (Table 1, Figure S4, Figure S5A). Additionally, dilp single mutants did not show increased glycogen or lipid storage, with the exception of dilp6 mutants, which had slightly increased lipid levels (Figure S5B, S5E). Whole body trehalose levels were increased in dilp2 mutants, but not changed in dilp3 or dilp5 mutants (Figure S5D), consistent with the previous suggestion that stored trehalose levels are specifically regulated by DILP2 [18].
dilp1–4 mutants were slightly more resistant to paraquat and starvation treatment than controls, and dilp2–3,5 mutants were highly resistant to oxidative stress, as demonstrated by their increased survival under both paraquat and hydrogen peroxide treatment (Figure S4). However, and in contrast to MNC ablated flies, they were not resistant to starvation (Figure S5), even though they stored more energy in the form of glycogen (Figure S5C) and lipids (Figure S5F). This finding suggests either that lack of DILPs other than DILP2, 3 or 5 is causal for the increased starvation resistance of MNC-ablated flies, or that MNCs mediate starvation resistance independent of DILP function. The latter is consistent with the proposed function of the dARC1 protein in MNCs, which has been suggested to control the behavioural response to starvation, the lack of which might induce starvation resistance [33].
Adult lifespan
MNC-ablation experiments have suggested a role for DILPs in the determination of lifespan [11],[34]. In particular DILP2 has been proposed by a number of studies to play an important role, because of its transcriptional down-regulation in mutant, long-lived flies [14]–[17]. However, this view has been challenged recently by the finding that RNAi-mediated knock-down of DILP2 is not sufficient to extend lifespan in flies [18].
We measured the lifespans of all seven dilp null mutants using female flies kept on standard food. We did not observe lifespan-extension in dilp1, 3, 4, 5, 6 or 7 mutants (Figure S6A). However, in contrast to dilp2 RNAi hypomorphs, dilp2 null mutants were significantly longer-lived than controls (Figure 5). An increase between 8% and 13% in median lifespan was observed in four independent trials, two genetic backgrounds and in dilp2 mutants originating from independent homologous recombination events tested individually or as transheterozygotes (Figure 5 and data not shown). Furthermore, a 9% extension of median lifespan was also observed for dilp2 mutant males, demonstrating that DILP2 is limiting for lifespan in both sexes. dilp2–3 mutants were also long-lived, although no more so than dilp2 mutants (Figure 5). Interestingly, while heterozygous dilp2–3,5 mutants were slightly long-lived, neither homozygous dilp2–3,5 mutants nor dilp1–4 mutants showed an increased median lifespan under standard food conditions (Figure 5, Figure S6A). However, maximum lifespan of homozygous dilp2–3,5 mutants was increased by 14%, as reported for the demographic aging of flies in which MNC were ablated early during development [35]. These findings might suggest that the strong reduction in insulin signalling in these mutants produced a general decrease in adult viability as well as a slowing down of the increase in death rates with age.
Fig. 5. dilp2 mutant flies are long-lived. 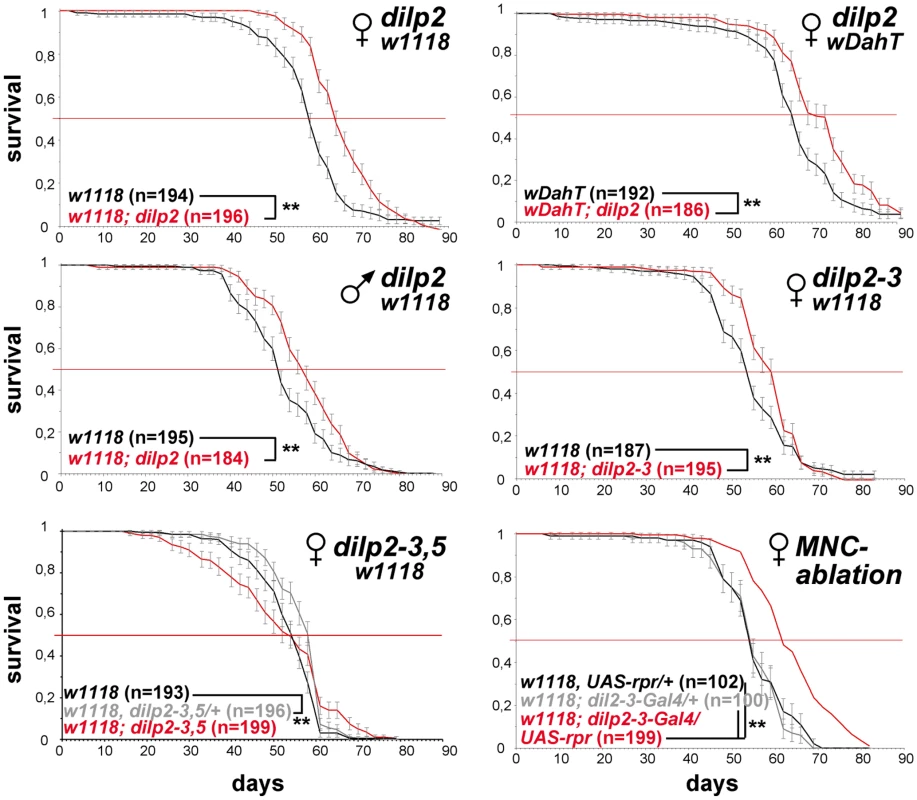
Survival curves of dilp mutant flies on standard food. Lack-of DILP2 extends median lifespan in two independent genetic backgrounds (w1118 and wDahT) and in both sexes. Combined knock out of DILP2 and 3 results in increased lifespan. In contrast to MNC-ablated flies, homozygous w1118; dilp2–3,5 mutants do not show increased median lifespan. However, median lifespan is slightly increased in dilp2–3,5 heterozygous flies. ** p<0.001, log-rank test. The intracellular symbiont Wolbachia pipientis, a maternally transmitted bacterium, has been shown to modulate longevity in wild type and mutant stocks of Drosophila [36],[37] and has recently been suggested to reduce the severity of IIS mutants by increasing IIS downstream of the DInR [38]. Therefore, we decided to test the influence of Wolbachia on lifespan and other fitness-related traits of dilp2–3,5 mutants (Figure 6A–6E). Intriguingly, Wolbachia-positive wDah;dilp2–3,5 mutants were extremely long-lived, showing an increase on standard food in both median and maximum lifespan of 29% and 22%, respectively, compared to wDah controls (Figure 6A). Lifespan extension was even more pronounced on a high yeast diet with an increase of up to 55% and 27% in medium and maximum lifespan, respectively (Figure 7G). In contrast, Wolbachia had no effect on the lifespan of wild type flies (Figure 6A), confirming that lifespan extension of dilp2–3,5 mutants is the result of a specific interaction between Wolbachia and the IIS pathway. However, Wolbachia did not affect IIS pathway activity as measured by 4E-BP expression. Although 4E-BP was up-regulated in dilp2–3,5 mutants (Figure 3B), there was no significant difference in 4E-BP expression between dilp2–3,5 mutants or wild type flies, respectively, with and without Wolbachia (Figure 6G). Some other consequence of insulin signaling must therefore mediate the effects of Wolbachia.
Fig. 6. Wolbachia-dependent lifespan extension of dilp2–3,5 mutants is correlated with xenobiotic resistance. 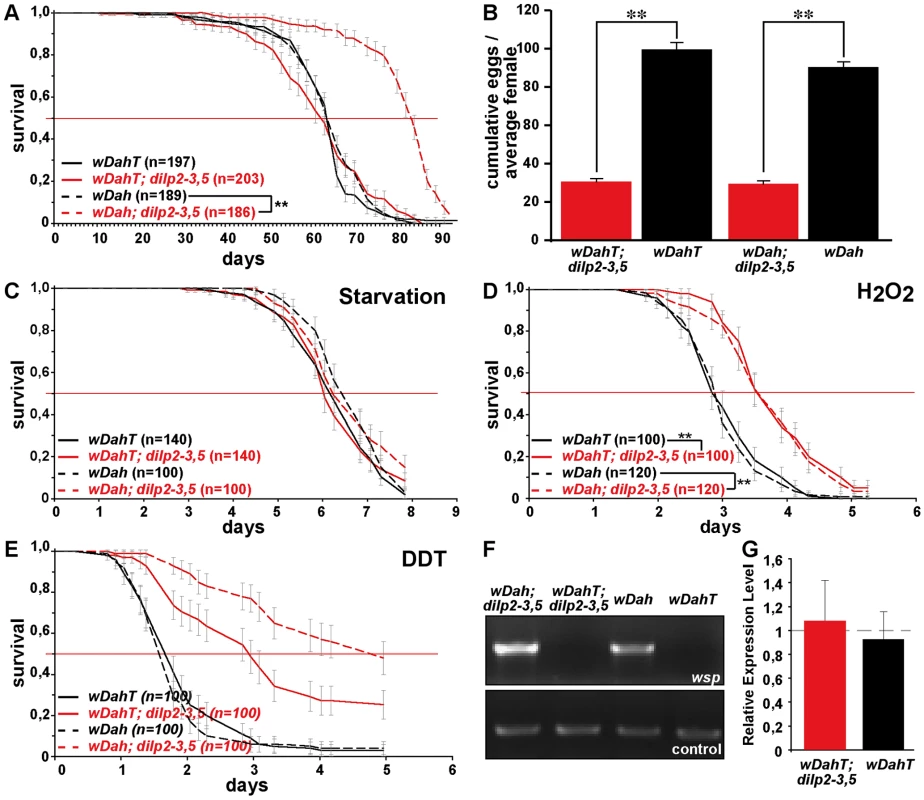
Wolbachia affects longevity (A) and xenobiotic resistance (E) of dilp2–3,5 mutants, but not fecundity (B), starvation (C) or oxidative stress resistance (D). (A) Median and maximum lifespan of wDah;dilp2–3,5 females (83/92,5 days) compared to wDah (64,5/76 days) control females is increased by 29% and 22%, respectively. ** p<0.001, log rank test. In contrast, wDahT;dilp2–3,5 (62,5/81 days) only show a small increase in maximum, but not in median lifespan compared to wDahT (64,5/74 days) females. (B) Index of lifetime fecundity of dilp2–3,5 females is reduced to 30% but not affected by Wolbachia infection. Shown is the cumulative number of eggs laid by an average female. ** p<0.01, Wilcoxon rank sum test. (C–E) Survival of dilp2–3,5 females with and without Wolbachia on (C) 1,5% agarose, (D) 5% hydrogen peroxide and (E) DDT (275 mg/l). ** p<0.01, log rank test. (F) PCR analysis confirms the presence of Wolbachia in wDah flies and the absence in Tetracycline-treated wDahT flies. (G) Q-RT-PCR analysis shows that 4E-BP expression is not affected by Wolbachia-infection status. 4E-BP transcript level in Wolbachia- flies was normalised to the corresponding Wolbachia+ sample, which by default was set to 1. Fig. 7. Dietary restriction in Drosophila is mediated by DILPs. 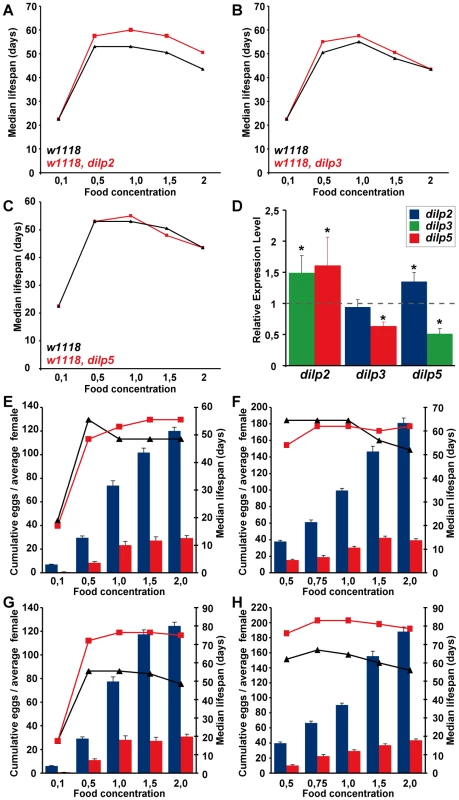
(A–C) dilp 2, 3 or 5 single mutants exhibit a normal response to DR compared to wild type controls. (D) Compensatory regulation among MNC-expressed DILPs on a high yeast diet. Expression levels of DILP2, 3 and 5 were measured by Q-RT-PCR on RNA extracted from heads of 10 day old dilp mutant females kept on 2.0x food. * p<0.05. (E–H) In two independent trials dilp2–3,5 mutants failed to show a normal response to DR. (E,F) wDahT; dilp2–3,5 mutants (Wolbachia-). (G,H) wDah; dilp2–3,5 mutants (Wolbachia+) Bars: index of lifetime fecundity±standard error of mean; connected points: median lifespan in days. dilp2–3,5 mutants in red, wDahT controls in black/blue. Intriguingly, although Wolbachia is well known to manipulate the reproductive system of its host, we did not detect significant differences in fecundity between mutants or controls with or without bacterial infection (Figure 6B), and nor did it have an effect on development time (data not shown) or energy storage (Figure S5C, S5F). Wolbachia did affect growth, but not by increasing IIS; flies without Wolbachia infection had significantly lower body weights than flies carrying the bacterium (Figure 4C), but this effect was seen in wild type and dilp2–3,5 mutants. Wolbachia infection status did not change survival of dilp2–3,5 mutants under starvation or hydrogen peroxide treatment (Figure 6C and 6D), suggesting that oxidative stress resistance of wDah;dilp2–3,5 mutants is not causal for their increased lifespan. In contrast, Wolbachia-positive dilp2–3,5 mutants were more resistant to DDT treatment than Wolbachia-free dilp2–3,5 mutants (Figure 6E), which suggests that xenobiotic resistance may contribute to their increased longevity.
Fecundity
IIS has been implicated in the maintenance of germ-line stem cells (GSC) in Drosophila and over-expression of DILP2 suppressed GSC loss in adult females [13]. Consistently, dilp2 mutant females exhibited a significantly reduced lifetime egg-production (−25%) compared to control flies (Figure S6B). However, fecundity was already reduced in young 3 day old dilp2 mutant females (data not shown), suggesting that at least part of the phenotype is caused by developmental defects, consistent with results from flies with ablated MNCs [34]. A small decrease in fecundity was also observed in dilp3 (−22%), dilp5 (−18%), dilp2–3 (−27%) and dilp1–4 (−14%) mutants (Figure S6B). In contrast, egg-production of dilp2–3,5 mutants was more severely reduced (−69%, Figure 6B), suggesting that DILP2, 3 and 5 can act redundantly in the control of egg-production. Notably, dilp2–3,5 females are not completely sterile like other strong IIS pathway mutants e.g. homozygous chico females [39]. Thus, the finding that dilp6 mutants exhibited the strongest reduction in fecundity of all dilp single mutant females (−46%, Figure S6B) suggests that egg-production is under the combined control of DILPs expressed in MNCs and the fat body. In contrast, fecundity was not significantly reduced in dilp1, 4 and 7 mutants, the latter contrary to the suggestion that DILP7 may be a Drosophila relaxin [7].
Increased lifespan and reduced fecundity in response to dietary restriction are mediated by DILPs
DR and IIS have been suggested to act via overlapping mechanisms to extend lifespan in flies [19],[22], and the abundance of dilp5 mRNA is reduced in DR flies [20], suggesting a possible role for DILP5 in the flies' responses to DR. To determine whether DILPs contribute to the DR response, we measured the lifespan of dilp2, 3 and 5 mutant females (Figure 7A–7C) under DR. The dilp single mutants showed a normal DR response, with a peak in lifespan on 1.0x food and the shortest lifespan on 2.0x food observed for both mutants and controls. Except for starvation conditions (0.1x), dilp2 mutants were long-lived on all food types, suggesting that the lack of DILP2 causes lifespan-extension independent of yeast concentration (Figure 7A). dilp5 mutants showed a normal DR response, demonstrating that DILP5 is not essential for DR mediated lifespan extension. However, lack of DILP5 may be compensated for by up-regulation of other DILPs on the higher yeast concentrations. We therefore measured DILP transcript levels in the dilp mutants on 2.0x food (Figure 7D). Whereas the transcriptional regulation of DILP transcripts in dilp2 and dilp3 mutants was similar on low (1.0x) and high (2.0x) yeast concentrations (compare Figure 7D to Figure 3C), in dilp5 mutants expression of DILP2 was up-regulated on 2.0x food (Figure 7D), suggesting that the lack of diet-dependent DILP5 expression was compensated for by up-regulation of DILP2. Thus, DILPs in the MNC can act redundantly to mediate the organismal response to DR.
To test for redundancy, we measured the DR response of dilp2–3,5 mutant females in two independent trials using flies with (Figure 7G and 7H) and without Wolbachia (Figure 7E and 7F). Wolbachia-free dilp2–3,5 mutants failed to show a normal response to DR. Instead, similar to chico1 mutants, their response was right shifted [22], with the flies shorter-lived compared to controls on low but longer-lived on high yeast concentrations (Figure 7E and 7F). The maximum lifespan of dilp2–3,5 mutants did not exceed the maximum that was achieved by DR alone, consistent with reduced IIS and DR extending lifespan through the same mechanisms. In addition, whereas wild-type flies showed a strong increase in egg-production between 1x and 2x food (62–81%), Wolbachia-free dilp2–3,5 mutants only laid 26% more eggs on the higher yeast concentration (Figure 7E and 7F). Absence of these three DILPs therefore strongly attenuated the response of fecundity to DR. As in wild type flies only DILP5 expression was nutritionally regulated, these results suggest that the normal response to DR is mainly mediated by DILP5.
Interestingly, infection with Wolbachia modified the DR response of dilp2–3,5 mutants. In contrast to Wolbachia-free dilp2–3,5 mutants, these mutants showed extended lifespan at all food concentrations except for starvation, and their maximum lifespan far exceeded that achieved by DR treatment alone (Figure 7G and 7H). In addition, the DR response of the Wolbachia-positive dilp2–3,5 mutant flies was greatly attenuated but not right shifted. Whereas control flies exhibited a DR-induced lifespan extension of 14–20% and an increase in egg production of 108–123% between 1x and 2x food, dilp2–3,5 mutants showed lifespan extension of only 2–6% and an increased egg production of 7–48%. Wolbachia infection status had no effect on the lifespan response to DR of wild type control flies, consistent with previous findings [40]. In conclusion, the DR response of Wolbachia-containing dilp2–3,5 mutants revealed both that these ligands mediate the responses to DR and that reduced IIS extends lifespan through mechanisms that both overlap with those of DR and through additional mechanisms that are independent of those at work in DR.
Discussion
We conducted a systematic mutational analysis of the 7 dilp genes of Drosophila melanogaster. Our results show that, while most dilp single mutants had only very mild or no obvious phenotypes, combinatorial lack of several DILPs resulted in more severe phenotypes, dependent upon the identity and number of DILPs knocked out, demonstrating that DILPs can act redundantly. Population-genetic theory suggests that newly duplicated gene copies can be evolutionarily unstable due to their functional overlap and that one copy may hence be rapidly lost. However, sufficiently rapid divergence in sequence and function can result in retention of partially redundant genes [23]. We have shown that the 7 dilp genes have been evolutionary conserved during the more than 40 million years of evolution of the genus Drosophila. Although this time span may seem to be small on a geological timescale, when generation time is taken into account the evolutionary divergence spanned by the genus Drosophila exceeds that of the entire mammalian radiation [26].
One hallmark of evolutionarily conserved redundancies is differential expression of redundant genes either spatially or temporally [24],[41]. Consistently, although dilp genes have overlapping expression domains, e.g. DILP1, 2, 3 and 5 are co-expressed in MNC during larval development [5],[10], each dilp gene also has its own spatio-temporal expression pattern [9] and thereby different DILPs may regulate the same process during different stages of the fly's lifecycle. This is supported by the consecutive activation of DILP2 (first instar), DILP5 (second instar) and DILP3 (mid-late third instar) expression in MNC during development, likely to reflect the requirement for higher DILP levels to support the extensive growth happening especially in later larval stages. Remarkably, only dilp2 single mutants showed developmental delay and reduced growth, suggesting that DILP3 and DILP5 cannot compensate fully for the lack of DILP2 expression. dilp2–3,5 triple mutants showed a much more severe delay in larval development and larval growth, suggesting that DILP2, 3, and 5 are the main DILPs controlling growth during larval stages, while the fat body expressed dilp6 gene has been shown to control growth specifically during pupal development [31]. Thus, individual dilp genes use specific enhancer elements resulting in specific spatio-temporal expression, which may explains why the redundant dilp genes are stable during the evolution of Drosophila flies.
A related explanation for the evolutionary stability of the dilp gene family could be that, while DILPs act in part redundantly, they are evolutionarily conserved due to diversification of some of their functions. In support, RNAi-mediated knock-down of DILP2 reduced stored trehalose levels to the same extent as in flies with ablated MNC [18]. We confirmed this finding using the dilp2 mutants and further showed that dilp5 and dilp3 single mutants have normal trehalose levels, suggesting that stored trehalose levels are specifically regulated by DILP2. However, we found no evidence for other specific functions of individual dilp genes, which was especially surprising for the evolutionarily more highly conserved DILP4 and DILP7 peptides. DILP7 is the only DILP with bona fide orthologues outside the Drosophila genus and does not seem to act redundantly with the other DILPs, suggesting that it has a specific and important function. Nevertheless, dilp7 mutants are viable, have a normal lifespan and are fertile, with no reduction in fecundity. DILP7 has been suggested to be a Drosophila relaxin based on the observation that DILP7 neurons project to the female reproductive tract and that silencing of these neurons results in sterile females that exhibit an egg-jamming phenotype [7]. However, our results demonstrate that the sterility of these flies is not due to the lack of DILP7 secretion. Thus, the function of DILP7 and its relatively high evolutionary conservation is currently unclear.
dilp4 null mutants showed no obvious phenotypes and we found no evidence that DILP4 acts redundantly to the other DILPs. Interestingly, however, by using a DILP4-specific antibody we detected expression of DILP4 in neurons within the brain. The DInR has been shown to be required for axon guidance of photoreceptor-cells during development of the visual system [42], however the ligands mediating this guidance are not known. The neuronal expression may indicate a possible function for DILP4 in axon guidance, which could be experimentally tested in future studies by using the dilp4 null mutants.
A hallmark of redundant genes is that they are typically cross-regulated by negative feedback [24]. Accordingly, we found up-regulation of DILP3 and DILP5 transcript levels in dilp2 mutants, consistent with results obtained upon RNAi-mediated knock-down of DILP2 [18]. We also found up-regulation of DILP2 and DILP3 transcript in dilp5 mutants. While expression of DILP4 and DILP7 was not significantly changed in dilp2–3,5 mutants (Figure 3B), which suggest that these two DILPs are not part of the MNC negative feedback system, the fat body expressed dilp6 gene was strongly up-regulated in the triple mutant flies (Figure 3B). Furthermore, the combined lack of DILP2, 3, 5 and 6 resulted in lethality, indicating that DILP6 can act redundantly to DILP 2, 3 and 5 and suggesting the existence of a negative feedback system that acts to coordinate DILP expression between MNC in the central nervous system and the peripheral fat body tissue (Figure 3E). The negative feedback system is not restricted to the compensatory regulation of DILP transcript levels, but also involves down-regulation of ImpL2 (Figure 3B), a negative regulator of IIS that directly binds to DILPs and inhibits their function [32].
Interestingly, we found evidence for synergy between the DILPs in the MNCs. DILP2 and DILP5 were both down-regulated in dilp3 mutants (Figure 3C and 3D), which is consistent with the decreased expression of DILP2 and DILP5 upon RNAi-mediated knock down of DILP3 [34]. In their study Buch et al. interpreted the combined down-regulation as an experimental artefact of the DILP3 RNAi construct due to high sequence homology among the three dilp genes [34]. However, the DILP3 RNAi-construct does not contain any off-targets for DILP2 or DILP5 (data not shown) suggesting that the down-regulation of DILP2 and DILP5 is not an experimental artefact but rather that the transcription of these two DILPs is positively regulated by DILP3. Thus, DILP3 might be part of a positive feedback system that acts in an autocrine/paracrine manner to regulate expression of DILPs in MNCs (Figure 3E).
Previous work has shown that the transcript levels of DILP3, but not of DILPs 2 and 5, are reduced in dFOXO null mutants [18]. In addition, putative FOXO bindings sites were found to be enriched in the DILP3 promotor region, suggesting that DILP3 levels are regulated by the IIS pathway itself [18]. Interestingly, a similar feed back system has been described for the mammalian analogue of MNC, the pancreatic beta cells, in which insulin regulates its own production [43],[44], suggesting that this feedback system has been evolutionary conserved from insects to mammals.
These findings suggest that autoregulation of DILP3 maintains a necessary minimal level of IIS pathway activity in MNC. In case IIS activity drops below a certain threshold, dFOXO is activated, which in turn upregulates DILP3 expression. The increase in DILP3 levels then causes an additional increase in transcript levels of DILPs 2 and/or 5 (Figure 3E).
Head fat body-specific over-expression of dFOXO has been shown to cause increased lifespan, systemic effects on lipid storage and reduced expression of DILP2 in MNCs [14]. However, molecules mediating the systemic effects of the tissue-specific dFOXO expression have not yet been identified. The findings that DILP6 is both part of a negative feedback system between the fat body and MNCs, which are involved in lifespan and lipid storage regulation, and a direct dFOXO target gene [31] makes DILP6 a good candidate to mediate systemic effects of dFOXO expression. dilp6 mutant flies were not long-lived, but they will allow testing of whether lifespan extension upon dFOXO over-expression is dependent on DILP6.
In conclusion, DILPs expressed in the MNC and the fat body act redundantly to regulate development, metabolism, reproduction and lifespan and their expression is tightly controlled by both negative and positive feedback mechanisms (Figure 3E). The fly may utilize the presence of redundant DILP copies to downplay stochastic variations in DILP expression or secretion in response to varying external conditions [24]. Synergy of expression, based upon autoregulation through IIS pathway activity, may enable rapid detection of and a systemic response to conditions that lower pathway activity. Because DILPs respond to nutritional changes this may also have helped Drosophila flies to adapt to new environments and food sources and thereby facilitated their evolution, generating flies with different feeding habits ranging from generalists like D. ananassae to specialists like D. sechellia. Although, except for DILP7, bona fide orthologues for the other DILPs could not be identified outside the Drosophilidae, most animals contain several ILPs, suggesting that, as in Drosophila, redundant ILPs may be of evolutionary advantage.
Specific interaction of Wolbachia and IIS in lifespan regulation
Wolbachia pipientis are maternally-inherited, obligate intracellular bacteria that are extremely widespread among wild and laboratory Drosophila populations [45] and their presence has been associated with parasitic and/or endosymbiontic modification of host fitness-related traits including lifespan [36],[37]. Interestingly, specific interaction between Wolbachia strain and host genotype have been demonstrated, e.g. Wolbachia can suppress the sterility phenotype of sex-lethal mutants or modify the longevity of a long-lived Drosophila strain [37],[46]. Here we show that longevity of dilp2–3,5 mutant flies is dependent on the presence of a maternally derived factor that can be removed by treatment with Tetracycline, most likely Wolbachia. The Tetracycline treatment itself is unlikely to have caused any negative effects because wild type flies with and without Wolbachia were phenotypically indistinguishable, except for their different body weight. Furthermore, dilp2–3,5 mutants had the same median lifespan as control flies in two Wolbachia-free genetic backgrounds, which demonstrates that the lack of lifespan extension is not specific to the outbred wDahT strain. Wolbachia-positive dilp2–3,5 mutants were extremely long-lived and had a prolonged survival under DDT treatment. Increased resistance to xenobiotic compounds has previously been associated with increased longevity [47]. For example, long-lived Little mice, mutant for the GH-releasing hormone receptor gene have been shown to be resistant to xenobiotic toxicity and show concerted up-regulation of xenobiotic detoxification genes [48],[49]. Furthermore, long-lived IIS mutant C. elegans and Drosophila also show increased expression of genes involved in xenobiotic metabolism [50], suggesting that xenobiotic resistance may contributes to the lifespan extension of wDah,dilp2–3,5 mutant flies. However, compared to control flies Wolbachia-free dilp2–3,5 mutants also showed increased survival under DDT treatment (Figure 6E), demonstrating that increased xenobiotic resistance alone was not sufficient to increase lifespan, suggesting that in addition other mechanisms contribute to the Wolbachia-dependent lifespan extension of dilp2–3,5 mutants. However, the molecular mechanisms by which Wolbachia influences its host are currently unknown.
Notably, lifespan extension of dilp2 and dilp2–3 double mutants was not dependent on the presence of Wolbachia (Figure 5), demonstrating that Wolbachia is not in general essential for lifespan extension due to reduced IIS in Drosophila. However, this finding raises the question why Wolbachia is essential for lifespan extension in one IIS mutant but not in another. There may exist an optimal range of down-regulation of IIS pathway activity in order to extend lifespan. In contrast to the relative mild phenotypes of dilp2 or dilp2–3 mutants, the combined loss of DILP2, 3 and 5 causes severe developmental and metabolic phenotypes, which may be detrimental to the flies, and Wolbachia may attenuate the expressivity of the dilp2–3,5 mutant phenotype. Recently, it has been suggested that Wolbachia acts to increase insulin signaling downstream of the DInR and thereby attenuates the phenotype of flies overexpressing a dominant negative DInR [38]. In contrast to these results we did not observe changes in IIS pathway activity when comparing expression of the IIS target 4E-BP in flies with or without Wolbachia. In addition, we found no difference in egg-to-adult survival, development time, energy storage, stress resistance or fecundity between dilp2–3,5 mutants with or without Wolbachia. This observation suggests either that lifespan, in contrast to the other traits, is very sensitive to even small changes in IIS activity or that Wolbachia mediates lifespan extension of dilp2–3,5 mutants by another mechanism.
The interaction between Wolbachia and its Drosophila host are complex and dependent both on the Wolbachia strain as well as the genetic background of the fly line. In addition, rapid co-evolution between Wolbachia and its host has been demonstrated [51]. In our study we analysed the interaction between IIS and one Wolbachia strain in the context of its natural host, the outbred wDahomey line. For future studies it will be interesting to determine whether this interaction is specific for Dahomey flies and its co-evolved Wolbachia strain, or whether other Wolbachia strains and/or other Drosophila wild type lines have the same effect on IIS.
DILPs mediate the response to dietary restriction in Drosophila
Dietary restriction, the reduced availability of nutrients without malnutrition, extends lifespan in a wide variety of organisms including worms, flies and mammals. However, the underlying molecular pathways mediating the effect of DR on lifespan are still elusive. In Drosophila, IIS and DR have been suggested to act through overlapping mechanisms, based on the DR response of chico mutants, which are short-lived on low food concentrations but long-lived on high food concentrations [22]. However recently the relevance of IIS for the response to DR in Drosophila has been challenged by the finding that flies mutant for the downstream target of IIS the transcription factor dFOXO are short-lived, but respond equally well to DR as control flies [19],[20]. Here we present evidence that DR in Drosophila is mediated by the up-stream ligands of IIS, DILPs, expressed in the MNCs. Although dilp single mutants responded as strongly to DR as did control flies, the DR response of dilp2–3,5 mutants was either severely attenuated or completely blocked, depending on the presence of Wolbachia, suggesting that DILPs can act redundantly in mediating the response to DR. Additionally, transcript levels of DILP5 were found to be regulated in a diet-dependent manner. Whereas DILP2 and DILP3 transcript levels remained constant across diets, the abundance of DILP5 mRNA was reduced in dietary restricted flies [20], suggesting that, under normal conditions, the response to DR is mediated by DILP5. However, when DILP5 is missing other DILPs may compensate the lack of diet-induced DILP5 expression, consistent with the up-regulation of DILP2 transcript in dilp5 mutants on high yeast food only. RNAi-mediated knock-down of DILP3 has been shown to reduce DILP5 transcript levels and to block diet-dependent changes in DILP5 transcription and these flies respond normally to DR treatment [20], which we could confirm using the dilp3 null mutant flies. However, this finding does not exclude a function for DILP5 in the response to DR, as DILP5 peptide is still present and could be regulated on the level of protein stability or secretion.
Thus, ligands of IIS mediate the changes in longevity seen under DR conditions in Drosophila, which raises the question why do mutants of the IIS downstream effector dFOXO show a normal response to DR. One explanation could be that dFOXO is involved in the response to DR under normal conditions but in its absence another pathway mediates the life span extension seen upon DR treatment. Fat body specific over-expression of dFOXO extends lifespan in a nutrient dependent manner, which would be consistent with a role of dFOXO in DR [19]. In C. elegans, the forkhead transcription factor pha-4, a Foxa orthologue, was shown to be required for lifespan extension under DR [52] and its fly orthologue would therefore be a candidate to mediate DR in the fly, redundant to IIS. The Target of Rapamycin (TOR) pathway has been linked to the determination of lifespan in flies and worms and lifespan-extension by decreased TOR signalling is dependent on nutritional conditions, suggesting a possible link between the TOR pathway and DR [53],[54]. The IIS pathway regulates TOR activity through the protein kinase AKT(PKB), an IIS component downstream of DILPs and Chico but upstream of dFOXO [1]. Thus, in Drosophila upstream IIS components such as DILPs and Chico may mediate the response to DR via the TOR pathway but not through the IIS downstream effector dFOXO.
Strong evolutionary conservation of dilp gene family membership and sequence differentiation has thus been accompanied by functional differentiation, redundancy and synergy between DILPs, and these features may themselves be of evolutionary advantage.
Materials and Methods
Ends-out homologous recombination
dilp mutants were generated by ends-out homologous recombination according to the methods described in [30],[55]. All fly stocks are summarized in Table S1. Donor constructs used for targeting dilp genes are summarized in Table S2. DNA fragments homologous to approximately 4 kb of dilp gene flanking sequences were amplified by PCR and subsequently cloned into the pW25 vector [55]. pw25 was obtained from the Drosophila Genomics Resource Center, (Bloomington, Indiana, USA). Long-range PCR was done using Takara LA Taq polymerase (Lonza, UK) on BAC clone DNA as template. BAC clones RP98-7A5 for dilp1–5 and RP98-32E2 for dilp7 were obtained from the BACPAC Resource Center (Oakland, California, USA). Primer sequences and restriction sites used for subcloning into pw25 are summarized in Table S3. DILP donor constructs were full-length sequenced and checked for base pair substitutions in gene coding sequences before used to generate transgenic flies. DNA constructs were transformed into the germline of Drosophila melanogaster by P-element-mediated germ line transformation using the Best Gene Drosophila Embryo Injection Services, (Chino Hills, California, USA). Crosses for ends-out homologous recombination were carried out according to the rapid targeting scheme [56]. Subsequently, the whitehs marker gene was genetically mapped and homologous recombination events were identified by PCR.
In order to generate dilp2–3,5 and dilp1–4,5 mutants, homologous recombination was done using flies carrying the dilp5 donor construct and hs-FLP, hs-SceI in the dilp2–31 and dilp1–41 mutant background, respectively.
Generation of dilp6 mutants
y1 P{y[+mDint2] w[BR.E.BR] = SUPor-P}KG04972 [57] flies that carry an KG-element transposon-construct integration in the 5′upstream region of the dilp6 gene at chromosome X position 2,229,002, corresponding to position -2831 relative to the putative start ATG in dilp6 exon 2, were obtained from the Bloomington Drosophila stock center, Indiana, USA. dilp6 deletion mutants were generated by conventional P-element imprecise excision resulting in the small dilp641 deletion and the large dilp668 deletion. Both dilp6 alleles contain residual P-element sequence, which impeded the exact mapping of the 3′ breakpoints at the sequence level. By PCR analysis the 3′ breakpoint in dilp641 could be mapped to the first dilp6 intron, i.e. dilp641 mutant flies lack the first dilp6 exon but contain the full dilp6 ORF. dilp668 mutants carry a deletion that covers at least 14 kb of genomic sequence including 5 annotated genes: dilp6, CG33218, CG2854, CG14050, CG34052.
Backcrossing and Wolbachia
dilp mutants were backcrossed for at least ten generations into two different wild-type stocks, the inbred lab strain w1118 and the outbred strain white DahomeyT (wDahT). dilp6 mutants were only backcrossed into the wDahT background. Naturally, Dahomey flies carry the intracellular bacterium Wolbachia. wDahT flies were generated by treating white Dahomey (wDah) flies with Tetracycline to remove Wolbachia. The presence of Wolbachia was checked by PCR using a Wolbachia specific primer combination (wsp-81F/wsp-691R, Table S3 and Figure 6F). w1118 flies do not contain Wolbachia and were therefore not treated with Tetracycline. Unless stated otherwise all experiments were done using Wolbachia-free flies. Flies carrying Wolbachia were generated by backcrossing dilp mutants into the original wDah background. All backcrossed stocks were maintained in large numbers in culture bottles on 1,0 SY-A medium at 25°C on a 12L∶12D cycle.
Generation of experimental flies
Experimental flies were generated by crossing age-matched heterozygous mutants and wild type, heterozygous and homozygous mutant progeny were collected from the same bottle based on eye colour. (Note: as homozygous dilp5 mutant males could not unambiguously be identified by their eye colour they were not used for experiments.) Eggs were collected during a six-hour time window and the same volume of embryos was transferred to each rearing bottles (1,0 SY-A) ensuring standard larval density. Flies eclosed during a 12 h time window were transferred to new bottles (1,0 SY-A) and left for 48 h to mate (once mated). Subsequently, flies were sorted under brief CO2 anaesthesia and transferred to vials. All experiments were conducted at 25°C on a 12 h∶12 h light:dark cycle at constant humidity (65%).
Lifespan, dietary restriction, and fecundity
For lifespan experiments, flies were maintained in vials at a density of 10 flies per vial on 1.0x SYA medium. Flies were transferred to new vials every two to three days and the number of dead flies was counted. DR experiments were done according to the optimised DR protocol described in [58]. Fly media used for DR experiments are summarized in Table S4. For fecundity assays mated females were kept at a density of 5 females per vial. Eggs were collected during 16–20 hour periods during the first 3–4 weeks. The number of eggs laid per vial at each time point was counted. Data are reported as the cumulative number of eggs laid per average female ± SEM over the whole period. For DR experiments egg numbers were collected from the lifespan vials.
Development time and body weight
Development time was analyzed by crossing heterozygous mutant flies and collecting eggs over a period of 3 h. Embryos were allowed to hatch and first instar larvae were hand picked and transferred 50 per vial on standard food. When adult flies started to hatch the number of eclosed homozygous, heterozygous and w control flies was counted in regular intervals.
For body weight determination flies were briefly anaesthetized on ice and weighted individually or in pairs on a ME235S analysis balance (Sartorius Mechantronics).
Stress test and energy storage
For stress tests 20 once mated females per vial were kept on 1x SY-A food for 8–10 days before the assay and transferred every other day. Media used for oxidative stress, starvation and DDT assays are summarized in Table S4. For DDT treatment time to knock-down was measured. Organismal lipid, glycogen and trehalose content of 8–10 day old females was quantified as described [18],[59].
Western blot and immunohistochemistry
DILP2 Western blots were done as described [18]. Immunohistochemistry on fly brains of 8–10 day old females was done according to the procedures described in [11]. The following primary antibodies were used: anti-DILP2 (rabbit), anti-DILP3 (rabbit), anti-DILP4 (rabbit) and anti-DILP5 (rat). Pictures were taken using an LSM 510 confocal microscope (Zeiss).
Quantitative RT–PCR
Total RNA was extracted from 25 fly heads or 25 bodies using Trizol (Gibco) according to the protocol of the manufacturer. cDNA was prepared using the Superscript II RT system (Invitrogen) and oligo(dT) primer. Quantitative RT-PCR was performed on a PRISM 7000 sequence detection system using SYBR green master mix (Applied Biosystems) and four independent RNA extractions per genotype. Results were calculated according to the standard curve method and normalized using act primers. Primers used for Q-RT-PCR are summarized in Table S3.
Data analyses
Lifespan and stress assays were recorded using Excel and were subjected to survival analysis (log rank test) and presented as survival curves. Fecundity data were analysed using the Wilcoxon rank test. Q-RT-PCR, glycogen, trehalose and lipid data were recorded in Excel and analysed using Student's t-test.
Supporting Information
Zdroje
1. EdgarBA
2006 How flies get their size: genetics meets physiology. Nat Rev Genet 7 907 916
2. BroughtonS
PartridgeL
2009 Insulin/IGF-like signalling, the central nervous system and aging. Biochem J 418 1 12
3. WuQ
BrownMR
2006 Signaling and function of insulin-like peptides in insects. Annu Rev Entomol 51 1 24
4. LiC
KimK
2008 Neuropeptides. WormBook 1 36
5. BrogioloW
StockerH
IkeyaT
RintelenF
FernandezR
2001 An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11 213 221
6. OkamotoN
YamanakaN
SatakeH
SaegusaH
KataokaH
2009 An ecdysteroid-inducible insulin-like growth factor-like peptide regulates adult development of the silkmoth Bombyx mori. Febs J 276 1221 1232
7. YangCH
BelawatP
HafenE
JanLY
JanYN
2008 Drosophila egg-laying site selection as a system to study simple decision-making processes. Science 319 1679 1683
8. Miguel-AliagaI
ThorS
GouldAP
2008 Postmitotic specification of Drosophila insulinergic neurons from pioneer neurons. PLoS Biol 6 e58 doi:10.1371/journal.pbio.0060058
9. IkeyaT
GalicM
BelawatP
NairzK
HafenE
2002 Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol 12 1293 1300
10. RulifsonEJ
KimSK
NusseR
2002 Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296 1118 1120
11. BroughtonSJ
PiperMD
IkeyaT
BassTM
JacobsonJ
2005 Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A 102 3105 3110
12. WangS
TulinaN
CarlinDL
RulifsonEJ
2007 The origin of islet-like cells in Drosophila identifies parallels to the vertebrate endocrine axis. Proc Natl Acad Sci U S A 104 19873 19878
13. HsuHJ
Drummond-BarbosaD
2009 Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci U S A 106 1117 1121
14. HwangboDS
GershmanB
TuMP
PalmerM
TatarM
2004 Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429 562 566
15. BauerJH
ChangC
MorrisSN
HozierS
AndersenS
2007 Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci U S A 104 13355 13360
16. WangMC
BohmannD
JasperH
2005 JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121 115 125
17. LeeKS
KwonOY
LeeJH
KwonK
MinKJ
2008 Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol 10 468 475
18. BroughtonS
AlicN
SlackC
BassT
IkeyaT
2008 Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One 3 e3721 doi:10.1371/journal.pone.0003721
19. GiannakouME
GossM
PartridgeL
2008 Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: not required, but its activity modulates the response. Aging Cell 7 187 198
20. MinKJ
YamamotoR
BuchS
PankratzM
TatarM
2008 Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell 7 199 206
21. LakowskiB
HekimiS
1998 The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A 95 13091 13096
22. ClancyDJ
GemsD
HafenE
LeeversSJ
PartridgeL
2002 Dietary restriction in long-lived dwarf flies. Science 296 319
23. NowakMA
BoerlijstMC
CookeJ
SmithJM
1997 Evolution of genetic redundancy. Nature 388 167 171
24. KafriR
SpringerM
PilpelY
2009 Genetic redundancy: new tricks for old genes. Cell 136 389 392
25. MaY
CreangaA
LumL
BeachyPA
2006 Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443 359 363
26. StarkA
LinMF
KheradpourP
PedersenJS
PartsL
2007 Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450 219 232
27. BendtsenJD
NielsenH
von HeijneG
BrunakS
2004 Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340 783 795
28. KriegerMJ
JahanN
RiehleMA
CaoC
BrownMR
2004 Molecular characterization of insulin-like peptide genes and their expression in the African malaria mosquito, Anopheles gambiae. Insect Mol Biol 13 305 315
29. RiehleMA
FanY
CaoC
BrownMR
2006 Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptides 27 2547 2560
30. GongWJ
GolicKG
2003 Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A 100 2556 2561
31. SlaidinaM
DelanoueR
GrönkeS
PartrigdeL
LeopoldP
2009 A Drosophila Insulin-like Peptide Promotes Growth during Nonfeeding States. Developmental Cell 17 874 884
32. HoneggerB
GalicM
KohlerK
WittwerF
BrogioloW
2008 Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol 7 10
33. MattalianoMD
MontanaES
PariskyKM
LittletonJT
GriffithLC
2007 The Drosophila ARC homolog regulates behavioral responses to starvation. Mol Cell Neurosci 36 211 221
34. BuchS
MelcherC
BauerM
KatzenbergerJ
PankratzMJ
2008 Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab 7 321 332
35. WessellsRJ
FitzgeraldE
CypserJR
TatarM
BodmerR
2004 Insulin regulation of heart function in aging fruit flies. Nat Genet 36 1275 1281
36. MinKT
BenzerS
1997 Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc Natl Acad Sci U S A 94 10792 10796
37. ToivonenJM
WalkerGA
Martinez-DiazP
BjedovI
DriegeY
2007 No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PLoS Genet 3 e95 doi:10.1371/journal.pgen.0030095
38. IkeyaT
Broughton1Susan
AlicNazif
GrandisonRichard
PartridgeLinda
2009 The endosymbiont Wolbachia increases Insulin/IGF-like signaling in Drosophila. Proceedings of the Royal Society B
39. BohniR
Riesgo-EscovarJ
OldhamS
BrogioloW
StockerH
1999 Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 97 865 875
40. GrandisonRC
WongR
BassTM
PartridgeL
PiperMD
2009 Effect of a standardised dietary restriction protocol on multiple laboratory strains of Drosophila melanogaster. PLoS One 4 e4067 doi:10.1371/journal.pone.0004067
41. KafriR
LevyM
PilpelY
2006 The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A 103 11653 11658
42. SongJ
WuL
ChenZ
KohanskiRA
PickL
2003 Axons guided by insulin receptor in Drosophila visual system. Science 300 502 505
43. KulkarniRN
BruningJC
WinnayJN
PosticC
MagnusonMA
1999 Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96 329 339
44. CantleyJ
ChoudhuryAI
Asare-AnaneH
SelmanC
LingardS
2007 Pancreatic deletion of insulin receptor substrate 2 reduces beta and alpha cell mass and impairs glucose homeostasis in mice. Diabetologia 50 1248 1256
45. ClarkME
AndersonCL
CandeJ
KarrTL
2005 Widespread prevalence of wolbachia in laboratory stocks and the implications for Drosophila research. Genetics 170 1667 1675
46. StarrDJ
ClineTW
2002 A host parasite interaction rescues Drosophila oogenesis defects. Nature 418 76 79
47. GemsD
PartridgeL
2008 Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab 7 200 203
48. Amador-NoguezD
DeanA
HuangW
SetchellK
MooreD
2007 Alterations in xenobiotic metabolism in the long-lived Little mice. Aging Cell 6 453 470
49. Amador-NoguezD
YagiK
VenableS
DarlingtonG
2004 Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell 3 423 441
50. McElweeJJ
SchusterE
BlancE
PiperMD
ThomasJH
2007 Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol 8 R132
51. WeeksAR
TurelliM
HarcombeWR
ReynoldsKT
HoffmannAA
2007 From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol 5 e114 doi:10.1371/journal.pbio.0050114
52. PanowskiSH
WolffS
AguilaniuH
DurieuxJ
DillinA
2007 PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447 550 555
53. KapahiP
ZidBM
HarperT
KosloverD
SapinV
2004 Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14 885 890
54. HansenM
TaubertS
CrawfordD
LibinaN
LeeSJ
2007 Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6 95 110
55. GongWJ
GolicKG
2004 Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics 168 1467 1476
56. RongYS
GolicKG
2001 A targeted gene knockout in Drosophila. Genetics 157 1307 1312
57. BellenHJ
LevisRW
LiaoG
HeY
CarlsonJW
2004 The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761 781
58. BassTM
GrandisonRC
WongR
MartinezP
PartridgeL
2007 Optimization of dietary restriction protocols in Drosophila. J Gerontol A Biol Sci Med Sci 62 1071 1081
59. GrönkeS
MildnerA
FellertS
TennagelsN
PetryS
2005 Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 1 323 330
Štítky
Genetika Reprodukční medicína
Článek Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the GenomeČlánek Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in MiceČlánek Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' DiseaseČlánek Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD TagsČlánek Wing Patterns in the Mist
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 2- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Scale of Population Structure in
- Allelic Exchange of Pheromones and Their Receptors Reprograms Sexual Identity in
- Genetic and Functional Dissection of and in Age-Related Macular Degeneration
- A Single Nucleotide Polymorphism within the Acetyl-Coenzyme A Carboxylase Beta Gene Is Associated with Proteinuria in Patients with Type 2 Diabetes
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Cdk2 Is Required for p53-Independent G/M Checkpoint Control
- Uncoupling of Satellite DNA and Centromeric Function in the Genus
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in the Clade
- Use of DNA–Damaging Agents and RNA Pooling to Assess Expression Profiles Associated with and Mutation Status in Familial Breast Cancer Patients
- Cheating by Exploitation of Developmental Prestalk Patterning in
- Replication and Active Demethylation Represent Partially Overlapping Mechanisms for Erasure of H3K4me3 in Budding Yeast
- Cdk1 Targets Srs2 to Complete Synthesis-Dependent Strand Annealing and to Promote Recombinational Repair
- A Genome-Wide Association Study Identifies Susceptibility Variants for Type 2 Diabetes in Han Chinese
- Genome-Wide Identification of Susceptibility Alleles for Viral Infections through a Population Genetics Approach
- Transcriptional Rewiring of the Sex Determining Gene Duplicate by Transposable Elements
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in
- Proteasome Nuclear Activity Affects Chromosome Stability by Controlling the Turnover of Mms22, a Protein Important for DNA Repair
- Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in Mice
- Structure, Function, and Evolution of the spp. Genome
- Human and Non-Human Primate Genomes Share Hotspots of Positive Selection
- A Kinase-Independent Role for the Rad3-Rad26 Complex in Recruitment of Tel1 to Telomeres in Fission Yeast
- Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' Disease
- Molecular Evolution and Functional Characterization of Insulin-Like Peptides
- Molecular Poltergeists: Mitochondrial DNA Copies () in Sequenced Nuclear Genomes
- Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags
- Wing Patterns in the Mist
- DNA Binding of Centromere Protein C (CENPC) Is Stabilized by Single-Stranded RNA
- Genome-Wide Association Study Reveals Multiple Loci Associated with Primary Tooth Development during Infancy
- Mutations in , Encoding an Equilibrative Nucleoside Transporter ENT3, Cause a Familial Histiocytosis Syndrome (Faisalabad Histiocytosis) and Familial Rosai-Dorfman Disease
- Genome-Wide Identification of Binding Sites Defines Distinct Functions for PHA-4/FOXA in Development and Environmental Response
- Ku Regulates the Non-Homologous End Joining Pathway Choice of DNA Double-Strand Break Repair in Human Somatic Cells
- Nucleoporins and Transcription: New Connections, New Questions
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Nucleoporins and Transcription: New Connections, New Questions
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



