-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Nucleoporins and Transcription: New Connections, New Questions
article has not abstract
Published in the journal: . PLoS Genet 6(2): e32767. doi:10.1371/journal.pgen.1000861
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1000861Summary
article has not abstract
It seems to make perfect sense that RNA, which must be exported from the nucleus to be translated, would be produced near or in association with nuclear pores. Indeed, recent reports proposed that Saccharomyces cerevisiae genes located close to the nuclear pore complex (NPC) tend to be highly transcribed [1],[2] and that, upon activation, some genes relocate from the nuclear interior to the nuclear periphery [3]. However, there is a critical difference in nuclear envelope organization between yeast and multicellular organisms. Yeast lacks lamins, a set of the structural proteins that coat the inner surface of the nuclear envelope, whereas multicellular organisms contain both lamins and NPCs. What is the relationship between the NPC and transcription in multicellular organisms? In this issue of PLoS Genetics, Vaquerizas and colleagues approach this issue [4], and in the process introduce an exciting set of new questions.
NPCs are gateways through which macromolecules are selectively imported from, or exported to, the cytoplasm. NPCs are large and highly structured protein assemblies built from more than 400 individual proteins (∼30 distinct subunits) called nucleoporins [5]. Nucleoporins and the structure of NPCs are highly conserved among eukaryotes from yeast to mammals. NPCs reside in the nuclear envelope, which is classically regarded to be associated with heterochromatin. For example, the nuclear lamina associates with transcriptionally silent regions in human and fly cells [6],[7], and artificial tethering of active genes to the nuclear lamina or to inner nuclear membrane proteins can cause transcriptional silencing [8],[9]. Indeed, electron microscopy and high-resolution light microscopy of mammalian cells clearly captures condensed heterochromatin at most of the nuclear envelope; however, heterochromatin is generally not localized at NPCs [3],[10]. So, there is an apparent paradox in the model. While the lamina interacts with transcriptionally silent loci, the NPCs, which are juxtaposed to the lamina, associate with active genes.
The Akhtar group performed chromatin immunoprecipitation coupled with genomic tiling microarrays (ChIP-chip) and identified genomic regions associated with two nucleoporins in the fruit fly Drosophila melanogaster—Nup153 or Megator (Mtor), a fly homolog of mammalian Tpr (Figure 1A) [4]. While the genomic binding profiles of these two nucleoporins were similar to each other, both patterns were very different from those of typical transcription factors. Instead of being localized at discrete loci, these nucleoporins are associated with large genomic domains spanning 10–500 kb in size. These regions, named Nucleoporin Associated Regions (NARs), contain predominantly actively transcribed genes. Concordantly, within NARs the authors found high levels of RNA polymerase II binding and histone H4 lysine 16 acetylation, a modification known to relax chromatin structure in vitro [11]. The results clearly demonstrate that at least a subset of nucleoporins associate with active genes in Drosophila.
Fig. 1. Nucleoporins Nup153 and Mtor are located at both NPCs and the nuclear interior, and associate with active transcription. 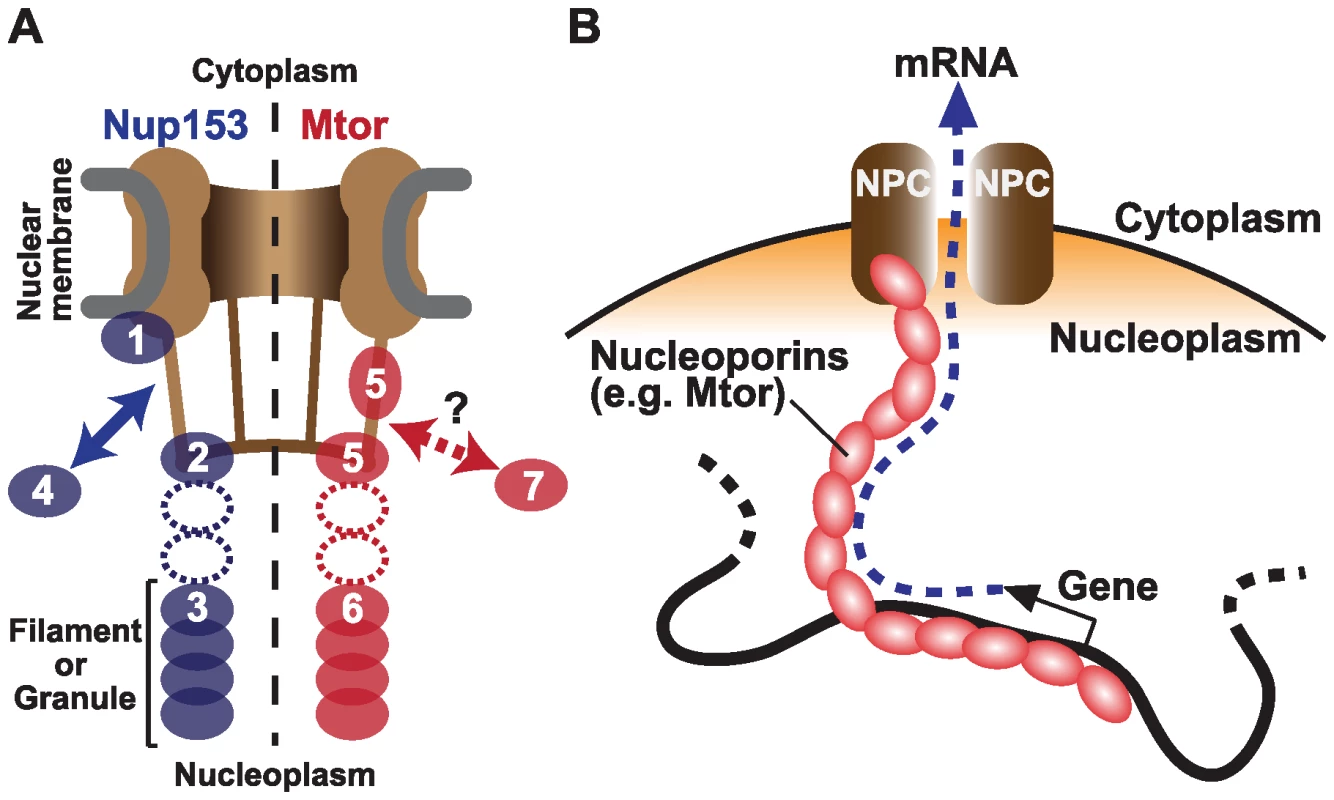
(A) Schematic representation of Nup153 (left) [13]–[15] and Mtor (right) [14],[16],[17] localization at the NPC and the nucleoplasm. Nup153 is proposed to be localized at the nuclear coaxial ring in proximity to the nuclear membrane (1); at the distal pore basket (2); as nucleoplasmic filaments (3); and shuttle between NPCs and the nucleoplasmic pool (arrow, 4). Mtor is proposed to constitute the pore basket (5) and nucleoplasmic filaments or granules (6). The mobile property of Mtor is unknown (7). It is still unclear whether the nucleoplasmic NUP153 and Mtor structures are extended from NPCs (dotted ovals). (B) Possible role of nucleoplasmic nucleoporins in transporting mRNA from the nuclear interior to NPCs. But do the nucleoporins associate with genes that are already active, or do they themselves promote transcription? The authors' current and previous experiments support a causal link between nucleoporins and transcriptional activation. They show that Nup153 and Mtor have a special relationship with the dosage compensation machinery. In Drosophila, unlike mammals and Caenorhabditis elegans, expression from genes on the single male X chromosome is doubled to balance expression with the two X chromosomes in female. The authors showed that NARs are over-represented on the X chromosome, but only in male cells, providing evidence for specific association between Nup153/Mtor and the active X chromosome [4]. Concordantly, these nucleoporins interact directly with a histone H4K16 acetyltransferase MOF (males absent on the first), which binds to dosage compensation complex MSL (male-specific lethal) proteins [12]. When Nup153 or Mtor were knocked down by RNAi, both MSLs and MOF dissociated from the X chromosome, resulting in reduced X-linked gene expression [12]. These findings suggest a mechanism wherein Nup153 and Mtor aid in directing the MSLs and MOF to the male X chromosome to facilitate transcription through H4K16 acetylation, further supporting an active role of nucleoporins in gene activation.
Perhaps the most surprising result is that the nucleoporins might carry out their function in transcription independent of their role in the NPC, and even independent of their localization to the nuclear envelope. By 3-D fluorescent in situ hybridization (3D-FISH), the authors determined the locations of NARs. While many NARs are found at the nuclear periphery, a subset of NARs are located at interior nuclear positions. Nup153 is known to be “mobile”, shuttling between NPCs and the nucleoplasm [13], and has been found in both the “basket” of the NPC and in filamentous structures in the nuclear interior (Figure 1A) [14],[15]. Likewise, while Mtor is localized to NPCs, it also constitutes granular or filamentous structures that extend into the nuclear interior. [16],[17]. Therefore, it is possible that the genome is associated with these internal structures to create the large NAR domains, rather than through association with the nuclear pore itself. An interesting model is that these interior nucleoporins serve as a physical route on which mRNAs are transported from deep in the nucleus to the NPC (Figure 1B) [16],[17]. This structure could be physically associated with nuclear territories or bodies to facilitate co-regulation of functionally linked genes.
Vaquerizas and colleagues clearly link a subset of nucleoporins to active gene expression and involvement with Drosophila dosage compensation, a chromosome-wide activation mechanism. Intriguing questions remain about how nucleoporins are targeted to specific genomic regions and the mechanism by which they affect RNA levels. The observation of both peripheral and non-peripheral NARs raises the question of whether nucleoporin-mediated regulation occurs at the NPC, in the nuclear interior, or at both locations. Finally, it remains unclear whether other nucleoporins, particularly those found exclusively as part of the NPCs, are associated with the genome or gene activity in multicellular organisms. Like any good study, this one has left us with new questions to explore.
Zdroje
1. CasolariJM
BrownCR
KomiliS
WestJ
HieronymusH
2004 Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117 427 439
2. BrownCR
SilverPA
2007 Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev 17 100 106
3. AkhtarA
GasserSM
2007 The nuclear envelope and transcriptional control. Nat Rev Genet 8 507 517
4. VaquerizasJM
SuyamaR
KindJ
MiuraK
LuscombeNM
2010 Nuclear pore proteins Nup153 and Megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 6(2) e1000846 doi:10.1371/journal.pgen.1000846
5. BrohawnSG
PartridgeJR
WhittleJR
SchwartzTU
2009 The nuclear pore complex has entered the atomic age. Structure 17 1156 1168
6. PickersgillH
KalverdaB
de WitE
TalhoutW
FornerodM
2006 Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38 1005 1014
7. GuelenL
PagieL
BrassetE
MeulemanW
FazaMB
2008 Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453 948 951
8. ReddyKL
ZulloJM
BertolinoE
SinghH
2008 Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452 243 247
9. FinlanLE
SproulD
ThomsonI
BoyleS
KerrE
2008 Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4 e1000039
10. SchermellehL
CarltonPM
HaaseS
ShaoL
WinotoL
2008 Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320 1332 1336
11. Shogren-KnaakM
IshiiH
SunJM
PazinMJ
DavieJR
2006 Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311 844 847
12. MendjanS
TaipaleM
KindJ
HolzH
GebhardtP
2006 Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21 811 823
13. RabutG
DoyeV
EllenbergJ
2004 Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6 1114 1121
14. KrullS
ThybergJ
BjorkrothB
RackwitzHR
CordesVC
2004 Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 15 4261 4277
15. BallJR
UllmanKS
2005 Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 114 319 330
16. ZimowskaG
ArisJP
PaddyMR
1997 A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci 110 927 944
17. ZimowskaG
PaddyMR
2002 Structures and dynamics of Drosophila Tpr inconsistent with a static, filamentous structure. Exp Cell Res 276 223 232
Štítky
Genetika Reprodukční medicína
Článek Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the GenomeČlánek Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in MiceČlánek Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' DiseaseČlánek Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD TagsČlánek Wing Patterns in the Mist
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Scale of Population Structure in
- Allelic Exchange of Pheromones and Their Receptors Reprograms Sexual Identity in
- Genetic and Functional Dissection of and in Age-Related Macular Degeneration
- A Single Nucleotide Polymorphism within the Acetyl-Coenzyme A Carboxylase Beta Gene Is Associated with Proteinuria in Patients with Type 2 Diabetes
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Cdk2 Is Required for p53-Independent G/M Checkpoint Control
- Uncoupling of Satellite DNA and Centromeric Function in the Genus
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in the Clade
- Use of DNA–Damaging Agents and RNA Pooling to Assess Expression Profiles Associated with and Mutation Status in Familial Breast Cancer Patients
- Cheating by Exploitation of Developmental Prestalk Patterning in
- Replication and Active Demethylation Represent Partially Overlapping Mechanisms for Erasure of H3K4me3 in Budding Yeast
- Cdk1 Targets Srs2 to Complete Synthesis-Dependent Strand Annealing and to Promote Recombinational Repair
- A Genome-Wide Association Study Identifies Susceptibility Variants for Type 2 Diabetes in Han Chinese
- Genome-Wide Identification of Susceptibility Alleles for Viral Infections through a Population Genetics Approach
- Transcriptional Rewiring of the Sex Determining Gene Duplicate by Transposable Elements
- Genomic Hotspots for Adaptation: The Population Genetics of Müllerian Mimicry in
- Proteasome Nuclear Activity Affects Chromosome Stability by Controlling the Turnover of Mms22, a Protein Important for DNA Repair
- Deletion of the Huntingtin Polyglutamine Stretch Enhances Neuronal Autophagy and Longevity in Mice
- Structure, Function, and Evolution of the spp. Genome
- Human and Non-Human Primate Genomes Share Hotspots of Positive Selection
- A Kinase-Independent Role for the Rad3-Rad26 Complex in Recruitment of Tel1 to Telomeres in Fission Yeast
- Analysis of the Genome and Transcriptome Uncovers Unique Strategies to Cause Legionnaires' Disease
- Molecular Evolution and Functional Characterization of Insulin-Like Peptides
- Molecular Poltergeists: Mitochondrial DNA Copies () in Sequenced Nuclear Genomes
- Population Genomics of Parallel Adaptation in Threespine Stickleback using Sequenced RAD Tags
- Wing Patterns in the Mist
- DNA Binding of Centromere Protein C (CENPC) Is Stabilized by Single-Stranded RNA
- Genome-Wide Association Study Reveals Multiple Loci Associated with Primary Tooth Development during Infancy
- Mutations in , Encoding an Equilibrative Nucleoside Transporter ENT3, Cause a Familial Histiocytosis Syndrome (Faisalabad Histiocytosis) and Familial Rosai-Dorfman Disease
- Genome-Wide Identification of Binding Sites Defines Distinct Functions for PHA-4/FOXA in Development and Environmental Response
- Ku Regulates the Non-Homologous End Joining Pathway Choice of DNA Double-Strand Break Repair in Human Somatic Cells
- Nucleoporins and Transcription: New Connections, New Questions
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study in Asian Populations Identifies Variants in and Associated with Systemic Lupus Erythematosus
- Nucleoporins and Transcription: New Connections, New Questions
- Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Genome
- The Genetic Interpretation of Area under the ROC Curve in Genomic Profiling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



