-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
Phylogenomics analysis indicates divergent mechanisms for host cell invasion across diverse species of obligate intracellular Rickettsia. For instance, only some Rickettsia species carry RalF, the rare bacterial Arf-GEF effector utilized by Legionella pneumophila to facilitate fusion of ER-derived membranes with its host-derived vacuole. For R. prowazekii (Typhus Group, TG), prior in vitro studies suggested the Arf-GEF activity of RalF, which is absent from Spotted Fever Group species, might be spatially regulated at the host plasma membrane. Herein, we demonstrate RalF of R. typhi (TG) and R. felis (Transitional Group) localizes to the host plasma membrane, yet R. bellii (Ancestral Group) RalF shows perinuclear localization reminiscent of RalF-mediated recruitment of Arf1 by L. pneumophila to its vacuole. For R. typhi, RalF expression occurs early during infection, with RalF inactivation significantly reducing host cell invasion. Furthermore, RalF co-localization with Arf6 and the phosphoinositide PI(4,5)P2 at the host plasma membrane was determined to be critical for R. typhi invasion. Thus, our work illustrates that different intracellular lifestyles across species of Rickettsia and Legionella have driven divergent roles for RalF during host cell infection. Collectively, we identify lineage-specific Arf-GEF utilization across diverse rickettsial species, previously unappreciated mechanisms for host cell invasion and infection.
Published in the journal: . PLoS Pathog 11(8): e32767. doi:10.1371/journal.ppat.1005115
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005115Summary
Phylogenomics analysis indicates divergent mechanisms for host cell invasion across diverse species of obligate intracellular Rickettsia. For instance, only some Rickettsia species carry RalF, the rare bacterial Arf-GEF effector utilized by Legionella pneumophila to facilitate fusion of ER-derived membranes with its host-derived vacuole. For R. prowazekii (Typhus Group, TG), prior in vitro studies suggested the Arf-GEF activity of RalF, which is absent from Spotted Fever Group species, might be spatially regulated at the host plasma membrane. Herein, we demonstrate RalF of R. typhi (TG) and R. felis (Transitional Group) localizes to the host plasma membrane, yet R. bellii (Ancestral Group) RalF shows perinuclear localization reminiscent of RalF-mediated recruitment of Arf1 by L. pneumophila to its vacuole. For R. typhi, RalF expression occurs early during infection, with RalF inactivation significantly reducing host cell invasion. Furthermore, RalF co-localization with Arf6 and the phosphoinositide PI(4,5)P2 at the host plasma membrane was determined to be critical for R. typhi invasion. Thus, our work illustrates that different intracellular lifestyles across species of Rickettsia and Legionella have driven divergent roles for RalF during host cell infection. Collectively, we identify lineage-specific Arf-GEF utilization across diverse rickettsial species, previously unappreciated mechanisms for host cell invasion and infection.
Introduction
Bacteria invading eukaryotic cells employ diverse strategies for successful entry, intracellular colonization and intercellular spread [1,2]. Whether facultative or obligate, intracellular species must either modify the phagocytic vacuole for survival or lyse the phagosome and live freely within the host cytoplasm (or invade other cellular organelles) [3–6]. Either strategy is delicately underpinned by bacterial secretion of effectors, which have a myriad of characterized functions: e.g., engaging host signaling pathways, rearranging the host cytoskeleton, polymerizing host actin, subverting host vesicular traffic, etc. [7–9]. It is well established that divergent effectors from distantly-related intracellular species can operate in similar processes [10]; e.g., actin nucleators from species of Shigella, Listeria and Rickettsia [11,12] and phospholipases from species of Pseudomonas and Legionella [13,14]. Conversely, the ability for highly similar effectors from distantly-related species to function differently in host cells is a phenomenon that is poorly known, probably reflective of effector repertoires being highly specific to bacterial genera [15–17].
Species of Rickettsia (Alphaproteobacteria: Rickettsiales) are Gram-negative obligate intracellular parasites of a wide range of eukaryotic species [18]. Rickettsiae bind to host cells and induce phagocytosis [19,20], with internalized bacteria released into the cytosol upon rapid escape from the phagocytic vacuole. Bacteria spread intercellularly upon death and lysis of host cells, though some species move intercellularly prior to host cell lysis via host actin polymerization [21–23]. Several surface proteins characterized for adhesion and/or entry of host cells (Sca5, Adr1, Adr2) [24–28] and activation of cytoskeletal vinculin (Sca4) [29] are conserved across sequenced Rickettsia genomes, as are several enzymes implicated in phagosomal lysis (TlyC, PLD, Pat1) [30–33]. In contrast, other characterized adhesins (Sca0, Sca1, Sca2) [34–38], proteins involved in Arp2/3-dependent (RickA) [39,40] and -independent (Sca2) [41,42] host actin polymerization, and another phospholipase (Pat2) [43,44] are sporadically encoded across rickettsial lineages. This suggests that, despite superficially similar infection strategies, diverse Rickettsia species employ distinct molecular mechanisms for successful colonization of host cells [45].
One such protein that is differentially encoded across Rickettsia genomes is a highly similar counterpart to the RalF protein of Legionella spp. Collectively, these proteins contain a Sec7-domain, which in eukaryotes functions as a guanine nucleotide exchange factor (GEF) of ADP-ribosylation factors (Arfs) [46]. Remarkably, bacterial Sec7-domain containing proteins are unknown from other bacteria [47]. Legionella RalF (RalFL) is a secreted effector, with its proximal C-terminal sequence mediating secretion through the dot/icm type IV secretion system (T4SS) [48]. RalFL activates and recruits host Arf1 to the Legionella-containing vacuole (LCV), which is a modification of the phagosome [49]. The structure of RalFL contains two distinct domains: an N-terminal Sec7 domain (S7D) and a C-terminal Sec7-capping domain (SCD) that regulates active site access to Arfs [50]. The S7D and SCD across RalFL and Rickettsia RalF (RalFR) share ~45% aa identity, though an extended variable region flanks the SCD of RalFR proteins at the C-terminus [51].
A comparative study of RalFL and RalFR determined similar GEF activities for both proteins, yet divergent subcellular localization patterns driven primarily by intrinsic characteristics of the SCD [52]. The RalFL SCD positions the protein at the endoplasmic reticulum for interception of host secretory vesicles, while the RalFR SCD targets the protein to the host plasma membrane. Furthermore, a proline-rich region within the extended variable region of RalFR interacts with components of the host actin cytoskeleton. Subsequently, membrane sensor regions were identified within the SCDs of RalFL and RalFR, with differential enrichments in aromatic and positively charged residues determining divergent lipid substrates that regulate Arf-GEF activities [53]. Collectively, these studies suggest that these distinguishing features (divergent SCD sensor regions, RalFR-specific cytoskeletal-binding domain) mediate the spatial regulation of RalF activity in two diverse intracellular species with very different lifestyles.
Despite tremendous insight on the possible function of RalFR during rickettsial host cell infection, important questions are left unanswered. As previous studies were performed in vitro [52,53], it still remains unknown if those Rickettsia species that carry ralF genes actually express RalFR during infection, and if so, at what time point. Furthermore, as GEFs confer the spatial regulation of different Arf classes at discrete cellular locales [54–57], the Arf(s) specificity of RalFR needs to be determined in light of the different subcellular localization of the protein compared to Legionella spp. Our work presented here addresses these unknowns by demonstrating RalF expression by R. typhi early during host cell invasion. Across several Rickettsia species, we identified the domain requirements for positioning RalF at host membranes, and for R. typhi, determined that RalF co-localization with Arf6 and PI(4,5)P2 at entry foci was critical for invasion. Altogether, our work identifies Arf-GEF utilization as a lineage-specific invasion mechanism, illuminating the variable strategies that drive Rickettsia infection of host cells.
Results
RalFRt interacts with the rvh T4SS coupling protein (RvhD4) via its proximal C-terminal sequence and is secreted during host cell infection
As predicted Arf-GEFs, we anticipated RalFR proteins to be secreted extracellularly into the host cell. Prior to invasion, L. pneumophila utilizes its dot/icm I-T4SS to translocate RalFL into host cells [48]. Like RalFL, R. typhi RalF (RalFRt) lacks a predicted N-terminal Sec secretion signal [58], trans-membrane spanning regions [59] and a β-barrel structure [60], suggesting its secretion via a Sec-independent pathway, possibly the Rickettsiales vir homolog (rvh) T4SS [61]. Accordingly, in order to determine if RalFRt interacts with the rvh T4SS, we performed a bacterial two-hybrid assay with full length RalFRt (RalFRtFL) and RvhD4, the rvh T4SS coupling protein. T4SS coupling proteins (VirD4 family) are ATPases that function as “gatekeepers” to regulate substrate entry into the T4SS channel [62,63]. Co-transformation of bait (encoding RvhD4) and prey (encoding RalFRtFL) plasmids in BacterioMatch II reporter electrocompetent cells resulted in bacterial growth on selective media (Fig 1A), indicating RalFRtFL and RvhD4 interact, and thus implicating RalFRt as an rvh T4SS effector.
Fig. 1. R. typhi RalFRt interacts with RvhD4 and is expressed early during host cell invasion. 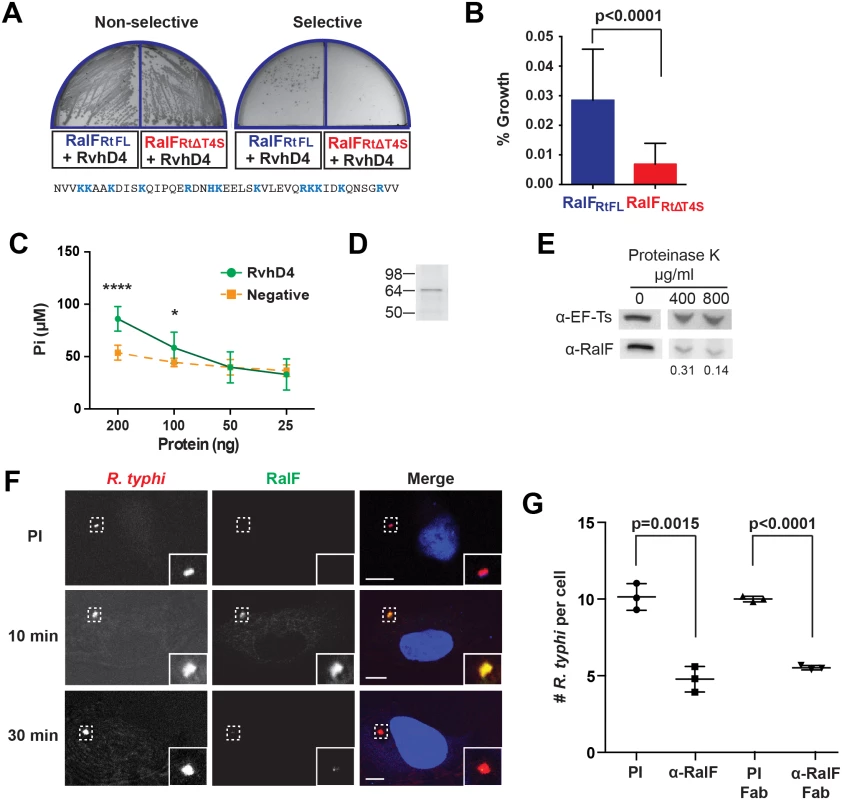
(A) Bacterial two-hybrid (B2H) assay reveals an interaction between RalFRt and RvhD4. ralFRtFL and ralFRtΔT4S were cloned into pTRG (prey) and rvhD4 was cloned into pBT (bait) of the B2H system. Constructed bait and prey plasmids were co-transformed into BacterioMatch II reporter electro-competent cells. Transformants were screened on non-selective plate (left) and positive interactions were identified on dual selective screening plate (right). The amino acid sequence deleted from ralFRtΔT4S (positively charged residues are colored blue) is shown at bottom. (B) Quantification of bacterial growth in the B2H assay described in panel A. Percent growth of CFUs of reporter cells harboring recombinant plasmids on dual selective screening medium was calculated relative to CFUs obtained on non-selective medium. Error bars represent mean ± SD of three independent experiments (Student’s two-sided t-test). (C) R. typhi RvhD4 exhibits ATPase activity. A series dilution of purified RvhD4 in assay buffer was incubated with reagent for 30 min at 21°C. The inorganic phosphate (Pi) released from ATP was quantified by measuring absorbance at OD 620 nm. As a negative control, a non-related R. typhi protein (RT0600) was assayed. Error bars represent mean ± SD of three independent experiments. * p = 0.01, **** p<0.0001; Student’s two-sided t-test. (D) Protein immunoblot of recombinant RvhD4 (~64 kDa) used in ATPase activity assays described in panel C. (E) RalFRt is surface exposed. Purified R. typhi was treated with 400 μg/mL or 800 μg/mL Proteinase K or in buffer alone for 1 hr. Lysates were resolved and immunoblotted for RalF or the R. typhi cytoplasmic control protein, elongation factor Ts (EF-Ts). Densitometry was performed using ImageJ and the intensity of RalF was normalized to EF-Ts. Representative image from two independent experiments is shown. Intensity of RalF normalized to EF-Ts and relative to untreated control is shown below the immunoblots. (F) RalF is expressed during early infection. HeLa cells infected with R. typhi for 10 and 30 min were fixed and R. typhi and RalF detected with rat anti-R. typhi (red) and affinity purified rabbit anti-RalFRt (green) antibodies, respectively. DAPI (blue) is shown in the merged image. Boxed regions are enlarged to show detail. Pre-immune (PI) cells were treated with rabbit PI serum in place of anti-RalFRt antibody. (Scale bar: 10 μm). (G) Anti-RalFRt IgG and Fab fragments inhibit R. typhi host cell infection. HeLa cells were infected with partially purified R. typhi pre-absorbed for 30 min with 20μg PI IgG serum, anti-RalFRt IgG, PI Fab fragments or anti-RalFRt Fab fragments. Cells were fixed 2 hrs post infection and R. typhi and the cell membrane detected with anti-R. typhi serum and Alexa Fluor 594 wheat germ agglutinin, respectively. The number of R. typhi per host cell was counted for 100 individual host cells in three independent experiments and normalized to PI serum. Error bars represent mean ± SD (Student’s two-sided t-test). Secretion of RalFL is dependent on hydrophobic residues within its C-terminal tail [48], while many other T4SS protein substrates have enrichments of positively charged residues at their C-termini that are important for secretion [64–66]. Accordingly, we evaluated RalFRt for the presence of a T4SS signal sequence (T4S) within its C-terminus. A T4S RalFRt truncation (RalFRtΔT4S) was generated and tested for its ability to bind RvhD4 via the bacterial two-hybrid assay (Fig 1A). The percent growth of colony forming units (CFUs) of reporter cells harboring recombinant plasmids on dual selective screening medium was calculated relative to percent growth of CFUs obtained on non-selective His dropout medium by drop plate method for counting. An approximately 77% decrease in CFUs on dual selective media was observed with RalFRtΔT4S compared to RalFRtFL, indicating that the RalF C-terminus is important for interacting with RvhD4 (Fig 1B).
The ATPase activity of T4SS coupling proteins is essential for substrate translocation [67]. To confirm functionality of RvhD4, recombinant RvhD4 was assayed for ATPase activity. RvhD4 was found to release inorganic phosphate (Pi) from ATP in a concentration dependent manner compared to a rickettsial protein that lacks predicted ATPase activity (RT0600) (Fig 1C and 1D). This indicates Rickettsia RvhD4 is a functional ATPase that likely regulates protein secretion through the rvh T4SS.
The interaction of RalFRt with machinery of the rvh T4SS implies extracellular secretion. Our prior report that characterized the R. typhi surface proteome demonstrated that RalFRt is expressed and surface exposed [68]. To further confirm RalFRt secretion, purified R. typhi was treated with proteinase K, on the premise that surface exposed protein would be degraded with proteinase K treatment while subsurface proteins would be protected. Protease treatment caused a dose-dependent degradation of RalFRt with respect to the R. typhi cytoplasmic control protein, elongation factor Ts (EF-Ts, Fig 1E).
RalFRt is expressed early during infection and is required for R. typhi invasion of host cells
To determine when RalFRt is expressed during R. typhi infection, a polyclonal antibody against RalFRt was generated, qualified (S1 Fig) and used for immunofluorescence assays (Fig 1F). During early infection of host cells (10 min), RalFRt expression is high and diminishes as internalization progresses (30 min). Given RalFRt expression during early infection, we assessed its role during R. typhi invasion of host cells. When R. typhi was pre-treated with the anti-RalFRt polyclonal antibody, the average number of R. typhi per host cell decreased by 52% from an average of 10 to 4.8 bacteria per host cell (Fig 1G), indicating a role for RalF during host cell invasion. To rule out possible steric hindrance induced by the Fc portion of the anti-RalFRt antibody inhibiting rickettsial-host cell interactions that promote entry, R. typhi was pre-absorbed with anti-RalFRt Fab fragments. The average number of R. typhi per host cell was significantly decreased by 45% from 10 to 5.5 bacteria per host cell (Fig 1G) further confirming the involvement of RalF in host cell invasion.
RalFR is divergent from RalFL within the SCD lipid sensor region and also contains a C-terminal extension that is highly variable across Rickettsia homologs
Utilizing over 60 Rickettsia genome sequences, phylogenomics analyses were carried out to provide further insight on the role of RalF in rickettsial biology and pathogenesis. While a key factor in R. typhi infection of host cells, RalF-mediated invasion is not a strategy employed by all Rickettsia species, as evident by ralF pseudogenization in all species of Spotted Fever Group (SFG) rickettsiae, as well as two other species (R. canadensis and R. helvetica) [45]. Still, the remaining species, including R. bellii and all species within the Typhus Group (TG) and Transitional Group (TRG) rickettsiae, contain genes encoding RalFR proteins that are highly conserved within the S7D and SCD as compared to RalFL (Fig 2). Specifically, and in agreement with previous studies [52,53], all RalFL and RalFR proteins contain a highly conserved Sec7 active site within the S7D (S2A and S2B Fig), with RalFR proteins having an enrichment of positively charged residues in the lipid sensor region of the SCD relative to RalFL proteins (S3C Fig). Thus, based on these characteristics, all RalFR proteins are predicted to spatially regulate their Arf-GEF activities at the host plasma membrane, where concentrated negatively charged phospholipids attract the RalFR SCD [53].
Fig. 2. Characteristics and comparative analysis of bacterial Sec7 domain-containing proteins (RalF). 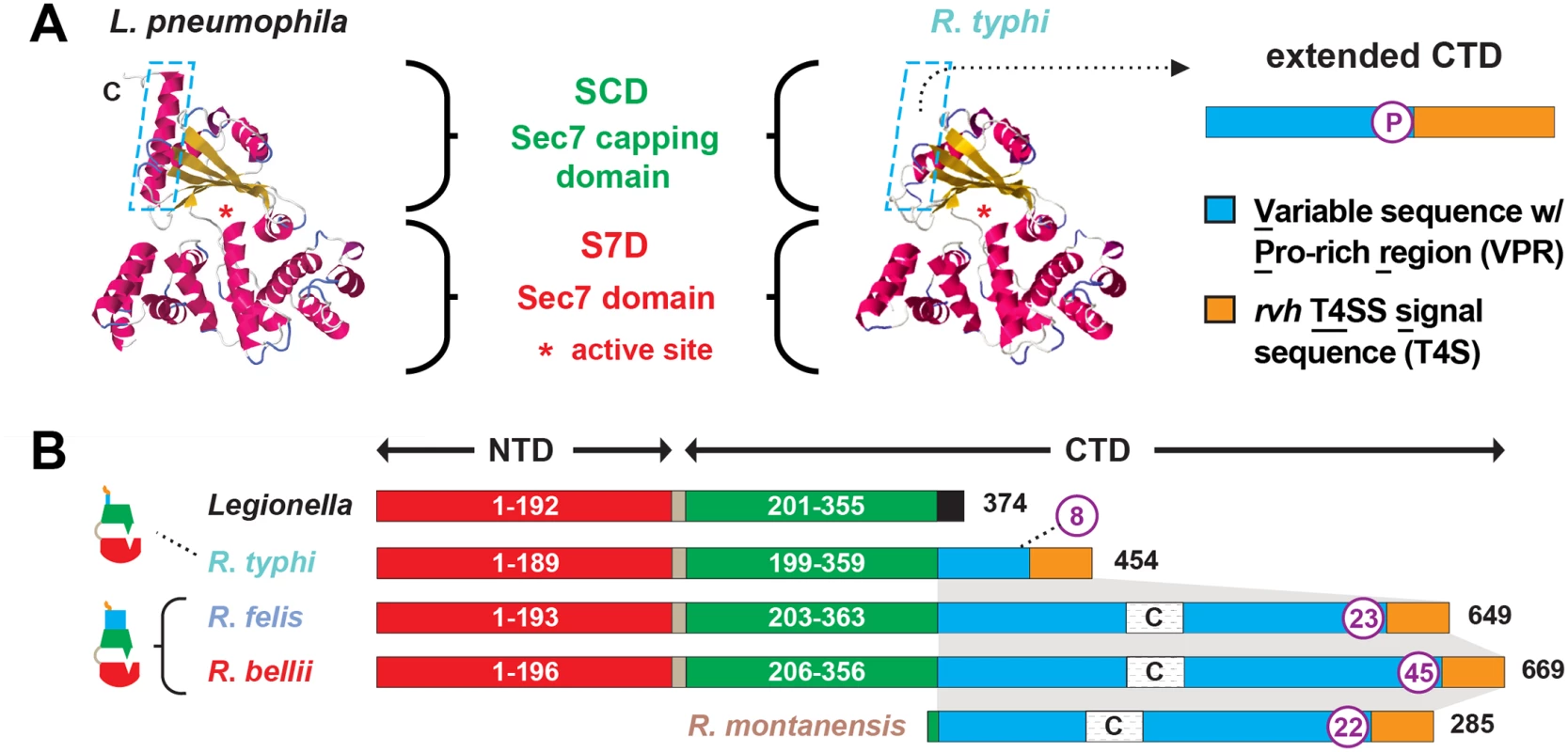
(A) Comparison of the crystal structure of Legionella pneumophila RalF (PDB 4C7P) [53] with the predicted structure of R. typhi RalF (RT0362). Modeling done with Phyre2 [69]. The delineation of the Sec7 domain (S7D, red) and Sec7-capping domain (SCD, green) is shown, with an approximation of the active site Glu (asterisk), which is essential for Arf recruitment to the Legionella containing vacuole [52]. The distinguishing feature of the otherwise highly similar proteins is the extended C-terminal domain in R. typhi RalF relative to L. pneumophila RalF. The blue dashed box depicts the extended C-terminal domain of Rickettsia RalF sequences, which can be delineated into a variable sequence with Pro-rich region (VPR) and an rvh T4SS signal sequence (T4S). (B) Domain organization of Legionella and Rickettsia RalF proteins. The structural conservation witnessed in panel A is encoded by conserved S7D (S2B Fig) and SCD (S3B Fig) sequences (~45% ID across Legionella and Rickettsia). Rickettsia RalF VPRs vary extensively across homologs; some Rickettsia RalF proteins contain only the VPR and T4S (S4A Fig). C, coiled-coil. Number of Pro residues within purple circles. NCBI GenBank accession numbers for all proteins are provided in the legend of S2 Fig. Relative to RalFL, the major distinguishing factor of RalFR proteins is the presence of a variable sequence with Pro-rich region (VPR) within the C-terminal domain (Fig 2). Pro-rich regions are a common characteristic of proteins that target the actin cytoskeleton [70], and are typically present in Arf-GEFs recruited to cytoskeletal/plasma membrane junctions [71,72]. Across RalFR proteins, the VPR is flanked by the SCD and T4S and is extraordinarily variable in sequence length and number of Pro residues across RalFR proteins (Fig 2B). Remarkably, many SFG rickettsiae species, e.g. R. montanensis, contain putative ORFs encoding complete VPRs. Alignment of these ORFs with VPRs from full-length RalFR proteins illustrates that a gene encoding RalFR was present in the Rickettsia ancestor, with pseudogenization purging the complete Arf-GEF from most Rickettsia genomes (S4A Fig). This conclusion is supported by genome synteny analysis across ralFR loci, which indicates a conserved position for ralF flanking the maeB gene in all sequenced Rickettsia genomes (S5A Fig). Thus, RalFL and RalFR proteins diversified early upon their establishment in ancestral Legionella and Rickettsia genomes, with the retention of VPRs within full-length RalFR proteins implying an important function.
The RalFR SCD regulates membrane localization while the VPR targets the host cytoskeleton
In light of the variability across the VPR of RalFR proteins, we determined the C-terminal domain (CTD) requirements for subcellular localization across RalFR proteins from several species (R. typhi, R. felis, R. montanensis and R. bellii) (Fig 3). The SCD - and VPR-mediated targeting to the host plasma membrane and actin cytoskeleton, respectively, for RalF of R. prowazekii (RalFRp) was used as a reference [52,53]. R. typhi and R. felis full-length RalF (RalFFL) proteins primarily had diffuse staining within the cytoplasm with some plasma membrane localization. However, RalFCTD (SCD-VPR-T4S) localized strongly to the plasma membrane and disrupted actin stress fibers. Additionally, R. typhi and R. felis RalFCTD induced membrane ruffling and microvilli-like protrusions suggesting that the CTD plays a role in cytoskeletal rearrangements, similar to the known Arf6-GEF, EFA6 [71,72]. Furthermore, RalFVPR (VPR-T4S), as well as the full-length VPR-containing ORF of R. montanensis, did not uniformly localize to the host plasma membrane, but instead were found strongly associated with intact actin stress fibers. Collectively, these results indicate that R. typhi and R. felis RalF proteins are similar to RalFRp, with both the SCD and VPR required to spatially regulate Arf-GEF activities at plasma membrane/actin cytoskeletal junctions.
Fig. 3. RalF subcellular localization and actin filament disruption mediated by the SCD and VPR. 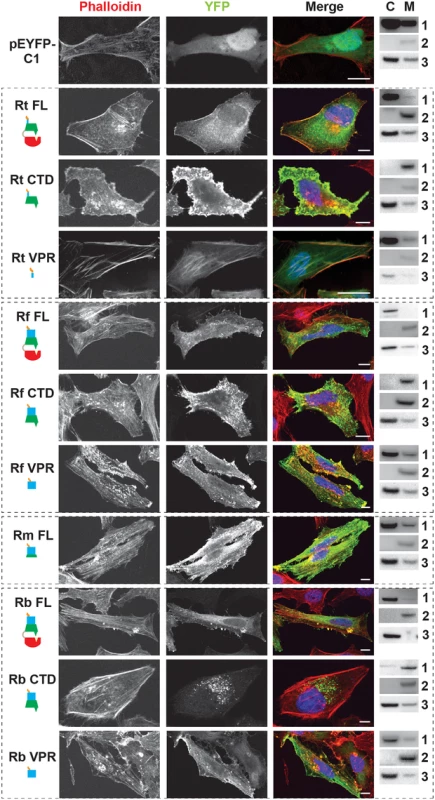
HeLa cells transfected with YFP tagged constructs (green, described in Fig 2B) were stained with Alexa Fluor 594 phalloidin to detect actin (red). DAPI (blue) is shown in the merged image. Cytoplasmic (C) and membrane (M) localization was confirmed via membrane fractionation of HEK293T cells Lipofectamine 2000 transfected with the indicated plasmids followed by immunoblotting. Immunoblot primary antibodies: 1, rabbit anti-GFP (Life Technologies); 2, membrane marker rabbit anti-Calnexin (Abcam); 3, cytoplasmic marker mouse anti-GAPDH (Abcam). Rt, R. typhi; Rf, R. felis; Rm, R. montanensis; Rb, R. bellii. (Scale bar: 10 μm). Remarkably, RalFFL and RalFCTD of R. bellii did not target the plasma membrane, yet instead showed perinuclear localization reminiscent of ectopically expressed RalFL. As R. bellii RalFVPR associated with intact actin stress fibers, these data collectively indicate that the SCD alone is sufficient to localize RalFRb to the host cytoplasm. Visualization of the SCD sequence alignment across all RalFL and RalFR proteins revealed that RalFRb lacks three separate insertions within the SCD that are conserved in all other RalFR proteins (S3B Fig). Thus, from a structural perspective, the SCD of RalFRb is more similar to RalFL proteins than RalFR proteins, which could explain why the SCD of R. bellii localizes to the perinuclear region of the cytoplasm. This is consistent with R. bellii sharing more genomic attributes with Legionella spp. [73], as well as being able to grow in various amoeba species unlike most other Rickettsia spp. (see Discussion).
RalF membrane localization was further confirmed using two independent approaches. First, membrane fractionation of HeLa cells transfected with RalF-expressing plasmids revealed that all RalFCTD proteins were predominately enriched in the membrane fraction, with RalFFL and RalFVPR proteins having less membrane enrichment (Fig 3 and S6 Fig). Second, RalF transfected cells were stained with Alexa Fluor 594 wheat germ agglutinin to detect plasma membrane or probed with anti-PDI (endoplasmic reticulum) or anti-GM130 (Golgi apparatus) antibodies (Fig 4 and S7 Fig), with the Pearson’s correlation coefficients calculated to measure co-localization with the respective membrane markers (S8 Fig). RalFCTD of R. typhi, R. felis, R. montanensis indicate localization to the plasma membrane, while R. bellii RalFCTD localized to the endoplasmic reticulum membrane recapitulating results observed with labeling host cell actin (Fig 3). Collectively, these data demonstrate the affinities of RalFR proteins for host membranes, identifying the SCD as the major determinant for membrane localization, combined with the targeting of actin cytoskeleton by the VPR (Table 1).
Fig. 4. Subcellular localization of rickettsial RalF proteins to host membranes. 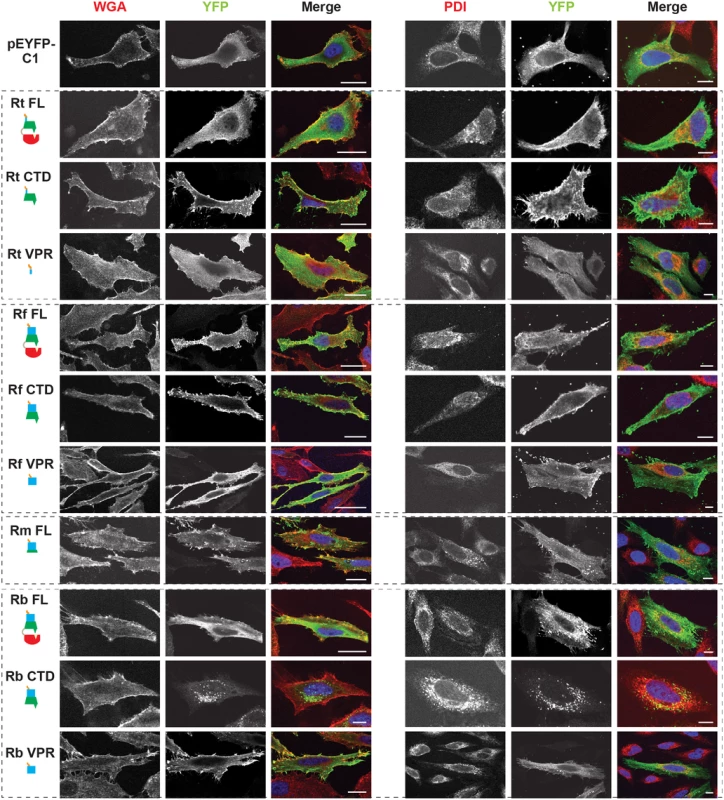
HeLa cells expressing YFP tagged RalF proteins (green, described in Fig 2) were fixed and stained with Alexa Fluor 594 wheat germ agglutinin (WGA) to detect the plasma membrane (left) or anti-PDI antibody to detect the endoplasmic reticulum (right). DAPI (blue) is shown in the merged image. (Scale bar: 10 μm). Tab. 1. Rickettsia RalF domain characterization. 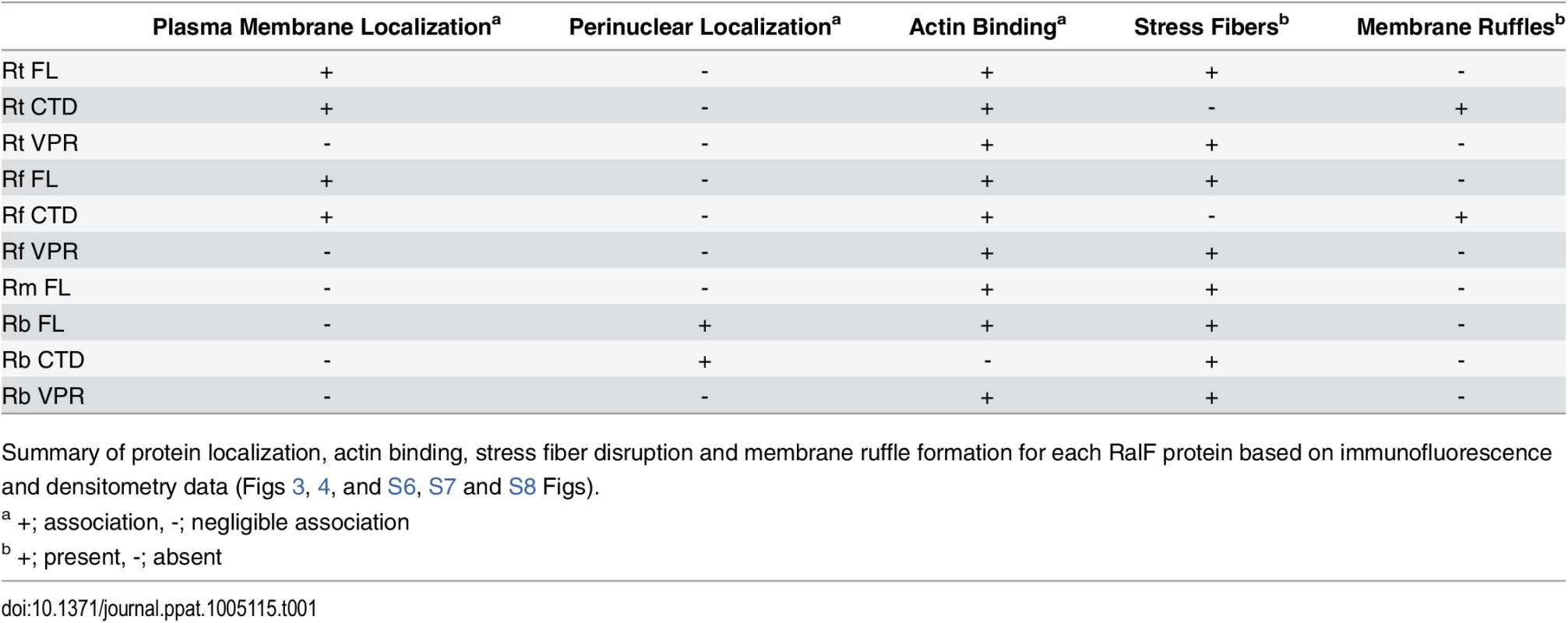
Summary of protein localization, actin binding, stress fiber disruption and membrane ruffle formation for each RalF protein based on immunofluorescence and densitometry data (Figs 3, 4, and S6, S7 and S8 Figs). Finally, for R. typhi, we monitored the subcellular localization of its RalFCTD construct lacking the T4S (RalFRtCTDΔT4S). We observed indistinguishable localization patterns between RalFRtCTD and RalFRtCTDΔT4S (S4C Fig), suggesting that the T4S has no effect on localization or stress fiber disruption. In conjunction with results above (Fig 1A and 1B), these data bolster the role of the T4S of RalFR proteins as an rvh T4SS translocation signal.
Plasma membrane localization of RalFRt is dependent on PI(4,5)P2
Previous studies showed the SCD of RalFRp has affinities for negatively-charged phospholipids; i.e., phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) and phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3) [52,53]. Given the enrichment of PI(4,5)P2 at host membranes during early stages of phagocytosis [74], we evaluated the role of PI(4,5)P2 in RalFR localization. As a baseline, we utilized the standard phospholipase C (PLC)-mediated catalyzation of PI(4,5)P2 within the IP3/DAG pathway of host cells [75]. Specifically, in the presence of ionomycin and Ca2+, PI(4,5)P2 is hydrolyzed to inositol 1,4,5-trisphosphate and diacylglycerol via PLC isozymes that regularly deplete the plasma membrane of PI(4,5)P2 following its role as a substrate in many signaling pathways [76]. To test PI(4,5)P2-dependent localization of RalFRt to the plasma membrane, HeLa cells ectopically expressing RalFRtCTD were treated with ionomycin and Ca2+, with the distribution pattern of RalFRtCTD monitored by immunofluorescence. With ionomycin and Ca2+ treatment, RalFRtCTD becomes cytosolic compared to plasma membrane localization in the presence of ionomycin alone (Fig 5A). Upon treatment with EGTA, which chelates Ca2+, PI(4,5)P2 accumulates and RalFRtCTD returns to the plasma membrane. HeLa cells expressing GFP-C1-PLCδ-PH, a biosensor of PI(4,5)P2, were used as a positive control to demonstrate the hydrolysis of PI(4,5)P2 in the presence of ionomycin and Ca2+. Additionally, RalFRbCTD subcellular localization was shown to be unaffected by PI(4,5)P2 hydrolysis, consistent with its perinuclear distribution in host cells. Collectively, these data indicate that PI(4,5)P2 enrichment at the host plasma membrane is a requirement for efficient recruitment of RalFRt, and probably also RalFRf, given its similar subcellular localization pattern.
Fig. 5. PI(4,5)P2 interacts with RalFRt and mediates R. typhi infection. 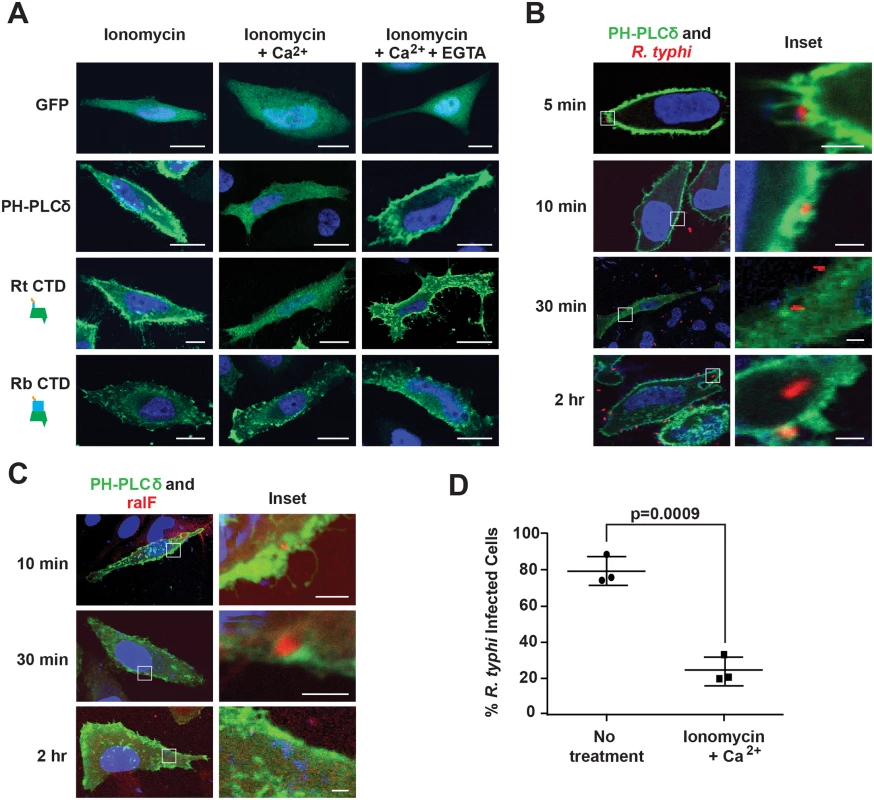
(A) RalFRtCTD co-localizes with PI(4,5)P2. HeLa cells transfected with pEYFP-C1 empty vector, GFP-C1-PLCδ-PH (a PI(4,5)P2 biosensor), EYFP–RalFRtCTD, or EYFP–RalFRbCTD were treated with 5 μM ionomycin alone, with Ca2+, or with Ca2+ and EGTA. Nuclei were stained with DAPI (blue). (Scale bar: 10 μm). (B) PI(4,5)P2 is recruited during R. typhi infection. HeLa cells transfected with GFP-C1-PLCδ-PH (green) were infected with R. typhi (MOI ~100:1) for indicated times. R. typhi was detected with rat anti-R. typhi serum and Alexa Fluor 594 anti-rat antibody (red). Nuclei were stained with DAPI (blue). Boxed regions are enlarged to show detail (inset). (Scale bar: 1 μm). (C) RalF localizes to PI(4,5)P2-enriched regions of the plasma membrane during R. typhi infection. HeLa cells transfected with GFP-C1-PLCδ-PH (green) were infected with R. typhi (MOI ~100:1) for indicated times. RalFRt was detected with rabbit anti-RalFRt and Alexa Fluor 594 anti-rabbit antibodies (red). Nuclei were stained with DAPI (blue). Boxed regions are enlarged to show detail (inset). (Scale bar: 1 μm). (D) Ionomycin and Ca2+ treatment decreases R. typhi infection. HeLa cells treated with 5 μM ionomycin and Ca2+ or no treatment were infected with R. typhi (MOI ~100:1) for 2 hrs. R. typhi was detected with rat anti-R. typhi serum and Alexa Fluor 488 anti-rat antibody. Cell membrane was stained with Alexa Fluor 594 wheat germ agglutinin. The number of infected host cells was counted, with percent infection of three independent experiments (100 host cells counted for each) plotted. Error bars represent mean ± SD (Student’s two-sided t-test). PI(4,5)P2 recruitment is critical for R. typhi invasion of host cells
Phosphatidylinositols enriched at the host plasma membrane often play a critical role in bacterial internalization [74]. Given that RalFRt is expressed early (Fig 1F) and required (Fig 1G) for host invasion, and its localization to the host plasma membrane requires PI(4,5)P2 enrichment (Fig 5A), we sought to determine if PI(4,5)P2 is recruited by RalFRt during R. typhi infection. PI(4,5)P2 localization during R. typhi invasion was analyzed using immunofluorescence microscopy with GFP-C1-PLCδ-PH as a biosensor of PI(4,5)P2 localization. During early infection (i.e. 5 and 10 min post infection), PI(4,5)P2 was highly localized to pseudopodia at the R. typhi entry foci (Fig 5B). As internalization progressed, R. typhi was surrounded by a vacuole with diminished PI(4,5)P2 localization. Once R. typhi detached from the membrane, it was no longer associated with PI(4,5)P2. Furthermore, detection of RalFRt during the infection process revealed co-localization of PI(4,5)P2 and RalFRt during early infection (Fig 5C). In agreement with RalFRt early expression, which diminished at later stages of infection (Fig 1F), PI(4,5)P2 recruitment decreased as infection progressed.
Finally, we evaluated whether or not the recruitment of PI(4,5)P2 to R. typhi entry foci is critical for R. typhi infection. Pretreatment of HeLa cells with ionomycin and Ca2+ to deplete PI(4,5)P2 from the membrane prior to infection resulted in a significant decrease in R. typhi infection (Fig 5D), strengthening the evidence that PI(4,5)P2 is a target molecule involved in RalFRt-associated host cell invasion.
RalFRt interacts with Arf6 but not Arf5
The Arf-GEF activity of RalFL is activated upon membrane binding, with Arf1 the preferred substrate [49,52]. Arf1 is predominantly localized to the Golgi apparatus and plays a role in intra-Golgi transport [77]. Given the association of RalFRt with the plasma membrane, we hypothesized that it might instead recruit Arf6, which is predominantly localized to the plasma membrane where it is involved in endocytosis, endosomal recycling and exocytosis of secretory granules [78–81]. Using immunofluorescence, RalFRtFL was found to recruit Arf6 but neither Arf5 (Fig 6A) nor Arf1 (S9 Fig) to the plasma membrane. Arf5 localizes primarily to the endoplasmic reticulum/Golgi intermediate compartment and the cis-Golgi, where it regulates endoplasmic reticulum to Golgi transport; therefore, Arf5 was used as a negative control [82]. Interestingly, RalFRbFL was similarly found to co-localize with Arf6 but not Arf5 or Arf1 in the perinuclear space.
Fig. 6. Arf6 is recruited by R. typhi RalF and is required for infection. 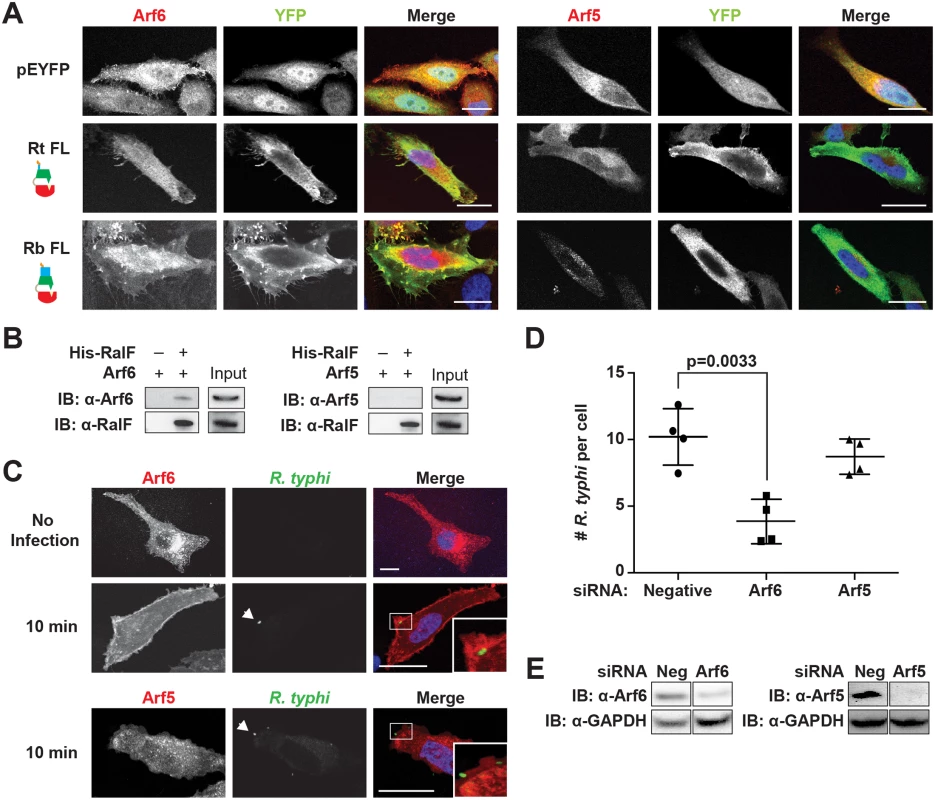
(A) Ectopically expressed RalFRtFL co-localizes with Arf6 but not Arf5. HeLa cells co-expressing EYFP, EYFP-RalFRtFL or EYFP-RalFRbFL and mRFP-Arf6 (left) or -Arf5 (right) were fixed with 4% para-formaldehyde. Nuclei were stained with DAPI (blue). (Scale bar: 10 μm). (B) RalFRtFL pull-down of Arf6. Lysates from HEK293T cells expressing mRFP-Arf5 or -Arf6 were incubated with HisPur Cobalt resin bound with rHis-RalFRtFL or resin alone. Bound proteins were eluted with imidazole and analyzed by protein immunoblot using antibodies as indicated. (C) Arf6 is recruited during R. typhi entry. HeLa cells expressing mRFP-Arf5 or -Arf6 (red) were infected with partially purified R. typhi (MOI ~100). Ten minutes post infection, cells were fixed and R. typhi detected with anti-R. typhi serum (green). DAPI (blue) is shown in the merged image. Boxed regions are enlarged to show detail. White arrowheads indicate R. typhi. (Scale bar: 5 μm). (D) Arf6 knockdown inhibits R. typhi infection. HeLa cells transfected with negative, Arf6, or Arf5 siRNA were infected with partially purified R. typhi (MOI ~100). At 2 hrs post infection, cells were fixed, plasma membrane stained with Alexa Fluor 594 wheat germ agglutinin, and R. typhi detected with rat anti-R. typhi serum and Alexa Fluor 488 anti-rat antibody. The number of R. typhi per host cell was counted for 100 host cells for three independent experiments. Error bars represent mean ± SD (Student’s two-sided t-test). (Scale bar: 5μm). (E) Confirmation of Arf6 and Arf5 knockdown. Arf6 and Arf5 knockdown, 80% and 96% respectively, was confirmed by western blot and densitometry analysis using ImageJ (NIH). To further confirm a RalFRtFL and Arf6 interaction, a protein pull-down assay was performed. Using rHis-RalFRtFL as bait and mRFP-Arf5 or mRFP-Arf6 as the prey, we confirmed that RalFRtFL interacted with Arf6 and not Arf5 (Fig 6B).
Arf6 recruitment is critical for R. typhi invasion of host cells
Activation of Arf6 at the plasma membrane drives the recruitment of phospholipase D and phosphatidylinositol 4-phosphate 5-kinase (PIP5K), which ultimately results in actin remodeling [83,84]. To determine if Arf6 is recruited during R. typhi entry, we used immunofluorescence microscopy to monitor Arf6 localization. As early as 10 min post infection, Arf6 was recruited to the plasma membrane at R. typhi entry foci, while Arf5 remained cytoplasmic (Fig 6C). Given that RalFRt localizes with Arf6 at plasma membrane (Fig 6A) and recruits Arf6 at the R. typhi entry foci (Fig 6C), we predicted that knockdown of Arf6 would decrease R. typhi infection. Indeed, siRNA-mediated Arf6 knockdown significantly decreased the number of R. typhi per cell, while Arf5 knockdown had no significant effect on R. typhi infection(Fig 6D and 6E). These results indicate that RalFRt recruits Arf6 at the plasma membrane during early infection, with spatially regulated Arf-GEF activity required for host cell invasion.
Discussion
Bacteria invading eukaryotic cells employ diverse strategies to subvert the host cellular actin cytoskeleton, allowing for internalization into normally non-phagocytic host cells [85]. For some bacterial species, surface proteins bind host cell receptors and trigger an “outside-in” signaling cascade, which induces cytoskeletal rearrangements and recruits the endocytic machinery to entry foci [86]. Such receptor-mediated induction of bacterial uptake is a strategy employed by Listeria monocytogenes, which utilizes two adhesins (InlA and InlB) to bind host proteins (E-cadherin, receptor gC1qR, proteoglycans) and activate the tyrosine kinase receptor Met [87]. Invasive species of Yersinia also employ two adhesins (invasin and YadA) to bind a subset of β1-integrin host receptors, facilitating invasion that is dependent on signaling from the Rho GTPase Rac1 and activation of the actin nucleating complex Arp2/3 [88,89]. Alternatively, other bacterial species translocate effectors into host cells to initiate actin remodeling and facilitate bacterial uptake. For example, Salmonella typhimurium utilizes its type III secretion system to inject host cells with the effector SopE, which stimulates GDP/GTP nucleotide exchange on Rho GTPases Rac1 and Cdc42, resulting in membrane ruffling and actin cytoskeleton rearrangement [90]. While a receptor-mediated process has been previously characterized for R. conorii invasion of mammalian cells (discussed below), to date no secreted effectors for any Rickettsia species have been characterized for their role in inducing uptake into host cells.
While most species of the order Rickettsiales encode the rvh T4SS [91], effectors have only been identified for some species of the family Anaplasmataceae. For Anaplasma phagocytophilum, rvh effectors are translocated to the mitochondria (Ats-1) and nucleus (AnkA) to inhibit etoposide-induced apoptosis and down-regulate host defense genes, respectively [92–94]. AM185, AM470, AM705 (AnkA), and AM1141 have been identified as putative rvh T4SS effectors of Anaplasma marginale using a heterologous T4SS (L. pneumophila dot/icm), yet none have been characterized for their roles in invasion [95]. Ehrlichia chaffeensis utilizes the rvh T4SS to translocate the effector ECH0825 into host mitochondria, resulting in inhibition of Bax-induced apoptosis [96]. Herein, we identified RalF as the first rvh T4SS effector for species in the family Rickettsiaceae. We provide evidence that R. typhi RalF interacts with RvhD4, the rvh T4SS coupling protein that presumably recognizes effectors and regulates their translocation similar to VirD4 proteins of other P-type T4SSs. Treatment of purified R. typhi with proteinase K degraded surface exposed RalF, providing further evidence that RalF is secreted. Furthermore, using immunofluorescence we show that RalFRt is expressed early during host cell invasion. Because RalF is expressed early during invasion, we hypothesized that RalF is critical for invasion, which was confirmed using antibody pretreatment assays.
Prior studies comparing the subcellular localization of ectopic RalFL and RalFRp determined that, despite strong conservation in the S7D and SCD across these proteins, cryptic signatures within the SCD targeted these proteins to different host membranes [52,53]. RalFRp localization to the plasma membrane, mediated by elevated positively charged residues within the lipid sensor of the SCD, was anticipated to be true for other RalFR proteins, given the strong sequence conservation within the SCD across RalFR homologs. Furthermore, despite extensive variation within the VPR across RalFR proteins, the presence of proline-rich regions in all proteins suggested that this region likely encodes a conserved motif that facilitates interaction with the host cytoskeleton, as was shown for RalFRp [52,53]. Indeed, our co-localization assays confirmed that, for RalF of R. typhi and R. felis, the SCD mediates interaction with the host plasma membrane, with the VPR facilitating interaction with the host cytoskeleton.
In contrast, the perinuclear localization of RalF of R. bellii, reminiscent of the localization of ectopic expressed RalFL proteins at the host secretory network, was unexpected. The VPR of RalFRb is similar in length to VPRs of RalF proteins from R. felis, R. akari and R. australis, with all of these proteins predicted to encode a coiled-coil motif typical of some eukaryotic Arf-GEFs; e.g., EFA6 [71]. While containing an extraordinary number of Pro residues, the VPR of RalFRb nonetheless targets the host cytoskeleton, suggesting that other characteristics of the protein mediate its localization to the cytoplasm. Indeed, in silico analyses revealed three conserved insertions within the SCD of RalFR proteins that are absent from RalFRb. Furthermore, relative to all RalF proteins, the S7D of RalFRb contains an odd insertion as well as a slightly less hydrophobic active site (S2 Fig), the significance of which is unknown. It is tempting to speculate that the perinuclear localization of RalFRb reflects a unique cytosolic lifestyle of R. bellii, an ancestral lineage with a different genomic repertoire relative to other Rickettsia species [51] and the unique ability to grow and survive in several species of amoeba [73]. As R. bellii has been observed invading nuclei of mammalian cells in vitro [73], RalF may play a role in this process, though other Rickettsia species that lack RalF also are known to invade host cell nuclei [97]. Notwithstanding, the SCD-driven perinuclear localization of RalFRb might thus be considered the retention of an ancestral role for RalFR proteins in targeting host vesicular trafficking, similar to RalFL proteins. Collectively, our detailed dissection of the domain requirements for subcellular localization strongly implies differential utilization of Arf-GEF activities for those species of Rickettsia that encode RalF.
RalFRb aside, the subcellular localization of other RalFR proteins to the plasma membrane suggested Arf6 might be their preferred host target, given the predominant localization of Arf6 to the plasma membrane [98] and a previous study showing that RalFRp can catalyze nucleotide exchange on Arf6 [52]. Arf6 activation by some intracellular pathogens (e.g., species of Salmonella, Yersinia and Chlamydia) is known to induce actin remodeling and mediate bacterial invasion via unique pathways. Salmonella enterica activates Arf6 to recruit the Arf-GEF ARNO, which in turn activates Arf1 to enable WASP family veroprolin homolog (WAVE) regulatory complex-dependent actin assembly [99]. Arf6 activation by species of Yersinia and Chlamydia leads to activated PIP5K, which converts PI(4)P to PI(4,5)P2 at the plasma membrane [100,101]. As PI(4,5)P2 enrichment at the host plasma membrane modulates many actin-binding proteins, including α-actinin, talin, vinculin, gelsolin, and the WASP-Arp2/3 complex [102–107], effector-driven accumulation of this phosphatidylinositide can be considered a strategy for induction of phagocytosis.
Given that R. typhi secretes RalF early during host cell invasion, we hypothesized that this Arf-GEF recruits Arf6 to entry foci, precipitating the enrichment of PI(4,5)P2 at the plasma membrane to facilitate bacterial invasion. Indeed, our in vitro and in vivo results confirm that Arf6 co-localizes with RalF and R. typhi at entry foci. Additionally, during R. typhi infection, PI(4,5)P2 noticeably accumulated in the membranes of pseudopodia, with a decreased concentration at the base of the phagocytic cup as internalization progressed. Furthermore, the role of PI(4,5)P2 in bacterial internalization was bolstered by the significant reduction in R. typhi invasion upon PI(4,5)P2 hydrolysis. Thus, an increase in PI(4,5)P2 induced by rickettsial RalF activation of Arf6 is predicted to initiate actin remodeling and ultimately facilitate bacterial invasion (Fig 7). Remarkably, this function for RalFR is markedly different than RalFL, which is utilized by L. pneumophila to recruit Arf1 to the LCV [49]. Unlike species of Legionella, Rickettsia species do not reside in vacuoles but rather lyse the phagosome and replicate within the host cytoplasm. Thus, despite carrying a similar Arf-GEF that is unknown from any other bacteria, different intracellular lifestyles across species of Rickettsia and Legionella have driven divergent roles for RalF during bacterial infection.
Fig. 7. Schematic of R. typhi entry. 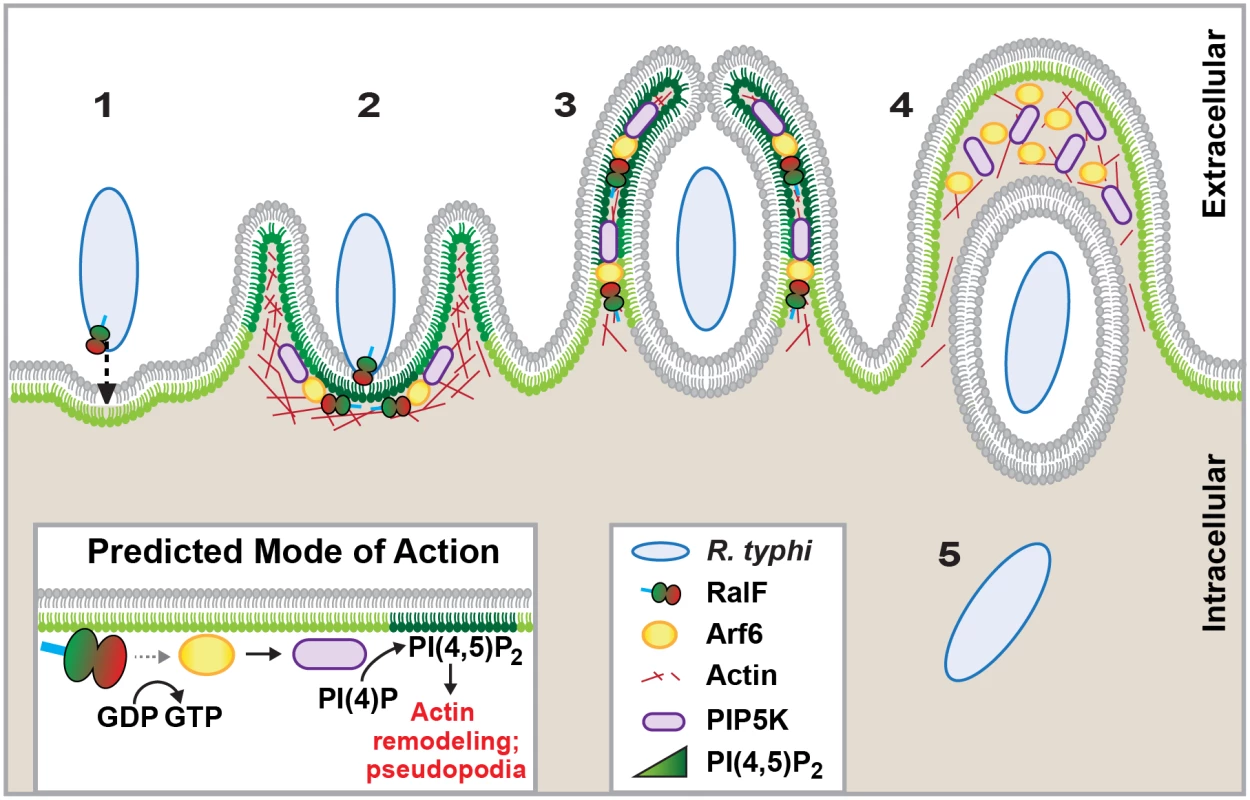
R. typhi entry has been broken down into five conceptual stages: binding (1); extension of pseudopodia (2); membrane fusion and internalization (3); formation of early endosome (4); bacterial escape from endosome (5). Schematic is a representation of micrographs from Figs 5B, 5C and 6C. Inset depicts hypothetical recruitment and activation of PIP5K via RalFRt-activated Arf6, which results in PI(4,5)P2 enrichment and actin rearrangement to facilitate for R. typhi entry. Currently, the predominant knowledge of rickettsia entry and invasion of host cells is based on studies of species from SFG rickettsiae, whereby the surface antigen Sca5 binds host receptor Ku70 to activate a signaling cascade leading to Arp2/3 activation and ultimately actin polymerization, membrane rearrangement and bacterial invasion [26,108,109]. The conservation of Sca5 across all Rickettsia species implies that this receptor-mediated mechanism for entry is likely conserved (Fig 8) [45]. However, depletion of host Rho family GTPases and nucleation-promoting factors that activate Arp2/3 has only a modest effect on rickettsial invasion, suggesting there are other bacterial or host proteins that activate Arp2/3 during infection [108]. Most species of SFG rickettsiae encode an Arp2/3 activating protein, RickA, which could potentially play this role, although its secretion during infection has yet to be demonstrated. Interestingly, genes encoding RickA are absent from species of TG rickettsiae (R. typhi and R. prowazekii); thus, if bacterial Arp2/3 activators are a requirement for invasion, factors other than RickA must be utilized for species of TG rickettsiae. Accordingly, we propose that RalF plays a role in host actin rearrangement and bacterial invasion.
Fig. 8. Model for the variable pathways utilized by divergent Rickettsia species for host cell entry. 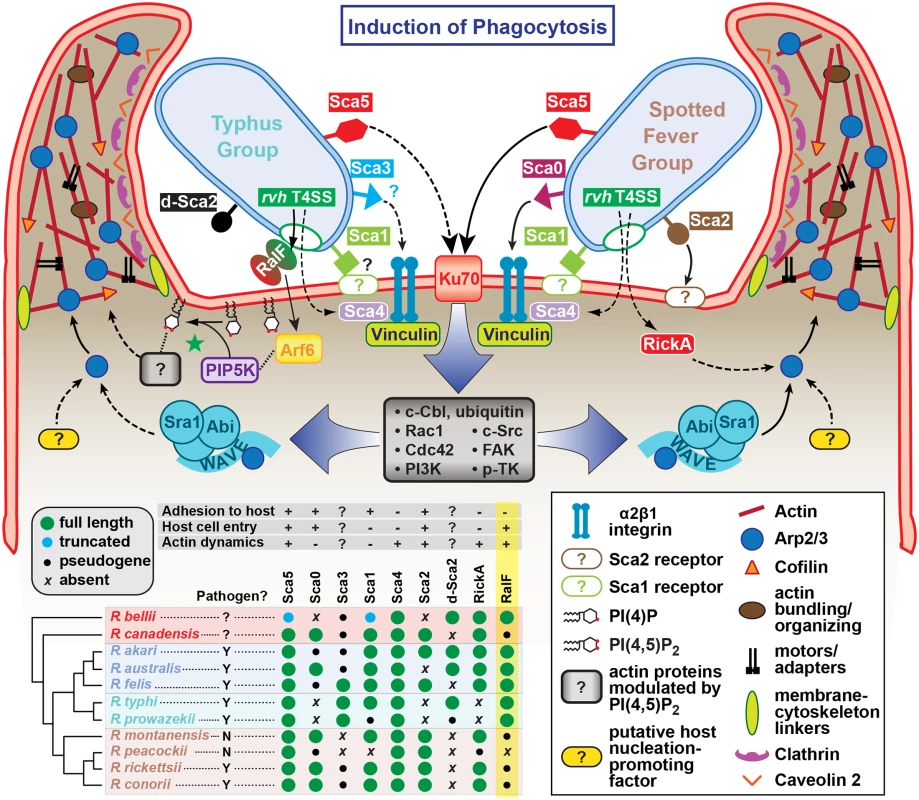
General pathways for Typhus Group (TG, left) and Spotted Fever Group (SFG, right) rickettsiae species are inferred primarily from previous work on SFG rickettsiae species R. conorii [26] and R. parkeri [108] or data from the present study (R. typhi). At center, a conserved proximal hub of the pathway commences with Sca5 binding to host receptor Ku70 [110], which triggers a host-signaling cascade (gray box) involving c-Cbl-mediated ubiquitination of Ku70, Rho-family GTPases Cdc42 and Rac1, phosphoinositide 3-kinase (PI3K) activity, and activation of tyrosine kinases (e.g., c-Src, FAK and p-TK) and their phosphorylated targets. The divergent distal arms of this pathway involve recruitment of factors for activating the actin nucleating complex (Arp2/3), which leads to host actin polymerization, extensive membrane ruffling and filopodia formation, and bacterial internalization in a clathrin and calveolin dependent process. For SFG rickettsiae, the WAVE complex recruits Arp2/3, with its activation via an unknown nucleation promoting factor (either host or bacterial; e.g., RickA). While these processes remain to be characterized for TG rickettsiae, our work suggests that secreted RalF recruits the GTPase Arf6, precipitating an accumulation of PI(4,5)P2 that modulates the activities of a range of actin-associated host proteins (green star). Additional bacterial proteins, some of which are known to facilitate host cell entry, have white lettering with colored boxed backgrounds. Known pathways for protein secretion and host cell receptor-binding, as recently reviewed [45], are shown with solid black lines; all other modeled pathways (shown with dashed lines) are either inferred by homology (e.g., Sca1 of TG rickettsiae as an adhesin based on characterization for Sca1 of R. conorii [36]) or estimated based on in silico analyses (e.g., Sca3 of TG rickettsiae as a putative analog to the α2β1 integrin-binding Sca0 of R. conorii [35]). A phylogenomics analysis across select Rickettsia species (bottom, left) illustrates the genomic variation underlying all of the bacterial components of the models. Adapted from our recent report on the Rickettsia secretome [45]. Red, ancestral group (AG); blue, transitional group (TRG); aquamarine, TG; brown, SFG. Aside from the Sca5-Ku70 interaction and subsequent downstream signaling cascade, other rickettsial adhesins have been characterized for facilitating host cell invasion (Fig 8). However, the lack of conservation of these adhesins (e.g., Sca0 and Sca2) across diverse Rickettsia species implies the existence of multiple mechanisms for rickettsial host cell invasion [45]. It is also probable that each species likely encodes redundancy for factors that facilitate entry, and that some factors may selectively operate for invasion of specific cells (arthropod versus mammalian) throughout the complex rickettsial lifecycle. Thus, it is probable that lineage-specific factors are employed by different species of Rickettsia to successfully invade and colonize diverse eukaryotic cells. Our identification of lineage-specific Arf-GEF utilization across diverse rickettsial species exemplifies this, and illuminates previously unappreciated mechanisms for host cell invasion and infection.
Materials and Methods
Bacterial strains, cell culture, and infection
Vero76 (African green monkey kidney, ATCC: CRL-1587), HEK293T and HeLa (ATCC: CCL-2) cells were maintained in minimal Dulbecco’s Modified Eagle’s Medium (DMEM with 4.5 gram/liter glucose and 480 L-glutamine; Mediatech, Inc.) supplemented with 10% heat inactivated fetal bovine serum (FBS) at 37°C with 5% CO2. R. typhi strain Wilmington (ATCC: VR-144) was propagated in Vero76 cells grown in DMEM supplemented with 5% heat inactivated fetal bovine serum at 34°C with 5% CO2. Rickettsiae were partially purified as previously described [111]. Infections with R. typhi were performed 18–24 hrs post transfection with a multiplicity of infection (MOI) of ~100 : 1. For antibody pretreatment experiments, partially purified R. typhi was incubated with 20 μg Melon Gel IgG (Thermo Scientific) purified rabbit pre-immune serum or anti-RalFRt polyclonal antibody or 20 μg purified rabbit pre-immune serum or anti-RalFRt Fab fragments for 30 min prior to infection. Fab fragments were purified using the Fab Purification Kit (Thermo Scientific) according to manufacture’s protocol.
Recombinant protein purification and antibody production
The expression and purification of recombinant proteins were performed as previously described [111]. Codon optimized (Life Technologies) R. typhi rvhD4 (RT0284) was cloned into pTrcHis2-TOPO vector under the control of the trc promoter (Life Technologies). Full-length R. typhi ralF was cloned into pEXP5-NT/TOPO (Life Technologies) and transformed into E. coli strain bl21-codonplus(de3)-ril (Stratagene). Primers used for cloning can be found in S1 Table. The expression of recombinant proteins was induced with 1 mM IPTG and recombinant proteins were purified by affinity chromatography under native conditions using nickel-nitrilotriacetic acid resin (Ni-NTA) superflow columns (Qiagen) according to manufacturer’s instructions. Polyclonal antibody was generated in rabbit using recombinant RalFRtFL (Alpha Diagnostic Intl. Inc).
Bacterial two-hybrid assay
R. typhi gene sequences (RT0362, GenBank accession no. YP_067323) encoding full-length RalF (RalFRtFL) and the rvh T4SS signal truncation (RalFRtΔT4S) were cloned into the pTRG “prey” plasmid (BacterioMatch II two-hybrid system; Stratagene). A codon optimized (Life Technologies) R. typhi rvhD4 gene (RT0284, YP_067246) was cloned into the pBT “bait” plasmid. Primers used for cloning can be found in S1 Table. The bait (pBT-RvhD4) and prey (pTRG-RalFRtFL or pTRG-RalFRtΔT4S) plasmids (100ng each) were co-transformed into BacterioMatch II reporter electrocompetent cells according to the manufacturer’s instruction (GenePulser Xcell, BioRad). The percent growth of CFUs of reporter cells harboring recombinant plasmids on dual selective screening medium were calculated relative to CFUs obtained on non-selective His dropout medium by a drop plate method for counting CFUs [112].
VirD4 ATPase assay
RvhD4 ATPase activity was monitored using a Quantichrom ATPase/GTPase assay kit (Bioassay Systems), according to the manufacturer’s instructions and as described previously [113]. Briefly, 200–12.5 ng/well of purified recombinant RvhD4 protein was incubated in the presence of 1 mM ATP for 30 min at 37°C. Generated free phosphate was quantified by measuring absorbance at OD 620 nm. All of the samples were measured in triplicate wells, and data are given as averages ± S.D. of three independent experiments.
Protease treatment of R. typhi
R. typhi was purified from heavily infected Vero76 cells. Briefly, infected cells were scrapped into media and spun at 12,000 x g for 10 min at 4°C. Cells were resuspended in ice cold PBS, pH 7.2 containing MgCl2 (PBS-Mg) and sonicated for 10 sec on ice using output 6 of a Sonic Dismembrator (Fisher Scientific). The lysate was filtered through a 5.0 μm filter (Millipore). The filtrate containing R. typhi was layered onto a 20% sucrose cushion at a 1 : 1 ratio and centrifuged at 16,000 x g for 15 min at 4°C to pellet R. typhi. R. typhi was resuspended in PBS-Mg and again purified with a 20% sucrose cushion. Purified R. typhi was treated with 400 μg/mL or 800 μg/mL Proteinase K (Sigma-Aldrich) for 1 hr at room temperature in PBS-Mg buffer as previously described [114]. Following incubation, Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) was added to the reaction, and bacteria centrifuged at 16,000 x g for 10 min at 4°C. R. typhi were washed with PBS-Mg and resuspended in PBS-Mg and NuPAGE LDS sample buffer and reducing reagent (Life Technologies). Lysates were separated on a NuPAGE Bis-Tris SDS-gel (Life Technologies) and immunoblotted with rabbit anti-RalF or anti-EF-Ts as the R. typhi cytoplasmic marker [43,115]. Densitometry was performed using ImageJ (NIH) and RalF intensity was normalized to EF-Ts.
Bioinformatics and phylogenomics analyses
Using RalFRt as a query, BLASTP searches were performed against the NCBI ‘Rickettsia’ database (taxid:780). Full length RalFR homologs were aligned with MUSCLE v3.6 [116] using default parameters. Initial domain characterization of RalFR proteins followed that previously described for R. prowazekii [52]. Using Phyre v.2.0 [69], RalFRt was modeled to the crystal structures of Legionella pneumophila RalF (PDB 1XSZ, 4C7P) [50,53] to confirm the boundaries of the Sec7 domain (S7D) and Sec7-capping domain (SCD). The S7D and SCD of Rickettsia and Legionella RalF homologs were aligned with MUSCLE, superimposing the secondary structure of RalFL over the alignment.
The divergent C-terminal domain (CTD) of RalFR proteins was further described based on distinct characteristics, i.e. a sequence of variable length that includes a Pro-rich tract, as well as a putative secretion signal sequence within the terminal 40 aa. Additional Rickettsia proteins that lack the S7D and SCD, mostly from SFG rickettsiae, were utilized to characterize this region. All full length and partial RalFR homologs were used to assess the synteny of the ralF locus across select Rickettsia genomes. For these genomes, gene neighborhood models were constructed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [117], with manual adjustment to gene annotations. Additional bioinformatics/phylogenomics methodologies are described in S2–S5 Figs.
Mammalian expression plasmids and transfections
Genomic DNA from R. bellii str. OSU 85–1299, R. felis str. Pedreira, R. montanensis str. M5/6, and R. typhi str. Wilmington was purified using DNeasy Blood and Tissue Kit (Qiagen). RalF constructs were amplified as EcoRI/BamHI fragments using primers in S1 Table, with the exception of RalFRbFL, which was cloned using Clontech InFusion technology. Amplicons were cloned into the pGEMT-Easy vector (Promega) and confirmed by sequencing (The Biopolymer/Genomics Core Facility, University of Maryland School of Medicine). Plasmids were digested with EcoRI and BamHI, with ralF fragments subcloned into the pEYFP-C1 vector (Clontech). All plasmids were transformed into Mix & Go Competent Cells—Strain Zymo 5α (Zymo Research). Plasmids pCDNA3-mRFP-Arf1, pCDNA3-mRFP-Arf5 and pCDNA3-mRFP-Arf6 were generous gifts from Prof. Vassilis Koronakis (University of Cambridge, UK). GFP-C1-PLCδ-PH (Addgene plasmid # 21179) was kindly gifted by Tobias Meyer [118]. All plasmids were purified using PerfectPrep EndoFree Plasmid Maxi Kit (5 Prime).
For transfections, HeLa cells seeded in 8-well chamber slides were transfected with 200ng plasmid per well using Xfect (Clontech) and HEK293T cells in T-75 flasks were transfected with 10μg plasmid using Lipofectamine 2000 (Life Technologies) according to manufactures’ protocols.
Immunofluorescence
Twenty-four hours post transfection or at indicated times post infection, cells were fixed with 4% PFA for 10 min at room temperature. Cells were washed three times with PBS and permeabilized in Blocking Buffer (0.2% saponin, 5% FBS in PBS) for 30 min. Primary antibodies mouse anti-PDI (clone RL90, BD Transduction Laboratories, diluted 1 : 200), mouse anti-GM130 (clone 610822, BD Transduction Laboratories, diluted 1 : 200), rat anti–R. typhi serum (1 : 500), rabbit anti-RalFRt (1 : 100), and rabbit anti-GFP (Life Technologies, diluted 1 : 1000) were diluted in blocking buffer and incubated with cells for 1 h. Cells were then washed with PBS and incubated with Alexa Fluor 594 or Alexa Fluor 488 secondary antibodies (Life Technologies) diluted 1 : 2000 in Blocking Buffer for 1 h or 30 min. Finally, cells were washed three times with PBS and mounted using ProLong Gold Anti-Fade mounting media with DAPI (Life Technologies). Actin was stained with Alexa Fluor 594 phalloidin (Life Technologies) and the plasma membrane stained with Alexa Fluor 594 wheat germ agglutinin (WGA, Life Technologies) according to manufacturer’s protocol. For confocal microscopy, cells were viewed under a Zeiss LSM 510 Meta Confocal Microscope (University of Maryland Baltimore Confocal Core Facility). For conventional fluorescence microscopy a Nikon Eclipse E600 fluorescent microscope with a Q Imaging Retiga 2000R camera was used to capture images with QCapture Pro software. Images were processed using ImageJ software (NIH). Co-localization analysis was performed using the CoLoc2 plugin in the ImageJ software program [119]. The Pearson’s correlation coefficient was calculated for 5–10 cells per condition from two independent experiments to measure the strength of association between each RalF protein and the cell organelle (i.e., plasma membrane, endoplasmic reticulum, or Golgi apparatus). Two-sided Student’s t-tests were performed to determine statistical significance for co-localization coefficients compared to control eYFP.
Cell fractionation
Cellular fractionation was completed as previously described [52]. Briefly, at 24 hrs post transfection, HEK293T cells were washed once with PBS and collected in 500 μL homogenization buffer (150 mM KCl, 20 mM HEPES pH 7.4, 2 mM EDTA) containing protease inhibitors and passed 30 times through a 27G-needle. The lysate was centrifuged at 2000 x g for 5 min at 4°C to remove the nuclear fraction. The supernatant was subsequently centrifuged at 100,000 x g for 1 h at 4°C to pellet the membrane fraction. The supernatant was removed (cytoplasmic fraction) and the pellet (membrane fraction) was resuspended in 80 μL of homogenization buffer. Twenty micrograms of cytoplasmic and membrane fractions were separated by SDS-PAGE and blotted with anti-GFP rabbit serum (Life Technologies). Rabbit anti-calnexin [clone ab13505] and mouse anti-GAPDH [clone 6C5] antibodies (Abcam) were used as markers of the membrane and cytosol fractions, respectively.
Ionomycin treatment
Transfected HeLa cells were washed with PBS and incubated in 100 μL of either phosphate buffered saline (PBS) or Krebs-Ringer solution (120 mM NaCl, 4.7 mM KCl, 1.1 mM CaCl2, 0.7 mM MgSO4, 10 mM glucose, 10 mM Na-HEPES, pH 7.4). Ionomycin (Sigma Aldrich) was added to a final concentration of 5 μM and cells were incubated for 10 min. EGTA was added to a final concentration of 2 mM and cells were incubated for 10 min. Cells were then infected with R. typhi (described above) or fixed and stained as described above.
Protein pull-down
RalFRt was cloned into the pTrcHisA vector (Life Technologies, see S1 Table for primer sequences) and transformed into Top10 E. coli cells (Life Technologies). Protein expression was induced with 1 mM IPTG overnight at 30°C. E. coli were lysed using Pierce Lysis Buffer in the presence of HALT Protease Inhibitors (Thermo Scientific) and imidazole added to a final concentration of 10 mM. Lysates were sonicated three times for 20 sec each using setting 6 of a Sonic Dismembranator (Fisher Scientific). mRFP-Arf5 and –Arf6 were expressed in HEK293T cells as described above. HEK293T cells were lysed using Pierce Lysis Buffer in the presence of HALT Protease Inhibitors (Thermo Scientific) and imidazole added to a final concentration of 10 mM.
Pull-down assays were performed using the Pierce Pull-Down PolyHis Protein:Protein Interaction kit according to manufacture’s protocol. Briefly, HisPur Cobalt Resin was incubated with rHis-RalFRt E. coli lysate or buffer alone for 1 hr. The resin was washed 5 times and then incubated with either mRFP-Arf5 or -Arf6 HEK293T lysate for 2 hr. Resin was again washed 5 times and bound proteins eluted with 290 mM imidazole elution buffer. Eluted proteins and 10% of the input protein were analyzed by protein immunoblot using the primary antibodies rabbit anti-RalFRt, rabbit anti-Arf5 (1 : 1000, Thermo Scientific, PA5-31432) and rabbit anti-Arf6 (1 : 1000, Thermo Scientific, PA1-093) and the secondary antibody HRP anti-rabbit IgG (1 : 2000, BioLegend, clone 6B9G9).
Gene knockdown
Negative and MISSION siRNAs against human Arf5 (SASI_Hs01_00024789) and Arf6 (SASI_Hs02_0033275) were obtained from Sigma Aldrich. All siRNA knockdowns were performed in HeLa cells using Lipofectamine 2000 (Life Technologies). Cells were used 24 hrs post transfection. Knockdowns were verified by western blot analysis using 1 : 1000 dilution of primary antibodies rabbit anti-Arf5 or anti-Arf6 (Thermo Scientific). As a loading control, membranes were re-probed with rabbit anti-GAPDH antibody (1 : 1000, Abcam).
Data analysis
Graphs show the mean ± SD of three independent experiments; 100 cells were counted for each condition in every experiment. Statistical analyses were performed using two-tailed equal variance Student’s t-test.
Supporting Information
Zdroje
1. Casadevall A. Evolution of intracellular pathogens. Annu Rev Microbiol. 2008;62 : 19–33. doi: 10.1146/annurev.micro.61.080706.093305 18785836
2. Hybiske K, Stephens RS. Exit strategies of intracellular pathogens. Nat Rev Microbiol. 2008;6 : 99–110. doi: 10.1038/nrmicro1821 18197167
3. Kumar Y, Valdivia RH. Leading a sheltered life: intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe. 2009;5 : 593–601. doi: 10.1016/j.chom.2009.05.014 19527886
4. Creasey EA, Isberg RR. Maintenance of vacuole integrity by bacterial pathogens. Current Opinion in Microbiology. 2014. pp. 46–52. doi: 10.1016/j.mib.2013.11.005
5. Garcia-del Portillo F, Finlay BB. The varied lifestyles of intracellular pathogens within eukaryotic vacuolar compartments. Trends Microbiol. 1995;3 : 373–380. 8564355
6. Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7 : 333–340. doi: 10.1038/nrmicro2112 19369949
7. Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiology Reviews. 2011. pp. 1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x 21517912
8. Mattoo S, Lee YM, Dixon JE. Interactions of bacterial effector proteins with host proteins. Current Opinion in Immunology. 2007. pp. 392–401. 17662586
9. Hicks SW, Galán JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11 : 316–26. doi: 10.1038/nrmicro3009 23588250
10. Galán JE. Common Themes in the Design and Function of Bacterial Effectors. Cell Host and Microbe. 2009. pp. 571–579. doi: 10.1016/j.chom.2009.04.008 19527884
11. Gouin E, Welch MD, Cossart P. Actin-based motility of intracellular pathogens. Current Opinion in Microbiology. 2005. pp. 35–45. 15694855
12. Welch MD, Way M. Arp2/3-mediated actin-based motility: A tail of pathogen abuse. Cell Host and Microbe. 2013. pp. 242–255. doi: 10.1016/j.chom.2013.08.011 24034611
13. Sato H, Frank DW, Hillard CJ, Feix JB, Pankhaniya RR, Moriyama K, et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22 : 2959–2969. 12805211
14. Vanrheenen SM, Luo ZQ, O’Connor T, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (Phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74 : 3597–3606. 16714592
15. Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444 : 567–573. 17136086
16. Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2004/03/24 ed. 2003;1 : 137–149.
17. Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Current Opinion in Microbiology. 2006. pp. 207–217. 16529981
18. Gillespie J, Nordberg E, Azad A, Sobral B. PHYLOGENY AND COMPARATIVE GENOMICS: THE SHIFTING LANDSCAPE IN THE GENOMICS ERA. In: Azad A.F. and Palmer G.H., editor. Intracellular Pathogens II: Rickettsiales. Boston: American Society of Microbiology; 2012. pp. 84–141.
19. Walker TS, Winkler HH. Penetration of cultured mouse fibroblasts (L cells) by Rickettsia prowazeki. Infect Immun. 1978;22 : 200–208. 215542
20. Walker TS. Rickettsial interactions with human endothelial cells in vitro: Adherence and entry. Infect Immun. 1984;44 : 205–210. 6425214
21. Heinzen RA, Hayes SF, Peacock MG, Hackstadt T. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect Immun. 1993;61 : 1926–1935. 8478082
22. Heinzen RA, Grieshaber SS, Van Kirk LS, Devin CJ. Dynamics of actin-based movement by Rickettsia rickettsii in Vero cells. Infect Immun. 1999;67 : 4201–4207. 10417192
23. Van Kirk LS, Hayes SF, Heinzen RA. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect Immun. 2000;68 : 4706–4713. 10899876
24. Uchiyama T, Kawano H, Kusuhara Y. The major outer membrane protein rOmpB of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes Infect. 2006;8 : 801–809. 16500128
25. Chan YGY, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol. 2009;11 : 629–644. doi: 10.1111/j.1462-5822.2008.01279.x 19134120
26. Martinez JJ, Seveau S, Veiga E, Matsuyama S, Cossart P. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123 : 1013–1023. 16360032
27. Renesto P, Samson L, Ogata H, Azza S, Fourquet P, Gorvel JP, et al. Identification of two putative rickettsial adhesins by proteomic analysis. Res Microbiol. 2006;157 : 605–612. 16574381
28. Vellaiswamy M, Kowalczewska M, Merhej V, Nappez C, Vincentelli R, Renesto P, et al. Characterization of rickettsial adhesin Adr2 belonging to a new group of adhesins in ??-proteobacteria. Microb Pathog. 2011;50 : 233–242. doi: 10.1016/j.micpath.2011.01.009 21288480
29. Park H, Lee JH, Gouin E, Cossart P, Izard T. The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem. 2011;286 : 35096–103. doi: 10.1074/jbc.M111.263855 21841197
30. Renesto P, Dehoux P, Gouin E, Touqui L, Cossart P, Raoult D. Identification and characterization of a phospholipase D-superfamily gene in rickettsiae. J Infect Dis. 2003;188 : 1276–1283. 14593584
31. Radulovic S, Troyer JM, Beier MS, Lau AOT, Azad AF. Identification and molecular analysis of the gene encoding Rickettsia typhi hemolysin. Infect Immun. 1999;67 : 6104–6108. 10531273
32. Whitworth T, Popov VL, Yu XJ, Walker DH, Bouyer DH. Expression of the Rickettsia prowazekii pld or tlyC gene in Salmonella enterica serovar typhimurium mediates phagosomal escape. Infect Immun. 2005;73 : 6668–6673. 16177343
33. Rahman MS, Gillespie JJ, Kaur SJ, Sears KT, Ceraul SM, Beier-Sexton M, et al. Rickettsia typhi Possesses Phospholipase A2 Enzymes that Are Involved in Infection of Host Cells. PLoS Pathog. 2013;9. doi: 10.1371/journal.ppat.1003399
34. Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog. 1998;24 : 289–298. 9600861
35. Hillman RD, Baktash YM, Martinez JJ. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with ??2??1 integrin. Cell Microbiol. 2013;15 : 727–741. doi: 10.1111/cmi.12068 23145974
36. Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YGY, Martinez JJ. The Rickettsia conorii autotransporter protein sca1 promotes adherence to nonphagocytic mammalian cells. Infect Immun. 2010;78 : 1895–1904. doi: 10.1128/IAI.01165-09 20176791
37. Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun. 2009;77 : 5272–5280. doi: 10.1128/IAI.00201-09 19805531
38. Cardwell MM, Martinez JJ. Identification and characterization of the mammalian association and actin-nucleating domains in the Rickettsia conorii autotransporter protein, Sca2. Cell Microbiol. 2012;14 : 1485–1495. doi: 10.1111/j.1462-5822.2012.01815.x 22612237
39. Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427 : 457–461. 14749835
40. Jeng RL, Goley ED, D’Alessio JA, Chaga OY, Svitkina TM, Borisy GG, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6 : 761–769. doi: 10.1111/j.1462-5822.2004.00402.x 15236643
41. Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010;78 : 2240–2247. doi: 10.1128/IAI.00100-10 20194597
42. Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12 : 1057–1063. doi: 10.1038/ncb2109 20972427
43. Rahman MS, Ammerman NC, Sears KT, Ceraul SM, Azad AF. Functional characterization of a phospholipase A2 homolog from Rickettsia typhi. J Bacteriol. 2010;192 : 3294–3303. doi: 10.1128/JB.00155-10 20435729
44. Housley N a, Winkler HH, Audia JP. The Rickettsia prowazekii ExoU homologue possesses phospholipase A1 (PLA1), PLA2, and lyso-PLA2 activities and can function in the absence of any eukaryotic cofactors in vitro. J Bacteriol. 2011;193 : 4634–42. doi: 10.1128/JB.00141-11 21764940
45. Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, et al. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev. 2014; 1–44. doi: 10.1111/1574-6976.12084
46. Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8 : 1476–85. doi: 10.1111/j.1600-0854.2007.00634.x 17850229
47. Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 2004;15 : 1487–1505. 14742722
48. Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A. 2005;102 : 826–831. doi: 10.1073/pnas.0406239101 15613486
49. Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295 : 679–682. doi: 10.1126/science.1067025 11809974
50. Amor JC, Swails J, Zhu X, Roy CR, Nagai H, Ingmundson A, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280 : 1392–1400. doi: 10.1074/jbc.M410820200 15520000
51. Gillespie JJ, Williams K, Shukla M, Snyder EE, Nordberg EK, Ceraul SM, et al. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One. 2008;3: e2018. doi: 10.1371/journal.pone.0002018 19194535
52. Alix E, Chesnel L, Bowzard BJ, Tucker AM, Delprato A, Cherfils J, et al. The Capping Domain in RalF Regulates Effector Functions. PLoS Pathog. 2012;8. doi: 10.1371/journal.ppat.1003012
53. Folly-Klan M, Alix E, Stalder D, Ray P, Duarte L V., Delprato A, et al. A Novel Membrane Sensor Controls the Localization and ArfGEF Activity of Bacterial RalF. PLoS Pathog. 2013;9. doi: 10.1371/journal.ppat.1003747
54. Paris S, Béraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, et al. Role of protein-phospholipid interactions in the activation of ARF1 by the guanine nucleotide exchange factor Arno. J Biol Chem. 1997;272 : 22221–22226. doi: 10.1074/jbc.272.35.22221 9268368
55. Bui QT, Golinelli-Cohen MP, Jackson CL. Large Arf1 guanine nucleotide exchange factors: Evolution, domain structure, and roles in membrane trafficking and human disease. Molecular Genetics and Genomics. 2009. pp. 329–350. doi: 10.1007/s00438-009-0473-3
56. Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12 : 362–375. doi: 10.1038/nrm3117 21587297
57. Aizel K, Biou V, Navaza J, Duarte L V., Campanacci V, Cherfils J, et al. Integrated Conformational and Lipid-Sensing Regulation of Endosomal ArfGEF BRAG2. PLoS Biol. 2013;11. doi: 10.1371/journal.pbio.1001652
58. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8 : 785–6. doi: 10.1038/nmeth.1701 21959131
59. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305 : 567–80. doi: 10.1006/jmbi.2000.4315 11152613
60. Berven FS, Flikka K, Jensen HB, Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32: W394–9. 15215418
61. Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, et al. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One. 2009;4: e4833. Available: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19279686 doi: 10.1371/journal.pone.0004833 19279686
62. Gomis-Rüth FX, Solà M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10 : 1551–65. 15134575
63. Llosa M, Gomis-Rüth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45 : 1–8. 12100543
64. Vergunst AC, van Lier MCM, den Dulk-Ras A, Stüve TAG, Ouwehand A, Hooykaas PJJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A. 2005;102 : 832–7. doi: 10.1073/pnas.0406241102 15644442
65. Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49 : 1699–713. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3882298&tool=pmcentrez&rendertype=abstract 12950931
66. Christie PJ, Cascales E. Structural and dynamic properties of bacterial type IV secretion systems (review). Mol Membr Biol. 22 : 51–61. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3921681&tool=pmcentrez&rendertype=abstract 16092524
67. Moncalián G, Cabezón E, Alkorta I, Valle M, Moro F, Valpuesta JM, et al. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J Biol Chem. 1999;274 : 36117–24. Available: http://www.ncbi.nlm.nih.gov/pubmed/10593894 10593894
68. Sears KT, Ceraul SM, Gillespie JJ, Allen ED, Popov VL, Ammerman NC, et al. Surface proteome analysis and characterization of surface cell antigen (Sca) or autotransporter family of Rickettsia typhi. PLoS Pathog. 2012;8: e1002856. doi: 10.1371/journal.ppat.1002856 22912578
69. Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4 : 363–371. doi: 10.1038/nprot.2009.2 19247286
70. Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14 : 231–41. Available: http://www.ncbi.nlm.nih.gov/pubmed/10657980 10657980
71. Derrien V, Couillault C, Franco M, Martineau S, Montcourrier P, Houlgatte R, et al. A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J Cell Sci. 2002;115 : 2867–79. Available: http://www.ncbi.nlm.nih.gov/pubmed/12082148 12082148
72. Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, et al. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. EMBO Press; 1999;18 : 1480–91.
73. Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, et al. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006;2: e76. doi: 10.1371/journal.pgen.0020076 16703114
74. Pizarro-Cerdá J, Kühbacher A, Cossart P. Phosphoinositides and host-pathogen interactions. Biochim Biophys Acta. 2014; doi: 10.1016/j.bbalip.2014.09.011
75. Rhee SG, Bae YS. Regulation of phosphoinositide-specific phospholipase C isozymes. J Biol Chem. 1997;272 : 15045–8. Available: http://www.ncbi.nlm.nih.gov/pubmed/9182519 9182519
76. Várnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium - and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143 : 501–10. 9786958
77. Stearns T, Willingham MC, Botstein D, Kahn RA. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc Natl Acad Sci U S A. 1990;87 : 1238–42. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=53446&tool=pmcentrez&rendertype=abstract 2105501
78. D’Souza-Schorey C, Li G, Colombo MI, Stahl PD. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267 : 1175–8. Available: http://www.ncbi.nlm.nih.gov/pubmed/7855600 7855600
79. D’Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140 : 603–16. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2140168&tool=pmcentrez&rendertype=abstract 9456320
80. Vitale N, Chasserot-Golaz S, Bailly Y, Morinaga N, Frohman MA, Bader M-F. Calcium-regulated exocytosis of dense-core vesicles requires the activation of ADP-ribosylation factor (ARF)6 by ARF nucleotide binding site opener at the plasma membrane. J Cell Biol. 2002;159 : 79–89. 12379803
81. Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139 : 49–61. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2139810&tool=pmcentrez&rendertype=abstract 9314528
82. Chun J, Shapovalova Z, Dejgaard SY, Presley JF, Melançon P. Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol Biol Cell. 2008;19 : 3488–500. doi: 10.1091/mbc.E08-04-0373 18524849
83. Massenburg D, Han JS, Liyanage M, Patton WA, Rhee SG, Moss J, et al. Activation of rat brain phospholipase D by ADP-ribosylation factors 1,5, and 6: separation of ADP-ribosylation factor-dependent and oleate-dependent enzymes. Proc Natl Acad Sci U S A. 1994;91 : 11718–22. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=45303&tool=pmcentrez&rendertype=abstract 7972129
84. Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99 : 521–32. Available: http://www.ncbi.nlm.nih.gov/pubmed/10589680 10589680
85. Stradal TEB, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. Elsevier; 2004;14 : 303–11. doi: 10.1016/j.tcb.2004.04.007
86. Alonso A, García-del Portillo F. Hijacking of eukaryotic functions by intracellular bacterial pathogens. Int Microbiol. 2004;7 : 181–91. Available: http://www.ncbi.nlm.nih.gov/pubmed/15492932 15492932
87. Cossart P, Pizarro-Cerdá J, Lecuit M. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 2003;13 : 23–31. Available: http://www.ncbi.nlm.nih.gov/pubmed/12480337 12480337
88. Alrutz MA, Srivastava A, Wong KW, D’Souza-Schorey C, Tang M, Ch’Ng LE, et al. Efficient uptake of Yersinia pseudotuberculosis via integrin receptors involves a Rac1-Arp 2/3 pathway that bypasses N-WASP function. Mol Microbiol. 2001;42 : 689–703. Available: http://www.ncbi.nlm.nih.gov/pubmed/11722735 11722735
89. Eitel J, Dersch P. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect Immun. 2002;70 : 4880–91. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=128239&tool=pmcentrez&rendertype=abstract 12183532
90. Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galán JE. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93 : 815–26. Available: http://www.ncbi.nlm.nih.gov/pubmed/9630225 9630225
91. Gillespie JJ, Brayton KA, Williams KP, Diaz MAQ, Brown WC, Azad AF, et al. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect Immun. 2010;78 : 1809–23. doi: 10.1128/IAI.01384-09 20176788
92. Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 2010;6: e1000774. doi: 10.1371/journal.ppat.1000774 20174550
93. Lin M, den Dulk-Ras A, Hooykaas PJJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9 : 2644–57. doi: 10.1111/j.1462-5822.2007.00985.x 17587335
94. Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect Immun. 2009;77 : 2385–91. doi: 10.1128/IAI.00023-09 19307214
95. Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, et al. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS One. 2011;6: e27724. doi: 10.1371/journal.pone.0027724 22140462
96. Liu H, Bao W, Lin M, Niu H, Rikihisa Y. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol. 2012;14 : 1037–50. doi: 10.1111/j.1462-5822.2012.01775.x 22348527
97. Schulz F, Horn M. Intranuclear bacteria: inside the cellular control center of eukaryotes. Trends Cell Biol. 2015; doi: 10.1016/j.tcb.2015.01.002
98. Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, et al. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128 : 1003–17. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2120412&tool=pmcentrez&rendertype=abstract 7896867
99. Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc Natl Acad Sci U S A. 2013;110 : 16880–5. doi: 10.1073/pnas.1311680110 24085844
100. Wong K-W, Isberg RR. Arf6 and phosphoinositol-4-phosphate-5-kinase activities permit bypass of the Rac1 requirement for beta1 integrin-mediated bacterial uptake. J Exp Med. 2003;198 : 603–14. 12925676
101. Balañá ME, Niedergang F, Subtil A, Alcover A, Chavrier P, Dautry-Varsat A. ARF6 GTPase controls bacterial invasion by actin remodelling. J Cell Sci. 2005;118 : 2201–10. doi: 10.1242/jcs.02351 15897187
102. Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150 : 1311–20. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2150692&tool=pmcentrez&rendertype=abstract 10995437
103. Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 1996;381 : 531–5. doi: 10.1038/381531a0 8632828
104. Hartwig JH, Kwiatkowski DJ. Actin-binding proteins. Curr Opin Cell Biol. 1991;3 : 87–97. doi: 10.1016/0955-0674(91)90170-4 1854489
105. Johnson RP, Craig SW. Actin activates a cryptic dimerization potential of the vinculin tail domain. J Biol Chem. 2000;275 : 95–105. Available: http://www.ncbi.nlm.nih.gov/pubmed/10617591 10617591
106. Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150 : 1299–310. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2150699&tool=pmcentrez&rendertype=abstract 10995436
107. Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10 : 102–111. doi: 10.1016/S0955-0674(98)80092-6 9484601
108. Reed SCO, Serio AW, Welch MD. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol. 2012;14 : 529–45. doi: 10.1111/j.1462-5822.2011.01739.x 22188208
109. Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci. 2004;117 : 5097–106. doi: 10.1242/jcs.01382 15383620
110. Chan YG-Y, Riley SP, Martinez JJ. Adherence to and invasion of host cells by spotted Fever group rickettsia species. Front Microbiol. 2010;1 : 139. doi: 10.3389/fmicb.2010.00139 21687751
111. Kaur SJ, Sayeedur Rahman M, Ammerman NC, Beier-Sexton M, Ceraul SM, Gillespie JJ, et al. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol. 2012;194 : 4920–4932. doi: 10.1128/JB.00793-12 22773786
112. Herigstad B, Hamilton M, Heersink J. How to optimize the drop plate method for enumerating bacteria. J Microbiol Methods. 2001;44 : 121–129. doi: 10.1016/S0167-7012(00)00241-4 11165341
113. Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I). J Biol Chem. 2011;286 : 27278–87. doi: 10.1074/jbc.M111.256974 21659521
114. El-Hage N, Babb K, Carroll JA, Lindstrom N, Fischer ER, Miller JC, et al. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology. 2001;147 : 821–30. Available: http://www.ncbi.nlm.nih.gov/pubmed/11283278 11283278
115. Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320 : 1651–4. doi: 10.1126/science.1158160 18566289
116. Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32 : 1792–1797. doi: 10.1093/nar/gkh340 15034147
117. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 1999. pp. 29–34. doi: 10.1093/nar/27.1.29 9847135
118. Suh B-C, Inoue T, Meyer T, Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314 : 1454–7. 16990515
119. Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. AJP Cell Physiol. 2011;300: C723–C742. doi: 10.1152/ajpcell.00462.2010
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type GenesČlánek Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion IsolatesČlánek -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated GlycolipidsČlánek Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- The Long and Winding Road (Apologies to the Beatles)
- The Ebola Virus: From Basic Research to a Global Health Crisis
- Riding the R Train into the Cell
- The Two-Phase Emergence of Non Pandemic HIV-1 Group O in Cameroon
- Tumor Progression Locus 2 Promotes Induction of IFNλ, Interferon Stimulated Genes and Antigen-Specific CD8 T Cell Responses and Protects against Influenza Virus
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
- The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type Genes
- Hyperinvasive Meningococci Induce Intra-nuclear Cleavage of the NF-κB Protein p65/RelA by Meningococcal IgA Protease
- Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
- HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation
- Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion Isolates
- Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion
- Inhibiting the Recruitment of PLCγ1 to Kaposi’s Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells
- Goblet Cell Derived RELM-β Recruits CD4 T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation
- HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses
- An siRNA Screen Identifies the U2 snRNP Spliceosome as a Host Restriction Factor for Recombinant Adeno-associated Viruses
- Extracellular Adenosine Protects against Lung Infection by Regulating Pulmonary Neutrophil Recruitment
- : Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
- Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
- Intracellular Uropathogenic . Exploits Host Rab35 for Iron Acquisition and Survival within Urinary Bladder Cells
- A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane
- Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect
- -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- The Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
- A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in
- Double-Edge Sword of Sustained ROCK Activation in Prion Diseases through Neuritogenesis Defects and Prion Accumulation
- The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in and Influences the Kinetics of Growth and Development
- Inhibits Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- Illuminating Targets of Bacterial Secretion
- Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment
- Interdisciplinarity and Infectious Diseases: An Ebola Case Study
- Fungi That Infect Insects: Altering Host Behavior and Beyond
- Plasticity and Redundancy in Proteins Important for Invasion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
- A Novel Virus Causes Scale Drop Disease in
- STAT2 Knockout Syrian Hamsters Support Enhanced Replication and Pathogenicity of Human Adenovirus, Revealing an Important Role of Type I Interferon Response in Viral Control
- Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
- Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative
- AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity
- Interferon-γ Promotes Inflammation and Development of T-Cell Lymphoma in HTLV-1 bZIP Factor Transgenic Mice
- Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie
- Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of . Virulence Pathways
- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors
- Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins the Activation of the MnK/eIF4E Pathway
- Nanoformulations of Rilpivirine for Topical Pericoital and Systemic Coitus-Independent Administration Efficiently Prevent HIV Transmission
- Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection
- sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic
- Evolutionary and Functional Analysis of Old World Primate TRIM5 Reveals the Ancient Emergence of Primate Lentiviruses and Convergent Evolution Targeting a Conserved Capsid Interface
- Hepcidin and Host Defense against Infectious Diseases
- Type I IFN Induction via Poly-ICLC Protects Mice against Cryptococcosis
- Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIV Rectal Challenge
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Illuminating Targets of Bacterial Secretion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



