-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklama: Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
In this work we performed a comprehensive evaluation of the infective potential of Leptomonas seymouri, repeatedly isolated from kala-azar patients infected by Leishmania donovani in India and neighboring countries, and have tested the capacity of this monoxenous trypanosomatid to utilize the sand fly vectors permissive for Leishmania donovani. We concluded that despite several genetic adaptations it has developed, Leptomonas seymouri remains a predominantly monoxenous species not able to infect mammalian macrophages either alone or in co-infection with Leishmania. Under certain circumstances it is able to infect mammals, but probably only when the host is immunocompromised by infection with another pathogen, such as Leishmania donovani or HIV.
Published in the journal: . PLoS Pathog 11(8): e32767. doi:10.1371/journal.ppat.1005127
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005127Summary
In this work we performed a comprehensive evaluation of the infective potential of Leptomonas seymouri, repeatedly isolated from kala-azar patients infected by Leishmania donovani in India and neighboring countries, and have tested the capacity of this monoxenous trypanosomatid to utilize the sand fly vectors permissive for Leishmania donovani. We concluded that despite several genetic adaptations it has developed, Leptomonas seymouri remains a predominantly monoxenous species not able to infect mammalian macrophages either alone or in co-infection with Leishmania. Under certain circumstances it is able to infect mammals, but probably only when the host is immunocompromised by infection with another pathogen, such as Leishmania donovani or HIV.
Introduction
Flagellates of the family Trypanosomatidae are single-celled obligatory parasites. They can be either dixenous (i.e. those with two hosts in their life cycle—Trypanosoma, Leishmania, and Phytomonas spp.) or monoxenous (i.e. those having only one host). For decades, monoxenous trypanosomatids of insects were effectively neglected. However, this situation is rapidly changing, as a remarkable diversity of these flagellates is being revealed within insects—a group which is known to be extraordinarily species rich [1,2]. In addition, the study of these parasites is expected to shed light on the origin of the dixenous life cycle (alternation of an insect vector and a vertebrate or plant host). It is generally accepted that the dixenous species have evolved from their monoxenous kins and that this transition has happened independently at least three times during the evolution of Trypanosomatidae, as the dixenous genera Trypanosoma, Leishmania, and Phytomonas are interspersed by the monoxenous genera Angomonas, Blastocrithidia, Blechomonas, Crithidia, Herpetomonas, Kentomonas, Leptomonas, Paratrypanosoma, Sergeia, Strigomonas, and Wallacemonas (S1 Fig) [3,4]. This suggests that some (presumably) monoxenous species may occasionally try switching to dixeny. Indeed, the presence of the monoxenous trypanosomatids in vertebrates has been noted already about 100 years ago [5]. More recently, several monoxenous flagellates belonging to the genera Herpetomonas, Crithidia, Leptomonas, and Blechomonas have been identified from human clinical isolates [6–8]. Importantly, most of them involved immuno-compromised individuals, leading to a hypothesis that these usually non-infectious species may explore new ecological niches in vertebrates that have their immune system suppressed [9,10]. Within this paradigm, about two dozen cases of monoxenous trypanosomatids co-infecting humans along with various Leishmania spp. have been reported almost exclusively from the Indian subcontinent. Most of them implicated causative agents of visceral leishmaniasis (kala-azar) of the L. donovani complex [11]. It was also demonstrated that both dixenous and monoxenous flagellates may be transmitted by the same Phlebotomus vector, yet the evidence is not very strong [12,13]. The cytochrome b and 18S rRNA-based PCR analyses were confined to the isolates from a small geographical area and the identity of non-Leishmania parasites could not be elucidated to the species level.
The species most often recovered from co-infections in leishmaniasis patients is Leptomonas seymouri Wallace, 1959 [14]. Together with all Leishmania spp. it belongs to the subfamily Leishmaniinae (S1 Fig) [15] and was originally isolated from a cotton stainer Dysdercus suturellus (Hemiptera: Pyrrhocoridae) [16]. Nonetheless, when a broad-scale survey of trypanosomatids parasitizing pyrrhocorids throughout the world was undertaken, none of the samples proved to contain L. seymouri [17]. So the question remains whether the original isolate was obtained from a specific host (e.g. species that is evolutionary adapted for parasite's life cycle). L. seymouri can even multiply in plants under experimental conditions [18] proving it to be non-fastidious and able to adapt to quite different environments.
Recent whole-genome analysis of kala-azar clinical isolates from splenic aspirates demonstrated heavy "contamination" with unidentified Leptomonas sp. [19]. This result is not so surprising provided that both parasites are almost indistinguishable by morphology and that Leptomonas outgrows Leishmania in culture [20].
We speculate that several species of monoxenous trypanosomatids are capable of surviving in the hostile environment of the vertebrate body. Molecular details of such adaptation are not worked out, yet it is clear that some monoxenous trypanosomatids must be able to tolerate heat shock up to the temperatures they might experience in warm-blooded vertebrates. Indeed, a number of representatives of the genera Crithidia and Herpetomonas can withstand elevated temperature reaching 37°C [21–23].
In this study we addressed the issue of Leishmania–Leptomonas co-infection from the point of view of the monoxenous partner. To understand molecular mechanisms and biochemical pathways responsible for survival within warm-blooded vertebrates, we have demonstrated that Leptomonas seymouri can withstand elevated temperatures in vitro, sequenced its genome, and assessed transcriptional profiles of cells cultivated in different conditions. Furthermore, we tested L. seymouri ability to survive in Phlebotomus argentipes and P. orientalis, two sand fly species implicated in Leishmania donovani transmission.
Results
Identification of Leptomonas seymouri in clinical kala-azar isolates
Whole genome sequencing of two clinical Indian kala-azar field isolates, a strain resistant to sodium antimony gluconate therapy (Ld 39, May 2000, Muzaffarpur, Bihar) and a strain sensitive to treatment (Ld 2001, February 2000, Balia, Uttar Pradesh), revealed numerous (over 95%) sequences apparently derived from Leptomonas sp. in addition to those of L. donovani [19]. These isolates were cultivated from splenic aspirates in frame of a large screen aimed to understand molecular differences between confirmed kala-azar cases. For precise identification of the co-infecting species we applied an arsenal of molecular tools developed over the years [24–27]. Three genetic loci, namely 18S rRNA, glycosomal glyceraldehyde-3-phosphate dehydrogenase (gGAPDH), and ITS regions were amplified, sequenced and compared with other representatives of the subfamily Leishmaniinae [15]. 18S rRNA sequences of the isolates Ld 39 and Ld 2001 (GenBank accession numbers KP717894 and KP717895, respectively) were identical and indistinguishable from the corresponding sequence of L. seymouri (GenBank accession number AF153040). gGAPDH sequences (GenBank accession numbers KP717896 and KP717897 for isolates Ld 39 and Ld 2001, respectively) were nearly identical with only 1 nt substitution in the coding sequence. They both were very similar (except for the degenerative primer sequences) to the gGAPDH sequence of L. seymouri (GenBank accession number AF047495). 18S rRNA and gGAPDH sequences are informative for higher level taxonomy, and are usually adequate for the genus (and up) level ranking [4,28]. For proper species identification we used other well-established markers, ITS1 and ITS2 [14,20,29]. Their sequences were identical with the exception of a 2 nt-long indel (GenBank accession numbers KP717898 and KP717899 for isolates Ld 39 and Ld 2001, respectively). BLAST search revealed 100% identity with the ITS1-5.8S rRNA region of L. seymouri (GenBank accession number JN848802).
The data presented above allowed us to conclude that the monoxenous co-infectant of the clinical kala-azar isolates Ld 39 and Ld 2001 is L. seymouri. We also would like to note that the cases of co-infections of Leishmania and Leptomonas are likely underreported in the literature, as several sequences attributed to L. donovani in GenBank do in fact belong to L. seymouri. Our analysis of the ITS-containing region, SL, gGAPDH, HSP70, HSP83, RNA polymerase II, α-tubulin and some mitochondrial genes (A6, cytb, COI, COII, COIII, NADH) revealed that 38 out of 170 (22%) and 3 out of 217 (1.4%) ITS sequences of L. seymouri were misidentified as Leishmania donovani and L. tropica, respectively (see S1 Table for GenBank accession numbers).
Leptomonas seymouri withstands elevated temperature typically associated with vertebrate infection
The presence of monoxenous L. seymouri in co-infections with dixenous L. donovani implies several adaptations to the environment of the human body. One of the important factors to be considered is temperature. Typical monoxenous trypanosomatids of the insect gut are temperature-sensitive and cannot withstand conditions of the warm-blooded vertebrates [6]. In order to investigate temperature resistance of several trypanosomatid species in vitro, we compared growth kinetics of two different Leptomonas species, L. seymouri ATCC 30220 (hereafter used as a proxy of filed isolated Ld 39 and Ld 2001, which were not available) and L. pyrrhocoris H10, under different experimental conditions. Parasites were incubated at temperatures 23°C, 29°C, and 35°C for up to 7 days. The highest temperature (35°C) approximately corresponds to that faced by the flagellates upon transfer from a sand fly into a vertebrate. To imitate the conditions of insect gut and vertebrate blood, SDM and two-phased blood-agar were used, respectively. No considerable difference was observed in growth kinetics of two trypanosomatid species incubated at 23°C in both media. Interestingly, increasing the cultivation temperature to 29°C and 35°C inhibited growth of L. pyrrhocoris, while growth of L. seymouri was not significantly affected (Fig 1). We concluded that L. seymouri is capable of withstanding the elevated temperature reaching that of the human body. In contrast, L. pyrrhocoris is temperature-sensitive and halts its cell division in non-optimal conditions. In all cases, cultivation on blood-agar medium resulted in higher cells density.
Fig. 1. Growth kinetics of Leptomonas pyrrhocoris and L. seymouri at 23°C, 29°C, and 35°C in SDM and blood-agar media. 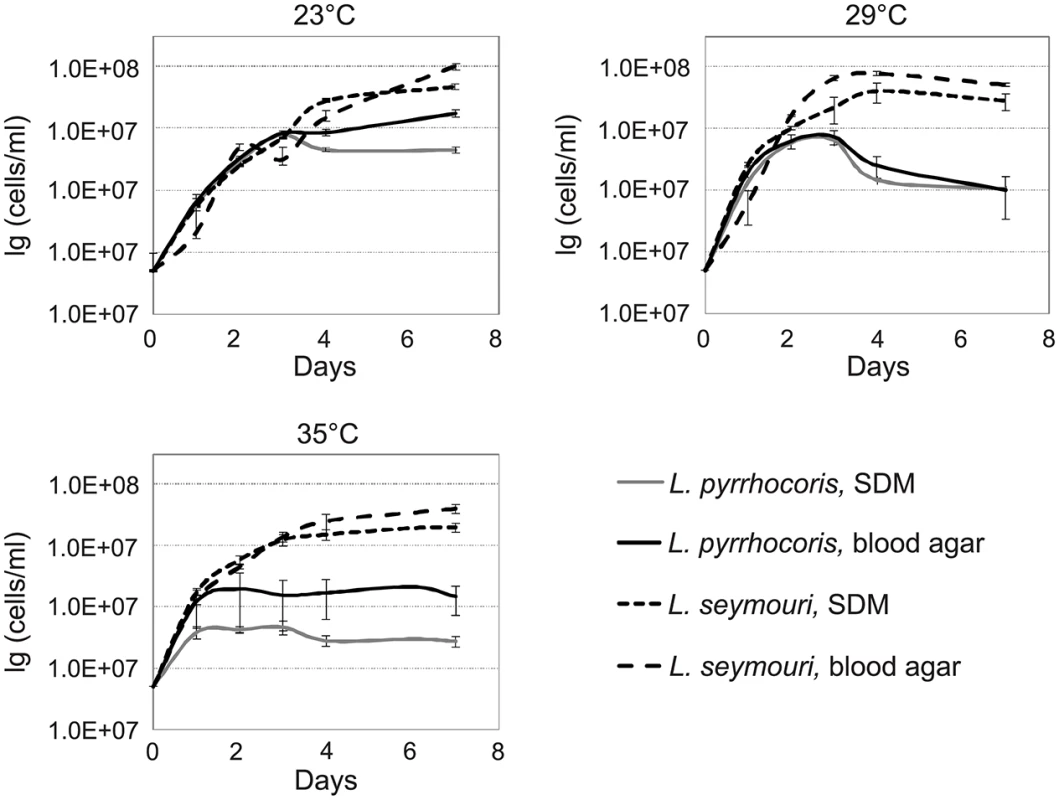
Data from 3 independent biological replicates are presented. Light microscopy of Giemsa stained smears of L. seymouri cultivated under different experimental conditions revealed statistically significant morphological changes (Fig 2). The most noticeable one was shortening of the free portion of the flagellum observed in cells cultivated at high temperature. This phenomenon was observed for both media used but it was more pronounced in blood-agar. Also elevated temperature resulted in more diverse body sizes and shapes with the most conspicuous feature being elongated and tapered posterior end of some cells.
Fig. 2. Morphology of Leptomonas seymouri cells cultivated in vitro in SDM and blood-agar media at low (23°C) and high (35°C) temperature. 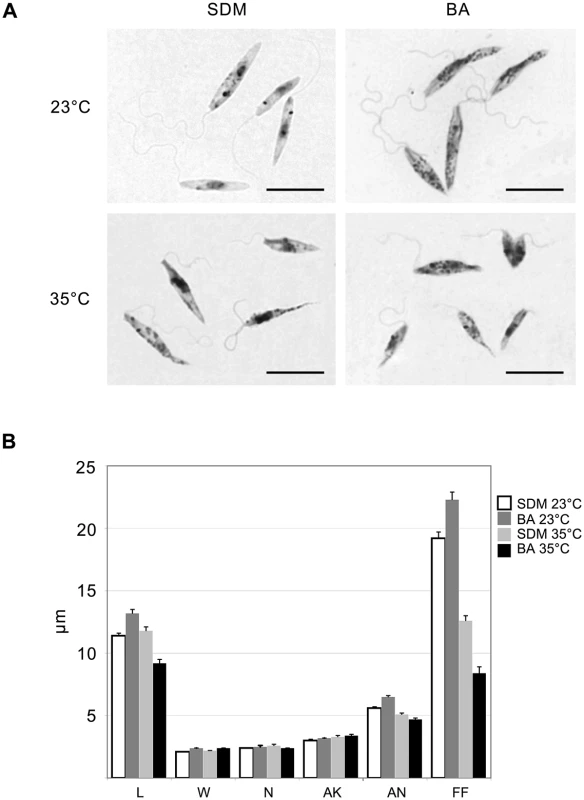
A, Giemsa-stained slides, scale bar is 10 μm. B, ANOVA statistical analysis of 100 cells, average and standard error are presented in micrometers (μm). L–length, W–width, N–length of the nucleus, AK–distance between the kinetoplast and anterior end of the cell, AN–distance between the nucleus and anterior end of the cell, FF–free flagellum length. Genome of Leptomonas seymouri
The genome of L. seymouri ATCC 30220 was assembled into 1,222 scaffolds (maximum length 326,845 bp) with N50 of 70,646 bp and a total assembly length of approximately 27.3 Mbp. This is a substantial improvement over the previously reported assembly of the unidentified Leptomonas sp. (14,518 contigs with maximum length of 26,366 and N50 of 3,370 bp) [19]. Both assemblies had almost the same total genome length (27.3 and 27.4 Mb). Importantly, over 85% of the reads could be cross-mapped (length fraction = 0.9; similarity fraction = 0.9) confirming identity of the L. seymouri isolates. The number of annotated protein-coding genes, 8,488, was also within the range of previously reported genomes (6,451 for Phytomonas sp. HART1; 8,309 for Leishmania major; 10,109 for Trypanosoma brucei) [30–32]. Consistent with other trypanosomatids, the protein-coding genes lack conventional introns. The only exceptions reported so far in Trypanosoma spp. and Leishmania spp. are poly(A) polymerase and DEAD/H RNA helicase [32,33]. Indeed, their L. seymouri orthologs also contain introns and thus require cis-splicing for proper expression.
A typical aspect of the L. seymouri genome is that it contains a relatively small number of genes that have undergone tandemly linked duplications. Using a cutoff value of 10−50, the number of genes present in two or more homologous copies has been estimated at about 9.9% in L. seymouri. Same numbers for Phytomonas sp., L. major, T. brucei, and C. fasciculata are 9.6%, 18.3%, 26.0%, and 40.2%, respectively. This is one of the major components determining differences in genome size among these species.
Metabolic pathways in L. seymouri
Genomic information was used to predict the metabolic pathways in L. pyrrhocoris and L. seymouri, two phylogenetic kins with different sensitivity to temperature and ability to co-infect vertebrate hosts (S2 Fig). In essence, the metabolism in these two species is very similar, with important features and differences highlighted below. A classical glycolytic pathway, partly inside glycosomes (as inferred from the presence of peroxisome targeting signals), is responsible for the metabolism of various exogenous sugars (S2 Table). Carbohydrate metabolism is characterized by an incomplete aerobic oxidation because one of the classical mitochondrial tricarboxylic acids (TCA) cycle enzymes (NAD-linked isocitrate dehydrogenase) is absent. However, the other TCA cycle enzymes can be used for the inter-conversion of metabolic building blocks required for gluconeogenesis and other biosynthetic purposes (S3 Table). While both L. pyrrhocoris and L. seymouri are able to synthesize their own pyrimidines, they depend on a supply of external purines. They lack the capacity to oxidize aromatic amino acids and require an external supply of most of the essential amino acids, cofactors and vitamins for growth (S4 Table). Both Leptomonas spp. have a fully developed mitochondrion with 9 of the 10 TCA cycle enzymes present, a complete respiratory chain with the respiratory complexes I—IV, and a fully functional mitochondrial F1-ATPase (S5 Table).
Although lacking the alternative oxidase found in many other trypanosomatids, L. seymouri possesses an alternative NADH dehydrogenase gene. Our analysis predicts that it is able to feed on a large variety of polysaccharides, carbohydrates, both hexoses and pentoses, with the anticipated end products of carbohydrate metabolism being acetate, succinate, carbon dioxide, ethanol, alanine, and D-lactate. L. seymouri has a complete set of β-oxidation enzymes, which are associated with the mitochondrion. A few additional lipid-metabolizing enzymes are present in the glycosomes. It appears that the analyzed flagellate does not possess a type-I system of fatty acid synthesis, but makes its fatty acids in the cytosol by the action of a series of elongases (S6 Table). It is able to oxidize 16 of the 20 amino acids, but the necessary enzymes for the metabolism of lysine and the three aromatic amino acids (phenylalanine, tyrosine and tryptophan) are lacking. The urea cycle is not functional since two mitochondrial enzymes of the cycle are missing (S7 Table). The remaining three cytosolic enzymes have all been acquired by lateral gene transfer and allow arginine to be utilized in polyamine biosynthesis. Surface proteins, previously identified in Trypanosoma, Leishmania and Crithidia spp., have also been found in Leptomonas (S8 Table). Homologues of GP63, amastin, 3’-nucleotidase, integral membrane protein, prohibitin, membrane-bound acid phosphatases MBPA1 and MBPA2 and tartrate-sensitive acid phosphatase, but not oligosaccharyl transferase, are present. Protection against oxidative stress in monoxenous trypanosomatids differs from their dixenous kins. In addition to the trypanothione system and the presence of many homologues of tryparedoxins and peroxiredoxins, all monoxenous species analyzed thus far have a bacterial-type catalase acquired by lateral gene transfer (S9 Table).
Enzymes of the RNA interference pathway, namely the homologs of the Argonaute (AGO1) and the two dicer proteins (DCL1 and DCL2) were not detected in L. seymouri (S10 Table). Importantly, they were found in the genome of L. pyrrhocoris arguing that these two closely related species differ in their ability to regulate gene expression by RNA interference.
Lateral gene transfer
In the evolution of Trypanosomatidae many events of lateral gene transfer (LGT) have taken place, since genes of bacterial origin are frequently encountered in all trypanosomatid lineages [34]. This suggests that an ancestral flagellate had already acquired such genes, which include a number of enzymes of glycolysis, pentose-phosphate shunt and pyrimidine biosynthesis, as well as trypanothione reductase and pterin transporters [35–37]. Some LGT events including genes involved in sucrose and pentose sugar metabolism, haem synthesis and urea cycle seem to be more recent and specific to the Leishmaniinae clade that comprises Leishmania, Crithidia and Leptomonas spp. [38–40]. Even more recent acquisitions, shared only among Crithidia spp. and Leptomonas spp. include catalase, the diaminopimelate-metabolizing enzymes and those of β-glucosidase, nitroalkane oxidase, phenolic acid dehydrogenase and glycerol dehydrogenase families (S11 Table). In total, 70 out of 586, or 12% of all the metabolic genes analyzed, have resulted from the events of lateral transfer.
Gene family analysis using the OrthoMCL approach
For this analysis full proteomes for 23 trypanosomatid species were downloaded from TriTrypDB v. 7.0 and combined with newly annotated proteins from L. seymouri, L. pyrrhocoris, B. ayalai and Paratrypanosoma confusum (S12 Table). Comprehensive characterization of L. seymouri gene family repertoire and its comparison to that of other trypanosomatids may help to shed light on possible adaptations of this species to the dixenous lifestyle. Recently, a comparative genomics approach was used to define a "gene kit" implicated in cell invasion and intracellular parasitism in Leishmania spp. and Trypanosoma cruzi [41]. Authors have found that despite substantial differences in mechanisms of host cell invasion and survival within the host cell, 3,340 orthologous gene clusters are exclusively shared between intracellular parasites when compared to extracellular T. brucei. Many proteins within these clusters were already proven to play a pivotal role in Leishmania and Trypanosoma virulence (e.g. GP63, amastin, ascorbate peroxidase), while functions of other proteins require further detailed investigation.
In our study we were aiming to identify candidate proteins in L. seymouri that may define its ability to occasionally infect warm-blooded organisms. For that purpose Orthologous Groups (OG) presence/absence patterns in L. seymouri were analyzed and compared to those of other trypanosomatids. In the reference dataset for OrthoMCL analysis several Leishmania spp. (medically and veterinary important dixenous species), along with C. fasciculata and L. pyrrhocoris (both never encountered in vertebrates) are of primary interest for comparison with L. seymouri. According to a widely accepted view of trypanosomatid phylogeny, Leptomonas spp. are most closely related to Crithidia spp., and together they form a clade that clusters as a sister group to the genus Leishmania [2,15] (S2 Fig). Firstly, OG content was compared in L. seymouri, C. fasciculata, and L. pyrrhocoris in order to exclude from the analysis OGs that are present in typical monoxenous trypanosomatids. Leptomonas pyrrhocoris has a typical promastigote morphology and dwells in insect species of the family Pyrrhocoridae [17,42], while C. fasciculata uses various culicids as hosts [43–45]. Notably, some representatives of the genus Crithidia (C. hutneri, C. luciliae thermophila) can survive at temperatures of the mammalian and avian bodies [21,22]. Therefore C. fasciculata may possess genes involved in survival at elevated temperatures, and in order to exclude possible biases caused by the presence of C. fasciculata genes in our OrthoMCL analysis, OG repertoire comparisons were performed twice: with and without C. fasciculata in the datasets being compared.
Out of 7,935 L. seymouri OGs, 79 OGs were absent in L. pyrrhocoris, and 26 OGs were absent from both L. pyrrhocoris and C. fasciculata (S13 Table and Fig 3). Our assumption is that among the genes belonging to the above-mentioned groups there are at least several that predispose L. seymouri metabolism to dixeny. Fifty five out of 79 OGs absent in L. pyrrhocoris do not have any functional annotation assigned and thus represent a broad field for further studies (S13 Table). Nevertheless, several genes identified by comparative genomics approach in our study were already proven to play a pivotal role in parasite survival and virulence (see below).
Fig. 3. Orthologous group presence (denoted by "+")/absence (denoted by "-") patterns for Leptomonas seymouri, Leptomonas pyrrhocoris, Crithidia fasciculata, and several Leishmania species. 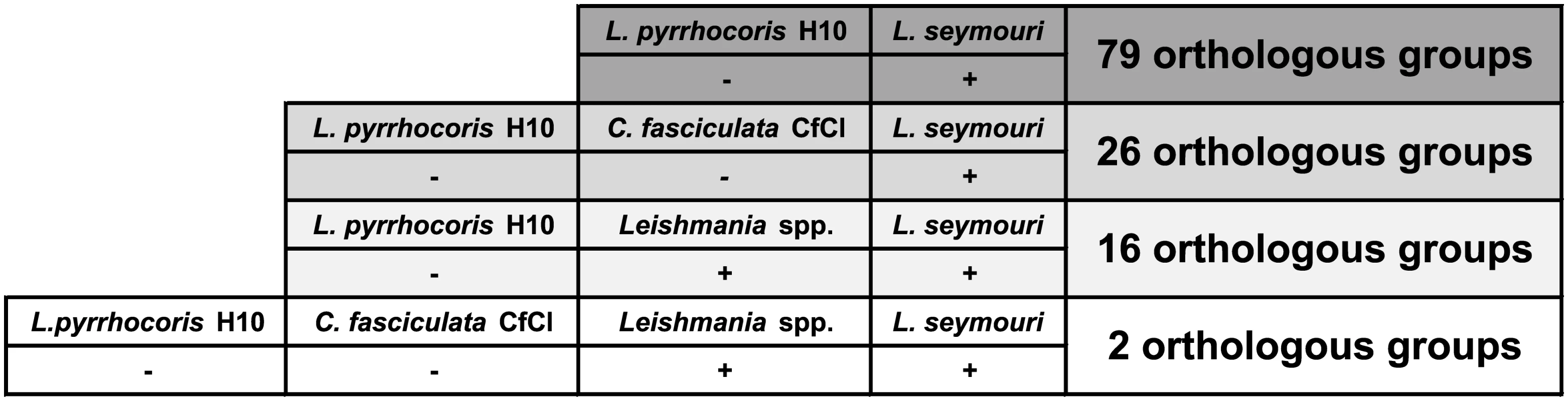
The number of present OGs is indicated on the right. In order to further narrow down the set of such genes we introduced one more condition into the comparison: gene family present in L. seymouri must be also present in all Leishmania species considered in the analysis (L. braziliensis MHOM/BR/75/M2903, L. braziliensis MHOM/BR/75/M2904, L. donovani BPK282A1, L. infantum MCAN/ES/98/JPCM5, L. major MHOM/IL/80/Friedlin, and L. mexicana MHOM/GT/2001/U1103). A reptile parasite L. tarentolae ParrotTarII was not included in the analysis due to its inability to infect warm-blooded organisms [46]. Additional BLASTP search (E-value ≤ 10−10) for proteins belonging to OGs and meeting the criteria stated above was performed in order to determine whether these OGs have related OGs with homologous proteins clustered separately by the sensitive OrthoMCL algorithm. Cases when related OGs have a presence/absence pattern which violates the abovementioned criteria are not discussed here since unambiguous conclusion cannot be made concerning the role of such proteins in L. seymouri thermotolerance.
Sixteen OGs absent from L. pyrrhocoris are shared by L. seymouri and Leishmania spp. Importantly, only 2 of them are absent from both L. pyrrhocoris and C. fasciculata (Fig 3). These two OGs represent a kinase-like protein and a ubiquinol-cytochrome c reductase-like protein. According to the results of additional BLASTP search, the latter protein OG does not have any related OGs and all of its orthologs in the TriTrypDB are annotated as ubiquinol-cytochrome c reductase-like proteins. Aiming to identify homologs of this protein in other species beyond the TriTrypDB, we conducted a BLAST search against the NCBI nr database and found a close homolog only in Strigomonas culicis (ubiquinol-cytochrome c reductase subunit 6, E-value ≤ 10−30, protein accession number: EPY16273.1). The kinase-like protein mentioned above has weak hits with E-value over 10−30 to several other OGs containing protein kinases. Due to the relatively high E-values of the BLAST hits and quite unspecific annotations of kinases within related OGs, this protein was not excluded from our analysis (although several related OGs have absence/presence patterns that differ from the required ones), and its possible role in L. seymouri thermotolerance cannot be ruled out.
Having excluded the requirement of OG being absent from C. fasciculata, the overlap mentioned above extends to 16 OGs (Fig 3), which include one more group with putative protein kinases as well as putative anaphase-promoting complex subunit, putative epsin and several hypothetical proteins with unknown functions. In order to obtain a global picture of corresponding OG distribution for subunits of the anaphase-promoting complex and putative epsin, we extended analysis of OG presence/absence patterns to the whole dataset of 27 trypanosomatid species. As expected, OGs containing these proteins have shown nearly omnipresent distribution (being absent from L. pyrrhocoris as required in our analysis and additionally missing in several Trypanosoma spp.). Additional BLAST search (with more relaxed parameters) for these proteins against proteins belonging to other OGs also did not return any hits. Such results can be explained assuming considerable sequence diversity in these proteins families. For epsins, a group of eukaryotic proteins broadly implicated in clathrin-mediated endocytosis, there is evidence for substantial sequence dissimilarities and lineage-specific protein architecture [47]. Anaphase-promoting complex is a multi-subunit E3 ubiquitin ligase that is necessary for proteolytic degradation of crucial cell cycle regulators, which causes segregation of sister chromatids [48]. Taking into account a universal role of the proteins mentioned above and their phyletic patterns (especially their presence in several monoxenous species) we conclude that they are unlikely to be involved in L. seymouri thermotolerance. Interestingly, 3 OGs containing hypothetical proteins (OG_09193, OG_10013, and OG_10042) within the group of 16 OGs fully satisfy the conditions applied in the study, including the absence of closely related OGs. Moreover, these groups of homologous proteins do not occur in any Trypanosoma spp. and in monoxenous trypanosomatids for which genome sequences are available (except for C. fasciculata). Proteins within these groups represent primary targets for additional studies aiming to reveal mechanisms contributing to L. seymouri thermotolerance.
Leptomonas seymouri harbors dsRNA viruses
Prompted by our observation that L. seymouri lacks RNAi machinery (see above) and by patterns of RNAi retention in Trypanosomatidae [49], we also examined L. seymouri for the presence of dsRNA viruses. Two complementary methods, the nuclease digestion assay and immunofluorescence microscopy, were used [50]. Indeed, the anti-dsRNA antibodies detected small sharp dots, which are reminiscent of those found in the virus-positive isolate of Leishmania guyanensis [51]. Importantly, these putative viral particles did not co-localize with the mitochondrion (Fig 4A). The nuclease digestion assay of L. seymouri RNA was performed in parallel with the virus-free Blechomonas pulexsimulantis used as a negative control [52]. It detected dsRNA bands resistant to DNase I and S1 nuclease, which were present in RNA preparations from L. seymouri (Fig 4B). Interestingly, this dsRNAs differ in size from that of the previously characterized LRV1 virus of Leishmania guyanensis (1.5 + 2.9 kb versus 5.3 kb, respectively) [53,54]. It remains to be investigated whether this reflects critical differences in genomic organization of viruses, such as segmented versus whole dsRNA genomes.
Fig. 4. Leptomonas seymouri harbors dsRNA viruses. 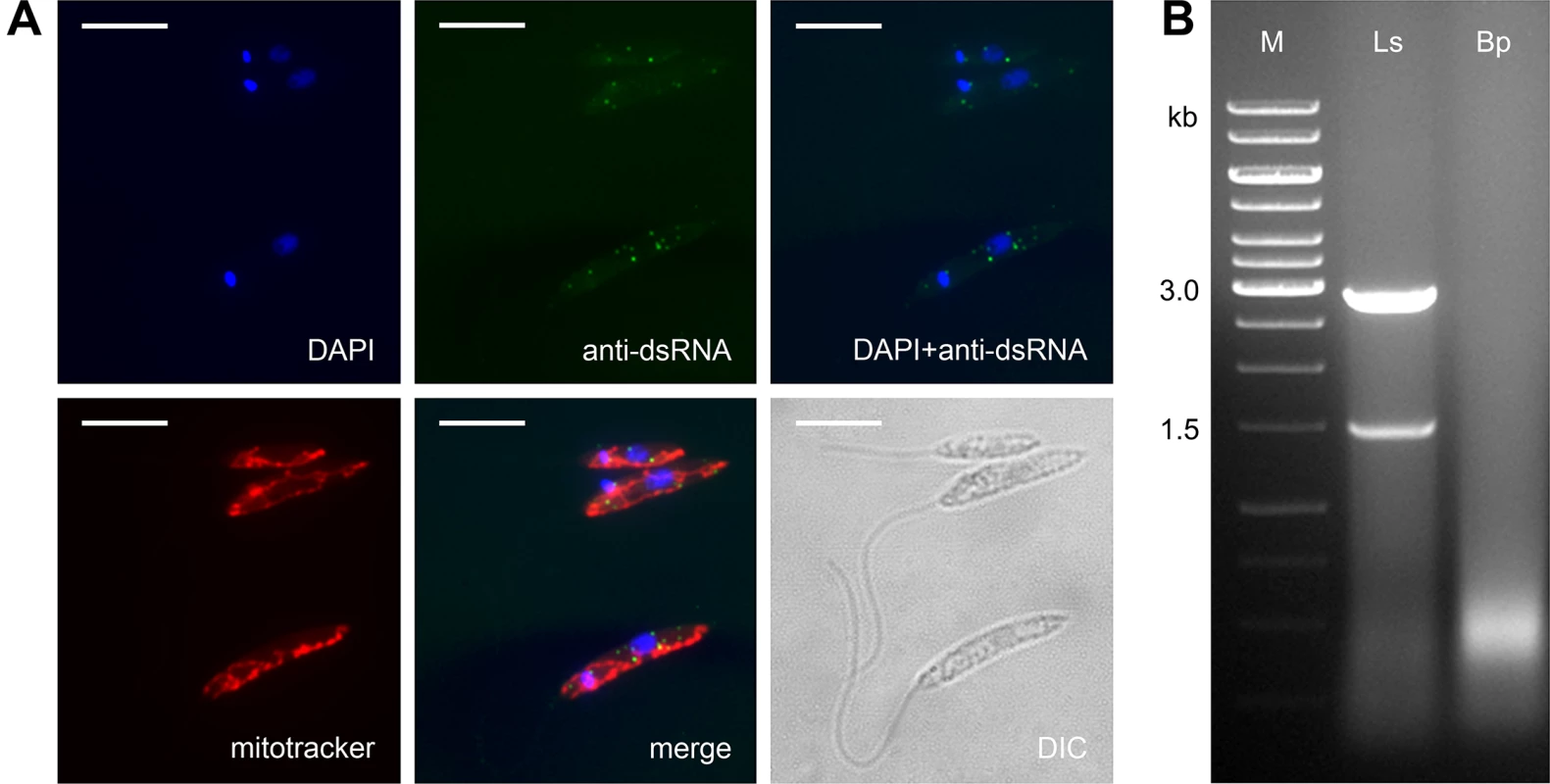
A, Cells were stained with DAPI, mitotracker Red and anti-dsRNA antibody for viral detection. Scale bar is 10 μm. B, Total RNA samples isolated from Leptomonas seymouri (Ls) and Blechomonas pulexsimulantis (Bp) were treated with DNase I and S1 nuclease and separated by gel electrophoresis. M– 1 kb ladder (Thermo Fisher Scientific). Whole transcriptome profiles of L. seymouri cultivated at different temperatures
To identify genes and/or pathways responsible for thermoresistance of L. seymouri, we profiled whole transcriptomes of the parasites cultivated at low (23°C) and high (35°C) temperature. We presumed that in addition to genetic factors (e.g. chromosome ploidy) regulation of gene expression may also be involved in adaptation to dixeny. Reads passing the filtering step (61.4; 52.5; 39.1 million reads for replicates at 23°C and 61.1; 58.9; 37.9 million reads for replicates at 35°C) were used in subsequent analyses. Out of 8,488 genes identified in the L. seymouri genome 8,482 genes were recovered in our analysis. Results of the FDR test are shown in S3 Fig. In total, 340 genes (4% of the total number) were shown to be differentially expressed at the elevated temperature (S14 Table). Expression of 139 genes (1.6% of the total number) was found to be down-regulated at 35°C, whilst 201 genes (2.4% of the total number) were upregulated at least 1.5 fold (p-value ≤ 0.05). Several interesting cases are discussed in detail below.
Synthesis of sterols
Sterols and related compounds are important membrane components of the living cells that define cell membrane's fluidity. They act as bidirectional regulators by stabilizing the membrane and raising its melting point at high temperature, and by preventing phospholipids from clustering together and stiffening at low temperature [55]. Sterol biosynthesis is a fairly conserved biochemical pathway in eukaryotes responsible for the production of cholesterol in animals and several C24-alkyl sterols (ergostane-based sterols) in fungi, plants, and trypanosomatids. In L. major, genetic ablation of C14α-demethylase (C14DM) results in a complete loss of ergostane-based sterols and accumulation of C14-methylated sterols. Genetically modified (c14dm-/-) parasites were viable but exhibited some remarkable defects including increased membrane fluidity, and hypersensitivity to heat stress [56]. In T. brucei the decrease in the levels of squalene synthase and squalene monooxidase led to the depletion of cellular sterol intermediates and end products, impaired cell growth and aberrant morphologies, DNA fragmentation and profound modification of mitochondrial structure and function [57].
In L. seymouri numerous enzymes implicated in biosynthesis of C24-alkyl sterols were down-regulated at elevated temperature. This list includes sterol C14DM, squalene monooxygenase, lanosterol synthase, C-5 sterol desaturase, and lathosterol oxidase.
Oxidative stress protection
As mentioned above, the protection against oxidative stress in monoxenous trypanosomatids is unique since in addition to the trypanothione/ tryparedoxins/peroxiredoxins systems, they heavily rely on a bacterial-type catalase. In L. seymouri, expression of the trypanothione reductase goes down upon cultivation at 35°C, but this decrease is compensated by overexpression of two other enzymes, namely catalase and ascorbate-dependent peroxidase. The expression of an enzyme responsible for superoxide anions detoxification (superoxide dismutase) is also upregulated at elevated temperature.
Carbohydrate and fatty acids metabolism
Several glycosomal components of the carbohydrate metabolism are significantly down-regulated in L. seymouri at elevated temperature. These are glucose-6-phosphate isomerase, ATP-dependent 6-phospho-1-fructokinase, glycosomal phosphoglycerate kinase, fumarate hydratase, fumarate reductase, and malate dehydrogenases. Conversely, expression of some genes, such as pyruvate phosphate dikinase and cytosolic fumarase, is up-regulated at 35°C.
Consistent with the above-mentioned observations, L. seymouri catabolism of fatty acids by β-oxidation is enhanced at high temperature. Several enzymes implicated in this reaction, namely enoyl-CoA hydratase, elongase 4, and several desaturases (delta-6 fatty acid desaturase, delta-5 fatty acid desaturase, stearic acid desaturase), are all upregulated at 35°C. Enhanced catabolism is accompanied by diminished de novo synthesis, as is evidenced by the inhibition of three consequent elongases (1 to 3) responsible for the synthesis of saturated fatty acids.
Experimental infection of Phlebotomus spp. with Leptomonas seymouri
Experimental infections of the two proven vectors of Leishmania donovani, Phlebotomus orientalis and P. argentipes, were compared side-by-side. Insects were fed on either blood or sugar meals to mimic the range of conditions which may favor infection (Fig 5A and 5B). On day 2 after infective sugar meal all females of P. orientalis were infected, while the infection rate of P. argentipes females was lower (59%) (Fig 5A). Intensity of infection was generally weak in both species tested. On day 6 p. i. percentages of infected sand flies decreased to 77% and 46% for P. orientalis and P. argentipes females, respectively. On day 9 every third female remained infected, yet most of them harbored only few flagellates.
Infection via the blood meal was less efficient when compared to the sugar meal. On day 2 less than half of blood-fed females of P. orientalis and P. argentipes were infected (47% and 32%, respectively). Freely moving promastigotes were found enclosed in the ingested blood. On days 6 and 9 L. seymouri promastigotes persisted only in a few females (Fig 5B).
Fig. 5. Experimental infection and co-infection of Phlebotomus spp. 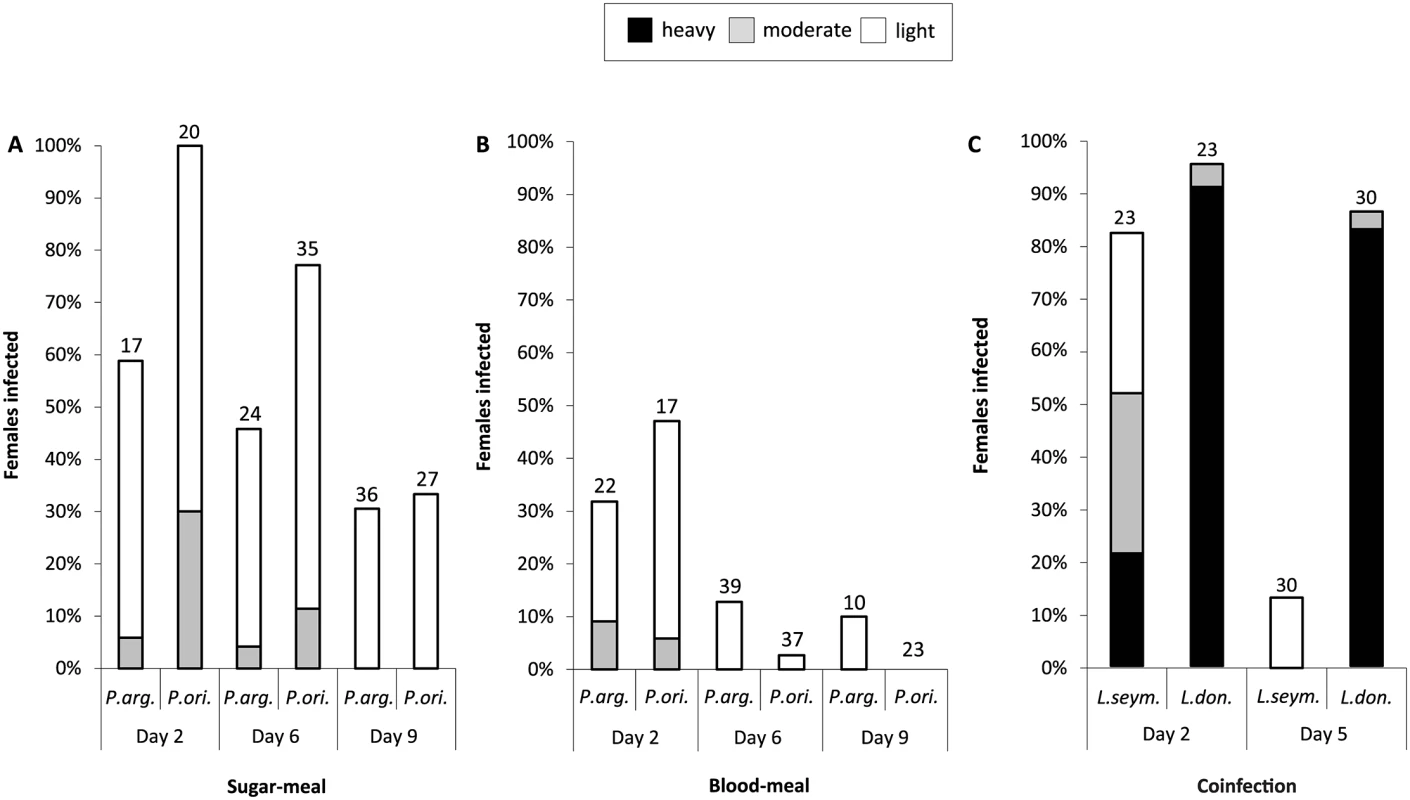
Phlebotomus argentipes and P. orientalis were infected with Leptomonas. seymouri using sugar- (A) or blood- (B) meal method. Intensity of infection was assayed on days 2, 6, and 9 post infection and defined as light (less than 100 promastigotes, white bar), moderate (100–1,000 promastigotes, grey bar), or heavy (over 1,000 promastigotes, black bar) depending on the number of parasites per gut. C, Experimental co-infection of P. argentipes with mCherry Leptomonas seymouri and GFP Leishmania donovani GR-374. Intensity of infection was assayed on days 2 and 5 post infection with blood meal and defined as light, moderate, or heavy as above. Numbers above each bar indicate the number of dissected females. Co-infection of sand flies with Leptomonas seymouri and Leishmania donovani
The experimental co-infection of sand fly females of P. argentipes were performed by blood meals containing either mCherry - (L. seymouri, ATCC-30220) and/or GFP-expressing (L. donovani, strain GR-374) flagellates. In the control dissection of five sand flies performed just a few hours p. i., both mCherry - and GFP-labeled cells were encountered at about 100 cells of each species per sand fly gut. On day 2 p. i. the infection rate of L. seymouri was lower than that of L. donovani (82.6% versus 95.7%, respectively), and also the intensity of infection with the former species was significantly weaker (Fig 5C). The differences between both parasite species became even more pronounced on day 5 p. i., when the percentage of infected sand flies remained unaltered for L. donovani (86.7%), while it markedly dropped for L. seymouri (13.3%). Moreover, the intensity of infection with L. donovani was high, whereas the few insects still infected with L. seymouri harbored only negligible number of free swimming mCherry-expressing cells (Fig 5C).
Infection of macrophages
Survival of parasites inside mammalian host cells J774 or BMMɸ was evaluated 3, 4, 5 and 6 days p. i. No viable L. seymouri cells were found in macrophages by fluorescent microscopy or in Giemsa-stained smears. In contrast, the control represented by L. donovani survived inside both J774 and BMMɸ cells. Similar results were obtained using peritoneal macrophages from BALB/c mice. The transformation assay has confirmed microscopic observations, as no L. seymouri cells were found after the lysis of macrophages. On the contrary, L. donovani propagated very well under the same conditions. Similar results were obtained when either J774 or BMMɸ macrophages were simultaneously co-infected with both parasites.
Discussion
Here we performed a multifarious evaluation of the infective potential of L. seymouri, repeatedly isolated from kala-azar patients infected by L. donovani in India and neighboring countries, and have tested the capacity of this monoxenous trypanosomatid to utilize the sand fly vectors permissive for L. donovani. Moreover, we attempted to find genetic and corresponding metabolic adaptations responsible for its survival at 35°C.
Firstly, we have sequenced the whole genome of L. seymouri and compared it with L. pyrrhocoris and C. fasciculata, the only monoxenous species for which high-quality assemblies are available. Twenty six OGs carried by the thermotolerant L. seymouri and absent in these closely related thermosensitive flagellates may potentially be associated with this adaptation. Including dixenous species into the comparative analysis narrowed down our search to just two OGs shared by L. seymouri and five Leishmania spp. and absent from L. pyrrhocoris and C. fasciculata, namely a kinase-like protein and a ubiquinol-cytochrome c reductase-like protein. It was shown previously that protein kinases are involved in amastigote differentiation in Leishmania spp. [58], a process in which temperature switch plays a decisive role [59]. Moreover, our search has identified a number of proteins with specific distribution among trypanosomatid lineages (e.g. absent in Trypanosoma spp. and/or Leishmania spp. but present in monoxenous flagellates) that are prime targets for functional analysis. In any case, the fact that the vast majority of genes within OGs with this phyletic distribution are annotated as hypothetical proteins with unknown function indicates our scarce knowledge of trypanosomatid metabolism.
A number of metabolic changes observed in L. seymouri exposed to elevated temperature are evocative of those in Leishmania amastigotes or T. brucei bloodstream forms in glucose-poor environment [60,61]. For example, inhibition of the de novo synthesis of sterols in L. seymouri resembles Leishmania amastigotes in which the relative abundance of C24-alkyl sterols was significantly decreased upon their differentiation from procyclics [62,63]. Similarly to their dixenous cousins, L. seymouri cells at high temperature reduce the uptake of glucose and shift their acetyl-CoA production in mitochondria from mainly pyruvate-based to the fatty acids-derived [64–66].
The detection of double-stranded viruses in L. seymouri is particularly relevant in the light of recent findings that their presence in Leishmania guyanensis correlates with its virulence and metastatic potential [51,67]. While molecular mechanisms of this phenomenon are just becoming to be understood, it is already clear that the host immune response is rewired [68,69]. We and others have detected dsRNA-containing viruses in several other monoxenous trypanosomatids parasitizing dipteran and heteropteran insects [27,51,70,71], but their relationships to the characterized viruses of Leishmania still remain a mystery. Two analyzed Leptomonas spp. differ in their acceptability for dsRNA viruses. This indicates fundamentally different mechanisms they may utilize to regulate their gene expression.
In summary, we conclude that although L. seymouri has developed several adaptations that allow it to grow well at 35°C, it remains a predominantly monoxenous species not able to infect mammalian macrophages either alone or in co-infection with Leishmania. This agrees with a recent report on selective elimination of Leptomonas from co-cultures with Leishmania [72]. Under certain circumstances it is able to infect mammals, but probably only when the host is immunocompromised by infection with another pathogen, such as L. donovani or HIV [14,73]. However, it is quite likely that such co-infections are much more frequent than the available literature suggests. This conclusion is further supported by our finding that L. seymouri can survive up to 9 days in the same sand fly species that is responsible for the transmission of pernicious Leishmania spp. Therefore, it will be important to analyze samples from patients suffering from visceral and other leishmaniases with primers specific for L. seymouri and related (presumably) monoxenous trypanosomatids to address the possibility that we see only the tip of the iceberg. In addition to the capacity to withstand elevated temperature, other factors, such as its ability to escape the host immune response, may likely play an important role in establishment of the Leptomonas infection in mammals. We cannot exclude the possibility that some isolates of L. seymouri may be exclusively transmitted by sandflies and spend part of their life cycle in vertebrates similar to their Leishmania spp. relatives.
Materials and Methods
Ethics statement
Animals were maintained and handled in the animal facility of Charles University in Prague in accordance with institutional guidelines and Czech legislation (Act Number 246/1992 and 359/2012 coll. on Protection of Animals against Cruelty in present statutes at large), which complies with all relevant European Union and international guidelines for experimental animals. The experiments were approved by the Committee on the Ethics of Animal Experiments of the Charles University in Prague (Permit Number 24/773/08-10001) and were performed under the Certificate of Competency (Registration Number CZU945/05 ext. CZ02573) and the Permission Number 31114/2013-MSMT-13 ext. 24115/2014-MZE-17214 of the Ministry of the Environment of the Czech Republic.
Origins of strains
Leptomonas seymouri isolate ATCC 30220 was obtained from the American Type Culture Collection (ATCC, Manassas, USA). It was isolated from the cotton stainer Dysdercus suturellus in the United States in 1959. Leptomonas pyrrhocoris isolate H10 [17], Blechomonas ayalai isolate B08-376 [52] and Leishmania donovani isolate MHOM/ET/2010/GR374 have originated from the research collections at Charles University in Prague, Institute of Parasitology in České Budějovice, and Life Science Research Centre in Ostrava.
In vitro cultivation and morphological analysis
Cultures of the monoxenous trypanosomatids were routinely maintained in the Schneider's Drosophila medium (SDM) (Thermo Fisher Scientific, Waltham, USA) supplemented with 10% Fetal Bovine Serum (FBS) (Thermo Fisher Scientific), 50 units/ml of penicillin, 50 μg/ml of streptomycin (both from Sigma-Aldrich, St. Louis, USA), and 10 μg/ml of hemin (Jena Bioscience GmbH, Jena, Germany) at 23°C. All isolates used in this work can also be cultivated in the Brain Heart Infusion (BHI) medium (Sigma-Aldrich) supplemented with 10% FBS and antibiotics as above, or in the two-phased blood-agar medium [74].
To estimate the dynamics of growth, 5 x 104 parasites were seeded into the SDM or the blood-agar medium. Cultures were incubated at 23°C, 29°C, and 35°C for 7 days. Cell numbers were counted using a hemocytometer and plotted in log scale. Morphology of the cells cultivated at low (23°C) and high (35°C) temperature, either in SDM or blood-agar media, was analyzed at day 4 (exponential phase) after staining cells with Giemsa as described previously [75,76]. One hundred cells per sample were measured and analyzed using ANOVA statistical models [77].
Leishmania donovani (MHOM/ET/2010/GR374) transfected with Green Fluorescent Protein (GFP) was cultured in M199 medium (Sigma) containing 20% heat-inactivated FBS (Thermo Fisher Scientific) supplemented with 1% BME vitamins (Sigma), 2% sterile urine, 50 units/ml penicillin, 250μg/ml amikacin (Bristol-Myers Squibb, New York, USA), and 150 μg/ml of geneticin, G418 (Sigma).
PCR amplification, cloning and sequencing
The internal transcribed spacer, ITS region of the rRNA locus was amplified using primers IAMWE and Tc5.8-rev and conditions described elsewhere [78]. Total genomic DNA samples of clinical Indian kala-azar field isolates Ld_39 and Ld_2001 were used as templates [19]. The 18S rRNA and gGAPDH genes were PCR-amplified, cloned into the pCR2.1 vector system (Thermo Fisher Scientific), sequenced and analyzed as described previously [79,80]. The obtained sequences were deposited to GenBank with the following accession numbers: KP717894, KP717895 (18S rRNA); KP717896, KP717897 (gGAPDH); KP717898, KP717899 (ITS1 + ITS2 regions).
Genome assembly and annotation
The Leptomonas seymouri ATCC 30220 genome was sequenced with 100 nt paired-end reads using the Illumina HiSeq 2000 platform (Macrogen, Seoul, South Korea). Prior to assembly, reads were subjected to trimming and filtering using CLC Genomics Workbench v. 7.0 (CLC Inc, Aarhus, Denmark): regions with Phred quality < 20 were trimmed, no more than one N was allowed in the remaining sequence, then TruSeq adapter trimming and a minimum length threshold of 75 nt were applied.
Draft genome of L. seymouri was assembled with the CLC Genomics Workbench v. 7.0 employing a De Bruijn graph-based algorithm with the average coverage of 180 x. Augustus v. 2.5.5 was used to annotate the draft genome of L. seymouri [81]. Prediction accuracy of Augustus was improved by retraining using a training set of L. seymouri conserved proteins. In brief, de novo assembled contigs were searched against proteins in the TriTrypDB v. 7.0 database [82] (BlastX E-value ≤ 10−5) and best BLAST hits were chosen based on the following criteria: a) E-value ≤ 10−30, b) hit length longer than 80 amino acids (aa), c) percent identity higher than 40. Subsequently, a non-redundant training set of 727 high-confidence gene models with unambiguous start site positions was created based on best BLAST hits to annotated proteins from TriTrypDB and RNA-seq coverage data. Non-redundancy of the training set was achieved by excluding genes with more than 70% identity at the amino acid level. Further analysis of the Augustus annotation included manual curation of predicted genes based on transcriptome sequencing data, e.g. removing start sites predicted in regions with no transcriptomic coverage and adding transcribed ORFs >200 aa in length not predicted by Augustus. For tRNA gene prediction tRNAscan-SE Search Server [83] was used with default parameters. For annotating other non-coding RNAs BlastN algorithm (E-value ≤ 10−10) was employed with subsequent manual inspection of BLAST results. As a result, 8,488 genes were annotated in the L. seymouri genome, which has been submitted to the NCBI (BioProject accession number PRJNA285179) and the TriTryp database, a part of the EuPathDB [84].
Gene family analysis using the OrthoMCL approach
Orthologous groups are the set of genes descended from a single common ancestral gene, containing both paralogs and orthologs. OGs for L. seymouri proteins were inferred using the OrthoMCL v.2.0 software [85]. Full proteomes for 23 trypanosomatid species were downloaded from the TriTrypDB v. 7.0 and combined with newly annotated proteins from L. seymouri and 3 other trypanosomatid species (Leptomonas pyrrhocoris, Blechomonas ayalai and Paratrypanosoma confusum). The reference protein dataset was subjected to removal of poor quality proteins (based on sequence length and percent of in-frame stop codons), all vs. all BLAST (E-value 10−10) and a clustering procedure implemented in the OrthoMCL algorithm. This resulted in 19,866 OGs, 7,935 of which contained proteins of L. seymouri.
Whole transcriptome data processing and analysis
L. seymouri was cultivated at 23°C and 35°C for 75 hrs. Total RNA was isolated from 2.5 x 107 cells using RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instruction. The mRNA-derived libraries were sequenced with 100 nt paired-end reads on the Illumina HiSeq 2000 platform (Macrogen). Total of 3 independent biological replicates were analyzed. The whole transcriptome data from this study have been submitted to TriTrypDB database [82].
Differential gene expression analysis was done using the RNA-Seq tool in CLC Genomics Workbench. Raw reads were subjected to quality-based trimming (regions with Phred quality < 20 were trimmed, no more than one N was allowed in the remaining sequence), adapter trimming, and a minimum length threshold of 30 bp. Processed reads were then mapped to the annotated L. seymouri genome with the following parameters: maximum number of mismatches, 2; minimum fraction of read length mapped, 0.8; minimum identity within the mapped sequence, 0.8; maximum number of best-scoring hits for a read, 30. All libraries were mapped as paired-end, and expression values (RPKM) for each gene were calculated. To identify gene sets that are differentially expressed between the two conditions, the FDR test was employed [86]. Genes with expression fold change ≥ 1.5 and FDR p-value ≤ 0.05 were chosen for further analyses.
Gene ontology (GO) terms for genes up - and down-regulated at high temperature were generated using the Blast2GO plugin in CLC Genomics Workbench [87]. Initially, BlastP search against the NCBI nr database was performed, GO terms associated with all the hits were retrieved, and most appropriate GO terms were selected according to the standard Blast2GO procedure. GO term enrichment was assessed using Fisher's exact test.
Detection of dsRNA viruses
For detection of dsRNA viruses, two complementary protocols were used. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI), mitotracker Red CMXRos (both from Thermo Fisher Scientific) and mouse monoclonal anti-dsRNA antibody (Scicons, Szirák, Hungary), followed by goat anti-mouse IgG–Alexa Fluor 488 (Thermo Fisher Scientific) antibody as described previously [50]. In addition, 50 μg of total RNA isolated using TRI reagent (Sigma-Aldrich) was treated with 1 unit of DNase I (New England Biolabs, Ipswich, USA) at 37°C for 1 hr, followed by digestion with 35 units of S1 nuclease (Sigma-Aldrich) for 45 min at the same temperature. Samples were analyzed on 0.8% native agarose in 1xTAE buffer [88].
Establishing a fluorescent strain of L. seymouri
A fragment encoding mCherry fluorescent protein was amplified with primers 5´-TTATCCATGGTTAGTAAAGGAGAA-3´ and 5´-TGTTAGCGGCCGCTTATGCGGTACCAGAACC-3´ using plasmid p2686 as a template [89]. The resulting 745 bp fragment was cloned into the pF4T7polNLS1.4sat vector digested with NcoI and NotI replacing the T7 polymerase ORF [90]. Log-phase L. seymouri cells (4 x 107) were transfected with 15 μg of SwaI-linearized pF4mCherry1.4sat as described before [91]. Recombinant clones were selected on agar—BHI growth medium supplemented with 10% FBS, 40mM HEPES, pH 7.4 and nourseothricin (Jena Bioscience) at final concentration of 250 μg/ml. Expression of mCherry was confirmed by fluorescence microscopy.
L. seymouri development in sandflies
Colonies of two sand fly species, Phlebotomus orientalis and P. argentipes, both representing major proven vectors for L. donovani, were maintained under standard conditions as described elsewhere [92]. Females of both colonies were fed either through a chick-skin membrane on suspension of heat-inactivated rabbit blood containing exponentially growing 1 x 107 promastigotes per ml of blood or on 20% sucrose solution containing 5 x 107 promastigotes per ml. In order to recognize sugar-fed females, the sucrose solution was stained by indigo carmin. Blood - and sugar-fed females were kept at 26°C with free access to 50% sucrose solution by day 1 post infection (p. i.).
Sand fly females were dissected at different intervals p. i. (1–2, 5–6 and 7–9 days). Numbers and location of flagellates in the sand fly gut were checked microscopically. Parasite loads were graded as previously described, i.e.: light (< 100 parasites/gut), moderate (100–1,000 parasites/gut) and heavy (> 1,000 parasites/gut) [93].
Leptomonas and Leishmania co-infection and development in sand flies
Females of P. argentipes were fed through a chick-skin membrane on suspension of heat-inactivated rabbit blood containing 1 x 106 per ml promastigotes of Leptomonas seymouri mCherry (passage 4) and 1 x 106 per ml promastigotes of Leishmania donovani (MHOM/ET/2010/GR374) GFP (passage 10) originating from exponentially growing cultures. Assorted blood-fed females were kept at 26°C with free access to 20% sucrose solution. Sand fly females were dissected on days 2 and 5 p. i., and the presence of parasites as well as other characteristics were analyzed as described previously [93].
In vitro infection of mouse macrophages with Leptomonas seymouri and Leishmania donovani
Macrophage cell line J774 was cultured in complete RPMI-1640 medium (Sigma) containing 10% FBS, 100 U/ml of penicillin, 100 μg/ml of streptomycin, 2mM of L-glutamine, and 0.05 mM of β-mercapto-ethanol (all from Sigma) at 37°C with 5% CO2. Bone marrow was obtained by flushing of tibias and femurs of BALB/c mice and flagellates were cultured in complete RPMI-1640 medium (Sigma) supplemented as above along with 20% of L929 fibroblast cell culture supernatant serving as a source of macrophage colony-stimulating factor at 37°C with 5% CO2. The differentiation from bone marrow precursor cells to bone marrow-derived macrophages proceeded for 7 to 8 days in sterile polystyrene Petri dishes. The bone marrow derived macrophages (BMMɸ) were washed and seeded into plates at density of 5 x 105 cells per ml. Consequently, stationary cultures of Leishmania donovani (GFP), Leptomonas seymouri (mCherry), alone or in combination were added in ratio of 8 : 1 (parasites: BMMɸ). Three days p. i. BMMɸ were extensively washed with pre-warmed RPMI-1640 to remove excess of parasites and the viability of trypanosomatids was monitored by fluorescence microscope Olympus CX-31 (Olympus, Tokyo, Japan) up to 6 day p. i. In addition, Giemsa staining was used to analyze intracellular forms in macrophages by light microscopy. All experiments were performed in two independent biological replicates.
To analyze survival of parasites, the transformation growth assay was used [94]. In brief, macrophages infected with Leishmania donovani, Leptomonas seymouri, alone or in combination for 96 hrs were extensively washed with RPMI-1640 and lysed with 0.016% SDS in RPMI-1640 for 7 min at room temperature to release their intracellular forms. The lysis reactions were neutralized by RPMI-1640 supplemented with 17% heat-inactivated FBS. Parasites were spun down at 3,200 rpm for 10 min at 4°C, washed in RPMI-1640, and re-suspended in a relevant promastigote medium (BHI or M199) supplemented with an appropriate selective antibiotic at 23°C. For macrophages co-infected with both parasites, two types of media and antibiotics were assessed. The status of viable parasites was checked for 6 consecutive days.
Supporting Information
Zdroje
1. Podlipaev SA (2001) The more insect trypanosomatids under study-the more diverse Trypanosomatidae appears. Int J Parasitol 31 : 648–652. 11334958
2. Maslov DA, Votýpka J, Yurchenko V, Lukeš J (2013) Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol 29 : 43–52. doi: 10.1016/j.pt.2012.11.001 23246083
3. Simpson AG, Stevens JR, Lukeš J (2006) The evolution and diversity of kinetoplastid flagellates. Trends Parasitol 22 : 168–174. 16504583
4. Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V (2014) Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol 195 : 115–122. doi: 10.1016/j.molbiopara.2014.05.007 24893339
5. Laveran A, Franchini G (1920) Infections experimentales de chiens et de cobayes a l'aide de cultures d'Herpetomonas d'insects. Bull Soc Pathol Exot 13 : 569–576.
6. McGhee RB, Cosgrove WB (1980) Biology and physiology of the lower Trypanosomatidae. Microbiol Rev 44 : 140–173. 6997722
7. Pacheco RS, Marzochi MC, Pires MQ, Brito CM, Madeira Md, et al. (1998) Parasite genotypically related to a monoxenous trypanosomatid of dog's flea causing opportunistic infection in an HIV positive patient. Mem Inst Oswaldo Cruz 93 : 531–537. 9711346
8. Morio F, Reynes J, Dollet M, Pratlong F, Dedet JP, et al. (2008) Isolation of a protozoan parasite genetically related to the insect trypanosomatid Herpetomonas samuelpessoai from a human immunodeficiency virus-positive patient. J Clin Microbiol 46 : 3845–3847. doi: 10.1128/JCM.01098-08 18832132
9. Ferreira MS, Borges AS (2002) Some aspects of protozoan infections in immunocompromised patients - a review. Mem Inst Oswaldo Cruz 97 : 443–457. 12118272
10. Dedet JP, Pratlong F (2000) Leishmania, Trypanosoma and monoxenous trypanosomatids as emerging opportunistic agents. J Eukaryot Microbiol 47 : 37–39. 10651294
11. Sundar S, Chakravarty J (2012) Recent advances in the diagnosis and treatment of kala-azar. Natl Med J India 25 : 85–89. 22686715
12. Wallace FG, Hertig M (1968) Ultrastructural comparison of promastigote flagellates (leptomonads) of wild-caught Panamanian Phlebotomus. J Parasitol 54 : 606–612. 5757733
13. Bhattarai NR, Das ML, Rijal S, van der Auwera G, Picado A, et al. (2009) Natural infection of Phlebotomus argentipes with Leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans R Soc Trop Med Hyg 103 : 1087–1092. doi: 10.1016/j.trstmh.2009.03.008 19345387
14. Ghosh S, Banerjee P, Sarkar A, Datta S, Chatterjee M (2012) Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J Clin Microbiol 50 : 2774–2778. doi: 10.1128/JCM.00966-12 22622439
15. Jirků M, Yurchenko VY, Lukeš J, Maslov DA (2012) New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J Eukaryot Microbiol 59 : 537–547. doi: 10.1111/j.1550-7408.2012.00636.x 22845426
16. Wallace FG (1977) Leptomonas seymouri sp. n. from the cotton stainer Dysdercus suturellus. J Protozool 24 : 483–484. 599500
17. Votýpka J, Klepetková H, Yurchenko VY, Horák A, Lukeš J, et al. (2012) Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist 163 : 616–631. doi: 10.1016/j.protis.2011.12.004 22341645
18. Conchon I, Campaner M, Sbravate C, Camargo EP (1989) Trypanosomatids, other than Phytomonas spp., isolated and cultured from fruit. J Protozool 36 : 412–414.
19. Singh N, Chikara S, Sundar S (2013) SOLiD sequencing of genomes of clinical isolates of Leishmania donovani from India confirm Leptomonas co-infection and raise some key questions. PLOS One 8: e55738. doi: 10.1371/journal.pone.0055738 23418454
20. Srivastava P, Prajapati VK, Vanaerschot M, Van der Auwera G, Dujardin JC, et al. (2010) Detection of Leptomonas sp. parasites in clinical isolates of Kala-azar patients from India. Infect Genet Evol 10 : 1145–1150. doi: 10.1016/j.meegid.2010.07.009 20633704
21. De Sa MF, De Sa CM, Veronese MA, Filho SA, Gander ES (1980) Morphologic and biochemical characterization of Crithidia brasiliensis sp. n. J Protozool 27 : 253–257. 7005431
22. Roitman I, Mundim MH, De Azevedo HP, Kitajima EW (1977) Growth of Crithidia at high temperature: Crithidia hutneri sp. n. and Crithidia luciliae thermophila s. sp. n. J Protozool 24 : 553–556.
23. McGhee RB (1959) The infection of avian embryos with Crithidia species and Leishmania tarentola. J Infect Dis 105 : 18–25. 13665046
24. Kostygov AY, Grybchuk-Ieremenko A, Malysheva MN, Frolov AO, Yurchenko V (2014) Molecular revision of the genus Wallaceina. Protist 165 : 594–604. doi: 10.1016/j.protis.2014.07.001 25113831
25. Maslov DA, Yurchenko VY, Jirků M, Lukeš J (2010) Two new species of trypanosomatid parasites isolated from Heteroptera in Costa Rica. J Eukaryot Microbiol 57 : 177–188. doi: 10.1111/j.1550-7408.2009.00464.x 20113381
26. Yurchenko V, Lukeš J, Jirků M, Zeledon R, Maslov DA (2006) Leptomonas costaricensis sp. n. (Kinetoplastea: Trypanosomatidae), a member of the novel phylogenetic group of insect trypanosomatids closely related to the genus Leishmania. Parasitology 133 : 537–546. 16834819
27. Yurchenko V, Votýpka J, Tesařová M, Klepetková H, Kraeva N, et al. (2014) Ultrastructure and molecular phylogeny of four new species of monoxenous trypanosomatids from flies (Diptera: Brachycera) with redefinition of the genus Wallaceina. Folia Parasitol 61 : 97–112. 24822316
28. Votýpka J, d'Avila-Levy CM, Grellier P, Maslov DA, Lukeš J, et al. (2015) New approaches to systematics of Trypanosomatidae: criteria for taxonomic (re)description. Trends Parasitol (in press).
29. Borghesan TC, Ferreira RC, Takata CS, Campaner M, Borda CC, et al. (2013) Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist 164 : 129–152. doi: 10.1016/j.protis.2012.06.001 22938923
30. El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, et al. (2005) Comparative genomics of trypanosomatid parasitic protozoa. Science 309 : 404–409. 16020724
31. Porcel BM, Denoeud F, Opperdoes FR, Noel B, Madoui M-A, et al. (2014) The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLOS Genet 10: e1004007. doi: 10.1371/journal.pgen.1004007 24516393
32. Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, et al. (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309 : 436–442. 16020728
33. Mair G, Shi H, Li H, Djikeng A, Aviles HO, et al. (2000) A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA 6 : 163–169. 10688355
34. Alves JM, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, et al. (2013) Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol 13 : 190. doi: 10.1186/1471-2148-13-190 24015778
35. Hannaert V, Bringaud F, Opperdoes FR, Michels PA (2003) Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol Dis 2 : 11. 14613499
36. Opperdoes FR, Coombs GH (2007) Metabolism of Leishmania: proven and predicted. Trends Parasitol 23 : 149–158. 17320480
37. Opperdoes FR, Szikora JP (2006) In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol Biochem Parasitol 147 : 193–206. 16546274
38. Alves JM, Voegtly L, Matveyev AV, Lara AM, da Silva FM, et al. (2011) Identification and phylogenetic analysis of heme synthesis genes in trypanosomatids and their bacterial endosymbionts. PLoS One 6: e23518. doi: 10.1371/journal.pone.0023518 21853145
39. Kořený L, Lukeš J, Oborník M (2010) Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? Int J Parasitol 40 : 149–156. doi: 10.1016/j.ijpara.2009.11.007 19968994
40. Kořený L, Oborník M, Lukeš J (2013) Make it, take it, or leave it: heme metabolism of parasites. PLoS Pathog 9: e1003088. doi: 10.1371/journal.ppat.1003088 23349629
41. Bartholomeu DC, de Paiva RM, Mendes TA, DaRocha WD, Teixeira SM (2014) Unveiling the intracellular survival gene kit of trypanosomatid parasites. PLoS Pathog 10: e1004399. doi: 10.1371/journal.ppat.1004399 25474314
42. Maslov DA, Westenberger SJ, Xu X, Campbell DA, Sturm NR (2007) Discovery and barcoding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J Eukaryot Microbiol 54 : 57–65. 17300521
43. Ibrahim EA, Molyneux DH (1987) Pathogenicity of Crithidia fasciculata in the haemocoele of Glossina. Acta Trop 44 : 13–22. 2884835
44. Schaub GA (1994) Pathogenicity of trypanosomatids on insects. Parasitol Today 10 : 463–468. 15275511
45. Alcolea PJ, Alonso A, Garcia-Tabares F, Torano A, Larraga V (2014) An insight into the proteome of Crithidia fasciculata choanomastigotes as a comparative approach to axenic growth, peanut lectin agglutination and differentiation of Leishmania spp. promastigotes. PLoS One 9: e113837. doi: 10.1371/journal.pone.0113837 25503511
46. Mizbani A, Taslimi Y, Zahedifard F, Taheri T, Rafati S (2011) Effect of A2 gene on infectivity of the nonpathogenic parasite Leishmania tarentolae. Parasitol Res 109 : 793–799. doi: 10.1007/s00436-011-2325-4 21442256
47. Gabernet-Castello C, Dacks JB, Field MC (2009) The single ENTH-domain protein of trypanosomes; endocytic functions and evolutionary relationship with epsin. Traffic 10 : 894–911. doi: 10.1111/j.1600-0854.2009.00910.x 19416477
48. Bessat M, Knudsen G, Burlingame AL, Wang CC (2013) A minimal anaphase promoting complex/cyclosome (APC/C) in Trypanosoma brucei. PLoS One 8: e59258. doi: 10.1371/journal.pone.0059258 23533609
49. Lye LF, Owens K, Shi H, Murta SM, Vieira AC, et al. (2010) Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6: e1001161. doi: 10.1371/journal.ppat.1001161 21060810
50. Zangger H, Ronet C, Desponds C, Kuhlmann FM, Robinson J, et al. (2013) Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis 7: e2006. doi: 10.1371/journal.pntd.0002006 23326619
51. Ives A, Ronet C, Prevel F, Ruzzante G, Fuertes-Marraco S, et al. (2011) Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 331 : 775–778. doi: 10.1126/science.1199326 21311023
52. Votýpka J, Suková E, Kraeva N, Ishemgulova A, Duží I, et al. (2013) Diversity of trypanosomatids (Kinetoplastea: Trypanosomatidae) parasitizing fleas (Insecta: Siphonaptera) and description of a new genus Blechomonas gen. n. Protist 164 : 763–781. doi: 10.1016/j.protis.2013.08.002 24113136
53. Weeks R, Aline RF Jr., Myler PJ, Stuart K (1992) LRV1 viral particles in Leishmania guyanensis contain double-stranded or single-stranded RNA. J Virol 66 : 1389–1393. 1738198
54. Salinas G, Zamora M, Stuart K, Saravia N (1996) Leishmania RNA viruses in Leishmania of the Viannia subgenus. Am J Trop Med Hyg 54 : 425–429. 8615459
55. Evans E, Rawicz W, Smith BA (2013) Back to the future: mechanics and thermodynamics of lipid biomembranes. Faraday Discuss 161 : 591–611. 23805759
56. Xu W, Hsu FF, Baykal E, Huang J, Zhang K (2014) Sterol biosynthesis is required for heat resistance but not extracellular survival in Leishmania. PLoS Pathog 10: e1004427. doi: 10.1371/journal.ppat.1004427 25340392
57. Perez-Moreno G, Sealey-Cardona M, Rodrigues-Poveda C, Gelb MH, Ruiz-Perez LM, et al. (2012) Endogenous sterol biosynthesis is important for mitochondrial function and cell morphology in procyclic forms of Trypanosoma brucei. Int J Parasitol 42 : 975–989. doi: 10.1016/j.ijpara.2012.07.012 22964455
58. Grant KM, Dunion MH, Yardley V, Skaltsounis AL, Marko D, et al. (2004) Inhibitors of Leishmania mexicana CRK3 cyclin-dependent kinase: chemical library screen and antileishmanial activity. Antimicrob Agents Chemother 48 : 3033–3042. 15273118
59. Bates PA, Rogers ME (2004) New insights into the developmental biology and transmission mechanisms of Leishmania. Curr Mol Med 4 : 601–609. 15357211
60. Lee SH, Stephens JL, Englund PT (2007) A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol 5 : 287–297. 17363967
61. Coombs GH, Craft JA, Hart DT (1982) A comparative study of Leishmania mexicana amastigotes and promastigotes. Enzyme activities and subcellular locations. Mol Biochem Parasitol 5 : 199–211. 6211617
62. Goad LJ, Holz GG Jr., Beach DH (1984) Sterols of Leishmania species. Implications for biosynthesis. Mol Biochem Parasitol 10 : 161–170. 6700638
63. Coppens I, Courtoy PJ (2000) The adaptative mechanisms of Trypanosoma brucei for sterol homeostasis in its different life-cycle environments. Annu Rev Microbiol 54 : 129–156. 11018126
64. Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, et al. (2008) Retooling Leishmania metabolism: from sand fly gut to human macrophage. Faseb J 22 : 590–602. 17884972
65. Saunders EC, Ng WW, Kloehn J, Chambers JM, Ng M, et al. (2014) Induction of a stringent metabolic response in intracellular stages of Leishmania mexicana leads to increased dependence on mitochondrial metabolism. PLoS Pathog 10: e1003888. doi: 10.1371/journal.ppat.1003888 24465208
66. Mottram JC, Coombs GH (1985) Leishmania mexicana: subcellular distribution of enzymes in amastigotes and promastigotes. Exp Parasitol 59 : 265–274. 3158538
67. Zangger H, Hailu A, Desponds C, Lye LF, Akopyants NS, et al. (2014) Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis 8: e2836. doi: 10.1371/journal.pntd.0002836 24762979
68. Ronet C, Beverley SM, Fasel N (2011) Muco-cutaneous leishmaniasis in the New World: the ultimate subversion. Virulence 2 : 547–552. doi: 10.4161/viru.2.6.17839 21971185
69. Hartley MA, Ronet C, Zangger H, Beverley SM, Fasel N (2012) Leishmania RNA virus: when the host pays the toll. Front Cell Infect Microbiol 2 : 99. doi: 10.3389/fcimb.2012.00099 22919688
70. Soares MJ, Motta MC, de Souza W (1989) Bacterium-like endosymbiont and virus-like particles in the trypanosomatid Crithidia desouzai. Microbios Lett 41 : 137–141.
71. Motta MC, de Souza W, Thiry M (2003) Immunocytochemical detection of DNA and RNA in endosymbiont-bearing trypanosomatids. FEMS Microbiol Lett 221 : 17–23. 12694905
72. Ahuja K, Arora G, Khare P, Selvapandiyan A (2015) Selective elimination of Leptomonas from the in vitro co-culture with Leishmania. Parasitol Int 64 : 1–5.
73. Alvar J, Aparicio P, Aseffa A, Den Boer M, Canavate C, et al. (2008) The relationship between leishmaniasis and AIDS: the second 10 years. Clin Microbiol Rev 21 : 334–359, table of contents. doi: 10.1128/CMR.00061-07 18400800
74. Svobodová M, Volf P, Votýpka J (2006) Experimental transmission of Leishmania tropica to hyraxes (Procavia capensis) by the bite of Phlebotomus arabicus. Microbes Infect 8 : 1691–1694. 16815725
75. Yurchenko V, Lukeš J, Xu X, Maslov DA (2006) An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae). J Eukaryot Microbiol 53 : 103–111. 16579812
76. Yurchenko V, Lukeš J, Jirků M, Maslov DA (2009) Selective recovery of the cultivation-prone components from mixed trypanosomatid infections: a case of several novel species isolated from Neotropical Heteroptera. Int J Syst Evol Microbiol 59 : 893–909. doi: 10.1099/ijs.0.001149-0 19329626
77. Huang S (2010) Statistical issues in subpopulation analysis of high content imaging data. J Comput Biol 17 : 879–894. doi: 10.1089/cmb.2009.0071 20632869
78. Dollet M, Sturm NR, Campbell DA (2012) The internal transcribed spacer of ribosomal RNA genes in plant trypanosomes (Phytomonas spp.) resolves 10 groups. Infect Genet Evol 12 : 299–308. doi: 10.1016/j.meegid.2011.11.010 22155359
79. Yurchenko V, Lukeš J, Tesařová M, Jirků M, Maslov DA (2008) Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus Crithidia. Protist 159 : 99–114. 17931968
80. Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, et al. (2014) Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist 165 : 825–838. doi: 10.1016/j.protis.2014.09.002 25460233
81. Stanke M, Keller O, Gunduz I, Hayes A, Waack S, et al. (2006) AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34: W435–439. 16845043
82. Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, et al. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38: D457–462. doi: 10.1093/nar/gkp851 19843604
83. Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–689. 15980563
84. Aurrecoechea C, Barreto A, Brestelli J, Brunk BP, Cade S, et al. (2013) EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res 41: D684–691. doi: 10.1093/nar/gks1113 23175615
85. Li L, Stoeckert CJ Jr., Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13 : 2178–2189. 12952885
86. Si Y, Liu P (2013) An optimal test with maximum average power while controlling FDR with application to RNA-seq data. Biometrics 69 : 594–605. doi: 10.1111/biom.12036 23889143
87. Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36 : 3420–3435. doi: 10.1093/nar/gkn176 18445632
88. Beiting DP, Peixoto L, Akopyants NS, Beverley SM, Wherry EJ, et al. (2014) Differential induction of TLR3-dependent innate immune signaling by closely related parasite species. PLoS One 9: e88398. doi: 10.1371/journal.pone.0088398 24505488
89. Kelly S, Reed J, Kramer S, Ellis L, Webb H, et al. (2007) Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol 154 : 103–109. 17512617
90. Kushnir S, Gase K, Breitling R, Alexandrov K (2005) Development of an inducible protein expression system based on the protozoan host Leishmania tarentolae. Protein Expres Purif 42 : 37–46.
91. Kraeva N, Ishemgulova A, Lukeš J, Yurchenko V (2014) Tetracycline-inducible gene expression system in Leishmania mexicana. Mol Biochem Parasitol (in press).
92. Volf P, Volfová V (2011) Establishment and maintenance of sand fly colonies. J Vector Ecol 36 Suppl 1: S1–9.
93. Myšková J, Votýpka J, Volf P (2008) Leishmania in sand flies: comparison of quantitative polymerase chain reaction with other techniques to determine the intensity of infection. J Med Entomol 45 : 133–138. 18283954
94. Rogers M, Kropf P, Choi BS, Dillon R, Podinovskaia M, et al. (2009) Proteophosophoglycans regurgitated by Leishmania-infected sand flies target the L-arginine metabolism of host macrophages to promote parasite survival. PLoS Pathog 5: e1000555. doi: 10.1371/journal.ppat.1000555 19696894
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type GenesČlánek Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion IsolatesČlánek -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated GlycolipidsČlánek Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
-
Všechny články tohoto čísla
- The Long and Winding Road (Apologies to the Beatles)
- The Ebola Virus: From Basic Research to a Global Health Crisis
- Riding the R Train into the Cell
- The Two-Phase Emergence of Non Pandemic HIV-1 Group O in Cameroon
- Tumor Progression Locus 2 Promotes Induction of IFNλ, Interferon Stimulated Genes and Antigen-Specific CD8 T Cell Responses and Protects against Influenza Virus
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Incomplete Neutralization and Deviation from Sigmoidal Neutralization Curves for HIV Broadly Neutralizing Monoclonal Antibodies
- E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3
- The Hos2 Histone Deacetylase Controls Virulence through Direct Regulation of Mating-Type Genes
- Hyperinvasive Meningococci Induce Intra-nuclear Cleavage of the NF-κB Protein p65/RelA by Meningococcal IgA Protease
- Active Transport of Phosphorylated Carbohydrates Promotes Intestinal Colonization and Transmission of a Bacterial Pathogen
- HTLV-1 Tax Stimulates Ubiquitin E3 Ligase, Ring Finger Protein 8, to Assemble Lysine 63-Linked Polyubiquitin Chains for TAK1 and IKK Activation
- Transgenic Mouse Bioassay: Evidence That Rabbits Are Susceptible to a Variety of Prion Isolates
- Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion
- Inhibiting the Recruitment of PLCγ1 to Kaposi’s Sarcoma Herpesvirus K15 Protein Reduces the Invasiveness and Angiogenesis of Infected Endothelial Cells
- Goblet Cell Derived RELM-β Recruits CD4 T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation
- HLA Class-II Associated HIV Polymorphisms Predict Escape from CD4+ T Cell Responses
- An siRNA Screen Identifies the U2 snRNP Spliceosome as a Host Restriction Factor for Recombinant Adeno-associated Viruses
- Extracellular Adenosine Protects against Lung Infection by Regulating Pulmonary Neutrophil Recruitment
- : Adaptations to the Dixenous Life Cycle Analyzed by Genome Sequencing, Transcriptome Profiling and Co-infection with
- Which Way In? The RalF Arf-GEF Orchestrates Host Cell Invasion
- Intracellular Uropathogenic . Exploits Host Rab35 for Iron Acquisition and Survival within Urinary Bladder Cells
- A Non-enveloped Virus Hijacks Host Disaggregation Machinery to Translocate across the Endoplasmic Reticulum Membrane
- Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect
- -Associated Polyomavirus Uses a Displaced Binding Site on VP1 to Engage Sialylated Glycolipids
- The Activation of Effector Avr3b by Plant Cyclophilin is Required for the Nudix Hydrolase Activity of Avr3b
- A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in
- Double-Edge Sword of Sustained ROCK Activation in Prion Diseases through Neuritogenesis Defects and Prion Accumulation
- The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in and Influences the Kinetics of Growth and Development
- Inhibits Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis
- Illuminating Targets of Bacterial Secretion
- Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment
- Interdisciplinarity and Infectious Diseases: An Ebola Case Study
- Fungi That Infect Insects: Altering Host Behavior and Beyond
- Plasticity and Redundancy in Proteins Important for Invasion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
- A Novel Virus Causes Scale Drop Disease in
- STAT2 Knockout Syrian Hamsters Support Enhanced Replication and Pathogenicity of Human Adenovirus, Revealing an Important Role of Type I Interferon Response in Viral Control
- Parsimonious Determination of the Optimal Infectious Dose of a Pathogen for Nonhuman Primate Models
- Twenty-Eight Years of Poliovirus Replication in an Immunodeficient Individual: Impact on the Global Polio Eradication Initiative
- AAV-Delivered Antibody Mediates Significant Protective Effects against SIVmac239 Challenge in the Absence of Neutralizing Activity
- Interferon-γ Promotes Inflammation and Development of T-Cell Lymphoma in HTLV-1 bZIP Factor Transgenic Mice
- Transgenic Rabbits Expressing Ovine PrP Are Susceptible to Scrapie
- Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of . Virulence Pathways
- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors
- Inhibition of mTORC1 Enhances the Translation of Chikungunya Proteins the Activation of the MnK/eIF4E Pathway
- Nanoformulations of Rilpivirine for Topical Pericoital and Systemic Coitus-Independent Administration Efficiently Prevent HIV Transmission
- Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection
- sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic
- Evolutionary and Functional Analysis of Old World Primate TRIM5 Reveals the Ancient Emergence of Primate Lentiviruses and Convergent Evolution Targeting a Conserved Capsid Interface
- Hepcidin and Host Defense against Infectious Diseases
- Type I IFN Induction via Poly-ICLC Protects Mice against Cryptococcosis
- Mucosal B Cells Are Associated with Delayed SIV Acquisition in Vaccinated Female but Not Male Rhesus Macaques Following SIV Rectal Challenge
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques
- Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria
- Illuminating Targets of Bacterial Secretion
- Are Human Intestinal Eukaryotes Beneficial or Commensals?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



