-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
article has not abstract
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1004923
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004923Summary
article has not abstract
Introduction
The principles of respiratory microbiology are being re-evaluated and re-written, starting with the debunked myth of lung sterility. The “terrain” of the respiratory ecosystem differs—anatomically and physiologically—from that of other mucosal sites, and changes dramatically in illness, when the dynamic homeostasis between host and microbiome is disrupted. Researchers are only just beginning to understand the contribution of viruses, phages, and fungi to the lung microbiome; thus, we have restricted our discussion to the bacterial microbiota of the lungs.
The Lungs Are Not Sterile
The notion that the lungs are sterile is still frequently stated in textbooks, virtually always without citation. This claim, if true, would be extraordinary. Bacteria are remarkably diverse and adaptable; as such, there is virtually no environmental niche on earth so extreme (in oxygen, pH, hydrophobicity, temperature, salinity, predators, nutrient scarcity, etc.) that bacterial communities cannot been found [1]. It would be remarkable if one of the rare bacteria-free environments on this planet was the warm, moist mucosa found inches below the oral cavity, a bacteria-rich environment over which there is a constant flow of bacteria-laden air, microaerosols, and fluids. Numerous studies, dating back nearly a century, have demonstrated that microaspiration is common in healthy, asymptomatic subjects [2–5], and knowledge of the bacterial content of inhaled air is as old as germ theory itself [6]. Since the first culture-independent report of the healthy lung microbiome [7], more than 30 published studies using molecular techniques for bacterial identification have found evidence of bacteria in the lower airways. No modern study has found evidence of their absence.
Mucosal Biology: The Lungs Are Not the Gut
While the gut and lungs are both mucosa-lined luminal organs with a shared embryological origin, their gross and micro-anatomical features are quite distinct, yielding marked differences in the composition and population dynamics of their microbiota. In the absence of vomiting or esophageal reflux, migration of microbes in the digestive tract is unidirectional (from the mouth to the anus), and is serially interrupted by widely varying physical and chemical barriers. In order for an orally introduced microbe to immigrate into the cecum, it must endure the acidic pH of the stomach (~2.0) and the alkaline pH of the duodenum (~8.0) and compete for resources with a densely populated resident microbiome. By contrast, movement of air, mucus, and microbes in the lung is bidirectional, with no physical barrier between the larynx and the most distal alveolus. Thus the microbiome of the lungs is more dynamic and transient than that of the lower gastrointestinal tract. While the gastrointestinal tract is of uniform temperature (37°C) throughout its entire 9 meters of length, the mucosa of the respiratory tract (a short half-meter in length) represents a gradient from ambient temperature at the point of inhalation to core body temperature in the alveoli [8]. Unlike the gut, the lung environment is oxygen-rich. Though the trachea and bronchi are, like the gut, lined with the heavily glycosylated proteins of secreted mucus, the vast majority of the lung’s surface area is lined with lipid-rich surfactant, which has bacteriostatic effects against select bacterial species [9]. Bacterial density in the airways is quite modest, comparable to that of the duodenum [7], orders of magnitude less than that of the large intestine; thus inter-bacterial metabolic interactions are markedly different. Finally, the gut and lungs differ in the character of host–bacterial interactions. Luminal IgA levels are far higher in the gut, while the lungs exhibit far more extraluminal interactions between bacteria and host leukocytes (alveolar macrophages). Together, these markedly divergent environmental conditions result in correspondingly divergent microbial communities.
The Lung Microbiome Is Determined by Three Ecological Factors
The composition of the lung microbiome is, by first principles, determined by the balance of three factors (Fig 1) [10]: (1) microbial immigration into the airways, (2) elimination of microbes from the airways, and (3) the relative reproduction rates of its community members, as determined by regional growth conditions. Any change in the microbiome—within an individual or across disease states—must be due to some perturbation in these factors. Sources of microbial immigration include the inhalation of air (which contains 104–106 bacteria/mm3 even before reaching the bacteria-dense upper airways [11]), subclinical microaspiration of upper respiratory tract contents [2–5], and direct dispersal along airway mucosa. Microbial elimination is driven by mucociliary clearance, cough (which is frequent even among healthy subjects [12]), and host immune defenses (both innate and adaptive). The environmental conditions that determine regional growth conditions in the lungs include both those common to all environmental niches (e.g., nutrient availability, temperature, pH, oxygen tension) as well as the abundance and activation state of host inflammatory cells. In health, these conditions are generally inhospitable for bacterial growth, resulting in relatively little bacterial reproduction; thus the primary determinant of the lung microbiome in health is the balance of immigration and elimination [13–15]. However, during disease, the regional growth conditions of the lungs change dramatically, creating permissive niches for selective bacterial reproduction. The long-recognized phenomenon of bacterial colonization in advanced lung disease reflects the enriched growth of species that are well-adapted to the specific environmental conditions of the injured respiratory tract. The selective reproductive advantage of the lung environment on these community members has overwhelmed the dynamic influence of immigration and elimination on the respiratory ecosystem.
Fig. 1. Ecological determinants of the respiratory microbiome. 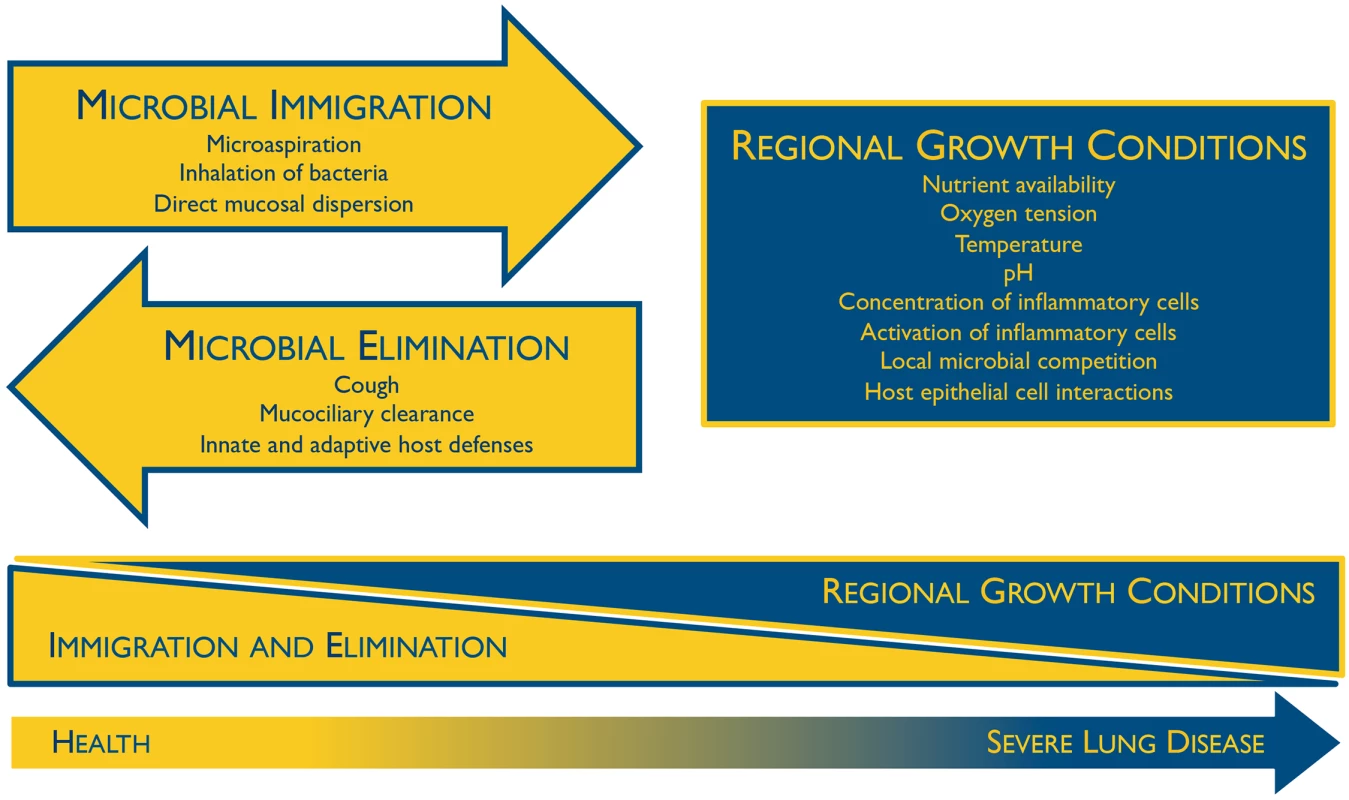
The constitution of the respiratory microbiome is determined by three factors: microbial immigration, microbial elimination, and the relative reproduction rates of its members. In health, community membership is primarily determined by immigration and elimination; in advanced lung disease, membership is primarily determined by regional growth conditions. Adapted from Dickson 2014 [10]. The Oral Microbiome Is the Primary Source of the Bacterial Microbiota in the Lungs During Health
The ubiquity of subclinical microaspiration of pharyngeal secretions among healthy subjects is a long-established and validated observation [2–5]. Numerous culture-independent studies have since confirmed that the microbiome of the lungs more closely resembles that of the oropharynx than it does competing source communities: inhaled air, the nasopharynx, or the lower gastrointestinal tract via hematogenous spread [15–18]. Both a direct study within individuals and a large population-based model have demonstrated that the nasal microbiome contributes little to lung communities in health [15,16]; the microbiome of the nose more closely resembles that of the skin than that of the lungs. Importantly, this similarity between lung and oral microbiota is evident even when the lung is sampled via a nasally introduced bronchoscope, demonstrating the minimal influence of upper respiratory tract contamination on bronchoscopically acquired specimens [10,19]. The oropharynx produces two liters of saliva per day, a far greater volume of secretions than is produced by the nasal mucosa in health. It is possible (but not proven) that lung and nasal microbial communities converge in times of increased rhinorrhea (e.g., acute viral infections or allergic rhinitis, both of which can provoke exacerbations of lung disease associated with nasal microbes such as Staphylococcus aureus and Moraxella catarrhalis).
The Lung Microbiome Changes during Disease
The ecological determinants of the lung microbiome—immigration, elimination, and regional growth conditions—all change dramatically during acute and chronic lung disease [10]. Consequently, the community membership of the lung microbiome is altered in disease states. Of the dozens of studies that have compared the microbiota of diseased lungs with those of healthy subjects, virtually all have found significant differences in community composition [20]. Many have described an increased community richness (number of species) in chronically diseased airways, often with a shift in community composition away from the Bacteroidetes phylum, which dominates the healthy lung microbiome, towards Proteobacteria, the phylum that contains many familiar lung-associated gram-negative bacilli. Baseline differences in lung microbiota have been associated with important clinical features of chronic lung disease: subsequent exacerbation frequency in bronchiectasis [21], mortality in idiopathic pulmonary fibrosis [22], and responsiveness to corticosteroids and antibiotics in asthma [23,24]. Active topics of investigation in the field include (1) whether an altered lung microbiome contributes to disease pathogenesis or is merely a marker of injury and inflammation, (2) whether the lung microbiome can be manipulated therapeutically to change exacerbation frequency or disease progression, and (3) how the diverse and dynamic homeostasis of the lung ecosystem collapses and is dominated by a single dominant pathogen in pneumonia [14,25].
Zdroje
1. Horikoshi K, Grant WD (1998) Extremophiles: Microbial Life In Extreme Environments: Wiley-Liss.
2. Gleeson K, Eggli DF, Maxwell SL Quantitative aspiration during sleep in normal subjects. Chest. 1997; 111 : 1266–1272. 9149581
3. Huxley EJ, Viroslav J, Gray WR, Pierce AK Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med. 1978; 64 : 564–568. 645722
4. Quinn LH, Meyer OO The relationship of sinusitis and bronchiectasis. Archives of Otolaryngology—Head & Neck Surgery. 1929; 10 : 152.
5. Amberson JB A clinical consideration of abscesses and cavities of the lung. Bull Johns Hopkins Hosp. 1954; 94 : 227–237. 13160680
6. Pasteur L Expériences relatives aux générations dites spontanées. Comptes Rendus Hebdomadaires des Séances de l'Académie des Sciences D: Sciences Naturelles. 1860; L: 303–307.
7. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010; 5: e8578. doi: 10.1371/journal.pone.0008578 20052417
8. Ingenito EP, Solway J, McFadden ER Jr., Pichurko B, Bowman HF, Michaels D, et al. Indirect assessment of mucosal surface temperatures in the airways: theory and tests. J Appl Physiol. 1987; 63 : 2075–2083. 3693240
9. Wu H, Kuzmenko A, Wan S, Schaffer L, Weiss A, Fisher JH, et al. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. 2003; 111 : 1589–1602. 12750409
10. Dickson RP, Martinez FJ, Huffnagle GB The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014; 384 : 691–702. doi: 10.1016/S0140-6736(14)61136-3 25152271
11. Lighthart B Mini-review of the concentration variations found in the alfresco atmospheric bacterial populations. Aerobiologia. 2000; 16 : 7–16.
12. Munyard P, Bush A How much coughing is normal? Arch Dis Child. 1996; 74 : 531–534. 8758131
13. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015.
14. Dickson RP, Erb-Downward JR, Huffnagle GB Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. 2014; 2 : 238–246. doi: 10.1016/S2213-2600(14)70028-1 24621685
15. Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. 2015; 6.
16. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015; 6.
17. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013; 187 : 1067–1075. doi: 10.1164/rccm.201210-1913OC 23491408
18. Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013; 1 : 19. doi: 10.1186/2049-2618-1-19 24450871
19. Dickson RP, Erb-Downward JR, Freeman CM, Walker N, Scales BS, Beck JM, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct pseudomonas species with distinct clinical associations. PLoS ONE. 2014; 9: e97214. doi: 10.1371/journal.pone.0097214 24831685
20. Dickson RP, Erb-Downward JR, Huffnagle GB The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013; 7 : 245–257. doi: 10.1586/ers.13.24 23734647
21. Rogers GB, Zain NM, Bruce KD, Burr LD, Chen AC, Rivett DW, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc. 2014;11 : 496–503. doi: 10.1513/AnnalsATS.201310-335OC 24592925
22. Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014; 190 : 906–913. doi: 10.1164/rccm.201403-0541OC 25184687
23. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011; 127 : 372–381 e371–373. doi: 10.1016/j.jaci.2010.10.048 21194740
24. Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013; 188 : 1193–1201. doi: 10.1164/rccm.201304-0775OC 24024497
25. Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014; 52 : 3605–3613. doi: 10.1128/JCM.01028-14 25078910
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



