-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
article has not abstract
Published in the journal: . PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1004936
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004936Summary
article has not abstract
Are Current Antifungal Drug Targets Sufficient for the Treatment of Systemic Mycosis?
Systemic and invasive mycoses caused by primary and opportunistic fungal pathogens have been emerging as global problems because of the increase in the number of immunocompromised individuals, due to solid-organ transplants, anti-cancer chemotherapy, and extended human lifespan. A recent report estimated that fungal pathogens, such as Cryptococcus neoformans, Candida albicans, and Aspergillus fumigatus, are responsible for more than 1 million deaths annually [1]. Despite this, the availability of antifungal drugs or targets for antifungal drug development are very limited. This is unlike the situation of bacterial pathogens and, to an extent, the eukaryotic parasites (which is serious enough) because the animals share a more recent common ancestor with the fungi than other pathogens. Ergosterol and its biosynthetic enzymes are the most popular antifungal drug targets because of the structural distinguishability of ergosterol from cholesterol in mammalian cell membranes. Polyene macrolides directly bind to ergosterol and generate lethal transmembrane channels that leak essential cellular ions and perturb osmotic balances, which leads to cell death [2]. Azole and allylamine derivatives are inhibitors of the ergosterol biosynthetic pathway that inhibit 14α-demethylase and squalene epoxidase, respectively, eventually leading to the accumulation of toxic precursors of ergosterol in the cell membrane and subsequent impairment of membrane integrity [3]. Another promising antifungal drug target is the fungal cell wall. Echinocandin inhibits β-1,3-glucan synthase and impairs cell wall integrity [4]. Nucleotide biosynthesis is also, somewhat unexpectedly, an appropriate antifungal drug target. For example, flucytosine itself does not have antifungal activity; however, after its uptake into cells, it is rapidly converted to 5-fluorouracil, which inhibits DNA and protein synthesis by cytosine deaminase, absent in humans [5]. However, all these antifungal drugs have problems, such as toxicity (e.g., hepatotoxicity and nephrotoxicity), frequent emergence of resistance, and a limited spectrum [2–5]. To overcome these problems, novel antifungal drug targets and drugs need to be discovered and developed.
Can Transcription Factors Serve As Novel Antifungal Drug Targets?
Transcription factors (TFs) are attractive as novel antifungal drug targets because they are evolutionarily divergent between fungi and humans (even among fungal species) and hence can be exploited as selective drug targets. In general, TFs have been considered poor drug targets because drugs that target them would have to specifically disrupt protein–nucleic acid or protein–protein interactions, rather than simply binding to an active pocket in an enzyme [6]. However, accumulated evidence thus far strongly demonstrates that TFs are chemically tractable [7–9]. Natural or synthetic chemicals or peptidomimetics have been identified based on their ability to inhibit hetero - or homo-dimerization of TFs, TF-binding DNA elements, DNA-binding domains of TFs, or the interaction between a TF and its essential modulating proteins, as summarized in Fig 1. Among the TF-targeting small molecules listed in Fig 1, nutlins, which are specific MDM2-p53 antagonists, are being evaluated in the early clinical trials for cancer treatment [10]. To this end, chemical biologists employ yeast two-hybrid or one-hybrid reporter assays; fluorescence resonance energy transfer (FRET) assays; small-molecule microarrays; or structure-based, computational, virtual, drug-protein docking simulations as screening tools [7–9]. Besides the approach using small molecules, other approaches using polyamides, small interfering RNAs, TF decoy oligonucleotides, and synthetic peptides have been employed to inhibit TFs [7–9]. Therefore, along with these recent technical breakthroughs in chemical biology fields, structural information and elucidation of the DNA-binding element and any co-regulators for a given virulence-regulating TF could pave the way for developing novel antifungal agents and therapeutic methods.
Fig. 1. Potential mechanisms for the chemical modulation of transcription factors. 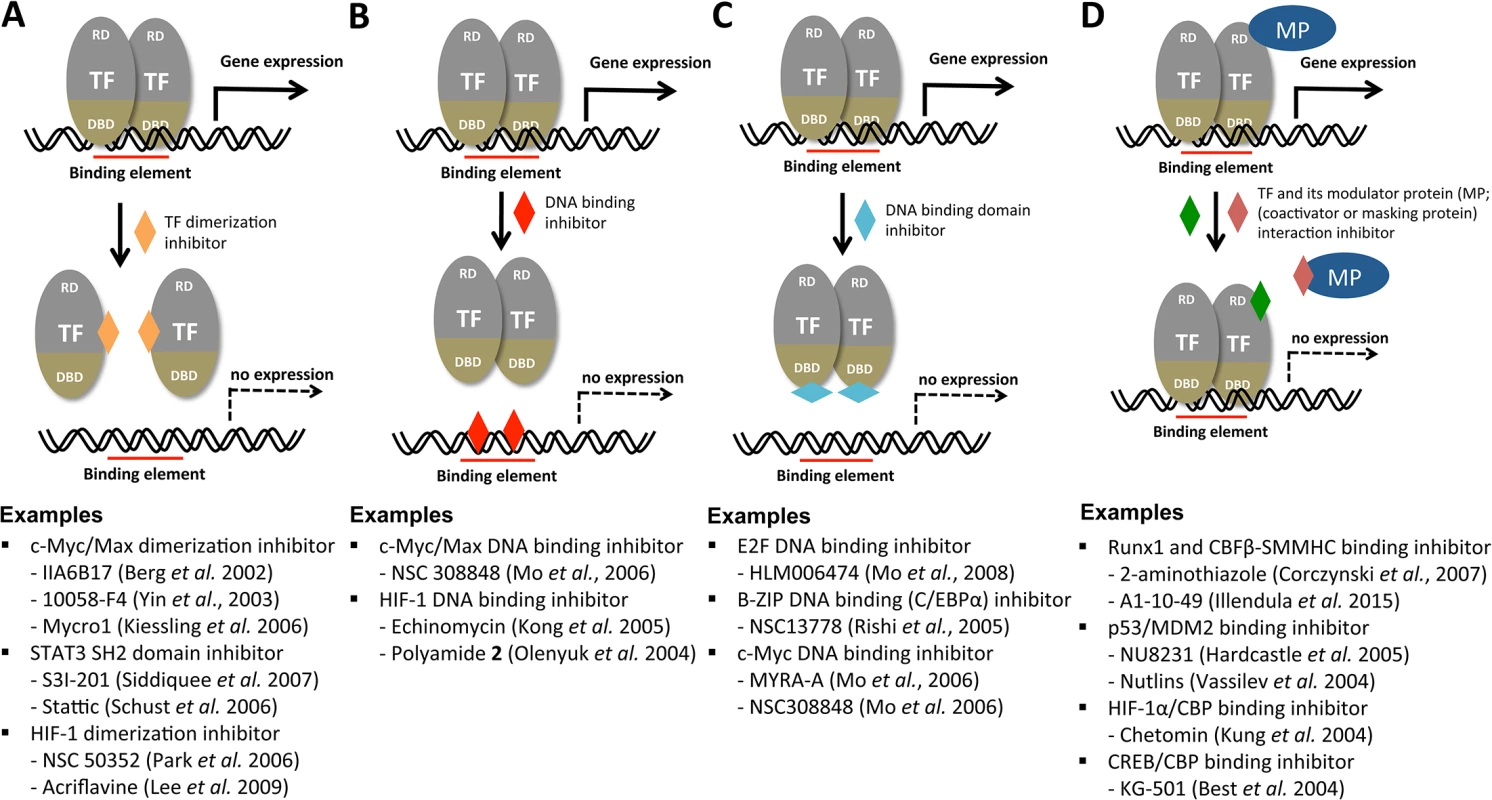
Transcription factors (TFs) are illustrated as a modular structure consisting of a DNA-binding domain (DBD) and regulatory domain (RD). MP indicates any modular protein (coactivator or masking protein). Although TFs are assumed to be homodimers here, they could have monomeric, multimeric, heterodimeric, or other structural configurations. In this overview, chromatin-remodeling enzymes, RNA polymerase and its multiple cofactors, all of which are required for general transcriptional induction of a gene, are not illustrated. The function of a TF can be perturbed by inhibiting TF dimerization (A), masking the DNA binding element (B), TF DNA binding domain (C), or inhibiting interaction between the TF and its modulator protein (D). Detailed reference information for each developing or developed TF inhibitor under each category can be obtained from the following review articles: [7–9]. How Much Have Transcription Factors Been Functionally Characterized in Human Fungal Pathogens?
Due to the reasons described above, the functional characterization of fungal TFs, particularly their role in pathogenicity, has been in high demand. To this end, several large-scale functional genomic analyses of fungal TFs have been independently performed in two major human fungal pathogens, C. albicans and C. neoformans. Nobile and Mitchell generated 83 TF mutants of C. albicans and addressed their roles in biofilm formation [11]. Homann et al. characterized the in vitro functions of 166 TFs under 50 different growth conditions [12]. However, neither study directly addressed their roles in the pathogenicity of C. albicans. Subsequently, Noble et al. generated homozygous mutant strains for 674 genes and discovered 115 infectivity-attenuated mutants [13]. Among these, 13 virulence-regulating TFs were discovered. In C. neoformans, three large-scale functional analyses of TFs have been performed thus far. Liu et al. generated gene-deletion mutants for 58 TFs as a part of large-scale gene deletion project and discovered 20 TFs that are involved in the lung infectivity of C. neoformans [14]. More recently, Jung et al. generated gene-deletion mutants for 155 non-essential, sequence-specific DNA-binding TFs and evaluated their virulence and infectivity potentials in both insect and murine models [15]. They discovered that 45 TFs are involved in either virulence or infectivity of C. neoformans. Furthermore, for the purpose of constructing model-driven comprehensive transcriptional networks for capsule biosynthesis in C. neoformans, Maier et al. analyzed 41 TF mutants with altered capsule production and found 10 infectivity-related TFs [16]. Besides these large-scale studies, a number of studies characterizing the function of individual TFs in the pathogenicity of C. albicans and C. neoformans have been performed. Therefore, a plethora of information is available for selecting potential TF targets that could be exploited for the development of TF-targeting antifungal drugs.
What Transcription Factors Could Be Broad-Spectrum Antifungal Drug Targets?
Comparison of functional TF analysis data of both C. albicans and C. neoformans provides an insight into what kinds of TFs could be exploited as broad - or narrow-spectrum antifungal drug targets. TFs that have been demonstrated to be involved in infectivity or virulence of C. albicans and C. neoformans are summarized in Fig 2. The following six TFs were found to be commonly involved in the virulence of both fungal pathogens: Crz1, Nrg1, Rim101, Bcr1/Usv101, Zap1/Zap104, and Brg1/Gat201.
Fig. 2. Virulence-regulating transcription factors in Candida albicans and Cryptococcus neoformans. 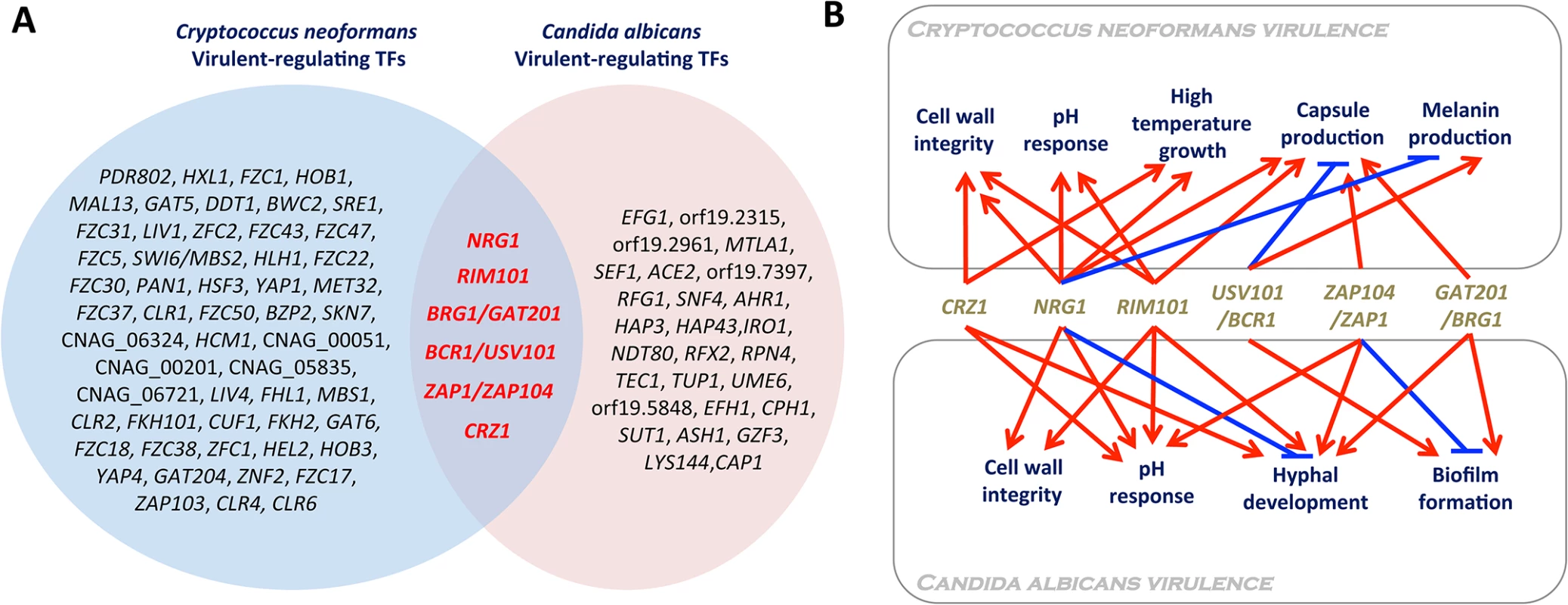
(A) The Venn diagram demonstrates TF genes whose deletion affects (either reduces or enhances) the pathogenicity (infectivity and/or virulence) of C. albicans and C. neoformans. List of virulence-regulating TFs was mainly retrieved from large-scale functional genetic studies of C. neoformans and C. albicans TFs [11,13–16] and a curated C. albicans genome database (http://www.candidagenome.org/). (B) Virulence-related cellular functions of TFs that could be exploited as broad-spectrum antifungal drug targets. Deletion of NRG1, BRG1/GAT201, BCR1/USV101, ZAP1/ZAP101, or CRZ1 is known to reduce the virulence of C. albicans and C. neoformans. Deletion of RIM101 enhances the virulence of C. neoformans but reduces the virulence of C. albicans. Interestingly, except Brg1/Gat201, Crz1, Nrg1, Rim101, Bcr1/Usv101, and Zap1/Zap104, all encode C2H2 zinc finger-type TFs. Crz1 is a direct downstream target of fungal calcineurin, a Ca2+/calmodulin-dependent protein phosphatase that modulates ion homeostasis, pH response, cell wall integrity, thermotolerance, developmental processes, and/or the virulence of a variety of human fungal pathogens including C. albicans, C. neoformans, and A. fumigatus [17]. The role of Rim101 and Nrg1 is similar to that of Crz1 in cell wall integrity; deletion of RIM101 and NRG1 alters cell wall integrity in both C. neoformans [18,19] and C. albicans [20,21]. Furthermore, Rim101 and Nrg1 are functionally well connected for modulating cellular pH responses in human fungal pathogens. Notably, however, deletion of RIM101 reduces the virulence of C. albicans [22] but increases the virulence of C. neoformans due to abnormal stimulation of immune responses [18,23], suggesting that Rim101 may not be a good broad-spectrum antifungal drug target. Rim101 negatively regulates Nrg1, which represses the expression of ENA1, a Na+/K+ transporter, in both C. albicans and C. neoformans [24,25]. In C. neoformans particularly, deletion of ENA1 completely abolishes virulence [24]. Therefore, their roles in pH response and cation homeostasis may also affect the virulence of fungal pathogens.
The function of Usv101 has been recently characterized in C. neoformans by Jung et al. [15] and Maier et al. [16]. The usv101∆ mutant had a reduced pulmonary infectivity, whereas it was shown to have an increased lung infectivity by Liu et al. [14]. This discrepancy seems to result from differences in the parental strains used for each study. The usv101∆ mutant exhibits increased capsule production, but decreased melanin synthesis, in the H99S and KN99α backgrounds. The latter phenotypic trait may be responsible for its reduced infectivity. By contrast, the CMO18 strain is inherently defective in melanin synthesis. Therefore, only capsule enhancement could be reflected in the increased lung infectivity of the usv101∆ mutant in the CMO18 strain. The closest C. albicans ortholog of Usv101 is Bcr1, which regulates biofilm formation and expression of cell-surface adherence genes [11]. Deletion of ZAP104 severely diminishes both capsule production and mating efficiency in C. neoformans [15], among which the former trait may affect the virulence of C. neoformans. The closest C. albicans ortholog of Zap104 is Zap1/Csr1. Zap1 is not only a regulator of zinc homeostasis but also regulates filamentous growth and biofilm maturation of C. albicans in positive and negative manners, respectively [26].
BRG1/GAT201 encodes a GATA-type TF. In C. neoformans, Gat201 regulates both capsule-dependent and-independent antiphagocytic mechanisms in C. neoformans [27]. Deletion of GAT201 severely reduces the lung infectivity of C. neoformans [14,15]. Surprisingly, Gat201 regulates about 16% of the C. neoformans genome, suggesting that it is one of the master regulators [27]. The closest C. albicans ortholog of Gat201 is Brg1 (also known as Gat2), which plays a major role in hyphal elongation in C. albicans by recruiting the histone deacetylase Hda1 to the promoters of hypha-specific genes, while the cAMP/PKA-dependent removal of Nrg1 is required for hyphal initiation [28]. Furthermore, Brg1 also promotes biofilm development [29]. The fact that both overexpression and deletion of BRG1 attenuated the virulence of C. albicans [30,31] suggests that orchestrated in vivo regulation of BRG1 is critical for its pathogenicity.
Notably, mutants of Crz1, Nrg1, Usv101, Gat201, and Zap104 exhibit increased susceptibility to polyene or azole drugs [15], suggesting that these broad-spectrum target TFs could also be exploited as combination therapeutic targets of antifungal drugs that are already clinically available. However, regardless of orthologous relationship for the common virulence-regulating TFs between C. neoformans and C. albicans, the homologous region is mainly restricted to the DNA binding region. Therefore, it will be still challenging to develop a broad-spectrum antifungal agent that specifically targets the TFs.
How About Exploiting Narrow-Spectrum Antifungal Drug Targets?
Although broad-spectrum antifungal drugs are generally favored from an industrial perspective, they have some potential drawbacks. For example, any known or unknown commensal or mutualistic fungal species in humans could be broadly affected, which may affect the balance of the normal microflora and cause secondary infection with other unwanted pathogens. In fact, C. albicans is part of the normal microflora in the gastrointestinal tract of healthy individuals. Therefore, if the identity of a fungal pathogen could be determined in the early stage of mycoses, pathogen-specific, narrow-spectrum targets could be even more optimal, and any drugs targeting such TFs would be expected to have less toxic effects (Fig 2). Several evolutionarily divergent, virulence-regulating TFs could be exploited as narrow-spectrum antifungal drugs.
Among many narrow-spectrum TF target candidates, Efg1 is the best characterized in C. albicans. Efg1, which is mainly controlled by the cAMP/PKA pathway, controls hyphal development and white-opaque phenotypic switching, both of which are critically involved in the pathogenicity of C. albicans [32]. In C. neoformans, HXL1, which is a bona fide downstream transcription factor of the Ire1 kinase in the unfolded-protein response pathway, is most notable, as its structure is evolutionarily divergent from its counterpart in humans (Xbp1) and its deletion completely abolishes the virulence of C. neoformans [33]. Another major benefit of targeting Efg1 or Hxl1 is that their inhibition strongly enhances the susceptibility to azole drugs [33,34], suggesting that they could be exploited as both single and combination therapeutic methods.
Perspectives
As large-scale functional genomics data of fungal TFs become more readily available and their roles in fungal pathogenicity are uncovered, chemical biologists will become more interested in developing TF-targeting antifungal agents. To provide an efficient drug screening system exploiting virulence-regulating TFs, the following data need to be accumulated: (1) identification of direct DNA-binding elements; (2) structural characterization and functional domain analysis; (3) identification of any essential interacting partners, particularly co-activators; and (4) identification of any upstream regulators and downstream effector proteins. The core consensus DNA-binding motif for Crz1, CDRE (calcineurin-dependent response element), has been shown to be present in some fungi [17]. Recently, Maier et al. identified DNA-binding motifs for Gat201, Nrg1, and Usv101 by chromatin immunoprecipitation-sequence analysis in C. neoformans [16]. This information will be useful in constructing any drug-screening system for identifying direct inhibitors of TF-DNA interactions. In conclusion, a further in-depth molecular and genetic analysis of the virulence-regulating TFs will provide new insight into developing novel classes of antifungal agents that could resolve the problems associated with the currently available drugs.
Zdroje
1. Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, et al. (2012) Hidden killers: human fungal infections. Science translational medicine 4 : 165rv113.
2. Kong D, Lee MJ, Lin S, Kim ES (2013) Biosynthesis and pathway engineering of antifungal polyene macrolides in actinomycetes. J Ind Microbiol Biotechnol 40 : 529–543. doi: 10.1007/s10295-013-1258-6 23515854
3. Geronikaki A, Fesatidou M, Kartsev V, Macaev F (2013) Synthesis and biological evaluation of potent antifungal agents. Curr Top Med Chem 13 : 2684–2733. 24083791
4. Zaas AK (2008) Echinocandins: a wealth of choice—how clinically different are they? Curr Opin Infect Dis 21 : 426–432. doi: 10.1097/QCO.0b013e328307c79c 18594297
5. Vermes A, Guchelaar HJ, Dankert J (2000) Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. J Antimicrob Chemother 46 : 171–179. 10933638
6. Imming P, Sinning C, Meyer A (2006) Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov 5 : 821–834. 17016423
7. Koehler AN (2010) A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol 14 : 331–340. doi: 10.1016/j.cbpa.2010.03.022 20395165
8. Berg T (2008) Inhibition of transcription factors with small organic molecules. Curr Opin Chem Biol 12 : 464–471. doi: 10.1016/j.cbpa.2008.07.023 18706517
9. Majmudar CY, Mapp AK (2005) Chemical approaches to transcriptional regulation. Curr Opin Chem Biol 9 : 467–474. 16122970
10. Duffy MJ, Synnott NC, McGowan PM, Crown J, O'Connor D, et al. (2014) p53 as a target for the treatment of cancer. Cancer Treat Rev 40 : 1153–1160. doi: 10.1016/j.ctrv.2014.10.004 25455730
11. Nobile CJ, Mitchell AP (2005) Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15 : 1150–1155. 15964282
12. Homann OR, Dea J, Noble SM, Johnson AD (2009) A phenotypic profile of the Candida albicans regulatory network. PLoS genetics 5: e1000783. doi: 10.1371/journal.pgen.1000783 20041210
13. Noble SM, French S, Kohn LA, Chen V, Johnson AD (2010) Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42 : 590–598. doi: 10.1038/ng.605 20543849
14. Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, et al. (2008) Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135 : 174–188. doi: 10.1016/j.cell.2008.07.046 18854164
15. Jung KW, Yang DH, Maeng S, Lee KT, So YS, et al. (2015) Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Comms 6 : 6757.
16. Maier EJ, Haynes BC, Gish SR, Wang ZA, Skowyra ML, et al. (2015) Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res. 25 : 690–700. doi: 10.1101/gr.184101.114 25644834
17. Thewes S (2014) Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot Cell 13 : 694–705. doi: 10.1128/EC.00038-14 24681686
18. O'Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA (2013) Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4: e00522–00512. doi: 10.1128/mBio.00522-12 23322637
19. Cramer KL, Gerrald QD, Nichols CB, Price MS, Alspaugh JA (2006) Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryot Cell 5 : 1147–1156. 16835458
20. Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, et al. (2008) Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol 10 : 2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x 18627379
21. Lotz H, Sohn K, Brunner H, Muhlschlegel FA, Rupp S (2004) RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryot Cell 3 : 776–784. 15189998
22. Davis D, Edwards JE Jr., Mitchell AP, Ibrahim AS (2000) Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun 68 : 5953–5959. 10992507
23. O'Meara TR, Norton D, Price MS, Hay C, Clements MF, et al. (2010) Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6: e1000776. doi: 10.1371/journal.ppat.1000776 20174553
24. Jung KW, Strain AK, Nielsen K, Jung KH, Bahn YS (2012) Two cation transporters Ena1 and Nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the HOG pathway. Fungal Genet Biol 49 : 332–345. doi: 10.1016/j.fgb.2012.02.001 22343280
25. Lamb TM, Mitchell AP (2003) The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol 23 : 677–686. 12509465
26. Ganguly S, Bishop AC, Xu W, Ghosh S, Nickerson KW, et al. (2011) Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell 10 : 1448–1454. doi: 10.1128/EC.05196-11 21890817
27. Chun CD, Brown JC, Madhani HD (2011) A major role for capsule-independent phagocytosis-inhibitory mechanisms in mammalian infection by Cryptococcus neoformans. Cell Host Microbe 9 : 243–251. doi: 10.1016/j.chom.2011.02.003 21402362
28. Lu Y, Su C, Liu H (2012) A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog 8: e1002663. doi: 10.1371/journal.ppat.1002663 22536157
29. Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, et al. (2013) Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLoS Pathog 9: e1003305. doi: 10.1371/journal.ppat.1003305 23637598
30. Cleary IA, Lazzell AL, Monteagudo C, Thomas DP, Saville SP (2012) BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol Microbiol 85 : 557–573. doi: 10.1111/j.1365-2958.2012.08127.x 22757963
31. Du H, Guan G, Xie J, Sun Y, Tong Y, et al. (2012) Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One 7: e29707. doi: 10.1371/journal.pone.0029707 22276126
32. Liu H (2002) Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol 292 : 299–311. 12452278
33. Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, et al. (2011) Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog 7: e1002177. doi: 10.1371/journal.ppat.1002177 21852949
34. Prasad T, Hameed S, Manoharlal R, Biswas S, Mukhopadhyay CK, et al. (2010) Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res 10 : 587–596 doi: 10.1111/j.1567-1364.2010.00639.x 20491944
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



