-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
The nuclear factor kappaB (NF-κB) signaling pathway is crucial for host defense, as it orchestrates both innate and adaptive immune responses. Beyond the best-studied Rel proteins (p65, RelB, c-Rel, p50 and p52), RPS3 has been recently identified as a “specifier” component of NF-κB, modulating the promoter selectivity and transcriptional specificity of NF-κB. In particular, the RPS3/p65-conferred signaling pathway was recently shown to play a critical role in host proinflammatory transcription and immune responses. Attaching and effacing (A/E) pathogens and others have acquired sophisticated mechanisms to modulate host NF-κB signaling pathways. We have found that NleC, a metalloprotease effector secreted by A/E pathogens, modulates host NF-κB signaling and inflammatory responses through a novel mechanism. NleC specifically recognizes and cleaves a small percentage of p65 and the generated N-terminal fragment of p65 interferes with the p65/RPS3 interaction, thereby amplifying the effect of cleaving only a small percentage of p65 molecules to selectively inhibit NF-κB gene expression. Our findings highlight a previously unappreciated mechanism through which pathogen-encoded proteases interfere with signaling cascades and inflammatory responses in host cells.
Published in the journal: . PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004705
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004705Summary
The nuclear factor kappaB (NF-κB) signaling pathway is crucial for host defense, as it orchestrates both innate and adaptive immune responses. Beyond the best-studied Rel proteins (p65, RelB, c-Rel, p50 and p52), RPS3 has been recently identified as a “specifier” component of NF-κB, modulating the promoter selectivity and transcriptional specificity of NF-κB. In particular, the RPS3/p65-conferred signaling pathway was recently shown to play a critical role in host proinflammatory transcription and immune responses. Attaching and effacing (A/E) pathogens and others have acquired sophisticated mechanisms to modulate host NF-κB signaling pathways. We have found that NleC, a metalloprotease effector secreted by A/E pathogens, modulates host NF-κB signaling and inflammatory responses through a novel mechanism. NleC specifically recognizes and cleaves a small percentage of p65 and the generated N-terminal fragment of p65 interferes with the p65/RPS3 interaction, thereby amplifying the effect of cleaving only a small percentage of p65 molecules to selectively inhibit NF-κB gene expression. Our findings highlight a previously unappreciated mechanism through which pathogen-encoded proteases interfere with signaling cascades and inflammatory responses in host cells.
Introduction
Foodborne diseases caused by enteric pathogens remain a significant and common health threat and an immense economic burden worldwide [1]. Among the causative agents of foodborne illness, diarrheagenic strains of Escherichia coli including enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC), typically cause diarrhea, hemorrhagic colitis, and pediatric renal failure [2]. EPEC, EHEC, and the rodent-specific pathogen Citrobacter rodentium produce characteristic attaching/effacing (A/E) lesions on the host intestinal epithelium after they adhere to these cells [3]. These pathogens translocate a variety of virulence proteins (effectors), through a conserved type III secretion system (T3SS), into intestinal epithelial cells (IECs) to modulate host cell functions to the pathogen’s advantage [4,5]. An ever-expanding repertoire of T3SS secreted effectors, termed non-LEE-encoded (Nle) effectors, was recently identified in A/E pathogens [6,7,8,9,10]. The target proteins of Nle effectors in host cells have started to be identified [11,12,13,14,15,16,17,18,19,20]; however, it remains largely unknown how Nle effectors interfere with cell signaling cascades and dampen the immune responses in host cells. The recognition of pathogens by host sensors activates multiple signaling pathways to induce inflammatory responses and eradicate the pathogens [21]. Among those, the NF-κB signaling pathway is crucial for host defense, as it orchestrates both innate and adaptive immune responses [21]. On the other hand, A/E bacteria, like other successful pathogens, have acquired sophisticated mechanisms to modulate host NF-κB signaling pathways [22,23,24,25,26,27,28]. Not surprisingly, a handful of the Nle effector target proteins within host cells have been revealed to be NF-κB signaling molecules [11,12,13,14,15,16,17,18,29,30]. Notably, however the molecular mechanisms through which each of these Nle effectors modulate NF-κB signaling have not been fully elucidated [25,31].
Besides the well-defined Rel family proteins (RelA/p65, RelB, c-Rel, p50 and p52) [32], RPS3 and Src-associated substrate during mitosis of 68kDa (Sam68) were recently identified as “specifier” components of NF-κB [33], where they modulate the promoter selectivity and transcriptional specificity of NF-κB [34,35]. The nuclear translocation and “specifier” function of RPS3 have been revealed to be tightly regulated by NF-κB signaling cascades [18]. Specifically, RPS3 is found in the cytoplasmic p65-p50-IκBα inhibitory complex in resting cells [34]. External stimuli activate the IκB kinase (IKK) complex, of which IKKβ phosphorylates IκBα resulting in its subsequent ubiquitination and degradation. IκBα removal unmasks a nuclear localization sequence (NLS), which allows nuclear import of p65 and p50 [36]. Likewise, IKKβ phosphorylates RPS3 at serine 209 (Ser209), independently enhancing the RPS3-importin-α interaction for nuclear translocation. Once in the nucleus, RPS3 cooperates with p65 to target NF-κB to select promoters and to trans-activate those genes [18]. Of note, the significance of RPS3/NF-κB signaling pathway has been highlighted in an increasing number of pathophysiological conditions [17,18,34,37,38,39], particularly in host proinflammatory transcription and immune responses against enteric pathogen infections [17,18]. More specifically, the EHEC NleH1 effector inhibits the nuclear translocation of RPS3, but not p65, during NF-κB activation by tempering RPS3 Ser209 phosphorylation [17,18]. As a consequence, NleH1 reduces the transcription of select, but not all, NF-κB target genes; most of the NleH1-attenuated RPS3/NF-κB-dependent genes encode proinflammatory cytokines/chemokines [17,18]. In support of the critical role of RPS3 in the transcriptional selectivity of NF-κB genes, we recently demonstrated that modulating the RPS3/p65 interaction by ectopic expression of an N-terminal fragment (amino acids 21–186) of p65 attenuates RPS3 nuclear translocation, without affecting p65, thus selectively blocking a subset of specific NF-κB gene transcription [40].
NleC, a zinc-dependent protease effector conserved among A/E pathogens, was recently identified as one of the key effectors that dampen the innate immune response in host cells, particularly the production of inflammatory cytokines including interleukin-8 (IL-8) as well as others [16,20,30]. Mutagenesis of the consensus zinc metalloprotease motif 183HEIIH187 abrogates the proteolytic activity of NleC and cleavage of host target proteins [16,29,30,41]. Although p300 [42], IκB [29], and the NF-κB Rel proteins p65 and p50 [16,29,30,41,42] have been reported as targets of NleC, it is largely understood that NleC cleaves and inactivates the NF-κB signaling pathway by primarily targeting the Rel proteins [16,29,30,41], in line with the known critical role of NF-κB in the transcription of inflammatory cytokine genes [43]. Previous studies have shown that ectopically expressed or T3SS-translocated NleC degrades p65, p50 and c-Rel, but not signal transducer and activator of transcription 1 (STAT1) or extracellular-signal-regulated kinases (ERKs), which indicates there may be cleavage specificity of NleC for the Rel subunits of NF-κB signaling pathway [30,41]. However, work by Yen et al. suggests that NleC could be specific for p65, as recombinant NleC could not digest p50 in cell lysates [16]. Moreover, two cleavage site(s) on p65 by NleC have been identified as between proline 10 and alanine 11 (P10/A11) [16] or cysteine 38 and glutamic acid 39 (C38/E39) [41,44,45], although none of these studies ruled out the other cleavage site experimentally. The detailed mechanisms on how NleC specifically recognizes and cleaves p65 remain poorly understood. Moreover, even though recombinant proteins and ectopic expression of NleC in cell lines were employed in previous studies, only a very small percentage of p65 was shown to be cleaved by NleC with a large portion of full-length p65 still present within the cells [16,41]. In contrast, it is puzzling that previous studies reported that NF-κB activity by luciferase assays and IL-8 production were markedly increased in HeLa cells infected with EPEC ΔnleC mutant, compared to wild-type EPEC [16,20,29,30,41,42].
Here we reveal that the N-terminus of p65 is specifically recognized by NleC, which is required for the subsequent NleC-mediated cleavage. Infection of mice with a C. rodentium mutant strain lacking NleC (ΔnleC) augmented the transcription of several proinflammatory cytokine genes including Cxcl1, Cxcl2, Il1b, Ifng, and Il22, and triggered more immune cell infiltration in the colon, compared to wild-type C. rodentium inoculation. Moreover, NleC primarily cleaves p65 at C38/E39 during C. rodentium infection, and we show that the generated p651–38 fragment binds to RPS3 in NF-κB complexes and selectively retards the nuclear translocation of RPS3, but not p65. While only a small percentage of molecules are cleaved, the association between the p651–38 fragment and RPS3 that interferes with RPS3/p65 interaction-mediated transcription amplifies the impact of NleC cleaving p65. Therefore our results reveal a novel mechanism by which A/E pathogens selectively dampen RPS3 signaling and ensure inhibition of the RPS3/NF-κB-dependent inflammatory responses in host cells.
Results
NleC specifically cleaves p65, but not other NF-κB subunits
The T3SS effector NleC from various A/E pathogens has been proposed to dampen the NF-κB-mediated proinflammatory responses in host cells by functioning as a metalloprotease that cleaves NF-κB, albeit through poorly-defined mechanism(s) [16,20,29,30,41,42]. Moreover, Rel subunits p65 [16,20,29,30,41,42], p50 [29], and c-Rel [16,20,29,30,41,42] were proposed to be NleC target proteins. Alignment of the N-terminal sequences of both human and mouse Rel family proteins reveals that the best-characterized cleavage site, C38/E39 within the Rel homology domain, in p65 is also conserved among all human and mouse Rel proteins (Fig. 1A and S1 Fig), indicating that NleC could potentially cleave all Rel proteins. To clarify which Rel protein(s) are NleC targets, we incubated whole cell lysates derived from HEK293T cells with recombinant NleC protein and examined the cleavage of endogenous Rel proteins using antibodies that specifically recognize the C-terminus of each Rel subunit. Consistent with previous studies [16,20,29,30,41,42], a percentage of endogenous p65 was cleaved by NleC at the N-terminus thus generating a large C-terminal fragment (Fig. 1B), whereas a catalytically inactive mutant of NleC, i.e. NleC (H117Y) with histidine 117 in the HExxH motif replaced by tyrosine [16], failed to do so (S2 Fig). Moreover, NleC cleaved p65 to a similar extent in the presence and absence of tumor necrosis factor (TNF), a strong NF-κB stimulus (S3 Fig), indicating that the NleC-mediated partial cleavage of p65 is not due to protection by other p65-binding proteins in the cytoplasm. In contrast to p65, the cleavage of other endogenous Rel proteins was not detectable, even when incubated with overwhelming amount (10 μg) of NleC recombinant protein (Fig. 1B). Moreover, recombinant NleC cleaved the N-terminally FLAG-tagged p65 protein, with the tag allowing for better resolution of the small N-terminal fragment (Fig. 1C). Of note, although recombinant NleC was able to cleave the ectopically expressed FLAG-tagged p50 protein, the product size marked by the N - and C-terminal FLAG tag indicates that cleavage occurs at the C-terminus of p50 rather than the conserved cysteine 62/glutamic acid 63 in the N-terminus (S4 Fig), which corresponds to C38/E39 in p65. In addition, recombinant NleC failed to cleave ectopically expressed RPS3, which is a non-Rel subunit of NF-κB that confers the promoter selectivity and transcriptional specificity [34], in the recombinant protease cleavage assay (S5 Fig). Therefore, NleC appears to selectively cleave p65, but not other NF-κB subunits.
Fig. 1. NleC specifically cleaves p65/RelA among the NF-κB Rel family proteins. 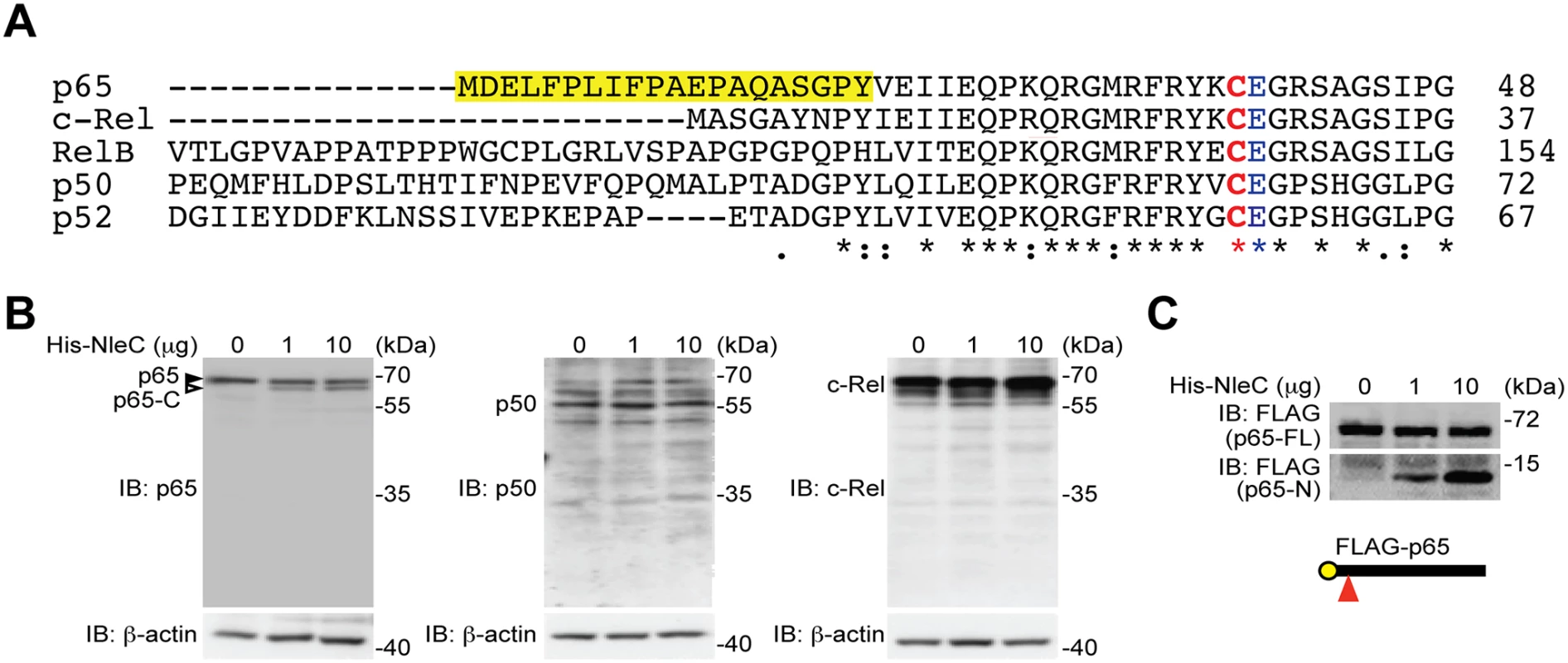
A. Sequence alignment of the human Rel family proteins. The conserved Cys/Glu residues in their respective Rel domains and the unique N-terminal 20 residues in p65/RelA are highlighted. The numbers at right show the position in the amino-acid sequence of the last residues depicted. B. Whole cell lysates derived from HEK293T cells were incubated with indicated amount of His-NleC recombinant protein, followed by SDS/PAGE separation and immunoblotted for indicated proteins with antibodies specifically recognizing the C-termini of p65, p50 and c-Rel, respectively. The full-length p65 and cleaved p65 C-terminal fragment are indicated by filled and open triangles, respectively. C. Whole cell lysates derived from HEK293T cells expressing N-terminally FLAG-tagged p65 were incubated with indicated amount of His-NleC recombinant protein. The cleavage of FLAG-tagged p65 was immunoblotted with anti-FLAG antibody, following SDS/PAGE separation. The NleC cleavage site in p65 is indicated by a red triangle. The N-terminal 20 amino acids of p65 are essential for NleC-mediated cleavage
Notably the first 20 amino acids of p65 are unique to this protein and not conserved among the Rel proteins (Fig. 1A and S1 Fig), providing a clue to why NleC cleaves p65 specifically, despite the fact that the C38/E39 cleavage site is shared by all other Rel subunits. Protease-substrate interaction-induced conformational changes are known to play an important role in the optimal cleavage of substrates by proteases [46]. We therefore hypothesized that the unique N-terminal sequence (a.a. 1–20) could be critical for the interaction between p65 and NleC, and be a prerequisite for NleC-mediated p65 cleavage. Using a library of C-terminal GFP-tagged p65 truncation constructs [34], we mapped the necessary region(s) of p65 for the NleC-conferred cleavage. Interestingly, recombinant NleC cleaved the p651–186 truncation as expected, whereas the p6521–186 truncation harboring the C38/E39 cleavage site but missing the first 20 amino acids was not cleaved by NleC, even in the presence of an overwhelming amount (10 μg) of the recombinant protease (Fig. 2A). To confirm this, we moved the GFP tag to the N-terminus of p6521–186 truncation to increase resolution of the cleavage product and we were still unable to detect cleavage (S6 Fig). These data suggest that the N-terminal 20 residues and C38/E39 cleavage site are both required for NleC-mediated p65 cleavage. In support of this notion, the full-length p65 containing both elements was cleaved by NleC, whereas no cleavage was detected in the p65186–311 and p65311–551 truncations as well as GFP vehicle control that do not contain either element (Fig. 2A, 3A-3B and S6 Fig). Together these results demonstrate that the first 20 amino acids are essential for NleC-mediated cleavage to occur, thus providing a rational for the specificity of NleC for p65 rather than other Rel homology proteins, despite a shared Rel homology domain sequence.
Fig. 2. The N-terminal 20 amino acids of p65 are required for NleC to bind and cleave p65. 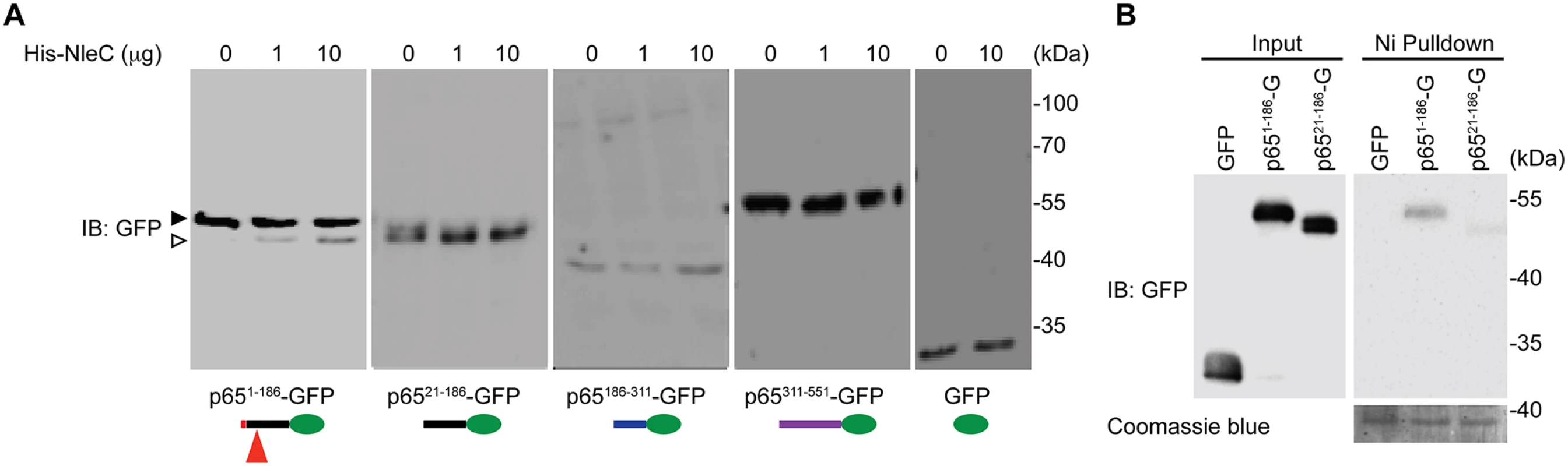
A. Various truncations of C-terminal GFP-tagged p65, as illustrated at the bottom, were transfected into HEK293T cells. Whole cell lysates were subjected to the His-NleC cleavage assay, and immunoblotted with anti-GFP antibody for NleC-cleaved fragments. The GFP-tagged p651–186 protein and cleaved fragment were labeled by filled and open triangles, respectively, and the NleC cleavage sites in p65 were indicated below. B. Whole cell lysates derived from HEK293T cells expressing GFP or the indicated GFP-tagged p65 proteins were incubated with the catalytic mutant His-NleC (H117Y) at 4°C. Nickel beads were added to pull-down His-NleC and associated proteins. Samples were separated by SDS/PAGE, followed by immunoblot for indicated proteins. Fig. 3. The dominant NleC cleave site on p65 is Cys38/Glu39. 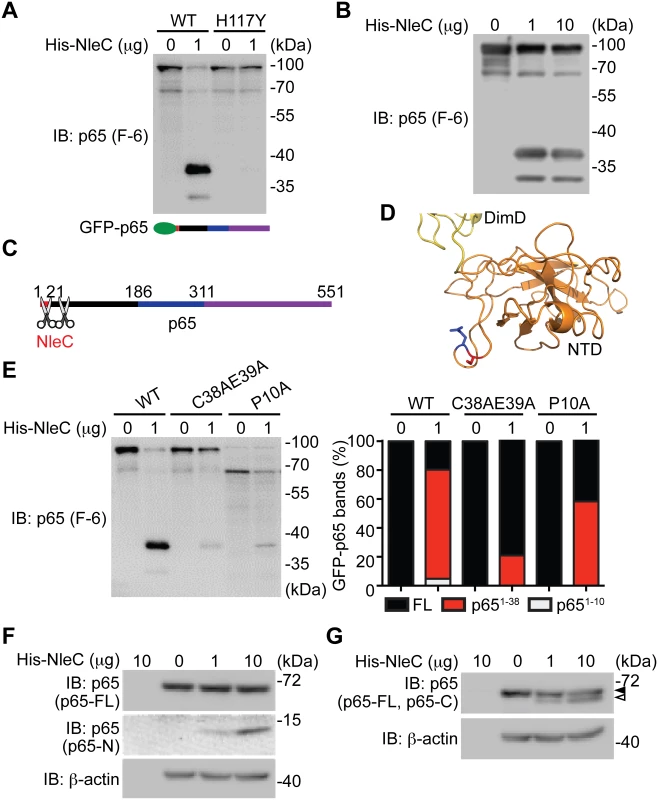
A-B. Whole cell lysates derived from HEK293T cells expressing N-terminal GFP-tagged p65 were subjected to the cleavage assay using wild-type His-NleC or the catalytic mutant His-NleC (H117Y), and immunoblotted with p65 N-terminus specific antibody for NleC-cleaved p65 fragments. C. Schematic of proposed NleC cleavage sites on p65. D. The structure of N-terminus of p65, with dimerization domain (DimD), N-terminal domain (NTD), Cys38, and Glu39 highlighted in yellow, orange, red, and blue, respectively. Image was created from PDB file 1VKX [47] using the Pymol software. E. Whole cell lysates derived from HEK293T cells expressing wild-type or mutant GFP-tagged p65 as indicated were subjected to the NleC cleavage assay and immunoblotted with p65 N-terminus specific antibody for NleC-cleaved p65 fragments. The percentage of GFP-tagged full-length p65, p651–38 fragment, and p651–10 fragment among the total GFP-tagged p65 proteins under each condition was quantified by densitometry. F-G. Whole cell lysates derived from HEK293T cells were subjected to the His-NleC cleavage assays, and immunoblotted with p65 N-terminus (F) and C-terminus (G) specific antibodies for NleC-cleaved p65 fragments. The N-terminal 20 amino acids of p65 are required for NleC recognition
It has been widely acknowledged that the recognition of a substrate by a protease is critical for subsequent conformational changes to form a stabilized tetrahedral intermediate and to initiate optimal cleavage [46]. To examine whether the N-terminal 1–20 residues of p65 are required for NleC to recognize and bind p65, we incubated the whole cell lysates from HEK293T cells expressing full-length or truncated p65, with catalytically inactive NleC (H117Y) mutant or wild-type NleC at 4°C and conducted pull-down assays using nickel beads. NleC associated with full-length p65, but did not interact with p65311–551 (S7 Fig), in line with the evidence that NleC cleaves full-length p65 rather than the truncated p65311–551 (Fig. 2A). Moreover, a significant interaction between NleC (H117Y) and p651–186 was detected, whereas there was little, if any, interaction between NleC (H117Y) and p6521–186 (Fig. 2B and S7 Fig.), suggesting that the N-terminal 20 amino acids could be the key targeting sequence for NleC to specifically recognize and bind to p65.
The major NleC cleavage site on p65 is between cysteine 38 and glutamic acid 39
Previous studies showed that C38/E39 is the NleC cleavage site in p65 [41,44,45], whereas P10/A11 was also reported as an NleC cleavage site [16]. We therefore conducted recombinant NleC cleavage assays using lysates containing p65 N-terminally tagged with GFP, which allowed us to further examine the cleavage location(s) on the N-terminus of p65. In agreement with our previous results (Fig. 2A), recombinant NleC cleaved p65 at the N-terminus generating two fragments that were N-terminally tagged with GFP; the cleaved products, which migrate at 37 kDa and 30 kDa, respectively, were verified using an antibody that specifically recognizes the N-terminus of p65 (Fig. 3A-3B). Of note, substantially more GFP-tagged p651–38 fragment was detected, indicating that NleC chiefly cleaves p65 at C38/E39. Therefore our results, in line with previous reports [16,41,44,45], demonstrate that the N-terminus of p65 harbors two NleC cleavage sites, P10/A11 and C38/E39, of which C38/E39 is the primary cleavage site whereas P10/A11 appears to be the secondary one (Fig. 3C). In support of this notion, the resolved p65 crystal structure [47] reveals that the C38/E39 residues are located in a surface loop of the protein, which facilitates protease access (Fig. 3D). Moreover, the NleC-generated p651–38 fragment is substantially reduced by an alanine substitution to C38/E39 (Fig. 3E). However, the NleC-cleaved p651–10 fragment was also abolished by the C38A/E39A mutation. In contrast, the NleC-generated p651–38 fragment is less profoundly impacted by the alanine substitution to P10, despite a complete inhibition of the NleC mediated cleavage of p65 at P10/A11 (Fig. 3E). These results therefore strengthen our conclusion that C38/E39 is the major NleC cleavage site within p65. Moreover, lysates from normal HEK293T cells were subjected to the NleC cleavage assays to examine how recombinant NleC cleaves endogenous p65. As detected by an antibody specific for the p65 N-terminus, a p65 fragment migrating as approximately 10 kDa on SDS/PAGE gels was generated by NleC in a dose dependent fashion (Fig. 3F). Because cleavage of p65 at P10/A11 would result in a fragment approximately 1 kDa in size that would likely be degraded, we therefore determined the cleaved p65 fragment is most likely the product of cleavage at C38/E39. Likewise, we observed a larger C-terminal fragment, migrating around 60 kDa on SDS/PAGE, using an antibody specific for the C-terminus of p65 (Fig. 3G). Together, our results suggest that C38/E39 is the major NleC cleavage site on p65, which generates a detectable p651–38 fragment.
NleC cleaves p65 and generates the p651–38 fragment during C. rodentium and EPEC infections
To further examine the pathophysiological relevance of NleC-mediated p65 cleavage, we employed the EPEC and C. rodentium infection models. After 3-hour infection of wild-type C. rodentium, we detected the p651–10 and p651–38 fragments in HEK293T cells expressing GFP-tagged p65 (Fig. 4A). The p651–10 and p651–38 cleavage was abolished in the cells infected with a C. rodentium mutant strain lacking NleC (ΔnleC) [20], compared to wild-type C. rodentium (Fig. 4A). Moreover, the attenuated p65 cleavage was robustly restored in the cells infected with a C. rodentium mutant strain that lacks NleC but was complemented with a HA-NleC plasmid (ΔnleC/pHA-NleC) [20] (Fig. 4A). Notably, we also observed the NleC-cleaved endogenous p651–38 fragment in Caco-2 cells, a human colon cancer cell line, and isolated mouse primary colon epithelial cells (CECs) infected by wild-type EPEC and C. rodentium, respectively (Fig. 4B and 4C). Strikingly, the p651–38 fragments were greatly diminished in cells infected with ΔnleC mutant bacteria; whereas the p65 cleavage was rescued in cells infected with complemented ΔnleC/pHA-NleC strains (Fig. 4B and 4C), suggesting that NleC cleaves p65 thus generating the p651–38 fragment during EPEC and C. rodentium infections in cell culture. In contrast to p65, the cleavage of p50 in Caco-2 cells and mouse CECs infected by EPEC and C. rodentium, respectively, was not detectable (S8 Fig), further supporting that NleC specifically cleaves p65 among NF-κB subunits. To examine the possibility that the abolished p65 cleavage in CECs infected with ΔnleC C. rodentium could be due to defective attachment to host cells, in comparison to wild-type and complemented strains, we measured the attachment of variant C. rodentium strains to CECs during infection. As shown in Fig. 4E, wild-type, ΔnleC, and ΔnleC/pHA-NleC C. rodentium attached to CECs in a similar pattern. Moreover, as assayed by enumeration of CEC-attached bacteria and immunoblot for marker proteins in mouse CECs (heat shock protein, Hsp90) and C. rodentium (lipopolysaccharides, LPS), the amount of ΔnleC and ΔnleC/pHA-NleC C. rodentium that attaches to CECs during infection was equal if not higher than the wild-type bacteria (Fig. 4D and 4F). These results rule out the possibility that NleC deletion affects the interaction between C. rodentium and CECs, and suggest that NleC executes the p65 cleavage during EPEC and C. rodentium infections, generating a small amount of p651–38 fragment and leaving most p65 intact.
Fig. 4. NleC cleaves p65 during C. rodentium and EPEC infections. 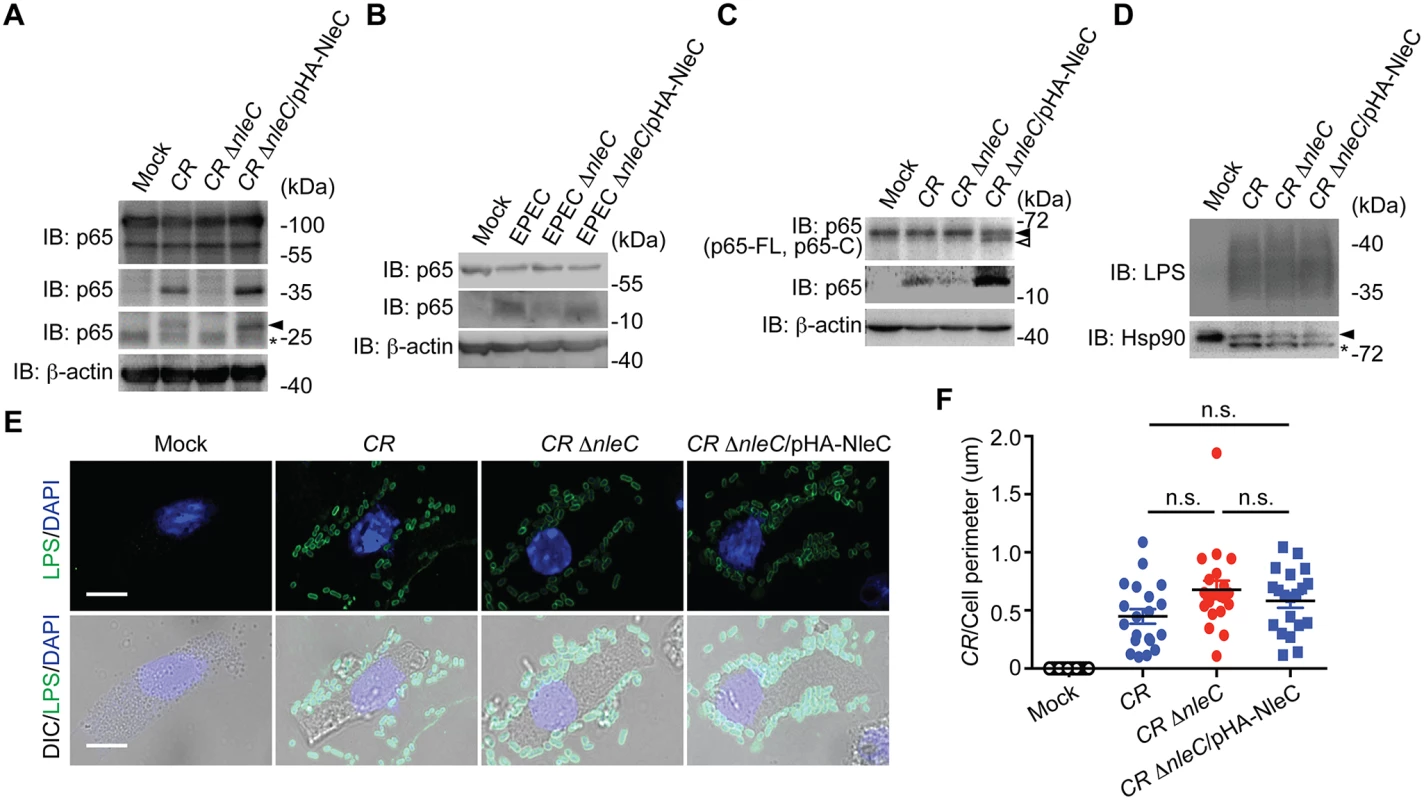
A. HEK293T cells expressing N-terminal GFP-tagged p65 were mock infected or infected with indicated C. rodentium strains at 100 MOI for 3 h. Whole cell lysates were derived, separated by SDS/PAGE, and immunoblotted for indicated proteins. The GFP-tagged p651–10 cleavage products are indicated by an arrow, and an asterisk labels nonspecific bands. B. Caco-2 cells were mock infected or infected with indicated EPEC strains at 100 MOI for 1 h. Whole cell lysates were derived, separated by SDS/PAGE, and immunoblotted for indicated proteins. C. Mouse colon epithelial cells (CECs) were mock infected or infected in suspension with indicated C. rodentium strains at 100 MOI for 3 h. Whole cell lysates were derived, separated by SDS/PAGE, and immunoblotted for indicated proteins. The full-length p65 and cleaved p65 C-terminal fragment are indicated by filled and open triangles, respectively. D. Mouse CECs infected as in C were separated from free C. rodentium by Percoll gradient centrifugation. Whole cell lysates from CECs and attached C. rodentium were derived, separated by SDS/PAGE, and immunoblotted for indicated proteins. E. Representative immunofluorescence micrographs of mouse CECs infected as in C that were centrifuged onto cover slips and stained for C. rodentium LPS, with nuclei counterstained by DAPI. Scale bars, 10 μm. F. The numbers of C. rodentium attached to mouse CECs as in E (from 6 random fields) were quantified and normalized to the perimeter of individual CEC. NleC affects host immune responses in mice infected by C. rodentium
Infection of C. rodentium in mice is known to cause colonic epithelial damage by acute inflammatory responses [48], therefore we examined the impact of NleC on colonic inflammatory response in mice inoculated with variant strains of C. rodentium. As we reported previously [20], the colonization of wild-type and ΔnleC mutant C. rodentium in the colon of infected mice was comparable at day 8 and day 10 (Fig. 5A and S9 Fig, respectively). In line with previous reports that NleC-mediated p65 cleavage plays a critical role in dampening the NF-κB signaling pathway and suppressing proinflammatory gene expression in EPEC-infected cells [16,30], we observed robust transcription of known NF-κB target genes Cxcl1 and Cxcl2 in the colon tissues removed from mice infected with ΔnleC C. rodentium, compared to wild-type bacterium at day 14 (Fig. 5B). We also detected significantly elevated levels of additional NF-κB target genes Ifng, Il1b, and Il22, thus highlighting the key function of NleC-mediated p65 cleavage in interfering with host NF-κB gene transcription (Fig. 5B). Consistent with the colonic expression of proinflammatory cytokine/chemokine genes, the amount of infiltrated CD11b+ myeloid cells in the colon was increased by two fold in mice orally inoculated with ΔnleC C. rodentium, compared to that from mice infected with wild-type bacterium (Fig. 5C-5D). As expected, wild-type C. rodentium-infected mice developed severe clinical symptoms characterized by crypt elongation, thickening of the mucosal surface, and goblet cell depletion in histological staining, in comparison to phosphate-buffered saline (PBS) inoculated animals (Fig. 5E-5F). By contrast, infection with ΔnleC C. rodentium induced more severe damage to colon epithelia and general enlargement of the colonic tissue (Fig. 5E-5F). Our results therefore demonstrate that the p65 cleavage by NleC during C. rodentium infection has a dramatic effect on NF-κB gene transcription and inflammatory response in the infected animals.
Fig. 5. NleC suppresses inflammatory responses in C. rodentium-infected mice. 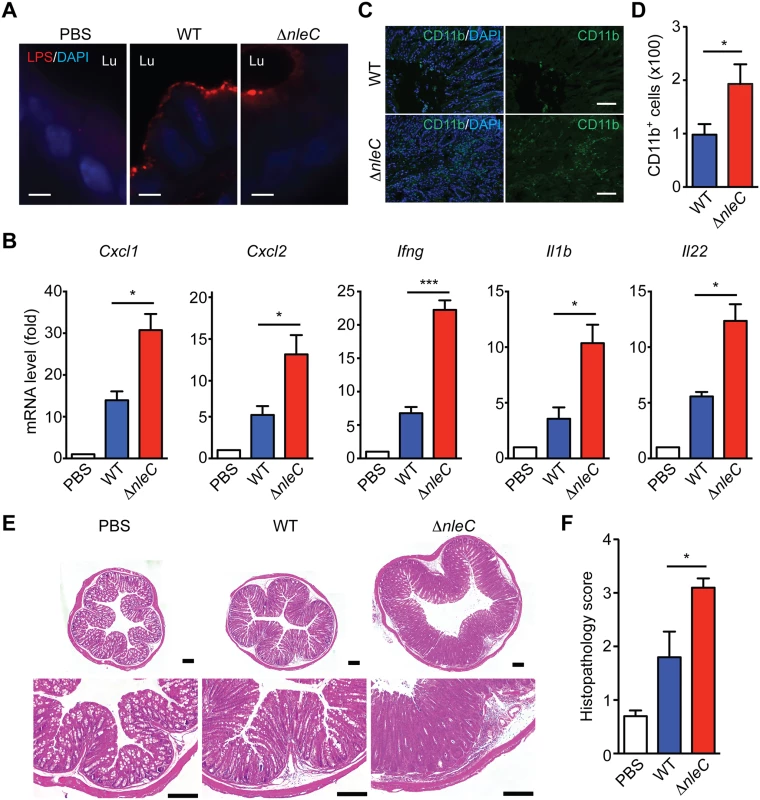
A. Representative immunofluorescence micrographs of C. rodentium LPS, with nuclei counterstained by DAPI, in colon sections derived from C57BL/6 mice 8 days post inoculation with the indicated strains of C. rodentium. Scale bars, 10 μm. Lu indicates the colon luminal space. B. Quantitative PCR (qPCR) was used to determine mRNA levels of indicated cytokine/chemokine genes relative to Actb (β-actin) in the colons collected from C57BL/6 mice 14 days post inoculation with wild-type or ΔnleC mutant strain of C. rodentium. C-D. Immunofluorescence micrographs of CD11b+ inflammatory immune cells in the colons collected from C57BL/6 mice as in B, with nuclei counterstained by DAPI. Scale bars, 10 μm. The colon-infiltrated inflammatory immune cells (from 5 random fields) with CD11b staining were quantified (D). E. Hematoxylin and eosin (H&E) staining of colons collected from C57BL/6 mice as in D. Scale bars, 200 μm. F. The histopathology scores of colon sections derived from mice infected as indicated as in D. Shown are mean ± s.e.m of 10 random fields from two independent experiments. The NleC-cleaved p651–38 interacts with RPS3
It is noteworthy that NleC cleaved only a small percentage of p65 during C. rodentium infection (Fig. 4A-4C) and even in the presence of an overwhelming amount (10 μg) of recombinant protease (Fig. 1B, 2A, 3A-3B, 3F-3G). In particular, the dramatic effect of NleC on the proinflammatory cytokine production and NF-κB activity was proposed to be mediated by the cleavage of p65 by NleC [16,20,29,30,41,42]; however, the large amount of full-length p65 resistant to NleC cleavage makes it difficult to explain the remarkable impact of NleC on dampening host NF-κB signaling and inflammatory response. Of note, RPS3, a non-Rel subunit of NF-κB, was revealed to confer the promoter selectivity and transcriptional specificity of NF-κB [34,49], in particular the RPS3/NF-κB-mediated transcription of a subset of proinflammatory genes is critical for host defense against A/E pathogens [17,18,19]. Moreover, our previous studies showed that interrupting the subcellular localization and function of RPS3 by small interfering RNA (siRNA) [34], bacterial effectors [17,18,19], and ectopic expression of an N-terminal truncated p6521–186 fragment [40], are able to selectively block NF-κB target gene transcription, without affecting the nuclear translocation of p65. We therefore hypothesized that the NleC-cleaved p651–38 product would execute a similar function as the p6521–186 fragment [40], which interferes with RPS3 signaling. To test this hypothesis, we examined the ability of GFP-tagged p651–38 and p6539–551 truncated proteins to alter NF-κB signaling in cultured cells. As demonstrated by subcellular fractionation, both p651–38 and p6539–551 truncated proteins were primarily located in the cytoplasm of transfected HEK293T cells, even following 30-min TNF treatment (Fig. 6A). This result is in agreement with previous reports that NleC inactivates p65 in the cytoplasm [16,41], and suggests that the NleC-cleaved products would mainly interfere with cytoplasmic NF-κB signaling in host cells. We further examined the interaction between RPS3 and the GFP-tagged p651–38 and p6539–551 fragments by immunoprecipitation. The RPS3-p651–38 interaction was comparable to, if not even stronger than, that of RPS3 and full-length p65, whereas the association between RPS3 and the p6539–551 fragment was barely detectable (Fig. 6B). Moreover, the p651–38 truncated protein was substantially enriched in the GST-RPS3 pulldown sample, compared to the GST vehicle control (S10 Fig), which independently verifies the interaction between p651–38 and RPS3. Our results therefore suggest that the NleC-cleaved p651–38 product, rather than the p6539–551 fragment, of p65 is able to interact with the NF-κB non-Rel subunit RPS3.
Fig. 6. The p651–38 fragment interferes with the RPS3-p65 interaction in the cytoplasm and selectively attenuates the nuclear translocation of RPS3. 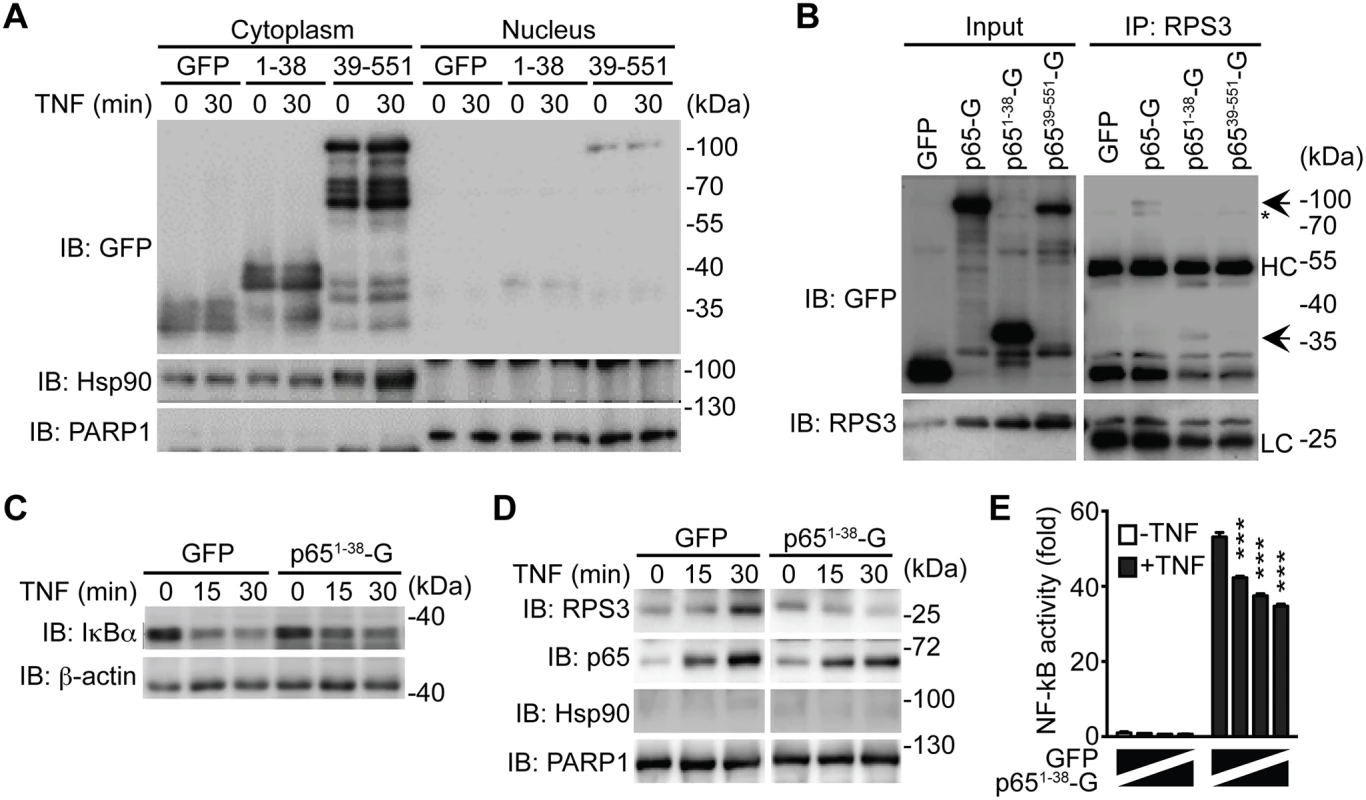
A. HEK293T cells were transfected with C-terminal GFP-tagged p651–38, p6539–551, or GFP control plasmids. 28 h later, the cytosolic and nuclear fractions were derived and immunoblotted (IB) for indicated proteins. Hsp90 and PARP1 served as loading controls and cytosolic and nuclear markers, respectively. B. HEK293T cells were transfected with indicated GFP-tagged p65 or GFP control plasmids. 28 h later, whole cell lysates (Input) were derived and directly IB, or after immunoprecipitation (IP) with RPS3 antibody, for indicated proteins. Non-specific bands and immunoprecipitated GFP-p65 proteins are indicated by asterisks and arrows, respectively. HC, heavy chain; LC, light chain. C. HEK293T cells expressing GFP-tagged p651–38 or GFP alone were stimulated with 50 ng ml-1 of TNF for indicated periods. Whole cell lysates were derived and IB for IκBα, with β-actin as a loading control. D. HEK293T cells were transfected and stimulated as in C, and nuclear fractions were derived and IB for indicated proteins. Hsp90 and PARP1 served as loading controls and cytosolic and nuclear markers, respectively. E. HEK293T cells were transfected with increasing amounts of GFP-p651–38, compensated with GFP control, together with 5 × κB-Luc reporter and pTKRL plasmids. After 28 h, the cells were stimulated in the presence or absence of TNF (50 ng ml-1) and analyzed for luciferase activity. The p651–38 product selectively attenuates the nuclear translocation of RPS3
Stimuli-triggered translocation from the cytoplasm to the nucleus is a prerequisite for RPS3 to facilitate NF-κB binding and transactivation of specific target genes [33,49]. We recently showed that ectopic expression of p6521–186 truncated protein competed RPS3 off endogenous full-length p65, thereby interfering with the nuclear translocation of RPS3 during the NF-κB response [40]. Ectopic expression of the p651–38 fragment, compared to the GFP control, did not alter TNF-stimulated IκBα degradation (Fig. 6C) and p65 nuclear translocation (Fig. 6D). By contrast, overexpression of p651–38 fragment remarkably attenuated TNF-triggered nuclear translocation of RPS3, which was induced normally in the GFP-expressing cells (Fig. 6D). To further assess the impact of p651–38 fragment on NF-κB activation, we examined the expression of an Ig-κB site-driven luciferase reporter gene, which was previously shown to be RPS3-dependent [18,34,40], in HEK293T cells expressing the p651–38 fragment. Indeed, in comparison to the GFP vehicle control, ectopic expression of the p651–38 fragment attenuated NF-κB reporter luciferase expression in a dose dependent manner (Fig. 6E). These results suggest that the p651–38 fragment is capable of interfering with the endogenous p65/RPS3 interaction and selectively attenuating the nuclear translocation of RPS3 rather than affecting other branches of NF-κB signaling (Fig. 7), which provides a novel mechanism for A/E pathogens to specifically modulate host NF-κB-mediated gene transcription and inflammatory responses.
Fig. 7. Schematic model of selective inhibition of NF-κB gene expression by the NleC-cleaved p651–38 fragment. 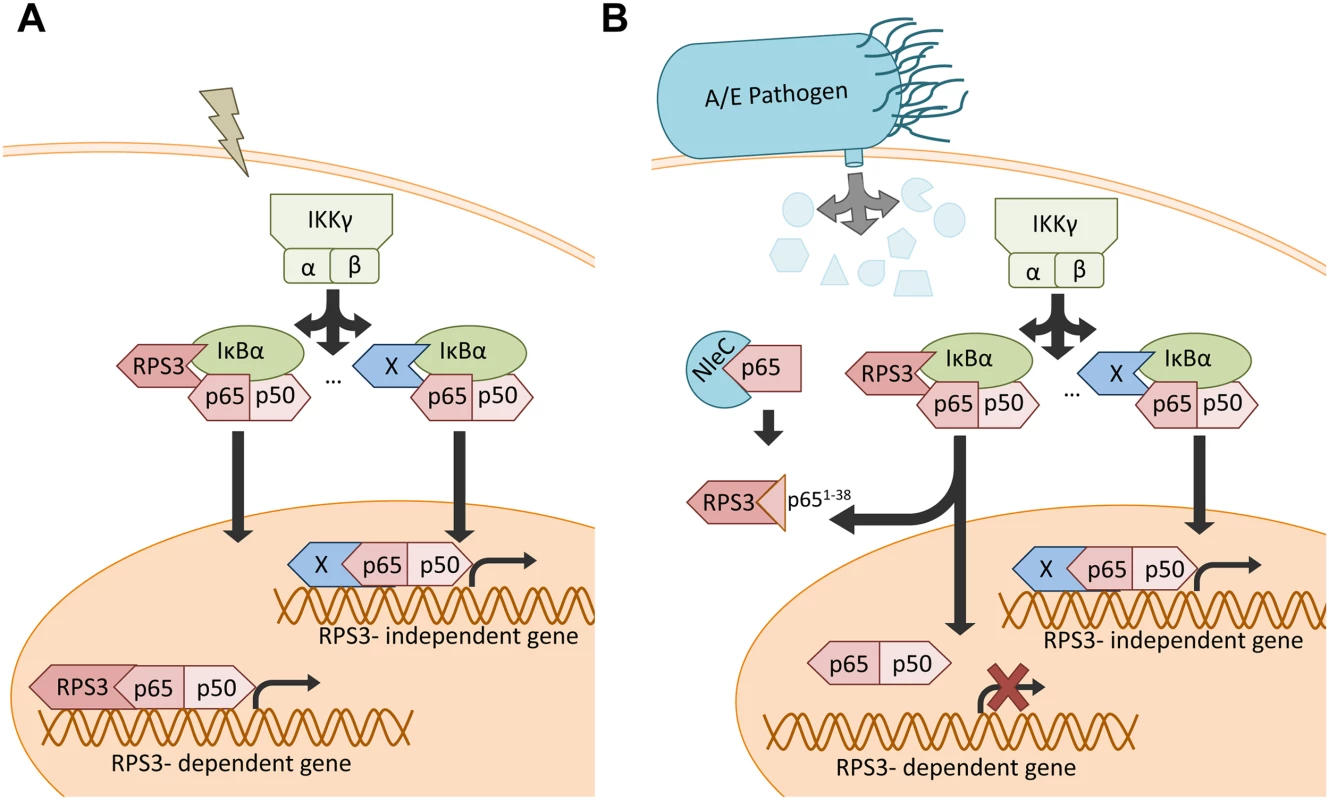
A. Under normal conditions, NF-κB stimuli activate both RPS3-dependent and-independent NF-κB signaling pathways. B. When injected into host cells via the T3SS during A/E pathogen infections, NleC cleaves a small percentage of p65, generating the p651–38 fragment that interferes with the RPS3-p65 interaction and sequestrates RPS3 in the cytoplasm. This leads to selective inhibition of the RPS3-dependent NF-κB gene transcription, without affecting the RPS3-independent gene transactivation. Discussion
The NF-κB signaling pathway orchestrates both innate and adaptive immune responses in host cells, thereby executing an important function in host defense to a variety of pathogens [21]. That said, the mechanisms controlling promoter selectivity and transcriptional specificity of NF-κB genes in acute pathogen-host interactions remain obscure. Beyond the well-characterized NF-κB Rel subunits, we recently identified two non-Rel subunits, i.e. RPS3 and Sam68, in the NF-κB DNA binding complexes that confer distinct transcriptional specificity of NF-κB [33,49]. The RPS3 - and Sam68-conferred NF-κB activation model suggests that the Rel subunits are required, but not the sole determinants, for the activation of NF-κB target genes. In contrast, the synergistic interactions between Rel and non-Rel subunits at the promoters are critical for the transactivation of certain NF-κB target genes [33,49]. An increasing number of studies [17,18,34,37,38,39] suggest that the RPS3/NF-κB signaling pathway is vital in the host proinflammatory transcription and immune responses against infection by A/E pathogens [17,18]. Specifically, the T3SS effector NleH1 from EPEC, EHEC, and C. rodentium attenuates the nuclear translocation of RPS3, but not p65, during NF-κB activation by inhibiting the IKKβ-mediated Ser209 phosphorylation of RPS3 [17,18]. This leads to reduced transcription of select, but not all, NF-κB target genes, most of which are RPS3/NF-κB-dependent proinflammatory cytokine genes [17,18]. In this work, we show that the T3SS effector protease NleC predominantly cleaves p65 at C38/E39. The NleC cleavage-generated p651–38 fragment interferes with the RPS3/p65 interaction in the cytoplasm, resulting in the attenuated nuclear translocation of RPS3, without affecting p65 nuclear translocation. Similar to what we have shown previously [17,18,34,40], cytoplasmic sequestering of RPS3 prevents the transactivation of the RPS3/p65-dependent proinflammatory cytokine genes and the infiltration of inflammatory immune cells to infected tissue. Of note, NleC cleaves a small percentage of p65 in C. rodentium-infected mouse CECs. In contrast, NleC cleavage has a substantial effect on proinflammatory cytokine gene transcription, as evidenced by infections using the EPEC and C. rodentium genetic mutants with disruptions in the NleC gene, compared to wild-type strains. We are aware that NleC, as a metalloprotease, cleaves other host proteins beyond p65, such as p300 [42], p38 [20], and others, which could also result in the observed striking effects of NleC on colonic cytokine gene expression and inflammatory responses in infected mice. However, our proposed “amplifying” mechanism through the RPS3-conferred NF-κB specific transcription provides a novel explanation for the disconnect regarding the extent of NleC-mediated NF-κB cleavage and the impact of NF-κB transactivation by such cleavage. In conjunction with our previous studies showing that NleH1 attenuates RPS3 Ser209 phosphorylation [17,18], our results highlight the critical role of the RPS3/NF-κB signaling pathway in the host immune response to A/E pathogen infections. Although NleC and NleH1 are both secreted by the A/E pathogen T3SS, the kinetics and amount of NleC and NleH1 within host cells during infection remains largely unknown. It has been well documented that certain successful pathogens have acquired sophisticated mechanisms to directly interfere with host NF-κB signaling through regulating or mimicking host proteins to their own advantage, and these co-opted strategies can operate at multiple levels of the sequential process of NF-κB signaling [11,12,13,14,15,16,17,18,29,30]. We therefore speculate that A/E pathogens utilize NleC and NleH1 to simultaneously (or sequentially) interfere with the RPS3/NF-κB signaling pathway through distinct strategies, therefore ensuring with this redundancy that the NF-κB signaling and inflammatory responses are dampened in host cells. Notably, the RPS3-p65 branch of NF-κB signaling has been associated with transcription of anti-apoptotic genes [39] and inflammatory genes [17,18] therefore selectively targeting this arm could prove advantageous to the pathogen. Given that the spatial and temporal coordination of effectors injected into host cells remains unclear [50,51], it is intriguing to consider that manipulating inflammatory gene transcription would be valuable early during infection and ensuring apoptosis would be an escape strategy allowing the pathogen to spread to the next host. While it has been documented that a relatively low amount of NleC is delivered into host cells during infection in cultured cells [9], the timing, longevity, and activity of the effectors within the host cell during an in vivo animal infection remains elusive. Greater resolution of the timing and amount of NleC’s introduction into the host cell and interaction with p65 is still needed.
The most abundant species among the NF-κB complexes in cells consists of p65, p50, and other proteins [34]. Moreover, p65 possesses a transactivation domain (TAD) that is essential to recruit general transcriptional machinery to transcribe target genes, whereas p50 lacks the TAD domain thereby normally suppressing gene expression [33]. These features make p65 unique within the NF-κB Rel subunits and a likely target for pathogen effectors. Our results show that NleC cleaves p65 more efficiently than other Rel family proteins and non-Rel subunits, RPS3 and Sam68. In further support of this notion, the N-terminal 20 residues of p65, which are not conserved among other Rel family proteins, play a critical role in the recognition and cleavage of p65 by NleC during A/E pathogen infection. Therefore it is not surprising that p65, the most important NF-κB Rel subunit, is a major target for a wealth of pathogens, allowing them to interfere with the NF-κB signaling pathway in host cells [16,20,29,30,41]. Indeed, previous studies showed that p65 was cleaved by a myriad of pathogen encoded proteases [52,53,54,55,56,57], although the direct consequence of cleaving p65 on NF-κB signaling has not been extensively studied. For instance, Chlamydia trachomatis, a gram-negative bacterium that causes urethritis, cervicitis, and other diseases, encodes a PDZ containing tail-specific protease [58]. The enzyme was proposed to suppress host NF-κB activity by cleaving p65 and generating two p65 fragments (approximately 40 kDa and 25 KDa, respectively) [59]. Moreover, the A-B toxin metalloprotease encoded by the fish pathogen Photobacterium damselae piscicida was recently shown to cleave p65 at C38/E39, similar to NleC [52]. It would be interesting to examine if p65 fragment interference is a previously unrecognized but possibly widespread mechanism of virulence for abrogating host NF-κB signaling, especially when pathogen encoded proteases are unable to cleave the majority or entire cellular source of p65. By selectively blocking NF-κB “specifiers”, as we demonstrated here for RPS3, pathogens could more acutely manipulate the host environment by regulating the timing and abundance of injected effectors, allowing collections of genes to be turned on and off to their advantage.
Materials and Methods
Ethics statement
All animal experiments were performed according to protocol number MO13-H349, approved by the Johns Hopkins University’s Animal Care and Use Committee and in direct accordance with the NIH guidelines for housing and care of laboratory animals.
Cell line, antibodies, and plasmids
HEK293T and Caco-2 cells (ATCC, Manassas, VA) were cultured in DMEM medium containing 10% fetal calf serum, 2 M glutamine, 100 U ml-1 penicillin, and 100 U ml-1 streptomycin. Antibodies used were: p65 (C-terminus, C-20, sc-372), p65 (N-terminus, F-6, sc-8008x), p50 (NLS, sc-114), c-Rel (C, sc-71) from Santa Cruz Biotechnology (Dallas, TX); β-actin (AC-15, A5441) and FLAG (M2, F1804) from Sigma-Aldrich (St. Louis, MO); PARP-1 (46D11, 9532) from Cell Signaling Technology (Danvers, MA); GFP (7.1 and 13.1, 11814460001) from Roche Applied Science (Indianapolis, IN); CD11b (M1/70, 101202) from BioLegend (San Diego, CA); Hsp90 (610418) from BD Biosciences (San Jose, CA); E. coli O152 LPS (81449) from Statens Serum Institut (Copenhagen, Denmark); RPS3 as previously described [34]. Tumor necrosis factor (TNF) was purchased from R&D System (Minneapolis, MN). The plasmids His-NleC [30], FLAG-p50, p50-FLAG [60], GFP-tagged full-length p65, p651–186, p6521–186, p651–311, p65186–311, and p65311–551 [34] were previously described. The GFP-p651–38 and GFP-p6539–551 were generated by inserting the appropriate fragments into the pEGFP-N1 vector (Clontech Laboratories, Mountain View, CA) using the InFusion Cloning System (Clontech Laboratories). The His-NleC (H117Y) and GFP-tagged p65 (C38A/E39A) and p65 (P10A) mutants were generated by site-directed mutagenesis using the Quick Change Kit (Stratagene, La Jolla, CA) with appropriate primers. All the plasmids were verified by DNA sequencing.
Transient transfection
DNA constructs were transfected into HEK293T cells using the TurboFect in vitro transfection reagent (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions, as described previously [40].
NleC protease digestion assays
Overnight cultures of BL21 (pET-NleC) were induced with1 mM IPTG and His-NleC proteins were purified by nickel affinity chromatography. The NleC protease digestion was conducted as previously described [30]. Briefly, cells were collected and lysed on ice with 0.4 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40 and 0.5% sodium deoxycholate, 1 × complete protease inhibitor cocktail [Roche Applied Science]) for 30 min. After centrifuge at 10,000 × g at 4°C for 10 min, 200 μl of supernatant was removed to a separate tube and incubated with indicated amount of His-NleC protein at 37°C for 3 h.
Nickel bead and GST pull-down assays
For the interactions between NleC and indicated proteins, the 200 μl of supernatant mixed with 1 μg of His-NleC protein were subjected to pull-down assays by adding 30 μl of Nickel beads (Qiagen, Germantown, MD), and rotating for 30 min at 4°C. The GST pulldown assays were conducted as previously described [35]. The pull-down proteins were washed at least four times with cold lysis buffer followed by separation with SDS-PAGE.
Isolation of primary colon epithelial cells
Colon epithelial cells (CECs) were isolated from C57BL/6J mice as previously described [61]. Briefly, after euthanizing mice, the entire colon was removed under aseptic conditions and washed twice with ice-cold PBS. After dividing the colon into 2–3 mm long fragments and transferring them into chelating buffer (27 mM trisodium cirtcrate, 5 mM Na2HPO4, 96 mM NaCl, 8 mM KH2PO4, 1.5 mM KCl, 0.5 mM DTT, 55 mM D-sorbitol, 44 mM sucrose, 6 mM EDTA, 5 mM EGTA [pH 7.3]) for 45 min at 4°C, CECs were then dislodged by repeated vigorous shaking. Tissue debris was removed by a 70-μm cell strainer (Fisher Scientific, Suwanee, GA) and CECs were harvested by centrifugation at 4°C. The viability of CECs was confirmed by trypan blue staining and isolated CECs were cultured at 37°C for 1 h for recovery, followed by infection.
Citrobacter rodentium and EPEC growth conditions and infection in cultured cells
Wild-type C. rodentium (DBS 100) and EPEC (E2348/69), as well as the NleC deletion mutant (ΔnleC) and the HA-NleC complemented (ΔnleC/pHA-NleC) strains [20] were grown from single colonies on Luria-Bertani (LB) plates in LB broth at 37°C overnight with shaking. Infection of EPEC in Caco-2 cells was performed as previously described [20]. Prior to infection experiments, C. rodentium was washed with ice-cold PBS and resuspended in pre-warmed corresponding media. Bacteria concentration was measured by absorbance at optical density 600, followed by a serial dilution and seeding on a MacConkey agar plate (VWR, Radnor, PA) to confirm the administered colony-forming units (CFU). The HEK293T cells and isolated CECs were infected with the indicated strains of C. rodentium at a multiplicity of infection (MOI) of 100 for 3 h, as described previously [20]. Cells were counted using a hemacytometer prior to each experiment and equal numbers of cells (1–2 × 106) were aliquoted into each infection condition. To determine bacterial attachment to CECs infected in suspension, cells and bacteria were passed through a Percoll gradient (40% and 60%) and CECs were collected from the top of half of the 40% gradients and overlay [62,63,64]. Cells were washed with PBS and placed on cover slips or lysed for immunofluorescence staining or immunoblot.
Immunofluorescence staining in colon epithelial cells
Post C. rodentium infection, mouse CECs were spun down to Poly-L-Lysine-coated coverslips, fixed with 4% PFA, and stained with appropriate primary antibodies and fluorescence dye-conjugated second antibodies. Following staining of nuclei with 1 μg ml-1 of DAPI (Sigma-Aldrich), coverslips were mounted onto slides using Fluoro-gel with Tris Buffer (Electron Microscopy Sciences, Hatfield, PA) and examined using an Axio Observer fluorescence microscope (Zeiss, Oberkochen, Germany). The numbers of C. rodentium that attached to mouse CECs were quantified using ImageJ software (NIH, Bethesda, MD) and normalized to cell perimeter.
Citrobacter rodentium infection in mice
Male C57BL/6 mice (6 to 8 weeks) purchased from the Jackson Laboratory (Bar Harbor, ME) were maintained in a specific pathogen-free facility and fed autoclaved food and water ad libitum. Food was withheld from the mice for 6–8 hours before they were orally inoculated with 200 μl of PBS containing 2 × 109 CFU of wild-type or ΔnleC mutant C. rodentium or PBS alone, and euthanized at the indicated time points post infection.
Colon tissue collection, histology, and immunofluorescence staining
After euthanizing mice, their colons were removed under aseptic conditions, washed once with ice-cold PBS, and the terminal 0.5-cm piece of the colon was frozen in optimal cutting temperature (O.C.T.) media (Tissue-Tek, Elkhart, IN) or incubated overnight in 4% PFA. 5-micron frozen sections were cut using a Microm HM 550 Cryostat (Thermo Scientific), collected on coated slides and processed for immunofluorescence staining. Frozen sections were fixed in paraformaldehyde, washed with PBS, and blocked with appropriate sera in PBS. After incubating with appropriate antibodies, sections were washed and incubated with fluorescence dye-conjugated second antibodies and 1 μg ml-1 of DAPI (Sigma-Aldrich). Stained sections were washed and mounted under a coverslip using Fluoro-gel with Tris Buffer (Electron Microscopy Sciences). For histological analysis, the colon tissue was embedded in paraffin and 5-micron sections were cut, collected on coated slides and processed for Hematoxylin and Eosin (H&E) staining. Stained sections were examined using an Axio Observer fluorescence microscope (Zeiss). Histopathology scores were determined in a blinded fashion using the following criteria as previously described by Qualls et al. [65]: 0, Normal tissue; Grade 1, mild inflammation was present containing mostly mononuclear cell infiltrate and little damage to the epithelia; Grade 2, inflammation greater than Grade 1 with mononuclear and polymorphonuclear infiltrate, mucin and Goblet cell depletion, and epithelium beginning to detach from basement membrane; Grade 3, inflammation and cellular infiltrate is greater than Grade 2 with cellular infiltrates reaching the submucosa, greater Goblet cell depletion, and greater epithelial disruption; Grade 4, severe inflammation containing mostly neutrophils, completely detached epithelium, and crypt destruction.
Quantitative real-time PCR
Total RNA was isolated from colon tissues using Trizol reagent (Life Technologies) and treated with the TURBO DNA-free Kit (Life Technologies) to remove residual genomic DNA. cDNA was synthesized using qScript cDNA SuperMix Kit (Quanta Biosciences, Gaithersburg, MD) according to the manufacturer’s instructions. Gene specific products were amplified using SsoAdvanced SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) with the following primers: Cxcl1-f, 5’-TGCACCCAAACCGAAGTCAT-3’; Cxcl1-r, 5’-TTGTCAGAA GCCAGCGTTCAC-3’; Cxcl2-f, 5’-CCTGCCAAGGGTTGACTTCA-3’; Cxcl2-r, 5’-TTCTGTCTGGGCGCA GTG-3’; Ifng-f, 5’-ATGAACGCTACACACTGCATC-3’; Ifng-r, 5’-CCATCCTTTTGCCAGTTCCTC-3’; Il1b-f, 5’-GAAATGCCACCTTTTGACAGTG-3’; Il1b-r, 5’-CTGGATGCTCTCATCAGGACA-3’; Il22-f, 5’-CAGAGGTAGACTTGATAACCAC-3’; Il22-r, 5’-GGTTATGGAAATGAAGTTACATAAGC-3’.
Subcellular fractionation
Subcellular fractionation was performed by differential centrifugation as previously described [35]. Briefly, cells were resuspended in Buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 0.4% NP-40, 0.5 mM PMSF, 1 × complete protease inhibitor cocktail [Roche Applied Science]) at 4°C for 5 min. Lysates were centrifuged at 4°C, 500 × g for 3 min, and supernatants were collected as cytosolic fractions. Pellets were incubated in Buffer C (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, 0.5 mM PMSF, 0.2 mM EDTA, 0.5 mM DTT, 1 × complete protease inhibitor cocktail) at 4°C for 10 min, followed by a centrifuge at 4°C, 13,000 × g for 10 min. Supernatants were collected as nuclear fractions.
Immunoprecipitation and immunoblot
The cells were harvested and lysed on ice with 0.4 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40 and 0.5% sodium deoxycholate, 1 × complete protease inhibitor cocktail) for 10 min. The lysates were centrifuged at 10,000 × g at 4°C for 10 min. The protein-normalized lysates were subjected to immunoprecipitation by adding 10 mg ml-1 of the appropriate antibody, 30 μl of protein G-agarose (Roche Applied Science), and rotating for more than 2 h in the cold room. The precipitates were washed at least four times with cold lysis buffer followed by a separation by SDS-PAGE under reduced and denaturing conditions. The resolved protein bands were transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA), probed as described previously [35], developed by the Super Signaling system (Thermo Scientific) according to the manufacturer’s instructions, and imaged using a FluorChem E System (Protein Simple, Santa Clara, CA).
Luciferase reporter gene assays
Luciferase reporter gene assays were performed as previously described [18,34,40]. Briefly, cells were cotransfected with 5 × Ig κB site-driven firefly luciferase constructs and the Renilla luciferase pTKRL plasmid (ratio 10 : 1), together with appropriate plasmids. Cells were cultured for 18 hours, stimulated in triplicate, and analyzed using the Dual-Luciferase Kit (Promega, Madison, WI).
Statistical analysis
All statistical analysis was performed using GraphPad Prism version 6.0 (GraphPad Software, San Diego, CA). The difference between treated and control groups were examined by unpaired Student’s t tests. Standard errors of means (s.e.m.) were plotted in graphs. n.s. means non-significant difference and significant differences were considered * at p < 0.05; ** at p < 0.01; and *** at p < 0.001.
Supporting Information
Zdroje
1. Clarke SC (2001) Diarrhoeagenic Escherichia coli—an emerging problem? Diagnostic microbiology and infectious disease 41 : 93–98. 11750160
2. Kaper JB, Nataro JP, Mobley HL (2004) Pathogenic Escherichia coli. Nature reviews Microbiology 2 : 123–140. 15040260
3. Donnenberg MS, Kaper JB, Finlay BB (1997) Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends in microbiology 5 : 109–114. 9080609
4. Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiology and molecular biology reviews: MMBR 62 : 379–433. 9618447
5. Coburn B, Sekirov I, Finlay BB (2007) Type III secretion systems and disease. Clinical microbiology reviews 20 : 535–549. 17934073
6. Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, et al. (2004) Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proceedings of the National Academy of Sciences of the United States of America 101 : 3597–3602. 14988506
7. Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, et al. (2006) An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proceedings of the National Academy of Sciences of the United States of America 103 : 14941–14946. 16990433
8. Garcia-Angulo VA, Deng W, Thomas NA, Finlay BB, Puente JL (2008) Regulation of expression and secretion of NleH, a new non-locus of enterocyte effacement-encoded effector in Citrobacter rodentium. Journal of bacteriology 190 : 2388–2399. doi: 10.1128/JB.01602-07 18223087
9. Deng W, Yu HB, de Hoog CL, Stoynov N, Li Y, et al. (2012) Quantitative Proteomic Analysis of Type III Secretome of Enteropathogenic Escherichia coli Reveals an Expanded Effector Repertoire for Attaching/Effacing Bacterial Pathogens. Molecular & cellular proteomics: MCP 11 : 692–709.
10. Deng W, de Hoog CL, Yu HB, Li Y, Croxen MA, et al. (2010) A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. The Journal of biological chemistry 285 : 6790–6800. doi: 10.1074/jbc.M109.086603 20034934
11. Nadler C, Baruch K, Kobi S, Mills E, Haviv G, et al. (2010) The type III secretion effector NleE inhibits NF-kappaB activation. PLoS pathogens 6: e1000743. doi: 10.1371/journal.ppat.1000743 20126447
12. Royan SV, Jones RM, Koutsouris A, Roxas JL, Falzari K, et al. (2010) Enteropathogenic E. coli non-LEE encoded effectors NleH1 and NleH2 attenuate NF-kappaB activation. Molecular microbiology 78 : 1232–1245. doi: 10.1111/j.1365-2958.2010.07400.x 21091507
13. Newton HJ, Pearson JS, Badea L, Kelly M, Lucas M, et al. (2010) The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65. PLoS pathogens 6: e1000898. doi: 10.1371/journal.ppat.1000898 20485572
14. Baruch K, Gur-Arie L, Nadler C, Koby S, Yerushalmi G, et al. (2011) Metalloprotease type III effectors that specifically cleave JNK and NF-kappaB. Embo J 30 : 221–231. doi: 10.1038/emboj.2010.297 21113130
15. Vossenkamper A, Marches O, Fairclough PD, Warnes G, Stagg AJ, et al. (2010) Inhibition of NF-kappaB signaling in human dendritic cells by the enteropathogenic Escherichia coli effector protein NleE. Journal of immunology 185 : 4118–4127. doi: 10.4049/jimmunol.1000500 20833837
16. Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, et al. (2010) NleC, a type III secretion protease, compromises NF-kappaB activation by targeting p65/RelA. PLoS pathogens 6: e1001231. doi: 10.1371/journal.ppat.1001231 21187904
17. Gao X, Wan F, Mateo K, Callegari E, Wang D, et al. (2009) Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS pathogens 5: e1000708. doi: 10.1371/journal.ppat.1000708 20041225
18. Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, et al. (2011) IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nature immunology 12 : 335–343. doi: 10.1038/ni.2007 21399639
19. Pham TH, Gao X, Tsai K, Olsen R, Wan F, et al. (2012) Functional differences and interactions between the Escherichia coli type III secretion system effectors NleH1 and NleH2. Infection and immunity 80 : 2133–2140. doi: 10.1128/IAI.06358-11 22451523
20. Sham HP, Shames SR, Croxen MA, Ma C, Chan JM, et al. (2011) Attaching and effacing bacterial effector NleC suppresses epithelial inflammatory responses by inhibiting NF-kappaB and p38 mitogen-activated protein kinase activation. Infection and immunity 79 : 3552–3562. doi: 10.1128/IAI.05033-11 21746856
21. Le Negrate G (2012) Subversion of innate immune responses by bacterial hindrance of NF-kappaB pathway. Cellular microbiology 14 : 155–167. doi: 10.1111/j.1462-5822.2011.01719.x 22044780
22. Santoro MG, Rossi A, Amici C (2003) NF-kappaB and virus infection: who controls whom. Embo J 22 : 2552–2560. 12773372
23. Mulhern O, Harrington B, Bowie AG (2009) Modulation of innate immune signalling pathways by viral proteins. Advances in experimental medicine and biology 666 : 49–63. 20054974
24. Rahman MM, McFadden G (2006) Modulation of tumor necrosis factor by microbial pathogens. PLoS pathogens 2: e4. 16518473
25. Rahman MM, McFadden G (2011) Modulation of NF-kappaB signalling by microbial pathogens. Nature reviews Microbiology 9 : 291–306. doi: 10.1038/nrmicro2539 21383764
26. O’Callaghan D, Stebbins CE (2010) Host-microbe interactions: bacteria. Current opinion in microbiology 13 : 1–3. doi: 10.1016/j.mib.2009.12.006 20044300
27. Lilic M, Galkin VE, Orlova A, VanLoock MS, Egelman EH, et al. (2003) Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms. Science 301 : 1918–1921. 14512630
28. Clements A, Young J, Constantinou N, Frankel G (2012) Infection strategies of enteric pathogenic E. coli. Gut microbes 3.
29. Muhlen S, Ruchaud-Sparagano MH, Kenny B (2011) Proteasome-independent degradation of canonical NFkappaB complex components by the NleC protein of pathogenic Escherichia coli. The Journal of biological chemistry 286 : 5100–5107. doi: 10.1074/jbc.M110.172254 21148319
30. Pearson JS, Riedmaier P, Marches O, Frankel G, Hartland EL (2011) A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-kappaB for degradation. Molecular microbiology 80 : 219–230. doi: 10.1111/j.1365-2958.2011.07568.x 21306441
31. Ruchaud-Sparagano MH, Maresca M, Kenny B (2007) Enteropathogenic Escherichia coli (EPEC) inactivate innate immune responses prior to compromising epithelial barrier function. Cellular microbiology 9 : 1909–1921. 17388785
32. Hayden MS, Ghosh S (2004) Signaling to NF-kappaB. Genes & development 18 : 2195–2224.
33. Wan F, Lenardo MJ (2009) Specification of DNA binding activity of NF-kappaB proteins. Cold Spring Harbor perspectives in biology 1: a000067. doi: 10.1101/cshperspect.a000067 20066093
34. Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, et al. (2007) Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131 : 927–939. 18045535
35. Fu K, Sun X, Zheng W, Wier EM, Hodgson A, et al. (2013) Sam68 modulates the promoter specificity of NF-kappaB and mediates expression of CD25 in activated T cells. Nature communications 4 : 1909. doi: 10.1038/ncomms2916 23715268
36. Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, et al. (1995) Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proceedings of the National Academy of Sciences of the United States of America 92 : 10599–10603. 7479848
37. Cadera EJ, Wan F, Amin RH, Nolla H, Lenardo MJ, et al. (2009) NF-kappaB activity marks cells engaged in receptor editing. The Journal of experimental medicine 206 : 1803–1816. doi: 10.1084/jem.20082815 19581408
38. Mokhtari D, Barbu A, Mehmeti I, Vercamer C, Welsh N (2009) Overexpression of the nuclear factor-kappaB subunit c-Rel protects against human islet cell death in vitro. American journal of physiology Endocrinology and metabolism 297: E1067–1077. doi: 10.1152/ajpendo.00212.2009 19706790
39. Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, et al. (2012) Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell 45 : 13–24. doi: 10.1016/j.molcel.2011.10.021 22244329
40. Wier EM, Neighoff J, Sun X, Fu K, Wan F (2012) Identification of an N-terminal truncation of the NF-kappaB p65 subunit that specifically modulates ribosomal protein S3-dependent NF-kappaB gene expression. The Journal of biological chemistry 287 : 43019–43029. doi: 10.1074/jbc.M112.388694 23115242
41. Baruch K, Gur-Arie L, Nadler C, Koby S, Yerushalmi G, et al. (2011) Metalloprotease type III effectors that specifically cleave JNK and NF-kappaB. The EMBO journal 30 : 221–231. doi: 10.1038/emboj.2010.297 21113130
42. Shames SR, Bhavsar AP, Croxen MA, Law RJ, Mak SH, et al. (2011) The pathogenic Escherichia coli type III secreted protease NleC degrades the host acetyltransferase p300. Cellular microbiology 13 : 1542–1557. doi: 10.1111/j.1462-5822.2011.01640.x 21812888
43. Vallabhapurapu S, Karin M (2009) Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27 : 693–733. doi: 10.1146/annurev.immunol.021908.132641 19302050
44. Li W, Liu Y, Sheng X, Yin P, Hu F, et al. (2014) Structure and mechanism of a type III secretion protease, NleC. Acta crystallographica Section D, Biological crystallography 70 : 40–47. doi: 10.1107/S1399004713024619 24419377
45. Turco MM, Sousa MC (2014) The structure and specificity of the type III secretion system effector NleC suggest a DNA mimicry mechanism of substrate recognition. Biochemistry 53 : 5131–5139. doi: 10.1021/bi500593e 25040221
46. Sorimachi H, Mamitsuka H, Ono Y (2012) Understanding the substrate specificity of conventional calpains. Biological chemistry 393 : 853–871. doi: 10.1515/hsz-2012-0143 22944687
47. Chen FE, Huang DB, Chen YQ, Ghosh G (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 391 : 410–413. 9450761
48. Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S (2005) Citrobacter rodentium of mice and man. Cellular microbiology 7 : 1697–1706. 16309456
49. Wan F, Lenardo MJ (2010) The nuclear signaling of NF-kappaB: current knowledge, new insights, and future perspectives. Cell research 20 : 24–33. doi: 10.1038/cr.2009.137 19997086
50. Shames SR, Finlay BB (2012) Bacterial effector interplay: a new way to view effector function. Trends in microbiology 20 : 214–219. doi: 10.1016/j.tim.2012.02.007 22425230
51. Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, et al. (2011) Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Molecular microbiology 80 : 1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x 21488979
52. Silva DS, Pereira LM, Moreira AR, Ferreira-da-Silva F, Brito RM, et al. (2013) The apoptogenic toxin AIP56 is a metalloprotease A-B toxin that cleaves NF-kappab P65. PLoS pathogens 9: e1003128. doi: 10.1371/journal.ppat.1003128 23468618
53. Doyle PS, Zhou YM, Hsieh I, Greenbaum DC, McKerrow JH, et al. (2011) The Trypanosoma cruzi protease cruzain mediates immune evasion. PLoS pathogens 7: e1002139. doi: 10.1371/journal.ppat.1002139 21909255
54. Christian J, Vier J, Paschen SA, Hacker G (2010) Cleavage of the NF-kappaB family protein p65/RelA by the chlamydial protease-like activity factor (CPAF) impairs proinflammatory signaling in cells infected with Chlamydiae. The Journal of biological chemistry 285 : 41320–41327. doi: 10.1074/jbc.M110.152280 21041296
55. Neznanov N, Chumakov KM, Neznanova L, Almasan A, Banerjee AK, et al. (2005) Proteolytic cleavage of the p65-RelA subunit of NF-kappaB during poliovirus infection. The Journal of biological chemistry 280 : 24153–24158. 15845545
56. Cameron P, McGachy A, Anderson M, Paul A, Coombs GH, et al. (2004) Inhibition of lipopolysaccharide-induced macrophage IL-12 production by Leishmania mexicana amastigotes: the role of cysteine peptidases and the NF-kappaB signaling pathway. Journal of immunology 173 : 3297–3304. 15322192
57. Coiras M, Lopez-Huertas MR, Mateos E, Alcami J (2008) Caspase-3-mediated cleavage of p65/RelA results in a carboxy-terminal fragment that inhibits IkappaBalpha and enhances HIV-1 replication in human T lymphocytes. Retrovirology 5 : 109. doi: 10.1186/1742-4690-5-109 19046417
58. Lad SP, Yang G, Scott DA, Wang G, Nair P, et al. (2007) Chlamydial CT441 is a PDZ domain-containing tail-specific protease that interferes with the NF-kappaB pathway of immune response. Journal of bacteriology 189 : 6619–6625. 17631635
59. Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, et al. (2007) Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proceedings of the National Academy of Sciences of the United States of America 104 : 2933–2938. 17301240
60. Dooher JE, Paz-Priel I, Houng S, Baldwin AS Jr, Friedman AD (2011) C/EBPalpha, C/EBPalpha oncoproteins, or C/EBPbeta preferentially bind NF-kappaB p50 compared with p65, focusing therapeutic targeting on the C/EBP:p50 interaction. Molecular cancer research: MCR 9 : 1395–1405. doi: 10.1158/1541-7786.MCR-11-0072 21813505
61. Flint N, Cove FL, Evans GS (1991) A low-temperature method for the isolation of small-intestinal epithelium along the crypt-villus axis. The Biochemical journal 280 (Pt 2): 331–334. 1747105
62. Childs WC 3rd, Gibbons RJ (1988) Use of Percoll density gradients for studying the attachment of bacteria to oral epithelial cells. Journal of dental research 67 : 826–830. 3163351
63. Colombo AV, Silva CM, Haffajee A, Colombo AP (2006) Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. Journal of medical microbiology 55 : 609–615. 16585650
64. Izhar M, Nuchamowitz Y, Mirelman D (1982) Adherence of Shigella flexneri to guinea pig intestinal cells is mediated by a mucosal adhesion. Infection and immunity 35 : 1110–1118. 7040246
65. Qualls JE, Kaplan AM, van Rooijen N, Cohen DA (2006) Suppression of experimental colitis by intestinal mononuclear phagocytes. Journal of leukocyte biology 80 : 802–815. 16888083
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



