-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
DENV causes widespread mosquito-borne viral infections worldwide and nearly 40% of the world’s population is at risk of being infected. Currently, no licensed vaccines or specific drugs are available to treat severe infections by DENV. NS5 is a large protein of 900 amino acids composed of two domains with several key enzymatic activities for viral RNA replication in the host cell and constitutes a prime target for the design of antiviral inhibitors. We succeeded in trapping a stable conformation of the full-length NS5 protein and report its crystal structure at a resolution of 2.3 Å. This conformation reveals the entire inter-domain region and clarifies the determinants of NS5 flexibility. The inter-domain interface is stabilized by several polar contacts between residues projecting from the MTase and RdRp domains of NS5. Several evolutionarily conserved residues at the interface play a crucial role for virus replication as shown by reverse genetics, although the analogous mutations mostly do not abolish the in vitro enzymatic activities of the recombinant proteins.
Published in the journal: . PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004682
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004682Summary
DENV causes widespread mosquito-borne viral infections worldwide and nearly 40% of the world’s population is at risk of being infected. Currently, no licensed vaccines or specific drugs are available to treat severe infections by DENV. NS5 is a large protein of 900 amino acids composed of two domains with several key enzymatic activities for viral RNA replication in the host cell and constitutes a prime target for the design of antiviral inhibitors. We succeeded in trapping a stable conformation of the full-length NS5 protein and report its crystal structure at a resolution of 2.3 Å. This conformation reveals the entire inter-domain region and clarifies the determinants of NS5 flexibility. The inter-domain interface is stabilized by several polar contacts between residues projecting from the MTase and RdRp domains of NS5. Several evolutionarily conserved residues at the interface play a crucial role for virus replication as shown by reverse genetics, although the analogous mutations mostly do not abolish the in vitro enzymatic activities of the recombinant proteins.
Introduction
Several flaviviruses such as Dengue virus (DENV), Japanese Encephalitis virus (JEV), West Nile virus (WNV), Yellow Fever virus (YFV) and Tick-Borne Encephalitis virus (TBEV) are major human pathogens. The mosquito-borne DENV serotypes 1–4 cause widespread epidemics and nearly 40% of the world’s population is at risk of being infected [1]. Infection by any of the four serotypes can lead to a broad spectrum of outcomes, ranging from asymptomatic infection, dengue fever, dengue hemorrhagic fever or dengue shock syndrome. A tetravalent vaccine is undergoing phase III of clinical trials, which requires three booster injections and only confers partial cross protection. No antivirals have been approved to treat Dengue so far, although availability of such molecules would be valuable to treat dengue infection.
Flavivirus RNA replication occurs within a multi-protein replication complex (RC), which assembles on ER-derived membranes and comprises both non-structural (NS) viral proteins and host cofactors [2–4]. With 900 amino acid residues, NS5 is the largest enzyme and the most conserved protein component of the flavivirus RC. Its N-terminal domain (residues 1–262 in DENV3) belongs to the S-adenosyl-L-methionine (SAM)-dependent methyltransferase (MTase) superfamily [5]. The MTase domain of NS5 caps the viral RNA genome, a step required for its stability and translation into viral polyproteins by host cell ribosomes [6]. Methylations of the N7 atom of Guanine-0, the 2’-O atoms of the ribose of Adenosine-1 and internal adenosines also contribute to viral escape from the host cell innate immune response [5] [7] [8] [9]. A putative guanylyltransferase activity (GTase) was also proposed for the N-terminal domain of NS5 [10,11] but this notion remains debatable. The C-terminal domain (residues 273–900) of NS5 contains the RNA-dependent RNA polymerase (RdRp) that synthesizes the anti-genome and progeny genome [12,13]. The RdRp domain is composed of the Finger, Thumb and Palm subdomains that are structurally conserved across viral RdRps [13]. Within the RdRp domain, residues 316–415 contain functional nuclear localization sequences that are hotspots for interactions with other viral and host proteins [14–17]. NS5 interacts with the NS3 protease-helicase and with several host proteins, including components of the ubiquitin proteasome pathway, NF90, and eEF1A [17–20]. In addition to its enzymatic functions required for RNA replication, NS5 acts as an antagonist of the host interferon response by interacting with and promoting the degradation of STAT2 [21]. In DENV, NS5 localizes to the nucleus of infected cells in a regulated and serotype-dependent manner that may modulate other host processes [22,23]. The importance of NS5 in viral replication and host immune response modulation makes it an ideal target for developing broad-acting antiviral inhibitors to treat diseases caused by flaviviruses [24–28]. A putative linker (or “inter-domain region” spanning residues 263–272) connects the two catalytic domains of NS5. Its sequence is relatively poorly conserved although its length has been preserved across flaviviruses.
Structural and functional studies of the complete NS5 polypeptide have been initiated by several research groups. Using in silico docking guided by reverse genetics, a first model for NS5 from WNV was proposed [12]. In this model, the MTase active site was placed in an orientation that would facilitate the capping of newly synthesized viral RNA exiting from the template-binding channel of the RdRp domain. The conformation of NS5 from DENV3 was also studied using small angle X-ray scattering (SAXS) [29] which suggested that the isolated NS5 protein from DENV may be flexible and could adopt a range of conformations, from compact (80% of the population) to more extended (20% of the population) structures in solution [29]. It was also reported that the MTase domain of NS5 does not affect its RdRp activity and that the two domains of NS5 responsible for RNA capping and synthesis respectively function essentially independently [30]. Recently, Lu and Gong reported a crystal structure for NS5 from JEV, in which the MTase and RdRp domains adopt a compact conformation stabilized by an interface dominated by hydrophobic interactions [31]. Moreover, a fragment of the RdRp from DENV3 NS5 containing extra residues from the linker has enhanced thermo-stability and polymerase activity, suggesting an impact of residues N-terminal to the polymerase domain on RdRp activity and stability [32]. This observation is supported by a recent study on viral RNA synthesis demonstrating that DENV2 NS5 has superior de novo and elongation activities than the NS5-RdRp domain and proposing that the MTase domain interacts with RdRp domain dynamically to regulate RNA synthesis during virus replication [33]. Clearly, more experimental data are needed to clarify the potential cross-regulatory effects between the two enzymatic domains of NS5, to reveal its distinct conformations, the molecular origin of its flexibility and to understand how its dynamic properties relate to the various steps required for RNA capping and synthesis. Here, we crystallized the full length NS5 protein from DENV3 and determined its crystal structure at 2.3 Å resolution, bound to SAH and also in the presence of GTP. Although the overall molecular shape of NS5 from DENV3 is similar to the NS5 protein from JEV, the relative orientation between their MTase and RdRp domains differs markedly between the two structures. The crystal structure of NS5 from DENV3 reveals a well-ordered linker region and a unique and mostly polar interface between the MTase and RdRp domains. The importance of selected interface residues in the structure and function of NS5 was addressed by site-directed mutagenesis for biochemical and virus replication studies. These studies corroborate the structural insight that inter-domain interactions are critical for viral RNA replication and infection.
Materials and Methods
Cells
BHK-21 cells (Baby Hamster Kidney fibroblast cells, ATCC) were maintained in RPMI 1640 media (Gibco), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S) at 37°C in 5% CO2.
Cloning, expression and purification of DENV3 NS5 for crystallization
To facilitate protein crystallization, we removed five flexible amino acids each from the N - and C - terminal ends of NS5, based on inspection of missing electron density in the structures of the individual MTase and RdRp domains from DENV3 (PDB codes 3P97 and 2J7U). The fragment corresponding to amino acid residues 6 to 895 of DENV3 NS5 (GenBank accession number AY662691.1) was amplified and cloned into pNIC28-Bsa4 plasmid using ligation independent cloning methods [34]. The expressed construct comprises the hexa-histidine tag and a Tobacco Etch Virus (TEV) protease cleavage site fused at the N terminus of DENV3 NS5. Transformed E. coli BL21-CodonPlus (Stratagene) was grown in LB medium containing 50 μg/ml kanamycin and 34 ug/ml chloramphenicol to an A600nm of 0.6–0.8 at 37°C. Expression of the recombinant proteins was induced by the addition of 0.5 mM isopropyl β-D-galactoside, and incubation was continued for a further 20 h at 18°C. The cells were rapidly cooled and harvested by centrifugation at 8000 g for 10 min at 4°C and stored at -20°C. Cells were resuspended in buffer A (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 5mM β-mercaptoethanol, 10% glycerol, 10 mM imidazole) and lysed by sonication. The lysate was clarified by centrifugation at 20,000 g for 60min at 4°C. The supernatant was purified by nickel-nitrilo-triacetic acid affinity chromatography by washing unbound protein with buffer A supplemented with 40 mM imidazole. The DENV3 NS5 protein was eluted using a linear gradient of imidazole ranging from 40 to 500 mM. Fractions containing His6-NS5 were dialyzed against buffer C (20 mM Na-Hepes, pH 7.5, 300 mM NaCl, 5 mM DTT, 10% (v/v) glycerol), and the proteins were cleaved by TEV (substrate-to-enzyme ratio of 40 : 1, w/w) at 4°C for overnight. NS5 was further purified by size-exclusion chromatography using a HiPrep Superdex-200 gel filtration column (Amersham Bioscience) in buffer C. Fractions containing NS5 were pooled and concentrated to ~10 mg/ml before storage in -80°C. SDS-PAGE analysis of the resulting NS5 protein indicated a purity of 95%.
Crystallization and data collection
Crystallization trials were set up at 20°C with a Phoenix crystallization robot using sitting drop vapour diffusion. After extensive robotic crystallization trials, small rhomboidal shaped crystals were obtained using precipitants containing divalent metal salts, including calcium acetate, magnesium acetate and magnesium formate. Crystallization conditions include conditions 18 and 46 (Crystal Screen HR2–110, Hampton Research), condition 20 (PEG/Ion Screen, HR2–126, Hampton Research) and condition 55 (Index screen HR2–144, Hampton Research). Larger crystals of rhombus or trapezoid shape were obtained over 2–5 days by mixing a volume of 1 μl of NS5 (6–895) at 4–6 mg/ml with 1 μl of precipitation solution (0.2 M calcium acetate or magnesium acetate, 0.1 M sodium cacodylate, pH = 6.4, 10–20% (w/v) PEG 8000). Crystals for the GTP complex were obtained by either co-crystallization of NS5 at a concentration of 6 mg/ml (~60 μM), with 0.4 mM GTP using a slightly different precipitating solution (0.1 M sodium cacodylate (pH 6.4), 0.2 M magnesium acetate or 0.2 M calcium acetate and 14% (w/v) polyethylene glycol 8000) at 18°C, or soaking of apo NS5 crystal in the precipitating solution supplemented with 5 mM GTP overnight. Although these crystals grow in slightly different conditions, they are isomorphous. Prior to data collection, crystals were soaked for a few seconds in a cryoprotecting solution containing 20% (v/v) glycerol before being mounted and cooled to 100 K in a nitrogen gas stream (Oxford Cryosystems). Diffraction intensities were collected at the PXIII (X10SA) beamline of the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland. Diffraction intensities for cocrystals with GTP were collected at the National Synchrotron Radiation Research Center, beamline 13B1 (Hsinchu, Taiwan). The best diffraction data were obtained for crystals of NS5 grown in the presence of calcium acetate. Integration, scaling, and merging of the intensities were carried out using programs MOSFLM and SCALA from the CCP4 suite [35]. The asymmetric unit contains one NS5 molecule with S-adenosyl-L-homocysteine (SAH) (copurified from E. coli) bound to the MTase domain. Crystal parameters and data collection statistics are summarized in Table 1.
Tab. 1. Data collection and refinement statistics. 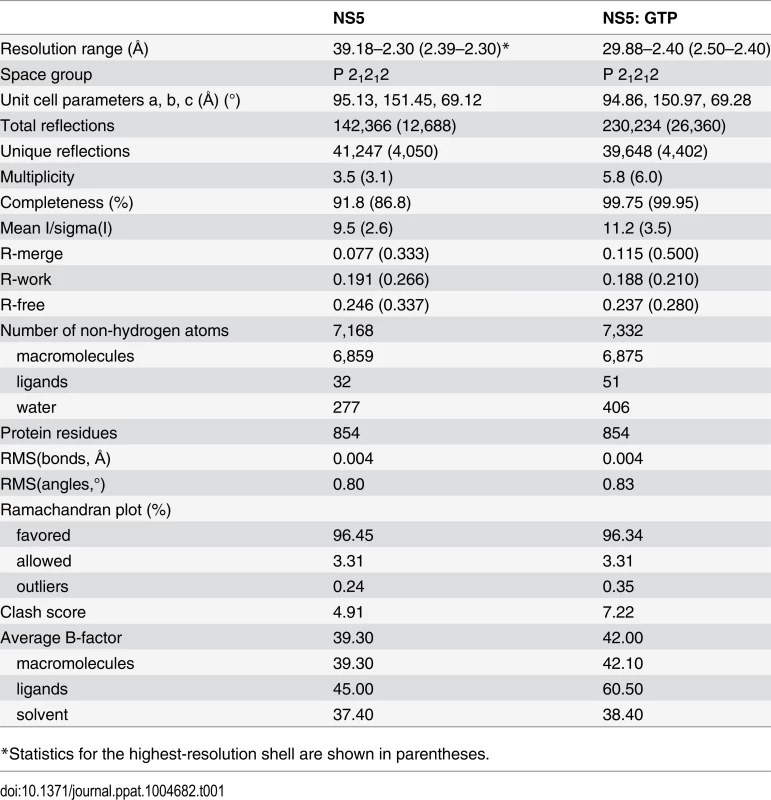
*Statistics for the highest-resolution shell are shown in parentheses. Structure solution and refinement
The crystals have an estimated solvent content of 50.2% based on a Matthews coefficient (Vm) value of 2.47 [36]. The structure was solved by molecular replacement with the program PHASER [37] using the DENV3 MTase (Protein Data Bank [PDB] accession code 3P97) [38] and DENV3 RdRp (PDB accession code 2J7U) [13] as search probes. Refinement cycles carried out using REFMAC5 [39] and PHENIX [40] were interspersed with manual model rebuilding sessions using Coot [41]. TLS refinement was introduced in the last refinement steps. The quality of the structure was analyzed using Molprobity [42]. A summary of structure refinement statistics is given in Table 1. Solvent-accessible surfaces areas were calculated using CCP4 program AREAIMOL with a 1.4-Å radius sphere as a probe. Superimpositions of structures were carried out using the program LSQKAB from the CCP4 suite. Figures were prepared using the program Pymol [43]. In the crystal structure of NS5 with GTP bound to the MTase domain (Table 1), the overall conformation of NS5 is very similar to the NS5: SAH structure: The root mean square deviation (r.m.s.d.) is 0.17 Å for a total of 719 Cα atoms superimposed. Buried surface areas were calculated with the PISA server (www.ebi.ac.uk/pisa) using the following domains definition: MTase residues 6–262 in DENV3 and 6–265 in JEV; RdRp residues 273–883 in DENV3 and 276–896 in JEV (S2 Table) [44]. The relative reorientation between domains was assessed using the program DynDom [45]. The refined coordinates were deposited in the PDB under accession codes 4V0Q (NS5: SAH) and 4V0R (NS5: SAH: GTP).
Hydrogen/Deuterium Exchange (HDX)
The buffer for the HDX reaction was of the same composition (20 mM Na-Hepes, pH 7.5, 300 mM NaCl, 5 mM DTT, 10% (v/v) glycerol) except that H2O was replaced with D2O (99.99%) and glycerol was omitted. Essentially the deuterium exchange reactions for NS5, or the individual MTase or RdRp domains was carried out by mixing a volume of 4 μl of the respective protein solutions (at a concentration of 18 μM), with 16 μl D2O or H2O buffer and incubated at 4°C for 0s, 30s, 60s, 600s, 1800s. The deuterium exchange (final D2O concentration 80%) for each sample at the various time points was quenched by the addition of 20 μl ice-cold solution consisting of 1 M guanidine hydrochloride and 1.5% (v/v) formic acid followed by rapid freezing in liquid nitrogen and storage at -80°C until HDX MS analysis.
In-column pepsin digestion, LCMS and HDX data processing
For capillary-flow LC, buffer A was H2O containing 0.3% (v/v) formic acid. Buffer B was acetonitrile containing 0.3% (v/v) formic acid. Deuterium exchanged protein samples from above were then digested online by passing through an immobilized pepsin-coupled column (2.1 mm i.d. × 30 mm) (Invitrogen) at 16°C and were de-salted for 3 min on a home-packed C4 trap (0.75 mm i.d. × 10 mm, C4 beads purchased from Michrom) with buffer A (H2O containing 0.3% (v/v) formic acid) at a flow rate of 150 μl min-1 driven by the LC loading pump (Dionex 3000 RSLC). The peptide samples were separated further on a home-packed C4 column (0.3 mm i.d. × 50 mm, C4 beads purchased from Michrom) using a 20 min gradient (5% to 35%) in buffer B (acetonitrile containing 0.3% (v/v) formic acid), followed by washing with 90% buffer B for 3 min and equilibration with 1% buffer B for 5 min) at a flow rate of 15 μl min-1 driven by LC NC pumps. MS raw data were acquired in the range of m/z 300–2000 for 30 min in positive mode on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with an ESI source (capillary temperature 275°C and spray voltage of 5 kV). All the HDX separations were performed at 0°C except for on-line pepsin digestion that was carried out at 16°C. Blank injections were made between runs to remove carry-over peptides. At least three independent deuterium exchange experiments were carried out for each time point. All HDX data were normalized to 100% D2O content, corrected for an estimated average deuterium recovery of 70%, and analyzed by the software HD Desktop [46]. Initial peptic peptide identifications were performed with the same HDX set up as described above. 4 μl of protein sample (20 μM) was injected into the HDX MS system. Product ion (MS/MS) spectra were acquired in linear ion trap LTQ with eight most abundant ions selected in the precursor (MS) scan with a 7.5 sec exclusion time. MS and tandem MS files were extracted and searched by using the Global proteome machine (http://www.thegpm.org) for high-confident peptide identification.
Generation of DENV4 NS5 mutant replicons
The K95A, Y119A, R263A (R262 in DENV3), E268A (E267 in DENV3), E270A (E269 in DENV3), and R353A (R352 in DENV3) mutants in the DENV4 NS5 sequence (GenBank accession number AF326825) were engineered into the subclone, pACYC-DENV4-F shuttle, using the QuikChange II XL site-directed mutagenesis kit according to the manufacturer’s protocol (Stratagene). This plasmid harbours nucleotides 7564–10653 (from NS3–3’UTR) from the DENV4, MY01–22713 strain, linked at the 3’ end to the Hepatitis D virus ribozyme (HDENVr) sequence [47]. Following sequence verification, the plasmids were digested with NotI and KpnI and inserted with a PCR product comprising the sequence spanning nucleotides 1–7563 under the control of the upstream T7 promoter in which the region from nucleotides 217–2291 in this cDNA had been replaced by renilla luciferase and foot-and-mouth disease virus 2A protease cDNAs [38]. A list of primers used for cloning and mutagenesis is available in S1A Table is S1 Text.
In vitro transcription, RNA transfection, and renilla luciferase (RenLUC) measurements
After linearization of the corresponding replicon cDNA plasmids with XhoI, in vitro transcription (IVT) was performed using a T7 mMESSAGE mMACHINE kit according to the manufacturer’s protocol (Ambion; Austin, Texas, USA). IVT RNAs (10 μg) were electroporated into 8×106 BHK-21 cells, after which the cells were re-suspended in a volume of 25 ml of DMEM with 10% FBS. Cell suspensions of 0.5 ml per well were seeded into 12-well plate; and the cells were assayed for luciferase activities at 4, 24, 48, 72, and 96 h post-transfection. Duplicate wells were seeded for each data point. Luciferase assays were performed using a RenLUC assay system following the manufacturer’s protocol (Promega, WI, USA).
Cloning and protein expression of DENV4 WT and mutant NS5 proteins
NS5 (serotype DENV4) WT and mutant cDNAs were amplified by PCR from the respective WT and mutant pACYC-DENV4-F shuttle plasmids and cloned into the pET28a vector (Stratagene) using the NheI and XhoI sites. Protein expression and heat stability analyses by thermo-fluoresence were performed as described previously [32].
DENV4 NS5 methyl-transferase in vitro assays
N-7 and 2’-O MTase assays were performed as described [38,48]. In general, the N7-MTase reaction comprised 25 nM protein, 240 nM biotinylated GpppA-DENV nt 1–110 in vitro transcribed RNA, 320 nM [3H-methyl]-SAM in 50 mM Tris-HCl, pH 7.5, 20 mM NaCl, and 0.05% (v/v) CHAPS. The 2’-O MTase reaction comprised 25 nM protein, 40 nM GpppA-7mer RNA (Trilink), 320 nM [3H-methyl]-SAM in 50 mM Tris-HCl, pH 7.5, 10 mM KCl, 2 mM MgCl2, and 0.05% CHAPS. Buffer, RNA substrate, and enzyme were first mixed in a single well in a 96-well half - area, white opaque plates (Corning Costar, Acton, MA), and the reaction was initiated by addition of [3H-methyl]-SAM. The N-7 and 2’-O reactions were incubated at RT for 15 min and 1 hr, respectively. The reaction was stopped with 25 μl of 2× stop solution (100 mM Tris/HCl, pH 7, 100 mM EDTA, 600 mM NaCl, 4 mg/ml streptavidin—SPA beads and 62.5 μM cold AdoMet) and shaken for 20 min at 750 rpm at room temperature followed by centrifugation for 2 mins at 1200 rpm. The plate was read in a Trilux microbeta counter (PerkinElmer, Boston, MA) with a counting time of 1 min per well. All data points were collected in duplicate wells.
DENV4 NS5 polymerase in vitro assays
The de novo initiation/elongation assay was described [28,49]. Briefly, the reaction comprised 100 nM DENV4 NS5, 100 nM in vitro transcribed DENV4 5´UTR-3´UTR RNA, 20 μM ATP, 20 μM GTP, 20 μM UTP, 5 μM Atto-CTP (Trilink Biotechnologies), in a volume of 15 μl of the assay buffer comprising 50 mM Tris-HCl, pH 7.5, 10 mM KCl, 1 mM MgCl2, 0.3 mM MnCl2, 0.001% Triton X-100 and 10 μM cysteine. The elongation assay reaction comprised 100 nM IVT 244 nt heteropolymeric RNA template, annealed with four primers (C1 primer 3’-AGTCAGTCAGTCAGTGT-biotin-5’, A1 primer 3’-GTCAGTCAGTCAGTCTC-biotin-5’, G1 primer 3’-TCAGTCAGTCAGTCACA-biotin-5’, T1 primer 3’-CAGTCAGTCAGTCAGAG-biotin-5’) [50], 2 μM ATP, 2 μM GTP, 2 μM UTP, 0.5 μM Atto-CTP, and 100 nM of DENV4 NS5 in 15 μl in assay buffer comprising 50 mM Tris-HCl at pH 7.5, 10 mM KCl, 0.5 mM MnCl2, 0.01% Triton X-100, and 10 μM cysteine. RNA was separately pre-annealed to the four different sets of primers at a ratio of 1 : 2 (w / w) by heating at 95°C for 3 min, and cooled to RT before mixing and used for the assay. All reactions were allowed to proceed for up to 3 hrs at RT. At the indicated time-points, 10 μl of 2.5× STOP buffer (200 mM NaCl, 25 mM MgCl2, 1.5 M DEA, pH 10; Promega) with 25 nM calf intestinal alkaline phosphatase (CIP; New England Biolabs) was added to the wells to terminate the reactions. The plate was shaken and centrifuged briefly at 1200 rpm, followed by incubation at RT for 60 min and the released AttoPhos was monitored by reading on a Tecan Saffire II microplate reader at excitationmax and emissionmax wavelengths 422 nm and 566 nm respectively. All data points were collected in triplicate wells in 384-well black opaque plates (Corning).
Construction of mutant DENV2 full-length infectious clone
Mutations of K95A, Y119A, E268A (E267 in DENV3) and R353A (R352 in DENV3) were constructed into full-length DENV2 cDNA clone (GenBank accession EU081177.1) using the subclone pWSK29 D2 fragment 3 as described [17]. Briefly, mutations were generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Primers used for mutagenesis are listed in S1D Table in S1 Text. Fragment 3 bearing the mutation was excised from the plasmid by XbaI and SacI, and inserted into subclone pWSK2 D2 fragment 1+2 that was similarly cut with XhaI and SacI.
In vitro transcription and replication profiling of mutants
The genome-length wild-type DENV2 infectious clone plasmid pWSK29 (GenBank accession EU081177.1) and the mutants were linearized with SacI and purified with phenol-chloroform. A quantity of ~2 μg linearized DNA was used for in vitro transcription with T7 mMESSAGE mMACHINE kit (Ambion). BHK-21 cells were trypsinized, washed twice with cold PBS and resuspended in Opti-MEM (Gibco) at a cell density of 1 × 107 cells/ml. 10 μg of in vitro transcribed RNA (wild-type DENV2 and mutants) were mixed with 800 μl cell suspension in a pre-chilled 0.4 cm cuvette, and electroporated at settings of 850 V and 25μ F, 2 pulses at 3 seconds intervals. Electroporated cells were allowed to recover at room temperature for 10 minutes prior to resuspending in complete RPMI 1640 media. 3 × 105 cells were then seeded into a 12-well plate and incubated at 37°C in the presence of 5% CO2. Media was changed to 2% FBS maintenance media after 6 hours post-transfection. Samples were harvested every 24 hours post-transfection until 120 hours. Supernatants were harvested and clarified for determination of virus titer by standard plaque assay on BHK-21 cells and extracellular viral RNA quantification by real time RT-PCR. Cells were then washed once with PBS before lysing with Trizol reagent (Invitrogen) for total intracellular viral RNA quantification.
Real time RT-PCR and Immunofluorescence assay
For extracellular viral RNA quantification, viral RNA was extracted from the supernatant using QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Real time RT-PCR was carried out in Bio-Rad Real time thermal cycler CFX96 by the use of iTaq Universal SYBR Green One-Step kit (Bio-Rad) with primers (forward, 5’ - CAGGCTATGGCACTGTCACGAT-3’; and reverse, 5’-CCATTTGCAGCAACACCATCTC-3’) targeting for DENV2 E region (adopted from [51]). A plasmid fragment containing genome sequences of the E region was used to establish a standard curve for quantification of viral genome copy number per ml of supernatant.
For cellular viral RNA quantification, total RNA was extracted using Trizol extraction method and 40 ng RNA was subjected to real time RT-PCR as described above. Absolute copies of the viral RNA genome was normalized to actin expression with β-actin primers (forward, 5’ - TCACCACACACTGTGCCCATCTACGA-3’ and reverse, 5’-CAGGGGAACCGCTCATTGCCAATGG-3’); and reported as viral RNA genome copy per μg RNA.
IFA against dsRNA with anti-dsRNA mAb J2 (Scicons) and NS5 protein with anti-NS5 hAb 5R3 were performed as described [52]. Images were captured on an inverted fluorescence microscope (Olympus IX71, Center Valley, USA) at 20× magnification and image analysis was done with ImageJ software.
Results
A compact conformation of NS5 from DENV3 with an inter-domain interface dominated by electrostatic interactions
Overall, the polypeptide chain of NS5 is well defined in the electron density map, including for inter-domain residues 262–273 (Fig. 1 and Table 1). The NS5 protein from DENV3 adopts a compact shape with overall dimensions 87 Å × 72 Å × 55 Å (Fig. 1A and B). The MTase domain is located above the fingers subdomain of the RdRp (Fig. 1A), such that the GTP binding pocket, the K61D146K180E216 catalytic tetrad and the SAH binding pockets of the MTase domain are positioned away from the inter-domain interface. In this conformation, the tunnels of the RdRp domain that permit the entry of NTPs and RNA template remain accessible. The MTase-RdRp inter-domain interface is comprised of two contact areas involving linker residues, the fingers subdomain from the RdRp and three segments (residues 63–69, 95–96 and 252–256) from the MTase domain (Fig. 1). The network of interactions stabilizing the interface mainly comprises charged residues (Fig. 1; S2 Table in S1 Text). In the first cluster, linker residues interact with both the RdRp and MTase domains. The side chain of E267 hydrogen bonds with residues Y119 and R262 from the MTase domain, while E269 forms a salt bridge with the guanidinium group of R361 from the RdRp domain (Fig. 1C). Several polar interactions are also formed between residues K95-K96 from the MTase domain and E296-K300 from the RdRp domain (Fig. 1C; S2 Table in S1 Text). The second cluster of inter-domain interactions is centered at helix α5 (residues F348-K357), which belongs to the evolutionary conserved bNLS motif of the RdRp domain, (Fig. 1B and D). Projecting from helix α5, the guanidinium side chain of R352 plays a central role via the formation of numerous contacts including electrostatic interactions with E67, E252 and Q63. Another salt bridge is formed between K357 and D256 (Fig. 1D). In addition, several water molecules trapped at the interface mediate H-bond interactions between residues projecting from both domains (Fig. 1C and D). In contrast to multiple non-polar contacts established between the MTase and RdRp domains of NS5 from JEV, the only hydrophobic contacts between the two domains of NS5 from DENV3 involve stacking interactions between W64, R68, F348 and P582 (Fig. 1D).
Fig. 1. Crystal structure of DENV3 NS5. 
(A) Overall structure of the NS5 protein from DENV3 in cartoon representation viewing from the bottom of RdRp. MTase is in yellow, RdRp fingers in green, palm in blue, thumb in salmon. The linker helix (residues 263–267) between the two domains is in orange. GTP and co-factor SAH are shown as sticks and labelled. Zinc ions are shown as spheres. (B) View of the NS5 molecule from the top of RdRp, which is rotated by 180° around a vertical axis as in (A). Interface regions are boxed. (C) and (D) Close-up views of the interface between the MTase domain and RdRp domain as indicated in (B). Key residues for inter-domain interactions are shown as sticks and labeled. (E) Multiple sequence alignment of flavivirus NS5 proteins. Interface residues are highlighted in gray. The linker residues (263–272 in DENV3 NS5) are boxed. List of accession numbers for genes and proteins used for alignment: DENV3: gi|50347097|gb|AAT75224.1|; DENV1: gi|194338413|gb|ACF49259.1|; DENV2: gi|266813|sp|P29990.1|; DENV4: gi|425895219|gb|AFY10034.1|; JEV: gi|4416167|gb|AAD20233.1|; WNV: gi|607369775|gb|AHW48802.1|; YFV: gi|27735297|ref|NP_776009.1|; TBEV: gi|1709707|sp|Q01299.1|. Despite an extensive inter-domain interface (Fig. 2), the MTase and RdRp domains of NS5 display only minimal differences at surface exposed regions compared to the individual enzymatic domains (S3 Fig in S1 Text). Superimposition with the NS5-MTase domain structures (PDB codes 3P97 and 1L9K) returns RMSD values of 0.42 Å and 0.40 Å for 221 Cα and 219 Cα atoms respectively [5,38]. In the MTase domain, the co-purified (SAH) molecule resides in the cofactor-binding pocket and is stabilized by a network of hydrogen bonds and van der Waals contacts (Fig. 1; S1 Fig in S1 Text) essentially identical to those reported earlier [5,53,54]. GTP is stabilized by base stacking with the aromatic ring of F25 and electrostatic interactions with residues L17, N18, and L20 of the MTase domain (S1 Fig in S1 Text) [5,54]. Likewise, superimposition with the DENV3 NS5-RdRp (PDB codes 2J7U and 4A11) yields RMSD values of 0.47 Å and 0.43Å for 492 and 495 Cα atoms respectively [13,32]. Like in the isolated RdRp domain structure reported earlier [13], however, two segments of the polypeptide chain are disordered in the present NS5 structure: residues 406–417 that harbor motif G, which regulate access of the ssRNA substrate to the template tunnel [13] and residues 455–468 which contain motif F. The latter were proposed to bind Stem Loop A, the viral promoter, prior to viral RNA replication [55]. Two Zinc ions bound to the RdRp domain are also observed as reported earlier [12,13].
Fig. 2. The dynamics of NS5 in solution probed by HDX-MS. 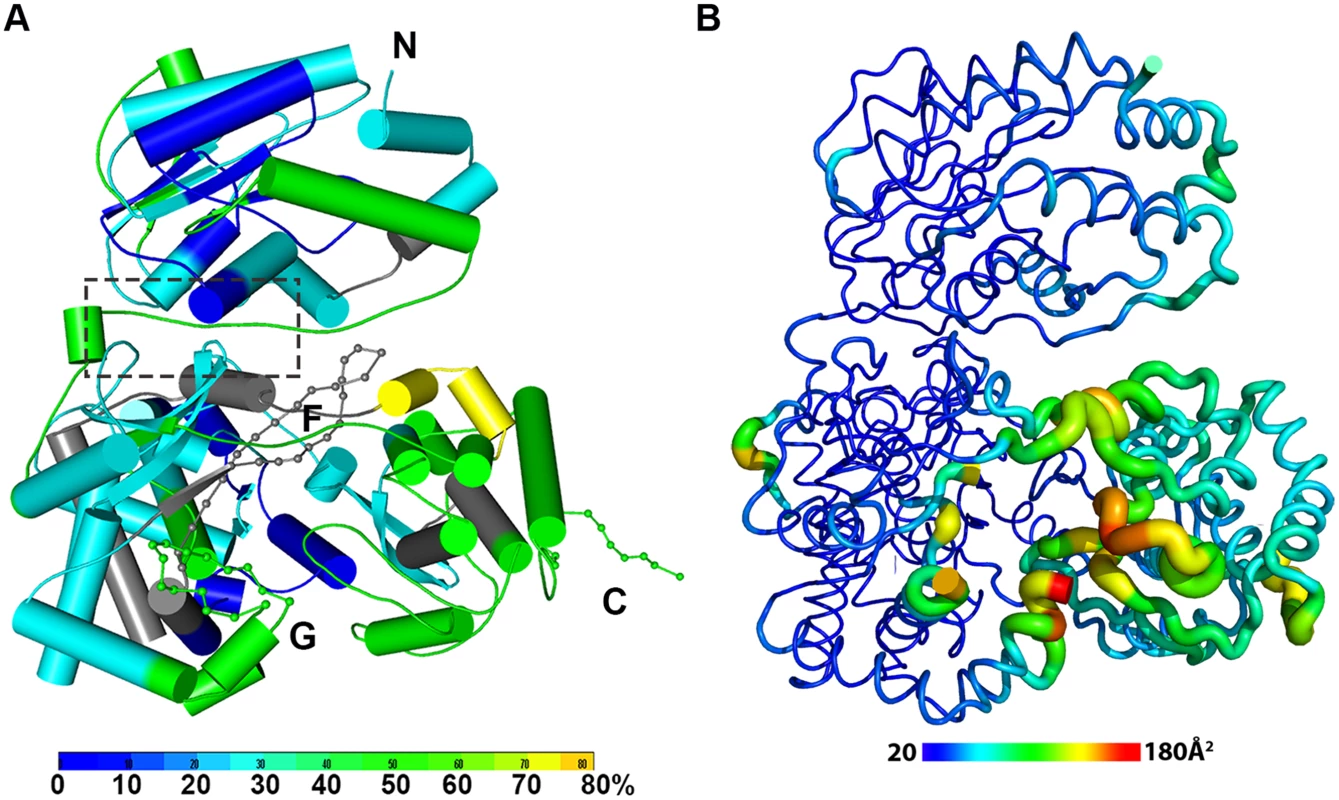
(A) Heat map data overlaid onto a crystal structure for the DENV3 NS5. The color key indicates the extent of the deuterium uptake (%), in which blue means the lowest deuterium uptake. Peptides in gray are regions where information of deuterium uptake is missing. F: motif F; G: motif G. (B) Putty cartoon view of temperature factor variation on the crystal structure of DENV3 NS5, colored from low to high values (20~180 Å2 as blue to red). Conformational dynamics of NS5 in solution
In order to determine whether the conformation of NS5 captured in the crystal state agrees with that in solution and also to better understand the dynamics of NS5, we performed Hydrogen/Deuterium Exchange Mass Spectrometry (HDX-MS) experiments using NS5, NS5-MTase or NS5-RdRp (Fig. 2A; S2 Fig in S1 Text). HDX-MS results are influenced by protein exposure to solvent and stability in solution, making this technique suitable to study solution-state protein dynamics [17,56–59]. The HDX-MS heat maps were overlaid onto the three respective crystal structures, with a color code indicating the extent of deuterium exchange with the protein backbone amide hydrogen atoms. The missing residues 406–417, 455–468, and 884–891 were modeled based on the homology to JEV NS5 structure. The corresponding HDX-MS profiles of peptic peptides across NS5 are listed in S2A Fig in S1 Text. The interface peptides from the MTase (residues 65–77 and 95–118) and the RdRp (residues 291–301 and 581–587) domains of NS5 display comparable deuterium exchange (Fig. 2A, boxed area; S2E and S2F Fig in S1 Text). However, the extent of deuterium exchange of these peptides (~ 20–30%) is not as low as the domain core region (~ 10% in blue), suggesting that the interface is solvent accessible as indicated by the crystal structure (Fig. 2A; S2 Fig in S1 Text).
Interestingly the HDX-MS also reveals the local protein dynamics of NS5. Peptides from the thumb subdomain (I735-L748, S781-M809, V812-L850, and L853-S895) and the middle finger (K311-M340) of the RdRp domain display higher deuterium-exchange (Fig. 2A; S2 Fig in S1 Text) in NS5 compared with the isolated NS5-RdRp domain. This suggests that in the context of the full length NS5 protein, the thumb subdomain of RdRp is more dynamic in solution than its fingers and palm subdomains. Remarkably the dynamics measurements with HDX-MS correlated very well with the distribution of crystallographic temperature factors observed for the RdRp domain of NS5, where higher temperature factors for the thumb subdomain than the fingers and palm subdomains are observed (Fig. 2B). Likewise, residues containing motifs F and G that are disordered in the crystal structure of NS5, also display a higher degree of amide hydrogen exchanges in HDX-MS (Fig. 2A; S2 Fig in S1 Text) [13,32]. It is worth noting that the conformation adopted by motifs F and G in the crystal structure of the isolated RdRp domain is different in the NS5 protein from JEV (S4 Fig in S1 Text). Overall, the HDX-MS experiment indicates that the conformation adopted by DENV3 NS5 in the crystal structure is also accessible in solution but that other conformations are possible.
Structure of the linker as the determinant of NS5 inter-domain flexibility
Despite a similar molecular shape maintained by intra-molecular interactions between the MTase and fingers subdomain of the RdRp, both the relative orientation and the interfaces between domains differ between the NS5 proteins from JEV and DENV3 (Fig. 3) [31]. Following superimposition of their RdRp domains, an additional rotation of 105° of their MTase domain is needed to bring them to coincidence (Fig. 3; S1 Movie). The inter-domain linker regions also differ markedly between the two proteins: In DENV3 NS5, the linker residues are well ordered (Fig. 3A and C), while in the JEV NS5 structure, residues A266-G270 adopt an extended conformation and residues E271-V272-H273 are disordered (Fig. 3B; S4 Fig in S1 Text) [31]. Since the linker residues are well ordered with clear electron density in the present DENV3 NS5 crystal structure (Fig. 3C), a precise definition of the structural determinants contributing to inter-domain flexibility is now possible. Earlier crystal structures of NS5 individual domains suggested that the evolutionarily conserved R262, whose guanidinium group hydrogen bonds with the main chain carbonyl oxygen of V97, defines the C terminus of the MTase domain (Fig. 1C; S3 Fig in S1 Text) [1,28,32,57,60]. Residues H263-V264-N265-A266, which are located after the C-terminal end of the MTase domain, are the least evolutionarily conserved in the interface region (Fig. 1E and 3D). Interestingly, these residues fold into a short 310-helix giving the polypeptide chain a compact conformation and allowing the two domains of NS5 to form a large interface leading to its observed globular shape (Figs. 1 and 2). Conversely, the segment E267-N272 forms extensive interactions with both the MTase and RdRp domains (Fig. 1C and 3C). The side chain of E267 projects towards the MTase domain forming hydrogen bonds with R262 and Y119, while the side chain of E269 projects towards the RdRp domain and interacts with R361 and K595. The two acidic residues E267 and E269 are highly conserved among all four dengue serotypes, but not in other flaviviruses (Fig. 1E and 3D). Likewise, T270 and N272 establish several polar contacts with the RdRp domain (Fig. 1C). To further delineate the boundaries of the RdRp domain of NS5, we observe that residues 268–271 adopt the same conformation in both the DENV3 NS5-RdRp and in the present NS5 structures (S3 Fig in S1 Text) [32]. Coincidentally, residues 268–272 enhance the thermostability of the truncated NS5-RdRp and also its polymerase activity [32] suggesting that they may be considered as an integral part of the RdRp domain of NS5 (Fig. 3D; S3 Fig in S1 Text). We conclude that DENV NS5 contains a short four-amino-acids long inter-domain linker (residues 263–266). Like a swivel, it is conceivable that structural transitions at this short linker—for instance a conversion from a helix to more extended conformations—are accompanied by important changes in the relative orientation between the two domains of NS5 [61].
Fig. 3. Comparison of conformations between DENV3 NS5 and JEV NS5 structures. 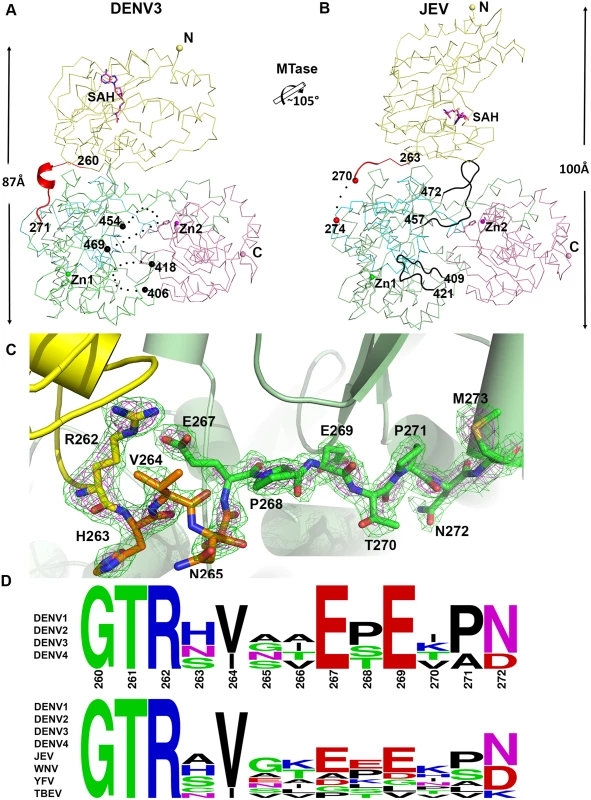
(A) and (B) Side-by-side view of the DENV3 NS5 and the JEV NS5 structures in thin ribbon style. Same color scheme as in Fig. 1: MTase is in yellow, RdRp fingers in green, palm in blue, thumb in salmon. The linker regions (260–271 in DENV3 NS5; 263–274 in JEV NS5) are shown as cartoon in red. Missing residues (407–417 and 455–468 in DENV3 NS5; 271–273 in JEV NS5) are indicated as dots. The rotation axis that relates both MTase domains is indicated with the corresponding angle. (C) The simulated annealing mFobs-DFcalc omit maps are in green, contoured at 3σ and in magenta, contoured at 5σ. (D) Sequence conservation of the linker region of DENV 1–4 and representative flaviviruses. Figure is generated with WebLogo [71]. Comparison of inter-domain interface
The total buried interdomain surface area calculated for DENV3 NS5 is 1502 Å2 which is only slightly larger than that for JEV NS5 (1464 Å2) (Domain definition: MTase residues 6–262 in DENV3 and 6–265 in JEV; RdRp residues 273–883 in DENV3 and 276–896 in JEV) (S2A Table in S1 Text) (http://www.ebi.ac.uk/pdbe/pisa/) [44]. These values suggest that both structures are relatively stable in solution [62]. This is consistent with the rather compact shapes adopted by NS5 with maximum molecular dimensions of approximately 87 Å (DENV3) and 100 Å (JEV) (Fig. 3A and B). Interestingly, major inter-domain interactions in DENV3 NS5 are of polar nature, occurring both directly between charged side-chains and through water-mediated contacts (Fig. 4A; S2B Table in S1 Text). In contrast, the inter-domain interactions in JEV NS5 are predominantly hydrophobic, including residues P113, L115, W121 from MTase and F467 (F464 in DENV3), F351(F348 in DENV3) and P585 (P582 in DENV3) from the RdRp domain (Fig. 4B; S2B Table in S1 Text) [31]. An analysis of interface residues reveals that residues in both interfaces were highly conserved during evolution (Fig. 4C and D) [63]. When superposing MTase domains of the two NS5 structures, the relative positions of the catalytic centers of the MTase domain (KDKE catalytic tetrad) and the RdRp domain (GDD, motif C) are far apart (Fig. 4E). It is thus evident that both DENV and JEV RdRp domains used the finger subdomains to interact with their respective MTase domains from two non-overlapping sides, thus establishing unique interfaces for DENV or JEV (Fig. 4).
Fig. 4. Comparison of interfaces between DENV3 and JEV NS5. 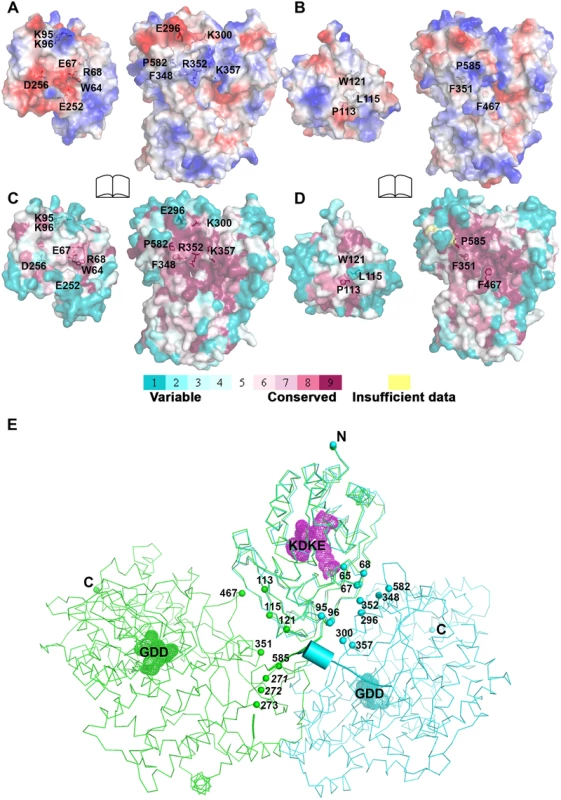
RdRp domains of the two structures have been displayed in the same orientation and the buried surfaces (indicated by the labeled interface residues) from the RdRp domains are relatively overlapping between the two NS5 structures. (A) and (B) Analysis of the electrostatic properties of the domain interfaces of the two NS5 structures. Positive surface charge is highlighted in red, negative charge in blue. (C) and (D) Analysis of the evolutionary conservation of the domain interfaces of the two NS5 structures. The color key indicates the degree of conservation, with cyan means highly variable and purple means highly conserved. (E) The relative orientations of the RdRp domain relative to the MTase domain in two structures in thin ribbon style. DENV3 NS5 is in blue, JEV NS5 is in green. “KDKE” highlights the MTase active site shown as mesh in purple and “GDD” for the RdRp active site shown as mesh. Residues at interface are highlighted and shown as dots. Linker (262–272) in DENV3 NS5 is shown as cartoon. Differential effects of interface mutations on in vitro enzymatic activities of NS5
To examine the functional importance of individual residues within the two clusters of interactions at the DENV NS5 inter-domain interface (Fig. 1C and D), we introduced single alanine mutations at residues K95, Y119, R263 (R262 in DENV3), and R353 (R352 in DENV3) into the DENV4 NS5 protein and compared their polymerase activities against the WT protein (Table 2). Note also that the residues selected for mutagenesis are conserved across the four DENV serotypes but not across other flaviviruses, except for R263 (R262 in DENV3) and R353 (R352 in DENV3) (Fig. 1E). All purified mutant NS5 proteins retained similar stability as WT as determined by a thermofluor assay, suggesting a native fold (Table 2). The RdRp activity of the mutant proteins compared to WT enzyme was assessed by both the de novo initiation/elongation assay and the elongation assay as reported [32]. The former uses the viral UTR sequence as template to start a new RNA chain while the latter employs an heteropolymeric RNA template annealed with four primers, which is extended during the course of the assay [28,49].
Tab. 2. Enzymatic activities and thermo-stabilities of DENV4 WT and mutant NS5 proteins. 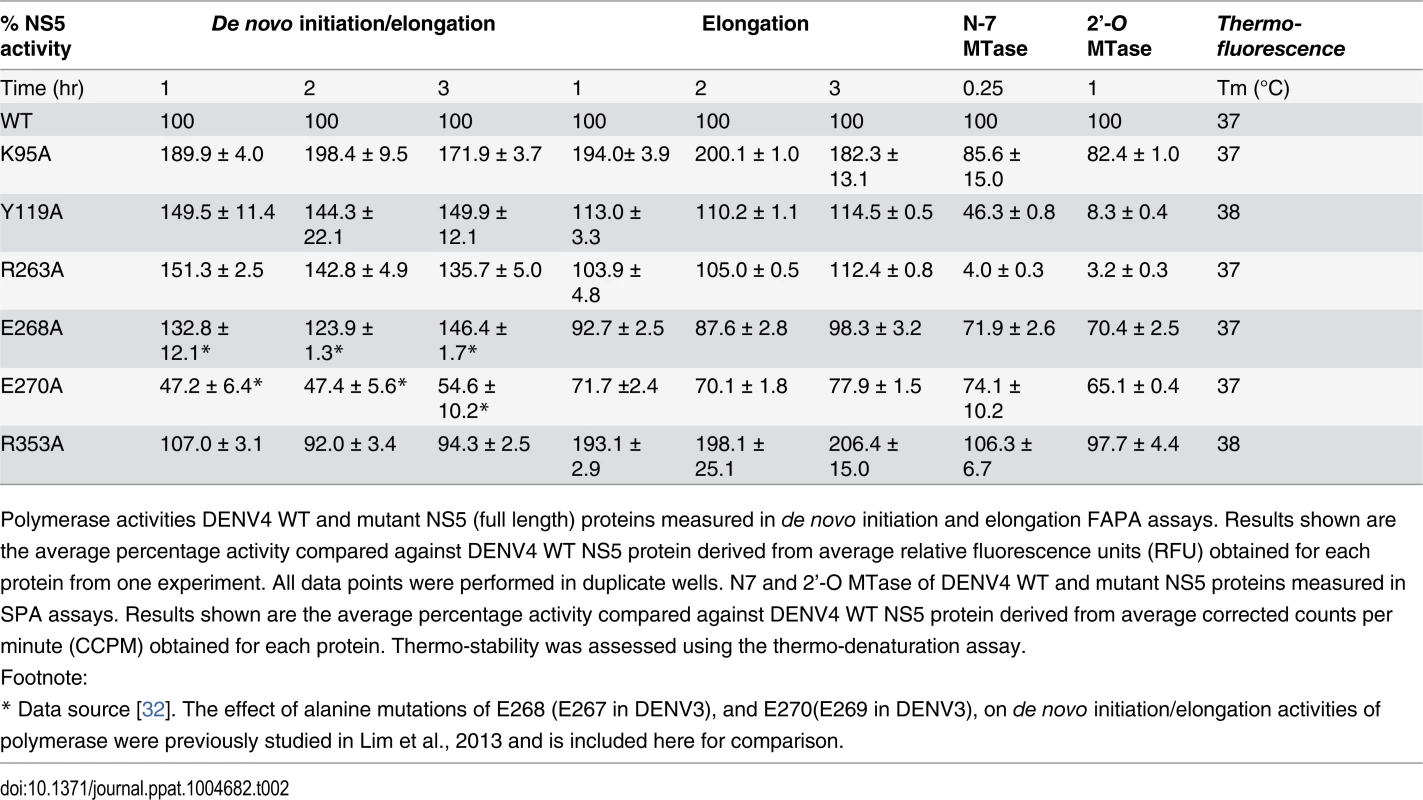
Polymerase activities DENV4 WT and mutant NS5 (full length) proteins measured in de novo initiation and elongation FAPA assays. Results shown are the average percentage activity compared against DENV4 WT NS5 protein derived from average relative fluorescence units (RFU) obtained for each protein from one experiment. All data points were performed in duplicate wells. N7 and 2’-O MTase of DENV4 WT and mutant NS5 proteins measured in SPA assays. Results shown are the average percentage activity compared against DENV4 WT NS5 protein derived from average corrected counts per minute (CCPM) obtained for each protein. Thermo-stability was assessed using the thermo-denaturation assay. Overall, with the exception of E270A (E269 in DENV3) that exhibits about 30–60% reduction in both polymerase activity, none of the NS5 mutants exhibit significant impairment in their de novo initiation or elongation activities. Mutants Y119A, R263A (R262 in DENV3), and E268A (E267 in DENV3) display a 24–50% increase in de novo initiation/elongation activities compared to WT NS5 (Table 2). The alanine mutation at K95 results in the most active RdRp so far with an almost 2 fold increase in both the de novo initiation/elongation and elongation activities, compared to WT. Residue R353 (R352 in DENV3) is located at the second interface cluster and belongs to a 20-amino-acids sequence conserved in all flavivirus NS5 proteins [15] and, as mentioned earlier, it forms part of the bNLS. The de novo initiation/elongation activity is 2-fold higher (similar to K95A) but the primer elongation activity is similar to WT (Table 2).
Next, we investigated the effect of the mutations on the N7 and 2’-O MTase activities of NS5 (Table 2). Mutation of residues Y119 and R263 (R262 in DENV3) to alanine strongly impacted both N7 and 2’-O MTase activities. Mutant Y119A exhibited less than 50% N7 activity compared to WT protein and minimal 2’-O activity. Both enzymatic activities were almost abolished in R263A (R262 in DENV3), the last residue of the MTase domain. On the other hand, E268A (E267 in DENV3) and E270A (E269 in DENV3)—mutants C-terminal to the inter-domain linker, retained 65–74% of both MTase activities, suggesting minor allosteric effects from the RdRp domain transmitted via the inter-domain interface. In contrast to the elevated polymerase activities, the K95A and R353A (R352 in DENV3) mutants that disrupt the transverse polar contacts appears to have little impact on the MTase activities (Table 2).
Essential role of the inter-domain interface in regulating virus replication
We next assessed whether residues involved in MTase-RdRp interactions are important for viral replication by introducing alanine mutations at positions K95, Y119, R263 (R262 in DENV3), E268 (E267 in DENV3), E270 (E269 in DENV3), and R353 (R352 in DENV3) in a DENV4 subgenomic RNA replicon, bearing a renilla luciferase reporter (Figs. 5A and 6) [32]. Both WT and mutant DENV4 replicon cDNAs were transcribed in vitro, electroporated into BHK-21 cells and luciferase activities were monitored at 4, 24, 48, 72 and 96 hr post-electroporation (Fig. 5A). The luciferase activity in cells harbouring WT DENV4 replicon showed a typical pattern, where a peak level is reached at approximately 24 to 48 hr post-electroporation (about 3320-fold above background levels), which steadily declined from 48–96 hr post-electroporation (Fig. 5A). Little or no luciferase activity was detected for DENV4 mutant replicons Y119A, R263A (R262 in DENV3), and E270A (E269 in DENV3), throughout the four days post-electroporation. Mutant replicons E268A (E267 in DENV3) and R353A (R352 in DENV3) exhibited significant luciferase activity that are almost comparable to WT at 48 hr point, although the mutants had some delay at the first 24 hrs (Fig. 5A).
Fig. 5. Replication profiles of NS5 interface mutants. 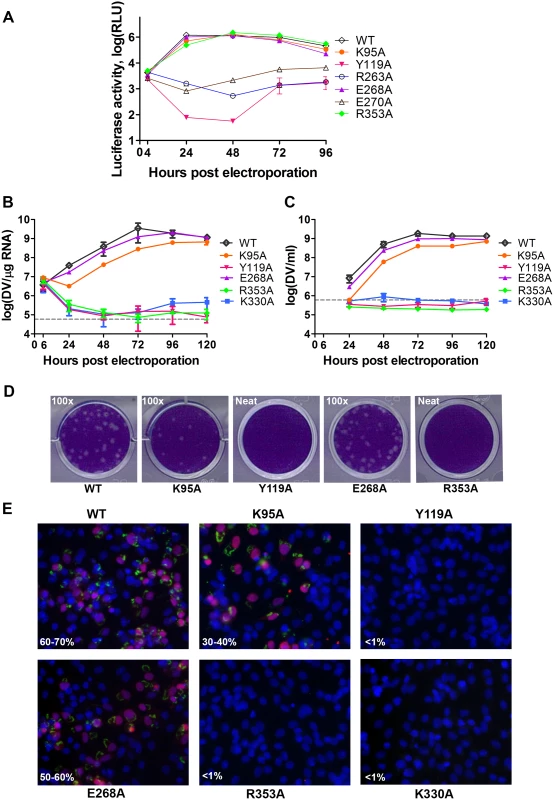
(A) Renilla luciferase activities of DENV4 WT and mutant replicons. Equal amounts of replicon RNA (WT or mutants) were electroporated into BHK-21 cells. At the indicated time points, the transfected cells were lysed and assayed for luciferase activities. The y axis shows the log10 value of Renilla luciferase activity (RLU). Each data point is the average for three replicates, and error bars show the standard deviations. (B) 10μg in vitro transcribed infectious clone RNA was electroporated into BHK-21 cells and viral replication was monitored over a course of 5 days. Intracellular viral RNA replication as detected by qRT-PCR. The grey dotted line represents the background detection of uninfected cells. (C) Extracellular viral RNA in the supernatants detected by qRT-PCR. The grey dotted line denotes background signal of uninfected supernatant. (D) Plaque morphologies of WT and the mutants at 72 hours post electroporation. (E) IFA images showing dsRNA and NS5 co-staining and percentage infection of cells at 72 hours post electroporation. Fig. 6. A schematic model for the divergent evolution of flaviviral NS5 proteins. 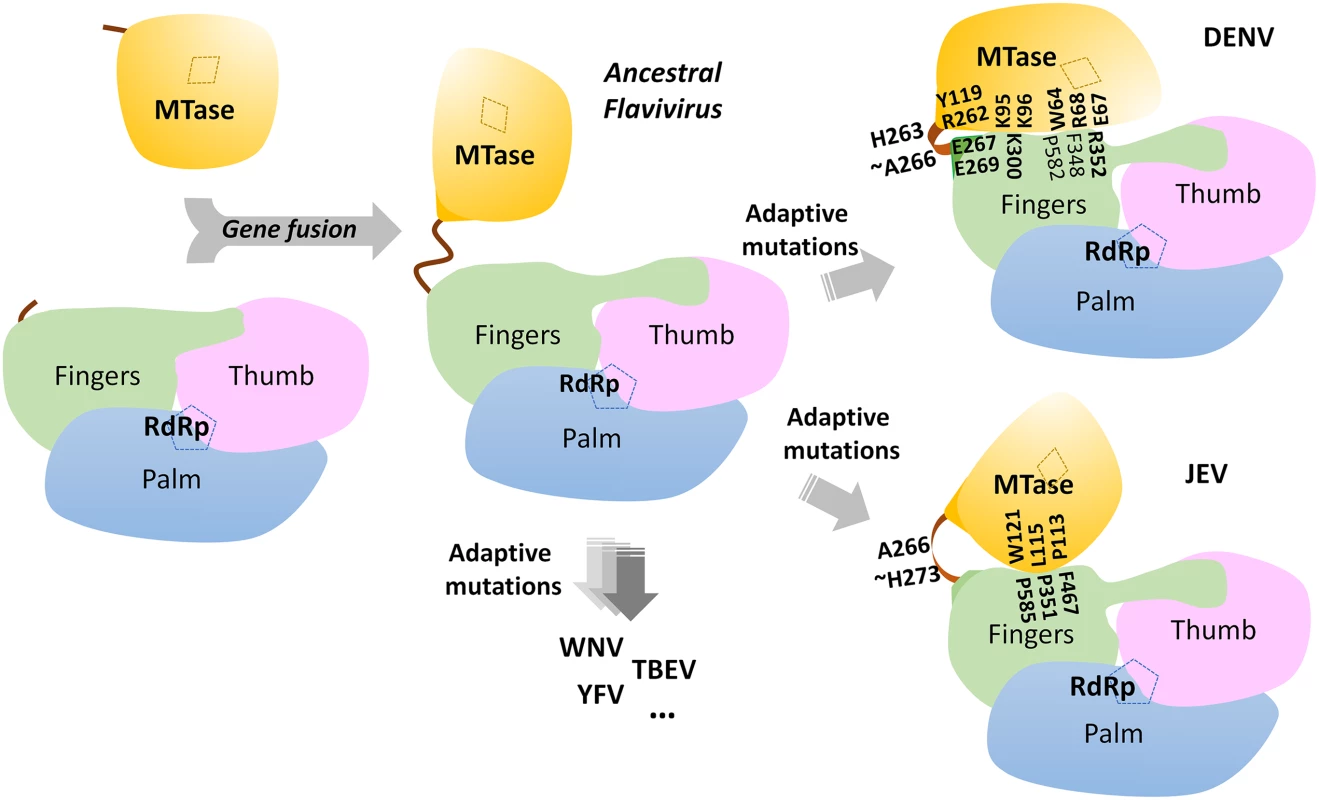
The same color scheme as in Fig. 1 is used: MTase is in yellow, RdRp fingers in green, palm in blue, thumb in salmon. The linker region 310 helix (residues 263–266) between the two domains is in orange. Active sites for MTase and RdRp are labelled with dotted tetragon and pentagon respectively. Linker residues and interface residues are labeled. A possible evolutionary pathway is presented: the MTase domain and RdRp domain originally existing as two separate proteins (left) became linked together to form the NS5 protein from an ancestral Flavivirus, possibly through gene fusion. This fusion promoted colocalization of both enzymatic activities and effectively increased the effective concentration of the proteins with respect to each other (middle panel). Following further (divergent) evolution, NS5 acquired different adaptive mutations and gave rise to the NS5 proteins now observed for various viruses, including DENV, JEV and possibly other flaviviruses) (right panel). Thus NS5 proteins from DENV and JEV may have different conformations and different allosteric mechanisms, in which the MTase and RdRp domain cross-talk to each other through unique interfaces specific to either DENV or JEV [72]. To further validate the biological significance of the interface, four residues including K95, Y119, E268 (E267 in DENV3), and R353 (R352 in DENV3) were selected and mutated into alanine in the DENV2 infectious clone (Fig. 5B, C, and D). Defective mutant K330A, which has previously been reported, was included as a negative control [14,17]. E268A mutant (E267 in DENV3) displayed similar growth kinetics, extracellular viral RNA level and infectious virus recovery compared to WT. Y119A mutation did not yield any viable virus or increased intracellular RNA quantified by qRT-PCR. This is likely due to the defective MTase activity (Fig. 5; Table 2). The K95A mutant virus replicated much slower than WT with a 10-fold lower intracellular and extracellular RNA level at 72 hr post electroporation. No RNA replication or virus recovery was detected for mutant R353A (R352 in DENV3). Immuno-fluorescence analyses at 72 hr using anti-dsRNA and anti-NS5 antibodies confirmed the intracellular levels for WT and mutant viruses (Fig. 5E). Taken together, these data demonstrated: (i) most interestingly, conserved residues that form the major inter-domain polar contacts, K95 and R353 (R352 in DENV3), play critical but non-enzymatic roles in virus RNA replication and infectivity; (ii) alanine mutation of E268 (E267 in DENV3) have no impact on the enzymatic activities of NS5 nor on virus RNA replication and infectivity (iii) Within the MTase domain, Y119, is important for the MTase enzymatic activity of NS5 and is therefore essential for the virus replication.
Discussion
A well-ordered linker region defines the boundaries for MTase and RdRp domains of DENV NS5
The present 3D structure of DENV3 NS5 at 2.3 Å provides the first view of a flavivirus NS5 protein showing how its MTase and RdRp domains are physically connected through a fully resolved linker region (Fig. 1 and 3). Based on the present study and recent work [32], we conclude that DENV NS5 contains a four amino-acids long inter-domain linker (residues 263–266), which is able to fold into a short 310-helix as observed in the present structure, but which can probably also adopt extended conformations (Fig. 6). In contrast, the JEV NS5 structure reveals a partially resolved linker region (266–275) which neither forms any secondary structure nor establishes interactions with either enzymatic domain (Fig. 3; S3 and S4 Fig in S1 Text) [31]. The low level of sequence conservation and the diverse conformations observed for the linker region strongly suggest conformational plasticity at the inter-domain interface for the flavivirus NS5 protein (Figs. 1 and 3D; S1 Movie). About 13 Å shorter in its largest dimension, DENV3 NS5 displays a more compact conformation than JEV NS5 (Fig. 3). This is reflected by the differences in the spatial arrangement of the two enzymatic domains (Fig. 4E). These different relative orientations are likely to result from different secondary structures adopted by the linker region that acts like a swivel, leading to the formation of different interfaces and enabling the two enzymatic domains to adopt various relative orientations during the transition from RNA synthesis to RNA capping (Figs. 3 and 6).
The MTase and RdRp domains engage in cross talk through a unique electrostatic-rich interface specific to DENV
Remarkably, the interface in DENV3 NS5 is mostly polar with several salt bridges and fewer hydrophobic stacking interactions than that of JEV NS5, which is mainly of hydrophobic nature (Fig. 4A and B) [60]. The interface peptides from MTase domain and the fingers subdomain of RdRp displayed similar hydrogen deuterium exchange patterns, which suggest that the crystal structure of NS5 represents a predominant conformation in solution (Figs. 1 and 2A). However, given the polar nature of the interface interactions, conformational flexibility and heterogeneity are likely. An earlier SAXS study also suggested that the interaction between the MTase and RdRp domains may be heterogeneous: approximately 80% of DENV3 NS5 were found to adopt a relatively compact structure in which the two domains interact with each other whilst the remaining 20% of the population were in more extended conformations [29]. Therefore the two enzymatic domains of DENV NS5 may be able to associate and dissociate from each other with a relatively low energy barrier, consistent with a limited inter-domain buried surface area and the predominance of polar interactions. The prevalence of a more compact conformation in solution is consistent with our observation of a relatively short but structured four-residue-linker (residues 263–266 in DENV3 NS5) that restricts inter-domain movements. Furthermore, the solution HDX-MS is consistent with the temperature factor analysis of the DENV3 NS5 structure, indicating that the RdRp domain of DENV3 NS5 adopts a more flexible conformation especially in its thumb sub-domain (Fig. 2). These observations rationalize the contributions of a covalently tethered MTase domain to the RNA synthesis activities by RdRp [33]. In addition, the dynamic nature of the DENV NS5 protein may also facilitate the recruitment of other viral or host proteins. In agreement with this proposal, we have shown previously that K330 within the α3-helix of RdRp is critical for binding to the NS3 helicase. Mutation of K330 to alanine did not inhibit the de novo initiation and elongation activities in RdRp nor the 2’O and N-7 MTase activities but completely abolished replication in an infectious clone [14].
The interface modulates virus replication
Using biochemical and reverse genetic tools—enzymology of MTase and RdRp activities for DENV4 NS5, DENV4 replicon and DENV2 infectious virus, we examined the functional relevance of the interface interactions to viral RNA replication and infectivity by structure-guided mutagenesis. When either the MTase (Y119A and R263A (R262 in DENV3) in DENV4) or the polymerase activities (E270A in DENV4 (E269 in DENV3)) were severely inhibited (over 50% reduction compared to WT), neither viral RNA nor infectious virus could be observed (Fig. 5A; Table 2), Mutation of key polar contacts at the interface (K95A and R353A (R352 in DENV3)), did not impair the enzymatic activities with a slightly increased RdRp activities (Table 2). Indeed regulation of the RdRp de novo initiation activity by the MTase domain was elegantly demonstrated using an array of enzyme assays by Selisko and colleagues [33]. However, this enhancement in enzymatic activity was not beneficial for the virus: we observed a slightly delayed RNA replication as indicated by the reporter activity in the first 24 hrs in the replicon assay (Fig. 5A) and more severely impaired virus replication and infectivity in the infectious clone (Fig. 5B, C and D). For K95A, the intracellular and extracellular RNA levels was about 1 log lower with <50% virus recovery measured by plaque assay compared to WT. In the case of R353A, which had a two-fold higher elongation activity, there was no detectable virus replication. It is possible that the enhanced polymerase activities of mutants K95A or R353A may promote the production of incomplete genomes, truncated viral RNA fragments or catastrophic mutations that abolish reinfection. These mutations may also alter the NS5 conformational dynamics and affect its ability to form competent virus replication complex with other cofactors from the virus and the host. Taken together we conclude that the inter-domain interface of DENV NS5 is essential for successful virus replication and infectivity.
An evolutionary view of the flavivirus NS5 MTase-RdRp structural heterogenity
Previous studies on WNV NS5 identified genetic interactions between residues K46A/R47A/E49A (MTase) and L512 (RdRp) from reverse genetic experiments, and a 3D model for WNV NS5 was built based on the hypothesis of direct molecular interactions [12]. However, mutagenesis studies targeting these residues do not affect the interaction between the two domains significantly using in vivo co-expression system [60]. The crystal structure of JEV NS5 revealed a hydrophobic inter-domain interface between the MTase and RdRp domains [31]. The follow-up mutagenesis study addressed the importance for JEV virus replication of residues P113 and L115 from the MTase domain and F351 (F348 in DENV3), F467 (F464 in DENV3) and P585 (P582 in DENV3) from the RdRp domain [64]. In the same study, corresponding mutations (P113D, P115D, F349D, F465D and to a lesser extent P583D) in DENV2 NS5 also yielded severely defective viruses [64]. The residues examined by mutagenesis involved drastic changes of the side-chain chemistry that may impact the folding and overall architecture of the mutants and limit the interpretations of these phenotypes. Subsequently in a follow-up work, the in vitro polymerase activity assays on JEV NS5 showed that proteins with mutations at the interface had only mildly impaired polymerase activities [65]. The authors thus suggested that the defect in virus replication was due to the amplifying effects of the defective enzymatic activity, the impact on the protein-protein interactions, or the combined effect of both [31,64,65].
Careful examination of the two available structures of NS5 revealed two non-overlapping buried surfaces on the MTase domain but one shared buried surface from the RdRp fingers subdomain (Figs. 1 and 3; S4 Fig in S1 Text) [31]. Indeed, F348 and P582 from DENV3 NS5 also form hydrophobic stacking interactions with the MTase domain (Fig. 1D and E). Mutations of these two residues, particularly F348, would disrupt the interface in both structures and the DENV3 NS5 structure explains very well the defective phenotypes of the mutant DENV2 viruses (F349D (F348 in DENV3) and P583D (P582 in DENV3)) (Figs. 1, 3 and 6) [64]. This further argues for the dynamic nature of NS5 and the differential impacts on virus replication within the replication complex machinery.
Based on the present report and comparable studies on NS5 from JEV and other flaviviruses, it is possible that the NS5 protein from different flaviviruses may have evolved to adopt different sets of conformations, stabilized by distinct interfaces and leading to different allosteric mechanisms to regulate their enzymatic activities (Figs. 3 and 6; S5 Fig in S1 Text). This hypothesis is detailed in Fig. 6. However, which molecular conformation(s) participates in the various stage(s) of the virus replication cycle remains elusive (Fig. 6). Thus, during virus replication, NS5 may adopt various conformations upon recruitment of and interaction with NS3, viral RNA and other viral and host cofactors, for instance in order to synthesize a fixed ratio of positive and negative RNA strands, for unwinding dsRNA intermediates and for the effective production of progeny viral genomic RNA. The short linker of DENV NS5 is likely to dictate the degree of freedom of the overall molecular conformation so that the necessary discrete functional conformations can be attained. Inspection of the sequence heterogeneity of the linker residues (Fig. 4E) suggests that the NS5 proteins from other flaviviruses may also be endowed with comparable (or even higher) degrees of freedom (Fig. 6), possibly reflecting the need to accommodate additional host factors that must be recruited to the RC for replication. This hypothesis is also indirectly supported by the disordered linker sequence of JEV NS5 (S3 Fig in S1 Text). Remarkably, the flaviviral NS3 protease-helicase also contains an inter-domain linker that is flexible and variable in sequence and which is able to adopt multiple conformations at different stages of the virus life cycle [66–70]. In this respect, the inter-domain linkers observed in the NS3 and NS5 proteins might have evolved to play a role for the assembly of the RC comparable to the extended and flexible arms that are needed for viral capsid proteins assembly. Further studies exploring possible connections between virus pathogenesis and the dynamics of viral replicative enzymes in various flaviviruses are now needed, which are likely to be of interest to both basic virology and immunology research and also to antiviral therapeutics development.
Supporting Information
Zdroje
1. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, et al. (2013) The global distribution and burden of dengue. Nature 496 : 504–507. doi: 10.1038/nature12060 23563266
2. Mackenzie J (2005) Wrapping things up about virus RNA replication. Traffic 6 : 967–977. 16190978
3. Miller S, Krijnse-Locker J (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6 : 363–374. doi: 10.1038/nrmicro1890 18414501
4. Salonen A, Ahola T, Kaariainen L (2005) Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol 285 : 139–173. 15609503
5. Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B (2002) An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21 : 2757–2768. 12032088
6. Ferron F, Decroly E, Selisko B, Canard B (2012) The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res 96 : 21–31. doi: 10.1016/j.antiviral.2012.07.007 22841701
7. Zhou Y, Ray D, Zhao Y, Dong H, Ren S, et al. (2007) Structure and function of flavivirus NS5 methyltransferase. J Virol 81 : 3891–3903. 17267492
8. Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, et al. (2006) West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80 : 8362–8370. 16912287
9. Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, et al. (2008) West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol 82 : 4295–4307. doi: 10.1128/JVI.02202-07 18305027
10. Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, et al. (2009) The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 15 : 2340–2350. doi: 10.1261/rna.1609709 19850911
11. Bollati M, Milani M, Mastrangelo E, Ricagno S, Tedeschi G, et al. (2009) Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in Flavivirus. J Mol Biol 385 : 140–152. doi: 10.1016/j.jmb.2008.10.028 18976670
12. Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, et al. (2007) Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J Biol Chem 282 : 10678–10689. 17287213
13. Yap TL, Xu T, Chen YL, Malet H, Egloff MP, et al. (2007) Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol 81 : 4753–4765. 17301146
14. Zou G, Chen YL, Dong H, Lim CC, Yap LJ, et al. (2011) Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem 286 : 14362–14372. doi: 10.1074/jbc.M110.214189 21349834
15. Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, et al. (2002) The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem 277 : 36399–36407. 12105224
16. Johansson M, Brooks AJ, Jans DA, Vasudevan SG (2001) A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J Gen Virol 82 : 735–745. 11257177
17. Tay MYF, Saw WG, Zhao Y, Chan WKK, Singh D, et al. (2014) The C-terminal 50 amino acid residues of Dengue NS3 protein are important for NS3-NS5 interaction and viral replication J Biol Chem In press. doi: 10.1074/jbc.M114.607341 25488659
18. Fernandez-Garcia MD, Meertens L, Bonazzi M, Cossart P, Arenzana-Seisdedos F, et al. (2011) Appraising the roles of CBLL1 and the ubiquitin/proteasome system for flavivirus entry and replication. J Virol 85 : 2980–2989. doi: 10.1128/JVI.02483-10 21191016
19. Gomila RC, Martin GW, Gehrke L (2011) NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS One 6: e16687. doi: 10.1371/journal.pone.0016687 21386893
20. Davis WG, Blackwell JL, Shi PY, Brinton MA (2007) Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol 81 : 10172–10187. 17626087
21. Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A (2009) NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83 : 5408–5418. doi: 10.1128/JVI.02188-08 19279106
22. Hannemann H, Sung PY, Chiu HC, Yousuf A, Bird J, et al. (2013) Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J Biol Chem 288 : 22621–22635. doi: 10.1074/jbc.M113.481382 23770669
23. Tay MY, Fraser JE, Chan WK, Moreland NJ, Rathore AP, et al. (2013) Nuclear localization of dengue virus (DENV) 1–4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res 99 : 301–306. doi: 10.1016/j.antiviral.2013.06.002 23769930
24. Keller TH, Chen YL, Knox JE, Lim SP, Ma NL, et al. (2006) Finding new medicines for flaviviral targets. Novartis Found Symp 277 : 102–114; discussion 114–109, 251–103. 17319157
25. Yin Z, Chen YL, Kondreddi RR, Chan WL, Wang G, et al. (2009) N-sulfonylanthranilic acid derivatives as allosteric inhibitors of dengue viral RNA-dependent RNA polymerase. J Med Chem 52 : 7934–7937. doi: 10.1021/jm901044z 20014868
26. Yin Z, Chen YL, Schul W, Wang QY, Gu F, et al. (2009) An adenosine nucleoside inhibitor of dengue virus. Proc Natl Acad Sci U S A 106 : 20435–20439. doi: 10.1073/pnas.0907010106 19918064
27. Noble CG, Shi PY (2012) Structural biology of dengue virus enzymes: towards rational design of therapeutics. Antiviral Res 96 : 115–126. doi: 10.1016/j.antiviral.2012.09.007 22995600
28. Lim SP, Wang QY, Noble CG, Chen YL, Dong H, et al. (2013) Ten years of dengue drug discovery: progress and prospects. Antiviral Res 100 : 500–519. doi: 10.1016/j.antiviral.2013.09.013 24076358
29. Bussetta C, Choi KH (2012) Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry 51 : 5921–5931. 22757685
30. Selisko B, Dutartre H, Guillemot JC, Debarnot C, Benarroch D, et al. (2006) Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351 : 145–158. 16631221
31. Lu G, Gong P (2013) Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog 9: e1003549. doi: 10.1371/journal.ppat.1003549 23950717
32. Lim SP, Koh JH, Seh CC, Liew CW, Davidson AD, et al. (2013) A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J Biol Chem 288 : 31105–31114. doi: 10.1074/jbc.M113.508606 24025331
33. Potisopon S, Priet S, Collet A, Decroly E, Canard B, et al. (2014) The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res.
34. Aslanidis C, de Jong PJ (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18 : 6069–6074. 2235490
35. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 : 760–763. 15299374
36. Matthews BW (1968) Solvent content of protein crystals. J Mol Biol 33 : 491–497. 5700707
37. Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr 60 : 432–438. 14993666
38. Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, et al. (2011) Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem 286 : 6233–6240. doi: 10.1074/jbc.M110.179184 21147775
39. Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 : 760–763. 15299374
40. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66 : 213–221. doi: 10.1107/S0907444909052925 20124702
41. Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132. 15572765
42. Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, et al. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66 : 12–21. doi: 10.1107/S0907444909042073 20057044
43. Schrödinger L (2010) The PyMOL Molecular Graphics System, Version 1.5.0.4
44. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372 : 774–797. 17681537
45. Hayward S, Lee RA (2002) Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model 21 : 181–183. 12463636
46. Pascal BD, Chalmers MJ, Busby SA, Griffin PR (2009) HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J Am Soc Mass Spectrom 20 : 601–610. doi: 10.1016/j.jasms.2008.11.019 19135386
47. Avis JM, Conn GL, Walker SC (2012) Cis-acting ribozymes for the production of RNA in vitro transcripts with defined 5′ and 3′ ends. Methods Mol Biol 941 : 83–98. doi: 10.1007/978-1-62703-113-4_7 23065555
48. Lim SP, Wen D, Yap TL, Yan CK, Lescar J, et al. (2008) A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase-kinetic and inhibition analyses. Antiviral Res 80 : 360–369. doi: 10.1016/j.antiviral.2008.08.005 18809436
49. Niyomrattanakit P, Chen YL, Dong H, Yin Z, Qing M, et al. (2010) Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J Virol 84 : 5678–5686. doi: 10.1128/JVI.02451-09 20237086
50. Hung M, Gibbs CS, Tsiang M (2002) Biochemical characterization of rhinovirus RNA-dependent RNA polymerase. Antiviral Res 56 : 99–114. 12367717
51. Johnson BW, Russell BJ, Lanciotti RS (2005) Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol 43 : 4977–4983. 16207951
52. Moreland NJ, Tay MY, Lim E, Rathore AP, Lim AP, et al. (2012) Monoclonal antibodies against dengue NS2B and NS3 proteins for the study of protein interactions in the flaviviral replication complex. J Virol Methods 179 : 97–103. doi: 10.1016/j.jviromet.2011.10.006 22040846
53. Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, et al. (2004) A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J Biol Chem 279 : 35638–35643. 15152003
54. Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, et al. (2007) Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol 372 : 723–736. 17686489
55. Iglesias NG, Filomatori CV, Gamarnik AV (2011) The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J Virol 85 : 5745–5756. doi: 10.1128/JVI.02343-10 21471248
56. Konermann L, Pan J, Liu YH (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev 40 : 1224–1234. doi: 10.1039/c0cs00113a 21173980
57. Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, et al. (2010) Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res 87 : 125–148. doi: 10.1016/j.antiviral.2009.11.009 19945487
58. Zhang X, Chien EY, Chalmers MJ, Pascal BD, Gatchalian J, et al. (2010) Dynamics of the beta2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal Chem 82 : 1100–1108. doi: 10.1021/ac902484p 20058880
59. Zheng J, Yong HY, Panutdaporn N, Liu C, Tang K, et al. (2014) High resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res Accepted.
60. Tan CS, Hobson-Peters JM, Stoermer MJ, Fairlie DP, Khromykh AA, et al. (2013) An interaction between the methyltransferase and RNA dependent RNA polymerase domains of the West Nile virus NS5 protein. J Gen Virol 94 : 1961–1971. doi: 10.1099/vir.0.054395-0 23740481
61. Vieira-Pires RS, Morais-Cabral JH (2010) 3(10) helices in channels and other membrane proteins. J Gen Physiol 136 : 585–592. doi: 10.1085/jgp.201010508 21115694
62. Jones S, Thornton JM (1996) Principles of protein-protein interactions. Proc Natl Acad Sci U S A 93 : 13–20. 8552589
63. Armon A, Graur D, Ben-Tal N (2001) ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol 307 : 447–463. 11243830
64. Li XD, Shan C, Deng CL, Ye HQ, Shi PY, et al. (2014) The interface between methyltransferase and polymerase of NS5 is essential for flavivirus replication. PLoS Negl Trop Dis 8: e2891. doi: 10.1371/journal.pntd.0002891 24852307
65. Wu J, Lu G, Zhang B, Gong P (2014) Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J Virol.
66. Li K, Phoo WW, Luo D (2014) Functional interplay among the flavivirus NS3 protease, helicase, and cofactors. Virologica Sinica: 1–12. doi: 10.1007/s12250-014-3437-7 24470264
67. Luo D, Lim SP, Lescar J (2012) The Flavivirus NS3 Protein: Structure and Functions. Molecular Virology and Control of Flaviviruses: 77.
68. Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, et al. (2010) Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. Journal of biological chemistry 285 : 18817–18827. doi: 10.1074/jbc.M109.090936 20375022
69. Assenberg R, Mastrangelo E, Walter TS, Verma A, Milani M, et al. (2009) Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol 83 : 12895–12906. doi: 10.1128/JVI.00942-09 19793813
70. Luo D, Xu T, Hunke C, Grüber G, Vasudevan SG, et al. (2008) Crystal structure of the NS3 protease-helicase from dengue virus. Journal of virology 82 : 173–183. 17942558
71. Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14 : 1188–1190. 15173120
72. Selisko B, Wang C, Harris E, Canard B (2014) Regulation of Flavivirus RNA synthesis and replication. Curr Opin Virol 9C: 74–83.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



