-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
Heterochromatin formation at specific genomic regions is critical for processes as diverse as gene expression and chromosome segregation. The formation of silent heterochromatin at repetitive DNA elements requires processing of transcripts by the RNA interference machinery. Curiously, factors involved in proper RNA splicing are required for heterochromatin assembly, and it was proposed that splicing factors provide a platform for the recruitment of RNAi complexes independently of their role in regulating splicing. In this study, we found several novel splicing factors involved in heterochromatin assembly. Our analysis of genome-wide splicing patterns by RNA sequencing showed that the mRNAs of RNAi factors are very sensitive to perturbations of RNA splicing machinery. Moreover, we found that splicing factors are critical for the production of a telomere shelterin component and proper telomeric heterochromatin assembly. Most importantly, we showed that introducing the cDNA versions of RNAi and shelterin components alleviates heterochromatin defects associated with splicing factor mutations. Thus splicing factors are required for heterochromatin assembly mainly by regulating the proper splicing of heterochromatin assembly factors.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004334
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004334Summary
Heterochromatin formation at specific genomic regions is critical for processes as diverse as gene expression and chromosome segregation. The formation of silent heterochromatin at repetitive DNA elements requires processing of transcripts by the RNA interference machinery. Curiously, factors involved in proper RNA splicing are required for heterochromatin assembly, and it was proposed that splicing factors provide a platform for the recruitment of RNAi complexes independently of their role in regulating splicing. In this study, we found several novel splicing factors involved in heterochromatin assembly. Our analysis of genome-wide splicing patterns by RNA sequencing showed that the mRNAs of RNAi factors are very sensitive to perturbations of RNA splicing machinery. Moreover, we found that splicing factors are critical for the production of a telomere shelterin component and proper telomeric heterochromatin assembly. Most importantly, we showed that introducing the cDNA versions of RNAi and shelterin components alleviates heterochromatin defects associated with splicing factor mutations. Thus splicing factors are required for heterochromatin assembly mainly by regulating the proper splicing of heterochromatin assembly factors.
Introduction
Eukaryotic genomic DNA associates with histone and non-histone proteins to form chromatin, which is necessary for the spatial and temporal organization of chromosomes. A distinction is commonly drawn between two types of chromatin: euchromatin and heterochromatin. Euchromatin is less condensed and often associated with genes that are actively transcribed. Heterochromatin is highly condensed and often forms over repetitive DNA elements such as transposons. The formation of heterochromatin prevents expression of transposons, improper recombination of repetitive genomic loci, and missegregation of chromosomes during mitosis and meiosis, thus maintaining genome stability [1], [2].
Histones within heterochromatin regions are usually hypo-acetylated and methylated at histone H3 lysine 9, which serves as a binding site for heterochromatin protein 1 (HP1) [3]–[5]. HP1 subsequently recruits diverse proteins to regulate cellular processes such as transcriptional silencing, recombination suppression, and chromosome segregation [1]. The mechanism that attracts histone methyltransferases and deacetylases to repetitive DNA elements is under intensive study. In certain cases, sequence-specific DNA binding proteins are directly involved in recruitment of these enzymes [6]–[9]. Alternatively, the repetitive nature may itself be sufficient to trigger heterochromatin assembly [10], [11]. Recent work has shown that DNA repeats are transiently transcribed, and the transcripts are processed by the RNA interference (RNAi) machinery into small interfering RNAs (siRNAs), which help target histone-modifying enzymes to repeat regions. However, the mechanistic details of RNAi-mediated heterochromatin assembly is not yet well understood [12]–[14].
The mechanisms by which heterochromatin is assembled and regulated have been extensively studied in the fission yeast S. pombe because it shares basic pathways of heterochromatin assembly with mammals, yet has the key advantages of facile genetics and single representative genes for most key families of mammalian chromatin-modifying factors. In this organism, heterochromatin is present mainly at pericentric regions, subtelomeres, and the silent mating-type region, all of which contain similar repeat sequences composed of dg and dh repeats [1]. These repeats are transcribed during S-phase of the cell cycle by RNA polymerase II. The transcripts are sliced by Ago1 and then reverse-transcribed by the RNA-directed RNA polymerase complex (RDRC: Rdp1, Cid12, and Hrr1) into double stranded RNAs, which are processed by Dcr1 into small interfering RNAs (siRNAs). These siRNAs are loaded onto Argonaute siRNA chaperone complex (ARC: Arb1, Arb2, and Ago1) and then onto the RNA-induced transcriptional silencing complex (RITS: Ago1, Chp1, and Tas3). RITS is targeted to the repeat regions through base pairing between siRNAs and the nascent transcripts and recruits the H3K9 methyltransferase complex CLRC, which contains SET domain protein Clr4 as its catalytic subunit. H3K9 methylation recruits HP1 family proteins Swi6 and Chp2, which in turn recruit histone deacetylases (HDACs) such as SHREC to further compact chromatin (see review [12]–[14]).
Surprisingly, in addition to these complexes, splicing factors are required for heterochromatin assembly at pericentric regions [15]–[17]. In fission yeast, 43% of genes contain introns [18], indicating the prevalence of splicing in this organism. The spliceosome and the splicing reactions of fission yeast are also highly conserved with those of higher eukaryotes, which utilize snRNAs U1, U2, U4, U5, and U6, as well as over one hundred protein components (see review [19]). The process of splicing starts when U1 and U2 snRNPs bind to the 5′ splice site and branch point on a pre-mRNA, respectively. The U4/U6.U5 snRNP and the Prp19 complex (also known as NineTeen Complex, or NTC) are then recruited to form the precatalytic spliceosome. After the release of U1 and U4, the spliceosome is activated and the 5′ splice site is cleaved and fused to the branch point to form a lariat structure. Next, 3′ intron cleavage is coupled to exon ligation in a post-spliceosomal complex of U2, U5, and U6. The mature mRNA is then released and the snRNPs recycled. Most notably, temperature-sensitive mutants prp10-1 (component of U2) and cwf10-1 (component of U5) exhibit silencing defects at pericentric regions [15]. Like dcr1Δ, these mutants lose most siRNAs derived from pericentric repeats [15]. Additionally, the spliceosome associates with RDRC component Cid12 [15], [20], indicating a possible direct role of splicing factors in connecting the nascent transcripts and the RNAi machinery during heterochromatin formation. It was hypothesized that splicing factors act in the RNAi pathway independently of RNA splicing because silencing defects are obvious when the splicing of a control tbp1 mRNA is intact and introducing cDNAs of ago1+ or hrr1+ was unable to rescue pericentric heterochromatin silencing defects of prp10-1 cells [15], [17]. However, the possibility that splicing factors regulate the proper processing of RNAi factors has not been rigorously tested. Notably, a number of recently identified factors involved in RNAi (arb1+, arb2+, ers1+, and dsh1+) contain introns and might require splicing factors for their proper expression [21]–[24].
In this study, we performed a screen of the S. pombe nonessential gene deletion strain library and discovered four new putative splicing factors involved in pericentric heterochromatin assembly. We demonstrated that the phenotype of the strongest of these, cwf14Δ, is similar to those of RNAi mutants in regulating pericentric heterochromatin assembly. RNA-seq analyses further found that cwf14Δ resulted in mis-splicing of a subgroup of genes, including a number of RNAi factors. Moreover, we showed that introducing the cDNAs of three RNAi factors, ago1+, arb2+, and ers1+, significantly alleviated silencing defects associated with cwf14Δ. Furthermore, we found that the mRNA of telomere shelterin protein Tpz1, which is involved in telomeric silencing, was also mis-spliced in splicing mutants and that introducing tpz1+ cDNA partially rescues telomeric silencing defects of splicing factor mutants. Thus splicing factors are involved in heterochromatin assembly mainly through regulating the proper splicing of heterochromatin assembly factors.
Results
A high-throughput screen for mutants affecting pericentric heterochromatin
To comprehensively identify factors required for pericentric heterochromatin assembly, we performed a screen of the fission yeast haploid deletion library for mutants that affect silencing of a reporter inserted into pericentric heterochromatin, otr::ade6+ (Figure 1A–C) [25]. In wild-type cells, otr::ade6+ is silenced, causing red colony color on low adenine medium. However, when heterochromatic silencing is lost at the pericentric region, otr::ade6+ is expressed, and colonies are white. Strains with intermediate silencing defects show variable degrees of pink/red color, allowing rough phenotypic quantification (Figure 1C). In order to eliminate strains that have inherent metabolic defects causing lighter-than-red color, we performed a control screen with an ade6-M210 query strain lacking the otr::ade6+ reporter (Figure 1B), which allowed us to filter false positives out of the screen. Our finalized list of hits is shown in Figure 1C. Each hit colony was assigned a score between 1 and 4, with 4 indicating the strongest silencing defects.
Fig. 1. A genetic screen for nonessential genes required for pericentric heterochromatin silencing in fission yeast. 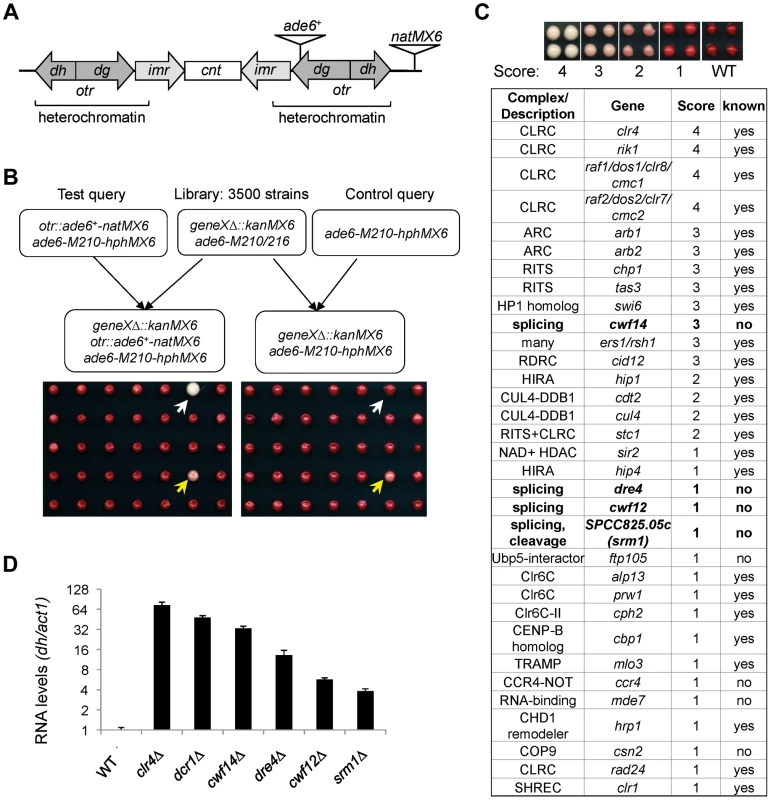
(A) Schematic diagram of the otr::ade6+-natMX6 reporter. (B) Workflow to introduce otr::ade6+ into the deletion library. A control screen was performed in parallel. The selection of only ade6-M210-hphMX6 progeny generates a uniform background for color development. White arrows denote a mutant that affects silencing, and yellows arrows denote a false positive that affects colony color independently of silencing. (C) List of mutants identified affecting pericentric silencing. Hit colonies were assigned color scores between 1 and 4, as indicated. (D) qRT-PCR analyses of transcripts derived from pericentric dh repeats, normalized to act1. Wild-type was set to 1. Error bars represent standard deviation of three experiments. Among the mutants identified, 25 were previously known to be required for pericentric heterochromatin assembly, validating the effectiveness of our screen. These were mutants in the complexes of CLRC, ARC, RITS, RDRC, CUL4-DDB1, HIRA, Clr6C, TRAMP, and SHREC, and in individual factors such as HP1 homolog Swi6, NAD+ histone deacetylase Sir2, CENP-B homolog Cbp1, and CHD1 remodeler Hrp1 (Figure 1C). There were also a number of previously reported heterochromatin mutants that are listed in the library but were missed in our screen, such as ago1Δ, rdp1Δ, and dcr1Δ. We confirmed by PCR that the null mutation was missing in these strains, indicating that false negatives are more likely the result of incorrect deletions present in the library than due to methodological bias.
Most interestingly, there were eight novel mutants identified in this screen, of which four are uncharacterized genes implicated in various steps of mRNA splicing: cwf14 (SPBC24C6.11), dre4 (SPAC13C5.02), cwf12 (SPBC32F12.05c), and the human SRRM1 homolog we named srm1 (SPCC825.05c). Each of these strains showed elevated levels of pericentric dh transcripts, a common phenotype of heterochromatin mutants, suggesting that these mutants indeed affected pericentric heterochromatin assembly (Figure 1D). As cwf14Δ showed the strongest phenotype among these four splicing mutants, we chose it as the focus of our subsequent experiments.
Cwf14 is required for RNAi-mediated pericentric heterochromatin assembly
We first confirmed by serial dilution analysis that cwf14Δ cells containing otr::ade6+ formed white colonies similar to those of dcr1Δ (Figure 2A). We also examined the effect of cwf14Δ on silencing of another reporter inserted at the same location, otr::ura4+ [26]. The silencing of this reporter gene in wild-type cells allows them to grow on counterselective medium containing 5-fluoroorotic acid (FOA). Serial dilution analyses showed that cwf14Δ has comparable silencing defects to dcr1Δ, as indicated by attenuated growth on FOA medium (Figure 2A). Moreover, chromatin immunoprecipitation (ChIP) analyses showed increased enrichment of RNA Polymerase II at pericentric regions in cwf14Δ cells, indicating that the effect on pericentric transcript levels was at least in part due to increased transcription (Figure 2B).
Fig. 2. Cwf14 is required for pericentric heterochromatin assembly through the RNAi pathway. 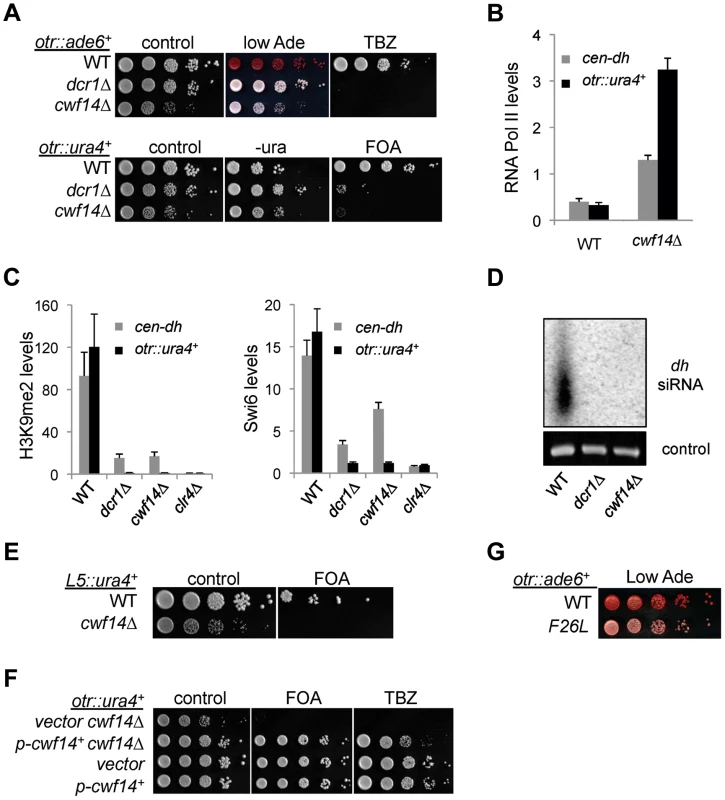
(A, E, F, and G) Serial dilution analyses to measure the expression of reporter genes and sensitivity to TBZ. (B and C) ChIP analyses of Pol II (Rpb1), H3K9me2, and Swi6 levels at pericentric dh repeats and otr::ura4+. Pol II ChIP was normalized to act1 gene, and H3K9me2 and Swi6 ChIP was normalized to act1 promoter. Error bars represent standard deviation of three experiments. (D) Northern blot analyses of siRNAs derived from pericentric dh repeats. Further ChIP analyses showed strong reduction in levels of heterochromatin hallmarks such as H3K9me2 and Swi6 at otr::ura4+ and to a lesser extent at the endogenous dh repeats in cwf14Δ cells (Figure 2C). This pattern is similar to RNAi mutants such as dcr1Δ, but less pronounced than that of clr4Δ, suggesting that Cwf14 might regulate RNAi-mediated heterochromatin assembly. Consistent with this idea, siRNAs derived from pericentric repeats were eliminated in cwf14Δ cells, similar to dcr1Δ cells (Figure 2D).
It was shown that a 1.6 kb fragment of pericentric dg repeats (termed L5) induces heterochromatin assembly and silencing of adjacent genes when inserted into an ectopic site in an RNAi-dependent manner [27], [28]. Silencing is lost in cwf14Δ cells as well, consistent with the idea that Cwf14 is involved in RNAi-mediated heterochromatin assembly (Figure 2E).
Since pericentric heterochromatin promotes proper loading of cohesin near centromeres to promote chromosome bi-orientation [29]–[31], most heterochromatin mutants show defects in chromosome segregation [26] and sensitivity to the microtubule-destabilizing drug thiabendazole (TBZ) [32]. As expected, cwf14Δ was also sensitive to TBZ (Figure 2A). Moreover, both the silencing and TBZ-sensitivity phenotypes were rescued by complementation of cwf14Δ with a plasmid containing an intact copy of cwf14+ under the control of its endogenous regulatory elements (Figure 2F). Since cwf14Δ cells had some growth defects, we also performed PCR-mediated random mutagenesis and isolated a mutant (cwf14-F26L) that resulted in silencing defects without significantly affecting growth (Figure 2G).
To test whether cwf14Δ affects H3K9 methyltransferase activity of CLRC, we used a system in which Clr4 is tethered via a fused Gal4 DNA binding domain (GBD) to an ectopically integrated ade6+ reporter adjacent to three copies of the Gal4 binding site (3xgbs-ade6+) [33] (Figure 3A). This tethering induces heterochromatin formation over a 6 kb locus, silencing transcription of the ade6+ reporter. RNAi factors are not required for this silencing, consistent with the idea that RNAi is required for the targeting of CLRC to DNA repeats [33]. Dilution analysis showed that cwf14Δ had no effect on Clr4-tethered silencing of 3xgbs-ade6+, and ChIP analyses showed that H3K9me2 was enriched at nearby loci 2 and 3 kilobases away in cwf14Δ cells, similar to wild-type cells (Figure 3B). Collectively, these data suggest that Cwf14 is involved in heterochromatin formation at pericentric repeats through the RNAi pathway.
Fig. 3. Cwf14 is not required for CLRC activity. 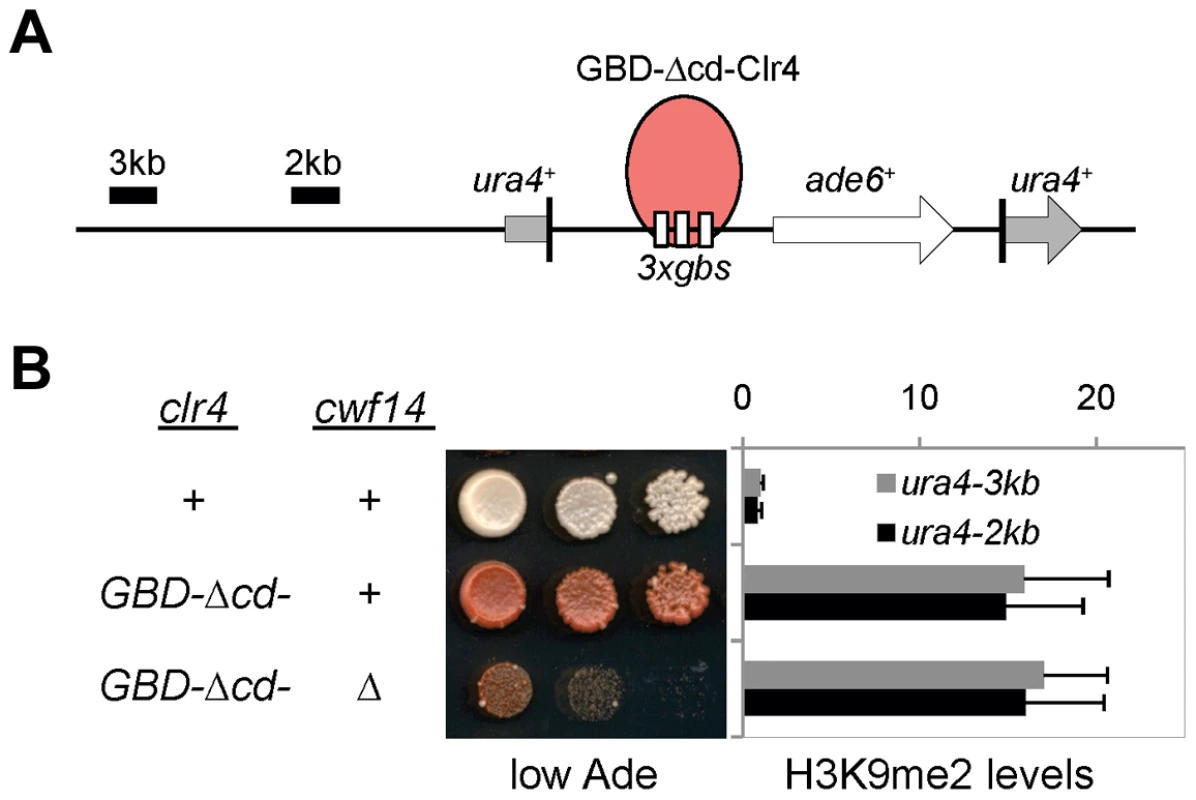
(A) Schematic diagram of the ura4::3xgbs-ade6+ reporter. (B) Left, serial dilution analysis to measure reporter gene expression. Right, ChIP analysis of H3K9me2 levels at the reporter, normalized to act1 promoter. Error bars represent standard deviation of three experiments. Cwf14 associates with the spliceosome
Cwf14 is highly conserved across species. Its budding yeast homolog, Bud31p, co-purifies with the spliceosome [34], and BUD31Δ causes mis-splicing of the mRNAs of ARP2 and SRC1, two factors required for proper budding [35]. Cwf14 was initially identified in a purification of splicing factor Cdc5, together with a number of spliceosome components [36]. However, whether Cwf14 is a stable component of the spliceosome and involved in splicing has not been tested directly. In order to further specify the mechanism of Cwf14 action, we constructed C-terminally tagged versions of cwf14 at its endogenous locus. Cwf14-GFP and Cwf14-myc were fully functional, as they did not show silencing defects of otr::ade6+ (Figure 4A). Imaging of Cwf14-GFP showed that Cwf14 localizes predominantly in the nucleus (Figure 4B). Western blot analysis showed that Cwf14-myc is a 40 kD protein. Moreover, the F26L mutation resulted in reduced Cwf14 protein levels, indicating that it is a partial loss-of-function allele (Figure 4C). We also performed immunoprecipitation of Cwf14-myc followed by MudPIT mass spectrometry to identify its interacting proteins. Most factors that co-immunoprecipitated with Cwf14, but not in a control purification, were components of the spliceosome, especially from subcomplexes NTC and U5 (Figure 4D and Table S1). This result corroborates data that Cwf14 co-precipitates with Prp17, Prp19, and Cwf2, all members of U5-associated NTC [19], [37], as well as a component of U5 snRNP, Cwf10 [38].
Fig. 4. Cwf14 associates with the spliceosome. 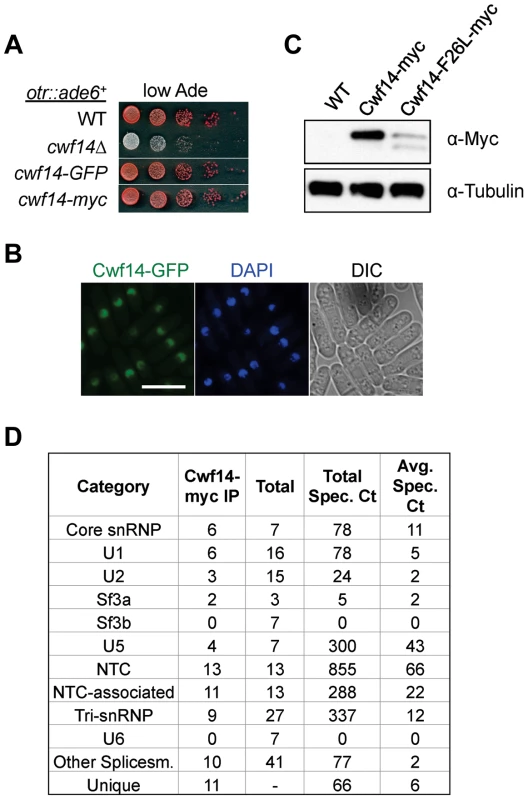
(A) Tagged versions of Cwf14 are functional as indicated by serial dilution analysis. (B) Imaging of cells expressing Cwf14-GFP, with DAPI to stain nucleus. Scale bar, 10 µm. (C) Western blot analysis of Cwf14-myc expression. (D) Mass spectrometry analyses of purified Cwf14-myc complex. Factors were binned into annotated subcomplexes. Cwf14 is involved in splicing of specific mRNAs
Previous work suggests that the spliceosome is involved in heterochromatin assembly through tethering RDRC to pericentric transcripts [15]. This is because RDRC component Cid12 associates with the spliceosome [15], [20], and no splicing defects were observed in the well-characterized splicing substrate tbp1 in splicing factor mutants when silencing defects were apparent [15], [17]. However, purifications of spliceosome components, including our purification of Cwf14, have not identified any other heterochromatin components [36], [37], [39] (Table S1). These results indicate either that the physical connection between the spliceosome and RDRC is very weak, or that Cid12 is present in two separate complexes, RDRC and spliceosome. It remains a possibility that splicing factors regulate the correct processing of mRNAs involved in RNAi-mediated heterochromatin assembly.
In order to test whether cwf14Δ has a general splicing defect, we performed RNA-seq analyses of total cellular RNAs from wild-type, dcr1Δ, and cwf14Δ cells. The gene expression profiles of cwf14Δ and dcr1Δ showed strong overlap of significantly up-regulated genes (Figure 5A and Tables S2 and S3), consistent with the idea that Cwf14 functions in the RNAi pathway. We found that cwf14Δ indeed resulted in intron retention of a portion of genes (Figure 5B and Table S4), consistent with the finding that it is associated with the spliceosome. The majority of introns were properly processed, which indicates that Cwf14 only moderately affected the activity of the spliceosome. Interestingly, many RNAi factors contain introns. Although unbiased ranking of exon-exon junction ratios between wild-type and cwf14Δ RNAs did not show preferential enrichment of RNAi or heterochromatin assembly factors (Table S4), significant intron retention was observed within mRNAs from ago1, arb2, ers1, and dsh1 in cwf14Δ cells as compared to those in wild-type cells, despite similar sequencing depths genome-wide and similar levels of each gene transcript in both samples (Figure 5C). RT-PCR analyses confirmed that these mRNAs were indeed spliced inefficiently in cwf14Δ as well as in cwf10-1 and prp10-1 cells (Figure 5D). Moreover, Western blot analysis showed strong reduction of protein levels of Flag-Ago1, the RNAi factor most severely affected by cwf14Δ, in both cwf14Δ and cwf10-1 strains (Figure S1), indicating that the mis-splicing of ago1 mRNA, and possibly other RNAi factors as well, resulted in altered protein levels.
Fig. 5. Cwf14 is required for the proper splicing of RNAi factors. 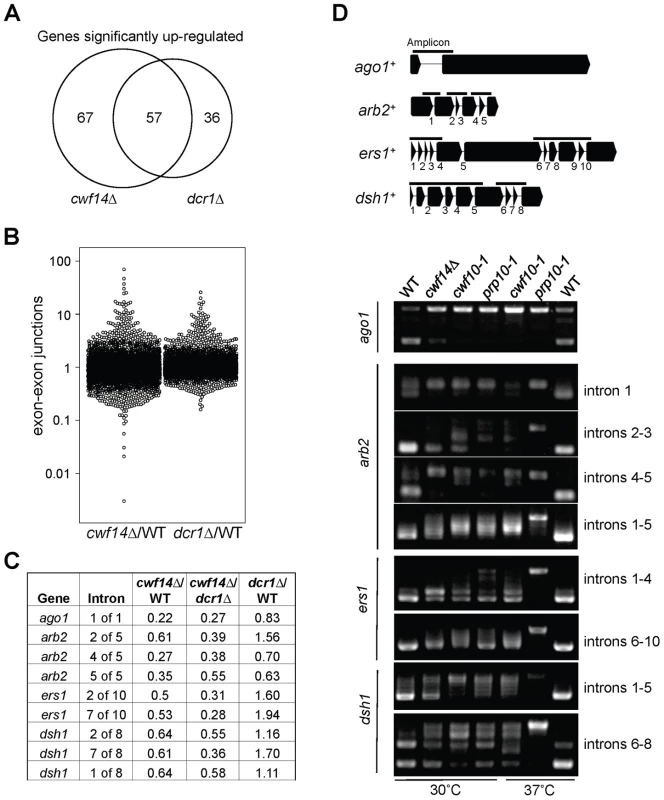
(A) Area-proportional overlap of genes up-regulated in cwf14Δ and dcr1Δ (each compared to wild type) at p<0.05. (B) Ratios of exon-exon junction counts from RNA-seq for each junction in the indicated samples. A ratio of 1 indicates no difference. The bottom tail of cwf14Δ/WT, which is less pronounced in dcr1Δ/WT, indicates mis-splicing. (C) RNA-seq analysis shows mis-splicing of a number of RNAi factor introns in cwf14Δ. Ratios are exon-exon junction counts from RNA-seq for each junction in the indicated samples. (D) Top, diagram of RNAi genes with intron positions indicated. Bars represent PCR fragments used to analyze intron retention. Bottom, RT-PCR analyses of RNA with primers flanking introns. Previously, it was shown that introducing cDNA versions of ago1+ or hrr1+ failed to rescue silencing defects of prp10-1 [15]. We also generated a cDNA version of ago1+ at its endogenous chromosomal location and under the control of its endogenous regulatory elements (ago1+::cDNA). This cDNA construct showed no defects in silencing of otr::ura4+, indicating that this replacement created a functional Ago1 protein (Figure S2). However, ago1+::cDNA was unable to rescue otr::ura4+ silencing defects and TBZ sensitivity of cwf14Δ (Figure 6A), even though it restored Flag-Ago1 protein levels (Figure S1). We reasoned that the inability of ago1+::cDNA to rescue cwf14Δ defects is because other RNAi factors are also mis-spliced. We thus generated cDNA versions of arb2+ and ers1+ at their endogenous chromosomal loci, which were both functional (Figure S2). Neither arb2+::cDNA nor ers1+::cDNA alone showed any effect on silencing of otr::ura4+ in cwf14Δ cells (Figure 6A). However, when combinations of two cDNAs were introduced together into cwf14Δ cells, there was a detectable rescue of silencing defects and TBZ sensitivity, and the effect was stronger when all three cDNAs were introduced (Figure 6A). Further ChIP analysis showed that both H3K9me and Swi6 protein levels were significantly increased at both otr::ura4+ and pericentric dh repeats in cwf14Δ cells supplemented with ago1+, arb2+, and ers1+ cDNAs (3cDNAs) (Figure 6B).
Fig. 6. Introducing cDNAs of RNAi factors rescues pericentric heterochromatin defects of cwf14Δ cells. 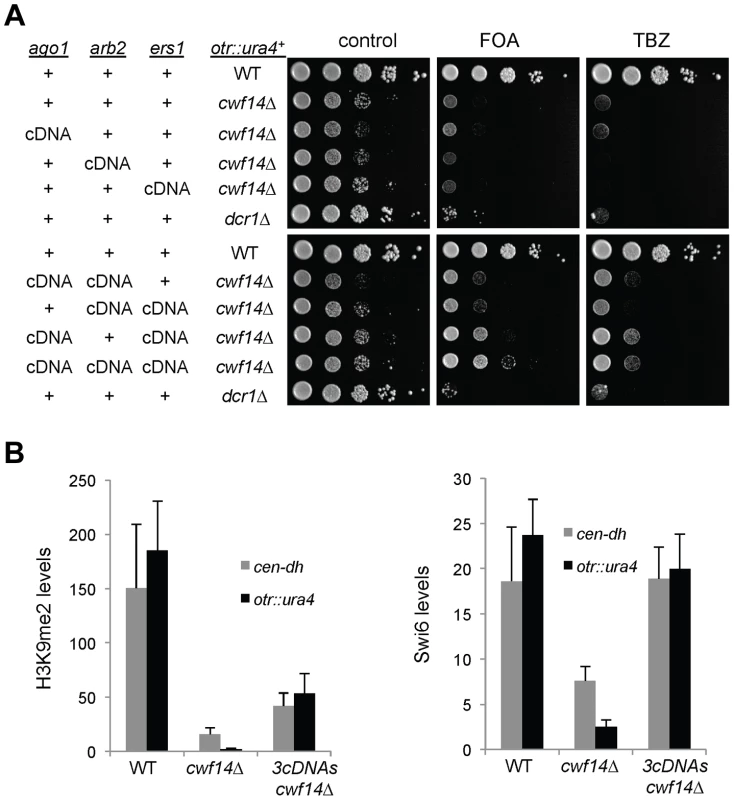
(A) Serial dilution analyses to measure the expression of otr::ura4+ and sensitivity to TBZ. (B) ChIP analyses of H3K9me2 and Swi6 levels at pericentric dh repeats and otr::ura4+, normalized to an act1 promoter fragment. Error bars represent standard deviation of three experiments. We also found that introducing ago1+::cDNA was sufficient to rescue the silencing defects of otr::ade6+ associated with cwf10-1 (Figure S3A). Moreover, there is a significant increase in both H3K9me and Swi6 levels at both otr::ade6+ and dh repeats in cwf10-1 ago1+::cDNA cells (Figure S3B). Altogether, these results suggest that mutations in different splicing factors affect the splicing of diverse RNAi factors to regulate heterochromatin assembly at pericentric regions.
Thus our results clearly demonstrated that splicing factors mainly exert their effects on pericentric heterochromatin assembly by promoting the proper splicing of RNAi factors. However, we caution that the rescue of heterochromatin silencing defects of cwf14Δ cells was incomplete even with ago1+, arb2+, and ers1+ cDNAs. This probably reflects the requirement of Cwf14 for the proper splicing of other RNAi factors such as dsh1+, rdp1+, arb1+, hrr1+, or some unidentified factors involved in heterochromatin assembly. Such an idea is supported by the fact that the rescue of cwf14Δ silencing defects correlated with the number of cDNA constructs that were introduced. It is also possible that splicing factors have a direct contribution in pericentric heterochromatin assembly. However, the strong rescue of pericentric silencing in cwf14Δ cells with cDNA constructs suggests that the regulation of RNAi factor splicing is a major role of splicing factors in this process.
Cwf14 regulates the proper splicing of tpz1 to control telomere silencing
We also found that telomere shelterin component tpz1+ showed a strong reduction in exon-exon junction sequencing reads in cwf14Δ cells relative to wild-type cells, and this phenotype was confirmed by RT-PCR analyses in cwf10-1 and prp10-1 as well (Figure 7A). A C-terminally Flag-tagged version of Tpz1 [40] affected silencing of a reporter gene inserted near telomere repeats (Figure S4), suggesting that Tpz1 is required for telomere silencing. We then analyzed the effect of splicing factor mutations on silencing of a reporter inserted near telomere repeats of chromosome two (Tel2::ura4+) [41]. Indeed, cwf14Δ resulted in silencing defects at this reporter gene (Figure 7B), accompanied by a reduction of H3K9me levels at tlh1, which is embedded at telomeric heterochromatin, as well as the accumulation of tlh1 transcripts (Figure 7C and 7D). Both cwf10-1 and prp10-1 cells showed increased tlh1 transcript levels, indicating that loss of telomeric silencing is a general feature of splicing mutants (Figure 7C and 7D). Because multiple DNA sequences contribute to heterochromatin assembly at telomeres, including tlh1+, telomere associated sequences (TAS), and terminal TEL repeats [8], we further tested silencing at TEL::ade6+, which is inserted on the mini-chromosome Ch16 adjacent to telomere repeats [42]. We found that cwf14Δ, cwf14-F26L, and cwf10-1 resulted in loss of silencing of this reporter (Figure 7E). Interestingly, replacement of tpz1+ with its cDNA partially rescued telomeric silencing phenotypes of the cwf14-F26L and cwf10-1 mutants (Figure 7E and 7F). We could only marginally rescue the telomere silencing defects of cwf14Δ cells with tpz1+::cDNA (Figure 7F and data not shown). Thus it is possible that additional factors involved in telomere silencing might also contain introns and depend on the spliceosome to properly regulate their splicing. Alternatively, splicing factors might affect telomere silencing through additional mechanisms. Nonetheless, these results demonstrate that inefficient splicing of tpz1 contributes to telomeric silencing defects in splicing mutants.
Fig. 7. Splicing factors regulate the proper splicing of shelterin component Tpz1. 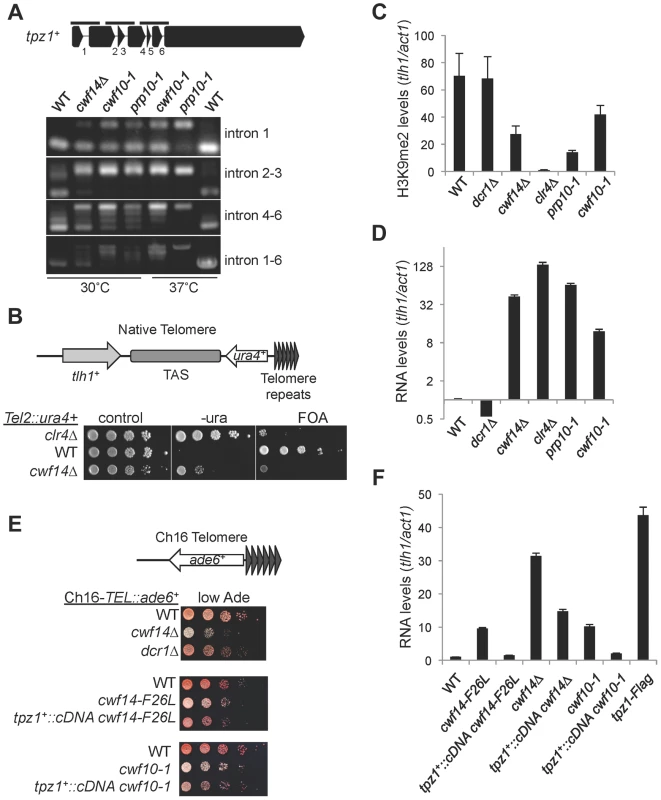
(A) RT-PCR analyses of RNA with primers flanking introns. (B, E) Serial dilution analyses to measure reporter gene expression. (C) ChIP analyses of H3K9me2 levels, normalized to act1 promoter. Error bars represent standard deviation of three experiments. (D, F) qRT-PCR analyses of transcripts derived from tlh1, normalized to act1 gene. Wild type was set to 1. Error bars represent standard deviation of three experiments. Discussion
The formation of heterochromatin requires RNAi-mediated processing of repeat-derived transcripts and the targeting of histone modifying activities to repeat regions, leading to H3K9me and the recruitment of HP1 proteins. It has been shown that splicing factors are required for RNAi-mediated heterochromatin assembly in fission yeast, although the mechanism by which they are involved is not well characterized. The previously prevailing model was that the spliceosome physically associates with RNAi factors to regulate heterochromatin assembly, rather than acting through its splicing activity [15], [16]. One of the main evidences for this idea is that splicing mutants show severe silencing defects even though the splicing of tbp1 mRNA was not affected [15], [17]. However, whether these splicing factor mutants selectively affect the splicing of RNAi factors has not been rigorously tested.
Our RNA-seq analyses showed prominent intron retention of a subgroup of mRNAs in cwf14Δ cells, even though the majority of introns (including those of tbp1) are still properly processed (Figure 5B and Table S4). Interestingly, a number of key RNAi factors were among the list of strongly mis-spliced introns, a result that is further corroborated by RT-PCR analyses of a selective set of RNAi factor mRNAs in other spliceosome mutants such as cwf10-1 and prp10-1 (Figure 5D). Most importantly, we found that introducing a combination of cDNAs of RNAi factors significantly alleviated pericentric heterochromatin defects associated with cwf14Δ and cwf10-1 (Figure 6 and S3). Thus splicing factors regulate the proper splicing of RNAi factors, which is a major, if not sole, contributor to heterochromatin assembly defects in splicing mutants. That introducing tpz1+::cDNA was able to partially rescue telomere silencing defects associated with splicing factors further supports the idea that mis-splicing of heterochromatin factors is the reason splicing factor mutants show heterochromatin assembly defects (Figure 7E and 7F).
It is noteworthy that cwf14Δ has only moderate splicing defects, with some introns show very strong sensitivity, whereas most others show little to no defects (Figure 5B and Table S4). This raises the question of whether specific intron sensitivity is a result of introns that are inherently difficult to splice. Consistent with this idea, our RT-PCR analyses showed prominent unspliced precursor mRNAs of RNAi factors even in wild-type cells (Figure 5D). It is also a striking pattern that heterochromatin factors in the same complexes tend to either have or not have introns. For example, many members of RNAi, such as ago1+, arb1+, arb2+, ers1+, dsh1+, rdp1+, and hrr1+, have introns, but none of CLRC (clr4+, rik1+, raf1+, raf2+, cul4+, stc1+), SHREC (clr1+, clr2+, clr3+, mit1+), or swi6+ have any introns, raising the possibility of selective regulation of the RNAi pathway through general or specific changes in splicing efficiency. In fact, an analysis of splicing changes during meiosis showed that one intron of arb1+ is induced to be spliced, while a different intron exhibits splicing repression [43]. Since splicing factors have been identified in screens that affect RNAi-based processes in worms, flies, and plants [44]–[48], it seems a conserved mechanism that proper splicing of the mRNAs of RNAi factors regulates the efficiency of RNAi inside the cell.
Materials and Methods
Genetic screens of the fission yeast deletion library
The Bioneer library is composed of strains of mixed endogenous ade6 alleles: ade6-M216, which forms pink colonies on low adenine medium, and ade6-M210, which give rise to red colonies, similar to ade6Δ. To avoid the complication of ade6-M216 alleles in our screen, we included in the query strain an ade6-M210-mCherry allele linked to a hphMX6 cassette, which confers resistance to the antibiotic hygromycin B, allowing us to generate a uniform ade6-M210 background. Screens were carried out according to previous protocols [49], with slight modifications. Yeast strains arrayed in 384 strains/plate format were pinned on YES agar medium containing 100 µg/ml G418, and otr::ade6+-natMX6 ade6-210-hphMX6 and ade6-210-hphMX6 strains were pinned on YES+100 µg/ml hygromycin B. After two days growth, strains were mated on SPA agar medium. Plates were incubated at 25°C for 3 days, then 42°C for 3 more days to kill vegetative cells. Strains were then germinated and correct genotype selected by pinning to YES+GNH (G418, Nat, and Hygromycin) or YES+GH (G418 and Hygromycin) medium, and then pinned to YE (low ade) medium for color readout.
Fission yeast strains and genetic analyses
Cwf14-GFP and Cwf14-13myc strains were constructed by a PCR-based module method. cwf14Δ, cwf12Δ, dre4Δ, and srm1Δ were derived from the Bioneer fission yeast deletion library, verified via PCR, and backcrossed. The p-cwf14+ plasmid was constructed by insertion of a PCR product containing cwf14+ promoter and coding region into SphI and XmaI sites of pREP41. The cwf14-F26L-myc mutant strain was constructed by integrating a cwf14+-myc-kanMX6 cassette amplified by error-prone PCR (high dNTP concentration) into the endogenous cwf14+ locus. ago1+::cDNA, arb2+::cDNA, and ers1+::cDNA strains were constructed by replacing the endogenous genes with intronless cDNA versions. All were sequenced to confirm full replacement and lack of mutation. cwf10-1 and prp10-1 strains were a kind gift from Robin Allshire. Genetic crosses were used to construct all other strains. The genotype of strains is provided in Table S5. For serial dilution plating assays, ten-fold dilutions of mid-log-phase culture were plated on the indicated medium and grown for 3 days at 30°C unless otherwise indicated.
RNA analyses
Total cellular RNA was isolated from log-phase cells using MasterPure yeast RNA purification kit (Epicentre) according to the manufacturer's protocol. Quantification with qRT-PCR was performed with Power SYBR Green RNA-to-CT one-step Kit (Applied Biosystems). RNA serial dilutions were used as templates to generate the standard curve of amplification for each pair of primers, and the relative concentration of target sequence was calculated accordingly. An act1 fragment served as reference to normalize the concentration of samples. Sequence of DNA oligos is provided in Table S6. For RNA-seq, purified RNA was prepared by TruSeq Stranded Total RNA Kit (Illumina), which includes rRNA depletion and chemical fragmentation. Index adapters were added to allow for multiplexing. Paired-end sequencing with 100 bp read lengths was performed on Illumina HiSeq. Mapping was performed with the Tuxedo Suite consisting of Bowtie, TopHat, and Cufflinks. For cwf14Δ, 57,521,513 reads were obtained, and 83% mapped to the genome via Bowtie. For WT, 47,636,518 reads were obtained, and 82% mapped. For dcr1Δ, 64,972,007 reads were obtained, and 83% mapped. RNA-seq data have been deposited to the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) with accession number SRP040479. For the dot plot, exon-exon junction ratios were filtered to remove several types: 1) junctions which mapped zero times in any sample (possible mapping noise), 2) junctions whose sum in WT, cwf14Δ, and dcr1Δ was less than 30 (to avoid randomness due to small sample sizes), and 3) ratios whose values were greater than 150 (to focus the diagram on splicing reduction). Northern blot of siRNAs was performed as described previously [50].
Chromatin immunoprecipitation (ChIP) analysis
ChIP analyses were performed as described previously [51]. Antibodies used were H3K9me2 (Abcam 1220), Swi6 [52], and RNA Pol II (Covance 8WG16). Quantification with qPCR was performed with Maxima SYBR Green/ROX qPCR Master Mix (Thermo). Enrichment was calculated as: relative levels in ChIP/relative levels in total DNA. An act1 promoter fragment was used as a control for normalization unless otherwise indicated. Sequence of DNA oligos was provided in Table S6.
Protein purification and mass spectrometry analysis
Exponentially growing yeast cells were harvested, washed with 2xHC buffer (300 mM HEPES-KOH pH 7.6, 2 mM EDTA, 100 mM KCl, 20% glycerol, 0.1% NP-40, 2 mM DTT, and protease inhibitor cocktail (Roche)) and frozen in liquid nitrogen. Crude cell extracts were prepared by vigorously blending frozen yeast cells with dry ice using a household blender, followed by sonication and incubation with 30 ml 1xHC buffer containing 250 mM KCl for 30 min. The lysate was cleared by centrifugation at 82,700×g for 3 hours. The supernatants were incubated with 50 µl of C-myc antibody (Sigma C3956) overnight, for three hours the next day with 50 µl protein G agarose beads, washed eight times with 1xHC containing 250 mM KCl, then two times with the same buffer without NP-40. For mass spectrometry analysis, bound proteins were eluted with 2×100 µl of 50 mM Tris pH 7.5, 5% SDS, 5% glycerol, 50 mM DTT. MudPIT mass spectrometry analysis was performed as described previously [53].
Supporting Information
Zdroje
1. GrewalSIS, JiaS (2007) Heterochromatin revisited. Nat Rev Genet 8 : 35–46 doi:10.1038/nrg2008
2. AlmouzniG, ProbstAV (2011) Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus 2 : 332–338 doi:10.4161/nucl.2.5.17707
3. ReaS, EisenhaberF, O'CarrollD, StrahlBD, SunZW, et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406 : 593–599 doi:10.1038/35020506
4. BannisterAJ, ZegermanP, PartridgeJF, MiskaEA, ThomasJO, et al. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 : 120–124 doi:10.1038/35065138
5. NakayamaJ, RiceJC, StrahlBD, AllisCD, GrewalSI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292 : 110–113 doi:10.1126/science.1060118
6. JiaS, NomaK-I, GrewalSIS (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304 : 1971–1976 doi:10.1126/science.1099035
7. KimHS, ChoiES, ShinJA, JangYK, ParkSD (2004) Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem 279 : 42850–42859 doi:10.1074/jbc.M407259200
8. KanohJ, SadaieM, UranoT, IshikawaF (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15 : 1808–1819 doi:10.1016/j.cub.2005.09.041
9. Bulut-KarsliogluA, PerreraV, ScaranaroM, la Rosa-Velazquez deIA, van de NobelenS, et al. (2012) A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol 19 : 1023–1030 doi:10.1038/nsmb.2382
10. DorerDR, HenikoffS (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77 : 993–1002.
11. GarrickD, FieringS, MartinDI, WhitelawE (1998) Repeat-induced gene silencing in mammals. Nat Genet 18 : 56–59 doi:10.1038/ng0198-56
12. MoazedD (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457 : 413–420 doi:10.1038/nature07756
13. LejeuneE, AllshireRC (2011) Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23 : 258–265 doi:10.1016/j.ceb.2011.03.005
14. CastelSE, MartienssenRA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. The EMBO Journal 14 : 100–112 doi:10.1038/nrg3355
15. BayneEH, PortosoM, KaganskyA, Kos-BraunIC, UranoT, et al. (2008) Splicing Factors Facilitate RNAi-Directed Silencing in Fission Yeast. Science 322 : 602–606 doi:10.1126/science.1164029
16. BernardP, DrogatJ, DheurS, GenierS, JaverzatJ-P (2010) Splicing factor Spf30 assists exosome-mediated gene silencing in fission yeast. Mol Cell Biol 30 : 1145–1157 doi:10.1128/MCB.01317-09
17. ChinenM, MoritaM, FukumuraK, TaniT (2010) Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J Biol Chem 285 : 5630–5638 doi:10.1074/jbc.M109.074393
18. WoodV, GwilliamR, RajandreamM-A, LyneM, LyneR, et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415 : 871–880 doi:10.1038/nature724
19. WahlMC, WillCL, LührmannR (2009) The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 136 : 701–718 doi:10.1016/j.cell.2009.02.009
20. MotamediMR, VerdelA, ColmenaresSU, GerberSA, GygiSP, et al. (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119 : 789–802 doi:10.1016/j.cell.2004.11.034
21. BukerSM, IidaT, BühlerM, VillénJ, GygiSP, et al. (2007) Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol 14 : 200–207 doi:10.1038/nsmb1211
22. RougemailleM, ShankarS, BraunS, RowleyM, MadhaniHD (2008) Ers1, a Rapidly Diverging Protein Essential for RNA Interference-dependent Heterochromatic Silencing in Schizosaccharomyces pombe. J Biol Chem 283 : 25770–25773 doi:10.1074/jbc.C800140200
23. RoguevA, BandyopadhyayS, ZofallM, ZhangK, FischerT, et al. (2008) Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322 : 405–410 doi:10.1126/science.1162609
24. KawakamiK, HayashiA, NakayamaJ-I, MurakamiY (2012) A novel RNAi protein, Dsh1, assembles RNAi machinery on chromatin to amplify heterochromatic siRNA. Genes Dev 26 : 1811–1824 doi:10.1101/gad.190272.112
25. EkwallK, OlssonT, TurnerBM, CranstonG, AllshireRC (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91 : 1021–1032.
26. AllshireRC, NimmoER, EkwallK, JaverzatJP, CranstonG (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9 : 218–233.
27. PartridgeJF, ScottKSC, BannisterAJ, KouzaridesT, AllshireRC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12 : 1652–1660.
28. SadaieM, IidaT, UranoT, NakayamaJ-I (2004) A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. The EMBO Journal 23 : 3825–3835 doi:10.1038/sj.emboj.7600401
29. BernardP, MaureJF, PartridgeJF, GenierS, JaverzatJP, et al. (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294 : 2539–2542 doi:10.1126/science.1064027
30. NonakaN, KitajimaT, YokobayashiS, XiaoG, YamamotoM, et al. (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4 : 89–93 doi:10.1038/ncb739
31. YamagishiY, SakunoT, ShimuraM, WatanabeY (2008) Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455 : 251–255 doi:10.1038/nature07217
32. EkwallK, NimmoER, JaverzatJP, BorgstrømB, EgelR, et al. (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109 (Pt 11) 2637–2648.
33. KaganskyA, FolcoHD, AlmeidaR, PidouxAL, BoukabaA, et al. (2009) Synthetic Heterochromatin Bypasses RNAi and Centromeric Repeats to Establish Functional Centromeres. Science 324 : 1716–1719 doi:10.1126/science.1172026
34. MasciadriB, ArecesLB, CarpinelliP, FoianiM, DraettaG, et al. (2004) Characterization of the BUD31 gene of Saccharomyces cerevisiae. Biochem Biophys Res Commun 320 : 1342–1350.
35. SahaD, BanerjeeS, BashirS, VijayraghavanU (2012) Context dependent splicing functions of Bud31/Ycr063w define its role in budding and cell cycle progression. Biochem Biophys Res Commun 424 : 579–585 doi:10.1016/j.bbrc.2012.06.156
36. OhiMD, LinkAJ, RenL, JenningsJL, McDonaldWH, et al. (2002) Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol 22 : 2011–2024 doi:10.1128/MCB.22.7.2011-2024.2002
37. RenL, McLeanJR, HazbunTR, FieldsS, Vander KooiC, et al. (2011) Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS ONE 6: e16719 doi:10.1371/journal.pone.0016719
38. LivesaySB, CollierSE, BittonDA, BählerJ, OhiMD (2013) Structural and functional characterization of the N-terminus of Schizosaccharomyces pombe Cwf10. Eukaryotic Cell 12 : 1472–89.
39. CarnahanRH, FeoktistovaA, RenL, NiessenS, YatesJRI, et al. (2005) Dim1p Is Required for Efficient Splicing and Export of mRNA Encoding Lid1p, a Component of the Fission Yeast Anaphase-Promoting Complex. Eukaryotic Cell 4 : 577 doi:10.1128/EC.4.3.577-587.2005
40. MoserBA, SubramanianL, KhairL, ChangY-T, NakamuraTM (2009) Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet 5: e1000622 doi:10.1371/journal.pgen.1000622
41. NimmoER, PidouxAL, PerryPE, AllshireRC (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392 : 825–828 doi:10.1038/33941
42. NimmoER, CranstonG, AllshireRC (1994) Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. The EMBO Journal 13 : 3801–3811.
43. WilhelmBT, MargueratS, WattS, SchubertF, WoodV, et al. (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453 : 1239–1243 doi:10.1038/nature07002
44. KimJK, GabelHW, KamathRS, TewariM, PasquinelliA, et al. (2005) Functional genomic analysis of RNA interference in C. elegans. Science 308 : 1164–1167 doi:10.1126/science.1109267
45. HerrAJ, MolnàrA, JonesA, BaulcombeDC (2006) Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA 103 : 14994–15001 doi:10.1073/pnas.0606536103
46. ZhouR, HottaI, DenliAM, HongP, PerrimonN, et al. (2008) Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Molecular Cell 32 : 592–599 doi:10.1016/j.molcel.2008.10.018
47. TabachY, BilliAC, HayesGD, NewmanMA, ZukO, et al. (2013) Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature 493 : 694–698 doi:10.1038/nature11779
48. AusinI, GreenbergMVC, LiCF, JacobsenSE (2012) The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics : official journal of the DNA Methylation Society 7 : 29–33 doi:10.4161/epi.7.1.18782
49. BaryshnikovaA, CostanzoM, DixonS, VizeacoumarFJ, MyersCL, et al. (2010) Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Meth Enzymol 470 : 145–179 doi:10.1016/S0076-6879(10)70007-0
50. PartridgeJF, DebeauchampJL, KosinskiAM, UlrichDL, HadlerMJ, et al. (2007) Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Molecular Cell 26 : 593–602 doi:10.1016/j.molcel.2007.05.004
51. HouH, WangY, KallgrenSP, ThompsonJ, YatesJR, et al. (2010) Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J Biol Chem 285 : 1909–1918 doi:10.1074/jbc.M109.058487
52. ReddyBD, WangY, NiuL, HiguchiEC, MargueratSB, et al. (2011) Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev 25 : 214–219 doi:10.1101/gad.1993611
53. WangY, ReddyB, ThompsonJ, WangH, NomaK-I, et al. (2009) Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Molecular Cell 33 : 428–437 doi:10.1016/j.molcel.2009.02.002
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



