Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
Primary open angle glaucoma is a blinding disease for which there is currently no cure, only treatments that may slow its progress. To help understand the mechanisms of this disease and to design more effective treatments, we identified previously a locus, SIX6, that increases the risk of glaucoma. This gene is involved in early eye development and helps to form the retina. In this paper, we test specific sequence variants in SIX6 that are found in glaucoma patients. We show that these variants have a reduced function that interferes with their ability to direct proper formation of the retina. One variant in particular is common, and may be the main reason that this gene is important in the glaucoma disease process. Patients who have two copies of this sequence variant show a change in the structure of their eye consistent with fewer neurons that carry the visual signal to the brain. These neurons typically die as people age, and people who begin life with fewer visual neurons may have an increased risk of glaucoma. Additional research in this topic may lead to new treatments that preserve sight.
Published in the journal:
. PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004372
Category:
Research Article
doi:
https://doi.org/10.1371/journal.pgen.1004372
Summary
Primary open angle glaucoma is a blinding disease for which there is currently no cure, only treatments that may slow its progress. To help understand the mechanisms of this disease and to design more effective treatments, we identified previously a locus, SIX6, that increases the risk of glaucoma. This gene is involved in early eye development and helps to form the retina. In this paper, we test specific sequence variants in SIX6 that are found in glaucoma patients. We show that these variants have a reduced function that interferes with their ability to direct proper formation of the retina. One variant in particular is common, and may be the main reason that this gene is important in the glaucoma disease process. Patients who have two copies of this sequence variant show a change in the structure of their eye consistent with fewer neurons that carry the visual signal to the brain. These neurons typically die as people age, and people who begin life with fewer visual neurons may have an increased risk of glaucoma. Additional research in this topic may lead to new treatments that preserve sight.
Introduction
Primary open-angle glaucoma (POAG) is the most common type of glaucoma, a group of diseases that affect approximately 60 million people worldwide and is a leading cause of blindness [1]. Glaucoma is characterized by the progressive death of retinal ganglion cells, leading to optic nerve atrophy and loss of vision. POAG is a complex inherited disorder for which an increasing number of genetic associations have been described, each contributing modestly to disease burden [2], [3].
A recent POAG genome-wide association study found a significant genetic association (rs10483727, odds ratio (OR) = 1.32, p = 3.87×10−11) at the SIX1/SIX6 locus [4]. Variants in the SIX1/SIX6 locus were first associated with quantitative optic nerve parameters in controls, including vertical cup-disc ratio (VCDR), which is used clinically to diagnose and monitor POAG progression [5], [6]. Several studies have independently confirmed the association of the SIX1/SIX6 locus with both VCDR and POAG [7]–[9].
The human SIX gene family consists of six members (SIX1–SIX6), all of which contain two shared protein domains; a DNA binding homeobox domain and a SIX domain, which binds downstream effector molecules [10], [11]. Members of this conserved gene family were originally identified through homology to the Drosophila melanogaster (Drosophila) sine oculis (so) gene, which is required for proper eye development [10], [11] and are thought to function as transcription factors, regulating key developmental steps through a complex regulatory network. During embryonic development, SIX1 is expressed broadly in multiple tissues, including the otic vesicle and the limb mesenchyme. However, expression of SIX6 is restricted to regions of the retina and the pituitary [10], [12]. Drosophila with null so alleles have restricted retinal development, while morpholino knockdown of six6b in Danio rerio (zebrafish) embryos was recently shown to result in a small eye phenotype [10], [13]. In humans, a large deletion on chromosome 14q22.3-q23 that includes SIX6 causes bilateral anophthalmia, the absence of both eyes, demonstrating the importance of SIX gene family members in ocular development and human disease [10], [11], [14]–[16].
In this study, we have extended the current understanding of the molecular contributions of SIX6 to POAG risk. First, we identified potential POAG risk alleles by sequencing the SIX6 gene in a case-control dataset; we found both common and rare coding changes within SIX6 in POAG cases, as well as sequence variants in the SIX6 enhancer. We then used the zebrafish system to demonstrate that these human coding variants have functional consequences in eye development. This analysis of 2–3 day old zebrafish embryos is not intended to fully recapitulate the glaucomatous phenotype; however, it provides in vivo data about the functional effects of human genetic variation on the human SIX6 protein in the context of eye development. We next used luciferase reporter assays, through which we show that a sequence variant found in the SIX6 enhancer of POAG patients may increase SIX6 expression. Finally, we demonstrate that POAG cases homozygous for the SIX6 risk allele rs33912345 have a significantly thinner retinal nerve fiber layer, suggesting a glaucomatous pathogenic mechanism driven by SIX6 dysfunction.
Results
Identification of SIX6 risk alleles in POAG cases
Sequencing of the SIX1 and SIX6 genes in Caucasian POAG cases and controls (262 cases, 256 controls) revealed 23 SNPs (Supplemental Table S1). Nine SNPs were identified in SIX1, but no nonsynonymous SNPs were present in the POAG cases. Sequencing of SIX6 yielded 14 variants including five rare nonsynonymous SNPs in POAG cases and controls, one common nonsynonymous SNP located in the homeobox of SIX6 (rs33912345, Asn141His), and five sequence variants within the SIX6 enhancer. All of these variants are conserved evolutionarily as shown by their positive Genomic Evolution Rate Profiling (GERP) scores (Table 1) [17].
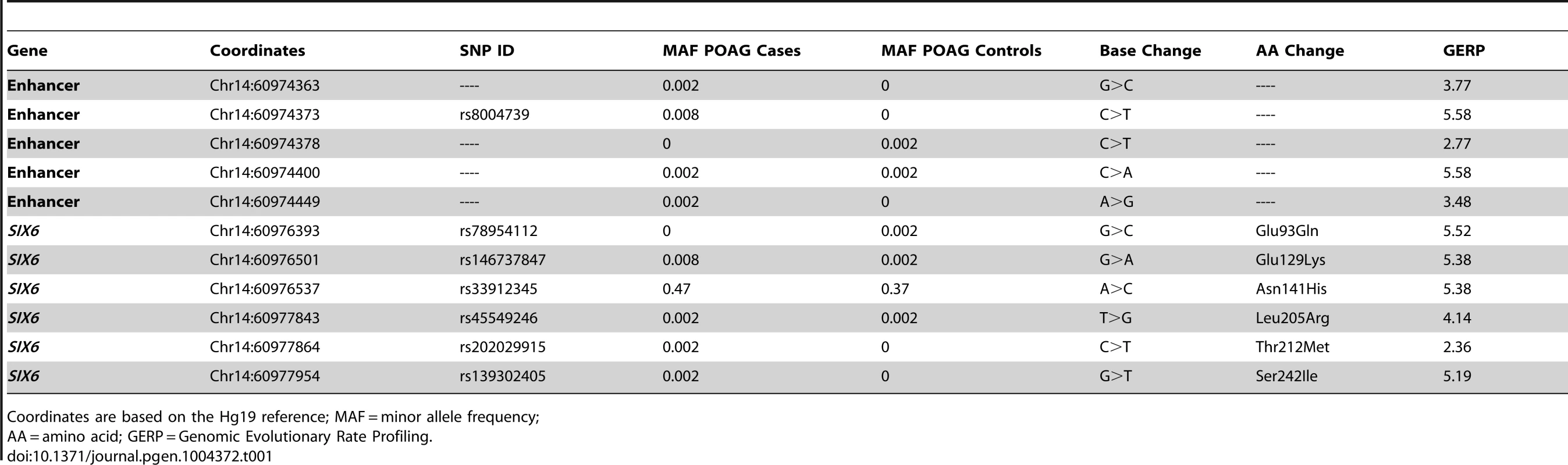
Genotyping of rs33912345 in the Duke POAG case-control dataset (482 cases, 433 controls) resulted in a significant association (OR = 1.40, p = 0.0005, POAG case minor allele frequency (MAF) = 0.47, POAG control MAF = 0.38) with POAG. This SNP is in high linkage disequilibrium (r2 = 0.95) with the intergenic SNP identified originally in POAG and VCDR genome-wide association studies (rs10483727) [4]–[6], [13]. As expected, meta-analysis of the imputed genotype data from the NEIGHBOR and GLAUGEN studies confirmed a significant association between POAG status and rs33912345 (OR = 1.27, p = 4.2×10−10) and other linked SNPs in the region (Supplemental Table S2). Further examination of this locus showed that the association signal includes both upstream and downstream regions of the SIX6 transcript, while remaining entirely downstream of SIX1 (Supplemental 1).
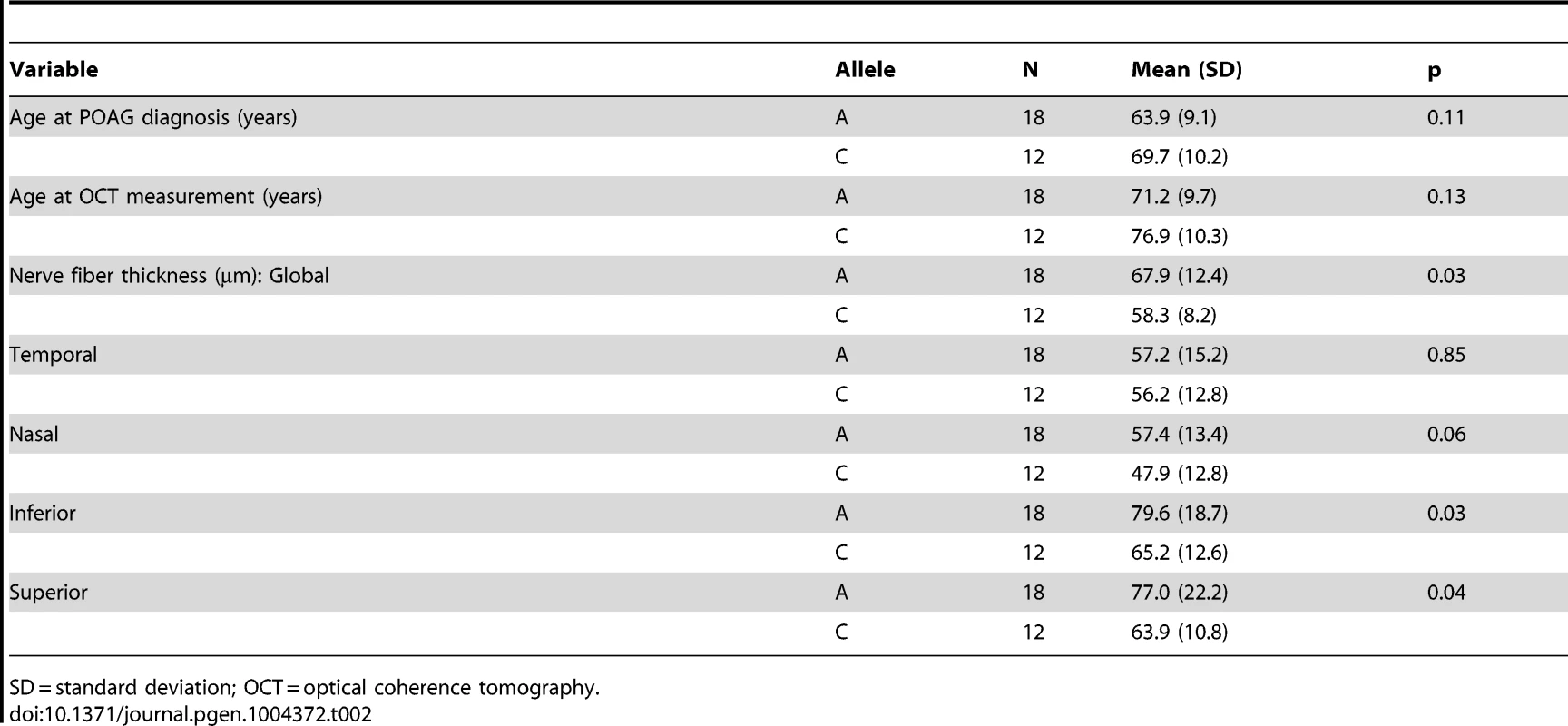
We next performed optical coherence tomography (OCT) to study the retinal characteristics of POAG cases possessing the SIX6 risk and non-risk variants (Table 2). OCT images were only available for POAG cases with the common SNP, rs33912345; no data were available for individuals with the rare SIX6 variants. We assessed retinal nerve fiber layer (RNFL) thickness in thirty POAG cases homozygous for the rs33912345 risk allele (C) or the non-risk allele (A) first by comparing age at disease diagnosis and age at OCT across the two genotypes, because age is known to influence retinal thickness and is thus a potential confounder. We observed no significant difference in age (p = 0.11, p = 0.14, respectively). Next, RNFL thickness was evaluated. The overall thickness (global RNFL) was reduced significantly in cases homozygous for the risk allele compared to cases with the non-risk allele (p = 0.03; mean (SD): C = 58.3 (8.2) µm, A = 67.9 (12.4) µm; Table 2), consistent with the hypothesis that SIX6 may increase POAG susceptibility via changes in the neural retina. To determine which quadrants might be driving this observation, we performed an exploratory, post-hoc comparison of RNFL thickness in the temporal, nasal, inferior, and superior regions. We found that RNFL thickness was reduced significantly in the inferior (p = 0.03) and superior (p = 0.04) quadrants, the two regions affecting directly VCDR measurements.
In vivo functional interrogation of SIX6 missense variants
Given 1) the observed association signal pattern; 2) the lack of coding changes identified in SIX1; 3) the presence of rare missense variants and a common, associated missense SNP in SIX6; 4) retinal nerve fiber layer thickness changes observed in POAG cases homozygous for the SIX6 risk allele; and 5) the localized expression of SIX6 in ocular tissues, we concluded SIX6 is a likely candidate gene in this region. We therefore evaluated the functional relevance of SIX6 and the potential burden of common and rare alleles in this locus in POAG using an in vivo zebrafish complementation assay.
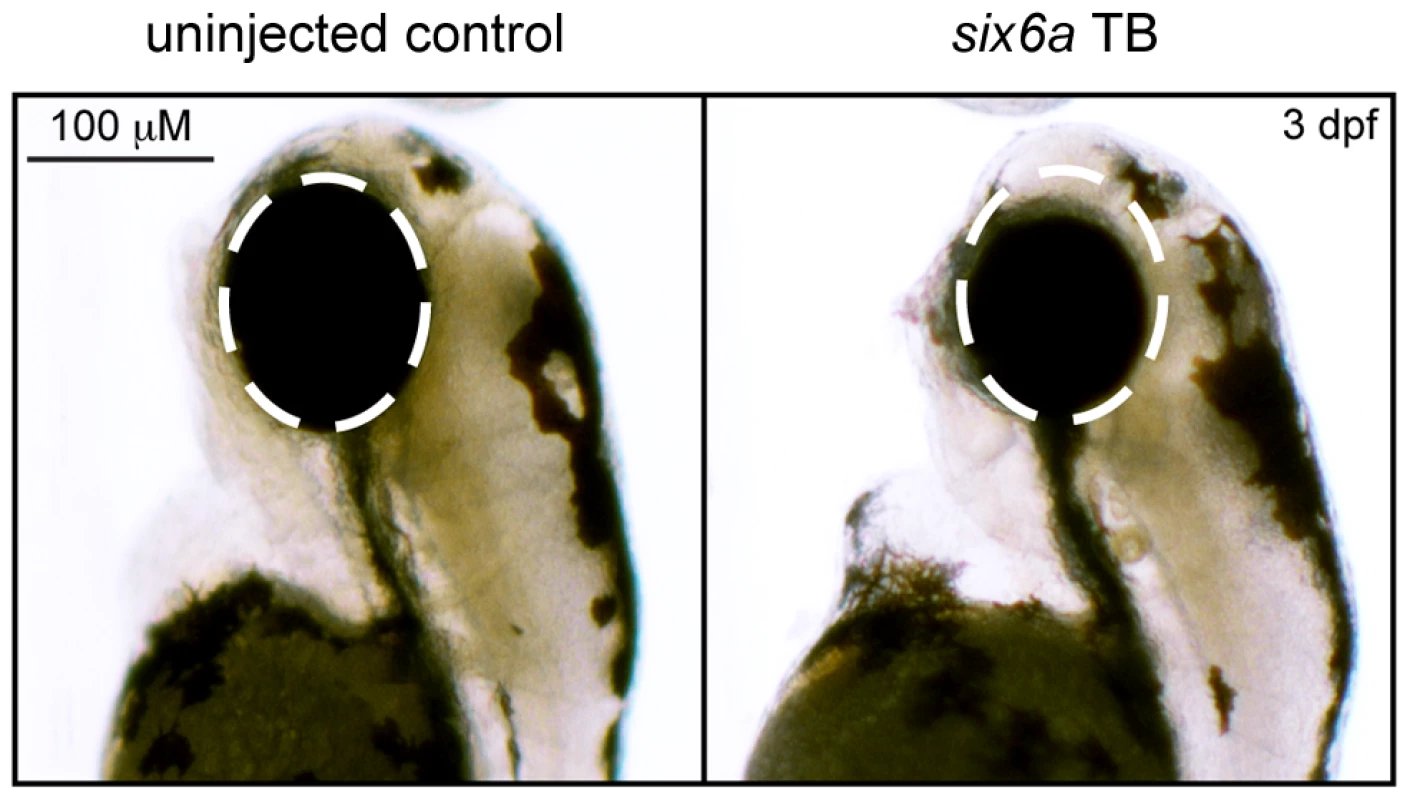
First, we performed a reciprocal BLAST analysis; we identified two orthologs of SIX6 in the zebrafish genome, Six6a and Six6b, both with 91% homology at the protein level (Supplemental Figure S2). Previous overexpression and loss of function studies of Six6 in Mus musculus (mouse) and Xenopus laevis (Xenopus) models reveal a role in regulating the proliferative state of retinal progenitor cells and the size of the eye [18], [19]; therefore, as a first test of whether the identified SIX6 variants are pathogenic and potentially relevant to POAG, we asked whether 1) morpholino-induced suppression of six6a or six6b leads to a reduced eye size; 2) expression of the human SIX6 non-risk allele rescues the morphant eye phenotype; and 3) expression of SIX6 alleles containing POAG risk variants rescues the morphant eye phenotype.
Using translation-blocking morpholinos (MOs) targeting zebrafish six6a and six6b, we injected 1–8 cell stage embryos (N = 50–150) and analyzed live embryos at 3 days post fertilization (dpf). We also tested a splice blocking MO. However, this induced non-specific toxicity, including the accumulation of pericardial fluid that could not be rescued with the human SIX6 transcript. This is not unexpected, as SIX6 is a two exon gene—splice-blocking MOs are generally not recommended for two exon genes; the targeted transcript will not be subject to nonsense mediated decay, possibly leading to the expression of a truncated protein and potential dominant-negative effects (described by the manufacturer: http://www.gene-tools.com/node/18). For these reasons, we used translation-blocking MOs for the remainder of our experiments. Masked scoring of both six6a and six6b morphants revealed ocular phenotypes consistent with loss of function, including a reduction in eye size in more than 80% of embryos (p<0.001, Figure 1). The specificity of the MO was tested by co-injection of 12.5 pg of the human SIX6 non-risk allele mRNA; we observed significant (p<0.001) rescue in six6a but not six6b morphant embryos (90% vs. 10% of embryos, respectively). Together, these data indicate that Six6a is the functional ortholog of human SIX6 and prompted subsequent evaluation of SIX6 variants using the six6a MO.
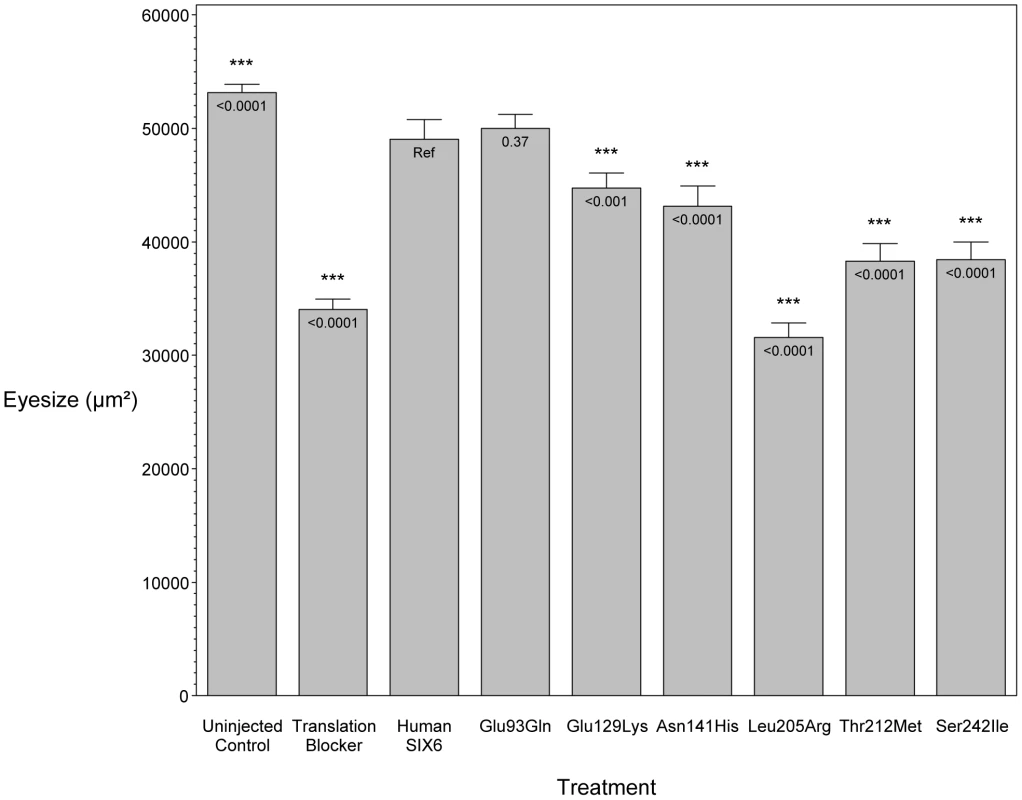
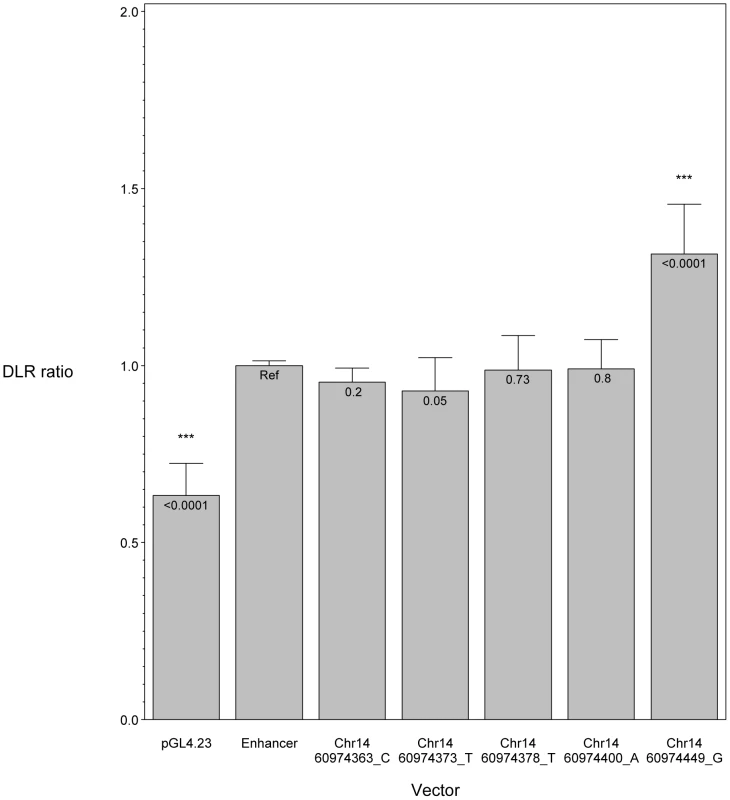
To investigate the pathogenic potential of all SIX6 variants, we used total eye size and the rescue of the morphant phenotype as the assay's phenotypic readout. We injected a mixture containing six6a MO and each of the human SIX6 alleles containing the coding variants identified via sequencing (Table 1). Subsequent to triplicate injections and masked scoring, these results were compared to the rescue condition of the human SIX6 non-risk allele. We found that five of the six variants tested were unable to fully rescue the small eye phenotype. Four of these alleles (Glu129Lys, Asn141His, Thr212Met, and Ser242Ile) resulted in an average eye size larger than the morpholino alone (p<0.001), but smaller than the rescue with the non-risk allele (p<0.001), indicating that these alleles are hypomorphic (Figure 2, Table 3). We also observed one variant (Leu205Arg) with an average eye size smaller than the morpholino alone (p = 0.002; mean (SD): MO = 34,042 (5,763) µm2, Leu205Arg = 31,568 (6,485) µm2; Figure 2), suggesting that it is functionally null. Finally, one allele (Glu93Gln) resulted in an eye size similar to the rescue with the non-risk allele (p = 0.37) and was determined to be benign. The benign allele was identified in one control individual, while the remaining hypomorphic and null alleles were identified either exclusively or primarily in POAG cases (Table 1). Injection of 12.5 pg of the human SIX6 risk mRNA into non-morphant zebrafish provided no evidence of a toxic gain of function compared to injection with the non-risk allele (data not shown).
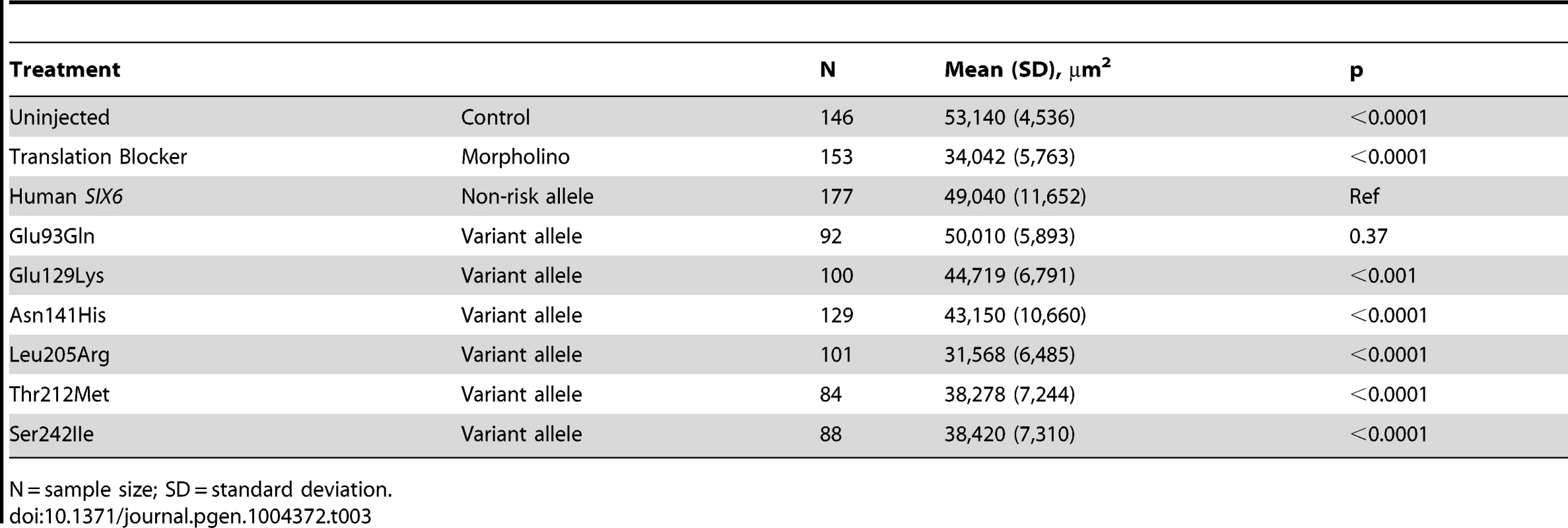
Given the reduction of the RNFL in cases homozygous for the risk allele, we next asked whether six6a and the identified SIX6 variants impacted the optic nerve, an anatomical site directly relevant to human POAG, in zebrafish. Using whole mount imaging of acetylated-tubulin expression in 2-dpf embryos injected with a control and six6a morpholino, we evaluated volumetric regions of interest (ROI) along the optic nerve (Supplemental Figure S3). Masked scoring of embryos revealed an approximately 3 fold reduction (p<0.001) in the volume of the optic nerve upon depletion of six6a (Figure 3 A–B). This was specific for the optic nerve as the volume of other axonal tracts in the brain were unaffected by six6a depletion (Figure 3A). Specificity of the volumetric measurements was demonstrated upon full rescue of the six6a morphant phenotype by co-injection of the non-risk allele (p<0.001) or a variant that scored as a benign allele in the eye size assay (Glu93Gln; p<0.001; Figure 3B). Both Leu205Arg and Asn141His variants performed as hypomorphic alleles (p<0.01), revealing concordance of our optic nerve assay with the OCT imaging findings in patients.
In vitro functional interrogation of SIX6 enhancer variants
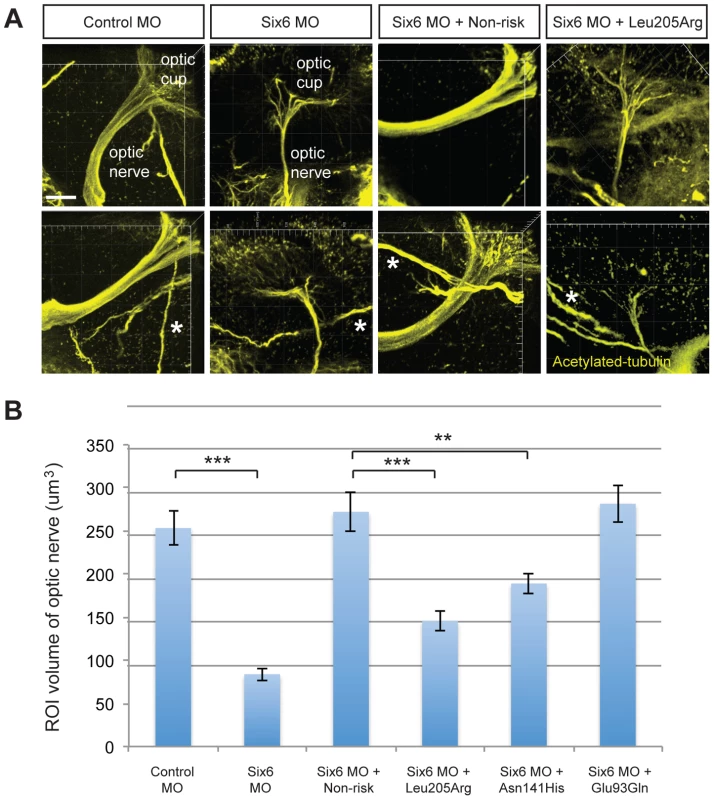
We hypothesized that POAG risk may be mediated not only by deficits in SIX6 protein function, but also by the level of SIX6 gene expression. To test this, we sequenced the SIX6 retinal specific enhancer element in 262 POAG cases and 256 POAG controls; we identified five variants (Chr14:60974363_C, Chr14:60974373_T, Chr14:60974378_T, Chr14:60974400_A, Chr14:60974449_G) (Table 1), and tested their effect on expression using an in vitro luciferase assay. We found that one of these variants (Chr14:60974449_G) resulted in a significant increase in expression compared to the reference enhancer (Figure 4). Activation of the SIX6 enhancer requires two cofactors, NeuroD and E47 (Supplemental Figure S4) [12]. Overexpression was observed with the Chr14:60974449_G variant even in the absence of these cofactors (Supplemental Figure S5), suggesting variants within the enhancer region may result in dysregulated protein expression.
Discussion
The SIX1/SIX6 locus has been shown to be associated significantly with POAG in several independent studies; however, the causal variant(s) driving this association have remained unknown [4], [6]–[9], [13], as has the direction of effect of these common alleles on protein function. We have demonstrated through several lines of evidence—the tissue specificity of the SIX gene family described in the literature; the identification of SIX6 missense variants in our POAG dataset; and the results of the in vivo and in vitro assays— that SIX6 is the most likely POAG susceptibility gene in this region. We identified both common and rare coding variants that alter the function of the SIX6 protein. We also identified a novel variant within the SIX6 enhancer that appears to disrupt the regulation of SIX6 expression, suggesting both regulatory and coding variants may influence POAG susceptibility at this locus. Finally, we identified hitherto unknown clinical features in POAG patients that may be dependent upon SIX6 genotype: patients homozygous for the SIX6 risk allele have a significantly thinner retinal nerve fiber layer than patients homozygous for the SIX6 non-risk allele.
The common variant, rs33912345 (Asn141His), which we showed has significantly reduced function in an in vivo model, was associated with POAG in our dataset and is in strong linkage disequilibrium with the originally identified GWAS SNP, rs10483727 [4]. This conserved amino acid is located within the alpha helix structure of the DNA homeobox domain of SIX6. Interestingly, the ancestral allele (C, His) is associated with POAG risk. The allele frequency of this variant differs markedly among populations (C allele frequency: YRI (0.99), ARF (0.96), ASN (0.76), and CEU (0.42); from 1000 Genomes release 14) [20]. This locus is associated significantly with an increase in vertical cup-disc ratio (VCDR) in population controls, indicating that it may be involved in the development of the optic nerve. VCDR is also a clinical measure used to track disease progression in POAG patients [5], [6], [9], [13], [21]. We note that African populations have larger VCDR and an increase in overall POAG prevalence [22]–[24] compared to CEU populations; in a recent study, the prevalence of POAG in Ghana exceeded 17%, the highest rate observed anywhere in the world [25]. While rs33912345 is not associated with POAG risk in a West African POAG case/control dataset from Ghana, the frequency of the ancestral (risk) allele is 99% in both cases and controls [26]. We hypothesize that differences in the structure of the optic nerve and the higher risk of POAG in individuals with African ancestry may, in part, arise from the fact that essentially all individuals in this population are homozygous for the rs33912345 ancestral risk allele.
Given the association of the SIX6 locus with neural retinal measurements such as VCDR, it is reasonable to hypothesize causal variants may function by inducing quantitative changes in cell populations in the retina. There is extensive evidence that SIX6 regulates early retinal progenitor cell proliferation during eye development [18], [19], [27]. Li et al. showed that Six6−/− mice display varying degrees of retinal hypoplasia that is due to a decrease in retinal ganglion cell proliferation arising from an early exit from the cell cycle during development, and results in a 20% decrease in the number of retinal ganglion cells by P35 [18]. This is consistent with functional studies of XOptx2, the Xenopus ortholog of SIX6 [19]. We have shown a reduction in eye size and in the volume of the optic nerve upon MO knockdown of zebrafish six6a, and we were able to rescue these phenotypes with co-injection of the human SIX6 non-risk allele, demonstrating that the zebrafish six6a gene is the likely functional ortholog to human SIX6. We identified five alleles that could not rescue the small eye and optic nerve phenotypes, and we observed a reduction in retinal nerve fiber layer thickness in POAG patients homozygous for the His141 SIX6 risk allele. Taken together with previously published findings, our results suggest that risk variants in human SIX6 increase POAG susceptibility by negatively affecting retinal ganglion cell development, likely leading to a reduction in the number of retinal ganglion cells in adulthood. Given that retinal ganglion cells are lost during the normal aging process, we speculate that this rate of loss could be increased by the presence of additional POAG risk alleles or other risk factors such as increased intraocular pressure [28]. The development of glaucomatous optic neuropathy and associated visual field loss would thus be hastened by a reduction in the initial number of retinal ganglion cells that an individual possesses. Future work will test the possibility that SIX6 variants also alter the rate of RGC death in the adult.
In summary, we have identified multiple common and rare SIX6 sequence variants in POAG cases, and used in vivo and in vitro assays to demonstrate that these variants have functional consequences on SIX6 expression and protein function. While other risk factors may be required for the onset of POAG, our data suggest that attenuation of SIX6 protein function increases an individual's susceptibility to developing the disease via changes to retinal development. Additional work is needed, possibly through the use of transgenic animal model studies, to fully understand the role of SIX6 in POAG.
Materials and Methods
Ethics statement
This research was approved by the Institutional Review Board of Duke University Medical Center and adheres to the tenets of the Declaration of Helsinki.
Subjects
Study subjects were unrelated patients from the Duke Eye Center and, after a comprehensive eye examine, were classified as either POAG cases or controls. POAG cases presented with glaucomatous optic neuropathy, defined as a cup-to-disc ratio greater than 0.7 and visual field loss in at least one eye. Patients with secondary forms of glaucoma or a history of ocular trauma were excluded from the study. POAG controls had no evidence of optic neuropathy, normal intraocular pressure (less than 22 mmHg in both eyes), and normal visual fields, assessed using standard automated perimetry.
DNA sequencing
Genomic DNA was extracted from patient blood samples using the PureGene chemistry following the manufacturer's standard protocol (Gentra, Minneapolis, MN). The coding portions (2 exons) of the SIX1 and SIX6 genes were sequenced in 518 Caucasian POAG cases and controls (262 cases, 256 controls) using a polymerase chain-reaction (PCR) containing 1× Qiagen PCR buffer (Tris·Cl, KCl, (NH4)2SO4, 15 mM MgCl2; pH 8.7); 200 µM each of dATP, dCTP, dGTP, and dTTP; 0.4 µM forward PCR primer; 0.4 µM reverse PCR primer; 3 µL of betaine, 10 ng genomic DNA; and 0.5 U HotStarTaq DNA polymerase (Qiagen, Venlo, Limburg) to a final volume of 25 µL. Primer sequences are available in Supplemental Table S3. The PCR was performed using a touchdown protocol (incremental lowering of annealing temperature) using the following thermocycler conditions: 94°C for 30 s, 65°C for 30 s, 72°C for 30 s with a 2°C decrease in the annealing temperature every two cycles until a final annealing temperature of 55°C was reached. The retinal specific SIX6 enhancer, previously described [12], was amplified a using a touchdown protocol with a final annealing temperature of 57°C. PCR products were sequencing using the BigDye chemistry on a 3730 DNA Analyzer (Applied Biosystems, Grand Island, NY).
Genotyping and imputation
The common missense single nucleotide polymorphism (SNP), rs33912345, was genotyped in the Duke POAG case-control dataset consisting of 482 POAG cases and 433 POAG controls using a TaqMan allelic discrimination assay according to the standard protocols from the manufacturer (Life Technologies, Grand Island, NY). For quality control purposes the following criteria were met: >95% genotyping efficiency, matching sample duplicates (two Centre d'Etude du Polymorphisme Humain samples per 96-well plate whose genotype data matched across all plates), and Hardy-Weinberg equilibrium assumptions. We tested for association of rs33912345 with POAG using an additive logistic regression model adjusted for age and sex using SAS [29].
Genome-wide genotype data were available from the NEI Glaucoma Human Genetics Collaboration (NEIGHBOR) and the Glaucoma Genes and Environment (GLAUGEN) consortia [4]. Chromosome 14 was imputed using IMPUTE2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html) with a global 1000 Genomes reference panel. We tested SNPs at the SIX1/SIX6 locus for association with POAG using an additive logistic regression model adjusted for age, sex, and four principal components (NEIGHBOR) or age, gender, study site, DNA extraction method, DNA specimen type and principal components 1–6 (GLAUGEN) implemented in PLINK and visualized using LocusZoom (http://csg.sph.umich.edu/locuszoom/) [30], [31]. A meta-analysis was performed in Plink using a random effects model. Linkage disequilibrium was calculated and visualized using Haploview (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview) [32].
Retinal nerve fiber layer thickness analysis
Optical coherence tomography (OCT) measurements of retinal nerve fiber layer (RNFL) by Spectralis (Heidelberg Engineering, Carlsbad, CA) spectral domain) and fundus photography were available from the Duke Eye Center. OCT images are not routinely performed in patients without ocular disease, so there was limited data available for controls. Therefore, the analysis was restricted to POAG cases homozygous for rs33912345. Thirty patients had both OCT measurements and SIX6 genotype data. RNFL thickness, age at POAG diagnosis, and the age at OCT measurement were compared between individuals homozygous for the risk or non-risk allele using a Student's t-test. Analyses were performed in SAS [29].
Microinjection of morpholino and mRNA
A vector containing human SIX6 was purchased from the CCSB Human ORFeome Collection that uses the Gateway technology system (Open Biosystems and Life Technologies, Grand Island, NY). SIX6 alleles identified by sequencing (Glu93Gln, Glu129Lys, Asn141His, Leu205Arg, Thr212Met, Ser242IIe), were created using the QuikChange II site-directed mutagenesis kit and protocols provided by the manufacturer (Agilent Technologies). SIX6 mRNA was in vitro transcribed using mMESSAGE mMACHINE SP6 Kit (Ambion, Life Technologies, Grand Island, NY). Translation blocker (TB) morpholinos against six6a (5′- CTGGAACATGGAGACTGTAATGTCT -3′) and six6b (5′ AATTGGCAACTGAAACATGAAGGCT 3′) were purchased from Gene Tools, LLC. Morpholino (2 ng) and mRNA (12.5 pg) were mixed and a volume of 0.5 nL was microinjected into each wild-type zebrafish embryo at one- to eight-cell stage as described previously (Stuart, McMurray et al. 1988).
Morphometric analyses of zebrafish
Morphometric analyses of eye size were conducted on zebrafish embryos at 3 days post fertilization, using a Nikon SMZ 745T microscope. Zebrafish were anesthetized in embryo medium containing 0.2 mg/ml tricaine (Ethyl 3-aminobenzoate methanesulfonate, Sigma, E10521). Lateral view images were captured with Nikon DS-Fi1 camera, and the size of eye was measured with Nikon NIS-Elements AR software. Analysis of the optic nerve was performed on 2 dpf embryos fixed in Dent's fixative (80% Methanol and 20% DMSO) overnight and stained with acetylated-tubulin (Sigma; T7451). Heads were isolated from stained embryos and oriented with the ventral aspect facing a coverslip on microscope cover glass. Image acquisition was performed on a Zeiss 710 inverted confocal microscope and ∼100 um optical sections were obtained and reconstructed. Volumetric measurements were calculated using Imaris software and 7.5 um×7.5 um×15 um ROIs along the optic nerve were analyzed between each condition. ROIs were restricted to portions of the optic nerve wherein all neurite processes coalesced to form the major aspect of the nerve.
In vitro luciferase assay
SIX6 enhancer alleles were tested using the dual-luciferase reporter assay system (Promega, Madison, WI). An experimental construct containing a minimal promoter (pGL4.23, firefly luciferase, Promega) was used to test the functional effect of the enhancer alleles identified by sequencing (Chr14:60974363_C, Chr14:60974373_T, Chr14:60974378_T, Chr14:60974400_A, Chr14:60974449_G) in the POAG case/control dataset. The experimental constructs (pGL4.23+Enhancer) were generated using a nested PCR protocol; the XhoI and HindII enzymes; the Quick Ligation kit (New England BioLabs, Ipswich, MA); and the QuikChange II site-directed mutagenesis kit (Agilent Technologies), following protocols provided by the manufacturers. Constructs were confirmed to be correct by sequencing. Hek293 cells were cultured according to the supplier's suggestions (ATCC, Manassas, VA). As described by Conte et al., co-transfection with NeuroD and E47 is required for SIX6 enhancer activation [12]. Therefore, cells were co-transfected with an experimental vector (pGL4.23+Enhancer), a control vector (pGL4.74, renilla luciferase, Promega), and vectors containing NeuroD and E47 (provided by the Center for Human Disease Modeling, Duke University) using a standard calcium phosphate transfection protocol. The experiment was performed three times in triplicate and the results were analyzed using the dual luciferase reporter (DLR) ratio (firefly luciferase sum: renilla luciferase sum) normalized by the reference SIX6 enhancer included on every plate. The data were analyzed using an ANOVA, adjusted for batch, and linear contrasts were used to determine the effect of each vector. Statistical analyses were performed in SAS [29].
Supporting Information
Zdroje
1. QuigleyHA, BromanAT (2006) The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol 90: 262–267.
2. BurdonKP (2012) Genome-wide association studies in the hunt for genes causing primary open-angle glaucoma: a review. Clin Experiment Ophthalmol 40: 358–363.
3. LiuY, AllinghamRR (2011) Molecular genetics in glaucoma. Exp Eye Res 93: 331–339.
4. WiggsJL, YaspanBL, HauserMA, KangJH, AllinghamRR, et al. (2012) Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet 8: e1002654.
5. MacgregorS, HewittAW, HysiPG, RuddleJB, MedlandSE, et al. (2010) Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet 19: 2716–2724.
6. RamdasWD, van KoolwijkLM, IkramMK, JansoniusNM, de JongPT, et al. (2010) A genome-wide association study of optic disc parameters. PLoS Genet 6: e1000978.
7. FanBJ, WangDY, PasqualeLR, HainesJL, WiggsJL (2011) Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest Ophthalmol Vis Sci 52: 1788–1792.
8. OsmanW, LowSK, TakahashiA, KuboM, NakamuraY (2012) A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet 21: 2836–2842.
9. RamdasWD, van KoolwijkLM, LemijHG, PasuttoF, CreeAJ, et al. (2011) Common genetic variants associated with open-angle glaucoma. Hum Mol Genet 20: 2464–2471.
10. KawakamiK, SatoS, OzakiH, IkedaK (2000) Six family genes–structure and function as transcription factors and their roles in development. BioEssays: news and reviews in molecular, cellular and developmental biology 22: 616–626.
11. KumarJP (2009) The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci 66: 565–583.
12. ConteI, Marco-FerreresR, BeccariL, CisnerosE, RuizJM, et al. (2010) Proper differentiation of photoreceptors and amacrine cells depends on a regulatory loop between NeuroD and Six6. Development 137: 2307–2317.
13. IglesiasAI, SpringelkampH, van der LindeH, SeverijnenLA, AminN, et al. (2013) Exome sequencing and functional analyses suggest SIX6 is a gene involved in an altered proliferation-differentiation balance early in life and optic nerve degeneration at old age. Hum Mol Genet 23: 1320–1332 doi:10.1093/hmg/ddt522
14. CheyetteBN, GreenPJ, MartinK, GarrenH, HartensteinV, et al. (1994) The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12: 977–996.
15. GallardoME, Lopez-RiosJ, Fernaud-EspinosaI, GranadinoB, SanzR, et al. (1999) Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics 61: 82–91.
16. KenyonKL, Yang-ZhouD, CaiCQ, TranS, ClouserC, et al. (2005) Partner specificity is essential for proper function of the SIX-type homeodomain proteins Sine oculis and Optix during fly eye development. Dev Biol 286: 158–168.
17. CooperGM, StoneEA, AsimenosG, GreenED, BatzoglouS, et al. (2005) Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15: 901–913.
18. LiX, PerissiV, LiuF, RoseDW, RosenfeldMG (2002) Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 297: 1180–1183.
19. ZuberME, PerronM, PhilpottA, BangA, HarrisWA (1999) Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell 98: 341–352.
20. AbecasisGR, AutonA, BrooksLD, DePristoMA, DurbinRM, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65.
21. CharlesworthJ, KramerPL, DyerT, DiegoV, SamplesJR, et al. (2010) The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci 51: 3509–3514.
22. GirkinCA (2008) Differences in optic nerve structure between individuals of predominantly African and European ancestry: Implications for disease detection and pathogenesis. Clin Ophthalmol 2: 65–69.
23. GirkinCA, SamplePA, LiebmannJM, JainS, BowdC, et al. (2010) African Descent and Glaucoma Evaluation Study (ADAGES): II. Ancestry differences in optic disc, retinal nerve fiber layer, and macular structure in healthy subjects. Arch Ophthalmol 128: 541–550.
24. KnightOJ, GirkinCA, BudenzDL, DurbinMK, FeuerWJ (2012) Effect of race, age, and axial length on optic nerve head parameters and retinal nerve fiber layer thickness measured by Cirrus HD-OCT. Arch Ophthalmol 130: 312–318.
25. BudenzDL, BandiJR, BartonK, NolanW, HerndonL, et al. (2012) Blindness and Visual Impairment in an Urban West African Population: The Tema Eye Survey. Ophthalmology 119: 1744–1753 doi:10.1016/j.ophtha.2012.04.017
26. LiuY, HauserMA, AkafoSK, QinX, MiuraS, et al. (2013) Investigation of known genetic risk factors for primary open angle glaucoma in two populations of African ancestry. Invest Ophthalmol Vis Sci 54: 6248–6254.
27. BernierG, PanitzF, ZhouX, HollemannT, GrussP, et al. (2000) Expanded retina territory by midbrain transformation upon overexpression of Six6 (Optx2) in Xenopus embryos. Mechanisms of development 93: 59–69.
28. CalkinsDJ (2013) Age-related changes in the visual pathways: blame it on the axon. Invest Ophthalmol Vis Sci 54: ORSF37–41.
29. SAS Institute Inc. (2002–2008) SAS 9.2 Software. Cary, NC: SAS Institute Inc.
30. PruimRJ, WelchRP, SannaS, TeslovichTM, ChinesPS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337.
31. PurcellS, NealeB, Todd-BrownK, ThomasL, FerreiraMA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81: 559–575.
32. BarrettJC, FryB, MallerJ, DalyMJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265.
Štítky
Genetika Reprodukční medicínaČlánek vyšel v časopise
PLOS Genetics
2014 Číslo 5
- Mateřský haplotyp KIR ovlivňuje porodnost živých dětí po transferu dvou embryí v rámci fertilizace in vitro u pacientek s opakujícími se samovolnými potraty nebo poruchami implantace
- Intrauterinní inseminace a její úspěšnost
- Akutní intermitentní porfyrie
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
Nejčtenější v tomto čísle
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
