-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
article has not abstract
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004330
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004330Summary
article has not abstract
Nature demonstrates many interesting variations in cell cycles and cell growth (Figure 1A). Some tissues in animals, such as skeletal muscle and syncytiotrophoblast cells of the placenta, arise through the fusion of post-mitotic diploid progenitor cells to form multinucleated cells. Multinucleated cells such as liver cells can also arise through endomitosis, in which nuclei that have replicated DNA undergo division, but it is not followed by cytokinesis. Perhaps the most interesting example is endoreduplication, a cell cycle in which rounds of DNA synthesis are not coupled with intervening mitoses, usually resulting in cells with enlarged cytoplasm volume. Endocycles are a curiosity because completion of mitosis is required in mitotic cells before another round of DNA replication can occur. However, endoreduplication is observed widely in plants, protozoa, insects, and higher animals, and many different mitotic cell cycle alterations have been defined [1]. In mammals, the best-studied endoreduplicating cell type is the trophoblast giant cells (TGC) of the rodent placenta. In the accompanying paper, Hannibal and colleagues show that mouse TGCs don't endoreduplicate their genomes evenly and have developmentally regulated, under-replicated domains, suggesting that under-replication may be a mechanism to regulate cell function [2].
Fig. 1. Modes of cell growth and polyploidy in mammals. 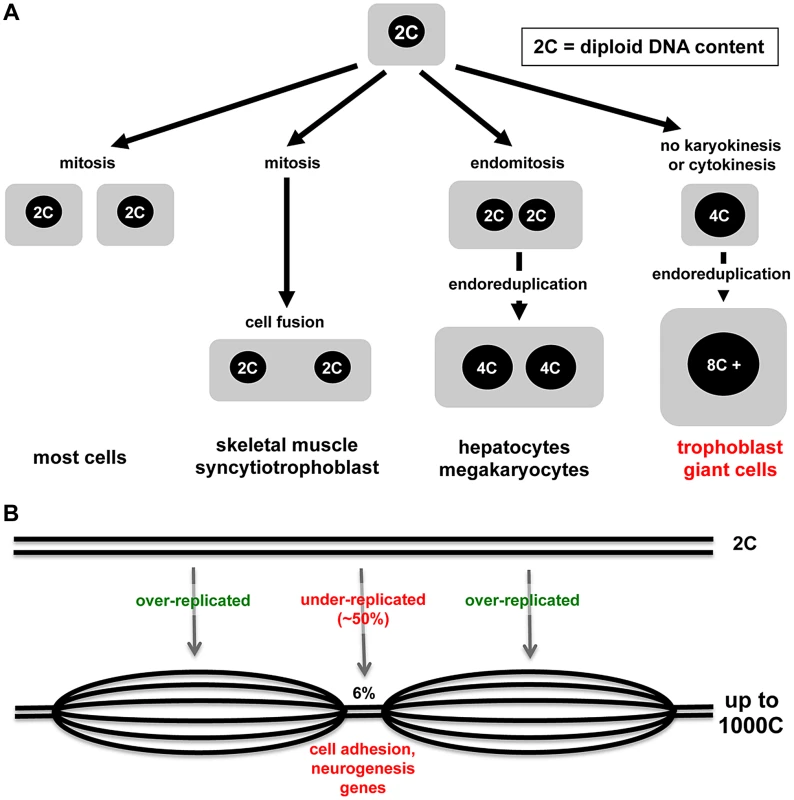
(A) Diagram depicting different mechanisms underlying the formation of polyploid cells ranging from fusion of post-mitotic diploid cells, endomitosis (nuclear but not cell division), and endoreduplication. (B) Nuclei of polyploid trophoblast giant cells show selected small regions of under-replication (based on results from Hannibal et al.). TGCs mediate uterine implantation of embryos, line the maternal blood space in the placenta, and secrete dozens of hormones thought to regulate maternal adaptations to pregnancy [3], [4]. Distinct TGC subtypes sit at different positions within the maternal blood space in the placenta [5], [6]. Parietal-TGCs, which form the interface with the maternal uterus, emerge first and achieve the highest ploidy [6]. After maternal circulation through the placenta is established, parietal-TGCs lie on the venous side as maternal blood leaves the placenta to enter uterine veins [5]. A distinct TGC subtype invades the maternal arteries that bring blood to the implantation site to replace the endothelial cells, while vascular spaces within the placenta itself are formed by morphogenesis of other TGC subtypes into tube-like structures [4].
The function of endoreduplication and polyploidy in TGCs remains a matter of debate, though it may be a way for the tissue to grow without the need to increase cell number [7], [8]—a matter of convenience at the maternal-fetal interface, which needs to develop rapidly. Several mouse mutants have defects in development and ploidy of TGCs [3], [8], but few of them cleanly distinguish the function of ploidy. Mutants in the E2F-7 and -8 cell cycle transcription factors show reduced TGC ploidy and cell size but, interestingly, have little change in TGC gene expression [9]–[11]. Functional models of TGC polyploidy are driven by the notion that over-replicated genes provide some advantage. In Drosophila polyploid cells, there are large regions of genome that are relatively over-replicated [1]. For many years it has been thought that TGCs endoreduplicate their entire genomes, but it would be hard to argue that the entire genome is important for TGC function. In vivo quantitation of TGC DNA content and of cells synchronously endoreduplicating in culture is consistent with a doubling of DNA content with each round [12]–[14]. In situ hybridization experiments using gene-specific probes suggest that TGC chromosomes are polytene due to the failure of replicated DNA strands to segregate [15]–[17]. In the 1990s, restriction landmark genomic scanning was developed to detect genome copy numbers and used to analyze CpG islands in rat placental TGCs [18]. At least 97% of the genome was similarly re-replicated, but the conclusion was limited by the technology of the day. In 2013, Sher et al. used array-based comparative genome hybridization to assess relative ploidies across the genome in TGCs dissected from mouse implantation sites and concluded that TGC genomes were uniformly duplicated [19]. This was also true for megakaryocytes and strikingly different than Drosophila polyploid cells [19].
With the advent of new technologies, it becomes possible to assess the genome with higher resolution and greater sensitivity, and so arrives the current paper from Julie Baker's lab [2]. Focusing on parietal-TGCs because of their high ploidy and using different technologies, Hannibal et al. demonstrate that some regions of the genome, though greater than diploid, are relatively under-replicated compared to the rest (Figure 1B) [2]. What accounts for the difference with the conclusions from Sher et al.? It may be that different TGC populations were sampled or that the approaches had different sensitivity. Comparative genome hybridization combined with whole genome sequencing showed that only 6% of the genome is under-replicated and, even in those regions, the under-replication is ∼50% compared to the rest of the genome [2]. Hannibal et al. show nicely that under-replicated regions are late replicating. Clearly the S-phase machinery must be kept away or inactivated in some regions. DNA synthesis in endoreduplicating TGCs is spread over ten to12 hours [12], [20], so there is ample opportunity to segregate regions of the genome.
The “if” and the “how” endoreduplication happens are only half of a good story; the “what” and the “why” are just as interesting. If under-replicated regions in TGCs were random, one could argue that under-replication was just a small error or a matter of convenience. What is intriguing about the data, however, is that the under-replication occurs in reproducible regions, detected both in TGCs in vivo and trophoblast progenitors differentiated in culture. Analysis of the 47 under-replicated regions shows that they are enriched for some classes of genes, including those involved in cell adhesion and development of the nervous system. Trophoblast progenitor cells reduce cell-cell adhesion as parietal-TGCs develop [21]–[23]. Microarray data from cultured mouse trophoblast stem cells (http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS3948) show expression of “nervous system genes” like the nerve guidance protein Slit. Human trophoblast cells also express Slit and it is overexpressed in the placenta in preeclampsia [24], a pregnancy complication associated with defects in trophoblast cell function. This new evidence might start to change the thinking about endoreduplication. Instead of thinking that more copies of genes that are good for TGC function is the goal, perhaps it is important to reduce duplication of genes that impair TGC function. Let the experiments begin.
Zdroje
1. EdgarBA, ZielkeN, GutierrezC (2014) Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol 15 : 197–210.
2. HannibalRL, ChuongEB, Rivera-MuliaJC, GilbertDM, ValouevA, et al. (2014) Copy number variation is a fundamental aspect of the placental genome. PLoS Genet 10: e1004290 doi:10.1371/journal.pgen.1004290
3. HuD, CrossJC (2010) Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol 54 : 341–354.
4. RaiA, CrossJC (2014) Development of the hemochorial maternal vascular spaces in the placenta through endothelial and vasculogenic mimicry. Dev Biol 387 : 131–141.
5. GasperowiczM, Surmann-SchmittC, HamadaY, OttoF, CrossJC (2013) The transcriptional co-repressor TLE3 regulates development of trophoblast giant cells lining maternal blood spaces in the mouse placenta. Dev Biol 382 : 1–14.
6. SimmonsDG, FortierAL, CrossJC (2007) Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol 304 : 567–578.
7. PanditSK, WestendorpB, de BruinA (2013) Physiological significance of polyploidization in mammalian cells. Trends Cell Biol 23 : 556–566.
8. FoxDT, DuronioRJ (2013) Endoreplication and polyploidy: insights into development and disease. Development 140 : 3–12.
9. PanditSK, WestendorpB, NantasantiS, van LiereE, TootenPC, et al. (2012) E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol 14 : 1181–1191.
10. OusephMM, LiJ, ChenHZ, PecotT, WenzelP, et al. (2012) Atypical E2F repressors and activators coordinate placental development. Dev Cell 22 : 849–862.
11. ChenHZ, OusephMM, LiJ, PecotT, ChokshiV, et al. (2012) Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol 14 : 1192–1202.
12. MacAuleyA, CrossJC, WerbZ (1998) Reprogramming the cell cycle for endoreduplication in rodent trophoblast cells. Mol Biol Cell 9 : 795–807.
13. ZybinaTG, ZybinaEV, ShteinGI (1985) DNA content of the nuclei of secondary giant cells of the rat trophoblast at different phases of the polytene nucleus cycle. Tsitologiia 27 : 957–960.
14. ZybinaEV, ZybinaTG (1996) Polytene chromosomes in mammalian cells. Int Rev Cytol 165 : 53–119.
15. VarmuzaS, PrideauxV, KotharyR, RossantJ (1988) Polytene chromosomes in mouse trophoblast giant cells. Development 102 : 127–134.
16. BowerDJ (1987) Chromosome organisation in polyploid mouse trophoblast nuclei. Chromosoma 95 : 76–80.
17. KeighrenM, WestJD (1993) Analysis of cell ploidy in histological sections of mouse tissues by DNA-DNA in situ hybridization with digoxigenin-labelled probes. Histochem J 25 : 30–44.
18. OhganeJ, AikawaJ, OguraA, HattoriN, OgawaT, et al. (1998) Analysis of CpG islands of trophoblast giant cells by restriction landmark genomic scanning. Dev Genet 22 : 132–140.
19. SherN, Von StetinaJR, BellGW, MatsuuraS, RavidK, et al. (2013) Fundamental differences in endoreplication in mammals and Drosophila revealed by analysis of endocycling and endomitotic cells. Proc Natl Acad Sci U S A 110 : 9368–9373.
20. HattoriN, DaviesTC, Anson-CartwrightL, CrossJC (2000) Periodic expression of the Cdk inhibitor p57Kip2 in trophoblast giant cells defines a G2-like gap phase of the endocycle. Mol Biol Cell 11 : 1037–1045.
21. ParastMM, AederS, SutherlandAE (2001) Trophoblast giant-cell differentiation involves changes in cytoskeleton and cell motility. Dev Biol 230 : 43–60.
22. WatsonED, HughesM, SimmonsDG, NataleDR, SutherlandAE, et al. (2011) Cell-cell adhesion defects in Mrj mutant trophoblast cells are associated with failure to pattern the chorion during early placental development. Dev Dyn 240 : 2505–2519.
23. El-HashashAH, KimberSJ (2006) PTHrP induces changes in cell cytoskeleton and E-cadherin and regulates Eph/Ephrin kinases and RhoGTPases in murine secondary trophoblast cells. Dev Biol 290 : 13–31.
24. LiaoWX, LaurentLC, AgentS, HodgesJ, ChenDB (2012) Human placental expression of SLIT/ROBO signaling cues: effects of preeclampsia and hypoxia. Biol Reprod 86 : 111.
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



