-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
Mutations can fix during evolution for two reasons:
they can be beneficial and fix for adaptive reasons, or they can be neutral or deleterious and fix solely by chance. Most studies focus on adaptation, where the evolving population is increasing in fitness due to a new selection pressure. Such studies have found an important evolutionary role for epistasis, the phenomenon where the effect of one mutation depends on another mutation. But adaptation only accounts for a fraction of overall evolutionary change. Here we investigate whether epistasis is as common during non-adaptive as adaptive evolution. We do this by comparing the same protein from human and swine influenza. Human influenza is constantly adapting to escape from the immunity that people acquire from previous influenza infections. But swine influenza is under less pressure to escape from acquired immunity since pigs have shorter lifetimes and are less likely to be infected with influenza multiple times. We find that epistasis is less common during the evolution of the swine influenza protein than its human influenza counterpart. Overall, our results suggest that mutations that interact via epistasis are more likely to fix during adaptive evolution.
Published in the journal: . PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004328
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004328Summary
Mutations can fix during evolution for two reasons:
they can be beneficial and fix for adaptive reasons, or they can be neutral or deleterious and fix solely by chance. Most studies focus on adaptation, where the evolving population is increasing in fitness due to a new selection pressure. Such studies have found an important evolutionary role for epistasis, the phenomenon where the effect of one mutation depends on another mutation. But adaptation only accounts for a fraction of overall evolutionary change. Here we investigate whether epistasis is as common during non-adaptive as adaptive evolution. We do this by comparing the same protein from human and swine influenza. Human influenza is constantly adapting to escape from the immunity that people acquire from previous influenza infections. But swine influenza is under less pressure to escape from acquired immunity since pigs have shorter lifetimes and are less likely to be infected with influenza multiple times. We find that epistasis is less common during the evolution of the swine influenza protein than its human influenza counterpart. Overall, our results suggest that mutations that interact via epistasis are more likely to fix during adaptive evolution.Introduction
Epistasis occurs when the effect of a change at one site in a genome depends on the presence or absence of a change at another site. Understanding epistasis is of profound importance in evolutionary biology, as epistasis can constrain evolutionary pathways and shape patterns of sequence change. As a result, epistasis has been extensively studied at an experimental level. Nearly all of these studies have focused on adaptive evolution, where the population is undergoing changes that improve its fitness in response to some new selection pressure. Examples include bacterial adaptation to new environmental conditions [1]–[3], the acquisition of drug resistance [4]–[7], and changes in enzyme activity or specificity [8]–[10]. These studies have almost universally emphasized a crucial role for epistasis in adaptive evolution.
But adaptive evolution accounts for only a portion of total evolutionary change, which can also be driven by stochastic forces such as genetic hitchhiking and drift [11]–[15]. In many cases, these stochastic forces probably drive a greater fraction of overall sequence change than does adaptive evolution [13]–[17]. Do insights about epistasis from studies of adaptive evolution also apply to evolutionary change by non-adaptive forces?
There are reasons to suspect that epistatically interacting substitutions may be more prevalent in adaptive than non-adaptive evolution. Two main mechanisms have been identified for the fixation of epistatically interacting mutations during adaptive evolution: compensatory mutations and permissive mutations. In the compensatory-mutation mechanism, selection favors an initial mutation that confers an overall adaptive benefit but also creates secondary defects, which are then remedied by a subsequent compensatory mutation. An example is the evolution of broad-spectrum antibiotic resistance, where an initial mutation that confers resistance to a new antibiotic but impairs protein stability is followed by a compensatory mutation that restores stability [5], [18], [19]. In this compensatory-mutation mechanism, both epistatic mutations are immediately beneficial.
In the permissive-mutation mechanism, an initially neutral or mildly deleterious [20] mutation that rises in frequency due to stochastic forces is essential for permitting the subsequent than adaptive mutation. An example is the evolution of steroid-receptor specificity, where initial neutral mutations modulate protein conformational stability in a way that permits subsequent adaptive mutations to alter specificity [8]. In this permissive-mutation mechanism, only the subsequent adaptive mutations are directly favored by selection – but selection for the adaptive mutations indirectly favors linked permissive mutations, leading to expansion of lineages carrying the combination of mutations and increasing their rate of fixation [21].
Crucially, in both the compensatory-mutation and the permissive-mutation mechanisms described above, adaptive evolution is ultimately responsible for driving fixation of the epistatic mutations. It is possible to imagine scenarios for the fixation of epistatic mutations by stochastic forces in the absence of adaptation – but it is not immediately obvious whether epistatic mutations would fix as commonly in the absence of a driving selective force. This idea that the frequency of epistatically interacting substitutions might differ between adaptive and non-adaptive evolution would be consistent with theoretical work suggesting that patterns of epistasis depends on the selective forces at play [22], [23].
Here we examine whether epistasis is more common during adaptive evolution by comparing a pair of protein homologs of which only one is subject to a known selection pressure for adaptation. Specifically, we compare nucleoprotein (NP) homologs from human and swine influenza. In both of these influenza lineages, NP has a highly conserved and essential function in the packaging and transcription of viral RNA, and this function is under strong stabilizing selection [24], [25].
Because human influenza circulates in a population of long-lived hosts that are infected with influenza repeatedly during their lifetimes, human influenza is also under constant diversifying selection for adaptive mutations that escape immune memory that accumulates in the host population [26]–[29]. A major way in which human immune memory targets NP is via cytotoxic T lymphocytes (CTLs), and mutations in CTL epitopes are therefore of adaptive value to human influenza [30]–[33]. We have previously shown that the evolution of NP from human influenza involves the fixation of mutations involved in strong epistatic interactions, and that these epistatic mutations occur in epitopes targeted by CTLs [34]. This prior work hints at an association between epistasis and adaptation.
To systematically test the hypothesis that epistasis is enriched during adaptation, here we compare human influenza NP with its swine influenza homolog. Swine influenza is not targeted by human CTLs (CTL epitopes are highly species specific [35], [36]) – so mutations in human CTL epitopes are not of any special significance to swine influenza. Furthermore, swine influenza is unlikely to be under strong diversifying selection even from swine CTLs. In contrast to human influenza, swine influenza circulates in a population of short-lived hosts that have much less opportunity to acquire anti-influenza immune memory before they are slaughtered [37]. As a result, swine influenza is under less pressure to escape from host immune memory. For example, the HA of classical swine influenza underwent minimal antigenic change from 1918 through the late 1990s [37]–[42] – a timeframe during which human influenza HA underwent extremely extensive antigenic change [43], [44]. Although reassortment events and swine vaccination may have recently somewhat increased antigenic change [38]–[40], overall antigenic change in swine influenza is clearly far less than in human influenza [40], [43], [44].
For this reason, the NPs from swine and human influenza represent an ideal pair of homologs for comparative studies of how adaptation affects patterns of epistasis during evolution. While both NPs are under strong stabilizing selection to maintain their essential and conserved biochemical functions [24], [25], only NP from human influenza is under substantial diversifying selection to change sequence epitopes recognized by CTLs. Comparison of the evolution of NPs from these two influenza lineages therefore provides a naturally occurring case study of how ongoing adaptation affects evolutionary patterns.
In the work described below, we first infer evolutionary trajectories for human and swine NP homologs. We then comprehensively mine existing experimental data to define sites in both NP homologs that are targeted by human CTLs. We show that the human NP homolog exhibits an increased frequency of substitutions in these sites relative to the swine NP homolog, a finding consistent with the expectation that mutations to these sites are adaptive only to human influenza. We then experimentally show that the swine NP homolog lacks the type of epistatic mutations that are fixed in the adaptively evolving human NP homolog. Finally, we use our comprehensive analysis of human CTL epitopes to systematically verify that epistatic interactions within the human NP homolog occur at sites that are targeted by CTLs, where mutations are of adaptive value. Overall, these results demonstrate that during NP evolution, epistatically interacting substitutions are enriched during adaptation.
Results
Evolutionary trajectories of NP homologs from human and swine influenza
We set out to compare the evolution of NP homologs from human and swine influenza. Figure 1 shows a phylogenetic tree of NP from human and swine influenza lineages that derive this gene from a common ancestor closely related to the viruses that caused concurrent human and swine pandemics in 1918 [45], [46]. The NP genes of the human influenza lineages in Figure 1 have circulated exclusively in humans since 1918 [45], [46], while the NP genes of the swine influenza lineages in Figure 1 have circulated exclusively in swine since 1918 [38], [47].
Fig. 1. Phylogenetic tree of human and swine NP homologs. 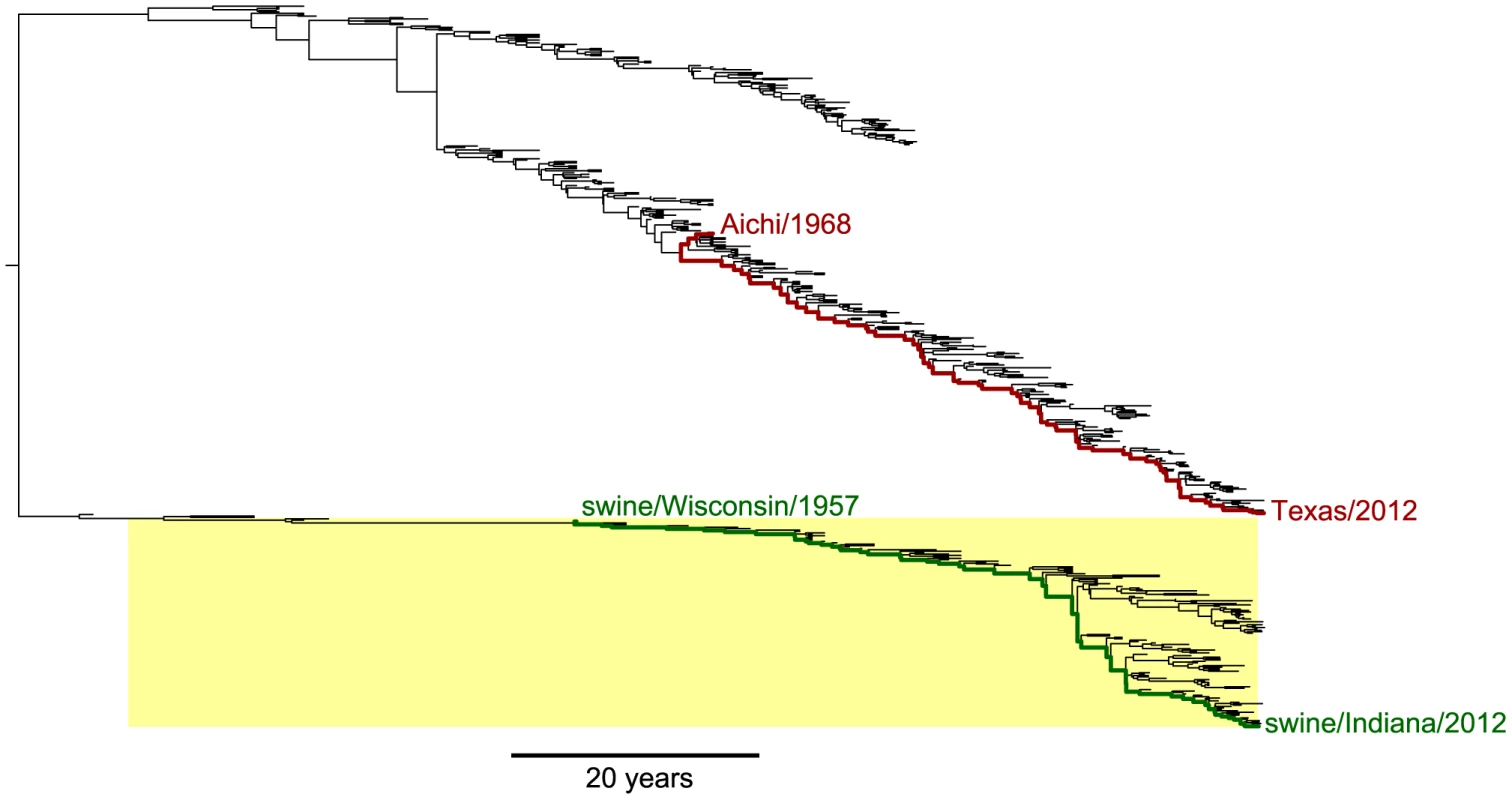
The human and swine NP lineages in this tree are descended from a virus closely related to the 1918 virus. Swine viruses are highlighted in yellow; all other viruses are human. In red are the lines of descent to the human H3N2 strains Aichi/1968 and Texas/2012 from their most-recent common ancestor. In green are the lines of descent to the swine H1N1 strains swine/Wisconsin/1957 and swine/Indiana/2012 from their most-recent common ancestor. Overall, this tree shows NPs from the following lineages: human seasonal H1N1, human H2N2, human H3N2, and North American swine viruses. The tree is a maximum clade credibility summary of a posterior distribution sampled from date-stamped protein sequences using BEAST [51] with a JTT [66] substitution model. See http://jbloom.github.io/mutpath/example_influenza_NP_1918_Descended.html for code, input data, and detailed documentation. Upon transfer into a new host, influenza undergoes a process of adaptation to the ecology, physiology, cell biology and innate immunology of the new host [48]. Because the details of this host adaptation are incompletely understood, we confined our studies to NP homologs that had already been circulating in their respective hosts for several decades. Our expectation is that during these decades of host-specific evolution, the NP homologs will have become highly adapted to the genetically encoded characteristics of their hosts – and that any further adaptation will be driven largely by non-genetic changes in the hosts, such as the acquisition of immune memory due to prior infections.
We therefore focused on the two evolutionary trajectories indicated in Figure 1. For human influenza, we examined the trajectory separating the H3N2 strains A/Aichi/2/1968 and A/Texas/JMM 49/2012. For swine influenza, we examined the trajectory separating the H1N1 strains A/swine/Wisconsin/1/1957 and A/swine/Indiana/A00968365/2012. In both cases, the starting strains for these trajectories meet the criterion specified in the previous paragraph – they are viruses with NPs that have had several decades to adapt to their respective hosts.
In order to map the mutations along these evolutionary trajectories, we utilized a previously described approach [34] for estimating the posterior distribution of mutational paths through protein sequence space by probabilistically placing mutations [49], [50] on trees sampled from a posterior distribution using BEAST [51]. The inferred mutational paths are shown in Figure 2. The human influenza NP accumulated 40 amino-acid mutations along the roughly 44-year trajectory, corresponding to 34 unique mutations relative to the initial Aichi/1968 NP (six mutations are reversions). The swine influenza NP accumulated 18 amino-acid mutations along the roughly 55-year trajectory, corresponding to 18 unique mutations relative to the initial swine/Wisconsin/1957 NP (there are no reversions).
Fig. 2. Evolutionary trajectories of human and swine NP. 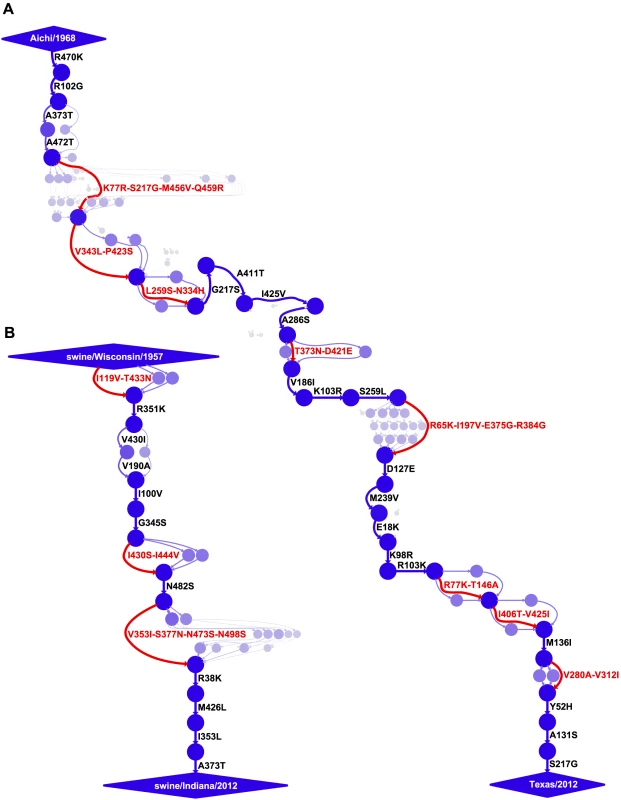
Mutational paths through protein sequence space along (A) the evolutionary trajectory from the human strain Aichi/1968 to Texas/2012 and (B) the evolutionary trajectory from swine/Wisconsin/1957 to swine/Indiana/2012. In the mutational paths, circles represent unique protein sequences, with areas and intensities proportional to the posterior probability that the sequence was part of the trajectory. Blue lines with black labels represent single mutations between sequences, with thicknesses and intensities proportional to the posterior probability that the mutational connection was part of the trajectory. When there is no single high-probability one-mutation connection between sequences, red lines and labels indicate that several mutations fixed in an unknown order. See http://jbloom.github.io/mutpath/example_influenza_NP_1918_Descended.html for code, input data, and detailed documentation. The trajectory in (A) is highly similar to that reported in [34], but is slightly longer and contains sequences from prior to 1968. The inclusion of these pre-1968 sequences is the reason why the first portion of the trajectory is slightly better resolved than that in [34]. We posit that two factors contribute to the slower rate of amino-acid substitution along the swine NP evolutionary trajectory relative to that of the human NP. First, as discussed in the previous section, the swine NP homolog is under less direct selection from immune memory than its human counterpart. Second, the strongest selection on influenza is from antibodies against the viral surface proteins, and so much of NP's sequence evolution is driven by stochastic genetic hitchhiking with adaptive antibody-escape mutations in these surface proteins [27], [52]. The reduced immune selection on these surface proteins in the swine lineage [37]–[43] probably curtails opportunities for similar genetic hitchhiking by mutations to the swine NP homolog. However, it is important to note that NP function is absolutely essential for viral replication in all strains of influenza [24], [25], and that decreases in NP function dramatically impair viral fitness [34]. Therefore, both adaptive and hitchhiking mutations in NP must first satisfy the stringent stabilizing selection for retention of protein function before they have an opportunity to fix.
Human and swine NP possess similar numbers of known human CTL epitopes
In order to examine the association between NP evolution and selection from CTLs, we comprehensively mapped human CTL epitopes in the human and swine influenza NP homologs. Numerous experimental studies have identified epitopes in NP that are targeted by human CTLs (see for example [30], [53]–[57] plus many others). The Immune Epitope Database [58] contains a comprehensive listing of such experimentally characterized epitopes. We created a software package (https://github.com/jbloom/epitopefinder) to systematically parse this database for MHC class I epitopes with an experimentally verified human T-cell response that are between 8 and 12 residues in length and align with no more than one mismatch to NP. We considered epitopes to be present in human influenza NP if they matched to either the Aichi/1968 or Texas/2012 NP, and to be present in swine influenza NP if they matched to either the swine/Wisconsin/1957 or swine/Indiana/2012 NP. We removed redundant epitopes from the same MHC class I gene allele group (see http://hla.alleles.org/nomenclature/naming.html) or from the same supertype [59] if the allele group was not specified.
Figure 3A shows the number of characterized epitopes that contain each site in NP. As can be seen from this figure, the distribution of CTL epitopes is non-uniform along NP's sequence, with some sites falling in many known epitopes and others falling in none. The distributions of epitopes along the NP sequence are highly similar for the human and swine NP homologs. Figure 3B shows the distribution of number of epitopes per site for the human and swine NP homologs. These distributions are nearly indistinguishable (see the Figure 3 legend for statistical testing). Overall, Figure 3 indicates that the human and swine NP homologs contain nearly identical numbers of known human CTL epitopes.
Fig. 3. Human and swine NP possess similar numbers of human CTL epitopes. 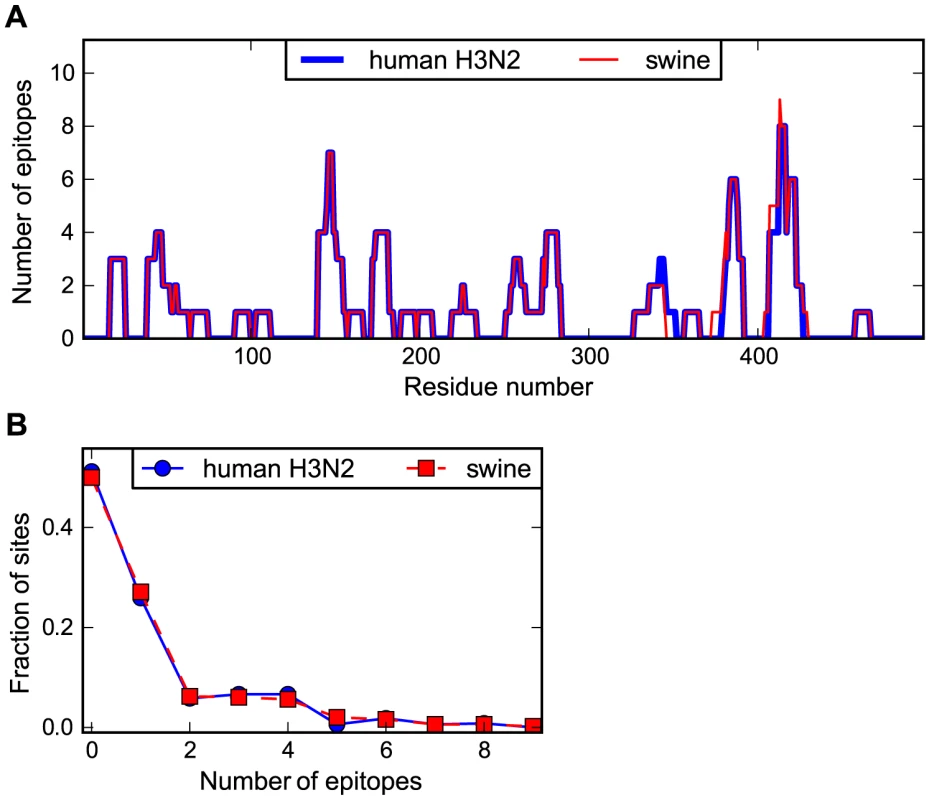
(A) The number of known human CTL epitopes for each residue for human and swine NP. (B) The distribution of number of epitopes per site. The curves in (B) are consistent with the null hypothesis that the human and swine per-site epitope counts are drawn from the same underlying distribution (Kolmogorov-Smirnov test, P = 1.00). The number of epitopes for each site was determined by downloading all human MHC class I epitopes with experimentally verified T-cell responses from the Immune Epitope Database [58], and identifying epitopes between 8 and 12 residues in length that aligned with Aichi/1968 or Texas/2012 (for human NP) or with swine/Wisconsin/1957 or swine/Indiana/2012 (for swine NP) with no more than one mismatch. Redundant epitopes for the same MHC allele were removed. The epitopes per site are listed in Table S1 and Table S2. See http://jbloom.github.io/epitopefinder/example_NP_CTL_epitopes_H3N2_and_swine.html for code, input data, and detailed documentation. Human NP exhibits increased evolution in CTL epitopes relative to swine NP
If the NP from human influenza is under selection from human CTLs, we might expect this to lead to an increased rate of fixation of mutations in CTL epitopes. No such selection is expected to occur for the NP from swine influenza, as swine influenza is definitely not under pressure from human CTLs, and is probably not under strong selection even from swine CTLs for the reasons discussed in the Introduction.
To compare the relative rate of substitution in known CTL epitopes for the two NP homologs, we determined the number of epitopes at the sites of the mutations that fixed along the evolutionary trajectories from Figure 2. As shown in Figure 4, for the human NP homolog, the typical fixed mutation falls in more epitopes than an average site – whereas for the swine NP homolog, the typical fixed mutation falls in fewer epitopes than an average site. We interpret these results as follows: the known epitopes in NP tend to involve sites that are less inherently mutationally tolerant than the average site, either due to a tendency of CTLs to target conserved regions or a bias towards the experimental discovery of epitopes in conserved regions of NP (the tendency of characterized CTL epitopes to fall in conserved regions of viral proteins has also been noted by others [60], [61]). This tendency for the epitopes to fall in less mutationally tolerant regions of NP means that in the absence of CTL selection, the site of the typical fixed mutation contributes to fewer epitopes than an average site – this is the case for the swine NP homolog. But for the human NP homolog, selection for adaptive mutations in sites targeted by CTLs is sufficient to cause the fixed mutations to fall in more epitopes than an average site – and in significantly more epitopes than mutations fixed in the swine NP homolog (P = 0.008, see the Figure 4 legend for statistical testing).
Fig. 4. Human NP exhibits increased evolution in CTL epitopes relative to swine NP. 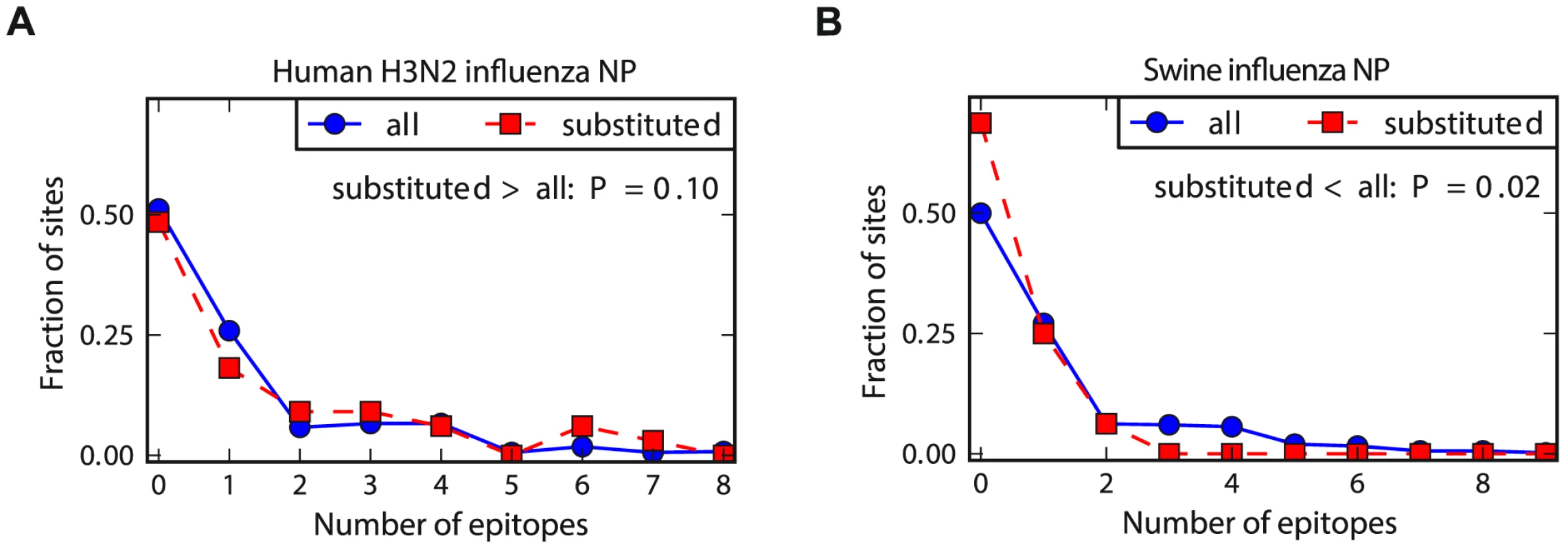
The number of CTL epitopes per site for all sites in NP versus those that substituted along the evolutionary trajectories for (A) human and (B) swine influenza. In human influenza, the substituted sites contain more epitopes than average sites – but in swine influenza, the substituted sites contribute to fewer epitopes than average sites. The P-values on the plots are the fraction of random subsets of all sites that contain as many (human NP) or as few (swine NP) total epitopes as the sites that actually substituted during the natural evolution of that homolog. The hypothesis of greatest interest is whether the substituted sites in the human NP contain more epitopes than do substituted sites in the swine NP. To test this hypothesis, we drew paired random subsets of sites from the human and swine NP homolog of the same size as the actual numbers of substituted sites for each homolog, and determined the fraction of these paired random subsets in which the number of epitopes for the human NP exceeded that for the swine NP by at least as much as for the actual data. This test gives a P-value of 0.008, supporting the hypothesis that human NP exhibits an increased rate of evolution in epitopes relative to swine NP. See http://jbloom.github.io/epitopefinder/example_NP_CTL_epitopes_H3N2_and_swine.html for code, input data, and detailed documentation. Epistatic interactions are fixed in human but not swine NP
The results in the previous section support the idea that there is pressure for adaptive change in human CTL epitopes for human influenza NP, but not for swine influenza NP. The facts discussed in the Introduction also strongly suggest that swine influenza NP is also under much less selection from swine CTLs than human influenza NP is from human CTLs. How do these differences in adaptive pressures influence the prevalence of epistasis during evolution?
We have previously performed a systematic test for a specific form of epistasis in the Aichi/1968 human influenza NP [34]. Specifically, we introduced all single mutations from the human NP evolutionary trajectory (Figure 2A) into the initial Aichi/1968 NP parent sequence, and quantified the effect of the mutations on total transcriptional activity by the influenza polymerase in transfected 293T cells. The previously described results from these experiments are shown in Figure 5A. Three of the 34 single mutations are highly deleterious as individual changes to the Aichi/1968 NP, despite the fact that they eventually fixed during the virus's evolution. We have previously shown that these three individually deleterious mutations were able to fix during NP's natural evolution due to epistatic interactions with other mutations that alleviated their deleterious effects [34].
Fig. 5. Epistatically constrained mutations are fixed in human but not swine NP. 
All single mutations that occurred along the evolutionary trajectories were introduced individually into the Aichi/1968 (human NP) or swine/Wisconsin/1957 (swine NP), and the impact of the mutation on the total transcriptional activity of the influenza polymerase was measured experimentally. (A) The effect of the mutations to human NP, as originally reported in [34]. (B) The effect of the mutations to swine NP. Individual mutations that are strongly deleterious are classified as “epistatically constrained,” since their fixation during natural evolution required additional secondary mutations to counteract the deleterious effects. Three epistatically constrained mutations fixed along the human NP trajectory, but no epistatically constrained mutations fixed along the swine NP trajectory. The epistatically constrained mutations are colored red in the plot. The numerical data in Figure 5A are in [34]; the numerical data in Figure 5B are in Table S3. Do similar epistatic interactions occur during the evolution of the swine influenza NP? To experimentally address this question, we introduced all of the single mutations from the swine NP evolutionary trajectory (Figure 2B) into the initial swine/Wisconsin/1957 NP parent sequence, and quantified the effect on transcriptional activity. These results are shown in Figure 5B. None of the mutations have a substantial deleterious effect as individual changes, indicating that none of them were dependent on epistatic interactions with other mutations. Therefore, while the 44-year evolutionary trajectory of the adaptively evolving human influenza NP involved the fixation of three mutations involved in strong epistatic interactions, we see no evidence of similar epistatically interacting substitutions along a 55-year evolutionary trajectory of the swine influenza NP. We acknowledge that the difference in the numbers of substitutions involved in epistatic interactions (3 out of 34 for human influenza NP, 0 out of 18 for swine influenza NP) is not statistically significant, and therefore merely provides anecdotal support for the idea that epistatically interacting substitutions are more common in the adaptively evolving human NP homolog. However, this anecdotal support becomes much more convincing when combined with the observations in the next section.
Epistasis in human NP occurs at sites enriched in CTL epitopes
Is the presence of epistasis in the human but not the swine influenza NP due to the fact that only the former is adaptively evolving to escape from CTL selection? One way to test this idea is to examine whether the epistatic mutations in the human NP are at sites that contribute disproportionately to CTL escape. We have previously noted that the three epistatically constrained mutations in human NP are in known CTL epitopes [34]. Here we use our new comprehensive mapping of CTL epitopes described above to more thoroughly test the hypothesis that epistasis in the human NP is associated with CTL escape. Figure 6 shows that the epistatic mutations occur at sites that contain significantly more CTL epitopes than either average sites in NP or the set of sites that actually substituted along the evolutionary trajectory. Therefore, not only are epistatically interacting substitutions enriched during the evolution of the adaptively evolving human influenza NP relative to its swine influenza homolog – furthermore, the epistasis involves mutations that play an especially important role in the protein's adaptive evolution.
Fig. 6. Epistasis in human NP occurs at sites enriched in CTL epitopes. 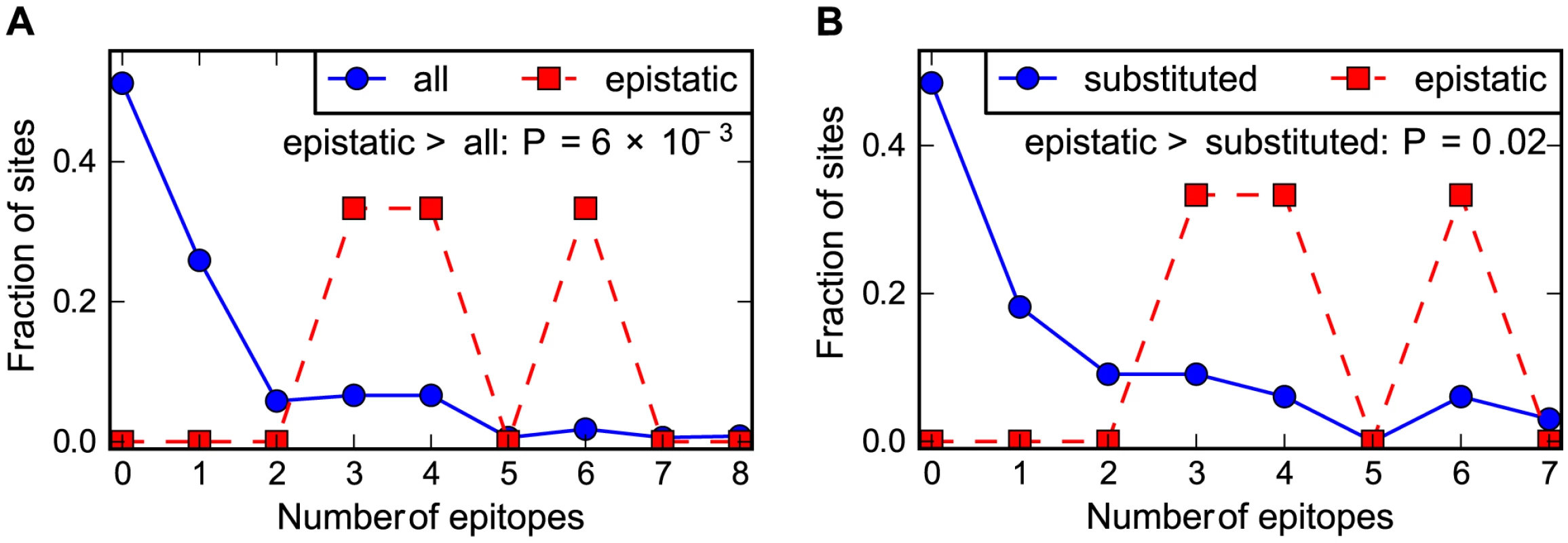
The number of CTL epitopes per site for the sites of the epistatically constrained substitutions in the human influenza NP versus (A) all sites or (B) the full set of sites that substituted along the evolutionary trajectory. The P-values shown on the plots represent the fraction of random subsets that contain as many total epitopes as the actual sites of the epistatically constrained substitutions. See http://jbloom.github.io/epitopefinder/example_NP_CTL_epitopes_H3N2_and_swine.html for code, input data, and detailed documentation. Discussion
We have used a combination of computational and experimental analyses to examine whether epistasis is more common during adaptive protein evolution. We did this by comparing the evolution of an adaptively evolving NP from human influenza with a closely related homolog from swine influenza that is not under similar pressure for adaptive change. Experimentally, we find that strong epistatic interactions are fixed only during the evolution of the adaptively evolving human influenza NP homolog. Our computational analyses strongly suggest that the different patterns of epistasis are due to the fact that only the human influenza NP homolog is undergoing continuing adaptive evolution. Specifically, mutations that fix in the human influenza NP are significantly more likely to be in sites targeted by human immune memory than are mutations in the swine influenza homolog – and the epistatic interactions all involve sites that are heavily targeted by such immune selection. Overall, these results suggest that epistatically interacting substitutions are significantly enriched in adaptive versus non-adaptive evolution.
Why are epistatically interacting substitutions more prevalent during adaptive evolution? Our experiments probe for epistatic interactions involving a mutation that is individually deleterious but becomes neutral or adaptive when paired with secondary mutations. As discussed in the Introduction, there are two mechanisms by which such epistatic interactions have been shown to fix during adaptive evolution: compensatory mutations and permissive mutations. Our prior work suggests that the epistatic mutations in human influenza NP fix primarily via the latter mechanism, although compensatory mutations may also play a lesser role [34]. Crucially, the driving force for both mechanisms is adaptation. For the compensatory-mutation mechanism, this driving force is obvious: an initial deleterious mutation is more likely to persist long enough to be paired with a compensatory mutation if the initial mutation also confers some adaptive benefit (although mildly deleterious mutations can also fix without compensation, albeit at a lower rate). Somewhat less obviously, a similar force drives the permissive-mutation mechanism: although the initial permissive change is stochastic, the fixation of its subsequent pairing with the mutation that it permits is more likely if the latter change is adaptive [21]. Although epistatically interacting mutations can fix during non-adaptive evolution by similar temporal mechanisms, there is no underlying force to favor these relatively rare epistatic combinations over more abundant and easily accessible non-epistatic mutations.
This explanation can be stated more succinctly in terms specific to the NP homologs studied here. In the absence of adaptation, evolution tends to fix easily accessible non-epistatic mutations that have no adverse effect – in other words, the evolution of the swine influenza NP is dominated by stabilizing selection for retention of function. The human influenza NP is also under strong stabilizing selection for retention of function, but in addition experiences diversifying selection for change in immune epitopes. Some of these adaptive immune-escape mutations have adverse effects on NP function, and so selection biases evolution towards epistatic combinations that enable the adaptive mutations to fix while retaining NP function.
Most experimental studies of epistasis have focused on its role in constraining adaptation [1]–[10]. Our results suggest that caution may be warranted in extrapolating findings about the frequency of epistatically interacting substitutions during adaptation to more general evolutionary scenarios, since such substitutions appear to be more common during adaptive than non-adaptive evolution.
Materials and Methods
Phylogenetic tree and mutational paths
The input sequences for construction of the phylogenetic tree (Figure 1) and mutational paths (Figure 2) were downloaded from the Influenza Virus Resource [62]. For human influenza, up to 5 sequences per year were retained from the following lineages: H1N1 (isolation dates from 1918 to 1957, and then from 1977 to 2008), H2N2 (isolation dates from 1957 to 1968), and H3N2 (isolation dates from 1968 to 2012). For swine influenza, up to 5 sequences per year and subtype were retained from North American swine influenza. For the human H1N1 isolated in 1977 or later, 24 years were subtracted from the isolation dates because these sequences are from an influenza lineage revived after being frozen for roughly 24 years [45]. We excluded sequences that were classified as mis-annotated by [63] or that are strong outliers from the molecular clock based on an analysis with RAxML [64] and Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/).
The sequences were translated, date-stamped, and used as input to BEAST [65] with a strict molecular clock, a JTT [66] model of substitution, and a relatively loose coalescent-based prior on the tree. Figure 1 shows a maximum clade credibility tree rendered with FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
The mutational paths in Figure 2 were constructed using the approach described in [34], and were rendered using GraphViz (http://www.graphviz.org/).
The source code, input data, and detailed documentation for the construction of the phylogenetic tree and the mutational paths can be accessed on GitHub via http://jbloom.github.io/mutpath/example_influenza_NP_1918_Descended.html
Mapping of CTL epitopes
The CTL epitopes were identified by downloading from the Immune Epitope Database [58] all epitopes with a positive T-cell response with source organism Influenza A virus and host Homo sapiens. We created a new software package, epitopefinder (https://github.com/jbloom/epitopefinder), to map specific epitopes to NP.
This mapping was done by parsing all MHC class I peptide epitopes of 8 to 12 residues, and removing as redundant any epitopes that overlapped by 8 or more residues and were from the same MHC class I allele group (see http://hla.alleles.org/nomenclature/naming.html) or from the same MHC class I supertype [59] if no allele group was specified. For redundant epitopes, the shortest epitope sequence was retained. The non-redundant epitopes were aligned to NP: if they aligned to Aichi/1968 or Texas/2012 with no more than one mismatch then they were considered to be present in the human NP homolog, and if they aligned with no more than one mismatch to swine/Wisconsin/1957 or swine/Indiana/2012 with no more than one mismatch then they were considered to be present in the swine NP homolog. The number of epitopes in which each site participates is listed in Tables S1 and S2.
The source code, input data, and detailed documentation for mapping the epitopes and for the computing the P-values can be accessed on GitHub via http://jbloom.github.io/epitopefinder/example_NP_CTL_epitopes_H3N2_and_swine.html
Experimental assays of NP function
We measured the function of the NP mutants by using flow cytometry to quantify the mean fluorescent intensity of 293T cells 20 hours after they had been transfected with plasmids encoding the NP variant in question, the three influenza polymerase proteins (PB2, PB1, PA), and the fluorescent reporter pHH-PB1flank-eGFP [67]. The data for the human NP homolog in Figure 5A were originally described in [34], and are reprinted here.
The data for the swine NP homolog in Figure 5B were generated by following the protocol described in [34] with the following modifications: the polymerase proteins were derived from the A/California/4/2009 swine-origin H1N1 strain, and the measured signal was normalized to that obtained using the wild-type swine/1957 NP. The polymerase plasmids (pHWCA09tc-PB2, pHWCA09tc-PB1, and pHWCA09tc-PA) have been described previously [68], while the insert for the swine/1957 NP plasmid (pHWswine57-NP) was synthesized commercially and cloned into pHW2000 [69]; the viral-RNA sequences for all four plasmids are in Dataset S1. The A/California/4/2009 swine-origin H1N1 polymerase proteins were chosen because the NP of this strain is closely related to NPs from the latter part of the swine influenza trajectory in Figure 1. We verified that the NP plasmid concentration used in [34] gave signal that was near the midpoint of the assay's dynamic range when using this combination of NP and polymerase genes (Figure S1). The data in Figure 5B represent the mean and standard error of at least three independent replicates; numerical values are in Table S3.
Supporting Information
Zdroje
1. ChouHH, ChiuHC, DelaneyNF, SegreD, MarxCJ (2011) Diminishing Returns Epistasis Among Beneficial Mutations Decelerates Adaptation. Science 332 : 1190–1192.
2. BlountZD, BorlandCZ, LenskiRE (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A 105 : 7899–7906.
3. KhanAI, DinhDM, SchneiderD, LenskiRE, CooperTF (2011) Negative Epistasis Between Beneficial Mutations in an Evolving Bacterial Population. Science 332 : 1193–1196.
4. SchenkMF, SzendroIG, SalverdaML, KrugJ, de VisserJA (2013) Patterns of Epistasis between beneficial mutations in an antibiotic resistance gene. Mol Biol Evol 30 : 1779–1787.
5. WeinreichDM, DelaneyNF, DepristoMA, HartlDL (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312 : 111–114.
6. BeadleBM, ShoichetBK (2002) Structural bases of stability-function tradeoffs in enzymes. J Mol Biol 321 : 285–296.
7. BershteinS, SegalM, BekermanR, TokurikiN, TawfikDS (2006) Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444 : 929–932.
8. OrtlundEA, BridghamJT, RedinboMR, ThorntonJW (2007) Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317 : 1544–1548.
9. BridghamJT, OrtlundEA, ThorntonJW (2009) An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461 : 515–519.
10. BloomJD, LabthavikulST, OteyCR, ArnoldFH (2006) Protein stability promotes evolvability. Proc Natl Acad Sci U S A 103 : 5869–5874.
11. BartonNH (2000) Genetic hitchhiking. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 355 : 1553–1562.
12. ChenRB, HolmesEC (2010) Hitchhiking and the Population Genetic Structure of Avian Influenza Virus. Journal of molecular evolution 70 : 98–105.
13. Kimura M (1983) The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge University Press.
14. KingJL, JukesTH (1969) Non-Darwinian evolution. Science 164 : 788–798.
15. LangGI, RiceDP, HickmanMJ, SodergrenE, WeinstockGM, et al. (2013) Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500 : 571–574.
16. LynchM (2007) The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A 104 Suppl 1 : 8597–8604.
17. NeiM (2005) Selectionism and neutralism in molecular evolution. Mol Biol Evol 22 : 2318–2342.
18. SiderakiV, HuangW, PalzkillT, GilbertHF (2001) A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci U S A 98 : 283–288.
19. WangX, MinasovG, ShoichetBK (2002) Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol 320 : 85–95.
20. CovertAW3rd, LenskiRE, WilkeCO, OfriaC (2013) Experiments on the role of deleterious mutations as stepping stones in adaptive evolution. Proc Natl Acad Sci U S A 110: E3171–3178.
21. DraghiJA, ParsonsTL, PlotkinJB (2011) Epistasis increases the rate of conditionally neutral substitution in an adapting population. Genetics 187 : 1139–1152.
22. DraghiJA, PlotkinJB (2013) Selection biases the prevalence and type of epistasis along adaptive trajectories. Evolution; international journal of organic evolution 67 : 3120–3131.
23. SzendroIG, SchenkMF, FrankeJ, KrugJ, de VisserJAGM (2013) Quantitative analyses of empirical fitness landscapes. J Stat Mech 2013: P01005 doi:10.1088/1742-5468/2013/01/P01005
24. PortelaA, DigardP (2002) The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol 83 : 723–734.
25. YeQ, KrugRM, TaoYJ (2006) The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444 : 1078–1082.
26. SmithDJ, LapedesAS, de JongJC, BestebroerTM, RimmelzwaanGF, et al. (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305 : 371–376.
27. RambautA, PybusOG, NelsonMI, ViboudC, TaubenbergerJK, et al. (2008) The genomic and epidemiological dynamics of human influenza A virus. Nature 453 : 615–619.
28. GerhardW, YewdellJ, FrankelME, WebsterR (1981) Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290 : 713–717.
29. WileyDC, WilsonIA, SkehelJJ (1981) Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289 : 373–378.
30. RimmelzwaanGF, BoonAC, VoetenJT, BerkhoffEG, FouchierRA, et al. (2004) Sequence variation in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. Virus Res 103 : 97–100.
31. BerkhoffEG, BoonAC, NieuwkoopNJ, FouchierRA, SintnicolaasK, et al. (2004) A mutation in the HLA-B*2705-restricted NP383-391 epitope affects the human influenza A virus-specific cytotoxic T-lymphocyte response in vitro. J Virol 78 : 5216–5222.
32. BerkhoffEG, Geelhoed-MierasMM, FouchierRA, OsterhausAD, RimmelzwaanGF (2007) Assessment of the extent of variation in influenza A virus cytotoxic T-lymphocyte epitopes by using virus-specific CD8+ T-cell clones. J Gen Virol 88 : 530–535.
33. ValkenburgSA, RutiglianoJA, EllebedyAH, DohertyPC, ThomasPG, et al. (2011) Immunity to seasonal and pandemic influenza A viruses. Microbes and infection/Institut Pasteur 13 : 489–501.
34. GongLI, SuchardMA, BloomJD (2013) Stability-mediated epistasis constrains the evolution of an influenza protein. eLife 2: e00631.
35. RenardC, HartE, SehraH, BeasleyH, CoggillP, et al. (2006) The genomic sequence and analysis of the swine major histocompatibility complex. Genomics 88 : 96-+.
36. AdamsEJ, ParhamP (2001) Species-specific evolution of MHC class I genes in the higher primates. Immunological Reviews 183 : 41–64.
37. SheerarMG, EasterdayBC, HinshawVS (1989) Antigenic conservation of H1N1 swine influenza viruses. J Gen Virol 70 (Pt 12) 3297–3303.
38. VincentAL, MaW, LagerKM, JankeBH, RichtJA (2008) Swine influenza viruses a North American perspective. Advances in virus research 72 : 127–154.
39. VincentAL, LagerKM, MaW, LekcharoensukP, GramerMR, et al. (2006) Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Veterinary microbiology 118 : 212–222.
40. GartenRJ, DavisCT, RussellCA, ShuB, LindstromS, et al. (2009) Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325 : 197–201.
41. LuohSM, McGregorMW, HinshawVS (1992) Hemagglutinin mutations related to antigenic variation in H1 swine influenza viruses. J Virol 66 : 1066–1073.
42. NobleS, McGregorMS, WentworthDE, HinshawVS (1993) Antigenic and genetic conservation of the haemagglutinin in H1N1 swine influenza viruses. J Gen Virol 74 (Pt 6) 1197–1200.
43. WeiCJ, BoyingtonJC, DaiK, HouserKV, PearceMB, et al. (2010) Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med 2 : 24ra21.
44. BedfordT, SuchardMA, LemeyP, DudasG, GregoryV, et al. (2013) Integrating influenza antigenic dynamics with molecular evolution. Elife 3: e01914 doi:10.7554/eLife.01914
45. dos ReisM, HayAJ, GoldsteinRA (2009) Using non-homogeneous models of nucleotide substitution to identify host shift events: application to the origin of the 1918 ‘Spanish’ influenza pandemic virus. Journal of molecular evolution 69 : 333–345.
46. MorensDM, TaubenbergerJK, FauciAS (2009) The persistent legacy of the 1918 influenza virus. N Engl J Med 361 : 225–229.
47. Brockwell-StaatsC, WebsterRG, WebbyRJ (2009) Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influenza and other respiratory viruses 3 : 207–213.
48. TaubenbergerJK, KashJC (2010) Influenza virus evolution, host adaptation, and pandemic formation. Cell host & microbe 7 : 440–451.
49. MininVN, SuchardMA (2008) Counting labeled transitions in continuous-time Markov models of evolution. Journal of mathematical biology 56 : 391–412.
50. O'BrienJD, MininVN, SuchardMA (2009) Learning to count: robust estimates for labeled distances between molecular sequences. Mol Biol Evol 26 : 801–814.
51. DrummondAJ, SuchardMA, XieD, RambautA (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29 : 1969–1973.
52. BhattS, HolmesEC, PybusOG (2011) The genomic rate of molecular adaptation of the human influenza A virus. Mol Biol Evol 28 : 2443–2451.
53. DiBrinoM, ParkerKC, MarguliesDH, ShiloachJ, TurnerRV, et al. (1995) Identification of the peptide binding motif for HLA-B44, one of the most common HLA-B alleles in the Caucasian population. Biochemistry 34 : 10130–10138.
54. VoetenJT, BestebroerTM, NieuwkoopNJ, FouchierRA, OsterhausAD, et al. (2000) Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J Virol 74 : 6800–6807.
55. AssarssonE, BuiHH, SidneyJ, ZhangQ, GlennJ, et al. (2008) Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol 82 : 12241–12251.
56. AlexanderJ, BilselP, del GuercioMF, Marinkovic-PetrovicA, SouthwoodS, et al. (2010) Identification of broad binding class I HLA supertype epitopes to provide universal coverage of influenza A virus. Human immunology 71 : 468–474.
57. CheungYK, ChengSC, KeY, XieY (2012) Human immunogenic T cell epitopes in nucleoprotein of human influenza A (H5N1) virus. Hong Kong medical journal 18 Suppl 2 : 17–21.
58. VitaR, ZarebskiL, GreenbaumJA, EmamiH, HoofI, et al. (2010) The immune epitope database 2.0. Nucleic Acids Res 38: D854–862.
59. SidneyJ, PetersB, FrahmN, BranderC, SetteA (2008) HLA class I supertypes: a revised and updated classification. BMC immunology 9 : 1.
60. da SilvaJ, HughesAL (1998) Conservation of cytotoxic T lymphocyte (CTL) epitopes as a host strategy to constrain parasite adaptation: evidence from the nef gene of human immunodeficiency virus 1 (HIV-1). Mol Biol Evol 15 : 1259–1268.
61. HertzT, NolanD, JamesI, JohnM, GaudieriS, et al. (2011) Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J Virol 85 : 1310–1321.
62. BaoY, BolotovP, DernovoyD, KiryutinB, ZaslavskyL, et al. (2008) The influenza virus resource at the National Center for Biotechnology Information. J Virol 82 : 596–601.
63. KrasnitzM, LevineAJ, RabadanR (2008) Anomalies in the influenza virus genome database: new biology or laboratory errors? J Virol 82 : 8947–8950.
64. StamatakisA (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 : 2688–2690.
65. DrummondAJ, RambautA (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology 7 : 214.
66. JonesDT, TaylorWR, ThorntonJM (1992) The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences : CABIOS 8 : 275–282.
67. BloomJD, GongLI, BaltimoreD (2010) Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328 : 1272–1275.
68. BloomJD, NayakJS, BaltimoreD (2011) A computational-experimental approach identifies mutations that enhance surface expression of an oseltamivir-resistant influenza neuraminidase. PLoS ONE 6: e22201.
69. HoffmannE, NeumannG, KawaokaY, HobomG, WebsterRG (2000) A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97 : 6108–6113.
Štítky
Genetika Reprodukční medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
- Prof. Petr Urbánek: Potřebujeme najít pacienty s nediagnostikovanou akutní intermitentní porfyrií
-
Všechny články tohoto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



