-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSuicidal Autointegration of and Transposons in Eukaryotic Cells
Transposons (“jumping genes”) are ubiquitous, mobile genetic elements that make up significant fraction of genomes, and are best described as molecular parasites. During ‘cut and paste’ transposition, the excised transposon relocates from one genomic location to another. Here we focus on the molecular events following excision of two eukaryotic DNA transposons, Sleeping Beauty and piggyBac. Both transposons are primarily used in a cellular environment that is different from their original hosts, thereby offering a new model to study host-parasite interaction in higher organisms. In the last decade, they have been developed into a technology platform for vertebrate genetics, including gene discovery, transgenesis, gene therapy and stem cell manipulation. Despite the wide range of their application, relatively little is known about their molecular mechanism in vertebrates. We show that these elements are not capable of self-avoidance, as a significant portion of the excised transposons integrates into its own genome in a suicidal process. Despite mechanistic differences, both transposons are affected similarly, and larger transposons are particularly vulnerable. We propose that transposons might recruit phylogenetically conserved cellular factors in a new host that protects against self-disruption. Suboptimal conditions in a new environment could generate abnormal, genotoxic transposition reactions, and should be monitored.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004103
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004103Summary
Transposons (“jumping genes”) are ubiquitous, mobile genetic elements that make up significant fraction of genomes, and are best described as molecular parasites. During ‘cut and paste’ transposition, the excised transposon relocates from one genomic location to another. Here we focus on the molecular events following excision of two eukaryotic DNA transposons, Sleeping Beauty and piggyBac. Both transposons are primarily used in a cellular environment that is different from their original hosts, thereby offering a new model to study host-parasite interaction in higher organisms. In the last decade, they have been developed into a technology platform for vertebrate genetics, including gene discovery, transgenesis, gene therapy and stem cell manipulation. Despite the wide range of their application, relatively little is known about their molecular mechanism in vertebrates. We show that these elements are not capable of self-avoidance, as a significant portion of the excised transposons integrates into its own genome in a suicidal process. Despite mechanistic differences, both transposons are affected similarly, and larger transposons are particularly vulnerable. We propose that transposons might recruit phylogenetically conserved cellular factors in a new host that protects against self-disruption. Suboptimal conditions in a new environment could generate abnormal, genotoxic transposition reactions, and should be monitored.
Introduction
Mobilization of transposable elements (TEs) is a DNA recombination reaction that can occur either via RNA (retroelement/retrovirus) or DNA intermediates (DNA transposon). In non-replicative, ‘cut and paste’ DNA transposition, the excised transposon relocates from one genomic location to another. In contrast, the ‘copy and paste’ mobilization of a retroelement/retrovirus does not include the excision step, but the downstream events of retroviral integration are highly similar to DNA transposition [1] Many DNA transposons are bracketed by terminal inverted repeats (IRs) that contain binding sites for the recombinase, the transposase. The transposition process is catalysed by the transposase, and can be divided into four steps: (i) the transposase recognizes and binds to the ends of the transposon; (ii) the transposase and two transposon ends form a complex called synaptic or paired end complex; (iii) the transposon is excised from the donor site; and (iv) the excised transposon is transferred to a new location by the transposase reviewed in [2].
TEs are ubiquitous components of both prokaryotic and eukaryotic genomes [3] Even though TEs are best viewed as molecular parasites that propagate themselves using resources of the host cells, their long-term coexistence with their host has provided ample examples of mutual adaptation. The mobility of TEs is regulated by diverse molecular mechanisms, and can be achieved by self-limiting regulatory features intrinsic to the TE itself [4] or mechanisms provided by the host cell. For example, the RNA interference (RNAi) machinery in eukaryotes is probably the best-known cellular mechanism that evolved to control transposition [5], [6]. Notably, generally little is known about the regulation of DNA transposons in eukaryotes. Indeed, our understanding of the mechanisms and the regulation of transposition in eukaryotes are mostly based on assuming analogies to bacterial transposons [2], [7], [8].
In the last decade, the DNA transposition of Sleeping Beauty (SB), a resurrected fish transposon [9] was intensively studied [10]–[13]. Using SB as a model to study host-transposon interaction in eukaryotic cells, a series of evolutionarily conserved (from fish to human) cellular determinants has been identified. HMGB1, a non-histone chromatin factor, is required for synaptic complex formation during SB transposition [11]. Factors of the non-homologous-end-joining (NHEJ) pathway of double strand DNA break (DSB) repair, including Ku70 and the DNA-dependent protein kinase (DNA-PKcs) are required for SB transposition by acting at repairing the transposon excision sites [10]. Through its association with Myc-interacting zinc finger protein 1 (ZBTB17 or Miz1), the SB transposase down-regulates cyclin D1 expression in human cells, resulting in a cell cycle slowdown [12]. A temporary G1 arrest enhances transposition, suggesting that SB transposition is favoured in the G1 phase of the cell cycle, where NHEJ is preferentially active [10]. The HMG-box transcription factor HMGXB4 (HMG2L1), a component of the Wnt-signaling pathway is involved in a feedback regulation of SB transposase expression [13]. These studies indicate that eukaryotic transposons can participate in a complex interactive regulatory platform involving evolutionary conserved cellular mechanisms.
Although, SB is a relatively well-characterised eukaryotic transposon, one part of the transposition reaction, the step following excision but prior reintegration, is yet unexplored. In the process of productive transposition, the excised molecule integrates into a new genomic location. However, in principle, the excised transposon molecule could reinsert, in a self-disruptive process, into its own genome. This suicidal transposition event is called autointegration, self-integration or intramolecular transposition, and is well characterized in prokaryotes [14]–[16]. The best-understood example in bacteria is Tn10 transposition, in which regulation of transposition is a delicate interplay between the transposon and host-encoded factors [17]–[19]. These host factors, namely IHF (integration host factor), HU (heat unstable nucleoid protein) and H-NS (nucleoid structuring protein) are among the most important regulatory factors in E. coli. IHF and HU stimulate the early steps of transposition prior to excision of Tn10 [19]. However, if they remain associated with the transpososome (a DNA-protein complex minimally containing the excised transposon and the transposase), they promote autointegration [7]. By opposing the effects of IHF [20] and HU, H-NS inhibits autointegration and promotes productive transposition [18], [19]. In eukaryotes, autointegration was reported in mariner transposition [21], [22] Curiously, one third of the autointegration events mediated by Mos1 (mariner) were recovered from non-canonical target sites [22]. Self-disruptive autointegration has also been observed during retroviral integration [23]–[25]. A host-encoded protein, barrier-to-autointegration factor (BANF1 or BAF) has been identified by its ability to protect retroviruses from autointegration [23].
Two observations suggest that, similarly to bacterial transposons and retroviruses, autointegration could be a significant factor affecting productive DNA transposition in eukaryotes as well. First, similarly to certain bacterial DNA transposons [26], [27], transposition of SB from a genomic locus frequently occurs into sites that are close to the donor locus [28]; this phenomenon is termed “local hopping”. Obviously, the transposon itself is the closest target to integrate. In Tn10 transposition, the host factor IHF promotes ‘target site channelling’ close to the IR of the transposon [19]. Second, larger transposons are expected to be particularly attractive targets for autointegration. Indeed, it has been observed that, similarly to certain bacterial TEs, longer elements of SB tend to transpose less efficiently [29], [30]. Thus, both ‘local hoping’ and size-sensitivity might be associated with vulnerability of SB transposition to self-integration.
In the present study, we investigated the post-excision fate of two DNA transposons, SB [9] and piggyBac (PB) [31] in vertebrate cells. Although, both SB and PB belong to the superfamily of DDE/D transposases, characterized by a highly conserved catalytic domain [1], they exhibit significant differences in their mechanisms of transposition [32], [33]. For example, the activity of SB is essentially restricted to vertebrates [29], [34], with the exception of a chordate, Ciona intestinalis [35]. By contrast, PB seems to have an extremely wide host range as it can transpose in insects as well as in human cells [36]–[38]. In comparison to PB, SB was reported to exhibit a much stronger ‘local hopping’ phenotype [39], [40]. Furthermore, SB, but not PB was reported to be sensitive to the size of the mobilized element. Specifically, the transposition of PB was reported to be independent on the size of the element below 14 kb [41].
Importantly, both SB and PB are valuable genomic tools for genome manipulation [42], and mostly used in heterologous cellular environments, thereby offering a unique opportunity to investigate various survival strategies of DNA elements in eukaryotes. Indeed, we can model how these elements behave in naïve genomes, and adapt to their new environment. We have used a simple experimental setup, i. e., transfection into cultured cells to monitor the process of establishing a host-parasite relationship in a heterologous environment. This strategy identified BANF1 as a host-encoded factor influencing this process. We propose that deciphering the mechanism and regulation of transposon reactions and translating this knowledge can be effectively used to derive transposon-based genetic tools for genome manipulation or for gene therapy.
Results
Self-destructive autointegration events mediated by the Sleeping Beauty transposase
To detect and characterise potential autointegration products, the following assay system was established. The test construct, SBrescue, is a plasmid comprising a replication origin (Ori) and an antibiotic resistance cassette for zeocin (Zeo) located between the IRs of the transposon (Figure 1A). Outside of the transposon SBrescue contains the rpsL gene rendering bacteria sensitive to streptomycin [43]. SBrescue and the helper plasmid encoding for the transposase are co-transfected into cells. Plasmid DNA is recovered from the cells two days post-transfection and transformed into E. coli. Bacteria are subjected to double antibiotic selection of zeocin and streptomycin (Figure 1B). Following transposon excision and circularization of the excised transposon, the rpsL is lost, thereby rendering bacteria StrepR (Figure 1B). Autointegrative transposition events can be rescued in the form of either two deletion circles or a single inversion circle, depending on the topology of the strand attack (Figure 1C). The assay can detect autointegration events occurring into regions designated A, B, C and IR (Figure 1A). In addition, integration events into the rpsL gene would render bacteria resistant to streptomycin and recovered by the assay. In contrast, autointegration events into Zeo or Ori would not be detectable with the assay system, because these regions are required for plasmid propagation and maintenance.
Fig. 1. Autointegration of SB transposon. 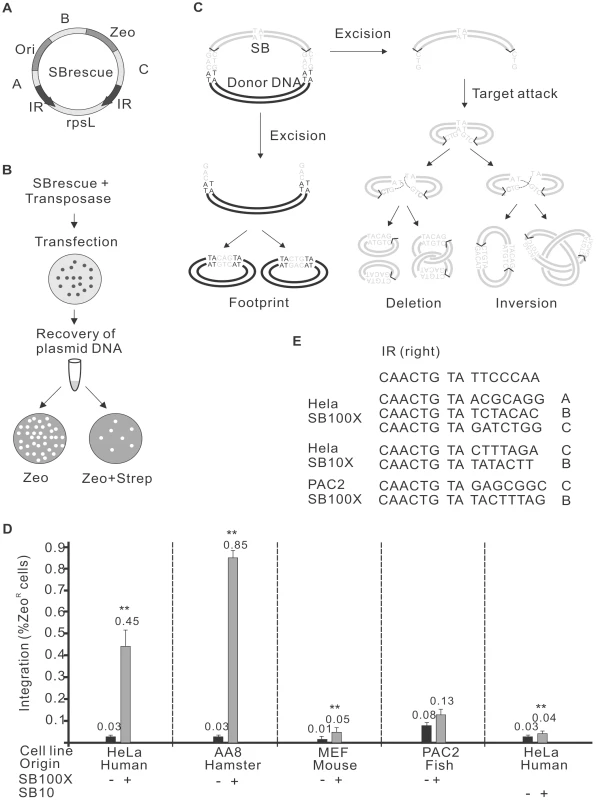
A. The structure of the SBrescue construct. The SBrescue contains an SB transposon carrying an origin of replication (Ori) and a zeocin gene (Zeo). The backbone DNA encodes a streptomycin sensitive gene rpsL (gray box). Bacteria carrying autointegration products with intact Ori and Zeo and integrations disrupting rpsL function (regions of A, B, C, IR and rpsL) could be rescued following double antibiotic selection of zeocin and streptomycin. Black arrow: inverted repeat (IR). B. Flowchart of the autointegration assay. SBrescue and helper plasmids encoding for the transposase are cotransfected into cells. Two days postransfection, low-molecular weight (plasmid) DNA is recovered from cells and transformed into bacteria. Bacteria were subjected to a selection of either zeocin or a double selection of zeocin/streptomycin. Frequency is calculated as zeoR/StrepR normalized by zeoR. The assay would capture circularized molecules generated in eukaryotic cells, while linear DNA degrades in bacteria. C. A model of SB autointegration. The excision and reintegration steps of autointegration are similar to canonical transposition. Excision; The transposition initiates with a staggered cut. The SB transposon (gray lines) is separated from the donor DNA (black lines) by the transposase. Autointegration; the transposon attacks a target site (TA) within the transposon. The autointegration products can be either two deletion circles (left) or inversion products, knotted or unknotted (right). For details see [26]. In the inversion products the orientation of the IRs would be different from the donor substrate and would contain two ends of the transposon and target site duplications. The host DNA repair machinery would repair the single stranded gaps at the integration site and the double-strand breaks at the excision site [10]. The excision site repair products (also called “footprints”) can be either CAG or CTG (gray). The backbone DNA does not have Ori and would exist only transiently in bacteria. D. Frequency of SB autointegration events in various vertebrate cells HeLa (human), AA8 (Chinese hamster), MEF (mouse embryonic fibroblast) and PAC2 (zebrafish) cells using either SB100X [44] or SB10 [9] transposases. The statistical significance of differences is shown by asterisk above the bars, **P<0.01. E. Confirmation of autointegration events by sequencing the de novo target sites. Sequences from the donor construct (bold) right-IR (6 bp), TA target sites (bold, italic) and 3′ flanking DNA (7 bp) are shown. The location of the targeted region is shown in the right side. To identify conditions affecting autointegration of SB, the following factors were considered: (a) cell type specificity; (b) transposase activity; (c) target site distribution; (d) the size of the transposon; (e) host-transposon interaction. First, SBrescue was introduced into human HeLa cells with or without a helper plasmid expressing the hyperactive SB100X transposase [44] (Figure 1B). Compared to the control (0.03%, 1.19×103/3.88×106), significantly elevated numbers (0.45%, 4×103/9.09×105) of ZeoR/StrepR bacterial colonies were observed when SB100X transposase was present in the experiments (Figure 1D). To characterize potential autointegration events and map the transposon insertion sites, the recovered products were subjected to DNA sequencing. Sequencing data confirmed that similarly to productive transposition, the autointegration events of SB transposition were targeted into TA dinucleotides within the mappable A, B, C and IR regions of the transposon (Figure 1E). To investigate if cellular factors in various vertebrate species might differentially promote or protect against autointegration of SB, the assay was performed in cultured cells of different origin, including AA8 (Chinese hamster, ovarian; 0.85% vs 0.03%, 1.51×103/1.77×105 vs 476/5.52×106), MEF (mouse, embryonic fibroblast; 0.05% vs 0.01%, 8.06×103/1.71×107 vs 332/9.54×105) and PAC2 (zebrafish, fibroblast; 0.13% vs 0.08%, 1.03×103/8.42×105 vs 770/9.69×105) cells (Figure 1D). Our results revealed that the SB-mediated autointegration events were detectable in all tested cell lines, including fish, the natural cellular environment of SB (Figure 1D). Similarly to productive transposition, the frequency of autointegration varied in the different cell types [29]. The highest frequencies of autointegration were detected in HeLa and AA8 cells that generally support efficient transposition [29], suggesting that the frequency of autointegration was primarily dependent on the activity of the transposase, rather than the cell type (Figure 1D). Indeed, compared to the original SB10 transposase [9], autointegration by the hyperactive SB100X transposase [44] was higher by one order of magnitude in human HeLa cells.
Remobilization of the SB transposon from a genomic donor site exhibits a significant bias toward the donor locus (local hopping) [32]. Similarly, the reintegration of Tn10 transposons is not unbiased and targeted to the IRs of the transposon during autointegration, referred as ‘target site channelling’ [19]. In contrast, when launched from an extrachromosomal donor molecule, the genomic distribution of SB insertion sites is fairly random [45]–[47]. Target site selection during transposition of SB from an extrachromosomal plasmid is primarily determined on the level of DNA structure, as insertion sites tend to have a palindromic pattern and a bendable structure [45]. Accordingly, the insertion profile of the SB transposon can be modelled by determining the DNA-deformability scores, called Vstep for each potential TA target site, using the software ProTIS [48]. To determine the autointegration profile of SB, Vstep values were generated for the mappable regions of SBrescue and the observed insertion frequencies were compared to the calculated Vstep values (Figure 2B). Altogether, 53 autointegration products were identified and mapped to the regions of IR, A, B and C. Most of the autointegration events occurred into region B that is farther away from the IRs, and relatively few into regions A and C that are closer to the transposon ends (Figure 2). In regions B and C, there was a correlation between insertion frequencies and Vstep scores (Figure 2B). These results suggest that similarly to transposition from an extrachromosomal donor, insertion site selection during autointegration of SB is largely independent from the donor site and did not exhibit ‘target site channelling’ close to the IRs of the transposon. On the contrary, despite of the predicted high Vstep score, only a single insertion event was recovered from the IRs (Figure 2), suggesting that the transposon ends of SB, embedded in a paired end complex are limited in their abilities to target the IRs or sites close to the IRs during autointegration. Due to the linkage, the autointegration of SB was primarily intramolecular, and no insertions were detected from the rpsL region. Thus, the transposon was fully excised from the flanking donor DNA prior its integration into a new site.
Fig. 2. Comparing autointegration profile to the predicted, close-to-random target site distribution of SB transposition. 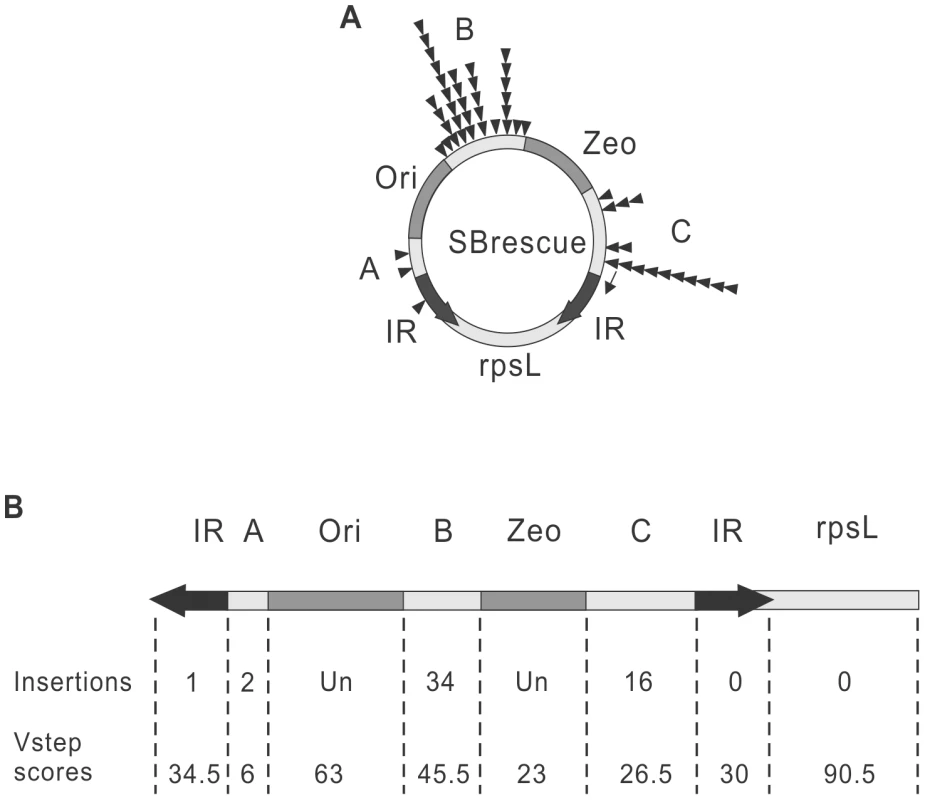
A. Distribution of 53 de novo autointegration events (triangles) detected by the assay shown in (Figure 1B). Autointegration products were isolated from individual bacterial clones, sequenced and mapped to the SBrescue construct. The thin arrow indicates the location of the sequencing primer on the left IR. B. Comparison of the predicted and experimental insertion events. The SBrescue construct is shown in a linear mode. The SB Vstep scores and experimental insertion events were shown below. Un, undetectable. Autointegration properties of the piggyBac transposon and single-ended transposition
Next, we tested whether self-destructive autointegration could also occur during PB transposition. We have used a transposon donor construct that is identical to SBrescue, except that the SB IRs were replaced by PB IRs [49] (PB2K in Fig. 3B), together with a mouse codon-optimized PB transposase (mPB) [50]. As shown in Figure 3A, autointegration of the PB transposon occurred at frequencies comparable to SB100X (0.49%, 3.2×104/6.4×106) in HeLa cells. As predicted and confirmed by DNA sequencing, autointegration of PB occurred into TTAA motifs, the canonical target site of PB [31] (Supporting Figure S1). Altogether, 23 integration sites were mapped and twelve were recovered from regions B and C (Figures 3B,C). However, unlike with SB, a significant number of integration events (48%, 11/23) mapped outside of the transposon, in the rpsL gene (Figures 3B,C). These non-canonical transposition events also targeted TTAA target sites, but involved only a single end of the transposon. The other IR was not separated from the donor molecule during the reaction. We refer to these non-canonical transposition events as single-ended transposition.
Fig. 3. Autointegration properties of PiggyBac transposition. 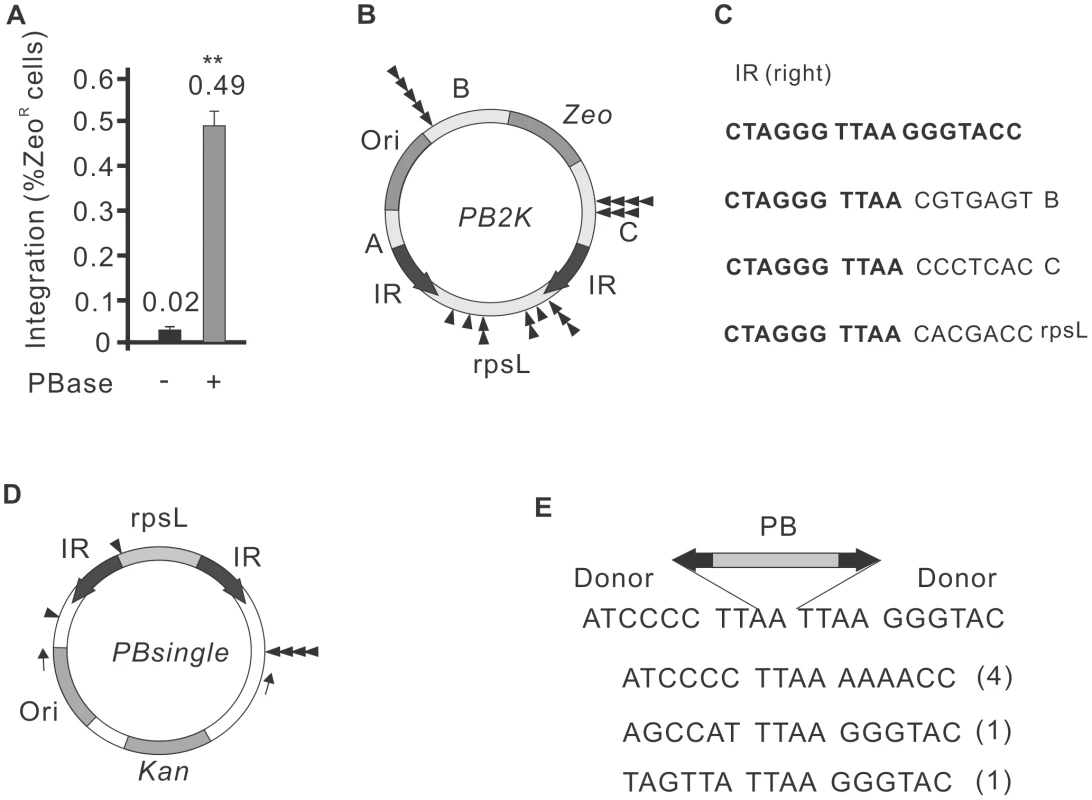
A. Frequency of PB autointegration events in HeLa cells using the PB2K construct. PBase, mPB transposase [50]. B. The structure of the PB2K construct. For explanation, see Figure 1A. Distribution of de novo PB insertions indicated by black triangles (n = 22) on the PB2K construct. C. Sequence of three (3/22) representative single-ended transposition events mapped to the B, C and rpsL regions of PB2K. Sequences flanking the right inverted repeat of the PB transposon in PB2K. Original sequences (bold); de novo integration events (normal); target site of PB transposition, TTAA (italic). D. Distribution of six single-ended transposition events on the PBsingle construct. Kan: kanamycin resistant gene (Kan). Dark bars indicate the control experiment with only transposon vector; light bars indicate the experiment with both transposon vector and transposase expressing vector. E. Sequence of the six individual single-ended transposition shown on Figure 3D. The PB transposon is shown as a two-headed arrow, representing the IRs (black). Frequencies are shown in parentheses. To investigate the phenomenon of single-ended transposition of PB further, a reciprocal construct, PBsingle was generated, where the PB transposon carried an rpsL gene (Figure 3D). In addition to single-ended transposition events detected by PB2K, the PBsingle assay system was suitable to capture various deletion products (Supporting Figure S2). Bacteria that gained StrepR could report on (i) double-ended excision products, (ii) single-ended integration events into either rpsL or (iii) the vector sequence flanking the transposon. The autointegration assay was performed as shown in Figure 1B, except bacteria were exposed to double selection of kanamycin and streptomycin. To capture single-ended events, 336 transposition products were pre-filtered by colony PCR, using primers flanking the PB excision site. Canonical excision products would appear as uniformly sized PCR products, while size difference would report on either single-ended transposition or non-transposase-mediated small deletions/insertion events generated by DNA repair. 31/336 pre-filtered PCR products were analysed further by DNA sequencing, and six out of 31 (19%) products were clearly generated by PB transposase-mediated, single-ended transposition that occurred into TTAA either inside or outside of the transposon (Figures 3D and 3E).
Bimolecular transposition
In the ‘single-ended’ transposition of PB, only one of the IRs was mobilized. Still, true single ended events, when the second IR is not involved in any of the steps of transposition, cannot be convincingly demonstrated. In fact, alternative mechanisms can generate similar, hard-to distinguish products. For example, the canonical transposition reaction might fail at the final step, and only one end of the transposon is transferred (lariat model), (Supporting Figure S2). Aberrant transposition might also occur by a mechanism that involves pseudo or cryptic sites mistakenly recognized as IRs. In addition, the ends of the transposon can also be derived from two separate molecules [51] (bimolecular transposition).
To explore the scenario of bimolecular transposition, truncated ‘solo’ transposons were generated. ‘Solo’ substrates, lacking either the left (PBΔleft; SBΔleft) or the right IRs (PBΔright; SBΔright) were tested in a cell culture-based transposition assay [9]. Molecular analysis of the resistant colonies revealed that neither PBΔright nor SBΔleft supported transposition (Table 1). In contrast, the analysis confirmed transposase-mediated transposition of the ‘solo’ substrates, PBΔleft (4.6%) and SBΔright (0.56%) [52] (Table 1), indicating that both transposases are capable of utilizing ‘solo’ substrates. In either cases, the IRs of the ‘solo’ transposons were properly integrated into respective target sites (Supporting Test S1). Notably, in clone PBΔleft#8, we have identified a second right IR integrated into a same genomic locus, confirming that the transposase used the two IRs from separate molecules (Supporting Text SF1). As ‘solo’ transposition occurred ∼8-fold more frequently for PB, we monitored the PB system further in the ‘solo-mixing’ experiments. In this strategy, the PBΔleft and PBΔright constructs were transfected either alone or mixed in equimolar ratios, and tested in the colony forming, transposition assay. If transposition utilizes the IRs from separate molecules, one would expect elevated colony numbers when either PBΔleft or both ‘solo’ substrates are present in the assay, compared to PBΔright that does not support transposition alone (Table 1). The higher number of resistant colonies in the respective experiments indicated that the transposase was able to utilize the IRs from different copies of the transposon, supporting the bimolecular model (Figure 4).
Fig. 4. Bimolecular transposition events generated by PB. 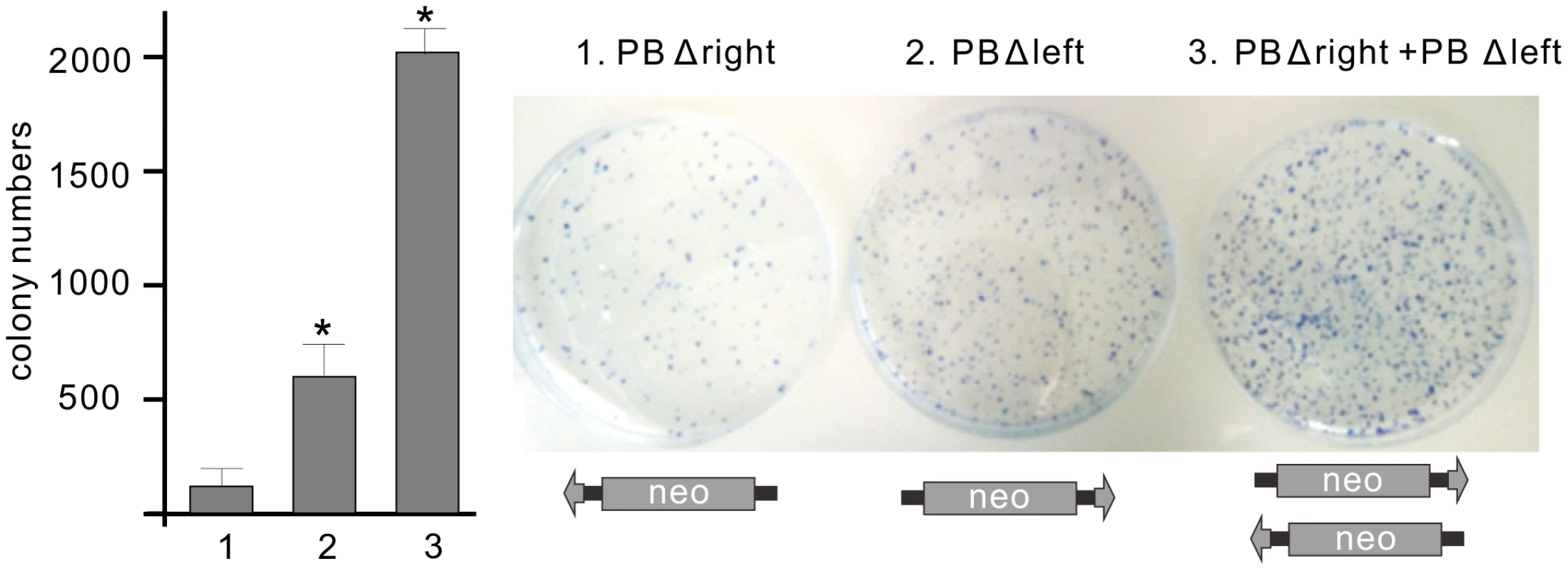
Transposition assay was performed by using ‘solo’ transposon substrates, either alone or mixed in equimolar ratios, in the present of the mPB transposase. The statistical significance of differences is shown by asterisk above the bars, *P<0.05. Molecular analysis identified no transposase-mediated integration events in the resistant colonies using PBΔright (background). See also Table 1. Tab. 1. Transposase-mediated integration events of ‘solo’ substrates. 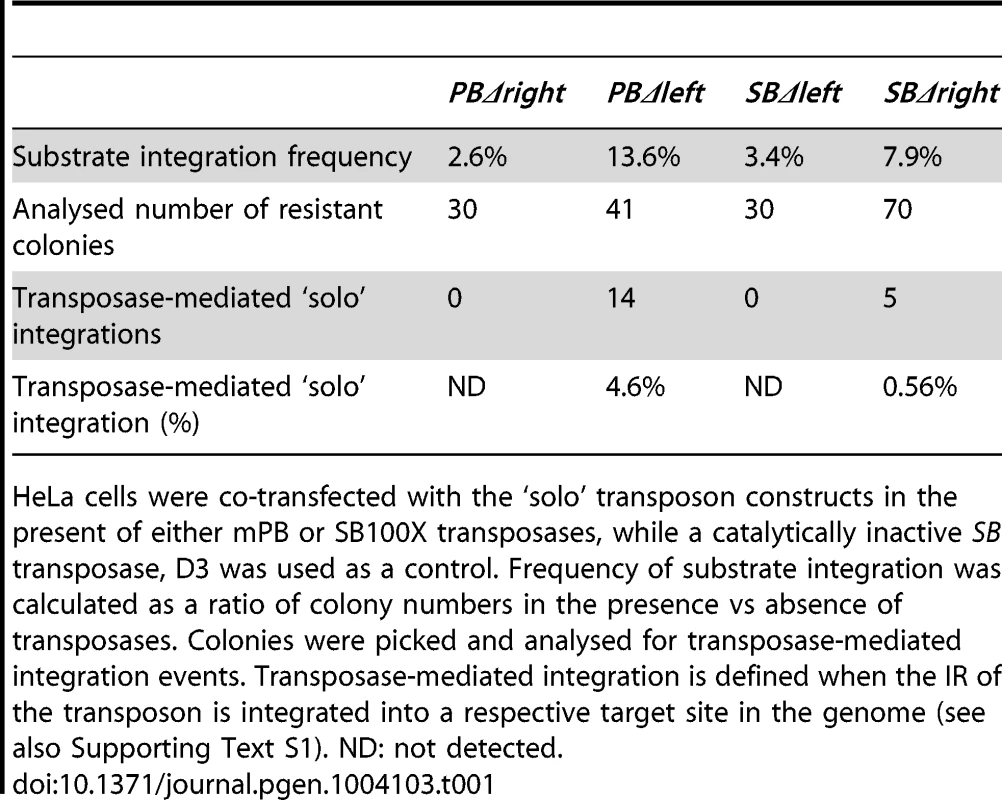
HeLa cells were co-transfected with the ‘solo’ transposon constructs in the present of either mPB or SB100X transposases, while a catalytically inactive SB transposase, D3 was used as a control. Frequency of substrate integration was calculated as a ratio of colony numbers in the presence vs absence of transposases. Colonies were picked and analysed for transposase-mediated integration events. Transposase-mediated integration is defined when the IR of the transposon is integrated into a respective target site in the genome (see also Supporting Text S1). ND: not detected. Both SB and PB transposons are sensitive to the size of the transposon
The efficacy of transposition was reported to depend on the size of the transposon [29], [30], [53]–[55]. One potential mechanism responsible for such size-dependence is that following transposon excision, self-disruptive autointegration competes with productive transposition. Since larger transposons have more target sites, they could be particularly attractive targets for autointegration. This hypothesis predicts that the size of the transposon does not affect the frequency of excision, but it shifts the ratio between autointegration and productive transposition. To test this assumption, a series of transposons of different size, ranging from 2679 bp to 7256 bp (SB2K, SB3K, SB4K, SB7K) and 2795 bp to 7319 bp (PB2K, PB3K, PB4K, PB7K) were generated for SB and PB, respectively. Frequencies of transposon excision, autointegration and productive transposition events were determined for the various transposons. Excision frequencies were estimated by quantitative PCR, autointegration was monitored as above. Productive transposition was determined in a cell culture-based assay [9]. Figure 5A shows that excision frequencies declined with increasing size, while autointegration frequencies elevated over 4 kb either moderately or sharply for SB and PB transposons, respectively (Figure 5A). Accordingly, productive transposition frequencies dropped with increasing size of both SB and PB transposons. These results indicated that the size of the transposon affected transposition already at the excision step, thereby arguing against the hypothesis of autointegration being the sole factor that compromises productive transposition with increasing transposon size. Nevertheless, autointegration contributes as an additive element to the less efficient transposition of long transposons. Surprisingly, the two transposons behaved similarly in all three assays (Figures 5A an 5B). Thus, in contrast to general assumptions, and similarly to SB, size affects PB transposition as well.
Fig. 5. Both SB and PB transposons are sensitive to the size of the transposon. 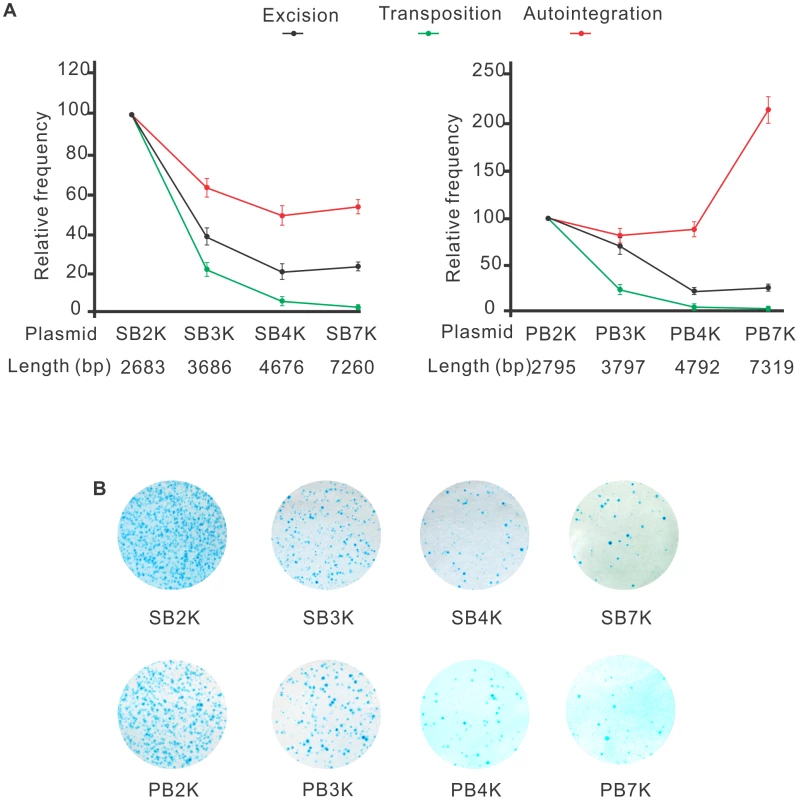
A. Excision, autointegration and transposition profiles of SB (left panel) and PB (right panel) transposons. The name and the size of the various constructs are shown below the plots. The values using the smallest constructs (SB2K or PB2K) were set to 100% (n = 3). B. Transposition assay performed by using SB (upper panel) and PB (lower panel) transposon constructs of various sizes. Inhibition of autointegration by a cellular, barrier-to-autointegration factor
A cellular protein, BANF1 (BAF) barrier-to-autointegration factor was identified by its ability to protect retroviruses from autointegration [23]. BANF1 binds to double-stranded DNA, including freshly transfected, extrachromosomal plasmid DNA [56], in a non-specific manner [57], [58]. Thus, in principle, BANF1 could affect DNA transposition as well, between the molecular steps of excision and reintegration, when the transposon exists as an extrachromosomal molecule in the cell. To test this assumption, we asked if BANF1 could protect DNA transposons from autointegration. We addressed this question by monitoring autointegration events in HeLa cells, where BANF1 was either knocked-down or transiently overexpressed (Figure 6A). When BANF1 expression was knocked-down by RNA interference (Supporting Figure S3), the frequency of autointegration of SB was increased by two-fold compared to the control (Figure 6A, left panel). In contrast, BANF1 overexpression decreased the frequency of autointegration to one third (Figure 6A, left panel). Similar results were obtained by using the PB transposon (Figure 6A, right panel). No significant effect of BANF1 was observed at the excision step of SB transposition (not shown), suggesting the BANF1 acted specifically following excision.
Fig. 6. The cellular factor of BANF1 interferes with autointegration. 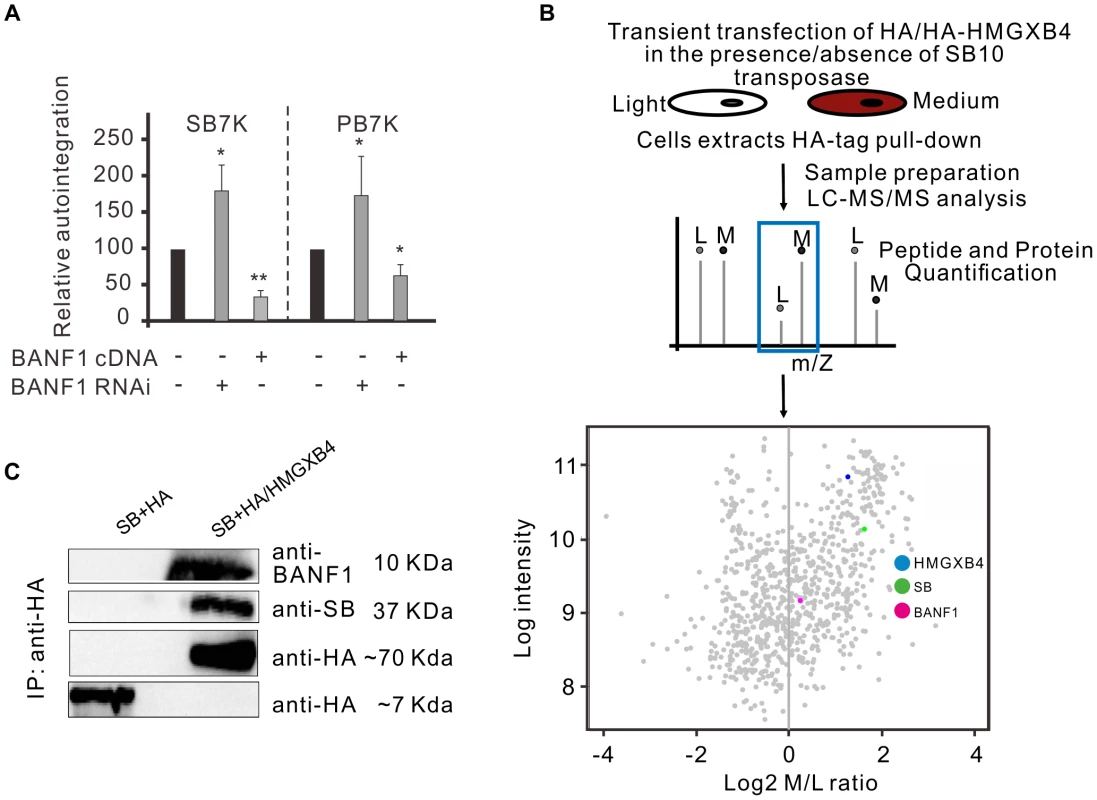
A. Relative autointegration frequencies of SB (SB7K, left panel) and PB (PB7K, right panel) in HeLa cells, where BANF1 was either knocked-down or overexpressed. Knocking down of BANF1 stimulated, whereas overexpressing of BANF1 inhibited autointegration of both SB and PB transposons (n = 3). The statistical significance of differences is shown by asterisk above the bars *P<0.05. B. A SILAC pull-down experiment using anti-HA resin to investigate interaction partners of HMGXB4 in the presence/absence of SB10 transposase in transiently transfected HEK293T cells. Schematic representation of the SILAC/pull-down experimental approach in which stable isotope labeled amino acids [Light (L) or Medium heavy (M)] are added in the form of medium supplement to culture HEK293T cells. Detection of interaction partners is performed by mass spectrometry. Scatter plot displays the normalized log2 SILAC ratio M/L values (X-axis) versus log2 intensity (Y-axis) of proteins detected in the interactome around HMGXB4− in presence of the SB transposase. Each dot represents an individual protein, while their position indicates their abundance in the complex pulled down by the bait of HMGXB4. Proteins with a positive log2 SILAC M/L ratio, including BANF1 and SB are enriched in the protein complex around HMGXB4. C. Co-immunoprecipitation assay to investigate the interaction partners of HMGXB4, a physical interaction partner of the Sleeping Beauty transposase, SB10 [13]. SB10 and HA-tagged HMGXB4 were transiently transfected into HEK293T cells (see Methods). In comparison to negative control, BANF1 and SB are enriched in the pull-down by HMGXB4-HA. In addition to BANF1, the effect of another host-encoded factor, the high-mobility group protein (HMGB1) was tested on autointegration. Similarly to BANF1, HMGB1 binds DNA in a non-specific manner [59]. In SB transposition, the transposase physically associates with HMGB1 and recruits it to the transposon DNA [11]. Autointegration was monitored in cells where HMGB1 was either transiently overexpressed or knocked-out [60]. Although, HMGB1 overexpression or deficiency was significantly affecting productive transposition [11], it had no detectable influence on autointegration (Supporting Figure S4). These results indicate that despite their similar non-specific DNA-binding activity, BANF1 and HMGB1 have a clearly distinct effect on DNA transposition.
Alternatively to a non-specific engagement, and similarly to retroviruses, BANF1 might be actively recruited to a preintegration complex of a transposon. In order to distinguish between these two scenarios, a high throughput immunoprecipitation experiment was designed to analyse a protein interactome forming around the SB transposase in mammalian cells. Affinity purification combined with mass spectrometry is a powerful strategy to detect protein-protein interactions among proteins in their native cellular environment [61]. This method is suitable to reveal the composition of entire protein complexes. If we use the analogy to retroviruses [62], one should keep in mind that even if BANF1 is recruited actively to the preintegration complex, it might not be recruited directly by the transposase. To distinguish true interaction partners from non-specific contaminants, we needed an easy-to-detect, confirmed interacting partner of the SB transposase as bait. We can readily monitor interactions of HMGXB4 (HMG2l1) with either the transposon or the transposase in vivo [13]. Thus, HMGXB4 was chosen as bait to analyse higher order complexes formed around SB. The experiments were run in parallel, in the presence and in the absence of the SB transposase. In the control experiment, it is not expected to detect interaction partners of the SB transposase. HEK293T cells were transiently transfected with HA-tagged HMGXB4 protein in the presence/absence of the SB10 transposase [9]. A SILAC pull-down experiment was performed. This experimental strategy identified BANF1 as an interaction partner of HMGXB4− in the presence, but not in the absence of the SB transposase (Figure 6B). The presence of BANF1 was also detectable when the bait, HMGXB4 was used in a co-immunoprecipitation assay (Figure 6C). This observation predicts that BANF1 can be actively recruited into a higher order protein complexes forming around the SB transposase in mammalian cells.
Discussion
Suicidal autointegration of Sleeping Beauty and piggyBac transposons
This study focuses on molecular events following the excision steps of two eukaryotic DNA transposons, SB and PB, derived from fish and insect genomes, respectively. The transposition reactions were performed in a heterologous host environment, phylogenetically distant from their natural hosts. The experimental setup mimics the scenario of introducing DNA transposons into a naïve eukaryotic host. We have shown that a significant portion of SB and PB transposon excision events is accompanied by suicidal integration into the transposon's own DNA. Although, different transposons may have different frequency of autointegration depending on the structure of the transpososome and the number of the integration target sites on the transposon, autointegration would influence the success of a transposon in a new environment. Neither SB nor PB was immune to the suicidal process of autointegration. Thus, in general, transposases/integrases in eukaryotes might not be able to distinguish between their own genome form foreign DNA. This would define autointegration as the lack of ability of self-avoidance upon integration. In contrast, certain prokaryotic transposons, including Tn7 and Mu exhibit ‘target immunity’ that prevents the transposon from transposing into its own genome [63], [64]. Both Tn7 and Mu avoid integration into DNA molecules that already have a copy of the transposon. As an alternative to self-encoded ‘target immunity’, some bacterial transposons and eukaryotic retroviruses recruit cellular host factors to protect against autointegration [19], [23]–[25]. In Tn10 transposition a host protein, histone-like nucleoid structuring (H-NS) plays a role in promoting intermolecular and supressing self-destructive intramolecular integration events [19]. Similarly, DNA transposons in eukaryotes might also capture cellular factors to protect their genome against autointegration. This strategy could defend the invading molecule and contribute establishing a stable host-transposon relationship.
BANF1 interferes with self-destructive autointegration of SB and PB transposons in eukaryotes
BANF1 is involved in several critical processes, including host defence [65], [66]. The usual mode of BANF1 is repressive, due to its propensity to coat DNA. For example, BANF1 acts as a potent inhibitor of virus replication, defending against poxvirus invasion [67]. Intriguingly, and in contrast to its original function in host defence, BANF1 is piggybacked by various retroviruses to protect their viral genome against autointegration. BANF1 inhibits autointegration of the Moloney Murine Leukemia retrovirus, MoMLV [23], [68], [69] or HIV-1 [62]. By physically protecting the retrovirus, BANF1 promotes productive viral integration into the host genome [62]. In our experimental setup, BANF1 was influencing the fate of the excised molecules of two DNA transposons of different origin, SB and PB. Thus, in addition to its reported activity to bind freshly transfected DNA [56] or retroviral cDNA [23], BANF1 might influence the fate of DNA transposons as well. An important ramification of utilizing phylogenetically conserved cellular proteins by transposons might be the ability to survive and establish stable host-parasite relationship in a heterologous host environment. Accordingly, in addition to its role in Tn10 transposition, H-NS was reported to selectively bind the transpososomes of Tn5, and is likely to modulate many other transposition processes in Gram-negative bacteria [70].
SB and PB are members of the superfamily of DDE/D transposases and retroviral integrases, utilizing the same strategy for target joining. Still, how reasonable it is to assume an interaction of BANF1 with both DNA transposons and retroviruses? In fact, BANF1 might be an ideal cellular factor for integrating elements in higher eukaryotes. Due to its non-specific DNA-binding activity to double-stranded DNA [58], a capacity to compact DNA and assemble higher-order nucleoprotein complexes, BANF1 could influence the fate of any extrachromosomal DNA molecule. As in retroviral integration [23], [69], BANF1 may compact the transposon genome to be a less accessible target for autointegration, and promote the integration step. Furthermore, similarly to retroviruses, BANF1 could be even actively recruited to preintegration complexes. The exact manner of recruitment might vary, providing specificity. BANF1 is recruited via physical interaction by the viral matrix protein gag to the retroviral preintegration complex of HIV-1 [62]. In SB transposition, BANF1 was enriched in a higher order complex containing the SB transposase and its interactor HMGXB4. Thus, the enrichment was mediated via protein-protein interaction. Since the experimental setup did not include the transposon DNA, we could not faithfully simulate preintegration complex formation. Nevertheless, HMGXB4 is a specific interaction partner of both the transposon and the transposase of SB [71]. Therefore, it might be reasonable to assume that BANF1 associates with the preintegration complex.
In sum, our strategy to model the process of establishing a host-transposon relationship in a naïve environment identified BANF1 as a host encoded factor influencing this process. Future work will have to clarify if a common role of BANF1 to protect integrating mobile elements in general exists.
Excision, a step prior to integration, is already affected by the size of SB and PB transposons
Traditional models predict that efficient integration must follow the excision of DNA elements. Strikingly, autointegration was estimated to be over 90% in mariner transposition in vitro, suggesting that under the standard reaction conditions, the vast majority of the excised transposon inserts into itself, rather than into another DNA molecule [21]. This high frequency would establish autointegration as a major factor affecting productive integration. Furthermore, as longer transposons present more potential target sites, autointegration would be a reasonable explanation for size-dependence of transposition, observed for both SB [29], [30] and PB (this work) transposition.
Still, the role of autointegration in counteracting productive transposition might be overestimated. We found that transposon excision, a step prior to integration, is already affected by the size of the transposons (Figure 5A), indicating that a larger transposon might have difficulty to form a synaptic complex. Our data argue that competition between self-integration and productive transposition is unlikely to be the only factor responsible for sensitivity to size. If we assume that unproductive transposition equals suicidal autointegration, the gap between transposon excision and productive transposition could be a good estimate for the effect, and was reported to be around 25% in SB transposition in vivo [32].
Excision, autointegration and transposition of SB and PB transposons are similarly affected by size
In contrast to an earlier report [41], we found that SB and PB transposons were affected similarly by the size of the transposon in three different assays (Figure 5). When the size of the transposon increased from 2683 to 7260 and 2795 to 7319 bps, the frequency of productive transposition dropped by 83% and 89.6% for SB and PB, respectively (Figure 4B). In addition, SB and PB behaved similarly in assays monitoring either excision or autointegration (Figure 5A). Therefore, our data argue against the general assumption that the PB transposon is not sensitive to size below 14 kb [41]. The different observation might be related to the fact that (i) the DNA fragment that Ding et al. used to increase the size of the transposon contained a higher density of TTAA target sites than the existing transposon. Actually, it is impossible to separate the true effects of length and numbers of target sites for a transposon that is highly specific in terms of integrating into a given sequence; (ii) Ding et al. estimated transposition frequencies in transgenic mouse experiments by counting transgenic embryos, regardless of the copy number of the integrated elements per embryo. Therefore, to compare productive transposition of SB and PB transposons, we have adjusted transgenic frequencies by the copy number of the integrated transgenes [72]. Importantly, small size does not seem to be an absolute requirement for mobilization in either case. Decreasing the distance outside the transposon ends of SB was reported to increase transpositional rates under experimental conditions [29]. Moreover, both PB and SB100X were reported to capable of mobilizing giant molecules of DNA, such as BACs (bacterial artificial chromosomes) [37], [73]. These reports indicate that in contrast to viruses, DNA transposons have no strict (if any) upper limit regarding their cargo capacity.
Autointegration of SB, likely due to physical constraints, avoided the IRs, suggesting that the captured events were rather intramolecular than intermolecular. Nevertheless, SB integration is not channelled to the terminal repeats of the transposon as it was observed for Tn10 [19]. Furthermore, the lack of linkage of autointegration sites to nearby regions at the donor DNA molecule would argue against an association between the ‘local hoping’ phenotype and autointegration.
Aberrant transposition events may pose a threat to genome stability
Our experimental approach gave us the opportunity to have a closer insight into the mechanism of both PB and SB transpositions. We have captured autointegration products at comparable frequencies for both SB and PB. We assume that the excision and reintegration steps of autointegration and canonical transposition are mechanistically not significantly different [32], [41], [74] (Figures 1 and S1).
In addition to the autointegration products, our assays detected aberrant, pseudo-transposition events. In the ‘single-ended’ transposition products of PB, one IR of the transposon was clearly separated from the donor site, without obvious involvement of the other IR in the reaction (Figure 3D). The liberated end of PB targeted either the transposon or the backbone DNA (Figures 3C and 3E). SB did not display this feature in a similar assay system. By contrast, both transposons were capable of mobilizing substrates, lacking one of the IRs from separate molecules ([52] and this work). These bimolecular transposition events were eight-fold more frequently detected for PB.
How could aberrant transposition events be generated? In fact, ‘true single ended’ transposition, when a transposase interacts with a single transposon end, performs the cleavage and integration steps without the involvement of another end has not been undoubtedly reported from any system. In fact, alternative mechanisms can generate hard-to distinguish, similar products. For example, the canonical transposition reaction could fail at the final step, and only one end of the transposon is transferred (lariat model). In addition, our ‘solo’ experimental data support the ‘bimolecular model’, when the ends of the transposon derive from separate molecules [51]. In addition to single ended events, small deletions at the donor sites of PB transposition are assumed to be associated with imprecise transposon excision, and involve non-homologous end joining [40]. These structures were reported following PB excision in Drosophila (4.3%), mouse (5%) and in human cells [38], [40].
Aberrant pseudo-transposition can be considered as a fidelity problem of the transposition reaction, and has been observed with P-element in Drosophila, Ds element in Arabidopsis, Ac/Ds elements in maize [51], [75] or Tam3 in Antirrhinum majus [76]–[80]. Small sequence variations generated by NHEJ at the excision sites are unlikely to cause genome rearrangements. By contrast, pseudo-transposition events can generate difficult-to-repair lesions and be genotoxic. Aberrant transposition events were reported to induce deletions, insertions, chromosome translocations and could initiate McClintock's chromosomal breakage-fusion-bridge cycles [51], [81]. Occasional mis-pairing between extrachromosomal molecules would not compromise the safety feature of a transposon-based transfer vector in a heterologous environment. However, fidelity problems could be problematic when the transposon is mobilized from the genome. Thus, cells subjected to PB-based genome manipulation techniques, e.g., transgene-free iPS cells generated by PB excision [82], should be carefully monitored for genome rearrangements.
Wide host range vs fidelity: A price to pay?
There seems to be a basic difference in the ways transposons in pro - and eukaryotes control their activity to minimize the potential genotoxicity generated by improper synapsis of the transposon ends. For all classical bacterial transposons characterized to date, including Tn5 transposition, the catalytic steps of the reaction are tightly coupled to the synapsis of the transposon ends [83]. In addition, the coupling of transcription and translation in bacteria also increases the probability of a proper synapsis as the transposase binds tightly to the first IR before searching for nearby ends. In contrast, eukaryotic transposases must search at random for transposon ends when they enter the nucleus. Therefore, regulatory mechanisms promoting accurate double-ended reactions from the same transposon molecule are crucial.
Tc1/mariner transpositions, including SB, might have invented novel “built in regulatory checkpoints” to enforce synapsis prior catalysis [21]. A simple topological filter could also suppress promiscuous synapses of distant ends of the transposon [84]. Furthermore, certain transposition-like reactions, including V(D)J recombination, are also capable of filtering out unpaired reaction products. This regulatory mechanism, assisted by a cellular factor, HMGB1, regulates a highly controlled, ordered assembly process [85], [86]. Similarly to V(D)J recombination, HMGB1 was reported to assist paired end complex formation of SB [11]. In addition to HMGB1, SB transposition requires various vertebrate-specific host factors [10], [11], [13], [29] that render SB transposition restricted to vertebrates. In contrast, PB has an incredibly wide host range (from yeast to human) that could be associated with loose or no host factors requirement.
In comparison to SB, PB transposition results in more frequent, aberrant transposition products in a heterologous environment. Why is it so? If PB does not use host factors to enforce fidelity of the end pairing before excision, the reaction might be less precise by its nature. Alternatively, PB might utilize a host factor in its endogenous host (insect) that guarantees precise regulation. However, this factor is diverged or not available in mammalian cells. Finally, both PB and SB transposons have “built in regulatory checkpoints” that are most effectively filter out aberrant products under optimal conditions and in appropriate hosts. Notably, aberrant transposition events, including single-ended transposition of the Mos1, mariner element were observed under suboptimal conditions [22]. In sum, when a transposon is transferred too far from its original host, the conditions in a new environment could be suboptimal, and the fidelity of the reaction could be compromised. The wide host range of PB can be explained by relative independence from host-encoded factors, perhaps a price to be paid for fidelity.
Materials and Methods
Plasmid constructs
The IRs of the transposons were identical to the versions published earlier [49], [87] and were not modified for the assays. All the primers used for construct cloning were listed in Supporting Table S1. SBrescue: XmnI/BsaI fragment (Klenow-filled) containing ampicillin gene on pUC19 was replaced by PstI and SalI fragment containing zeocin gene from vector pZEO (isolate SV1, Invitrogen) resulting in pUC19-zeo. Klenow-filled SapI/SspI fragment containing zeocin gene and replication origin was inserted into EcoRI site of PT2/HB to get PT2/SBzeo. The transposon was PCR-amplified with primer AATASB-IR from PT2/SBzeo and ligated to rpsL gene fragment, which was PCR-amplified with primers rps1F/rpslR from nNG639 [43]. SB2K: BspHI/EcoRI fragment containing zeocin gene on SBrescue was replaced by BsaI/BglII fragment containing zeocin and promoter sequences from pFP-Zeo [88]. SB3K, SB4K and SB7K: DNA fragments were PCR-amplified from bacteriophage lamda DNA, using primers lam1kF/lam1kR, lam1kF/lam2kR and lam1kF/lam6kR, respectively, and were inserted into XbaI site (Klenow filled) of SB2K. PB2K: Klenow-filled NotI/HindIII fragment containing zeocin gene from SBrescue was inserted into SpeI site of pUC19PBneo [72] resulting in PUC19XLzeo. PvuII fragment containing PB transposon was ligated to rpsL gene PCR-amplified with primers rps1F/rpslR from nNG639. PB3K, PB4K, PB7K: The AatII/BglII fragments containing lamda DNA from SB3K, SB4K and SB7K were inserted into AatII/BglII sites of PBPr respectively. pcDNA3.1BANF1 (BANF1 gene expressing vector): BANF1 coding sequence was PCR-amplified from pcDNA3.1/HiscBANF1 (a gift from Katherine Wilson, Johns Hopkins University) with primers BAFF/BAFR and cloned into EcoRV site of pcDNA3.1/Zeo (+) (Invitrogen). BAF-RNAi: Oligos of BAF96F/BAF96R were annealed together and cloned into BglII/HindIII site of pFP-Neo-H1 [88]. To generate ‘solo’ substrates PB pUC19XLneo [69] was digested with BamHI to delete the right IR (PBΔright) or with KpnI to remove the left IR (PBΔleft). For “solo” SB, pTneo was digested EcoRI to generate SBΔleft, while the digestion with BamHI yielded SBΔright.
Cell culture maintenance and transfections
HeLa, AA8 and mouse MEF cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Gibco/Invitrogen) supplemented with 10% fetal calf serum (FCS, PAA). The zebrafish PAC2 cells were grown at room temperature and atmospheric CO2 concentrations in Leibovitz L15 medium (Gibco/Invitrogen) supplemented with 15% FCS. Cells were transfected at 50–80% confluence with QIAGEN-purified plasmid DNA using jetPEI (Polyplus transfection, for mammalian cells) or FuGene6 (Roche, for fish cells) according to instructions of manufacture. Transfection efficacy of a ∼3 kb and a ∼7 kb plasmid containing GFP cassette was monitored and compared by FACS analysis, but no significant difference was found (not shown).
Autointegration assay
Cell culture and transfection was done as described [9]. Typically, 1.5×105 cell were subjected to transfection with plasmids containing the transposon (500–1000 ng) and the transposase (60–100 ng). Two days post transfection plasmid DNA was recovered and transformed into bacteria (Invitrogen, ElectroMAX DH10B Cells, Cat. No. 18290-015, Genotype: F – mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu) 7697 galU galK rpsL nupG λ–). Bacteria were subjected to either zeocin (to determine total number of plasmids) or zeocin/streptomycin double selection (to determine autointegration events). The number of autointegration events was normalized by total number of plasmids. To confirm autointegration events, individual bacterial colonies were cultured and recovered plasmid DNA was subjected to DNA sequencing using primers of psbLacR3 and PB-F or PB-R for SB- and PB transposon, respectively. For BANF1 overexpression or knockdown experiments, 300 ng of pcDNA3.1BANF1 or BAF-RNAi plasmid was cotransfected with the transposon and helper constructs.
Transposition assay
Cell culture and transfection was done as described [9]. Two days post transfection 105 cells were plated on 10 cm dishes and exposed to antibiotic selection (100 ng/ml zeocin, for two weeks). Resistant colonies were visualized by methylene blue staining [9]. Transgene copy number was normalized by using qPCR specific to zeocin.
Excision assay
The plasmid DNA was prepared as described in autointegration assay and dissolved in 50 µl water. Excision frequencies of eight transposon plasmid constructs of various sizes (four SB and four PB) were estimated by using a quantitative, real-time PCR (7700 sequence detection system from ABI, Applied Biosystems, Foster City, CA). To determine the total number of parental plasmid DNA molecules, a ‘parental’ titration curve was established. PCR primers of rpsL-F/rpsL-probe/rpsL-R were used to amplify the rpsL gene on the construct of SBrescue. For the curve, dilutions of 10−2, 10−3, 10−4, 10−5, 10−6 ng of SBrescue plasmid DNA were subjected to a PCR reaction to amplify the rpsL gene (rpsL-F/rpsL and probe/rpsL). To quantify the total number of parental plasmid molecules, total DNA extract was used (3 µl, diluted by 2000-fold, rpsL-F/rpsL-probe/rpsL-R). The excision products were PCR-amplified from the total extract DNA using nested PCR (1st round, primers of rpslexciF1/rpslexciR1, 94°C for 30 s and 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s; 2nd round, rpslexciF2/rpslexciR2, 1 µl, diluted by 100-fold, 94°C for 30 s and 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s). The amplified products (10−2, 10−3, 10−4, 10−5, 10−6 ng) were used to establish a second titration curve, specific for the excision products. To quantify excision products, primers of SB-F/SB-probe/SB-R and PB-F/PB-probe/PB-R were used on a total DNA extract (5 µl), for SB and for PB, respectively. The excision frequency was calculated as the ratio of excision products normalized by the total number of parental plasmid molecules. qPCR was performed for each experimental sample in triplicates. Ct values were determined following recommendations by the manufacturer.
Colony PCR
Briefly, bacteria were picked by a pipette tip and directly subjected to a PCR assay using primers of PB-F and PB-R (5 pmol of each, Supporting Table S1) and Taq polymerase (Takara) in a total volume of 20 µl. PCR program: 94°C for 1 min; 30 cycles of 94°C for 30 s, 58°C for 30 s, and 2°C for 30 s; and 72°C for 2 min.
Protein-protein interaction studies using the SILAC/pull-down assay
A triple SILAC pull-down experiment was performed using anti-HA resin. HEK293T cells were transiently transfected with HA-tagged wild type or mutant HMGXB4 (HMG2l1) [13] and SUMO1 in the presence/absence of Sleeping Beauty, SB10 [9] using Polyplus-transfection jetPEI transfection reagent with 3 µg of plasmids each. We compared proteins co-purifying with HA in cells expressing the empty vector (“light”), HA-tagged HMGXB4− with mutated sumoylation site (“medium”) and HA-tagged wild-type HMGXB4 (“heavy”). The cells were plated on a 15-cm dish and harvested 48 h post-transfection. Two dishes were used for each condition. Detection of interaction partners is performed by mass spectrometry and the results obtained were analyzed by MaxQuant computational platform [89]. Results presented show protein abundance ratios between cells transfected with HMGXB4− and the empty vector control.
Co-immunoprecipitation, immunoblotting and antibodies
Whole-cell extracts were prepared using extraction buffer (Tris-HCl 50 mM at pH 8.0, NaCl 150 mM, 0.1% SDS (Na-dodecylsulphate) Triton X-100 1% and Na-deoxycholate 0.5%) supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany). For immunoprecipitations, equal amounts of lysate (containing 5 mg of total cellular protein from HEK293 cells) were pre cleared with protein G-agarose beads (Sigma, St Louis, MO). Pre-cleared extracts were incubated with EZview Red Anti-HA Affinity Gel (Sigma-Aldrich, USA) for 1 h at 4°C. Precipitates were washed extensively in extraction buffer. Bound complexes were eluted with 2× SDS–PAGE sample buffer and resolved by 7.5–15% SDS–PAGE. Immunoblotting was performed according to standard procedures and proteins detected with the indicated antibodies. Antibodies were detected by chemiluminescence using ECL Advance Western Blotting Detection Kit (Amersham Bioscience).
Supporting Information
Zdroje
1. CraigNL (1995) Unity in transposition reactions. Science 270 : 253–254.
2. GueguenE, RousseauP, Duval-ValentinG, ChandlerM (2005) The transpososome: control of transposition at the level of catalysis. Trends Microbiol 13 : 543–549.
3. AgrenJA, WrightSI (2011) Co-evolution between transposable elements and their hosts: a major factor in genome size evolution? Chromosome Res 19 : 777–786.
4. LeungPC, TeplowDB, HarsheyRM (1989) Interaction of distinct domains in Mu transposase with Mu DNA ends and an internal transpositional enhancer. Nature 338 : 656–658.
5. KettingRF, HaverkampTH, van LuenenHG, PlasterkRH (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99 : 133–141.
6. TabaraH, SarkissianM, KellyWG, FleenorJ, GrishokA, et al. (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 : 123–132.
7. ChalmersR, GuhathakurtaA, BenjaminH, KlecknerN (1998) IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93 : 897–908.
8. Claeys BouuaertC, LipkowK, AndrewsSS, LiuD, ChalmersR (2013) The autoregulation of a eukaryotic DNA transposon. Elife 2: e00668.
9. IvicsZ, HackettPB, PlasterkRH, IzsvakZ (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91 : 501–510.
10. IzsvakZ, StuweEE, FiedlerD, KatzerA, JeggoPA, et al. (2004) Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell 13 : 279–290.
11. ZayedH, IzsvakZ, KhareD, HeinemannU, IvicsZ (2003) The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res 31 : 2313–2322.
12. WaliskoO, IzsvakZ, SzaboK, KaufmanCD, HeroldS, et al. (2006) Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc Natl Acad Sci U S A 103 : 4062–4067.
13. WaliskoO, SchornA, RolfsF, DevarajA, MiskeyC, et al. (2008) Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther 16 : 359–369.
14. ChiangSJ, JordanE, ClowesRC (1982) Intermolecular and intramolecular transposition and transposition immunity in Tn3 and Tn2660. Mol Gen Genet 187 : 187–194.
15. BenjaminHW, KlecknerN (1989) Intramolecular transposition by Tn10. Cell 59 : 373–383.
16. TomcsanyiT, BergCM, PhadnisSH, BergDE (1990) Intramolecular transposition by a synthetic IS50 (Tn5) derivative. J Bacteriol 172 : 6348–6354.
17. SignonL, KlecknerN (1995) Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. Genes Dev 9 : 1123–1136.
18. WardleSJ, O'CarrollM, DerbyshireKM, HanifordDB (2005) The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev 19 : 2224–2235.
19. SinghRK, LiburdJ, WardleSJ, HanifordDB (2008) The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. J Mol Biol 376 : 950–962.
20. HanifordDB (2006) Transpososome dynamics and regulation in Tn10 transposition. Crit Rev Biochem Mol Biol 41 : 407–424.
21. Claeys BouuaertC, ChalmersR (2010) Transposition of the human Hsmar1 transposon: rate-limiting steps and the importance of the flanking TA dinucleotide in second strand cleavage. Nucleic Acids Res 38 : 190–202.
22. SinzelleL, JegotG, BrilletB, Rouleux-BonninF, BigotY, et al. (2008) Factors acting on Mos1 transposition efficiency. BMC Mol Biol 9 : 106.
23. LeeMS, CraigieR (1998) A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci U S A 95 : 1528–1533.
24. LiY, KappesJC, ConwayJA, PriceRW, ShawGM, et al. (1991) Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol 65 : 3973–3985.
25. GarfinkelDJ, StefaniskoKM, NyswanerKM, MooreSP, OhJ, et al. (2006) Retrotransposon suicide: formation of Ty1 circles and autointegration via a central DNA flap. J Virol 80 : 11920–11934.
26. KlecknerN, ChalmersRM, KwonD, SakaiJ, BollandS (1996) Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr Top Microbiol Immunol 204 : 49–82.
27. GreenblattIM (1984) A Chromosome Replication Pattern Deduced from Pericarp Phenotypes Resulting from Movements of the Transposable Element, Modulator, in Maize. Genetics 108 : 471–485.
28. HorieK, YusaK, YaeK, OdajimaJ, FischerSE, et al. (2003) Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol 23 : 9189–9207.
29. IzsvakZ, IvicsZ, PlasterkRH (2000) Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol 302 : 93–102.
30. KarsiA, MoavB, HackettP, LiuZ (2001) Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Mar Biotechnol (NY) 3 : 241–245.
31. FraserMJ, SmithGE, SummersMD (1983) Acquisition of Host Cell DNA Sequences by Baculoviruses: Relationship Between Host DNA Insertions and FP Mutants of Autographa californica and Galleria mellonella Nuclear Polyhedrosis Viruses. J Virol 47 : 287–300.
32. LuoG, IvicsZ, IzsvakZ, BradleyA (1998) Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc Natl Acad Sci U S A 95 : 10769–10773.
33. MitraR, Fain-ThorntonJ, CraigNL (2008) piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J 27 : 1097–1109.
34. LoboNF, FraserTS, AdamsJA, FraserMJJr (2006) Interplasmid transposition demonstrates piggyBac mobility in vertebrate species. Genetica 128 : 347–357.
35. HozumiA, MitaK, MiskeyC, MatesL, IzsvakZ, et al. (2013) Germline transgenesis of the chordate Ciona intestinalis with hyperactive variants of sleeping beauty transposable element. Dev Dyn 242 : 30–43.
36. WangW, BradleyA, HuangY (2009) A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res 19 : 667–673.
37. RostovskayaM, FuJ, ObstM, BaerI, WeidlichS, et al. (2012) Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res 40: e150.
38. KimH, KimK, KimJ, KimSH, YimJ (2012) Mutagenesis by imprecise excision of the piggyBac transposon in Drosophila melanogaster. Biochem Biophys Res Commun 417 : 335–339.
39. KengVW, YaeK, HayakawaT, MizunoS, UnoY, et al. (2005) Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods 2 : 763–769.
40. WangW, LinC, LuD, NingZ, CoxT, et al. (2008) Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A 105 : 9290–9295.
41. DingS, WuX, LiG, HanM, ZhuangY, et al. (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122 : 473–483.
42. IvicsZ, LiMA, MatesL, BoekeJD, NagyA, et al. (2009) Transposon-mediated genome manipulation in vertebrates. Nat Methods 6 : 415–422.
43. NairJ, RouseDA, BaiGH, MorrisSL (1993) The rpsL gene and streptomycin resistance in single and multiple drug-resistant strains of Mycobacterium tuberculosis. Mol Microbiol 10 : 521–527.
44. MatesL, ChuahMK, BelayE, JerchowB, ManojN, et al. (2009) Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet 41 : 753–61.
45. VigdalTJ, KaufmanCD, IzsvakZ, VoytasDF, IvicsZ (2002) Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol 323 : 441–452.
46. YantSR, WuX, HuangY, GarrisonB, BurgessSM, et al. (2005) High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol 25 : 2085–2094.
47. AmmarI, Gogol-DoringA, MiskeyC, ChenW, CathomenT, et al. (2012) Retargeting transposon insertions by the adeno-associated virus Rep protein. Nucleic Acids Res 40 (14) 6693–712.
48. GeurtsAM, HackettCS, BellJB, BergemannTL, CollierLS, et al. (2006) Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res 34 : 2803–2811.
49. FraserMJ, BruscaJS, SmithGE, SummersMD (1985) Transposon-mediated mutagenesis of a baculovirus. Virology 145 : 356–361.
50. CadinanosJ, BradleyA (2007) Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res 35: e87.
51. EnglishJJ, HarrisonK, JonesJ (1995) Aberrant Transpositions of Maize Double Ds-Like Elements Usually Involve Ds Ends on Sister Chromatids. Plant Cell 7 : 1235–1247.
52. IzsvakZ, KhareD, BehlkeJ, HeinemannU, PlasterkRH, et al. (2002) Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem 277 : 34581–34588.
53. FischerSE, van LuenenHG, PlasterkRH (1999) Cis requirements for transposition of Tc1-like transposons in C. elegans. Mol Gen Genet 262 : 268–274.
54. LampeDJ, GrantTE, RobertsonHM (1998) Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149 : 179–187.
55. UrasakiA, MorvanG, KawakamiK (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174 : 639–649.
56. IbrahimN, WicklundA, WiebeMS (2011) Molecular characterization of the host defense activity of the barrier to autointegration factor against vaccinia virus. J Virol 85 : 11588–11600.
57. UmlandTC, WeiSQ, CraigieR, DaviesDR (2000) Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39 : 9130–9138.
58. BradleyCM, RonningDR, GhirlandoR, CraigieR, DydaF (2005) Structural basis for DNA bridging by barrier-to-autointegration factor. Nat Struct Mol Biol 12 : 935–936.
59. TangD, KangR, ZehHJ3rd, LotzeMT (2010) High-mobility group box 1 and cancer. Biochim Biophys Acta 1799 : 131–140.
60. CalogeroS, GrassiF, AguzziA, VoigtlanderT, FerrierP, et al. (1999) The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet 22 : 276–280.
61. VermeulenM, HubnerNC, MannM (2008) High confidence determination of specific protein-protein interactions using quantitative mass spectrometry. Curr Opin Biotechnol 19 : 331–337.
62. MansharamaniM, GrahamDR, MonieD, LeeKK, HildrethJE, et al. (2003) Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J Virol 77 : 13084–13092.
63. StellwagenAE, CraigNL (1997) Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J 16 : 6823–6834.
64. AdzumaK, MizuuchiK (1989) Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell 57 : 41–47.
65. CaiM, HuangY, GhirlandoR, WilsonKL, CraigieR, et al. (2001) Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. Embo J 20 : 4399–4407.
66. MargalitA, BrachnerA, GotzmannJ, FoisnerR, GruenbaumY (2007) Barrier-to-autointegration factor–a BAFfling little protein. Trends Cell Biol 17 : 202–208.
67. WiebeMS, TraktmanP (2007) Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe 1 : 187–197.
68. LeeMS, CraigieR (1994) Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci U S A 91 : 9823–9827.
69. SuzukiY, CraigieR (2002) Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J Virol 76 : 12376–12380.
70. WhitfieldCR, WardleSJ, HanifordDB (2009) The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. Nucleic Acids Res 37 : 309–321.
71. WaliskoO, SchornA, RolfsF, DevarajA, MiskeyC, et al. (2007) Transcriptional Activities of the Sleeping Beauty Transposon and Shielding Its Genetic Cargo With Insulators. Mol Ther
72. GrabundzijaI, IrgangM, MatesL, BelayE, MatraiJ, et al. (2010) Comparative analysis of transposable element vector systems in human cells. Mol Ther 18 : 1200–1209.
73. LiMA, TurnerDJ, NingZ, YusaK, LiangQ, et al. (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res 39: e148.
74. LiangQ, KongJ, StalkerJ, BradleyA (2009) Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 47 : 404–408.
75. HuangJT, DoonerHK (2012) The spectrum and frequency of self-inflicted and host gene mutations produced by the transposon Ac in maize. Plant Cell 24 : 4149–4162.
76. HuetF, LuJT, MyrickKV, BaughLR, CrosbyMA, et al. (2002) A deletion-generator compound element allows deletion saturation analysis for genomewide phenotypic annotation. Proc Natl Acad Sci U S A 99 : 9948–9953.
77. PrestonCR, SvedJA, EngelsWR (1996) Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144 : 1623–1638.
78. PageDR, KohlerC, Da Costa-NunesJA, BarouxC, MooreJM, et al. (2004) Intrachromosomal excision of a hybrid Ds element induces large genomic deletions in Arabidopsis. Proc Natl Acad Sci U S A 101 : 2969–2974.
79. MartinC, MackayS, CarpenterR (1988) Large-Scale Chromosomal Restructuring Is Induced by the Transposable Element Tam3 at the Nivea Locus of Antirrhinum Majus. Genetics 119 : 171–184.
80. AvilaP, GrinstedJ, de la CruzF (1988) Analysis of the variable endpoints generated by one-ended transposition of Tn21. J Bacteriol 170 : 1350–1353.
81. GrayYH (2000) It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet 16 : 461–468.
82. StadtfeldM, HochedlingerK (2009) Without a trace? PiggyBac-ing toward pluripotency. Nat Methods 6 : 329–330.
83. DaviesDR, GoryshinIY, ReznikoffWS, RaymentI (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289 : 77–85.
84. Claeys BouuaertC, LiuD, ChalmersR (2011) A simple topological filter in a eukaryotic transposon as a mechanism to suppress genome instability. Mol Cell Biol 31 : 317–327.
85. van GentDC, HiomK, PaullTT, GellertM (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J 16 : 2665–2670.
86. AgrawalA, SchatzDG (1997) RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89 : 43–53.
87. GeurtsAM, YangY, ClarkKJ, LiuG, CuiZ, et al. (2003) Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther 8 : 108–117.
88. KaufmanCD, IzsvakZ, KatzerA, IvicsZ (2005) Frog Prince transposon-based RNAi vectors mediate efficient gene knockdown in human cells. J RNAi Gene Silencing 1 : 97–104.
89. CoxJ, MannM (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26 : 1367–1372.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



