-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
Tumour cells overexpress ribosomal RNA (rRNA), which is required for ribosome assembly and cell growth. rRNA gene repression is mediated by the chromatin remodeling complex (NoRC) and a non-coding RNA that binds to this enzyme. This study addresses the mechanism of nucleosome positioning by NoRC and the functional role of the non-coding RNA, which is termed pRNA because it corresponds to the promoter sequence. NoRC recognises the promoter nucleosome in a chromatin array with high affinity and uses a release mechanism to position the nucleosome over the transcription initiation site. The pRNA binds specifically to NoRC and inhibits its ATPase activity. We suggest that the RNA retains NoRC at the gene promoter after remodeling, linking its chromatin modification and scaffolding activity to inactive rDNA copies.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004157
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004157Summary
Tumour cells overexpress ribosomal RNA (rRNA), which is required for ribosome assembly and cell growth. rRNA gene repression is mediated by the chromatin remodeling complex (NoRC) and a non-coding RNA that binds to this enzyme. This study addresses the mechanism of nucleosome positioning by NoRC and the functional role of the non-coding RNA, which is termed pRNA because it corresponds to the promoter sequence. NoRC recognises the promoter nucleosome in a chromatin array with high affinity and uses a release mechanism to position the nucleosome over the transcription initiation site. The pRNA binds specifically to NoRC and inhibits its ATPase activity. We suggest that the RNA retains NoRC at the gene promoter after remodeling, linking its chromatin modification and scaffolding activity to inactive rDNA copies.
Introduction
Nucleosomes present a major obstacle for the binding of sequence-specific DNA-binding factors, the interaction of positively charged histone tails with DNA and the masking of DNA binding sites that face in towards the histone octamer surface [1], [2]. As a result, all DNA-dependent processes, such as transcription, replication, repair and recombination, are affected by the positioning of nucleosomes on regulatory sites. ATP-dependent chromatin remodeling enzymes, which use energy from ATP hydrolysis to slide, evict or replace histones within nucleosomes, are key modulators of chromatin structure and DNA-dependent processes [3]. Thus, it is of particular importance to reveal their molecular mechanism of nucleosome remodeling, how these enzymes are targeted to their genomic loci and their role in defining nucleosome positions in vivo [4]–[10].
In mammalian cells, there are numerous types of remodeling enzymes that associate with different subunits to form remodeling complexes with distinct biological functions. Due to the high combinatorial complexity, it is estimated that several hundred different chromatin remodeling complexes exist in humans. These remodeling enzymes comprise several groups of ATPases classified into the Snf2, ISWI, Mi-2, Chd1, Ino80, ERCC6, ALC1, CHD7, Swr1, RAD54 and Lsh subfamilies [9], [11]. In addition to their diversity, chromatin remodeling enzymes are highly abundant, with approximately one enzyme for every 10 nucleosomes in yeast and human cells [5], [10].
Remodeling enzymes preferentially localise to specific genomic regions, raising the questions of which signals target the enzymes to these locations and what their functions are at these sites [12], [13]. Recently, the continuous sampling model was suggested for the abundant ISWI type remodeling enzymes. According to this model, the enzyme continuously samples all nucleosomes by transiently binding and dissociating without translocation. Only upon introducing additional signals, such as the direct interaction with sequence-specific DNA-binding factors, histone modifications and altered DNA/nucleosome structures, do these nucleosomes become marked as “to be translocated” by converting them to high-affinity substrates [13]. However, there is still a lack of mechanistic proof for the continuous sampling model.
Active rRNA genes cover the promoter-bound nucleosomes from −157 to −2 (relative to the transcription start site), compatible with the binding of the UBF and TIF-IB/SL1 factors required for transcription initiation [8]. On repressed genes, the nucleosome is shifted 24 nt downstream, occluding the TIF-IB binding site [8], [14]. NoRC (nucleolar remodeling complex), which is an ISWI type remodeling enzyme that consists of two subunits, Tip5 (TTFI interacting protein 5) and the Snf2H ATPase, is required to establish the repressed rRNA genes and initiate heterochromatin formation [15], [16]. NoRC is recruited to the rRNA gene by the Transcription Termination Factor-I (TTF-I), which binds upstream of the gene promoter [17]. Recent studies have revealed that NoRC also interacts with the pRNA (promoter-associated RNA), a 150–200 nt long non-coding RNA that is complementary to the rRNA gene promoter sequences and is required for efficient rRNA gene silencing and subsequent DNA methylation [18], [19].
We addressed whether NoRC affects the architecture of the repressed rRNA gene, its mechanism of nucleosome positioning and how the enzyme is targeted to the promoter nucleosome. We demonstrate that, within arrays of nucleosomes, NoRC is capable of recognising the rRNA gene promoter nucleosome with a higher affinity than that for other nucleosomes and that it specifically repositions the nucleosome to the site that was observed in vivo. We show that the mechanism of positioning corresponds to a release model of nucleosome positioning, in which NoRC has a reduced affinity for the remodeled substrate. We further studied the role of the pRNA-NoRC interaction and observed that this RNA serves as a negative regulator of NoRC activity, indicating that tight regulation of these enzymes reduces the wasteful turnover of ATP when maintained within chromatin.
Results
NoRC requires linker DNA for nucleosome binding and remodeling
The remodeling complex NoRC, consisting of the Snf2H and Tip5 subunits, was expressed using the baculovirus system and purified to apparent homogeneity (Figure 1A). The activity of NoRC was tested on mononucleosomal substrates reconstituted on the 601 nucleosome positioning sequence in the centre or at the border of the DNA fragment ([20], [21], Figure 1B and S1). The end-positioned nucleosomes were repositioned by NoRC to the central locations in an ATP-dependent remodeling reaction (Figure 1B, upper panel). In contrast, when the nucleosomes were located at the centre of the DNA fragment, only minor ATP-dependent effects were detected (Figure 1B, lower panel). The initial analysis indicated that the recombinant NoRC complex was active but required a specific nucleosomal substrate for its activity.
Fig. 1. Analysis of NoRC activity and nucleosome binding. 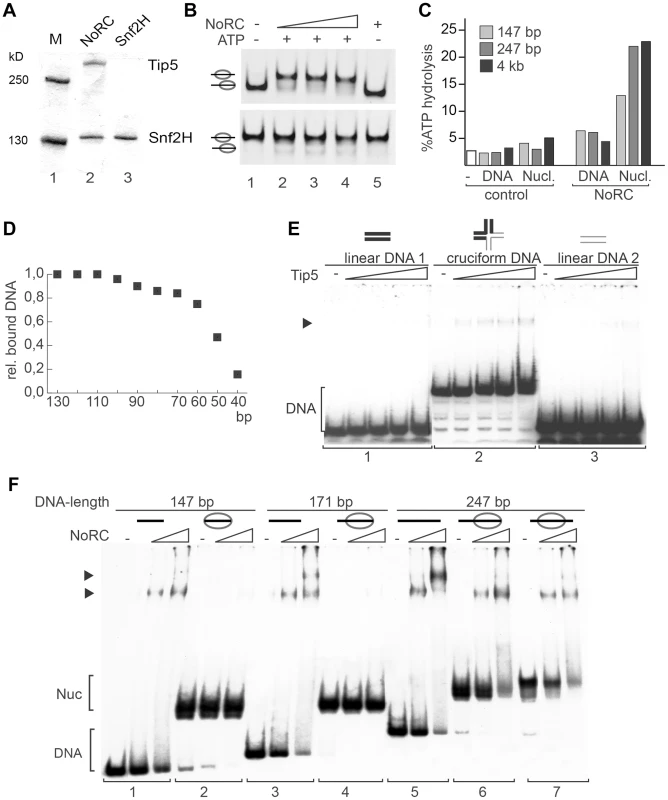
(A) Purified recombinant NoRC and Snf2H proteins were analysed by SDS-PAGE and Coomassie blue staining. Molecular weight markers are shown on the left and arrows on the right point to the recombinant proteins. (B) Remodeling activity of NoRC was tested on nucleosomes reconstituted on 601 DNA [20]. Nucleosomes positioned at the border (upper panel), or at the center of the DNA fragment (lower panel) were incubated with increasing amounts of NoRC and ATP as indicated. After the remodeling reaction the nucleosome positions were analysed on native PAA gels. (C) The ATPase activity of NoRC in the presence of DNA and nucleosomes exhibiting different linker lengths. ATP hydrolysis was measured using radioactive ATP as a tracer and the hydrolysed phosphate was separated via thin layer chromatography. Total ATP hydrolysis was quantified and plotted. (D) The binding affinity of NoRC to DNA molecules of different length was quantified and plotted. NoRC was incubated with a mixture of DNA molecules of different length and analysed by EMSA. The graph shows a quantification of the relative binding of the individual DNA fragments by NoRC. (E) Analysis of Tip5 binding to cruciform DNA. Radioactive labelled cruciform DNA (panel 2) and the two 40 bp DNA controls (panel 1 and 3), which cover the same nucleotide sequence, were incubated with increasing amounts of recombinant Tip5. DNA binding was analysed on native PAA gels. The structure of the annealed oligonucleotides is given on top. (F) Binding of NoRC to DNA and nucleosomes with different lengths of linker DNA. Purified nucleosomes, assembled on a 247 bp rDNA fragment, are either positioned at the border or the center of the DNA fragment. The reconstituted nucleosomes contained either no linker DNA (146 bp fragment), ∼25 bp linker DNA (171 bp fragment), ∼50 bp linker DNA (247 bp, middle position) or ∼100 bp linker DNA (end-positioned nucleosome). A scheme of the nucleosomes is shown on the top. Arrowheads indicate the DNA/NoRC or nucleosome/NoRC complexes. One of the features of the nucleosomal substrate is the linker DNA. To test whether linker DNA is required for NoRC function, we analysed the ATPase activity of NoRC in the presence of nucleosomal arrays and mononucleosomes with and without linker DNA (Figure 1C and S1D). Interestingly, mononucleosomes lacking linker DNA stimulated the ATPase activity of NoRC significantly less than the linker-containing mononucleosomes or nucleosomal arrays. This experiment suggests that recognition of the nucleoprotein structures in the core nucleosome by NoRC activates its ATPase activity but that linker DNA is required for full stimulation.
Next, to determine the minimal length of DNA required for NoRC binding, we carried out DNA-binding experiments using a mixture of DNA molecules with different lengths (from 10 to 130 bp in 10 bp increments, Figure 1D). Quantification of DNA:NoRC complexes in a competitive assay revealed that the DNA-binding affinity of NoRC strongly decreases with DNA lengths below 60 bp and that the remodeler does not significantly bind to DNA of 40 bp or shorter.
Initial experiments did not demonstrate that Tip5 or NoRC have any sequence-specific DNA binding activity (data not shown). However, NoRC may recognise DNA with a particular structure. Therefore, initial binding of Tip5 to cruciform DNA was analysed. Cruciform DNA and two linear, double-stranded 40 bp DNA fragments (‘DNA sequence controls’) were prepared as described [22]. Increasing amounts of Tip5 were incubated with either the cruciform DNA or the linear DNA and analysed in an electromobility shift assay (EMSA). No binding of Tip5 to either of the linear DNA fragments was visible under the experimental conditions (Figure 1E, panels 1 and 3). In contrast, the incubation of Tip5 with the cruciform DNA resulted in the formation of protein/cruciform DNA complexes (panel 2). The experiment shows preferential binding of NoRC to structured DNA.
To test whether linker DNA is required for a stable interaction of NoRC with the nucleosomes, EMSAs using reconstituted mononucleosomes containing linker DNA of 0 bp (146 bp template), ∼25 bp (171 bp template), ∼50 bp (247 bp template, centrally positioned nucleosome) and ∼100 bp (247 bp template, end-positioned nucleosome) and increasing amounts of NoRC were performed (Figure 1F). NoRC bound with similar affinity to the DNA molecules ranging in length from 146 bp to 247 bp, forming discrete NoRC:DNA complexes as expected from the previous experiment. However, when this DNA was reconstituted into nucleosomes, NoRC failed to form a stable complex with the nucleosomes containing 0 bp and 25 bp of linker DNA but formed discrete NoRC-nucleosome complexes with nucleosomes bearing 50 or 100 bp of linker DNA (Figure 1F). Thus, NoRC has a higher binding affinity for free DNA than nucleosomal cores, which suggests that linker DNA is required for efficient targeting of NoRC to remodeling sites.
NoRC interacts symmetrically with the nucleosomal edges and the linker DNA
To determine the relative orientation of NoRC when bound to the nucleosome, we performed DNase I footprinting experiments. Nucleosomes were reconstituted on the central position of the radioactively end-labelled 247 bp mouse rDNA promoter fragment, a known target site of NoRC [15]. Free DNA, nucleosomes and NoRC-nucleosome complexes were incubated with DNase I, the reaction was stopped by the addition of EDTA and the reaction products were resolved by EMSA (Figure 2A, B). Free DNA, nucleosomes and the corresponding NoRC-nucleosome complexes were gel-purified and further analysed on sequencing gels. When compared to free DNA, DNase I digestion of the nucleosomal DNA resulted in a characteristic cleavage pattern, revealing sites of protection and a repeated pattern of DNase I-sensitive sites with a distance of approximately 10 bp, indicating a nucleosome positioned in the centre of the rDNA fragment (Figure 2C). Because a natural DNA sequence was used in this study, the nucleosome lacked precise positioning and a mixture of rotationally phased nucleosomes broadened the protected region [23]. To avoid the formation of multimeric complexes or template precipitation, NoRC was incubated with the nucleosomal substrates at concentrations that result in 50–70% complex formation. NoRC significantly protected the borders of the nucleosome and the adjacent linker DNA from DNase I digestion (Figure 2D). Our data suggest that the binding of NoRC to the nucleosome is bilateral, interacting with both exit and entry sites of the nucleosome, and confirms that NoRC binds to the linker DNA.
Fig. 2. NoRC binds to the entry/exit sites of the nucleosome. 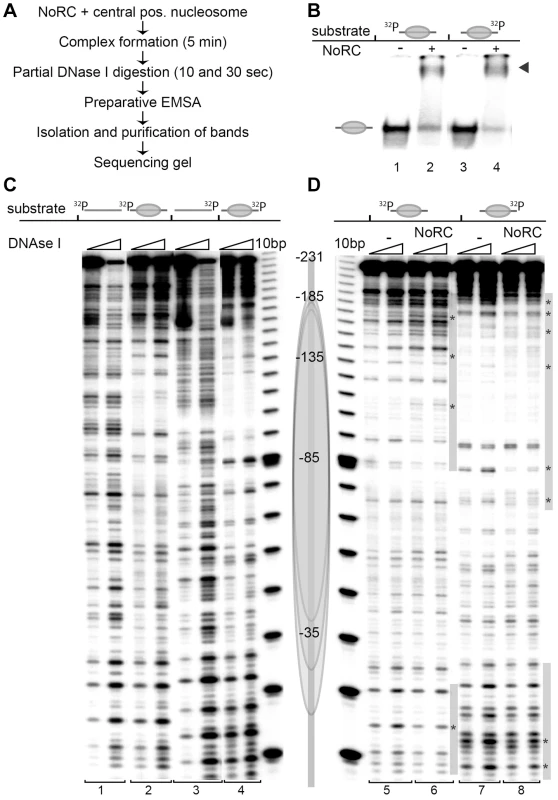
(A) Overview of the experimental approach. (B) Analytical EMSA of the DNase I footprinting reaction. For further analysis of the DNase I digestion pattern the nucleosome and nucleosome/NoRC complexes were isolated from the gel. The arrow indicates the NoRC/nucleosome complexes. (C) DNase I footprinting of DNA and centrally positioned nucleosomes. A 247 bp rDNA promoter fragment (−231 to +16 respective to the start site) was radioactively labelled either at the 5′ or 3′ end. The free DNA (bar) and the centrally positioned nucleosome (gray ellipse) were treated with DNase I and after 10 sec and 30 sec the reactions were stopped with EDTA. Nucleosomes and DNA were resolved by EMSA and the bands were isolated. Purified DNA was subsequently analysed on 7% sequencing gels. A scheme of the central positioned nucleosome is shown on the right. (D) Recombinant NoRC was incubated with a purified nucleosome positioned at the center of the DNA fragment and partially digested with DNase I (10 and 30 sec). The reaction was stopped by the addition of EDTA and the nucleoprotein complexes were separated by native gel electrophoresis. Nucleosomes and NoRC/nucleosome complexes were isolated, DNA purified and analysed on 7% sequencing gels. The nucleosome position (gray ellipse) and the radioactive end-labeling (32P) are indicated. Changes in the digestion pattern upon NoRC treatment are marked with a gray bar, significant changes are highlighted with stars. NoRC determines the nucleosome positions at the rRNA gene promoter
To examine the ability of NoRC to reposition nucleosomes on its target site, we reconstituted mononucleosomes on a DNA fragment containing the rRNA gene promoter sequence in vitro (position −190 to +90, relative to the transcription start site). Nucleosomes reconstituted on the rDNA promoter region occupied multiple positions on the DNA, as demonstrated by native gel electrophoresis (Figure 3A, lane 1). NoRC dependent remodeling establishes a preferential nucleosome position that is located close to the center of the DNA (Figure 3A). This nucleosome position was characterised by Exonuclease III footprinting, showing that it protected the DNA from positions −120 to +27 (Figure S2). This position correlates well with the nucleosome position of the repressed rRNA genes in vivo [8]. This suggests that NoRC recognises specific DNA sequences or structures on the nucleoprotein complex that allow site-specific positioning.
Fig. 3. NoRC targeting and remodeling mechanism. 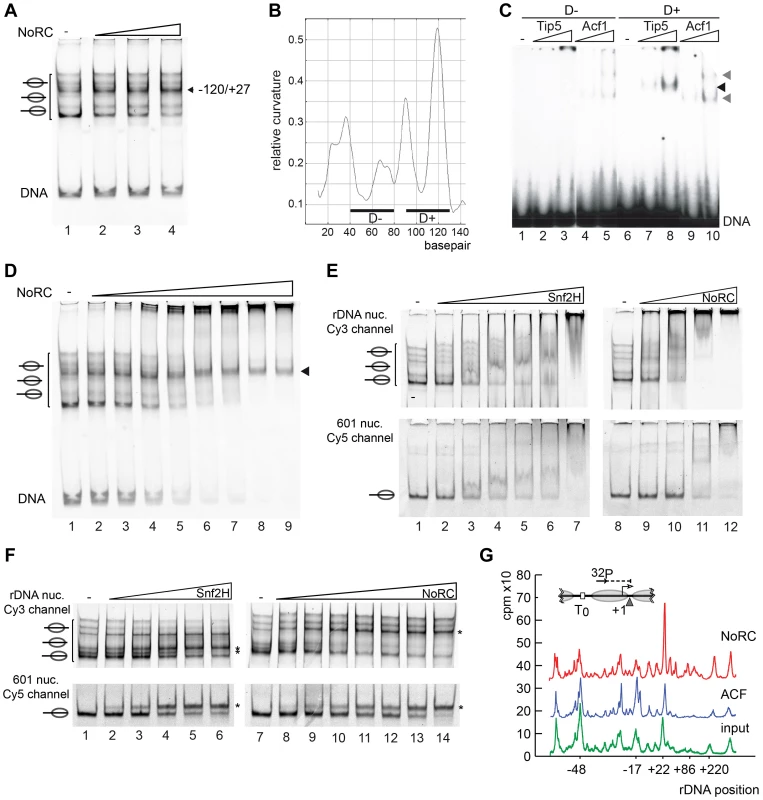
(A) NoRC repositions nucleosomes reconstituted on the rDNA promoter. Mononucleosomes assembled on the rDNA promoter (DNA from position −190 to +90) were incubated with NoRC (20 to 100 nM) and ATP. The reaction was stopped by addition of competitor DNA and samples were analysed by EMSA. The position of the remodelled nucleosome is shown on the left. (B) DNA curvature prediction of the murine rDNA promoter sequence (−231 bp to −95 bp, relative to transcription start site). Curvature was calculated using a DNA curvature prediction program (Bolshoy algorithm/bandit program [26]). The locations and curvatures of the 40 bp DNA fragments used in EMSA are shown: D− (−192 bp to −153 bp) contains nearly no curvature, whereas D+ (−137 bp to −98 bp) oligonucleotide is strongly bent. (C) Analysis of Tip5 binding to structured DNA. Increasing amounts of Tip5 and Acf1 were incubated with the radioactively labelled DNA fragments D− and D+ and the complex formation was monitored by EMSA. (D) NoRC exhibits a reduced affinity for the remodeled nucleosome. Nucleosomes reconstituted on the rDNA promoter were incubated with increasing NoRC concentrations in the absence of ATP and analysed by EMSA. The arrowhead indicates the nucleosome position −120/+27, the final position of the NoRC dependent remodeling reaction. (E) NoRC preferentially binds the rDNA promoter nucleosomes. In the same reaction Cy3-labelled rDNA promoter (upper panel) and Cy5-labelled 601 nucleosomes (lower panel) were incubated with increasing NoRC or Snf2H concentrations in the absence of ATP. Reactions were analysed by EMSA and imaged for the Cy5 and Cy3 channel, respectively. The positions of the nucleosomes are indicated. (F) NoRC preferentially remodels the rDNA promoter nucleosomes. Reactions were performed essentially as shown in (E), but in the presence of 1 mM ATP. The reactions were stopped with competitor DNA and analysed by EMSA. The respective remodeling reaction is visualized by scanning the Cy3 or the Cy5 channel. The positions of the remodeled nucleosomes are indicated. (G) NoRC repositions specifically the promoter nucleosome on an array of nucleosomes. Chromatin was reconstituted on a plasmid DNA containing the rRNA gene promoter and incubated with NoRC or ACF, followed by partial digestion by MNase. The DNA was isolated and analysed by primer extension footprint and denaturing gel electrophoresis. The input chromatin (green), ACF (blue) and NoRC reactions (red) are shown. The relative positions of the peaks to the transcription start site are given. A common feature of ribosomal gene promoters is that they lack sequence homology but retain structural similarity and contain intrinsically distorted regions [24]. The relative DNA curvature of the mouse rDNA promoter was calculated with the Bolshoy algorithm using the ‘bandit’ program (Figure 3B, [25], [26]). The mouse rRNA gene promoter contains a region of high local DNA curvature ([25]; at about position −110) that is specifically bound by Tip5 (Figure 3C). This result agrees with the results of the previous experiment, which demonstrated the preferential binding of Tip5 to cruciform DNA (Figure 1E). Thus, these data indicate the specific recognition of structured DNA by the remodeling enzyme, suggesting a potential mechanism for targeting NoRC to the rRNA gene promoter.
NoRC remodels nucleosomes according to the release model
Two kinetic models were proposed to explain how chromatin remodelers are able to direct the nucleosome to a specific position on DNA [9]. The release model implies that remodelers bind with high affinity to nucleosomes positioned at the “wrong” sites and remodel the nucleosome until it reaches the final (correct) position. The nucleosome at the final position exhibits the lowest affinity for the remodeling enzyme and is thus the worst substrate for the remodeling enzyme. In contrast, the arrest model postulates that the nucleosome exhibits a much higher affinity for the remodeling enzyme at the final position, locking it on the nucleosome and reducing the catalytic conversion rate [9], [27]. To assign one of the kinetic models for a particular remodeler, the binding and remodeling of nucleosomes must be compared. Thus, we compared the differential binding affinities of NoRC to the individual nucleosome positions by EMSA. The incubation of rDNA −190/+90 reconstituted into nucleosomes with increasing concentrations of NoRC resulted in a stepwise binding of the different nucleosome species (Figure 3D). Free DNA and most of the nucleosomes were bound with similar affinities and retarded in the gel. However, the nucleosome occupying the −120/+27 position bound with the lowest affinity. This nucleosome position is the final position of the NoRC-dependent remodeling reaction (Figure 3A), revealing that NoRC has the lowest binding affinity for the “remodeled” nucleosome, therefore suggesting that NoRC remodels nucleosomes according to the release model.
Tip5 targets NoRC to the rDNA promoter
Differential local binding affinities are required to position nucleosomes on DNA. However, on a more global scale, differential binding affinities could also serve to target the remodeling enzymes to specific genes and regulatory regions. To test how NoRC and Snf2H select their remodeling targets, we used competitive binding and remodeling assays. Nucleosomes were reconstituted on a fluorescently labelled rRNA gene promoter fragment (Cy5 labelled) and the 601 nucleosome positioning sequence ([20], Cy3 labelled). Nucleosomes were mixed and binding or remodeling reactions were performed with increasing amounts of remodelers. Snf2H bound with similar affinity to both nucleosome substrates, and remodeled them with similar efficiency (Figure 3E, F). In contrast, NoRC showed preferential binding to the nucleosomes reconstituted on the rRNA gene promoter, preferentially binding the DNA and nucleosomes at lower NoRC concentrations when compared to the 601 substrate (Figure 3E, lanes 8 to 12). Binding with higher affinity was mirrored in the remodeling assay where NoRC was remodeling the rRNA gene promoter nucleosomes prior to the 601 nucleosomes (Figure 3F and Figure S3).
NoRC selectively remodels the promoter nucleosome within a nucleosomal array
As cellular nucleosomes are arranged in arrays, we tested whether NoRC is also capable of selectively recognising and repositioning the rRNA gene promoter nucleosome within nucleosomal arrays. Chromatin was reconstituted using the salt dialysis method on a circular DNA containing the rRNA gene promoter and incubated with NoRC or ACF in the presence of ATP. A partial MNase digestion of the nucleosomal DNA was performed and analysed in a primer extension reaction (Figure 3G). ACF did not qualitatively change the distribution of the nucleosomes within the analysed region of the rRNA gene promoter. However, NoRC induced a specific relocalisation of the promoter nucleosomes, placing the 3′ end of the nucleosome at position +22. NoRC-dependent nucleosome positioning at +22 perfectly corresponds to the cellular nucleosomal configuration of the repressed rRNA gene [8]. The 5 bp discrepancy between the mononucleosome remodeling and array remodeling assay could arise from internucleosomal interactions that influence the remodeling outcome. Our data strongly support the hypothesis that nucleosome remodeling complexes determine nucleosome positioning in vivo, thereby directly affecting gene expression.
Previous studies have revealed a specific interaction between TTF-I and NoRC, suggesting that TTF-I recruits NoRC to the rRNA gene promoter [15], [17]. The results described here reveal an additional targeting signal, encoded by the high affinity of NoRC for nucleosomes positioned at “wrong” sites of the rDNA promoter. TTF-I improves the efficiency of NoRC recruitment to the rRNA gene promoter without affecting the outcome of the NoRC-dependent nucleosome remodeling reaction (Figure S4).
pRNA switches off the remodeling activity of NoRC
Recent studies have demonstrated that NoRC binds to a non-coding RNA, which is initiated upstream of the rRNA gene promoter and contains promoter sequences in the sense orientation. It was suggested that promoter RNA (pRNA) is required to tether NoRC to inactive rRNA genes, where it establishes repressive epigenetic marks [18], [19], [28]. We studied two pRNA constructs that exhibit strong and weak binding affinities for Tip5, pRNA−143/−39 and pRNA−113/−39, respectively [19]. pRNAs were generated by in vitro transcription, re-natured and added to the remodeling reactions (Figure 4). First, the presence of the pRNAs did not influence the nucleosome positioning behaviour of NoRC. Second, we observed specific inhibition of the NoRC-dependent remodeling reaction with increasing levels of pRNA−143/−39 (Figure 4A). In contrast, Snf2H was similarly inhibited by both pRNAs, suggesting that the Tip5 subunit determines RNA-binding specificity and activity. Moreover, NoRC recognises the secondary structure of the pRNA, as inhibition of its nucleosome-remodeling activity was lost when the stem-loop structure was mutated (Figure 4B). We identified a regulatory role of the pRNA, demonstrating that the non-coding RNA serves as an inhibitor of the remodeling enzyme.
Fig. 4. pRNA inhibits the activity of NoRC. 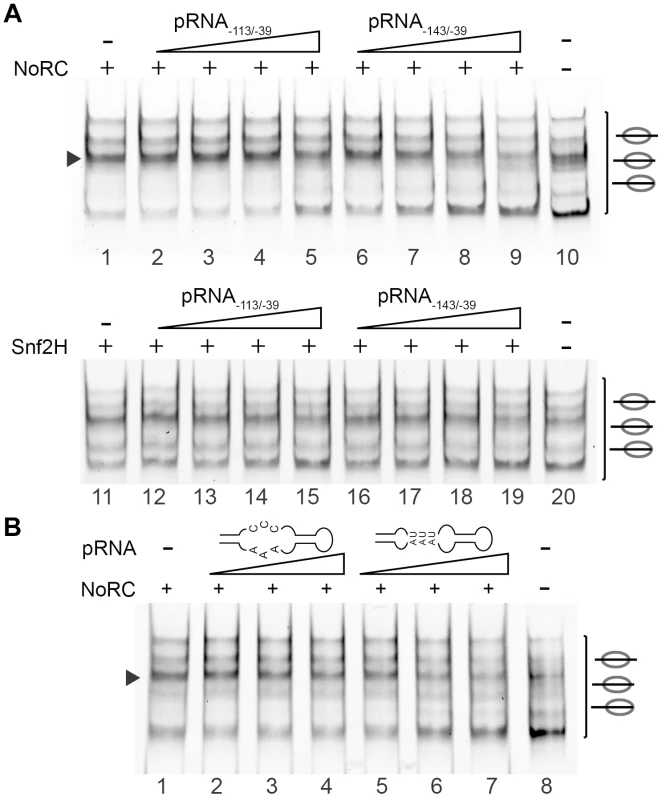
(A) Nucleosomes assembled on the −190 to +90 rDNA DNA fragment were incubated with NoRC or Snf2H, ATP and increasing concentrations of pRNA−143/−39 and pRNA−113/−39 (5 to 200 nM). The remodeling reactions were analysed by EMSA. The arrowhead indicates the nucleosome at the −120/+27 position. (B) NoRC specifically recognizes the hairpin-loop structure of the pRNA. Mononucleosomes assembled on −190 to +90 rDNA promoter region were incubated with NoRC and increasing concentrations of pRNA−127/−39 (20 to 80 nM) or the mutated pRNA missing the hairpin-loop structure in the presence of ATP. Remodeling reactions were stopped after 45 min and analysed by EMSA. The arrowhead indicates the nucleosome at the −120/+27 position. To gain more insight into the inhibitory mechanism of pRNA, we investigated the effect of pRNA on NoRC ATPase activity. The incubation of NoRC with an increasing amount of DNA or pRNA only modestly stimulated the NoRC ATPase activity (Figure S5), whereas the presence of nucleosomes considerably accelerated ATP/ADP exchange. The incubation of NoRC with nucleosomes and increasing amounts of pRNA−143/−39 or pRNA−113/−39 resulted in a RNA concentration-dependent inhibition of the ATPase activity (Figure 5A). As in the remodeling reaction, pRNA−143/−39 inhibited the NoRC-dependent ATPase activity more efficiently than pRNA−113/−39, confirming the higher binding affinity of the remodeling complex for this RNA and explaining the inhibition of the nucleosome remodeling reaction.
Fig. 5. Nucleosomes and pRNA compete for the binding of NoRC. 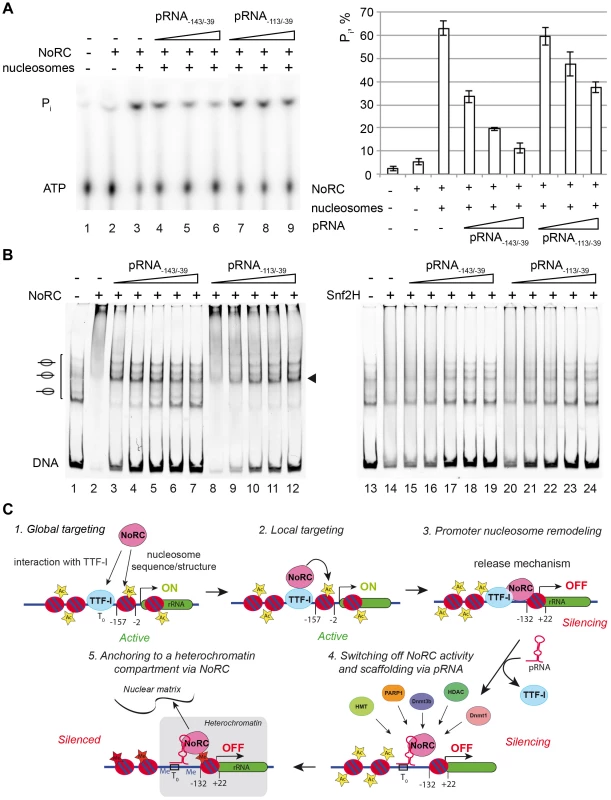
(A) ATPase assay. NoRC was incubated with the indicated pRNAs and radioactive ATP as a tracer. Hydrolysed phosphate was separated via thin layer chromatography and analysed on a PhosphoImager. The quantification of three independent reactions is plotted. Error bars show the standard deviations. (B) Competitive binding assays using NoRC, pRNA and nucleosomes. Nucleosomes assembled on the rDNA promoter (−190 to +90) (lane 1) were incubated with NoRC (lanes 2 to 12), resulting in quantitative complex formation (lanes 2 and 8). These complexes were incubated with increasing concentrations of pRNA as indicated and analysed by EMSA. The nucleosome occupying the position −120/+27 is indicated. Lanes 13 to 24 shows the experiment, but performed with Snf2H. (C) Model describing the putative roles of NoRC and pRNA in rRNA gene silencing. To reveal the mode of RNA-dependent inhibition, we studied the binding of NoRC to nucleosomes in the presence of RNA (Figure 5B). A competitive EMSA revealed that the pRNA competes with nucleosomes for NoRC binding, indicating that only exclusive NoRC:pRNA or NoRC:nucleosomes complexes exist. Again, competition of nucleosomes from the NoRC:nucleosome complex required less pRNA−143/−39 than pRNA−113/−39, indicating the higher binding affinity of pRNA−143/−39 for NoRC (Figure 5B). Both RNA species competed similarly with Snf2H, pointing to the specific role of Tip5 in NoRC (lanes 13 to 24). In summary, our data demonstrate that pRNA competes with nucleosomes for NoRC binding and therefore directly interferes with its ATPase activity and the nucleosome remodeling reaction.
Discussion
NoRC is an ISWI type remodeling enzyme that requires linker DNA for nucleosome binding and efficient activation of its ATPase activity and remodeling. The complex recognises structured and non-structured DNA with a minimal length of 30 bp, and the same length of linker DNA is required for stable interactions with nucleosomes. Our data suggest that the most stable interactions are formed with the linker DNA rather than the nucleosome core, as we were not able to detect interactions between NoRC and the nucleosome core in electromobility shift assays. Reduced binding affinities to the nucleosome core potentially explain the reduced ATPase activity observed with NoRC using nucleosome cores. However, binding to the linker DNA and the orientation of the complex with respect to the nucleosome core are not random, as specific interactions with the DNA entry/exit sites of the nucleosome were visible in DNase I footprinting experiments. NoRC was specifically aligned adjacent to the nucleosome, giving rise to symmetrical DNase I protected and enhanced cleavage sites, a pattern reminiscent of ACF binding to nucleosomes [23], [29].
Recognition and remodeling of the rRNA gene promoter
Ribosomal genes present an ideal model system for studying the dynamics and mechanism of chromatin remodeling, as the epigenetic marks, the chromatin structure of the active and repressed genes and the factors involved are well characterised. Active rRNA genes contain a nucleosome covering the gene promoter from positions −157 to −2, allowing the binding of UBF and TIF-IB/SL1 to their recognition sites at the nucleosomal borders [8]. In contrast, repressed genes have a nucleosome covering the positions from −132 to +22 relative to the transcription start site, masking the binding site of TIF-IB. The repression of rRNA genes is intimately linked with the recruitment of NoRC, which induces nucleosome remodeling, gene repression and the acquisition of heterochromatic marks [16]. We show that the activity of NoRC is sufficient for recognition of the promoter structure and nucleosome positioning in vivo. Nucleosomal arrays are required to establish the cellular nucleosome positioning pattern, suggesting that internucleosomal interactions influence the activity of remodeling enzymes. Our results are in good agreement with data demonstrating that ISWI machines are molecular rulers and potentially act in the context of di-nucleosomes [30], [31]. Although NoRC does not serve as a sequence-independent spacing factor, it is capable of recognising sequence features of the rRNA gene promoter, which serve as positioning signals.
Several studies have demonstrated the importance of positioned nucleosomes in the genome [32]. However, irrespective of the ability of many sequences to position nucleosomes in vitro they fail to do so in vivo [33], [34], suggesting that there are additional mechanisms that structure chromatin. We show that NoRC positions nucleosomes according to the release mechanism [9], [13]. The enzyme binds with high affinity to nucleosomes positioned at “wrong” sites, which is the recruitment signal. The remodeling reaction is highly processive, with ACF moving a nucleosome for approximately 200 bp without leaving the nucleosomal substrate [35]. After initiation of the remodeling reaction, the endpoint of the translocation reaction is determined by a reduced affinity of the remodeler for the nucleosome at this site. As any remodeler with distinct binding affinities to nucleosomes at different but close positions on DNA could position nucleosomes, we suggest that chromatin remodeling enzymes serve to organise chromatin structure with respect to the underlying DNA sequence. The concentration and composition of the remodeling enzymes in combination with the specific targeting of those complexes to chromatin would determine a specific chromatin architecture and specify accessible regulatory sequences that determine the activity of DNA-dependent processes. We suggest that the combinatorial aspect of remodeling enzymes and complex constitution may determine cell types and their responses to the environment.
There are a multitude of signals targeting remodeling enzymes to specific genomic regions, including direct recruitment by proteins, protein modifications, histone variants, coding and non-coding RNAs, as well as nucleosomes at “wrong” positions [13]. The continuous sampling model for chromatin remodeling enzymes suggests that high concentrations of remodeling enzymes and low binding affinities towards the non-signalling nucleosomes allow for efficient screening of the genome for signals that attract remodeling enzymes [10]. Here, we provide evidence for the continuous sampling mechanism of NoRC, where the remodeling enzyme selectively remodels the promoter nucleosome within an array of nucleosomes. Differential binding affinities guide the remodeling enzyme to these sites of action. However, on the genomic scale additional targeting signals help to further increase the local concentration of the remodeling enzymes at their sites of action. In the case of NoRC, interaction with TTF-I directly recruits NoRC and thereby improves the efficiency of the remodeling reaction, but does not influence the remodeling outcome [14], [17].
Effect of pRNA on NoRC-dependent remodeling
Previous studies have shown that the TAM domain in the Tip5 subunit interacts with pRNA and that this interaction is a prerequisite for maintaining NoRC in the nucleolus [18]. We show that pRNA competes with nucleosomes for NoRC binding, specifically inhibiting its ATPase activity. Therefore, we suggest that a ternary complex consisting of NoRC, nucleosomes and RNA does not exist, despite the fact that NoRC contains several DNA/nucleosome-binding domains and an RNA-binding TAM domain [36].
We suggest, that the pRNA serves three functions (Figure 5C). First, after replacing TTF-I at the rRNA gene promoter, it serves to maintain NoRC localisation at the promoter. Due to the release mechanism of nucleosome positioning, NoRC has a low affinity for the remodeled chromatin structure and most likely would dissociate from the promoter. Given, that Grummt and colleagues have shown that the 5′-end of the pRNA forms a triplex with the T0 site at the promoter region and that the 3′-end interacts with Tip5, we propose a tethering function for the pRNA. Switching off the ATPase activity of NoRC ensures that the nucleosome is stably maintained in the OFF position and that the enzyme does not waste ATP. The pRNA and NoRC recruit DNA methyltransferases, histone deacetylases and histone methyltransferases to silence the rRNA genes [37]–[39] and recruit the silenced genes to the heterochromatin environment of the nuclear matrix [36].
Materials and Methods
Proteins
The proteins were expressed in SF21 cells. N-terminally His tagged Snf2H with or without Tip5 was purified via Ni-NTA (Qiagen) chromatography. Flag tagged Snf2H and Acf1 were purified using M2 beads (Sigma) [14].
DNA and RNA preparation
Murine rRNA gene promoter fragments of 146 bp (−231 to −86; positions relative to the transcription start site), 171 bp (−231 to −61), 247 bp (−231 to +16) and 280 bp (−190 to +90) were amplified by PCR from a plasmid containing the genomic DNA isolated from the NIH3T3 cell line (genbank access #KC202874.1). To radioactively label the DNA fragments, α-32P dCTP was added to the PCR reaction mix. The 601 DNA and the pRNA were prepared by PCR as described [19], [40]. PCR products were used for nucleosome assembly reactions as described [23].
Nucleosome assembly
Nucleosomes were assembled according to Rhodes and Laskey using the salt gradient dialysis technique [41]. A typical assembly reaction (50 µl) contained 5 µg DNA, varying amounts of histone octamer, 200 ng BSA/ml, and 250 ng competitor DNA in high salt buffer (10 mM Tris, pH 7.6, 2 M NaCl, 1 mM EDTA, 0.05% NP-40, 2 mM β-mercaptoethanol). The salt was continuously reduced to 200 mM NaCl during 16–20 h. The quality of the assembly reaction was analysed on a 5% PAA gel in 0.4× TBE followed by ethidium bromide staining. Nucleosomes reconstituted on the 247 bp rDNA promoter fragment display two distinct positions that can be separated by native gel electrophoresis [21].
Nucleosome remodeling assay
Nucleosome mobility was assayed as described [42]. Briefly, reactions contained 4 nM Cy5 labelled DNA reconstituted into nucleosomes, 1 mM ATP, 100 ng/µl BSA, 1 mM DTT, 70 mM imidazole in Ex80 buffer (20 mM Tris pH 7.6, 80 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 1 mM β-mercaptoethanol, 10% glycerol, 200 ng/µl BSA) and recombinant remodeling enzymes. Nucleosomes were incubated with NoRC for 45 min at 30°C. The reactions were stopped by the addition of 1700 ng CMV14 plasmid DNA and incubated for 15 min on ice. The nucleosome positions were analysed by electrophoresis on 5% PAA gels in 0.4× TBE and fluorescence scanning.
DNA and nucleosome binding assays
Tip5 binding to cruciform DNA was performed as described [22]. NoRC binding to the DNA and nucleosomes was studied by electromobility shift assays (EMSA). The substrates used in the assay were either radioactively or fluorescently labelled as indicated in the legends. Reactions were performed in Ex80 buffer and the indicated amounts of NoRC. Reactions were incubated for 45 min at 30°C and then analysed by native PAGE. Competitive titration experiments were performed using identical reaction conditions, containing 25 nM NoRC, 4 nM fluorescently labelled mononucleosomes and the indicated amounts of the indicated pRNA constructs. The reactions were analysed on 5% polyacrylamide gels in 0.4× TBE and subsequent fluorescence scanning.
DNaseI footprinting assay
NoRC/nucleosome and nucleosome DNase I footprinting experiments were performed as described [29]. Essentially, radioactively end-labelled DNA was reconstituted into nucleosomes and incubated with NoRC using the same experimental conditions as in the remodeling reactions. DNase I digestions were stopped by the addition of EDTA to a final concentration of 5 mM. The complexes were resolved on native PAA gels and the DNA, nucleosome and NoRC/nucleosome complexes were excised from the gel. DNA was purified and analysed on 7% sequencing gels. Mapping nucleosomal boundaries on nucleosomal arrays before, or after remodelling with NoRC or ACF was performed as described [14].
ATPase assay
An ATPase reaction contained 150 ng of DNA or chromatin in 10 µl of Ex75 buffer, 10 µM ATP and γ32P-ATP (0.1 µl; 3000Ci/mmol, Hartmann Analytic), the indicated amounts of pRNA−143/−39 or pRNA−113/−39 and 10 units RNasin. The reactions were initiated by the addition of the remodeling enzyme and incubated for 60 min at 30°C. Aliquots of 1 µl were spotted on thin layer cellulose chromatography plates (Merck) and air-dried. The hydrolyzed phosphate was separated from unreacted ATP by thin layer chromatography in 0.5 M LiCl/acetic acid buffer. The plates were dried at 65°C for 5 min and exposed to Phospho Imager plates (FujiFilm BAS-1500). ATP and hydrolyzed phosphate spots were quantified using the Multigauge software (Fuji). The percentage of hydrolyzed ATP was calculated according to the following equation: Pi/(ATP+Pi)×100%, where Pi: amount of hydrolyzed radioactive phosphate; ATP: amount of left γ32P-ATP.
Exonuclease III mapping of nucleosome boundaries
Nucleosome positioning on the Cy5 5′ end-labelled mouse rDNA fragment (from positions −190 to +90 relative to the transcription start site) was determined with Exo III mapping. Reactions were carried out in an initial volume of 50 µl with 30 nM nucleosomes and 2 U/µl of Exo III (NEB) in 10 mM Tris, 90 mM KCl,1 mM MgCl2, and 1 mM DTT at 16°C. At different time points 7 µl of the reaction mix were removed and the reaction was stopped by the addition of EDTA (final concentration of 50 mM). Proteins were digested with Proteinase K after the addition of SDS to a final concentration of 1% and the DNA was subsequently purified by ethanol precipitation. DNA samples were analysed on 6% sequencing gels. The DNA ladder was prepared with the DNA Cycle Sequencing Kit (Jena Bioscience) using a Cy5 labelled oligonucleotide and the mouse rDNA promoter fragment (−190 to +90), with either ddTTP or ddCTP in the reaction mix. Results were imaged with a FLA-5000 imager (Fujifilm). As control, we carried out Exo III digestions with naked DNA in order to discriminate nucleosome positions from exonuclease pause sites on free DNA. To map NoRC dependent positions a remodeling reaction was performed prior to Exo III analysis. Remodeling was performed with 7.4 ng/µl of NoRC and Cy5 labelled nucleosomes in the presence or absence of 1 mM ATP for 60 min at 30°C. The reaction was stopped with competitor plasmid DNA and used for native gel analysis and Exo III footprinting.
Supporting Information
Zdroje
1. WorkmanJL, BuchmanAR (1993) Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem Sci 18 : 90–95 doi:10.1016/0968-0004(93)90160-O
2. WolffeAP (1994) Transcriptional activation. Switched-on chromatin. Curr Biol 4 : 525–528.
3. ClapierCR, CairnsBR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78 : 273–304 doi:10.1146/annurev.biochem.77.062706.153223
4. DeindlS, HwangWL, HotaSK, BlosserTR, PrasadP, et al. (2013) ISWI Remodelers Slide Nucleosomes with Coordinated Multi-Base-Pair Entry Steps and Single-Base-Pair Exit Steps. Cell 152 : 442–452 doi:10.1016/j.cell.2012.12.040
5. FlausA, Owen-HughesT (2011) Mechanisms for ATP-dependent chromatin remodelling: the means to the end. FEBS J 278 : 3579–3595 doi:10.1111/j.1742-4658.2011.08281.x
6. RandoOJ, ChangHY (2009) Genome-wide views of chromatin structure. Annu Rev Biochem 78 : 245–271 doi:10.1146/annurev.biochem.78.071107.134639
7. ZhangZ, WippoCJ, WalM, WardE, KorberP, et al. (2011) A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332 : 977–980 doi:10.1126/science.1200508
8. LiJ, LängstG, GrummtI (2006) NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J 25 : 5735–5741 doi:10.1038/sj.emboj.7601454
9. RippeK, SchraderA, RiedeP, StrohnerR, LehmannE, et al. (2007) DNA sequence - and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci USA 104 : 15635–15640 doi:10.1073/pnas.0702430104
10. ErdelF, SchubertT, MarthC, LängstG, RippeK (2010) Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci USA 107 : 19873–19878 doi:10.1073/pnas.1003438107
11. FlausA, MartinD, BartonG, Owen-HughesT (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34 : 2887–2905 doi:10.1093/nar/gkl295
12. YadonAN, SinghBN, HampseyM, TsukiyamaT (2013) DNA looping facilitates targeting of a chromatin remodeling enzyme. Mol Cell 50 : 93–103 doi:10.1016/j.molcel.2013.02.005
13. ErdelF, KrugJ, LängstG, RippeK (2011) Targeting chromatin remodelers: signals and search mechanisms. Biochim Biophys Acta 1809 : 497–508 doi:10.1016/j.bbagrm.2011.06.005
14. StrohnerR, NémethA, NightingaleKP, GrummtI, BeckerPB, et al. (2004) Recruitment of the nucleolar remodeling complex NoRC establishes ribosomal DNA silencing in chromatin. Mol Cell Biol 24 : 1791–1798.
15. StrohnerR, NémethA, JansaP, Hofmann-RohrerU, SantoroR, et al. (2001) NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20 : 4892–4900 doi:10.1093/emboj/20.17.4892
16. GrummtI, LängstG (2013) Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta 1829 : 393–404 doi:10.1016/j.bbagrm.2012.10.004
17. NémethA, StrohnerR, GrummtI, LängstG (2004) The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res 32 : 4091–4099 doi:10.1093/nar/gkh732
18. MayerC, SchmitzK-M, LiJ, GrummtI, SantoroR (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22 : 351–361 doi:10.1016/j.molcel.2006.03.028
19. MayerC, NeubertM, GrummtI (2008) The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 9 : 774–780 doi:10.1038/embor.2008.109
20. ThåströmA, et al. (1999) ThåströmA, LowaryPT, WidlundHR, CaoH, et al. (1999) Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol 288 : 213–229 doi:10.1006/jmbi.1999.2686
21. FelleM, ExlerJH, MerklR, DachauerK, BrehmA, et al. (2010) DNA sequence encoded repression of rRNA gene transcription in chromatin. Nucleic Acids Res 38 : 5304–5314 doi:10.1093/nar/gkq263
22. BianchiME, BeltrameM, PaonessaG (1989) Specific recognition of cruciform DNA by nuclear protein HMG1. Science 243 : 1056–1059.
23. LängstG, BonteEJ, CoronaDFV, BeckerPB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97 : 843–852.
24. SchrothGP, SiinoJS, CooneyCA, Th'ngJP, HoPS, et al. (1992) Intrinsically bent DNA flanks both sides of an RNA polymerase I transcription start site. Both regions display novel electrophoretic mobility. J Biol Chem 267 : 9958–9964.
25. LängstG, SchätzT, LangowskiJ, GrummtI (1997) Structural analysis of mouse rDNA: coincidence between nuclease hypersensitive sites, DNA curvature and regulatory elements in the intergenic spacer. Nucleic Acids Res 25 : 511–517.
26. SchätzT, LangowskiJ (1997) Curvature and sequence analysis of eukaryotic promoters. J Biomol Struct Dyn 15 : 265–275 doi:10.1080/07391102.1997.10508191
27. ErdelF, Müller-OttK, BaumM, WachsmuthM, RippeK (2011) Dissecting chromatin interactions in living cells from protein mobility maps. Chromosome Res 19 : 99–115 doi:10.1007/s10577-010-9155-6
28. SantoroR, SchmitzK-M, SandovalJ, GrummtI (2010) Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep 11 : 52–58 doi:10.1038/embor.2009.254
29. StrohnerR, WachsmuthM, DachauerK, MazurkiewiczJ, HochstatterJ, et al. (2005) A “loop recapture” mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol 12 : 683–690 doi:10.1038/nsmb966
30. RichmondTJ (2012) Nucleosome recognition and spacing by chromatin remodelling factor ISW1a. Biochem Soc Trans 40 : 347–350 doi:10.1042/BST20110748
31. YangJG, MadridTS, SevastopoulosE, NarlikarGJ (2006) The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol 13 : 1078–1083 doi:10.1038/nsmb1170
32. StruhlK, SegalE (2013) Determinants of nucleosome positioning. Nat Struct Mol Biol 20 : 267–273 doi:10.1038/nsmb.2506
33. LiQ, WrangeO, ErikssonP (1997) The role of chromatin in transcriptional regulation. Int J Biochem Cell Biol 29 : 731–742.
34. PeralesR, ZhangL, BentleyD (2011) Histone occupancy in vivo at the 601 nucleosome binding element is determined by transcriptional history. Mol Cell Biol 31 : 3485–3496 doi:10.1128/MCB.05599-11
35. BlosserTR, YangJG, StoneMD, NarlikarGJ, ZhuangX (2009) Dynamics of nucleosome remodelling by individual ACF complexes. Nature 462 : 1022–1027 doi:10.1038/nature08627
36. ZillnerK, FilarskyM, RachowK, WeinbergerM, LängstG, et al. (2013) Large-scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res 41 : 5251–5262 doi:10.1093/nar/gkt218
37. ZhouY, SantoroR, GrummtI (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21 : 4632–4640.
38. SantoroR, LiJ, GrummtI (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32 : 393–396 doi:10.1038/ng1010
39. SchmitzK-M, MayerC, PostepskaA, GrummtI (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24 : 2264–2269 doi:10.1101/gad.590910
40. FelleM, HoffmeisterH, RothammerJ, FuchsA, ExlerJH, et al. (2011) Nucleosomes protect DNA from DNA methylation in vivo and in vitro. Nucleic Acids Res 39 : 6956–6969 doi:10.1093/nar/gkr263
41. RhodesD, LaskeyRA (1989) Assembly of nucleosomes and chromatin in vitro. Meth Enzymol 170 : 575–585.
42. LängstG, BeckerPB (2001) ISWI induces nucleosome sliding on nicked DNA. Mol Cell 8 : 1085–1092.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



