-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
The genetic association with clearance of Hepatitis C virus (HCV) is one of the strongest and most elusive known associations with disease. The genetic variant more strongly associated with improved HCV clearance inactivates the recently discovered IFNL4 gene, which encodes for antiviral IFN-λ4 protein, and turns it into a polymorphic pseudogene. We show that functional IFN-λ4 is conserved and functionally important in mammals. In humans though the inactivating mutation appeared in Africa just before the out-of-Africa migration and quickly became advantageous, with the strength of selection (the degree of advantage) varying across human groups. In particular, selection became stronger out of Africa and was strongest in East Asia, raising the frequency of the pseudogene and resulting in the virtual loss of functional IFN-λ4 protein in several Asian populations. Although the environmental force driving selection is unknown, this process resulted in variable clearance of HCV in modern human populations. The complex selective history of IFNL4-inactivating allele has thus shaped present-day heterogeneity across populations not only in genetic variation, but also in relevant phenotypes and susceptibility to disease.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004681
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004681Summary
The genetic association with clearance of Hepatitis C virus (HCV) is one of the strongest and most elusive known associations with disease. The genetic variant more strongly associated with improved HCV clearance inactivates the recently discovered IFNL4 gene, which encodes for antiviral IFN-λ4 protein, and turns it into a polymorphic pseudogene. We show that functional IFN-λ4 is conserved and functionally important in mammals. In humans though the inactivating mutation appeared in Africa just before the out-of-Africa migration and quickly became advantageous, with the strength of selection (the degree of advantage) varying across human groups. In particular, selection became stronger out of Africa and was strongest in East Asia, raising the frequency of the pseudogene and resulting in the virtual loss of functional IFN-λ4 protein in several Asian populations. Although the environmental force driving selection is unknown, this process resulted in variable clearance of HCV in modern human populations. The complex selective history of IFNL4-inactivating allele has thus shaped present-day heterogeneity across populations not only in genetic variation, but also in relevant phenotypes and susceptibility to disease.
Introduction
Interferon-lambda (IFN-λ) proteins induce antiviral effectors in host target cells and have a crucial role in immune defense against pathogens [1]. The IFNL family classically included three genes (IFNL1, IFNL2, and IFNL3; formerly IL29, IL28A, IL28B, respectively) located within a 50 kb region of chromosome 19 [2], [3]. Several intergenic variants within the IFNL cluster had been identified as showing remarkable association with clearance of hepatitis C virus (HCV) [4]–[6], which is worldwide responsible for ∼170 million infections and over 350,000 deaths per year [7], [8]. The underlying functional basis of this association remained unclear despite numerous efforts to identify functional consequences of these variants [9]–[16].
An additional member of the IFN-λ family has recently been discovered: IFN-λ4, which bears only 30% amino acid identity with the other IFN-λs and is encoded by the IFNL4 gene, also located within the IFNL locus [2], [17]. IFN-λ4 shows similar antiviral activity like IFN-λ3 but as it shows limited secretion it might also act intracellularly, unlike the other IFN-λs [18]. A compound di-nucleotide exonic variant (rs368234815, ΔG>TT) in IFNL4 causes a frame-shift of its open reading frame and results in the polymorphic pseudogenization of IFNL4 - the polymorphic loss of IFN-λ4 protein [17]. The existence of IFNL4 was not even computationally predicted because the human reference genome contains the TT allele and lacks the IFNL4 open reading frame [17]. Remarkably, the derived TT allele not only eliminates IFN-λ4, but it also shows the strongest genetic association reported to date with improved spontaneous and treatment-induced HCV clearance [17], [19], [20].
The function of IFN-λ proteins is crucial for response to pathogens and this locus has evolved under natural selection, with signatures of positive selection being described in the three classical IFNL genes (IFNL1-3) [21]. However, that analysis did not cover the IFNL4 gene, nor the frame-shift rs368234815 variant, which were then unknown [21]. Therefore, the evolutionary history of this interesting functional variant and its influence on the local signatures of selection remained unknown.
Here we report an in-depth comparative and population genetic analysis that focuses on IFNL4 and the rs368234815 polymorphism. We show that the functional IFN-λ4 protein is under purifying selection in mammals, while in humans the IFNL4 pseudogenizing TT allele carries strong signatures of positive selection. We use a new Approximate Bayesian Computation (ABC) approach [22], [23] to provide evidence of a complex selective history of the TT allele, which involves changes in selective strength across human populations. This selective process had important implications in present-day phenotypic diversity and susceptibility to disease.
Results
Functional IFN-λ4 is strongly conserved in mammals
The IFNL4 gene is present in most mammals analyzed, although it is absent in mouse and rat (Methods). To understand the evolutionary conservation of IFNL4 we performed a comparative analysis of the IFNL4 coding sequences from a representative set of mammals (N = 12). The overall dN/dS (non-synonymous to synonymous substitution ratio) is 0.23 across mammals and 0.22 across primates (Figure 1), indicative of purifying selection maintaining the sequence and function of the protein. Notably, all individual branches except squirrel monkey have dN/dS<1 and no model of protein evolution supported dN/dS>1 in specific branches or sites (Table S1). This reveals strong evolutionary conservation of IFN-λ4 in mammals, reflecting its functional relevance.
Fig. 1. Phylogenetic tree showing the dN/dS ratio of each lineage analyzed. 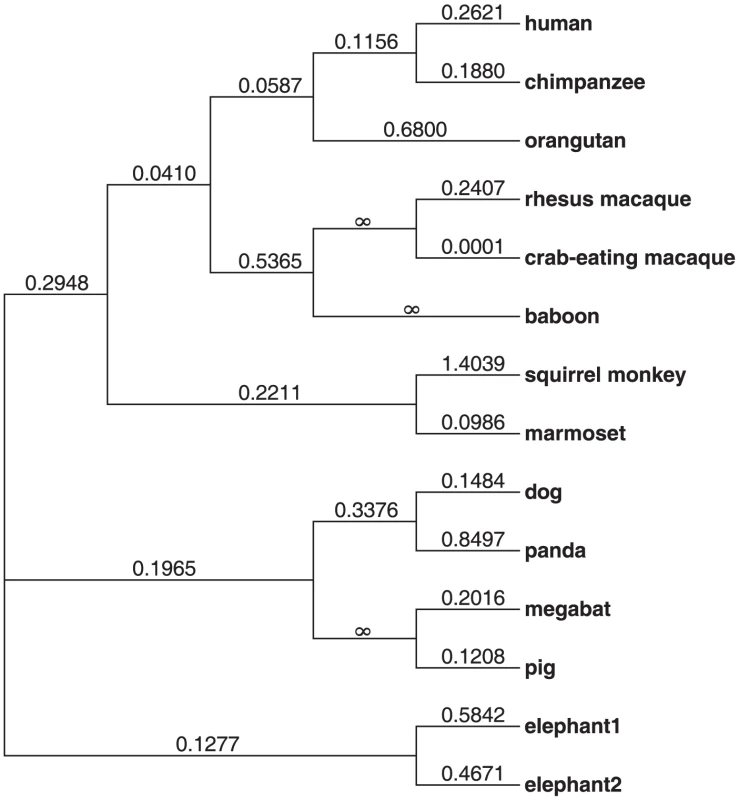
Strong population differentiation for the TT allele
The selective constraint on IFN-λ4 contrasts with the pseudogenization of the gene in humans through the derived TT allele [17]. The multiple-species alignment shows that ΔG is the conserved, ancestral allele and TT is the derived human-specific allele. The mutational process from ΔG to TT in humans is unclear, but only these two forms have been observed, so they should be considered as two alleles of a di-nucleotide variant (Methods). The TT allele shows considerable frequency variation across human groups. The 1000 Genomes data [24] reveals a gradient in frequency that rises from Africa (0.29–0.44) to Europe (0.58–0.77) and the New World (0.51–0.65), and reaches near fixation in East Asia (0.94–0.97) (Figure 2, Table S2, full population names in Methods).
Fig. 2. Allele frequency of rs368234815 - ΔG allele (blue) and TT allele (green) for each population from the 1000 Genomes dataset. 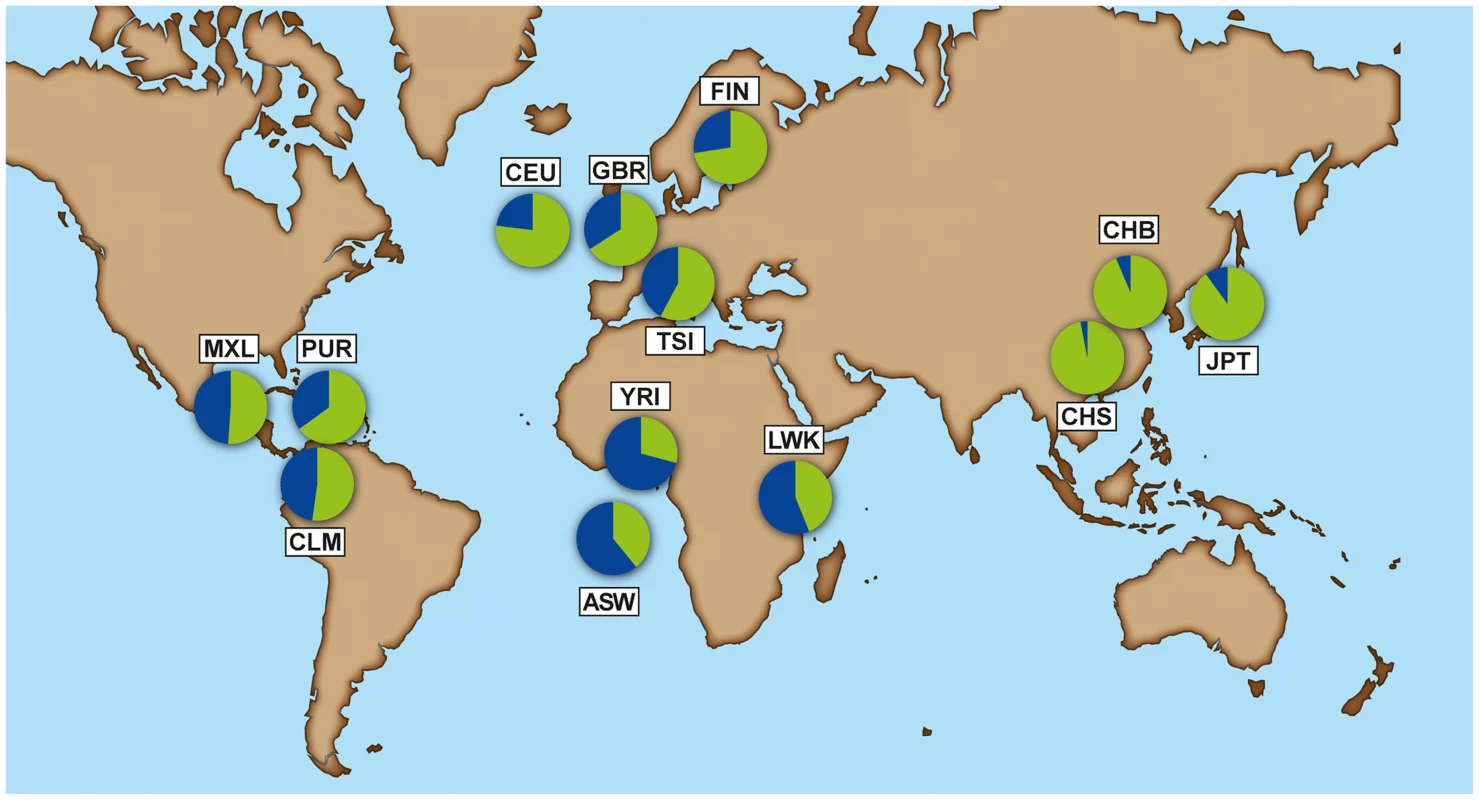
American populations of European and African origin (CEU, ASW) are placed near the geographic area of origin. For full population names see Methods. Population differentiation can be quantified with the fixation index FST [25], a measure of the pairwise level of differentiation in allele frequencies. We used Yoruba (YRI) as the background population because it has the lowest frequency of the derived TT allele in Africa. To put these values in the context of genome-wide population differences, FST was also calculated for every SNP in the 1000 Genomes dataset. For the TT allele the largest FST, 0.63, corresponds to Southern Han Chinese (CHS) versus YRI, which places the TT allele in the 0.5% tail of the empirical genomic distribution of CHS-YRI FST (Fig. 3A, Table 1). FST is also in the 0.8% tail of the genomic distribution for the other East Asian populations (CHB, JPT, Fig. 3B and C, Table 1), and in the 4% tail for Europeans (CEU) and one African population, Luhya (LWK) (Table 1, Fig. S1). These results remain significant when other populations were used as background and in continental comparisons, and when the genome-wide distribution was restricted to SNPs with the lowest frequency in Yoruba (Table S3). Therefore, rs368234815 is among the 0.8% most differentiated SNPs between African and East Asians, and among the 12% most differentiated SNPs between African and European populations.
Fig. 3. Empirical P-values of the FST and XP-EHH analysis (depicted as dots or diamonds, respectively) in the 30 Kb genomic locus around IFNL4 (chr19:39724153–39754153) for (A) CHS, (B) CHB, and (C) JPT using YRI as background. 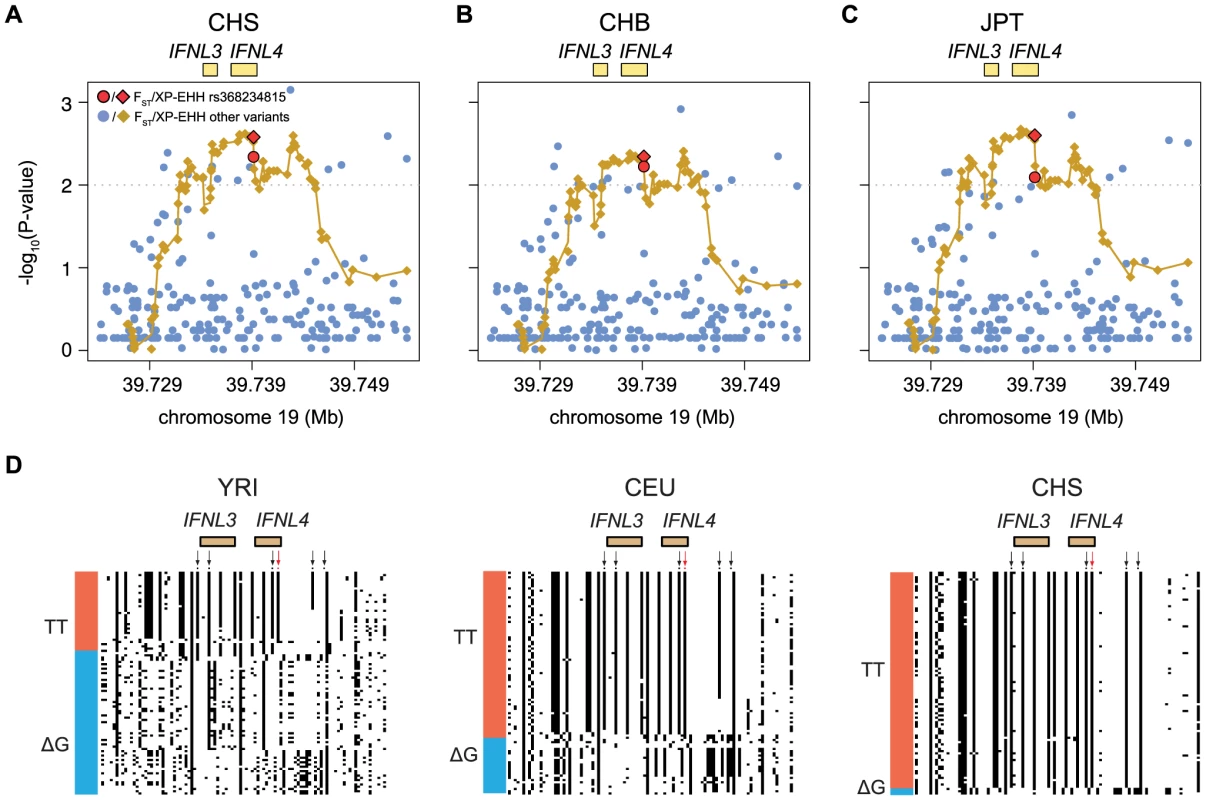
All XP-EHH values are connected by a fitting curve (yellow line). The 1% tail of the genomic empirical distribution is indicated by the horizontal, dashed line. (d) Haplotype structure in the same region as above, for an African (YRI), European (CEU), and East Asian (CHS) population. Columns represent SNPs with a derived allele frequency >5% in at least one population (n = 99 SNPs), with the ancestral allele in white, and the derived allele in black. Lines represent the haplotypes they fall in, as inferred with SHAPEIT by the 1000 Genomes consortium [24]. Haplotypes were sorted based on rs368234815 (red arrow) and SNPs in perfect LD with it in CHS (black arrows); see also Table 2 and Figure 4. The bar on the left-hand side of each plot indicates the haplotypes that carry the TT allele (red) or the ΔG allele (blue). Tab. 1. FST values and for FST, XP-EHH, iHS and Fay and Wu's H (FW) the empirical P-values are shown for rs368234815 in every population. 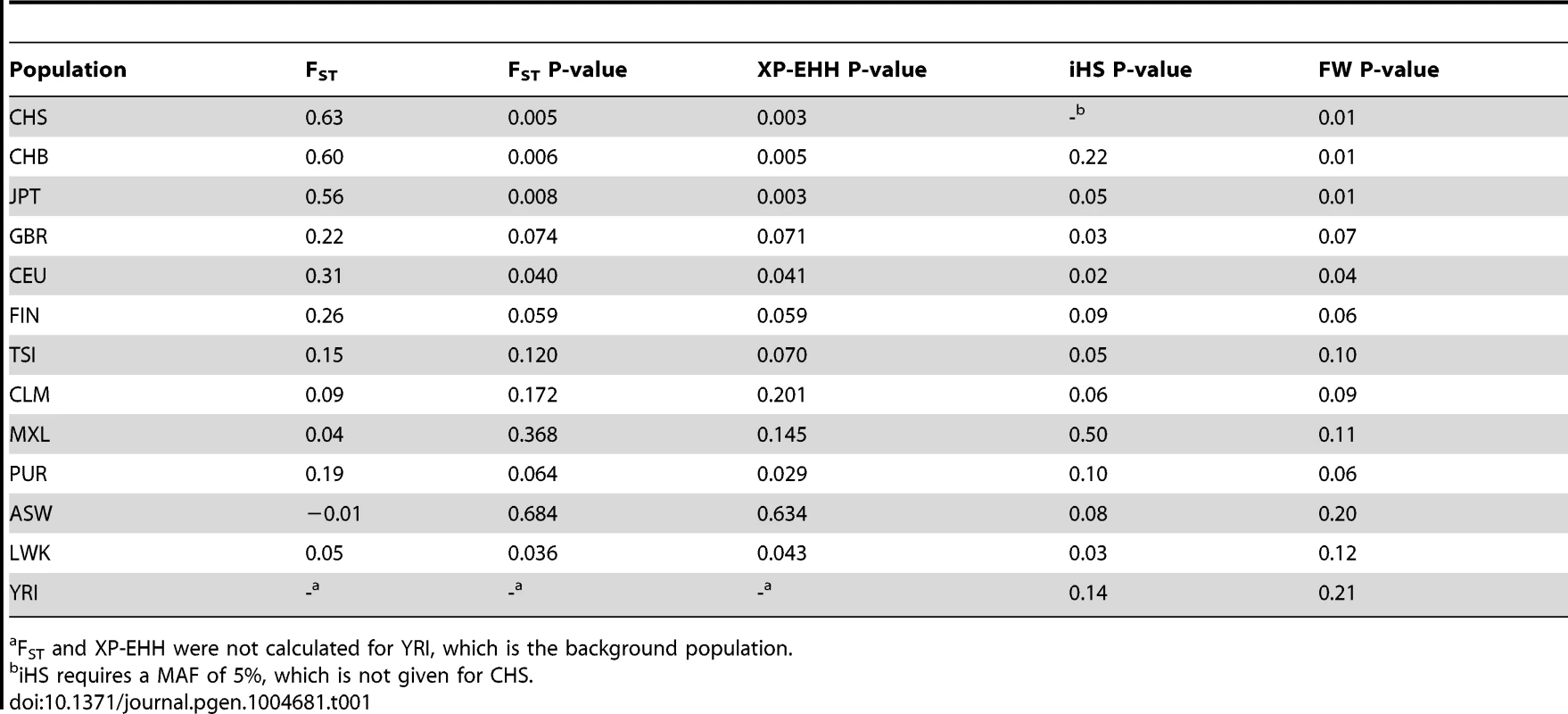
FST and XP-EHH were not calculated for YRI, which is the background population. The TT allele resides in a population-specific extended haplotype
The unusually high population differentiation of the TT allele is compatible with a scenario of recent population-specific natural selection. Under certain selection models such high differentiation should be accompanied by extended haplotype homozygosity in the populations experiencing selection, but not in other populations. We evaluated such a signature with the cross population extended haplotype homozygosity test [26] (XP-EHH), which was calculated across the genome relative to the Yoruba population. The XP-EHH value for the TT allele is in the 0.5% tail of the empirical distribution for East Asian populations (p = 0.003, 0.005, and 0.003, for CHS, CHB, and JPT, Fig. 3A–C, Table 1), and the signal remains significant when calculated relative to a European population (GBR) and in analyses at the continental level (Table S4). In addition, some non-Asian populations show marginally significant signatures of positive selection too (CEU, PUR, LWK, Table 1, Fig. S1). Similar results were obtained with iHS, a statistic that explores haplotype homozygosity within a single population [27] (Table 1) (although iHS lacks power when population frequency is very high, like in Asia). The unusual allele-specific haplotype homozygosity is evident in Figure 3D, which shows the haplotype structure of the locus in one African, one European, and one East Asian population (for all populations see Fig. S3). We note that FST shows very weak correlation with both XP-EHH and iHS in the genome (r = 0.12 and r = −0.08 Spearman rank test, respectively, although the large number of data points makes these weak correlations significant, P-value<2.2e-16). Therefore, the FST and XP-EHH/iHS observations can be considered largely independent.
Finally, not only rs368234815 itself but also its genetic locus shows signatures of recent positive selection, with significant Fay and Wu's H test [28] (FW), which detects an excess of high-frequency derived alleles in the region (Table 1). Together, the combined signatures of FST, XP-EHH, iHS and FW provide strong evidence for the action of natural selection rapidly increasing the frequency of the TT allele in East Asia. The signature outside Asia is less clear, with most populations showing significant signatures of selection for a subset of the tests performed.
Cumulative evidence that TT allele drives the signatures of selection
A classical problem in population genetics is the identification of the genetic variant responsible for a selection signal. High linkage disequilibrium (LD) in the region surrounding IFNL4 (Table 2, Fig. 3D, Fig. 4, Fig. S7) hampers the distinction of signatures across all the linked variants, making it difficult to identify the causal variant. We conclude that rs368234815 is the most likely variant driving the signatures of selection, based on three lines of evidence: (1) its functionality and phenotypic consequences, (2) its genetic association with viral clearance, which reflects its effect on fitness, and (3) its signatures of selection.
Fig. 4. Map of the <i>IFNL</i> locus with locations of relevant SNPs (from <em class="ref"><b>Table 2</b></em>) and the inferred recombination hotspot based on recombination rates from <em class="ref">[60]</em>. ![Map of the <i>IFNL</i> locus with locations of relevant SNPs (from <em class="ref"><b>Table 2</b></em>) and the inferred recombination hotspot based on recombination rates from <em class="ref">[60]</em>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/5838c46a589bfcb64ebfc0ecefede8a9.png)
Tab. 2. Derived allele frequency (DAF), LD to rs368234815, and signatures of selection (empirical P-values for FST and XP-EHH) for other relevant SNPs across the IFNL-locus. 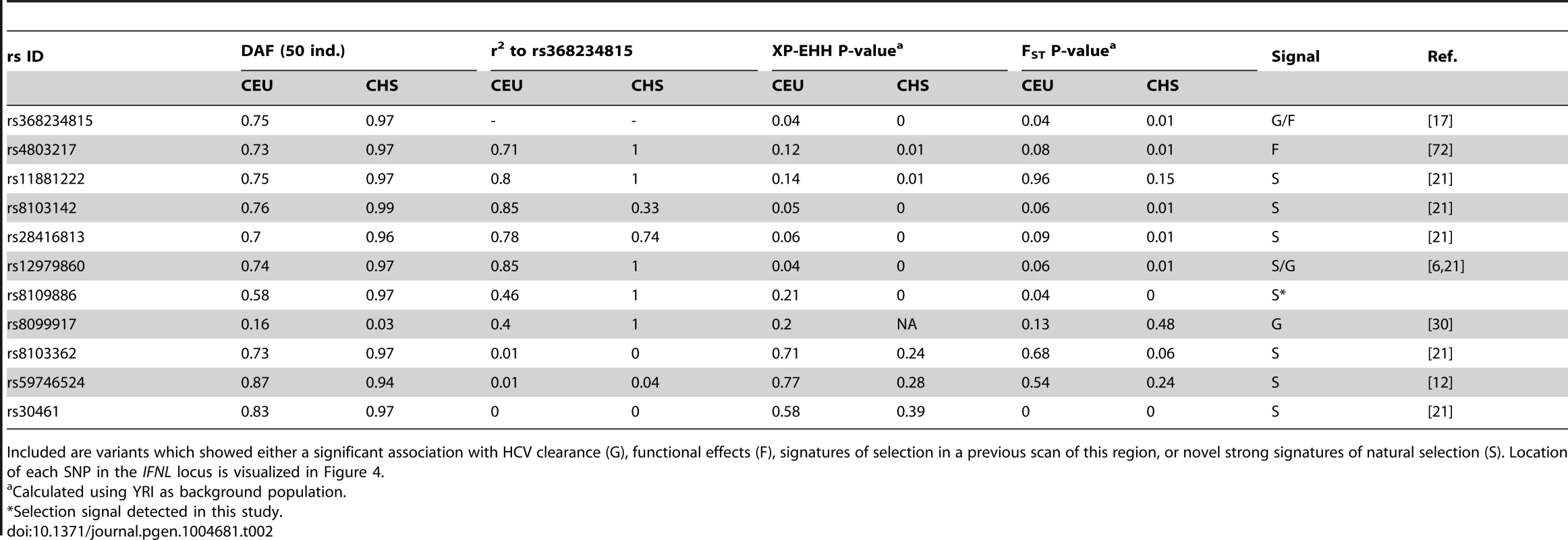
Included are variants which showed either a significant association with HCV clearance (G), functional effects (F), signatures of selection in a previous scan of this region, or novel strong signatures of natural selection (S). Location of each SNP in the IFNL locus is visualized in Figure 4. First, the TT allele has a clear phenotypic consequence as it leads to abrogation of IFN-λ4. This is in contrast with other variants in the locus for which no conclusive functional data has been reported despite numerous efforts [9]–[14]. Second, of all variants in the IFNL region, rs368234815 shows the strongest genetic association with spontaneous and treatment-induced HCV clearance in African Americans [17], [19]; in Europeans and Asians the strong LD across the region results in comparable associations for many variants [15]–[17], [20], [29] (Table 2, Fig. S2, Fig. S7). Third, of all protein-coding or HCV-associated variants in this locus, rs368234815 shows the strongest combined signatures of positive selection in East Asians (Fig. 3A–C, Fig. S1 and S2, Table 2). Only one other polymorphism (intergenic rs8109886, located upstream of IFNL4, Fig. 4), shows signals of selection comparable to rs368234815 (Fig. S2, Table 2). No function has been ascribed to this variant despite a moderate HCV association that is likely due to linkage to TT [6], [17], [30] (Table 2 and Fig. 4), making it a priori a less likely candidate for selection. Indeed, simulations of the evolutionary process showed that the large frequency change of rs8109886 can be explained by linkage to the TT allele alone (Note S1).
We also put rs368234815 in the context of the signatures of selection in the larger genomic region. IFNL4 is located upstream of IFNL3 in a region of moderate LD that is separated from the IFNL1/IFNL2 locus by a recombination hotspot (Fig. S7, Table 2). Manry et al. [21] identified signatures of recent positive selection in all three original IFNL genes (IFNL1-3) but neither IFNL4 nor the rs368234815 variant were known at that time and thus they were not considered. The recombination hotspot breaks LD between the IFNL1/INFL2 locus and the IFNL3/IFNL4 locus (Fig. S7, Table 2), showing that these signatures are in all likelihood independent, as suggested by Manry et al. [21]. There is moderate LD between IFNL4 and IFNL3, with an average r2 between rs368234815 and IFNL3 SNPs of 0.18 in CEU and of 0.44 in CHS (see also Fig. S7). So, the selection signatures in IFNL3 and IFNL4 may not be independent. In fact, the seven SNPs identified by Manry et al. [21] (detailed in Table 2 and Fig. 4) have (1) weaker signatures of selection; (2) unclear functional effects, and (3) weaker association with HCV clearance than the TT allele in Africa (Table 2, Figure 4). Also, those that show some signatures of selection have high to moderate LD with rs368234815 (Table 2), with LD broken mostly by a few recombination events in the ancestral haplotype (Note S1). Taken together, these lines of evidence confirm that IFNL1/2 and IFNL3/IFNL4 have likely been independently targeted by positive selection in recent human history, as suggested by Manry et al. [21], and highlight rs368234815 TT as the most likely selected allele in its region.
Mode and tempo of positive selection on the TT allele
The classical model of positive selection involves selection on a de novo mutation (SDN), a so-called hard sweep, where a new mutation immediately becomes beneficial and selected (reviewed in [31]). This scenario is difficult to reconcile with our observations, because unequivocal signatures of selection are observed only in East Asians but the TT allele is common worldwide. The TT-carrying haplotype harbors the highest genetic diversity in Africa indicating that it arose there before the out-of-Africa dispersion (Note S2, Table S5), a result that is consistent with the IFNL4 haplotype network (Fig. S4). Under SDN, only a model where selection begins weak in Africa and becomes stronger outside of Africa could explain our observations (Fig. 5A). An alternative model is selection from standing variation (SSV), also known as a soft sweep (reviewed in [31]). In this scenario an existing neutral or nearly neutral allele becomes advantageous, for example upon environmental change (Fig. 5A).
Fig. 5. (A) Graphical representation of the different models of selection tested in the ABC analysis (NTR - neutral, SDN - selection on a de novo mutation, and SSV - selection on standing variation). 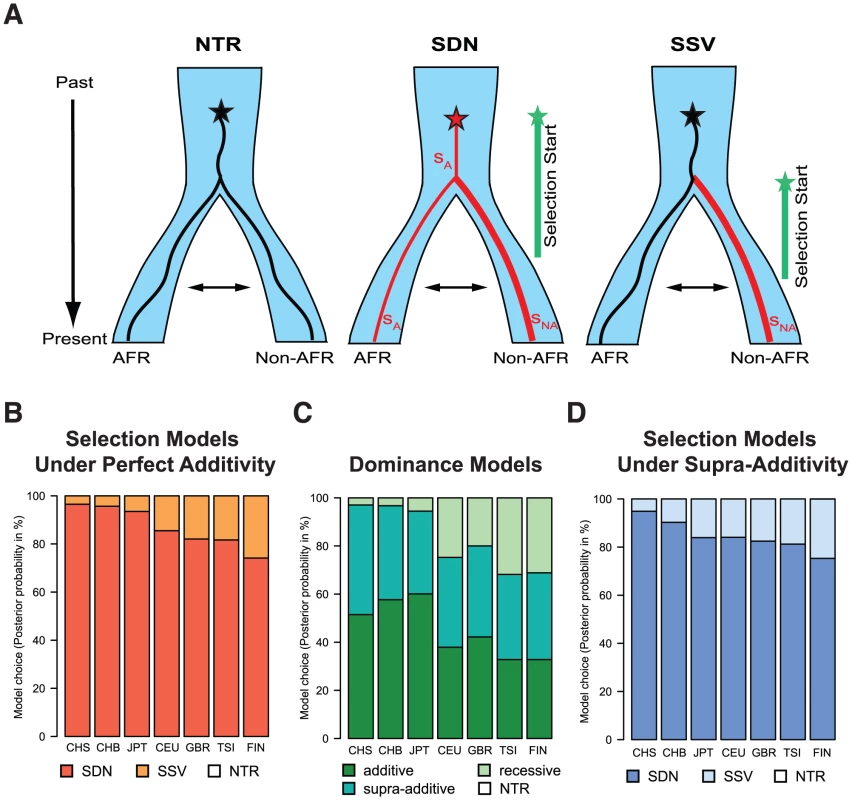
We simulated one ancestral population that splits at the out-of-Africa event (at 51,000 years ago) into the African (AFR) and the non-African (non-AFR) populations, which experience subsequent migration. The star indicates the appearance of the focal mutation. In the first case the neutral (black) mutation appeared and evolved under neutrality in both populations. In the SDN model the advantageous mutation (red) is immediately under positive selection with strength sA, and time when selection started tmut (the prior parameter space for tmut is indicated by a green line); selection strength is allowed to change in the non-African population to sNA. In the SSV model the neutral (black) mutation appeared and evolved under neutrality, becoming advantageous in the non-African population (red line) at time tmut. Prior parameter spaces can be found in methods. (B) Posterior probabilities of the model choice for the different selection models under perfect additivity. (C) Posterior probabilities of the model choice for the different dominance models (and neutrality, NTR). For all models except NTR the posterior probability represent the sum for the SDN and SSV selection models. (D) Posterior probabilities of the model choice for the different selection models under the supra-additive model. In (B), (C), and (D), NTR has negligible posterior probability and is therefore not visible. To disentangle the most likely model of selection for the TT allele we applied a modified version of a recently published ABC approach [23], which we extended to be able to analyze two-population models. In brief, we match millions of simulations under the different models to a summary of the observed genetic data in the IFNL4 region, and use the best matching simulations for further inferences. Under reasonable assumptions we expect the most realistic selection model to produce the closest simulations to real data, and thus simulations can be used to make inferences about the selective history of the allele [23] including the model of selection and relevant parameters (Note S3). While the method relies on some assumptions (e.g. correct demographic and dominance models) this approach has been shown to be robust and to have high power to recover the correct selection scenario [23]. We assess that we overall have high power to recover the correct model, with 76% of the SSV and 95% of the SDN simulations being assigned correctly under the East Asian demographic model, and 70% of the SSV and 97% of the SDN simulations being assigned correctly under the European demographic model (Note S3). The slight bias observed was considered when interpreting the results. For our analysis we consider three models: neutrality (no selection), selection from a de novo mutation (SDN) and selection from standing variation (SSV) (Fig. 5A).
In East Asian populations we obtain negligible support for neutrality and very strong support for the SDN model (Fig. 5B, Table 3, Table S6). Results in Europeans are also consistent with the SDN model, although the weaker signals of selection and the slight bias observed above make these results less conclusive (Table 3, Note S3, Fig. S5, Table S6). The posterior probability for the SDN model is ∼95% in East Asia and ∼80% in Europe, corresponding to Bayes factors (Bayesian measures of relative model support [32]) of ∼10 and ∼4, respectively. This provides substantial and robust evidence for the SDN model, compared to the SSV and NTR models for East Asian and European populations according to Jeffrey's interpretation [33]. Therefore, we conclude that the TT allele was likely positively selected upon appearance. The ABC-based parameter estimates are less reliable than the model choice [23] because they always have large credible intervals (Bayesian measures of confidence). However, the posterior distributions have modes that differ from the modes of the prior distributions, indicating that they are determined by information from the data and not by the prior (Fig. S5). Also, the estimates are quite concordant within and between continental groups (Fig. S5, Table 3). So while they should be interpreted with appropriate caution, the estimates do provide additional useful information about the model and timing of selection. We infer that the TT allele emerged before the out-of-Africa migration (estimated tmut≈55,900 years ago (41,360–68,640)) and was immediately, or shortly thereafter, targeted by moderate positive selection (selection coefficient, sA, ≈0.58% (0.17–1.23)); we estimate that selection intensified substantially outside of Africa, with the selection strength nearly quadrupling in Europe and in Asia (sNA≈2.6% (0.6–4.8); Table 3, Fig. S5).
Tab. 3. ABC results and inferred parameter estimates for the SDN model. 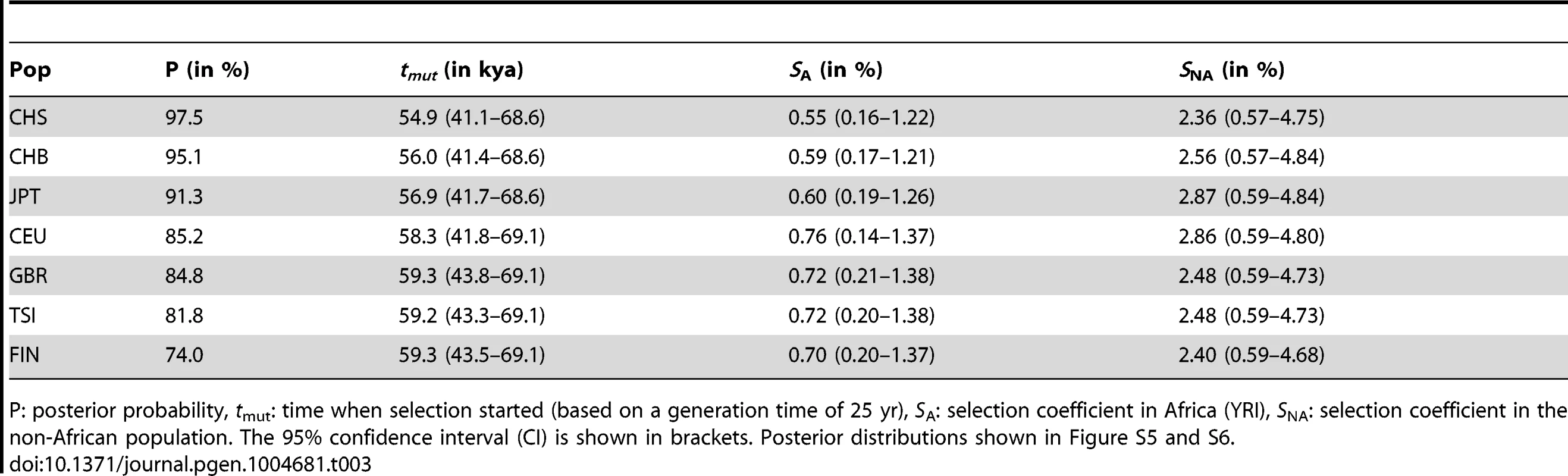
P: posterior probability, tmut: time when selection started (based on a generation time of 25 yr), SA: selection coefficient in Africa (YRI), SNA: selection coefficient in the non-African population. The 95% confidence interval (CI) is shown in brackets. Posterior distributions shown in Figure S5 and S6. One important aspect of the simulations is the mode of dominance (also known as the genetic model), and the ABC analysis above was performed on simulations under a perfectly additive model where heterozygotes have half the fitness effect of homozygotes (dominance coefficient h = 0.5). This model is reasonable because in TT/ΔG heterozygotes only one IFNL4 copy is truncated, and because genetic studies show that the odds ratios (ORs) for HCV clearance in heterozygotes are intermediate to those in the two homozygotes [17]. These two arguments argue strongly against a model of complete dominance for TT as realistic, but other models are more difficult to discard a priory. We thus compare three dominance models: (1) a fully recessive model for the TT allele (h = 0), (2) the perfectly additive model used above (h = 0.5), and (3) a supra-additive model where the additive effect is non-linear and heterozygotes are closest in fitness to ΔG homozygotes. This model has been proposed based on the ORs for the intronic IFNL4 variant, rs12979860 which is in high LD with rs368234815 and is thus a good proxy for the dominance effects of TT (Table 2) [34]. Based on those results we use a dominance coefficient h = 0.38 (see Note S4). When we compare the three dominance models in East Asia, regardless of the selection model, the fully recessive model has marginal support (4%), with the two additive models showing similar posterior probabilities (slightly higher for additive: 56%, than supra-additive: 44%, Fig. 5C and Note S4). When we compare the ABC results in the two additive models, they both strongly support the SDN model over the SSV model (95% in the additive model and 90% in the supra-additive model, corresponding to a Bayes factor of ∼12), and both models provide virtually no support for the neutral model (Figure 5B–D and Note S4). Parameter estimates also agree well among these two models (Fig. S5, Fig. S6 and Note S4). Therefore our results in East Asia validate the use of an additive model and show that the ABC inferences are not sensitive to the particularities of the additive model used. In European population the results are less clear, just as in the original ABC analysis and as expected given the weaker signatures of selection. Still, these results also support the two additive models (36% support for additive and 38% for supra-additive; Fig. 5C) as well as the SDN model (∼81% support for SDN in both the additive and the supra-additive, corresponding to a Bayes factor ∼4, Fig. 5B and D, Note S4).
These results show a complex selection history for the TT allele, with selection starting upon appearance of the allele but with intensity changing over time and geographic range. The model is consistent with all our observations, including the marginal evidence for selection observed in non-Asian populations (Table 1). It is interesting that we infer selection on the TT allele even in Yoruba, where the signature is undetectable with classical methods likely because of weak selection and lower frequency although the TT allele shows clear signatures of homozygosity (Fig. 3D). Interestingly, and in agreement with this model, we do observe some signatures of positive selection in another African population, the Luhya. It remains possible that the advantage of the TT allele was counteracted by additional selective forces in Africa that maintained the TT allele at an intermediate frequency, such as balancing selection, although we note that the locus lacks classical signatures of long-standing balancing selection (Note S5, Table S7).
Discussion
Here we show that functional IFN-λ4 is under purifying selection throughout the mammal clade while positive selection has favored the elimination of IFN-λ4 through pseudogenization in humans. Selection on the TT allele has been particularly strong in specific populations, leading to extremely high frequency of the pseudogene and subsequent virtual loss of IFN-λ4. This event is phenotypically relevant: not only is IFN-λ4 biologically important [17], [18] and evolutionary conserved, but the loss of IFN-λ4 through pseudogenization shows remarkable association with improved HCV clearance [17], [19], [20].
The precise reason behind the advantage of IFN-λ4 elimination is unknown, but its immunological role and clear antiviral activity against HCV [17] make exposure to pathogens (and in particular viral agents) the most likely selective force. However, due to its slow progression into fatal disease [35] HCV is unlikely to have exerted such strong selective pressure, although we cannot completely discard this possibility. Besides HCV, it has been shown that functional IFN-λ4 has antiviral activity against coronaviruses [18], while the IFN-λ4 pseudogene increases susceptibility to cytomegalovirus retinitis among HIV-infected patients [36]. Suggesting that IFN-λ4 pseudogenization is likely associated with several phenotypic traits. It is perhaps surprising that suppression of an antiviral protein results in improved viral clearance, although it has for example been shown that during chronic infection blockage of persistent signaling of IFN I (a different type of interferon) can improve viral clearance [37], [38].
We showed that a complex selective regime, with variation in selection strength in different geographical areas, best explains the history of the IFNL4 locus. Signatures of non-neutral evolution have been detected in other interferons, including at least one other IFNL family member (IFNL1 or IFNL2) [21]. Although the mode and tempo of selection in these other IFNL genes are not well understood, together these observations suggest that IFN-λ proteins have played an important role in recent human adaptation, probably as a consequence of their role in individuals' constant fight with pathogens. It is likely, though, that only the selective history of the IFNL4-TT allele had a strong influence in the rate of clearance of some viruses, at least HCV, across human groups.
It has been proposed that gene loss may exert an important role in evolution, including human evolution [39], and the loss of otherwise conserved regulatory elements may play a role in the acquisition of human-specific phenotypes [40]. Loss-of-function mutations show global signatures of purifying selection [41]–[43] and tend to carry detrimental effects [44]. A few exceptions exist, though, where truncating polymorphisms show signatures of positive or balancing selection [45]–[50]. Still, as with other targets of selection, most of these cases lack biological interpretation. In fact, IFNL4 joins a small group of known genes where a striking signature of local adaptation is coupled with a clear molecular phenotype (e.g. [46], [47], [51]), which in this case is also associated with disease risk. As such, it contributes to our understanding of how recent human evolution has shaped genetic and phenotypic human diversity, including present-day heterogeneity in susceptibility to disease.
Materials and Methods
Molecular Evolution of IFNL4 across species
In order to explore the level of functional constraint in IFNL4, we estimated the level of protein conservation in primate and non-primate mammals. Specifically, we assessed the ratio (dN/dS) of non-synonymous substitutions per non-synonymous site (dN) to synonymous substitutions per synonymous site (dS) across gene orthologs. Since purifying selection eliminates deleterious protein-coding changes, dN/dS decreases with negative selection and increases with relaxed constraint and positive selection.
We used human IFNL4 reference sequence NM_001276254.2 to BLAT genomes of other species and generate multiple-species sequence alignment of IFNL4 coding exons 1 through 5 (Table S8). The panda-predicted IFNL4 ortholog was subsequently used as BLAT query to extract coding exons for additional non-primate species (Table S8). Further, we sequenced IFNL4 (exons and introns) in genomic DNA and reconstructed complete IFNL4 cDNA sequences of chimpanzee (Genbank accession JX867772), baboon (Genbank accession KC525947) and crab-eating macaque (Genbank accession KC525948). The whole IFNL4 genomic region is absent in mouse or rat. All discovered functional IFNL4 sequences (Table S8) where used for a multiple-sequence alignment which was created using ClustalW [52] and annotated with Jalview [53].
The alignment was analyzed with codeml (part of PAML4 [54]) to test various models of selection. We estimated the overall dN/dS for the complete tree and compared likelihoods for models that allowed: i) free dN/dS for each branch (i.e., lineage heterogeneity); ii) a primate-specific dN/dS; and iii) a human-specific dN/dS. Additionally, we performed tests aimed to detect site-specific signatures of positive selection across the phylogeny (branch models): i) model 1a (neutral) vs. model 2 (positive selection); ii) model 7 (neutral) vs. model 8 (with dN/dS>1); and iii) model 8a (with dN/dS = 1) vs. model 8 (with dN/dS>1).
Human population genetic data
We analyzed genome-wide data from the 1000 Genomes release (2010/11/23; phase I) [24]. We considered (1) autosomal variants detected in the low coverage sequencing, and (2) populations with information for at least 50 unrelated individuals, which was met by 13 populations from four different continents [African ancestry: YRI (Yoruba in Ibadan, Nigeria), LWK (Luhya in Webuye, Kenya), ASW (African Ancestry in Southwest US); European ancestry: GBR (British from England and Scotland), CEU (Utah residents (CEPH) with Northern and Western European ancestry), FIN (Finnish from Finland), TSI (Toscani in Italia); East Asian ancestry: CHS (Han Chinese South), CHB (Han Chinese in Beijing, China), JPT (Japanese in Toyko, Japan); American ancestry: MXL (Mexican Ancestry in Los Angeles, CA), CLM (Colombian in Medellin, Colombia), PUR (Puerto Rican in Puerto Rico)]. As an exception PUR (Puerto Rico) was analyzed although it contains only 44 individuals. Some analyses were performed both by population and by continent; in these cases the continental groups contain 150 randomly selected, unrelated individuals (America 144) with an equal contribution from each population within the continent.
For the rs368234815 ΔG/TT frameshift-substitution variant the 1000 Genomes dataset only contains the T insertion/deletion variant rs11322783 (-/T, chr19 : 39739154, dbSNP b138), while the substitution rs74597329 (G/T, chr19 : 39739155, dbSNP b138) is absent. This is due to the automatic variant caller failing to correctly identify an insertion and a substitution in the same genomic position. We sequenced an amplicon containing rs368234815 in 153 individuals included both in the 1000 Genomes and HapMap sets (CEU, YRI and CHB/JPT). Sequencing confirmed the presence of only two alleles (ΔG and TT) and showed good concordance with the 1000 Genomes data between our ΔG/TT genotypes and 1000 Genomes genotypes for the overlapping insertion/deletion variant rs11322783 (4 individuals of 153 tested were discordant, providing an estimated 97.4% genotype and 98.7% allele concordance rate). This validated the use of 1000 Genomes dataset for our subsequent analyses. We used the ancestral allelic state annotated in the 1000 Genomes data, which is based on the Ensembl 59 comparative 32 species alignment [55]; only SNPs with a high-confidence ancestral inference were used, and indels were excluded due to their cryptic variation patterns [56].
Signatures of selection
We used FST, iHS and XP-EHH to explore the signatures of selection of rs368234815 TT allele. FST is a measure of population differentiation and unusually high FST can indicate population-specific positive selection that drastically increases allele frequency in the population under selection [57]. To calculate FST we used the Weir and Cockerham [25] estimator implemented in vcf-tools [58].
Positively selected alleles rapidly increase in frequency with recombination having little chance to break their association with nearby variants. If the selected allele was originally in few haplotype backgrounds and it has not reached fixation, it will be associated with extended haplotype homozygosity (EHH), a pattern that will be absent for the non-selected allele. We used two statistics to explore this expectation. First, iHS [27] measures the allele-specific decay of EHH within a population by comparing the associated EHH of ancestral and derived alleles. Second, XP-EHH [26] that detects alleles that are under selection in one population only, by comparing EHH patterns both among allelic types and across populations; as such XP-EHH has higher power to detect population-specific selection. Low frequency variants break the EHH signal, so following [59] we considered only SNPs with derived allele frequency ε 5% for XP-EHH or minor allele frequency ε 5% for iHS. Local recombination rate estimates were obtained from a combined recombination map based on HapMap data [60] from Africa, European, and Asian populations. Both statistics were standardized to a mean of zero and a standard deviation of one; for iHS, scores were then binned by frequency (1%) as previously suggested [27]. Correlation of FST with XP-EHH (CHS vs. YRI) or iHS (CHS) was calculated for all variants present in the respective dataset with Spearman's rank correlation test implemented in R [61].
We used each of these statistics to analyze every non-African population; for between-population comparisons we used Yoruba as background, unless noted otherwise. To assess the putative effects of this choice of populations we repeated the analyses for continental groups, for different background populations, and for SNPs that have their lowest allele frequency in Yoruba. In all cases the empirical P-values were obtained by comparing the score for rs368234815 to the whole-genome empirical distribution of the respective statistic. Since this is a hypothesis-driven analysis with a single variant analyzed within a single locus, no multiple testing or genome-wide corrections are needed.
We also applied tests that analyze the signatures of selection in the IFNL4 genetic region (∼2.5 kb). Here we show results for Fay and Wu's H test [28], which detects the excess of high-frequency derived alleles expected after a recent sweep with recombination. Significance was estimated using 10,000 standard neutral coalescent simulations [62]. Because demography affects the SFS and can cause spurious results if not properly accounted for, our simulations are run under a demographic model which includes inferred parameters for populations of African [63], European [63], Asian [63] and American [64] ancestry. A custom made perl program (Neutrality Test Pipeline) was used to calculate the statistic and corresponding P-value.
ABC analysis
To infer the model of selection that best fits IFNL4 data and estimate the timing and selection strength of the TT allele, we used an Approximate Bayesian Computation (ABC) approach [22]. In particular, we followed a published approach [23], which has been previously shown to discriminate well between SDN, SSV and neutrality (NTR) [23]. In brief, this approach is based on performing a large number of simulations under different selection models, with random parameters drawn from some probability distribution (called the prior distribution). Real data and simulations are compared based on summary statistics, and through a rejection scheme the simulations that most closely resemble real data help inform inferences about the best-fitting model. The parameter values that generate these simulations are then used to obtain the posterior distribution of each parameter, whose mean and standard deviation are used to perform the parameter inferences. We extended the method to consider more than one population, since two-population statistics are most informative in our case.
Specifically, the approach uses msms [65] to simulate data, custom python scripts to calculate all summary statistics, and ABCtoolbox [66] for all ABC inferences. Under both selection models, we started with uniform priors with a range as follow (see Fig. 5A):
-
SDN model - selection strength in Africa sA∼U(0,1.5%); selection strength in non-Africa sNA∼U(0.5,5%); time when selection started tmut∼U(40,70kya)
-
SSV model - selection strength in non-Africa sNA∼U(>0,5%); frequency of the allele when selection started f0∼U(0,20%); time when selection started tmut∼U(21,51kya)
-
NTR model - time when mutation appears t∼U(40,70kya)
Because simulations with the selected allele fixed are likely to be very different from the observed data, we conditioned on the selected allele segregating in both populations. This resulted in non-uniform prior distributions presented in Figure S5 and S6. We used 104 simulations to distinguish between the neutral model and the two selection models, and a larger set of 8×105 simulations for the more subtle distinction between the two selection models and for parameter estimation. For the simulations, we used the population history model estimated by Gravel et al. [63] and assumed a constant recombination rate of 1.76 cm/Mb throughout the region (average recombination rate in the IFNL locus [60]), and a perfectly additive model of dominance (h = 0.5). Lack of an appropriate demographic model for American and non-Yoruba African populations precludes analysis for those populations. The following single-population statistics were calculated: the average number of pairwise differences π, Watterson's θ, Fay and Wu's H [28] and Tajima's D [67], all for both 4 kb around the site and a 8 kb (6 kb upstream and 2 kb downstream of the site) interval around the TT allele. The between-population statistics employed were: FST [68] for the selected site, FST in 4 kb around the site, FST for the whole region, and XP-EHH on the selected site [26]. In addition, we also included the frequency of the selected allele in both populations. This resulted in a set of 16 summary statistics, which, following Wegmann et al. [69] and Peter et al. [23], was reduced to seven summary statistics using PLS-DA [70] for model choice and regular PLS for parameter inference [71]. Performance of the ABC model choice and parameter distribution for the SDN model has been assessed for each particular model (Note S3). Confidence in the choice of selection models has been supported with Bayes factors.
In addition, we investigated the influence of the dominance model in our inferences. We analyzed a recessive model for TT (h = 0), the perfectly additive model above (h = 0.5), and a supra-additive model (h = 0.38), using 500,000 simulations for each model. We run an ABC analysis for model selection with all simulations (from all three dominance models and the three selection models NTR, SDN, and SSV). We then assess the posterior probability of each dominance model regardless of selection model, and the posterior probability (and parameter estimates) of each selection model for the additive and supra-additive dominance models (see Note S4).
URLs
1000 Genomes, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/; GENCODE, http://pseudogene.org/psidr/; HapMap, http://hapmap.ncbi.nlm.nih.gov; XP-EHH and iHS executables, http://hgdp.uchicago.edu/Software/; VCFtools, http://vcftools.sourceforge.net; ABCtoolbox: http://www.cmpg.iee.unibe.ch/content/softwares__services/computer_programs/abctoolbox/index_eng.html; msms: http://www.mabs.at/ewing/msms/
Supporting Information
Zdroje
1. DonnellyRP, KotenkoSV (2010) Interferon-Lambda: A New Addition to an Old Family. Journal of Interferon & Cytokine Research 30 : 555–564.
2. SheppardP, KindsvogelW, XuW, HendersonK, SchlutsmeyerS, et al. (2002) IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4 : 63–68.
3. KotenkoSV, GallagherG, BaurinVV, Lewis-AntesA, ShenM, et al. (2002) IFN-θs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4 : 69–77.
4. TanakaY, NishidaN, SugiyamaM, KurosakiM, MatsuuraK, et al. (2009) Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet 41 : 1105–1109.
5. ThomasDL, ThioCL, MartinMP, QiY, GeD, et al. (2009) Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461 : 798–801.
6. GeD, FellayJ, ThompsonAJ, SimonJS, ShiannaKV, et al. (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461 : 399–401.
7. HanafiahKM, GroegerJ, FlaxmanAD, WiersmaST (2013) Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57 : 1333–1342.
8. PerzJF, ArmstrongGL, FarringtonLA, HutinYJ, BellBP (2006) The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 45 : 529–538.
9. DillMT, DuongFH, VogtJE, BibertS, BochudP, et al. (2011) Interferon-Induced Gene Expression Is a Stronger Predictor of Treatment Response Than {IL28B} Genotype in Patients With Hepatitis C. Gastroenterology 140 : 1021–1031.e10.
10. FukuharaT, TaketomiA, MotomuraT, OkanoS, NinomiyaA, et al. (2010) Variants in {IL28B} in Liver Recipients and Donors Correlate With Response to Peg-Interferon and Ribavirin Therapy for Recurrent Hepatitis C. Gastroenterology 139 : 1577–1585.e3.
11. LanghansB, KupferB, BraunschweigerI, ArndtS, SchulteW, et al. (2011) Interferon-lambda serum levels in hepatitis C. J Hepatol 54 : 859–865.
12. SugiyamaM, TanakaY, WakitaT, NakanishiM, MizokamiM (2011) Genetic variation of the IL-28B promoter affecting gene expression. PLoS One 6: e26620.
13. HondaM, SakaiA, YamashitaT, NakamotoY, MizukoshiE, et al. (2010) Hepatic {ISG} Expression Is Associated With Genetic Variation in Interleukin 28B and the Outcome of {IFN} Therapy for Chronic Hepatitis C. Gastroenterology 139 : 499–509.
14. UrbanTJ, ThompsonAJ, BradrickSS, FellayJ, SchuppanD, et al. (2010) IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 52 : 1888–1896.
15. de CastellarnauM, AparicioE, PareraM, FrancoS, TuralC, et al. (2012) Deciphering the interleukin 28B variants that better predict response to pegylated interferon-α and ribavirin therapy in HCV/HIV-1 coinfected patients. PLoS One 7: e31016.
16. SmithKR, SuppiahV, O'ConnorK, BergT, WeltmanM, et al. (2011) Identification of improved IL28B SNPs and haplotypes for prediction of drug response in treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome medicine 3 : 57.
17. Prokunina-OlssonL, MuchmoreB, TangW, PfeifferRM, ParkH, et al. (2013) A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45 : 164.
18. HammingOJ, Terczyńska-DylaE, VieyresG, DijkmanR, JørgensenSE, et al. (2013) Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J
19. AkaP, KuniholmMH, PfeifferRM, WangAS, TangW, et al. (2013) Association of the IFNL4-ΔG Allele with Impaired Spontaneous Clearance of Hepatitis C Virus. Journal of Infectious Diseases 32 : 3055–65 doi: 10.1038/emboj.2013.232
20. BibertS, RogerT, CalandraT, BochudM, CernyA, et al. (2013) IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med 210 : 1109–1116.
21. ManryJ, LavalG, PatinE, FornarinoS, ItanY, et al. (2011) Evolutionary genetic dissection of human interferons. J Exp Med 208 : 2747–2759.
22. BeaumontMA, ZhangW, BaldingDJ (2002) Approximate Bayesian computation in population genetics. Genetics 162 : 2025–2035.
23. PeterB, Huerta-SanchezE, NielsenR (2012) Distinguishing between Selective Sweeps from Standing Variation and from a De Novo Mutation. PLoS Genet 8: e1003011.
24. McVeanGA, Altshuler (Co-Chair)DM, Durbin (Co-Chair)RM, AbecasisGR, BentleyDR, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491 : 56–65.
25. WeirS, CockerhamCC (1984) Estimating F-Statistics for the Analysis of Population Structure. Evolution 38 : 1358.
26. SabetiPC, VarillyP, FryB, LohmuellerJ, HostetterE, et al. (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449 : 913–918.
27. VoightBFAK, WenSA, PritchardXA, KJ (2006) A Map of Recent Positive Selection in the Human Genome. PLoS Biol 4: e72.
28. FayJC, WuCI (2000) Hitchhiking under positive Darwinian selection. Genetics 155 : 1405.
29. di IulioJ, CiuffiA, FitzmauriceK, KelleherD, RotgerM, et al. (2011) Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology 53 : 1446–1454.
30. SuppiahV, MoldovanM, AhlenstielG, BergT, WeltmanM, et al. (2009) IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat Genet 41 : 1100–1104.
31. FuW, AkeyJM (2013) Selection and Adaptation in the Human Genome. Annu Rev Genomics Hum Genet 14 : 467–89 doi: 10.1146/annurev-genom-091212-153509
32. KassRE, RafteryAE (1995) Bayes factors. Journal of the american statistical association 90 : 773–795.
33. Jeffreys H (1998) The theory of probability. Oxford University Press.
34. SheblFM, PfeifferRM, BuckettD, MuchmoreB, ChenS, et al. (2011) IL28B rs12979860 genotype and spontaneous clearance of hepatitis C virus in a multi-ethnic cohort of injection drug users: evidence for a supra-additive association. Journal of Infectious Diseases 204 : 1843–1847 jir647.
35. FreemanA (2001) Estimating progression to cirrhosis in chronic hepatitis C virus infection. Hepatology 34 : 809–816.
36. BibertS, WojtowiczA, TafféP, ManuelO, BernasconiE, et al. (2014) The IFNL3/4 [DELTA]G variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS [In Press].
37. TeijaroJR, NgC, LeeAM, SullivanBM, SheehanKCF, et al. (2013) Persistent LCMV Infection Is Controlled by Blockade of Type I Interferon Signaling. Science 340 : 207–211.
38. WilsonEB, YamadaDH, ElsaesserH, HerskovitzJ, DengJ, et al. (2013) Blockade of Chronic Type I Interferon Signaling to Control Persistent LCMV Infection. Science 340 : 202–207.
39. OlsonMV (1999) When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet 64 : 18.
40. McLeanCY, RenoPL, PollenAA, BassanAI, CapelliniTD, et al. (2011) Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471 : 216.
41. MacArthurDG, BalasubramanianS, FrankishA, HuangN, MorrisJ, et al. (2012) A Systematic Survey of Loss-of-Function Variants in Human Protein-Coding Genes. Science 335 : 823–828.
42. MontgomerySB, GoodeD, KvikstadE, AlbersCA, ZhangZ, et al. (2013) The origin, evolution and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res 23 : 749–61 doi: 10.1101/gr.148718.112
43. YngvadottirB, XueY, SearleS, HuntS, DelgadoM, et al. (2009) A Genome-wide Survey of the Prevalence and Evolutionary Forces Acting on Human Nonsense {SNPs}. Am J Hum Genet 84 : 224–234.
44. FrischmeyerPA, DietzHC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8 : 1893–1900.
45. AndrésAM, DennisMY, KretzschmarWW, CannonsJL, Lee-LinSQ, et al. (2010) Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet 6: e1001157.
46. HamblinMT, RienzoAD (2000) Detection of the Signature of Natural Selection in Humans: Evidence from the Duffy Blood Group Locus. Am J Hum Genet 66 : 1669–1679.
47. WangXAG, ZhangWEA, Jianzhi (2006) Gene Losses during Human Origins. PLoS Biol 4: e52.
48. XueY, DalyA, YngvadottirB, LiuM, CoopG, et al. (2006) Spread of an inactive form of caspase-12 in humans is due to recent positive selection. The American Journal of Human Genetics 78 : 659–670.
49. MacArthurDG, SetoJT, RafteryJM, QuinlanKG, HuttleyGA, et al. (2007) Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat Genet 39 : 1261–1265.
50. SeixasS, IvanovaN, FerreiraZ, RochaJ, VictorBL (2012) Loss and Gain of Function in SERPINB11: An Example of a Gene under Selection on Standing Variation, with Implications for Host-Pathogen Interactions. PLoS One 7: e32518.
51. TishkoffSA, ReedFA, RanciaroA, VoightBF, BabbittCC, et al. (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39 : 31–40.
52. ThompsonJD, GibsonT, HigginsDG (2002) Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics Chapter 2. Unit 2.3. doi: 10.1002/0471250953.bi0203s00
53. WaterhouseAM, ProcterJB, MartinDM, ClampM, BartonGJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 : 1189–1191.
54. YangZ (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24 : 1586–1591.
55. PatenB, HerreroJ, FitzgeraldS, BealK, FlicekP, et al. (2008) Genome-wide nucleotide-level mammalian ancestor reconstruction. Genome Res 18 : 1829–1843.
56. KvikstadEM, DuretL (2014) Strong Heterogeneity in Mutation Rate Causes Misleading Hallmarks of Natural Selection on Indel Mutations in the Human Genome. Mol Biol Evol 31 : 23–36.
57. NielsenR (2005) Molecular signatures of natural selection. Annu Rev Genet 39 : 197–218.
58. DanecekP, AutonA, AbecasisG, AlbersCA, BanksE, et al. (2011) The variant call format and VCFtools. Bioinformatics 27 : 2156–2158.
59. GrossmanS, AndersenK, ShlyakhterI, TabriziS, WinnickiS, et al. (2013) Identifying Recent Adaptations in Large-Scale Genomic Data. Cell 152 : 703–713.
60. FrazerKA, BallingerDG, CoxDR, HindsDA, StuveLL, et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449 : 851–861.
61. Team RC (2011) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. Open access available at: http://cran.r-project.org
62. HudsonRR (2002) Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18 : 337–338.
63. GravelS, HennBM, GutenkunstRN, IndapAR, MarthGT, et al. (2011) Demographic history and rare allele sharing among human populations. Proceedings of the National Academy of Sciences 108 : 11983–11988.
64. GutenkunstRNAH, WilliamsonRDA, BustamanteSHA, CarlosC (2009) Inferring the Joint Demographic History of Multiple Populations from Multidimensional SNP Frequency Data. PLoS Genet 5: e1000695.
65. EwingG, HermissonJ (2010) MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26 : 2064–2065.
66. WegmannD, LeuenbergerC, NeuenschwanderS, ExcoffierL (2010) Abctoolbox: a versatile toolkit for approximate bayesian computations. BMC Bioinformatics 11 : 116.
67. TajimaF (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 : 585.
68. ReynoldsJ, WeirBS, CockerhamCC (1983) Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105 : 767–779.
69. WegmannD, LeuenbergerC, ExcoffierL (2009) Efficient approximate Bayesian computation coupled with Markov chain Monte Carlo without likelihood. Genetics 182 : 1207–1218.
70. TenenhausM (1998) La régression PLS: théorie et pratique. Editions Technip
71. BoulesteixA-L, StrimmerK (2007) Partial least squares: A versatile tool for the analysis of high-dimensional genomic data. Brief Bioinform 8 : 32–44.
72. McFarlandAP, HornerSM, JarretA, JoslynRC, BindewaldE, et al. (2013) The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus–induced microRNAs. Nat Immunol 15 : 72–79.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



