-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
Plasmids are extrachromosomal, autonomously replicating units that are harbored by many bacteria. Many plasmids encode transfer function allowing them to be transferred into plasmid-free bacteria by a process named conjugation. Since many of them also carry antibiotic resistance genes, plasmid-mediated conjugation is a major mechanism in the dissemination of antibiotic resistance. In depth knowledge on the regulation of conjugation genes is a prerequisite to design measures interfering with the spread of antibiotic resistance. pLS20 is a conjugative plasmid of the soil bacterium Bacillus subtilis, which is also a gut commensal in animals and humans. Here we describe in detail the molecular mechanism by which the key transcriptional regulator tightly represses the conjugation genes during conditions unfavorable to conjugation without compromising the ability to switch on accurately the conjugation genes when appropriate. We found that conjugation is subject to the control of a unique genetic switch where at least three levels of regulation are integrated. The first level involves overlapping divergent promoters of different strengths. The second layer involves a triple function of the transcriptional regulator. And the third level of regulation concerns formation of a DNA loop mediated by the transcriptional regulator.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004733
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004733Summary
Plasmids are extrachromosomal, autonomously replicating units that are harbored by many bacteria. Many plasmids encode transfer function allowing them to be transferred into plasmid-free bacteria by a process named conjugation. Since many of them also carry antibiotic resistance genes, plasmid-mediated conjugation is a major mechanism in the dissemination of antibiotic resistance. In depth knowledge on the regulation of conjugation genes is a prerequisite to design measures interfering with the spread of antibiotic resistance. pLS20 is a conjugative plasmid of the soil bacterium Bacillus subtilis, which is also a gut commensal in animals and humans. Here we describe in detail the molecular mechanism by which the key transcriptional regulator tightly represses the conjugation genes during conditions unfavorable to conjugation without compromising the ability to switch on accurately the conjugation genes when appropriate. We found that conjugation is subject to the control of a unique genetic switch where at least three levels of regulation are integrated. The first level involves overlapping divergent promoters of different strengths. The second layer involves a triple function of the transcriptional regulator. And the third level of regulation concerns formation of a DNA loop mediated by the transcriptional regulator.
Introduction
Bacteria exchange genetic material at high rates by different processes, which are collectively named Horizontal Gene Transfer (HGT). HGT can be beneficial for bacteria because the newly acquired DNA may endow them with novel features enabling them to adapt to changing conditions in the environment, i.e. rapid evolution. On the other hand, HGT is notorious for its role in the dissemination of virulence/pathogenicity determinants and antibiotic resistance. The main mechanisms responsible for HGT are transformation mediated by natural competence, transduction and conjugation [1]–[6]. The latter mechanism, -conjugation-, concerns the transfer of a DNA element from a donor to a recipient cell. Conjugative elements containing all the information required for DNA transfer of a donor to a recipient cell are often found on plasmids, but they can also be embedded within a bacterial chromosome. These latter forms are generally named integrative and conjugative elements (ICE).
Some basic features of the conjugation process are conserved among plasmids [for review see], [ 7]–[10]. In most cases, a single-stranded DNA (ssDNA), which is generated by a rolling circle-like mode of DNA replication, is transferred into the recipient cell through a membrane-associated intercellular mating channel, named transferosome, which is a form of type IV secretion system. Conjugative plasmids can be exploited for the construction of tools to genetically modify bacteria of clinical or industrial relevance that are reluctant to genetic manipulation by other ways. Besides its intrinsic scientific interest, a detailed understanding about how conjugation genes are regulated is crucial to design strategies helping to interfere with the rapid spread of antibiotic resistance, and for the construction of genetic tools based upon conjugative plasmids.
Various conjugative plasmids have been studied in considerable detail [for review see], [ 7]–[10]. Although most of the well-studied conjugative plasmids replicate in Gram-negative bacteria, an increasing interest in conjugative plasmids of Gram-positive bacteria has resulted in the recent analysis of conjugative plasmids from for instance streptococci, enterococci, staphylococci and clostridia [11]–[17]. However, conjugation systems present on the Gram-positive soil bacterium Bacillus subtilis had not been reported until recently. This is most remarkable taking into account that (i) it is one of the best-studied Gram-positive bacteria; (ii) it has important industrial applications; and (iii) it is closely related to pathogenic and fastidious bacilli [for review see], [ 18,19]. Moreover, several B. subtilis strains are gut commensals in animals including humans [20]. B. subtilis plasmids may therefore play an important role in HGT in different environments. We chose the B. subtilis plasmid pLS20 for our studies. Originally, this 65 kb plasmid was identified in the Bacillus subtilis natto strain IFO3335 that is used in the fermentation of soybeans to produce “natto”, a dish that is popular in South Asia [21]. Previous studies on pLS20 have shown that it is conjugative in liquid media as well as on solid media [22], [23]. The presence of pLS20 has a broad impact on the physiology of the host, and the localization of some components of the conjugation machinery has been determined [24], [25]. The replication region of pLS20 has been characterized, and it has been demonstrated that it uses a dedicated segregation mechanism involving the actin-like Alp7A protein [26], [27]. pLS20 encodes a protein, RokLS20, that suppresses the development of natural competence of B. subtilis [28].
Recently, we have reported a global view of the regulatory circuitry of the pLS20 conjugation genes. A conjugation operon encompassing more than 40 genes is located next to a divergently oriented single gene, rcoLS20, which encodes the master regulator of conjugation responsible for keeping conjugation in the default “OFF” state. Activation of conjugation requires an anti-repressor, RapLS20, that belongs to the family of Rap proteins. Inactivation of the rapLS20 gene on pLS20 severely compromises conjugation, and conjugation was enhanced when rapLS20 was expressed from an ectopic locus. The activity of RapLS20, in turn, is regulated by a signaling peptide, Phr*LS20. The small phrLS20 gene, located immediately downstream of rapLS20, encodes a pre-protein. After being secreted, PhrLS20 can be processed by a second proteolytic cleavage, resulting in generation of the functional pentapeptide, Phr*LS20, corresponding to the five C-terminal residues of PhrLS20. When (re)imported, this peptide inactivates RapLS20. Therefore, activation of conjugation is ultimately regulated by the Phr*LS20 signaling peptide. The Phr*LS20 concentration will be relatively high or low when donor cells are predominantly surrounded by donor or recipient cells, respectively. Hence, conjugation will become activated particularly under conditions in which recipient cells are potentially present. In addition, Phr*LS20 has a crucial role in returning conjugation to the default “OFF” state [29].
Despite identification of the players involved in regulation of the conjugation genes, our knowledge on regulation of the genetic switch responsible for activating conjugation is still very limited. Using a combination of various in vitro and in vivo approaches, we show that the genetic switch controlling pLS20 conjugation involves at least three layers of regulation. Together, they tightly repress the main conjugation promoter under conditions that do not favor conjugation, while maintaining the ability to accurately switch on the conjugation genes when appropriate conditions occur. The three layers involve coinciding or overlapping divergent promoters of different strengths, autoregulated expression of RcoLS20, which turns out to be a tri-functional transcriptional regulator, and formation of RcoLS20-mediated DNA looping. The sophisticated regulatory mechanism that combines three layers of control into a single switch is novel for plasmids of Gram-positive bacteria. The implications of the uncovered regulation mechanisms for conjugation are discussed in the context of regulatory systems present on other HGT elements and with other regulatory systems involving DNA looping.
Results
Promoters Pc and Pr
The rcoLS20-gene 28 intergenic region contains the strong main conjugation promoter, Pc, which is under the negative control of the master regulator of conjugation RcoLS20
According to our standard presentation (Fig. 1A), the transcription of pLS20cat gene 27, encoding the main repressor of conjugation genes, RcoLS20, reads leftwards. Flanking genes 28 to 74, which are all transcribed in the opposite direction, probably constitute a large conjugation operon [29]. To test whether a promoter that would drive expression of this operon is located upstream of gene 28 we cloned the ∼600 bp intergenic rcoLS20-gene 28 region in the appropriate orientation in front of a promoterless lacZ reporter, and subsequently placed a single copy of this cassette at the B. subtilis chromosomal thrC locus (strain PKS3). Transcriptional fusions to several sub-fragments of this region were also constructed. In addition, all the fragments were cloned in the opposite orientation to analyze the divergent promoter of the rcoLS20 gene (see below). For simplicity, the cloned fragments are indicated with Roman letters. Fragments cloned in the orientation to analyze the conjugation or the rcoLS20 promoter are indicated with the extension “c” or “r”, respectively. The entire intergenic region is referred to as Fragment I (or F_I). A schematic representation of the different strains and fusions described in this work is given in Figs. 1B–C.
Fig. 1. Genetic map of the pLS20cat conjugation region and summary of the transcriptional lacZ fusions used in this study. 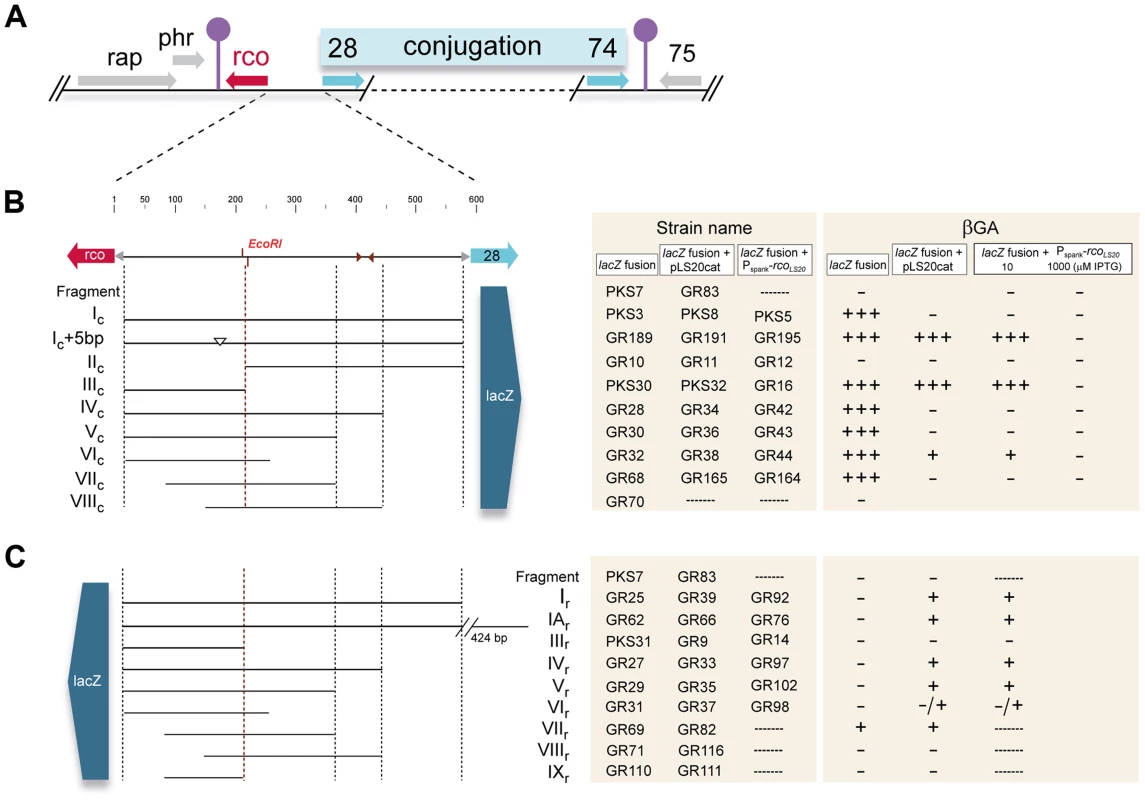
A. Map of the conjugation region of plasmid pLS20cat. Positions and directions of the genes and the positions of the predicted transcriptional terminators are indicated with arrows and lollipop symbols, respectively. Panels B and C show a blow-up of the 600 bp rcoLS20 gene 28 intergenic region and the different fragments fused to lacZ. Fusions in (B) and (C) were used to study activities of the promoters Pc and Pr, respectively. Features of the intergenic region are given on the top line. Numbers correspond to the bp position in this region. Names of the fragments cloned are indicated. The small triangles indicate position of an inverted repeated sequence (see text). Strains containing Pc-lacZ fusions in combination with the Pspank-rcoLS20 cassette were grown on plates containing 10 µM or 1 mM IPTG. The symbols “+++”, “+”,and “−” reflect intense blue, pale blue, and white colonies after growth on X-gal containing plates. Colors of the colonies were observed after 16 and 48 hours of incubation at 37°C for strains containing pLS20cat or the Pspank-rcoLS20 cassette, respectively. Colonies of strain PKS3 containing F_Ic-lacZ fusion were blue when grown overnight on Luria-Bertani (LB) agar plates supplemented with the chromogenic substrate 5-bromo-4-chloro-indolyl-β-D-galactopyranoside (Xgal; Fig. S1), demonstrating that the rcoLS20-gene 28 intergenic region contains a promoter, which we named Pc. Analysis of PKS3 samples gave relatively high levels of β-galactosidase (βG) activities that were in the range of 300 and 500 Miller Units (MU) during mid-exponential and stationary phase, respectively. These results indicate that Pc is a rather strong promoter that does not seem to be regulated by host-encoded factors when grown under these conditions.
Under our laboratory conditions, efficient conjugation is limited to a narrow time window near the end of the exponential growth phase [29]. If Pc is the main conjugation promoter it is expected that (i) its activity would generally be lower in the presence of pLS20cat and (ii) there would be a correlation between promoter Pc activity and the efficiency of conjugation. The following results show that this is indeed the case. Thus, we introduced pLS20cat into strain PKS3, and colonies of the resulting strain, PKS8, were white after overnight growth on Xgal-containing plates (Fig. S1). In addition, when we used PKS8 as donor strain and simultaneously determined the kinetics of conjugation and promoter Pc activity we found that promoter Pc is only active during a rather short window of time near the end of the exponential growth phase, which coincides with the period of high conjugation efficiency (Fig. 2).
Fig. 2. Correlation between the kinetics of Pc promoter activity and conjugation efficiencies of pLS20cat. 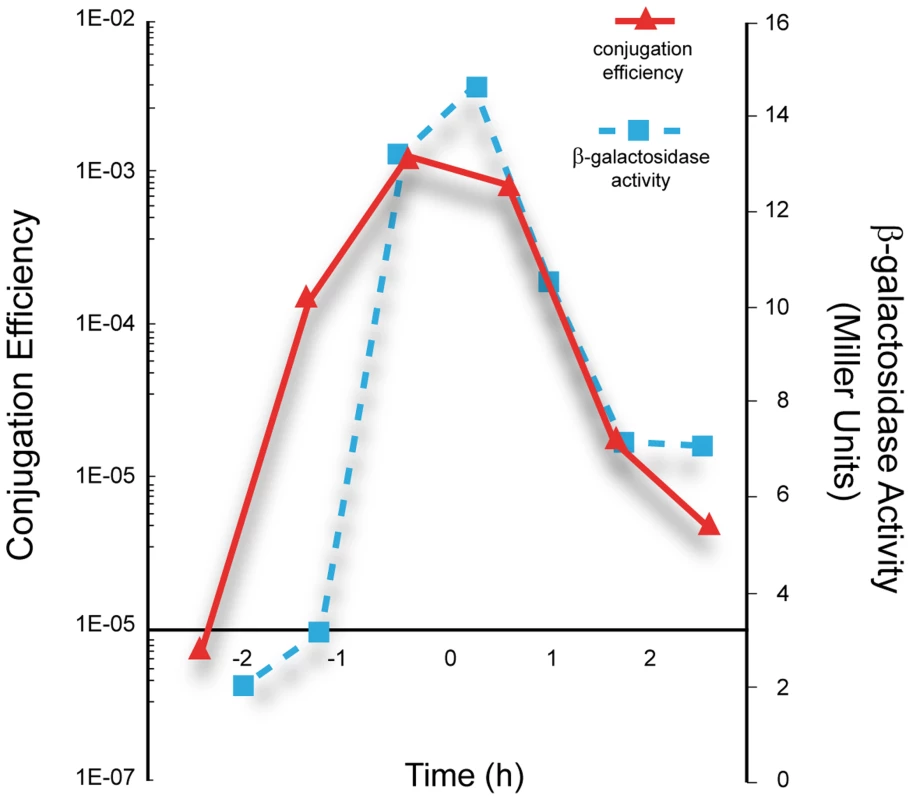
Overnight cultures of the strain PKS8 (F_Ic-lacZ, pLS20cat) and recipient strain PS110 were diluted to an OD600 of 0.05. Next, samples taken at different times were used to determine conjugation efficiency of pLS20cat by a standard conjugation protocol (continuous line), and the promoter Pc activity by measuring β-galactosidase activity (broken line). T = 0 corresponds to the end of the exponential growth phase. The presented graph corresponds to a representative experiment. The experiment was carried out three times and the corresponding values differed by less than 10%. Next we tested whether RcoLS20, encoded by the pLS20cat gene 27 (rcoLS20), is responsible for repression of the Pc promoter. To this end, we placed the rcoLS20 gene under the control of the isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible Pspank promoter and introduced this cassette at the amyE locus of strain PKS3 (harboring the F_Ic-lacZ fusion at the thrC locus). Colonies of this strain, PKS5, were blue when grown on Xgal-supplemented LB agar plates, but white when the plates contained IPTG (Fig. S1). Together, these results indicate that promoter Pc located upstream of gene 28 constitutes the main conjugation promoter that is negatively controlled by RcoLS20.
Interestingly, PKS5 colonies were white when plates contained as little as 10 µM of IPTG (colonies shown in Fig. S1). Taking also into account that Pspank is a relatively weak promoter, these results indicate that the Pc promoter is very sensitive to RcoLS20.
Promoter Pc is located at an unusually large distance upstream of the first gene of the conjugation operon
We next set out to delineate the position of the Pc promoter. As a first approach, we constructed strains containing lacZ fused to different subregions of Fragment Ic. Surprisingly, whereas no significant promoter activity was obtained with the strain having lacZ fused to Fragment IIc (strain GR10), the βG activities obtained with strains harboring lacZ fused to Fragment IIIc, IVc, Vc or VIc were very similar to those obtained when lacZ was fused to Fragment Ic. These results show that promoter Pc is located at an unusually large distance of at least 350 bp upstream of gene 28.
Analyses of strains containing lacZ fused to Fragment VIIc (GR68) or VIIIc (GR70) revealed that promoter activity was sustained only by Fragment VIIc (Fig. 1B), showing that the 5′-located 63 bp region of Fragment VIIc contains (at least part of) the Pc promoter. This 63 bp region contains the sequence 5′ - ttaaaaatttcactgaaatac-TTtACA-gttaaaaaaatgtc-TATctT-3′, which constitutes a putative σA-dependent promoter endorsing several features characteristic for a strong promoter. First, the hexamer sequences 5′-TTtACA-3′ and 5′-TATctT-3′ are very similar to the consensus −35 (5′-TTGACA-3′) and −10 (5′-TATAAT-3′) sequences recognized by σA. Second, an optimal spacer length of 17 bp separates the putative −35 and −10 boxes. Third, the spacer contains the extended −10 motif (5′-TGn-3′, double underlined). Fourth, AT-rich tracts are located directly upstream of the predicted −35 box which are likely binding sites for the C-terminal domain of the RNA polymerase α-subunit. Additional evidence that this sequence constitutes the Pc promoter was obtained by primer extension analysis to determine the transcription start site. The detected extension product is shown in Fig. 3B. The position of the deduced transcription start site is located 6 bp downstream of the Pc core promoter sequences mentioned above (see Fig. 3A). The position of the transcription start site corroborates with our RNAseq data, which provides a good estimation of the position of the transcriptional start site. Thus, total RNA isolated from pLS20cat-harboring cells was processed as described in Materials and Methods after which it was employed to generate cDNA libraries using a “directional RNA-seq” procedure that preserves information about the transcript's direction. The schematic representation of the distribution and directionality of the reads presented in Figure 3C shows that the rightward-oriented transcripts, driving expression of gene 28 and downstream genes (shown in green), start close to the divergently oriented rcoLS20 gene (shown in red).
Fig. 3. Promoter Pc is located 461 bp upstream of the start codon of gene 28 and overlaps with the divergently oriented Pr promoter. 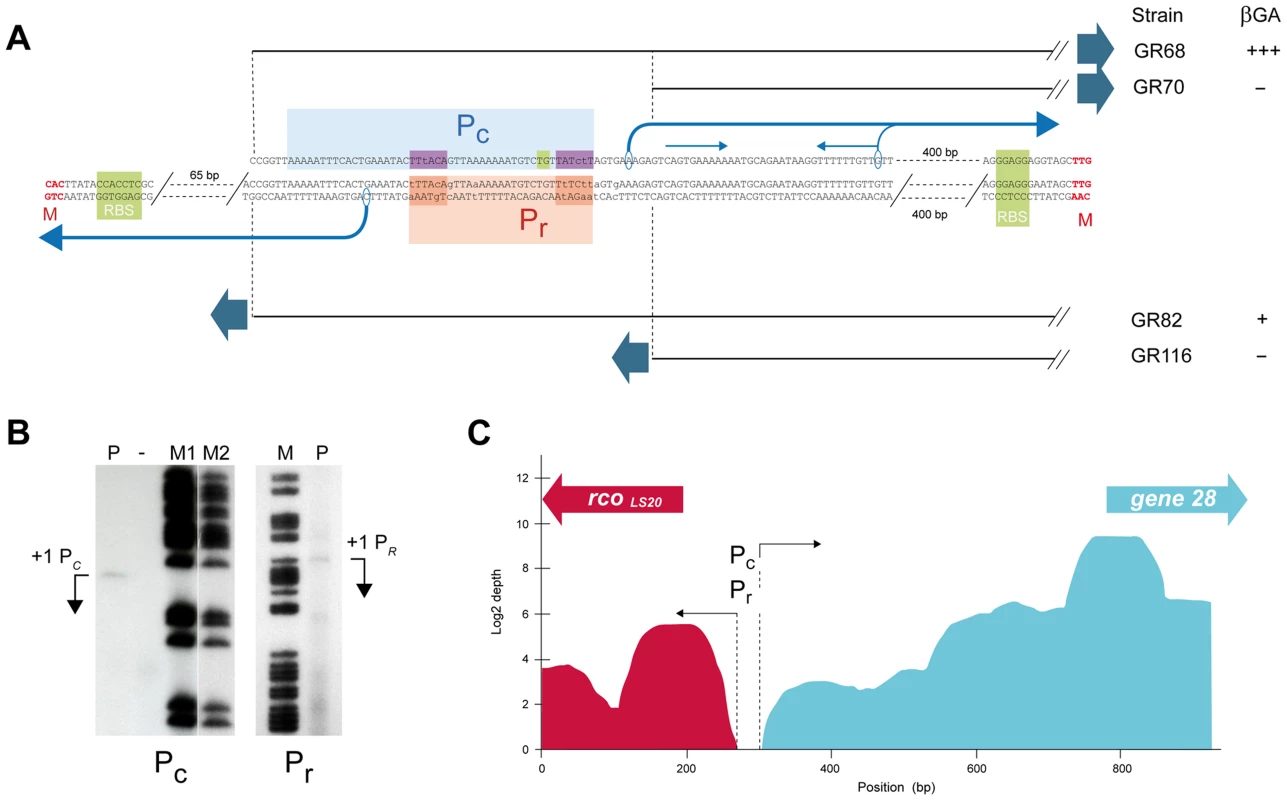
A. Determination of promoter Pc and Pr sequences by deletion analysis and primer extension. pLS20cat containing cells harvested at the end of the exponential growth phase were processed to isolate their total RNA, which was used in primer extension assays as described in Materials and Methods. Features of the promoter Pc are shown above the sequence. The dotted vertical lines and black straight lines indicate the 5′ end points of the transcriptional lacZ fusions present in strains GR68 and GR70, displaying and not displaying promoter activity, respectively. The core promoter and the putative upstream UP element is indicated by a light blue box; the −35 and −10 hexamers, and the extended −10 motif are indicated with dark blue and green boxes, respectively. The transcription start site determined by primer extension and the direction of transcription are indicated with the corresponding encircled base and a black bent arrow. The thin grey bent arrow corresponds to the 3′ end point of the smaller extension product that coincides with the start of an inverted repeat which is marked with a pair of thin blue arrows above the sequence. Features of promoter Pr are shown below the sequence. The dotted vertical lines and the black straight lines indicate the 3′ end points of the transcriptional fusions with lacZ reporter present in strains GR82 and GR116, displaying and not displaying promoter activity, respectively. The deduced position of the Pr core promoter, and the −35 and −10 boxes are indicated with orange and red boxes, respectively (see text). The transcription start site determined by primer extension and the direction of transcription are indicated with the corresponding encircled base and a black bent arrow. B. Primer extension to determine the transcription start sites of promoters Pc (left panel) and Pr (right panel). The cDNA products of the primer extension reactions are indicated with bent arrows (lane P). Free lane in which no sample was run is indicated with “−”. Lanes M, M1 and M2 correspond to [G+A] chemical sequencing reactions of a short 230 bp DNA fragment corresponding to the studied pLS20cat region obtained by PCR amplification as described in Materials and Methods. In the case of the Pc promoter, a smaller extension product with a relatively strong signal was observed 37 bp downstream of the extension product shown. The longer extension product most likely reflects the correct transcription start site based on the following arguments. First, it is known that AMV reverse transcriptase prematurely terminates cDNA synthesis when reaching a stem loop in the RNA, and that the prematurely terminated molecules map at the bottom of the secondary structure [71], [72]. The position of the strong signal coincides with the 3′ end of an inverted repeat (indicated in Fig. 3A). Second, no putative core promoter sequences are evident upstream of the 5′ position of the shorter extension product. Third, if the stronger signal corresponds to the transcription start site, the responsible promoter would be present on Fragment VIIIc used for the transcriptional lacZ fusion in strain GR70. However, no promoter activity was observed with this strain (see text). And fourth, the transcription start site based on the longer extension product corroborates the RNAseq data. C. Schematic overview of RNAseq expression data of pLS20cat genes rcoLS20 and 28 under conditions with (top panel) and without overexpression of rcoLS20 (lower panel). The amount of right and leftward “reads”, given in green and red, respectively, are presented on a log2 scale. The positions of the divergently oriented genes rcoLS20 and 28 are indicated on the top with a red and green arrow, respectively. Dotted lines and black arrows indicate the approximate start sites of the divergent transcripts driven by the Pc and Pr promoters. The rcoLS20-28 intergenic region contains the weak Pr promoter that is activated and repressed at low or high RcoLS20 concentrations, respectively
As for Pc, we constructed lacZ fusion strains to characterize the divergently oriented Pr promoter responsible for expression of RcoLS20. Surprisingly, no promoter activity was observed when lacZ was fused to the 570 bp Fragment Ir (strain GR25, Fig. 1C). One possibility could be that promoter Pr is located even further upstream. This does not seem to be the case however, because Fragment IAr, corresponding to the 1,014 bp region upstream of rcoLS20 (strain GR62), also did not provide detectable levels of promoter activity. We then introduced pLS20cat into these strains to study whether it encoded a protein that might be required to activate promoter Pr. Colonies of the resulting pLS20cat-harboring strains GR39 (F_Ir-lacZ) and GR66 (F_IAr-lacZ) turned pale blue when grown on Xgal-containing plates (Fig. 1C), consistent with pLS20cat providing a protein that activates the Pr promoter. In addition, the results show that the Pr promoter is located on Fragment Ir.
RcoLS20 might be responsible for activating its own promoter. To test this possibility, we engineered strain GR92 that contains the F_Ir-lacZ fusion combined with the cassette in which expression of rcoLS20 is under the control of the IPTG-inducible Pspank promoter. Colonies of strain GR92 were white when grown on agar plates containing only Xgal, but turned pale blue when the plates contained also low levels of IPTG. These results demonstrate that RcoLS20 activates its own promoter. In addition, the fact that colonies only developed a pale blue color suggests that promoter Pr is weaker than Pc. To test this more directly, we measured Pr promoter activities at late-exponential growth phase using strain GR92 grown at different levels of RcoLS20 induction (Table 1). Interestingly, maximum Pr promoter activity was obtained when cells were grown in the presence of 50 µM IPTG. Promoter Pr activity decreased at higher IPTG concentrations and equaled background levels in the presence of 1 mM of IPTG, indicating that RcoLS20 represses its own promoter at higher concentrations.
Tab. 1. Pr promoter activity at different induction levels of RcoLS20. 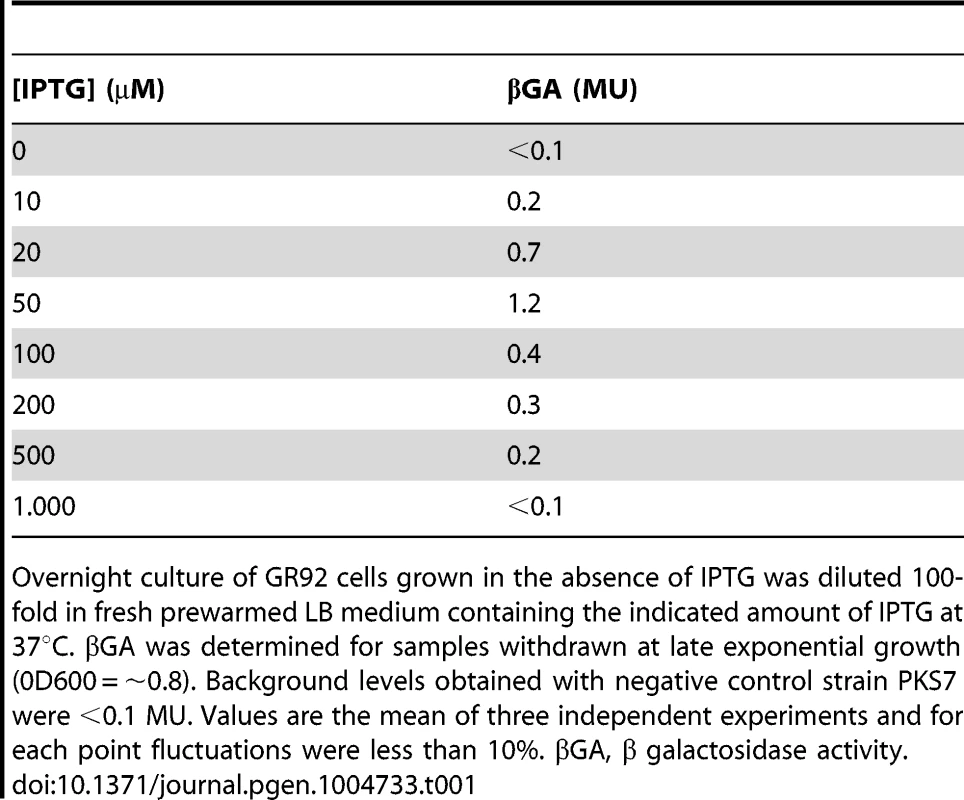
Overnight culture of GR92 cells grown in the absence of IPTG was diluted 100-fold in fresh prewarmed LB medium containing the indicated amount of IPTG at 37°C. βGA was determined for samples withdrawn at late exponential growth (0D600 = ∼0.8). Background levels obtained with negative control strain PKS7 were <0.1 MU. Values are the mean of three independent experiments and for each point fluctuations were less than 10%. βGA, β galactosidase activity. Together, these results show that Pr is a weak promoter whose strength is several hundred folds weaker than that of Pc. The results also show that RcoLS20 has a triple function. First, low levels of RcoLS20 are required to activate its own promoter Pr; second, at higher concentrations RcoLS20 represses its own promoter; and third, RcoLS20 is responsible for repression of the oppositely oriented Pc promoter. This triple function of RcoLS20 is likely to have important consequences for regulation of the conjugation genes (see discussion). It is worth mentioning that whereas maximum activation of the Pr promoter was achieved when rcoLS20 was induced from the Pspank promoter at 50 µM IPTG, efficient repression of the Pc promoter was observed by inducing rcoLS20 with as low as 10 µM IPTG. Finally, the results obtained show that RcoLS20 is the only pLS20cat protein required for activation and repression of the Pr and Pc promoters.
The divergent Pr and Pc promoters overlap
As a first approach to determine the position of the Pr promoter we constructed strains containing lacZ gene preceded by different subregions of Fragment Ir combined with pLS20cat to provide RcoLS20 in trans. The transcriptional regulator RcoLS20 is a DNA binding protein (see below). Therefore, a lack of Pr promoter activity in the reporter assay can be due to the absence of (part of) the Pr promoter or the RcoLS20 binding sites required for activation of Pr. Since activator proteins generally bind upstream of promoters, we tested constructs having deletions at the 3′ end of Fragment Ir (i.e. flanking the rcoLS20 gene). Promoter Pr activity was detected when lacZ was fused to Fragment VIIr (strain GR82), but not when it was fused to Fragment VIIIr (strain GR116) (Figs. 1C and 3A). Interestingly, these results suggested that promoter Pr would be (partially) located on the 63 bp 5′ region of Fragment VII on which the divergently oriented Pc promoter is also located (see above, Fig. 3A). In a complementary approach, we determined the transcriptional start site of promoter Pr by primer extension (Fig. 3B). The determined transcription start site of promoter Pr is positioned 6 bp upstream of the −35 box of the Pc promoter (see Fig. 3A). This implies that promoter Pr overlaps with the Pc promoter.
Pr is a weak promoter whose activity requires RcoLS20 (see above). It is therefore unlikely that the −35 and −10 boxes will be very similar to the consensus sequences. The following two sequences that may constitute a σA-dependent promoter are located upstream of the determined Pr transcription start site: (i) [5′-aaGAtA - 17 bp -TgTAAa-3′] and (ii) [5′-aTaACA-18bp-aAgtAT-3′] (mismatches with respect to consensus −35 (5′-TTGACA-3′) and −10 boxes (5′-TATAAT-3′) given in lower case, see Fig. 3A). The position of the determined transcription start is optimally spaced with respect to the first but not the second possible promoter sequence. Therefore, we favor the first sequence to correspond to the Pr promoter. Interestingly, this would imply that the positions of the −10 and −35 boxes correspond exactly to the −35 and −10 boxes, respectively, of the divergently oriented Pc promoter.
The results of the RNAseq experiments presented in Figure 3C (see also above) support the conclusion that the Pr and Pc promoters overlap. RNA transcripts mapped against the entire intergenic region except for a small region that is located near the start of the rcoLS20 gene. The divergent promoters Pr and Pc, responsible for the left - (red) and rightward (green) oriented transcripts, respectively, must both be located in the small nontranscribed region which corresponds to the position of the Pr/Pc promoters according to their transcriptional start sites determined by primer extension.
In summary, results obtained by a combination of different approaches demonstrate that divergent Pc and Pr promoters overlap, if not coincide.
RcoLS20 operator sites
In vivo evidence that RcoLS20 binds to two operator sites; one of them, -located more than 85 bp downstream of Pc-, is required for efficient regulation of promoters Pc and Pr
RcoLS20 belongs to the Xre-family of transcriptional regulators and is predicted to contain a Helix-Turn-Helix (HTH) DNA binding motif in its N-terminal region [29]. It is therefore likely that RcoLS20 will exert its transcriptional regulatory effects on Pr and Pc by binding to DNA sequences. We employed the following in vivo approach to gain insights into the location of the RcoLS20 binding sites. Either pLS20cat or the Pspank-rcoLS20 cassette was introduced into the various lacZ fusion strains (see Fig. 1). The resulting strains were then grown on Xgal containing LB plates, -supplemented with or without 10 µM of IPTG for strains containing the Pspank-rcoLS20 cassette-, and expression of the different lacZ fusions in response to RcoLS20 was screened by the color of their colonies.
A schematic summary of the results obtained for promoter Pc is given in Figure 1B. In agreement with results presented above, the strain harboring lacZ fused to Fragment Ic (PKS3) displayed high Pc promoter activity, but no promoter activity was detected when RcoLS20 was provided in trans (strains PKS5 and PKS8). Efficient RcoLS20-mediated repression of the Pc promoter was lost however when lacZ was fused to Fragment IIIc (strains GR16 and PKS32). This strongly indicates that an RcoLS20 operator site is located on the 368 bp Fragment IIc and that this operator, which would be located at least 85 bp downstream of the Pc promoter, is crucial for efficient repression of the Pc promoter. Fragment IIc contains an inverted repeated sequence (5′-ATCAAAATCAtgctgcaactTGGTTTTGAT-3′). To test whether this region constitutes an RcoLS20 operator site we constructed lacZ fusions to Fragments IVc or Vc, and also engineered derivatives of these two strains containing pLS20cat or Pspank-rcoLS20. The 5′ ends of these Fragments are located up - or downstream of the inverted repeat (see Fig. 1B). The Pc promoter present on Fragment IVc and Vc was efficiently repressed by RcoLS20, indicating that the RcoLS20 operator site is present on Fragment V and not on the 212 bp region immediately upstream of gene 28 containing the mentioned inverted repeat. Efficient RcoLS20-mediated repression of promoter Pc was not observed for the lacZ fusion based on Fragment IIIc (see above). Together these in vivo results strongly indicate that an ∼160 bp region, located 85 bp downstream of Pc, contains an RcoLS20 operator site that is required for efficient repression of this promoter. We name this operator site OI.
Results described above show that promoter Pc was not repressed by RcoLS20 when the lacZ fusion was based on Fragment IIIc (strain GR16) and cells were grown in the presence of 10 µM IPTG. Interestingly though, promoter Pc in strain GR16 was efficiently repressed when the concentration of IPTG was increased to 1 mM (see Fig. 1B). This indicates that another RcoLS20 operator site is present on the 201 bp Fragment IIIc. We name this operator site OII.
Next, we used the same strategy to delineate the regions required for activation of the divergent Pr promoter. The results of these analyses are summarized in Figure 1C. Interestingly, the region required for efficient repression of Pc by RcoLS20, is also required for RcoLS20-mediated activation of promoter Pr. Thus, RcoLS20 activated the Pr promoter when lacZ was fused to Fragments IVr or Vr (strains GR97/GR33 and GR102/GR35, respectively) but not when it was fused to Fragment IIIr (strains GR14/GR9, Fig. 1C). In summary, the in vivo results obtained provide strong evidence that one RcoLS20 operator, OI, is located in an ∼160 bp region located 85 bp downstream of promoter Pc, and that this operator is crucial for proper repression and activation of promoters Pc and Pr, respectively. In addition, the results indicate the presence of another RcoLS20 operator, OII, which would be located near promoters Pc and Pr.
In vitro approaches show that RcoLS20 binds cooperatively to multiple binding sites present in operators OI and OII
To study the position of the RcoLS20 binding sites in more detail we purified RcoLS20 and used it in Electrophoretic Mobility Shift Assays (EMSA). To facilitate purification, we constructed an E. coli strain that expresses an RcoLS20-His(6) tagged fusion protein. The his(6)-tag was placed at the C-terminus because RcoLS20 contains a predicted Helix-Turn-Helix DNA binding motif close to its N-terminus. The following result demonstrates that the RcoLS20-His(6) protein is functional in vivo. We constructed B. subtilis strain GR90 in which the expression of rcoLS20-his(6) gene is placed under the control of the inducible Pspank promoter, and which also contains the F_Ic-lacZ reporter fusion. The activity of promoter Pc in strain GR90 was repressed in an IPTG-dependent manner similar to that observed for strain PKS5 containing an inducible copy of native rcoLS20 (not shown).
The in vivo transcriptional fusion results presented above indicated the presence of two operators. One of them, operator OII, located near promoters Pc/Pr, and another one, operator OI, present in an ∼160 bp region about 85 bp downstream of Pc. In addition, this analysis indicated that the ∼200 bp region immediately upstream of gene 28 does not contain RcoLS20 binding sites. Accordingly, we began analyzing binding of RcoLS20 to Fragments X (200 bp region upstream gene 28), III (expected to contain operator OII) and XII (expected to contain operator OI) (see Fig. 4). Independent of the concentrations used, RcoLS20 did not bind to Fragment X (Fig. 4B). Together with the in vivo data presented above, this provides strong evidence that this region does not contain RcoLS20 binding sites. Also in agreement with the in vivo data, RcoLS20 bound to both Fragment III and Fragment XII (Fig. 4B). Interestingly, the retardation patterns obtained for these fragments were similar, and resulted in the appearance of a maximum of two retarded species. The observation that the two retarded species were already present at low RcoLS20 concentrations when the majority of the DNA migrated to the position of unbound DNA, strongly indicates that RcoLS20 binds cooperatively to at least two binding sites present in each operator. In addition, the observation that DNA fragments entered the gel even at very high protein concentrations indicates that RcoLS20 binds to specific sites and that it does not spread along the DNA. To delineate the OI and OII regions further we used overlapping and subregions of Fragments III and XII as probes. Fragment IIIA (130 bp containing promoters Pc/Pr) and Fragment XIIA (125 bp) both produced up to two shifts, and RcoLS20 did not bind to the 46 bp region that separates these two fragments. This latter conclusion is based on comparison of gel retardations obtained with fragments XI and XII.
Fig. 4. Analysis of the RcoLS20 binding sites in the rcoLS20 - gene 28 intergenic region by EMSA. 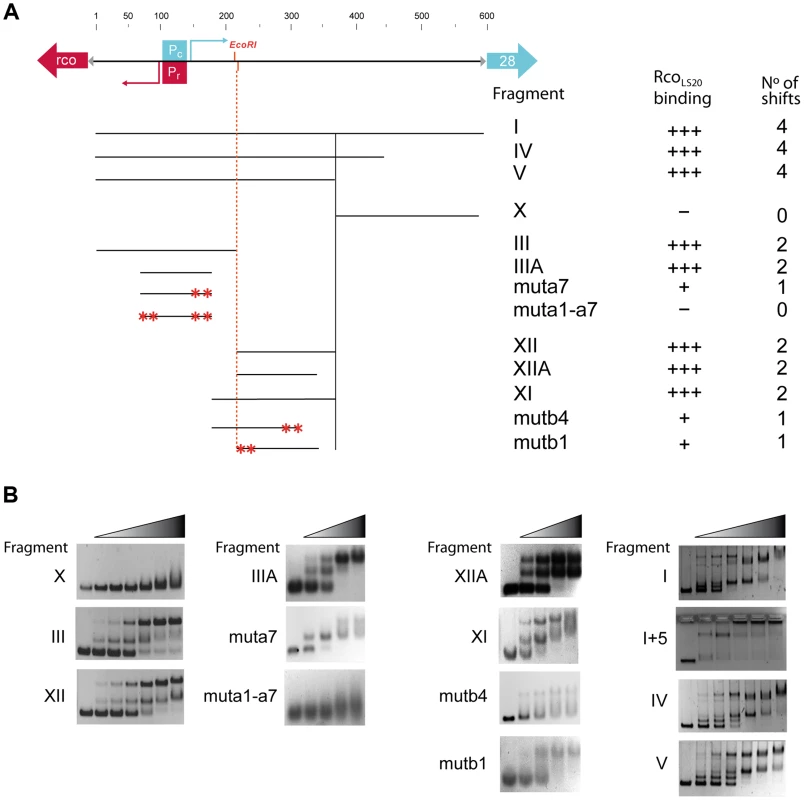
A. Schematic representation of the DNA fragments used as probes and a summary of the EMSA results. Top panel. A map of the 600 bp rcoLS20-gene 28 intergenic region is shown as a black line alongside with the positions of the characteristics in this region. The divergently oriented genes rcoLS20 and 28 are indicated with a red and blue arrow, respectively. The triangles indicate ribosomal binding sites. The position of the Pc and the Pr promoter (boxed) and their transcription start sites (bent arrows) are indicated in blue and red, respectively. The position of the unique EcoRI site in this region is also indicated. Lower panel shows the different fragments used in EMSA; the names of the DNA fragments, the levels of RcoLS20 binding and the numbers of shifts are indicated on the right. Asterisks indicate positions of introduced mutations. B. Examples of the EMSA results obtained using DNA fragments described in panel A. The left-most lane of each panel corresponds to samples loaded without protein. Increasing concentrations of RcoLS20 were prepared using a two-fold dilution method, and ranged from 0.212–6.8 (7 lane panels) or 0.95–7.3 µM (5 lane panels). We next analyzed binding of RcoLS20 to Fragments I, IV and V that encompass both operators. These fragments gave similar retardation patterns. Interestingly, in these cases, RcoLS20 binding resulted in the appearance of four retarded species. All four of these retarded species could be detected at low RcoLS20 concentrations when most of the fragment had not bound RcoLS20, indicating that RcoLS20 binds cooperatively to multiple sites on these fragments.
To search for the presence of conserved motifs in the two RcoLS20 operators we used the motif-identification programs MEME [30] and BIOPROSPECTOR [31]. These analyses revealed the identification of an 8 bp conserved motif that is present seven times in OII (Fragment III_A), and four times in OI (Fragment XII_A). We named the seven motifs identified in the OII operator a1–a7, and the four motifs in the OI operator b1–b4 (see Fig. 5). Whereas motifs b1 to b4 are all located on the lower strand, motifs a1–a7 are located on the upper strand, except motif a3. It is worth mentioning some characteristics of motifs a1 to a7. First, motif a5 overlaps with the Pc/Pr core promoter sequences, and motifs a1–a4 and a6–a7 flank them. Second, motifs a1 and a7 form part of a 13 bp direct repeat. Third, motifs a1 and a3 form an inverted repeat. Fourth, the oppositely oriented motifs a3 and a4 overlap in a region that has an inverted repeat (5′TTTCAgTGAAA-3′).
Fig. 5. Identification of a conserved motif constituting (part of) the RcoLS20 binding site. 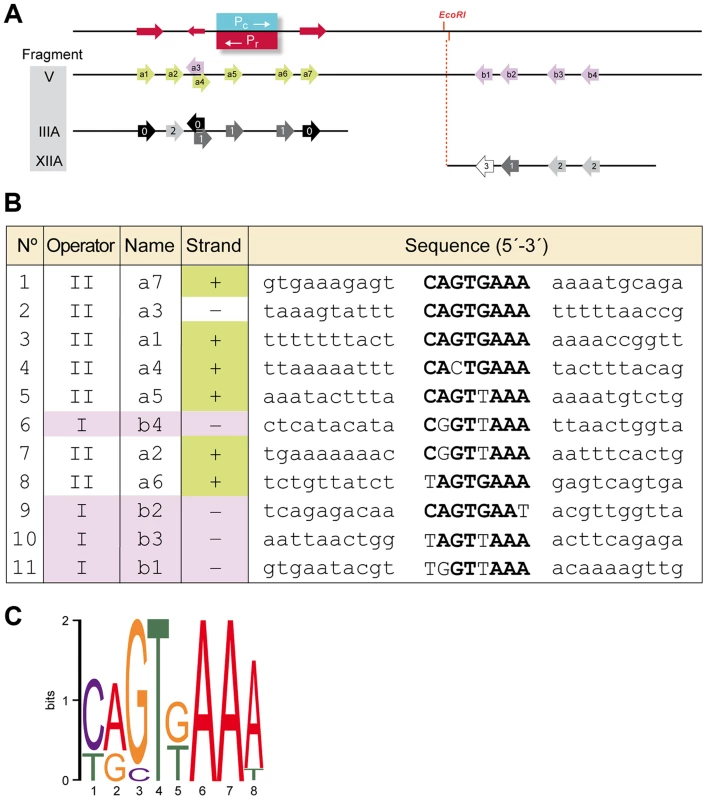
A. Schematic representation of the region corresponding to Fragment V, which encompasses promoters Pr/Pc and the identified repeated motif that form (part of) the binding site for RcoLS20. Top line. Position of promoters Pc and Pr are indicated in blue and red, respectively. The position of the unique EcoRI site is indicated. A 13 bp long direct repeat (5′-TCAGTGAAAAAAA-3′) is indicated with rightward directed red arrows. The leftward-directed arrow indicates the position of the complementary 9 bp sequence 5′-TTTCACTGA-3′. Second line (Fragment V). Arrows indicate the positions of the identified motifs a1–a7 and b1–b4. Motifs present on the upper and the lower strand are shown in green and purple, respectively. Third and fourth line show identified motifs present on Fragment III_A and XII_A, respectively. Black, dark grey, grey and light grey indicate motifs identical to the consensus sequence or deviating at one, two or three positions, respectively. B. An alignment of the nucleotide sequences of the eleven identified motifs and their flanking sequences. Names according to the nomenclature in “A” are given together with information on the strand and the region. Sequences corresponding to the consensus sequence of the motif are given in bold. C. A representation of the consensus motif generated by Weblogo [73]. The size of each nucleotide corresponds to the frequency with which that nucleotide is observed in that position. Evidence that the identified motif constitutes (part of) the binding site for RcoLS20 was obtained by DNase I footprinting (see below) and mutational analysis. Thus, gel retardation assays showed that binding of RcoLS20 is affected in probes containing alterations in one or two motifs in either operator. For instance, RcoLS20 did not bind to a derivative of Fragment III_A containing mutations in both motifs a1 and a7; and binding was affected when only motif a7 was mutated. Similarly, mutation of motif b1 or b4 resulted in the appearance of only one retarded species instead of two observed for corresponding fragments without mutations (Fig. 4B). In summary, the results obtained show that the intergenic rcoLS20-gene 28 region contains two RcoLS20 operators that are separated by 75 bp. Operator OII overlaps with promoters Pr/Pc and the other region is located 75 bp towards the direction of gene 28. Each region contains repeats of a motif whose consensus sequence is 5′-CAGTGAAA-3′ and which forms (part of) the binding site of RcoLS20. Motifs in OI are located on the lower strand, and except for one, motifs in OII are located on the upper strand.
Binding of RcoLS20 to operators OI and OII was confirmed by DNase I footprinting. The results presented in Figure 6 confirm that RcoLS20 binds to a region that overlaps with the Pr/Pc promoters and to another region located about 75 bp downstream of the Pc promoter. The combined in vitro results are in line with the in vivo results presented above.
Fig. 6. Footprint analyses of the binding of RcoLS20 to the rcoLS20 - -gene 28 intergenic region. 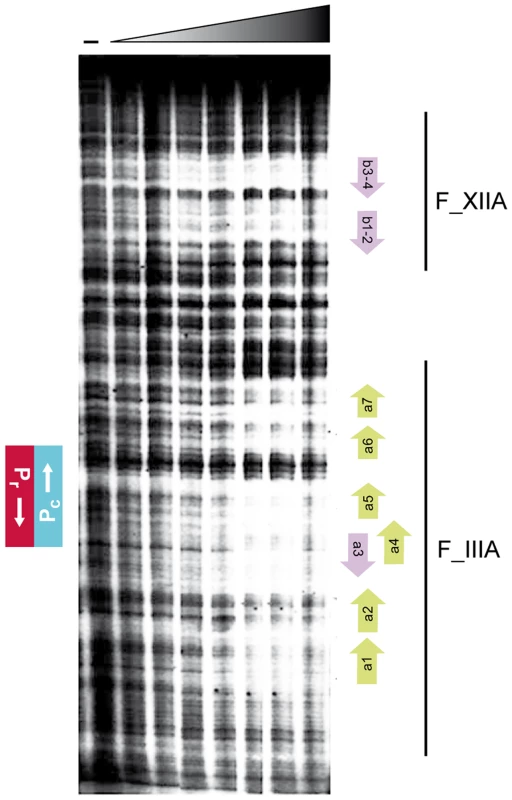
Fragment V, end-labeled at the Pc template strand, was analyzed for binding of RcoLS20-His. First lane (−) was not incubated with protein. Concentrations of RcoLS20-His, increasing by two-folds, ranged from 0.11 to 7.04 µM. The positions of the Pc and Pr promoters are indicated on the left. Bars on the right reflect the regions covered by Fragments IIIA (F_IIIA) and XIIA (F_XIIA). Positions of motifs a1–a7 and b1–b4 are indicated with green or purple arrows at the right. Evidences that proper regulation of the Pr/Pc promoters involves DNA looping
Operator OI, -located at a distance of more than 75 bp from Pr/Pc-, is needed for proper regulation of these promoters. This and other data presented above, suggest that proper regulation of the Pr/Pc promoters involves DNA looping mediated by RcoLS20 bound to operators OI and OII. Due to the intrinsic stiffness of DNA, loops are generally longer than 90 bp because the curvature energy required to make smaller loops is too large, unless the DNA region separating the two operator sites is bent [32]. Operators OI and OII are separated by only 75 bp. Several periodically spaced A/T tracts can result in formation of a static bent [33]. The spacer region contains periodically spaced A/T tracts, and computer-assisted analysis predicts that the spacer region forms a static curve (see Fig. S2). These data prompted us to perform circular permutation assays. Thus, three overlapping fragments of identical size (314 bp) were generated in which the predicted static curve is located at different positions (see Fig. 7A). As expected, these fragments migrated to identical positions when run on a 2% agarose gel (Fig. 7B). However, when run on a native 8% PAA gel the fragments migrated differently and all of them run slower than expected for their size, with the fragment containing the predicted bent in the middle of the fragment migrating slowest (Fig. 7C). These results show that the 75 bp spacer contains a static bent.
Fig. 7. The 75 bp spacer region separating operators OI and OII contains a static bent. 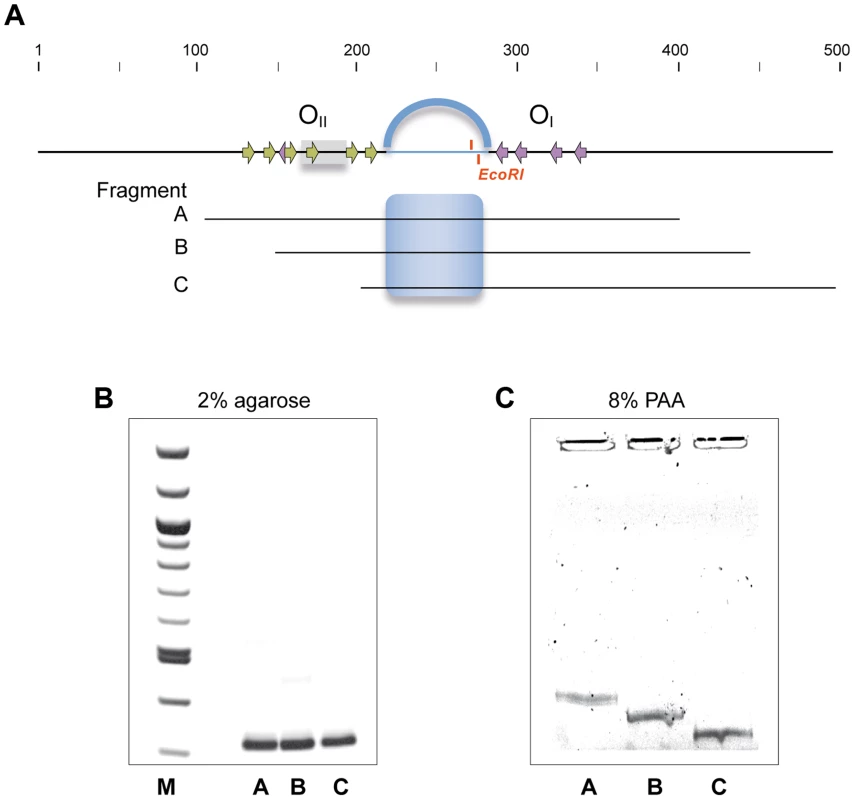
A. Schematic representation of the region encompassing operators OI and OII and the regions corresponding to the DNA fragments amplified by PCR that were subjected to 2% agarose (B) and 8% native PAA (C). Position of the Pc/Pr promoters and the RcoLS20 binding motifs within the operators are indicated with grey rectangles and arrows, respectively. The 75 bp region separating OI and OII is shown as an interrupted line and the position of the unique EcoRI site is given. The predicted curvature in this region is represented by the blue arc above the top line, and by a blue shading in the equivalent region in fragments A–C. Fragments A–C were run on 2% agarose or on 8% native PAA gel followed by ethidium bromide staining. If RcoLS20-mediated DNA looping occurs then it is expected (i) that RcoLS20 will form oligomers thereby creating a DNA binding unit able to bind simultaneously to OI and OII, and (ii) that the two operators are in phase such that the RcoLS20 binding sites have a spatial orientation that is optimal for RcoLS20 binding. We tested both predictions. The oligomerization state of RcoLS20 was studied by two complementary analytical ultracentrifugation approaches (Fig. 8). In sedimentation velocity experiments, RcoLS20 was observed as a single species with an experimental sedimentation coefficient of 3.8 S. This value corrected to standard conditions (s20,w = 4.1S) was compatible with an elongated protein tetramer (Fig. 8A). To confirm this result, sedimentation equilibration experiments were carried out within the concentration range from 10 to 30 µM. The calculated average molecular mass obtained was 85,200 Da±1,700, which corresponds to the tetrameric form of RcoLS20 (Fig. 8B).
Fig. 8. RcoLS20 forms tetramers in solution. 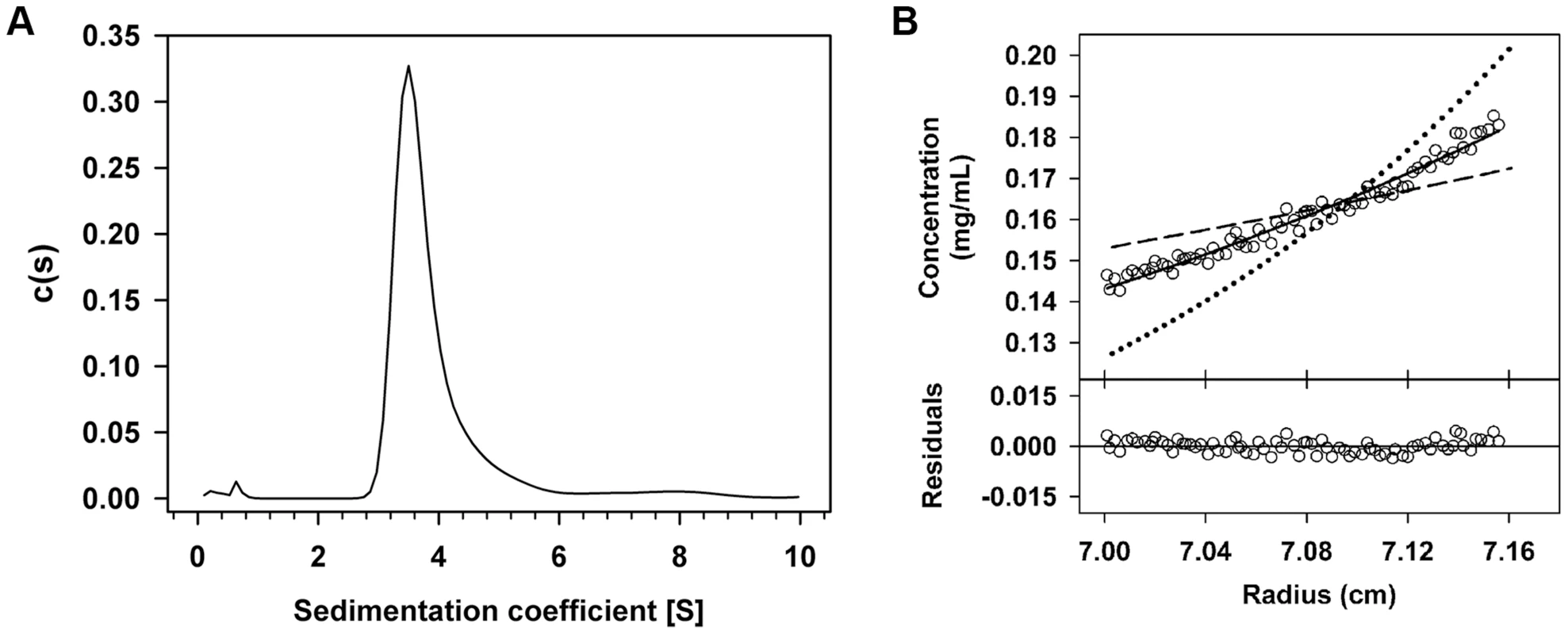
The oligomerization state of RcoLS20 protein in solution was studied by two complementary analytical ultracentrifugation assays. A. Sedimentation velocity assay. Sedimentation coefficient distribution c(s) profile corresponding to 10 µM purified RcoLS20. B. Sedimentation equilibrium assay. Upper part: Sedimentation equilibrium data for RcoLS20 (empty circles) are presented together with best-fit analysis assuming protein dimer (dashed line), tetramer (black line), or octamer (dotted line) species. The data indicate that RcoLS20 is a tetramer at 10 µM. Lower part: The difference between estimated values and experimental data for protein tetramers (residuals). To test if a specific phasing between OI and OII is important for RcoLS20 to carry out its regulatory role we constructed a derivative of Fragment I, I+5, in which we enlarged the spacer half a helical turn by inserting 5 bp and cloned this fragment in front of lacZ (see Fig. 1B). Next, we tested the responsiveness of promoter Pc to RcoLS20 using strains containing either Fragment F_Ic or F_Ic +5 fused to lacZ. As expected, RcoLS20, which was provided in trans by pLS20cat, efficiently repressed promoter Pc when lacZ was fused to Fragment Ic (strain PKS8). Promoter Pc was not efficiently repressed by RcoLS20 however, when lacZ was fused to Fragment Ic+5 (strain GR191). Thus, colonies of pLS20cat-harboring cells were blue when grown on Xgal-containing plates (see Fig. S3). These results show that enlarging the distance between OI and OII with half a helical turn destroys proper regulation of promoter Pc by RcoLS20. Besides affecting the phasing, the 5 bp insertion might also affect the static curvature of the spacer region. Regardless whether the loss of RcoLS20-mediated regulation is due to phasing and/or altered curvature, the results provide compelling evidence that RcoLS20 mediates its regulatory effect through DNA looping.
Next, we analyzed by EMSA if the 5 bp insertion between operators OI and OII affects RcoLS20 binding. As described above, even in the presence of the highest RcoLS20 concentration applied, DNA fragments F_I, F_IV and F_V containing operators OI and OII entered the gel migrating to distinct positions, indicating multiple intramolecular RcoLS20 binding events (Fig. 4B, right column, first, third and fourth panel). Interestingly, however, whereas Fragment I+5 entered the gel at low RcoLS20 concentrations, most of the DNA did not enter the gel at medium or high RcoLS20 concentrations (Fig. 4B, right column, second panel). One possible explanation is that dephasing between the two operators allows RcoLS20 to bind intermolecularly resulting in the formation of high molecular weight nucleoprotein complexes that do not enter the gel. Together, these results support the view that the phasing between OI and OII is crucial for proper RcoLS20-mediated regulation of transcription.
Discussion
Conjugation is a complex and energy consuming process, involving the generation and transfer of ssDNA, synthesis and assembly of a sophisticated type IV secretion system, and establishment of specific contacts with the recipient cell. Hence, the process of conjugation and expression of the genes involved are strictly controlled. Analysis of the regulation of conjugation genes present on ICEs in bacteria and those on plasmids of Gram-negative bacteria indeed indicates that this is the case [for review see], [ 5,7]. In our previous studies, we have sequenced and annotated plasmid pLS20cat of the Gram-positive bacterium B. subtilis and identified a large conjugation operon. We have also identified rcoLS20 as the gene encoding the master regulator of conjugation, RapLS20 as the anti-repressor required to activate the conjugation genes, and we showed that the activity of RapLS20 is in turn regulated by the signaling peptide Phr*LS20. In this study, we analyzed the underlying molecular mechanism of how the pLS20 conjugation genes are regulated. The results obtained provide compelling evidence that the conjugation genes of pLS20 are controlled by a complex genetic switch, which is composed of at least three intertwined layers. A scheme of the three layers is shown in Figure 9. One of the levels results from the relative positioning of the main conjugation promoter, Pc, and the divergently oriented promoter Pr, driving expression of the rcoLS20 gene (Fig. 9A). The presence of divergently oriented promoters is a common form of gene organization in bacteria, and the (likely) role of this organization in transcriptional regulation has long been recognized [34]. Nevertheless, direct proof for and detailed analysis of the implications on transcriptional regulation are restricted to only a minor fraction of the divergently oriented transcriptional units detected. Here, we identified the conjugation promoter Pc and showed that it is a relatively strong promoter, which is repressed by the master regulator of conjugation RcoLS20. Importantly, the position of promoter Pc coincides, or at least partially overlaps, with the divergently oriented weak Pr promoter. It has been demonstrated that an RNA polymerase can bind only to one of two overlapping promoters [35], [36]. Thus, in the special configuration of overlapping promoters the RNA polymerase may itself act as a transcriptional regulator. Recently, Bendtsen et al. [37] described theoretical scenarios backed up by experimental data that overlapping promoters indeed can result in a transcriptional switch, provided that they have different activities in the absence of the regulatory protein, combined with a regulator that has a strong differential effect on the regulation of both promoters. This is exactly the case for the Pc/Pr promoter pair; in the absence of the regulator promoter Pc is several hundred folds stronger than Pr, and the presence of the regulator strongly represses the Pc promoter while activating the Pr promoter.
Fig. 9. Model of the different layers contributing to the genetic switch controlling expression of the pLS20 conjugation genes. 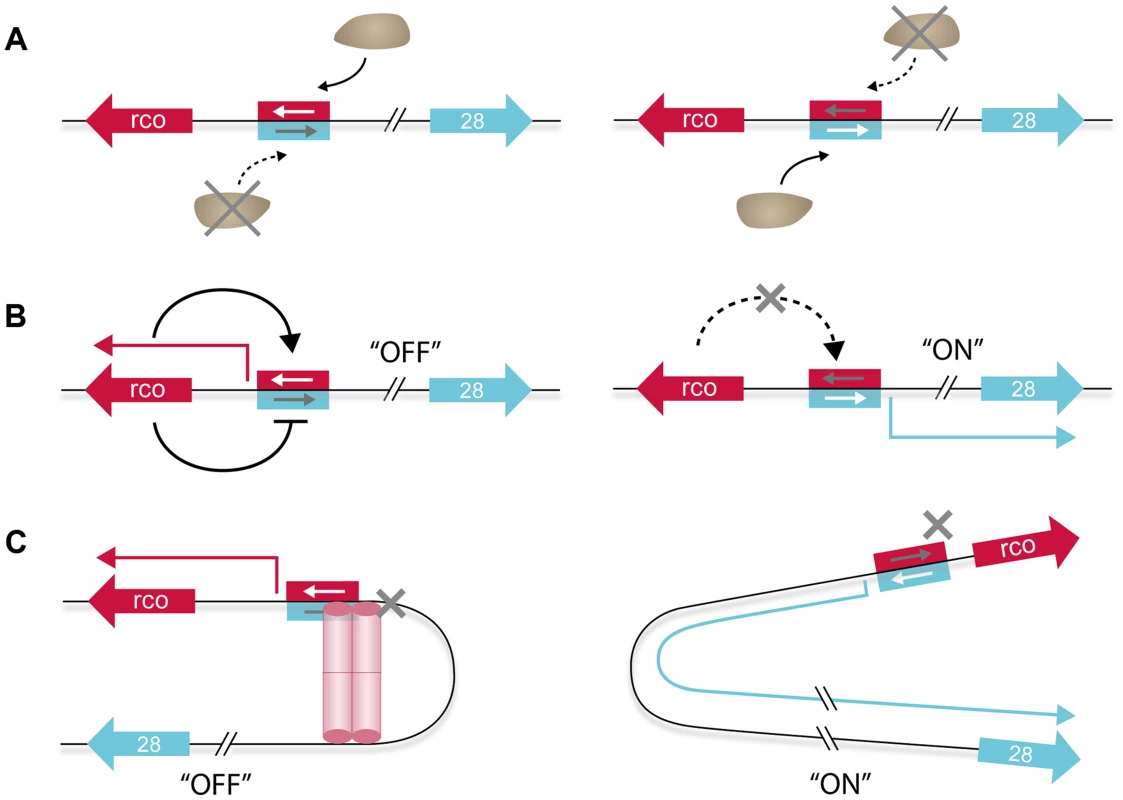
A. RNA polymerase acts itself as a switch because it is unable to bind simultaneously to both of the two overlapping and divergently oriented promoters. Consequently, RNA polymerase (the brown ellipse shaped form) binds only one promoter at a time resulting in transcription of only the gene(s) controlled by this promoter. B. RcoLS20 generates a self-sustaining positive feedback loop by activating transcription from its own promoter (Pr) (left panel). This, combined with the simultaneous repression of the divergent conjugation promoter (Pc), results in conjugation being maintained effectively in the “OFF” state. Relief of RcoLS20-mediated repression of the Pc promoter results in activation of the conjugation genes (right panel). In addition, this interrupts the auto-stimulation of the Pr promoter, preventing further synthesis of RcoLS20, which in turn will contribute in pushing and maintaining conjugation in the “ON” state. The negative auto-regulatory loop of RcoLS20 that probably functions to keep RcoLS20 within a low concentration range (see text) is not presented. C. DNA looping results in a high local concentration of RcoLS20, increasing specificity and affinity that dampens transcriptional fluctuations between and within individual cells (left panel). This would contribute to tight repression of the Pc promoter, keeping conjugation in the “OFF” state under conditions antithetic to conjugation without compromising the ability to switching rapidly to a high expression state (i.e. “ON”, right panel) of the conjugation genes when appropriate conditions occur. rcoLS20 and gene 28, -the first gene of the conjugation operon-, are indicated with large red and blue arrows, respectively. The same coloring scheme is used for the corresponding promoters (rectangular) and transcripts (thin broken arrows). Activation and repression of transcription are indicated with continuous black lines ending in an arrow and a “T” shape, respectively. The red cylindrical structures, which may reflect one or two RcoLS20 tetramers, represent the RcoLS20 oligomer mediating DNA looping. The second level of regulation contributing to the genetic switch concerns the multiple roles that RcoLS20 plays in the Pc/Pr regulation (Fig. 9B). We showed that, on the one hand, RcoLS20 activates transcription of its own weak promoter, Pr, thereby generating a self-sustaining positive feedback loop. On the other hand, RcoLS20 functions simultaneously as an efficient repressor of the Pc promoter. The dual effect that RcoLS20 has on Pc and Pr maintains conjugation effectively in the “OFF” state. We also showed that the level of rcoLS20 induction from an inducible promoter required for efficient repression of the Pc promoter was about ten-fold lower than that required for maximum auto-activation of the Pr promoter. These differential effects of RcoLS20 on repressing and activating the Pc/Pr promoters will also contribute towards maintaining conjugation stably in the “OFF” state under conditions when conjugation should not be activated. Interestingly, we found that at elevated concentrations RcoLS0 inhibits its own transcription. This negative autoregulation probably functions to keep RcoLS20 within a low concentration range in order to respond accurately to the anti-repressor RapLS20 to activate the conjugation genes. The triple effects RcoLS20 has on the regulation of the Pc/Pr promoters will also play an important role when RapLS20 induces the system to switch to the “ON” state. In addition to relieving repression of the strong conjugation Pc promoter, this will simultaneously annihilate autostimulation of the Pr promoter, preventing further synthesis of RcoLS20, which in turn will contribute in pushing and maintaining conjugation in the “ON” state.
A third level contributing to the genetic switch to activate the conjugation genes involves the DNA looping mediated by simultaneous binding of RcoLS20 to operators OI and OII (Fig. 9C). DNA looping mediated by a transcriptional regulator has been reported for several other regulatory systems in prokaryotes and their analyses have revealed that several features are conserved and necessary for DNA looping to occur [for review see, 38]. Our results showed that the properties of RcoLS20 and the DNA in the Pc/Pr region comply with the necessary features for RcoLS20-mediated loop formation. First, using different techniques, we show that RcoLS20, -predicted to contain a helix-turn-helix DNA binding motif in its N-terminal region [29]-, is a DNA binding protein and that it binds specifically to two operators, OI and OII. Second, operator OI, which is located more than 85 bp away from promoters Pc and Pr, is required for efficient regulation of both promoters. Third, RcoLS20 binds cooperatively to both operators. Fourth, dephasing the positions of the two operators by inserting 5 bp in the spacer region destroys proper regulation of the conjugation genes. And fifth, we showed that RcoLS20 forms tetramers in solution. This will create a unit containing multiple DNA binding motifs, facilitating cooperative binding to multiple sites within the two operators.
The DNA loop in the Pc/Pr region of pLS20 is characterized by a small spacer region that separates RcoLS20 operators OI and OII. The spacer length can be used to classify DNA loops into two categories: short or energetic loops, and long or entropic ones. Due to intrinsic stiffness and torsional rigidity of the DNA, loop formation is normally unfavorable for those with spacer lengths shorter than the DNA persistence length (approximately 150 bp), because the curvature energy required for forming such small loops becomes too great. For such short loops to occur specific features like intrinsic static bending or binding of an additional protein inducing bending are required. In the case of pLS20, in which the operators OI and OII are separated by only 75 bp, we show that the spacer region contains a static bent.
The first experimental demonstration that a DNA loop can play a crucial role in transcriptional regulation was reported for the E. coli ara operon in 1984 [39]. Since then, some other operons have been shown to be also regulated by transcriptional regulator-mediated DNA looping [for reviews see], [ 38], [40]–[43], though the actual number of transcriptional systems for which DNA looping has been conclusively demonstrated is remarkably low. In the case of plasmids, reports demonstrating DNA looping systems are limited to only few cases. One of these includes regulation of initiation of DNA replication at the beta origin of the E. coli R6K plasmid [44]; and in the case of Enterococcus faecalis plasmid pCF10 it has been proposed that regulation of its conjugation system involves DNA looping mediated by the pheromone-responsive transcriptional regulator PrgX [for review see, 45]. Bio-informatic analyses suggest that DNA looping mediated regulation of transcription is likely to be more common than the few cases for which this has been demonstrated so far. For instance, Cournac and Plumbridge [38] have screened the E. coli genome for the presence of putative “simple DNA looping systems” in which looping would involve a single regulator (i.e., this analysis included only transcriptional regulators for which the operator sequence is known, and did not take into account the putative loops that would involve heterologous proteins and/or global transcriptional regulators). Under these restrictive settings, this survey identified 48 genes/operons in which DNA looping mediated regulation is likely to play a role. Interestingly, fourteen of them involve divergently oriented promoters. In the context of our studies, it is worth mentioning the regulation of the conjugation genes located on the integrative and conjugative element ICEBs1 that is present in several B. subtilis strains. The gene encoding the transcriptional regulator ImmR, and the excision and conjugation genes are expressed from two divergently oriented promoters that are separated by ∼130 bp. At low concentrations, the ImmR protein can bind to six regions, three being proximal to each promoter. It has been suggested that repression of the immR promoter might involve cooperative interactions between ImmR molecules bound to binding sites proximal to both promoters, i.e. DNA looping [46]. Based on the distribution of the operator sites, DNA looping could also be involved in the transcriptional regulation of the Gram-negative plasmids Ti or IncP-plasmids, where divergent promoters have been shown to be involved in controlling both the replication and transfer functions [47], [48].
What are the benefits of DNA looping in general and for the regulation of the conjugation genes of pLS20 in particular? A major consequence of DNA looping is that it results in a high local concentration of the transcriptional regulator at the right place, which would increase its specificity and affinity [for recent review see, 49]. Often, -and RcoLS20 is not an exception-, transcriptional regulators are produced in limited amounts per cell. Low numbers of regulators enhance the possibility of transcriptional fluctuations between individual cells within a population. In addition, the intrinsic stochasticity of transcription, -also referred to as noise-, affects the temporal effectiveness of transcriptional regulation; again this is especially prominent when the number of regulatory proteins involved is low. Recent evidences indicate that DNA looping contributes importantly to controlling temporal transcriptional noise, as well as dampening transcriptional fluctuations between cells within a population [50], [51]. Thus, DNA looping contributes to the tight regulation of promoters especially when levels of transcriptional regulators are low by diminishing stochastic fluctuations in transcription.
For some differentiation processes, cell-to-cell or stochastic variability in levels of transcriptional regulators form the basis for activation of these processes, resulting in different behavior of genetically identical cells within a population [52]–[54]. Examples of these processes are the formation of persister cells, development of natural genetic competence, spore formation and swimming/chaining. It is believed that such a bet-hedging strategy is beneficial for the fitness of the species because there will always be some cells that are prepared to cope with a deteriorating environmental condition that may arise in the near future. However, for other processes, there may not be such an advantage and it would then be important to tightly repress the process at times when conditions for that process are not apt. Conjugation probably is such a process because there is no benefit in activating the conjugation genes when there is no recipient present to receive the plasmid. The fact that the efficiency of pLS20 transfer during growth conditions antithetic to conjugation is below the detection limit (at least six orders of magnitude lower than those observed during optimal conjugation conditions) strongly indicates that conjugation genes are tightly repressed under such conditions. However, the tight repression of conjugation should not compromise the ability of rapidly switching to high expression of the conjugation genes when appropriate conditions occur. In pLS20 this is achieved by the constellation of DNA looping combined with autoregulated expression of RcoLS20 and overlapping divergent promoters of different strength.
A well-studied genetic switch involving DNA looping is the one that governs the switch from the lysogenic to the lytic state of the Escherichia coli phage λ [for review see], [ 55,56]. In the lysogenic or prophage state, phage λ replicates passively with the host while the lytic genes are repressed. This prophage state is extremely stable and can be maintained for many generations. Upon induction of the SOS response, however, a switch is made to the lytic cycle resulting in excision of the phage genome, followed by its amplification and eventually lysis of the cell and release of phage progeny. The early lytic phage λ genes are located in two divergently oriented operons, which are controlled by the lytic promoters PR and PL. A third operon, which encodes amongst others the CI transcriptional regulator, is located in between the two early lytic operons such that the promoter of gene cI, PRM, flanks the divergently oriented PR promoter driving expression of one of the two early operons. In several aspects, functional analogies exist between CI and RcoLS20 although they share only 16% of identity at their primary protein sequence level. Both RcoLS20 and CI stimulate and repress their own promoter at low and high concentrations, respectively, resulting in a self-sustaining positive feedback loop while keeping the transcriptional regulator in a low concentration range. Above, arguments have been given that for pLS20 this situation, together with the effects of the DNA loop, is important for the tight repression of the Pc promoter during conditions in which conjugation is not favourable, while maintaining the sensitivity to be able to respond rapidly to switch on the conjugation genes when appropriate conditions occur. The transcriptional regulation of λ appears to serve a similar purpose. Thus, on the one hand the lytic genes are tightly repressed since spontaneous switching to the lytic cycle occurs less than once every 108 generations [57]. On the other hand, mutations that specifically eliminate the negative autoregulation of cI expression impair prophage induction [58], [59]. Another analogy between the pLS20 and λ systems is that both the regulators RcoLS20 and CI, can form higher order oligomers, permitting them to bind cooperatively to multiple sites distributed in two operators, effectively resulting in DNA looping which plays an important role in the genetic regulation of the conjugation and the lytic operon, respectively. Taking the analogy further, it is interesting to note that these regulatory systems both control a process of horizontal gene transfer.
However, there are also several differences between the two systems. For instance, whereas regulation of pLS20 conjugation genes involves a short loop of 75 bp, regulation of the λ lytic genes involves a long loop of 2.3 kb. A second difference is that CI protein forms dimers in solution. A pair of CI dimers tetramerizes when binding to the binding sites in one operator and another dimer pair does the same when binding to the other operator. Upon DNA looping, interaction between the two tetramers constitutes a functional octamer. In addition, when a loop is formed another pair of dimers may bind to additional binding sites present in both operators, and this additional bridge is responsible for repressing PRM promoter. At present, we do not have such detailed insights in transcriptional regulations at the molecular level for RcoLS20. However, instead of dimers, RcoLS20 forms tetramers in solution, which probably means that the molecular mechanism by which the pLS20 promoters Pr and Pc are regulated is distinct from the way CI regulates λ promoters PR and PRM. Another argument supporting this assumption is the different configuration of the divergent promoters and the binding sites for the regulator protein. In pLS20, the position of promoters Pc/Pr overlaps and the RcoLS20 binding sites in OII overlap and flank these core promoters. In λ the binding sites for CI regulator in one operator overlap the PR promoter and are located upstream of the PRM core promoter sequences. Finally, a major difference between the DNA looping involved systems of pLS20 and λ is how the switches are induced. In λ, the switch is induced by an SOS response which results in RecA-mediated CI autocleavage. In the case of pLS20, the switch is dictated ultimately by intercellular quorum sensing signaling involving the signaling peptide Phr*LS20 that regulates the activity of RapLS20, the anti-repressor of RcoLS20 [29]. This quorum sensing system will lead to activation of the conjugation genes when donor cells are surrounded by recipient cells. However, high levels of Phr*LS20 will build up when the majority of the cells that surround a donor cell already contain pLS20, and this will inactivate RapLS20 and hence block activation of the conjugation genes.
Besides those described here, it is possible that the pLS20 conjugation genes are regulated by additional mechanism(s). For example, the conspicuously long 5′ untranslated region upstream of gene 28 is predicted to form complex secondary structures, which might modulate expression of the downstream genes in a variety of scenarios. Currently, a study to elucidate a possible role of this long 5′ untranslated region is carried out in our laboratory.
In summary, in this work we have provided evidence that regulation of the conjugation genes present on pLS20 is based on a unique genetic switch that combines at least three levels of control. These include (i) overlapping divergent promoters of different strengths, (ii) auto-stimulation and repression of the weak Pr promoter by the transcriptional regulator at low and elevated concentrations, respectively, combined with simultaneous repression of the divergent strong conjugation promoter, and (iii) DNA looping mediated by binding of RcoLS20 regulator to two operators separated by a short loop. Most likely, the combination of these different layers causes tight repression of the main conjugation promoter Pc when conditions for conjugation are not optimal, while allowing the system to switch rapidly to high expression of the conjugation genes when appropriate conditions occur.
Materials and Methods
Bacterial strains, plasmids, media and oligonucleotides
Bacterial strains were grown in LB liquid medium or on 1.5% LB agar plates [60]. When appropriate, the following antibiotics were added to media or plates: ampicillin (100 µg/ml), erythromycin (1 and 150 µg/ml in B. subtilis and E. coli, respectively), chloramphenicol (5 µg/ml), spectinomycin (100 µg/ml), and kanamycin (10 µg/ml). Table S1 lists the B. subtilis strains used. All of them are isogenic with B. subtilis strain 168. Plasmids and oligonucleotides used are listed in Table S2 and S3, respectively. All oligos were purchased from Isogen Life Science, The Netherlands.
Transformation
E. coli cells were transformed using standardized methods as described in Singh et al [61]. For standard B. subtilis transformations, competent cells were prepared as described by Bron [62]. Transformants were selected on LB agar plates with appropriate antibiotics.
Construction of plasmids and strains
Standard molecular methods were used to manipulate DNA [60]. Sequence analysis was used to verify the correctness of all constructs. The same strategy was used to construct B. subtilis strains containing a copy of lacZ fused to the entire or part of the rcoLS20-gene 28 intergenic DNA region. First, the region of DNA to be cloned was amplified using appropriate primers (see Table S3), purified, and digested with the appropriate restriction enzymes. Next, the fragment was used to prepare a ligation mixture together with the integration vector pDG1663 digested with the same enzymes. The ligation mixture was transformed into E. coli XL1-blue cells. The plasmid content of several ampicillin resistant transformants was checked and clones containing the insert with appropriate size and orientation were subjected to DNA sequencing to verify the absence of mutations. The names of the pDG1663 derivatives and their characteristics are listed in Table S2. Plasmid DNA of each pDG1663 derivative was used to transform competent B. subtilis 168 cells. Transformants were initially selected for resistance to erythromycin. Next, double cross-over events were distinguished from single cross-over events by selecting transformants sensitive to spectinomycin. The resulting B. subtilis strains containing a single copy of lacZ preceded by different regions of the rcoLS20-gene 28 region at the thrC locus of the B. subtilis chromosome are listed in Table S1. Next, plasmid pLS20cat was introduced into the different lacZ fusion strains by conjugation. B. subtilis strain PKS9 contains a single copy of the rcoLS20 gene under the control of the IPTG-inducible Pspank promoter at its amyE locus and this cassette is linked to the spectinomycin gene. Chromosomal DNA of strain PKS9 was used to transform competent cells of the various lacZ fusion strains in order to construct derivatives of the lacZ fusion strains containing the Pspank-rcoLS20 cassette.
The following strategy was used to construct a translational fusion of rcoLS20 with his(6). The rcoLS20 gene was amplified from pLS20cat by PCR using primers oPKS14N and oPKS8. The purified PCR product was digested with NcoI and SalI and cloned into the vector pET28b+ digested with the same restriction enzymes to produce plasmid pRcoLS20-His. B. subtilis strain GR90 contains the rcoLS20-his(6) under the control of the Pspank promoter at the amyE locus. To construct this strain rcoLS20-his(6) was amplified from pRcoLS20-His by PCR using primers oGR3 and oGR4. The PCR product was digested with NheI and SphI and cloned into the vector pDR110 digested with the same enzymes to generate pPspankrcoLS20-His. This plasmid was used to transform competent B. subtilis cells selecting for spectinomycin resistance. Double cross-over events were selected by loss of amylase gene.
β-Galactosidase activity assays
β-galactosidase activities were determined as described previously [63]. Overnight grown cultures were diluted 100 times into fresh prewarmed medium and samples were taken every 45 min.
Conjugation assays
Conjugation was carried out in liquid medium as described previously [29]. The effect of ectopic expression on conjugation of a gene controlled by the IPTG-inducible Pspank promoter was studied as follows. Overnight cultures were diluted in prewarmd LB supplemented with IPTG at the indicated concentrations to an OD600 of ∼0.05. Next, samples were taken at regular intervals to determine OD600 and were subjected to matings with proper recipient cells.
RNA isolation and RNA sequencing
Preparation of total RNA samples, RNA sequencing and Bioinformatic analysis of RNAseq data was done as described previously [29].
RcoLS20-His(6) purification
E. coli BL21 (DE3) cells carrying plasmid prcoLS20-His were used to inoculate 1 litre of fresh LB medium supplemented with 30 mg/ml kanamycin and grown at 37°C with shaking. At an OD600 of 0.4, expression of rcoLS20-his(6) was induced by adding IPTG to a final concentration of 1 mM and growth was continued for 2 h. Cells were further processed as described previously [28]. Purified protein (>95% pure) was dialysed against buffer B (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 250 mM NaCl, 10 mM MgCl2, 7 mM β-mercaptoethanol, 50% v/v glycerol) and stored in aliquots at −80°C. Bradford assay was used to determine the protein concentrations.
Gel retardation
In essence, the gel retardation assays were carried out as described before [28]. Thus, different fragments of intergenic regions between gene 28 and rcoLS20 were amplified by PCR using pLS20cat as template. The resulting PCR fragments were purified and equal concentrations (300 nM) were incubated on ice in binding buffer [20 mM Tris HCl pH 8, 1 mM EDTA, 5 mM MgCl2, 0.5 mM DTT, 100 mM KCl, 10% (v/v) glycerol, 0.05 mg ml−1 BSA] without and with increasing amounts of purified RcoLS20His(6) in a total volume of 16 µl. After careful mixing, samples were incubated for 20 min at 30°C, placed back on ice for 10 min, then loaded onto 2% agarose gel in 0.5XTBE. Electrophoresis was carried out in 0.5X TBE at 50 V at 4°C.Finally, the gel was stained with ethidium bromide, destained in 0.5XTBE and photographed with UV illumination.
Primer extension experiments
Determination of the transcription start sites by primer extension was performed essentially as described [64]. In brief, total RNA (30 µg) was mixed with 4 pmol of end-labeled oligonucleotide that served as primer; the mixture was heated at 70°C for 5 min and allowed to anneal for 5 min at 23°C. The annealed RNA was ethanol precipitated, resuspended and primer extension was performed with 30 U of AMV reverse transcriptase (Promega) at 42°C, as recommended by the supplier. The extended cDNA products were analysed by electrophoresis on a denaturing 6% urea-polyacrylamide gel, in parallel with a DNA sequence ladder performed by chemical sequencing [65] of a DNA fragment encompassing the mapped promoters (see below). The primer used to map promoter Pc was 5′-ttctagttctttttacac, while that used for promoter Pr was 5′-tctctattgcccacttat. Oligonucleotides were end-labeled with [γ-32P]-ATP and T4 polynucleotide kinase as recommended by the supplier (New England Biolabs). The 186 bp DNA fragment that served as sequence ladder was PCR amplified with primers 5′-acggtctagcgcttacaat and 5′-ttctagttctttttacac, the last one labeled at its 5′ end.
DNase I footprinting
DNaseI footprinting assay was carried out as described [66]. The Pc/pr promoter encompassing region was amplified by PCR using primers p28_Δ16 and Prom28UpBam, and pLS20cat as template. One of the ends was radio-labeled by digesting the fragment with BamHI and subsequently filling in the end with exo− Klenow fragment in the presence of [α-32P]-ATP.
Computer-assisted analysis
Presence of conserved motifs was searched by using motif-identification programs MEME [30] and BIOPROSPECTOR [31]. Prediction of the static bending properties of DNA sequences was carried out by calculating the global 3D structure according to the dinucleotide wedge model [67]. All graphics work was done by using Adobe Photoshop CS2 and adobe illustrator. Graphs were plotted using Excel program.
Ultracentrifugation
Sedimentation velocity assay. Samples in 20 mM Tris-HCl, 250 mM NaCl, 10 mM MgCl2, 1 mM EDTA and 100 mM glycerol, pH 7.4, were loaded (320 µL) into analytical ultracentrifugation cells. The experiments were carried out at 43–48 krpm in an XL-I analytical ultracentrifuge (Beckman-Coulter Inc.) equipped with UV-VIS absorbance and Raleigh interference detection systems. Sedimentation profiles were recorded at 280 nm. Sedimentation coefficient distributions were calculated by least-squares boundary modelling of sedimentation velocity data using the continuous distribution c(s) Lamm equation model as implemented by SEDFIT 14.1 [68]. Experimental s values were corrected to standard conditions (water, 20°C, and infinite dilution) using the program SEDNTERP [69] to get the corresponding standard s values (s20,w).
Sedimentation equilibrium assay. Using the same experimental conditions as in the SV experiments, short columns (90 µL) SE experiments were carried out at speeds ranging from 7,000 to 10,000 rpm and at 280 nm. After the last equilibrium scan, a high-speed centrifugation run (48,000 rpm) was done to estimate the corresponding baseline offsets. Weight-average buoyant molecular weights of protein were determined by fitting a single species model to the experimental data using the HeteroAnalysis program [70], and corrected for solvent composition and temperature with the program SEDNTERP [69].
Supporting Information
Zdroje
1. OchmanH, LawrenceJG, GroismanEA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405 : 299–304.
2. FrostLS, LeplaeR, SummersAO, ToussaintA (2005) Mobile genetic elements: the agents of open source evolution. Nat Rev Micobiol 3 : 722–732.
3. ThomasCM, NielsenKM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3 : 711–721.
4. NovickRP, ChristieGE, PenadesJR (2010) The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol 8 : 541–551.
5. WozniakRA, WaldorMK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8 : 552–563.
6. AuchtungJM, LeeCA, MonsonRE, LehmanAP, GrossmanAD (2005) Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102 : 12554–12559.
7. FrostLS, KoraimannG (2010) Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol 5 : 1057–1071.
8. SmillieC, Garcillán-BarciaMP, FranciaMV, RochaEPC, De la CruzF (2010) Mobility of plasmids. Microbiol Mol Biol Rev 74 : 434–452.
9. Alvarez-MartinezCE, ChristiePJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73 : 775–808.
10. FronzesR, ChristiePJ, WaksmanG (2009) The structural biology of type IV secretion systems. Nat Rev Microbiol 7 : 703–714.
11. Goessweiner-MohrN, GrumetL, ArendsK, Pavkov-KellerT, GruberCC, GruberK, Birner-GruenbergerR, Kropec-HuebnerA, HuebnerJ, GrohmannE, KellerW (2013) The 2.5 A structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem 288 : 2018–2028.
12. LiJ, AdamsV, BannamTL, MiyamotoK, GarciaJP, UzalFA, RoodJI, McClaneBA (2013) Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77 : 208–233.
13. LiuMA, KwongSM, JensenSO, BrzoskaAJ, FirthN (2013) Biology of the staphylococcal conjugative multiresistance plasmid pSK41. Plasmid 70 : 42–51.
14. CarylJA, O'NeillAJ (2009) Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the Staphylococci. Plasmid 62 : 35–38 S.
15. ClewellDB (2011) Tales of conjugation and sex pheromones: A plasmid and enterococcal odyssey. Mob Genet Elements 1 : 38–54.
16. DunnyGM, JohnsonCM (2011) Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr Opin Microbiol 14 : 174–180.
17. ChatterjeeA, CookLC, ShuCC, ChenY, ManiasDA, RamkrishnaD, DunnyGM, HuWS (2013) Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110 : 7086–7090.
18. Sonenshein, A. L., Hoch, J. A., and Losick, R. (1993) Bacillus subtilis and other Gram-positive bacteria; Biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology. 987 p.
19. Sonenshein, A. L., Hoch, J. A., and Losick, R. (2001) Bacillus subtilis and its closest relatives: from genes to cells. ASM Press.
20. CuttingSM (2011) Bacillus probiotics. Food Microbiol 28 : 214–220.
21. TanakaT, KoshikawaT (1977) Isolation and characterization of four types of plasmids from Bacillus subtilis (natto). J Bacteriol 131 : 699–701.
22. KoehlerTM, ThorneCB (1987) Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J Bacteriol 169 : 5271–5278.
23. ItayaM, SakayaN, MatsunagaS, FujitaK, KanekoS (2006) Conjugational transfer kinetics of pLS20 between Bacillus subtilis in liquid medium. Biosci Biotechnol Biochem 70 : 740–742.
24. BauerT, RoschT, ItayaM, GraumannPL (2011) Localization pattern of conjugation machinery in a Gram-positive bacterium. J Bacteriol 193 : 6244–6256.
25. RöschTC, GolmanW, HucklesbyL, Gonzalez-PastorJE, GraumannPL (2014) The presence of conjugative plasmid pLS20 affects global transcription of Its Bacillus subtilis host and confers beneficial stress resistance to cells. Appl Environ Microbiol 80 : 1349–1358.
26. MeijerWJJ, de BoerA, van TongerenS, VenemaG, BronS (1995) Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res 23 : 3214–3223.
27. DermanAI, BeckerEC, TruongBD, FujiokaA, TuceyTM, ErbML, PattersonPC, PoglianoJ (2009) Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 73 : 534–552.
28. SinghPK, RamachandranG, Duran-AlcaldeL, AlonsoC, WuLJ, MeijerWJ (2012) Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ Microbiol 14 : 2812–2825.
29. SinghPK, RamachandranG, Ramos-RuizR, Peiro-PastorR, AbiaD, WuLJ, MeijerWJ (2013) Mobility of the Native Bacillus subtilis Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling. PLoS Genet 9: e1003892.
30. BaileyTL, ElkanC (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 : 28–36.
31. LiuX, BrutlagDL, LiuJS (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138.
32. HagermanPJ (1988) Flexibility of DNA. Annu Rev Biophys Biophys Chem 17 : 265–286.
33. HaranTE, MohantyU (2009) The unique structure of A-tracts and intrinsic DNA bending. Q Rev Biophys 42 : 41–81.
34. BeckCF, WarrenRA (1988) Divergent promoters, a common form of gene organization. Microbiol Rev 52 : 318–326.
35. WangP, YangJ, LawleyB, PittardAJ (1997) Repression of the aroP gene of Escherichia coli involves activation of a divergent promoter. J Bacteriol 179 : 4213–4218.
36. WangP, YangJ, IshihamaA, PittardAJ (1998) Demonstration that the TyrR protein and RNA polymerase complex formed at the divergent P3 promoter inhibits binding of RNA polymerase to the major promoter, P1, of the aroP gene of Escherichia coli. J Bacteriol 180 : 5466–5472.
37. BendtsenKM, ErdossyJ, CsiszovszkiZ, SvenningsenSL, SneppenK, KrishnaS, SemseyS (2011) Direct and indirect effects in the regulation of overlapping promoters. Nucleic Acids Res 39 : 6879–6885.
38. CournacA, PlumbridgeJ (2013) DNA looping in prokaryotes: experimental and theoretical approaches. J Bacteriol 195 : 1109–1119.
39. DunnTM, HahnS, OgdenS, SchleifRF (1984) An operator at −280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A 81 : 5017–5020.
40. AdhyaS (1989) Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet 23 : 227–250.
41. MatthewsKS (1992) DNA looping. Microbiol Rev 56 : 123–136.
42. SchleifR (1992) DNA looping. Annu Rev Biochem 61 : 199–223.
43. HochschildA, LewisM (2009) The bacteriophage lambda CI protein finds an asymmetric solution. Curr Opin Struct Biol 19 : 79–86.
44. MukherjeeS, EricksonH, BastiaD (1988) Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell 52 : 375–383.
45. DunnyGM (2013) Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47 : 457–482.
46. AuchtungJM, LeeCA, GarrisonKL, GrossmanAD (2007) Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis. Mol Microbiol 64 : 1515–1528.
47. ZatykaM, Jagura-BurdzyG, ThomasCM (1997) Transcriptional and translational control of the genes for the mating pair formation apparatus of promiscuous IncP plasmids. J Bacteriol 179 : 7201–7209.
48. LangJ, FaureD (2014) Functions and regulation of quorum-sensing in Agrobacterium tumefaciens. Front Plant Sci 5 : 14.
49. OehlerS, Muller-HillB (2010) High local concentration: a fundamental strategy of life. J Mol Biol 395 : 242–253.
50. VilarJM, SaizL (2005) DNA looping in gene regulation: from the assembly of macromolecular complexes to the control of transcriptional noise. Curr Opin Genet Dev 15 : 136–144.
51. SaizL, VilarJM (2006) DNA looping: the consequences and its control. Curr Opin Struct Biol 16 : 344–350.
52. KorobkovaE, EmonetT, VilarJM, ShimizuTS, CluzelP (2004) From molecular noise to behavioural variability in a single bacterium. Nature 428 : 574–578.
53. DubnauD, LosickR (2006) Bistability in bacteria. Mol Microbiol 61 : 564–572.
54. VeeningJW, SmitsWK, KuipersOP (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62 : 193–210.
55. DoddIB, ShearwinKE, EganJB (2005) Revisited gene regulation in bacteriophage lambda. Curr Opin Genet Dev 15 : 145–152.
56. OppenheimAB, KobilerO, StavansJ, CourtDL, AdhyaS (2005) Switches in bacteriophage lambda development. Annu Rev Genet 39 : 409–429.
57. LittleJW, ShepleyDP, WertDW (1999) Robustness of a gene regulatory circuit. EMBO J 18 : 4299–4307.
58. DoddIB, PerkinsAJ, TsemitsidisD, EganJB (2001) Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev 15 : 3013–3022.
59. DoddIB, ShearwinKE, PerkinsAJ, BurrT, HochschildA, EganJB (2004) Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev 18 : 344–354.
60. Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
61. SinghPK, Ballestero-BeltranS, RamachandranG, MeijerWJ (2010) Complete nucleotide sequence and determination of the replication region of the sporulation inhibiting plasmid p576 from Bacillus pumilus NRS576. Res Microbiol 161 : 772–782.
62. Bron S (1990) Plasmids. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester, UK: John Wiley & Sons Ltd. pp. 75–174.
63. Miller, J. H. (1982) Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
64. MorenoR, FonsecaP, RojoF (2012) Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol Microbiol 83 : 24–40.
65. MaxamAM, GilbertW (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 65 : 499–560.
66. MeijerWJJ, Castilla-LlorenteV, VillarL, MurrayH, ErringtonJ, SalasM (2005) Molecular basis for the exploitation of spore formation as survival mechanism by virulent phage φ29. EMBO J 24 : 3647–3657.
67. BolshoyA, McNamaraP, HarringtonRE, TrifonovEN (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci U S A 88 : 2312–2316.
68. SchuckP (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78 : 1606–1609.
69. Laue TM, Shah BD, Ridgeway TM, Pelletier SL (1992) Interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, UK: Royal Society of Chemistry. pp. 90–125.
70. ColeJL (2004) Analysis of heterogeneous interactions. Methods Enzymol 384 : 212–232.
71. ShimomayeE, SalvatoM (1989) Use of avian myeloblastosis virus reverse transcriptase at high temperature for sequence analysis of highly structured RNA. Gene Anal Tech 6 : 25–28.
72. LoreauN, BoiziauC, VerspierenP, ShireD, ToulmeJJ (1990) Blockage of AMV reverse transcriptase by antisense oligodeoxynucleotides. FEBS Lett 274 : 53–56.
73. CrooksGE, HonG, ChandoniaJM, BrennerSE (2004) WebLogo: a sequence logo generator. Genome Res 14 : 1188–1190.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



