-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
The development of a living organism is influenced by the environmental conditions such as nutrient availability. Under starvation conditions, the C. elegans larvae will enter a special developmental stage called dauer larva. An insulin-like signaling pathway controls dauer formation as well as adult lifespan by inhibiting the activity of FOXO transcription factor DAF-16 that regulates expression of stress-resistant genes. Here we isolate a new gene called daf-31; this gene encodes a protein that regulates C. elegans larval development, metabolism and adult lifespan. This protein has been found in other species to be part of an enzyme that functions to modify other proteins. We show that overexpression of our newly discovered protein stimulates the transcriptional activity of DAF-16. Interestingly, abnormal regulation of human proteins similar to DAF-31 results in tumor formation. It is known that human FOXO proteins prevent tumorigenesis. Therefore, it is possible that abnormal DAF-31 activity may lead to tumor growth by reducing DAF-16 activity. Thus, the present study may not only contribute to understanding the role of a universal enzyme in controlling development, metabolism and lifespan in other organisms besides worms but may also shed light on the mechanisms of tumorigenesis in humans.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004699
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004699Summary
The development of a living organism is influenced by the environmental conditions such as nutrient availability. Under starvation conditions, the C. elegans larvae will enter a special developmental stage called dauer larva. An insulin-like signaling pathway controls dauer formation as well as adult lifespan by inhibiting the activity of FOXO transcription factor DAF-16 that regulates expression of stress-resistant genes. Here we isolate a new gene called daf-31; this gene encodes a protein that regulates C. elegans larval development, metabolism and adult lifespan. This protein has been found in other species to be part of an enzyme that functions to modify other proteins. We show that overexpression of our newly discovered protein stimulates the transcriptional activity of DAF-16. Interestingly, abnormal regulation of human proteins similar to DAF-31 results in tumor formation. It is known that human FOXO proteins prevent tumorigenesis. Therefore, it is possible that abnormal DAF-31 activity may lead to tumor growth by reducing DAF-16 activity. Thus, the present study may not only contribute to understanding the role of a universal enzyme in controlling development, metabolism and lifespan in other organisms besides worms but may also shed light on the mechanisms of tumorigenesis in humans.
Introduction
Animal development is a complex process that involves hierarchical gene regulatory networks and is influenced by environmental conditions. When food is abundant, the post-embryonic development of C. elegans consists of four larval stages (L1–L4) and the adult. During the L1 stage, environmental factors determine whether C. elegans molts to an L2 larva or a pre-dauer L2d larva [1]. At least three environmental cues have been defined: food supply, temperature, and a constitutively secreted dauer-inducing pheromone that signals population density [2]. The L2 larva is developmentally committed to continued growth, whereas the L2d larva can molt to a dauer larva if food is scarce and the animals are overcrowded, or to an L3 larva should conditions improve.
Mutations affecting dauer larval development include dauer-defective (daf-d) mutations that prevent entry into the dauer stage, and dauer-constitutive (daf-c) mutations that mandate entry into the dauer stage [2]. Based on epistatic relationships between daf-c and daf-d mutations, more than twenty genes controlling dauer formation have been ordered in a genetic pathway [2] representing generation of the pheromone signal [3], response by chemosensory neurons [4], [5] and transduction of the signal to other cells. Three functionally overlapping neural pathways control the developmental response to environmental cues. They involve DAF-7/TGF-ß [6], [7], DAF-11/cyclic GMP [8], and DAF-2/insulin-like [9], [10] pathways, which relay the environmental signals to a nuclear hormone receptor, DAF-12 [11], to control dauer versus non-dauer morphogenesis.
Mutations in two genes, daf-9 and daf-15, lead to non-conditional formation of detergent-sensitive dauer-like larvae [12]. These mutants form dauer larvae constitutively and display some characteristics of dauer larvae formed under starvation, such as a high density of intestinal and hypodermal storage granules. daf-9 encodes a cytochrome P450 related to those involved in the biosynthesis of steroid hormones in mammals [13], [14]; it was found to specify a step in the biosynthetic pathway for a DAF-12 steroid ligand called dafachronic acid [15]–[17]. daf-15 encodes the C. elegans ortholog of Raptor [18] that is proposed to interact with C. elegans target-of-rapamycin kinase (LET-363/CeTOR) to control C. elegans larval development [18]. Both daf-9 and daf-15 also regulate fat metabolism and adult lifespan [13], [14], [18].
The dauer-like mutants represent a mutant class distinct from the previously defined daf-c and daf-d mutants. Unlike most daf-c mutants, the dauer-like mutants are not ts, and they do not complete dauer morphogenesis. The daf-d genes such as daf-12 have non-conditional alleles and fail to respond to pheromone [1], but unlike the dauer-like mutants they can execute non-dauer development. The dauer-like mutants define a third class of mutants, one in which the animals are incapable of executing either complete dauer or non-dauer development.
The daf-31 mutant was isolated in genetic screens to identify genes similar to daf-9 and daf-15 [17]. The overall aim of the present study was to clone the daf-31 gene and characterize the DAF-31 function. Our genetic epistasis analysis suggests daf-31 functions downstream of or in parallel to daf-3, daf-12 and daf-16 dauer-defective genes, and acts upstream of or in parallel to daf-15/raptor. We cloned the daf-31 gene by positional cloning and showed that it encodes an ortholog of arrest-defective-1 protein (ARD1), the catalytic subunit of the major N alpha-acetyltransferase (NatA). Moreover, our data reveal that daf-31 has an essential role in controlling C. elegans larval development, metabolism and adult longevity.
Results
daf-31 mutations cause developmental larval arrest
Entry into the dauer stage is determined by the pheromone/food ratio, with high pheromone and low food supply favoring dauer formation [2]. Dauer larva is considered as an alternative L3 larval stage. Compared to L3 larva (Figure 1A), the dauer larva has a constricted pharynx (Figure 1B) and a special cuticle with dauer alae (Figure 1C). In the presence of dauer-inducing pheromones, daf-31 mutants cannot form SDS-resistant dauer larvae [17]. In order to determine whether daf-31 mutants enter the dauer stage in response to starvation, we examined the progeny of strain unc-24daf-31/nT1 under starvation conditions and observed uncoordinated (Unc) dauer larvae. These dauer larvae showed normal dauer features, such as a dark body, fully constricted pharynx (Figure 1D), and a cuticle with dauer alae (Figure 1E). However, daf-31 dauer larvae were not SDS-resistant like normal dauer larvae. Furthermore, daf-31 dauer larvae could not resume development when food was provided, dying shortly thereafter. Therefore, daf-31 mutants could not complete dauer morphogenesis under starved conditions, and those incomplete dauer larvae could not finish reproductive development after food was provided.
Fig. 1. Characteristics of daf-31 mutant dauer larvae. 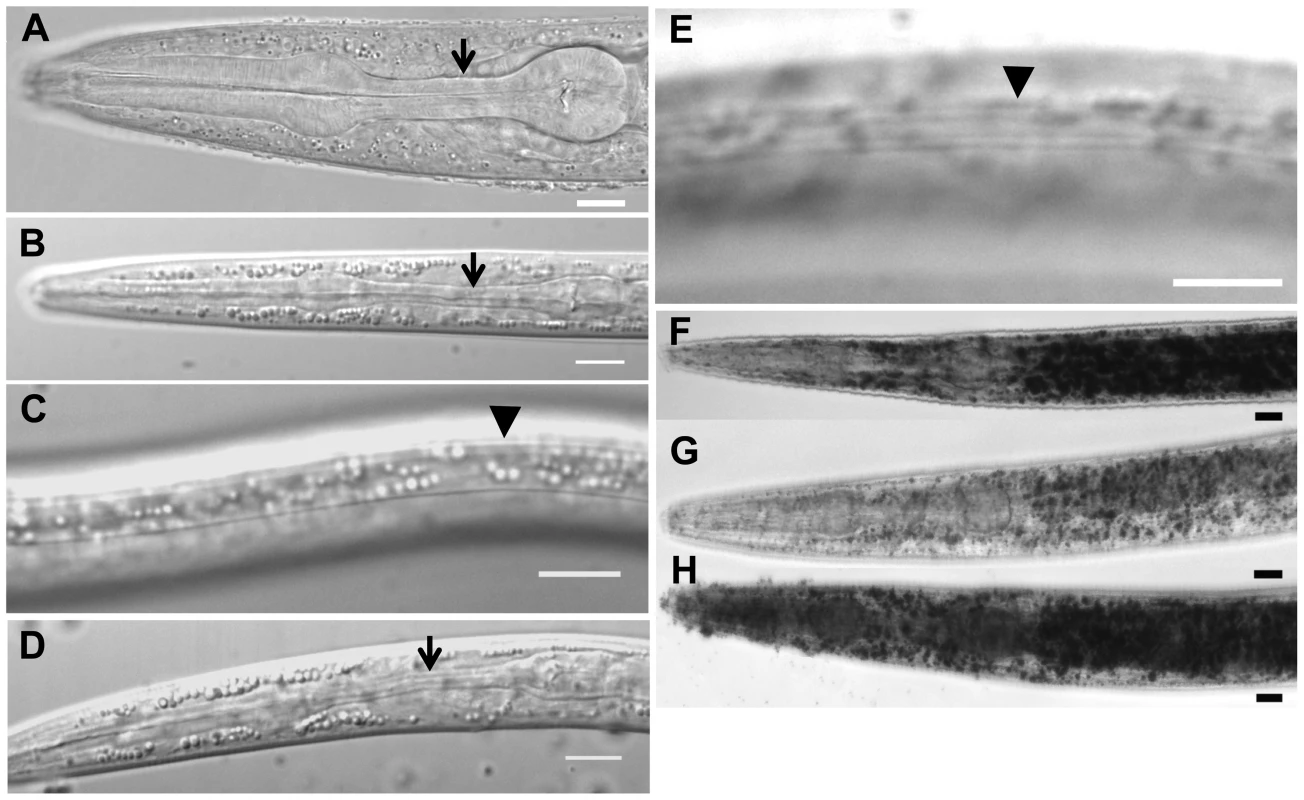
(A) N2 L3 larva pharynx. (B) N2 dauer larva with fully constricted pharynx. (C) N2 dauer larva with dauer alae along the lateral surface of the cuticle (body beneath focal plane). (D) unc-24(e138)daf-31(m655) dauer larva with fully constricted pharynx. (E) unc-24(e138)daf-31(m655) dauer larva with dauer alae (body beneath focal plane). Arrows indicate isthmus of pharynx in panels A, B and D. Arrowheads indicate dauer alae in panels C and E. N2 and unc-24daf-31/nT1 animals were grown on NG agar plates at 20°C. Unc dauer larvae (unc-24daf-31) were identified after animals were starved. (F–H) Representative pictures showing fat accumulation detected by Sudan Black B in daf-2 mutants (F), N2 (G) and daf-31 mutants (H). N2, daf-2(e1370) and daf-31(m655)IV/nT1[unc-?(n754) let-?](IV;V) synchronized L1 larvae were placed on NG agar plates, incubated at 20°C until they entered L3 or dauer-like stages, then collected for staining. Scale bars: 10 µm. Fat accumulation is one characteristic of C. elegans dauer larvae. We examined fat accumulation in daf-31 homozygous mutants using Sudan Black B staining. As shown in Figure 1F, daf-2 mutant dauer larvae accumulate fat as described previously [9]. The daf-31 mutant worms also accumulate more fat droplets than wild-type worms and fat droplets in the daf-31 mutants are larger than those in wild-type worms (Figure 1G and 1H). To confirm this phenotype, Nile red was used to stain fixed worms; this approach has been reported to reliably detect fat droplets in C. elegans [19]. Similar to Sudan Black staining, Nile red also detected fat accumulation in daf-31 mutant worms (Figure S1). Therefore, daf-31 mutants shift metabolism to fat accumulation.
daf-31 is epistatic to daf-d genes but acts upstream of or in parallel to daf-15/raptor
To position daf-31 in the dauer formation pathway, we examined the epistatic relationship between daf-31 and daf-d genes including daf-3, daf-12 and daf-16. The daf-31 mutation is epistatic to all three daf-d mutations as judged by the ratio of progeny (1∶2∶1) (Table 1). For the epistasis analysis with daf-16, the ratio of progeny is 1∶2 because nT1 homozygous animals are lethal. We repeated the epistasis analysis of daf-31 and daf-12 by using the daf-12(rh61rh411) null allele [11] and obtained a similar result (Table 1). These epistatic relationships suggest that daf-31 functions downstream of or in parallel to daf-3, daf-12 and daf-16 in dauer formation.
Tab. 1. Epistatic tests between daf-31 and daf-d mutations for dauer formation. 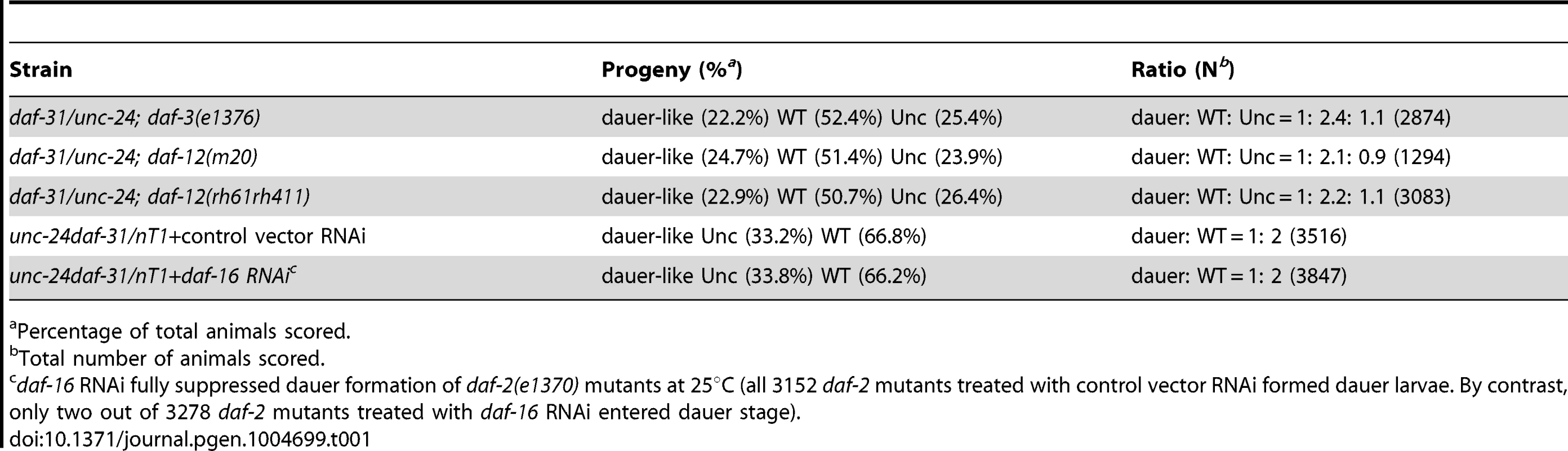
Percentage of total animals scored. To examine the epistatic relationship of daf-31 and daf-15, we injected dsRNA of daf-15 into unc-24daf-31/nT1 young adult worms. Wild-type (N2) animals were treated equally and used as controls. We examined the phenotype of progeny reproduced at various time periods after injection. The progeny reproduced between seven and eighteen hours after injection arrested development at a dauer-like stage three days after egg lay (Table 2). These dauer-like animals have a similar phenotype to daf-15 mutants. Thus, regarding dauer entry, it appears that the daf-15 mutation is epistatic to the daf-31 mutation. We scored the recovery of both N2 and unc-24daf-31 dauer-like animals two days after dauer-like arrest. 48% of N2 dauer-like worms remained at dauer-like stage and the rest of the animals recovered and grew to L4 larval or adult size (Table 2). By contrast, 100% of unc-24daf-31 animals stayed at dauer-like stage (Table 2). For unc-24daf-31 dauer-like larvae without daf-15 RNAi treatment, the majority of these animals died within five days. However, surviving animals all grew to L4 larval or adult size (n = 52). Taken together, these data indicate that daf-15 is epistatic to daf-31 as unc-24daf-31 mutants treated by daf-15 RNAi form dauer-like larvae similar to daf-15 mutants. Moreover, these two mutants have a synergistic effect on C. elegans development because no unc-24daf-31 dauer-like larvae treated by daf-15 RNAi recovered. Thus, these two genes may function in the same pathway and daf-31 is upstream of daf-15. However, the possibility that these two genes act in parallel cannot be excluded.
Tab. 2. Epistatic test between daf-31 and daf-15 mutations for dauer recovery. 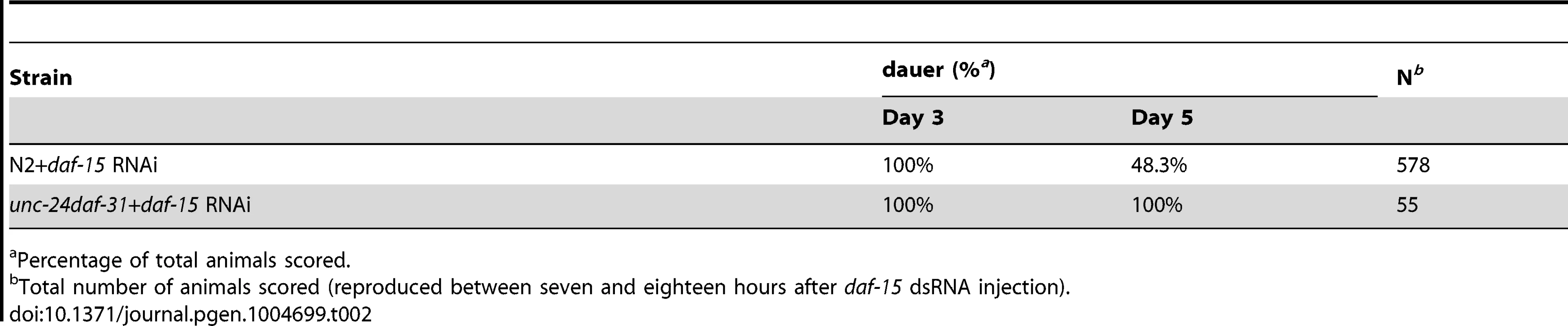
Percentage of total animals scored. daf-31 encodes an ortholog of ARD1
A positional cloning strategy was used to identify the daf-31 gene on chromosome IV between unc-24 and fem-3 (Figure S2A). daf-31 was found to lie between the physical SNP markers T09A12 and F17E9 (Figure S2A). A genomic fragment corresponding to the K07H8.3 open reading frame fully rescued the genetic daf-31 null mutant [daf-31(m655)] phenotype, i.e. the transgenic animals did not form dauer-like larvae, but grew to fertile adults. Sequence analysis of the daf-31 gene in the mutant revealed a 393 bp deletion which removed 151 bp of promoter upstream of the ATG start codon and 242 bp of daf-31 coding region downstream of the ATG start codon, which may completely block daf-31 transcription as both the essential promoter region and the N-terminal portion of the gene were deleted (Figure S2B). Primers were designed to flank the deletion region and PCR analysis of mutant worms' genomic DNA detected a 1,449 bp band (393 bp smaller than the wild-type band) in both homozygous and heterozygous daf-31(m655) mutant worms (Figure S2C).
The daf-31 gene encodes the ortholog of ARD1 with a predicted molecular weight of 21.2 kDa. ARD1 is the catalytic subunit of NatA that catalyzes the acetylation of proteins beginning with Met-Ser, Met-Gly and Met-Ala [20]. Amino acid sequence alignment showed that DAF-31 shares 75% identity with human ARD1, 77% identity with mouse ARD1, 72% identity with Drosophila melanogaster ARD1 and 46% identity with yeast ARD1 (Figure S2D).
Given that there is only a single mutant allele of daf-31, we used RNAi to inhibit daf-31 in the N2 background to confirm the daf-31 mutant phenotype. Inhibition of daf-31 by feeding animals with E. coli that express daf-31 dsRNA did not induce a dauer-like phenotype. From our previous work, dsRNA injection can create a stronger mutant phenotype similar to that of a genetic null mutant [18]. In vitro synthesized daf-31 dsRNA was injected into gonads of N2 young adult worms and the progeny displayed a dauer-like phenotype similar to the daf-31(m655) mutants. The starvation-induced dauer morphology of daf-31(m655) mutants, such as dauer alae formation and contrasted pharynx (described in Figure 1) could not be examined using this RNAi method. Therefore, we examined fat accumulation in daf-31 RNAi-treated animals. Similar to the daf-31(m655) mutant, daf-31 RNAi-treated animals accumulated fat as detected by both Sudan Black and Nile red staining of fixed animals (Figure S3). Based on these results, we conclude that the daf-31 mutant phenotypes described in this study most likely resulted from daf-31 mutation instead of secondary mutations in the background.
daf-31 is expressed in multiple tissues
In order to characterize the daf-31 expression pattern, we constructed a daf-31 promoter::gfp reporter construct. In N2 animals, GFP expression was detected from L1 to the adult stages in multiple tissues including the hypodermis, pharynx, intestine, and neurons (Figure 2). To confirm the GFP expression pattern of daf-31 promoter fusion reporter, we constructed daf-31 translational fusion reporter genes in which the GFP open reading frame was fused to the full-length daf-31 genomic DNA in frame either at the N-terminus or at the C-terminus. Both translation fusion reporter genes fully rescued the dauer-like phenotypes of daf-31 mutants. However, we did not observe GFP expression in the rescued daf-31 mutant worms, a phenomenon previously reported with other GFP fusion gene mutant rescues [21]. Thus, our observations of daf-31 expression pattern were limited to the daf-31 promoter fusion, which may not represent the endogenous expression pattern of the entire daf-31 gene if enhancer elements are present in introns or in 3′ untranslated sequences.
Fig. 2. daf-31 expression pattern in wild-type N2 animals. 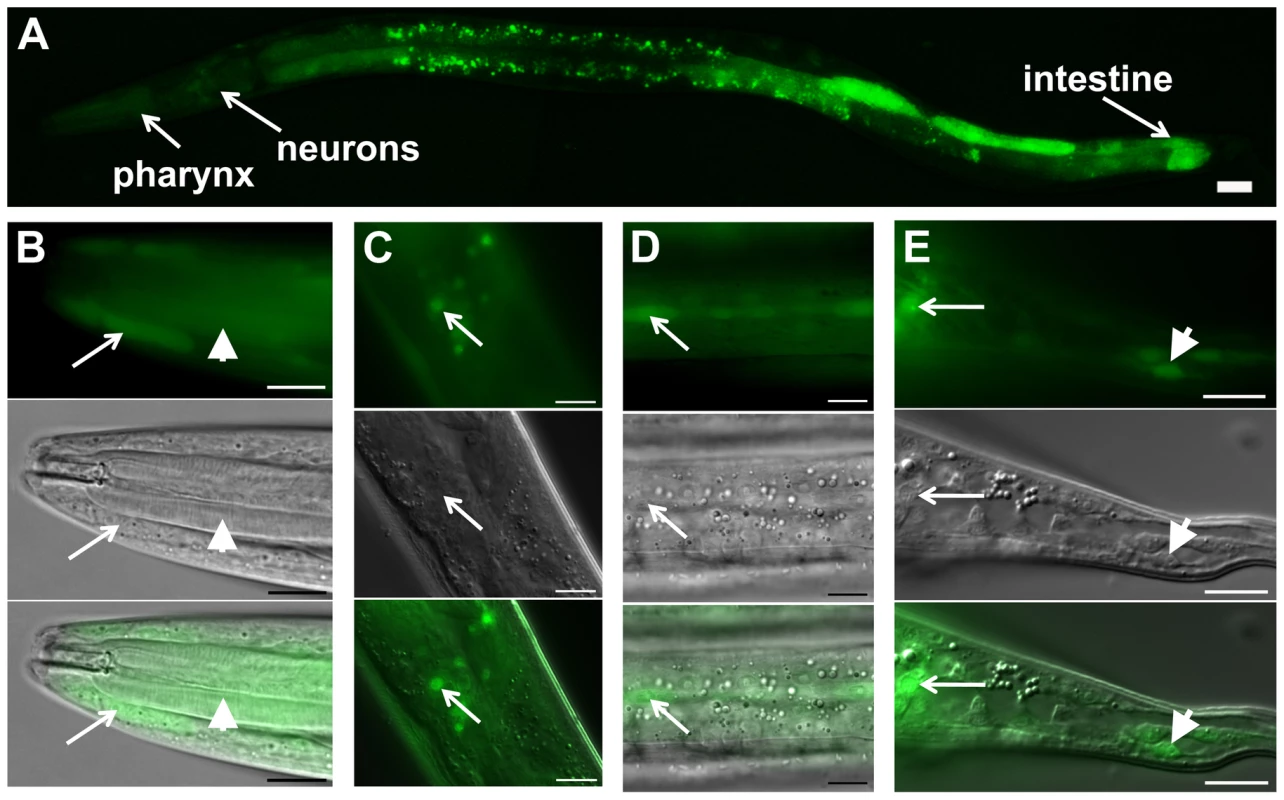
GFP expression is under the control of the same 760 bp daf-31 promoter that successfully drove daf-31 gene expression and rescued the dauer-like larval arrest phenotype of daf-31 mutants. (A) daf-31 expression in multiple tissues including pharynx, hypodermis, neurons and intestine. High magnification pictures showing the expression of daf-31 in pharynx (B, indicated by arrowheads), head hypodermal cells (B, indicated by arrows), head neurons (C), hypodermal seam cells (D), tail neurons (E, indicated by arrows) and tail hypodermal cells (E, indicated by arrowheads). Photos in B though E: the upper panels show the GFP signal, the middle panels show the same animals in the same focal planes under Nomarski optics, and the bottom panels show the merged images from the upper and middle panels. For all pictures, the left is anterior and the right is posterior. Scale Bars: 10 µM. daf-31 influences C. elegans lifespan
Increased adult longevity is a phenotype associated with many dauer mutants. Since daf-31 homozygous mutants arrest development at L4 stage, we inhibited the daf-31 gene by feeding RNAi. The RNAi treatment successfully reduced daf-31 mRNA level as measured by qRT-PCR (Figure S4). However, daf-31 RNAi treatment had no obvious effect on the lifespan of wild-type worms as the daf-31 RNAi-treated worms had similar mean and maximum lifespans as control vector RNAi-treated worms (Figure 3A and Table S1). When RNAi-sensitive rrf-3(pk1426) mutants were treated with daf-31 RNAi, their lifespans were significantly decreased (p<0.0001, log-rank test) (Figure 3B and Table S1). Compared to controls, the mean lifespan of rrf-3 mutants treated with daf-31 RNAi was shortened by four days (Figure 3B and Table S1).
Fig. 3. Influence of daf-31 on C. elegans lifespan. 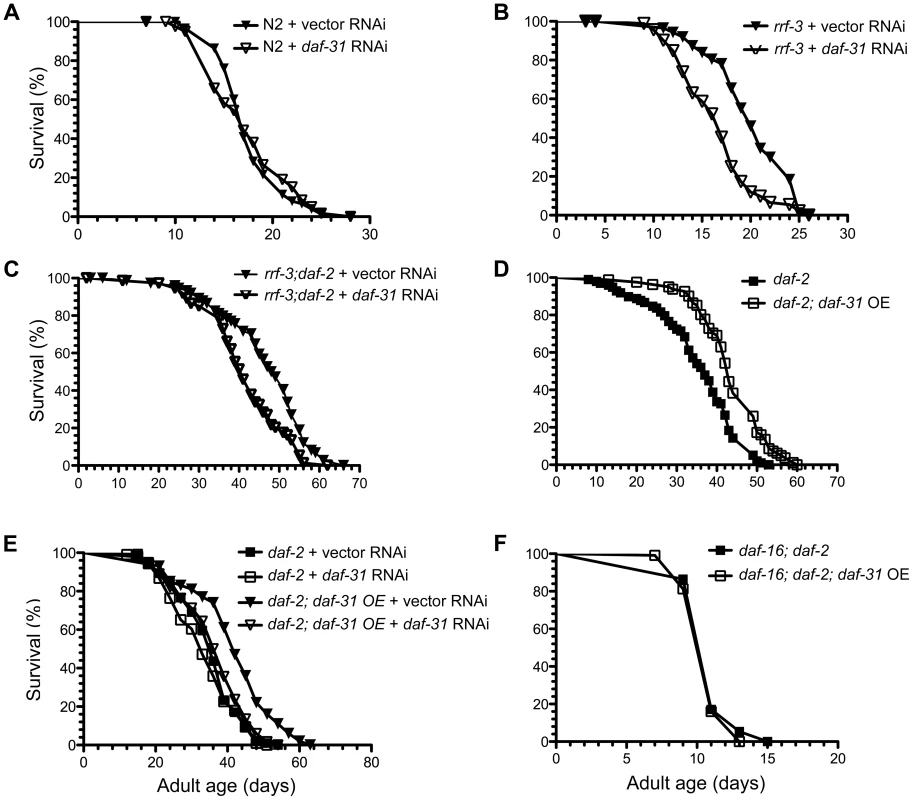
(A) N2 animals treated with daf-31 RNAi have a similar lifespan to those treated with the vector RNAi control. daf-31 RNAi significantly decreases the lifespan of RNAi-sensitive rrf-3 mutants (B) and RNAi-sensitive daf-2 mutants (rrf-3;daf-2) (C). (D) daf-31 overexpression enhances the longevity phenotype of daf-2 mutants. (E) daf-31 RNAi abrogates the effect of daf-31 overexpression on the daf-2 mutant lifespan. (F) daf-16 mutations block the further lifespan extension of daf-2 mutants conferred by daf-31 overexpression. To test if daf-31 mutations influence the longevity phenotype of daf-2 mutants, we treated the rrf-3(pk1426);daf-2(e1370) mutant with daf-31 RNAi. The mean lifespan of daf-31 RNAi-treated rrf-3;daf-2 mutants was five days shorter compared to that of control animals (p = 0.0005, log-rank test) (Figure 3C and Table S1). Thus, inhibition of daf-31 partially suppressed the longevity phenotype of daf-2 mutants. Based on this result, we postulate that overexpression of daf-31 may further increase the lifespan of daf-2 mutants.
To test this, daf-2(e1370) and daf-16(mgDf47);daf-2(e1370) mutants overexpressing daf-31 were constructed. The overexpression of daf-31 was confirmed by qRT-PCR (Figure S4). As shown in Figure 3D, daf-31 overexpression increased the lifespan of daf-2 mutant worms (p<0.0001, log-rank test) (Figure 3D and Table S1). The mean lifespan was increased by eight days and the maximum lifespan was extended by seven days (Figure 3D and Table S1). This increased lifespan was due to daf-31 overexpression as daf-31 RNAi treatment completely abrogated it (Figure 3E and Table S1). Interestingly, daf-31 overexpression failed to extend the lifespan of daf-16;daf-2 double mutants (Figure 3F and Table S1), indicating that DAF-16 is required for daf-31 overexpression to enhance the daf-2 longevity phenotype. We also measured the lifespan of N2 animals overexpressing the daf-31 gene. As shown in Figure S5 and Table S1, daf-31 overexpression did not extend the lifespan of N2 worms. In fact, it slightly decreased the lifespan of N2 worms. Finally, to confirm daf-31 functions through daf-16 in C. elegans lifespan regulation, we tested if daf-31 RNAi can further decrease the lifespan of RNAi-sensitive daf-16 mutants (daf-16;rrf-3). We found daf-31 RNAi had no obvious effect on the lifespan of daf-16;rrf-3 mutants (Figure S6 and Table S1).
daf-31 overexpression stimulates daf-16 transcriptional activity
The forkhead transcription factor DAF-16/FOXO controls the transcription of an array of genes essential for lifespan extension and oxidative stress resistance including the antioxidant enzyme superoxide dismutase (sod)-3 gene and beta-carotene 15,15′-monooxygenase gene (bcmo-2) [22]. We used qRT-PCR to measure the expression level of sod-3 and bcmo-2 in daf-31 overexpression strains and control animals. As shown in Figure 4A, the expression level of sod-3 was significantly increased when daf-31 was overexpressed in the N2 background and in daf-2 mutants. Similar to sod-3, the expression of bcmo-2 is also significantly increased when daf-31 is overexpressed in the daf-2 mutant background (Figure 4B). Taken together, these data indicate overexpression of daf-31 stimulates the transcriptional activity of DAF-16.
Fig. 4. daf-31 overexpression stimulates the transcriptional activity of DAF-16 without influencing the subcellular localization of DAF-16. 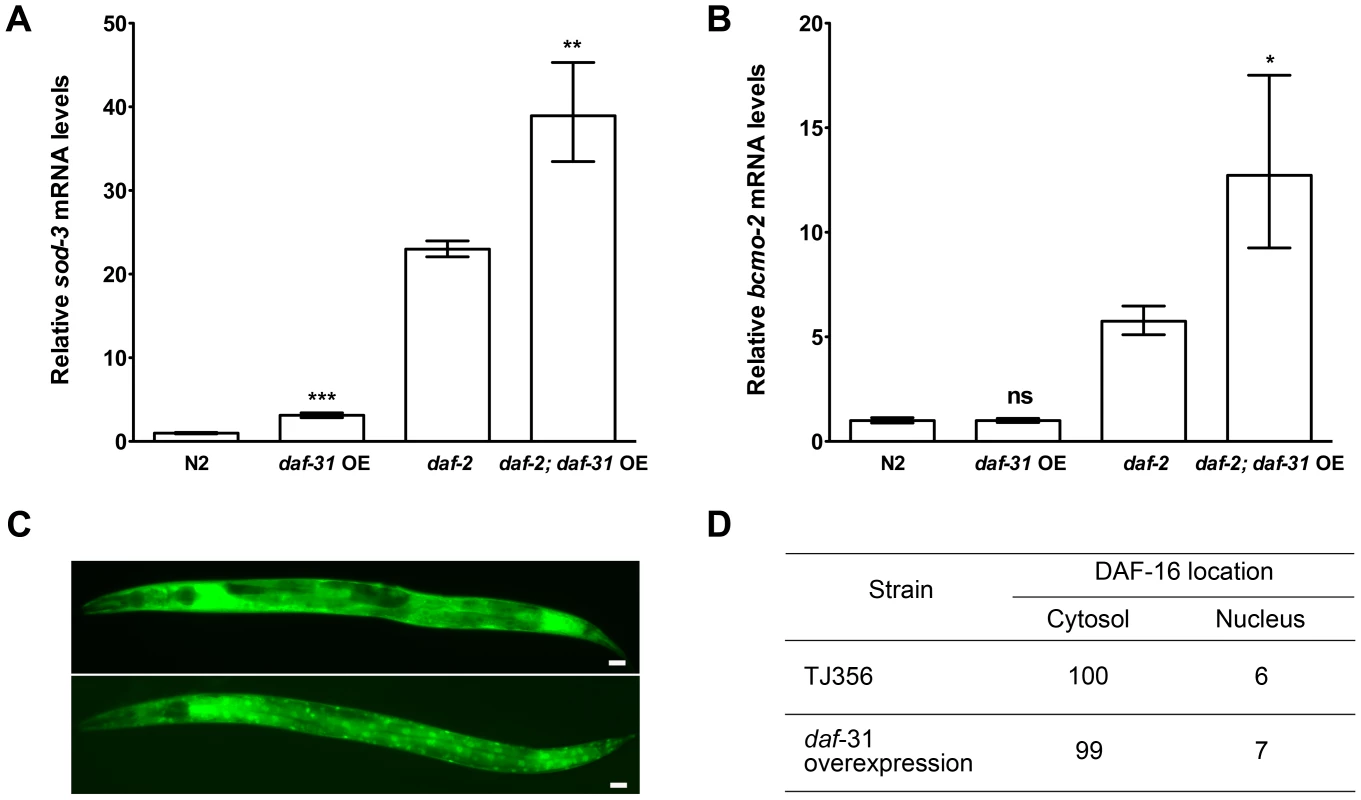
qRT-PCR was performed to measure the mRNA expression level of two DAF-16 target genes, sod-3 and bcmo-2, in indicated strains. Y-axis stands for relative mRNA levels. Daf-31 overexpression up-regulates the expression of sod-3 mRNA in both N2 worms and daf-2 mutants (A), and up-regulates the expression of bcmo-2 mRNA in daf-2 mutants (B) **, P<0.01, ***, P<0.001 (t-test). (C) Representative pictures showing the cytosolic localization of DAF-16 (upper panel) and nuclear localization of DAF-16 (lower panel). Scale Bars: 10 µM. (D) The percentage of worms showing the nuclear localization of DAF-16 in wild-type background (TJ356) is similar to that in daf-31 overexpression animals. Reduction of daf-2 insulin-like signaling activity increases C. elegans lifespan by promoting nuclear localization of DAF-16 [23], [24]. We crossed the DAF-16::GFP reporter gene into daf-31 overexpressing animals to examine if daf-31 overexpression influences the subcellular localization of DAF-16. We found the percentage of animals showing DAF-16 nuclear localization was not significantly different between daf-31 overexpressing animals and control worms (Figure 4C and D).
daf-31 overexpression does not confer stress resistance
daf-2 mutants are resistant to environmental stresses such as high temperature [25], [26]. We examined if daf-31 overexpression could enhance the thermotolerance of daf-2 mutants. As reported previously [25], the survival rate of daf-2 mutants at 35° is doubled compared to N2 worms (Figure S7 and Table S2). However, the mean survival for N2 and N2 overexpressing daf-31 was similar (9.8 hours vs. 10 hours) (p = 0.2420, log-rank test) (Figure S7 and Table S2). Similarly, daf-31 overexpression did not increase the survival of daf-2 mutants at 35° (p = 0.4623, log-rank test) (Figure S7 and Table S2).
daf-31 overexpression enhances reproduction
We tested the influence of daf-31 overexpression on the reproduction of N2 and daf-2 mutant animals. daf-31 overexpression increased the brood size of N2 animals and daf-2 mutants by about 23% and 30%, respectively (Figure S8). While daf-31 overexpression increased the daf-2 mutant lifespan in a daf-16 dependent manner, daf-31 overexpression increased the reproduction of daf-2 mutants significantly in a daf-16 independent way. Overexpression of daf-31 increased the brood size of daf-16;daf-2 mutants by 40% (P<0.01, t-test) (Figure S8).
Discussion
We demonstrated that daf-31 mutants form dauer-like larvae that share some characteristics of wild-type dauer larvae such as fat accumulation. Many daf genes, especially those from the insulin-like signaling pathway, are involved in the regulation of lifespan [27]. Mutations in daf-2, which encodes an insulin/IGF-1 receptor [9], convey a temperature-sensitive Daf-c phenotype, and the adults live twice as long as wild-type animals [9], [28], [29]. Mutations in some genes downstream of daf-2, such as age-1 and pdk-1, also extend lifespan [30], [31]. Conversely, mutations in other downstream genes, including daf-18 and daf-16, shorten lifespan [32]–[34].
We examined whether daf-31 is also involved in aging and found that daf-31 partially mediates the effect of reduced daf-2/IGF signaling pathway on C. elegans lifespan. Moreover, overexpression of daf-31 enhances the longevity phenotype of daf-2 mutants depending on the activity of DAF-16. Supporting this lifespan data, the expression levels of sod-3 and bcmo-2, the transcriptional targets of the DAF-16 FOXO3 transcription factor, are up-regulated in the daf-31 overexpression strains. Thus, it is reasonable to argue that DAF-31 regulates C. elegans lifespan by influencing DAF-16 transcriptional activity and daf-31 overexpression stimulates DAF-16 activity. Indeed both DAF-31 and DAF-16 are expressed in neurons and intestine, two major tissues essential for regulation of C. elegans lifespan by the DAF-2/IGF signaling pathway [35]–[37]. However, overexpression of daf-31 has no influence on the subcellular localization of DAF-16. It is consistent with the lifespan data that overexpression of daf-31 does not increase the lifespan of N2 animals. Thus, overexpression of DAF-31 only extends C. elegans lifespan in the daf-2 mutant background in which DAF-16 has entered the nucleus due to inhibition of the IGF signaling.
Previous studies show that the stress-resistance phenotype can be uncoupled from the longevity phenotype [38]. Indeed, although daf-31 overexpression further increases the long-lived lifespan of daf-2 mutants, it has no effect on the thermotolerance of daf-2 mutants. Interestingly, daf-31 overexpression increases the reproduction of both wild-type animals and daf-2 mutants, which is not dependent on DAF-16, suggesting DAF-31 functions through DAF-16 for lifespan regulation but not for reproduction. Since DAF-16 is required for the stress resistance of daf-2 mutants, it is likely that daf-31 overexpression extends the daf-2 mutant lifespan through DAF-16-dependent mechanisms other than increasing stress-resistance. DAF-31 is found in multiple tissues including neurons. It is known that many C. elegans neurons are refractory to RNAi treatment in wild-type background [39]. It is possible that neuronal DAF-31 activity is more important for lifespan regulation because daf-31 RNAi treatment only shows influence on lifespan of RNAi-sensitive mutants. Supporting this assumption, it has been reported that daf-16/FOXO activity in neurons accounted for only 5–20% of the lifespan extension seen in daf-2 mutants [37]. Since DAF-31 may only influence DAF-16 activity in neurons, its overexpression only increases the daf-2 mutant lifespan modestly.
We cloned the daf-31 gene and sequence analysis indicates DAF-31 is a worm ortholog of ARD1 that was first identified in yeast [40]. ARD1 is the catalytic subunit of the major NatA that transfers an acetyl group from acetyl coenzyme A to the N-terminal of nascent polypeptides. Yeast ARD1 mutants fail to enter stationary phase and sporulate during nitrogen deprivation [40]. The yeast stationary phase is comparable to C. elegans dauer stage and is essential for survival when nutrients are limited. C. elegans enters dauer stage during starvation or under high concentration of pheromone. Our data show daf-31 mutants could not complete dauer morphogenesis in response to pheromone and starvation, which indicates daf-31 is required for dauer formation. Thus, both yeast ARD1 and worm DAF-31 play an important role in the developmental switch in response to the environmental nutrient limitation. Additionally, daf-31 mutants could not grow to fertile adults in an environment with abundant food suggesting its essential role in normal development. Similar to our observation, it has been reported that loss of Ard1 is lethal for D. melanogaster and affects cell survival or proliferation, indicating ARD1 is required for D. melanogaster development [41]. In addition to developmental arrest, the daf-31 mutants shift metabolism to fat accumulation. Interestingly, yeast ARD1 mutants not only fail to enter stationary phase but also do not accumulate as much carbohydrates as wild-type yeast strains [40]. Thus, the function of ARD1 in regulating development and metabolism appears conserved from yeast to C. elegans.
N-terminal acetylation is one of the most common posttranslational protein modifications. It is estimated to occur on 50% of yeast proteins [20], 71% of D. melanogaster cytosolic proteins [20] and 84% of human proteins [42]. NatA plays the most prominent role in N-terminal acetylation. It would be interesting to know whether the pleiotropic phenotypes of ard1 mutants result from global changes of protein N-acetylation or from acetylation status of specific protein substrates. It has been reported that human ARD1 directly acetylates β-catenin and enhances its transcriptional activity [43]. We show that overexpression of DAF-31 stimulates the transcriptional activity of DAF-16. It would be interesting to examine if DAF-31 overexpression acetylates DAF-16. Alternatively, a suppressor screening of daf-31 mutants may help to identify the essential substrates of the DAF-31 acetyltransferase and contribute to understanding the mechanisms by which ARD1 influences development, metabolism and aging. Moreover, emerging evidence has revealed that abnormal regulation of ARD1 is associated with tumorigenesis and ARD1 represents a novel cancer drug target [44], [45]. Identification of DAF-31 substrate proteins may uncover new therapeutic targets of cancer diseases.
Materials and Methods
Culture conditions and C. elegans strains
All strains were grown on NG agar plates seeded with E. coli strain OP50 [46]. Mutations are listed by linkage groups as follows: LG I: daf-16(mgDf47); LG II: rrf-3(pk1426); LG III: daf-2(e1370); LG IV: unc-24(e138), daf-31(m655); LG X: daf-3(e1376), daf-12(m20), daf-12(rh61rh411). All mutants are derived from the wild-type Bristol N2 strain.
To make the daf-16(mgDf47)I; daf-2(e1370)III double mutant, daf-2(e1370) males were mated with daf-16(mgDf47) hermaphrodites. Ten F1 adults (daf-16/+; daf-2/+) were incubated at 25°C. F2 dauer larvae (either +/+; daf-2 or daf-16/+; daf-2) were transferred to a fresh plate at 15°C for recovery. Then adults were shifted to 25°C. Since daf-16(mgDf47) can suppress the daf-2(e1370) Daf-c phenotype, non-dauer adults from the next generation were daf-16(mgDf47); daf-2(e1370) double mutants.
Double mutants were constructed for epistatic tests between daf-31 and daf-d mutants at 20°. However, daf-16 RNAi was used for epistasis analysis between daf-31 and daf-16. daf-16 RNAi fully suppressed dauer formation of daf-2(e1370) mutants. daf-12(m20) and daf-12(rh61rh411) mutations were used to construct the strain+daf-31(m655)/unc-24(e138)+; daf-12(m20) and +daf-31(m655)/unc-24(e138)+; daf-12(rh61rh411) using standard genetic methods. The daf-12(rh61rh411) mutation was confirmed by sequencing. +daf-31(m655)/unc-24(e138)+; daf-3(e1376) was constructed to determine the epistatic relationship between daf-31 and daf-3. As the daf-31(m655) mutation was not marked by a genetic mutation in daf-31;daf-3 and daf-31;daf-12 mutant worms, the daf-31(m655) deletion mutations were confirmed by using single worm PCR. Representative gel pictures are shown in Figure S9. Injection of RNAi was used to inhibit the daf-15 gene in unc-24(e138)daf-31(m655)/nT1 to examine the epistatic relationship between daf-15 and daf-31.
To construct the daf-31 overexpressing strains, the full-length daf-31 genomic DNA, including its native 760-bp promoter and 3′-UTR was cloned into pGEM-T (Promega); this construct successfully rescued the daf-31 dauer-like mutant phenotype. Multiple copies of the construct were integrated into chromosomal DNA by γ-irradiation to make an N2 transgenic line overexpressing daf-31. Then the daf-31 overexpressing chromosome was introduced into both daf-2(e1370) and daf-16(mgDf47);daf-2(e1370) mutants by genetic crosses.
To make a daf-31 promoter-GFP transcriptional fusion, the 760-bp daf-31 promoter was inserted into the GFP vector pPD95.70 (a gift from Dr. Andrew Fire at Stanford University) between the PstI and BamHI sites. The construct was injected into N2 adults at a concentration of 100 µg/ml. pRF4, which encodes a mutant collagen and induces a dominant roller (Rol) phenotype, was co-injected at the same concentration as a transformation marker. Rol animals were selected from the F2 generation and used to establish stable transgenic lines.
To evaluate the subcellular location of DAF-16, TJ356 (zIs356 [daf-16p::daf-16a/b::GFP+rol-6]) males were crossed to daf-31 overexpressing hermaphrodites. The roller progeny were mounted on 2% agar pads to examine DAF-16::GFP subcellular localization.
Fat staining
To stain fat using Sudan Black B, N2, daf-2(e1370) and daf-31(m655)IV/nT1[unc-?(n754) let-?](IV;V) synchronized L1 larvae were placed on NG agar plates, incubated at 20°C until they entered L3 or L4 stages, collected and washed two to three times with M9 buffer. Paraformaldehyde stock solution (10%) was added to a final concentration of 1%. The samples were frozen in dry ice/ethanol and then thawed under a stream of warm water. After a total of three freeze-thaw cycles, the worms were stained with Sudan Black B as described by Kimura et al. [9].
Nile red staining of fixed worms was performed as described by Pino et al. [47]. Worm samples were collected and washed twice with M9 buffer. After the final wash, worms were fixed in 40% isopropanol at room temperature for three minutes. The fixed worms were stained in Nile red/isopropanol solution for 30 minutes at room temperature with gentle rocking. The stained worms were washed once with 1 ml M9 buffer and mounted on a 2% agarose pad for microscopy under the fluorescence channel. In order to compare the fat content in different strains, the pictures were taken at the same camera setting under 20× magnification.
daf-31 cloning
Three-factor-mapping with SNP markers and cosmid rescue were performed as previously described [18]. To determine the mutation in daf-31(m655), the K07H8.3 gene (GenBank accession # NM_068991.4) was amplified using primers 5′ - GTG AGT CGA AAC CCA TTT TG -3′ and 5′ - GAA TGA ACC AGT TGG AAA AGG -3′ from both N2 and daf-31(m655) mutant homozygotes. PCR products were cloned into the pGEM-T vector (Promega) following the manufacturer's instructions. T7 primer 5′ - GTA ATA CGA CTC ACT ATA GGG -3′ and SP6 primer 5′ - TAC GAT TTA GGT GAC ACT ATA G -3′ were used in DNA sequencing reactions.
RNAi
Part of the daf-31 coding region was amplified from C. elegans genomic DNA using primers 5′ - CGG GAT CCA TTC GTT GTG CTC GCG TG -3′ and 5′ - CCC AAG CTT GCA GTG GTA TAG GCC TC -3′. The PCR products were then purified and cloned into the feeding RNAi vector L4440 (Addgene) between the BamHI and HindIII sites. The RNAi construct was transformed into E. coli HT115 (DE3) and RNAi feeding was performed as previously described [48].
To inhibit daf-15 and daf-31 genes by injection of RNAi, a 1 kb daf-15 cDNA fragment and the full-length daf-31 cDNA were cloned into pGEM-T vector (Promega), respectively. The gene identity was confirmed by sequencing. The Riboprobe Combination System-SP6/T7 (Promega) was used to transcribe RNA in vitro according to the manufacturer's protocol. Double-stranded RNA was synthesized and injected as described by Fire et al. [49].
qRT-PCR
Synchronized N2 L1 larvae were treated with either control (empty) vector or daf-31 RNAi by feeding as previously described [48]. Day 1 adult animals were collected for total RNA extraction using the Trizol kit (Zymo). Synchronized L1 larvae of daf-31 overexpressing strains were allowed to grow on OP50 food plates. Day 1 adult animals were collected for total RNA extraction using the Trizol reagent (Zymo). The first strand cDNA was synthesized using the ImProm-II reverse transcription system (Promega). SYBR green dye (Quanta) was used for qRT-PCR to measure the expression level of daf-31, sod-3 and bcmo-2 in corresponding worm samples. Reactions were performed in triplicate on an ABI Prism 7000 real-time PCR machine (Applied Biosystems). Relative-fold changes were calculated using the 2−ΔΔCT method. The primers used for qRT-PCR were: daf-31, 5′ - GAA GAT CAC AAG GGA AAT GTT G -3′ and 5′ - CTC TTG CGG TCT GAT CCA TC -3′; act-1, 5′ - CAA TCC AAG AGA GGT ATC CTT ACC CTC -3′ and 5′ - GAG GAG GAC TGG GTG CTC TTC -3′; bcmo-2, 5′ - GCC GAT TTA GAG AAC GGA GAT CAC -3′ and 5′ - TGA GAA TTC CGT CAT CTT CCC GA -3′; sod-3, 5′ - GGA ATC TAA AAG AAG CAA TTG CTC -3′ and 5′ - CGC GCT TAA TAG TGT CCA TCA G -3′.
Adult lifespan, thermotolerance and reproduction
About 120–150 L4 larvae raised at 20°C were transferred to ten NG agar plates (twelve to fifteen animals per plate spread with either OP50 or RNAi food) and incubated at 25°C. The first day of adulthood is day 1 in the survival curves. During the reproductive period, adult animals were transferred daily to fresh plates. Thereafter, animals were transferred every ten days (OP50 food) or every six days (RNAi food). Animals were scored as alive, dead, or lost every other day. Animals that do not move in response to touching were scored as dead. Animals that died from causes other than aging, such as sticking to the plate walls, internal hatching or bursting in the vulval region, were scored as lost. GraphPad Prism was used for statistical analysis and generation of survival curves. For the thermotolerance experiment, day 1 adult animals were incubated at 35°C and survival was scored as described above. To measure reproduction of worms, L4 larvae growing at 20°C were transferred daily to fresh plates and the progeny were counted.
Supporting Information
Zdroje
1. GoldenJW, RiddleDL (1984) A pheromone-induced developmental switch in Caenorhabditis elegans: Temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc Natl Acad Sci U S A 81 : 819–823.
2. Riddle DL, Albert PS (1997) Genetic and environmental regulation of dauer larva development. In C elegans II (ed D L Riddle, T Blumenthal, B J Meyer and J R Priess), pp 739–768 NY: Cold Spring Harbor Laboratory Press.
3. GoldenJW, RiddleDL (1985) A gene affecting production of the Caenorhabditis elegans dauer-inducing pheromone. Mol Gen Genet 198 : 534–536.
4. AlbertPS, BrownSJ, RiddleDL (1981) Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol 198 : 435–451.
5. BargmannCI, HorvitzHR (1991) Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251 : 1243–1246.
6. RenP, LimCS, JohnsenR, AlbertPS, PilgrimD, et al. (1996) Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274 : 1389–1391.
7. SchackwitzWS, InoueT, ThomasJH (1996) Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17 : 719–728.
8. BirnbyDA, LinkEM, VowelsJJ, TianH, ColacurcioPL, et al. (2000) A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155 : 85–104.
9. KimuraKD, TissenbaumHA, LiuY, RuvkunG (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 : 942–946.
10. PierceSB, CostaM, WisotzkeyR, DevadharS, HomburgerSA, et al. (2001) Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev 15 : 672–686.
11. AntebiA, YehWH, TaitD, HedgecockEM, RiddleDL (2000) daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev 14 : 1512–1527.
12. AlbertPS, RiddleDL (1988) Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev Biol 126 : 270–293.
13. GerischB, WeitzelC, Kober-EisermannC, RottiersV, AntebiA (2001) A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell 1 : 841–851.
14. JiaK, AlbertPS, RiddleDL (2002) DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129 : 221–231.
15. MotolaDL, CumminsCL, RottiersV, SharmaKK, LiT, et al. (2006) Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124 : 1209–1223.
16. RottiersV, MotolaDL, GerischB, CumminsCL, NishiwakiK, et al. (2006) Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell 10 : 473–482.
17. Caldicott IM (1995) Non-conditional dauer and dauer-like mutants of Caenorhabditis elegans [PhD dissertation]. Columbia (Missouri): Division of Biological Sciences, University of Missouri, Columbia.
18. JiaK, ChenD, RiddleDL (2004) The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131 : 3897–3906.
19. YenK, LeTT, BansalA, NarasimhanSD, ChengJX, et al. (2010) A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS ONE 5: e12810.
20. PolevodaB, ShermanF (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol 325 : 595–622.
21. GuntherCV, GeorgiLL, RiddleDL (2000) A Caenorhabditis elegans type I TGF beta receptor can function in the absence of type II kinase to promote larval development. Development 127 : 3337–3347.
22. LiJ, TewariM, VidalM, LeeSS (2007) The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol 301 : 82–91.
23. HendersonST, JohnsonTE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11 : 1975–1980.
24. LinK, HsinH, LibinaN, KenyonC (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28 : 139–145.
25. LithgowGJ, WhiteTM, MelovS, JohnsonTE (1995) Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A 92 : 7540–7544.
26. HondaY, HondaS (1999) The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J 13 : 1385–1393.
27. KenyonC (2005) The plasticity of aging: insights from long-lived mutants. Cell 120 : 449–460.
28. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464.
29. GemsD, SuttonAJ, SundermeyerML, AlbertPS, KingKV, et al. (1998) Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150 : 129–155.
30. MorrisJZ, TissenbaumHA, RuvkunG (1996) A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382 : 536–539.
31. ParadisS, AilionM, TokerA, ThomasJH, RuvkunG (1999) A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev 13 : 1438–1452.
32. OggS, RuvkunG (1998) The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell 2 : 887–893.
33. OggS, ParadisS, GottliebS, PattersonGI, LeeL, et al. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 : 994–999.
34. LinK, DormanJB, RodanA, KenyonC (1997) daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 : 1319–1322.
35. ApfeldJ, KenyonC (1998) Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95 : 199–210.
36. WolkowCA, KimuraKD, LeeMS, RuvkunG (2000) Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290 : 147–150.
37. LibinaN, BermanJR, KenyonC (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115 : 489–502.
38. FujiiM, TanakaN, MikiK, HossainMN, EndohM, et al. (2005) Uncoupling of longevity and paraquat resistance in mutants of the nematode Caenorhabditis elegans. Biosci Biotechnol Biochem 69 : 2015–2018.
39. TimmonsL, CourtDL, FireA (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 : 103–112.
40. WhitewayM, SzostakJW (1985) The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 43 : 483–492.
41. WangY, MijaresM, GallMD, TuranT, JavierA, et al. (2010) Drosophila variable nurse cells encodes arrest defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Dev Dyn 239 : 2813–2827.
42. ArnesenT, Van DammeP, PolevodaB, HelsensK, EvjenthR, et al. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci U S A 106 : 8157–8162.
43. LimJH, ParkJW, ChunYS (2006) Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res 66 : 10677–10682.
44. ArnesenT, ThompsonPR, VarhaugJE, LillehaugJR (2008) The protein acetyltransferase ARD1: a novel cancer drug target? Curr Cancer Drug Targets 8 : 545–553.
45. KuoHP, HungMC (2010) Arrest-defective-1 protein (ARD1): tumor suppressor or oncoprotein? Am J Transl Res 2 : 56–64.
46. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
47. PinoEC, WebsterCM, CarrCE, SoukasAA (2013) Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J Vis Exp 73 : 50180.
48. KamathRS, Martinez-CamposM, ZipperlenP, FraserAG, AhringerJ (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002.
49. FireA, XuS, MontgomeryMK, KostasSA, DriverSE, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 : 806–811.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



