-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
High salinity, as a world-wide abiotic stress, restricts root water uptake, damages cell physiology, and limits the productivity of agricultural crops. Ethylene is a major phytohormone that regulates plant development in response to adverse environments, including high salt stress. However, the molecular mechanisms of how ethylene signal exerts its effect and how ethylene signaling is modulated upon salt stress remain to be explored. Here, we report that high salinity induces EIN3/EIL1 protein accumulation and EBF1/2 protein degradation in an EIN2-independent manner. Moreover, the activated EIN3 deters excess ROS accumulation and increases salt tolerance. Transcriptome analysis and functional studies reveal an EIN3-directed gene network in salt stress response. Functional studies of 114 SIED (Salt-Induced and EIN3/EIL1-Dependent) genes identify a novel regulator of ROS dismissal and salt tolerance. This new understanding of ethylene/salt mutual regulation would allow a better manipulation and engineering of EIN3 and its downstream SIED genes to enhance plant tolerance and adaption to salt stress, particularly in those economically important crops in the future.
Published in the journal: . PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004664
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004664Summary
High salinity, as a world-wide abiotic stress, restricts root water uptake, damages cell physiology, and limits the productivity of agricultural crops. Ethylene is a major phytohormone that regulates plant development in response to adverse environments, including high salt stress. However, the molecular mechanisms of how ethylene signal exerts its effect and how ethylene signaling is modulated upon salt stress remain to be explored. Here, we report that high salinity induces EIN3/EIL1 protein accumulation and EBF1/2 protein degradation in an EIN2-independent manner. Moreover, the activated EIN3 deters excess ROS accumulation and increases salt tolerance. Transcriptome analysis and functional studies reveal an EIN3-directed gene network in salt stress response. Functional studies of 114 SIED (Salt-Induced and EIN3/EIL1-Dependent) genes identify a novel regulator of ROS dismissal and salt tolerance. This new understanding of ethylene/salt mutual regulation would allow a better manipulation and engineering of EIN3 and its downstream SIED genes to enhance plant tolerance and adaption to salt stress, particularly in those economically important crops in the future.
Introduction
Soil salinity is a major abiotic stress that reduces plant growth and limits the productivity of agricultural crops. The detrimental effects of salt on plants are a consequence of both a water deficit resulting in osmotic stress and the effects of excess sodium ions imposed on critical biochemical processes [1]. The sessile nature of plants has favored the evolution of mechanisms to cope with various environmental stresses. One of these mechanisms is the release and utilization of a multitude of phytohormones, including a gaseous molecule ethylene [2].
Ethylene can trigger multiple physiological and morphological responses, including inhibition of cell expansion, induction of fruit ripening and abscission, and adaptation to stress conditions [3]. One of the well documented ethylene responses is the so-called “triple response” of etiolated seedlings, i.e. short, thickened root and hypocotyl, as well as exaggerated curvature of the apical hook [4]. Based on this highly reproducible and specific phenotype, a largely linear ethylene signal transduction pathway has been established [5]. In Arabidopsis, ethylene is perceived by a family of membrane-associated receptors [6], [7], [8], which are negative regulators of the signaling pathway, and ethylene binding leads to functional inactivation of the receptors [9]. In the absence of ethylene, the active receptors recruit CTR1 (CONSTITUTIVE TRIPLE RESPONSE1) to associate with the membrane and thus become activated [10], which subsequently represses the downstream signaling pathway mediated by ETHYLENE INSENSITIVE2 (EIN2) and EIN3. EIN2 is a central component of the ethylene signaling transduction pathway, and its null mutant ein2 is completely insensitive to ethylene [11]. EIN2 is shown to locate in endoplasmic reticulum membrane [12], and undergoes a hormone-induced cleavage and translocation event that is controlled by CTR1-directed phosphorylation of its carboxyl-terminus [13], [14], [15]. As the requisite component for ethylene signaling, EIN2 positively regulates the functions of EIN3/EIL1 transcription factors, which results in the activation of transcription of ERF1 and other downstream genes [16], [17]. EIN3/EIL1 are short-lived proteins, which are quickly stabilized and accumulate in the nucleus in the presence of ethylene. Genetic and biochemical studies revealed that EIN3/EIL1 are subject to ubiquitin/proteasome-mediated proteolysis that requires two F-box proteins, EBF1/EBF2 [16], [18], [19], [20]. Recently, our studies have demonstrated that ethylene stabilizes EIN3/EIL1 at least partly by promoting the proteasomal degradation of EBF1/EBF2, and that EIN2 is indispensable for mediating ethylene-induced EIN3/EIL1 accumulation and EBF1/2 degradation [18], highlighting the importance of EIN2 in the control of EIN3/EIL1 abundance.
In addition to its role in regulating plant growth and development, ethylene also plays a key role in plant responses to biotic and abiotic stresses [21]. Recently, the functions of components of ethylene signaling in salt stress response were investigated. The ctr1-1 mutant exhibited increased salt tolerance and the germination rate and post-germination development of ctr1-1 were more tolerant under salt and osmotic stress treatments, especially under high concentration of salt [22]. Under salt stress, the ein2-1 mutant was severely affected in both seedling growth and seed germination processes, suggesting that EIN2 is required for salt stress tolerance [23], [24]. The ein3-1eil1-1 double mutant exhibited remarkably reduced tolerance to high concentration of salt [22], [23], [24]. Recently, Jiang et al. reported that salinity-induced ethylene promotes Arabidopsis soil-salinity tolerance by enhancing Na/K homeostasis [25].
Despite such clear demonstration of a vital role of ethylene in salt stress response, the molecular mechanisms of how the ethylene signaling is modulated under salt stress condition and how ethylene signaling increases salinity tolerance are poorly understood. In this study, we demonstrated that plants pretreated with ethylene exhibited increased tolerance to salt stress, and that EIN3/EIL1 are both necessary and sufficient for salt tolerance. Interestingly, we found that salt stabilized EIN3/EIL1 protein by promoting EBF1/EBF2 proteasomal degradation in an EIN2 independent manner. Microarray analysis identified a large number of EIN3/EIL1-regulated genes (SIEDs) that participate in salt stress response, including many genes encoding reactive oxygen species (ROS) scavengers. A novel EIN3 target gene, SIED1, was functionally studied and defined as an important mediator of ethylene-evoked salt tolerance.
Results
ACC/Ethylene Pretreatment or Activated Ethylene Signaling Increases Salt Tolerance
Previous studies investigating the effect of ethylene in salt stress were conducted in conditions where ethylene and salt stress were simultaneously applied [23]. Because several salt-induced seedlings responses, such as leaf epinasty, chlorophyll loss and growth retardation, are also regulated by ethylene [4], [5], it is sometimes not clear how the final morphological output is the result of an altered salt or ethylene response. To specifically ascertain the role of ethylene in salt response, we pretreated Arabidopsis seedlings with ethylene or its biosynthesis precursor ACC and then transferred to MS medium supplemented with 200 mM NaCl alone. Upon ACC pretreatment, wild-type Col-0 displayed enhanced tolerance to salt compared with untreated control, with higher survival rate and lower relative electrolyte leakage (an indicator for the salt stress damage) [26] (Figure 1A–C). By comparison, ebf1-1 mutant showed slightly lower survival rate, whereas ebf2-1, an ethylene hypersensitive mutant [16], was more tolerant to salt than Col-0 upon ACC pretreatment (Figure 1A–C). Consistent with their respective ethylene response phenotype, ctr1-1, EIN3ox (a transgenic plant overexpressing EIN3) as well as EIL1ox displayed constitutively enhanced salt tolerance, whereas ACC pretreatment had virtually no effect on the salt tolerance of ethylene insensitive mutants, etr1-1, ein2-5 and ein3-1eil1-1 (Figure 1A–C; Figure S1). To further examine the effect of ACC pretreatment on salt tolerance, 5-day-old seedlings of wild-type, ein3-1eil1-1 and EIN3ox were also transferred onto MS medium supplemented with serial concentrations of NaCl (0, 50, 100, 150 and 200 mM). Similarly, ACC pretreatment significantly increased the survival rate, fresh weight as well as root length of wild-type but not ein3-1eil1-1 compared with ACC-untreated plants when 100 mM or higher concentrations of NaCl were applied (Figure S2E vs S2B, S2F vs S2C and S2G vs S2D). Consistently, EIN3ox seedlings showed constitutively increased salt tolerance in terms of survival rate, fresh weight and root length (Figure S2E vs S2B, S2F vs S2C and S2G vs S2D). Together, these results demonstrate that ACC pretreatment or overexpression of EIN3 leads to increased tolerance to salt stress, which depends on the canonical ethylene signaling pathway.
Fig. 1. ACC/Ethylene pretreatment or enhanced ethylene signaling increases salt tolerance. 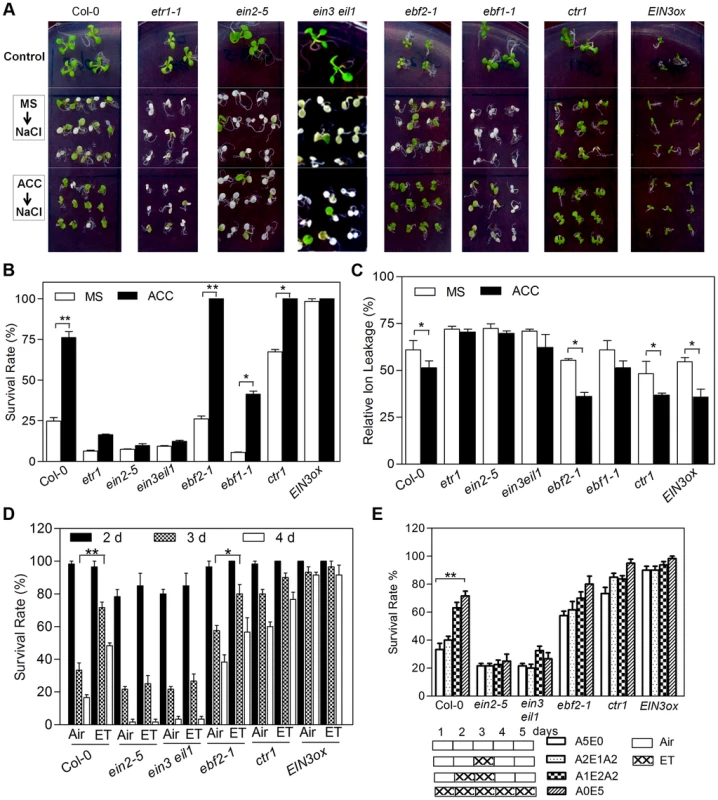
(A) Plants were grown on MS medium with or without 10 µM ACC for 5 d and then transferred onto MS medium supplemented with 200 mM NaCl for 3 d. Plants were also transferred onto MS medium as controls. (B) Survival rate of plants shown in (A). Seedling death was scored as complete bleaching of cotyledons and leaves. Values are mean ± SD from 25 seedlings per replicate (n = 3 replicates). (Student's t test, *P<0.05 and **P<0.01). (C) Relative electrolyte leakage of plants shown in (A). Values are mean ± SD from 50 seedlings per replicate (n = 3 replicates). (Student's t test, *P<0.05 and **P<0.01). (D) Survival rate of plants pretreated with air (Air) or 20 ppm ethylene (ET) for 5 d and then transferred onto MS medium supplemented with 200 mM NaCl. Survival rates were calculated on the second, third and fourth day. Values are mean ± SD from 20 seedlings per replicate (n = 4 replicates). (Student's t test, *P<0.05 and **P<0.01). (E) Survival rate of plants pretreated with air or 20 ppm ethylene for indicated time and then transferred onto medium supplemented with 200 mM NaCl. Survival rates were calculated on the third day after transfer. Values are mean ± SD from 20 seedlings per replicate (n = 4 replicates). A5E0: 5 d of air treatment. A2E1A2: 2 d of air followed by 1 d of ethylene then 2 d of air treatment. A1E2A2: 1 d of air followed by 2 d of ethylene then 2 d of air treatment. A0E5: 5 d of ethylene treatment. ET: ethylene. (Student's t test, *P<0.05 and **P<0.01). To exclude the possibility that the observed effect of ACC pretreatment was due to the residual ACC remained in the pretreated seedlings, the experiment was repeated using ethylene gas, which was quickly diffusing away. After 5 days of 10 ppm ethylene gas treatment, seedlings were transferred onto MS medium supplemented with 200 mM NaCl in the air, and survival rates were calculated after two, three and four days, respectively. We found that ethylene pretreatment effectively increased the tolerance to salt in wild-type, ebf2-1 and ctr1-1, evidenced by higher survival rates after three or four days of salt treatment, but had little effect on ein2-5 and ein3-1eil1-1 (Figure 1D). Ethylene pretreatments with different lengths of time were also investigated. Seedlings pretreated with ethylene for 2 or 5 days exhibited increasingly enhanced survival rate in Col-0, ebf2-1, but not in ein2-5 and ein3-1eil1-1, while 1 day of ethylene pretreatment had only marginal effect (Figure 1E). Together with the ACC pretreatment experiments, these results support that exogenous ethylene application beforehand effectively increases salt tolerance.
To further study the function of EIN3/EIL1 in ethylene-mediated salt response, a transgenic line expressing estradiol-inducible EIN3-FLAG in the ein3 eil1 ebf1 ebf2 quadruple mutant (iE/qm) was investigated [18]. Previous studies demonstrated that the iE/qm seedlings were completely insensitive to exogenously applied ethylene, and the accumulation of EIN3-FLAG fusion protein can be induced by estradiol (but not by ethylene) in a dose-dependent manner [18]. As reported, the EIN3-FLAG protein was undetected in estradiol-untreated iE/qm, and it was evidently induced in iE/qm upon estradiol treatment, but ACC treatment did not further increase its protein accumulation (Figure S3A, Lane 1, 2, 8, or Lane 7, 9). We also found that salt treatment did not affect EIN3 protein level in estradiol-treated iE/qm regardless of treatment time (Figure S3A, Lane 3–5, or Lane 2, 6). These results demonstrated that the estradiol-induced EIN3 protein accumulation in iE/qm seedlings is not altered by ethylene or salt treatment.
Next, we investigated whether the estradiol-induced EIN3 protein in iE/qm seedlings effectively increased the tolerance to salt stress. In the absence of estadiol, where EIN3 protein was not detectable, the cotyledons and leaves of iE/qm seedlings treated with 200 mM NaCl were severely bleached after 3 days (Figure S3B), and the survival rate was declined to less than 30% (Figure S3C). In contrast, in the presence of serial concentrations of estradiol (from 0.1 to 20 µM), iE/qm seedlings treated with 200 mM NaCl for 3 days appeared largely green and healthy (Figure S3B), and the survival rates remained over 90% in all cases (Figure S3C). Despite all concentrations of estradiol were sufficient for conferring salt tolerance, we noted that iE/qm seedlings supplemented with lower concentrations (0.1 or 1 µM) grew better on salt medium (Figure S3B). Therefore, all subsequent physiological experiments were performed with 1 µM estradiol. In line with cotyledon yellowing phenotype and survival rate, the leaf chlorophyll content and fresh shoot weight were significantly higher in estradiol-treated iE/qm seedlings than the untreated control under salt stress condition (Figure S3D, S3F). Conversely, upon salt treatment, the ion leakage was evidently lower in estradiol-treated iE/qm plants than the untreated control (Figure S3E). Taken together, these results indicate that loss of EIN3/EIL1 function leads to hypersensitivity to salt stress whereas accumulation of EIN3 alone results in enhanced salt tolerance, highlighting the requirement and sufficiency of EIN3/EIL1 for salt tolerance in Arabidopsis.
High Salinity Enhances EIN3 Protein Accumulation and Transcriptional Activity in Both EIN2-Dependent and EIN2-Independent Manners
Given that EIN3 is a critical regulator of plant salt responses, we next determined whether, and if so, how EIN3 is modulated by salt stress. We first monitored the level of endogenous EIN3 protein using an anti-EIN3 antibody [16], [18] in response to salt treatment. We found that the levels of EIN3 protein started to increase after 3 h of salt treatment and dramatically accumulated after 6 h of treatment in wild-type Col-0 (Figure 2A). We also checked the levels of EIN3 mRNA and found no obvious change after 6 h of salt treatment (Figure S4), suggesting that salt regulates EIN3 accumulation at the protein level.
Fig. 2. Salt treatment promotes protein accumulation and transcriptional activity of EIN3 in both EIN2-dependent and EIN2-independent manners. 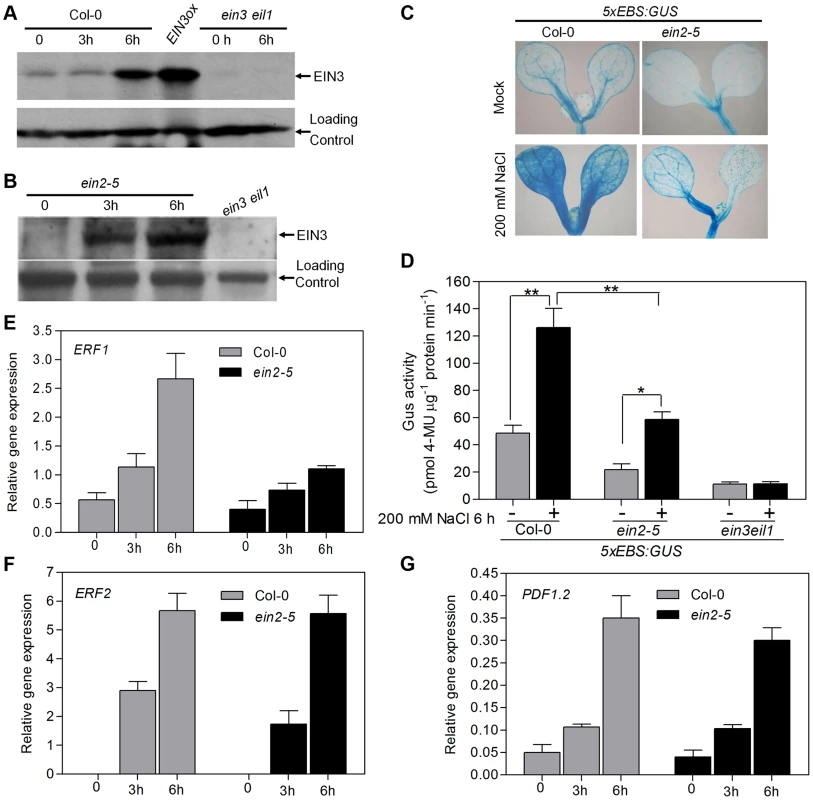
(A) Salt treatment promotes EIN3 protein accumulation in wild type. 5-d-old seedlings were treated with 200 mM NaCl for 3 h and 6 h. Protein was extracted and subjected to immunoblots using anti-EIN3 antibody. A nonspecific band was used as a loading control. (B) Salt treatment promotes EIN3 protein accumulation in ein2-5 mutant. Experiments were repeated three times with similar results. (C) Histochemical analysis of 5xEBS:GUS transgenic plants. (D) 5xEBS:GUS activity in Col-0 or ein2-5 background was measured. Two biological replicates and three technical replicates were performed (Student's t test, *P<0.05 and **P<0.01). (E–G) Real-time RT-PCR analysis of gene expression of ERF1 (E), ERF2 (F) and PDF1.2 (G). Previous study indicated that EIN2 is absolutely required for ethylene-induced EIN3/EIL1 accumulation, as no EIN3 or EIL1 protein can be detected in ein2 mutant [16], [18]. To determine whether EIN2 is required for salt-induced EIN3/EIL1 protein accumulation, we detected the EIN3/EIL1 protein level in ein2-5 mutant under salt treatment. Surprisingly, we found that salt treatment (but not mock treatment, Figure S5A) promoted EIN3 and EIL1 proteins accumulation in the ein2-5 mutant background, although not as dramatic as in wild-type (Figure 2B and Figure S6). These results indicate that the salt stress signal is able to promote EIN3/EIL1 proteins accumulation in an EIN2-independent manner. In addition, we also analyzed the EIN3 protein levels in an ethylene receptor mutant etr1-1 [9] upon treatment with 200 mM NaCl for 3 h and 6 h using anti-EIN3 antibody. We found that EIN3 protein was evidently induced in etr1-1 mutants upon salt treatment for 6 h (Figure S5B), suggesting that salt induced EIN3 protein accumulation does not require the canonical ethylene perception.
We next investigated whether salt-induced EIN3 protein is transcriptionally functioning. A transgenic reporter line that harbors the GUS report gene driven by five tandem repeats of the EIN3 binding site (EBS) followed by the minimal 35S promoter, 5xEBS:GUS, has been previously used to monitor the transcriptional activity of EIN3 [27], [28]. Upon salt treatment, GUS staining became overly intensified in the cotyledons and hypocotyls of 5xEBS:GUS/Col-0 plants (Figure 2C), indicative of elevated levels of EIN3 activity under this condition, which was further supported by a quantification assay (Figure 2D). Compared to that in Col-0 background, GUS activity was also evidently up-regulated in 5xEBS:GUS/ein2-5 plants upon salt treatment, although to a lesser extent (Figure 2C and 2D). By contrast, GUS activity in 5xEBS:GUS/ein3 eil1 plants did not increase upon salt stress (Figure 2D), suggesting that salt-increased 5xEBS:GUS activity is EIN3/EIL1-dependent. We also observed that the expression levels of several ethylene responsive genes, including ERF1, ERF2 and PDF1.2, were up-regulated by salt in both wild type and ein2-5 mutants (Figure 2E–G). In keeping with the results of EIN3 accumulation and GUS expression (Figure 2B–D), the expression level of ERF1, a direct target gene of EIN3 [17], was also lower in salt-treated ein2-5 mutant compared with that in wild-type (Figure 2E). Thus, although salt treatment did promote EIN3 protein accumulation in ein2-5, the relative lower level and activity of EIN3 might be inadequate to compensate for the loss of EIN2 that could elicit additional pathways contributing to salt tolerance. Taken together, our results suggest that, in addition to the canonical EIN2-dependent pathway, there exists a new pathway independent of EIN2 to mediate the salt stress signal to promote EIN3/EIL1 protein accumulation.
Salt Promotes EBF1/EBF2 Protein Degradation in an EIN2-Independent Manner
It has been established that the stability of EIN3 is controlled by two F-box proteins, EBF1/EBF2, and that ethylene-induced EIN3 stabilization is at least partly mediated by the destabilization of EBF1/EBF2 proteins in an EIN2-dependent manner [16], [18], [19], [20]. To further characterize how EIN3 accumulation is enhanced by salt, we examined the levels of EBF1/EBF2 protein after salt treatment. Our initial effort to produce polyclonal antibodies recognizing endogenous EBF1 or EBF2 protein in plant tissues was unsuccessful. Therefore, two transgenic lines, 35S:EBF1-MYC/Col-0 and 35S:EBF2-MYC/Col-0 [18], were used to detect the EBF1 and EBF2 protein levels. Immunoblot analysis showed that the protein levels of EBF1-MYC and EBF2-MYC markedly decreased upon salt treatment (Figure 3A). We also noted that ACC pretreatment seemed to reinforce the destruction of EBF1/EBF2 proteins, as seedlings pretreated with ACC accumulated less EBF1/EBF2 proteins after salt application (Figure 3A). Together with the finding that EIN3 protein level is not altered by salt in iE/qm seedlings (Figure S3A), these results suggest that the salt-induced accumulation of EIN3 protein is due to reduced levels of EIN3-targeting F-box proteins.
Fig. 3. Salt treatment promotes EBF1/EBF2 protein degradation in an EIN2-independent manner. 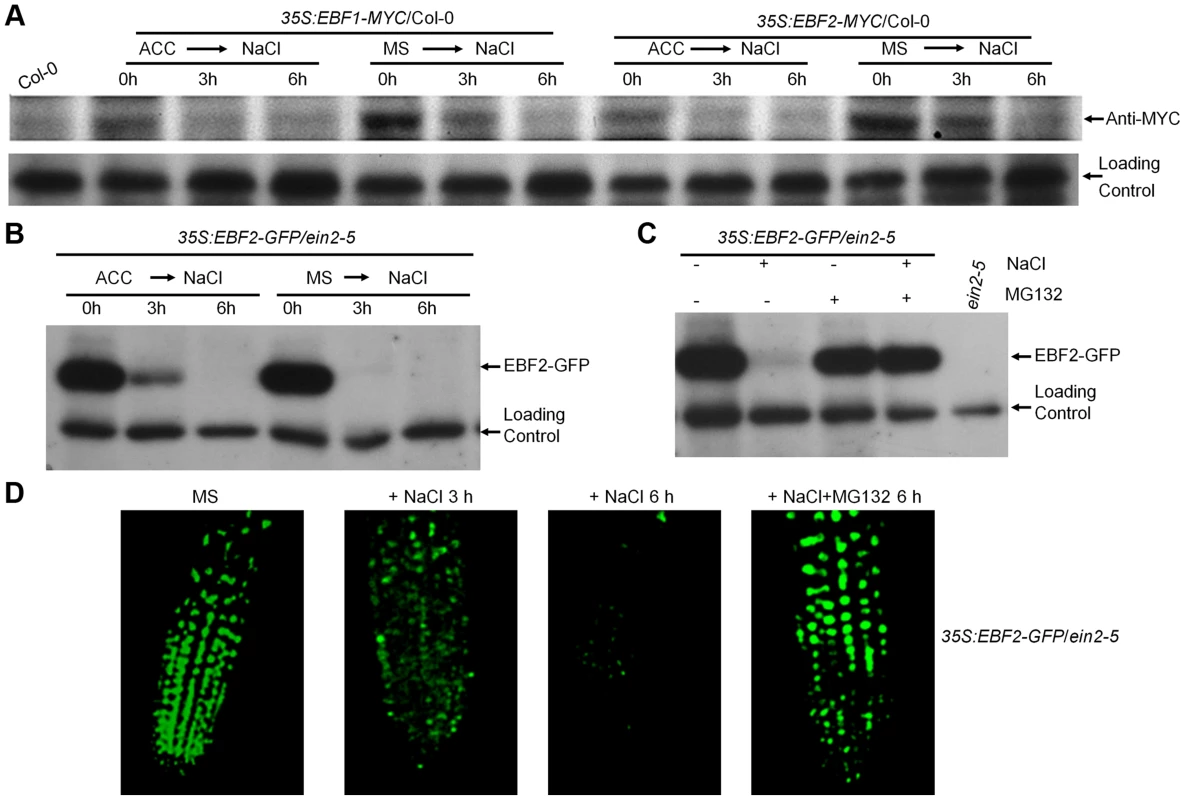
(A) Immunoblot assays of EBF1/2-MYC protein in Col-0. Transgenic seedlings overexpressing EBF1/2-MYC in Col-0 grown on MS medium supplemented with or without 10 µM ACC for 5 d were subjected to 200 mM NaCl for 3 h and 6 h. A nonspecific band was used as a loading control. (B) Immunoblot assay of EBF2-GFP protein in ein2-5 background. Transgenic seedlings overexpressing EBF2-GFP in ein2-5 background grown on medium supplemented with or without 10 µM ACC for 5 d were subjected to 200 mM NaCl for 3 h and 6 h. Experiments were repeated three times with similar results. (C) Salt induced EBF2 protein degradation was inhibited by MG132. 5-d-old plants were treated with 200 mM NaCl or/and 50 µM MG132 for 6 h. Experiments were repeated three times with similar results. (D) GFP fluorescence of 35S:EBF2-GFP in the roots of ein2-5 mutant. The seedlings grown on MS medium for 5 d were treated with 200 mM NaCl and/or 50 µM MG132 for 6 h. Our above data showed that salt induced EIN3 protein accumulation in ein2-5 mutant, so we asked whether salt-induced destruction of EBF1/EBF2 proteins also take place in the absence of EIN2 function. To address this question, a previously generated transgenic line, 35S:EBF2-GFP/ein2-5, which showed high level of EBF2-GFP accumulation [18], was used. Immunoblot analysis showed that the protein levels of EBF2-GFP markedly decreased upon salt treatment, regardless of ACC pretreatment (Figure 3B). By contrast, treatment with MG132, a 26S proteasome inhibitor, promoted a dramatic accumulation of EBF2-GFP and reversed the salt-induced EBF2-GFP degradation (Figure 3C). Similarly, GFP fluorescence was dramatically reduced in the 35S:EBF2-GFP/ein2-5 after salt treatment, but MG132 treatment effectively reversed the salt effect and stabilized EBF2-GFP protein (Figure 3D). We further examined the effect of other salt ions on EBF2 stability, and found that, as NaCl, treatments of KCl, NaNO3 and KNO3 all similarly led to the destruction of EBF2-GFP protein in ein2-5 mutant background (Figure S7A). We also excluded the involvement of osmotic stress in the control of salt-induced EBF protein degradation, as high dose of mannitol treatment (200 mM) had no effect on EBF2-GFP stability (Figure S7B). Collectively, our data clearly demonstrated that salt stress leads to the proteasome-mediated degradation of EBF1/EBF2 proteins independent of the upstream ethylene signaling components, such as EIN2.
Transcriptome Profiling Analyses Identify Salt-Regulated EIN3/EIL1-Dependent Genes
Our above data indicated that EIN3/EIL1 are both necessary and sufficient for conferring enhanced salt tolerance. To elucidate the molecular network underlying EIN3/EIL1-induced salt tolerance, we performed transcriptome profiling of EIN3ox, ein3eil1 and wild type Col-0. For this analysis, 5-day-old light-grown seedlings treated with or without 200 mM NaCl for 6 h were used. This design enabled us to compare the transcriptional profiles among plants with different levels of EIN3 activity, as well as to identify salt-regulated and EIN3/EIL1-dependent genes.
Treatment with high salt for 6 h resulted in the induction of 1482 transcripts while the repression of 1745 transcripts (using both q value<0.05 and 2-fold as a cutoff) in wild-type Col-0 (Figure 4E, 4F). Applying a q value<0.05 and 5-fold as a cutoff, 509 transcripts were induced while 209 transcripts were repressed in wild type by salt treatment, which were arbitrarily defined as salt-regulated genes in this study (Figure 4A and 4B). Using the same cutoff, 365 and 74 transcripts in EIN3ox while 281 and 98 transcripts in ein3eil1 were induced and repressed by salt treatment, respectively (Figure 4A, 4B). Of the 509 salt-induced genes, 162 were also elevated in salt-treated EIN3ox and ein3eil1 mutant (Figure 4A). Conversely, 36 out of 209 salt-repressed genes were also down-regulated in salt-treated EIN3ox and ein3eil1 mutant (Figure 4B). To investigate how EIN3 activation leads to increased salt tolerance, we were particularly interested in two classes of genes: salt-induced EIN3/EIL1-dependent (SIED) genes and salt-repressed EIN3/EIL1-dependent (SRED) genes (Figure 4C, 4D). The former class includes those genes whose levels are induced by salt at least 5-fold in Col-0 (P value 0.0041), plus that salt induction is more pronounced in EIN3ox (P value 0.00099) but less in ein3 eil1 (P value 0.0094) (i.e. salt-induced gene expression is at least partly dependent on EIN3 activity) (Table S1). The latter class includes those genes whose levels are repressed by salt at least 5-fold in Col-0 (P value 0.0016), plus that salt repression is more pronounced in EIN3ox (P value 0.00093) but less in ein3 eil1 (P value 0.0081) (Table S2).
Fig. 4. Transcriptome profiling analyses identify salt-regulated EIN3/EIL1-dependent genes. 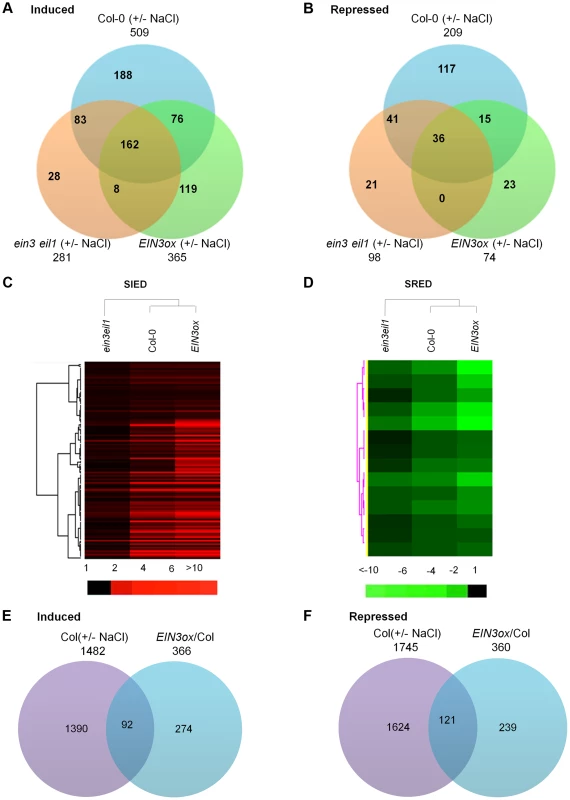
(A) and (B) Venn diagrams showing the overlaps among transcripts induced or repressed (q<0.05 and 5-fold change as a cutoff) by salt in Col-0, ein3eil1 and EIN3ox plants. (C) and (D) Hierarchical clusters displaying the salt-induced expression of those SIED (Salt-Induced EIN3/EIL1-Dependent) and SRED (Salt-Repressed EIN3/EIL1-Dependent) genes in Col-0, ein3eil1 and EIN3ox plants. A total of 114 SIED genes (C) and 14 SRED genes (D) were included in the cluster (Table S1 and S2). (E) and (F) Venn diagrams showing the overlaps between transcripts induced or repressed (2-fold cutoff) by salt in Col-0 and transcripts induced or repressed (2-fold cutoff) by overexpression of EIN3 (comparing transcriptome of EIN3ox versus Col-0 under unstressed condition). Based on these criteria, 114 SIED genes and 14 SRED genes were identified (Table S1 and S2). The drastic difference on the number of SIED and SRED genes suggested that EIN3/EIL1 might enhance salt tolerance mainly through inducing genes or pathways that participate in plant survival, rather than repressing genes or pathways that lead to plant death under salinity stress. In support of this speculation, 18 out of 114 SIED genes (∼16%) are defense-related genes that function to enhance plant tolerance or resistance to abiotic or biotic stresses. Several ERF (11) and JAZ (3) genes were found to be SIED genes, implying that the signaling pathways of ethylene and jasmonic acid (JA), two stress hormones, have been preferentially activated by salt stress, which is consistent with previous studies [23], [29], [30]. The considerable enrichment of ERF genes, many of which are direct target genes of EIN3 [17], [31], suggests that the identification of SIED genes is biologically relevant. Furthermore, when compared the SIEDs with ethylene-regulated EIN3-target genes identified by ChIP-Sequencing [32], we found that 15 out of 114 SIEDs (Highlighted in Table S1), such as At5g22270 and At5g59820 (ZAT12), are the direct targets of EIN3. However, most of SIEDs are not the target genes of EIN3 identified by Chang et al. [32]. This could be due to that EIN3 preferentially binds to specific subsets of target promoters dependent on the initial treatment/stimulus. Alternatively, it is also possible that the majority of SIED genes are indirectly induced by EIN3, for instance, via the ERF transcription factors. The SIED genes were further analyzed using the gene ontology (GO) enrichment tool Gorilla [33]. We found that, in terms of molecular function category, there were notable enrichments for metabolic processes, as well as transcription, DNA binding, and oxidoreductase activity (Figure S8). For instance, of 114 SIED genes, we found 9 genes encoding oxidoreductases and 4 genes involved in electron transport or energy pathways, suggesting that modulation of oxidative/reductive status under salt stress might be an important mechanism of EIN3/EIL1 action to enhance plant survival.
In this study, we demonstrated that pretreatment with ethylene conferred increased salt tolerance, which depends on the action of EIN3/EIL1 (Figure 1). One explanation for this priming effect of ethylene is that EIN3/EIL1 activation in advance alters the expression of genes that ultimately leads to salt tolerance. To test this possibility, we compared the salt-regulated transcriptome (salt-treated versus untreated Col-0) and EIN3-regulated transcriptome (EIN3ox versus Col-0 without salt treatment). By a 2-fold cutoff, 366 genes (P value 0.0096) were identified as EIN3-induced while 360 genes (P value 0.0099) were EIN3-repressed based on transcriptome profiling (Figure 4E, 4F). We found that 92 out of 366 EIN3-induced genes (∼25%) and 121 out of 360 EIN3-repressed genes (∼34%) were also induced and repressed by salt stress, respectively (Figure 4E, 4F). By a 5-fold cutoff, 17 out of 62 genes (∼27%) vastly up-regulated by EIN3 (EIN3ox versus Col-0) (P value 0.0095), were also highly induced by salt, including several ERFs, defense genes, and biosynthetic process and metabolism genes (Table S3). Six genes were selected to further verify the microarray data using qRT-PCR, which showed largely similar expression patterns (Figure S9). These results indicated that overexpression of EIN3 activated the expression of a number of stress-responsive defense and metabolism genes even under unstressed conditions. Therefore, the priming effect of ACC/ethylene pretreatment could be attributed to altered expression of numerous salt-responsive genes, which subsequently increases tolerance when salt stress is encountered. Further investigation on the functionality of these stress-responsive defense and metabolism genes in salt tolerance is needed to test this hypothesis.
Functional Studies of SIED Genes Identify a Novel Regulator of Salt Tolerance
To further investigate the roles of the SIED genes in salt tolerance, Salk T-DNA insertion lines of SIED genes were ordered from ABRC, and the homozygous lines of 47 insertion mutants were obtained and verified by genotyping (Table S4). Characterization of salt stress phenotype showed that, while 41 mutants were indistinguishable from wild type, 6 mutants, namely zat12, azf2, cni1, szf2, phil and SALK_067396 (hereafter designated as sied1, salt-induced and EIN3/EIL1-dependent gene 1), exhibited a salt-hypersensitivity phenotype similar to ein3 eil1, which showed low survival rate under salt stress (Figure 5A, 5B). PCR genotyping assays showed that the six mutant lines were knockout alleles in their corresponding genes (Figure S10), suggesting that these genes are positive regulators of salt tolerance.
Fig. 5. Functional characterization of SIED genes identifies a novel regulator of salt tolerance. 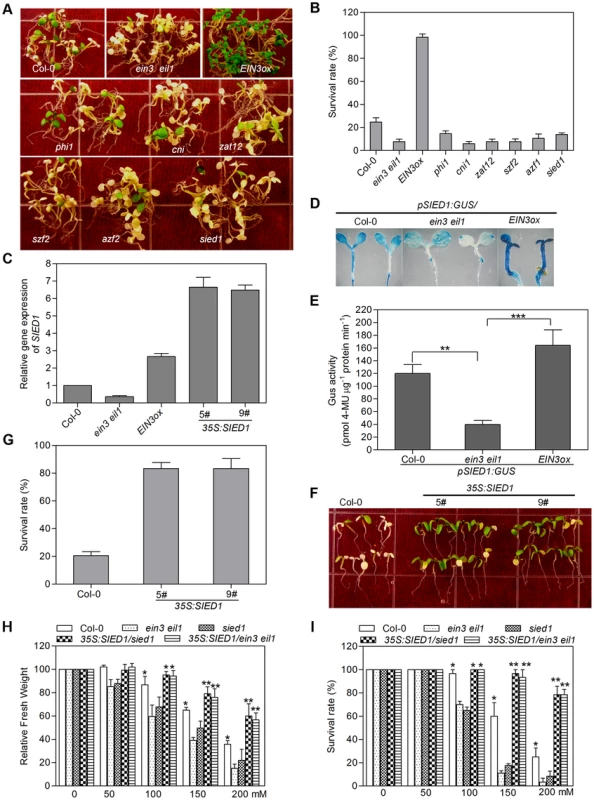
(A) Plants were grown on MS medium for 5 d and then transferred onto MS medium supplemented with 200 mM NaCl for 4 d. Experiments were repeated three times with similar results. (B) Survival rate of plants shown in (A). Values are mean ± SD from at least 50 seedlings per replicate (n = 4 replicates). (C) qRT-PCR analysis of SIED1 expression. (D) Histochemical analysis of SIED1 expression in Col-0, ein3eil1 and EIN3ox plants. (E) pSIED1:GUS activity in Col-0, ein3eil1 or EIN3ox background (Student's t test, **P<0.01 and ***P<0.001). (F) Overexpression of SIED1 in wild-type enhanced salt tolerance. Seedlings were grown on MS medium for 5 d and then transferred onto MS medium supplemented with 200 mM NaCl for 4 d. Experiments were repeated three times with similar results. (G) Survival rate of plants shown in (F). Values are mean ± SD from at least 50 seedlings per replicate (n = 3 replicates). (H) and (I) Overexpression of SIED1 in ein3eil1 or sied1 backgrounds enhanced salt tolerance. Fresh weight (H) and survival rate (I) were measured (Student's t test, **P<0.01 and ***P<0.001). Interestingly, five of these SIEDs, ZAT12 [34], [35], [36], AZF2 [37], SZF2 [38], CNI1 [39] and PHI1 [40] have been previously demonstrated to modulate various abiotic stresses. For instance, transgenic plants overexpressing ZAT12 were more tolerant to osmotic stress, while zat12 knockout mutants were more sensitive to osmotic and salt stress [34]. Overexpression of CNI1 (Carbon/Nitrogen Insensitive 1), a RING-type ubiquitin ligase, caused a hyposensitivity to C/N stress, and cni1 knockout mutants resulted in hypersensitivity to C/N stress and salt treatment [39]. Of the five genes, three genes encode zinc-finger transcription factors (ZAT12, AZF2, SZF2). In fact, we have identified at least 9 genes encoding zinc-finger transcriptional regulators as SIED (Table S1). It thus remains interesting to determine whether all other identified zinc-finger proteins are involved in salt tolerance. Together, our results supported the idea that many EIN3/EIL1 target genes identified in this analysis are involved in various stress responses, including salt stress.
The sixth salt-hypersensitivity mutant, sied1, corresponds to At5g22270, a functionally unknown gene encoding a 93-amino acid polypeptide. Microarray data and qRT-PCR analysis showed that SIED1 had evidently higher expression level in EIN3ox and lower level in ein3 eil1 compared with that of wild type (Figure 5C). To confirm its EIN3-induced expression pattern, we generated a transgenic reporter line that harbors the β-glucuronidase (GUS) gene driven by the promoter of SIED1. Consistent with the gene expression data, GUS staining was weaker in ein3 eil1 but stronger in EIN3ox than that in wild type (Figure 5D, 5E). Moreover, we found that EIN3 directly binds to the promoter of SIED1, as well as other SIED genes including ZAT12, SZF2, PHI and AZF2, but does not bind to the promoter region of CNI1 (Figure S11E–F), indicating that EIN3 selectively binds to the promoters of many SIED genes in vivo.
Since loss-of-function SIED1 mutation led to salt hypersensitivity, we next generated transgenic plants constitutively overexpressing SIED1 in wild type to further investigate its role in salt tolerance. qRT-PCR analysis revealed that higher levels of SIED1 mRNA were detected in two independent overexpression lines 5# and 9# compared with Col-0 (Figure 5C). Phenotypic analysis showed that overexpression of SIED1 effectively enhanced salt tolerance and greatly increased survival rate upon salt treatment for 4 days (Figure 5F, 5G). Since SIED1 is a direct target of EIN3, we then determined whether overexpression of SIED1 could repress the salt-hypersensitivity phenotype of ein3eil1. Toward this end, we generated the 35S:SIED1/ein3eil1 transgenic plants. Compared with ein3eil1, the seedlings of 35S:SIED1/ein3eil1 showed significantly increased survival rate and fresh weight upon salt treatment (Figure 5H, 5I), indicating that SIED1 acts genetically downstream of EIN3 and overexpression of SIED1 is sufficient to suppress the salt-hypersensitivity phenotype of ein3eil1. As expected, overexpression of SIED1 also repressed the salt-hypersensitivity phenotype of sied1 mutant (Figure 5H, 5I). Taken together, these results identify SIED1, acting downstream of EIN3, as a novel component that plays a positive role in salt tolerance.
EIN3 Induces the Transcription of Peroxidases (POD) and Increases POD Activity and Diminishes ROS Accumulation
The transcriptome profiling analysis revealed that genes encoding oxidoreductase activity are enriched in SIED, suggesting that EIN3 might modulate the oxidative/reductive status under salt stress condition. Interestingly, we found that the expression of many genes encoding peroxidases (PODs) was induced in wild type by salt treatment, whose expression was also elevated in EIN3ox but reduced in ein3eil1 under salt stress (Table S1 and Figure S12). These observations were further confirmed by qRT-PCR results with six selected POD genes (Figure 6A). In the meanwhile, we did not find evident differences in the expression levels of genes encoding superoxide dismutases (SOD), catalases (CAT1, CAT2 and CAT3) and NADPH oxidases (AtRobhA-F) among the three genotypes upon salt treatment (Figure S13A–C). These results suggest that EIN3 selectively induces the expression of genes encoding peroxidases. Peroxidase activity assay also showed that POD activity was significantly higher in EIN3ox seedlings than that in wild type (P<0.05) or ein3eil1 (P<0.01) under salt stress condition (Figure 6B).
Fig. 6. EIN3 increases activity of peroxidases through transcriptional regulation of POD genes directly. 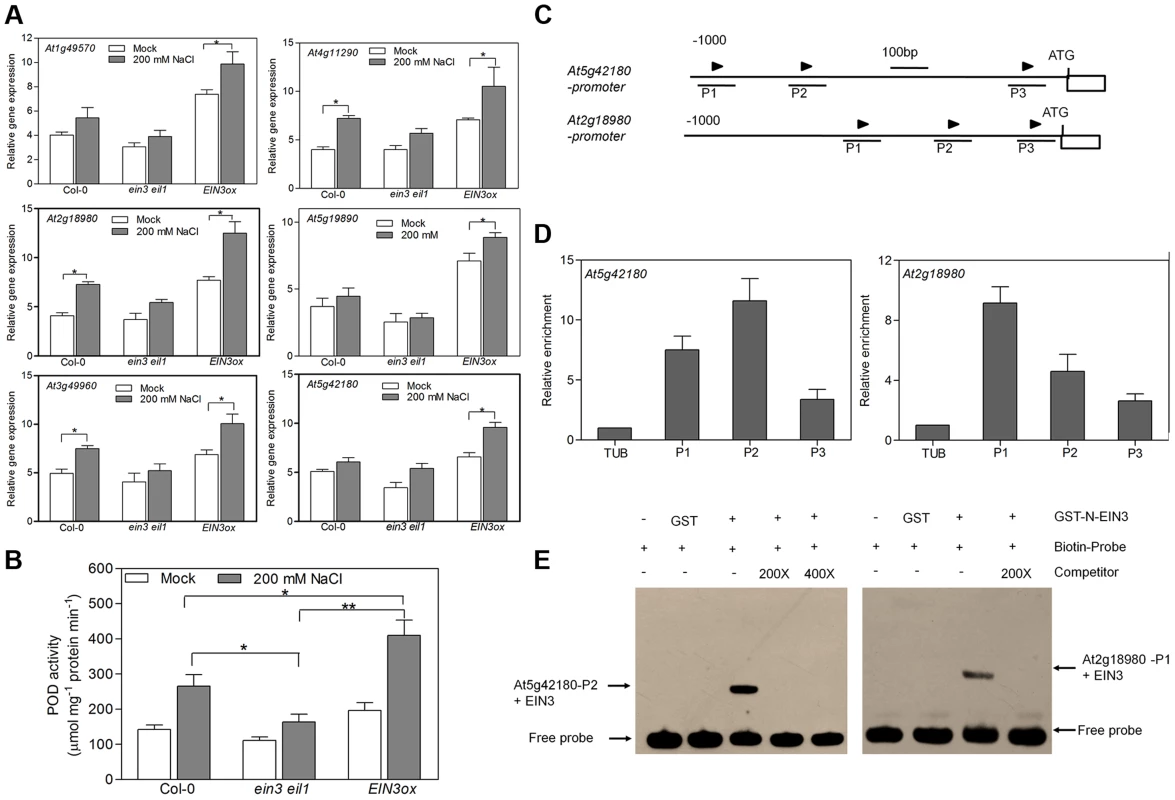
(A) qRT-PCR analysis of PODs expression. (B) Measurement of POD activity. The treated seedlings in (A) were also used (Student's t test, *P<0.05 and **P<0.01). (C) Schematic diagrams of putative EIN3 Binding Site (EBS) (arrows) in the promoters of two POD genes. The 1 kb upstream sequences are shown, and the translational start sites (ATG) are shown at position +1. (D) ChIP-qPCR assays of the promoter regions of POD genes from DNA of Col-0 seedlings with anti-EIN3 antibody. A Tubulin 8 fragment was amplified as control. Three biological replicates and two technical replicates were performed with similar results. (E) EMSA showing the interaction between the EBS containing region of POD genes and EIN3 protein. GST-tagged EIN3 N-terminus fusion protein was incubated with biotin-labeled DNA fragment. Competition for the biotin-labeled promoter region was done by adding an excess of unlabeled wild-type probe (competitor) or mutated probe (mutant competitor). Two biological replicates and two technical replicates were performed with similar results. We next analyzed the promoter regions of two selected POD genes (At5g42180 and At2g18980) and found three EBSs in each promoter (Figure 6C). Chromatin immunoprecipitation (ChIP) assay using wild-type seedlings showed that the anti-EIN3 antibody bound strongly to the P2 fragment of At5g42180 and the P1 fragment of At2g18980 (Figure 6D) respectively, suggesting that EIN3 binds directly to the promoter regions of these genes in vivo. Furthermore, the promoter sequences of At5g42180 (P2) and At2g18980 (P1) were used for electrophoresis mobility shift assay (EMSA), showing that EIN3 can bind to these promoter sequences in vitro (Figure 6E). These results indicate that EIN3 increases peroxidase activity through the direct transcriptional regulation of PODs expression.
Peroxidases have been shown to participate in plant response against abiotic stresses as key scavengers of ROS [41]. The induction of several POD genes by EIN3 suggested that activation of EIN3 might facilitate the scavenging of ROS when plants are stressed with high salinity, thus leading to enhanced salt tolerance. To test this possibility, we determined the endogenous ROS accumulation and H2O2 content of wild-type Col-0, ein3eil1 and EIN3ox upon salt treatment. Salt treatment evidently increased ROS accumulation and H2O2 content in the cotyledons of Col-0, indicated by H2DCFA fluorescence [42], [43] and DAB (3,3-diaminobenzidine) staining [44], respectively (Figure 7A–C). We further found higher level of ROS and H2O2 accumulation in ein3eil1 while lower level of ROS and H2O2 production in EIN3ox than that of Col-0 upon salt treatment (Figure 7A–C), in accordance with the salt tolerance phenotypes of these genotypes (Figure 1). Additionally, we found that upon salt treatment, a higher ROS accumulation (indicated by DAB staining and H2O2 contents) was found in sied1 mutant plants, while a significant decrease was observed in SIED1ox seedlings (Figure S14), suggesting that SIED1 acts to enhance salt tolerance also via reducing ROS accumulation.
Fig. 7. ROS accumulation in salt-treated Col-0, ein3eil1, EIN3ox and ebf2 plants. 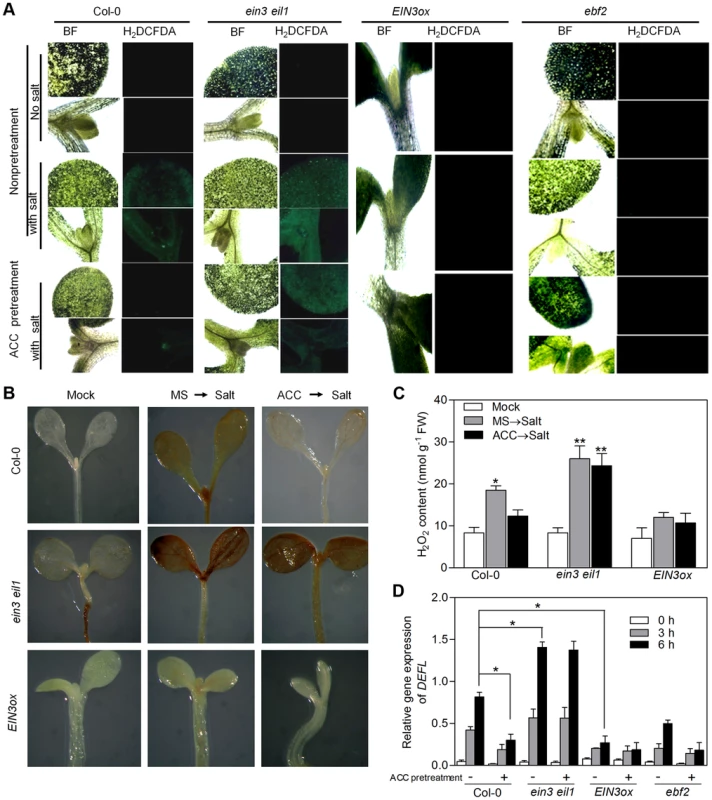
(A) Fluorescence microscopy images of ROS (indicated by H2DCFDA fluorescence). Seedlings grown on MS medium supplemented with or without 10 µM ACC for 5 d were subjected to salt treatment. BF: bright field. Experiments were repeated three times with similar results. (B) DAB staining of seedlings under normal conditions or salt treatment. Seedlings grown on MS medium supplemented with or without 10 µM ACC for 5 d were treated with 200 mM NaCl for 6 h, and used for DAB staining. (C) H2O2 content in the seedlings in (B) (Student's t test, *P<0.05 and **P<0.01). (D) qRT-PCR analysis of DEFL expression (a ROS marker gene) (Student's t test, *P<0.05 and **P<0.01). Since ACC-pretreated plants exhibited tolerance to salt stress, we next examined the effect of ACC pretreatment on ROS accumulation and H2O2 production in salt-treated plants. Compared with non-pretreated seedlings, ACC-pretreated Col-0 showed evident lower levels of ROS accumulation and H2O2 generation when stressed with high salinity (Figure 7A–C). Meanwhile, ROS level and H2O2 content remained constantly high in ein3eil1 and low in EIN3ox, and no obvious changes were found in these two genotypes upon ACC pretreatment (Figure 7A–C). These results are also in good correlation with their respective salt stress phenotypes with or without ACC pretreatment (Figure 1). We next examined the expression levels of a well-established ROS marker gene, DEFL (defensin-like), which was shown to be induced by various ROS [45]. In agreement with the histochemical observations of ROS accumulation, ACC-pretreated Col-0 seedlings accumulated less DEFL mRNA than non-pretreated Col-0 upon 3 h and 6 h of salt applications (Figure 7D). By contrast, DEFL expression was highly induced by salt stress in ein3 eil1 regardless of ACC pretreatment, whereas its expression levels remained constantly lower in salt-stressed EIN3ox and ebf2-1 than that in Col-0 (Figure 7D), further supporting the importance of EIN3 action in the modulation of salt-evoked ROS accumulation.
Taken together, our results indicate that activation of EIN3, either by EIN3 overexpression, ACC pretreatment, or EBF2 mutation, induces the expression of numerous POD and SIED genes, which arguably contributes to the decreased accumulation of ROS, and consequently the detoxification of salt stress-elicited damages.
Discussion
Salt Stress Stabilizes EIN3/EIL1 Proteins and Destabilizes EBF1/EBF2 Proteins in Both EIN2-Dependent and EIN2-Independent Manners
Genetic and biochemical studies revealed that EIN3/EIL1 proteolysis is mediated by two F-box proteins, EBF1/EBF2. Upon ethylene treatment, the levels of EBF1/EBF2 proteins are down-regulated through 26S proteasome pathway, which leads to the accumulation of EIN3 and EIL1 proteins [16], [18], [19], [20]. Moreover, our previous work indicated that EIN2 is indispensable for ethylene-induced EIN3 and EIL1 stabilization and degradation of EBF1/EBF2 proteins, because no EIN3 or EIL1 protein can be detected in ein2 but EBF1/EBF2 proteins are constitutively accumulated in the ein2 background [18]. In this study, we found that the level of EIN3 and EIL1 proteins was remarkably up-regulated by salt treatment in wild type, and also elevated in ein2-5 background, although to a lesser extent. Conversely, we further found that the levels of EBF1/EBF2 proteins were evidently down-regulated by salt treatment in both Col-0 and ein2-5 background, which is the result of 26S proteasome-executed EBF1/EBF2 degradation. These findings indicate that EIN2 is dispensable for salt-induced EIN3/EIL1 accumulation and EBF1/EBF2 degradation, which is distinct from the regulatory mechanism in ethylene signaling that fully depends on EIN2. Thus, we propose that salt treatment promotes EBF1/EBF2 protein degradation, which consequently induces EIN3 protein accumulation in both EIN2-dependent and EIN2-independent pathways.
These findings for the first time report the existence of an alternative pathway that is distinct from the canonical ethylene signaling pathway to modulate the protein stability of EBF1/EBF2 and EIN3/EIL1. In addition to NaCl, we found that treatments with equal concentration of KCl, NaNO3 or KNO3 salt also effectively induced EBF2-GFP protein degradation in ein2-5 mutant (Figure S7A), suggesting that this regulation is a general salt stress response rather than the specific effect caused by sodium chloride. High salt condition often affects osmotic homeostasis and causes osmotic stresses. However, treatment with 200 mM mannitol did not affect EBF2-GFP protein level, suggesting that the ionic stress but not osmotic stress regulates EBF1/EBF2 protein turnover (Figure S7B). Further investigation is needed to elucidate the regulatory mechanism behind the salt-induced proteasomal degradation of EBF1/EBF2.
In this study, we demonstrate that the protein stability of EBF1/EBF2 and EIN3/EIL1 is regulated by ethylene and salt stress in different ways. Recently, increasing body of evidence indicates that EIN3/EIL1 might act as a signaling hub that integrates multiple hormone and stress signals [18], [31], [43], [46], [47], [48], [49], [50]. Our previous works and other studies reported that EIN3 protein stability is also controlled by light irradiation [43], auxin and its biosynthesis inhibitor, Kyn [28], and glucose [51]. These studies collectively indicate that EIN3/EIL1 are not limited to the ethylene signaling, but rather participating in the regulation of myriad processes, whose function and/or abundance are also modulated by various signals besides the ethylene gas. Given that only a subset of abiotic stresses (e.g. high salt, high sucrose, freezing, but not osmosis) alter the stability of EIN3 or EBF proteins, it is thus interesting to ascertain whether, and if so, how other types of abiotic stresses, such as drought, chilling, heat, and heavy metals, affect the stabilization of EBF1/EBF2 and EIN3/EIL1 proteins. The elucidation of differential regulation of EIN3 and/or EBF protein stability by environmental and stress signals would provide insights into how EIN3 exerts its unique effect in plant adaptation to various growth conditions.
EIN3 Enhances Salt Tolerance by Decreasing ROS Accumulation
Previous studies revealed a number of salt-tolerance genes that might be EIN3 target genes, such as ESE1 (an AP2/EREBP transcription factor) [52] and JERF3 [53]. However, a systematic analysis of EIN3-regulated genes in salt tolerance is lacking. In this study, we conducted a genome-wide transcriptome profiling in combination with genetic approach to dissect and identify numerous EIN3-regulated genes and pathways that might contribute to ethylene-directed salt tolerance (Table S1).
One of the pathways is the scavengers of ROS, in which EIN3 up-regulates the expression of numerous peroxidases. One important cause of high salinity-imposed damage is ROS generated by salt stress. ROS plays a dual role in plants, as actors or modulators of cellular signaling pathways on one hand, and as oxidative agents or toxic products elicited by cellular stresses on the other hand [54]. ROS is tightly regulated by the equilibrium between production and scavenging. Transgenic plants overexpressing enzymes involved in oxidative protection, such as glutathione peroxidase (GPX) [55], superoxide dismutase (SOD) [56], ascorbate peroxidase (APX) [57], exhibited enhanced salt tolerance. In this study, based on the transcriptome analysis, we found that the expression of several ROS scavenger peroxidases (PODs) was notably elevated in EIN3ox under salt condition compared with that in wild type or ein3 eil1 (Table S1 and Figure 6). Accordingly, the overall activity of POD enzymes was significantly higher in EIN3ox than that in wild type or ein3 eil1 upon salt stress (Figure 6B). The transcript level of a zinc-finger transcription factor, ZAT12, which has been shown to induce the expression of APX1 [35], was also up-regulated in EIN3ox (Table S1). Consistently, we observed lower levels of H2O2 accumulation and ROS marker gene expression in EIN3ox but higher levels in ein3 eil1 compared with wild type upon salt treatment (Figure 7). Thus, elimination of excessive ROS accumulation under salt stress through inducing the expression of peroxidases is one contributing mechanism behind EIN3-mediated salt tolerance.
In addition, we found that EIN3/EIL1 enhanced salt tolerance through regulating a myriad of SIED genes. We provided genetic evidence to indicate that a portion of these SIED genes participate in salt tolerance, including 5 genes previously known to be induced by and/or involved in various abiotic stresses, and a novel gene (SIED1) whose function is previously unknown. Further biochemical studies uncovered that 5 of these 6 SIED genes, including SIED1, are direct target genes of EIN3. We found that overexpression of SIED1 also decreased ROS accumulation upon salt treatment (Figure S14). Nevertheless, the lack of salt stress phenotype for other SIED knockout mutants does not necessarily mean that these genes are not involved in salt tolerance. For instance, it is well known that ERF family transcription factors and JAZ family transcriptional regulators possess tremendous functional redundancy within the family members [31], [58], [59]. In addition, we cannot completely rule out the possibility that the lack of salt stress phenotype is simply because the T-DNA insertions in many SIED genes may have a marginal effect on gene expression/function (Table S4). Additional mutant alleles, gain-of-function studies or multigenic mutants analysis would help clarify whether those SIED genes play a role in EIN3-induced salt tolerance. Based on our genomic and genetic studies, the identification of numerous SIED and SRED genes would thus serve as a proper starting point to further dissect the complicated signaling network that is directed by EIN3/EIL1 in plant adaptation to salt stress.
Intracellular K+/Na+ homeostasis is crucial for cell metabolism and is considered to be a key component of salinity tolerance in plants. Jiang et al. recently reported that salinity-induced ethylene is a potent promoter of salt tolerance through enhancing Na+/K+ homeostasis in Arabidopsis [25]. In fact, we also found that salt-stressed EIN3ox seedlings had accumulated more K+ and less Na+, whereas ein3-1 eil1-1 seedlings had higher Na+ but lower K+ content compared with wild type (data not shown). These results indicate that the modulation of intracellular K+/Na+ homeostasis serves as another contributing mechanism for ethylene-induced tolerance to high salt stress.
In summary, our study provides new insights into how ethylene enhances plants' tolerance to high salinity. We propose that EIN3 and EIL1 play a central role in conferring salt tolerance via at least two mechanisms, one is to modulate intracellular K+/Na+ homeostasis, the other is to deter ROS accumulation by inducing SIEDs and PODs gene expression. Our study also provides a possible explanation for the priming effect of ethylene pretreatment, as ethylene treatment decreases EBF1/2 stability and increases EIN3/EIL1 abundance, which enhances the sensitivity of this pathway to salt stress signal. Once the EIN3 pathway is activated in advance (by ethylene/ACC), it will alter the expression of downstream SIED genes, many of which are direct target genes of EIN3, and induce a myriad of defense pathways, and switch plants to a more resistant state.
Materials and Methods
Plant Material, Growth Conditions and Salt Stress Experiments
Arabidopsis thaliana ecotype Col-0 was the parent strain for all mutants and transgenic lines used in this study. Surface-sterilized seeds were plated on MS medium supplemented with or without 10 µm ACC and imbibed for 4 d in 4°C to improve germination uniformity. For phenotypic analysis under salt stress, seedlings were transferred onto MS agar plates containing 200 mM NaCl and their subsequent appearance was recorded photographically 3 days after transfer. The salt stress of seedlings was indicated by visibly bleached leaves.
Measurements of ROS, Relative Electrolyte Leakage and Chlorophyll Content
Reactive oxygen species (ROS) accumulation in seedlings was detected using the cell-permeable fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH2-DA; Molecular Probes) according to Schopher et al. [42]. 5-day-old seedlings grown on MS with or without 10 µM ACC were treated with 200 mM NaCl for 6 h. Then, seedling were incubated in 100 µM DCFH2-DA in 1% ethanol for 20 min, and washed with distilled H2O to remove the dye before the observation of ROS accumulation under the confocal microscope. Confocal images were obtained after excitation at 488 nm and emission at 522 nm. H2O2 production was detected in seedlings using 3, 3-diaminobenzidine (DAB) as substrate. Relative electrolyte leakage and chlorophyll content were measured as described previously [60].
Assay for GUS Activity
Histochemical and Fluorimetric GUS assays were performed using the method described by Jefferson [61].
Confocal Laser Microscopy
A Leica TCS SP2 inverted confocal laser microscope with ×40 objectives was used to detect GFP fluorescence. The excitation wavelength was 488 nm, and a bandpath filter of 510 to 525 nm was used for emission.
Protein Extract and Western Blotting
Plant samples were ground in liquid N2 and soluble protein extracts were made by homogenization in 50 mM Tris–HCl (pH 8.0), 10 mM NaCl, 0.1 M PMSF, and 0.1 M DTT, with subsequent centrifugation at 13.000× g for 30 min at 4°C. The protein in the supernatants was quantified by Bradford's assay (Bradford, 1976). Western blot analysis was performed as described previously [18], [60] with anti-GFP (Invitrogen), anti-FLAG (Sigma-Aldrich), anti-MYC (AbChem), or anti-EIN3 antibodies [16].
qRT-PCR Assay
Total RNA was extracted from seedlings and analyzed as described previously [62]. First strand cDNA samples were generated from total RNA samples by reverse transcription using an AMV reverse transcriptase 1st strand cDNA synthesis kit (Life Sciences, Promega) and were used as templates for qPCR-based gene expression analysis as described previously [60]. The oligonucleotide primer sequences used to amplify specific cDNAs were described in Table S5.
Whole-Genome Transcriptome Profiling Analysis
Microarray experiments were performed using Arabidopsis Affymetrix chips (Santa Clara, CA). Total RNA was extracted from 5-d-old post germinated seedlings grown on MS medium, which were subsequently subjected to 200 mM NaCl for 6 h. Each experiment used two biological replicates, and each represented a pool of around 200 seedlings from two individual plates. The expression data were analyzed using Gene Spring version 4.2.1 (Silicon Genetics Inc., Red - wood City, CA, USA). A q value<0.05 and fold change >2 between control and treatment samples were considered as a cutoff.
Gene Ontology (GO) Enrichment Analysis
GO enrichment analysis on SIED genes was performed using the software GOrilla (Gene Ontology enRIchmentanaLysis and visuaLizAtion tool) as described previously [33].
ChIP and EMSA Assays
10 g of 5-d-old Col-0 seedlings were prepared for ChIP assays using anti-EIN3 antibody [16], and the enriched DNA fragments were measured by qPCR as previously described [60]. All assays were performed with two biological replicates and three technical replicates. EMSA assay was performed as described previously [60]. The N-terminus DNA binding domain of EIN3 protein (amino acids 141 to 352) that is sufficient for DNA binding [31] was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli and purified, and used for EMSA experiments.
Generation of Transgenic Plants
Plants overexpressing SIED1 were generated by Agrobacterium tumefaciens strain GV3101 -mediated transformation into Arabidopsis Col-0 by floral dip [63], using a construct that contained the full-length coding region of SIED1 (At5g22270) in the PBI121vector. To generate pSIED1:GUS construct, a 2.2-kb SIED1 promoter region was amplified from genomic DNA and inserted into PBI101 vector, introduced into GV3101, and transformed into Col-0, ein3 eil1 and EIN3ox plants [63]. To generate EIL1pro:EIL1-GFP construct, the promoter region and the full-length coding region of EIL1 was amplified from genomic DNA and inserted into pCHF3 vector, introduced into GV3101, and transformed into wild-type Col-0 and ein2-5 plants.
Statistical Analysis
The values we obtained in the figures were expressed as the means (SD). Two-tailed Student's t tests were used.
Accession Numbers
The microarray data reported in this paper have been deposited at NASC (The Nottingham Arabidopsis Stock Centre) database under accession number NASCARRAYS-659.
Supporting Information
Zdroje
1. ApseMP, AharonGS, SneddenWA, BlumwaldE (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 : 1256–1258.
2. BradfordKJ, YangSF (1980) Stress-induced Ethylene Production in the Ethylene-requiring Tomato Mutant Diageotropica. Plant Physiol 65 : 327–330.
3. BleeckerAB, KendeH (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16 : 1–18.
4. EckerJR (1995) The ethylene signal transduction pathway in plants. Science 268 : 667–675.
5. ChenYF, EtheridgeN, SchallerGE (2005) Ethylene signal transduction. Ann Bot 95 : 901–915.
6. ChenYF, RandlettMD, FindellJL, SchallerGE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277 : 19861–19866.
7. ChangC, KwokSF, BleeckerAB, MeyerowitzEM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262 : 539–544.
8. HuaJ, ChangC, SunQ, MeyerowitzEM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269 : 1712–1714.
9. HuaJ, MeyerowitzEM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94 : 261–271.
10. KieberJJ, RothenbergM, RomanG, FeldmannKA, EckerJR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72 : 427–441.
11. AlonsoJM, HirayamaT, RomanG, NourizadehS, EckerJR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 : 2148–2152.
12. BissonMM, BleckmannA, AllekotteS, GrothG (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424 : 1–6.
13. JuC, YoonGM, ShemanskyJM, LinDY, YingZI, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci U S A 109 : 19486–19491.
14. QiaoH, ShenZ, HuangSS, SchmitzRJ, UrichMA, et al. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338 : 390–393.
15. WenX, ZhangC, JiY, ZhaoQ, HeW, et al. (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22 : 1613–1616.
16. GuoH, EckerJR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 : 667–677.
17. SolanoR, StepanovaA, ChaoQ, EckerJR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12 : 3703–3714.
18. AnF, ZhaoQ, JiY, LiW, JiangZ, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22 : 2384–2401.
19. GagneJM, SmalleJ, GingerichDJ, WalkerJM, YooSD, et al. (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci U S A 101 : 6803–6808.
20. PotuschakT, LechnerE, ParmentierY, YanagisawaS, GravaS, et al. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115 : 679–689.
21. van LoonLC, GeraatsBP, LinthorstHJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11 : 184–191.
22. AchardP, ChengH, De GrauweL, DecatJ, SchouttetenH, et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311 : 91–94.
23. CaoWH, LiuJ, HeXJ, MuRL, ZhouHL, et al. (2007) Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol 143 : 707–719.
24. LeiG, ShenM, LiZG, ZhangB, DuanKX, et al. (2011) EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ 34 : 1678–1692.
25. JiangC, BelfieldEJ, CaoY, SmithJA, HarberdNP (2013) An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25 : 3535–3552.
26. VersluesPE, AgarwalM, Katiyar-AgarwalS, ZhuJ, ZhuJK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45 : 523–539.
27. StepanovaAN, YunJ, LikhachevaAV, AlonsoJM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19 : 2169–2185.
28. HeW, BrumosJ, LiH, JiY, KeM, et al. (2011) A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23 : 3944–3960.
29. IsmailA, RiemannM, NickP (2012) The jasmonate pathway mediates salt tolerance in grapevines. J Exp Bot 63 : 2127–2139.
30. ZhangH, HuangZ, XieB, ChenQ, TianX, et al. (2004) The ethylene-, jasmonate-, abscisic acid - and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220 : 262–270.
31. ZhuZ, AnF, FengY, LiP, XueL, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci U S A 108 : 12539–12544.
32. ChangKN, ZhongS, WeirauchMT, HonG, PelizzolaM, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife 2: e00675.
33. EdenE, NavonR, SteinfeldI, LipsonD, YakhiniZ (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10 : 48.
34. DavletovaS, SchlauchK, CoutuJ, MittlerR (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139 : 847–856.
35. RizhskyL, DavletovaS, LiangH, MittlerR (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279 : 11736–11743.
36. VogelJT, ZarkaDG, Van BuskirkHA, FowlerSG, ThomashowMF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41 : 195–211.
37. SakamotoH, MaruyamaK, SakumaY, MeshiT, IwabuchiM, et al. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136 : 2734–2746.
38. SunJ, JiangH, XuY, LiH, WuX, et al. (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48 : 1148–1158.
39. SatoT, MaekawaS, YasudaS, SonodaY, KatohE, et al. (2009) CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J 60 : 852–864.
40. KrepsJA, WuY, ChangHS, ZhuT, WangX, et al. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130 : 2129–2141.
41. HiragaS, YamamotoK, ItoH, SasakiK, MatsuiH, et al. (2000) Diverse expression profiles of 21 rice peroxidase genes. FEBS Lett 471 : 245–250.
42. SchopferP, PlachyC, FrahryG (2001) Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid. Plant Physiol 125 : 1591–1602.
43. ZhongS, ZhaoM, ShiT, ShiH, AnF, et al. (2009) EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci U S A 106 : 21431–21436.
44. ZhouF, AndersenCH, BurhenneK, FischerPH, CollingeDB, et al. (2000) Proton extrusion is an essential signalling component in the HR of epidermal single cells in the barley-powdery mildew interaction. Plant J 23 : 245–254.
45. GadjevI, VanderauweraS, GechevTS, LaloiC, MinkovIN, et al. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 : 436–445.
46. ShiY, TianS, HouL, HuangX, ZhangX, et al. (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24 : 2578–2595.
47. ChenH, XueL, ChintamananiS, GermainH, LinH, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21 : 2527–2540.
48. LingamS, MohrbacherJ, BrumbarovaT, PotuschakT, Fink-StraubeC, et al. (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23 : 1815–1829.
49. ZhaoQ, GuoHW (2011) Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol Plant 4 : 626–634.
50. LiuZ, WuY, YangF, ZhangY, ChenS, et al. (2013) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A 110 : 6205–6210.
51. YanagisawaS, YooSD, SheenJ (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425 : 521–525.
52. ZhangL, LiZ, QuanR, LiG, WangR, et al. (2011) An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol 157 : 854–865.
53. WuL, ZhangZ, ZhangH, WangXC, HuangR (2008) Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol 148 : 1953–1963.
54. ApelK, HirtH (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55 : 373–399.
55. RoxasVP, LodhiSA, GarrettDK, MahanJR, AllenRD (2000) Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol 41 : 1229–1234.
56. TsengMJ, LiuCW, YiuJC (2007) Enhanced tolerance to sulfur dioxide and salt stress of transgenic Chinese cabbage plants expressing both superoxide dismutase and catalase in chloroplasts. Plant Physiol Biochem 45 : 822–833.
57. BadawiGH, KawanoN, YamauchiY, ShimadaE, SasakiR, et al. (2004) Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol Plant 121 : 231–238.
58. De BoerK, TillemanS, PauwelsL, Vanden BosscheR, De SutterV, et al. (2011) APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J 66 : 1053–1065.
59. ShyuC, FigueroaP, DepewCL, CookeTF, SheardLB, et al. (2012) JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24 : 536–550.
60. LiZ, PengJ, WenX, GuoH (2013) Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25 : 3311–3328.
61. JeffersonRA, KavanaghTA, BevanMW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 : 3901–3907.
62. PengJY, LiZH, XiangH, HuangJH, JiaSH, et al. (2005) Preliminary studies on differential defense responses induced during plant communication. Cell Res 15 : 187–192.
63. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
Štítky
Genetika Reprodukční medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



