-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
Predicting organismal phenotypes from genotype data is important for plant and animal breeding, medicine, and evolutionary biology. Genomic-based phenotype prediction has been applied for single-nucleotide polymorphism (SNP) genotyping platforms, but not using complete genome sequences. Here, we report genomic prediction for starvation stress resistance and startle response in Drosophila melanogaster, using ∼2.5 million SNPs determined by sequencing the Drosophila Genetic Reference Panel population of inbred lines. We constructed a genomic relationship matrix from the SNP data and used it in a genomic best linear unbiased prediction (GBLUP) model. We assessed predictive ability as the correlation between predicted genetic values and observed phenotypes by cross-validation, and found a predictive ability of 0.239±0.008 (0.230±0.012) for starvation resistance (startle response). The predictive ability of BayesB, a Bayesian method with internal SNP selection, was not greater than GBLUP. Selection of the 5% SNPs with either the highest absolute effect or variance explained did not improve predictive ability. Predictive ability decreased only when fewer than 150,000 SNPs were used to construct the genomic relationship matrix. We hypothesize that predictive power in this population stems from the SNP–based modeling of the subtle relationship structure caused by long-range linkage disequilibrium and not from population structure or SNPs in linkage disequilibrium with causal variants. We discuss the implications of these results for genomic prediction in other organisms.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002685
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002685Summary
Predicting organismal phenotypes from genotype data is important for plant and animal breeding, medicine, and evolutionary biology. Genomic-based phenotype prediction has been applied for single-nucleotide polymorphism (SNP) genotyping platforms, but not using complete genome sequences. Here, we report genomic prediction for starvation stress resistance and startle response in Drosophila melanogaster, using ∼2.5 million SNPs determined by sequencing the Drosophila Genetic Reference Panel population of inbred lines. We constructed a genomic relationship matrix from the SNP data and used it in a genomic best linear unbiased prediction (GBLUP) model. We assessed predictive ability as the correlation between predicted genetic values and observed phenotypes by cross-validation, and found a predictive ability of 0.239±0.008 (0.230±0.012) for starvation resistance (startle response). The predictive ability of BayesB, a Bayesian method with internal SNP selection, was not greater than GBLUP. Selection of the 5% SNPs with either the highest absolute effect or variance explained did not improve predictive ability. Predictive ability decreased only when fewer than 150,000 SNPs were used to construct the genomic relationship matrix. We hypothesize that predictive power in this population stems from the SNP–based modeling of the subtle relationship structure caused by long-range linkage disequilibrium and not from population structure or SNPs in linkage disequilibrium with causal variants. We discuss the implications of these results for genomic prediction in other organisms.
Introduction
Most efforts to understand the genetic architecture of quantitative traits have focused on mapping the variants causing phenotypic variation in quantitative trait locus (QTL) mapping populations derived from crosses between lines genetically divergent for the trait, or in association mapping populations, with the goal of understanding the biological underpinnings of trait variation [1]. However, the ability to accurately predict quantitative trait phenotypes from information on genotypic variation in the absence of knowledge of causal variants will revolutionize evolutionary biology, medicine and human biology, and breeding of agriculturally important plant and animal species. The premise of personalized medicine is based on prediction of individual genetic risk to disease from genome-wide association studies [2], [3], and the ability to select individuals or lines in animal and plant breeding programs based on genotypic information circumvents the costly process of progeny testing and reduces the generation interval in applied breeding programs, leading to greater efficiency [4], [5].
In classical animal and plant breeding, the genetic quality of individuals or lines is predicted from phenotypic values of selection candidates and their relatives. The widely used Best Linear Unbiased Prediction (BLUP, [6]) method models the covariance structures between individuals via the numerator relationship matrix, which is constructed from known pedigree information and thus reflects expected relationships between individuals (i.e. the proportion of shared alleles of identical ancestral origin) given the pedigree. The advent of high-throughput genotyping platforms for many agronomic species [7] enabled genotyping large numbers of individuals for dense panels of single nucleotide polymorphisms (SNPs) spanning the genome. The expected, pedigree-based numerator relationship matrix can then be replaced by a realized, genome-based relationship matrix (often called the “genomic” relationship matrix, [8]). This approach is equivalent to a random regression approach in which all SNP genotypes are simultaneously accounted for as explanatory variables in a multiple regression model [9]. In animal and plant breeding, selection based on genome-based predictions of genetic values is expected to massively increase genetic progress [4], [10] and has quickly found its way into widespread practical application (see [4], [5] for reviews).
Genome based-prediction follows a different paradigm than genome wide association studies (GWAS). GWAS identify single molecular variants associated with phenotypic variability using individual statistical tests for significance of each variant. Genome-based prediction uses the entire genomic variability captured by the available marker set to explain the observed phenotypic variation, and does not rely on selection of single loci based on significance tests. Standard prediction methods are thought to work for traits with a highly polygenic or even infinitesimal [11] genetic architecture, where the effect of a single variant is too small to be captured by a statistical test in a GWAS. There is strong empirical evidence that many quantitative traits have such a highly polygenic genetic architecture in farm animals [12], agriculturally used plants [13], model organisms and humans [14], [15].
With the advent of next generation sequencing technologies, it is now feasible to implement genomic prediction based on complete genome sequences of higher organisms. While these techniques have only been applied to individuals or cohorts of limited size [16] to date, initiatives to sequence larger panels are under way [17], [18], and genotyping by whole genome resequencing will become a standard technology in the foreseeable future.
The accuracy of prediction methods based on marker data depends on the heritability of the trait, its genetic architecture (number of loci affecting trait variation, mode of inheritance, and distribution of allelic effects, [19]), the LD reflecting effective population size, the size of the genome, the marker density and the sample size used in the statistical analysis [20]. Various methods of prediction incorporating genomic information have been studied on real and simulated data, including Genomic Best Linear Unbiased Prediction (GBLUP) approaches with genomic relationship matrices [8], Random Regression BLUP (RRBLUP), Bayesian linear regression methods [10], [21] or fully non-parametric approaches [22]–[25].
GBLUP approaches are based on a linear model for the phenotypic values, which encompasses a vector of random genetic values of individuals whose covariance structure is inferred from genomic data. The linear model underlying the RRBLUP approach includes a vector of random marker effects (instead of a vector of genetic values) which are assumed to be drawn from the same normal distribution and uncorrelated. The model primarily provides estimates of SNP effects, but estimated genetic values of individuals can be derived as linear combinations of the estimated SNP effects, yielding the same predictions of individual genotypic or phenotypic values as GBLUP. The BayesB method [10], on the other hand, fits only a small fraction of the available markers to conform with the assumption that most loci are expected to have zero effect on the phenotype, and the remaining non-zero marker effects are drawn from normal distributions with random variances.
It has been suggested [26] that differences between prediction methods will become more pronounced with the availability of full genome sequence data. According to a study with simulated data [26], RRBLUP and equivalent GBLUP procedures do not take full advantage of high-density marker data if the number of causal SNPs is small, while approaches with an implicit feature selection such as BayesB might be more accurate. If, on the other hand, the number of causal loci is large, RRBLUP or GBLUP methods may yield accurate predictions because the assumption that every SNP has an effect is closer to reality.
Implementing genomic prediction with full genome sequence data raises a number of questions. What is the most efficient way to incorporate the complete genomic information in prediction? How much predictive ability is gained by using whole genome sequence data compared to high density SNP panels? Is it possible to increase predictive ability by a pre-selection of SNPs or models with an internal feature selection? How comparable are the results of genomic prediction and genome wide association? Here, we address these questions empirically based on full genomic sequences of a population of Drosophila melanogaster inbred lines. The inbred lines have been sequenced, and constitute the Drosophila Genetics Reference Panel (DGRP), a new community resource for genetic studies of complex traits [27].
We report the results of a full sequence based genomic prediction for two quantitative traits, starvation stress resistance and locomotor startle response, both of which display considerable genetic variation in natural populations and respond rapidly to artificial selection [28]–[30]. We used whole-genome sequences determined on the Illumina platform for DGRP-lines for starvation resistance (startle response) [27]. Our reference method is a GBLUP approach in which ∼2.5 million polymorphic SNPs are used to derive a genomic relationship matrix [8]. We evaluated predictive ability via cross-validation (CV), and compared prediction within vs. across sexes, various SNP densities, and training set sizes. We assessed whether BayesB is superior over GBLUP given full genome sequence data [26], and compared our genomic prediction results with those of GWAS conducted on the same DGRP lines [27].
To our knowledge, this is the first application of genomic prediction on empirical whole genome sequence in a substantial sample of a higher organism. However, this study, as well as all previous association studies, only assesses the effects of common SNPs, since the effects of rare alleles cannot be estimated due to the small sample of sequenced lines. The results illustrate both the potential of the approach and challenges to be addressed in the future.
Results
Genomic Best Linear Unbiased Prediction (GBLUP)
We constructed a genomic relationship matrix [8] from ∼2.5 million SNPs for which the minor allele was present in at least four of the DGRP lines [27]. A histogram of the off-diagonal elements of this matrix for DGRP lines used in the GBLUP analyses (Figure 1) and a corresponding heatmap (Figure 2) show that there were no large blocks of high genomic relationship among the lines. The average genomic relationship is close to zero, as expected, but there is considerable variance around this average (Figure 1), as indicated by two block of lines with average genomic relationships within each block of and (Figure 2). We performed genomic prediction for starvation stress resistance and locomotor startle response. The phenotypes used were the medians of many () individually tested males and females for each line, or the average of the male and female medians (Table S1). We used several cross-validation (CV) procedures for each trait (Table 1). In the -fold CV, predictive ability was for starvation resistance and for startle response. In human studies the efficiency of a predictor is reported as the squared correlation rather than [31], so that in terms of variance explained the estimates were for starvation resistance and for startle response. The observed accuracy depends on the size of the training set (Figure 3), with decreasing accuracies obtained with smaller training sets. Predictive abilities are roughly halved for both traits when using only instead of of the data to train the model. Maximum likelihood estimates of narrow-sense heritabilities based on the GBLUP model using the genomic relationship matrix were in all analyses (Table S2), reflecting the fact that phenotypes are averages over many replicates and thus residual variance is minimal. Hence, the phenotypes used represent the line genotypes with maximum accuracy, which is the ideal case for training the genomic model.
Fig. 1. Histogram of the offdiagonal elements of the genomic relationship matrix . 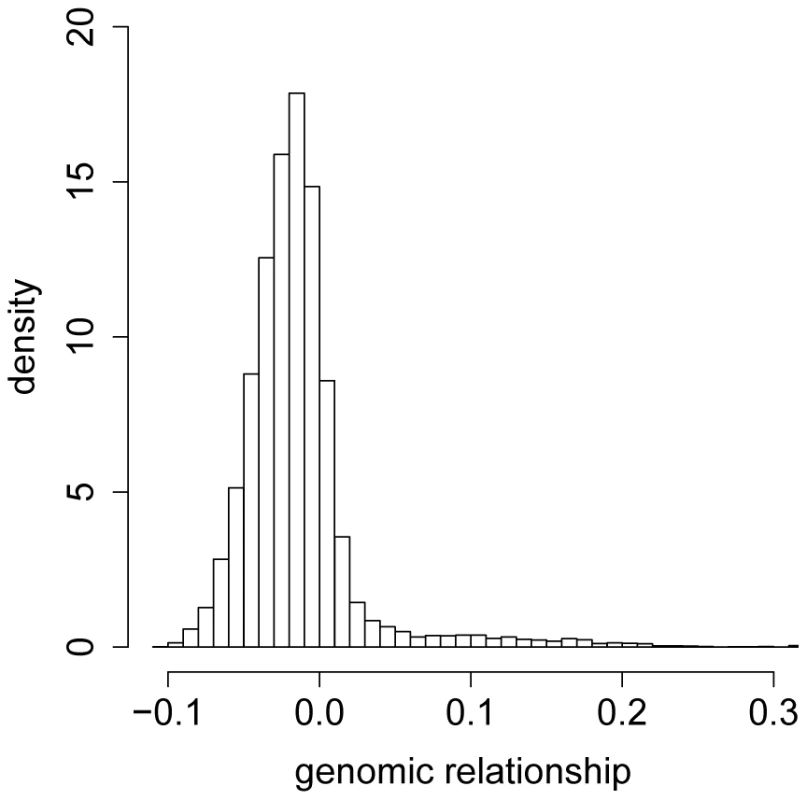
The genomic relationship matrix was calculated according to [8] using 157 lines and 2.5 million SNPs. Fig. 2. Heatmap of the genomic relationship matrix . 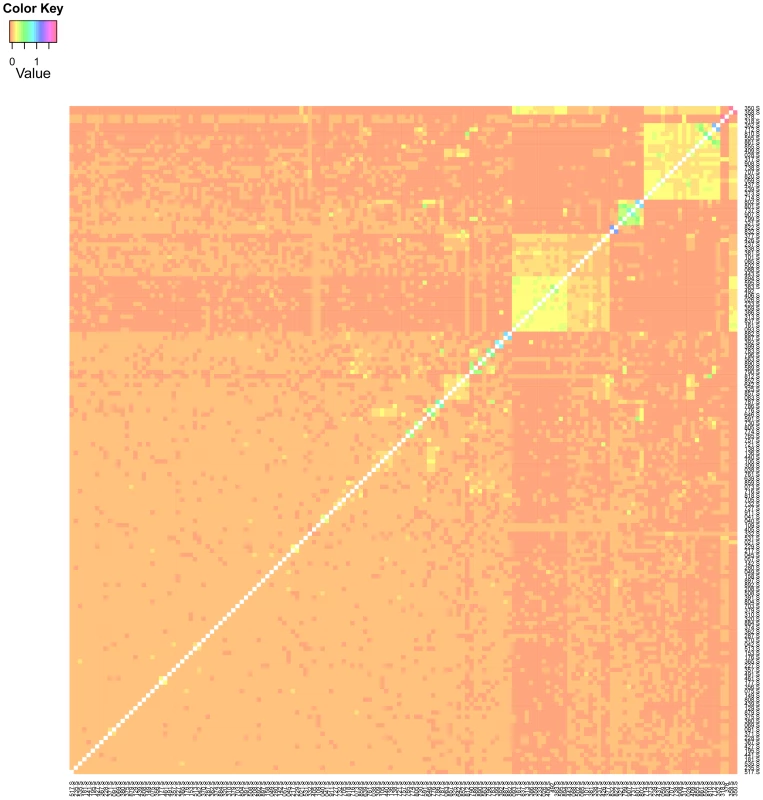
The genomic relationship matrix was calculated according to [8] using 157 lines and 2.5 million SNPs. The “S” after the line-ID indicates that the line belongs to the set of lines for which phenotypic records for startle response were also available (in addition to the phenotypic records of starvation resistance). Fig. 3. Accuracy of prediction of GBLUP for CVs with different numbers of lines in the training set. 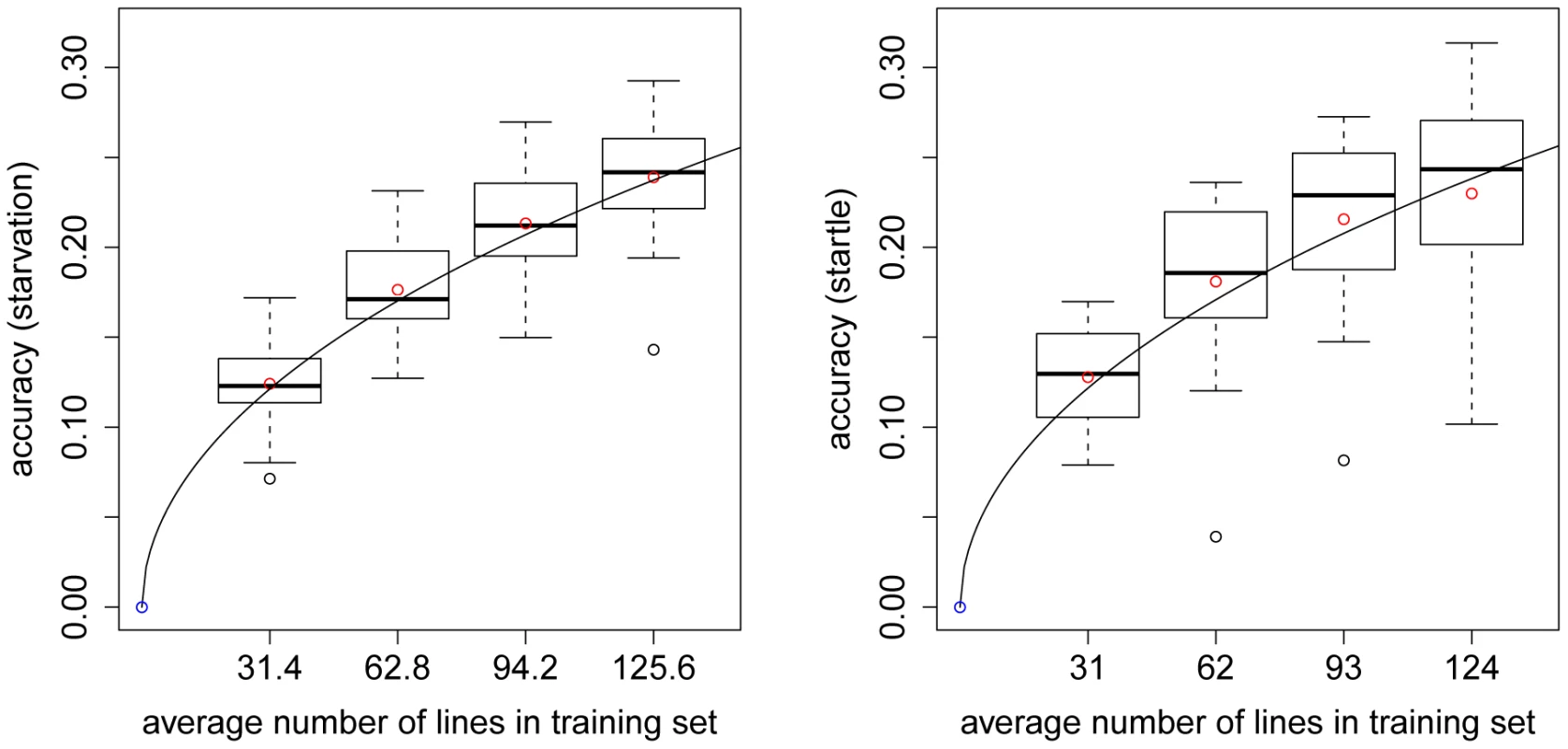
Each boxplot illustrates the average accuracies for 20 replicates of the CV procedure using GBLUP. The left (right) plot shows accuracies for starvation resistance (startle response). The solid line is the curve of [20] fitted to the empirical data, which results in estimates of and for starvation resistance and startle response. All 2.5 million SNPs were used to construct the genomic relationship matrix in the GBLUP model. Tab. 1. Average correlations between predicted genetic values and observed phenotypes for different CV procedures with GBLUP and different traits. 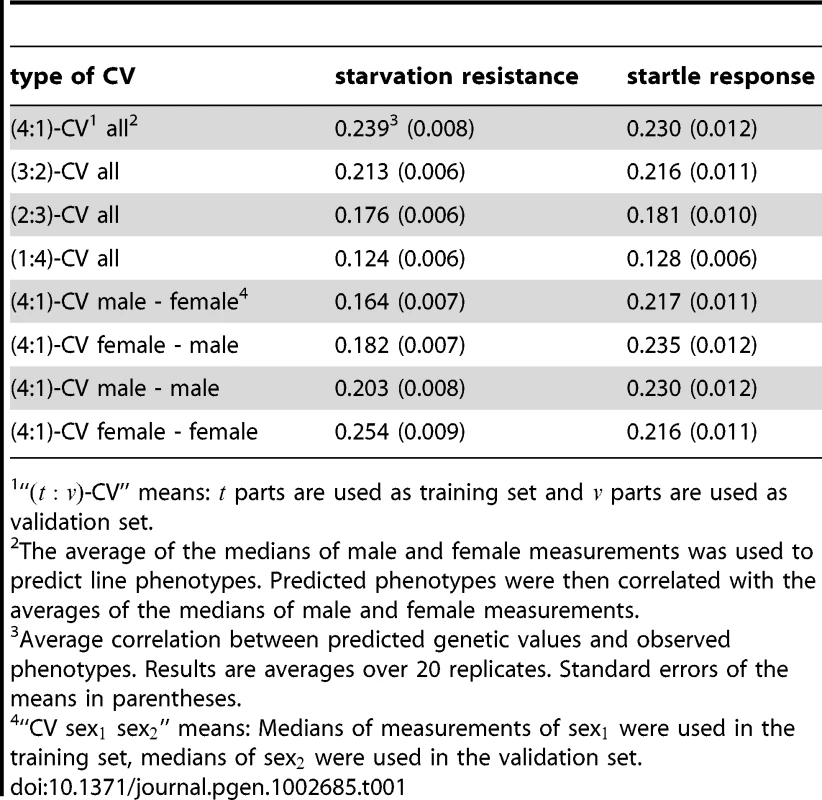
“-CV” means: parts are used as training set and parts are used as validation set. Using male performance data to train the model and using the results to predict the female performance (or vice versa) does not affect the predictive ability for startle response, but substantially reduces the predictive ability for starvation resistance, reflecting a higher degree of genotype by sex interaction in this trait ([27], and see below). Prediction is more accurate in females than in males ( vs. ) for starvation resistance, while there is little difference for startle response.
A series of -fold CVs for starvation resistance using different SNP densities showed that predictive ability remained almost constant if every SNP (∼150,000 SNPs) was used to construct the genomic relationship matrix (Figure 4). The predictive ability began to deteriorate when fewer than SNPs were used, but only vanished completely when as few as ∼2,500 SNPs (every SNP) were used. The corresponding LD distribution for SNP neighbors for different SNP densities is shown in Figure 5, illustrating the extreme short-range extent of LD in the D. melanogaster genome. The average LD between SNPs (after imputation) whose distance lay in the interval bp was for the autosomes and for the X-chromosome. Long-range LD between pairs of loci at the opposite ends of chromosome arms or across different chromosome arms was on average both for the autosomes and the X-chromosome.
Fig. 4. Predictive ability of 5-fold CV with GBLUP for starvation resistance using different numbers of SNPs. 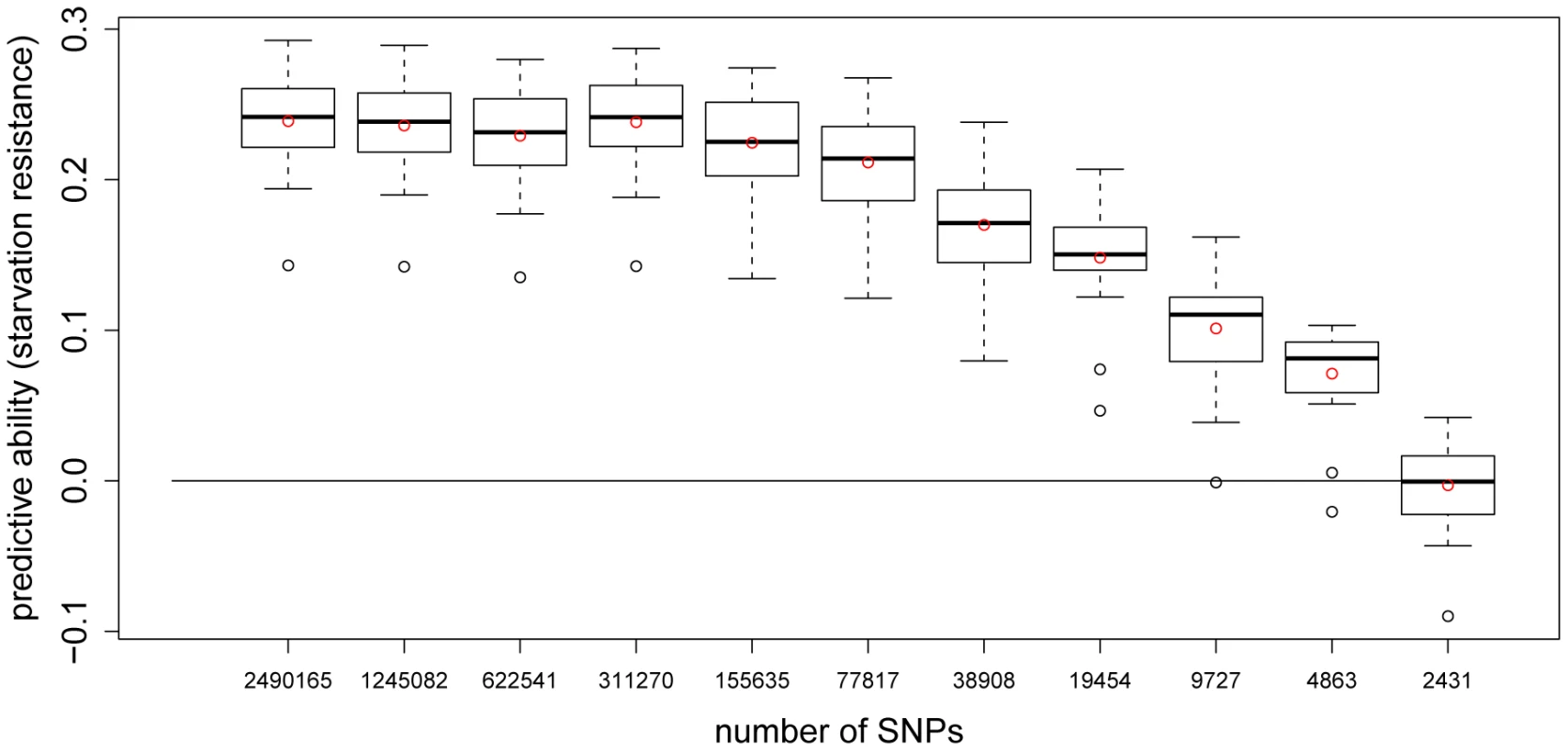
Each boxplot shows the average predictive abilities for 20 replicates of 5-fold CV using GBLUP. For the CVs leading to the -th boxplot, every 2-th SNP was used to build the genomic relationship matrix according to [8]. This was done for the thinning factors 010. The red dots indicate the average predictive abilities. Fig. 5. The distribution of between SNP neighbors for different SNP densities. 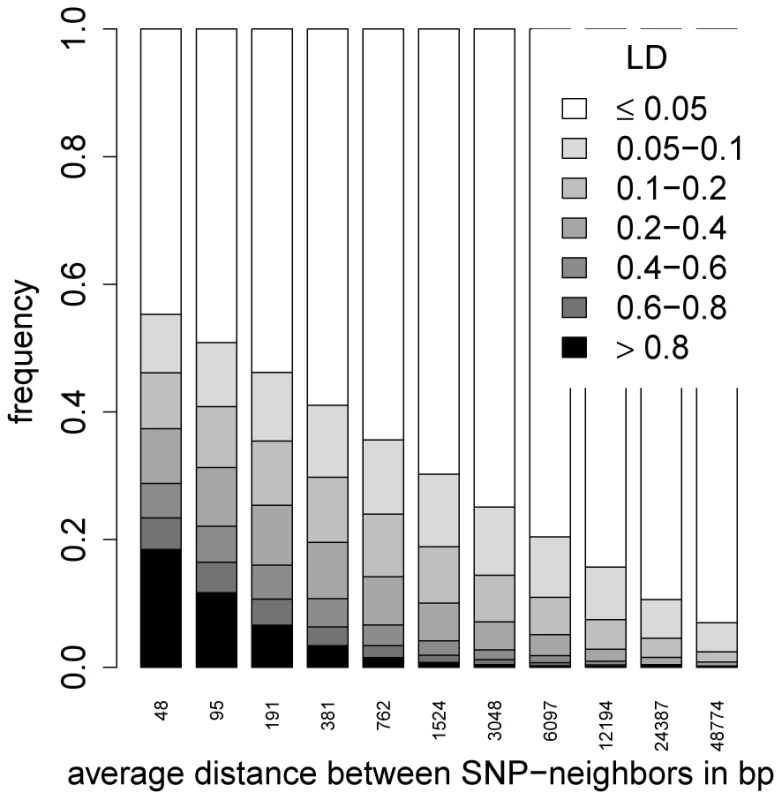
For the -th stacked bar, every -th SNP was used, 010. Then, the distribution of for the resulting SNP neighbors was calculated. For starvation resistance, the influence of the minor allele frequency of the SNPs used on the predictive ability was assessed with a series of 5-fold CVs using SNP sets with different average minor allele frequency. We find that the variability of the predictive ability increases when the average minor allele frequency of the SNPs used to construct the genomic relationship matrix is decreased (Figure S1). In replicates of an additional 5-fold CV, in which we randomly chose SNPs to build the genomic relationship matrix, an average predictive ability of was obtained, which is in the range obtained when every SNP (∼77,817 SNPs) was used (, Figure 4). Running replicates of a 5-fold CV using randomly chosen blocks of adjacent SNPs (each block consisting of SNPs) led to an average predictive ability of .
To analyze whether the predictive ability is due to lines which are more highly related, we ran an additional 5-fold CV with replicates in which the two groups of higher overall relatedness (Figure 2) were excluded. Here we found an average predictive ability of for starvation resistance, which is larger than the average predictive ability we obtained using all lines (). For startle response, excluding the two groups led to a decrease in predictive ability ( in comparison to ).
Effective population size derived from empirical accuracies of genomic prediction
The accuracy of genomic prediction is a function of a number of quantities, including the size of the training set and the effective population size [20]. has an effect on the number of independently segregating chromosome segments, , in a population (the larger , the larger ); and the predictive ability of GBLUP is higher when the number of segments is small. By varying the size of the training set in a series of CVs, we can estimate by fitting a curve through the empirical accuracies obtained (Figure 3).
We estimated for starvation resistance and for startle response. The coefficient of determination of the fitted curve was for starvation resistance (startle response). The bias corrected empirical confidence intervals for the estimates obtained with bootstrapping [32] were for starvation resistance and for startle response.
The effective population size in the Raleigh population (from which the DGRP-lines were drawn) was estimated to be ∼19,000 in , with a massive fluctuation between years [33]. Our estimates of correspond to independently segregating chromosome segments. In this formula is the length of the female genome in Morgans (there is no recombination in male Drosophila). Since the sequenced animals resulted from generations of full sib mating following the original sampling from the Raleigh population, the DGRP lines are not expected to have the same as the original population and are consequently expected to have a different .
We can use the curves fitted through the empirical accuracies (Figure 3), to predict the expected accuracy of prediction for an arbitrarily large size of the training set: If lines were available in the training set, the curve would predict accuracies of ∼0.58 for starvation resistance and startle response. This value was obtained by using and as well as and in the modified formula of [20].
Effective population size derived directly from linkage disequilibrium
We also estimated the effective population size based on LD directly. For a distance bin of Morgan we obtained average LD-values of for chromosome 2L (2R, 3L, 3R, X). These values correspond to an estimated effective population size of , approximately generations ago. The average estimated effective population size is , which is in the range of the estimates based on the observed accuracies.
Genomic prediction with SNP selection
Genomic prediction might be improved if we only fit SNPs which are associated with variance in a trait, because we then concentrate on the biologically relevant genomic regions, and excluding SNPs which are not associated with the trait reduces statistical noise. We tested this hypothesis using the starvation resistance data. We identified the SNPs with the highest absolute estimated effect or the highest estimated genetic variance, respectively, in the training set of the respective of the folds in a -fold CV. We then used these subsets of selected SNPs to predict the phenotype in the remaining of the fold. Predictive ability was improved by over the reference scenario when using the SNPs with largest effects (average predictive ability of in comparison to ). Using the SNPs with greatest variance explained, predictive ability was improved by (average predictive ability of ). In both cases, the improvement is marginal and provides little support for the idea of SNP pre-selection.
We also compared our GBLUP results to those from a method which does not assume that all SNP effects are drawn from the same normal distribution and carries out an internal feature selection. We ran replicates of a -fold CV for starvation resistance using BayesB [10]. In each round of the Markov Chain Monte Carlo based procedure (see Methods), of the SNPs were assumed to have no effect and the effects of the remaining of the SNPs were drawn from normal distribution with random variances. In most folds of each single CV and for all replicates of CV, the observed predictive abilities differed only marginally between BayesB and GBLUP (Figure 6). The average predictive ability obtained with BayesB was which is not appreciably different from the result obtained with GBLUP ().
Fig. 6. Predictive ability for GBLUP versus BayesB using phenotypic values of starvation resistance. 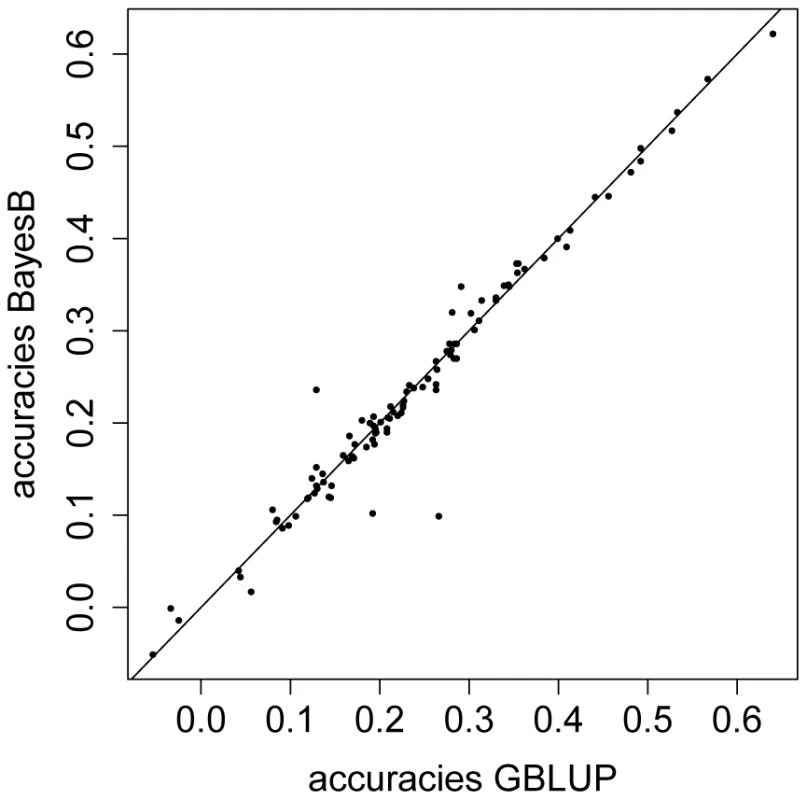
Predictive abilities are plotted for 20 replicates of a 5-fold CV, each replicate consisting of 5 corresponding folds of CV. Genomic prediction versus GWAS
Although genomic prediction follows a different paradigm than genome-wide association studies, it is informative to compare significant SNP positions from the GWAS to areas of large estimated SNP effects resulting from the GBLUP model. Previously [27], a GWAS of DGRP lines (of which the material used here is a subset) identified SNPs associated with starvation resistance and SNPs associated with startle response at a nominal p-value in the analyses of sex-averaged data. We estimated SNP effects using RRBLUP and compared them to the significant SNPs from the GWAS study (Figure S2, Figure S3). There is excellent concordance of signals from both approaches in some regions (e.g. the genome-wide largest SNP effects on chromosome 3L for starvation resistance and 2L for startle response), while concordance is poor in other regions, especially on the X chromosome.
We further investigated whether the most significant SNPs detected in the GWAS are reflected by large SNP effects in the GBLUP study using a different approach. For each significant SNP position from the GWAS we took the neighboring SNPs ( on each side) and calculated the sum of the absolute values of their estimated effects using the GBLUP model. To avoid an effect of different sample size, we used the most significant loci from the GWAS for both traits. We compared these sums to the sums of the absolute values of estimated SNP effects in sliding windows spanning the whole genome (with each window containing neighboring SNPs). We observed a clear separation of the density functions of these sums for both startle response and starvation resistance (Figure 7).
Fig. 7. Distribution of absolute SNP effects. 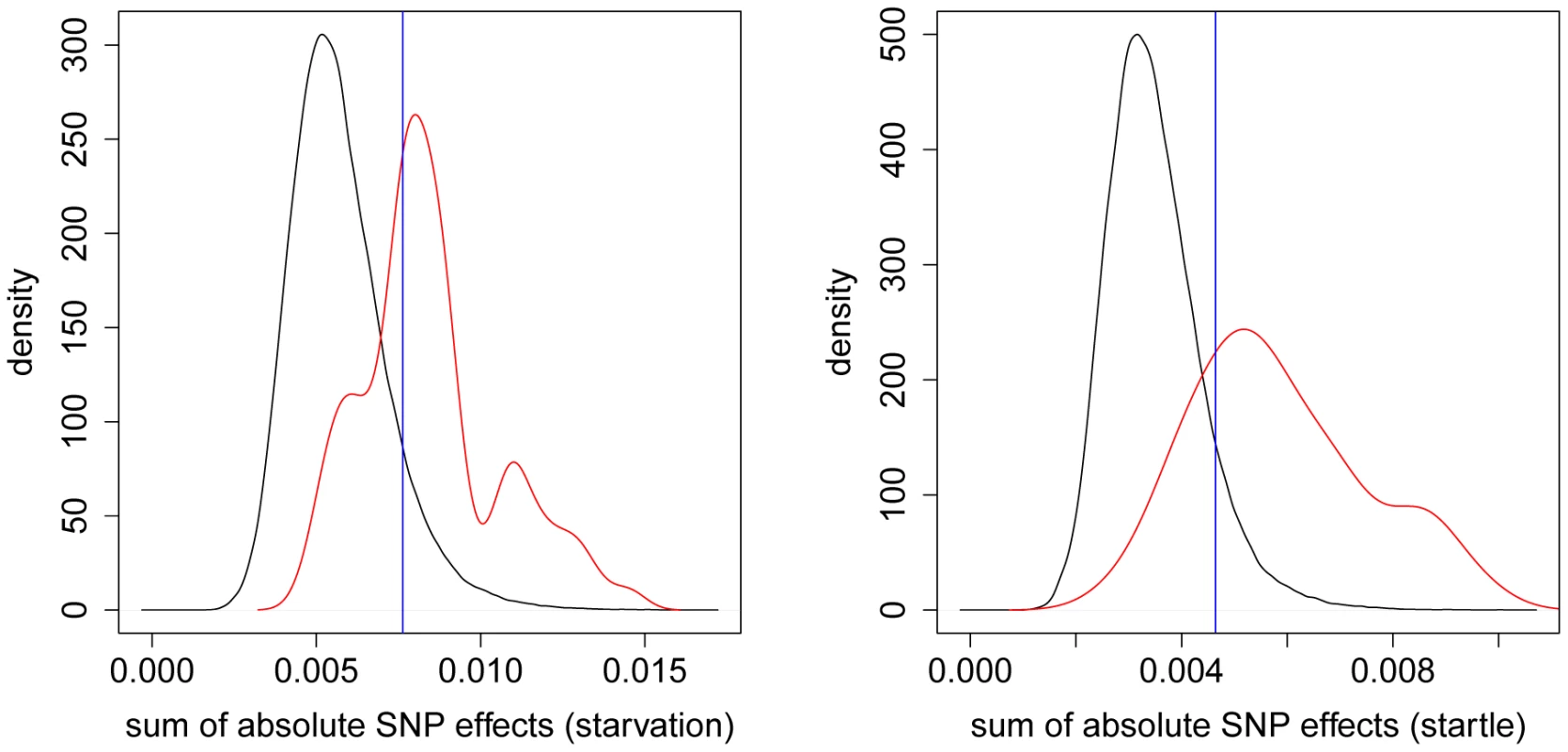
The density of the sum of the absolute SNP effects from GBLUP is plotted for sliding windows of 100 adjacent SNPs covering the whole genome (black) and for windows around the 75 most significant SNPs (red) according to the GWAS of [27]. The left (right) plot shows the densities for starvation resistance (startle response). The blue vertical line indicates the 90% quantile of the black density function. The density resulting from the sliding window approach reflects the overall distribution of the suggested statistic in the sample. For starvation resistance (startle response) a threshold value of , cf. Figure 7, cuts off the upper of the respective distribution. Applying the same threshold with the density function reflecting the statistic for the significant GWAS positions, of the distribution exceeds the threshold, indicating that signals found in the GWAS are also associated with large estimates of the SNP effects in the genomic model.
Analyses of individual trait data
In addition to the line means we also analyzed individual records ( individual flies per line tested for starvation resistance and for startle response) to assess whether the variance between lines can be fully explained by additive gene effects or if non-additive mechanisms have an impact. This was done by modeling the covariance structure between lines based on the additive and additiveadditive genomic relationship matrix and testing the goodness of fit of the respective models. Most applications of genomic prediction are for outbred populations, for which the additive genetic variance and corresponding narrow-sense heritability determine the extent to which phenotypes in the next generation can be predicted from information obtained on the current generation. However, the variance among DGRP lines is the total genetic variance, and is possibly inflated by additive by additive epistatic variance [34]. Therefore, we performed several analyses on measurements of individual flies to determine the nature of the total genetic variance, especially to what extent the presence of non-additive genetic variance might have affected predictive abilities. We fitted three different models to the individual phenotype data: Model 1 contained a random line effect, and lines were assumed to be unrelated. In Model 2, a random additive line effect was added, whose covariance structure was modeled via the genomic relationship matrix . In Model 3, an additional random additiveadditive epistatic effect was included, whose covariance structure was modeled via the Hadamard product . Since the between line variance relates to inbred lines, while the additive and additiveadditive variance component pertain to the non-inbred base population (or a hypothetical random mating produced from the inbred lines), the variance between inbred lines in Model 1 is expected to be twice the additive genetic variance in Model 2 or 3 under a fully additive model.
We estimated variance components for all three models pooled across sexes and separately for males and females (Table S3, Table S4). We find little evidence for non-additive genetic variance for these traits. The estimate of from Model 2 is from Model 1, and Model 2 gave a significantly better fit than Model 1 when applying the likelihood ratio test, again indicating that the observed between line variance is due to additive gene action. Inclusion of the component was not significant for either of the traits. We found significant sex by line interaction variance for starvation resistance, but not for startle response (Tables S3, S4), which is in accordance with the findings of the genomic prediction across sexes (Table 1) and previous analyses of these data [27].
Discussion
We report the first (to our knowledge) application of genomic prediction to a real set of full genomic sequencing data in a eukaryotic organism. Although predictive abilities obtained with starvation resistance and startle behavior are only moderate to low, and although we limited our analysis to SNPs that are common due to the small sample size of lines, this study can be seen as a proof of concept for this approach. There are several reasons for the limited predictive ability obtained in this study. First, the training set is small, with a maximum of observations in the -fold CV, and the accuracy of genomic prediction is a function of the size of the training set [20]. Using the curves fitted through the empirical accuracies (Figure 3), we predict accuracies of for starvation resistance and startle response, if sequenced lines were available for the training set.
The second important factor affecting accuracy of prediction is the number of independently segregating chromosome segments, [20]. In our study we obtained . This is larger than usually observed for Holstein cattle ( with and genome length Morgans [35]), but is smaller than the corresponding value in the human genome ( with Morgans, [36]). (Note that in mammalian species, there is recombination in both sexes and [9].)
Accuracy of genomic prediction is thought to come from two sources: (i) SNPs in useful LD with causal loci; and (ii) SNPs reflecting the relationship structure between the training set and the set to be predicted [37]. Due to the very fast decay of LD in the D. melanogaster genome, few SNPs are in useful LD with any causal polymorphism. Even if we define “useful LD” very conservatively as , then on average only a region of bp around a causal polymorphism was in useful LD on an autosome ( bp on the X chromosome). This means that on average () SNPs were in useful LD with a causal autosomal (X-linked) polymorphism, as the average distance between neighboring SNPs was bp ( bp) on an autosome (X chromosome). If predictive ability was mainly driven by SNPs in LD with causal polymorphisms, reducing the SNP density should lead to a massive decay of predictive ability of the models, which was not observed. Little decrease in accuracy was seen, even if every SNP was used in the model, in which case hardly any SNP would be in useful LD with causal polymorphisms. The underlying mechanism therefore seems to depend on a sufficient number of SNPs being in low LD with causal polymorphisms, rather than few SNPs in close physical association and high LD. In the DGRP population, LD approaches a small but positive baseline level with increasing physical distance [27], so that even with large physical distances a minimum level of LD is maintained, which was on average with being the sample size.
The number of SNPs for maximal accuracy of genomic prediction with unrelated individuals has been estimated as [38], corresponding to SNPs in the present study.
For starvation resistance, we find that the empirical accuracy levels off when approximately every SNP is used, which is equivalent to or SNPs. Adding more SNPs beyond this value does not lead to any improvement in the genomic prediction of starvation resistance, but also does not reduce accuracy, which one might expect when using more SNPs than actually needed. While fitting large numbers of “superfluous” SNPs may be considered as noise in the RRBLUP model, these SNPs can also be seen to provide a better basis to estimate the realized relationship matrix in the GBLUP model, which leads to a higher accuracy of the estimated realized relationships. Since both models are fully equivalent [9] no penalty is expected in the prediction of genomic values.
Since pedigree information for the founders of the inbred lines was not available, our estimates of heritability and genomic prediction are based on the actual degree of identity-by-descent sharing between relatives [39]. There is little pedigree structure in the DGRP lines, with the exception of two distinct blocks of higher relatedness, comprising and lines, respectively, with a genomic relationship within blocks of and . When these blocks were excluded from the data, predictive accuracy in a -fold CV increased (decreased) for starvation resistance (startle response), suggesting that prediction in the DGRP population does not rely on distinct family structures. Given this together with the short-range extent of LD in the D. melanogaster genome and the robustness of the accuracy of genomic prediction with reduced marker density, we conclude that the observed accuracy of prediction for starvation resistance and startle response is primarily due to the long-range LD in the population, or equivalently, the subtle relationship structure as reflected by the genomic relationship matrix.
We restricted our analyses to SNPs for which the minor allele was present in at least four DGRP lines (a minor allele frequency of ). We applied this threshold to avoid computational limitations, especially when applying the BayesB method; and for consistency with the GWAS in the DGRP [27], which used the same filtering criterion. Thus, we did not utilize the million SNPs with minor allele frequencies less than this, nor did we take other forms of molecular variation into account.
Structural variations such as transposable elements have been repeatedly reported to be associated with phenotypic variation [40], therefore we must consider to what extent not including these variants in the models affected prediction accuracy. Given that we do not observe an increase in accuracy when increasing the number of SNPs from to million, we do not expect that increasing the marker density by adding more SNPs and other variants will have a significant effect on predictive ability. Additionally, SNPs with low minor allele frequencies were shown to be highly variable in predictive ability, so that the potential amount of information possibly added by the million low frequency SNPs is limited. However, accounting for all polymorphisms in the model means that some fraction of the genetic variants must causally affect the trait. Simulations [26] including the causal polymorphism in the model improves the predictive ability over models based only on neutral SNPs in LD with the causal variants. Further research is needed to understand these mechanisms in the context of genomic prediction based on empirical data.
The accuracy of BayesB has outperformed that of GBLUP in several simulation studies [10], [37]. Simulation results have suggested that GBLUP did not take full advantage of genome sequence data, suggesting that Bayesian methods are needed to obtain maximum accuracy [26]. The superiority of BayesB over GBLUP is expected to increase with marker density, and decrease when the size of the training data set is increased [38]. However, we did not find that BayesB yielded a significantly higher predictive ability than GBLUP in the replicates of -fold CV with starvation resistance implemented in the present study. We used a very high marker density and a small training set, and yet GBLUP performed as well as BayesB. These conclusions should be taken with caution, since the available size of the training set was extremely small in our study due to the limited availability of fully sequenced lines. In [20], BayesB yielded a higher accuracy than GBLUP, when the number of simulated QTL was low; but GBLUP slightly outperformed BayesB, when the number of QTL became large, since the GBLUP model is equivalent to RRBLUP, in which all SNPs are assumed to have an effect drawn from the same normal distribution. Although this model may not seem biologically plausible, it performed as well as BayesB in the present study, consistent with several studies on real data from dairy cattle for different traits [4], [41].
The finding that BayesB did not outperform GBLUP in the present study is consistent with a quasi-infinitesimal genetic architecture; and results indicate that starvation resistance and startle response are complex traits with a highly polygenic genetic architecture rather than being driven by a few major causal genes. This is in agreement with previous studies stating that starvation resistance and startle response can be considered to be model traits with a complex (i.e. quasi-infinitesimal) genetic background [28]–[30]; and it is also in line with the results from the GWAS [27]. One reasonable conclusion might be that there are so many causal polymorphisms, each with a small effect, that the effective chromosome segments are saturated with causal variants and the effects of segments follow a normal distribution. Under this circumstance, GBLUP is expected to perform as well as BayesB. However, these hypotheses clearly need further investigation. More systematic model comparisons based on the available data were not considered here due to the prohibitive computing time required for BayesB.
Previously, gene centered multiple regression and partial least square (PLS) regression models were used to predict starvation resistance and startle response phenotypes from genotypic data [27]. In both cases only SNPs that had nominal significance levels of from the GWAS were used. The gene centered prediction models found that a few SNPs explained a large fraction of the genetic and phenotypic variance of the traits, while the PLS models found that the significant SNPs explained a high fraction of the phenotypic variance. The purpose of these studies was a comparison with human association studies, in which the faction of the variance explained by significant variants in the entire sample is commonly quoted. These approaches are fundamentally different from the BLUP approach used in this study. The BLUP approach includes random components and their covariance structure in the model, whereas regression models do not incorporate random terms except from the residuals; and the BLUP approach does not rely on a pre-selection of SNPs based on a GWAS. Most critically, we evaluated the robustness of the BLUP predictions using -fold cross-validation; whereas the previous analyses only tested the explanatory power of the most significant associated SNPs using the entire sample. Had we done the same analysis using GBLUP, we would be able to predict of the variance.
The imperfect concordance of the positions of the most significant SNPs from the GWAS and the largest estimates of SNP effects from RRBLUP is a consequence of the different objectives of the two approaches. A sequence-based GWAS is conducted to identify causal polymorphisms and provide estimates of allelic effects and frequencies. Also, the GWAS suffers from estimating one effect at a time and so does not necessarily position the QTL accurately. The goal of RRBLUP is to predict the phenotype using all available SNP information simultaneously. Here, estimated SNP effects are a by-product and mapping causal variants is not the primary objective. Given that the number of SNP effects to estimate is much larger than the number of observations, effects are estimated using penalized multiple regression approaches, shrinking estimated effect sizes towards zero. In addition, the magnitude of estimated SNP effects from RRBLUP is a function of the marker density. The higher the marker density, the more SNPs will be in LD with a causal mutation; therefore, the true allele substitution effect of a causal polymorphism will be split up and assigned in parts to a series of SNPs in the respective haplotype block. This can mask both the effect size, because one large effect may come in many small pieces; and the mapping position, because any SNP in LD with the causal polymorphism may have a substantial estimated effect. Nevertheless, some of the largest SNP effects from RRBLUP are in the proximity of prominent SNPs identified in the GWAS, so that to some extent positional information can still be retrieved from the RRBLUP results.
A methodology combining the strengths of both approaches – unbiased effect estimates and high positional resolution of GWAS with the simultaneous analysis of all SNPs, high predictive power and quality control via CV of genomic approaches – still needs to be developed. Results obtained in our study cannot be directly compared to predictive abilities in human studies due to the extremely small training set size ( in CV), and Drosophila has much larger and rapid decline of LD compared to humans. When genomic prediction in human studies was based on large training sets (thousands), substantial SNP panels (k) and a highly heritable trait (), predictive ability of genomic models was found to exceed what has been previously reported using a reduced number of markers pre-selected based on GWAS [31] and genomic prediction based on pre-selected SNPs was found to be of limited use in human studies of height [42].
In the near future individual whole genome sequences will become increasingly available for large numbers of individuals in many species [17], [18]. Sequence-based predictions will therefore be relevant for prediction of risk disease and individualized medicine in humans, and for genome-based selection in farm animals and crops. The main findings of our study are: (i) genomic prediction can be efficiently implemented via GBLUP with full genome sequence data; (ii) there is little, if any, gain in predictive ability if the number of SNPs is increased above (equivalent to in Holstein cattle and in humans); and (iii) approaches based on external or internal (BayesB) selection of subsets of SNPs were not found to provide a substantial gain in accuracy of prediction compared to GBLUP. All findings must be seen against the background of the small sample size and the specific genetic constellation, with almost unrelated inbred lines and highly accurate phenotypes. Nevertheless, these results provide a realistic assessment of the potential benefits of sequenced-based prediction applied to non-model organisms and indicate avenues for future research.
Materials and Methods
The Drosophila Genetic Reference Panel (DGRP)
The full Drosophila Genetic Reference Panel (DGRP) [27], a recently developed new community resource for genetic studies of complex traits, consists of D. melanogaster lines derived by generations of full sib mating from wild-caught females from the Raleigh, North Carolina population. Whole genome sequence data of DGRP lines (Freeze 1.0) have been obtained using a combination of Illumina and next generation sequencing technology, which are available from the Baylor College of Medicine, http://www.hgsc.bcm.tmc.edu/project-species-i-DGRP_lines.hgsc. We used the Illumina sequences for DGRP lines in this study.
Data preprocessing
SNPs were called from the raw sequence data as described previously [27]. We used SNPs with a coverage greater than 2X but less than 30X, for which the minor allele frequency was present in at least four lines, and for which SNPs were called in at least 60 lines. This series of filters gave a total of SNPs for this analysis; on 2L, on 2R, on 3L, on 3R and on the X chromosome. We did not consider the few SNPs on the very short chromosome 4. In total there were missing SNP genotypes (), which we imputed using Beagle Version 3.3.1 software [43].
Phenotypic values
Phenotypic measurements for starvation resistance were available for all DGRP lines, and for startle response on lines [27]. We used the average of the medians of measurements for each trait in males and females as the phenotypic value of the line, i.e. , where and are the medians of the measurements for female and male individuals of the line. We used medians because of the skewed distribution of traits; however, medians are highly correlated with line means. For starvation resistance (startle response) there were on average measurements for females, and measurements for males (Table S1). Measurements were taken in several replicates for each trait [27].
Cross-validation
We used different cross-validation (CV) procedures [44]–[46] to assess the predictive ability of different methods. In one replicate of a CV, the lines are randomly divided into a training set, which is used for parameter estimation; and a validation set, for which genetic values are predicted. The CV procedures differ in the ratios of the numbers of lines belonging to the training and validation sets: In a -CV (with integers and ), the lines are randomly divided into groups. The groups build the training set, and the remaining groups build the validation set. For this classification, there are possibilities. For each of these possibilities (“folds”), total genetic values for the lines of the validation set are predicted and the corresponding predictive ability is calculated. The predictive abilities are then averaged to obtain one average correlation per CV replicate. For example, one (3∶2)-CV, consists of CV folds, over which predictive abilities are averaged. A -CV is also called -fold CV.
We used (4∶1)-, (3∶2)-, (2∶3) - and (1∶4)-CVs to analyze the effect of decreasing training set size. The CVs also differed in the constellations of phenotypic records used for the training and validation set. For example, the notation “(4∶1) male – female” indicates that only the medians of male records were used in the training set, and that the predicted genetic values were correlated with the medians of female records of the validation set to obtain the predictive ability in a (4∶1)-CV. CVs were also run for different marker densities, using every -th SNP (). Additionally, 5-fold CVs using only the SNPs with the largest absolute values of estimated effects (obtained in the training set), or using only the SNPs with the largest SNP variances (obtained in the training set) were performed. The additive genetic variance marked by the SNP was calculated as with allele frequency and estimated SNP effect . In another series of 5-fold CVs we randomly chose SNPs to build the genomic relationship matrix or we randomly chose blocks of adjacent SNPs (each block consisting of SNPs). In an additional 5-fold CV we excluded the lines in the two blocks of higher relatedness (Figure 2) from the data. Each type of CV was replicated times, resulting in average predictive abilities.
We also analyzed the influence of minor allele frequency on the predictive ability by another series of 5-fold CV. For this, we sorted all SNPs by their minor allele frequency and divided the sorted vector into blocks. For each block we ran replicates of a 5-fold CV using GBLUP and the corresponding SNPs.
Predictive ability and accuracy
Predictive ability was measured in terms of correlation between predicted genetic values and observed phenotypic values. The corresponding accuracy , defined as the correlation between true and predicted genetic value, was obtained by dividing the observed predictive ability by the square root of the observed heritability [47]. The heritability was based on the GBLUP model (see below).
Genomic prediction with GBLUP
The underlying statistical model is(1)In this model, the component of the -vector is the phenotypic value of the line that is used for prediction, i.e. the average of the medians of the phenotypic measurements for males and females for this line. Moreover, is the overall mean; is assumed to be multivariate normal, with the genomic relationship matrix of all lines [8] and the additive genetic variance among lines. The matrix is an -incidence matrix, whose rows consist of unit vectors with one component being and all the others zero, indicating the respective positions of lines used for prediction in the -vector of genetic values of all lines. is the residual term, where is the residual variance. Following the approach of [8], was defined aswhere is the -matrix of SNP genotype vectors for the lines with the SNPs coded as and the column of is , where is the frequency of the second allele at locus .
Variance components were estimated via maximum likelihood (ML) using the R-package “RandomFields”, Version 2.0.46 (http://CRAN.R-project.org/package=RandomFields), and its function “fitvario”. The BLUP approach to obtain the vector of genetic values is equivalent to solving the so-called Mixed Model Equations (MME):A narrow-sense heritability based on the GBLUP model (1) was calculated as
Estimation of SNP effects
The GBLUP model (1) is equivalent to the following linear model in which all SNPs are assumed to have an effect drawn from the same normal distribution [9]:where and are as described above and is the vector of SNP effects with . Using this equivalence, the SNP effects can be predicted asTo estimate the SNP effects resulting from GBLUP for a single trait, we used all of the available lines, i.e. in model (1) contained the phenotypic values of all lines so that in the corresponding formulas. Note that only the inversion of a matrix of size equal to the number of sequenced lines is required.
Distribution of linkage disequilibrium
We used [48] as a measure of LD between a pair of loci. With two biallelic loci and with alleles and and frequencies and , we denote the frequencies of the genotypes and as and respectively. Then,We performed the LD analyses using the imputed SNP matrix of million SNPs for the lines. We calculated the distribution of LD between all pairs of neighboring SNPs for different marker densities, using every -th SNP (). The extent of long-range LD was calculated for pairs of SNPs randomly sampled from the first and the last SNPs per chromosome arm. Moreover, the average LD was calculated between SNPs on different chromosome arms, by sampling pairs of SNPs for each combination of chromosome arms.
Effective population size derived from empirical accuracies of genomic prediction
We modified the formula [20] for the expected accuracy, of GBLUP given different population parameters (see Text S1 for more details on the derivation in the case of D. melanogaster):(2) is the effective population size, is the size of the training set, is the length of the female genome in Morgans and is the narrow-sense heritability of the trait estimated from model (1). The term describes the number of independently segregating genome segments [9].
We ran CVs with different numbers of lines ( for starvation resistance and for startle response) in the training set ( replicates each). Average numbers of lines in the training set are reported, which are non-integer values for starvation resistance because in a -CV, division of lines into groups may give unequal numbers of lines in the different partitions. Given the corresponding average accuracies for the CV replicates, we estimated by fitting a curve to the points . To fit the curve, we chose such that the sum of the squared differences of the observed accuracies and the accuracies obtained by (2) was minimized:using and Morgan. We calculated the length of the female genome in Morgans by summing the lengths of the chromosomes in base-pairs ( (, , , ) Mbp for chromosome 2L (2R, 3L, 3R, X), [49]) and multiplying by the average recombination rates of females for the different chromosomes in Morgans per base-pair [50].
After performing bootstrapping ( replicates), the bias corrected empirical confidence intervals ( error in each tail) for the estimates [32], [51] were calculated aswhere is the -percentile of the bootstrap cumulative distribution function, is the -percentile of the standard normal distribution function , and .
Effective population size derived directly from linkage disequilibrium
To estimate the effective population size based on LD, the following formula was used [52]:where is the number of lines and is the recombination rate in female individuals, cf. Text S1 for more details on this formula.
Genomic prediction with BayesB
The underlying model for the Markov Chain Monte Carlo based BayesB [10] method iswhere and are as defined previously and is the vector of normally distributed and independent SNP effects. The variance of the SNP effect, , is assigned an informative prior. The prior distribution of the genetic variances aims to resemble a situation where there are many loci with zero variance and only some loci with variance not equal to zero. Therefore, the prior distribution of the variance of a marker effect is a mixture of distributions which is given byNote that this implies that the unconditional distribution of each single marker effect is a mixture of a point mass at (with probability ) and of a t-distribution with zero mean, degrees of freedom and scale parameter [21], i.e. BayesB assigns the same unconditional prior distribution to each marker effect.
In our studies, we used and the scale parameter was calibrated asWe chose , such that approximately markers were contributing to the additive genetic variance. For the residual variance, , the prior distribution was with andValues for and were chosen in the order of magnitude of the variance components of the GBLUP model (1), which were estimated using all lines and “fitvario”. The BayesB procedure is described in detail in [10]. It consists of running a Gibbs chain, where additionally a Metropolis-Hastings algorithm ( iterations) is used to sample from , where denotes the data corrected for the mean and all genetic effects other than the marker effect . Following graphical inspection, we ran BayesB with a chain length of iterations including a burn in of iterations that were discarded. To perform the BayesB approach, we used the software “GenSel”, Version 2.36, by R. Fernando and D. Garrick (cf. http://taurus.ansci.iastate.edu/Site/Welcome.html), which is implemented in C++. BayesB is computationally very intensive. The analyses were run on a Mac Pro 2× 2.93 GHz 6-Core Intel Xeon with 64 GB RAM running Mac OS X Server 10.6.7. One fold of a -fold CV for starvation resistance took approximately hours.
Comparing areas with large SNP effects with significant SNP positions
A genome-wide association study (GWAS) revealed significant SNP positions for starvation resistance (startle response) [27], where a SNP position was considered significant if at least one of the three p-values, obtained using only male, only female or sex-pooled phenotypic records, was . We considered the subset of SNPs for which p-values of SNP effects of pooled data were , to be more conservative and to be consistent with the previous analyses, leading to significant SNPs for starvation resistance (startle response).
We compared genomic regions for which GBLUP estimated large SNP effects to these significant SNP positions of the GWAS. To avoid an effect of different sample sizes, we chose the most significant SNPs from the GWAS analysis for each trait. For each of these SNPs, we chose the closest (neighboring) SNPs ( on each side) and calculated the sums of absolute values of the corresponding SNP effects (resulting from the GBLUP model). We compared the distribution of these sums to the distribution of the sums of the absolute values of estimated SNP effects in windows of neighboring SNPs covering the whole genome by plotting the corresponding density functions. To obtain the sums of the absolute values of estimated SNP effects covering the whole genome, the windows were overlapping, displaced by SNP positions. If the genomic regions for which GBLUP estimated large SNP effects coincide with the significant SNP positions of the GWAS, we expect the density functions to be separated.
Variance component estimation using ASReml and individual trait records
For each trait, we fitted three different models using individual trait records. The first model included a fixed sex effect, a random line effect, a random line-sex-interaction term and a random term accounting for the different replicates in which measurements of the traits were taken:In the second model, an additional random genetic effect was added for each line. The variance-covariance matrix of the vector of these genetic effects was assumed to be given by the genomic relationship matrix of [8]:In the third model, an additional random additiveadditive epistatic effect was included for each line. The variance-covariance matrix of the vector of these genetic effects was given by the Hadamard product [53] of the genomic relationship matrix of [8]:Other two-way epistatic interactions, like additivedominance or dominancedominance, should not exist in inbred lines, provided inbreeding is complete. Variance components and their standard errors were estimated using ASReml 2.0 [54]. The analyses were done pooled across sexes as well as separately for males and females. The analyses of separate sexes did not include the sex term, and the replicate(sexline) term was reduced to replicate(line).
Heritabilities
The broad-sense heritability for Model 1 was calculated ascf. [28]. Narrow sense heritabilities for Models 2 and 3 were calculated asandThese heritabilities are based on individual trait records.
Unless stated otherwise, all statistical analyses were performed using R software [55]. The R-package “ff”, Version 2.2-1 (http://CRAN.R-project.org/package=ff), was used to handle the large amount of SNP data efficiently in terms of memory capacity.
Supporting Information
Zdroje
1. MackayTFCStoneEAAyrolesJF 2009 The genetics of quantitative traits: Challenges and prospects. Nat Rev Genet 10 565 577 doi:10.1038/nrg2612
2. WrayNRGoddardMEVisscherPM 2007 Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res 17 1520 1528
3. de los CamposGGianolaDAllisonDB 2010 Predicting genetic predisposition in humans: The promise of whole-genome markers. Nat Rev Genet 11 880 886 doi:10.1038/nrg2898
4. HayesBJBowmanPJChamberlainAJGoddardME 2009 Genomic selection in dairy cattle: Progress and challenges. J Dairy Sci 92 433 443
5. LorenzAJChaoSAsoroFGHeffnerELHayashiT 2011 Genomic selection in plant breeding: Knowledge and prospects. Adv Agron 110 77 123
6. HendersonCR 1973 Sire evaluation and genetic trends. J Anim Sci 1973 10 41
7. RanadeKChangMSTingCTPeiDHsiaoCF 2001 High-throughput genotyping with single nucleotide polymorphisms. Genome Res 11 1262 1268
8. VanRadenPM 2008 Efficient methods to compute genomic predictions. J Dairy Sci 91 4414 4423
9. GoddardM 2009 Genomic selection: Prediction of accuracy and maximisation of long-term response. Genetica 185 1021 1031
10. MeuwissenTHEHayesBJGoddardME 2001 Prediction of total genetic value using genomewide dense marker maps. Genetics 157 1819 1829
11. FisherRA 1918 The correlation between relatives under the supposition of mendelian inheritance. Trans Roy Soc Edinburgh 52 399 433
12. PimentelEErbeMKoenigSSimianerH 2011 Genome partitioning of genetic variation for milk production and composition traits in Holstein cattle. Front Gene 2 doi:10.3389/fgene.2011.00019
13. SchönCCUtzHFGrohSTrubergBOpenshawS 2004 Quantitative trait locus mapping based on resampling in a vast maize testcross experiment and its relevance to quantitative genetics for complex traits. Genetics 167 485 498 doi:10.1534/genetics.167.1.485
14. MackayTFC 2004 The genetic architecture of quantitative traits: Lessons from Drosophila. Curr Opin Genetics Dev 14 253 257
15. FlintJMackayTFC 2009 Genetic architecture of quantitative traits in mice, ies, and humans. Genome Res 19 723 733 doi:0.1101/gr.086660.108
16. EckSHBenet-PagèsAFlisikowskiKMeitingerTFriesR 2009 Whole genome sequencing of a single Bos taurus animal for single nucleotide polymorphism discovery. Genome Biol 10 doi:10.1186/gb-2009-10-8-r82
17. The 1000 Genomes Project Consortium 2010 A map of human genome variation from populationscale sequencing. Nature 467 1061 1073
18. ElshireRJGlaubitzJCSunQPolandJAKawamotoK 2011 A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6 e0019379 doi:10.1371/journal.pone.0019379
19. HayesBJPryceJChamberlainAJBowmanPJGoddardME 2010 Genetic architecture of complex traits and accuracy of genomic prediction: Coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet 6 e1001139 doi:10.1371/journal.pgen.1001139
20. DaetwylerHDPong-WongRVillanuevaBWoolliamsJA 2010 The impact of genetic architecture on genome-wide evaluation methods. Genetics 185 1021 1031
21. GianolaDde los CamposGHillWGManfrediEFernandoR 2009 Additive genetic variability and the Bayesian alphabet. Genetics 183 347 363
22. GianolaDvan KaamJBCHM 2008 Reproducing kernel Hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 178 2289 2303
23. de los CamposGGianolaDRosaGJM 2009 Reproducing kernel Hilbert spaces regression: A general framework for genetic evaluation. J Anim Sci 87 1883 1887
24. LongNGianolaDRosaGJMWeigelKAKranisA 2010 Radial basis function regression methods for predicting quantitative traits using SNP markers. Genet Res 92 209 225
25. OberUErbeELongNPorcuESchlatherM 2011 Predicting genetic values: A kernelbased best linear unbiased prediction with genomic data. Genetics 188 695 708
26. MeuwissenTGoddardM 2010 Accurate prediction of genetic values for complex traits by wholegenome resequencing. Genetics 185 623 631
27. MackayTFCRichardsSStoneEABarbadillaAAyrolesJF 2012 The Drosophila Genetic Reference Panel. Nature 482 173 178 doi:10.1038/nature10811
28. AyrolesJFCarboneMAStoneEAJordanKWLymanRF 2009 Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41 299 307
29. HarbisonSTYamamotoAHFanaraJJNorgaKKMackayTFC 2004 Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics 166 1807 1823
30. JordanKWCarboneMAYamamotoAMorganTJMackayTFC 2007 Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol 8 doi:10.1186/gb-2007-8-8-r172
31. MakowskyRPajewskiNMKlimentidisYCVazquezAIDuarteCW 2011 Beyond missing heritability: Prediction of complex traits. PLoS Genet 7 e1002051 doi:10.1371/journal.pgen.1002051
32. EfronBTibshiraniR 1986 Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Statist Sci 1 54 75
33. KusakabeSYamaguchiYBabaHMukaiT 2000 The genetic structure of the Raleigh natural population of Drosophila melanogaster revisited. Genetics 154 679 685
34. FalconerDSMackayTFC 1996 Introduction to quantitative genetics Harlow, England Pearson
35. QanbariSPimentelETetensJThallerGLichtnerP 2010 The pattern of linkage disequilibrium in german Holstein cattle. Anim Genet 41 346 356 doi:10.1111/j.1365-2052.2009.02011.x
36. TenesaANavarroPHayesBJ 2007 Recent human effective population size estimated from linkage disequilibrium. Genom Res 17 520 526
37. HabierDFernandoRLDekkersJCM 2007 The impact of genetic relationship information on genome-assisted breeding values. Genetics 177 2389 2397
38. MeuwissenTHE 2009 Accuracy of breeding values of ‘unrelated’ individuals predicted by dense SNP genotyping. Genet Sel Evol 41 doi:10.1186/1297-9686-41-35
39. VisscherPMMedlandSEFerreiraMARMorleyKIZhuG 2006 Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full sublings. PLoS Genet 2 e0020041 doi:10.1371/journal.pgen.0020041
40. GonzálezJPetrovDA 2009 The adaptive role of transposable elements in the Drosophila genome. Gene 448 124 133
41. VanRadenPMVan TassellCPWiggansGRSonstegardTSSchnabelRD 2009 Reliability of genomic predictions for North American Holstein bulls. J Dairy Sci 92 16 24
42. AulchenkoYSStruchalinMVBelonogovaNMAxenovichTIWeedonMN 2009 Predicting human height by Victorian and genomic methods. Eur J Human Genet 17 1070 1075
43. BrowningBLBrowningSR 2009 A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84 210 223
44. StoneM 1974 Cross-validation choice and assessment of statistical predictions. J Roy Statist Soc B 36 111 147
45. StoneM 1977 An aymptotic equivalence of choice of model by cross-validation and Akaike's criterion. J Roy Statist Soc B 39 44 47
46. AllenD 1977 The relationship between variable selection and data augmentation and a method of prediction. Technometrics 16 125 127
47. LegarraARobert-Grani_eCManfrediEElsenJM 2008 Performance of genomic selection in mice. Genetics 180 611 618
48. HillWGWeirBS 1995 Maximum likelihood estimation of gene location by linkage disequilibrium. Am J Hum Genet 54 704 714
49. AdamsMDCelnikerSEHoltRAEvansCAGocayneJD 2000 The genome sequence of Drosophila melanogaster. Science 287 2185 2195 doi:10.1126/science.287.5461.2185
50. Fiston-LavierASSinghNDLipatovMPetrovDA 2010 Drosophila melanogaster recombination rate calculator. Gene 463 18 20
51. EfronB 1987 Better bootstrap confidence intervals. J Am Stat Assoc 82 171 185
52. SvedJA 1971 Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor Popul Biol 2 125 141
53. HendersonCR 1984 Applications of Linear Models in Animal Breeding Guelph, Canada University of Guelph
54. GilmourARGogelBJCullisBRThompsonR 2006 ASReml user guide release 2.0 Hemel Hempstead, UK VSN International Ltd.
55. IhakaRGentlemanR 1996 R: A language for data analysis and graphics. J Comput Graph Statist 5 299 314
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Pánevní endometrióza spojená s volnou tekutinou v peritoneální dutině snižuje úspěšnost otěhotnění po intrauterinní inseminaci
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



