-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
Epigenetic modifications influence gene expression and provide a unique mechanism for fine-tuning cellular differentiation and development in multicellular organisms. Here we report on the biological functions of UTX-1, the Caenorhabditis elegans homologue of mammalian UTX, a histone demethylase specific for H3K27me2/3. We demonstrate that utx-1 is an essential gene that is required for correct embryonic and postembryonic development. Consistent with its homology to UTX, UTX-1 regulates global levels of H3K27me2/3 in C. elegans. Surprisingly, we found that the catalytic activity is not required for the developmental function of this protein. Biochemical analysis identified UTX-1 as a component of a complex that includes SET-16(MLL), and genetic analysis indicates that the defects associated with loss of UTX-1 are likely mediated by compromised SET-16/UTX-1 complex activity. Taken together, these results demonstrate that UTX-1 is required for many aspects of nematode development; but, unexpectedly, this function is independent of its enzymatic activity.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002647
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002647Summary
Epigenetic modifications influence gene expression and provide a unique mechanism for fine-tuning cellular differentiation and development in multicellular organisms. Here we report on the biological functions of UTX-1, the Caenorhabditis elegans homologue of mammalian UTX, a histone demethylase specific for H3K27me2/3. We demonstrate that utx-1 is an essential gene that is required for correct embryonic and postembryonic development. Consistent with its homology to UTX, UTX-1 regulates global levels of H3K27me2/3 in C. elegans. Surprisingly, we found that the catalytic activity is not required for the developmental function of this protein. Biochemical analysis identified UTX-1 as a component of a complex that includes SET-16(MLL), and genetic analysis indicates that the defects associated with loss of UTX-1 are likely mediated by compromised SET-16/UTX-1 complex activity. Taken together, these results demonstrate that UTX-1 is required for many aspects of nematode development; but, unexpectedly, this function is independent of its enzymatic activity.
Introduction
The proper development of multicellular organisms requires strict regulation of cell-specific gene expression to ensure appropriate cell fate specification, cellular differentiation, and organogenesis. In addition to transcription factors, gene expression is controlled by chromatin organization, which is regulated by chromatin-remodelling factors and the post-translational modifications of histone proteins [1]–[3].
An important post-translational modification is the mono - (me), di - (me2), or tri - (me3) methylation of lysine residues (K) on the tail of histone 3 (H3). Specifically, the methylation of specific lysine residues plays a major role in the maintenance of active and silent gene expression states. The combination of H3 K4, K36, and K79 tri-methylation generally marks transcriptionally active regions, whereas H3 K9 and K27 tri-methylation marks regions of transcriptionally silenced genes [2]. The levels of methylation are modulated by the action of histone methyltransferases (HMTs) and histone demethylases (HDMs). The largest group of histone demethylases contains a Jumonji C-domain (JmjC) that catalyzes the demethylation of specific lysine and arginine residues by an oxidative reaction requiring iron [Fe(II)] and α-ketoglutarate (αKG) as cofactors [4]. There are 28 JmjC-containing proteins in humans, grouped in different families, and the majority of these are evolutionarily conserved [5]. The KDM6 subfamily (UTX/UTY/JMJD3) was shown to catalyze the demethylation of H3K27me2/3 [6]–[11], and the individual members were shown to regulate differentiation in several cellular systems [6], [7], [10]. In C. elegans, there are four KDM6 family members: jmjd-3.1, jmjd-3.2, jmjd-3.3, closely related to JMJD3, and utx-1, the unique homologue of the human UTX/UTY. The functional role of these proteins in nematodes is not well defined. jmjd-3.1 has been reported to regulate somatic gonadal development [6], while utx-1 has been implicated in vulva differentiation and aging [12]–[14].
In this report, we have analyzed the developmental functions of UTX-1. We show that utx-1 plays a vital role during embryogenesis and acts in several aspects of nematode postembryonic development. Surprisingly, we found that the catalytic activity of UTX-1 is not of critical importance for UTX-1 function in development. Genetic and biochemical analyses indicate that UTX-1 acts through a SET-16(MLL)/UTX-1 complex and that the primary role of UTX-1 resides in the regulation of the activity of this complex.
Results
Loss of utx-1 results in reduced fertility and lethality
C. elegans D2021.1 encodes for a predicted protein of 134 kDa that has high homology and co-linearity with the mammalian UTX/UTY proteins (Figure 1A); thus we named this gene and its product utx-1 and UTX-1, respectively. UTX-1 is expressed in most, if not all, nuclei of early and late stage embryos (Figure 1B) as well as during all of the larval stages and into adulthood (Figure 1C), suggesting that UTX-1 could have a functional role throughout C. elegans development. To determine the biological function of UTX-1 two deletion mutant strains, utx-1(tm3136) and utx-1(tm3118), were analyzed (Figure 1A). The tm3136 allele is a 236 bp deletion that creates a premature stop codon, potentially encoding a truncated protein of only 28 amino acids, and very likely producing a null mutant. The tm3118 allele is an out–of-frame deletion of 547 bp. The truncated protein potentially retains the first 620 amino acids, but is lacking the JmjC domain and catalytic activity. The two alleles have similar phenotypes suggesting that they are both loss of function mutants.
Fig. 1. UTX-1 expression and utx-1 embryonic phenotypes. 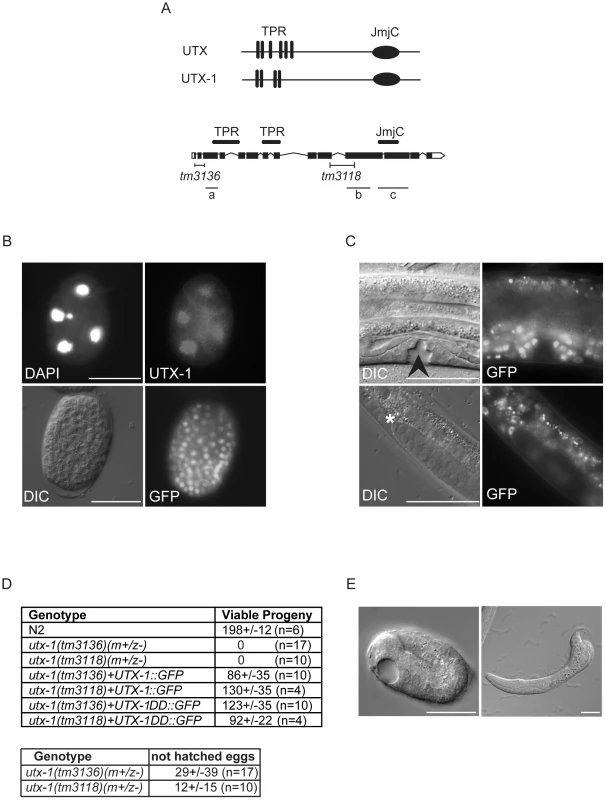
(A) Top: Human UTX and the C. elegans homologue UTX-1. TPRs, tetratricopeptide repeats; JmjC, Jumonji C domain. Bottom: Genomic organization of utx-1. Black H-shaped lines indicate the position of the tm3136 and tm3118 deletions. Black lines indicate the position of the different RNAi fragments (a, b, and c) used in this study. (B) UTX-1 expression during embryogenesis analyzed by immunostaining with an anti-UTX-1 antibody (top panel, right) and by epifluorescence (bottom panel, right). DAPI staining and Nomarski (DIC) images are also shown on the left. (C) UTX-1 expression by epifluorescence (right panels) during larval development. Nomarski (DIC) images are shown on the left panels. Asterisks indicate the distal tip cell, arrow head the forming vulva. Animals are oriented anterior to the left. (D) Brood size of wild type, utx-1 mutant worms and rescued utx-1 lines. Progeny is given as the average number of viable progeny per worm ± SD. The number of laid, not hatched, eggs counted in utx-1 (m+/z−) mutants is reported in the lower table. utx-1+UTX-1::GFP and utx-1+UTX-1DD::GFP, are utx-1 transgenic lines expressing wild-type or catalytically inactive mutant of UTX-1, respectively, as extrachromosomal arrays. (E) Representative Nomarski (DIC) images of a utx-1(tm 3136) mutant embryo and escaper L1 larva. Similar phenotypes are observed in utx-1(tm 3118) (not shown). Bars in B and E are 20 µm, in C 50 µm. Animals are oriented anterior to the left. Homozygous utx-1 mutant worms that are derived from heterozygous mothers providing maternal UTX-1, utx-1(m+/z−), are viable and reach adulthood. However, they produce only a few, mostly unviable, utx-1(m−/z−) eggs (Figure 1D and 1E), suggesting that UTX-1 is required for embryogenesis and that the lack of UTX-1 can be overcome by maternal contribution. Analysis of the dead embryos revealed that mutant utx-1 animals mainly arrested as late embryos (Figure 1E). Dead L1 larvae, with misshapen bodies (Figure 1E), were rarely observed (5%, n>200). A putx-1::UTX-1::GFP (UTX-1::GFP) translational reporter as extra-chromosomal array was able to rescue the embryonic lethal phenotype observed in heterozygous utx-1(m−/z−) progeny from mothers carrying either the tm3136 or tm3118 allele (Figure 1D) in several transgenic lines, leading us to conclude that UTX-1 is essential for embryogenesis and that the zygotic expression of UTX-1 is sufficient to restore embryonic viability. Indeed, progeny that did not receive the transgene from the mother, died as late stage embryos (not shown) or malformed L1 larvae (Figure S1A), suggesting that UTX-1 is not required for very early embryogenesis. In agreement with this, analysis of epithelial junctions using an AJM-1::GFP translational reporter [15] suggests that the morphology of utx-1(m−/z−) embryos, that did not inherit the transgene, was normal at early stages and progressively deteriorated throughout development (Figure S1B). Analysis of markers for intestinal (elt-2::GFP), muscular (hlh-1::GFP and myo-2::GFP), and hypodermal (dpy-7::GFP) cells revealed that these cell lineages are correctly established in utx-1(tm3118) mutant worms (Figures S2, S3, S4, S5). However, a progressive loss of myo-3::GFP transgene expression during embryogenesis was observed and little GFP signal was detected in L1 escapers (Figure S6), suggesting that defects in muscle function might account, at least in part, for the lethality of utx-1 null animals (Figure 2B).
Fig. 2. utx-1 postembryonic phenotypes. 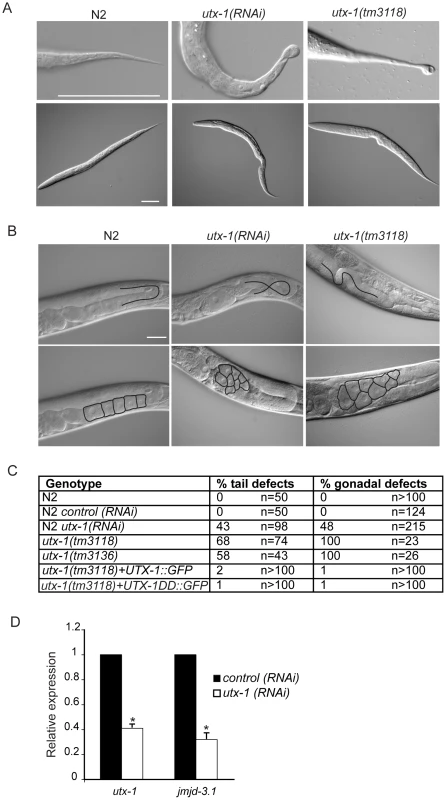
(A) Representative DIC images of L1 larvae tails (upper panel) and L1 larvae (lower panel) of N2, utx-1(RNAi) and utx-1(tm3118) animals. Animals are oriented anterior to the left. Scale bar is 50 µm. (B) Representative DIC images of gonads in adults N2, utx-1(RNAi) and utx-1(tm3118) animals. Scale bar is 25 µm. In the upper panels, blacks lines indicate the migration of the gonad arm. In the lower panels, black lines indicate the contours of the oocytes. Animals are oriented anterior to the left. (C) Percentages of posterior (% tail defects) and gonad (% gonadal defects) defects in the indicated strains are shown. For RNAi, F1 larvae and adults from at least three independent experiments were analyzed. (D) utx-1 and jmjd-3.1 mRNA levels in embryos after control (black bars) or utx-1 (white bars) RNAi treatment as measured by qPCR, using rpl-26 mRNA as internal control. *P<0.01 (Student's t-test). Reduction of utx-1 results in posterior defects and in aberrant gonad migration and organization
To determine the function of UTX-1 at later developmental stages, we analyzed both the utx-1(m+/z−) mutant worms, in which the zygotic contribution of UTX-1 is lost, and wild-type worms in which utx-1 expression was downregulated by feeding RNA interference (RNAi). RNA interference, with constructs targeted to three different regions of utx-1 (Figure 1A), resulted in an approximately 60% reduction of utx-1 mRNA in F1 progeny and in a significant reduction of UTX-1 protein expression (Figure S7A). In agreement with the phenotype of utx-1(m+/z−) mutant animals, the utx-1(RNAi) F1 animals had reduced fertility (Figure S7B). Furthermore, variable defects, often located posteriorly, were observed in about 40% of the utx-1(RNAi) worms (Figure 2A and 2C). The posterior defects observed in animals treated with utx-1(RNAi) were very similar to the defects observed in utx-1(m−/z−) dead larvae (Figure 2A, 2C and Figure S8). This demonstrates that RNA interference can be used to efficiently analyze the postembryonic roles of UTX-1, and that the posterior phenotype in utx-1(m−/z−) is due to the loss of utx-1. Importantly, transgenic expression of wild-type utx-1 fully rescued the posterior defects in larvae.
The reduced fertility observed in utx-1(m+/z−) animals might be due to a regulatory role for UTX-1 in either somatic gonad or germline development. Homozygous mutant utx-1(m+/z−) animals from heterozygous animals generally develop germlines with correct proliferation and differentiation patterns (not shown), as demonstrated by the fact that oocytes are formed (Figure 2B) and by an ability to lay a few dead embryos, suggesting that the sterility is not related to a germline defect. However, animals lacking UTX-1 activity have defects in gonad migration and oocyte organization. The shape of the gonad is dictated by the coordinated migration of two distal tip cells (DTCs), which are part of the somatic gonad structure and move away from the gonad primordium during postembryonic development, leading to two consecutive turns forming the U-shaped gonad arms observed in adult animals. Using transgenic animals carrying a distal tip cell marker, lag-2::GFP [16], we observed aberrant gonadal migration in 42% (n = 176) of the utx-1(RNAi) animals. Morphological analysis by DIC of utx-1(RNAi) animals further confirmed that 48% (n = 215) of the animals showed a failure to turn or abnormal turning of at least one gonad arm (Figure 2B and 2C), and these animals often (41%, n = 137) developed misshapen gonads, with an enlargement of the proximal end of the gonad arms and a misorganization of oocytes (Figure 2B). These gonad phenotypes were also identified in utx-1(m+/z−) mutant animals (Figure 2B, 2C and Figure S9), and they were efficiently rescued by the UTX-1::GFP transgene, reinforcing that these aberrations are caused by the loss of utx-1 (Figure 2C). The fact that the transgenic expression of wild-type utx-1 is able to rescue the sterility and the gonadal phenotypes suggests that utx-1 has a role in the somatic gonad rather than in the germline, where transgenes are normally silenced. Consistent with this, GFP-tagged UTX-1 is expressed in the distal tip cells during migration (Figure 1C) and other tissues of the somatic gonad, such as the sheath cells and the spermatheca (not shown) and not in the germline.
The aberrant migration and oocyte organization defects are similar those we reported for jmjd-3.1 loss-of-function mutants, which encodes one of the C. elegans homologues of the JMJD3 family [6]. To determine if there is a link between these two observations, we tested if UTX-1 affected the expression of jmjd-3.1 by performing quantitative PCR on utx-1(RNAi) animals. As shown in Figure 2D, utx-1(RNAi) animals have reduced levels of jmjd-3.1, suggesting that UTX-1 may, directly or indirectly, regulate jmjd-3.1 expression. Additionally, an enhancement of the phenotype was not observed when utx-1 was reduced in a jmjd-3.1 mutant genetic background (see below), suggesting that both genes are acting in the same genetic pathway to regulate somatic gonadal development.
UTX-1 demethylates H3K27me2/3, but the catalytic activity is not required for proper development
UTX-1 belongs to the KDM6 family, of which members have been shown to catalyze the demethylation of H3K27me3 and H3K27me2 [17]. Several observations indicate that this role is conserved throughout the C. elegans life cycle. First, loss of the zygotic and maternal contributions of utx-1 results in increased global levels of H3K27me2/3 at the embryonic stage (Figure 3A). Second, reduction of UTX-1 by RNA interference results in a significant increase of H3K27me3 levels at different larval stages (data not shown, [12], [18]). Third, exogenous expression of wild-type UTX-1 in utx-1 null animals restores H3K27me3 to wild-type level (Figure 3A). Fourth, over-expression of UTX-1 in wild-type animals results in a significant reduction of global H3K27me3 levels (Figure 3B). Finally, the decreased level of H3K27me3 observed in N2 worms overexpressing UTX-1 (Figure 3B) is well correlated with the degree of UTX-1::GFP overexpression, as shown in Figure S10.
Fig. 3. UTX-1 is an H3K27me2/3 demethylase. 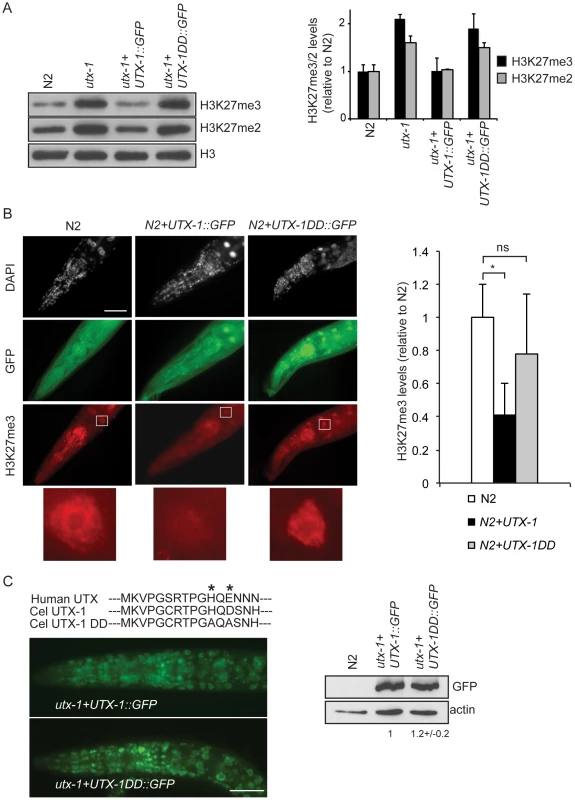
(A) Protein lysates from embryos of the indicated strains were probed with antibodies against H3K27me3 and H3K27me2. H3 was used as loading control. Quantification of the western blot is shown in the graphic on the right. Error bars indicate the standard deviation calculated using at least 2 replicates. The signals were quantified using ImageJ software and normalized to H3. The values are relative to N2 levels. Similar results were obtained with at least two different transgenic lines and in the two utx-1 genetic backgrounds (tm3118 and tm3136). (B) Representative images of N2 expressing a translational construct for wild-type (N2+UTX-1::GFP) and catalytically inactive UTX-1 (N2+UTX-1DD::GFP) GFP fusion and GFP-negative siblings, fixed and stained with H3K27me3 antibody and DAPI. The white square encloses an intestinal cell, used for the H3K27me3 quantification. Enlargement of the white square is shown at the bottom of the panel. Quantification of H3K27me3 levels is shown in the graphic on the right. At least 25 cells for each genotype were quantified as described in Materials and Methods. Mean fluorescence + s.e.m. values of two independent experiments are displayed. *P<0.01. (Student's t-test). Animals are oriented anterior to the left. (C) Top: Alignment of a part of the Jumonji C domain of human UTX with UTX-1 and with the catalytically inactive UTX-1DD (DD = Demethylase Dead). Asterisks indicate two of the three conserved amino acids in the iron-binding domain (HXD/EXnH) of the JmjC-domain, modified in the UTX-1DD. Bottom: Epifluorescence of utx-1 mutant animals, carrying a translational GFP fusion of wild-type UTX-1 (utx-1+UTX-1::GFP) or catalytically inactive UTX-1 (utx-1+UTX-1DD::GFP). Anterior parts of the animals are shown, with anterior to the left. On the right, lysates from L1 carrying the two transgenes were probed with GFP antibody. Actin was used as loading control. The signal was quantified using ImageJ program and normalized to actin. Next, we tested if the catalytic activity of UTX-1 is responsible for the phenotype observed in utx-1 null mutants and utx-1(RNAi) animals. To this end, we expressed in utx-1 mutants a GFP-tagged mutated form of the UTX-1 protein (for simplicity called UTX-1DD::GFP, DD = Demethylase Dead), carrying mutations in two of the three conserved amino acids in the iron-binding motif (HXD/EXnH) of the JmjC-domain (indicated by asterisks in Figure 3C). Several reports have shown that these amino acids are required for iron binding and thus for the catalytic activity of all JmjC-domain containing demethylases characterized so far [6]–[11], [19]–[21]. All UTX-1::DD::GFP transgenic lines generated (8/8) showed expression at levels similar to wild-type UTX-1 (Figure 3C) and were fertile and able to produce viable progeny (Figure 1D). Importantly, re-expression of catalytically inactive UTX-1 did not restore the wild-type level of H3K27me3 in utx-1 null animals (Figure 3A) and did not influence the H3K27me3 level when overexpressed in wild-type animals (Figure 3B), thus confirming that the amino acids substitutions affected UTX-1 enzymatic activity. This unexpected result strongly indicates that the demethylase activity of UTX-1 is not important for either embryonic development or animal viability. Subsequently, we tested if the other observable phenotypes were dependent on UTX-1 enzymatic activity. Tail and gonadal defects were also efficiently rescued (Figure 2C) in 50% (4/8) of the transgenic lines, indicating that UTX-1, but not its catalytic activity, is required for correct posterior and gonadal development.
C. elegans KDM6 family members do not compensate for the reduced H3K27me2/3 demethylase activity in utx-1 mutant worms
jmjd-3.1, jmjd-3.2, and jmjd-3.3 (Figure 4A) are C. elegans KDM6 family members closely related to human JMJD3. Animals carrying mutations in one of these genes are viable, fertile (not shown), and do not show up-regulated levels of H3K27me3 by western blot analysis (Figure 4B and Figure S11B). However, triple mutant worms carrying deletions in all three JMJD3-like genes showed increased global levels of H3K27me3 (Figure 4B and Figure S11B), suggesting that these proteins are H3K27me3 demethylases and might act redundantly with UTX-1. Several lines of evidences indicate that the JMJD3-like genes do not function redundantly with UTX-1. Analysis of the transcriptional expression levels of the JMJD3-like genes in wild-type worms indicated that only jmjd-3.1 is expressed at levels comparable to utx-1, while jmjd-3.2 and jmjd-3.3 are only weakly expressed, in particular during larval stages (Figure S11A). Furthermore, the transcriptional expression pattern of the JMJD3-like genes appeared generally restricted to specific tissues or, as in the case for jmjd-3.2, even to few cells (Figure S11C); this is in contrast to the broad expression pattern of UTX-1. In addition, the triple mutant lacking the JMJD3-like genes is viable and fertile, with no defects in the posterior region of the body (Figure 4C) and with only minor gonadal defects (Figure 4C), most likely due to the absence of jmjd-3.1. Importantly, the down-regulation of utx-1 by RNA interference in the triple mutant genetic background did not exacerbate the posterior or the gonadal defects associated with utx-1 reduction in wild-type animals (Figure 4C). Taken together, these results imply that the members of the KDM6 class do not act redundantly.
Fig. 4. KDM6 family in C. elegans. 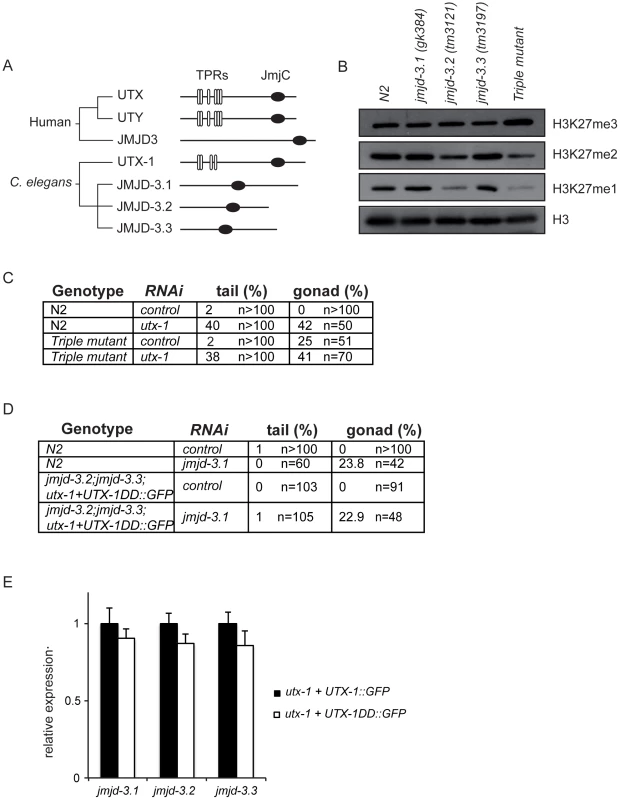
(A) Phylogenetic cluster of human UTX, UTY, JMJD3 and homologous proteins in C. elegans. TPRs, tetratricopeptide repeats; JmjC, Jumonji C domain. (B) Protein lysates from eggs of the indicated strains were analyzed by western blot using the indicated antibodies. H3 was used as loading control. Quantification of the western blot is shown in Figure S11. (C) Percentage of tail and gonad defects observed in N2 and the triple mutant (jmjd-3.1;jmjd-3.2;jmjd-3.3) after treatment with control or utx-1(RNAi). F1 animals from at least two independent experiments were scored. (D) Percentage of tail and gonad defects observed in N2 and the triple mutant (jmjd-3.2; jmjd-3.3;utx-1+UTX-1DD::GFP) after treatment with control or jmjd-3.1(RNAi). F1 animals from at least two independent experiments were scored. (E) Levels of expression of the JMJD3-like genes in eggs derived from the utx-1(tm3136) mutant strain rescued with the catalytically inactive utx-1 (utx-1+UTX-1DD::GFP) relative to utx-1(tm3136) rescued with utx-1 wild-type (utx-1+UTX-1::GFP). rpl-26 mRNA was used as internal control for normalization. However, in light of the unexpected results obtained with the catalytically inactive UTX-1 mutant, it is important to take into consideration the possibility that JMJD3-like genes could, nevertheless, compensate for the lack of UTX-1 activity in utx-1 mutant worms expressing the catalytically inactive form of UTX-1. In this case, we would expect that the loss or reduction of other H3K27me3 demethylases in the utx-1 mutant rescued with the catalytically inactive UTX-1 would result in utx-1-specific abnormalities (posterior defects and aberrant gonadal migration). To test this hypothesis, we generated a triple mutant jmjd-3.2; jmjd-3.3;utx-1+UTX-1DD::GFP in which the fourth member of the KDM6 family, jmjd-3.1, was down-regulated by RNA interference. In this genetic background, no posterior defects were observed and the degree of gonadal defects was similar to those observed in wild-type animals under the same conditions (Figure 4D). Furthermore, quantitative PCR showed no increased expression levels of the JMJD3-like genes in the rescued transgenic line utx-1+UTX-1DD::GFP compared to utx-1+UTX-1::GFP (Figure 4E). These results, together with the fact H3K27me3 levels are still up-regulated in utx-1 expressing the catalytically inactive form of UTX-1 (Figure 3A), strongly indicate that the JMJD3-like proteins do not compensate for the lack of UTX-1 catalytic activity and that the catalytic activity of UTX-1 is not required for proper development.
UTX-1 acts in the SET-16/MLL complex
The mammalian UTX is part of the MLL3/MLL4 H3K4me3 methyltransferase complex [22]–[24] that also includes the specific component PTIP, and WDR5, ASH2L, and RbBP5 as core components, which are also shared by other complexes [25]. The high conservation of these proteins in nematodes (WDR5/tag-125/wdr-5.1, ASH2L/ash-2, RbBP5/F21H12.1/rbbp-5, MLL3-4/set-16, UTX/utx-1, and PTIP/pis-1), suggests that a similar complex could also exist in C. elegans. To test if an MLL3-4/UTX-like complex (SET-16/UTX-1) is present in C. elegans, we purified GFP-tagged UTX-1 and associated proteins from a mixed population of transgenic animals, enriched with embryos (Figure 5A). The identities of the interacting proteins were determined by mass spectrometry and are listed in the Table S1. As a control, N2 lysates were subject to the same procedure and the recovered proteins (listed in Table S2) were considered contaminants and used to confirm the specificity of the identified interacting proteins. Strikingly, all of homologous components of the mammalian MLL3/4 complex were identified as UTX-1 partners in C. elegans (Figure 5B). As further verification, we utilized a transgenic line carrying HA-tagged WDR-5.1 [26], the most prominent WDR5-like protein recovered by mass spectrometry, in which we expressed UTX-1::GFP. As shown in Figure 5C, in lysates derived from embryos, both UTX-1::GFP and endogenous UTX-1 were found associated with WDR5.1, further supporting the existence of a SET-16/UTX-1 complex in C. elegans. Importantly, the catalytically inactive mutant UTX-1DD::GFP was also recovered by WDR-5.1 immunoprecipitation (Figure 5C). Gel filtration analysis of lysates from transgenic lines carrying either the wild-type or the catalytically inactive forms of UTX-1 further confirmed that both UTX-1 and UTX-1DD are engaged in large complexes (Figure 5D), further supporting that a functional JmjC domain is not required for the association with the complex.
Fig. 5. UTX-1 is part of a MLL-like complex. 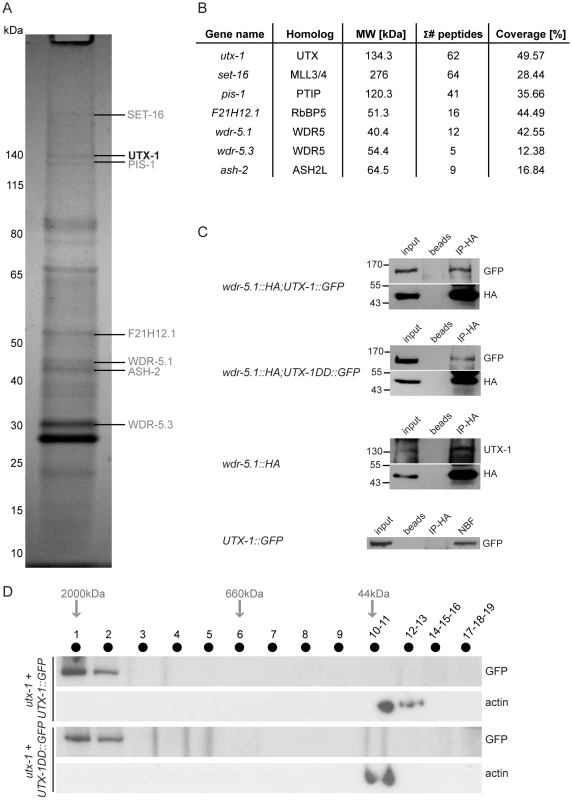
(A) Immunoprecipitation of GFP tagged UTX-1 from a mixed population (eggs and adults) of utx-1(tm3118) rescued with UTX-1::GFP. Affinity purified proteins were resolved by SDS-PAGE and stained with colloidal Coomassie. Homologues of the mammalian UTX-MLL complex co-eluted with the bait and identified by LC-MS/MS are listed in grey. Position of the bait protein is shown in black. Molecular weight markers are indicated to the left of the gel. (B) Table summarizing the identified homologues of the components of the mammalian UTX-MLL complex. Gene names, molecular weight in kDa, number of unique peptides and sequence coverage in percentage are reported. (C) Co-immunoprecipitations of WDR-5.1::HA and UTX-1. Total protein extracts from eggs of the indicated strains were immunoprecipitated using anti-HA affinity gel beads. The precipitates were analyzed by SDS-PAGE followed by western blotting using antibodies against HA, GFP or endogenous UTX-1. Input = 30 µg of protein extract. NBF = non bound fraction. (D) UTX-1-associated protein complex assessed by size exclusion chromatography. Superose 6 gel filtration of total protein extracts derived from UTX-1 mutant rescued with wild-type (utx-1+UTX-1::GFP) and catalytically inactive UTX-1 (utx-1+UTX-1DD::GFP). Fractions were analyzed by western blotting using GFP and actin antibodies. We then verified the functional correlation of the SET-16/UTX-1 complex components by testing if their loss or downregulation could result in phenotypes similar to those observed in the utx-1 mutant. Loss of set-16 results in embryonic and early larval lethality [27]. The analysis of set-16(n4526) young larvae revealed the presence of posterior defects similar to those identified in utx-1 null animals (Figure 6A and Figure S8), and set-16(RNAi) animals that escaped embryonic and early larval lethality, often had abnormal gonad migration and enlargement (Figure 6A, 6B and Figure S9), which phenocopied the effect of the loss of utx-1. Similarly, in pis-1(ok3720) mutants and pis-1(RNAi) animals, posterior and gonadal defects were observed, although at a lower degree (Figure 6A and 6B, Figures S8 and S9). RNA interference of the core components of the complex (F21H12.1, wdr-5.1, and ash-2) also resulted in posterior and gonadal defects similar to the ones observed in utx-1 mutants (Figure 6C and Figure S9). It should be noted, that enlargement of the proximal gonad was never observed after the reduction by RNAi of F21H12.1 and ash-2 and was rarely observed in wdr-5 (RNAi) animals (Figure S9). We then tested the effects of simultaneously downregulating specific components of the complex by RNAi. As shown in Figure 6B, the concurrent knockdown of utx-1 and set-16 or pis-1 did not enhance the phenotypes; similar results were obtained with concomitant silencing of pis-1 and set-16. The high degree of phenotypic similarity and the absence of redundancy are evidence that these genes are acting in the same genetic pathways to regulate posterior patterning and somatic gonadal development. Along the same line, qPCR analysis revealed that set-16 downregulation by RNA interference results in a reduction of jmjd-3.1 mRNA (about 60% decrease compared to control RNAi, data not shown), further supporting the notion that UTX-1 and SET-16 act in the same complex.
Fig. 6. Functions of the SET-16/UTX-1 complex. 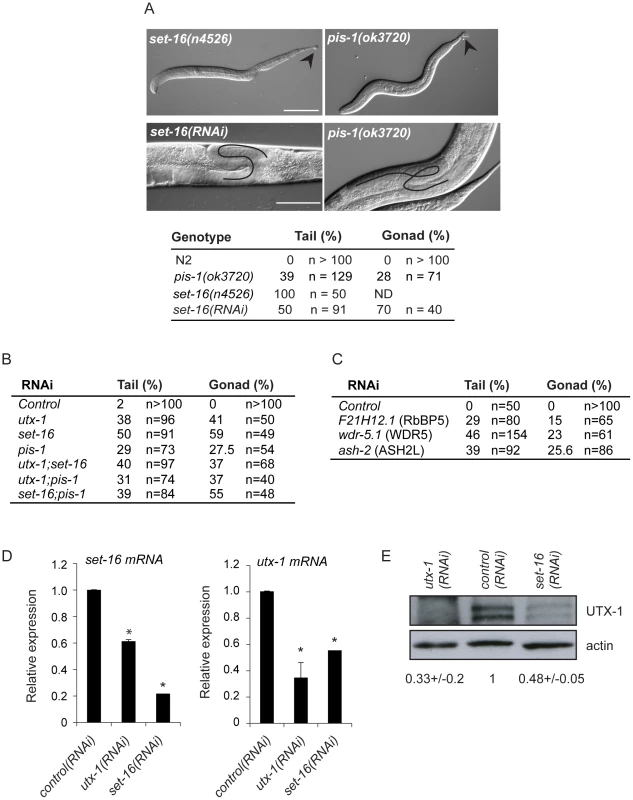
(A) (Upper panel) DIC images of set-16(n4526) and pis-1(ok3720) mutant larvae. Arrowhead indicates misshapen tails. Scale bar is 50 µm. (Lower panel) DIC images of set-16(RNAi) and pis-1(ok3720) adults. Gonadal migration defects are shown. The black line indicates the aberrant gonadal migration. Scale bar is 20 µm. Animals are oriented anterior to the left. The percentage of tail and gonadal defects associated to loss or reduction of set-16 and pis-1 are reported on the right. (B) Percentage of tail and gonad defects after RNAi of the indicated genes. F1 L1 larvae and adult animals from at least two independent experiments were scored. (C) Percentage of tail and gonad defects after RNAi of the indicated genes. F1 or F2 L1 larvae and adult animals from at least two independent experiments were scored. (D) utx-1 and set-16 mRNA levels in embryos of worms treated with control, utx-1 or set-16(RNAi). *P<0.01 (Student's t-test). (E) Protein lysates of embryos from worms treated with control, utx-1 or set-16(RNAi) were probed with an antibody against UTX-1. Actin was used as loading control. The signal was quantified using ImageJ program and normalized to actin. Indicated values are relative to control (RNAi) and derive from two independent experiments. Since the catalytic activity of UTX-1 is not necessary to rescue the developmental defects observed both in utx-1 mutants and in animals in which different factors of the complex were lost or down-regulated, we hypothesized that UTX-1 might regulate the expression of other components of the complex. In support of this, we found that the levels of set-16 mRNA were reduced in utx-1(RNAi) animals (Figure 6D). Interestingly, downregulation of set-16 also results in decreased expression of utx-1 mRNA and protein (Figure 6D and 6E), suggesting an interdependent regulation of, at least, these two members of the complex.
Taken together the data demonstrate that the SET-16/UTX-1 complex is present in C. elegans, and it is required for development. That the loss or downregulation of single components of the complex results in similar phenotypes as those observed in utx-1 null mutants, indicates that each component is required for the complex to function normally and that the defects associated with the loss of UTX-1 are likely the result of compromised SET-16/UTX-1 complex activity.
Discussion
We have demonstrated that C. elegans UTX-1 is an H3K27me2/3 demethylase that is essential for development during embryonic and larval stages of the nematode, independently of its demethylase activity. Animals lacking the maternal and zygotic contribution of UTX-1 arrest during late embryogenesis. Although, analyses of reporter genes revealed no major defects in lineage specifications, a reduction of myo-3::GFP, expression, but not hlh-1::GFP, was observed in utx-1 mutant animals, suggesting that utx-1 might regulate genes involved in muscle function. In agreement, mammalian UTX has been implicated in terminal differentiation of muscle cells [28]. The maternal contribution of UTX-1 allows utx-1(m+/z−) worms to reach adulthood, but defects arise at different stages of development, including abnormal gonad migration and oocyte misorganization. This latter phenotype could explain, at least in part, the reduced fertility of utx-1(m+/z−) animals. We have previously shown that proper gonad migration partly depends on another H3K27me3 demethylase, jmjd-3.1 [6]. The expression level of jmjd-3.1 is significantly reduced in utx-1(RNAi) animals. However, it should be noted that the loss of utx-1 leads to a more severe phenotype than the loss of jmjd-3.1, which only influences gonadal processes at high temperature and moderately reduces fertility. These results suggest that utx-1, in addition to jmjd-3.1, modulate additional genes required for establishing the correct developmental program of gonads.
While utx-1 represents the unique UTX/UTY homologue, C. elegans has three other genes with homology to the single mammalian JMJD3 gene (jmjd-3.1, jmjd-3.2 and jmjd-3.3). We generated mutant animals carrying mutations in all three JMJD3-like genes and, unexpectedly, we did not detect any additional phenotypes in the triple mutants, other than the phenotypes already reported for jmjd-3.1 [6]. While it is possible that residual gene function remains in these mutants, the global level of H3K27me3 was significantly increased in the triple knockout worms, whereas no increase was observed in the jmjd-3.1 mutant strain alone ([6]; Figure 4B). This data suggests that the JMJD3-like demethylases might regulate the expression of restricted sets of genes or that they have overlapping functions. Our analysis of the global levels of H3K27me2/3 also suggests that UTX-1 is the most important demethylase for the removal of the H3K27me3 mark among the members of the KDM6 family. Accordingly, the loss of utx-1 results in sterility (in m+/z− worms) and in embryonic lethality (in m−/z− worms) while animals lacking the three JMJD3 homologues are fertile and viable. This result indicates that utx-1 plays unique and essential roles during embryonic and postembryonic development and suggests that the JMJD3-like proteins, like the human homologues [10], [29], are mainly required for regulating cellular responses under particular conditions, such as stress or aging.
Strikingly, we found that the catalytic activity of C. elegans UTX-1 is not required for the function of the protein in the developmental processes analyzed. This is at odds with a previous report describing the role of utx1 genes in D. rerio, in which human wild-type, but not the catalytically inactive mutant, partially rescued the defects in UTX morphant animals [9]. We do not know if this apparent dissimilarity is due to an organismal difference, as suggested by the fact that C. elegans UTX-1 does not regulate HOX genes (data not shown) as it does in zebrafish [9] and that zebrafish has two UTX homologues, or to the different experimental approaches. Interestingly, recent results also suggest a catalytic-independent role for human JMJD3 and UTX in chromatin remodeling in a subset of T-box target genes [30]. Quantitative PCR and analysis of reporter genes failed, however, to identify any regulation of selected C. elegans T-box genes by UTX-1 (not shown).
The demonstration that UTX, which mediates H3K27me2/3 demethylase activity, is part of the MLL3/4 complex, which also has H3K4 methyltransferase activity [6], [7], suggests a model in which the coordinated removal of repressive marks (H3K27me3) and the deposition of activating marks (H3K4me3) fine-tune transcription during differentiation. We have shown that a similar complex is present in C. elegans, and that it is required to achieve proper development. Indeed, loss or reduction of each component of the complex results in phenotypes similar to those we observed in utx-1 mutants. The lack of synergistic effects in double RNAi experiments further supports the notion that the components of the complex act in the same pathway(s) to regulate posterior body and somatic gonad development. Surprisingly, utx-1 phenotypes are rescued by catalytically inactive UTX-1. The catalytically inactive mutant binds WDR-5.1 similarly to the wild-type protein, and it was identified in gel filtration experiments in a large complex, similarly to its wild-type counterpart. WDR-5.1 is also a component of other complexes and we cannot exclude at this time that the UTX-1/WDR-5.1 interaction might take place in the context of another complex. However, the components of other complexes with which WDR-5.1 is involved have, thus far, not been recovered by our mass spectrometry analysis. For example we did not identify the known WDR-5.1 binding partner SET-2 (the main H3K4me3 methyltransferase in C. elegans) [26], [31], [32], suggesting that UTX-1 is specifically recruited in the SET-16(MLL)-like complex.
Taken together these results strongly suggest that UTX-1 acts through a SET-16/UTX-1 complex and indicate that the primary role of UTX-1 in C. elegans development is independent of the demethylase activity, possibly through the regulation of expression of the complex components. This is suggested by our results showing that UTX-1 is, at least, required for the proper expression of set-16, and that SET-16 is required for the expression of utx-1, suggesting a positive feed forward mechanism for retaining the activity of the SET-16/UTX-1 complex. It is possible that there are additional functions for UTX-1; UTX-1 may be required for targeting the complex to specific genomic regions or it might play a role in the stability of the complex. To correctly address these possibilities, chromatin immunoprecipitation and mass spectrometry analysis must be performed in the context of utx-1 null mutants. Unfortunately, these experiments are currently unfeasible since the utx-1 mutant is unviable. It should be mentioned, however, that downregulation of the human UTX does not interfere with MLL complex formation (Agger K., Helin K., unpublished data), at least in mammals.
Finally, we do not know if UTX-1 works exclusively in association with the SET-16 complex or if it has additional roles as a single protein or in association with other complexes. The results obtained by mass spectrometry analysis suggest that this latter hypothesis might be correct. UTX-1 immunoprecipitates with other proteins involved in distinct chromatin complexes, such as HDA-1 and LIN-53, components of the NuRD complex [33], and MIX-1, which functions in the dosage compensation complex (DCC) [34]. While these interactions await further validation, it is worth noting that elements of the NuRD complex have been involved in vulva formation [33], a postembryonic event in which both UTX-1 and SET-16 have been implicated ([12] and data not shown), and that the DCC has been recently shown to interact with ASH-2, a member of the complex that we describe here. Therefore, it is conceivable that UTX-1 works in diverse chromatin complexes to accomplish functions required at different stages of development or under specific conditions. A MLL complex-independent role for UTX is also supported by the fact that a substantial amount of mammalian UTX is bound to promoter regions depleted of H3K4 methyl marks [35].
We did not detect reduced global levels of H3K4me3 in utx-1 mutant animals (data not shown), which could be expected if UTX-1 regulates the function of a complex having H3K4 methyltransferase activity. This is in agreement with previously reported result in mammals and C. elegans [7], [12] and it is consistent with the fact that inactivation or downregulation of set-16 only results in a minor, if any, reduction in global levels of H3K4me3 [12], [35], [36]. Indeed, similar to mammals, H3K4me3 deposition in C. elegans is mainly regulated by the other H3K4me3 methyltransferase, set-2 [26], [36]. This observation suggests that the SET-16/UTX-1 complex regulates the mark deposition only for a subset of genes, and, consequently, complex impairment does not impact the global levels H3K4me3.
Our analysis failed to uncover a role for UTX-1 catalytic activity during development, and a major question is therefore whether this activity is required for any biological function in C. elegans. Since this work focused on the role of UTX-1 during development, the catalytic activity might be required for other processes that are not implicated in developmental programs and are dispensable for viability. Indeed, recent reports implicate the catalytic activity of UTX-1 in aging [13], [18]. Moreover, UTX-1 activity could act during germline formation to counteract the well-established role of the PRC2/MES complex [37]–[43]. However, we have thus far not been able to establish a function of UTX-1 during germline formation neither alone nor in synthetic interaction with components of the MES complex (data not shown).
In summary, we have shown that UTX-1 plays an essential role in several developmental processes in C. elegans. Surprisingly, the catalytic activity is dispensable for proper development, and our data suggest that UTX-1 acts, instead, through a SET-16/UTX-1 complex. Future studies will be directed at identifying the specific target genes regulated by the complex and the possible role that UTX-1 might play in the stability of the complex and in its recruitment to the target genes.
Materials and Methods
Genetics and strains
C. elegans strains were cultured using standard methods [44]. Strains used were as follows: wild-type Bristol (N2), utx-1(tm3118)X, utx-1(tm3136)X, jmjd-3.1(gk384)X, jmjd-3.2(tm3121)X, jmjd-3.3(tm3197)X, JK2049 qls19 V, set-16(n4526)III, pis-1(ok3720)IV, AZ217(myo-2::GFP), MS438(elt-2::GFP), GS3798(dpy-7::YFP), OP64(hlh-1::GFP), PS3729(ajm-1::GFP). The strain wdr-5.1/tag-125::HA and OE4201(myo-3::GFP) were generous gifts from Francesca Palladino and Thomas Bürglin, respectively. Transgenic animals with specific genetic backgrounds were generated by standard crossing procedure. The C. elegans utx-1 sequence is located on chromosome X and the transcript encompasses 14 exons coding for a predicted protein of 1168 amino acids. The ATG of the gene is located at position 13888 bp of the D2021 cosmid (U23513) and a TAG terminator codon at position 19549 bp. Two alleles of utx-1 were identified at the National BioResource Project (NBRP), Japan. Both alleles were backcrossed three times with N2 before the phenotypic analysis and maintained in culture as heterozygotes. The tm3136 allele lacks 236 bp and the deletion is found at position 13920–14155 bp of the Genbank entry U23513. This deletion creates a premature stop codon and the deleted gene could potentially encode for a truncated protein of 28 amino acids. The tm3118 is an out–of-frame deletion of 547 bp situated at position 17361–17907 bp of the Genbank entry U23513. The truncated protein potentially retains the first 620 amino acids and lacks the JmjC domain. Phenotypic analyses of utx-1 mutant animals (tm3136 and tm3118) were done in blind, before genotyping. jmjd-3.2(tm3121)X and jmjd-3.3(tm3197)X were backcrossed three times before analysis and their deletions, described in Wormbase, were confirmed by sequencing. KDM6 members are all positioned on the X-chromosome, with utx-1 located very closely to jmjd-3.1, thus precluding the generation of a quadruple mutant lacking all the four H3K27me3 demethylases. The triple mutant with deletions in jmjd-3.1(gk384)X, jmjd-3.3(tm3197)X and jmjd-3.2(tm3121)X was generated using standard crossing methods. The triple mutant jmjd-3.2(tm3121);jmjd-3.3(tm3197);utx-1(tm3118)+UTX-1DD::GFP (expressing UTX-1DD as an extrachromosomal array) was generated using standard crossing methods.
Brood size
Single fourth-stage (L4) larvae were plated in agar plates with OP50 bacteria and moved to a new plate every 24 h. Viable progeny were counted every day, for 4 days at 25°C. The average number of progeny produced by a single animal is reported.
RNA interference (RNAi)
RNAi was performed by feeding and carried out as described previously [45]. For UTX-1, a clone (X-4I10) containing the region from 18167 bp to 19214 bp of the GenBank entry U23513 (c in Figure 1A), was obtained from the C. elegans RNAi feeding library (J. Ahringer's laboratory, Wellcome Trust/Cancer Research UK Gurdon Institute, University of Cambridge, Cambridge, UK). Two other clones (a and b in Figure 1A), spanning the regions 14109–14331 bp and 17512–18014 bp of the GenBank entry U23513, respectively, were constructed by PCR. We generated RNAi clones for set-16, ash-2, pis-1, tag-125, and F21H12.1 by amplifying cDNA fragments (approximately 500 pb) before cloning in L4440 plasmid using EcoRI restriction sites (all primer sequences available upon request). Eggs, prepared by hypochlorite treatment, were added onto RNAi bacteria-seeded NMG plates and cultivated at 25°C. Control animals were fed with bacteria carrying the control vector (L4440). Generally, F1 progeny was scored for phenotypes.
Real-time quantitative PCR (RT–qPCR)
Total RNA was isolated from eggs using TRIzol reagent (Invitrogen) and RNAeasy Minikit (Qiagen). cDNA was synthesized using reagents from the TaqMan Reverse Transcription kit (Applied Biosystems). qPCR was performed using SYBR Green 2× PCR Master mix (Applied Biosystems) in an ABI Prism 7300 Real Time PCR system (Applied Biosystems). The measures were normalized to ribosomal protein (rpl-26) RNA levels. All reactions were performed in triplicate, in at least three independent experiments. All primer sequences are available upon request.
Quantitative Western blot analysis
For mutant strains, total protein extracts were prepared from eggs obtained by hypochlorite treatment of adults grown on OP50 at 25°C. For RNAi-treated animals, extracts were prepared from eggs obtained by hypochlorite treatment of adults grown on HT115 containing either the empty feeding vector, or specific RNAi. Protein concentration was estimated using the modified micro-Lowry assay and equal amounts of protein were loaded. The following antibodies were used: polyclonal anti-H3 (Abcam 1791, lot GR9204-1) 1∶30000; polyclonal anti-H3K27me1 (Upstate 07-448, lot DAM1598790) 1∶5000, polyclonal anti-H3K27me2 (Abcam 24684, lot 956943) 1∶2000; polyclonal anti-H3K27me3 (Upstate 07-449, lot 701050758) 1∶2000; monoclonal anti-actin (Chemicon International MAB1501) 1∶10000; peroxidase-labeled anti-rabbit and anti-mouse secondary antibodies (Vector). The specificity of H3K27 antibodies has been tested as shown in Figure S12. Polyclonal C. elegans UTX-1 antibodies were obtained through the Eurogentec polyclonal antibody production service. To generate a specific UTX-1 antiserum, rabbits were immunized with two UTX-1 peptides (MDESEPLPEERHPGNC and SYRRSYKDDANRLDHC). Antibodies were purified using affinity columns coupled with the same peptides and used at 1∶500 dilution. The antibody recognizes in the lysate of wild-type animals a specific band of the predicted size of 134 kDa, absent in the lysates obtained from utx-1 mutant alleles (Figure S12D). Western blots were quantified using ImageJ program (National Institutes of Health).
Construction of tagged UTX-1 and UTX-1DD
For the UTX-1::GFP construct, a 6956-bp fragment of utx-1 including 1290 bp of promoter region and the entire coding region was PCR-amplified from N2 genomic DNA. The resulting fragment was inserted in the multiple cloning site of the pPD95.75 vector (Fire lab).
For the UTX-1DD::GFP construct, the UTX-1::GFP construct was mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Specifically, the DNA sequence was mutated so that the histidine at position 914 (H914) and the aspartic acid at position 916 (D916) were changed to alanine. The DNA sequences of both constructs were verified by sequencing.
Microinjection and production of transgenic lines
To obtain lines carrying extra-chromosomal arrays, 20 ng/µl of UTX-1::GFP and UTX-1 DD::GFP constructs were each co-injected with 100 ng/µl pRF4(rol-6(su1006)) or ttx-3p::RFP in wild-type N2 worms (N2+UTX-1::GFP and N2+UTX-1DD::GFP). Transgenic lines in utx-1(tm3136 and tm3118) genetic backgrounds (utx-1+UTX-1::GFP and utx-1+UTX-1DD::GFP) were generated by crossing.
Microscopy, image processing, quantification, and statistical analysis
Fluorescence microscope and DIC pictures were acquired using an Axiovert 135, Carl Zeiss, Inc. with a 63× Plan Apochrome objective with a NA of 1.4 in immersion oil and a 40× Plan NEOFLUAR with a NA of 0.75, respectively. Pictures were taken at room temperature with a CoolSNAP cf2; Photometrics camera. All pictures were exported in preparation for printing using Photoshop (Adobe). MetaMorph software (MDS Analytical Technologies) was used to quantify the mean and s.e.m. of integrated intensities per cell as described in [46]. The 2–4 most anterior intestinal cells were used for the quantification of H3K27me3/GFP and more than 20 cells from at minimum of 10 animals for each genotype (N2+UTX-1::GFP , N2+UTX-1DD::GFP and GFP-negative siblings) were analyzed in two independent experiments. Only animals showing good H3K27me3 signal in the gonads, as indication of successful immunostaining, were used for quantification. Statistical calculations were performed using the Graphpad Prism software package (GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego California USA, www.graphpad.com). Distribution of data was assessed using three different normality tests: KS normality test, D'agostino & Pearson Normality test and Shapiro-Wilk normality test. When data were normally distributed according to these tests, parametric statistics were applied (t-test), otherwise non-parametrical statistical analysis (Mann-Whitney U test) was performed. When comparing more than two groups ANOVA tests were applied. For all tests p≤0.05 was considered significant.
Immunofluorescence
For immunostaining, animals were fixed and permeabilized as described [47]. Polyclonal anti-H3K27me3 (Upstate 07-449) and polyclonal anti-UTX-1 (Eurogentec, clone 3917, this study) were used. Secondary antibodies were: goat anti-mouse IgG (Alexafluor 488); goat anti-rabbit IgG (Alexafluor 594), both purchased from Invitrogen. DAPI (Sigma, 2 ug/ul) was used to counter-stain DNA. Eggs immunofluorescence was performed by freeze crack method, adding eggs to polylysine treated slides. After freezing at −80°C for 30 minutes, the cover slip was removed and embryos were fixed in methanol at −20°C for 10 min. Primary antibody was incubated overnight at 4°C in a humid chamber and secondary antibody was incubated 1 h at room temperature. Washes were in PBS/tween 0.2%. Mounting medium for fluorescence with DAPI (Vectashield H1200) was used.
GFP pulldown
Generation of transgenic strain utx-1+UTX-1::GFP has been described earlier in Materials and Methods. Total protein extracts was obtained by grinding a frozen pellet of mixed eggs and adults with a mortar and pestle into powder, the latter was resuspended in IP buffer containing 300 mM KCl, 0.1% Igepal, 1 mM EDTA, 1 mM MgCl2, 10% glycerol, 50 mM Tris HCl (pH 7.4) and protease inhibitors. GFP-Trap beads (Chromotek) were used to precipitate GFP-tagged proteins from this lysate. Approximately 200 mg of total proteins was used for the pulldown in IP buffer. Following incubation and washes with the same buffer, proteins were eluted with acidic glycine (0.1 M [pH 2.5]), resolved on a 4–12% NuPage Novex gel (Invitrogen), and stained with Imperial Protein Stain (Thermo Scientific). The gel was sliced into 21 bands across the entire separation range of the lane. Cut bands were reduced, alkylated with iodoacetamide, and in-gel digested with trypsin (Promega) as described previously [48], prior to LC/MS-MS analysis.
Mass spectrometry of proteins
Peptide identification was performed on an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Germany) coupled with an EASY-nLC nanoHPLC (Proxeon, Odense, Denmark). Samples were loaded onto a 100 µm ID×17 cm Reprosil-Pur C18-AQ nano-column (3 µm; Dr. Maisch GmbH, Germany). The HPLC gradient was from 0 to 34% solvent B (A = 0.1% formic acid; B = 95% MeCN, 0.1% formic acid) over 30 minutes and from 34% to 100% solvent B in 7 minutes at a flow-rate of 250 nL/min. Full-scan MS spectra were acquired with a resolution of 60,000 in the Orbitrap analyzer. For every full scan, the seven most intense ions were isolated for fragmentation in the LTQ using CID. Raw data were viewed using the Xcalibur v2.1 software (Thermo Scientific). Data processing was performed using Proteome Discoverer beta version 1.3.0.265 (Thermo Scientific). For database search we included both Mascot v2.3 (Matrix Science) and SEQUEST (Thermo Scientific) search engines. Database of C. elegans protein sequences was downloaded from Uniprot. Trypsin was selected as digestion enzyme and two missed cleavages were allowed, carbamidomethylation of cysteines was set as fixed modification and oxidation of methionine as variable modification. MS mass tolerance was set to 10 ppm, while MS/MS tolerance was set to 0.6 Da. Peptide validation was performed using Percolator and peptide false discovery rate (FDR) was set to 0.01. For additional filtering, maximum peptide rank was set to 1 and minimum number of peptides per protein was set to 2. Protein grouping was performed, in order to avoid presence of different proteins identified by non-unique peptides. We manually investigated whether the protein listed to represent the protein group was the most characterized in terms of sequence coverage and number of peptides identified.
Protein interaction assay
For co-immunoprecipitation assays, frozen eggs (prepared by hypochlorite treatment) were reduced into powder using a mortar and pestle. The powder was resuspended in IP buffer (described in GFP pulldown section) and 5–10 mg was incubated with Protein G agarose beads (Upstate) overnight at 4°C. Soluble fraction was collected and incubated with EZview anti-HA affinity gel beads (Sigma Aldrich) during 2 h at 4°C. The immunoprecipitates and the protein G beads were washed five times in IP buffer, boiled in SDS-sample buffer and analyzed by SDS-PAGE followed by western blotting. Antibodies used in those experiments were: anti-HA (Covance HA.11, clone 16B12), anti-GFP (Roche, 11814460001) and anti-UTX-1 (Eurogentec, clone 3917, this study). Quantification of western blots was performed using ImageJ program (National Institutes of Health, USA).
Analytical gel filtration chromatography
Eggs from indicated strains were grinded to powder, resupended in IP buffer (50 mM Tris-HCl pH 7.4, 300 mM KCl, 1 mM MgCl2, 1 mM EDTA, 0.1% Igepal and complete protease inhibitors [Roche]) and incubated on wheel for 30 min at 4°C. Protein extracts were recovered by centrifugation at 20,000 g, 30 min at 4°C and clarified by ultracentrifugation at 627,000 g for 30 min at 4°C. Fresh extracts were fractionated on a Superose 6 HR 10/300 GL column (GE Healthcare) equilibrated in IP buffer. Size exclusion chromatography was performed on a fast protein liquid chromatography (FPLC) system and an ÄKTA purifier (GE Healthcare). Elution profiles of blue dextran (2,000 kDa), thyroglobulin (660 kDa) and bovine serum albumin (66 kDa) were used for calibration. Fractions of 1 ml were collected and precipitated with 25% trichloroacetic acid and then centrifuged at 20,000 g for 10 min at 4°C. Pellets were washed two times in cold acetone, air dried, and resuspended in loading buffer for Western blot analysis.
Supporting Information
Zdroje
1. StrahlBDAllisCD 2000 The language of covalent histone modifications. Nature 403 41 45
2. KouzaridesT 2007 Chromatin modifications and their function. Cell 128 693 705
3. BergerSL 2007 The complex language of chromatin regulation during transcription. Nature 447 407 412
4. TsukadaYFangJErdjument-BromageHWarrenMEBorchersCH 2006 Histone demethylation by a family of JmjC domain-containing proteins. Nature 439 811 816
5. CloosPAChristensenJAggerKHelinK 2008 Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22 1115 1140
6. AggerKCloosPAChristensenJPasiniDRoseS 2007 UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449 731 734
7. LeeMGVillaRTrojerPNormanJYanKP 2007 Demethylation of H3K27 Regulates Polycomb Recruitment and H2A Ubiquitination. Science 318 447 450
8. SmithERLeeMGWinterBDrozNMEissenbergJC 2007 Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28 1041 1046
9. LanFBaylissPERinnJLWhetstineJRWangJK 2007 A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449 689 694
10. De SantaFTotaroMGProsperiniENotarbartoloSTestaG 2007 The Histone H3 Lysine-27 Demethylase Jmjd3 Links Inflammation to Inhibition of Polycomb-Mediated Gene Silencing. Cell 130 1083 1094
11. HongSChoYWYuLRYuHVeenstraTD 2007 Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA 104 18439 18444
12. FisherKSouthallSMWilsonJRPoulinGB 2010 Methylation and demethylation activities of a C. elegans MLL-like complex attenuate RAS signalling. Developmental biology 341 142 153
13. MauresTJGreerELHauswirthAGBrunetA 2011 The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging cell 10 980 990
14. JinCLiJGreenCDYuXTangX 2011 Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell metabolism 14 161 172
15. MohlerWASimskeJSWilliams-MassonEMHardinJDWhiteJG 1998 Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Current biology : CB 8 1087 1090
16. HendersonSTGaoDLambieEJKimbleJ 1994 lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120 2913 2924
17. SwigutTWysockaJ 2007 H3K27 demethylases, at long last. Cell 131 29 32
18. JinCLiJGreenCDYuXTangX 2011 Histone Demethylase UTX-1 Regulates C. elegans Life Span by Targeting the Insulin/IGF-1 Signaling Pathway. Cell metabolism 14 161 172
19. CloosPAChristensenJAggerKMaiolicaARappsilberJ 2006 The putative oncogene GASC1 demethylates tri - and dimethylated lysine 9 on histone H3. Nature 442 307 311
20. KloseRJYamaneKBaeYZhangDErdjument-BromageH 2006 The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442 312 316
21. ChangBChenYZhaoYBruickRK 2007 JMJD6 is a histone arginine demethylase. Science 318 444 447
22. IssaevaIZonisYRozovskaiaTOrlovskyKCroceCM 2007 Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27 1889 1903
23. ChoYWHongTHongSGuoHYuH 2007 PTIP associates with MLL3 - and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282 20395 20406
24. PatelSRKimDLevitanIDresslerGR 2007 The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 13 580 592
25. ShilatifardA 2008 Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Current opinion in cell biology 20 341 348
26. SimonetTDulermoRSchottSPalladinoF 2007 Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Developmental biology 312 367 383
27. AndersenECHorvitzHR 2007 Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134 2991 2999
28. SeenundunSRampalliSLiuQCAzizAPaliiC 2010 UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. The EMBO journal 29 1401 1411
29. AggerKCloosPARudkjaerLWilliamsKAndersenG 2009 The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene - and stress-induced senescence. Genes Dev 23 1171 1176
30. MillerSAMohnSEWeinmannAS 2010 Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Molecular cell 40 594 605
31. GreerELMauresTJHauswirthAGGreenEMLeemanDS 2010 Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466 383 387
32. XiaoYBedetCRobertVJSimonetTDunkelbargerS 2011 Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proceedings of the National Academy of Sciences of the United States of America 108 8305 8310
33. SolariFAhringerJ 2000 NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Current biology : CB 10 223 226
34. LiebJDAlbrechtMRChuangPTMeyerBJ 1998 MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92 265 277
35. WangJKTsaiMCPoulinGAdlerASChenS 2010 The histone demethylase UTX enables RB-dependent cell fate control. Genes & development 24 327 332
36. LiTKellyWG 2011 A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet 7 e1001349 doi:10.1371/journal.pgen.1001349
37. CapowskiEEMartinPGarvinCStromeS 1991 Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics 129 1061 1072
38. PaulsenJECapowskiEEStromeS 1995 Phenotypic and molecular analysis of mes-3, a maternal-effect gene required for proliferation and viability of the germ line in C. elegans. Genetics 141 1383 1398
39. KellyWGFireA 1998 Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125 2451 2456
40. HoldemanRNehrtSStromeS 1998 MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development 125 2457 2467
41. KorfIFanYStromeS 1998 The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development 125 2469 2478
42. BenderLBCaoRZhangYStromeS 2004 The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol 14 1639 1643
43. BenderLBSuhJCarrollCRFongYFingermanIM 2006 MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133 3907 3917
44. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
45. TimmonsLCourtDLFireA 2001 Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103 112
46. KragCMalmbergEKSalciniAE 2010 PI3KC2alpha, a class II PI3K, is required for dynamin-independent internalization pathways. Journal of cell science 123 4240 4250
47. FinneyMRuvkunG 1990 The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63 895 905
48. VandammeJVolkelPRosnobletCLe FaouPAngrandPO 2011 Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Molecular & cellular proteomics : MCP 10 M110 002642
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



