-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Recombination Drives Vertebrate Genome Contraction
Selective and/or neutral processes may govern variation in DNA content and, ultimately, genome size. The observation in several organisms of a negative correlation between recombination rate and intron size could be compatible with a neutral model in which recombination is mutagenic for length changes. We used whole-genome data on small insertions and deletions within transposable elements from chicken and zebra finch to demonstrate clear links between recombination rate and a number of attributes of reduced DNA content. Recombination rate was negatively correlated with the length of introns, transposable elements, and intergenic spacer and with the rate of short insertions. Importantly, it was positively correlated with gene density, the rate of short deletions, the deletion bias, and the net change in sequence length. All these observations point at a pattern of more condensed genome structure in regions of high recombination. Based on the observed rates of small insertions and deletions and assuming that these rates are representative for the whole genome, we estimate that the genome of the most recent common ancestor of birds and lizards has lost nearly 20% of its DNA content up until the present. Expansion of transposable elements can counteract the effect of deletions in an equilibrium mutation model; however, since the activity of transposable elements has been low in the avian lineage, the deletion bias is likely to have had a significant effect on genome size evolution in dinosaurs and birds, contributing to the maintenance of a small genome. We also demonstrate that most of the observed correlations between recombination rate and genome contraction parameters are seen in the human genome, including for segregating indel polymorphisms. Our data are compatible with a neutral model in which recombination drives vertebrate genome size evolution and gives no direct support for a role of natural selection in this process.
Published in the journal: . PLoS Genet 8(5): e32767. doi:10.1371/journal.pgen.1002680
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002680Summary
Selective and/or neutral processes may govern variation in DNA content and, ultimately, genome size. The observation in several organisms of a negative correlation between recombination rate and intron size could be compatible with a neutral model in which recombination is mutagenic for length changes. We used whole-genome data on small insertions and deletions within transposable elements from chicken and zebra finch to demonstrate clear links between recombination rate and a number of attributes of reduced DNA content. Recombination rate was negatively correlated with the length of introns, transposable elements, and intergenic spacer and with the rate of short insertions. Importantly, it was positively correlated with gene density, the rate of short deletions, the deletion bias, and the net change in sequence length. All these observations point at a pattern of more condensed genome structure in regions of high recombination. Based on the observed rates of small insertions and deletions and assuming that these rates are representative for the whole genome, we estimate that the genome of the most recent common ancestor of birds and lizards has lost nearly 20% of its DNA content up until the present. Expansion of transposable elements can counteract the effect of deletions in an equilibrium mutation model; however, since the activity of transposable elements has been low in the avian lineage, the deletion bias is likely to have had a significant effect on genome size evolution in dinosaurs and birds, contributing to the maintenance of a small genome. We also demonstrate that most of the observed correlations between recombination rate and genome contraction parameters are seen in the human genome, including for segregating indel polymorphisms. Our data are compatible with a neutral model in which recombination drives vertebrate genome size evolution and gives no direct support for a role of natural selection in this process.
Introduction
A link between the dynamics of intron evolution and recombination has been found in form of a negative relationship between recombination rate and intron size, seen in Drosophila [1], [2], humans [2] and chicken [3]. Two hypotheses based on natural selection have been proposed to explain this relationship. First, insertion mutations increasing intron length, which may confer higher energy cost for transcription or replication and thus be mildly deleterious, may be more efficiently removed by purifying selection in regions with high recombination rate where Hill-Robertson interference is reduced [1]. Second, insertion mutations increasing intron length may be favored in regions with low recombination rate because large introns reduce the effect of Hill-Robertson interference [4].
The negative relationship between intron length and recombination could also be possible to explain under a neutral scenario if recombination itself, either directly (by being mutagenic) or indirectly (by affecting other genomic features) affects the direction or magnitude of changes in intron length. More generally, a mutational bias associated with recombination that leads to increases or decreases in sequence length all over the genome (and not only in introns) will have implications to the overall DNA content, i.e., the evolution of genome size. Several models for genome size evolution have been presented. Broadly speaking they can be defined as adaptive [5]–[8], non-adaptive [9] or neutral [10], [11]. Short deletions are almost ubiquitously found to outnumber short insertions in eukaryotic genomes and it has been proposed that the degree of deletion bias is a main factor for variation of genome size under a neutral model [10], [11]. Recombination-associated processes can potentially provide a mechanistic explanation to the deletion bias, which remains to be tested. Taking possible mutagenic effect of recombination into account is clearly necessary before inference on selection from correlation between recombination rate and sequence length is made.
In this study we address the underlying evolutionary forces that contribute to a negative relationship between recombination rate and sequence length by focusing on three sequenced and annotated vertebrate genomes from two major lineages, mammals (human) and birds (chicken and zebra finch). Detailed recombination rate maps are available for all these species [12]–[14]. Avian genomes are typically smaller than mammalian genomes; 75% of >400 characterized bird species have a haploid DNA content of 1.2–1.6 pg, whereas 75% of >600 characterized mammalian species have 2.5–4.3 pg [15]. A focus on avian genomes is of particular interest in the context of genome size evolution in relation to recombination because both chicken and zebra finch display an unusual heterogeneity in the rate of recombination, including recombination-prone microchromosomes [3] and a stronger “telomere-effect” (elevated recombination rates toward chromosome ends) than so far seen in any other species [13], [14]. Potentially, such heterogeneity can increase the power in detecting correlations between recombination rate and other genomic parameters. In addition, because avian genomes show a high degree of karyotype and synteny conservation [16], [17], genomic correlates may be less affected by noise following from frequent chromosomal rearrangements.
Using comparative genomics to analyze structural variation in non-coding DNA is usually limited by the problem of aligning sequences evolving under low or no constraint in other than closely related species. Moreover, unless sequence data can be aligned from three or more species, it is impossible to distinguish insertions from deletions. Furthermore, if insertions or deletions occur in genomic regions containing functional elements [18], selection may act differently on the two types of structural changes. Here we circumvent these problems by using transposable elements contained within non-coding DNA to measure insertion and deletion rates in individual lineages. Specifically, we infer insertion and deletion events from alignments of repeat elements with their ancestral master sequence, as introduced by Petrov et al. [19]. Our main observation, consistent across all three species, is that loss of DNA is most pronounced in regions of high recombination. This is compatible with a neutral model of genome evolution where recombination drives genome contraction.
Results
Transposable elements, sequence length, and recombination rate in avian genomes
We identified Long Interspersed Elements (LINEs) from pre-masked genome assemblies of chicken and zebra finch using Repeatmasker. A total of 239,812 (chicken) and 169,576 (zebra finch) LINEs were found, the far most abundant type being the well-known CR1 retroposon [20]–[23]. Using data from 1 Mb non-overlapping windows across the genome, we found a significant negative correlation between recombination rate and intron length in both species (chicken, τ = −0.18, p<0.001; zebra finch τ = −0.14, p<0.001; Kendall's rank test) (Table 1), and this was also the case when only first introns were considered in chicken (τ = −0.12, p<0.001) but not in zebra finch (τ = −0.04, p = 0.120). Moreover, there was a significant negative relationship between recombination rate and the length of individual LINE sequences located within introns (chicken, τ = −0.32, p<0.001; zebra finch, τ = −0.40, p<0.001) as well as between recombination rate and the length of intronic sequence that is not LINE sequence (chicken, τ = −0.16, p<0.001; zebra finch, τ = −0.13, p<0.001) (Table 1). This shows that if the rate of LINE integration is higher in regions with low recombination rate, it cannot fully explain the negative relationship between intron length and recombination rate. Furthermore, there was a significant negative correlation between recombination rate and the length of intergenic sequence (intergenic spacer) in both species (chicken, τ = −0.32, p<0.001; zebra finch, τ = −0.15, p<0.001) and a positive correlation between recombination rate and gene density (chicken, τ = 0.30, p<0.001; zebra finch τ = 0.16, p<0.001) (Table 1). All these observations point at a pattern of more condensed structure in regions of high recombination in avian genomes.
Tab. 1. Strength (correlation coefficient, τ) and statistical significance (<i>p</i>) of Kendall's rank correlations between recombination rate and various genomic parameters in non-overlapping 1 Mb windows. 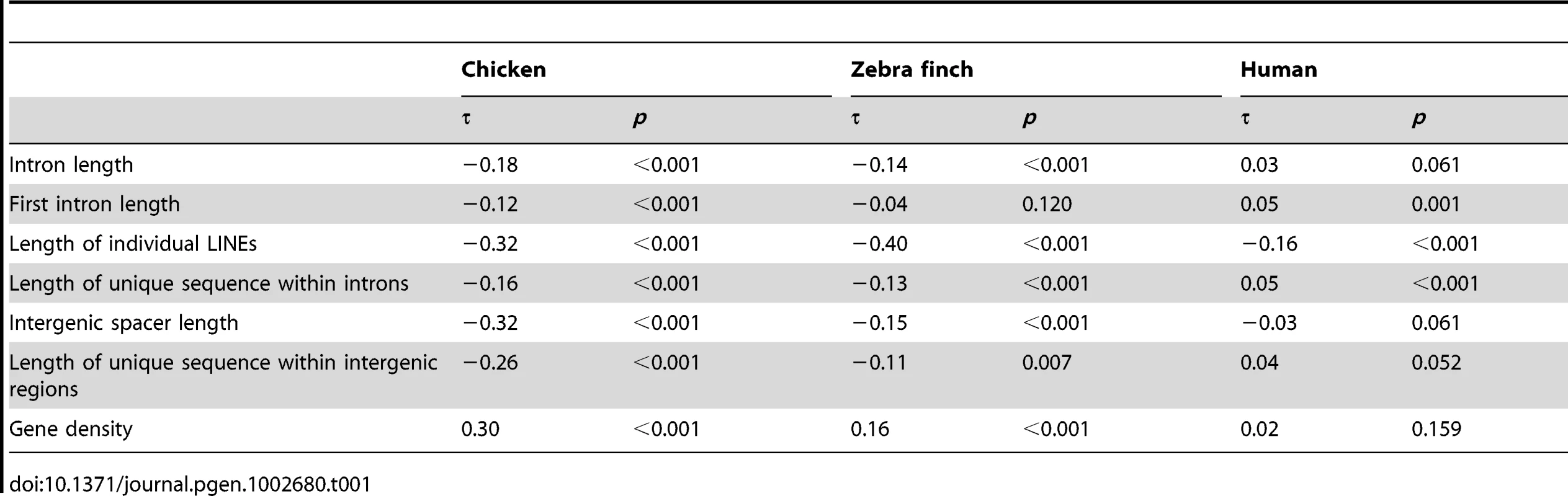
Rates of insertion and deletion and their relationship with recombination rate
We estimated insertion and deletion rates by aligning repeat elements with their master sequence and by inferring events of small insertion and deletion from gaps in master and repeat element sequence, respectively (see Figure S1 for distribution of the length of insertions and deletions). The rate of deletion defined as the number of bp deleted per bp repeat (LINE) sequence was consistently higher than the rate of insertion, giving a deletion bias of 3.24 and 3.45 in chicken and zebra finch, respectively (Table 2). Using data from 1 Mb windows, there was a significant positive correlation between recombination rate and deletion rate (chicken, τ = 0.23, p<0.001; zebra finch, τ = 0.32, p<0.001) (Figure 1), but no correlation between recombination rate and insertion rate (chicken, τ = −0.01, p = 0.776; zebra finch, τ = −0.03, p = 0.323). There was also a positive correlation between recombination rate and the number of deletion events (chicken, τ = 0.37, p<0.001; zebra finch, τ = 0.38, p<0.001).
Fig. 1. The relationship between recombination rate (x-axis, fourth-root) and deletion rate, insertion rate, deletion bias (deletion rate/insertion rate), and rate of net sequence length change (insertion rate – deletion rate). 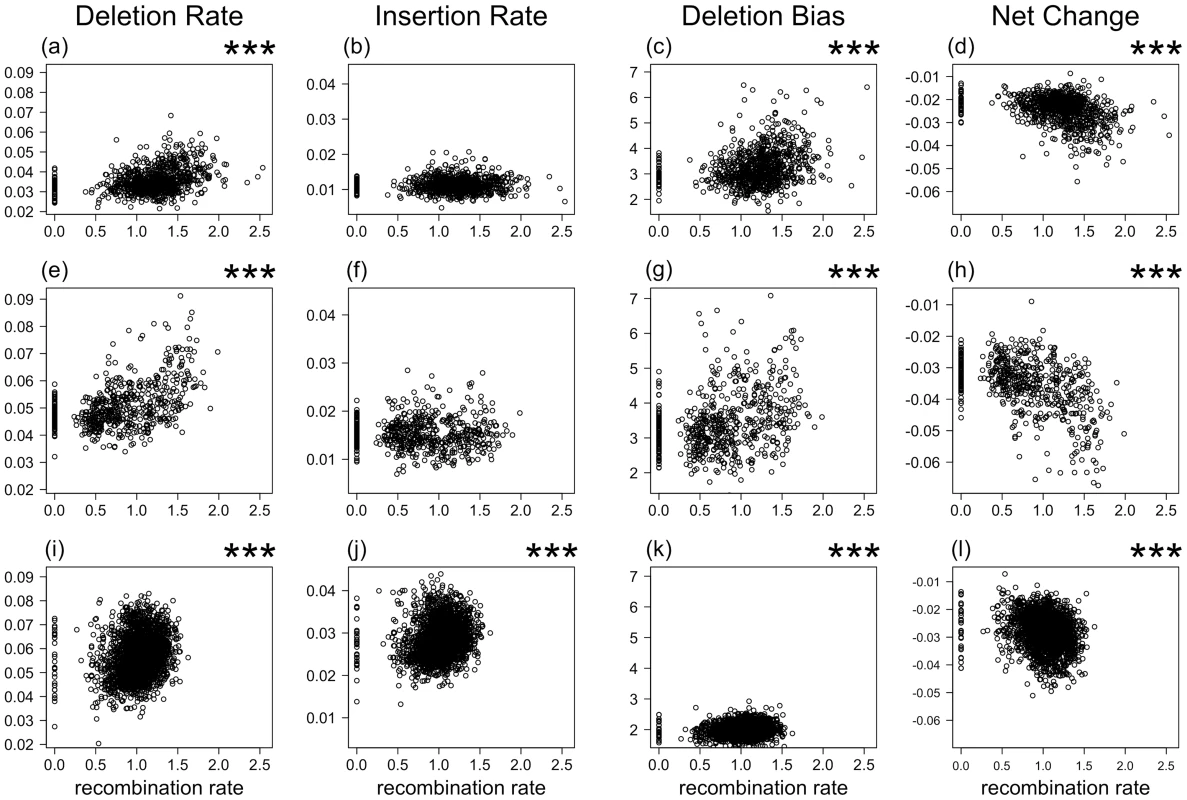
In chicken (a, b, c, and d), zebra finch (e, f, g, and h), and human (i, j, k, and l). p<0.001 in Kendall's rank correlation test is depicted by ***. Tab. 2. The mean rate of substitution, insertion, and deletion (as the number of bp inserted or deleted per bp repeat sequence) for LINEs in the genomes of chicken, zebra finch, and human. 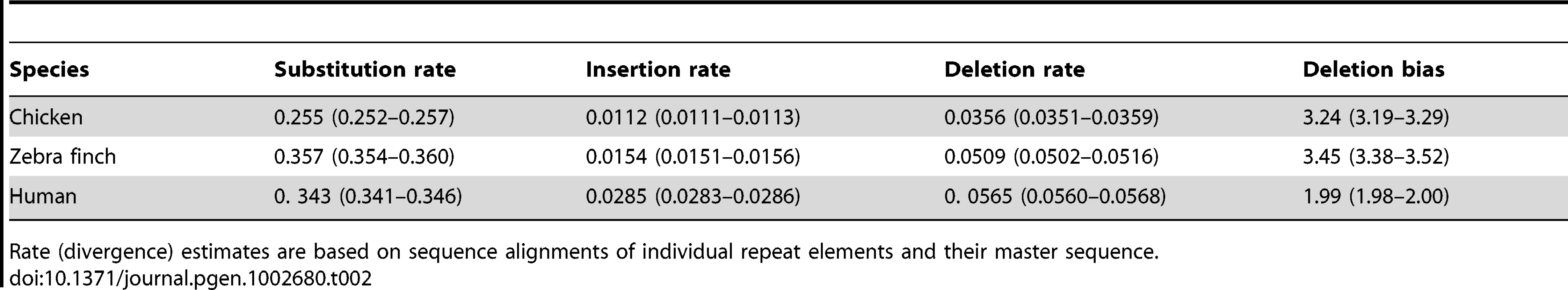
Rate (divergence) estimates are based on sequence alignments of individual repeat elements and their master sequence. Insertions and deletions taken together, and not surprisingly given the above correlations, there was a positive relationship between recombination rate and the deletion bias (chicken, τ = 0.19, p<0.001; zebra finch, τ = 0.24, p<0.001) (Figure 1). While this suggests genomic contraction in high recombination regions, such trend could in theory be mitigated by relatively short insertion and deletion events (although the deletion bias being high) in high recombination regions. However, recombination rate was as strongly correlated with the net change in sequence length (amount of sequence deleted minus amount of sequence inserted; chicken, τ = 0.24, p<0.001; zebra finch, τ = 0.33, p<0.001) (Figure 1) as it was to the deletion bias calculated on basis of rates of insertion and deletion. We thus conclude that there has been a process of genomic contraction in high recombination rate regions during avian evolution. This process is manifested in a more condensed present-day genomic structure in regions with high recombination.
In the above we have assumed a star phylogeny for the relationship among repeat copies and their master sequence. To exclude the possibility that violation of this assumption would affect our conclusions we repeated the analyses only using insertions and deletions that were seen once. This replicated the correlations seen with the whole data set (deletion bias: chicken, τ = 0.09, p<0.001; zebra finch, τ = 0.08, p<0.001; net change in sequence length: chicken, τ = −0.23, p<0.001; zebra finch, τ = −0.22, p<0.001).
All relationships reported above are for 1 Mb genomic regions, which is the smallest window size for which we have resolution in data on the regional recombination rate. Since recombination in at least mammalian genomes is often concentrated to narrow hot spot regions [24], more fine-scale recombination maps could potentially have given stronger correlations between recombination and parameters of genome contraction. On the other hand, primate recombination hot spots tend to be ephemeral with a rapid turnover rate [25], [26] and this may obscure correlations with genomic parameters representing mutational events that have accumulated over long evolutionary time scales. While we are not able to analyze smaller windows we repeated all analyses using 5 Mb windows. Interestingly, most parameters related to genome contraction showed stronger correlation with recombination rate for this window size than for 1 Mb windows (Table S1). For example, Kendall's τ was as high as −0.43 and −0.53 for the correlation with the length of individual LINEs and 0.34 and 0.47 for the correlation with the net change in sequence length in chicken and zebra finch, respectively.
Comparison between sex chromosome and autosomes
As a specific test of the role of recombination in affecting genome size evolution we compared sequences on autosomes and sex chromosomes. Birds have female heterogamety (males ZZ, females ZW) so the Z chromosome does only recombine in males and thus have a lower recombination rate than autosomes. If recombination drives genome contraction we expect the deletion bias to be higher on autosomes than on the Z chromosome and the mean length of LINEs to be longer on the Z chromosomes than on autosomes. Data from both chicken and zebra finch meet these expectations. The deletion bias was significantly lower on the Z chromosome than on the autosomes (chicken, 3.23 vs. 3.32, p = 0.045; zebra finch, 3.19 vs. 3.74, p<0.001, 10,000 times of non-parametric bootstrapping based on stratified sampling) and the mean length of LINEs was longer on the Z chromosome than on autosomes (chicken, 419.8 bp vs. 314.5 bp; zebra finch 384.6 bp vs. 266.6 bp, p<0.001 in both species).
The pattern for the non-recombining, female-specific W chromosome should be expected to differ even more from that of autosomes. The W chromosome is not included in the zebra finch assembly and the amount of W-linked sequence in the chicken assembly is limited (0.26 Mb). However, the chicken W chromosome had the longest mean length of LINEs (446.7 bp, significantly different from autosomes, p<0.001) and the least pronounced deletion bias (2.38, p<0.001).
Chromosome size per se does not explain the relationship between recombination rate and genome contraction parameters
Recombination rate is closely correlated with chromosome size in both chicken [3] and zebra finch [27]. In theory, it is possible that some other genomic parameter that also correlates with chromosome size causes the observed relationships between recombination rate and different attributes of genome contraction (e.g., intron length, deletion rate and deletion bias). To test this possibility we used a mixed model with chromosome identity as a random variable. However, the majority of the observed correlations between recombination rate and parameters associated with genome contraction remained statistically significant when chromosome identity was controlled for (Table S2).
Another way of excluding possible effects of chromosome identity is to study the relationship between recombination rate and deletion rate/deletion bias for individual chromosomes (Table S3). For the microchromosomes the number of available windows is not sufficient for this analysis and we thus restricted the analysis to chromosomes with at least 20 windows (i.e. chromosomes >20 Mb in size). Nine out of 11 such chromosomes in chicken showed a positive correlation (mean Kendall's τ = 0.11) between recombination rate and deletion rate (randomization test with 106 replicates, p = 0.033) and 10 out of 11 chromosomes showed a positive correlation (mean τ = 0.11) between recombination rate and the deletion bias (p = 0.006). In zebra finch, eight out of eight chromosomes had a positive correlation (mean τ = 0.22) between recombination rate and deletion rate (p = 0.004), and seven out of eight had a positive correlation (mean τ = 0.12) between recombination rate and the deletion bias (p = 0.035). The genome-wide relationships between recombination rate and genome contraction parameters can thus also be seen within individual chromosomes.
The impact on avian genome size variation
We simulated the impact of deletion-biased length mutations on avian genome size evolution over time by fitting an exponential decay function based on the assumptions of a constant rate of sequence loss and neutral evolution. We used sequence divergence (rather than years) as a time scale to avoid uncertainties associated with rate calibration of the molecular clock; this is particularly warranted given apparent heterotachy in avian substitution rates [28]. From 8,328 ancestral CR1 sequences identified in whole-genome alignment of chicken, turkey, and zebra finch [29], we estimated rates of sequence evolution in the chicken branch as follows: substitution rate 4.21% (95% confidence interval, CI: 4.01–4.45%), deletion rate 2.61% (1.99–3.33%), and insertion rate 0.58% (0.52–0.65%). Combining these three estimates, this translates into a loss of 0.48 (0.34–0.60) nucleotides per nucleotide substitution. The rate parameter of this exponential decay was 0.489 (0.347–0.664; see Methods).
To get an idea of the estimated effect of the deletion bias on avian genome size evolution we note that lineage-specific divergence (nucleotide substitutions) in the chicken lineage subsequent to the split between birds and lizards has been estimated to 0.411 [30]. If we assume a constant rate parameter of sequence loss (0.489), the chicken genome has lost sequences corresponding to 18.2% of the total DNA content (95% CI: 13.3–23.9%; Figure S2) due to small insertions and deletions since the common ancestor of birds and lizards. This assumes that the rate and pattern of indel mutations observed within transposable elements are representative for the whole genome. In the comparison of short (1–2 bp), intermediate (3–20 bp) and large (>20 bp) indel events in our data, the intermediate size category has had the largest influence on genome size change (Figure S3). Note that the estimated loss of DNA may at least in part have been balanced by gain of DNA due to large-scale insertions.
Recombination also correlates with the deletion bias in the human genome
To test if genome compaction driven by recombination is widespread among vertebrates we analyzed data from a total of 1,724,413 LINEs in the human genome (Table 1). Similar to what was seen in birds, the deletion bias in the human genome was positively correlated with recombination rate (τ = 0.07, p<0.001; τ = 0.07, p = 0.001 using only unique events), although the bias was less pronounced (1.98). Recombination rate was positively correlated with deletion rate (τ = 0.20, p<0.001; Figure 1) and in this case also with insertion rate (τ = 0.16, p<0.001). The net change of sequence length was negatively correlated with recombination rate (τ = −0.17, p<0.001; τ = 0.03, p = 0.047 using only unique events). As for the avian data, these correlations remained statistically significant when chromosome identity was controlled for (Table S2). Moreover, when individual chromosomes were analyzed separately, 19 out of 22 chromosomes showed a positive correlation (mean Kendall's τ = 0.13) between recombination rate and deletion rate (randomization test with 106 replicates, p<0.001) and 17 out of 22 chromosomes showed a positive correlation (mean τ = 0.06) between recombination rate and the deletion bias (p = 0.001) (Table S3). In summary, the patterns of insertion and deletion seen in the two avian genomes were largely replicated by data from the human genome.
Human polymorphism data give no support that selection would explain the link between recombination and the deletion bias
All the observations made above are consistent with a neutral model in which recombination promotes deletion. Could they also be compatible with a model invoking a role of selection? Selection is more efficient in regions of high recombination and slightly deleterious alleles are therefore expected to accumulate at a lower rate (and advantageous alleles at a higher rate) in such regions. However, it may be difficult to imagine a scenario where recombination rate would correlate positively with the deletion bias due to an increased fixation probability of deletions within transposable elements, or decreased fixation probability of insertions, in high recombination regions. This would require that small indels within LINEs are not selectively neutral (or that there is differential selection for insertions and deletions; see below) but Lunter et al. [31] showed that the distribution of insertions and deletions in ancestral repeats shared between human and mouse is consistent with a neutral model and Petrov and colleagues [19], [32] have convincingly argued against purifying selection acting on indels in dead-on-arrival elements in Drosophila.
If recombination promotes deletions by being mutagenic, rather than via selection and altered fixation probabilities of indels, we should expect to see a correlation between recombination rate and the deletion bias in within-species polymorphism data. There is no large-scale data on polymorphic indels in birds but Mills et al. [33] reported nearly 2 million segregating indels in the human genome. These polymorphisms are mostly from unique sequence given the difficulty to confidently map short next-generation sequencing reads to repeat elements. Insertions were distinguished from deletions by comparison to chimpanzee outgroup sequence. We found that there was a significant positive correlation between recombination rate and the deletion bias among polymorphic human indels (τ = 0.08, p<0.001), and this holds true also when introns (τ = 0.06, p<0.001) and intergenic sequence (τ = 0.06, p<0.001) were analyzed separately.
As mentioned above, for a positive correlation between recombination rate and deletion bias to be seen under a selection model is required that purifying selection against insertions is more effective than purifying selection against deletions in high recombination regions. Put in other words, the deleterious effects of insertions have to be larger than those of deletions. For indels in functional regions of the genome, like protein-coding sequence, the opposite is observed in mammals and Drosophila [11], [18], [34]. We used allele frequency data from 10,003 human indels [33] to see if the site frequency spectrum differs between insertions and deletions in the genome. The spectrum is expected to be biased towards rare alleles in the presence of purifying selection, and increasingly so as the intensity of selection increases [35]. However, we found no evidence for that segregating rare alleles (minor allele frequency, MAF, <0.05) would occur more frequently among insertions than among deletions (proportion of loci with MAF<0.05 in intergenic sequence: 0.229 vs. 0.210, chi-square = 2.72, p = 0.099; in LINEs: 0.146 vs. 0.231, chi-square = 1.97, p = 0.160) (Table S4). For intronic sequence, where functional elements are more likely to be present, deletions were significantly more biased towards rare alleles than insertions (chi-square = 12.14, p<0.001)
Discussion
Petrov and colleagues [10], [11] have hypothesized that the extent to which small deletions outnumber small insertions, the deletion bias, is a main factor determining genome size. This hypothesis comes mainly from the observation that in species with small genomes, the deletion bias is more pronounced than in species with larger genomes. A genomic parameter that affects the magnitude of this mutational bias could then be a driving force of the evolution of genome size under a neutral model. The same could apply to variation in compactness and chromosome size within genomes. Our data suggest that the rate of recombination represents such a parameter. Using data from two avian genomes where recombination is highly heterogeneous we find that recombination rate correlates (a) negatively with the length of introns as well as intergenic regions and with the inverse of gene density, (b) positively with the rate of deletion but negatively with the rate of insertion, and (c) positively with the deletion bias as well as the net change in sequence length. We make similar observations for the human genome, including for polymorphism data, indicating that recombination is a general factor modulating genome size variation in vertebrates. This conclusion is in line with the observation that, across species, mammalian genome size is negatively correlated with recombination rate [36].
A main criticism against the idea that the deletion bias affects genome size evolution is that the number of small deletions is too small to impact on genome size [37]. Our simulations suggest a loss of nearly 20% of the DNA content in the chicken lineage since the common ancestor of birds and lizards due to small insertions and deletions. This may very well have been sufficient to counteract genome expansion due to the spread of interspersed repeats during this period of time. Less than 10% of the chicken genome consists of recognizable transposable elements [3] and although ancient elements that have mutated beyond recognition may add to this proportion, it is clear that transposable element activity has been low in the avian lineage [3], [23]. Using bone-cell size as an indirect measure of genome size, Organ et. al [38] showed that the small genome size typical for contemporary birds was present already in the saurischian dinosaur lineage 230–250 million years ago [39]. We suggest that this apparent stasis of genome size through the evolution of non-avian dinosaurs and modern birds relates to a balance between moderate repeat expansion and DNA loss from the deletion bias.
Avian genomes differ from mammalian genomes in several respects, notably by being much smaller and therefore more condensly organized with shorter introns and shorter intergenic distances [40], [41]. Another avian characteristic is the significant within-genome variation in chromosome size with numerous small microchromosomes (<20 Mb). The origin and evolution of microchromosomes remains to be an enigmatic issue [42]. Although fissions and fusions are likely to be involved in generating variation in chromosome size, our results point at an interesting model for the maintenance and perhaps even further diminutivization of microchromosomes. Recombination rate correlates closely with chromosome size in avian genomes [3], a situation that follows from an obligate crossing-over per chromosome (arm) [43]. Given the observation that recombination rate correlates with the deletion bias, we propose, inspired by Burt [42], that there is a vortex where high recombination rates in small chromosomes make them even smaller due to the deletion bias, in turn leading to even higher recombination rates, etc. However, and as suggested by Petrov [11], as genome structure becomes more condensed, the likelihood for deletion events to involve functionally important sequences will increase. As a consequence, at some point selection against deleterious deletion events will occur sufficiently often to counteract quantitatively the mutational deletion bias.
Our results are compatible with that recombination by some mechanism introduce deletion mutations. While the often seen (e.g. humans, Drosophila) positive correlation between recombination rate and levels of within-species genetic diversity [44]–[47] could potentially be interpreted to reflect that recombination is mutagenic also for point mutations, recombination reduces the effect of selection at linked loci thereby acting towards maintenance of genetic variation. On the other hand, support for a neutral link between recombination and nucleotide substitution has been provided by the observation in humans and Drosophila that regions of the genome with low recombination rate also show reduced rates of between-species divergence [45], [48], [49]. However, this remains a contentious issue because several contradictory conclusions have been claimed [50]–[54].
With these uncertainties about recombination and point mutation in mind, we may ask if there is any mechanistic support for recombination being mutagenic for deletion. DNA polymerases δ and ε are key enzymes for eukaryotic DNA replication, including in connection with homologous recombination (reviewed in [55]). Both enzymes tend to cause deletions more often than insertions [56]–[59], a situation that is likely to explain the general phenomenon of deletion bias. Possibly, proofreading is less efficient to correct for unpaired bases in the primer strand than in the template strand [57]. Important in this context, DNA polymerase δ is preferentially used to promote heteroduplex extension during recombination [60]. DNA polymerase δ has lower fidelity than DNA polymerase ε, and this difference is especially pronounced for deletions. Fortune et al. analyzing Saccharomyces cerevisiae found that DNA polymerase δ has a 30-fold lower accuracy for large deletions and a 13-fold lower accuracy for single nucleotide deletions compared to DNA polymerase ε [57]. This may point at a mechanistic link between recombination and the rate of small deletions.
The model of recombination driving genome compactization, if correct, can explain another observation made for most investigated eukaryotic genomes: a positive correlation between GC content and gene density [61]–[67]. In both mammals and birds, GC content is one of the strongest predictors of recombination rate [28], [36]. It has been suggested that this is due to recombination driving GC-biased biased gene conversion (gBGC), a process of segregation distortion favoring the fixation of G and C nucleotides, leading to increased GC content in regions with high recombination rates [68]–[71]. If the deletion bias is more pronounced in these high recombining regions, as our data suggest, they will come to have a more compact structure with less intergenic DNA and thereby giving rise to a correlation between GC and gene density.
A general caveat in studies of the relationship between recombination and genomic parameters is that while estimates of recombination rates reflect the contemporary situation, most genomic parameters (substitution rates, base composition, chromosomal organization) are the result of long-term evolutionary processes. It follows that if regional recombination rates vary over time [72], this may obscure correlations between recombination rate and genomic parameters. However, it seems plausible that this would mostly lead to weakened correlations, not cause spurious correlations. Importantly, the recombinational landscape in birds of more conserved than in other vertebrate groups; we recently found that the recombination rate measured in 1 Mb windows are highly correlated (Spearman's rho = 0.50) between chicken and zebra finch despite these two lineages diverged 60–80 million years ago [14]. The unusually stable karyotype of birds [16], [17] is likely to contribute to this conservation.
There are at least two ways to study mutation processes using divergence data from transposable repeat elements spread across the genome. First, divergence can be estimated from alignments of ancestral (orthologous) repeats (ARs) shared by species; when AR data is available for three or more species, lineage-specific divergence can be estimated. Second, divergence can be estimated by alignments of master (consensus) and “offspring” sequences, like in the present study. Using ARs shared by human, chimpanzee and macaque, Kvikstad et al. [73] found that the rate of insertion, but not the rate of deletion, was dependent on recombination rate. They also reported that the deletion bias was not significantly correlated with recombination rate, observations that are at odds with our findings from the human genome. In Text S1 we show that primate ancestral repeats have a lower deletion rate and a lower deletion bias than more recently evolved repeats in the human lineage. We hypothesize that this is because of an ascertainment bias in the analysis of ARs since sequences that can be aligned over large evolutionary distances are less likely to harbor deletions. Moreover, since ancestral LINEs shared by human, chimpanzee, and macaque comprise less than 10% of total amount of LINEs in the human genome, they will have relatively limited influence on overall patterns inferred from analyses of present-day repeats.
Although transposable elements have emerged as a widely used sequence category for inferences of neutral rates and patterns of nucleotide substitution (e.g. [74]), as well as of insertion and deletion [75], a final cautionary note could be added. For example, it might be argued that the presence of undetected and active subfamilies originating from a single master sequence would violate the assumption of independent divergence of individual elements from the presumed master sequence. This could inflate estimates of divergence within individual LINE subfamilies. However, our results were not affected by restricting the analyses to indel events that were only seen once. This also excludes the possibility of concerted evolution from frequent gene conversion affecting our results. Moreover, unless the genomic distribution of repeats spreading from incorrectly inferred dead-on-arrival elements would be non-random with respect to recombination, the occurrence of undetected subfamilies is anyway unlikely to affect our conclusions. Finally, we note that the chronological order of activity of different LINE subfamilies as revealed by patterns of nested LINEs is entirely congruent with the relative age of subfamilies as revealed by divergence between individual elements and master sequences (Figure S4).
Methods
Sequence data
Sequence alignments of LINEs and their master sequences from zebra finch (taeGut1), chicken (galGal3), and human (hg18) were downloaded from the Repeatmasker homepage (http://www.repeatmasker.org/PreMaskedGenomes.html) [76]. These repeat elements had been identified using Repeatmasker 3.2.7 or 3.2.8 with the reference sequences and annotations of Repbase update 20090604 [77]. We excluded repeats located within exons and repeats of unassigned contigs (contigs with an unknown location in the genome). Data on sex-averaged recombination rates were obtained from [12] for human, from [13] for chicken, and from [14] for zebra finch.
Since SINEs constitute only a small proportion of all transposable elements in avian genomes [3], we limited the study to LINEs. We did not include DNA transposons since their cut-and-paste mechanism for transposition prohibits an unbiased analysis of insertion and deletion events within repeats. LTR retrotransposons were also excluded because solo-LTR elements, the product of intra-strand recombination, can bias divergence estimates.
Data analysis
We concatenated all LINEs together with their aligned master sequences within 1 Mb windows. The insertion and deletion rates within transposable elements were calculated by dividing the length sum of insertions or deletions (in bp) by the length sum of transposable elements within the window in question. The deletion bias was calculated by dividing the number of deleted nucleotides by the number of inserted nucleotides within each window. Substitution rate of LINEs was calculated by using the baseml program in PAML4.4 [78]. To be able to take possible biased distribution of different LINE subfamilies (with different age profiles) across the genome into account, divergence was normalized by the relative age of each subfamily using the TinT program which counts the frequency of nested transposable elements [79]. Divergence of each window was normalized by the following equation:where is the divergence, is the relative time of the maximal activity of subfamily i (note that a low ti value indicates a high age), is number of LINEs in a window, and is the mean value of ti for all subfamilies in the genome.
It might be argued that analyses of repeat elements from two avian genomes cannot be seen as independent samples if elements inserted before the split of chicken and zebra finch. We therefore made separate analyses involving recombination rate using galliform-specific (chicken) and passeriform-specific (zebra finch) subfamilies of repeats, respectively. The results from these lineage-specific analyses were very similar to the full data set and are not reported.
Since several parameters were not normally distributed we used Kendall's rank tests for correlation analyses. All statistical analyses were performed in the R platform (http://www.r-project.org). Mixed model analysis was performed in order to control for chromosome identity using the lme4 package [80]. We then used the pvals.fnc function that calculated p-values based on the t statistic, with the upper bound for the number of degrees of freedom.
Comparison of autosomes and sex chromosomes
Non-parametric bootstrapping was performed in order to compare the sequence length of LINEs between sex chromosome and autosomes. The sequence length of each LINE was collected based on stratified random sampling and the difference in the mean LINE length between pairs of randomly grouped samples was used to test the null hypothesis. Bootstrapping was performed 10,000 times and significance level (p value) was obtained by calculating the proportion of replicates that had higher mean length difference between random categories than the real categories.
Comparison of the deletion bias between sex chromosome and autosomes was also tested using non-parametric bootstrapping and stratified random sampling. The difference in mean deletion bias between two categories of replicates was calculated by:where D* and I* are the respective number of deleted and inserted sites from a randomly chosen LINE using stratified sampling, a is the number of LINEs in a single category, and n is the number of LINEs in both categories. Bootstrapping was performed 10,000 times and significance level (p value) was obtained by calculating the proportion of replicates that had higher (or lower) than the difference of the mean deletion bias from the real dataset.
Modeling of the effect of the deletion bias on genome size
Change in sequence length can be expressed by the exponential decay function:where f(x) is the length of neutrally evolving sequence, r is the rate parameter for an exponential decay function, and t is time. r was calculated from the change in sequence length over a given time period defined by the substitution rate using:where D, I and S are deletion, insertion and substitution rates, respectively. This gives:D, I and S of the chicken lineage after the split between chicken and turkey were calculated from 8,328 ancestral CR1 sequences identified in whole-genome alignment of chicken, turkey and zebra finch [29]. This identification was based on the Repeatmasker output file (http://www.repeatmasker.org/genomes/galGal3/galGal3.fa.out.gz) of the chicken genome, using in house perl programs. We excluded alignments where sequence length of turkey or zebra finch was shorter than 80% of the alignment length in order to minimize the effect from spurious sequence originating from non-repetitive CR1 flanking sequences. Ancestral CR1 elements were concatenated within each window, followed by the estimation of divergence in the chicken lineage. Genome-wide divergence was then estimated from the weighted divergences of each window according to the length of alignments. Confidence intervals were calculated from bootstrapping with 1,000 replicates.
Supporting Information
Zdroje
1. CarvalhoABClarkAG 1999 Intron size and natural selection. Nature 401 344
2. ComeronJMKreitmanM 2000 The correlation between intron length and recombination in Drosophila: Dynamic equilibrium between mutational and selective forces. Genetics 156 1175 1190
3. ICGSC 2004 Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432 695 716
4. ComeronJMKreitmanM 2002 Population, evolutionary and genomic consequences of interference selection. Genetics 161 389 410
5. VinogradovAE 2004 Compactness of human housekeeping genes: selection for economy or genomic design? Trends in Genetics 20 248 253
6. VinogradovAE 2006 “Genome design” model: Evidence from conserved intronic sequence in human-mouse comparison. Genome Research 16 347 354
7. GregoryTR 2002 A bird's-eye view of the C-value enigma: Genome size, cell size, and metabolic rate in the class aves. Evolution 56 121 130
8. AndrewsCBMackenzieSAGregoryTR 2009 Genome size and wing parameters in passerine birds. Proceedings of the Royal Society B-Biological Sciences 276 55 61
9. LynchMConeryJS 2003 The origins of genome complexity. Science 302 1401 1404
10. PetrovDASangsterTAJohnstonJSHartlDLShawKL 2000 Evidence for DNA loss as a determinant of genome size. Science 287 1060 1062
11. PetrovDA 2002 Mutational equilibrium model of genome size evolution. Theoretical Population Biology 61 531 544
12. KongAGudbjartssonDFSainzJJonsdottirGMGudjonssonSA 2002 A high-resolution recombination map of the human genome. Nature Genetics 31 241 247
13. GroenenMAMWahlbergPFoglioMChengHHMegensHJ 2009 A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Research 19 510 519
14. BackstromNForstmeierWSchielzethHMelleniusHNamK 2010 The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Research 20 485 495
15. GregoryTR 2011 Animal Genome Size Database. http://www.genomesize.com
16. EllegrenH 2010 Evolutionary stasis: the stable chromosomes of birds. Trends in Ecology & Evolution 25 283 291
17. GriffinDKRobertsonLBWTempestHGSkinnerBM 2007 The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenetic and Genome Research 117 64 77
18. SjödinPBataillonTSchierupMH 2010 Insertion and deletion processes in recent human history. PLoS One 5 e8650
19. PetrovDALozovskayaERHartlDL 1996 High intrinsic rate of DNA loss in Drosophila. Nature 384 346 349
20. AbrusanGKrambeckHJJunierTGiordanoJWarburtonPE 2008 Biased distributions and decay of long interspersed nuclear elements in the chicken genome. Genetics 178 573 581
21. LiuGEJiangLTianFZhuBSongJZ 2009 Calibration of mutation rates reveals diverse subfamily structure of galliform CR1 repeats. Genome Biology and Evolution 1 119 130
22. VandergonTLReitmanM 1994 Evolution of chicken repeat 1 (CR1) elements: evidence for ancient subfamilies and multiple progenitors. Molecular Biology and Evolution 11 886 898
23. WickerTRobertsonJSSrSFeltusFAVM 2005 The repetitive landscape of the chicken genome. Genome Research 15 126 136
24. JeffreysAJNeumannR 2009 The rise and fall of a human recombination hot spot. Nature Genetics 41 625 629
25. PtakSEHindsDAKoehlerKNickelBPatilN 2005 Fine-scale recombination patterns differ between chimpanzees and humans. Nature Genetics 37 429 434
26. WincklerWMyersSRRichterDJOnofrioRCMcDonaldGJ 2005 Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308 107 111
27. WarrenWCClaytonDFEllegrenHArnoldAPHillierLW 2010 The genome of a songbird. Nature 464 757 762
28. NabholzBKunstnerAWangRJarvisEEllegrenH 2011 Dynamic evolution of base composition: causes and consequences in avian phylogenomics. Molecular Biology and Evolution 28 2197 2210
29. DalloulRALongJAZiminAVAslamLBealK 2010 Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biology 8 e1000475
30. NamKMugalCNabholzBSchielzethHWolfJB 2010 Molecular evolution of genes in avian genomes. Genome Biology 11 R68
31. LunterGPontingCPHeinJ 2006 Genome-wide identification of human functional DNA using a neutral indel model. PLoS Computational Biology 2 e5
32. PetrovDAHartlDL 1998 High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Molecular Biology and Evolution 15 293 302
33. MillsREPittardWSMullaneyJMFarooqUCreasyTH 2011 Natural genetic variation caused by small insertions and deletions in the human genome. Genome Research 21 830 839
34. TaylorMSPontingCPCopleyRR 2004 Occurrence and consequences of coding sequence insertions and deletions in mammalian genomes. Genome Research 14 555 566
35. TajimaF 1989 Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585 595
36. RomiguierJRanwezVDouzeryEJGaltierN 2010 Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Research 20 1001 1009
37. GregoryTR 2003 Is small indel bias a determinant of genome size? Trends in Genetics 19 485 488
38. OrganCLShedlockAMMeadeAPagelMEdwardsSV 2007 Origin of avian genome size and structure in non-avian dinosaurs. Nature 446 180 184
39. ShedlockAMBotkaCWZhaoSShettyJZhangT 2007 Phylogenomics of nonavian reptiles and the structure of the ancestral amniote genome. Proceedings of the National Academy of Sciences of the United States of America 104 2767 2772
40. BurtDW 2005 Chicken genome: Current status and future opportunities. Genome Research 15 1692 1698
41. EllegrenH 2005 The avian genome uncovered. Trends in Ecology & Evolution 20 180 186
42. BurtDW 2002 Origin and evolution of avian microchromosomes. Cytogenetic and Genome Research 96 97 112
43. QumsiyehMB 1994 Evolution of number and morphology of mammalian chromosomes. Journal of Heredity 85 455 465
44. BegunDJAquadroCF 1992 Levels of naturally-occurring DNA polymorphism correlate with recombination rates in Drosophila-melanogaster. Nature 356 519 520
45. LercherMJHurstLD 2002 Human SNP variability and mutation rate are higher in regions of high recombination. Trends in Genetics 18 337 340
46. NachmanMW 2001 Single nucleotide polymorphisms and recombination rate in humans. Trends in Genetics 17 481 485
47. NachmanMWBauerVLCrowellSLAquadroCF 1998 DNA variability and recombination rates at X-linked loci in humans. Genetics 150 1133 1141
48. HellmannIEbersbergerIPtakSEPaaboSPrzeworskiM 2003 A neutral explanation for the correlation of diversity with recombination rates in humans. American Journal of Human Genetics 72 1527 1535
49. KulathinalRJBennetttSMFitzpatrickCLNoorMAF 2008 Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proceedings of the National Academy of Sciences of the United States of America 105 10051 10056
50. CutterADMosesAM 2011 Polymorphism, divergence, and the role of recombination in Saccharomyces cerevisiae genome evolution. Molecular Biology and Evolution 28 1745 1754
51. HuangSWFriedmanRYuNYuALiWH 2005 How strong is the mutagenicity of recombination in mammals? (vol 22, pg 426, 2005). Molecular Biology and Evolution 22 1157 1157
52. KeinanAReichD 2010 Human population differentiation is strongly correlated with local recombination rate. PLoS Genetics 6 e1000886
53. NoorMA 2008 Mutagenesis from meiotic recombination is not a primary driver of sequence divergence between Saccharomyces species. Molecular Biology and Evolution 25 2439 2444
54. StevisonLSNoorMAF 2010 Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. Journal of Molecular Evolution 71 332 345
55. BebenekKKunkelTA 2004 Functions of DNA polymerases. DNA Repair and Replication 69 137 165
56. AlbertsonTMOgawaMBugniJMHaysLEChenY 2009 DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proceedings of the National Academy of Sciences of the United States of America 106 17101 17104
57. FortuneJMPavlovYIWelchCMJohanssonEBurgersPMJ 2005 Saccharomyces cerevisiae DNA polymerase delta - High fidelity for base substitutions but lower fidelity for single - and multi-base deletions. Journal of Biological Chemistry 280 29980 29987
58. HicksWM 2010 Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329 82 85
59. SchmittMWMatsumotoYLoebLA 2009 High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie 91 1163 1172
60. MaloiselLFabreFGangloffS 2008 DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Molecular and Cellular Biology 28 1373 1382
61. OliverJLBernaola-GalvanPCarpenaPRoman-RoldanR 2001 Isochore chromosome maps of eukaryotic genomes. Gene 276 47 56
62. BernardiGCostantiniMAulettaF 2007 Isochore patterns and gene distributions in fish genomes. Genomics 90 364 371
63. CarelsNBernardiG 2000 The compositional organization and the expression of the Arabidopsis genome. FEBS Letters 472 302 306
64. AdamsMDCelnikerSEHoltRAEvansCAGocayneJD 2000 The genome sequence of Drosophila melanogaster. Science 287 2185 2195
65. VersteegRvan SchaikBDCvan BatenburgMFRoosMMonajemiR 2003 The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Research 13 1998 2004
66. DuretLMouchiroudDGautierC 1995 Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. Journal of Molecular Evolution 40 308 317
67. BernardiG 1995 The human genome: Organization and evolutionary history. Annual Review of Genetics 29 445 476
68. DuretLGaltierN 2009 Biased gene conversion and the evolution of mammalian genomic landscapes. Annual Review of Genomics and Human Genetics 10 285 311
69. MaraisG 2003 Biased gene conversion: implications for genome and sex evolution. Trends in Genetics 19 330 338
70. MeunierJDuretL 2004 Recombination drives the evolution of GC-content in the human genome. Molecular Biology and Evolution 21 984 990
71. GaltierNPiganeauGMouchiroudDDuretL 2001 GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics 159 907 911
72. SmukowskiCSNoorMAF 2011 Recombination rate variation in closely related species. Heredity 107 496 508
73. KvikstadEMTyekuchevaSChiaromonteFMakovaKD 2007 A macaque's-eye view of human insertions and deletions: Differences in mechanisms. PLoS Computational Biology 3 1772 1782
74. PollardKSHubiszMJRosenbloomKRSiepelA 2010 Detection of nonneutral substitution rates on mammalian phylogenies. Genome Research 20 110 121
75. MakovaKDYangSChiaromonteF 2004 Insertions and deletions are male biased too: A whole-genome analysis in rodents. Genome Research 14 567 573
76. SmitAHubleyRGreenP 1996–2004 (1996–2004) RepeatMasker http://www.repeatmasker.org
77. JurkaJ 2000 Repbase Update - a database and an electronic journal of repetitive elements. Trends in Genetics 16 418 420
78. YangZH 2007 PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24 1586 1591
79. ChurakovGGrundmannNKuritzinABrosiusJMakalowskiW 2010 A novel web-based TinT application and the chronology of the Primate Alu retroposon activity. BMC Evolutionary Biology 10 376
80. BatesDMSarkarD 2007 lme4: Linear mixed-effects models using S4 classes. pp. R package version 0.9975-9912
Štítky
Genetika Reprodukční medicína
Článek Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication inČlánek Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the TranscriptomeČlánek Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells inČlánek Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing BoneČlánek Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Slowing Replication in Preparation for Reduction
- Chromosome Pairing: A Hidden Treasure No More
- Loss of Imprinting Differentially Affects REM/NREM Sleep and Cognition in Mice
- Six Novel Susceptibility Loci for Early-Onset Androgenetic Alopecia and Their Unexpected Association with Common Diseases
- Regulation by the Noncoding RNA
- UDP-Galactose 4′-Epimerase Activities toward UDP-Gal and UDP-GalNAc Play Different Roles in the Development of
- Deletion of PTH Rescues Skeletal Abnormalities and High Osteopontin Levels in Mice
- Karyotypic Determinants of Chromosome Instability in Aneuploid Budding Yeast
- Genome-Wide Copy Number Analysis Uncovers a New HSCR Gene:
- MicroRNA-277 Modulates the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- Functional Centromeres Determine the Activation Time of Pericentric Origins of DNA Replication in
- Dynamic Deposition of Histone Variant H3.3 Accompanies Developmental Remodeling of the Transcriptome
- Scientist Citizen: An Interview with Bruce Alberts
- YY1 Regulates Melanocyte Development and Function by Cooperating with MITF
- Congenital Heart Disease–Causing Gata4 Mutation Displays Functional Deficits
- Recombination Drives Vertebrate Genome Contraction
- KATNAL1 Regulation of Sertoli Cell Microtubule Dynamics Is Essential for Spermiogenesis and Male Fertility
- Re-Patterning Sleep Architecture in through Gustatory Perception and Nutritional Quality
- Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in
- Genome-Wide Analysis of GLD-1–Mediated mRNA Regulation Suggests a Role in mRNA Storage
- Meiotic Chromosome Pairing Is Promoted by Telomere-Led Chromosome Movements Independent of Bouquet Formation
- LINT, a Novel dL(3)mbt-Containing Complex, Represses Malignant Brain Tumour Signature Genes
- The H3K27 Demethylase UTX-1 Is Essential for Normal Development, Independent of Its Enzymatic Activity
- Suppresses Senescence Programs and Thereby Accelerates and Maintains Mutant -Induced Lung Tumorigenesis
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
- Identification of Genes That Promote or Antagonize Somatic Homolog Pairing Using a High-Throughput FISH–Based Screen
- Principles of Carbon Catabolite Repression in the Rice Blast Fungus: Tps1, Nmr1-3, and a MATE–Family Pump Regulate Glucose Metabolism during Infection
- Integrin α PAT-2/CDC-42 Signaling Is Required for Muscle-Mediated Clearance of Apoptotic Cells in
- Histone H3 Localizes to the Centromeric DNA in Budding Yeast
- Collapse of Telomere Homeostasis in Hematopoietic Cells Caused by Heterozygous Mutations in Telomerase Genes
- Hypersensitive to Red and Blue 1 and Its Modification by Protein Phosphatase 7 Are Implicated in the Control of Arabidopsis Stomatal Aperture
- Extent, Causes, and Consequences of Small RNA Expression Variation in Human Adipose Tissue
- TBC-8, a Putative RAB-2 GAP, Regulates Dense Core Vesicle Maturation in
- Regulating Repression: Roles for the Sir4 N-Terminus in Linker DNA Protection and Stabilization of Epigenetic States
- Common Genetic Determinants of Intraocular Pressure and Primary Open-Angle Glaucoma
- Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone
- Fitness Landscape Transformation through a Single Amino Acid Change in the Rho Terminator
- Repeated, Selection-Driven Genome Reduction of Accessory Genes in Experimental Populations
- Allelic Variation and Differential Expression of the mSIN3A Histone Deacetylase Complex Gene Promote Mammary Tumor Growth and Metastasis
- DNA Demethylation and USF Regulate the Meiosis-Specific Expression of the Mouse
- Knowledge-Driven Analysis Identifies a Gene–Gene Interaction Affecting High-Density Lipoprotein Cholesterol Levels in Multi-Ethnic Populations
- A Duplication CNV That Conveys Traits Reciprocal to Metabolic Syndrome and Protects against Diet-Induced Obesity in Mice and Men
- EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice
- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women
- Stratifying Type 2 Diabetes Cases by BMI Identifies Genetic Risk Variants in and Enrichment for Risk Variants in Lean Compared to Obese Cases
- New Insight into the History of Domesticated Apple: Secondary Contribution of the European Wild Apple to the Genome of Cultivated Varieties
- Activated Cdc42 Kinase Has an Anti-Apoptotic Function
- The Region Is Critical for Birth Defects and Electrocardiographic Dysfunctions Observed in a Down Syndrome Mouse Model
- COP9 Signalosome Integrity Plays Major Roles for Hyphal Growth, Conidial Development, and Circadian Function
- Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest
- Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing
- The Number of X Chromosomes Causes Sex Differences in Adiposity in Mice
- Target Gene Analysis by Microarrays and Chromatin Immunoprecipitation Identifies HEY Proteins as Highly Redundant bHLH Repressors
- Acquisition Order of Ras and p53 Gene Alterations Defines Distinct Adrenocortical Tumor Phenotypes
- ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes
- Histone H1 Depletion Impairs Embryonic Stem Cell Differentiation
- IDN2 and Its Paralogs Form a Complex Required for RNA–Directed DNA Methylation
- Separation of DNA Replication from the Assembly of Break-Competent Meiotic Chromosomes
- Genomic Hypomethylation in the Human Germline Associates with Selective Structural Mutability in the Human Genome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Inactivation of a Novel FGF23 Regulator, FAM20C, Leads to Hypophosphatemic Rickets in Mice
- Genome-Wide Association of Pericardial Fat Identifies a Unique Locus for Ectopic Fat
- Slowing Replication in Preparation for Reduction
- An Essential Role for Katanin p80 and Microtubule Severing in Male Gamete Production
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



