-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
Transportin-SR (TRN-SR) is a member of the importin-β super-family that functions as the nuclear import receptor for serine-arginine rich (SR) proteins, which play diverse roles in RNA metabolism. Here we report the identification and cloning of mos14 (modifier of snc1-1, 14), a mutation that suppresses the immune responses conditioned by the auto-activated Resistance (R) protein snc1 (suppressor of npr1-1, constitutive 1). MOS14 encodes a nuclear protein with high similarity to previously characterized TRN-SR proteins in animals. Yeast two-hybrid assays showed that MOS14 interacts with AtRAN1 via its N-terminus and SR proteins via its C-terminus. In mos14-1, localization of several SR proteins to the nucleus was impaired, confirming that MOS14 functions as a TRN-SR. The mos14-1 mutation results in altered splicing patterns of SNC1 and another R gene RPS4 and compromised resistance mediated by snc1 and RPS4, suggesting that nuclear import of SR proteins by MOS14 is required for proper splicing of these two R genes and is important for their functions in plant immunity.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002159
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002159Summary
Transportin-SR (TRN-SR) is a member of the importin-β super-family that functions as the nuclear import receptor for serine-arginine rich (SR) proteins, which play diverse roles in RNA metabolism. Here we report the identification and cloning of mos14 (modifier of snc1-1, 14), a mutation that suppresses the immune responses conditioned by the auto-activated Resistance (R) protein snc1 (suppressor of npr1-1, constitutive 1). MOS14 encodes a nuclear protein with high similarity to previously characterized TRN-SR proteins in animals. Yeast two-hybrid assays showed that MOS14 interacts with AtRAN1 via its N-terminus and SR proteins via its C-terminus. In mos14-1, localization of several SR proteins to the nucleus was impaired, confirming that MOS14 functions as a TRN-SR. The mos14-1 mutation results in altered splicing patterns of SNC1 and another R gene RPS4 and compromised resistance mediated by snc1 and RPS4, suggesting that nuclear import of SR proteins by MOS14 is required for proper splicing of these two R genes and is important for their functions in plant immunity.
Introduction
In eukaryotes, the nuclear envelope forms a barrier between the cytoplasm and the nucleus. Trafficking of macromolecules across the nuclear envelope occurs through the nuclear pore complex (NPC) [1]. Previous studies on MOS3 [2], MOS6 [3], MOS7 [4] and MOS11 [5] have revealed the importance of nucleocytoplasmic trafficking in plant immunity. Mutations in MOS3, MOS6, MOS7 and MOS11 suppress the constitutive defense responses of snc1 (suppressor of npr1-1, constitutive 1), a gain-of-function mutant carrying a mutation in a Toll/interleukin-1 receptor-Nucleotide Binding-Leucine Rich Repeat (TIR-NB-LRR) R protein [6]. MOS3 encodes the nucleoporin Nup96 [2], whereas MOS11 encodes a putative RNA binding protein [5]. Both MOS3 and MOS11 are required for mRNA export. MOS6 encodes a putative importin-α [3], whereas MOS7 encodes another nucleoporin, Nup88, which is required for nuclear accumulation of snc1 and two general defense regulators, Enhanced Disease Susceptibility 1 (EDS1) and Nonexpresser of PR genes 1 (NPR1) [4].
Nuclear import receptors play essential roles in transferring proteins from the cytoplasm to the nucleus. The largest group of nuclear import receptors belong to the importin-β super-family. Members of the importin-β super-family have rather low overall sequence similarity but they all have a conserved N-terminal RAN-binding domain [7], [8]. The import receptors recognize the nuclear localization sequence (NLS) of target proteins to facilitate their transport through the NPC. Upon RAN-GTP binding to importin-β, the importin-β complex is dissociated and the cargo is released into the nucleus.
The importin-β super-family can be divided into several sub-families according to the direction and the cargo type they transport [9]. Among them, the transportin-SR (TRN-SR) subfamily functions as nuclear import receptors for serine-arginine rich (SR) proteins. TRN-SR was originally identified as an interactor of SR domains of ASF/SF2 [10] and papillomavirus E2 [11]. In humans, the C-terminus of TRN-SR interacts with SR proteins and the interaction can be disrupted upon RAN-binding to its N-terminus [10].
SR proteins are a highly conserved family of nuclear proteins that play important roles in splicing [12]–[14]. They contain RNA recognition motifs (RRM) at the N-terminus and an arginine-serine rich (RS) domain at the C-terminus. The NLS is located in the RS domain. SR proteins not only function as splicing factors for constitutive splicing [15], [16], they also regulate alternative splicing through splice site selection in a concentration-dependent manner [17], [18].
Several plant R genes including the tobacco N gene [19], the barley Mla6 [20], Arabidopsis SNC1 [21] and RPS4 [22]–[24] are alternatively spliced. For example, six transcript variants (TV) have been identified for RPS4 [23], [24]. Compromised RPS4-mediated resistance resulting from a lack of the TVs suggests that alternative splicing of RPS4 is required for its function [23]. However, it is unclear how alternative splicing of these R genes is controlled and why it is necessary for immunity. In this study, we report that Arabidopsis MOS14 encodes a TRN-SR that is required for proper splicing of SNC1 and RPS4, suggesting that SR proteins may play important roles in the control of the splicing of these two R genes.
Results
Identification of mos14-1 snc1 npr1
Arabidopsis snc1 constitutively activates defense responses and displays enhanced resistance to pathogens. snc1 mutant plants exhibit dwarf morphology with curly leaves. Suppressor screens of snc1 have previously been carried out using fast neutron and T-DNA insertional mutagenesis [2], [25]. To identify additional suppressor mutants of snc1, we treated snc1 npr1 seeds with ethane methyl sulfonate (EMS) and screened the M2 plants for mutants that suppressed snc1 dwarfism. From this population, we identified mos14-1 snc1 npr1 (Figure 1A).
Fig. 1. Constitutive immune responses in snc1 are suppressed by mos14-1. 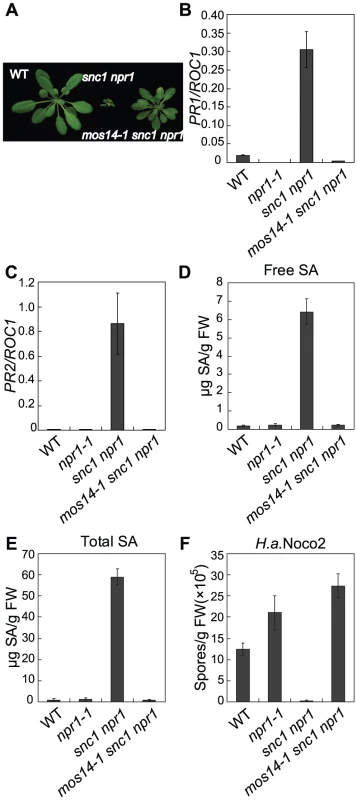
(A) Morphology of five-week-old soil-grown plants of the indicated genotypes. (B–C) PR1 (B) or PR2 (C) expression in the indicated genotypes. RNAs were extracted from two-week-old plants grown on MS media and reverse transcribed to obtain cDNA for real-time PCR analysis. Values were normalized to the expression of the reference gene Cyclophilin (ROC1). Error bars represent ±SD of three replicates. (D–E) Free SA (D) and total SA (E) levels in the indicated genotypes. SA was extracted from five-week-old soil-grown plants and measured by HPLC. Error bars represent ±SD of three replicates. (F) Growth of H. a. Noco2 on the indicated genotypes. Plants were inoculated by H. a. Noco2 at 5×104 spores/ml and spores were collected and counted seven days later. Error bars represent ±SD of three replicates. In snc1 npr1, defense marker gene PR1 and PR2 are constitutively expressed. As shown in Figure 1B and 1C, constitutive activation of PR1 and PR2 is suppressed in mos14-1 snc1 npr1. Analysis of SA levels also showed that the elevated SA levels in snc1 npr1 are suppressed by mos14-1 (Figure 1D and 1E). To test whether enhanced pathogen resistance in snc1 npr1 is affected by mos14-1, mos14-1 snc1 npr1 seedlings were challenged with the virulent oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) Noco2. As shown in Figure 1F, resistance to H. a. Noco2 is lost in mos14-1 snc1 npr1.
Map-based cloning of mos14-1
To map the mos14-1 mutation, we crossed mos14-1 snc1 npr1 (in the Columbiaecotype background) with Landsberg erecta (Ler)-snc1 [2]. In the F2 mapping population, about a quarter of the progeny showed morphology similar to the triple mutant. Crude mapping using 24 F2 plants revealed that mos14-1 is linked to the lower arm of chromosome 5 (Figure 2A). Further analysis indicated that mos14-1 is flanked by marker MMN10 and MUB3. Fine mapping using about 1200 F2 plants narrowed mos14-1 to a 60 kb region between marker K19B1 and MRG21. To identify the mos14-1 mutation, PCR fragments covering this 60 kb region was amplified directly from mos14-1 snc1 npr1 and sequenced. A single G to A mutation was found in At5g62600 (Figure 2B), which is located at the junction of the 13th intron and 13th exon of the gene. RT-PCR analysis using primers flanking the mutation showed that splicing of At5g62600 was affected by the mutation (Figure 2C). The RT-PCR fragments were cloned into the pGEM-T vector. Subsequent sequence analysis of cDNA clones from mos14-1 revealed that they fell into six different classes. All of them represent transcript variants that were incorrectly spliced. An alignment of wild type cDNA and the cDNA variants from mos14-1 are shown in Figure S1.
Fig. 2. Map-based cloning of mos14-1. 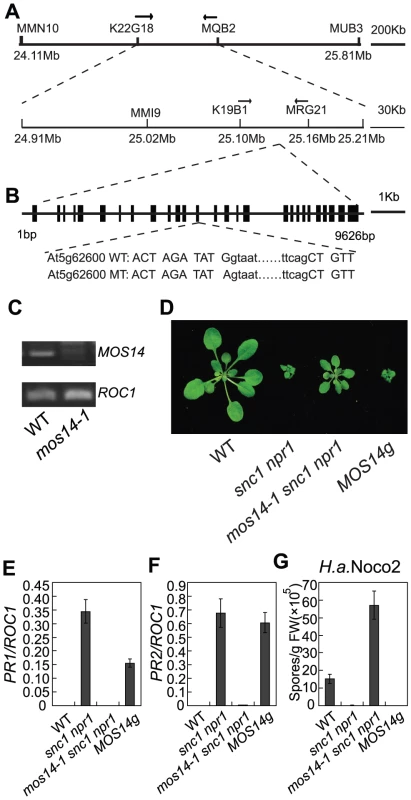
(A) Mapping of mos14-1. BAC clones and markers are indicated. The mos14-1 mutation is flanked between markers K19B1 and MRG21. (B) Gene structure of MOS14 and the mutation site in mos14-1. The exons are indicated with boxes and introns with lines. The mutation site is located at the junction between the 13th intron and 13th exon. The lower case letters represent nucleotides in the intron and the uppercase letters represent nucleotides in the exon. (C) Expression of MOS14 in mos14-1. Primers (F47 and SNPR, Table S1) flanking the mutation site were used to amplify MOS14 from wild type and mos14-1 cDNA. ROC1 was used as loading control. (D) Morphology of mos14-1 snc1 npr1 carrying the MOS14 transgene. Five-week-old soil-grown plants were photographed. “MOS14g” stands for “mos14-1 snc1 npr1 containing the MOS14 transgene under its native promoter”. (E–F) Restoration of PR1 (E) and PR2 (F) expression in mos14-1 snc1 npr1 by the MOS14 transgene. (G) Restoration of resistance to H. a. Noco2 in mos14-1 snc1 npr1 by the MOS14 transgene. To confirm that the mutation in At5g62600 is responsible for the suppression of snc1 npr1 mutant phenotypes, a genomic clone containing At5g62600 was constructed and transformed into mos14-1 snc1 npr1. Transgenic plants from five independent lines carrying the wild type At5g62600 displayed snc1-like morphology (Figure 2D). Further analysis of a representative transgenic line showed that the expression of PR1 and PR2 was similar to snc1 npr1 (Figure 2E and 2F). In addition, resistance to H. a. Noco2 was also restored in the transgenic line (Figure 2G), confirming that At5g62600 complemented mos14-1 and MOS14 is At5g62600.
To obtain the mos14-1 single mutant, we backcrossed mos14-1 snc1 npr1 with wild type plants. The mos14-1 single mutant was obtained by genotyping the F2 plants. The mos14-1 single mutant flowers late and has reduced fertility. Besides, it exhibits small stature (Figure S2). When the genomic clone of At5g62600 was introduced into the mos14-1 single mutants, it reverted the size and fertility of the mutant to wild type-like and also suppressed the late flowering phenotype, showing that the developmental phenotypes observed in mos14-1 are caused by the mos14-1 mutation.
MOS14 encodes a transporter for SR proteins
MOS14 is a single copy gene in Arabidopsis. It encodes a protein with 25% identity and 45% similarity to the TRN-SR in Drosophila, suggesting that MOS14 may be a transporter for SR proteins. MOS14 and its animal homologs are highly conserved at their N-terminus (Figure S3), which contain the importin-β N-terminal domains.
To determine the subcellular localization of MOS14, transgenic plants expressing MOS14 under its native promoter with a C-terminal GFP tag were generated in both wild type and mos14-1 backgrounds. Expression of MOS14-GFP in mos14-1 suppresses the developmental phenotypes of mos14-1 (Figure S4), suggesting that the fusion protein is functional. Confocal fluorescence microscopy analysis of transgenic plants expressing the MOS14-GFP fusion protein showed that the GFP signal is found exclusively in the nucleus (Figure 3), indicating that MOS14 is a nuclear protein. In the nuclei of root cells, GFP fluorescence was excluded from a large part of the nucleus, probably the nucleolus. We did not observe similar exclusion of MOS14-GFP from parts of the nuclei in epidermal cells, probably because these nuclei are much smaller than those in root cells.
Fig. 3. MOS14 localizes to the nucleus. 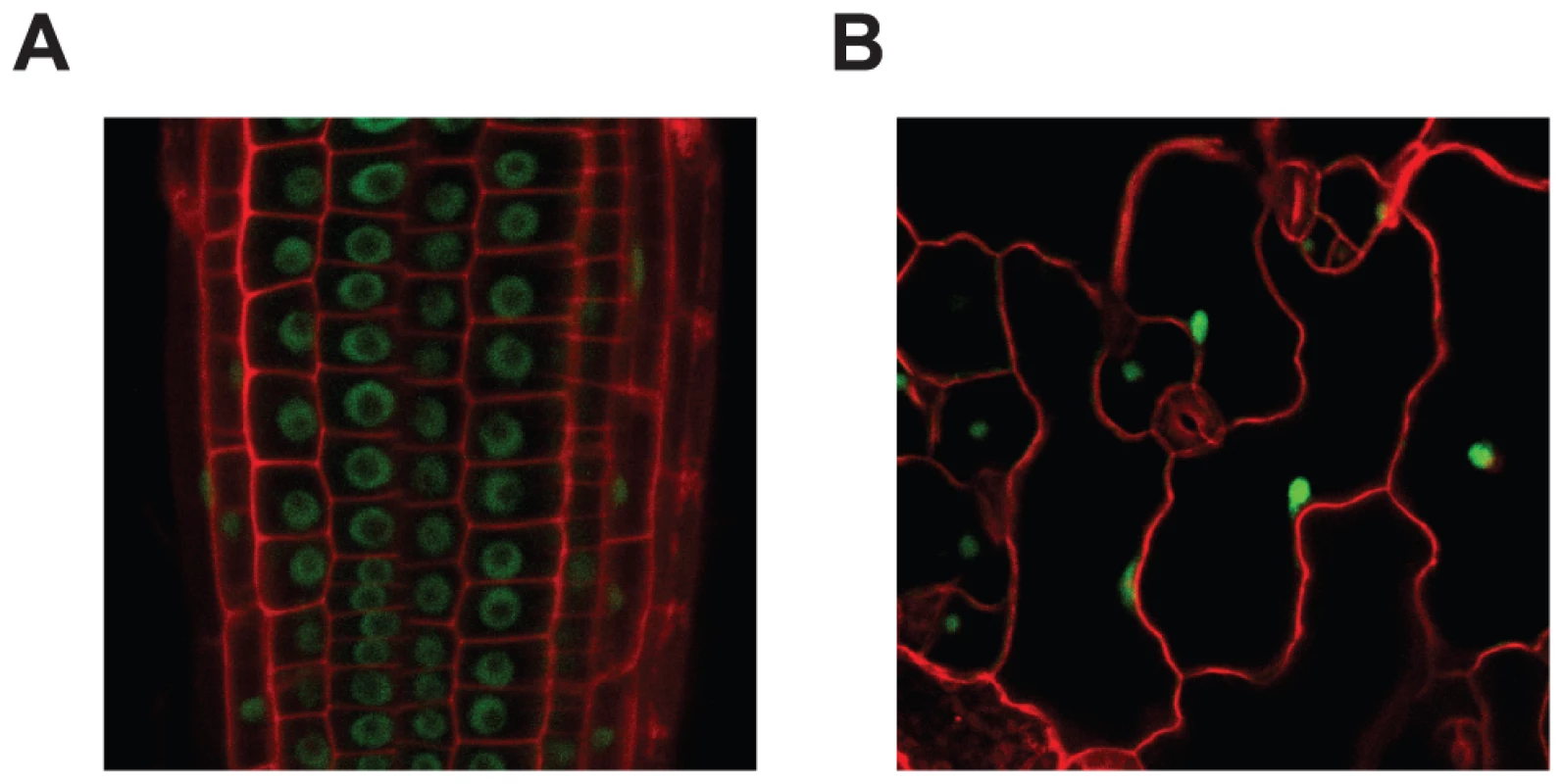
GFP fluorescence in root (A) and epidermal (B) cells from transgenic plants expressing MOS14-GFP under its own promoter in Col-0 (WT). Cell walls were stained with PI before confocal microscopy, which fluoresces red. In animals, TRN-SR binds SR proteins via its C-terminus and transport SR proteins through the nuclear envelope. Binding of RAN-GTP to the N-terminus of TRN-SR in nucleus results in the release of SR proteins. To test whether MOS14 is able to interact with SR proteins, the N-terminus (1–281) and C-terminus (282–958) of MOS14 were expressed in the bait vector and four selected Arabidopsis SR proteins (AtRS2Z33, AtRSZ21, AtRS31 and AtSR34) were expressed in the prey vector for yeast two-hybrid assays. As shown in Figure 4A, the C-terminus, but not the N-terminus of MOS14 interacts with the SR proteins. We also tested the interactions between MOS14 and AtRAN1. As shown in Figure 4B, the N-terminus, but not the C-terminus of MOS14 interacts with AtRAN1.
Fig. 4. MOS14 interacts with AtRAN1 and SR proteins in yeast two-hybrid assays. 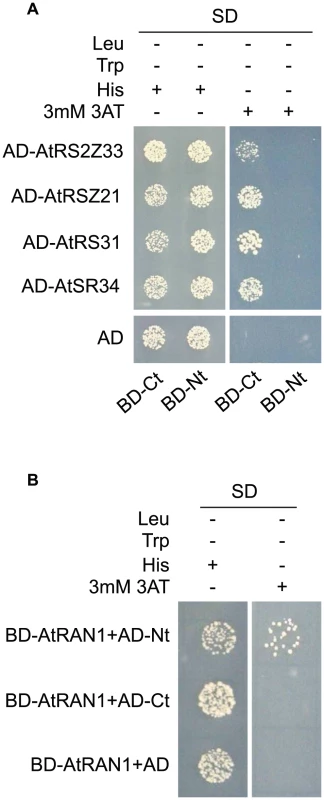
(A) Interaction between the C-terminus (Ct, amino acid 282–958) of MOS14 and the indicated SR proteins. (B) Interaction between the N-terminus (Nt, amino acid 1–281) of MOS14 and AtRAN1. 3 mM 3-AT was included in the medium to increase the stringency of selection. SD, synthetic dropout medium. To test whether the mos14-1 mutation affects the nuclear import of Arabidopsis SR proteins, we made constructs expressing four SR genes AtRS2Z33, AtRSZ21, AtRS31 and AtSR34 with a C-terminal GFP tag. These constructs were transformed into protoplasts of wild type and mos14-1 plants to check for the localization of the SR-GFP proteins. A construct expressing the SARD1-GFP fusion protein was included as the control [26]. As shown in Figure 5A, in both wild type and mos14-1 protoplasts, SARD1 was localized in the nucleus. Consistent with previous studies [27], the SR-GFP proteins were clearly localized in the nucleus of wild type protoplasts. However, in mos14-1 protoplasts, the SR-GFP proteins were mainly localized in the cytoplasm (Figure 5A and Table 1), suggesting that MOS14 is required for the nuclear localization of SR proteins.
Fig. 5. Localization of SR proteins in wild type and mos14-1. 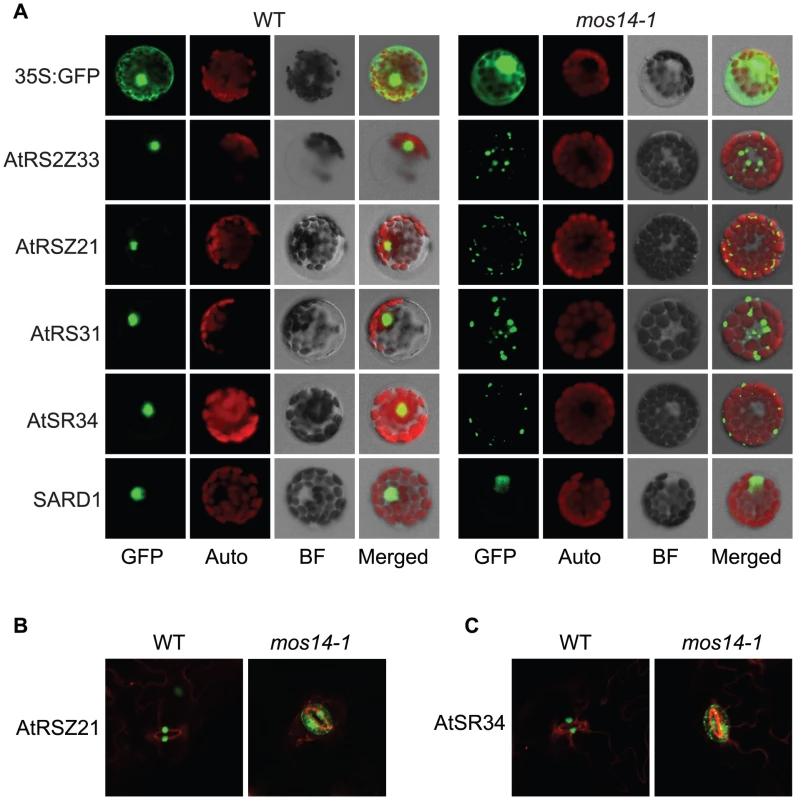
Localization of SR proteins in wild type and mos14-1 protoplasts. Protoplasts were prepared from four-week-old soil-grown plants and transformed with 10 µg of plasmid expressing the indicated SR protein with a C-terminal GFP tag or GFP alone . All constructs were under the control of a 35S promoter. GFP (GFP fluorescence); Auto (chloroplast autofluorescence); BF (bright field). (B–C) Localization of AtRSZ21-GFP and AtSR34-GFP in transgenic plants expressing the GFP fusion proteins under the control of 35S promoter in Col-0 (WT) or mos14-1 background. Tab. 1. Analysis of SR protein and SARD1 localization in protoplasts of Col-0 and mos14-1. 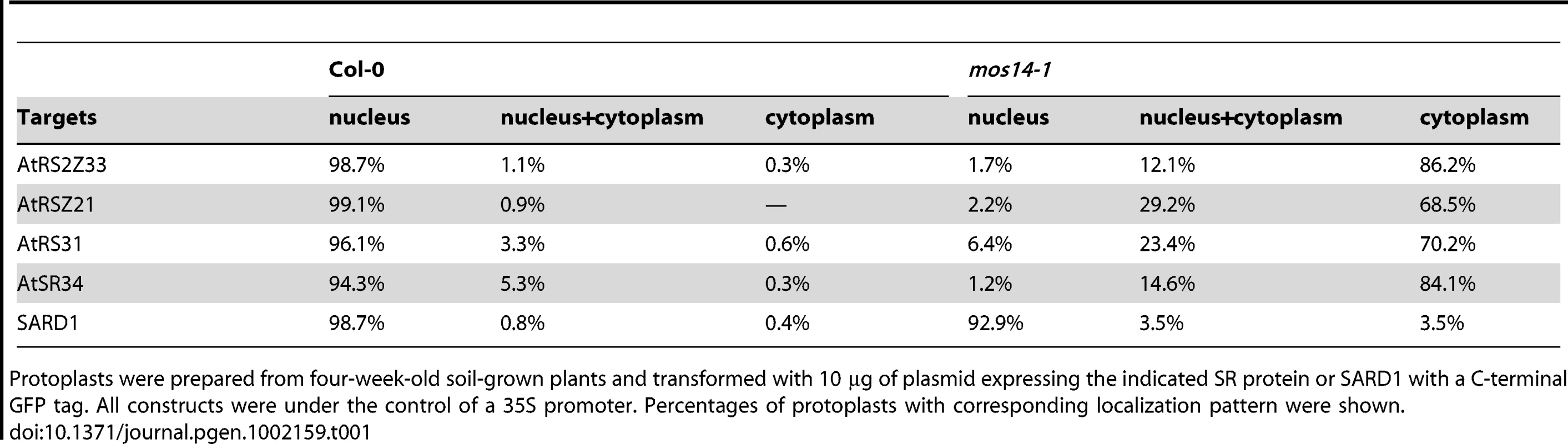
Protoplasts were prepared from four-week-old soil-grown plants and transformed with 10 µg of plasmid expressing the indicated SR protein or SARD1 with a C-terminal GFP tag. All constructs were under the control of a 35S promoter. Percentages of protoplasts with corresponding localization pattern were shown. Unlike GFP expressed under 35S promoter which is distributed throughout the whole cell, the SR-GFP proteins were localized to discrete foci in the cytoplasm of mos14-1 protoplasts. The pattern of these foci resembles that of P-bodies, which are distinct foci in the cytoplasm of eukaryotic cells containing many enzymes involved in mRNA turnover. Because of the diverse roles of SR proteins in RNA metabolism, it would not be surprising if they also function in P-bodies. The effect of mos14-1 on the localization of AtRSZ21 and AtSR34 was further confirmed in transgenic plants expressing the AtRSZ21-GFP and AtSR34-GFP fusion proteins. As shown in Figure 5B and 5C, AtRSZ21-GFP and AtSR34-GFP were localized in discrete foci in the cytoplasm of guard cells in mos14-1 background. The GFP fusion proteins were also observed in the cytoplasm of leaf pavement cells in mos14-1. Taken together, these experiments indicate that MOS14 is a transporter for SR proteins.
mos14-1 affects splicing of SNC1 and RPS4
Multiple SNC1 transcripts with intron 2 and intron 3 removed or retained have previously been detected [21]. Because none of the transgenic plants expressing the snc1 cDNA exhibit dwarf morphology like snc1 mutant plants (Figure S5), alternative splicing is probably required for the function of SNC1. Since mos14-1 affects the nuclear localization of SR proteins and SR proteins participate in pre-mRNA splice site recognition and spliceosome assembly, we tested whether splicing of SNC1 was affected in mos14-1. Primers flanking the introns of SNC1 were designed to evaluate its splicing pattern of SNC1 (Figure 6A). Consistent with the previous report, we detected transcripts with either intron 2 or 3 retained (Figure S6). As shown in Figure 6B, we detected another transcript that contains both intron 2 and 3 (TV1) in addition to the regular transcripts with both intron 2 and 3 removed (TV4) in mos14-1 snc1 npr1. In wild type plants, the amount of TV2 and TV3 is small compared to that of TV4. Both TV2 and TV3 increased dramatically in the mos14-1 snc1 npr1 mutant plants (Figure 6B). Similar alteration of SNC1 transcript patterns was also observed in the mos14-1 single mutant (Figure S8B). Since PCR reaction using the RNA samples showed no amplification, the DNA fragments from RT-PCR represent SNC1 transcripts rather than genomic DNA contamination. Further analysis of SNC1 transcript variants in mos14-1 and mos14-1 snc1 npr1 lines carrying the wild type MOS14 transgene showed that the splicing patterns of SNC1 in the transgenic lines are similar to those in the wild type plants (Figure S8A and S8B). These data indicate that mos14-1 affects the splicing of the SNC1 transcript.
Fig. 6. mos14-1 affects the splicing pattern and causes reduced expression of SNC1 and RPS4. 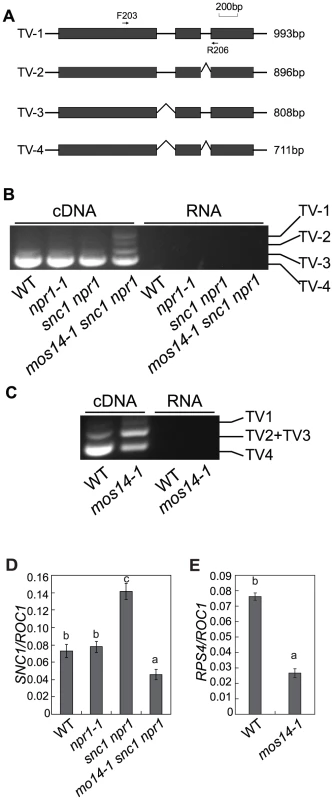
(A) Gene structure of 5′ end of SNC1. Exons are indicated with boxes and introns are indicated with lines. Locations of the primers used to amplify the transcript variants (TV) are indicated. The lengths of different transcript variants amplified by F203 and R206 are indicated on the right. (B) Transcription patterns of SNC1 in wild type (WT), npr1-1, snc1 npr1 and mos14-1 snc1 npr1. PCR was performed on cDNA (left) or RNA samples (right) using primer F203 and R206 (Table S1). RNAs incubated in reverse transcription reaction without RTase M-MLV were used as the negative control to ensure that genomic DNA contamination did not occur. (C) RPS4 alternative transcription patterns in wild type (WT) and mos14-1. PCR was performed on cDNA (left) or RNA samples (right). TV1 represents transcripts with both intron 2 and intron 3 retained. TV2 and TV3 represent two transcripts with similar size which retained either intron 2 or intron 3. TV4 is the regular transcript with intron 2 and intron 3 removed. (D) SNC1 expression in mos14-1 snc1 npr1. Values are normalized to the expression of ROC1. Bars represent ±SD of three replicates. Statistical differences among the samples are labeled with different letters (P<0.01). (E) RPS4 expression in mos14-1. Values are normalized to the expression of ROC1. Bars represent ±SD of three replicates. Statistical differences among the samples are labeled with different letters (P<0.01). The R gene RPS4 was also reported to be alternatively spliced [23]. We designed primers to detect the transcript variants for RPS4 by RT-PCR. As shown in Figure 6C, the levels of TV1 are similar in wild type and mos14-1. However, TV2+TV3 increased considerably and TV4 was significantly reduced in mos14-1, indicating that mos14-1 also affects the splicing pattern of RPS4 transcripts. The altered RPS4 transcript patterns in mos14-1 snc1 npr1 and mos14-1 can be complemented by the MOS14 transgene (Figure S8C and S8D).
To determine whether MOS14 has a general role in RNA splicing, we analyzed splicing of two housekeeping genes Actin1 and β-tubulin4 by RT-PCR using primers that flank introns. ROC1 was used as the control because it contains no intron. We found that splicing of Actin1 and β-tubulin4 was not affected in mos14-1 (Figure S7). We also analyzed the splicing patterns of U1-70K, AtSR30 and AtSR34, three genes reported to be alternatively spliced [28], [29]. As shown in Figure S7, the splicing of AtSR30 and AtSR34, but not U1-70K was clearly affected by mos14-1. Alteration of the transcription patterns of AtSR30 and AtSR34 in mos14-1 further supports the role of MOS14 in alternative splicing. Since the splicing of Actin1, β-tubulin4 and U1-70K is not affected by mos14-1, there may be a certain level of specificity in MOS14-mediated pre-mRNA processing.
To test whether the splicing defect in mos14-1 leads to a decrease in snc1 and RPS4 transcripts, real-time RT-PCR was carried out using primers to amplify an unspliced region at the 3′ end of the two genes. As shown in Figure 6D and 6E, expression levels of both snc1 and RPS4 decreased significantly in the presence of the mos14-1 mutation.
RPS4-mediated immunity and basal resistance are compromised in mos14-1
Since mos14-1 altered the splicing pattern of RPS4 and reduced its expression, we tested whether RPS4-mediated immunity is affected by mos14-1. As shown in Figure 7A, growth of Pseudomonas syringae pv. tomato (P.s.t.) DC3000 avrRps4 in mos14-1 is about ten-fold higher than that in wild type, suggesting RPS4-mediated immunity is compromised in mos14-1. We also tested whether MOS14 is required for basal resistance by challenging the mos14-1 plants with the virulent pathogen P.s.t. DC3000. As shown in Figure 7B, bacterial growth is about ten-fold higher in mos14-1 compared to wild type, indicating that MOS14 is also required for basal resistance.
Fig. 7. mos14-1 compromises RPS4-mediated immunity and basal resistance. 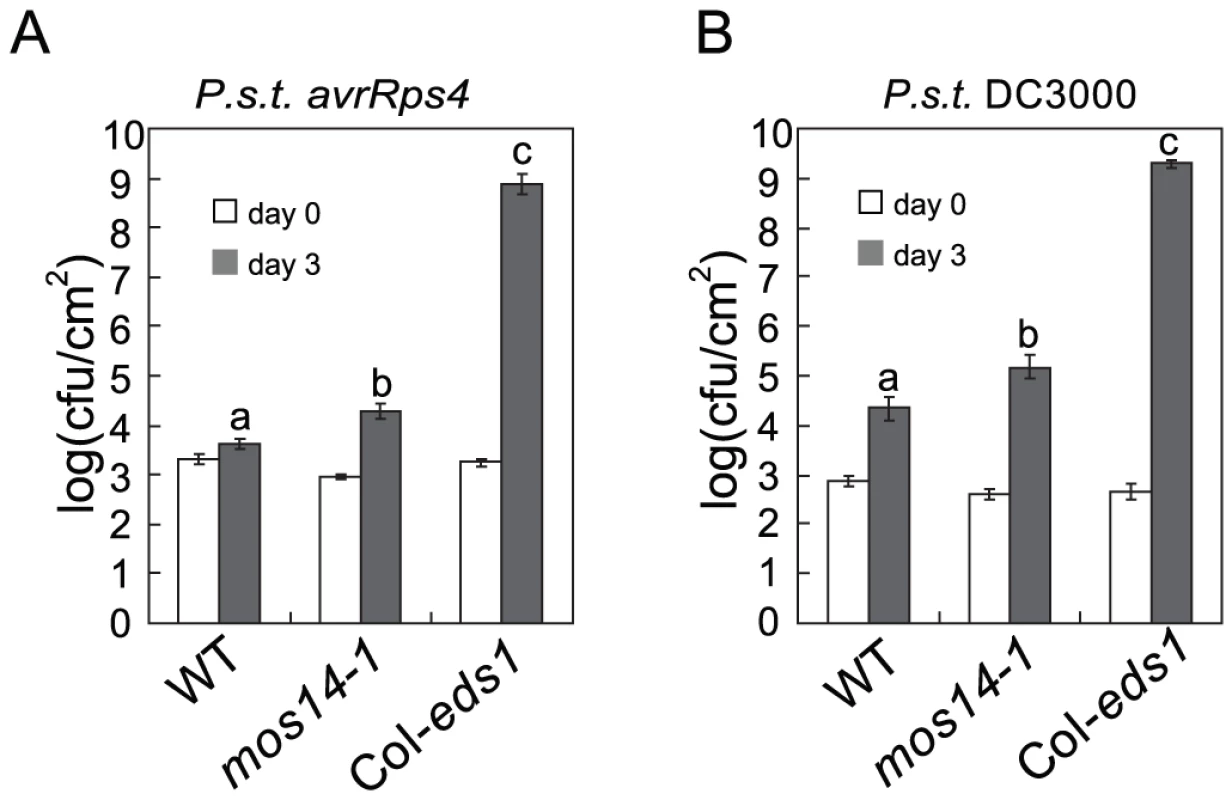
(A) Growth of P.s.t. DC3000 avrRps4 on wild type (WT), mos14-1 and eds1-2 (Col). (B) Growth of P.s.t. DC3000 on wild type (WT), mos14-1 and eds1-2 (Col). Error bars represent ±SD of six replicates. Statistical differences among the samples are labeled with different letters (P<0.01). Discussion
Previous studies on snc1 suppressor mutants revealed that multiple components are involved in the regulation of plant immunity. In particular, pathways involved in mRNA export, protein import and protein export were found to contribute to immune responses. Here we report the identification of MOS14 as a novel component of nucleocytoplasmic trafficking required for plant immunity. Loss of MOS14 function suppresses the constitutive defense responses of snc1, compromises resistance mediated by RPS4 and impairs basal resistance against P.s.t. DC3000. These findings show that MOS14 plays a critical role in plant immunity.
MOS14 encodes a nuclear protein with high sequence similarity to TRN-SR proteins in animals. TRN-SR proteins have been shown to function as nuclear import receptors for both phosphorylated SR proteins as well as the splicing repressor protein RSF1 which antagonizes SR proteins in the nucleus [11], [30]. Since their discovery, TRN-SR proteins have not been extensively studied [10]. MOS14 is a single-copy gene, while the Arabidopsis genome has 18 genes belonging to six subfamilies of SR proteins, of which three are plant-specific [31]. There is no close homolog of RSF1 in Arabidopsis. Like the TRN-SR proteins in animals, the N-terminus of MOS14 interacts with AtRAN1 and the C-terminus interacts with SR proteins. In addition, localization of several SR proteins to the nucleus was impaired by mos14-1. These data indicate that the mechanism of nuclear import of SR proteins is conserved between plants and animals.
Very limited studies have been performed on the genetic characterization of TRN-SR proteins. In C.elegans, RNAi of the MOS14 homolog Transporter of SR-1 (TSR-1) leads to embryonic lethality, suggesting TRN-SR proteins can be essential for viability [32]. Intriguingly, the mos14-1 mutation is not lethal, although it does cause multiple development phenotypes such as reduced stature and fertility. In addition to its functions in development, our genetic analysis of MOS14 revealed that it plays important roles in both R gene-mediated resistance as well as basal defense, suggesting that nuclear import of SR proteins is important for plant immunity. The reasons why mos14-1 leads to these pleiotropic defects and not lethality awaits further investigation.
SR proteins play important roles in general RNA splicing, alternative splicing, as well as other processes of RNA metabolism. Consistent with the function of MOS14 in the nuclear import of SR proteins, the mos14-1 mutation affects the splicing of SNC1 and RPS4. Several R genes including SNC1, RPS4 and tobacco N gene are alternatively spliced, and alternative splicing of RPS4 and N gene are required for their function [23], [33]. In mos14-1, alternative splicing of both SNC1 and RPS4 are altered. This effect probably contributes to the suppression of snc1 mutant phenotypes by mos14-1 and compromised RPS4 function in the mos14-1 single mutant. In addition to the altered ratio of transcript variants, the expression levels of snc1 and RPS4 were also reduced. The reduced expression of snc1 and RPS4 is probably caused by splicing defects resulting from the reduced nuclear localization of SR proteins.
In mos14-1 snc1 npr1, the SNC1 TV-4 transcript level is only modestly reduced, suggesting that reduced accumulation of TV-4 may not be the only factor that contributes to the complete suppression of snc1 mutant phenotype. In addition to reduced accumulation of TV-4, levels of SNC1 TV-1, TV-2 and TV-3 are considerably increased in mos14-1 snc1 npr1. These transcripts are predicted to produce truncated snc1 proteins because of introduction of early stop codons. It is possible that these truncated proteins may interfere with the function of the full-length snc1. Because snc1 and RPS4 are not the only genes whose splicing are affected by mos14-1, altered splicing of one or more unknown positive regulators of plant defense could also contribute to the suppression of snc1 mutant phenotypes.
In addition to the compromised resistance responses mediated by snc1 and RPS4, basal resistance against P.s.t. DC3000 is also compromised in mos14-1. It remains to be determined how mos14-1 affects basal resistance. One possibility is that MOS14 is required for the splicing of one or more R genes that contribute to basal resistance against P.s.t. DC3000. Alternatively, mos14-1 may cause splicing defects in defense regulators required for basal resistance.
In summary, we have identified MOS14 as a nuclear transporter of SR proteins. Our data suggest that regulation of R gene splicing by SR proteins is critical for plant immunity. Future studies on individual SR proteins will help us better understand how SR proteins regulate the splicing of R genes.
Materials and Methods
Plant growth conditions and mutant screen
All plants were grown at 23°C under 16 hr light/8 hr dark in plant growth rooms or chambers, if not specifically mentioned. To identify mutations that suppress the mutant phenotypes of snc1, snc1 npr1 seeds were mutagenized with EMS. About 30,000 M2 plants representing about 1,500 M1 families were screened for suppression of the dwarf morphology of snc1 npr1-1. Mutants lacking the dwarf phenotype were further analyzed for suppression of the constitutive defense responses in snc1 npr1.
Gene expression analysis
About 0.1 g tissue was collected and RNA was extracted by Takara RNAiso reagent. The RNA was treated with Promega RQ1 RNase-Free DNase to remove contaminating genomic DNA. Reverse transcription was subsequently carried out using oligo-dT and the M-MLV RTase cDNA synthesis kit from Takara. About 200 ng of total RNA was included in each RT reaction. For semi-quantitative and real-time PCR , one fiftieth of the cDNA was used in each reaction. A total of 40 cycles were performed for semi-quantitative RCR except 28 cycles for ROC1. Real-time PCR was carried out using Takara SYBR® Premix Ex Taq™ II. The primers for real-time PCR analysis of PR1, PR2, SNC1 [34] and ROC1 (also called cyclophilin) [35] were described previously. ROC1 is a housekeeping gene without introns. The sequences of primers used for SNC1 and RPS4 transcript variants analysis are shown in Table S1. Primers to amplify U1-70K [28], AtSR30 and AtSR34 [29] were described previously.
Pathogen infections and SA measurements
For infections with H. a. Noco2, three-week-old soil-grown plants were sprayed with H. a. Noco2 at 5×104 spores/ml. The inoculated seedlings were subsequently kept in a growth chamber with high humidity (>80%) at 18°C under 12 hr light/12 hr dark cycle for seven days before growth of H. a. Noco2 was quantified, as previously described [36].
For infections with P.s.t. DC3000 or P.s.t. DC3000 avrRps4, five-week-old soil-grown plants were infiltrated with bacterial suspensions (OD600 = 0.001) in 10 mM MgCl2. Samples were taken at day 0 and day 3.
To analyze the SA levels in the mutant plants, SA was extracted using a previously described procedure [37] and measured by high-performance liquid chromatography.
Construction of plasmids
For the transgenic complementation test, three PCR fragments, F12R37 (3.9K), F14R38 (3.8K) and F23R19 (2.9K) covering the 11 kb region where MOS14 is located were amplified from wild type genomic DNA. The primers used for amplification of F12R37, F14R38 and F23R19 are F12, R37, F14, R38, F23 and R19 respectively, and their sequences are provided in Table S1. These fragments were sequentially sub-cloned into pBluescript SK+. The complete 11 kb fragment was subsequently cloned into a modified pGreen0229 vector containing the NOS terminator to obtain the construct pMOS14:MOS14. The final construct containing MOS14 was transformed into mos14-1 snc1 npr1 through Agrobacterium-mediated transformation.
For the subcellular localization study of MOS14, PCR fragments F12R37 (3.9K), F14R38 (3.8K) and F23R20 (2.9K) were sequentially sub-cloned into pBluescript SK+. The primers used for amplification of F23R20 are F23 and R20 and their sequences are listed in the Table S1. The 11 kb fragment described above was cloned into a modified pCambia1305 vector expressing C-terminal tagged GFP to obtain pMOS14:MOS14-GFP.
For transient expression of SR proteins in protoplasts, full-length cDNAs of AtRS2Z33, AtRSZ21, AtRS31 and AtSR34 without the stop codons were amplified by PCR and cloned into the modified pUC19 vector pUC19-35S-cmGFP4 that expresses GFP under the 35S promoter.
To obtain transgenic plants expressing snc1 cDNA, full-length snc1 cDNA was amplified from total cDNA of snc1 and cloned into a modified pGreen0229 vector. The cDNA clone was sequenced to make sure the sequence is correct and no intron was retained.
To obtain transgenic plants expressing AtSR34-GFP and AtRSZ21-GFP, full-length cDNAs of AtSR34 and AtRSZ21 without the stop codons were amplified by PCR and cloned into a modified pCambia1300 vector expressing C-terminal tagged GFP under 35S promoter. The constructs were transformed into Col-0 and mos14-1 through Agrobacterium-mediated transformation.
Yeast two-hybrid analysis
To make constructs for the yeast two hybrid assays, an SfiI restriction site was first introduced to the multiple cloning site of pGBKT7 and pGADT7 to obtain pGBKT7a and pGADT7a, respectively. cDNA expressing the N-terminal or C-terminal region of MOS14 and AtRAN1 were amplified by PCR and cloned into pGBKT7a. Full-length cDNAs of AtRS2Z33, AtRSZ21, AtRS31 and AtSR34 were amplified by PCR and cloned into pGADT7a. cDNA expressing the N-terminal or C-terminal region of MOS14 were also cloned into pGADT7a. The plasmids expressing the MOS14 fragments were co-transformed with the vectors expressing AtRAN1 or one of the SR proteins into the yeast strain PJ694α for yeast two-hybrid analysis.
Confocal fluorescence microscopy of MOS14-GFP localization
For confocal fluorescence microscopy analysis of MOS14-GFP, the roots or leaves of six-day-old seedling grown on MS plates were first stained with propidium iodide (PI) for 1 min and then washed in ddH2O for at least three times. The concentration of PI used for staining the roots was 10 µg/ml, whereas the concentration of PI used for the leaves is 10 mg/ml. The stained sample was observed using a Zeiss Meta 510 confocal microscope. Excitation wavelengths for GFP and PI were 488 nm and 543 nm, respectively. For root samples, the emission filter used for PI was LP560 nm. For leaf samples, the emission filter used for PI was BP560 nm-615 nm. For both root and leaf samples, the emission filter for GFP was BP505 nm-530 nm.
Confocal fluorescence microscopy of localization of SR-GFPs
Plasmids used for protoplast transfections were purified with Invitrogen PureLink™ HiPure Plasmid Filter Purification Kit. Transformation of wild type or mos14-1 protoplasts was performed as previously described [38]. After transformation, protoplasts were kept in the dark for about 16 hours. The transformed protoplasts were examined using a Zeiss Axiovert 200 fluorescence microscope. The pictures of representative protoplasts were taken using confocal fluorescence microcopy. For autofluorescence, the emission filter used was 650 nm-740 nm. Confocal fluorescence microscopy analysis of transgenic plants expressing AtSR34-GFP and AtRSZ21-GFP was performed on three-week-old seedlings using a procedure described in the analysis of MOS14-GFP localization.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g62600 (MOS14), At2g14610 (PR1), At3g57260 (PR2), At4g38470 (ROC1), At2g37620 (Actin1), At5g44340 (β-tubulin4 ), AAD38537 (hTRN-SR1), CAB42634 (hTRN-SR2), NP608708 (dTRN-SR), AF025464 (TSR1) and CAA99366 (MTR10a).
Supporting Information
Zdroje
1. MerkleT 2003 Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr Genet 44 231 260
2. ZhangYLiX 2005 A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17 1306 1316
3. PalmaKZhangYLiX 2005 An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol 15 1129 1135
4. ChengYTGermainHWiermerMBiDXuF 2009 Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21 2503 2516
5. GermainHQuNChengYTLeeEHuangY 2010 MOS11: a new component in the mRNA export pathway. PLoS Genet 6 e1001250 doi:10.1371/journal.pgen.1001250
6. ZhangYGoritschnigSDongXLiX 2003 A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 2636 2646
7. GorlichDDabrowskiMBischoffFRKutayUBorkP 1997 A novel class of RanGTP binding proteins. J Cell Biol 138 65 80
8. HarelAForbesDJ 2004 Importin beta: conducting a much larger cellular symphony. Mol Cell 16 319 330
9. StromACWeisK 2001 Importin-beta-like nuclear transport receptors. Genome Biol 2 REVIEWS3008
10. KataokaNBachorikJLDreyfussG 1999 Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol 145 1145 1152
11. LaiMCLinRIHuangSYTsaiCWTarnWY 2000 A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem 275 7950 7957
12. BartaAKalynaMLorkovicZJ 2008 Plant SR proteins and their functions. Curr Top Microbiol Immunol 326 83 102
13. ReddyAS 2004 Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci 9 541 547
14. LongJCCaceresJF 2009 The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417 15 27
15. LopatoSWaigmannEBartaA 1996 Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell 8 2255 2264
16. LopatoSGattoniRFabiniGSteveninJBartaA 1999 A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol Biol 39 761 773
17. LazarGSchaalTManiatisTGoodmanHM 1995 Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc Natl Acad Sci U S A 92 7672 7676
18. LopatoSKalynaMDornerSKobayashiRKrainerAR 1999 atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev 13 987 1001
19. WhithamSDinesh-KumarSPChoiDHehlRCorrC 1994 The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78 1101 1115
20. HaltermanDZhouFWeiFWiseRPSchulze-LefertP 2001 The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J 25 335 348
21. YiHRichardsEJ 2007 A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19 2929 2939
22. GassmannWHinschMEStaskawiczBJ 1999 The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20 265 277
23. ZhangXCGassmannW 2003 RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 15 2333 2342
24. ZhangXCGassmannW 2007 Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 145 1577 1587
25. ZhuZXuFZhangYChengYTWiermerM 2010 Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A 107 13960 13965
26. ZhangYXuSDingPWangDChengYT 2010 Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci U S A
27. LorkovicZJHilscherJBartaA 2004 Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol Biol Cell 15 3233 3243
28. GolovkinMReddyAS 1996 Structure and expression of a plant U1 snRNP 70K gene: alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8 1421 1435
29. Savaldi-GoldsteinSAvivDDavydovOFluhrR 2003 Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15 926 938
30. AllemandEDokudovskayaSBordonneRTaziJ 2002 A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1. Mol Biol Cell 13 2436 2447
31. BartaAKalynaMReddyAS 2010 Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22 2926 2929
32. LongmanDJohnstoneILCaceresJF 2000 Functional characterization of SR and SR-related genes in Caenorhabditis elegans. Embo J 19 1625 1637
33. Dinesh-KumarSPBakerBJ 2000 Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci U S A 97 1908 1913
34. LiYTessaroMJLiXZhangY 2010 Regulation of the expression of plant Resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 153 1425 1434
35. PalusaSGAliGSReddyAS 2007 Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J 49 1091 1107
36. BiDChengYTLiXZhangY 2010 Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol 153 1771 1779
37. LiXZhangYClarkeJDLiYDongX 1999 Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329 339
38. YooSDChoYHSheenJ 2007 Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Pronto 2 1565 1572
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



