-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
SKN-1, the Caenorhabditis elegans Nrf1/2/3 ortholog, promotes both oxidative stress resistance and longevity. SKN-1 responds to oxidative stress by upregulating genes that detoxify and defend against free radicals and other reactive molecules, a SKN-1/Nrf function that is both well-known and conserved. Here we show that SKN-1 has a broader and more complex role in maintaining cellular stress defenses. SKN-1 sustains expression and activity of the ubiquitin-proteasome system (UPS) and coordinates specific protective responses to perturbations in protein synthesis or degradation through the UPS. If translation initiation or elongation is impaired, SKN-1 upregulates overlapping sets of cytoprotective genes and increases stress resistance. When proteasome gene expression and activity are blocked, SKN-1 activates multiple classes of proteasome subunit genes in a compensatory response. SKN-1 thereby maintains UPS activity in the intestine in vivo under normal conditions and promotes survival when the proteasome is inhibited. In contrast, when translation elongation is impaired, SKN-1 does not upregulate proteasome genes, and UPS activity is then reduced. This indicates that UPS activity depends upon presence of an intact translation elongation apparatus; and it supports a model, suggested by genetic and biochemical studies in yeast, that protein synthesis and degradation may be coupled processes. SKN-1 therefore has a critical tissue-specific function in increasing proteasome gene expression and UPS activity under normal conditions, as well as when the UPS system is stressed, but mounts distinct responses when protein synthesis is perturbed. The specificity of these SKN-1–mediated stress responses, along with the apparent coordination between UPS and translation elongation activity, may promote protein homeostasis under stress or disease conditions. The data suggest that SKN-1 may increase longevity, not only through its well-documented role in boosting stress resistance, but also through contributing to protein homeostasis.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002119
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002119Summary
SKN-1, the Caenorhabditis elegans Nrf1/2/3 ortholog, promotes both oxidative stress resistance and longevity. SKN-1 responds to oxidative stress by upregulating genes that detoxify and defend against free radicals and other reactive molecules, a SKN-1/Nrf function that is both well-known and conserved. Here we show that SKN-1 has a broader and more complex role in maintaining cellular stress defenses. SKN-1 sustains expression and activity of the ubiquitin-proteasome system (UPS) and coordinates specific protective responses to perturbations in protein synthesis or degradation through the UPS. If translation initiation or elongation is impaired, SKN-1 upregulates overlapping sets of cytoprotective genes and increases stress resistance. When proteasome gene expression and activity are blocked, SKN-1 activates multiple classes of proteasome subunit genes in a compensatory response. SKN-1 thereby maintains UPS activity in the intestine in vivo under normal conditions and promotes survival when the proteasome is inhibited. In contrast, when translation elongation is impaired, SKN-1 does not upregulate proteasome genes, and UPS activity is then reduced. This indicates that UPS activity depends upon presence of an intact translation elongation apparatus; and it supports a model, suggested by genetic and biochemical studies in yeast, that protein synthesis and degradation may be coupled processes. SKN-1 therefore has a critical tissue-specific function in increasing proteasome gene expression and UPS activity under normal conditions, as well as when the UPS system is stressed, but mounts distinct responses when protein synthesis is perturbed. The specificity of these SKN-1–mediated stress responses, along with the apparent coordination between UPS and translation elongation activity, may promote protein homeostasis under stress or disease conditions. The data suggest that SKN-1 may increase longevity, not only through its well-documented role in boosting stress resistance, but also through contributing to protein homeostasis.
Introduction
Maintenance of protein homeostasis is critical for organismal health, and protection against environmental challenges. Protein homeostasis depends upon the balance among the processes of protein synthesis, folding, and degradation. Disruptions in this balance result in accumulation of abnormal proteins, which over time leads to deterioration of cellular functions, and ultimately to cell death [1], [2]. Imbalances in proteostasis are central to progression of numerous disorders, including some cancers, neurodegenerative and alcoholic liver disease, and type 2 diabetes [3], [4].
Most intracellular proteolysis is mediated by the 26S proteasome, a multicatalytic protease that degrades polyubiquitinated proteins [5]. The ubiquitin-proteasome system (UPS) regulates the stability of proteins involved in a wide range of cellular processes [6]. The 26S proteasome is composed of two subcomplexes: a barrel-shaped 20S catalytic core structure, and a 19S regulatory particle that caps it at either or both ends. The 19S regulatory particle facilitates the entry of polyubiquitinated proteins, and is composed of base and lid subcomplexes [6], [7]. It is a major challenge to understand how the levels and activity of the proteasome are regulated to maintain the balance of protein synthesis and degradation.
Several lines of evidence indicate that the proteasome associates with the mRNA translation machinery, and that the processes of protein synthesis and degradation may be linked. Proteins are synthesized through the steps of translation initiation, elongation, and termination. The elongation cycle adds amino acids to a growing polypeptide chain, and requires a set of translation elongation factors (TEFs) (Figure S1; Table S1). The elongation process is regulated through phosphorylation of TEFs in response to growth and nutrient availability signals [8]. In addition, some TEFs are involved in functions besides translation. The elongation factor eEF1A binds to proteasome subunits and ubiquitinated proteins, and thereby seems to promote degradation of damaged nascent proteins [7], [9]–[11]. Given that up to 30% of nascent polypeptides may be degraded cotranslationally [12], [13], this interaction could be important for protein quality control and homeostasis. Consistent with this idea, in S. pombe translation initiation factors (TIFs), TEFs, and the proteasome associate together within a “translasome” supercomplex that is proposed to facilitate degradation of defective newly-synthesized proteins [14].
Nrf1/2/3 (NF-E2-related factor) proteins defend against oxidative and xenobiotic stress by regulating transcription of numerous cytoprotective genes [15]. Recent evidence indicates that Nrf proteins also promote proteasome gene expression in some cellular contexts. Proteasome activity is increased in many cancers, and it has been shown that in colon cancer cells Nrf2 upregulates proteasome expression and activity, and thereby seems to protect against apoptosis [16]. In cultured cell lines, Nrf1 and possibly Nrf2 mobilize a compensatory “bounce-back” response in which proteasome subunit genes are upregulated when the proteasome is inhibited [17]–[19]. These findings have important implications for development of cancer therapeutics that target the proteasome, because concomitant inhibition of Nrf proteins might enhance their effectiveness [18]. Nrf1 seems to have a relatively minor role in steady state proteasome gene expression, however, raising the question of how proteasome activity might normally be fine-tuned by Nrf proteins or other mechanisms in vivo.
In C. elegans, the Nrf1/2/3 ortholog SKN-1 defends against various stresses, and upregulates expression of a wide range of cellular defense, metabolism, and repair genes under either normal or stress conditions [20], [21]. Several proteasome subunit genes are among those that appear to be regulated by SKN-1 [20], [21], and a recent genome-scale chromatin immunoprecipitation (ChIP) analysis detected SKN-1 at the promoters of most proteasome genes under non-stressed conditions during the L1 larval stage [22]. Taken together, these findings raise the possibility that SKN-1/Nrf proteins might have a conserved and essential function in regulating proteasome synthesis in vivo, even under normal conditions. Furthermore, in a recent screen our lab identified genes for which RNA interference (RNAi) resulted in constitutive expression of stress-inducible SKN-1 targets [23]. These genes include several involved in protein folding or degradation, the TEF eef-1B.1, and some TIFs. RNAi against multiple TIFs resulted in a SKN-1-dependent transcriptional response that increased stress resistance and lifespan [23]. Together, these results suggest that SKN-1 might defend against perturbations in either protein synthesis or degradation.
In this study, we have investigated how impairment of translation elongation influences the activity of SKN-1 and the proteasome, and how SKN-1 and the translation machinery affect the proteasome. We show that distinct but overlapping sets of SKN-1 target genes are induced when translation initiation or elongation is inhibited. In the intestine, which is the C. elegans counterpart to the gut, liver, and adipose tissue, SKN-1 mediates a bounce-back response to proteasome gene inhibition, and also maintains UPS activity in vivo under normal conditions. Importantly, impairment of translation elongation does not induce this bounce-back response, and instead reduces intestinal UPS activity. The data reveal a remarkable degree of complexity in how SKN-1/Nrf proteins respond to different stresses, and suggest that the functional relationships between the translation elongation apparatus, SKN-1/Nrf proteins, and the proteasome are important for protein homeostasis.
Results
Induction of distinct SKN-1–mediated stress responses by inhibition of translation initiation or elongation
To investigate whether SKN-1 activity is generally influenced by translation elongation, we performed RNAi against 5 of the 7 predicted C. elegans TEFs (Table S1). We first monitored expression of a transgene in which the promoter for the SKN-1 target gene gcs-1 (γ-glutamyl cysteine synthetase) is fused to green fluorescent protein (GFP) (Figure S2A) [24]. RNAi against each TEF upregulated gcs-1p::GFP in the anterior intestine (Figure 1A) and increased expression of endogenous gcs-1 mRNA (Figure 1B). Mutation of an important SKN-1 binding site (Figure S2A) [24] diminished gcs-1 promoter induction (Figure S2B), and upregulation of endogenous gcs-1 mRNA was eliminated in a skn-1 mutant (Figure 1C), indicating that SKN-1 was required for gcs-1 induction in response to TEF RNAi.
Fig. 1. Induction of a SKN-1–mediated stress response by interference with translation elongation. 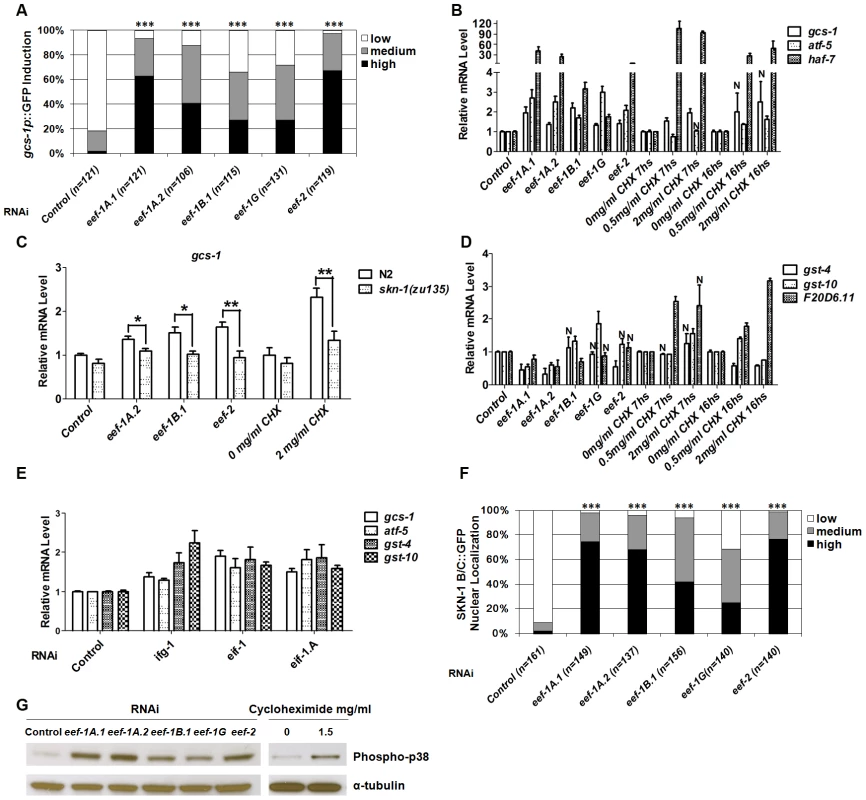
(A) The SKN-1-regulated reporter gcs-1p::GFP is activated by TEF RNAi. GFP levels were scored in the anterior intestine by a system similar to that we have described previously [24], with unambiguously bright levels defined as “High”, barely-detectable GFP levels scored as “Low”, and medium expression being intermediate between “Low” and “High”. Here and in (F), P values were derived from a chi2 test, *** P<0.0001. (B) Induction of endogenous SKN-1 target genes by interference with translation elongation. mRNA levels relative to control are shown, detected by quantitative Real Time PCR (qRT-PCR) after TEF RNAi or CHX treatment (for 7 or 16 hours (hs)). Here and in (E), P<0.05 compared to control except where not significant is indicated by N. Error bars indicate SEM. (C) SKN-1-dependence of target gene induction. Endogenous gcs-1 mRNA was detected by qRT-PCR in wild-type (N2) or skn-1(zu135) animals that had been fed with TEF RNAi bacteria, or treated with CHX for 18 hr at 15°C. Differences between wild-type (N2) and skn-1(zu135) animals were analyzed by paired t test (two-tailed). In all qRT-PCR experiments where asterisks are shown, ***P<0.001, **P<0.01, *P<0.05. An unpaired t test (two-tailed) was used to compare effects of TEF RNAi or CHX treatment with the corresponding N2 control. For each comparison to N2, P<0.001. (D) Interference with translation elongation does not upregulate all SKN-1 target genes. Endogenous SKN-1 target mRNA levels were assayed by qRT-PCR after TEF RNAi or CHX treatment. (E) Induction of SKN-1 target genes by TIF RNAi, assayed by qRT-PCR. (F) SKN-1 accumulates in intestinal nuclei in response to TEF RNAi. Examples of nuclear SKN-1 B/C::GFP scoring are shown in Figure S2E. (G) Activation of p38 MAPK signaling in response to inhibition of translation elongation. Lysates from worms that were exposed to TEF RNAi or CHX (13 hs) were analyzed by Western blotting for phosphorylated (active) p38 kinase. We next investigated how TEF knockdown influences expression of other SKN-1 target genes. The SKN-1-dependent genes atf-5 and haf-7 [20], [23] were upregulated in a manner that was either partially or completely dependent upon skn-1 (Figure 1B and Figure S2C). In contrast, the SKN-1 targets gst-4, gst-10 and F20D6.11 were generally not induced in response to TEF RNAi (Figure 1D and Figure S2D). This was surprising, because gst-4 is upregulated by SKN-1 under normal conditions, in response to various stresses, and after inhibition of insulin-like signaling (IIS) or translation initiation [20], [23], [25]–[27]. Similarly, gst-10 and F20D6.11 are induced by SKN-1 in response to reduced IIS and TIF RNAi, respectively [23], [25]. We further compared effects of translation initiation and elongation by analyzing animals subjected to RNAi against the TIFs ifg-1 (eIF4G), eif-1 (eIF-1) and eif-1.A (eIF-1A). In contrast to the effects of TEF RNAi, RNAi against these TIFs consistently upregulated endogenous gst-4 and gst-10, along with gcs-1 and atf-5 (Figure 1E and Figure S2D). Taken together, our data indicate that SKN-1 upregulates overlapping but distinct sets of target genes in response to inhibition of translation initiation or elongation.
Cycloheximide (CHX) blocks translation elongation by competing with the binding of ATP to the 60S ribosomal subunit, and inhibiting eEF2-mediated translocation (Figure S1) [28]. Treatment with CHX generally mimicked the effects of TEF RNAi on SKN-1 target gene expression, except that F20D6.11 was also upregulated (Figure 1B and 1D). This suggests that a SKN-1-dependent stress response is induced by inhibition of the translation elongation process per se, not simply by a lack of TEFs.
We next investigated how TEF knockdown influences the levels of SKN-1 in intestinal nuclei. A transgenic protein that includes two SKN-1 isoforms fused to GFP (SKN-1 B/C::GFP) readily accumulates in intestinal nuclei in response to various stresses, or reductions in IIS [24]–[26]. TEF knockdown also dramatically increased SKN-1 accumulation in intestinal nuclei, without upregulating endogenous skn-1 transcripts, indicating that elongation inhibition increases SKN-1 nuclear accumulation post-transcriptionally (Figure 1F, Figure S2E and S2F). In striking contrast, TIF RNAi does not detectably increase the overall levels of nuclear SKN-1 [23]. TIF inhibition therefore appears to upregulate SKN-1 target genes through a different mechanism, and may act on processes that cooperate with SKN-1 but do not influence its nuclear accumulation.
The evolutionarily conserved p38 mitogen-activated protein kinase (MAPK) signaling pathway is required for oxidative stress to induce SKN-1 nuclear accumulation and target gene activation [29]. The activity of this pathway can be assessed in C. elegans by Western blotting for the dually phosphorylated, active form of p38 kinase [29], [30]. We observed that both TEF RNAi and CHX treatment dramatically elevated the levels of phospho-p38 (Figure 1G). This signal and gcs-1 promoter induction were markedly reduced in the MAPKK and MAPKKK null mutants sek-1(km4) and nsy-1(ok593) respectively, indicating that the canonical p38 pathway was required (Figure S2G and S2B, respectively). With the exception of ifg-1, RNAi against TIFs did not robustly activate sek-1-dependent p38 MAPK activity, further supporting the idea that TIFs and TEFs influence SKN-1 activity largely through distinct mechanisms (Figure S2H).
Impaired translation elongation does not induce stress defenses non-specifically
It is an important question whether the SKN-1-mediated response to reduced translation elongation might derive simply from a non-specific activation of multiple stress defenses. To test this idea, we investigated how other stress responses involved in protein homeostasis are influenced by TEF RNAi. An accumulation of misfolded proteins in the endoplasmic reticulum (ER) or mitochondria triggers the ER unfolded protein response (UPRer), or mitochondrial UPR (UPRmt), respectively. To investigate whether the UPRer or UPRmt is activated in response to inhibition of TEFs, we examined transcriptional levels of hsp-4, an indicator of the UPRer [31], along with the UPRmt indicators hsp-6 and hsp-60 [32]. In general, TEF RNAi did not robustly increase the levels of hsp-4, hsp-6, or hsp-60 mRNAs (Figure 2A and 2B). Heat shock proteins (HSPs) act as chaperones that cope with misfolded cytoplasmic proteins during multiple stresses. As an indicator of effects on this network, we assayed for induction of genes representing four major classes of heat shock proteins: small HSPs (hsp-16.2), DnaJ/HSP40s (dnj-19, dnj-12), Hsc/HSP70s (hsp-70), and HSP90s (T05E11.3, daf-21) [33]. These HSP genes were also not upregulated in response to inhibition of most TEFs (Figure 2C, Figure S3A and S3B). Taken together, the data indicate that RNAi against TEFs does not broadly activate stress responses involved in proteostasis.
Fig. 2. TEF inhibition does not globally induce stress responses. 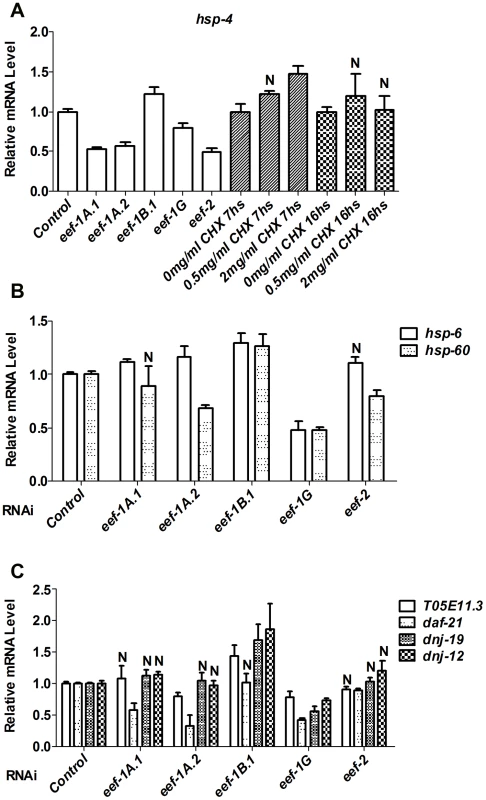
In (A–C), Relative levels of the indicated endogenous mRNAs were assayed by qRT-PCR after RNAi against the indicated TEF, or CHX treatment. N = not significant. In (A), all other P<0.025; in (B), all other P<0.05; in (C), all other P<0.02. In C. elegans, interference with translation initiation or elongation decreases brood size (Figure S3C) [34]–[36], raising the concern of whether the activation of SKN-1 that results from TEF RNAi might derive in part from reduction in germline proliferation. Interference with germ cell proliferation stimulates translocation of the transcription factor DAF-16/FOXO into intestinal nuclei, resulting in increased DAF-16 target gene expression and a daf-16-dependent increase in longevity [37], [38]. In contrast, TEF RNAi only minimally affected either DAF-16 nuclear levels, or expression of the DAF-16 target sod-3 (Figure S3D and S3E). Interference with germ cell proliferation also dramatically upregulated expression of the SKN-1 target gst-4 (Blackwell lab, unpublished), which is not induced by TEF RNAi (Figure 1D and Figure S2D). Together, these results suggest that the effects of translation elongation inhibition on SKN-1 activity do not derive from either a non-specific stress response, or indirect effects mediated by the germline.
SKN-1 increases stress resistance in response to reduced translation elongation
RNAi against TIF or ribosomal protein genes increases resistance to various environmental stresses [23], [34]–[36]. We therefore examined whether TEF knockdown affects resistance to two different sources of oxidative stress, the organic hydroperoxide tert-butyl hydrogen peroxide (TBHP), and the metalloid sodium arsenite (As) [20]. TBHP resistance was dramatically increased after knockdown of multiple TEFs in wild type animals (Figure 3A; Table S2). In contrast, RNAi against eef-2 or eef-1G did not robustly increase oxidative stress resistance in skn-1(zu135) mutants, indicating that skn-1 is essential for the TBHP resistance that derives from TEF knockdown (Figure 3B; Table S3). TEF inhibition also increased resistance to As (Figure 3C; Table S4). We conclude that the SKN-1-mediated transcriptional response to impaired translation elongation increases oxidative stress resistance.
Fig. 3. TEF knockdown increases oxidative stress resistance. 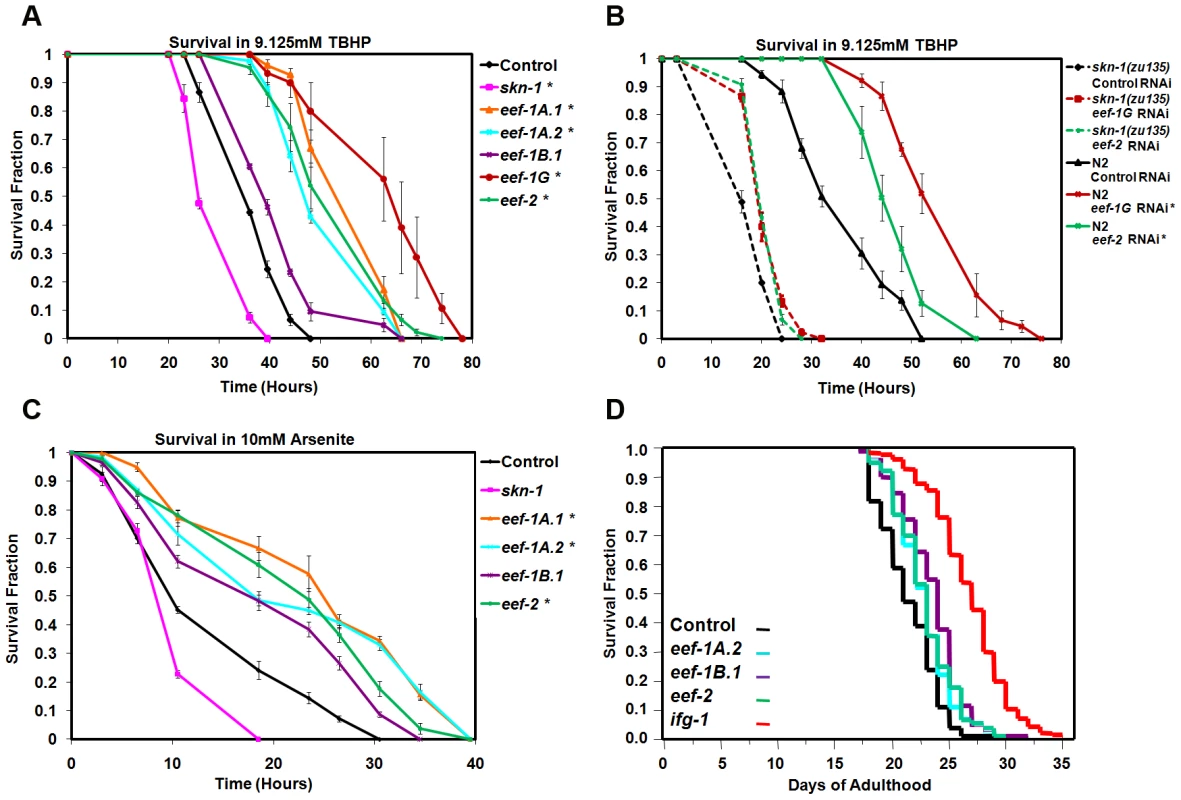
(A) Increased TBHP resistance after TEF RNAi. In (A–C), data were analyzed by JMP and plotted with EXCEL. Representative experiments are shown, with replicates, statistics, and percent changes in survival time provided in Table S2 (A), Table S3 (B), and Table S4 (C). Error bars represent the SEM, and P values were calculated by log-rank. *P<0.0001. (B) skn-1 contributes to TBHP resistance deriving from TEF RNAi. (C) Resistance to Arsenite resulting from TEF RNAi. (D) Lifespan analysis of worms fed TEFs or TIF (ifg-1) RNAi bacteria. A composite of three replicates (Table S5A) is shown. C. elegans lifespan is increased by mutation or adulthood knockdown of several TIFs, ribosomal proteins, or other translation regulators [23], [34], [35]. TIF and TEF mRNAs are expressed at lower levels in the long-lived IIS mutant daf-2, also consistent with an opposing correlation between protein synthesis and longevity [39]. However, when we performed TEF RNAi by feeding during adulthood, lifespan was increased slightly by knockdown of eef-1A.2, eef-1B.1 and eef-2, but not by eef-1A.1 or eef-1G (Figure 3D; Table S5A; Figure S3F; Table S5B). This failure of TEF RNAi to increase lifespan robustly could arise from TEF RNAi having more pleiotropic effects on the animal than TIF knockdown, or could be related to the differences in gene expression responses that result from interference with translation elongation and initiation.
SKN-1 mediates a proteasome bounce-back response and maintains UPS activity tissue-specifically in vivo
For multiple reasons, we examined the involvement of SKN-1 in proteasome gene regulation and activity. Firstly, our microarray-based expression profiling suggested that SKN-1 contributes to transcription of 14 proteasome subunit genes (44% of the apparent total), under both normal and oxidative stress conditions [20]. Secondly, a transgenic SKN-1::GFP fusion protein was detected with high confidence at the promoter regions of 25 proteasome genes (78% of the apparent total) during the L1 larval stage [22]. These included all of the proteasome genes that expression profiling suggested are regulated by SKN-1, with only a single exception (rpt-5). In addition, some SKN-1 target genes are induced by RNAi knockdown of proteasome genes [23], [26], [27]. Finally, as suppression of translation elongation might increase the fraction of incompletely translated proteins, it seemed possible that SKN-1 might increase proteasome gene expression and activity in response to interference with translation elongation.
We first investigated the extent to which skn-1 is required for proteasome gene expression under normal conditions. The 26S proteasome consists of at least 32 subunits in C. elegans, including 19S ATPases involved in substrate unfolding (rpt-1∼6), other 19S subunits (rpn-1∼12), 20S α-rings (pas-1∼7) and 20S β-rings (pbs-1∼7) [11]. We examined how skn-1 RNAi affected the expression of the endogenous proteasome subunit genes rpt-3, rpn-12, pas-4 and pbs-6, which represent the four subunit classes above. Each of these genes is a predicted SKN-1 target at which at least four canonical SKN-1 binding sites lie within 1 kb upstream of the translation initiation codon, and SKN-1::GFP was detected by ChIP [20], [22] (data not shown). In whole animals skn-1 RNAi slightly decreased the expression of each gene, except for rpt-3 (Figure 4A). We also examined expression of transcriptional reporters in which proteasome promoters are fused to GFP. RNAi against skn-1 slightly decreased expression of reporters for rpt-5, rpn-11 and pas-5, particularly in the intestine, but did not detectably affect rpn-2 or pbs-4 (Figure 4B and 4C, Figure S4A and S4B, Table S6). The data suggest that under normal conditions SKN-1 contributes to but is apparently not essential for the expression of many proteasome subunit genes.
Fig. 4. Importance of SKN-1 for the proteasome bounce-back response. 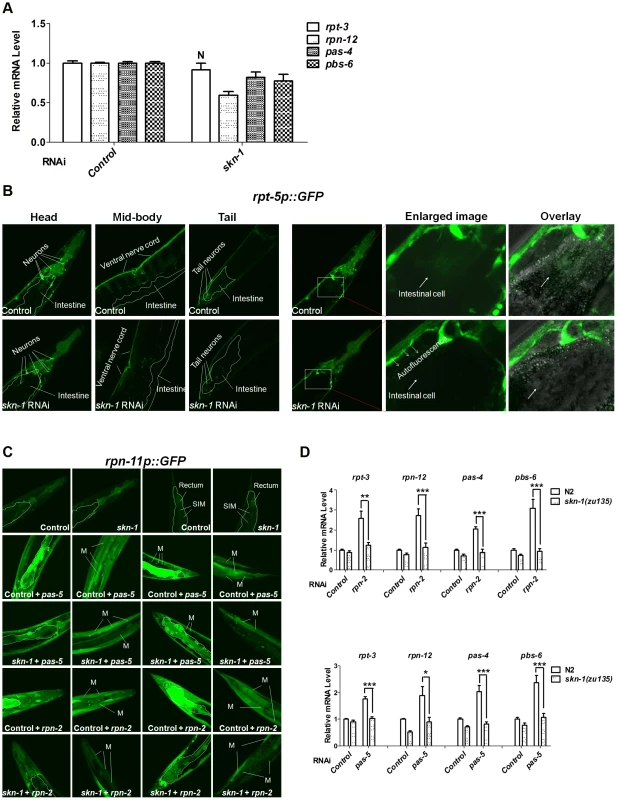
(A) Relative mRNA levels of endogenous proteasome subunit mRNAs in N2 animals fed with control (L4440) or skn-1 RNAi bacteria. All P<0.005 compared to control, except where N = not significant. Error bars indicate SEM. (B) skn-1 RNAi slightly decreases rpt-5p::GFP expression, particularly in the intestine, under normal conditions. Representative projection images show all z-stacks in the left panels, and z-stacks through the intestine on the right. Here and in (C), 2-day-old adults were used for imaging, boundaries of the intestine are indicated by dashed lines, and quantification and statistics are listed in Table S6. (C) skn-1 RNAi blocks the bounce-back response to proteasomal subunit knockdown. Representative confocal projection images of pas-5 or rpn-2 RNAi effects on the rpn-11p::GFP transcriptional reporter are shown, with z-stacks through the intestine or muscles displayed for double RNAi experiments. Abbreviations: M, body-wall muscle; SIM, stomatointestinal muscle. (D) Impaired bounce-back response in skn-1 mutants. Endogenous proteasome subunit mRNAs were detected by qRT-PCR in wild-type (N2) or skn-1(zu135) animals that had been fed with proteasome subunit RNAi bacteria. A paired t test (two-tailed) was employed to compare wild-type (N2) and skn-1(zu135) animals. An unpaired t test (two-tailed) was used to compare proteasome subunit vs control RNAi in N2 animals. Compared to N2 control, all P<0.05. mRNA levels were normalized to tba-1 (α-tubulin). In mammalian cells, Nrf1 and Nrf2 have been implicated in the “bounce-back” response whereby inhibition of the proteasome results in a compensatory upregulation of proteasome subunit gene expression [17]–[19]. To test this model in C. elegans tissues in vivo, we blocked proteasome activity by performing RNAi against an essential proteasome subunit gene, then examined expression of other proteasome genes. Knockdown of pas-5 or rpn-2 resulted in dramatic upregulation of the pbs-4, rpt-5 and rpn-11 transcriptional reporters, as well as an RPN-11::GFP translational fusion protein (Figure 4C, Figure S4A and S4B, Table S6). These increases in proteasome gene expression were largely dependent upon skn-1 in the intestine, where SKN-1 is prominently expressed [24], as well as in some muscles. Additionally, pas-5 or rpn-2 knockdown increased endogenous proteasome subunit mRNA levels in a skn-1-dependent manner (Figure 4D). In certain tissues, therefore, SKN-1 is required in vivo for the compensatory induction of proteasome gene upregulation that occurs in response to proteasome inhibition.
As our data suggested that skn-1 contributes to proteasome gene expression, particularly when proteasome activity is impaired, we used a novel in vivo assay to investigate whether SKN-1 is important for UPS activity under normal conditions [40]. We generated a strain (Pvha-6::UbG76V-Dendra2) in which the intestine-specific promoter vha-6 drives expression of a photoswitchable green-to-red fluorescent protein (Dendra2) that is fused to a non-hydrolyzable ubiquitin moiety (UbG76V) [40], [41]. By monitoring this fusion protein after photoconversion, we could assess ubiquitin-dependent protein degradation activity in living animals [40]. In control RNAi animals, at 9 hours after photoconversion the levels of red-fluorescing intestinal UbG76V-Dendra2 had been reduced to 40% of that present just after photoconversion, but a control Dendra2 that lacked UbG76V was still stable (Figure 5A, upper left panels). UbG76V-Dendra2 degradation was dramatically inhibited by RNAi against the proteasome genes pbs-5, rpn-2, or rpt-4, indicating that this degradation required the proteasome (Figure 5B and S4C). Together, the data show that this intestinal UbG76V-Dendra2 protein is degraded by the UPS.
Fig. 5. Knockdown of SKN-1 or TEFs, but not TIFs, reduces UPS activity in the intestine. 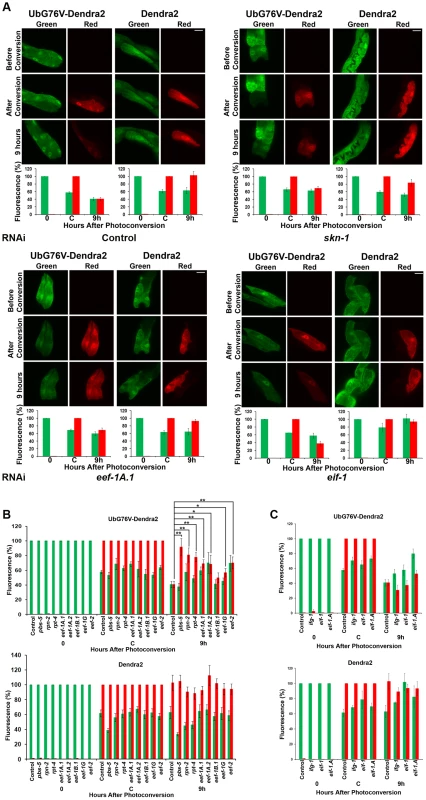
(A) Representative images of Pvha-6::UbG76V-Dendra2 and Pvha-6::Dendra2 reporters in animals exposed to control, skn-1, eef-1A.1 (TEF) or eif-1(TIF) RNAi. Red fluorescence derives from substrate that was present at the time of photoconversion. Bar: 20 µm. In (A–C), bar graphs depict the percentages of green and red fluorescence compared to either the initial value (t = 0 for green) or point of photoconversion (t = C for red) for UbG76V-Dendra2 and Dendra2, respectively (± SEM). (B,C) Summary of UbG76V-Dendra2 and Dendra2 imaging in intestinal cells after 9 hours in response to TEF and TIF RNAi, respectively. Significant differences relative to control RNAi (red fluorescence after 9 hours) are indicated with **P<0.01, *P<0.05. Degradation of intestinally-expressed UbG76V-Dendra2 was also markedly reduced by skn-1 RNAi, indicating that UPS activity in the intestine depends upon SKN-1 (Figure 5A, upper panels). In contrast, skn-1 RNAi did not impair degradation of UbG76V-Dendra2 that was expressed specifically in the body-wall muscle, and slightly increased its degradation in dopaminergic neurons (Figure S4D and S4E, respectively). skn-1 RNAi also decreased the total proteasome activity in the animal under normal conditions, as detected in vitro by a proteasome in-gel activity assay (Figure S5A). Treatment with the proteasome inhibitor MG132 was more toxic for animals in which skn-1 had been knocked by RNAi than for control animals (Figure S5B), further supporting the idea that SKN-1 is important for proteasome gene expression and activity. We conclude that SKN-1 functions tissue-specifically to maintain UPS activity in the intestine under normal conditions, and that a significant proportion of total C. elegans UPS activity is skn-1-dependent.
Dependence of the UPS on translation elongation factors
Having determined that SKN-1 is important for proteasome gene expression and UPS activity in the intestine, and that RNAi against TEFs induces SKN-1 to upregulate particular target genes, we wanted to investigate whether interference with translation elongation might direct SKN-1 to increase proteasome expression and degradation activity. This seemed like a plausible model, because it might be advantageous for proteasome activity to be increased upon interference with elongation, in order to ensure that any incompletely translated proteins are degraded. Surprisingly, however, the levels of endogenous mRNAs encoding four proteasome subunits decreased slightly in response to RNAi against each TEF that we examined, with the exception of eef-1B.1 (Figure S6A). Intestinal fluorescence from the pas-5p::GFP reporter was not increased by either eef-1A.1 or eef-2 RNAi, but was slightly decreased by eef-2 knockdown (Figure S6B; Table S6). Furthermore, in whole animals the levels of proteasome 20S α subunits were decreased slightly in response to RNAi against each TEF (Figure S6C). Expression of proteasome subunits was similarly reduced slightly by RNAi against TIFs (Figure S6A and S6D). We conclude that whereas SKN-1 directly upregulates proteasome gene transcription under normal conditions, and particularly after depletion of individual proteasome subunits, it does not do so after inhibition of translation elongation or initiation.
In yeast, eEF1A interacts with proteasome subunits and may escort incompletely translated proteins to the proteasome, thereby facilitating their degradation [7], [9], [10]. This raised an alternative possibility, that the translation elongation apparatus might be important for proteasome activity. Consistent with this notion, in C. elegans EEF-1A.1 interacts with proteasome subunits RPN-2 and RPT-4 [11] and inhibition of three TEFs resulted in the premature aggregation of transgenic proteins, suggesting a possible downregulation of proteasome activity [42]. When we monitored UbG76V-Dendra2 degradation in the intestine, we observed that its degradation was significantly impaired by knockdown of multiple different TEFs, but not TIFs (Figure 5; Table S7). Having observed that proteasome gene expression is affected similarly by TEF and TIF RNAi (Figure S6A–S6D), this suggests that UPS activity may be mechanistically dependent upon the translation elongation machinery.
These findings raised an unexpected model for why SKN-1 target genes are induced by RNAi against TEFs: that the resulting reduction in proteasome activity might stimulate a SKN-1-dependent stress response. However, several observations argue against this interpretation. In contrast to the effects of TEF RNAi, knockdown of proteasome subunit genes induced skn-1-dependent expression of other proteasome genes, and did not increase p38 MAPK signaling or SKN-1 nuclear occupancy (Figure 4C, 4D and S6E–S6G; Table S6). Also different from TEF RNAi effects, proteasome gene RNAi activated the skn-1-regulated gst-4p::GFP reporter [26], [27], and knockdown of pas-5, rpn-2 or rpt-4 induced skn-1-dependent endogenous gst-4 and gst-10 expression (Figure S6G and S6H). Finally, proteasome subunit but not TEF RNAi activated heat shock promoters hsp-70 and hsp-16.2 (Figure S3A and S3B). Induction of a SKN-1-mediated stress response by TEF RNAi therefore does not derive from an indirect effect on the proteasome, and may result directly from signals associated with slowed translation elongation.
Discussion
We have determined that interference with either mRNA translation or proteasome integrity results in induction of SKN-1-mediated stress responses. These responses are remarkably specific, in that SKN-1 upregulates distinct suites of target genes in response to impairment of translation initiation, translation elongation, or proteasome activity (Figure 6). When protein synthesis is inhibited, SKN-1 increases oxidative stress resistance. If proteasome subunit expression is blocked, SKN-1 attempts to compensate by upregulating proteasome genes in multiple tissues. In contrast, proteasome gene expression is not increased when translation is impaired, and proteasome activity is actually decreased in response to reduced translation elongation, suggesting that the specificity of these SKN-1-mediated functions may be important for maintaining protein homeostasis.
Fig. 6. Specific SKN-1–mediated responses to impaired protein synthesis and degradation. 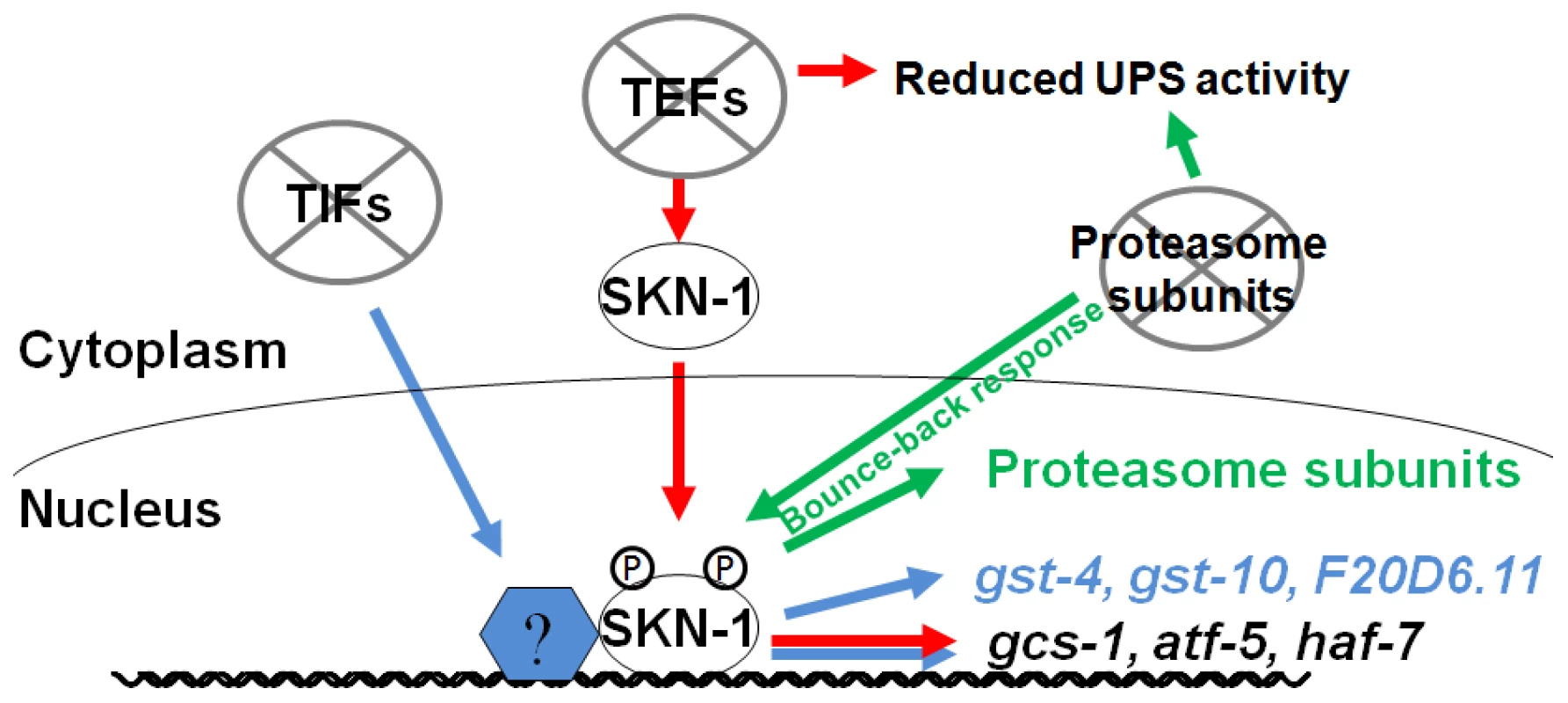
SKN-1 target genes are activated through distinct mechanisms by interference with translation initiation or elongation, so that TEF RNAi (red) induces a more restricted set of SKN-1 target genes than TIF RNAi (blue). TEF RNAi also impairs UPS activity, supporting the idea that proteasome activity and translation elongation are mechanistically coupled processes. In the intestine, SKN-1 is also important for the “bounce-back” response to proteasome inhibition, and for UPS activity under normal conditions (green). It is intriguing that different mechanisms seem be involved when SKN-1 is directed to activate target genes in response to inhibition of translation initiation or elongation. It is unlikely that the differences between these SKN-1-dependent responses derive simply from different degrees of translation activity or stress, because these responses are qualitatively distinct. Whereas TIF but not TEF RNAi upregulates transcription of the SKN-1 target genes gst-4 and gst-10 (Figure 1B, 1D and 1E), TEF but not TIF RNAi leads to accumulation of SKN-1 in intestinal nuclei (Figure 1F) [23]. In addition, TEF RNAi increases p38 pathway signaling more robustly (Figure 1G and Figure S2G). One possible model is that interference with translation initiation might upregulate SKN-1-dependent gene expression by acting on transcription factors that cooperate with SKN-1. Consistent with this idea, several mRNAs are translated preferentially when translation initiation is inhibited, including some that encode stress response factors [43]. It may be important to increase oxidative and xenobiotic stress resistance when either translation initiation or elongation is impaired, because broad reductions in protein synthesis could disrupt cellular metabolism or redox buffering, particularly in a key metabolic and synthetic tissue like the C. elegans intestine [23]. In addition, oxidizing conditions facilitate IIS, suggesting that under conditions of growth and high translation rates it could be advantageous to suppress SKN-1-regulated oxidative stress defenses [44].
In addition to its well-documented role in small molecule detoxification, we have found that SKN-1 is also important for regulating proteasome gene expression and sustaining UPS activity, particularly in the intestine. The SKN-1 orthologs Nrf1 or Nrf2 have been reported to induce compensatory proteasome gene expression when proteasome activity is impaired in cultured mammalian cells [17]–[19]. We have shown that this SKN-1/Nrf function is both important in vivo and evolutionarily conserved, and involves each class of proteasome genes. We also obtained the novel finding that SKN-1 orchestrates this response in multiple post-mitotic tissues, including the intestine. It will be important to investigate the extent to which the proteasome bounce-back response might rely on different Nrf1/2/3 isoforms or other mechanisms in various mammalian cell types, particularly the gut, liver, and adipose tissues, which are counterparts to the C. elegans intestine.
Under non-stressed conditions, lack of SKN-1 or Nrf1 decreased proteasome gene expression only modestly in C. elegans and mammalian cells, respectively (Figure 4A and 4B) [18], [20]. However, this seemingly small effect of SKN-1 evidently has substantial consequences, because we determined that under normal conditions SKN-1 has a major effect on UPS activity in vivo in the intestine (Figure 5A, upper panels), and contributes to the total proteasome activity in the animal (Figure S5A). Taken together with recent ChIP data indicating that SKN-1 occupies the promoters of most proteasome genes under non-stressed conditions [22], our findings suggest that SKN-1/Nrf proteins are critical regulators of proteasome genes even under normal circumstances. Perhaps the “bounce-back” function of SKN-1 is needed for fine-tuning the levels of proteasomal subunits in the intestine, so that proteasome assembly can proceed efficiently.
Our observation that animals fed skn-1 RNAi bacteria are sensitized to treatment with a proteasome inhibitor (Figure S5B) suggests that SKN-1 is critical for sustaining proteasomal defenses against proteotoxicity in vivo. The involvement of SKN-1/Nrf proteins in regulating proteasome gene expression might be important not only under acute stress conditions, but also in situations of chronic proteotoxic stress such as alcoholic liver and neurodegenerative diseases. Interestingly, in mice liver-specific inactivation of Nrf1 results in non-alcoholic steatohepatitis and hepatic cancer [45]. This syndrome is associated with oxidative damage in hepatocytes, but our results suggest that impaired proteasome activity might also be involved. SKN-1/Nrf proteins have been shown to increase longevity in both C. elegans and Drosophila [15], [25]. Our new results predict that this effect may derive not only from their function in protecting against reactive small molecules, but also may involve their role in sustaining proteasome expression and activity, and thereby helping to maintain protein homeostasis. Consistent with this idea, a recent study showed that skn-1 is required for C. elegans lifespan to be extended by an amyloid-binding compound that suppresses toxicity deriving from misfolded proteins [46].
We also observed that UPS activity is dependent upon the translation elongation machinery. A conclusive assessment of how TEF RNAi affected total proteasome activity in the animal, as measured in vitro, was problematic because translation inhibition reduced the total amount of protein present (data not shown). However, our in vivo assay [40] demonstrated clearly that intestinal UPS activity was reduced by TEF but not TIF RNAi (Figure 5). Previous work in yeast had noted that TEFs interact with the proteasome, and that eEF1A may facilitate degradation of defective newly synthesized proteins by escorting them to the proteasome (see Introduction). Working in a metazoan, we have now obtained support for the idea that UPS-mediated protein degradation and translation elongation are mechanistically coupled processes. We determined that inhibition of translation elongation but not initiation reduced UPS activity in the intestine in vivo, an effect that seems unlikely to be mediated by the modest decline in proteasome gene expression seen after RNAi of either TEFs or TIFs (Figure 5 and Figure S6A–S6D). It also seems unlikely that this effect derived simply from the UPS being swamped by incompletely translated proteins arising from inhibition of elongation, because we did not see simultaneous upregulation of proteasome genes. Interestingly, our assay measured degradation of fluorescent and presumably folded UbG76V-Dendra2, suggesting that the translation elongation apparatus may promote UPS-mediated degradation of complete polypeptides that are no longer associated with the translation apparatus. The physical interactions that have been described between the proteasome and elongation factors [6], [14] therefore may be generally important for UPS activity.
It is an intriguing question why SKN-1 does not increase proteasome gene expression and activity when translation elongation is inhibited, particularly when it appears to be present at most proteasome gene promoters constitutively [22]. Perhaps it would be deleterious for SKN-1 to do so, because if translation elongation were to slow in response to limited nutrients or other conditions, an inappropriate increase in proteasome activity might prematurely degrade nascent polypeptides, and thereby could globally impair protein synthesis. This could provide a rationale not only for the failure of TEF RNAi to induce proteasome gene upregulation, but also for the apparent dependence of UPS activity on translation elongation but not initiation factors. Taken together, our findings indicate that SKN-1 plays an important role in sensing and maintaining protein homeostasis, by mobilizing distinct responses to perturbations in polypeptide chain elongation and proteasomal degradation (Figure 6). They also indicate that the stress defenses that are regulated by SKN-1/Nrf proteins are not controlled in unison through a simple on/off switch, but are remarkably customized for specific conditions. This raises important questions concerning how these stresses are sensed at the molecular level, and how different stress signals are integrated by SKN-1/Nrf proteins to achieve specificity in their responses.
Materials and Methods
RNAi
RNAi was performed by feeding essentially as described [23], except that L3 and (or) early L4-stage worms were fed RNAi bacteria for 3 days at 20°C unless otherwise indicated. Bacteria carrying the vector plasmid L4440 were used as the control. RNAi constructs were taken from the Vidal ORFeome-Based RNAi library [47] and confirmed by sequencing. In all double RNAi experiments, RNAi and/or control bacteria were mixed at a 1∶1 volume ratio. The wild-type strain is N2.
RNA isolation and quantitative RT-PCR
Animals subjected to RNAi were collected and washed 3 times in M9, then total RNA was extracted from approximately 60 animals for each treatment. RNA was extracted using the TRI Reagent (Sigma) and cDNA synthesized using the SuperScript First-Strand Synthesis Kit (Invitrogen). SYBR Green Real Time Quantitative PCR was carried out using the ABI 7900 and analyzed using the Standard Curve method [48]. For all RNAi experiments, qRT-PCR data were derived from 3–4 independent biological replicates. In CHX experiments, the values presented were derived from 2–3 independent PCR analyses of one biological experiment. Results were graphed so that the level of each mRNA that was seen in N2 animals fed with control (L4440) RNAi bacteria was set as 1. Unless otherwise indicated, act-1 (β-actin) was used for normalization and P values were derived from an unpaired t test (two-tailed). Primer sequences are listed in Table S8.
Western blotting
L2/L3 stage larvae were fed RNAi bacteria for two days, then collected and washed in M9 buffer, and snap frozen in liquid nitrogen. Worms were lysed in RIPA buffer (50 mM Tris [pH = 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with 0.2 mM sodium vanadate, 50 mM sodium fluoride, 0.1 mM PMSF and protease inhibitor cocktail (Roche). Supernatant was quantitated by the BCA protein assay kit (Pierce). Western blots were performed with antibodies specific for phospho-p38 (Cell signaling #9211), proteasome 20S α subunits (BIOMOL #8195), and α-tubulin (Sigma-Aldrich #9026).
Oxidative stress resistance assays
Analyses of oxidative stress resistance were performed essentially as described [23]. To assay TBHP resistance, L3/L4 stage animals were fed with RNAi bacteria for three days, then transferred to plates that contained 9.125 mM TBHP (Sigma-Aldrich) in the agar and E. coli OP50 food. Fresh TBHP plates were prepared fresh two hours before transfer. Animals that bagged, crawled off the plates and exploded were censored. As resistance assays were performed by essentially the same method, using freshly prepared plates that contained 10 mM NaAsO2 (Sigma-Aldrich) in the agar and E. coli OP50 food.
Lifespan analysis
Lifespan analyses were conducted at 20°C, with RNAi treatments performed only during adulthood. N2 animals were synchronized by timed egglay for 2–4 hours on plates seeded with control RNAi bacteria. Synchronized one-day-old adults were transferred to lifespan plates seeded with gene-specific or control RNAi bacteria. 2′ fluoro-5′ deoxyuridine (FUDR) was present (0.1 mg/ml) to prevent progeny development. The first day of adulthood was used as t = 0, with animals scored each day after the sixteenth day of adulthood. Those that crawled off the plate, exploded, or bagged were censored. JMP version 7, was used for statistical analyses, and P values were calculated using the log-rank method.
In vivo proteasome activity assay
L2/L3 larvae were placed on RNAi feeding plates, then exposed to photoconversion after 72 h (muscle cell imaging) or 48 h (all others). Photoconversion and image analysis were performed as described [40]. Only gravid adults were imaged, and worms were maintained on RNAi plates between time points. P values were determined by Student's t-test (homoscedastic).
Quantification and imaging of GFP reporters
For proteasome reporters, L2/L3 larvae were fed RNAi bacteria for 3 days, then normal-appearing worms that developed into gravid adults were analyzed. Animals were mounted on 5% agarose pads, immobilized in 1 mM levamisole and imaged with a Zeiss Axioplan 2 microscope. Confocal microscopy was used to generate z-stack projections for a representative subset of animals (LSM 510 Meta, 40× plan-neofluar objective, Zeiss, Germany; z-stacks with 0.5 µm interval). At least two stable transgenic lines for each proteasome reporter strain were examined. Fluorescent images were analyzed by the MCID system (Imaging Research) to measure the average fluorescence level of the entire worm, or particular regions. The average value of controls for each experiment was set as 100%, with values obtained in parallel from RNAi-treated worms converted to the relative fluorescence level. Data obtained from several experiments were pooled for statistical analysis.
For other reporters, an AxioVision (Zeiss) microscope was used to acquire imaging and fluorescence was scored by eye as Low, Medium, or High as described for each experiment.
Additional Materials and Methods are provided in Text S1.
Supporting Information
Zdroje
1. GoldbergAL 2003 Protein degradation and protection against misfolded or damaged proteins. Nature 426 895 899
2. MorimotoRICuervoAM 2009 Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci 64 167 170
3. BalchWEMorimotoRIDillinAKellyJW 2008 Adapting proteostasis for disease intervention. Science 319 916 919
4. Bousquet-DubouchMPNguenSBouyssieDBurlet-SchiltzOFrenchSW 2009 Chronic ethanol feeding affects proteasome-interacting proteins. Proteomics 9 3609 3622
5. CiechanoverA 2005 Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol 6 79 87
6. GuerreroCMilenkovicTPrzuljNKaiserPHuangL 2008 Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A 105 13333 13338
7. VermaRChenSFeldmanRSchieltzDYatesJ 2000 Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11 3425 3439
8. BrowneGJProudCG 2002 Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem 269 5360 5368
9. ChuangSMChenLLambertsonDAnandMKinzyTG 2005 Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol 25 403 413
10. GonenHSmithCESiegelNRKahanaCMerrickWC 1994 Protein synthesis elongation factor EF-1 alpha is essential for ubiquitin-dependent degradation of certain N alpha-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci U S A 91 7648 7652
11. DavyABelloPThierry-MiegNVaglioPHittiJ 2001 A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep 2 821 828
12. SchubertUAntonLCGibbsJNorburyCCYewdellJW 2000 Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404 770 774
13. TurnerGCVarshavskyA 2000 Detecting and measuring cotranslational protein degradation in vivo. Science 289 2117 2120
14. ShaZBrillLMCabreraRKleifeldOScheligaJS 2009 The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol Cell 36 141 152
15. SykiotisGPBohmannD 2010 Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal 3 re3
16. ArltABauerISchafmayerCTepelJMuerkosterSS 2009 Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2). Oncogene 28 3983 3996
17. KraftDCDeocarisCCWadhwaRRattanSI 2006 Preincubation with the proteasome inhibitor MG-132 enhances proteasome activity via the Nrf2 transcription factor in aging human skin fibroblasts. Ann N Y Acad Sci 1067 420 424
18. RadhakrishnanSKLeeCSYoungPBeskowAChanJY 2010 Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38 17 28
19. SteffenJSeegerMKochAKrugerE 2010 Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell 40 147 158
20. OliveiraRPPorter AbateJDilksKLandisJAshrafJ 2009 Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8 524 541
21. ParkSKTedescoPMJohnsonTE 2009 Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell 8 258 269
22. NiuWLuZJZhongMSarovMMurrayJI 2011 Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21 245 254
23. WangJRobida-StubbsSTulletJMRualJFVidalM 2010 RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet 6 e1001048 doi:10.1371/journal.pgen.1001048
24. AnJHBlackwellTK 2003 SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17 1882 1893
25. TulletJMHertweckMAnJHBakerJHwangJY 2008 Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 1025 1038
26. KahnNWReaSLMoyleSKellAJohnsonTE 2008 Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem J 409 205 213
27. ChoeKPPrzybyszAJStrangeK 2009 The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol 29 2704 2715
28. Schneider-PoetschTJuJEylerDEDangYBhatS 2010 Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6 209 217
29. InoueHHisamotoNAnJHOliveiraRPNishidaE 2005 The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19 2278 2283
30. KimDHFeinbaumRAlloingGEmersonFEGarsinDA 2002 A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297 623 626
31. CalfonMZengHUranoFTillJHHubbardSR 2002 IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415 92 96
32. HaynesCMYangYBlaisSPNeubertTARonD 2010 The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 37 529 540
33. MorleyJFMorimotoRI 2004 Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15 657 664
34. PanKZPalterJERogersANOlsenAChenD 2007 Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6 111 119
35. HansenMTaubertSCrawfordDLibinaNLeeSJ 2007 Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6 95 110
36. TohyamaDYamaguchiAYamashitaT 2008 Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans. FASEB J 22 4327 4337
37. LinKHsinHLibinaNKenyonC 2001 Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28 139 145
38. YamawakiTMArantes-OliveiraNBermanJRZhangPKenyonC 2008 Distinct activities of the germline and somatic reproductive tissues in the regulation of Caenorhabditis elegans' longevity. Genetics 178 513 526
39. Halaschek-WienerJKhattraJSMcKaySPouzyrevAStottJM 2005 Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15 603 615
40. HamerGMatilainenOHolmbergCI 2010 A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat Methods 7 473 478
41. DantumaNPLindstenKGlasRJellneMMasucciMG 2000 Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18 538 543
42. NollenEAGarciaSMvan HaaftenGKimSChavezA 2004 Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A 101 6403 6408
43. SonenbergNHinnebuschAG 2009 Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136 731 745
44. LohKDengHFukushimaACaiXBoivinB 2009 Reactive oxygen species enhance insulin sensitivity. Cell Metab 10 260 272
45. XuZChenLLeungLYenTSLeeC 2005 Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci U S A 102 4120 4125
46. AlavezSVantipalliMCZuckerDJSKlangIMLithgowGJ 2011 Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature
47. RualJFCeronJKorethJHaoTNicotAS 2004 Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14 2162 2168
48. Glover-CutterKKimSEspinosaJBentleyDL 2008 RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15 71 78
49. ProudCG 1994 Peptide-chain elongation in eukaryotes. Mol Biol Rep 19 161 170
50. PrahladVCorneliusTMorimotoRI 2008 Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320 811 814
51. LinkCDCypserJRJohnsonCJJohnsonTE 1999 Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones 4 235 242
52. PispaJPalmenSHolmbergCIJanttiJ 2008 C. elegans dss-1 is functionally conserved and required for oogenesis and larval growth. BMC Dev Biol 8 51
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



