-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Revisiting Heterochromatin in Embryonic Stem Cells
article has not abstract
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002093
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002093Summary
article has not abstract
It is widely believed that chromatin in embryonic stem (ES) cells exists in a unique “open” conformation, characterized by sparse, disorganized heterochromatin and prevalent global transcription. Upon differentiation, this “blueprint” of pluripotent state is thought to undergo dramatic remodelling. In this issue of PLoS Genetics, Lienert and colleagues [1] revisit heterochromatin and transcription in pluripotent and terminally differentiated cells to demonstrate that neither the abundance of repressive histone H3 lysine 9 dimethylation (H3K9me2) nor the net transcriptional output of the genome discriminate these two very different cell states.
Pluripotent ES cells, derived from the inner cell mass of developing mammalian blastocyst, have the distinctive ability to self-renew in culture and differentiate into multiple lineages when exposed to appropriate signals. The self-organizing regulatory network of transcription factors and the epigenetic mechanisms that are involved in maintenance of pluripotent state and self-renewal are actively debated and intensively studied by many laboratories [2], [3]. When induced to differentiate, ES cells respond by changes in gene expression, cell morphology, and chromatin structure, which may collectively contribute to a reduction in developmental plasticity [4], [5].
Several lines of evidence have suggested that DNA in stem cells is packaged into an unusually dynamic form of chromatin that carries ES cell–specific patterns of histone modifications. Thus, in ES cells, histone H3 and H4 tend to be hyperacetylated; constitutive heterochromatin foci, marked by histone H3 lysine 9 trimethylation (H3K9me3), are fewer and less well organized; and histone and non-histone chromatin-bound proteins, such as heterochromatin protein 1 (HP1), are more mobile [4], [6], [7]. In addition, a substantial number of gene promoters in ES cells is marked by closely juxtaposed active (H3K4me3) and repressive (H3K27me3) chromatin modifications [8], [9]. This so-called bivalent or poised chromatin is resolved into a monovalent state at most, but not all, loci upon differentiation [9], [10]. However, repressive chromatin marks come in several “flavours”. Of those, H3K9me2 is a relatively abundant modification associated with facultative heterochromatin that covers large, gene-poor regions of the genome [11]. It has been reported that these H9K9me2 domains are “minimally present” in ES cells, but undergo substantial expansion and stabilization in differentiated tissues, such as liver and brain, resulting in transcriptional silencing of genes residing in these domains [11], [12]. Further studies have found that chromatin regions marked by other repressive modifications, such as H3K9me3 and H3K27me3, are also larger in lineage-restricted human lung fibroblasts IMR90 when compared to human ES cells. These regions undergo remodelling and reduction in size upon reprogramming of IMR90 cells into induced pluripotent stem cells (iPSCs) [10]. Collectively, these observations suggest that lineage commitment and differentiation are accompanied by expansion and stabilization of repressive chromatin.
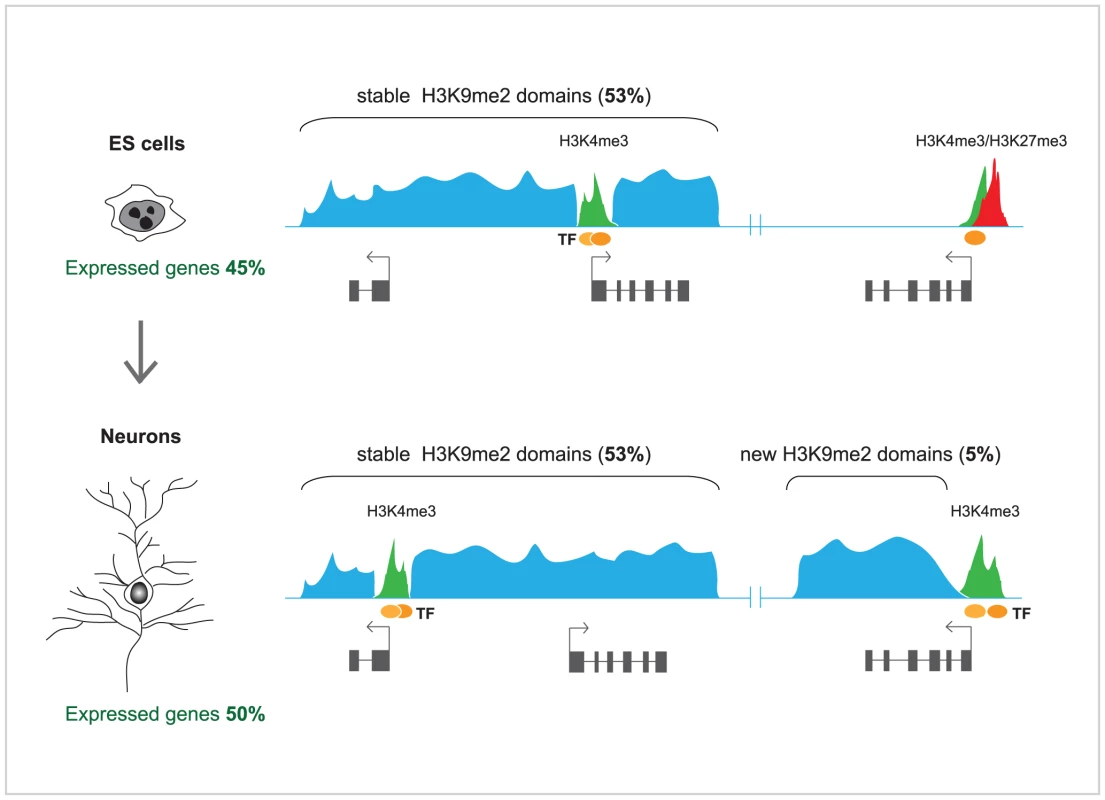
In order to investigate in detail the changes in H3K9me2-marked heterochromatin domains during terminal differentiation, Lienert et al. [1] used a robust in vitro neurogenesis system to differentiate ES cells into postmitotic pyramidal neurons [13]. Profiles of H3K9me2, representing ∼10% of the genome, including the entire chromosome 19, were generated for both cell types and compared to each other. Surprisingly, it was found that these profiles showed high degree of correlation between ES cells and neurons. In both cell types, H3K9me2 covered ∼50% of chromosome 19, and a very modest increase in H3K9me2 (5%) was observed in terminally differentiated neurons. In agreement with an earlier study [11], H3K9me2 was enriched at large chromosomal domains, but those were generally invariable in median size and distribution between ES cells and neurons, and mutually exclusive with active (H3K4me2) and other repressive chromatin marks (H3K27me3). Some discrete differences were observed; those included gain of H3K9me2 over new large domains in neurons, mostly over the bodies of transcribed genes, as well as loss of H3K9me2 from much smaller regions (Figure 1). Furthermore, high throughput sequencing of RNA (RNA-seq) from ES cells, neurons, and, additionally, mouse embryonic fibroblasts, showed well defined cell type–specific expression, but no significant overall difference in the transcribed portion of the genome, including most repetitive sequences. Although the findings of Lienert et al. [1] seem to disagree with previous studies [4], [11], these discrepancies could be largely explained by methodological differences in the analyses of H3K9me2 genomic microarray data [12] and the accuracy in discriminating between low and absent transcription by microarrays, which may suffer from crosshybridization, versus unambiguous direct counting of RNA sequence reads [1]. As both Effroni et al. [4] and Lienert et al. [1] have measured the abundance of polyadenylated RNAs, reflecting mostly the productive transcription, it might be interesting to employ global nuclear run-on coupled with high throughput sequencing (NRO-seq) [14] in order to explore whether the extent of non-productive transcription differs significantly between ES cells and terminally differentiated neurons.
In summary, the observations of Lienert et al. [1] highlight the remarkable conservation of the facultative heterochromatin domains and the global transcriptional output of the genome between ES cells and terminally differentiated neurons. They also suggest that genome reprogramming during lineage commitment and differentiation is largely achieved by developmental cues and strong transcription factors, which induce localized and highly specific changes in heterochromatin rather than promote genome-wide build up of H3K9me2 and suppression of global low-level transcription. Such a model is further supported by findings that differentiation of ES cells into neuronal progenitors and then into astrocytes is accompanied by focal, localized rearrangements in chromatin-nuclear lamina interactions, while the overall architecture of lamina-associated chromosomal domains remains largely preserved [15].
It cannot be completely ruled out that, although quantitatively similar, heterochromatin is qualitatively different, more fluid and, perhaps, less essential in ES cells than in terminally differentiated cells and tissues. Such plasticity could be mediated by chromatin remodelling ATPases, histone acetyltransferases, and histone demethylases, some of which are highly expressed in stem cells and essential for pluripotency [4], [16]–[18]. Is heterochromatin then functional in ES cells?
The vast majority of H3K9me2 in the genome is established by the euchromatic histone methylases EHMT2 and EHMT1, also known as G9a and GLP, respectively. Similar to the knockouts of DNA methyltransferases [19], ES cells lacking either G9a or GLP are viable and morphologically normal, but G9a−/− and Glp−/− embryos die in midgestation (E9–9.5) [20], [21]. This suggests that, although DNA methylation and G9a/GLP-dependent H3K9me2 are dispensable for self-renewal in ES cells, they become vital during differentiation and embryonic development. Unfortunately, the differentiation potential of G9a−/− and Glp−/− ES cells has never been investigated in detail. Nevertheless, these cells form embryonic bodies upon induction with retinoic acid, but fail to terminally silence OCT3/4 [22], indicating that G9a/GLP-dependent heterochromatin formation may safeguard rather than actively channel differentiation.
Despite the overwhelming evidence that heterochromatin is present, but somewhat “wimpy” in stem cells, it was reported that H3K9me2 - and H3K9me3-specific histone demethylases JMJD1A and JMJD2C, respectively, are directly regulated by OCT3/4 transcription factor and are essential for maintenance of pluripotency [18]. Depletion of these enzymes by small interfering RNAs (siRNAs) leads to accumulation of H3K9me and unscheduled differentiation. However, it was also clearly shown that JMJD1A and JMJD2C action is restricted to specific loci and does not lead to ubiquitous removal of H3K9me from the genome. Taken together with the studies of Lienert et al. [1], these findings firmly indicate that heterochromatin is functional in ES cells and has to be actively remodelled in order to allow the self-organizing network of transcription factors to prevent differentiation and promote self-renewal. The same general principle of local heterochromatin removal by lineage-specific transcriptional regulators may operate during differentiation.
Zdroje
1. LienertFMohnFTiwariVKBaubecTRoloffTC
2011
Genomic prevalence of heterochromatic H3K9me2 and transcription
do not discriminate pluripotent from terminally differentiated
cells.
PLoS Genet
7
e100290
doi:10.1371/journal.pgen.1002090
2. HannaJHSahaKJaenischR
2010
Pluripotency and cellular reprogramming: facts, hypotheses,
unresolved issues.
Cell
143
508
525
3. YoungRA
2011
Control of the embryonic stem cell state.
Cell
144
940
954
4. EfroniSDuttaguptaRChengJDehghaniHHoeppnerDJ
2008
Global transcription in pluripotent embryonic stem
cells.
Cell Stem Cell
2
437
447
5. MeshorerEMisteliT
2006
Chromatin in pluripotent embryonic stem cells and
differentiation.
Nat Rev Mol Cell Biol
7
540
546
6. MeshorerEYellajoshulaDGeorgeEScamblerPJBrownDT
2006
Hyperdynamic plasticity of chromatin proteins in pluripotent
embryonic stem cells.
Dev Cell
10
105
116
7. KeohaneAMO'NeillLPBelyaevNDLavenderJSTurnerBM
1996
X-Inactivation and histone H4 acetylation in embryonic stem
cells.
Dev Biol
180
618
630
8. BernsteinBEMikkelsenTSXieXKamalMHuebertDJ
2006
A bivalent chromatin structure marks key developmental genes in
embryonic stem cells.
Cell
125
315
326
9. MeissnerAMikkelsenTSGuHWernigMHannaJ
2008
Genome-scale DNA methylation maps of pluripotent and
differentiated cells.
Nature
454
766
770
10. HawkinsRDHonGCLeeLKNgoQListerR
2010
Distinct epigenomic landscapes of pluripotent and
lineage-committed human cells.
Cell Stem Cell
6
479
491
11. WenBWuHShinkaiYIrizarryRAFeinbergAP
2009
Large histone H3 lysine 9 dimethylated chromatin blocks
distinguish differentiated from embryonic stem cells.
Nat Genet
41
246
250
12. FilionGJvan SteenselB
2010
Reassessing the abundance of H3K9me2 chromatin domains in
embryonic stem cells.
Nat Genet
42
4; author reply 5–6
13. BibelMRichterJSchrenkKTuckerKLStaigerV
2004
Differentiation of mouse embryonic stem cells into a defined
neuronal lineage.
Nat Neurosci
7
1003
1009
14. CoreLJWaterfallJJLisJT
2008
Nascent RNA sequencing reveals widespread pausing and divergent
initiation at human promoters.
Science
322
1845
1848
15. Peric-HupkesDMeulemanWPagieLBruggemanSWSoloveiI
2010
Molecular maps of the reorganization of genome-nuclear lamina
interactions during differentiation.
Mol Cell
38
603
613
16. NiwaH
2007
Open conformation chromatin and pluripotency.
Genes Dev
21
2671
2676
17. SchnetzMPHandokoLAkhtar-ZaidiBBartelsCFPereiraCF
2010
CHD7 targets active gene enhancer elements to modulate ES
cell-specific gene expression.
PLoS Genet
6
e1001023
doi:10.1371/journal.pgen.1001023
18. LohYHZhangWChenXGeorgeJNgHH
2007
Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate
self-renewal in embryonic stem cells.
Genes Dev
21
2545
2557
19. GollMGBestorTH
2005
Eukaryotic cytosine methyltransferases.
Annu Rev Biochem
74
481
514
20. TachibanaMSugimotoKNozakiMUedaJOhtaT
2002
G9a histone methyltransferase plays a dominant role in
euchromatic histone H3 lysine 9 methylation and is essential for early
embryogenesis.
Genes Dev
16
1779
1791
21. TachibanaMUedaJFukudaMTakedaNOhtaT
2005
Histone methyltransferases G9a and GLP form heteromeric complexes
and are both crucial for methylation of euchromatin at
H3-K9.
Genes Dev
19
815
826
22. FeldmanNGersonAFangJLiEZhangY
2006
G9a-mediated irreversible epigenetic inactivation of Oct-3/4
during early embryogenesis.
Nat Cell Biol
8
188
194
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání