-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
The mammalian ortholog of yeast Slx4, BTBD12, is an ATM substrate that functions as a scaffold for various DNA repair activities. Mutations of human BTBD12 have been reported in a new sub-type of Fanconi anemia patients. Recent studies have implicated the fly and worm orthologs, MUS312 and HIM-18, in the regulation of meiotic crossovers arising from double-strand break (DSB) initiating events and also in genome stability prior to meiosis. Using a Btbd12 mutant mouse, we analyzed the role of BTBD12 in mammalian gametogenesis. BTBD12 localizes to pre-meiotic spermatogonia and to meiotic spermatocytes in wildtype males. Btbd12 mutant mice have less than 15% normal spermatozoa and are subfertile. Loss of BTBD12 during embryogenesis results in impaired primordial germ cell proliferation and increased apoptosis, which reduces the spermatogonial pool in the early postnatal testis. During prophase I, DSBs initiate normally in Btbd12 mutant animals. However, DSB repair is delayed or impeded, resulting in persistent γH2AX and RAD51, and the choice of repair pathway may be altered, resulting in elevated MLH1/MLH3 focus numbers at pachynema. The result is an increase in apoptosis through prophase I and beyond. Unlike yeast Slx4, therefore, BTBD12 appears to function in meiotic prophase I, possibly during the recombination events that lead to the production of crossovers. In line with its expected regulation by ATM kinase, BTBD12 protein is reduced in the testis of Atm−/− males, and Btbd12 mutant mice exhibit increased genomic instability in the form of elevated blood cell micronucleus formation similar to that seen in Atm−/− males. Taken together, these data indicate that BTBD12 functions throughout gametogenesis to maintain genome stability, possibly by co-ordinating repair processes and/or by linking DNA repair events to the cell cycle via ATM.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002094
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002094Summary
The mammalian ortholog of yeast Slx4, BTBD12, is an ATM substrate that functions as a scaffold for various DNA repair activities. Mutations of human BTBD12 have been reported in a new sub-type of Fanconi anemia patients. Recent studies have implicated the fly and worm orthologs, MUS312 and HIM-18, in the regulation of meiotic crossovers arising from double-strand break (DSB) initiating events and also in genome stability prior to meiosis. Using a Btbd12 mutant mouse, we analyzed the role of BTBD12 in mammalian gametogenesis. BTBD12 localizes to pre-meiotic spermatogonia and to meiotic spermatocytes in wildtype males. Btbd12 mutant mice have less than 15% normal spermatozoa and are subfertile. Loss of BTBD12 during embryogenesis results in impaired primordial germ cell proliferation and increased apoptosis, which reduces the spermatogonial pool in the early postnatal testis. During prophase I, DSBs initiate normally in Btbd12 mutant animals. However, DSB repair is delayed or impeded, resulting in persistent γH2AX and RAD51, and the choice of repair pathway may be altered, resulting in elevated MLH1/MLH3 focus numbers at pachynema. The result is an increase in apoptosis through prophase I and beyond. Unlike yeast Slx4, therefore, BTBD12 appears to function in meiotic prophase I, possibly during the recombination events that lead to the production of crossovers. In line with its expected regulation by ATM kinase, BTBD12 protein is reduced in the testis of Atm−/− males, and Btbd12 mutant mice exhibit increased genomic instability in the form of elevated blood cell micronucleus formation similar to that seen in Atm−/− males. Taken together, these data indicate that BTBD12 functions throughout gametogenesis to maintain genome stability, possibly by co-ordinating repair processes and/or by linking DNA repair events to the cell cycle via ATM.
Introduction
SLX1 and SLX4 were identified, together with MUS81 and MMS4 (Eme1 in mammals), in a S. cerevisiae screen for genes required for the viability of sgs1-deficient cells [1]. Slx1 is the founding member of a family of proteins with a predicted URI nuclease domain whose activity is enhanced 500-fold by its interaction with Slx4 [2]. Slx4 can also form complexes with Rad1-Rad10 [3], [4] to effect DSB repair during single-strand annealing in yeast [3], [5]. However, Slx4 can also act independently of both Slx1 and Rad1-Rad10 [2], [6], and is phosphorylated by Mec1 and Tel1, the yeast orthologs of ATR and ATM, respectively, in response to DNA damage [4].
Human, C. elegans and D. melanogaster orthologs of SLX4 were described recently [7]–[10] and named BTBD12 (for BTB domain-containing protein-12), Him-18, and Mus312, respectively. These proteins are considerably diverged from their yeast counterpart; BTBD12 encodes a 1834 amino acid protein, approximately 2.5-times larger than the yeast protein, and resembles its lower eukaryotic orthologs mostly in its C-terminal SAP and CCD domains [7]. Like the yeast ortholog, the human protein is a substrate of the ATM/ATR kinases [11] and its depletion also results in DNA damage sensitivity [8]. Recently, a subset of Fanconi anemia (FA) patients were found to have biallelic mutations in BTBD12, making this gene a novel complementation group for this disorder [12]. A complex of BTBD12 and SLX1 displays robust Holliday Junction (HJ) resolvase and 5′ flap endonuclease activity in vitro, and mammalian BTBD12 also binds to, and enhances the activity of, several DNA repair proteins including MUS81 [7], [10] and the MSH2-MSH3 heterodimer of the DNA mismatch repair (MMR) family [8], suggesting a role for this protein as a docking platform for structure-specific endonucleases.
Recent reports in D. melanogaster and C. elegans indicate that the Btbd12 orthologs, Mus312 and Him-18, respectively are essential for normal meiotic progression [13], [14]. In addition, Him-18 appears to function pre-meiotically in the germ line, being required for repair at stalled replication forks [13], suggesting that Him-18 functions throughout germ cell development to maintain genomic integrity. Given these data, the primary goal of the current studies was to understand the function of BTBD12 in the germ line of mice, with the hypothesis that BTBD12 may be critical for the processing of homologous recombination intermediates, whether as the result of replication errors during pre-meiotic proliferation, or during the repair of double strand breaks (DSBs) that underlie meiotic recombination. For the latter, our studies were aimed at investigating the role of BTBD12 in the regulation of meiotic recombination events during prophase I in mammals, particularly those that ensure accurate segregation of maternal and paternal chromosomes at the first meiotic division. Principal amongst these is the formation of DNA crossovers between the homologs, as initiated by DSB induction through the activity of the SPO11 endonuclease [15], [16]. The resolution of DSBs can be achieved through the recruitment of various repair pathway complexes, to produce crossovers (CO) or noncrossovers (NCO). The fact that CO formation occurs with tight precision, coupled with the observation that only a small subset of total DSBs will become COs, suggests that orchestration of DSB repair events, and the various repair pathways that give rise to either CO or NCO events, is highly regulated at the molecular level (reviewed by [17]).
Two pathways have been defined for CO formation, the first involving the so-called Class I or “ZMM” pathway (for ZIP3, MSH4/5 and MER3), and the second, Class II pathway, involving the Mus81 endonuclease, which functions as a heterodimer with EME1 (Mms4 in yeast) [18]–[21]. In S. cerevisiae, and possibly in the mouse, this Class II pathway appears to be restricted to a subset of DSBs that may be aberrant in structure and/or that may be processed initially by the RecQ helicase, Sgs1/BLM (yeast/mouse ortholog; [20]–[23]). These aberrant DSB repair intermediates (or joint molecules, JMs) include a variety of structures that result from secondary strand invasion events, usually involving independent activities of each end of the DSB, and/or from closely spaced DSBs. These aberrant JMs have been demonstrated biochemically in S. cerevisiae [20]–[23], but not yet in mammals. Under wildtype conditions in budding yeast, Sgs1 can disassemble and/or process many of these aberrant DSB repair intermediates towards NCO or Class I CO fates [20], [21]. However, a small proportion of them cannot be processed in this manner and thus become the target of MUS81-driven crossing over.
In the mouse, the class I pathway accounts for some 90–95% of COs, and the major intermediate marker for these events is the accumulation, in pachynema, of the MutL homolog heterodimer, MLH1 and MLH3 [24]–[27]. The remaining events are processed via the MUS81-dependent Class II pathway [27], [28]. Interestingly, however, our studies in mouse have demonstrated that the loss of Mus81 results in the recruitment of 5–10% additional MLH1/MLH3 foci during pachynema, and that these additional foci may act to maintain CO rates at normal levels in the absence of MUS81 [28]. This suggests that a unique, possibly mammalian-specific, level of integration exists between the two crossover pathways. Importantly, BTBD12 interacts with many of the key players in both CO pathways, including BLM (reviewed by [29]), leading us to hypothesize that BTBD12 may functionally integrate the different CO pathways during mammalian meiosis.
We obtained a mouse line from the European Conditional Mouse Mutagenesis Program (EUCOMM), harboring a Frt-flanked βGeo cassette upstream of LoxP-flanked exon 3 of Btbd12 gene, and as initially described by Crossan et al [30]. We call this conditional genetrap allele Btbd12βGeoFlox. In wildtype mice, BTBD12 protein localizes to spermatogonial and spermatocyte populations of the adult testis and is dramatically down-regulated at these sites in the mutant animals. Weak staining against BTBD12 persists, however, in the Btbd12βGeoFlox/βGeoFlox testes, but the protein fails to be recruited to meiotic chromosome cores to detectable levels. Despite residual protein, the mice are sub-fertile as a result of a reduced spermatogonial population coupled with failure to progress normally through meiotic prophase I resulting in less than 15% normal numbers of spermatozoa. Importantly, BTBD12 localization to meiotic chromosomes appears to be dependent on ATM, since the presence of BTBD12 protein is lost in Atm−/− males, and Btbd12βGeoFlox/βGeoFlox mice show genomic instability similar to that seen in the absence of ATM. Taken together, our data suggest that BTBD12 is a key substrate of ATM in the mammalian germ line, playing a role in the spermatogonial stages of gametogenesis, as well as in the entry into, and progression through, prophase I.
Results
BTBD12 protein localizes to spermatogonia and spermatocytes of the mouse testis
Wildtype testis sections were stained with a commercial anti-BTBD12 antibody from Novus Biologicals that was raised against amino acids 1650–1700 of human BTBD12 at the c-terminus of the protein (Figure 1A, 1B). BTBD12 is expressed ubiquitously in the mouse, and is found in both adult testis and fetal ovary [8]. In adult testis of wildtype mice, the protein was found to localize strongly to spermatogonia and spermatocytes (Figure 1A, black arrowheads and arrows, respectively). At earlier stages of spermatogenesis, prior to entry into meiosis, the signal for BTBD12 protein appears to localize through the nucleus, with increased intensity of signal around the nuclear periphery. Upon entry into prophase I, the BTBD12 signal becomes more punctate in nature, associating with the increasingly condensed chromatin, but continuing to occupy the majority of the nuclear space. Additionally, BTBD12 protein localized to bivalent chromosomes on the meiotic spindle during metaphase of meiosis I (Figure 1A, white arrowheads and inset panel).
Fig. 1. BTBD12 localization in wild type and Btbd12βGeoFlox/βGeoFlox spermatogenesis. 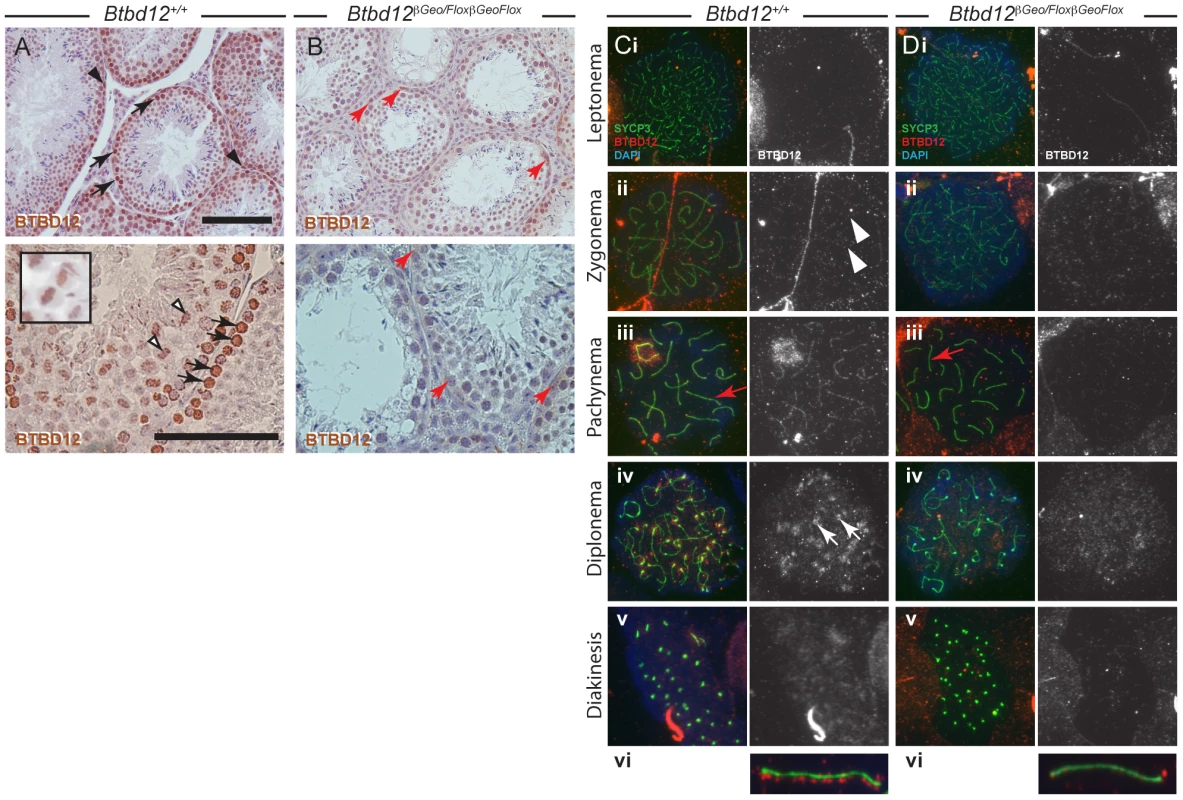
(A, B) Testis sections from wild type (A) or Btbd12βGeoFlox/βGeoFlox mice (B) were stained with an antibody against BTBD12 (brown). BTBD12 is localized to spermatogonia (black arrowheads), prophase I spermatocytes (black arrows) and metaphase I spermatocytes (white arrowheads, and inset) in wild type sections, however only a small amount of residual staining is seen in the Btbd12βGeoFlox/βGeoFlox sections, in cells along the basal membrane of the tubule (red arrows). Scale bars are 100 µm. (C, D) BTBD12 localization in prophase I substages in wild type (Ci-vi) and Btbd12βGeoFlox/βGeoFlox (Di-vi) spermatocytes. Cells were stained with antibodies against SYCP3 (green) and BTBD12 (red), while DNA was stained with DAPI (blue). BTBD12 staining is shown alone in the grey panels. BTBD12 accumulates along chromosome cores during zygonema in wild type cells (Cii, white arrowheads), persists through pachynema (Ciii, vi) and remains only at the centromeres in diplonema (Civ, white arrows). Single pachytene chromosomes stained with SYCP3 (green) and BTBD12 (red) from wild type (Cvi) and Btbd12βGeoFlox/βGeoFlox (Dvi) mice are shown, with the red channel slightly offset from the green channel to better visualize the BTBD12 localization on the chromosome cores. The same chromosomes are shown on the original images (Ciii, Diii, red arrows). BTBD12 localization was evaluated on chromosome spread preparations of prophase I spermatocytes. In all chromosome “spread” preparations presented herein, we utilize the major protein component of the axial element of the synaptonemal complex (SC, the meiosis specific structure that assembles along and between each homologous chromosome pair), SYCP3, to visualize meiotic chromosome cores. We utilized an affinity-purified antibody that targets amino acids 1–350 of murine BTBD12 (antibody “NT”). The chromosome spreads from wildtype adult males (Figure 1 Ci-vi) showed accumulation of BTBD12 protein in a punctate staining pattern along the chromosome cores in zygonema (Figure 1 Cii), becoming more intense by pachynema (Figure 1Ciii). In some cases, increased staining was observed within the sex body (94% of wildtype cells having BTBD12 in this domain, compared to 0% in the Btbd12 mutants; n of 33 and 31, respectively), in line with the fact that BTBD12 is a target of ATM, which plays a significant role in the formation of this sub-nuclear domain during mid-prophase I [31], [32]. One autosomal chromosome from a pachytene cell of each genotype is shown, enlarged, with the BTBD12 staining offset from the SYCP3 staining to facilitate visualization of the punctate pattern of the Btbd12 signal (Figure 1Cvi). By diplonema, BTBD12 was no longer present on the cores, but remained associated with the centromeres (Figure 1Civ), and had disappeared from the nucleus by diakinesis (Figure 1Cv).
Btbd12βGeoFlox/βGeoFlox mutant testes show some residual BTBD12 protein and an absence of detectable BTBD12 on meiotic chromosome cores
The Btbd12βGeoFlox allele contains an Frt-flanked βGeo insertion into the Btbd12 locus upstream of LoxP-flanked exon 3. The expectation, therefore, is that this allele would act as a gene trap, reducing expression of wildtype Btbd12 mRNA. Consistent with this, in the Btbd12βGeoFlox/βGeoFlox mutant testis sections, we observe weak BTBD12 protein staining persisting in some cells, particularly those close to the basal membrane of the tubules, which are presumptive spermatogonia (Figure 1B, red arrows). This persistent staining, however, was much fainter than that seen in the wildtype testis sections, indicating a significantly lower abundance of BTBD12 protein in the mutant animals, and suggesting a splicing event around the βGeo cassette to produce BTBD12 protein. Importantly, there was no staining evident on metaphase chromosomes in the Btbd12βGeoFlox/βGeoFlox testes.
In spermatocytes from Btbd12βGeoFlox/βGeoFlox mutant littermates, BTBD12 did not appear to localize to chromosome cores with as intense a signal as in wildtype cells (Figure 1Di-vi), although faint staining was observed at pachynema in the Btbd12βGeoFlox/βGeoFlox mutants. This staining, however, was barely detectable above background, even at higher magnifications (Figure 1Diii and vi). Thus, the persistent BTBD12 signal observed in testis sections from Btbd12βGeoFlox/βGeoFlox males is not associated with prophase I chromosome cores, or is present at levels that are undetectable on chromosome spreads.
Altered BTBD12 protein expression/stability in Btbd12βGeoFlox/βGeoFlox testis
To confirm the presence of BTBD12 protein in wild type testes, and to explore the status of BTBD12 protein in the homozygous mutant animals, western blots were performed using whole testis protein extracts from adult and juvenile animals (Figure 2). Two antibodies were utilized, one C-terminal and one N-(not shown) terminal, as described above. Both antibodies produced a band of similar size in protein extracts from wild type adult testis (lane 1), with decreasing amounts observed in Btbd12βGeoFlox/+ heterozygotes (lane 2) and further reduced expression observed in Btbd12βGeoFlox/βGeoFlox mutant protein extracts (lane 3). Quantitation of BTBD12 protein levels in testis extracts from all three genotypes revealed a decrease from wildtype levels of 23.5% and 45.6% for heterozygotes and homozygous mutant samples, respectively (Figure 2 graph). The presence of residual 130 kDa protein in the homozygous mutant animals is in line with the persistent protein signal observed in immunohistochemical staining of testis sections from these animals (Figure 1B), and suggests that the mutant allele is transcribed at a reduced level compared to the wildtype allele. As expected, the lanes containing Btbd12βGeoFlox/+ and Btbd12βGeoFlox/βGeoFlox protein (lanes 2 and 3 in Figure 2) also show positive staining for β-galactosidase at approximately the same size. No β-galactosidase band is observed in the wildtype testis protein lane. Taken together, these results show that the BTBD12 protein present in testis extracts from Btbd12βGeoFlox/+ and Btbd12βGeoFlox/βGeoFlox males encompasses some portions of both the N-terminus and C-terminus of the protein, and most likely arises as a result of splicing around the FRT-flanked cassette, but that expression of this protein is dramatically reduced by the presence of the βGeo cassette. Moreover, βGeo cassette itself is also transcribed from the targeted allele, either as a distinct protein and/or as a fusion with BTBD12. We favor the former option given that the size of the BTBD12 and βGeo bands are similar.
Fig. 2. BTBD12 protein is down regulated in the Btbd12βGeoFlox/βGeoFlox mutant and the Atm null mice. 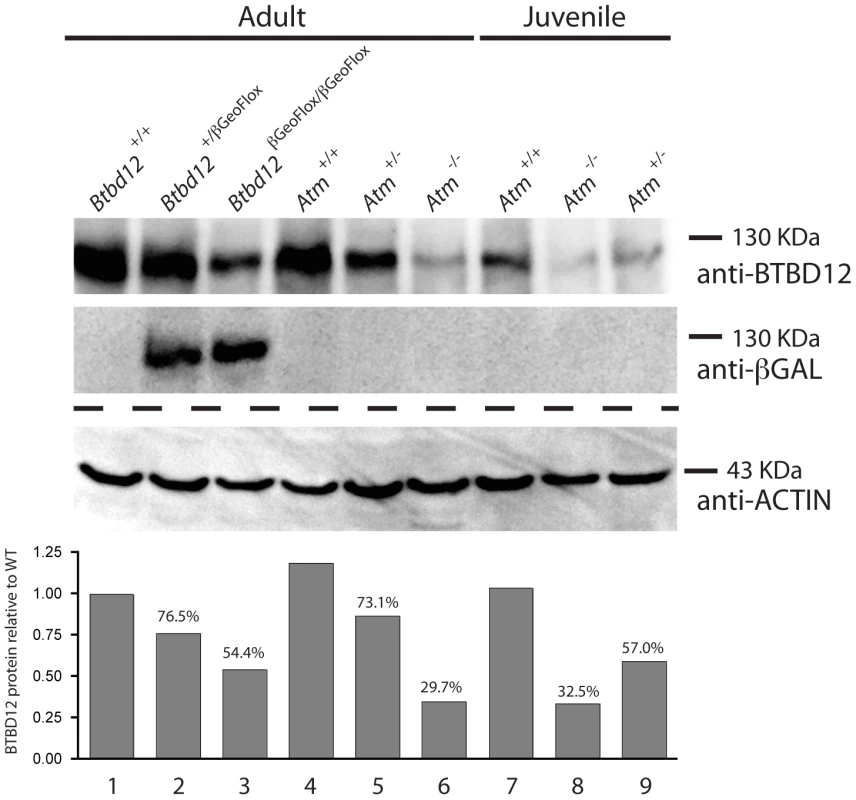
Western blot analysis of whole testis protein from adult Btbd12+/+, Btbd12+/βGeoFlox, Btbd12βGeoFlox/βGeoFlox mice (lanes 1–3), Atm+/+, Atm+/−, Atm−/− adult mice (lanes 4–6) and Atm+/+, Atm−/−, Atm+/− juvenile mice (lanes 7–9) using antibodies against BTBD12, β-Galactosidase and Actin (protein control). Protein levels for BTBD12 were normalized against Actin, and are shown as a percentage of the BTBD12 found in each wild type control. Btbd12βGeoFlox/βGeoFlox mice are viable, comparable in body weight to wildtype littermates, and are prone to anophthalmia and microphthalmia
Btbd12βGeoFlox/βGeoFlox mutant mice are viable and are born at slightly lower than expected Mendelian rates. Out of a total of 395 pups born in 65 litters from heterozygote breedings, 66 of them were mutants, compared with an expected frequency of 98.75 (Table S1). These data are significantly different, at the p<0.05 level, from the expected percentages mutant or wildtype numbers (P = 0.012, χ2 test). This rate is slightly higher than that seen in the previous report describing these mice [30]. Occasionally we observe that mutants exhibit lower birth weights than their wildtype litter mates (not shown) but, by adulthood, these animals have regained weights comparable to their siblings. Thus, Btbd12βGeoFlox/βGeoFlox adult mice were not significantly smaller than wildtype littermates (average weights 19.5 and 21.3 g, respectively P = 0.25, unpaired T-test) but exhibited varying degrees of anophthalmia and microphthalmia from birth. 33 out of 66 (50%) mutants showed defects of one or both eyes. No such deformities were apparent in wildtype litter mates indicating a possible role for BTBD12 in eye development and in line with previous reports describing this EUCOMM mouse line [30].
Btbd12βGeoFlox/βGeoFlox mutants have a higher level of genomic instability than their wildtype littermates
It is well known that mammalian species with mutations in Atm show an increase in genomic instability (GIN), including defects in cell cycle regulation, sensitivity to DNA damage-inducing agents, and chromosomal aberrations (reviewed by [33]). Since BTBD12 is believed to be a direct target for phosphorylation by ATM/ATR, we assessed both wild type and Btbd12βGeoFlox/βGeoFlox mutant mice for GIN. A micronucleus formation assay was performed on peripheral blood from both Btbd12βGeoFlox/βGeoFlox mutants and wild type littermate controls [34], [35]. The Btbd12βGeoFlox/βGeoFlox mutant animals showed over a two-fold increase in micronucleus formation compared with wild type mice, which was statistically significant (Figure S1; p<0.0001). This result indicates that these mutants have a high level of GIN in their somatic cells approaching, though not as great, as that seen in Atm null mice [36].
Male Btbd12βGeoFlox/βGeoFlox mice show reduced fertility associated with a paucity of spermatogenic cells in the testis
Btbd12βGeoFlox/βGeoFlox mutant mice showed significantly decreased testis size when compared to wildtype littermates (approximately 25%, Figure 3A, 3B, p<0.0001). Sperm counts performed on the two cohorts of mice revealed that Btbd12βGeoFlox/βGeoFlox mice have only about 10% of the amount of epididymal sperm found in wildtype littermates (Figure 3C), and exhibit dramatically reduced fertility, with only 3 litters born to two different Btbd12βGeoFlox/βGeoFlox males over the period of 9 months, with the youngest male to sire a litter being approximately 7 weeks old. Female mutants were sterile, with two mutant females bred to a fertile male, over a period of a year, yielding no pregnancies, while wildtype cage mates produced healthy offspring from the same male.
Fig. 3. Btbd12βGeoFlox/βGeoFlox mutants have reduced testicular germ cell proliferation. 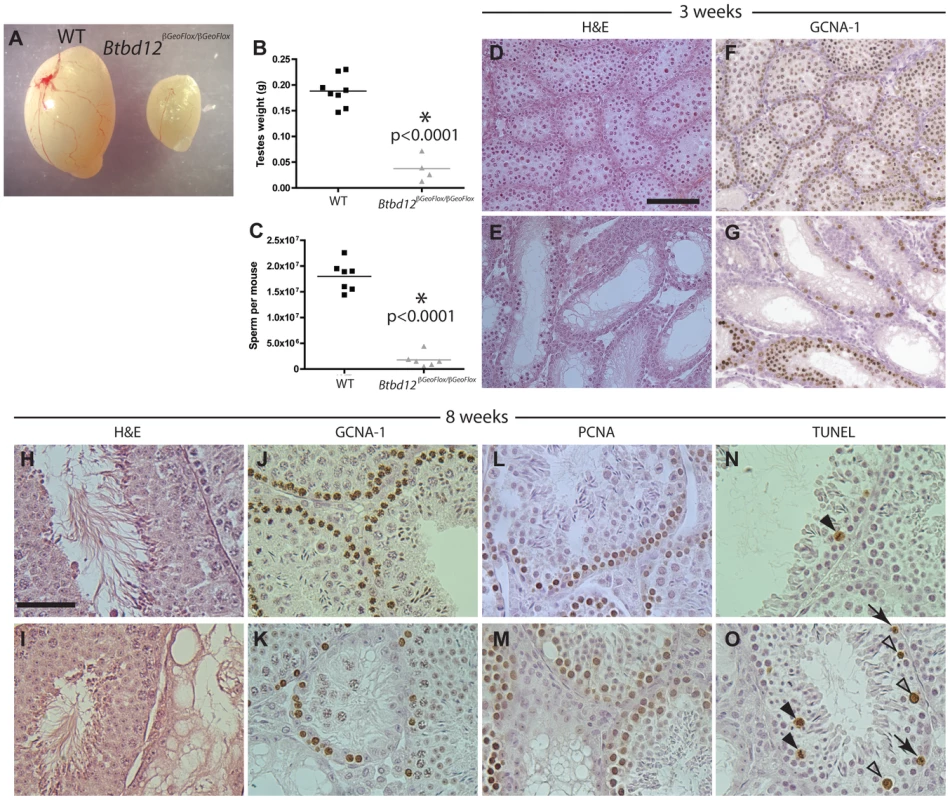
(A) Testes from Btbd12βGeoFlox/βGeoFlox mice are severely reduced in size compared with wild type littermates. (B) Testes weights of wild type (black) and Btbd12βGeoFlox/βGeoFlox mutant (grey) mice (P<0.0001). (C) Eipdidymal sperm numbers in wild type (black) and Btbd12βGeoFlox/βGeoFlox mutant (grey) mice (<0.0001). (D–G) Testis sections from wild type (D, F) or Btbd12βGeoFlox/βGeoFlox mutant (E, G) 3 week old mice, stained with either H&E (D–E) or an antibody against GCNA-1 (F–G, brown). (H–O) Testis sections from wild type (H, J, L, N) or Btbd12βGeoFlox/βGeoFlox mutant (I, K, M, O) 8 week old mice, stained with H&E (H–I), anti-GCNA-1 (J–K), anti-PCNA1 (L–M) or TUNEL (N–O). TUNEL stained apoptotic cells (N, O, arrowheads) were staged as either spermatocytes prior to metaphase I (white arrowheads) or at metaphase I (black arrowheads). D–G Scale bar is 50 µm. H–O scale bar is 100 µm. H&E staining of testis sections from both wildtype and Btbd12βGeoFlox/βGeoFlox mutant mice at both 3 and 8 weeks of age (Figure 3D, 3E, 3H, 3I) showed that the seminiferous tubules of Btbd12βGeoFlox/βGeoFlox males are extremely variable in their cell density and also in the progression through meiosis. For example, in 8-week old Btbd12βGeoFlox/βGeoFlox mutant mice, neighboring tubules showed almost normal tubule morphology, juxtaposed to almost empty, abnormal tubules (Figure 3I). Immunohistochemical staining with the spermatogonial and early spermatocyte cell marker, GCNA-1, showed a severe depletion of early germ cells within the tubules of the Btbd12βGeoFlox/βGeoFlox compared with those within the wildtype litter mate mice, at both 3 week and 8 weeks of age (Figure 3F, 3G, 3J, 3K). By contrast, the proliferating cell marker, PCNA, showed a similar staining pattern in the majority of tubules in both wildtype and Btbd12βGeoFlox/βGeoFlox mice (Figure 3L, 3M) suggestive of normal progression through spermatogonial divisions and in self-renewal capabilities. The maintainenance of PCNA signal in spite of reduced PGC pool is suggestive of prolonged S-phase. In line with this, we observed increased TUNEL labeling of apoptotic cells in testis sections of Btbd12βGeoFlox/βGeoFlox, mostly during meiosis I. The majority of these cells are undergoing apoptosis at around the time of exit from prophase I, but some also appear to be in mid-prophase (Figure 3N, 3O, arrowheads). In a few instances, some TUNEL-positive cells appear to be in pre-meiotic stages (Figure 3O, arrows), but these are clearly fewer in number than those apoptotic cells in prophase I. Collectively, these results demonstrate a loss of germ cells from the testis of Btbd12βGeoFlox/βGeoFlox males, starting as early as the first wave of meiotic entry within the first three weeks of postnatal life.
Male Btbd12βGeoFlox/βGeoFlox mice show declining primordial germ cell numbers within their testes from embryonic day 18 onwards
Given the apparently normal proliferative capacity of spermatogonia in testes of 8-week old Btbd12βGeoFlox/βGeoFlox mice, we questioned why the tubules of 3 week-old mice were so heterogeneous with respect to cellular density. If a failure of spermatogonial proliferation is not the cause of the lack of cellularity of certain tubules in the Btbd12βGeoFlox/βGeoFlox males, then a second possibility is that the testis of these mice fail to be populated with appropriate numbers of spermatogonial precursors, known as pro-spermatogonia or gonocytes, during development. To investigate this option, testes were obtained for both wildtype and Btbd12βGeoFlox/βGeoFlox males between embryonic (e) day 18 and day 3 post-partum (pp), and prospermatogonia were visualized with antiserum against GCNA-1. In wildtype males, the G0-arrested prospermatogonia population is established around embryonic day 12.5–16.5 following migration of the primordial germ cells (PGC) to the genital ridge [37], [38], the exact timing being somewhat controversial, and remain quiescent until just prior to birth [39]. During the period between e18 and day 3pp, a large number of prospermatogonia are lost by apoptosis. During that time, approximately 1 to 7 prospermatogonia may be observed within the seminiferous cords of the developing testis, and these cells then start to proliferate from day 4 pp onwards [39]. In Btbd12+/+ males, these large round cells appeared separated from the basement membrane by the Sertoli cells, which are more columnated in appearance, and they stained readily with numerous markers including GCNA-1 (Figure 4A–4C, brown cells) and mouse Vasa homolog (MVH; not shown) from e16 onwards. By day 3 pp, every testis cord section contains on average 2.08±0.19 prospermatogonia per cord, representing a range of 1 to 10 cells (Figure 4C, 4M; Table S2), having declined dramatically at around the time of birth. In Btbd12βGeoFlox/βGeoFlox males at e16, normal numbers of GCNA-1 positive prospermatogonia are observed (Figure 4D), but by e18, their numbers have declined significantly (Figure 4E, 4M; Table S2). By d3 pp, the majority of testis cords contain no prospermatogonia, with a mean prospermatogonia content of 0.92±0.28 (Figure 4F, 4M; Table S2). The Sertoli cell populations in both Btbd12+/+ and Btbd12βGeoFlox/βGeoFlox males appeared normal throughout (Figure 4B, 4D, 4F). In line with this reduced cellularity within the testicular cords, we observed a marked increase in apoptosis, as measured by TUNEL labeling of testis sections from wildtype (Figure 4G–4I, 4N) and mutant (Figure 4J–4L, 4N, arrows) animals, particularly at e16 and e18 (Figure 4J, 4K, 4N; Table S2). These data demonstrate that the population of spermatogonia within the testes of Btbd12βGeoFlox/βGeoFlox males is markedly lower than that seen in wildtype as a result of early loss of these cells after arriving at the genital ridge, and suggesting that the proliferation of PGCs in the developing testis is dependent on BTBD12.
Fig. 4. Spermatogonial germ cell proliferation in prepubescent males. 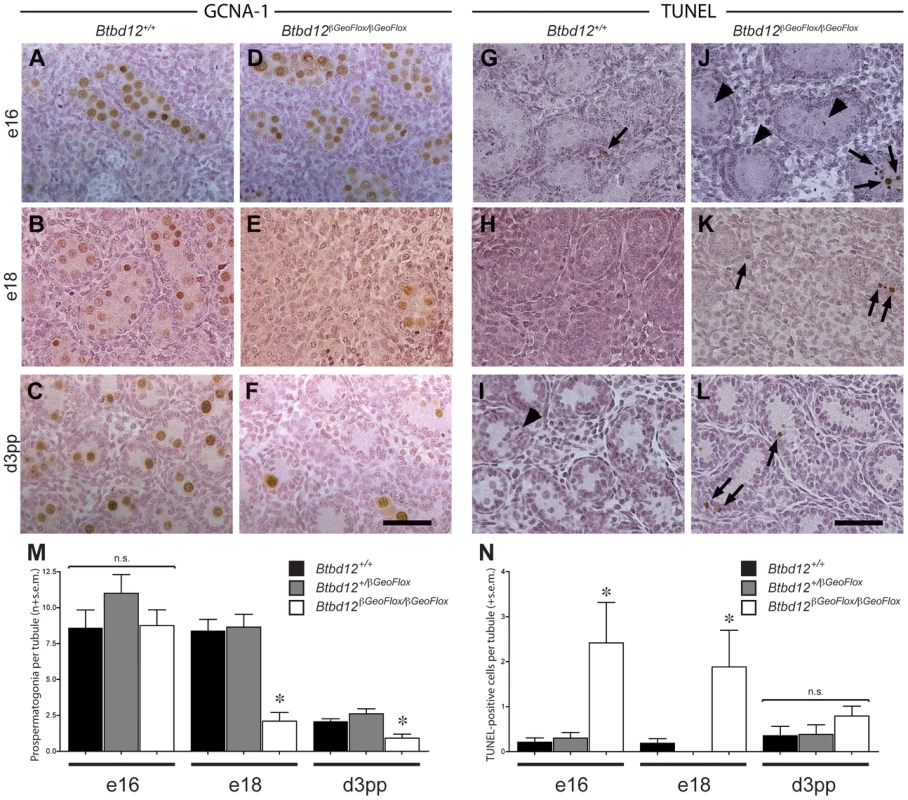
Testes sections from either wildtype (A–C, G–I) or Btbd12βGeoFlox/βGeoFlox (D–F, J–L) mice from three different ages (day e16, day e18, day 3 pp) were stained with either GCNA-1 (A–F) or TUNEL labeled (G–L). TUNEL labeled apoptotic spermatogonia are shown by the arrows. Arrowheads mark mitotic TUNEL-positive cells. Scale bar is 50 µm. Quantitation of the GCNA-1 labeled (M) and TUNEL-positive (N) cells reveals a statistically significant decrease in prospermatogonia, accompanied by an increase in apoptotic cells (asterisks). Male Btbd12βGeoFlox/βGeoFlox exhibit altered progression through prophase I
To examine meiotic prophase I progression, chromosome spreads from both Btbd12βGeoFlox/βGeoFlox and wildtype spermatocytes were stained with antibodies against SYCP3 and γH2AX (Figure 5A–5H), as a marker for DNA DSBs during prophase I. γH2AX accumulated on leptotene spermatocytes similarly in both wildtype and Btbd12βGeoFlox/βGeoFlox cells (Figure 5A, 5E), demonstrating the appearance and processing of DSB events. By zygonema, however, γH2AX localization began to diminish on the chromosome cores of wildtype spermatocytes (Figure 5B), coincident with the onset of DSB repair processes. By pachynema, and into diplonema, γH2AX localization was only restricted to a strongly-stained domain coincident with the sex body (Figure 5C, 5D and [40], [41]). By contrast, γH2AX staining is apparent at zygonema in the Btbd12βGeoFlox/βGeoFlox spermatocytes (Figure 5F), but persists along the autosomes well into pachynema, at which time this staining is limited to the sex body in wildtype cells (Figure 5C, 5G; ). γH2AX staining remains apparent well into diplonema in the Btbd12 mutants (Figure 5D, 5H; Figure S2).
Fig. 5. Btbd12βGeoFlox/βGeoFlox spermatocytes show altered meiotic progression. 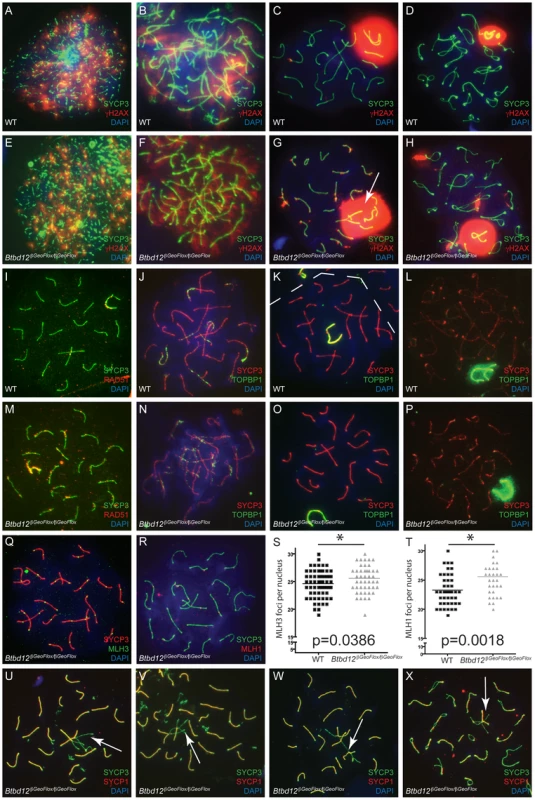
(A–H) Meiotic chromosome spreads from wild type (A–D) and Btbd12βGeoFlox/βGeoFlox mutants (E–H) were stained with antibodies against SYCP3 (green) and γH2AX (red), and DNA was stained with DAPI (blue). Abnormal sex body morphology in pachynema in the Btbd12βGeoFlox/βGeoFlox mutant is shown by the white arrow. (I, M) Meiotic chromosome spreads from wild type (I) and Btbd12βGeoFlox/βGeoFlox mutants (M) were stained with antibodies against SYCP3 (green) and RAD51 (red), and DNA was stained with DAPI (blue). Meiotic chromosome spreads from wild type (J–L) and Btbd12βGeoFlox/βGeoFlox mutants (N–P) were stained with antibodies against SYCP3 (red) and TOPBP1 (green), and DNA was stained with DAPI (blue). The dashed line represents the boundary between two cells (K). (Q) Meiotic chromosome spreads Btbd12βGeoFlox/βGeoFlox mutants were stained with antibodies against SYCP3 (red) and MLH3 (green) and DAPI (blue). (R) Meiotic chromosome spreads Btbd12βGeoFlox/βGeoFlox mutants were stained with antibodies against SYCP3 (green) and MLH1 (red) and DAPI (blue). (S) MLH3 focus counts per cell nucleus in wild type (black) and Btbd12βGeoFlox/βGeoFlox mutants (grey). (T) MLH1 focus counts per cell nucleus in wild type (black) and Btbd12βGeoFlox/βGeoFlox mutants (grey). (U–X) Meiotic chromosome spreads Btbd12βGeoFlox/βGeoFlox mutants were stained with antibodies against SYCP3 (green) and SYCP1 (red) and DAPI (blue). Regions of asynapsis are shown by the white arrows. To investigate the progression of DSB repair, we assessed RAD51 distribution along SCs during prophase I. Specifically, we were interested in observing persistence of RAD51 signal in Btbd12βGeoFlox/βGeoFlox spermatocytes as an indication of unrepaired, or delay in repair of, DSBs. As expected, we observed progressive loss of RAD51 from SCs in wildtype spermatocytes entering pachynema (Figure 5I; Table S3), whereas RAD51 focus numbers remained elevated through pachynema in Btbd12βGeoFlox/βGeoFlox spermatocytes (Figure 5M; Table S3). This difference between wildtype and mutant spermatocytes in terms of pachytene RAD51 foci (means of 26.2 and 45.1, respectively) was statistically significant.
The BRCT domain-containing protein, TOPBP1, functions in replication and DNA damage checkpoint processes, and in meiosis, it localizes to sites of DNA damage in response to DSBs [42], [43]. TOPBP1 is also known to be required for ATR binding/activation in a number of organisms [44]–[46] and, along with ATM/ATR kinases, may be a part of the machinery that monitors recombination during prophase I and activates the meiotic checkpoint. Indeed, TOPBP1 localizes exclusively to sites of SPO11-induced DSB, as demonstrated by co-localization with γH2AX [47].
Despite the massive increase in γH2AX staining, Btbd12βGeoFlox/βGeoFlox spermatocytes showed no difference in the TOPBP1 localization pattern compared to that seen in chromosome spreads from wild type spermatocytes (Figure 5J–5P). TOPBP1 accumulated on synapsed chromosomes during zygonema, and gradually decreased until it remained only at the sex chromosomes during pachynema, indicating that this signaling pathway is not affected by the loss of BTBD12 from the chromosome cores, in contrast to the persistent RAD51 observed on SCs from Btbd12βGeoFlox/βGeoFlox spermatocytes.
MLH1 and MLH3 localization was used to examine the progression of DSB repair events via the “ZMM”, Class I CO pathway (Figure 5Q–5T), which is overseen by key members of the DNA mismatch repair (MMR) family: MSH4, MSH5, MLH1 and MLH3. MLH1 and MLH3 form a heterodimer that binds to the MSH4/MSH5 heterodimer in pachynema [48]–[52]. MSH4/MSH5 assemble on DSB repair sites in zygonema in numbers that, in mice at least, far exceed the final tally of chiasmata [53], [54]. The number of these foci is then pared down through prophase I, but still maintains levels that are approximately two-fold higher than the final chiasmata count [53], [54]. Association of MLH1/MLH3 with a subset of these sites is thought to stabilize these events, resulting in the resolution of these structures via the class I CO pathway [24]–[26].
In spermatocyte spreads from both wildtype and Btbd12βGeoFlox/βGeoFlox males, MLH1 and MLH3 foci arise at pachynema, at frequencies of 1–2 foci per chromosome, which is comparable to that seen previously in wildtype [25], [26]. The temporal and spatial dynamics of MLH1 and MLH3 association with the SYCP3-positive chromosome core was similar for wild type and Btbd12βGeoFlox/βGeoFlox spermatocytes (Figure 5Q–5T), suggesting similar progression of class I CO events. When foci were counted and compared between wildtype and mutants, however, we observed a slight, yet statistically significant, increase in foci number for both MLH1 and MLH3 in Btbd12βGeoFlox/βGeoFlox spermatocytes, equating to approximately 1 additional focus per nucleus for each (Figure 5S, 5T, p = 0.0018 for MLH1 and p = 0.0389 for MLH3, unpaired T test). For MLH1, the mean number of foci was 23.28 and 25.56 for wildtype and Btbd12βGeoFlox/βGeoFlox males, respectively (Table S3). For MLH3, the mean number of foci was 24.62 and 25.57 for wildtype and Btbd12βGeoFlox/βGeoFlox males, respectively. MLH1 foci were also significantly elevated in meiotic spreads from female day e19 embryos, with average MLH1 focus numbers of 22.29 and 24.00 in wildtype and Btbd12βGeoFlox/βGeoFlox, respectively (p = 0.02, data not shown). The earlier recombination intermediate MSH4 remained the same in both wildtype and Btbd12βGeoFlox/βGeoFlox spermatocytes (data not shown).
To assess synapsis, chromosome spreads from Btbd12βGeoFlox/βGeoFlox spermatocytes were stained with an antibody against the central element component, SYCP1 (Figure 5U–5X). Interestingly, at pachynema, approximately 10% of cells harboring the mutant Btbd12 locus showed abnormal synapsis, compared to less than 1% for wildtype cells (not shown), characterized by frequent pairing between more than two chromosomes, incomplete synapsis at pachynema, and synapsis between chromosomes of differing lengths (Figure 5S–5U). In some cases, these synaptic errors appeared to persist into diplonema without first resulting in apoptosis (Figure 5X).
Btbd12βGeoFlox/βGeoFlox chromosomes form chiasmata at normal rates and show no defects in chromosome segregation at meiosis I in males and females
To assess the impact of loss of BTBD12 on the first meiotic division, we prepared air-dried diakinesis chromosome spreads and stained them with Giemsa. Chiasmata formation occurred in both wild type (not shown) and Btbd12βGeoFlox/βGeoFlox spermatocytes (Figure 6A). The number of diakinesis stage cells was severely depleted in Btbd12βGeoFlox/βGeoFlox mice, suggesting loss of these cells prior to completing prophase I and/or delayed progression through to diplonema. Of the diakinesis cells that we obtained from Btbd12βGeoFlox/βGeoFlox males, however, none showed any changes in chiasmata counts compared to that seen in wildtype litter mates (Figure 6B).
Fig. 6. Crossovers form in Btbd12βGeoFlox/βGeoFlox mutants and there is no evidence of spindle defects. 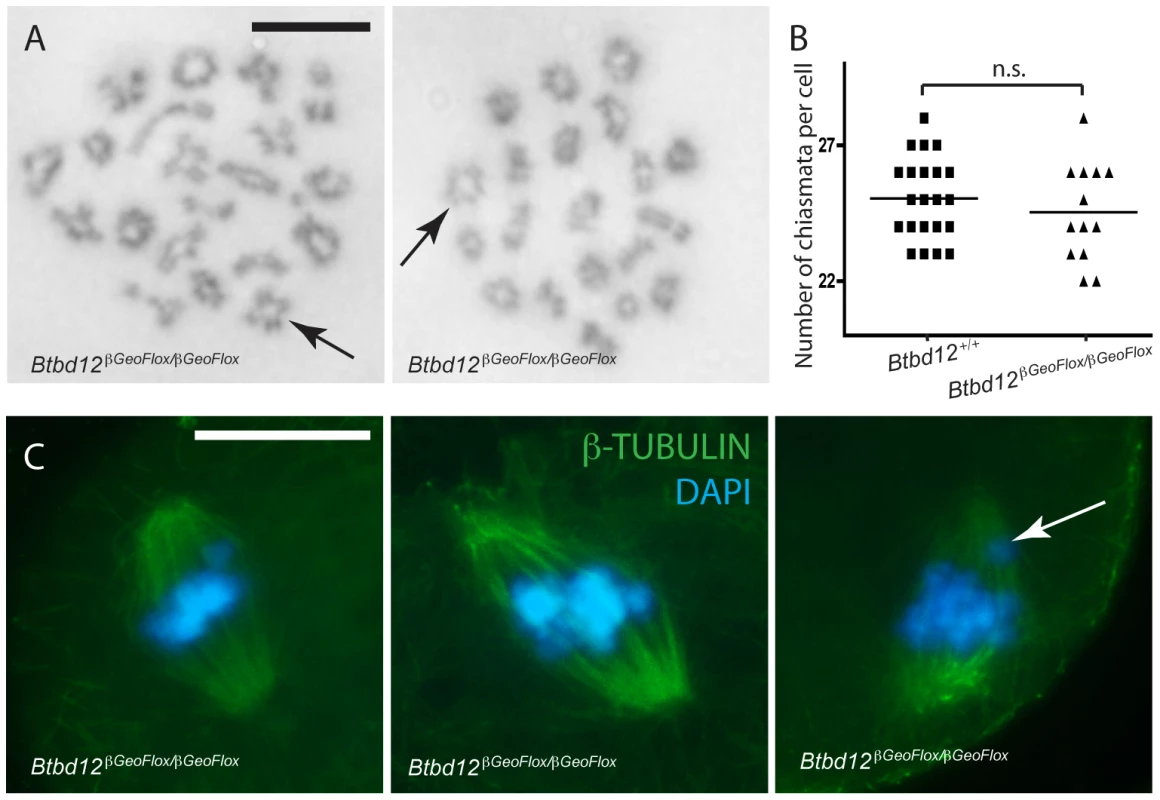
(A) Diakinesis preps from Btbd12βGeoFlox/βGeoFlox spermatocytes show evidence of chiasmata (black arrows). Scale bar is 10 µm. (B) Chiasmata counts from Btbd12+/+ (black squares) and Btbd12βGeoFlox/βGeoFlox (black triangles) diakinesis preparations show no significant difference (p = 0.2) (C) Btbd12βGeoFlox/βGeoFlox oocytes at metaphase I show no irregularities in chromosome alignment on the metaphase spindle. Although misaligned chromosomes were observed on occasion (white arrow), these were seen in wild type oocytes at the same frequency. Scale bar is 20 µm. To ask whether the number of chiasmata present in Btbd12βGeoFlox/βGeoFlox mutant animals is sufficient to cause appropriate separation of chromosomes during the first meiotic division, an examination of oocytes undergoing metaphase I to anaphase I progression was undertaken. Oocytes from wildtype and Btbd12βGeoFlox/βGeoFlox mice were stained with an antibody against β-tubulin to show the meiotic spindle and DAPI to stain the DNA (Figure 6C). The arrangement of chromosomes on the meiotic spindle in Btbd12βGeoFlox/βGeoFlox mutant oocytes was similar to wild type controls, with 53 cells examined from each genotype. In both cases, occasional cells appear to show one or two misaligned chromosomes, but this occurs at similar rates in oocytes from wild type and Btbd12βGeoFlox/βGeoFlox females (Figure 6C, arrow; 3/53 cells for both wildtype and mutant). Taken together, these results indicate that, despite elevated MLH1/MLH3 focus numbers, chiasmata counts are unaffected in Btbd12βGeoFlox/βGeoFlox mutant animals. Moreover, these chiasmata can, and do, result in normal metaphase I progression, resulting in appropriate chromosome segregation at the first meiotic division. However, the fact that very few diakinesis cells are obtained suggests either a delay in prophase I completion or loss of cells through prophase I prior to diakinesis.
BTBD12 localization to meiotic chromosome cores is disrupted in Atm null testes
BTBD12 was first identified as a potential kinase target of ATM [11]. To investigate the functional interaction between these two proteins, we examined the localization of BTBD12 on meiotic chromosomes in the absence of ATM. When co-immunostaining for BTBD12 and SYCP3 was performed on spread preparations from Atm null spermatocytes, we observed a complete absence of BTBD12 protein on chromosome cores (Figure 7A), compared to the punctate pattern of BTBD12 staining observed on chromosome cores wild type spermatocytes (Figure 1C). TOPBP1, however, localizes normally to the cores in Atm null cells (Figure 7B) in line with the demonstration in budding yeast that Mec1(ATR) activation is dependent on Dpb11(TOPBP1), rather than vice versa, and that Mec1, in turn, mediates only the functional interaction between Slx4(BTBD12) and Dpb11(TOPBP1), rather than regulating directly the localization of TOPBP1 [44], [55], [56].
Fig. 7. BTBD12 is down regulated in Atm null mice. 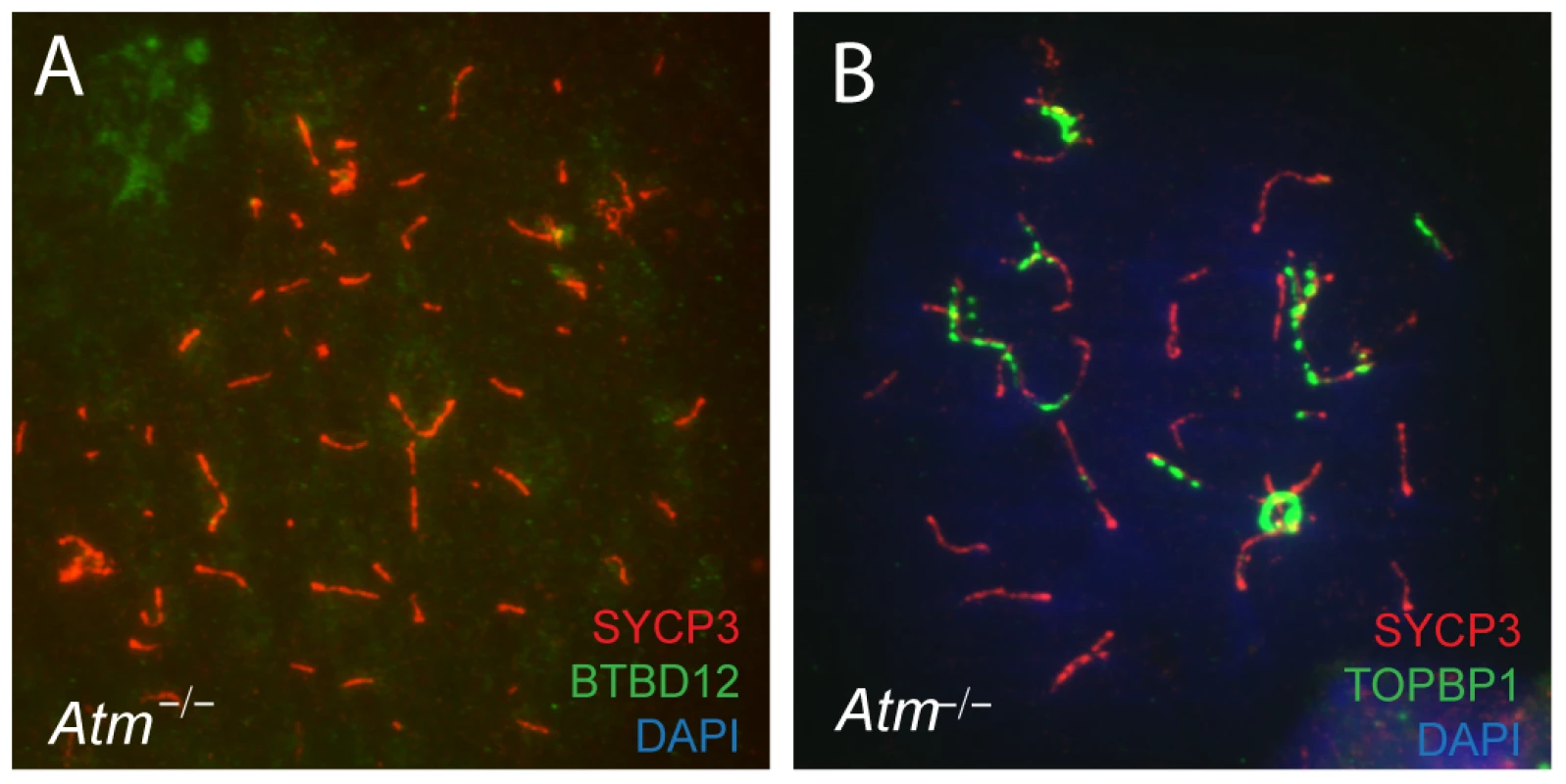
(A) Spermatocyte from an Atm−/− null mouse stained with anti-SYCP3 (red), anti-BTBD12 (green) and DAPI (blue). (B) Spermatocyte from an Atm−/− null mouse stained with anti-SYCP3 (red), anti-TOPBP1 (green) and DAPI (blue). BTBD12 protein is also down-regulated in Atm−/− males, when compared to the wildtype littermate controls, with Atm+/− males showing intermediate levels of BTBD12 protein (Figure 2, lanes 4–6). Testis protein extracts from Atm null mice show a decrease in BTBD12 protein to 70.3% of that seen in wildtype males, compared to a decrease to 26.9% in Atm heterozygotes (Figure 2 graph). Since ATM deletion results in pachytene meiotic failure in mice [57], [58], western blots were also performed on juvenile testis extracts at day 17 post-partum to ensure that all protein extracts from Atm+/+, Atm+/−, and Atm−/− contained equivalent cell populations. Indeed, even with higher proportions of leptotene and zygotene cells present in the day 17 extracts, protein from Atm−/− males showed a depletion of BTBD12 by 67.5% compared to Atm+/+ males (Figure 2, lanes 7 and 8), with Atm+/−protein extracts showing an intermediate decrease of 43% compared to wildtype levels (lane 9). Thus, even when taking into account the earlier meiotic failure of Atm−/− males, BTBD12 protein is severely reduced in the absence of ATM.
Discussion
The results presented herein describe, for the first time, the role of BTBD12 (SLX4) in mammalian gametogenesis. Our data show that BTBD12 plays dual roles in gametogenesis, firstly in facilitating primordial germ cell proliferation and establishment of the spermatogonial pool, possibly by ensuring genome stability, and secondly, in meiotic recombination events. These studies demonstrate that BTBD12 protein localizes to spermatogonia and spermatocytes of the testis. In the latter, BTBD12 is found along chromosome cores during prophase I, accumulating as early as zygonema and persisting through until late pachynema.
To explore the role of BTBD12 in mammalian gametogenesis, we obtained the Btbd12βGeoFlox/βGeoFlox mutant mouse line from the European Conditional Mouse Mutagenesis program (EUCOMM). The genetic disruption at the Btbd12 allele results in residual protein that appears on western blots and, to a lesser extent, on immunohistochemical sections. The detected protein could reflect either a truncated fusion protein consisting of BTBD12 and βGeo, or intact BTBD12 protein generated by a splicing event that removes the βGeo cassette, retaining separate and identiable β-galactosidase protein expression. We favor this latter possibility because the BTBD12 protein detected in the mutant animals can be detected with antibodies against either the N - or the C-terminus. Further, cDNA analysis reveals the presence of every exon of the mouse gene in reverse transcribed RNA from Btbd12βGeoFlox/βGeoFlox testis (data not shown). The reduced intensity of BTBD12 signal in the Btbd12βGeoFlox/βGeoFlox testis extracts compared to wildtype extracts indicates that the presence of the βGeo cassette dramatically reduces the efficiency of BTBD12 protein production and/or reduces the stability of the protein. Importantly, the residual BTBD12 protein does not localize appropriately to meiotic chromosome cores, or localizes at levels that are undetectable using standard chromosome spreads, leading us to conclude that the functional activity of BTBD12 is abnormal in these mice, at least in the context of prophase I.
Micronucleus formation in Btbd12βGeoFlox/βGeoFlox mice is very much elevated compared to wildtype litter mates, in line with a recent report that first described this mouse line [30]. Crossan et al report that the phenotype of Btbd12βGeoFlox/βGeoFlox mice bears some resemblance to human Fanconi anemia (FA), including blood cell cytopenia and numerous developmental deformaties such as the anophthalmia reported herein. This report is in line with another recent publication demonstrating that human SLX4 mutations are also found in a subset of FA patients [12]. Crossan et al also describe gonadal defects and subfertility in Btbd12βGeoFlox/βGeoFlox mice [30], but did not explore the origins of these phenotypes. They report that the histological appearance of the testes is consistent with a defect in meiosis, but they did not document pre-meiotic defects in these animals. They did, however, allude to similarities between the testicular phenotype of Btbd12βGeoFlox/βGeoFlox males and that of other FA-associated DNA repair proteins with which BTBD12 interacts, including Ercc1, Fancd2, Fancl, and Fanca [59]–. These latter mutations result in spermatogenic phenotypes ranging from spermatogonial proliferation defects, to meiotic defects, to defects in spermatozoa morphology. As will be discussed, results presented herein suggest that the phenotypic defects observed in the testes of Btbd12βGeoFlox/βGeoFlox males may be quite distinct from these other mutations.
Given the dramatic loss of testis weight we observe in the adult Btbd12βGeoFlox/βGeoFlox males, coupled with the severe paucity of cells in many seminiferous tubules as early as the period of the first wave of meiosis (days 13–26 pp), we reasoned that a large proportion of germ cells must be lost prior to entry into meiotic prophase I. Thus, these mice suffer from multiple germ cell defects, one involving spermatogonial proliferation and the other involving meiotic progression. The combined effect is a drop in testis size of 75% and a depletion of epididymal spermatozoa by about 90% of wildtype numbers. Given that we have observed only 3 viable pregnancies from females mated to Btbd12βGeoFlox/βGeoFlox males over a 9-month period, we conclude that these defects result in sub-fertility. Breeding data from Btbd12βGeoFlox/βGeoFlox females shows a more severe phenotype, with no pregnant females after 1 year of breeding. These data are more severe than the original description of these mice by the Crossan et al [30], but it should be noted that their analysis included many more mice over a much longer time period.
As predicted, histological examination of testis sections from e16 onwards revealed a dramatic decrease in the numbers of spermatogonial precursors from late gestation, known as prospermatogonia or gonocytes, within the developing seminiferous cords of Btbd12βGeoFlox/βGeoFlox male pups compared to their wildtype littermates. These data suggest that BTBD12 may function to promote DNA repair mechanisms during early proliferation of the PGC population within the developing gonad. PGCs originate at the posterior end of the primitive streak at e7, numbering around 45 [62]. They then migrate to the genital ridge, during which time they undergo a rapid cell proliferation to achieve a cell number of 3000 by e11.5 [63]. They continue to proliferate within the developing gonad until e13 when the testis/seminiferous cord structures form, trapping the now-mitotically arrested prospermatogonia within them [63]. They remain arrested until just prior to birth when they resume proliferation to provide the full complement of prospermatogonia to the post-natal testis. This late gestational wave of proliferation is also associated with increased germ cell apoptosis, even in testes from wildtype males. While the molecular pathways responsible for maintenance of genome integrity during this period are largely un-documented, it is likely that surveillance mechanisms exist similar to those found in somatic cells. Indeed, analysis of Atm mutant males reveals a requirement for ATM function in pre-meiotic spermatogonia [64]. Given the severe consequences of genomic instability within the PGC population for propagation of genetic mutations to offspring, it is plausible that particularly stringent DNA repair mechanisms must exist in these cells. Our results demonstrate that the PGCs arriving in the embryonic gonad appear normal in distribution and number since the testis of e16 mutants is similar in cellularity to that of wildtype littermates. From this time onwards, however, the numbers of prospermatogonia begin to decline rapidly in the testes of Btbd12βGeoFlox/βGeoFlox males such that the final number of these germ cells is dramatically lower in mutant males at e18 and at day 3 pp compared to wildtype males. This wave of apoptosis appears to coincide with the wave of proliferation that occurs around the time of birth because a large number of mitotic figures are observed in the testes of both wildtype and mutant males at this time (Figure 4, arrowheads).
Crossan et al have suggested that the germ cell defects present in Btbd12βGeoFlox/βGeoFlox mice may be similar to that of mutant mice for other key DNA repair genes, particularly those of various FA complementation groups [30]. Indeed, there are substantial similarities between these phenotypes described herein and those for proteins that have been shown to interact with BTBD12. For example, a targeted nullizygous mutation of Fancl also results in limited PGC populations in the embryonic gonad, similar to that seen in Btbd12βGeoFlox/βGeoFlox male embryos [65]. Interestingly, however, the residual PGCs can repopulate the testis gradually in the postnatal mouse such that, by 12 weeks of age, fertility is restored [65]. Thus, while the initial PGC defect may be similar in Btbd12βGeoFlox/βGeoFlox and Fancl−/− male embryos [65], the outcomes in terms of adult fertility are very distinct. In contrast to the age-related increase in fertility in Fancl nullizygous mice, and also to Fanca nullizygous mice, which show declining fertility with age [60], we see limited fertility in the Btbd12βGeoFlox/βGeoFlox males from 7 weeks of age onwards, with no subsequent restoration. Thus, it appears that mouse mutants for the FA complementation groups, while all showing similar anemia phenotypes, represent a spectrum of defects with respect to germ cell migration, proliferation, and differentiation. These differences underscore the importance of this family in genome stabilization within the germ line at all points in their development.
The pre-meiotic phenotype we observe in Btbd12βGeoFlox/βGeoFlox males is also temporally similar to that seen in Ercc1 nullizygous animals [66] in that both mutations result in significant loss of germ cells prior to entry into meiosis. Indeed, the testicular phenotype of Ercc1−/− males at day 3 pp is remarkably similar, if not identical, to that presented herein [66]. In the case of Ercc1, however, no restoration of fertility with age has been reported for nullizygous animals and, in fact, the mice have reduced numbers of epididymal spermatozoa throughout reproductive life, all of which show distinct morphological defects [66]. The limited spermatozoa observed in Btbd12βGeoFlox/βGeoFlox males appear to be morphologically normal and capable of fertilization, and could be a result of the fact that these mice retain residual BTBD12 protein.
Beyond spermatogonial stages, we present evidence of a role for BTBD12 in prophase I progression in the mouse, congruent with the localization of BTBD12 protein on synapsed meiotic chromosome cores. The early localization of BTBD12 at these sites implies an early function in recombination events. Accordingly, in the Btbd12βGeoFlox/βGeoFlox animals, there is persistence in the signal for γH2AX across the autosomes beyond pachynema in the absence of BTBD12. While some 10% of cells show synapsis errors in Btbd12βGeoFlox/βGeoFlox spermatocytes, synapsis is largely unaffected in these mutants, which is in contrast to that seen for FancD2 mutants in which spermatocytes show high levels of asynapsis in a subset of spermatocytes [59]. These data suggest a failure to repair DSBs in a timely fashion, leading to the persistent γH2AX and RAD51 observed at pachynema. However, we cannot rule out the possibility that the additional γH2AX signal is associated with persistent DNA damage arising during pre-meiotic replication events. Indeed, we did observe, by TUNEL labeling, a small fraction of spermatogonia undergoing apoptosis prior to meiotic entry, and so it is possible that if excessive DNA damage persists in the absence of BTBD12, then a proportion of these cells could avoid apoptosis and enter prophase I. Such a possibility can only be addressed through the use of prophase I-specific conditional knock-out strategies. Importantly, however, regardless of the timing of DNA damage induction (pre-meiotic or meiotic), these cells do not succumb to the pachytene checkpoint, as would be predicted from other knockout studies of early prophase I genes, including Msh4/Msh5 and Dmc1 [53], [54], [67]. Moreover, we do not see an overt increase in localization of TOPBP1, nor of MSH4 and MSH5 (data not shown), suggesting that the persistent DSBs observed at pachynema arise out of SPO11-induced events in early prophase I, and not as a result of DNA repair errors prior to meiosis.
Studies by the Sekelsky group were the first to indicate a role for vertebrate SLX4 orthologs in meiosis. These authors showed that the Drosophila Slx4 ortholog, MUS312, is essential for ensuring genomic stability and interacts with the MEI-9(XPF)/ERCC1 nuclease to produce meiotic COs [9], [14], [68], [69]. Thus mus312 mutant flies exhibit >90% reduction in meiotic crossovers [14], but this is unrelated to Mus81 events since Drosphila Mus81 does not participate in CO regulation during meiosis [70]. Similarly, Saito et al describe a role for the C. elegans Slx4 ortholog, named HIM-18, in meiotic recombination, since him-18 mutants exhibit reductions in crossing over of the order of 30–50%, depending on the chromosomal context [13]. Importantly, while Mus-81 does not appear to play a critical role during meiosis in worms, double mus-81;him-18 mutants show a more severe reduction in crossing over than him-18 alone, suggesting co-operative roles for MUS-81 and HIM-18 in meiotic recombination in C. elegans. It is interesting to note that a pre-meiotic function was also described for him-18 mutants, analogous to our observations for mouse BTBD12.
The role of BTBD12 in meiotic recombination is unclear at the current time, and it is interesting to note that the yeast ortholog, Slx4, does not appear to function in meiosis [71]. Given that BTBD12 interacts with components of both the Class I and Class II crossover pathways, as well as with BLM, it is well placed to play an important role in integrating CO decisions in mouse germ cells (Figure 8A). The loss of functional BTBD12 on meiotic chromosomes results in an increase in MLH1/MLH3 foci at mid-pachynema (Figure 5Q, 5R), similar to that seen in Mus81 nullizygous males [28]. Also similar to the Mus81 knockout mouse, we observe no increase in chiasmata at diakinesis in the Btbd12βGeoFlox/βGeoFlox males (Figure 6A, 6B). For Mus81 nullizygous mice, we hypothesize that the additional MLH1/MLH3 foci can restore/maintain chiasmata numbers in the absence of a functional Class II pathway (Figure 8C). By contrast, loss of MLH1 or MLH3, the major Class I mediators at pachynema, cannot be compensated for by MUS81 or any other pathway (Figure 8B). Given the similarity of the Btbd12 phenotype, described herein, to that of the Mus81 nullizygous phenotype, our data suggest that BTBD12 may drive recombination intermediates towards Class II events, thereby promoting MUS81-mediated crossing over. In the absence of either MUS81 or BTBD12 (Figure 8D), however, CO numbers are maintained because of a compensatory increase of MLH1/MLH3 foci, possibly suggesting that BTBD12 does not mediate this switch between the two pathways, but can promote Class II pathway choices under certain conditions. In this regard, it is possible that BTBD12 acts in concert with BLM helicase.
Fig. 8. Model for BTBD12 involvement in meiotic crossover control in the Class I and Class II pathways. 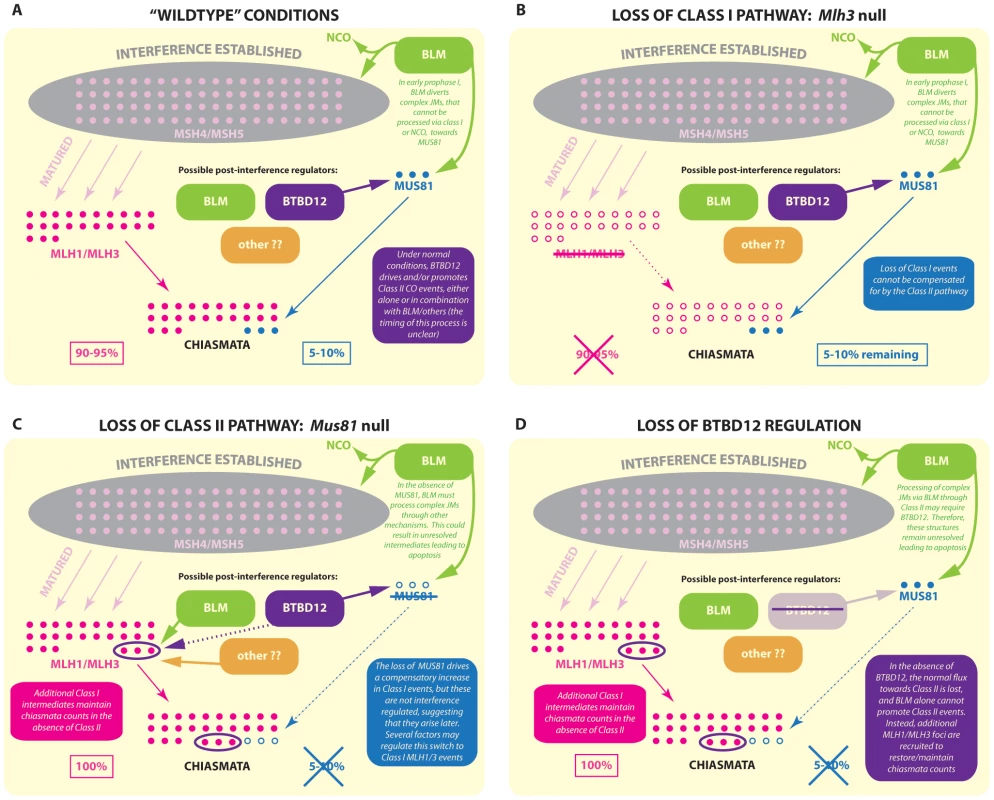
In wild type mice (A) MSH4/MSH5 foci (pale pink) accumulate on meiotic chromosomes during zygonema, and are subject to interference regulation (grey area). A subset of MSH4/MSH5 bound structures are then stabilized/matured by the accumulation of MLH1/MLH3 at pachynema, and these are designated as sites of Class I CO, which account for 90–95% of total crossovers. The remaining 5–10% of crossovers are generated by the Class II pathway, a major component of which is MUS81. It is thought that the precursors of these Class II COs may be aberrant JMs that, ordinarily, would be disassembled and/or processed towards NCO or Class I fates but that, by the complexity of their structure, avoid this fate. Thus, MUS81 processes those complex intermediates that “evade” BLM processing. (B) In the absence of MLH3, Class I COs cannot be generated, however a 5–10% subset of (presumably) Class II COs remain. These COs show CO breakpoints that are distal to the hotspot center and are more heterogeneic than MLH3 derived COs. (C) In the absence of MUS81, the full quota of Class I COs can be processed by MLH1/MLH3. However, the complex JMs that evade BLM processing cannot now be processed via Class II COs by MUS81. Instead, these structures are processed by some other mechanism or are left unresolved, increasing the apoptosis rate during prophase I. Meanwhile, however, through related or unrelated mechanisms, the Class I pathway upregulates acquisition of MLH1/MLH3 on MSH4/MSH5 sites, possibly taking advantage of the fact that there are excess numbers of the latter to which MLH1/MLH3 can bind and stabilize. Thus, the increase in MLH1/MLH3 foci by 5–10% does maintains the correct CO count. (D) The loss of BTBD12 strongly mirrors the loss of MUS81 in mammalian meiosis. An increase in Class I COs is indicated by a 5–10% increase in MLH1/MLH3 foci, whereas the final tally of COs in the form of chiasmata remains the same, indicating a loss of Class II events. Studies in yeast have suggested that the BLM ortholog, Sgs1, acts to limit the formation of aberrant JMs that arise from strand invasion events that involve both ends of the DSB (as just one example), instead producing substrates for the ZMM crossover pathway or instead resulting in a NCO fate, whilst Mus81-Mms4 can process those events that are not efficiently processed by Sgs1 even in wildtype situations [20], [21]. The absence of Sgs1, therefore, results in an overload of substrates for the Mus81 pathway [20], [21].
Given the model that emerges from the yeast data, together with the phenotypic characterization of Btbd12βGeoFlox/βGeoFlox mice described herein, we propose that the function of BTBD12 is to drive events towards Class II COs, possibly in a BLM-dependent fashion. This raises the question of the fate of those aberrant JMs that are not directed towards Class I or NCO pathways by the actions of BLM; those intermediates that, ordinarily, would be the substrates for MUS81 processing but which, in the absence of Class II-promoting BTBD12, still require resolution. Since it is unlikely that these aberrant structures are responsible for the additional MLH1/MLH3 sites that (presumably) maintain the normal chiasmata count, this suggests two important points. Firstly, the additional MLH1/MLH3 sites must be generated through some other mechanism, perhaps taking advantage of the fact that there is already an excess pool of MSH4/MSH5 foci from which to select for subsequent MLH1/MLH3-driven maturation/stabilization (see model Figure 8). Secondly, those BLM-processed JMs that fail to be diverted towards other recombination fates may remain un-repaired into late pachynema, as suggested by the persistent γH2AX and RAD51 on meiotic SCs in Blm conditional knockouts [72] and in Btbd12βGeoFlox/βGeoFlox males, and could well account for the gradual loss of spermatocytes via apoptosis leading up to the first meiotic division. Conflicting with these suggestions are our previous data showing increased chiasmata-like structures in the absence of Blm, that appear to be MLH1/MLH3-independent [72]. In addition, it should be noted that the complex intermediate structures reported for budding yeast have not yet been demonstrated in mammalian meiosis and, as such, our models, by necessity, relies on extrapolation from a number of organisms, most notably budding yeast.
BTBD12 and its orthologs have been shown to interact with a number of key players in the meiotic machinery, including Rad1-Rad10 [73], ERCC-1 [9], [14], [30], XPF (and its ortholog in Drosophila, MEI-9) [7], [9], [10], as well as with MUS81 and SLX1 [8], [10]. Studies by Svendson et al further demonstrated interactions between human BTBD12 and components of the DNA mismatch repair (MMR) family, including MSH2 [8]. Thus, a model emerges by which BTBD12 and its orthologs can mediate DNA repair and/or modification by directing the activities of certain nucleases in a substrate and context-specific manner. While the precise role of BTBD12 in mammalian meiosis remains to be further clarified, therefore, and its interactions with these various partners in mammalian germ cells have yet to be described, it is tempting to speculate on the function of BTBD12 based on localization data presented herein, on the phenotype of these mice, and based on previous studies of BTBD12 orthologs in other species. For example, studies in vitro have revealed distinct cutting activities for mammalian BTBD12 that are specific to its interactions with MUS81-EME1 (flaps and replication forks) and with SLX1 (HJ cleavage) [8], [10], [29]. Interestingly, the cleavage of HJs in vitro by BTBD12-SLX1 occurs in a near-symmetrical fashion, and in a manner similar to that seen for GEN1 [74], and it is plausible that such an activity may well confer at least limited HJ resolvase activity on the BTBD12-SLX1 complex in mammalian cells. This is in contrast to that seen for yeast Slx4, which cleaves HJ structures asymmetrically [2], [75] thereby reducing the likelihood of an in vivo resolvase role for the yeast protein. This difference in the cutting symmetry between the yeast and mammalian orthologs may explain why slx4 mutants in yeast have no meiotic phenotype, while we observe a distinct meiotic phenotype in the Btbd12βGeoFlox/βGeoFlox mice.
BTBD12 was first identified as a candidate target of the ATM kinase [11] and, in line with this, Slx4 is a substrate of the yeast ATM/ATR orthologs, Mec1 and Tel1 [5]. The function of Slx4 in replication fork repair is dependent on this phosphorylation [55]. In line with this, our western blot analysis reveals a depletion of BTBD12 protein within the testis of adult and juvenile Atm−/− males compared to that seen in wildtype litter mates. BTBD12 protein is also lost from the chromosome cores of Atm−/− spermatocytes, while pre-meiotic spermatogonial proliferation appears to require both BTBD12 and ATM [64]. Thus, while it remains to be seen how ATM and BTBD12 co-ordinate their pre-meiotic functions in spermatogonia, we conclude that ATM activity is essential for the normal loading of BTBD12 onto meiotic chromosomes during prophase I and/or for the stabilization of BTBD12 at these sites. Interestingly, the increased loading of MLH1/MLH3 observed in Btbd12βGeoFlox/βGeoFlox spermatocytes mirrors the phenotype seen in Spo11+/−Atm−/− animals, the Spo11 heterozygosity in this case rescuing the zygotene loss of spermatocytes in Atm single nulls and allowing progression to metaphase [31], [32], again pointing to a functional interaction between the ATM and BTBD12. Spo11+/−Atm−/− spermatocytes also show defects in the dynamics of sex chromosome synapsis and in the formation of the obligate crossover on the pseudoautosomal region (PAR) of the XY [31]. This phenotype is not shared with the Btbd12βGeoFlox/βGeoFlox males, suggesting that BTBD12 is not involved in this aspect of ATM function.
The Btbd12βGeoFlox/βGeoFlox strain of mice we have used herein do not, in all likelihood, represent a complete null allele, due to the presence of residual BTBD12 protein, as measured by both western blot and by immunohistochemistry. However, given the failure of this residual protein to accumulate on meiotic chromosome cores, at least to detectable levels, we consider this mouse to represent a significant impairment in BTBD12 function in germ cells. The use of the Btbd12βGeoFlox/βGeoFlox mouse, while not removing all of the endogenous protein, and while not permitting a focus on meiotic events alone, has been fortuitous in this instance, as it has highlighted the primordial germ cell proliferation defect, which otherwise would not have been evident using a meiosis-specific conditional null mouse. Clearly, any future meiosis-specific research should utilize a conditional approach, which would ensure BTBD12 absence only in meiotic cells, and allow much easier characterization of any defects as a result of loss of BTBD12 protein. Such studies are currently ongoing but will, by necessity, be lengthy.
As discussed, BTBD12 has many potential roles in DNA repair in mammals, from its function in somatic cell repair in human cell lines [7], [8], [10], [29], to its roles in pre-meiotic PGC proliferation and meiotic crossover formation demonstrated herein. The ability of BTBD12 to process not only HJs robustly, but other DNA structures, such as 3′ and 5′ flaps, as well [7], [8], [10], [29], indicates it has the potential to have numerous roles in DNA repair, both in mitosis and meiosis. Moreover, the evidence of functional interactions with a myriad of DNA repair proteins such as MUS81, BLM, XPF-ERCC1, MSH2-MSH3, and the Fanconi anemia genes, along with its well established binding partner SLX1, show that BTBD12 may integrate with several DNA repair complexes to effect its HJ (and other) processing abilities [7], [8], [10], [29]. These interactions no doubt contribute to the similarities between phenotypes at discrete stages of spermatogenesis for Btbd12, Fancl, Ercc1, and other mutant mouse models. Thus a clearer understanding of the function of mammalian BTBD12, both in the context of its multiple roles in gamete formation, and in its function in general genome stability/DNA repair will require more detailed knowledge of these key interactions.
Materials and Methods
Ethics statement
All animals used in this work were handled under strict guidelines imposed by Cornell Veterinary staff and by the Institutional Care and Use Committee (IACUC) under an approved protocol.
Animals and genotyping
We obtained a line of Btbd12 mice with a cassette inserted into the Btbd12 gene, containing a β-galactosidase fused with a neomycin resistance gene, flanked by FRT sites, and LoxP sites flanking exon 3 of the Btbd12 gene from the European Conditional Mouse Mutagenesis (EUCOMM) program (EPD0028_7_A08; Btbd12tm1a(EUCOMM)Wtsi). We termed these mice Btbd12βGeoFlox, to indicate that the FRT flanked and LoxP flanked regions are intact. Mice were genotyped using primers Btbd12_F 5′ CACTGAGCCATCTCACCAGC 3′ and Cas_R1 5′ TCGTGGTATCGTTATGCGCC 3′ to amplify the mutant allele (Btbd12βGeoFlox), and Btbd12_F 5′ CACTGAGCCATCTCACCAGC 3′ and Btbd12_R2b 5′ GGAGCCCAGTCTGGGACTCTG 3′ to amplify the wildtype allele (Btbd12+).
Antibody preparation
The anti Rabbit polyclonal BTBD12 antibodies were against recombinant His-tagged murine BTBD12 peptide comprising amino acid residues 1–350 (“NT”) and 750–1100 (“CT”). For that, the corresponding cDNA fragment was cloned in pET-28 expression vector (Novagen) and recombinant proteins fused to a histidine tag were purified using Ni-NTA resin (Qiagen) following the manufacturer's instructions.
Sperm counts
Epidiymides were removed from either Btbd12βGeoFlox/βGeoFlox or Btbd12+/+ adult mice, placed in human tubule fluid (HTF) culture medium containing BSA (Specialty Media, Millipore), ripped open using micro forceps and the contents squeezed into the medium. The spermatozoa were cultured for 20 minutes at 32°C, then a 20 µl aliquot was removed and fixed in 480 µl 10% formalin. The fixed cells were gently mixed then intact spermatozoa counted using a hemocytometer.
Histology
Testes were removed from pre-pubertal or adult mice and fixed either in Bouins fixative or 10% buffered formalin for 2–12 hours. Paraffin-embedded tissue was sectioned at 4 µm and processed for Hematoxylin and Eosin staining or immunohistochemical analyses using standard methods.
Chromosome spread analysis
Testes were removed from adult Btbd12βGeoFlox/βGeoFlox or Btbd12+/+ mice for the meiotic spread analysis, as well as for the focus counts, and processed as previously described [26]. Briefly, testes were removed and decapsulated into hypotonic sucrose extraction buffer (HEB, containing 1.7% sucrose) and left on ice for 60 minutes. Tubules were macerated on glass depression slides in a bubble of 0.03% sucrose and added to slides coated in 1% paraformaldehyde. The slides were dried slowly in a humidified chamber for 3 hours and washed in PBS containing Photoflo (Kodak, EMS). Ovaries were removed from day e19 female embryos and incubated in HEB for 20 minutes, before being macerated on a depression slide in 0.03% sucrose and added to a bubble of 1% PFA on a well slide, before drying as above.
Immunofluorescence and immunohistochemistry
Slides were processed as described previously [76] using antibodies generated in this lab [26], generously donated by colleagues and available commercially. Immunohistochemistry was performed on formalin-fixed sections using rat monoclonal hybridoma supernatant against germ cell nuclear antigen-1 (GCNA-1), 10D9G11, for staining of germ cells [77], rabbit anti-BTBD12 antibody, rabbit anti-β-galactosidase or TUNEL staining (Chemicon) to detect cells undergoing apoptosis. γH2AX staining was described as either “normal” or “abnormal”, in both pachytene and diplotene cells from Btbd12βGeoFlox/βGeoFlox or Btbd12+/+ spermatocytes. Abnormal cells were classified by >1 γH2AX focus per homologous chromosome core in pachynema, and 1 or more SC-associated γH2AX focus per nucleus in diplonema.
Meiotic preparations of oocyte spindles
Oocyte spindles were prepared using a modification of techniques described previously [78], [79] and used subsequently in our laboratory [80]. Briefly, ovaries were removed by puncturing ovaries from unstimulated females at 24–26 days of age, and placed in Waymouth's media (GIBCO, Invitrogen Corporation, Carlsbad, CA) supplemented with 100 units of penicillin (base) and 10 µg of streptomycin (base)/ml, 10% fetal bovine serum, and 0.23 mmol/l sodium pyruvate. Primary oocytes at germinal vesicle (GV) stage were cultured in 20 µl drops of KSOM (Millipore Corporation, Bedford, MA) overlaid with mineral oil (Chemicon, Millipore Corporation, Bedford, MA) and incubated at 37°C in an atmosphere of 5% CO2. After 2.5 hrs in culture, oocytes were transferred to fresh KSOM drops and scored for germinal vesicle break down (GVBD). In order to observe meiotic division at metaphase I, oocytes were cultured in KSOM for 8∼10 h and >18 h, respectively, and fixed in fibrin clots (below). Oocytes were fixed in fibrin clots, according to published methodology [78], prior to staining for β-tubulin (1∶500 mouse monoclonal antibody; Sigma-Aldrich, St. Louis, MO). Tubulin staining was visualized using a FITC-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), and counterstaining for DNA was achieved using 4′,6-Diamidino-2-phenylindole (DAPI).
Statistical analyses
Testis weights, spermatozoa numbers, TUNEL analysis, immunofluorescent focus counts and diakinesis spread counts were all analyzed for statistical significance by using an unpaired t-test using Prism 4.0 software.
Micronucleus assay material and methods
Analysis of micronucleus formation in peripheral blood cells was performed as previously described [35]. Briefly, peripheral blood was collected from the retro-orbital sinus, fixed in methanol, and incubated in bicarbonate buffer containing RNase A and anti-CD71: FITC antibody (Biodesign International). After washing and staining with propidium iodide, the cells were analyzed on a FACSCalibur flow cytometer (Becton-Dickinson, San Jose, CA).
Supporting Information
Zdroje
1. MullenJRKaliramanVIbrahimSSBrillSJ 2001 Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157 103 118
2. FrickeWMBrillSJ 2003 Slx1–Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev 17 1768 1778
3. LiFDongJPanXOumJHBoekeJD 2008 Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Mol Cell 30 325 335
4. FlottSAlabertCTohGWTothRSugawaraN 2007 Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol Cell Biol 27 6433 6445
5. FlottSRouseJ 2005 Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem J 391 325 333
6. ChangMBellaouiMBooneCBrownGW 2002 A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A 99 16934 16939
7. FekairiSScaglioneSChahwanCTaylorERTissierA 2009 Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138 78 89
8. SvendsenJMSmogorzewskaASowaMEO'ConnellBCGygiSP 2009 Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138 63 77
9. AndersenSLBergstralhDTKohlKPLaRocqueJRMooreCB 2009 Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol Cell 35 128 135
10. MunozIMHainKDeclaisACGardinerMTohGW 2009 Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35 116 127
11. MatsuokaSBallifBASmogorzewskaAMcDonaldER3rdHurovKE 2007 ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316 1160 1166
12. StoepkerCHainKSchusterBHilhorst-HofsteeYRooimansMA 2011 SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet
13. SaitoTTYoudsJLBoultonSJColaiacovoMP 2009 Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet 5 e1000735 doi:10.1371/journal.pgen.1000735
14. YildizOMajumderSKramerBSekelskyJJ 2002 Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell 10 1503 1509
15. KeeneySGirouxCNKlecknerN 1997 Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375 384
16. NealeMJPanJKeeneyS 2005 Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436 1053 1057
17. HandelMASchimentiJC 2010 Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11 124 136
18. de los SantosTHunterNLeeCLarkinBLoidlJ 2003 The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164 81 94
19. BornerGVKlecknerNHunterN 2004 Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 29 45
20. OhSDLaoJPTaylorAFSmithGRHunterN 2008 RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell 31 324 336
21. JessopLLichtenM 2008 Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell 31 313 323
22. OhSDLaoJPHwangPYTaylorAFSmithGR 2007 BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130 259 272
23. JessopLRockmillBRoederGSLichtenM 2006 Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet 2 e155 doi:10.1371/journal.pgen.0020155
24. EdelmannWCohenPEKaneMLauKMorrowB 1996 Meiotic pachytene arrest in MLH1-deficient mice. Cell 85 1125 1134
25. LipkinSMMoensPBWangVLenziMShanmugarajahD 2002 Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31 385 390
26. KolasNKSvetlanovALenziMLMacalusoFPLipkinSM 2005 Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol 171 447 458
27. SvetlanovABaudatFCohenPEde MassyB 2008 Distinct Functions of MLH3 at Recombination Hot Spots in the Mouse. Genetics 178 1937 1945
28. HollowayJKBoothJEdelmannWMcGowanCHCohenPE 2008 MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4 e1000186 doi:10.1371/journal.pgen.1000186
29. SvendsenJMHarperJW 2010 GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution. Genes Dev 24 521 536
30. CrossanGPvan der WeydenLRosadoIVLangevinFGaillardPH 2011 Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet
31. BarchiMRoigIDi GiacomoMde RooijDGKeeneyS 2008 ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet 4 e1000076 doi:10.1371/journal.pgen.1000076
32. BellaniMARomanienkoPJCairattiDACamerini-OteroRD 2005 SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/ − spermatocytes. J Cell Sci 118 3233 3245
33. DerheimerFAKastanMB 2010 Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett 584 3675 3681
34. DertingerSDTorousDKTometskoKR 1996 Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat Res 371 283 292
35. LevittPSZhuMCassanoAYazinskiSALiuH 2007 Genome maintenance defects in cultured cells and mice following partial inactivation of the essential cell cycle checkpoint gene Hus1. Mol Cell Biol 27 2189 2201
36. ShimaNHartfordSADuffyTWilsonLASchimentiKJ 2003 Phenotype-based identification of mouse chromosome instability mutants. Genetics 163 1031 1040
37. WesternPSMilesDCvan den BergenJABurtonMSinclairAH 2008 Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 26 339 347
38. OrthJM 1982 Proliferation of Sertoli cells in fetal and postnatal rats: a quantitative autoradiographic study. Anat Rec 203 485 492
39. OrthJMJesterWFLiLHLaslettAL 2000 Gonocyte-Sertoli cell interactions during development of the neonatal rodent testis. Curr Top Dev Biol 50 103 124
40. MahadevaiahSKTurnerJMBaudatFRogakouEPde BoerP 2001 Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27 271 276
41. Fernandez-CapetilloOMahadevaiahSKCelesteARomanienkoPJCamerini-OteroRD 2003 H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4 497 508
42. HondaYTojoMMatsuzakiKAnanTMatsumotoM 2002 Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem 277 3599 3605
43. YamaneKWuXChenJ 2002 A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol 22 555 566
44. MordesDAGlickGGZhaoRCortezD 2008 TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22 1478 1489
45. Parrilla-CastellarERKarnitzLM 2003 Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J Biol Chem 278 45507 45511
46. BurrowsAEElledgeSJ 2008 How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev 22 1416 1421
47. PereraDPerez-HidalgoLMoensPBReiniKLakinN 2004 TopBP1 and ATR colocalization at meiotic chromosomes: role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell 15 1568 1579
48. WangTFKlecknerNHunterN 1999 Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci U S A 96 13914 13919
49. HollingsworthNMPonteLHalseyC 1995 MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev 9 1728 1739
50. HunterNBortsRH 1997 Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev 11 1573 1582
51. Santucci-DarmaninSWalpitaDLespinasseFDesnuelleCAshleyT 2000 MSH4 acts in conjunction with MLH1 during mammalian meiosis. Faseb J 14 1539 1547
52. Ross-MacdonaldPRoederGS 1994 Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79 1069 1080
53. EdelmannWCohenPEKneitzBWinandNLiaM 1999 Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nature Genetics 21 123 127
54. KneitzBCohenPEAvdievichEZhuLKaneMF 2000 MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 14 1085 1097
55. OhouoPYBastos de OliveiraFMAlmeidaBSSmolkaMB 2010 DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell 39 300 306
56. Navadgi-PatilVMBurgersPM 2008 Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283 35853 35859
57. BarlowCLiyanageMMoensPBTarsounasMNagashimaK 1998 Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 125 4007 4017
58. AshleyTWestphalCPlug-de MaggioAde RooijDG 2004 The mammalian mid-pachytene checkpoint: meiotic arrest in spermatocytes with a mutation in Atm alone or in combination with a Trp53 (p53) or Cdkn1a (p21/cip1) mutation. Cytogenet Genome Res 107 256 262
59. HoughtalingSTimmersCNollMFinegoldMJJonesSN 2003 Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev 17 2021 2035
60. ChengNCvan de VrugtHJvan der ValkMAOostraABKrimpenfortP 2000 Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet 9 1805 1811
61. AgoulnikAILuBZhuQTruongCTyMT 2002 A novel gene, Pog, is necessary for primordial germ cell proliferation in the mouse and underlies the germ cell deficient mutation, gcd. Hum Mol Genet 11 3047 3053
62. LawsonKAHageWJ 1994 Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp 182 68 84; discussion 84–91
63. WilhelmDPalmerSKoopmanP 2007 Sex determination and gonadal development in mammals. Physiol Rev 87 1 28
64. TakuboKHiraoAOhmuraMAzumaMAraiF 2006 Premeiotic germ cell defect in seminiferous tubules of Atm-null testis. Biochem Biophys Res Commun 351 993 998
65. LuBBishopCE 2003 Late onset of spermatogenesis and gain of fertility in POG-deficient mice indicate that POG is not necessary for the proliferation of spermatogonia. Biol Reprod 69 161 168
66. HsiaKTMillarMRKingSSelfridgeJRedheadNJ 2003 DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development 130 369 378
67. PittmanDLCobbJSchimentiKJWilsonLACooperDM 1998 Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1 697 705
68. RadfordSJGoleyEBaxterKMcMahanSSekelskyJ 2005 Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170 1737 1745
69. RadfordSJMcMahanSBlantonHLSekelskyJ 2007 Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics 176 63 72
70. TrowbridgeKMcKimKBrillSJSekelskyJ 2007 Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176 1993 2001
71. RouseJ 2009 Control of genome stability by SLX protein complexes. Biochem Soc Trans 37 495 510
72. HollowayJKMorelliMABorstPLCohenPE 2010 Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol 188 779 789
73. LyndakerAMGoldfarbTAlaniE 2008 Mutants defective in Rad1-Rad10-Slx4 exhibit a unique pattern of viability during mating-type switching in Saccharomyces cerevisiae. Genetics 179 1807 1821
74. IpSCRassUBlancoMGFlynnHRSkehelJM 2008 Identification of Holliday junction resolvases from humans and yeast. Nature 456 357 361
75. CoulonSGaillardPHChahwanCMcDonaldWHYatesJR3rd 2004 Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell 15 71 80
76. EdelmannWCohenPEKneitzBWinandNLiaM 1999 Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 21 123 127
77. WangDEndersGC 1996 Expression of a specific mouse germ cell nuclear antigen (GCNA1) by early embryonic testicular teratoma cells in 129/Sv-Sl/+ mice. Cancer Lett 100 31 36
78. WoodsLMHodgesCABaartEBakerSMLiskayRM 1999 Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. Journal of Cell Biology 145 1395 1406
79. HodgesCALeMaire-AdkinsRHuntPA 2001 Coordinating the segregation of sister chromatids during the first meiotic division: evidence for sexual dimorphism. J Cell Sci 114 2417 2426
80. KanRSunXKolasNKAvdievichEKneitzB 2008 Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol Reprod 78 462 471
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



