-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Survival and Growth of Yeast without Telomere Capping by Cdc13 in the Absence of Sgs1, Exo1, and Rad9
Maintenance of telomere capping is absolutely essential to the survival of eukaryotic cells. Telomere capping proteins, such as Cdc13 and POT1, are essential for the viability of budding yeast and mammalian cells, respectively. Here we identify, for the first time, three genetic modifications that allow budding yeast cells to survive without telomere capping by Cdc13. We found that simultaneous inactivation of Sgs1, Exo1, and Rad9, three DNA damage response (DDR) proteins, is sufficient to allow cell division in the absence of Cdc13. Quantitative amplification of ssDNA (QAOS) was used to show that the RecQ helicase Sgs1 plays an important role in the resection of uncapped telomeres, especially in the absence of checkpoint protein Rad9. Strikingly, simultaneous deletion of SGS1 and the nuclease EXO1, further reduces resection at uncapped telomeres and together with deletion of RAD9 permits cell survival without CDC13. Pulsed-field gel electrophoresis studies show that cdc13-1 rad9Δ sgs1Δ exo1Δ strains can maintain linear chromosomes despite the absence of telomere capping by Cdc13. However, with continued passage, the telomeres of such strains eventually become short and are maintained by recombination-based mechanisms. Remarkably, cdc13Δ rad9Δ sgs1Δ exo1Δ strains, lacking any Cdc13 gene product, are viable and can grow indefinitely. Our work has uncovered a critical role for RecQ helicases in limiting the division of cells with uncapped telomeres, and this may provide one explanation for increased tumorigenesis in human diseases associated with mutations of RecQ helicases. Our results reveal the plasticity of the telomere cap and indicate that the essential role of telomere capping is to counteract specific aspects of the DDR.
Published in the journal: . PLoS Genet 6(8): e32767. doi:10.1371/journal.pgen.1001072
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001072Summary
Maintenance of telomere capping is absolutely essential to the survival of eukaryotic cells. Telomere capping proteins, such as Cdc13 and POT1, are essential for the viability of budding yeast and mammalian cells, respectively. Here we identify, for the first time, three genetic modifications that allow budding yeast cells to survive without telomere capping by Cdc13. We found that simultaneous inactivation of Sgs1, Exo1, and Rad9, three DNA damage response (DDR) proteins, is sufficient to allow cell division in the absence of Cdc13. Quantitative amplification of ssDNA (QAOS) was used to show that the RecQ helicase Sgs1 plays an important role in the resection of uncapped telomeres, especially in the absence of checkpoint protein Rad9. Strikingly, simultaneous deletion of SGS1 and the nuclease EXO1, further reduces resection at uncapped telomeres and together with deletion of RAD9 permits cell survival without CDC13. Pulsed-field gel electrophoresis studies show that cdc13-1 rad9Δ sgs1Δ exo1Δ strains can maintain linear chromosomes despite the absence of telomere capping by Cdc13. However, with continued passage, the telomeres of such strains eventually become short and are maintained by recombination-based mechanisms. Remarkably, cdc13Δ rad9Δ sgs1Δ exo1Δ strains, lacking any Cdc13 gene product, are viable and can grow indefinitely. Our work has uncovered a critical role for RecQ helicases in limiting the division of cells with uncapped telomeres, and this may provide one explanation for increased tumorigenesis in human diseases associated with mutations of RecQ helicases. Our results reveal the plasticity of the telomere cap and indicate that the essential role of telomere capping is to counteract specific aspects of the DDR.
Introduction
The ends of linear chromosomes pose two major threats to the proliferative potential and genetic stability of eukaryotic cells: inappropriate activation of the DNA damage response (DDR) and progressive shortening of the chromosome ends due to the end replication problem. The telomere, a specialised structure consisting of G-rich repetitive DNA and associated protein complexes at the end of linear chromosomes helps to overcome both of these problems by recruiting telomere capping proteins and telomerase to telomeres [1]. In metazoan organisms, maintenance of telomere integrity is critical for protecting against the processes of cancer and ageing [2], [3].
The mechanisms of chromosome end protection by telomere capping proteins are conserved in eukaryotes. The telomeric DNA of most eukaryotes consists of G-rich repetitive DNA with a 3′ single stranded DNA (ssDNA) overhang. In mammalian cells, telomeric DNA is bound by the shelterin complex [4], [5]. Two proteins of shelterin, TRF1 and TRF2, bind to the double stranded telomeric repeat and recruit TIN2, Rap1, TPP1 and POT1 to the telomere and help to cap the chromosome end [4], [5]. In addition to the shelterin complex, another conserved telomere capping complex, the CST complex, which consists of CTC1, STN1 and TEN1, has been recently described in mammal and plant cells [6]–[8]. In Saccharomyces cerevisiae, telomeric ssDNA is capped by the essential Cdc13-Stn1-Ten1 complex, analogous to the CST complex in other cell types [7], [9].
ssDNA binding proteins like POT1 and Cdc13 bind to the telomeric 3′ ssDNA overhang and play multiple roles to protect and maintain the chromosome end [4], [5]. Deletion of POT1 or CDC13 results in lethality in both mammalian and yeast cells [10]–[12]. Conditional inactivation of POT1 in mammalian cells leads to telomeric ssDNA generation, ATR-dependent checkpoint activation, deregulation of telomerase, telomere recombination and telomere fusion [4]. Similarly, acute inactivation of Cdc13 by the temperature sensitive cdc13-1 allele in budding yeast induces telomeric ssDNA generation, recombination and Mec1 (ATR orthologue) dependent cell cycle arrest [10], [13].
The RecQ helicases are a family of highly conserved proteins involved in the maintenance of genome stability and at telomeres [14]. There are five RecQ helicases in humans. Loss of function of three of these results in cancer predisposition disorders Bloom's syndrome (BS, defective in BLM), Werner's syndrome (WS, defective in WRN) and Rothmund Thomson syndrome (RTS, defective in RECQ4) [14]. WS and RTS are also characterised by various features of premature ageing. There is only one RecQ helicase in Saccharomyces cerevisiae - SGS1. RecQ helicases are 3′-5′ DNA helicases that unwind a variety of DNA replication and recombination structures. It is believed that in the absence of RecQ helicases, genomic rearrangements that arise due to a failure to unwind pathological DNA structures formed at stalled replication forks are a major cause for genetic instability and increases in tumourigenesis associated with these syndromes [14]. The RecQ helicases also play roles in maintaining telomeres and cells derived from WS patients show premature entry into telomere dysfunction-induced senescence, which has been proposed to be a cause for the premature ageing phenotype of WS [15]. Similarly, in yeast, deletion of SGS1 induces rapid senescence in yeast cell lacking telomerase and Sgs1 is required for the generation of certain type of recombination-dependent ALT (Alternative Lengthening of Telomere) like survivors [16]–[18]. Interestingly, the RecQ helicase in fission yeast, Rqh1 acts on dysfunctional telomeres to promote telomere breakage and entanglement [19].
Several recent studies have identified important roles for the RecQ helicase family in resection of DNA double strand breaks (DSBs) to generate ssDNA, and in generating 3′ overhangs at shortened telomeres [20]–[24]. These resection activities were partially redundant with the 5′ to 3′ exonuclease, Exo1. Since cdc13-1 induced telomere uncapping leads to telomere resection controlled by Exo1 and other, as yet unidentified, nuclease activities [25], we speculated that Sgs1 might contribute to resection and response to telomere uncapping in cdc13-1 mutants.
Here we examined the role of Sgs1 in responding to telomere capping defects in cdc13-1 mutants. We show that although Sgs1 contributes to the good growth of cdc13-1 mutants, it strongly inhibits the growth of cdc13-1 exo1Δ rad9Δ mutants. Our experiments reveal important and yet complex functions of Sgs1 in regulating the growth of budding yeast cells with telomere capping defects. Our results have implications for the role of mammalian RecQ helicases at dysfunctional telomeres.
Results
Sgs1, Exo1, and Rad9 affect cell proliferation of cdc13-1 strains
To begin to test the effect of Sgs1 on the response to telomere uncapping, we first deleted SGS1 in a cdc13-1 background. Cdc13-1 becomes increasingly non-functional at temperatures above 23°C, which leads to telomere uncapping, checkpoint activation and temperature sensitive growth [10]. We found that in marked contrast to deletion of EXO1, deletion of SGS1 made cdc13-1 cells more temperature sensitive at 26°C (Figure 1A). This suggests that Sgs1 contributes to the stability of uncapped telomeres in cdc13-1 mutants, and that the effect of Sgs1 differs from the effect of Exo1 which attacks uncapped telomeres, generates ssDNA and inhibits growth (Figure 1A) [25]. The role of Sgs1 in stabilising telomeres is consistent with other experiments in budding yeast and mammalian cells [16], [26].
Fig. 1. Sgs1, Exo1, and Rad9 affect cell proliferation of cdc13-1 strains. 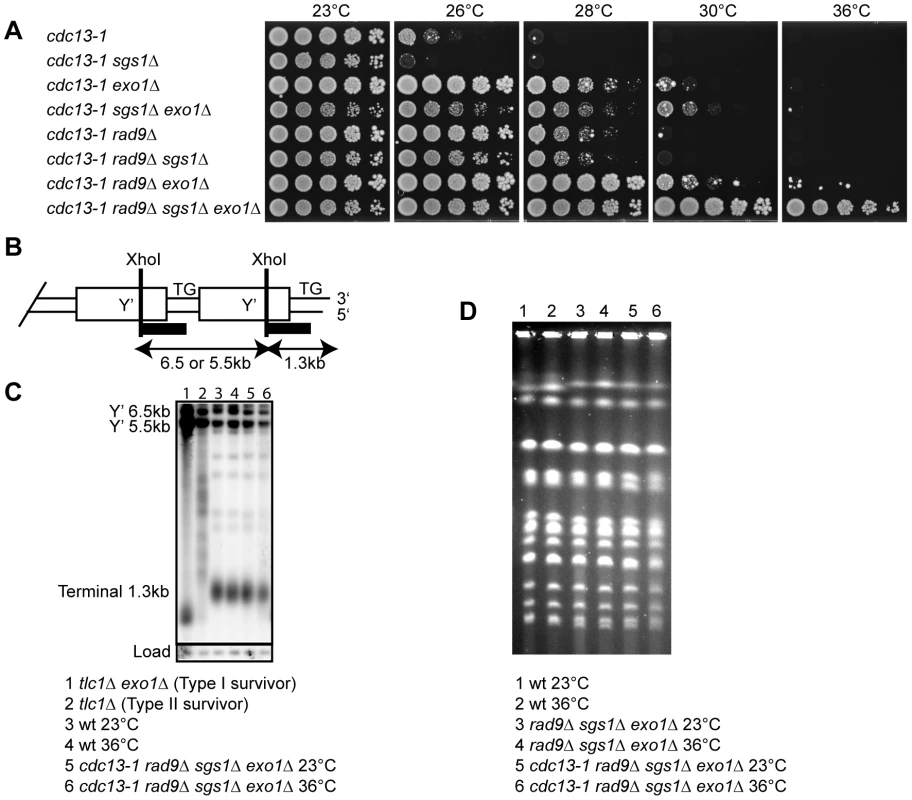
(A) Serial dilutions of yeast strains with the indicated genotypes and growing at 23°C were spotted onto YPD agar plates and incubated at the indicated temperatures for four days before being photographed. (B) Schematic representation of the end of a Y' repeat-containing yeast chromosome with position of XhoI sites and Southern blot hybridisation probe (black bar) indicated. (C) DNA was purified from the indicated strains following incubation in liquid culture overnight at 23°C or 36°C. The DNA was cut with XhoI and hybridised with a Y'+TG probe as described previously [31]. The membrane was stripped and reprobed with a CDC15 probe as described previously [31]. (D) Yeast strains with the indicated genotypes were grown in liquid cultures at 23°C or 36°C overnight before subjected to pulsed-field gel electrophoresis as described previously [42]. Since Sgs1 functions redundantly with Exo1 to generate ssDNA at DSBs and shortened telomeres [20]–[22], [24], we examined the effect of deleting both Sgs1 and Exo1 activities on growth of cdc13-1 mutants. We found that sgs1Δ exo1Δ mutants grew slightly poorer at all temperatures in both CDC13 and cdc13-1 backgrounds (Figure 1A, Figure S1). The poor growth of CDC13 sgs1Δ exo1Δ mutants has been reported previously [20], [21]. Despite the poor growth of sgs1Δ exo1Δ mutants, deletion of SGS1 allowed cdc13-1 exo1Δ strains to grow slightly better at high temperature, since more cells appeared able to divide and grow at 30°C (Figure 1A). This suggests that in the absence of Exo1, Sgs1 may play a role inhibiting cell division when telomeres uncap.
Checkpoint pathways, as well as nucleases, inhibit the growth of cdc13-1 cells at high temperature [25], [27]. We have previously shown that simultaneous deletion of EXO1 and the checkpoint gene RAD9 allow better suppression of cdc13-1 temperature sensitivity than either single deletion [25]. Strikingly, deletion of SGS1 fully suppressed the temperature sensitivity of cdc13-1 rad9Δ exo1Δ strains and allowed cdc13-1 rad9Δ sgs1Δ exo1Δ strains to form colonies at 36°C, a temperature at which Cdc13 is expected to be completely non-functional (Figure 1A, Figure S2) [10]. This suggests that deletion of SGS1, EXO1 and RAD9 allows budding yeast cells to grow in the absence of any Cdc13 activity.
It was possible that growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C was due to second site mutations/changes since we previously showed that at low rates (5×10−5) cdc13-1 exo1Δ rad9Δ strains could generate clones that can grow at 36°C [28]. Therefore to test the possibility that a second site mutation was responsible for the growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C, we performed a backcross with an independent cdc13-1 strain. We found that all cdc13-1 rad9Δ sgs1Δ exo1Δ segregants derived from the cross were able to grow at 36°C (Figure S3). This demonstrated that no independently segregating mutation contributed to growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at high temperature, and suggests that Sgs1, Exo1 and Rad9-dependent activities combine to inhibit the growth of telomere capping defective cdc13-1 mutants at 36°C.
At low rates a small fraction of telomerase-null ALT survivors that maintain telomeres by homologous recombination have also been shown to be able to survive without Cdc13 [29]. Therefore to test whether alterations in telomere structure were responsible for the good growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C, we examined telomere structure by Southern blot. As positive controls, we examined both Type I and Type II telomerase-null ALT survivors which have amplified the Y' region and the TG repeats respectively. We found that cdc13-1 rad9Δ sgs1Δ exo1Δ strains had telomere structures that looked similar to wild-type strains at both 23°C and 36°C (Figure 1B, Figure 1C and data not shown). Furthermore, we found that cdc13-1 rad9Δ sgs1Δ exo1Δ strains can grow and maintain normal telomere structure at 36°C without Rad52, suggesting that homologous recombination is not required for the survival of these strains following telomere uncapping (Figure S4). Therefore we conclude that cdc13-1 rad9Δ sgs1Δ exo1Δ strains are not ALT survivors and the growth at 36°C is independent of major alterations in telomeric DNA structure.
In Schizosaccharomyces pombe, inactivation of the telomere capping protein Pot1 results in growth crisis and generation of survivors with circular chromosomes [30]. To test whether cdc13-1 rad9Δ sgs1Δ exo1Δ strains survive growth at 36°C by circularising their chromosomes, we used pulsed-field gel electrophoresis to examine chromosome structure. Circular chromosomes do not enter pulsed field gels. We found that at 23°C, cdc13-1 rad9Δ sgs1Δ exo1Δ strains contain linear chromosomes that were indistinguishable from wild-type or rad9Δ sgs1Δ exo1Δ strains (Figure 1D). Interestingly, the chromosomes of cdc13-1 rad9Δ sgs1Δ exo1Δ strain remain linear at 36°C (Figure 1D). We conclude that the ability of cdc13-1 rad9Δ sgs1Δ exo1Δ strains to grow without telomere capping by Cdc13 is not due to chromosome circularisation.
The Mre11-Rad50-Xrs2 (MRX) complex and the nuclease Sae2 can resect DSBs in an Sgs1 and Exo1-independent pathway [20], [22]. To test whether inhibition of this pathway also has an effect on response to telomere uncapping and ability of cdc13-1 cells to grow at 36°C, we examined the effect of deleting SAE2 on growth of cdc13-1 mutants. We found that like the MRX complex [31], Sae2 has a role in the protection of uncapped telomeres as deletion of SAE2 renders cdc13-1 cells slightly more temperature sensitive at 26°C (Figure S5). Furthermore, deletion of SAE2, in combination with deletion of SGS1 or EXO1 in the presence or absence of RAD9 does not allow cdc13-1 cells to grow at 36°C (Figure S5). We were unable to analyse sgs1Δ sae2Δ exo1Δ triple mutant cells as simultaneous deletion of these genes is lethal, as reported previously [22], [24] (Figure S6). We conclude that Sae2 has a protective role at uncapped telomeres and that unlike Sgs1, deletion of SAE2 does not allow cdc13-1 rad9Δ exo1Δ cells to grow at 36°C.
Checkpoint response to telomere uncapping in the absence of Sgs1, Exo1, and Rad9
To further address the surprising ability of cdc13-1 rad9Δ sgs1Δ exo1Δ strains to grow at 36°C, we used synchronous cultures to examine two cellular responses to telomere uncapping: checkpoint activation and telomeric ssDNA generation. To examine checkpoint activation, we first quantified the fraction of cells arrested at medial nuclear division induced by cdc13-1 mutation at 36°C. A cdc15-2 mutation, which inhibits mitotic exit, was used to inhibit further cell cycle progression of any cdc13-1 cells that failed to arrest at medial nuclear division or escaped arrest and therefore progressed to reach the late nuclear division stage [27]. As expected, control cdc13-1 cells were largely arrested at medial nuclear division 80 minutes after release from G1 arrest to 36°C and the cells remained arrested for up to 4 hours (Figure 2A and 2B) [25]. A cdc13-1 sgs1Δ strain showed a similar cell cycle arrest to the cdc13-1 strain (Figure 2A and 2B). cdc13-1 exo1Δ strains showed no defect in the activation of checkpoint as most cdc13-1 exo1Δ cells were arrested at medial nuclear division after 80 minutes, but some cells escaped from the arrest at later time points, as previously reported (Figure 2A and 2B) [25]. We found that deletion of both SGS1 and EXO1 slightly reduced the number of cells arrested at medial nuclear division (to approximately 60%) after 80 minutes (Figure 2A). This is partly due to a slower cell cycle progression and also due to a mild defect in checkpoint activation as approximately 15% of cdc13-1 sgs1Δ exo1Δ cells had entered late nuclear division by 80 minutes (Figure 2B, Figure S7). At later time points, increasing fractions of cdc13-1 sgs1Δ exo1Δ cells passed through medial nuclear division and accumulated at the late nuclear division stage, presumably due to a defect in either activation or maintenance of checkpoint (Figure 2B). As expected, deletion of RAD9 completely eliminated checkpoint arrest in all these backgrounds (Figure 2A and 2B). We conclude that in the absence of both Sgs1 and Exo1, checkpoint activation/maintenance following telomere uncapping in cdc13-1 mutants is partially defective.
Fig. 2. Checkpoint response to telomere uncapping in the absence of Sgs1, Exo1, and Rad9. 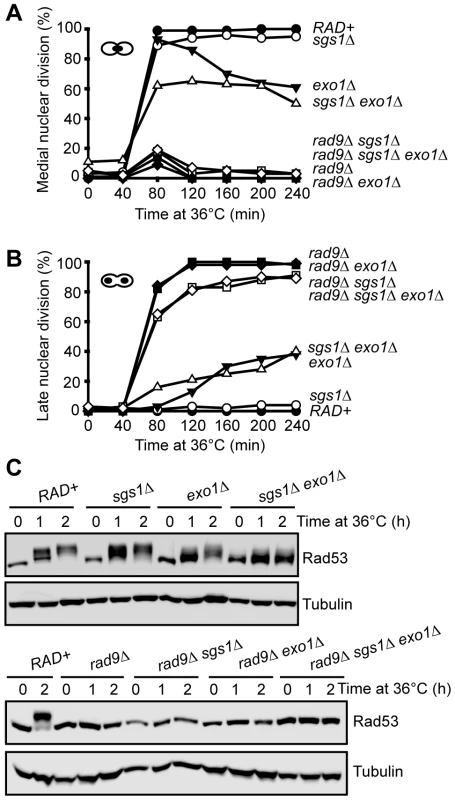
(A) Yeast strains of the indicated genotypes (all with cdc13-1 cdc15-2 bar1 mutations) were arrested in G1 at 23°C with α factor and released into 36°C, cells were collected at the indicated time points, and scored for the percentage of cells arrested in medial nuclear division using DAPI staining. (B) As in (A) but the percentage of cells arrested in late nuclear division were quantified. (C) Yeast strains of the indicated genotypes (all with cdc13-1 cdc15-2 bar1 mutations) were arrested in G1 at 23°C and released into 36°C for up to two hours. Western blot were first probed with anti-Rad53 antibody, membranes were stripped and reprobed with anti-tubulin antibody. We next examined Rad53 mobility shifts, caused by checkpoint-kinase-dependent phosphorylation, as a biochemical marker for checkpoint activation in cdc13-1 strains [32]. It has been shown that Rad53 is strongly phosphorylated two hours following telomere uncapping in cdc13-1 mutants [32]. Consistent with this finding, we observed a strong Rad53 phosphorylation shift two hours after cdc13-1 cells were shifted to 36°C (Figure 2C). Deletion of SGS1 in cdc13-1 strains had little effect on this Rad53 phosphorylation but deletion of EXO1 mildly reduced Rad53 phosphorylation (Figure 2C). Interestingly, we found that deletion of both SGS1 and EXO1 caused a more severe defect on Rad53 phosphorylation, although some residual Rad53 phosphorylation could be detected (Figure 2C). As expected, Rad53 phosphorylation was abolished when RAD9 was deleted in all strains (Figure 2C). Taken together, the cell cycle arrest and Rad53 phosphorylation experiments suggest that Sgs1 and Exo1 control parallel pathways to activate the checkpoint induced by uncapped telomeres. However, the presence of residual Rad53 phosphorylation and checkpoint arrest in cdc13-1 sgs1Δ exo1Δ strains indicates that there is at least one additional pathway to activate the checkpoint cascade after telomere uncapping in the absence of Sgs1 and Exo1.
Resection of uncapped telomeres in the absence of Sgs1, Exo1, and Rad9
The major stimulus for checkpoint cascades after telomere uncapping is ssDNA accumulation caused by 5′ to 3′ resection, generating ssDNA specifically on the TG rich, 3′ strand at telomeres [25], [33]. To study the role of Sgs1 in resection, we used Quantitative amplification of ssDNA (QAOS) [25], [33] to examine ssDNA production at two repetitive loci in subtelomeric regions and three single copy loci near the right telomere of chromosome V. We first examined the Y' repeats which are found in two thirds of yeast telomeres. We measured ssDNA accumulation at two different Y' loci, Y'600 and Y'5000, located approximately 600 bp and 5000 bp from many chromosome ends. As previously reported, we detected accumulation of ssDNA specifically on the TG strands and this ss DNA was partly dependent on Exo1 (Figure 3C and 3D, Figure S8) [25]. This ssDNA was also partly dependent on Sgs1, this was particularly clear at the Y'5000 locus (Figure 3C). The difference between ssDNA accumulation in cdc13-1 and cdc13-1 sgs1Δ cells was perhaps not as significant at Y'600 as some standard deviations overlapped (Figure 3D). Interestingly, in the absence of both Sgs1 and Exo1, telomere resection was not detectable at Y'5000, but was still detectable at Y'600 (Figure 3C and 3D, Figure S9). This suggests that Sgs1 and Exo1 independently regulate 5′-3′ resection at cdc13-1-induced uncapped telomeres, especially further away from the telomere ends at Y'5000 loci. This resection defect likely provides an explanation to the weak checkpoint activation observed in cdc13-1 sgs1Δ exo1Δ strains (Figure 2, Figure S7). At Y'600, the ssDNA generated in cdc13-1 sgs1Δ exo1Δ cells was not significantly different from that seen in cdc13-1 exo1Δ cells (Figure 3D). The presence of ssDNA at Y'600 loci in cdc13-1 sgs1Δ exo1Δ strains suggests that there is a third pathway that can resect uncapped telomeres near the telomere ends (Figure 3D). The small residual amounts of telomeric ssDNA likely provide the stimulus for checkpoint activation in cdc13-1 sgs1Δ exo1Δ strains.
Fig. 3. Resection of uncapped telomeres in the absence of Sgs1, Exo1, and Rad9. 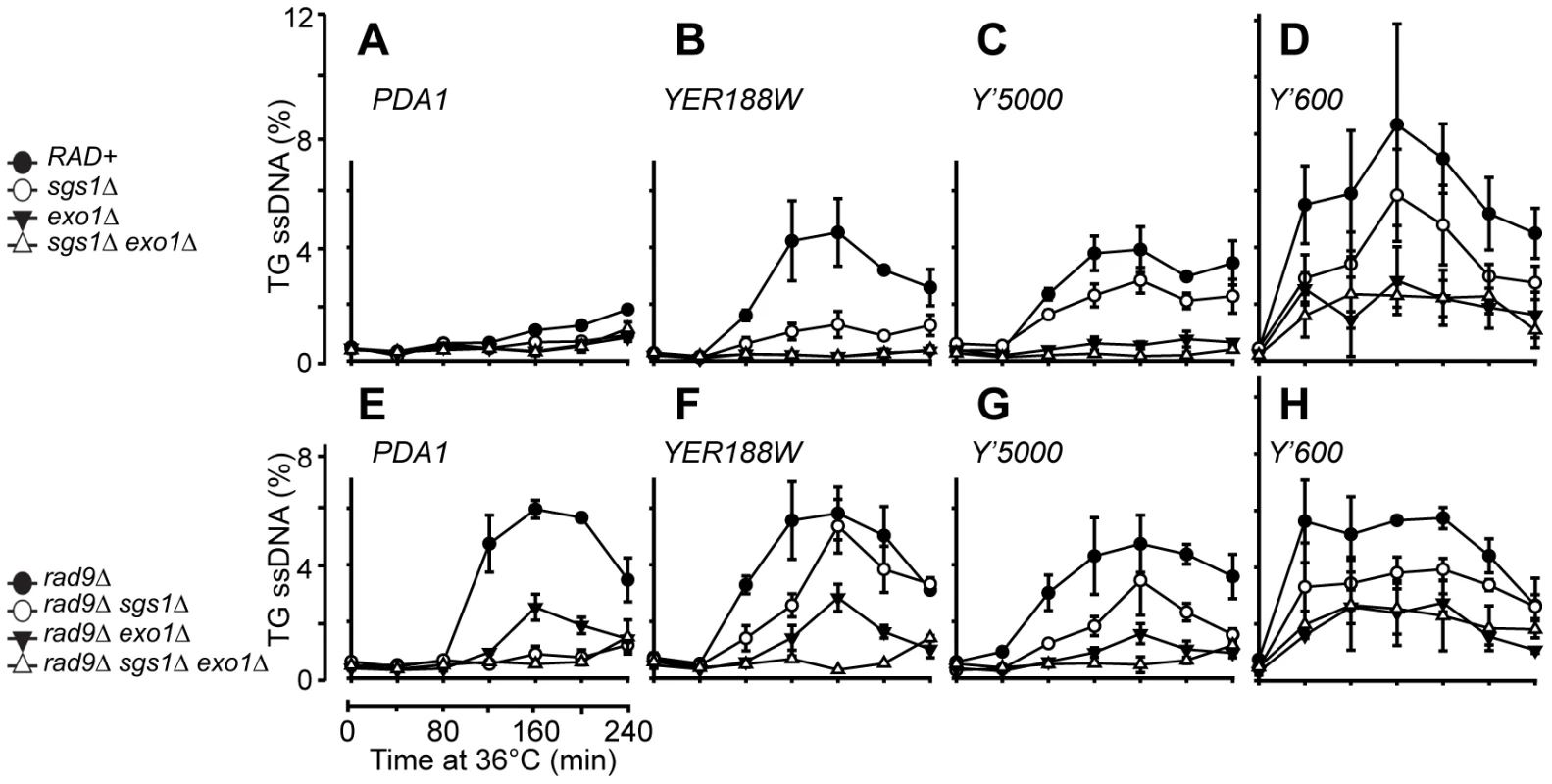
(A–H) A series of yeast strains with the indicated genotypes (all with cdc13-1 cdc15-2 bar1 mutations) were arrested in G1 at 23°C and released into 36°C to induce telomere uncapping, the amount of ssDNA at the TG strands at two repetitive telomeric locus, Y'600 and Y'5000 and two single copy loci, YER188W and PDA1 were measured by QAOS as described previously [41]. The values plotted are the mean value ± SD. It has been shown that following telomere uncapping in cdc13-1 mutants, the resection of uncapped telomeres extends into the single copy regions of chromosomes [10]. To test whether Sgs1 affects accumulation of ssDNA in single copy regions, we examined ssDNA accumulation further away from the telomeres at three single copy loci, YER188W, YER186C and PDA1 located at 8.5 kb, 14.5 kb and 29.7 kb from the right end of Chromosome V (Figure 3A and 3B, Figure S8). As previously observed, we detected ssDNA in cdc13-1 cells at both the YER188W, YER186C loci, but not at the PDA1 locus which is located further away from the telomeres (Figure 3A and 3B, Figure S8). We found that the resection at these loci were highly dependent on EXO1 as previously reported [25], but considerably less dependent on Sgs1 (Figure 3A and 3B, Figure S8). Thus our data suggests that Sgs1 and Exo1 regulate different types of nuclease activities in cdc13-1 mutants.
We next examined resection in the absence of RAD9 because Rad9 plays an important role in the inhibition of nuclease activities at uncapped telomeres [25], [27], [34]. Specifically, deletion of RAD9 allows nuclease(s) activities to generate ssDNA further away from the telomere ends, even in the absence of Exo1, which results in the resection of DNA up to 30 kb away from the telomeres [25]. We first examined ssDNA production at the Y' repeats and found that as in RAD9+ background, DNA resection of uncapped telomeres at the Y'5000 loci was totally dependent on the combined activities of Sgs1 and Exo1 as little ssDNA was detected at these loci following telomere uncapping (Figure 3G, Figure S8). This observation again supports the idea that Sgs1 and Exo1 act in two alternative pathways to resect the DNA at these loci. Consistent with what was observed in RAD9+ background, there appears to be another nuclease that can resect the uncapped telomeres near the telomere end at Y'600 in the absence of both Sgs1 and Exo1 (Figure 3H). As previously reported, we found that deletion of RAD9 allows ssDNA generation at single copy loci up to 30 kb away from the telomere ends even in the absence of Exo1 (compare cdc13-1 exo1Δ in Figure 3A and 3B and cdc13-1 rad9Δ exo1Δ in Figure 3E and 3F). Interestingly, deleting SGS1 abolished this Exo1-independent ssDNA generation (compare cdc13-1 rad9Δ exo1Δ and cdc13-1 rad9Δ sgs1Δ exo1Δ) (Figure 3E and 3F). This suggests that Rad9 inhibits an Sgs1-dependent, but Exo1-independent pathway of generating ssDNA further away from the uncapped telomere ends.
The results described above suggest that Sgs1 and Exo1 contribute to two distinct pathways of resection near the telomeres of cdc13-1 strains. Our data suggests that attenuation of telomeric DNA resection pathways by deletion of both SGS1 and EXO1, coupled with inactivation of checkpoint machinery by deletion of RAD9, is sufficient to allow cells to grow in the absence of telomere capping by Cdc13.
Cellular response to long term absence of Cdc13
Cdc13 plays at least two important roles at telomeres, one capping chromosome ends, the other recruiting telomerase [35]. cdc13-1 strains grown at 36°C would be expected to be defective in both activities. Therefore to test whether cdc13-1 rad9Δ sgs1Δ exo1Δ strains can maintain telomeres and/or tolerate long term absence of Cdc13, we grew cells for many passages at 36°C by repeatedly restreaking them onto plates which were incubated at 36°C (Figure 4A). We found that cdc13-1 rad9Δ sgs1Δ exo1Δ strains showed progressively slower growth with repeated passage at 36°C, but that some strains eventually accumulated faster growing colonies (Figure 4A). This pattern of growth is similar to telomerase deficient tlc1Δ cells which enter growth senescence due to shortened telomeres, and eventually accumulate post senescent survivors which survive by utilising recombination-dependent mechanisms to maintain their telomeres (Figure 4A) [36], [37].
Fig. 4. Cellular response to long term absence of Cdc13. 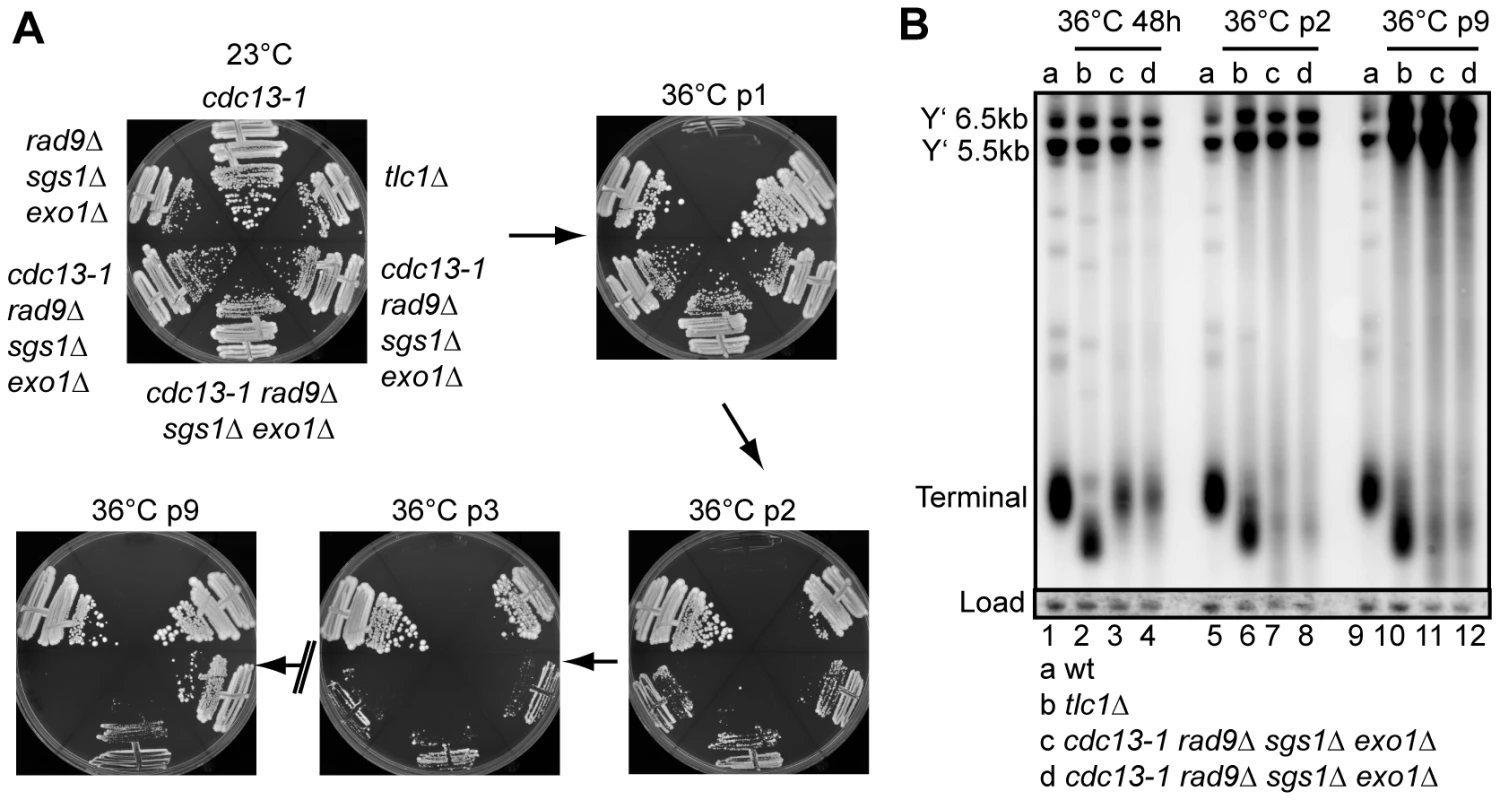
(A) Yeast strains of the indicated genotype were streaked onto YPD agar plates and the plates were incubated at 23°C or 36°C for three days. The strains were then photographed and restreaked onto another YPD plate. This cycle is repeated for nine times (p = passage). (B) DNA was purified from the strains in (A) or a wild-type strain following further incubation in liquid culture for 48 hours at 36°C. Southern blots were performed as in Figure 1C. To test whether cdc13-1 rad9Δ sgs1Δ exo1Δ strains growing at 36°C were behaving like telomerase deficient strains, we examined the telomere structure by Southern blot. At early passage, cdc13-1 rad9Δ sgs1Δ exo1Δ strains grown at 36°C had a comparatively normal telomeric DNA structure, but the terminal telomere fragments were weak (Figure 4B, Figure 1C). This confirms that growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C does not depend on gross changes in telomere structure. By passage 2, the terminal telomere fragments of these strains had become shorter and weaker, similar to but distinct from the telomeres in a tlc1Δ strain, possibly explaining the poor growth of these strains at this passage at 36°C (Figure 4A and 4B). At passage 9, when some of the cdc13-1 rad9Δ sgs1Δ exo1Δ strains grew well they showed alternative telomere structures similar to that in a Type I tlc1Δ survivor and had amplified the 5.5 and 6.5 kb Y' repeats (Lanes 10-12, Figure 4B) [36]. Unlike the Type I tlc1Δ survivor shown, the terminal fragments in cdc13-1 rad9Δ sgs1Δ exo1Δ survivors remained very weak. We conclude that initial growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C does not depend on gross alteration in telomere structure. However, continued growth without functional Cdc13 results in telomere shortening, cellular senescence and generation of post-senescence survivors with altered telomere structures.
Cdc13 is dispensable for cell viability in the absence of Sgs1, Exo1, and Rad9
The growth of cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C suggested that CDC13 might not be essential in cells lacking SGS1, EXO1 and RAD9. To directly test this hypothesis, we created diploid strains heterozygous for sgs1Δ, exo1Δ, rad9Δ and cdc13Δ and dissected tetrads following sporulation (Figure 5A, Figure S10). We found that cdc13Δ rad9Δ sgs1Δ exo1Δ spores were viable and formed colonies at a comparable frequency (average 89%) to CDC13 rad9Δ sgs1Δ exo1Δ spores, whereas other cdc13Δ genotypes were unviable (Figure 5A, Figure S10, Table S1). This result shows that deleting SGS1, EXO1 and RAD9 genes from yeast is sufficient to allow yeast cells to grow and divide in the absence of the usually essential telomere capping protein Cdc13. We found that the telomeres of cdc13Δ rad9Δ sgs1Δ exo1Δ strains shortened very quickly and the terminal fragments of fresh cdc13Δ rad9Δ sgs1Δ exo1Δ strains (marked with asterisk) were already shortened when first examined (Figure 5B). Similar to cdc13-1 rad9Δ sgs1Δ exo1Δ strains at 36°C, these terminal fragments of cdc13Δ rad9Δ sgs1Δ exo1Δ strains were very weak (Figure 5B). We found that these cdc13Δ rad9Δ sgs1Δ exo1Δ cells show variable rates of senescence, and faster growing ALT survivors that can grow for at least eight passages without Cdc13 eventually emerged (Figure S11, Figure S12). We conclude that Cdc13 is not an essential telomere capping protein in rad9Δ sgs1Δ exo1Δ strains, but that such strains eventually maintain telomeres like telomerase deficient cells.
Fig. 5. Cdc13 is dispensable for cell viability in the absence of Sgs1, Exo1, and Rad9. 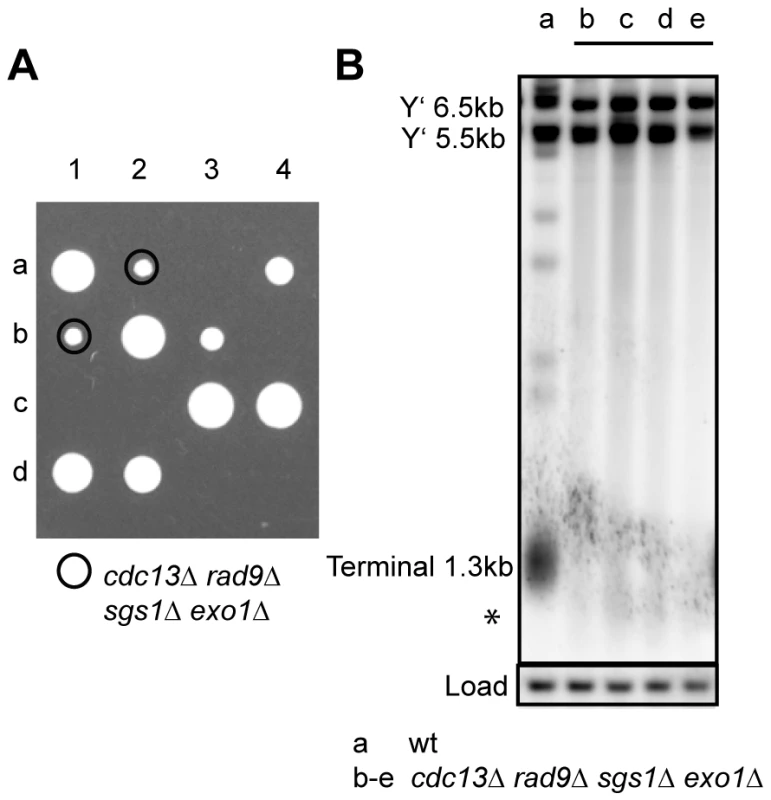
(A) Spores from a yeast strain heterozygous for cdc13Δ, sgs1Δ, exo1Δ and rad9Δ were dissected onto YPD agar plate and the plates were incubated at 23°C for five days before being photographed. Four tetrads (labelled 1–4) are shown (see Figure S10 for full genotype). (B) DNA was purified from fresh cdc13Δ sgs1Δ exo1Δ rad9Δ strains or control strains following further incubation in liquid culture for 48 hours at 23°C. Southern blots were performed as in Figure 1C. Discussion
Telomere capping protects the ends of linear chromosomes from the activation of DDR. However, cell division-induced telomere shortening in human somatic cells is believed to lead to progressive loss of telomere capping, and causes chromosome ends to be recognised as DNA breaks, which consequently activate a DDR and cell cycle arrest [4]. This telomere dysfunction-induced cell cycle arrest is thought to contribute to the establishment of replicative cellular senescence and the ageing process, and represent an important barrier to tumour formation [2], [38], [39]. Thus it is important to understand the cellular response to telomere uncapping.
Budding yeast temperature sensitive cdc13-1 mutants have been an informative model to understand the eukaryotic cell response to telomere uncapping. It was shown previously that at low rates cdc13-1 rad9Δ exo1Δ cells (5×10−5) or telomerase deficient survivors (8×10−5 to 4×10−2) were able to grow without telomere capping by Cdc13, but the nature of any other genetic or other change necessary to permit growth was unclear [28], [29]. Here we show that the simultaneous deletion of SGS1, EXO1 and RAD9 is sufficient to make Cdc13 dispensable for cell proliferation, as most cdc13Δ rad9Δ sgs1Δ exo1Δ cells are viable. This indicates that the essential function of telomere capping in yeast is to counteract the proliferative barrier exerted by specific components of the DDR. Our data suggests that attenuation of resection stimulating activities of Sgs1 and Exo1, together with DNA damage checkpoint inactivation is sufficient to allow yeast cells to grow in the absence of telomere capping by Cdc13. This suggests that in the absence of these proteins, either telomere capping is not required or that an alternative capping strategy can function in the absence of Cdc13. Intriguingly, the telomeres in cdc13-1 rad9Δ sgs1Δ exo1Δ strains (at 36°C) and cdc13Δ rad9Δ sgs1Δ exo1Δ strains look different to those in wild-type strains, as the telomeres have very weak terminal restriction fragments. Even though we showed that cdc13-1 rad9Δ sgs1Δ exo1Δ strains do not require Rad52 to survive telomere uncapping at 36°C, we cannot rule out a role for homologous recombination proteins at telomeres in cdc13Δ rad9Δ sgs1Δ exo1Δ strains.
Besides capping telomeres, Cdc13 is also required for the recruitment of telomerase [35], and as expected we found that in the absence of Cdc13, telomeres eventually shorten and are then maintained by the alternative lengthening of telomere (ALT) mechanisms, usually seen in telomerase deficient cells. Our work highlights the plasticity of the telomere cap and shows how modulation of the DDR can provide an important mechanism to overcome the proliferative barrier induced by dysfunctional telomeres. As telomere uncapping-induced cellular senescence represents an important proliferative barrier to prevent cancer formation, the mechanisms described here could be relevant to understanding the malignant transformation of human cells [2], [3].
The RecQ helicases, including Sgs1, are a family of highly conserved protein involved in the maintenance of genome stability and suppression of cancer formation in humans [14]. It is believed that in the absence of RecQ helicases, genome rearrangements that arise at stalled replication forks are a major cause for the increased tumorigenesis [14]. RecQ helicases also participate in the maintenance of telomeres, for example, deletion of SGS1 induces rapid senescence in yeast cells lacking telomerase and cells derived from WS patients show premature entry into telomere dysfunction-induced senescence [15], [16]. Here we report a similar role for Sgs1 in the protection of uncapped telomeres as deleting SGS1 makes cdc13-1 cells more temperature sensitive. However, paradoxically, we also show that Sgs1 plays a role in the resection of uncapped telomeres, an activity that is modulated by the checkpoint protein Rad9. This Sgs1-dependent resection activity leads to a critical role of Sgs1 in the inhibition of cell proliferation when telomeres uncap in the absence of Exo1 and Rad9. It is tempting to speculate that in certain genetic contexts RecQ helicase may also inhibit growth of mammalian cells with telomere capping defects, and a defect in this role could contribute to increased levels of tumorigenesis in BS, WS and RTS patients.
Previous studies have suggested that following telomere uncapping in cdc13-1 mutants, the telomeres were resected by three somewhat redundant nuclease activities, Exo1, ExoX and ExoY [25]. ExoX was defined as a Rad9-inhibited nuclease that was strongly dependent on Rad24 [25]. ExoY was a nuclease that acts on uncapped telomeres in the absence of Exo1 and Rad24 (ExoX). In this study, we have identified Sgs1 as a protein that shares similar properties with ExoX, because it is required for the resection of uncapped telomeres in an Exo1-independent pathway and its activity is inhibited by Rad9. Since the helicase Sgs1 acts with a nuclease Dna2 in the resection of DSBs and at shortened telomeres [20], [24], we speculate that Sgs1 and Dna2 might work together to regulate resection at uncapped telomeres (may contribute to ExoX). We also found that in the absence of Sgs1 and Exo1, ssDNA still accumulated at cdc13-1 induced uncapped telomeres, consistent with the existence of another nuclease (possibly ExoY). It is interesting to note that the Mre11-Rad50-Xrs2 (MRX) complex and the nuclease Sae2 can resect DSBs in an Sgs1 and Exo1-independent pathway [20], [22]. Thus a candidate for ExoY is MRX/Sae2. However, we have previously found that the MRX complex plays a capping role at telomeres, rather than contributing to resection [31]. Similarly, we found that Sae2 also has a protective role at uncapped telomeres. Therefore we believe that like the MRX complex, Sae2 is unlikely to be ExoY. Thus although uncapped telomeres share significant similarities with DSBs, there are also clearly significant differences. Future studies will be required to resolve the roles of the different types of nucleases and nuclease regulators at uncapped telomeres and DSBs.
Materials and Methods
Yeast strains
All experiments were performed using Saccharomyces cerevisiae W303 strains as listed in Table S2. Gene disruptions of SGS1 and CDC13 were constructed by a one step PCR-mediated method using kanMX and hphMX cassettes respectively [40]. All the yeast strains were generated by standard genetic crosses.
Spot tests
Single colonies were inoculated into 2 ml YPD and incubated at 23 °C overnight until saturation. Five-fold serial dilution of the cultures were spotted onto agar plates using a 48 or 96-prong replica plating device. Plates were incubated for 2–4 days at different temperatures before being photographed using a SPimager (S&P Robotics).
Protein extraction and western blot analysis
Protein extracts were prepared by using TCA extraction method, as previously described [32]. Briefly, cells were collected, washed in water, resuspended in 10% TCA and broken using glass beads. Protein suspensions were then resuspended in Laemmli buffer. Samples were boiled for three minutes, centrifuged for 10 minutes and the supernatant used as the protein extract. For Western blotting, proteins were separated on 7.5% SDS-PAGE. Anti-Rad53 and anti-tubulin antibodies were from Dan Durocher, Toronto and Keith Gull, Oxford respectively.
Analysis of telomere structure
DNA was purified from yeast strains following incubation in liquid culture for 24 or 48 hours. The DNA was cut with XhoI, run on a 0.8% (0.5xTBE) gel at 1V/cm overnight and transferred to a Magna nylon membrane. The membrane was then hybridised with a Y'+TG probe (synthesized using DIG-High Prime Labelling and Detection Kit (Roche)) as described previously [31]. The membrane was stripped and reprobed with a CDC15 probe as described previously [31].
Cell cycle analysis and single-stranded DNA measurement
Yeast strains with cdc13-1 cdc15-2 bar1 mutations were arrested in G1 at 23°C and released into 36°C to induce telomere uncapping, cell cycle position were scored using DAPI staining on a Nikon Eclipse 50i microscope. The amount of ssDNA at the TG and the AC strands at telomeric and single copy loci were measured by QAOS (quantitative amplification of single-stranded DNA) as described previously [41].
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis were carried out as described previously with the following modification: gels were run at 6.0 V/cm with a switch time of 70 seconds for 15 hours and 120 seconds for 11 hours [42].
Supporting Information
Zdroje
1. VerdunRE
KarlsederJ
2007
Replication and protection of telomeres.
Nature
447
924
931
2. d'Adda di FagagnaF
2008
Living on a break: cellular senescence as a DNA-damage response.
Nat Rev Cancer
8
512
522
3. StewartSA
WeinbergRA
2006
Telomeres: cancer to human aging.
Annu Rev Cell Dev Biol
22
531
557
4. PalmW
de LangeT
2008
How shelterin protects mammalian telomeres.
Annu Rev Genet
42
301
334
5. de LangeT
2009
How telomeres solve the end-protection problem.
Science
326
948
952
6. MiyakeY
NakamuraM
NabetaniA
ShimamuraS
TamuraM
2009
RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway.
Mol Cell
36
193
206
7. WellingerRJ
2009
The CST complex and telomere maintenance: the exception becomes the rule.
Mol Cell
36
168
169
8. SurovtsevaYV
ChurikovD
BoltzKA
SongX
LambJC
2009
Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes.
Mol Cell
36
207
218
9. LydallD
2009
Taming the tiger by the tail: modulation of DNA damage responses by telomeres.
Embo J
28
2174
2187
10. GarvikB
CarsonM
HartwellL
1995
Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint.
Mol Cell Biol
15
6128
6138
11. HockemeyerD
DanielsJP
TakaiH
de LangeT
2006
Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres.
Cell
126
63
77
12. WuL
MultaniAS
HeH
Cosme-BlancoW
DengY
2006
Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres.
Cell
126
49
62
13. GrandinN
DamonC
CharbonneauM
2001
Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination.
Embo J
20
6127
6139
14. ChuWK
HicksonID
2009
RecQ helicases: multifunctional genome caretakers.
Nat Rev Cancer
9
644
654
15. OpreskoPL
2008
Telomere ResQue and preservation–roles for the Werner syndrome protein and other RecQ helicases.
Mech Ageing Dev
129
79
90
16. LeeJY
KozakM
MartinJD
PennockE
JohnsonFB
2007
Evidence that a RecQ helicase slows senescence by resolving recombining telomeres.
PLoS Biol
5
e160
doi:10.1371/journal.pbio.0050160
17. HuangP
PrydeFE
LesterD
MaddisonRL
BortsRH
2001
SGS1 is required for telomere elongation in the absence of telomerase.
Curr Biol
11
125
129
18. JohnsonFB
MarciniakRA
McVeyM
StewartSA
HahnWC
2001
The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase.
Embo J
20
905
913
19. RogO
MillerKM
FerreiraMG
CooperJP
2009
Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres.
Mol Cell
33
559
569
20. ZhuZ
ChungWH
ShimEY
LeeSE
IraG
2008
Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends.
Cell
134
981
994
21. GravelS
ChapmanJR
MagillC
JacksonSP
2008
DNA helicases Sgs1 and BLM promote DNA double-strand break resection.
Genes Dev
22
2767
2772
22. MimitouEP
SymingtonLS
2008
Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing.
Nature
455
770
774
23. NimonkarAV
OzsoyAZ
GenschelJ
ModrichP
KowalczykowskiSC
2008
Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair.
Proc Natl Acad Sci U S A
105
16906
16911
24. BonettiD
MartinaM
ClericiM
LucchiniG
LongheseMP
2009
Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres.
Mol Cell
35
70
81
25. ZubkoMK
GuillardS
LydallD
2004
Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants.
Genetics
168
103
115
26. CrabbeL
VerdunRE
HaggblomCI
KarlsederJ
2004
Defective telomere lagging strand synthesis in cells lacking WRN helicase activity.
Science
306
1951
1953
27. LydallD
WeinertT
1995
Yeast checkpoint genes in DNA damage processing: implications for repair and arrest.
Science
270
1488
1491
28. ZubkoMK
LydallD
2006
Linear chromosome maintenance in the absence of essential telomere-capping proteins.
Nat Cell Biol
8
734
740
29. LarriveeM
WellingerRJ
2006
Telomerase - and capping-independent yeast survivors with alternate telomere states.
Nat Cell Biol
8
741
747
30. BaumannP
CechTR
2001
Pot1, the putative telomere end-binding protein in fission yeast and humans.
Science
292
1171
1175
31. FosterSS
ZubkoMK
GuillardS
LydallD
2006
MRX protects telomeric DNA at uncapped telomeres of budding yeast cdc13-1 mutants.
DNA Repair (Amst)
5
840
851
32. MorinI
NgoHP
GreenallA
ZubkoMK
MorriceN
2008
Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response.
Embo J
27
2400
2410
33. BoothC
GriffithE
BradyG
LydallD
2001
Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction.
Nucleic Acids Res
29
4414
4422
34. LazzaroF
SapountziV
GranataM
PellicioliA
VazeM
2008
Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres.
Embo J
27
1502
1512
35. PennockE
BuckleyK
LundbladV
2001
Cdc13 delivers separate complexes to the telomere for end protection and replication.
Cell
104
387
396
36. TengSC
ZakianVA
1999
Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae.
Mol Cell Biol
19
8083
8093
37. LundbladV
BlackburnEH
1993
An alternative pathway for yeast telomere maintenance rescues est1 - senescence.
Cell
73
347
360
38. GorgoulisVG
VassiliouLV
KarakaidosP
ZacharatosP
KotsinasA
2005
Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions.
Nature
434
907
913
39. HerbigU
FerreiraM
CondelL
CareyD
SedivyJM
2006
Cellular senescence in aging primates.
Science
311
1257
40. GoldsteinAL
McCuskerJH
1999
Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae.
Yeast
15
1541
1553
41. ZubkoMK
MaringeleL
FosterSS
LydallD
2006
Detecting repair intermediates in vivo: effects of DNA damage response genes on single-stranded DNA accumulation at uncapped telomeres in budding yeast.
Methods Enzymol
409
285
300
42. MaringeleL
LydallD
2006
Pulsed-field gel electrophoresis of budding yeast chromosomes.
Methods Mol Biol
313
65
73
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 8- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- A Model for Damage Load and Its Implications for the Evolution of Bacterial Aging
- Mutation in the Gene Encoding Ubiquitin Ligase LRSAM1 in Patients with Charcot-Marie-Tooth Disease
- Identification of the Bovine Arachnomelia Mutation by Massively Parallel Sequencing Implicates Sulfite Oxidase (SUOX) in Bone Development
- Did Genetic Drift Drive Increases in Genome Complexity?
- The 5p15.33 Locus Is Associated with Risk of Lung Adenocarcinoma in Never-Smoking Females in Asia
- An Alpha-Catulin Homologue Controls Neuromuscular Function through Localization of the Dystrophin Complex and BK Channels in
- Epigenetically-Inherited Centromere and Neocentromere DNA Replicates Earliest in S-Phase
- Survival and Growth of Yeast without Telomere Capping by Cdc13 in the Absence of Sgs1, Exo1, and Rad9
- Tuberous Sclerosis Complex 1 Regulates dE2F1 Expression during Development and Cooperates with RBF1 to Control Proliferation and Survival
- Disease-Associated Mutations That Alter the RNA Structural Ensemble
- The Transcriptomes of Two Heritable Cell Types Illuminate the Circuit Governing Their Differentiation
- Inactivation of VCP/ter94 Suppresses Retinal Pathology Caused by Misfolded Rhodopsin in
- Multiple Independent Loci at Chromosome 15q25.1 Affect Smoking Quantity: a Meta-Analysis and Comparison with Lung Cancer and COPD
- Transcriptional Regulation by CHIP/LDB Complexes
- Conserved Role of in Ethanol Responses in Mutant Mice
- A Global Overview of the Genetic and Functional Diversity in the Pathogenicity Island
- Common Inherited Variation in Mitochondrial Genes Is Not Enriched for Associations with Type 2 Diabetes or Related Glycemic Traits
- Extracellular Dopamine Potentiates Mn-Induced Oxidative Stress, Lifespan Reduction, and Dopaminergic Neurodegeneration in a BLI-3–Dependent Manner in
- Genetic Analysis of Baker's Yeast Msh4-Msh5 Reveals a Threshold Crossover Level for Meiotic Viability
- Genome-Wide Association Studies of Serum Magnesium, Potassium, and Sodium Concentrations Identify Six Loci Influencing Serum Magnesium Levels
- Something New: An Interview with Radoje Drmanac
- The Extinction Dynamics of Bacterial Pseudogenes
- Microtubule Actin Crosslinking Factor 1 Regulates the Balbiani Body and Animal-Vegetal Polarity of the Zebrafish Oocyte
- Consistent Association of Type 2 Diabetes Risk Variants Found in Europeans in Diverse Racial and Ethnic Groups
- Transmission of Mitochondrial DNA Diseases and Ways to Prevent Them
- Telomere Disruption Results in Non-Random Formation of Dicentric Chromosomes Involving Acrocentric Human Chromosomes
- Chromosome Axis Defects Induce a Checkpoint-Mediated Delay and Interchromosomal Effect on Crossing Over during Drosophila Meiosis
- Dynamic Chromatin Organization during Foregut Development Mediated by the Organ Selector Gene PHA-4/FoxA
- Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction
- A Wnt-Frz/Ror-Dsh Pathway Regulates Neurite Outgrowth in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Identification of the Bovine Arachnomelia Mutation by Massively Parallel Sequencing Implicates Sulfite Oxidase (SUOX) in Bone Development
- Common Inherited Variation in Mitochondrial Genes Is Not Enriched for Associations with Type 2 Diabetes or Related Glycemic Traits
- A Model for Damage Load and Its Implications for the Evolution of Bacterial Aging
- Did Genetic Drift Drive Increases in Genome Complexity?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



