-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Identification of Effective Subdominant Anti-HIV-1 CD8+ T Cells Within Entire Post-infection and Post-vaccination Immune Responses
Attempts to develop an HIV vaccine that elicits potent cell-mediated immunity have so far been unsuccessful. This is due in part to the use of immunogens that appear to recapitulate responses induced naturally by HIV that are, at best, partially effective. We previously showed that the capacity of CD8+ T cells from patients to block HIV replication in culture is strongly correlated with HIV control in vivo, therefore, we investigated the virological determinants of potent CD8+ T cell inhibitory activity. We observed that CD8+ T cells from patients with naturally low plasma viral loads (viremic controllers) were better able to inhibit the replication of diverse HIV strains in vitro than CD8+ T cells from HIV-noncontroller patients. Importantly, we also found that the potency of the antiviral activity in the latter group was strongly correlated with recognition of selected regions across the viral proteome that are critical to viral fitness. Vaccines that encode full-length viral proteins rarely elicited responses to these vulnerable regions. Taken together, our results provide insight into the characteristics of effective cell-mediated immune responses against HIV and how these may inform the design of better immunogens.
Published in the journal: . PLoS Pathog 11(2): e32767. doi:10.1371/journal.ppat.1004658
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004658Summary
Attempts to develop an HIV vaccine that elicits potent cell-mediated immunity have so far been unsuccessful. This is due in part to the use of immunogens that appear to recapitulate responses induced naturally by HIV that are, at best, partially effective. We previously showed that the capacity of CD8+ T cells from patients to block HIV replication in culture is strongly correlated with HIV control in vivo, therefore, we investigated the virological determinants of potent CD8+ T cell inhibitory activity. We observed that CD8+ T cells from patients with naturally low plasma viral loads (viremic controllers) were better able to inhibit the replication of diverse HIV strains in vitro than CD8+ T cells from HIV-noncontroller patients. Importantly, we also found that the potency of the antiviral activity in the latter group was strongly correlated with recognition of selected regions across the viral proteome that are critical to viral fitness. Vaccines that encode full-length viral proteins rarely elicited responses to these vulnerable regions. Taken together, our results provide insight into the characteristics of effective cell-mediated immune responses against HIV and how these may inform the design of better immunogens.
Introduction
Only two HIV vaccines designed to elicit protective T cell responses have reached clinical efficacy testing, both with disappointing results [1][2][3]. The reasons for this are not completely understood, despite much accumulated knowledge regarding the characteristics of cell-mediated immune responses associated with HIV and SIV control. The limited magnitude and breadth of vaccine-induced T cell responses, particularly when compared with responses to similar vaccines in non-human primate models, the modest cytotoxic capacity of CD8+ T cells, waning of responses over time, bias towards targeting of more variable regions of the viral proteome and the modest immunogenicity of the vaccine vector regimens are all likely contributing factors [2][4][5][6][7][8]. A critical first step towards addressing this is to determine whether the antiviral efficacy of CD8+ T cells is a function of their specificity.
The HVTN 502 (Step) and 503 (Phambili) trials were a test-of-concept for induction of protective T cell responses that collectively evaluated Merck’s trivalent adenovirus type 5 HIV-1 Gag/Pol/Nef vaccine in ∼3800 subjects at high risk of HIV acquisition [1][9]. Post-hoc analyses of HVTN 502 have shown that individuals in whom vaccine-induced responses targeted ≥3 epitopes in Gag achieved a lower viral load after HIV infection than subjects without Gag responses; it is striking, however, that these subjects were a small minority among the vaccinees (<7%) [6]. While this confirms several observational studies that showed an association between HIV control and preferential recognition of Gag epitopes [10][11], the question remains as to why vaccines that express full-length Gag proteins have so far failed to induce responses that can impact on HIV replication after infection. The answer may be two-fold: first, immunodominance hierarchies of the T cell responses elicited by these vaccines often mimic those of natural infection, with ‘hotspots’ in variable and least vulnerable regions of the viral proteome [12]; second, even within Gag and other conserved proteins, not all epitopes are equal in terms of vulnerability to immune pressure, or ‘fragility’, which is defined by the capacity to maintain function in the face of genetic mutations [13]. Thus, the efficacy of cell-mediated immune responses may depend on the specific epitopes targeted, both within and outside Gag. This was demonstrated in an observational study of 950 clade B - and C-infected individuals, in whom responses to overlapping peptides (OLP) spanning the entire viral proteome were systematically analysed [14]. A ‘protective ratio’ (PR) was calculated for each OLP from the ratio of the median viral load in subjects who failed to respond to the OLP to responders. OLP with a protective ratio >1 were defined as ‘beneficial’. Of note, Gag proteins contained the majority of the beneficial regions, though not all of them, and also contained regions that were not targeted by protective responses. Together, these data support the ‘decoy’ hypothesis, which proposes that certain epitopes within the viral proteome elicit dominant yet irrelevant responses that serve to undermine effective targeting of regions of vulnerability [15]. This question will only be adequately addressed by clinical testing of rationally designed immunogens based on ‘beneficial’ regions, as proposed by Rolland et al. and Mothe et al. [15][14].
Aside from identifying specific beneficial targets, the precise mechanisms and effector functions of antiviral T cell responses that underlie heterogeneity in HIV control among infected individuals need to be defined. We showed in a prospective study that CD8+ T cell viral inhibitory activity in vitro strongly correlated with HIV control in vivo, reflected in both viral load set-point and CD4+ cell decline over time [16]. This indicates that CD8+ T cell viral inhibitory activity is expressed on a continuum and is not a discrete function that is unique to HIV controllers with protective HLA alleles, providing scope for induction of effective CD8+ T cell responses by vaccination of subjects who do not have a favourable genotype. Viral inhibition assays that use polyclonal T cell populations provide a composite measure of lytic and non-lytic activity of all circulating HIV-specific CD8+ T cells, which may be heterogeneous in their functional capacity [17][18][19][20][21][22]. This activity is detectable in acute infection in a minority but rapidly wanes, likely as a result of viral escape and / or functional impairment [23][24][21][25]. Low level activity has also been detected in HIV-naïve recipients of DNA and adenovirus type 5-vectored vaccines encoding full-length HIV proteins even though such vaccines are capable of eliciting substantial numbers of Gag - and Pol-specific cytokine-secreting T cells [23][26][3]. These observations underscore the need for better understanding of the factors that determine the potency of CD8+ T cell viral inhibitory activity.
We also showed previously that CD8+ T cell viral inhibition in chronically infected individuals did not correlate with the total magnitude of IFN-γ-positive T cell response to any single HIV protein, including Gag [16]. This was surprising, given the known associations between Gag responses and HIV control, and led us to propose the hypothesis that potent viral inhibition depends on preferential targeting of selected regions that are not limited to Gag nor predicted by conservation score alone. We hypothesised that responses to such critical regions are generally subdominant and that this may explain the lack of efficacy of T cell-inducing vaccines. To this end, we investigated CD8+ T cell-mediated inhibitory activity in a subset of HIV-positive HVTN 502 and 503 vaccine trial participants. This comprised recipients of both the vaccine and placebo who were sampled at the same time during early HIV infection (1 year). They were naïve to antiretroviral therapy (ART), with CD4 cell counts >350 cells/μl, and were not selected for low virus loads or protective HLA class I alleles. In parallel, we studied ART-naïve subjects who showed spontaneous long-term control of HIV, with plasma viral loads consistently <5000 copies/ml (viremic controllers, VC). They were sampled later in infection (median 4.5 years) and were included as a reference cohort, as potent CD8+ T cell antiviral activity has been reported in such individuals [23][26][16].
Results
Limited potency and breadth of CD8+ T cell antiviral inhibitory activity in the majority of HIV-positive vaccine and placebo recipients
CD8+ T cell antiviral activity was measured in 34 HIV-positive HVTN 502 & 503 trial participants, who were infected with clade B and C viruses respectively. They were aligned for duration of infection, early post-infection viral load and CD4+ cell counts. Only a minority had either a protective HLA class I allele (n = 7, 20%) or evidence of spontaneous viremia control, indicated by plasma viral loads consistently below 5000 copies/ml (n = 5, 15%) (Table 1). We included both vaccinees and placebos in order to maximise the number of subjects with samples available for analysis. Fourteen VC with viral loads <5000 copies/ml were studied in parallel as a reference cohort. The estimated duration of HIV infection in latter ranged from 1–11 years. Six (43%) had a protective HLA class I allele and all were presumed clade B-infected (Table 2). The inclusion of clade B and C cohorts enabled us to ascertain whether the association between CD8+ T cell inhibitory activity and HIV control was clade-independent, as suggested by our previous results [16]. However, a major goal of this study was to explore the extent of cross-clade inhibition (breadth) using a panel of laboratory-adapted and primary HIV isolates representing clades A, B and C strains, as this had not been systematically examined in HIV-positive individuals before. CD8+ T cells from HIV-positive HVTN 502 & 503 participants were tested according to PBMC availability, using at least one clade B and one clade C virus, while all VC were tested against five viral isolates (S1 Table). Among the HIV-positive trial participants in whom viral inhibitory activity against a clade-matched virus was analysed at a CD8+/CD4+ cell ratio of 2 : 1, it was not significantly different between vaccinees (n = 20) and placebo (n = 8) recipients (ranges 0–87% vs. 0–93%, p = 0.32; Fig. 1A). Because no difference was observed, analyses presented in the main were performed by combining data from both the vaccinee and placebo groups. However, the vaccinees were also analysed independently as they accounted for two-thirds of the HVTN cohorts. The data are shown in Supplementary Results, S1 Text. Whether data were combined or independent, the results were similar. Inhibition of a clade-matched virus was significantly higher among VC at CD8+/CD4+ T cell ratios of both 2 : 1 (medians 85% and 37% respectively, p <0.0001) (Fig. 1B) and 1 : 10 (medians 61% and 0% respectively, p <0.0001 (Fig. 1C). VC also showed more potent cross-clade inhibition than HVTN 502 participants when tested using a clade C virus (CD8+/CD4+ T cell ratio of 2 : 1—medians 60% vs. 14%, p = 0.002) (Fig. 1D). These differences remained significant when the placebos were excluded from the analyses (Supplementary Results, S1 Text). Cross-clade activity was analysed further using at least 3 viruses in 14 HVTN 502 & 503 participants and 14 VC. Differences in the potency and breadth of CD8+ T cell-mediated inhibitory responses in these groups are highlighted in the heatmap (Fig. 1E).
Fig. 1. CD8+ T cell antiviral inhibitory activity in HIV-positive HVTN subjects and HIV viremic controllers. 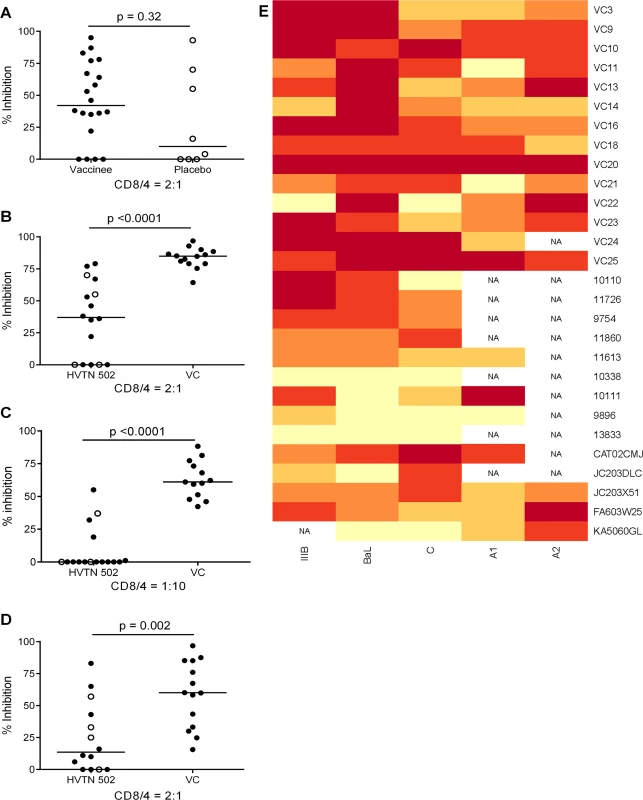
A. CD8+ T cell mediated-inhibition of a clade-matched virus was measured in 28 HIV-positive HVTN 502 & 503 subjects at a CD8+/CD4+ cell ratio of 2:1. Eight subjects are not shown as cell recoveries allowed testing at a CD8+/CD4+ cell ratio of 1:1 only (n = 6, see S1 Table) or CD4+ cells did not survive HIV superinfection (n = 2). Data are stratified by vaccine or placebo allocation. B-D. CD8+ T cells from 16 HVTN 502 participants and 14 VC (all infected with clade B viruses) were tested for inhibition of a clade B HIV isolate at CD8+/CD4+ cell ratios of 2:1 (B) and 1:10 (C) or inhibition of a clade C isolate, ES X-1936 (CD8+/CD4+ = 2:1) (D). Placebos are indicated by open circles. In all graphs, horizontal lines indicate medians. E. Heatmap showing potency and breadth of CD8+ T cell-mediated inhibition (CD8+/CD4+ cells = 2:1) among 14 viremic controllers and 14 HIV-positive HVTN 502 and 503 participants for whom at least 3 virus isolates were tested. Viral inhibition was measured on day 6 for all viruses except A2 (RW93024), for which it was measured on day 3 due its different replication kinetics (see Methods). Darker colour indicates higher inhibition; scale 0–100%, grading—20%. Tab. 1. Characteristics of HVTN 502 and 503 trial participants. 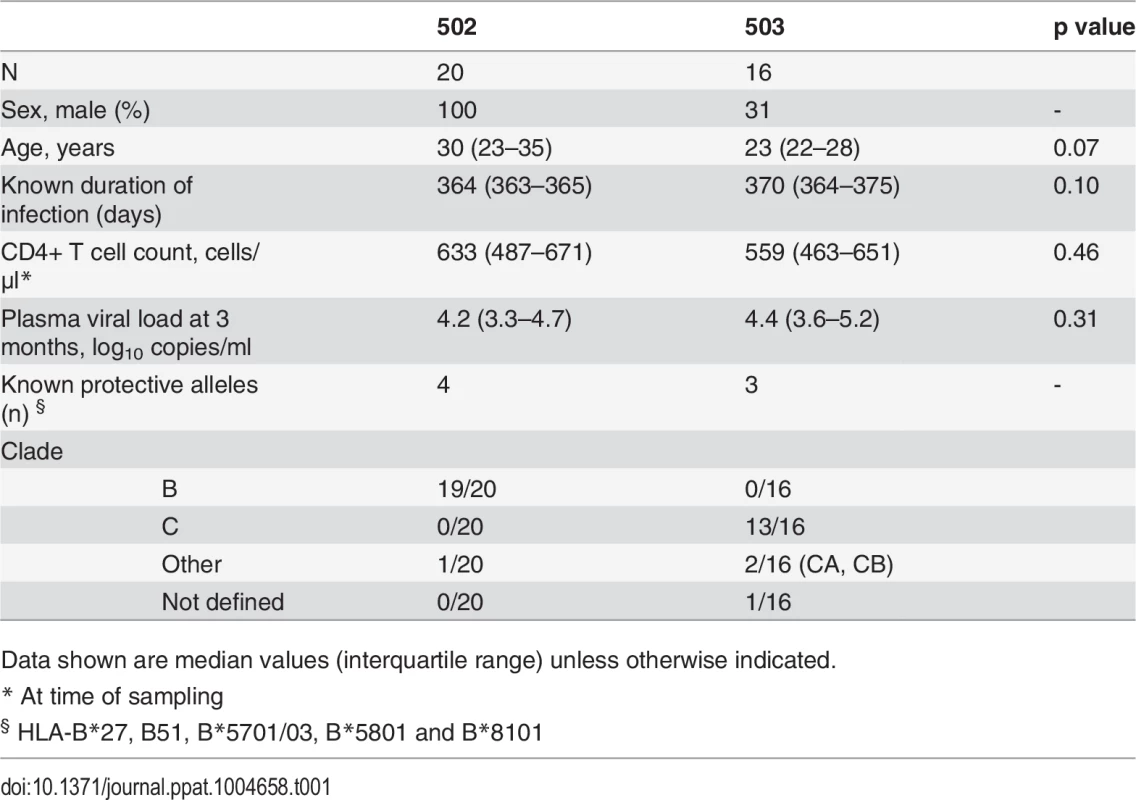
Data shown are median values (interquartile range) unless otherwise indicated. Tab. 2. Characteristics of Viremic Controller subjects. 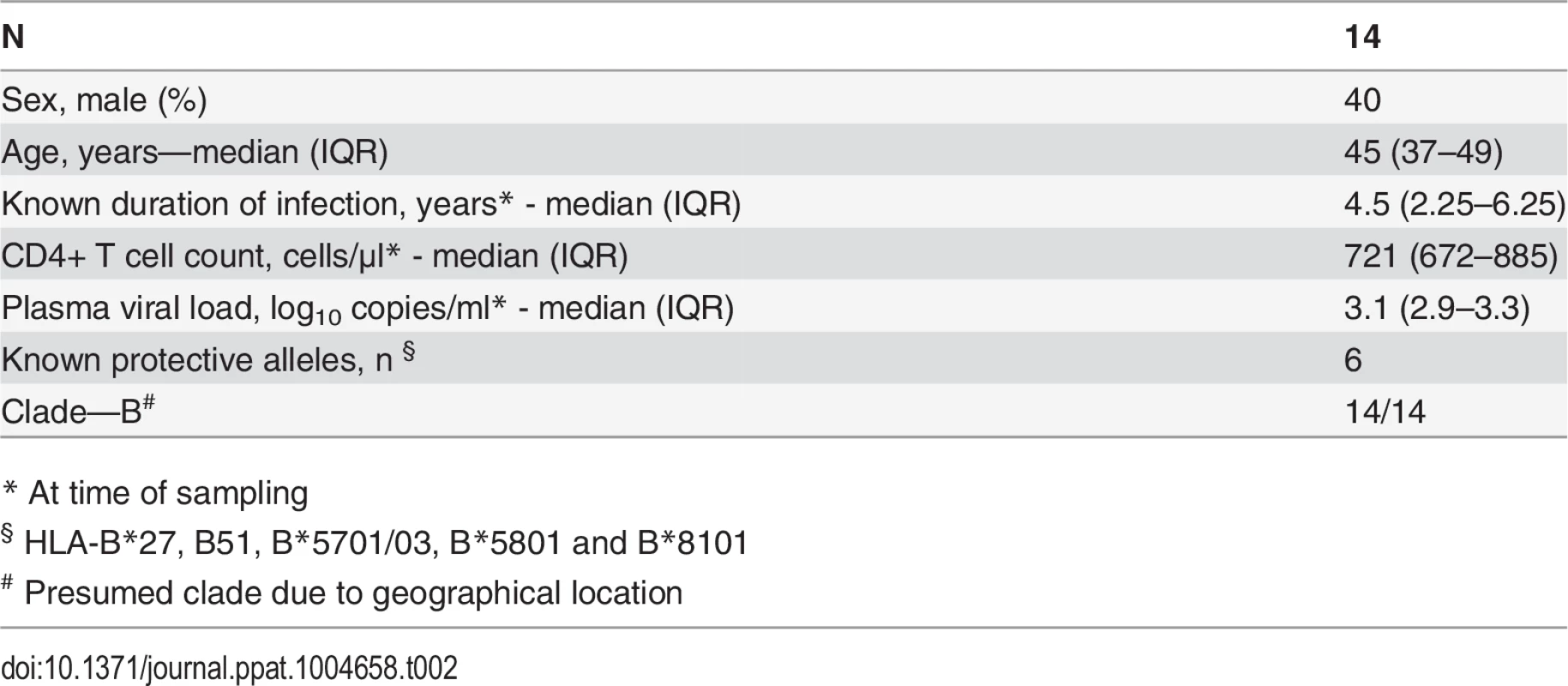
* At time of sampling We have previously reported a significant inverse relationship between CD8+ T cell antiviral activity measured 6 months post-infection in a primary HIV infection cohort and viral load set-point, a known predictor of the rate of progression to AIDS [16]. In the present study, CD8+ T cell inhibitory activity was measured later. Nevertheless, there was still a significant inverse correlation between CD8+ T cell inhibition of a clade-matched virus and viral load set-point (which was attained within 100 days of infection in the HVTN trial participants) or current viral load in the VC (r = -0.49, p = 0.0009, S1 Fig.).
CD8+ T cell antiviral potency is strongly associated with targeting of known ‘beneficial’ regions
The finding that HIV-positive trial participants showed less potent inhibition of a clade-matched virus isolate than VC was consistent with results from previous studies of early infected individuals [16][24]. Here, we extended these observations to clade-mismatched viruses. The broader CD8+ T cell inhibitory responses in VC suggested that they preferentially recognised conserved viral epitopes. However, when examining responses within the groups, we observed more potent inhibition of clade-matched than mismatched viruses in VC and HIV-positive trial participants alike. This indicated that CD8+ T cells targeting clade-specific viral epitopes must contribute to the overall potency of the response. To investigate this further, we used ex vivo IFN-γ Elispot assays to measure the magnitude of responses to two sets of overlapping 15-mer peptides. The first corresponded to the beneficial regions that were defined by Mothe et al. in clade B and clade C-infected populations (S2 and S3 Tables) and the second to a set of ‘conserved elements’ (CE) peptides that were originally defined by Rolland et al. and consisted of 7 regions in Gag p24 (S4 Table) [14][27][28]. The peptides representing beneficial regions were constituted in pools according to their previously defined protective ratio, with the first pool of each protein containing the peptides with the highest protective ratio (higher number indicating lower viral load in responders compared with non-responders) [14]. CE peptides were divided into pools A & B, also in accordance with previously observed associations with low virus loads [29] (S4 Table). To match their infecting clade, VC and HVTN 502 participants were tested with a peptide set representing beneficial regions in Clade B and HVTN 503 subjects were tested with the Clade C beneficial peptide set. All three groups were tested with the same CE peptide set. For all Elispot assays, CD8+ T cells were obtained from the same sample as that used in the viral inhibition assay (except for 2 VC in whom it was necessary to use an additional sample obtained within 1 year of the original bleed). Summed frequencies of IFN-γ-producing CD8+ T cells targeting the beneficial and CE peptides are shown in Fig. 2. The median response to beneficial peptides was 190 and 262 SFU/million CD8+ T cells for HVTN 502 and 503 groups respectively and 210 SFU/million CD8+ T cells for the VC (Fig. 2A). The median response to the CE peptides was 60 SFU/million CD8+ T cells for the combined HVTN groups and 35 SFU/million CD8+ T cells for the VC (Fig. 2B). These differences were not statistically significant, nor were there significant differences between vaccinees and placebos in terms of the magnitude of response to either beneficial (medians 198 vs. 415 SFU/million CD8+ T cells, p = 0.99) or CE peptides (medians 55 vs 60 SFU/million CD8+ T cells, p = 0.6).
Fig. 2. CD8+ T cell inhibitory activity and targeting of beneficial regions and conserved elements within the HIV proteome. 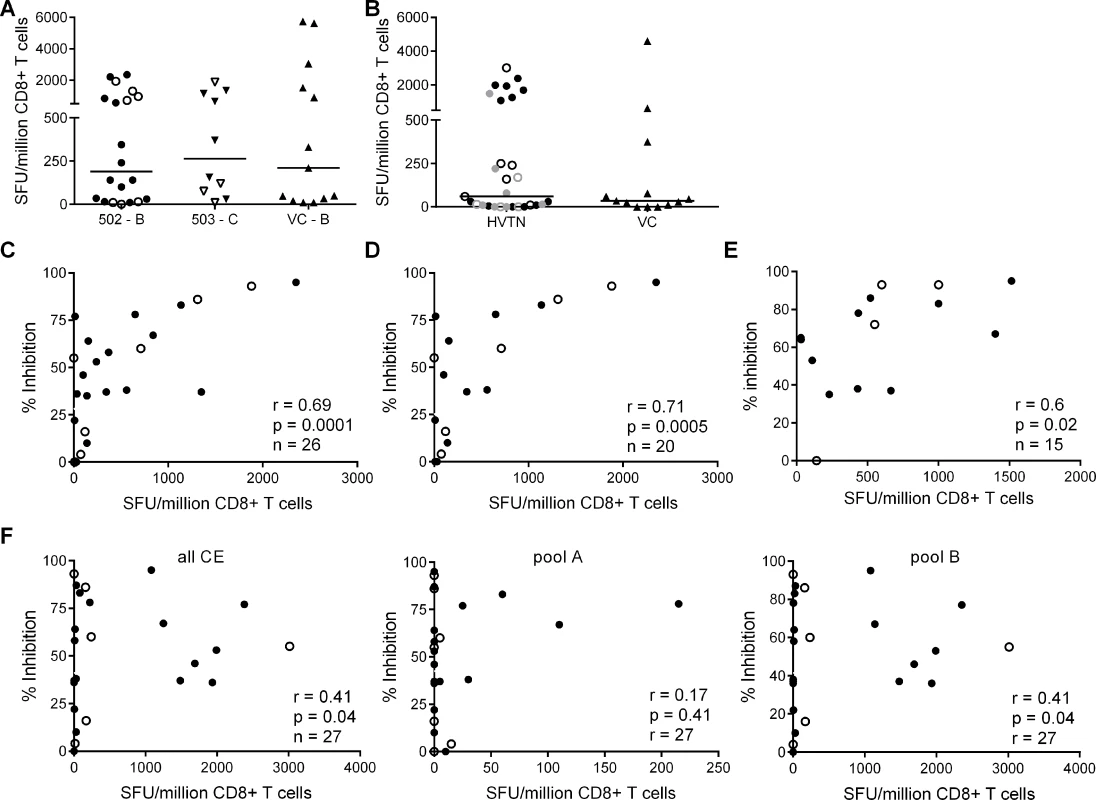
CD8+ T cell responses to peptides based on (A) clade-specific ‘beneficial’ regions and (B) Gag ‘conserved elements’ were measured by IFN-γ Elispot assays. Net responses (background subtracted) are shown; values for negative controls were median (IQR)– 10 (0–15) SFU/million CD8+ T cells. Horizontal lines indicate median values. HVTN vaccinees and placebos are shown as closed and open symbols respectively in A. In B, HVTN subjects are grouped together and represented as follows: HVTN 502—vaccinees, black closed circles, placebos, black open circles; HVTN 503—vaccinees, grey closed circles, placebos, grey open circles. VC are shown as triangles in A & B. Six HVTN 503 subjects were excluded as viral subtype data were not confirmed at the time of the analysis. One VC subject was excluded as no sample was available for Elispot assay. C. Correlation between CD8+ T cell inhibition of a clade-matched virus (CD8+/CD4+ cell ratio = 2:1) and magnitude of CD8+ T cell responses to beneficial peptides (summed) in 26 HVTN subjects. D. The analysis was repeated after removal of subjects with protective HLA class I alleles and (E) with short-term cell lines expanded from CD8+ T cells recovered from Elispot assays in 15 subjects that were then tested with individual peptides from the pools which elicited a response in the ex vivo Elispot assay. For C-F: closed circles—502 and 503 vaccinees; open circles—502 and 503 placebos. F. Correlation between CD8+ T cell inhibition (2:1 ratio) of a clade-matched virus and magnitude of CD8+ T cell responses to conserved elements peptides in 27 HVTN subjects (left panel—sum of all CE peptides, middle—CE pool A, right—CE pool B). This group of VC did not show significantly higher responses to beneficial or CE peptides than the HVTN subjects. This was unexpected in the light of previous reports but likely reflected the longer duration of infection (median 4.5 years vs. 1 year), which may be associated with loss of responses to epitopes within the regions studied, due to mutational escape [14][29][30][31][32]. For example, the two VC who were HLA-B*5701-positive did not make detectable responses to the beneficial or CE peptide pools that contained immunodominant Gag epitopes restricted by this allele (TW10 and KF11).
We next explored the relationship between virus inhibition and the magnitude of CD8+ T cell responses to the beneficial and CE regions in the HVTN subjects. We observed a strong correlation between the magnitude of T cell responses to beneficial regions and CD8+ T cell-mediated inhibition of a clade-matched virus (r = 0.69, p = 0.0001 for a CD8+/CD4+ cell ratio of 2 : 1) (Fig. 2C). This relationship was also confirmed using a lower CD8+/CD4+ cell ratio of 1 : 1 (r = 0.5, p = 0.01) and importantly, was maintained after removal of subjects with protective HLA class I alleles (HLA-B*27, B51, B*5701/03, B*5801, B*81) (r = 0.71, p = 0.0005) (Fig. 2D). Furthermore, these correlations remained statistically significant after exclusion of placebos (Supplementary Results, S1 Text). Taken together, these analyses suggested that CD8+ T cell viral inhibition of >85% (i.e. the median response in VC) was associated with a beneficial peptide response threshold of ∼1300 SFU/million CD8+ T cells. Additional support for the relationship between CD8+ T cell viral inhibition and magnitude of T cell responses to beneficial regions was obtained in a subset of subjects (n = 15) in which individual peptides were tested in cultured Elispot assays. The highest viral inhibition also correlated with the higher magnitude T cell responses to individual beneficial peptides (r = 0.61, p = 0.02, Fig. 2E). Unexpectedly, there was a weaker association between the magnitude of T cell responses to the conserved elements pools and CD8+ T cell viral inhibition (r = 0.41, p = 0.04) (Fig. 2F). This positive relationship was also maintained after exclusion of placebos (Supplementary Results, S1 Text) and was largely driven by responses to the conserved elements pool B, containing peptides spanning CE 4, 5 and 6.
We also analysed the frequency of T cell responses to the total HIV proteome as these had been measured previously by intracellular staining for IFN-γ (at a median of 5 weeks after HIV infection) after stimulation of PBMC with clade B consensus potential T cell epitope (PTE) peptide sets [2][30]. These were selected to optimise the detection of CD8+ T cell responses to circulating viruses and thus ensure accurate measurement of the maximum response [33]. The total proteome response (median) was 1.81% CD8+ T cells (Fig. 3A), with no significant difference between HVTN 502/503 vaccinees and placebos (median 2.1% and 1.5% of CD8+ T cells respectively, p = 0.23), which is similar to data obtained from chronic infection cohorts [31]. There was no correlation between CD8+ T cell antiviral activity and responses to the whole proteome, either for HVTN subjects as a whole (r = 0.14, p = 0.5) (Fig. 3B) or for the vaccinees only (Supplementary Results, S1 Text).
Fig. 3. CD8+ T cell responses to PTE peptides based on to the entire HIV proteome were measured in HIV-positive HVTN participants (502—vaccinees, black closed circles; placebos, black open circles) and (503—vaccinees, grey closed circles; placebos, grey open circles) at week 5 post-infection by intracellular staining for IFN-γ. 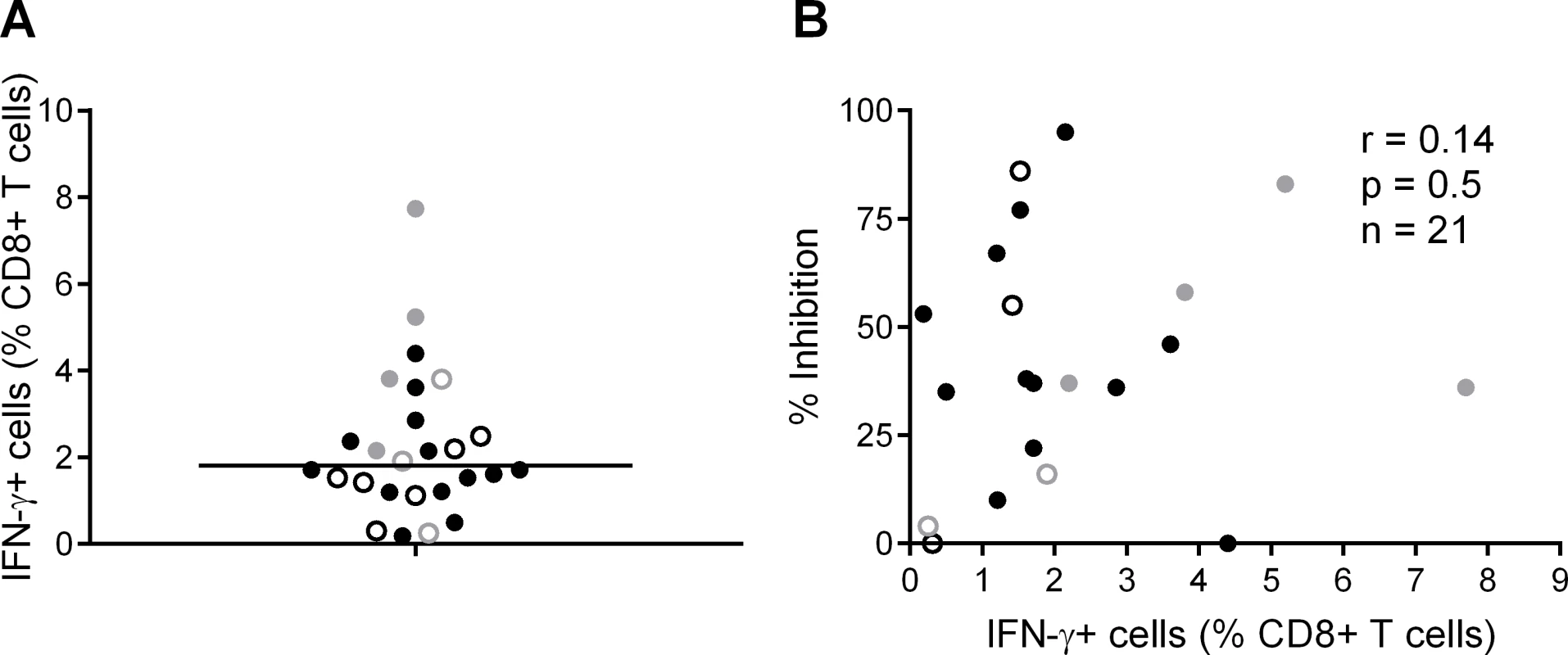
A. B. Correlation between CD8+ T cell inhibition (CD8+/CD4+ cell ratio = 2:1) of a clade-matched virus and magnitude of CD8+ T cell response to the entire HIV proteome, colour-coded as indicated in A. Five subjects are not shown in B, due to lack of viral inhibition data for the 2:1 ratio. Variation in CD8+ T cell antiviral activity is best explained by the magnitude of responses to beneficial peptides
In view of the strong correlation between CD8+ T cell antiviral activity and recognition of beneficial peptides, we explored this relationship further using a series of univariate and multivariable regression models, with CD8+ T cell antiviral activity as the dependent variable. We investigated associations with the following independent variables: 1) the Shannon entropy score for each beneficial region as a measure of its variability at the population level; 2) the magnitude of responses to beneficial regions (‘total beneficial’ response); 3) the magnitude of the Gag component of the beneficial regions (‘beneficial Gag’ response), in order to ascertain how much this contributed to the total beneficial response; 4) the magnitude of responses to CE peptides; 5) the ratio of magnitude of responses to beneficial regions to the total proteome response (relative magnitude or immunodominance) and 6) the presence of protective (‘good’) or non-protective (‘bad’) HLA class I alleles.
For our first set of models, entropy was used as the primary independent (or predictor) variable of interest since both the beneficial regions and CE regions were largely derived from conserved, i.e. low entropy regions in the viral proteome [14][28]. Thus, our first regression model included entropy as the only independent variable. Total beneficial responses, beneficial Gag responses or CE responses were then each added separately to this baseline model to ascertain whether they improved the fit of the model (as captured by a change in the model r2) and, thus, whether they were independently associated with CD8+ T cell activity. Entropy alone explained 13.5% of the variance in inhibition. Addition of the total beneficial response or the beneficial Gag response each improved the fit of the model (by 46% and 24% respectively) and the contribution of each of these was statistically significant (Table 3). By contrast, addition of the CE response had no effect (increase in model r2 of 0.1%).
Tab. 3. Univariate linear regression models to investigate associations between entropy, beneficial and conserved elements responses and % inhibition (dependent variable). 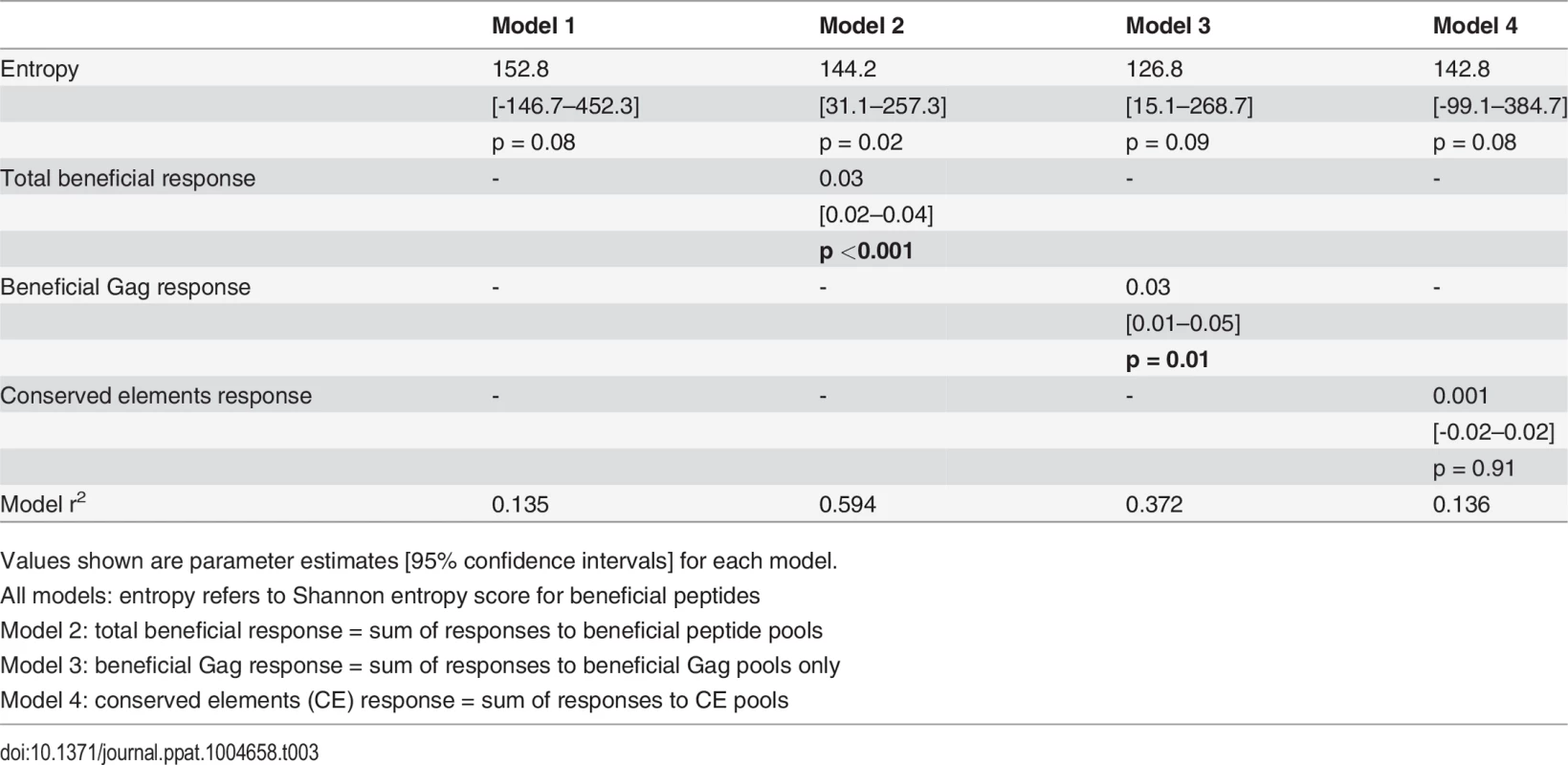
Values shown are parameter estimates [95% confidence intervals] for each model. We also constructed three multivariable regression models that included various combinations of the following factors: magnitude of total beneficial responses, relative magnitude or entropy of beneficial regions and good or bad HLA class I alleles. The combinations of covariates for these models were chosen to allow us to investigate several potential pathways for any associations, based on hypothesised interactions between absolute and relative magnitude of responses and between certain HLA alleles and entropy of epitopes restricted by these alleles. These models explained 39–49% of the variance in CD8+ T cell inhibition and all were significant as a whole. However, in each case the magnitude of the response to beneficial regions made the strongest unique contribution whereas the contribution of the other variables was not statistically significant (Table 4).
Tab. 4. Multivariate linear regression models to investigate associations between beneficial responses, entropy, HLA alleles and % inhibition (dependent variable). 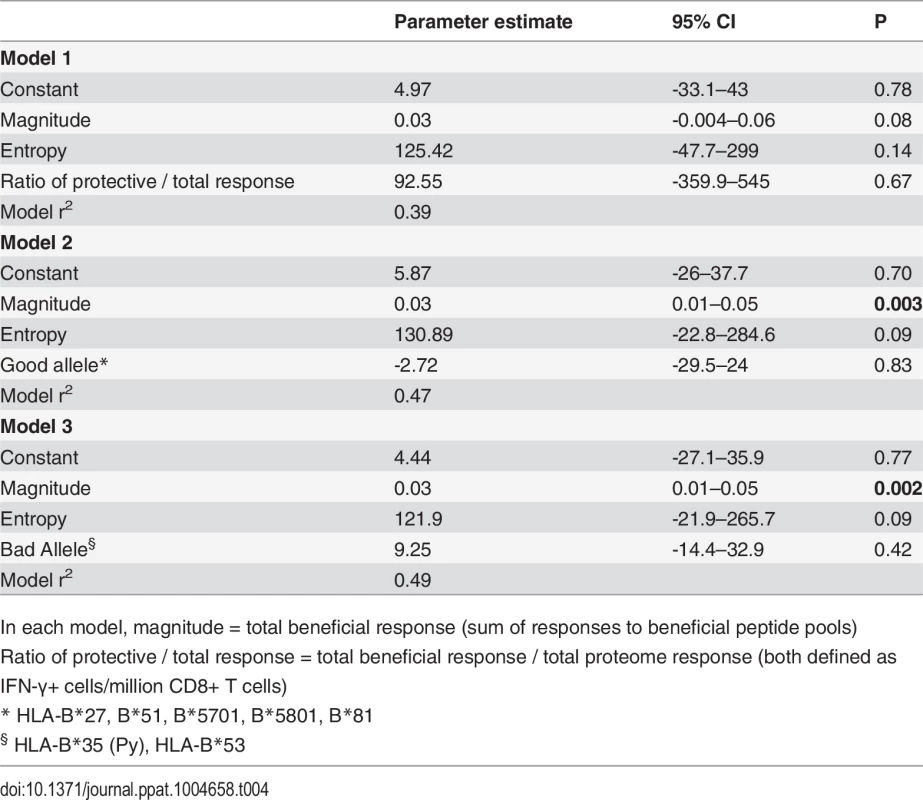
In each model, magnitude = total beneficial response (sum of responses to beneficial peptide pools) Responses to vaccines encoding full-length HIV proteins are skewed towards non-beneficial regions of the viral proteome
Given that responses to beneficial regions were subdominant in HIV-infected individuals, we next investigated whether this was also the case for responses that are primed in HIV-naïve individuals by vaccines encoding full-length HIV proteins. Data on responses that developed post-vaccination and prior to HIV acquisition were available for 13/20 of the HVTN 502 trial participants in this analysis (sampled 4 weeks after the second vaccination) [6]. We compared the magnitudes of vaccine-induced responses to peptides spanning the entire Gag/Pol/Nef immunogen with beneficial and CE regions. Vaccination induced responses to beneficial regions in 5/13 patients and to CE regions 3/13 patients, while no response to any of these regions was detected in 5 subjects. Overall, vaccine-induced responses to beneficial regions accounted for a median (range) of 0% (0–43%) of the response to the entire immunogen in these subjects, despite representing 36% of the immunogen sequence (Fig. 4A).
Fig. 4. Vaccines based on full-length protein immunogens elicit subdominant responses to beneficial regions and / or conserved elements within the HIV proteome. 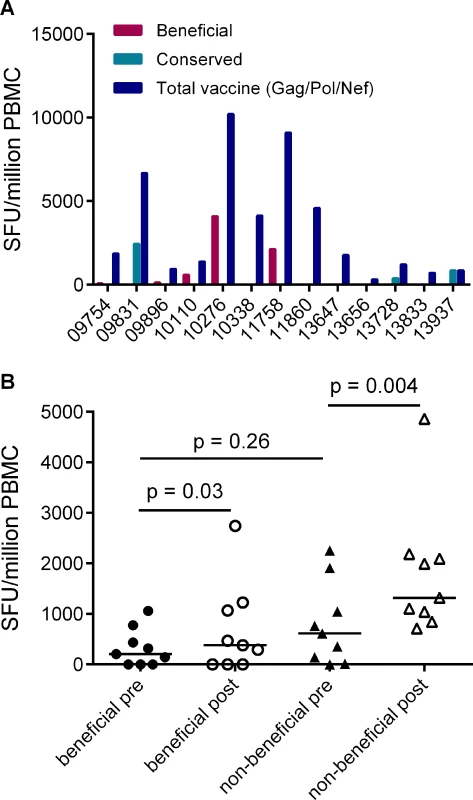
A. Post-vaccination, pre-infection CD8+ T cell responses to overlapping peptides spanning the MRK Ad5 Gag/Pol/Nef immunogen determined by IFN-γ Elispot assay in 13 HVTN 502 participants (dark blue bars) (4 weeks post-second vaccination). Responses to beneficial regions and Gag conserved elements within the immunogen are indicated by magenta and turquoise bars respectively. B. Pre- and post-vaccination responses to beneficial and non-beneficial regions within Gag determined by IFN-γ Elispot assay in 9 HIV-positive subjects after therapeutic vaccination with MVA.HIVA. Finally, we investigated whether natural immunodominance hierarchies were maintained or altered following the administration of a Gag immunogen as a therapeutic vaccine in chronic HIV infection. We mapped T cell responses to beneficial and non-beneficial regions before and after vaccination with an immunogen, ‘HIVA’ comprising full-length Gag p24/p17 sequences fused to a multiepitope string, delivered as a modified vaccinia virus Ankara-vectored vaccine to chronic ART-treated HIV-positive subjects with suppressed viremia [34][35]. Epitope mapping was performed in 9 subjects using overlapping 15-mers spanning p24 and p17, together with optimal 8–10-mer peptides for epitopes that had been defined previously (Table 5) [36]. We confined our analysis to responses to the Gag component of the immunogen, since the epitope string was, by definition, designed to focus responses on selected regions of the proteome. Prior to vaccination, the magnitude of summed responses to beneficial regions was lower than for non-beneficial Gag regions, although the difference was not statistically significant (median 205 and 615 SFU/million PBMC respectively, p = 0.27). MVA.HIVA vaccination significantly boosted T cell responses to the beneficial Gag regions (median change +150 SFU/million PBMC, p = 0.03). However, responses to non-beneficial Gag regions were preferentially expanded (median change +845 SFU/million PBMC, p = 0.004) (Fig. 4B, Table 5). Taken together, these data suggest that vaccines encoding full - or near full-length HIV proteins mimic natural HIV infection by eliciting responses that are biased towards non-beneficial targets, regardless of whether they are administered to HIV-naïve or primed individuals.
Tab. 5. Distribution of responses to Gag peptides in HIV-positive MVA.HIVA vaccinees. 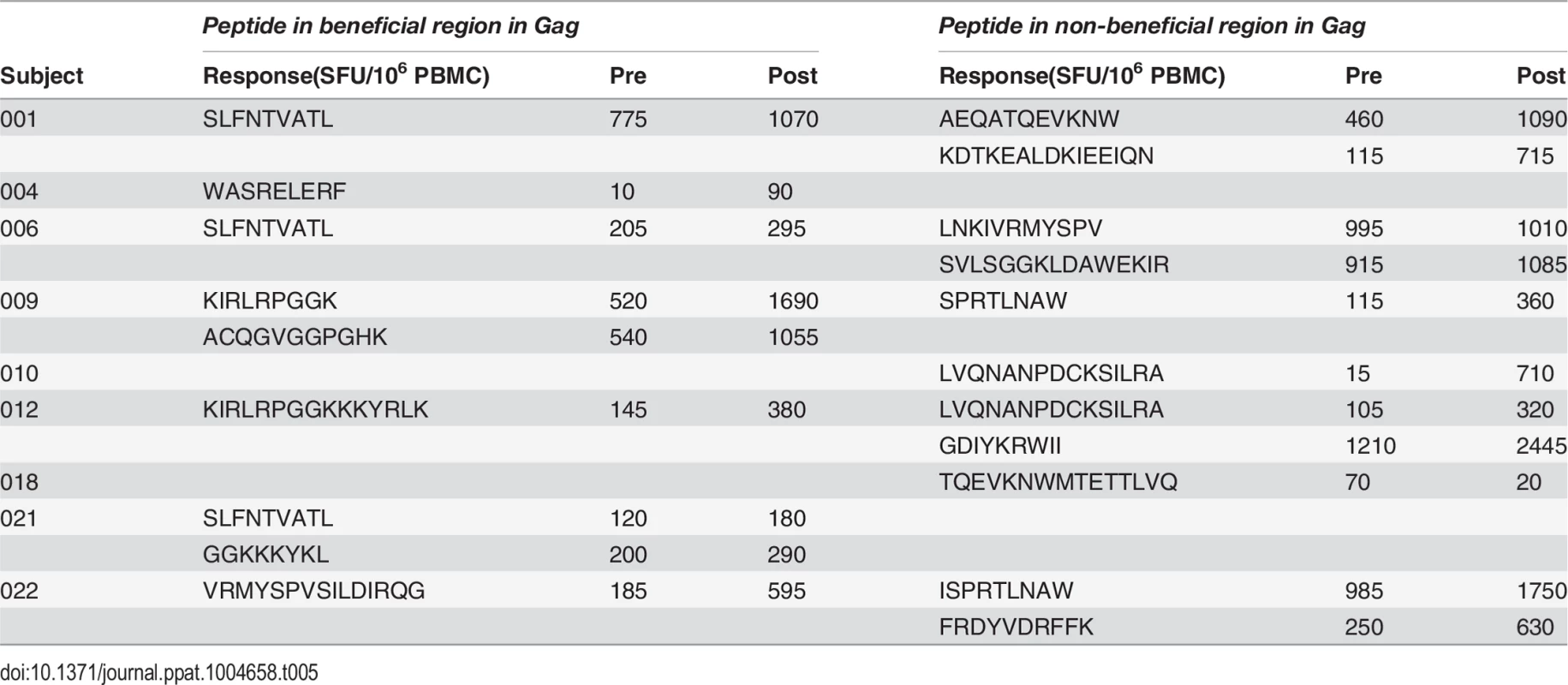
Discussion
The lack of a reliable correlate of protective immunity against HIV is a significant obstacle to systematic evaluation of vaccine candidates. Consequently, efforts to develop a T cell-based vaccine have focused broadly on recapitulating the immunological phenotype of HIV controllers, using immunogens incorporating near-complete gene sequences for many proteins. Recently, there has been greater emphasis on rationally designed immunogens, in particular, those that aim to maximise coverage of variable viral epitopes (mosaics) or avoid them altogether (conserved regions) [15][37][38][39][8]. CD8+ T cell-mediated viral inhibition was found to correlate with the frequency of T cells targeting conserved epitopes in HIV-uninfected vaccinees [8][40]. However, no vaccine candidate has yet been shown to elicit viral inhibitory activity of similar potency to that observed in HIV controllers. Here, we report that the total viral inhibitory capacity of anti-HIV CD8+ T cells is highly dependent on their specificity and we provide a mechanism to explain why conventional HIV immunogens elicit largely ineffective CD8+ T cell responses.
We reported previously that ex vivo CD8+ T cell-mediated viral inhibitory activity is inversely correlated with viral load set-point; we confirmed this finding here in genetically unrelated cohorts infected with different viruses [16]. While this is consistent with well-established associations between primary CD8+ T cell responses to HIV-1 and control of acute viraemia [41][42][32][43], the time interval between attainment of viral load set-point and sampling for the viral inhibition assay was longer in the present study, thus we cannot rule out the possibility that early control of viraemia was the cause rather than the consequence of the level of antiviral activity. It is also conceivable that a viral inhibition ‘set-point’ is attained soon after infection; this could explain the findings of Lecuroux et al., who reported that most HIV-infected individuals showed modest CD8+ T cell inhibitory activity throughout acute and early infection [24]. Nevertheless, our data give insight into the level of inhibitory activity that might be used as a benchmark to assess vaccine candidates: for example, inhibition of a clade-matched virus by ≥ 85% (observed in 50% of VC subjects but only 7% of HVTN trial participants) was associated with a median viral load of ∼ 2000 copies/ml. This suggests that the bar must be set very high if such assays are to be used to identify vaccine strategies that could clear HIV infection or reduce viral loads to undetectable levels [44].
We report for the first time, to our knowledge, that the breadth of inhibitory activity, indicated by inhibition of clade-mismatched viruses, was significantly greater in VC than subjects with uncontrolled viraemia. This suggested two non-mutually exclusive explanations: enrichment of the HIV-specific repertoire in VC for T cells recognising conserved epitopes and / or high frequencies of circulating cross-reactive CD8+ T cells that can tolerate epitope variation. However, potent clade-specific viral inhibitory activity, together with differential inhibition of diverse viruses was evident in both study groups. This led us to hypothesise that factors other than epitope conservation must play a role in the control of viral replication. We found that CD8+ T cell antiviral activity in HVTN subjects was highly correlated with the frequency of CD8+ T cells targeting selected peptides that had been shown in an independent study of two large cohorts to associate with control of viraemia [14]. This correlation was independent of protective HLA class I alleles, which suggests that effective CD8+ T cell responses may be restricted by a broader range of HLA class I alleles than previously suspected, as was also proposed by Mothe et al [14]. While the viral regions that were defined as beneficial were predominantly of low entropy, our regression analysis indicated that the magnitude of these responses accounted for a significantly greater proportion of the variation in viral inhibition than entropy alone. The Gag component of these regions explained nearly two-thirds of the effect. Interestingly, T cell responses to conserved elements peptides were weakly correlated with viral inhibition and this effect was driven by only three of the seven conserved regions tested. This is consistent with other studies showing that high population-level conservation per se does not necessarily predict viral fitness and may reflect the presence of invariant regions that are immunologically inert [27][45]. Collectively, these observations are not only reconcilable with previously described associations between broad Gag-specific T cell responses and reduced viral loads at the population level but also point to a mechanism that could explain them with greater precision [10][14][6]. The greater the breadth of responses to Gag, the higher the probability of targeting the most vulnerable epitopes, even though there is also the possibility of targeting the non-beneficial regions. The lack of responses to beneficial regions in some of the VC studied is quite likely explained by the small sample size studied and / or the extended time of untreated HIV infection which may have led to elimination of some of these T cell responses, or possibly that these VC made responses to other critical epitopes that were not represented in our peptide sets [32][46][47]. However, this does raise questions as to how long the effect of responses to beneficial regions lasts, in the face of ongoing viral escape. The rate of escape from CD8+ T cell responses is determined by the net effect on viral fitness of all escape mutations and is significantly slower in chronic than acute infection [48]. The association between the prevalence of T cell responses to beneficial regions and population-level viral load was made in chronically infected cohorts and suggests, therefore, that even though these beneficial responses may drive viral escape, the net effect is an overall impairment of viral fitness. This is consistent with observations made by Boutwell et al. who showed that CD8+ T cell escape mutations in HIV-1 Gag frequently impair viral fitness; many of the susceptible epitopes in their study were located in the beneficial regions [49].
It is possible that we have overlooked functional characteristics of Gag-specific CD8+ T cells such as the capacity to produce multiple cytokines simultaneously, as these have also been associated with control of viraemia [50][51]. However, viral inhibition assays arguably provide the most direct and complete measure of antiviral function, whereas the cytokines that are typically detected in assays of T cell polyfunctionality provide an indirect assessment. Our analysis indicated that individuals with potent viral inhibitory responses are rare, as was reported by others [24], and furthermore highlighted that responses to beneficial regions within the HIV proteome are both infrequent and subdominant. This is consistent with a previous study that showed infrequent targeting of epitopes in these regions in acute infection [32]. As spontaneous control of viraemia is itself a rare event, this provides further evidence that viral inhibitory activity in vitro accurately reflects immune control in vivo. It also raises questions as to whether long-term control or even clearance of infection can be achieved by vaccines that mimic priming by HIV. Responses elicited by the Ad5-HIV vaccine in HVTN 502 trial participants were shown previously to be limited in breadth, with a bias towards variable regions [2][7]. Our retrospective analysis of a subset of HVTN 502 vaccinees indicated preferential targeting of non-beneficial regions, which was concerning given that the Gag/Pol/Nef immunogen contained the majority of the previously described beneficial regions [14]. We observed a similar skewing of responses in HIV-positive subjects who received a therapeutic MVA vaccine encoding the immunogen, HIVA, which included 9 of the identified beneficial regions within Gag. Newer vaccine candidates such as Ad35-GRIN and Ad35-ENV, which comprise Gag, Reverse transcriptase, Integrase and Nef and Env sequences, induced responses to a median of one Gag epitope in HIV-uninfected healthy volunteers [40]. The common factor among these immunogens is the inclusion of full or near-full-length Gag sequences. A non-human primate study showed that full-length HIV immunogens induced responses to conserved regions that were of similar breadth to those elicited by non-native conserved region immunogens [52]; by contrast, Kulkarni et al. compared vaccination with p55 Gag and a conserved elements-only immunogen and showed better recognition of conserved elements epitopes with the latter approach [28,53]. Taken together, these observations highlight the need for vaccines to overcome natural immunodominance hierarchies in humans through the development of immunogens that focus responses on specific critical regions of the viral proteome. Additional refinements, such as inclusion of sequences that pre-empt predictable escape mutations, should also be considered [54]. Vaccine-mediated clearance of an AIDS virus infection in the non-human primate model was recently demonstrated for the first time with a persistent rhesus CMV SIV vaccine [55,56][57]. It is noteworthy that the responses elicited were unique in terms of their unprecedented breadth, absence of immunodominance and specificity for non-canonical viral epitopes, although the immunogen comprised entire proteins. While this may reflect unusual properties of the CMV vector and the specific mechanisms that contributed to virus eradication have yet to be resolved, such studies may provide vital lessons for human vaccine development.
In summary, these data provide several new insights that should inform HIV vaccine design. First, they suggest that induction of effective anti-HIV CD8+ T cell responses could be achieved with an immunogen comprising only a few selected regions of the viral proteome. In addition to the regions defined by Mothe et al., which were identified in chronically infected individuals, comprehensive analyses of responses that arise during acute / early HIV infection may yield viral targets that are critical to early and sustained control [32][58]. Secondly, we have identified a possible threshold for the magnitude of responses to these critical regions that should be attained in order to have a meaningful impact on viral replication. Our analysis of responses to vaccination with Ad5 Gag/Pol/Nef in a small subset of HVTN 502 subjects prior to HIV infection, together with other post-hoc studies, suggests that this is extremely unlikely to be achieved using immunogens that comprise full-length proteins. Exclusion of irrelevant decoy regions that when present, often induce immunodominant T cell responses, may be essential to prevent the development of such non-protective responses. Finally, our previous experience with potent heterologous viral vector combinations has shown that it is feasible to induce HIV-specific T cell responses in human subjects of the order of magnitude that we have proposed here [8]; rationally designed immunogens that exploit these vectors should be prioritised for clinical development.
Methods
Ethics statement
Approval was obtained from the Oxford Tropical Research Ethics Committee for analysis of anonymised PBMC samples that were made available to University of Oxford, UK by Fred Hutchinson Cancer Research Center via a Material Transfer Agreement (‘HVTN 502/Merck 023—HVTN 503 Ancillary Study’) following approval of the study by HVTN Protocol Committee. The PBMC samples were gathered and obtained from a collection held by HVTN. Viremic controllers (VC) were recruited at Duke University Medical Center with IRB approval and after obtaining written informed consent.
Study participants
The HVTN 502 and 503 studies have been described previously [1][9]. PBMC sampled from 36 HIV-positive HVTN 502 and 503 participants who were still naïve to ART 12 months after HIV acquisition, with CD4+ cell counts >350 cells/μl, were provided through the HVTN 502 Oversight Committee. Plasma viral load data were provided by SCHARP and set-point was determined using the method described by Fellay et al. [59]. Participants’ characteristics are given in Table 1. Criteria for enrolment of VC were plasma viremia consistently <5000 copies/ml for at least one year and a CD4+ cell count >400 cells/μl in the absence of ART. However, one subject was included despite a CD4+ cell count <400 cells μl because of viral loads consistently <2280 copies/ml for 5 years prior to enrolment; this individual maintained viral loads <448 copies/ml during the study. Two subjects had transient viraemia >5000 copies/ml which was subsequently spontaneously controlled. Patients’ characteristics are given in Table 2. All VC had presumed clade B infection, due to the geographical location. Therapeutic vaccine trial participants were patients with chronic HIV infection, receiving effective ART for at least 12 months, with CD4+ cell counts >350 cells/μl, who received two intramuscular immunisations of MVA.HIVA 5x107 pfu 4 weeks apart [34][60]. HLA typing was performed as described previously [6].
Virus subtyping
Virus subtyping was performed by near full-length genome sequencing, as described previously [61] or by bulk sequencing of p17 Gag and analysis using REGA HIV-1 & 2 Automated Subtyping Tool (Version 2.0) [62][63].
Virus isolates
HIV-1 isolates were obtained from the Programme EVA Centre for AIDS Reagents, National Institute for Biological Standards and Control (NIBSC), a centre of the Health Protection Agency, UK. The virus panel comprised two laboratory-adapted clade B isolates, BaL (CCR5-tropic) and IIIB (CXCR4-tropic) and three primary isolates, ES X-1936 (clade C, CCR5-tropic), 92UG029 (clade A, CCR5 / CXCR4 dual-tropic) and RW93024 (clade A, CXCR-tropic). All virus propagation was performed using primary CD4+ cells and 50% tissue culture infectious doses (TCID50) for each virus was calculated as described previously [64].
Peptides
Clades B and C consensus peptides spanning the entire HIV proteome (15-mers overlapping by 11 amino acids) were obtained from the NIH Aids Reagent Programme. 10mg/ml stocks were stored at -80°C until required, then were diluted to generate working stocks. One or more 15-mer peptides that matched most closely the beneficial OLP described by Mothe et al. and the CE peptides described by Kulkarni et al. were selected for use in Elispot assays [14][28] (Tables 3–5).
Viral inhibition assay
The viral inhibition assay has been described in detail elsewhere [16,65]. Briefly, CD8+ T cells were isolated from cryopreserved PBMC by magnetic bead selection (Miltenyi Biotec) and retained for use in IFN-γ Elispot assays. CD8-depleted cells (hereafter referred to as CD4+ T cells) were stimulated with PHA (5 μg/mL) in RPMI 1640 medium supplemented with 10% fetal calf serum (R10) for 3 days, washed, and infected with HIV-1 isolates at pre-determined optimal MOI (National Institute for Biological Standards and Control, United Kingdom). To assess viral inhibition, HIV-superinfected CD4+ T cells (5 × 104) were cultured in triplicate in R10 with interleukin 2 (20 IU/mL) in 96-well round-bottomed plates, alone or together with unstimulated ex vivo CD8+ T cells, obtained by positive bead selection of PBMCs from a second freshly thawed vial on day 3. CD8+ T cells were confirmed as >98% pure by staining for CD3, CD8, and CD56. CD8+ and CD4+ T cells were co-cultured for 6 days for all virus isolates except clade A2, for which the peak of virus replication is attained after 3 days [65]. CD8+/CD4+ ratios of 2 : 1, 1 : 1 and 1 : 10 were tested, according to cell availability. On the day of harvest, cells were stained first with Aqua Live/Dead Fixable stain (Invitrogen), fixed with 1% paraformaldehyde/20 μg/mL lysolecithin at RT, permeabilized with cold 50% methanol followed by 0.1% Nonidet P-40, and finally stained with p24 antibody (KC-57-FITC; Beckman Coulter) and antibodies to CD3, CD4, and CD8 (conjugated to APC-Cy7, PerCP, and APC, respectively; BD Biosciences). Samples were acquired on a CyAn flow cytometer. Data were analyzed using FlowJo software. Antiviral suppressive activity was expressed as percentage inhibition and determined as follows: [(fraction of p24 + cells in CD4 + T cells cultured alone)–(fraction of p24 + in CD4+ T cells cultured with CD8+ cells)]/(fraction of p24 + cells in CD4 + T cells cultured alone) × 100.
IFN-γ Elispot assay
Purified CD8+ T cells from the PBMC sample that was used to isolate CD4+ T cells for the viral inhibition assay were tested in IFN-γ Elispot assays with pools of beneficial or CE peptides (final concentration 2μg/ml) as described previously [16]. Mapping of responses to epitopes in the Gag component of the HIVA immunogen was performed using PBMC sampled pre - and 2 or 4 weeks post-vaccination, with overlapping 15-mer peptides (final concentration 4μg/ml) spanning the entire immunogen sequence, with confirmation using optimal 8–10-mer peptides where available [60]. Elispot assays with CD8-depleted PBMC were performed to confirm that these responses were CD8+ T cell-mediated. In selected assays, CD8+ T cells were recovered from the Elispot plate after overnight incubation with peptides, washed and cultured (2x106/ml) in R10 medium (RPMI with 10% fetal calf serum) plus IL-7 (25ng/ml). Cultures were supplemented with IL-2 (1.8 x103 units/ml) on day 3 and R10/IL-7/IL-2 medium was replaced on day 7. Cells were starved of IL-2 for 30 hours on day 10 and then used in cultured IFN-γ Elispot assays with individual peptides (2μg/ml).
Intracellular cytokine assay
Intracellular cytokine staining was performed as described previously, typically at the second visit after HIV infection had been confirmed [66][2].
Statistical analysis
Group comparisons were performed using the Mann Whitney test and correlations were investigated by determination of Spearman’s rank coefficient, using Graphpad Prism software, version 6. Models to explore predictors of inter-subject variation in viral inhibition by CD8+ T cells were tested using univariate and multivariable linear regression. Analyses were performed using SPSS version 22.
Supporting Information
Zdroje
1. Buchbinder SP, Mehrotra D V, Duerr A, Fitzgerald DW, Mogg R, et al. (2008) Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372 : 1881–1893. doi: 10.1016/S0140-6736(08)61591-3 19012954
2. McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, et al. (2008) HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372 : 1894–1905. doi: 10.1016/S0140-6736(08)61592-5 19012957
3. Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, et al. (2013) Efficacy Trial of a DNA/rAd5 HIV-1 Preventive Vaccine. N Engl J Med 369 : 2083–2092. doi: 10.1056/NEJMoa1310566 24099601
4. Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, et al. (2009) Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol 83 : 6508–6521. doi: 10.1128/JVI.00272-09 19403685
5. Migueles SA, Rood JE, Berkley AM, Guo T, Mendoza D, et al. (2011) Trivalent adenovirus type 5 HIV recombinant vaccine primes for modest cytotoxic capacity that is greatest in humans with protective HLA class I alleles. PLoS Pathog 7: e1002002. doi: 10.1371/journal.ppat.1002002 21383976
6. Janes H, Friedrich DP, Krambrink A, Smith RJ, Kallas EG, et al. (2013) Vaccine-Induced Gag-Specific T Cells Are Associated With Reduced Viremia After HIV-1 Infection. J Infect Dis 208 : 1231–1239. doi: 10.1093/infdis/jit322 23878319
7. Li F, Finnefrock AC, Dubey SA, Korber BT, Szinger J, et al. (2011) Mapping HIV-1 vaccine induced T-cell responses: bias towards less-conserved regions and potential impact on vaccine efficacy in the Step study. PLoS One 6: e20479. doi: 10.1371/journal.pone.0020479 21695251
8. Borthwick N, Ahmed T, Ondondo B, Hayes P, Rose A, et al. (2014) Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther 22 : 464–475. doi: 10.1038/mt.2013.248 24166483
9. Gray GE, Allen M, Moodie Z, Churchyard G, Bekker L-G, et al. (2011) Safety and efficacy of the HVTN 503/Phambili Study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect Dis 11 : 507–515. doi: 10.1016/s1473–3099(11)70098-6 21570355
10. Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, et al. (2007) CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13 : 46–53. 17173051
11. Zuniga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, et al. (2006) Relative Dominance of Gag p24-Specific Cytotoxic T Lymphocytes Is Associated with Human Immunodeficiency Virus Control. J Virol 80 : 3122–3125. 16501126
12. Hertz T, Ahmed H, Friedrich DP, Casimiro DR, Self SG, et al. (2013) HIV-1 Vaccine-Induced T-Cell Reponses Cluster in Epitope Hotspots that Differ from Those Induced in Natural Infection with HIV-1. PLoS Pathog 9: e1003404. doi: 10.1371/journal.ppat.1003404 23818843
13. Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, et al. (2013) Extreme Genetic Fragility of the HIV-1 Capsid. PLoS Pathog 9: e1003461. doi: 10.1371/journal.ppat.1003461 23818857
14. Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, et al. (2011) Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med 9 : 208. doi: 10.1186/1479-5876-9-208 22152067
15. Rolland M, Nickle DC, Mullins JI (2007) HIV-1 Group M Conserved Elements Vaccine. PLoS Pathog 3: e157. 18052528
16. Yang H, Wu H, Hancock G, Clutton G, Sande N, et al. (2012) Antiviral Inhibitory Capacity of CD8+ T cells Predicts the Rate of CD4+ T-Cell Decline in HIV-1 Infection. J Infect Dis 206 : 552–561. doi: 10.1093/infdis/jis379 22711904
17. Saez-Cirion A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, et al. (2007) HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A 104 : 6776–6781. 17428922
18. Julg B, Williams KL, Reddy S, Bishop K, Qi Y, et al. (2010) Enhanced anti-HIV functional activity associated with Gag-specific CD8 T-cell responses. J Virol 84 : 5540–5549. doi: 10.1128/JVI.02031-09 20335261
19. Payne RP, Kløverpris H, Sacha JB, Brumme Z, Brumme C, et al. (2010) Efficacious Early Antiviral Activity of HIV Gag - and Pol-Specific HLA-B*2705-Restricted CD8+ T Cells. J Virol 84 : 10543–10557. doi: 10.1128/JVI.00793-10 20686036
20. Buckheit RW, Salgado M, Silciano RF, Blankson JN (2012) Inhibitory Potential of Subpopulations of CD8+ T Cells in HIV-1-Infected Elite Suppressors. J Virol 86 : 13679–13688. doi: 10.1128/jvi.02439-12 23055552
21. Freel SA, Picking RA, Ferrari G, Ding H, Ochsenbauer C, et al. (2012) Initial HIV-1 Antigen-Specific CD8+ T Cells in Acute HIV-1 Infection Inhibit Transmitted/Founder Virus Replication. J Virol 86 : 6835–6846. doi: 10.1128/JVI.00437-12 22514337
22. Tomaras GD, Lacey SF, McDanal CB, Ferrari G, Weinhold KJ, et al. (2000) CD8+ T cell-mediated suppressive activity inhibits HIV-1 after virus entry with kinetics indicating effects on virus gene expression. Proc Natl Acad Sci 97 : 3503–3508. doi: 10.1073/pnas.97.7.3503 10725407
23. Freel SA, Lamoreaux L, Chattopadhyay PK, Saunders K, Zarkowsky D, et al. (2010) Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J Virol 84 : 4998–5006. doi: 10.1128/JVI.00138-10 20200250
24. Lécuroux C, Girault I, Chéret A, Versmisse P, Nembot G, et al. (2013) CD8 T-Cells from Most HIV-Infected Patients Lack Ex Vivo HIV-Suppressive Capacity during Acute and Early Infection. PLoS One 8: e59767. doi: 10.1371/journal.pone.0059767 23555774
25. Streeck H, Brumme ZL, Anastario M, Cohen KW, Jolin JS, et al. (2008) Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med 5: e100. doi: 10.1371/journal.pmed.0050100 18462013
26. Spentzou A, Bergin P, Gill D, Cheeseman H, Ashraf A, et al. (2010) Viral inhibition assay: a CD8 T cell neutralization assay for use in clinical trials of HIV-1 vaccine candidates. J Infect Dis 201 : 720–729. doi: 10.1086/650492 20132004
27. Rolland M, Manocheewa S, Swain JV, Lanxon-Cookson EC, Kim M, et al. (2013) HIV-1 Conserved-Element Vaccines: Relationship between Sequence Conservation and Replicative Capacity. J Virol 87 : 5461–5467. doi: 10.1128/JVI.03033-12 23468488
28. Kulkarni V, Rosati M, Valentin A, Ganneru B, Singh AK, et al. (2013) HIV-1 p24gag Derived Conserved Element DNA Vaccine Increases the Breadth of Immune Response in Mice. PLoS One 8: e60245. doi: 10.1371/journal.pone.0060245 23555935
29. Mothe B, Llano A, Ibarrondo J, Zamarreno J, Schiaulini M, et al. (2012) CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PLoS One 7: e29717. doi: 10.1371/journal.pone.0029717 22238642
30. Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, et al. (2008) Marked Epitope - and Allele-Specific Differences in Rates of Mutation in Human Immunodeficiency Type 1 (HIV-1) Gag, Pol, and Nef Cytotoxic T-Lymphocyte Epitopes in Acute/Early HIV-1 Infection. J Virol 82 : 9216–9227. doi: 10.1128/JVI.01041-08 18614631
31. Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, et al. (2009) HLA-B57/B*5801 Human Immunodeficiency Virus Type 1 Elite Controllers Select for Rare Gag Variants Associated with Reduced Viral Replication Capacity and Strong Cytotoxic T-Lymphotye Recognition. J Virol 83 : 2743–2755. doi: 10.1128/JVI.02265-08 19116253
32. Streeck H, Jolin JS, Qi Y, Yassine-Diab B, Johnson RC, et al. (2009) Human Immunodeficiency Virus Type 1-Specific CD8+ T-Cell Responses during Primary Infection Are Major Determinants of the Viral Set Point and Loss of CD4+ T Cells. J Virol 83 : 7641–7648. doi: 10.1128/JVI.00182-09 19458000
33. Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, et al. (2006) Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine 24 : 6893–6904. doi: http://dx.doi.org/10.1016/j.vaccine.2006.06.009 16890329
34. Dorrell L, Yang H, Ondondo B, Dong T, de Gleria K, et al. (2006) Expansion and diversification of virus-specific T cells following immunisation of HIV-1-infected individuals with a recombinant modified vaccinia virus Ankara / HIV-1 gag vaccine. J Virol 80 : 4705–4716. 16641264
35. Hanke T, McMichael AJ (2000) Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat Med 6 : 951–955. 10973301
36. Llano A, Williams A, Overa A, Silva-Arrieta S, Brander C (2013) HIV Molecular Immunology 2013. Yusim K, Korber B, Brander C, Barouch D, de Boer RJ, et al., editors Los Alamos National Laboratory, Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, New Mexico. doi: 10.1016/j.jfo.2013.06.003 25632406
37. Korber BT, Letvin NL, Haynes BF (2009) T-Cell Vaccine Strategies for Human Immunodeficiency Virus, the Virus with a Thousand Faces. J Virol 83 : 8300–8314. doi: 10.1128/JVI.00114-09 19439471
38. Barouch DH, O’Brien KL, Simmons NL, King SL, Abbink P, et al. (2010) Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med 16 : 319–323. doi: 10.1038/nm.2089 20173752
39. Letourneau S, Im EJ, Mashishi T, Brereton C, Bridgeman A, et al. (2007) Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2: e984. doi: 10.1371/annotation/fca26a4f-42c1-4772-a19e-aa9d96c4eeb2 17912361
40. Kopycinski J, Hayes P, Ashraf A, Cheeseman H, Lala F, et al. (2014) Broad HIV Epitope Specificity and Viral Inhibition Induced by Multigenic HIV-1 Adenovirus Subtype 35 Vector Vaccine in Healthy Uninfected Adults. PLoS One 9: e90378. doi: 10.1371/journal.pone.0090378 24609066
41. Koup RA, Safrit JT, Cao Y, Andrews C, McLeod G, et al. (1994) Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol 68 : 4650–4655. 8207839
42. Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, et al. (1999) Control of Viremia in Simian Immunodeficiency Virus Infection by CD8+ Lymphocytes. Science (80-) 283 : 857–860.
43. Goonetilleke N, Liu MKP, Salazar-Gonzalez JF, Ferrari G, Giorgi E, et al. (2009) The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206 : 1253–1272. doi: 10.1084/jem.20090365 19487423
44. Shapiro SZ (2014) Clinical Development of Candidate HIV Vaccines: Different Problems for Different Vaccines. AIDS Res Hum Retroviruses 30 : 325–329. doi: 10.1089/aid.2013.0114 24168166
45. Manocheewa S, Swain JV, Lanxon-Cookson E, Rolland M, Mullins JI (2013) Fitness Costs of Mutations at the HIV-1 Capsid Hexamerization Interface. PLoS One 8: e66065. doi: 10.1371/journal.pone.0066065 23785468
46. Streeck H, Lu R, Beckwith N, Milazzo M, Liu M, et al. (2014) Emergence of Individual HIV-Specific CD8 T Cell Responses during Primary HIV-1 Infection Can Determine Long-Term Disease Outcome. J Virol 88 : 12793–12801. doi: 10.1128/JVI.02016-14 25165102
47. Pereyra F, Heckerman D, Carlson JM, Kadie C, Soghoian DZ, et al. (2014) HIV Control Is Mediated in Part by CD8+ T-Cell Targeting of Specific Epitopes. J Virol 88 : 12937–12948. doi: 10.1128/JVI.01004-14 25165115
48. Ganusov V V, Goonetilleke N, Liu MKP, Ferrari G, Shaw GM, et al. (2011) Fitness Costs and Diversity of the Cytotoxic T Lymphocyte (CTL) Response Determine the Rate of CTL Escape during Acute and Chronic Phases of HIV Infection. J Virol 85 : 10518–10528. doi: 10.1128/JVI.00655-11 21835793
49. Boutwell CL, Carlson JM, Lin T-H, Seese A, Power KA, et al. (2013) Frequent and Variable Cytotoxic-T-Lymphocyte Escape-Associated Fitness Costs in the Human Immunodeficiency Virus Type 1 Subtype B Gag Proteins. J Virol 87 : 3952–3965. doi: 10.1128/JVI.03233-12 23365420
50. Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, et al. (2006) HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107 : 4781–4789. 16467198
51. Riou C, Burgers WA, Mlisana K, Koup RA, Roederer M, et al. (2014) Differential Impact of Magnitude, Polyfunctional Capacity, and Specificity of HIV-Specific CD8+ T Cell Responses on HIV Set Point. J Virol 88 : 1819–1824. doi: 10.1128/JVI.02968-13 24227857
52. Stephenson KE, SanMiguel A, Simmons NL, Smith K, Lewis MG, et al. (2012) Full-Length HIV-1 Immunogens Induce Greater Magnitude and Comparable Breadth of T Lymphocyte Responses to Conserved HIV-1 Regions Compared with Conserved-Region-Only HIV-1 Immunogens in Rhesus Monkeys. J Virol 86 : 11434–11440. doi: 10.1128/JVI.01779-12 22896617
53. Kulkarni V, Valentin A, Rosati M, Alicea C, Singh AK, et al. (2014) Altered Response Hierarchy and Increased T-Cell Breadth upon HIV-1 Conserved Element DNA Vaccination in Macaques. PLoS One 9: e86254. doi: 10.1371/journal.pone.0086254 24465991
54. Valkenburg SA, Gras S, Guillonneau C, Hatton LA, Bird NA, et al. (2013) Preemptive priming readily overcomes structure-based mechanisms of virus escape. Proc Natl Acad Sci 110 : 5570–5575. doi: 10.1073/pnas.1302935110 23493558
55. Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, et al. (2011) Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature: Epub ahead of print.
56. Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, et al. (2013) Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science (80-) 340 : 1237874. doi: 10.1126/science.1237874 23704576
57. Hansen SG, Piatak M Jr., Ventura AB, Hughes CM, Gilbride RM, et al. (2013) Immune clearance of highly pathogenic SIV infection. Nature 502 : 100–104. doi: 10.1038/nature12519 24025770
58. Liu MKP, Hawkins N, Ritchie AJ, Ganusov V V, Whale V, et al. (2013) Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest 123 : 380–393. doi: 10.1172/JCI65330 23221345
59. Fellay J, Shianna K V, Ge D, Colombo S, Ledergerber B, et al. (2007) A Whole-Genome Association Study of Major Determinants for Host Control of HIV-1. Science (80-) 317 : 944–947.
60. Yang H, Dong T, Turnbull E, Ranasinghe S, Ondondo B, et al. (2007) Broad TCR Usage in Functional HIV-1-Specific CD8+ T Cell Expansions Driven by Vaccination during Highly Active Antiretroviral Therapy. J Immunol 179 : 597–606. 17579081
61. Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, et al. (2011) Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med 17 : 366–371. doi: 10.1038/nm.2316 21358627
62. De Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, et al. (2005) An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinforma 21 : 3797–3800. doi: 10.1093/bioinformatics/bti607.
63. Alcantara LCJ, Cassol S, Libin P, Deforche K, Pybus OG, et al. (2009) A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 37: W634–W642. doi: 10.1093/nar/gkp455 19483099
64. Koup RA, Ho DD, Poli G, Fauci AS (2001) Isolation and quantitation of HIV in peripheral blood. Curr Protoc Immunol 5 : 2.1–2.11. doi: 10.1002/0471142735.im1202s05.
65. Yang H, Yorke E, Hancock G, Clutton G, Sande N, et al. (2013) Improved quantification of HIV-1-infected CD4+ T cells using an optimised method of intracellular HIV-1 gag p24 antigen detection. J Immunol Methods 391 : 174–178. doi: 10.1016/j.jim.2013.03.001 23500782
66. Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, et al. (2007) Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods 323 : 39–54. doi: http://dx.doi.org/10.1016/j.jim.2007.03.002 17451739
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek 2014 Reviewer Thank YouČlánek Characterization of Metabolically Quiescent Parasites in Murine Lesions Using Heavy Water LabelingČlánek High Heritability Is Compatible with the Broad Distribution of Set Point Viral Load in HIV Carriers
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 2- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- 2014 Reviewer Thank You
- A Case for Two-Component Signaling Systems As Antifungal Drug Targets
- Prions—Not Your Immunologist’s Pathogen
- Telomeric ORFS in : Does Mediator Tail Wag the Yeast?
- Livestock-Associated : The United States Experience
- The Neurotrophic Receptor Ntrk2 Directs Lymphoid Tissue Neovascularization during Infection
- The Intracellular Bacterium Uses Parasitoid Wasps as Phoretic Vectors for Efficient Horizontal Transmission
- CD200 Receptor Restriction of Myeloid Cell Responses Antagonizes Antiviral Immunity and Facilitates Cytomegalovirus Persistence within Mucosal Tissue
- Phage-mediated Dispersal of Biofilm and Distribution of Bacterial Virulence Genes Is Induced by Quorum Sensing
- CXCL9 Contributes to Antimicrobial Protection of the Gut during Infection Independent of Chemokine-Receptor Signaling
- Mitigation of Prion Infectivity and Conversion Capacity by a Simulated Natural Process—Repeated Cycles of Drying and Wetting
- Approaches Reveal a Key Role for DCs in CD4+ T Cell Activation and Parasite Clearance during the Acute Phase of Experimental Blood-Stage Malaria
- Revealing the Sequence and Resulting Cellular Morphology of Receptor-Ligand Interactions during Invasion of Erythrocytes
- Crystal Structures of the Carboxyl cGMP Binding Domain of the cGMP-dependent Protein Kinase Reveal a Novel Capping Triad Crucial for Merozoite Egress
- Non-redundant and Redundant Roles of Cytomegalovirus gH/gL Complexes in Host Organ Entry and Intra-tissue Spread
- Characterization of Metabolically Quiescent Parasites in Murine Lesions Using Heavy Water Labeling
- A Working Model of How Noroviruses Infect the Intestine
- CD44 Plays a Functional Role in -induced Epithelial Cell Proliferation
- Novel Inhibitors of Cholesterol Degradation in Reveal How the Bacterium’s Metabolism Is Constrained by the Intracellular Environment
- G-Quadruplexes in Pathogens: A Common Route to Virulence Control?
- A Rho GDP Dissociation Inhibitor Produced by Apoptotic T-Cells Inhibits Growth of
- Manipulating Adenovirus Hexon Hypervariable Loops Dictates Immune Neutralisation and Coagulation Factor X-dependent Cell Interaction and
- The RhoGAP SPIN6 Associates with SPL11 and OsRac1 and Negatively Regulates Programmed Cell Death and Innate Immunity in Rice
- Lymph-Node Resident CD8α Dendritic Cells Capture Antigens from Migratory Malaria Sporozoites and Induce CD8 T Cell Responses
- Coordinated Function of Cellular DEAD-Box Helicases in Suppression of Viral RNA Recombination and Maintenance of Viral Genome Integrity
- IL-33-Mediated Protection against Experimental Cerebral Malaria Is Linked to Induction of Type 2 Innate Lymphoid Cells, M2 Macrophages and Regulatory T Cells
- Evasion of Autophagy and Intracellular Killing by Human Myeloid Dendritic Cells Involves DC-SIGN-TLR2 Crosstalk
- CD8 T Cell Response Maturation Defined by Anentropic Specificity and Repertoire Depth Correlates with SIVΔnef-induced Protection
- Diverse Heterologous Primary Infections Radically Alter Immunodominance Hierarchies and Clinical Outcomes Following H7N9 Influenza Challenge in Mice
- Human Adenovirus 52 Uses Sialic Acid-containing Glycoproteins and the Coxsackie and Adenovirus Receptor for Binding to Target Cells
- Super-Resolution Imaging of ESCRT-Proteins at HIV-1 Assembly Sites
- Disruption of an Membrane Protein Causes a Magnesium-dependent Cell Division Defect and Failure to Persist in Mice
- Recognition of Hyphae by Human Plasmacytoid Dendritic Cells Is Mediated by Dectin-2 and Results in Formation of Extracellular Traps
- Essential Domains of Invasins Utilized to Infect Mammalian Host Cells
- High Heritability Is Compatible with the Broad Distribution of Set Point Viral Load in HIV Carriers
- Yeast Prions: Proteins Templating Conformation and an Anti-prion System
- A Novel Mechanism of Bacterial Toxin Transfer within Host Blood Cell-Derived Microvesicles
- A Wild Strain Has Enhanced Epithelial Immunity to a Natural Microsporidian Parasite
- Control of Murine Cytomegalovirus Infection by γδ T Cells
- Dimorphism in Fungal Pathogens of Mammals, Plants, and Insects
- Recognition and Activation Domains Contribute to Allele-Specific Responses of an Arabidopsis NLR Receptor to an Oomycete Effector Protein
- Direct Binding of Retromer to Human Papillomavirus Type 16 Minor Capsid Protein L2 Mediates Endosome Exit during Viral Infection
- Characterization of the Mycobacterial Acyl-CoA Carboxylase Holo Complexes Reveals Their Functional Expansion into Amino Acid Catabolism
- Prion Infections and Anti-PrP Antibodies Trigger Converging Neurotoxic Pathways
- Evolution of Genome Size and Complexity in the
- Antibiotic Modulation of Capsular Exopolysaccharide and Virulence in
- IFNγ Signaling Endows DCs with the Capacity to Control Type I Inflammation during Parasitic Infection through Promoting T-bet+ Regulatory T Cells
- Identification of Effective Subdominant Anti-HIV-1 CD8+ T Cells Within Entire Post-infection and Post-vaccination Immune Responses
- Viral and Cellular Proteins Containing FGDF Motifs Bind G3BP to Block Stress Granule Formation
- ATPaseTb2, a Unique Membrane-bound FoF1-ATPase Component, Is Essential in Bloodstream and Dyskinetoplastic Trypanosomes
- Cytoplasmic Actin Is an Extracellular Insect Immune Factor which Is Secreted upon Immune Challenge and Mediates Phagocytosis and Direct Killing of Bacteria, and Is a Antagonist
- A Specific A/T Polymorphism in Western Tyrosine Phosphorylation B-Motifs Regulates CagA Epithelial Cell Interactions
- Within-host Competition Does Not Select for Virulence in Malaria Parasites; Studies with
- A Membrane-bound eIF2 Alpha Kinase Located in Endosomes Is Regulated by Heme and Controls Differentiation and ROS Levels in
- Cytosolic Access of : Critical Impact of Phagosomal Acidification Control and Demonstration of Occurrence
- Role of Pentraxin 3 in Shaping Arthritogenic Alphaviral Disease: From Enhanced Viral Replication to Immunomodulation
- Rational Development of an Attenuated Recombinant Cyprinid Herpesvirus 3 Vaccine Using Prokaryotic Mutagenesis and In Vivo Bioluminescent Imaging
- HITS-CLIP Analysis Uncovers a Link between the Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Protein and Host Pre-mRNA Metabolism
- Molecular and Functional Analyses of a Maize Autoactive NB-LRR Protein Identify Precise Structural Requirements for Activity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Control of Murine Cytomegalovirus Infection by γδ T Cells
- ATPaseTb2, a Unique Membrane-bound FoF1-ATPase Component, Is Essential in Bloodstream and Dyskinetoplastic Trypanosomes
- Rational Development of an Attenuated Recombinant Cyprinid Herpesvirus 3 Vaccine Using Prokaryotic Mutagenesis and In Vivo Bioluminescent Imaging
- Telomeric ORFS in : Does Mediator Tail Wag the Yeast?
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



