-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Five Questions about Viruses and MicroRNAs
article has not abstract
Published in the journal: . PLoS Pathog 6(2): e32767. doi:10.1371/journal.ppat.1000787
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1000787Summary
article has not abstract
MicroRNAs (miRNAs) are ∼22-nt regulatory RNAs expressed by all multicellular eukaryotes [1]. Humans encode >700 miRNAs and similar numbers are likely to exist in other mammalian species. Almost all cellular miRNAs are initially transcribed by RNA polymerase II (Pol II) as part of a long, capped, polyadenylated primary miRNA (pri-miRNA) precursor. The miRNA forms part of one arm of an RNA stem-loop that consists of an ∼32-bp imperfect stem flanked by unstructured RNA sequences. This stem-loop is recognized by the nuclear RNase III enzyme Drosha, which cleaves the stem to liberate an ∼60-nt pre-miRNA hairpin. The pre-miRNA is then transported to the cytoplasm where it is cleaved by a second RNase III enzyme, called Dicer, which removes the terminal loop to generate the miRNA duplex intermediate. One strand of this duplex is incorporated into the RNA-induced silencing complex (RISC), where it acts as a guide RNA to direct RISC to complementary mRNA species [1]. Depending on the level of complementarity, RISC can either cleave bound mRNAs and/or inhibit their translation. Inhibition of mRNA translation generally requires full complementarity of the mRNA to nucleotides 2 through 7 or 8 from the miRNA 5′ end—the miRNA seed region. The primary, and possibly sole, function of mammalian miRNAs is therefore to act as specific post-transcriptional inhibitors of mRNA function.
Do All Viruses Encode miRNAs?
Because miRNAs are potent regulators of gene expression that are both small and non-antigenic, they would seem ideal tools to alter the cellular environment in ways that favor virus replication. While numerous virally encoded miRNAs have, in fact, been described for members of the herpesvirus family, few other viral miRNAs have been identified (Table 1). In general, we can divide mammalian viruses into three categories, i.e., the herpesviruses, which encode multiple viral miRNAs; other nuclear DNA viruses, which may encode one or two miRNAs; and the RNA viruses and cytoplasmic DNA viruses, which appear to lack any miRNAs [1]. While this generalization is clearly subject to revision as more viruses are analyzed, viruses that currently appear to lack any miRNAs include retroviruses (e.g., human immunodeficiency virus type 1 [HIV-1]), flaviviruses (e.g., yellow fever virus), hepatitis C virus (HCV), and papillomavirus [2],[3]. In addition, deep sequencing of small RNAs isolated from cells infected with cowpox virus or influenza A virus has also failed to identify any virally encoded miRNAs (J. L. Umbach and B. R. Cullen, unpublished data).
Tab. 1. MicroRNAs Encoded by Mammalian Viruses. 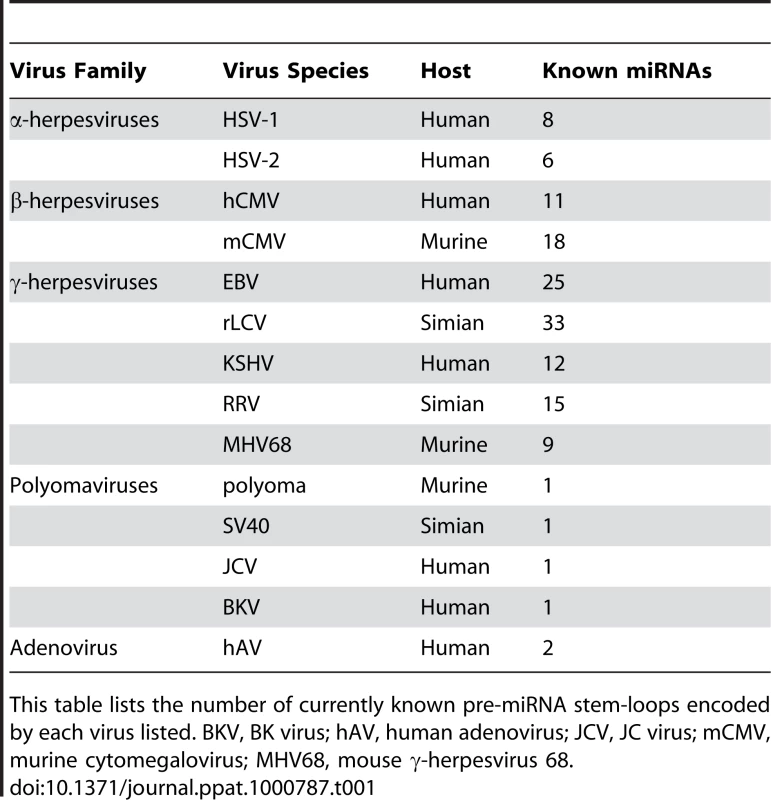
This table lists the number of currently known pre-miRNA stem-loops encoded by each virus listed. BKV, BK virus; hAV, human adenovirus; JCV, JC virus; mCMV, murine cytomegalovirus; MHV68, mouse γ-herpesvirus 68. Although the reason for this pattern of viral miRNA expression remains unknown, one can speculate that viruses that replicate in the cytoplasm (e.g., flaviviruses, HCV, and cowpox) might lack miRNAs because they never encounter the nuclear enzyme Drosha. On the other hand, nuclear RNA viruses (e.g., influenza and HIV-1) may avoid encoding miRNAs because excision of a genomically encoded pre-miRNA would induce the cleavage and degradation of the viral RNA genome. Finally, one can speculate that viruses that establish long-term, latent infections (e.g., herpesviruses) would derive more benefit from the use of non-antigenic miRNAs to inhibit cellular antiviral responses than would viruses that have short lytic replication cycles.
What Do Viral miRNAs Do?
Only a small number of mRNA targets of viral miRNAs have been defined, so a complete answer to this question is currently not possible. However, the emerging picture [4] suggests that viral miRNAs may serve two major functions. On the one hand, several viral miRNAs have been shown to inhibit the expression of cellular factors that play a role in cellular innate or adaptive antiviral immune responses. Examples of the inhibition of host cell factors involved in mediating immune responses include the natural killer cell ligand MICB, which is downregulated by miRNAs encoded by human cytomegalovirus (hCMV), Kaposi's sarcoma-associated herpesvirus (KSHV), and Epstein-Barr virus (EBV) [5], and the pro-apoptotic protein PUMA, which is downregulated by an EBV miRNA [6]. The potential selective advantage conferred on a virus by this downregulation is obvious. Secondly, several viral miRNAs downregulate the expression of viral proteins, including key viral immediate-early or early regulatory proteins. For example, herpes simplex virus 1 (HSV-1) downregulates the immediate-early transactivators ICP0 and ICP4 in latently infected cells by using viral miRNAs that are expressed at high levels during latency, but not during productive viral replication. This action is thought to stabilize the latent state [7]. Conversely, several polyomaviruses downregulate the expression of the viral T antigens, key early transcription factors, at late stages in the viral replication cycle [8],[9],[10]. It has been proposed that this reduces the killing of infected cells by cytotoxic T lymphocytes specific for T antigen epitopes [8].
Although it is therefore widely believed that viral miRNAs play an important role in the replication and pathogenesis of the viruses that encode them, little evidence in support of this hypothesis currently exists. Indeed, a mutant polyomavirus lacking the only polyomavirus-encoded miRNA replicated indistinguishably from wild-type in infected mice [9]. Whether herpesviruses, which encode more viral miRNAs, do depend on these small RNAs will no doubt soon be revealed.
Are Viral miRNAs Conserved between Related Viral Species?
If viral miRNAs do play an important role in viral replication and pathogenesis, then they should be under purifying selection. Indeed, comparison of the genomic sequences of diverse KSHV isolates reveals that the miRNAs encoded by KSHV are, in general, highly conserved, although isolates bearing mutations that block production of individual KSHV miRNAs have been described [11]. A similar high level of conservation also appears to be characteristic of miRNAs encoded by other human herpesviruses, including EBV and hCMV.
In contrast, a comparison between distinct but related herpesvirus species reveals far less evidence of miRNA sequence conservation, although the genomic localization of virally encoded miRNAs is often conserved. In the case of HSV-1, which encodes eight known viral miRNAs, and the related but distinct HSV-2, which encodes at least six miRNAs, four miRNAs are partially conserved in terms of sequence, and fully conserved in terms of genomic localization [12]. Similarly, 22 of the 25 miRNAs encoded by EBV are at least partially conserved in the related γ-herpesvirus rhesus lymphocryptovirus (rLCV), which encodes a remarkable 33 miRNAs [13]. In contrast, none of the 12 miRNAs expressed by KSHV are conserved in the somewhat more distantly related rhesus rhadinovirus (RRV), although all virally encoded miRNAs are found in a single cluster located adjacent to the viral ORF71 gene in both virus species [2],[13],[14]. It is therefore likely that viral miRNAs evolve through a combination of sequence evolution of pre-existing miRNA stem-loops combined with an occasional stem-loop duplication event [13]. As miRNAs are short, readily mutable sequences, any degree of evolutionary conservation implies that miRNAs can confer a significant selective advantage. Presumably, sequence conservation of viral miRNAs also implies the existence of cellular, rather than viral, mRNA targets, as the latter might be expected to rapidly co-evolve with viral miRNAs.
Do Viruses Regulate miRNA Transcription, Processing, or Function?
Viral infection represents a cataclysmic event in the life of a cell that generally results in dramatic changes in cellular mRNA expression. These may include the induction of cellular factors that have the potential to resist or promote viral replication. As cellular miRNAs are also transcribed by Pol II, it is expected that viral infection would also affect their pattern of expression. The question then is whether these changes are evolutionarily selected to favor or inhibit virus replication. At present, there is no evidence indicating that viruses modify the cellular environment to selectively favor the processing and expression of viral miRNAs, although viral miRNAs may be expressed at such high levels that they have the potential to competitively inhibit cellular miRNA function.
While it has been proposed that HIV-1 infection induces the expression of specific cellular miRNAs [15], the best-characterized example of a virus inducing cellular miRNAs that may favor virus infection occurs in the case of EBV, which induces the expression of several cellular miRNAs, including miR-155, in infected B cells [16]. In contrast, while KSHV does not induce miR-155 expression upon infection of B cells, it does encode a viral miRNA that functions as an ortholog of miR-155 [17],[18]. As miR-155 can promote the oncogenic transformation of B cells when overexpressed in vivo, it is likely that miR-155 induction by EBV, or expression of a miR-155 mimic by KSHV, plays a role not only in promoting the establishment of EBV or KSHV latency, but also in viral transformation of B cells [16],[17],[18].
While viruses may therefore enhance cellular miRNA expression, the more commonly observed effect is inhibition [15],[19]. Moreover, there have been several reports demonstrating that cellular miRNAs can inhibit virus replication and/or that artificial inhibition of miRNA production or function can enhance virus replication in culture [15],[20],[21],[22],[23]. Many viruses globally repress Pol II transcription in infected cells, and this would also repress miRNA biogenesis. In addition, adenoviruses produce the viral VA1 non-coding RNA, which acts as a potent inhibitor of both pre-miRNA nuclear export and pre-miRNA processing by Dicer [24]. The HIV-1 Tat protein has also been proposed to inhibit miRNA function, although this appears to require very high levels of Tat expression [19].
Can Viruses Use Cellular miRNAs to Promote Their Replication?
At least one clear-cut example of a cellular miRNA that facilitates virus replication is known, i.e., activation of HCV replication by the liver-specific miRNA miR-122 [25]. The HCV RNA genome contains two miR-122 binding sites in its 5′ untranslated region that are required for activation of viral genomic RNA replication, although the underlying mechanism remains obscure. At present, this result remains unique to HCV; i.e., no other RNA virus has been shown to rely upon direct binding of a cellular miRNA to its RNA genome for any aspect of its replication cycle.
An interesting and largely unresolved aspect of the replication cycle of RNA viruses is that their RNA genome must serve several essential yet mutually exclusive roles. Thus, the HCV genome is not only the mRNA that encodes all the viral proteins but also the substrate for the viral RNA-dependent RNA polymerase and the genomic RNA that is packaged into progeny virions. It is interesting to speculate that cellular miRNAs could play a role in retargeting viral RNAs away from translation and towards one of the latter two fates. Of note, RISC is believed to direct bound mRNAs into cytoplasmic processing bodies (P bodies), where they may be translationally repressed and/or degraded, yet emerging evidence suggests that P bodies can also function as sites of viral RNA synthesis [26]. Currently, it remains unclear how viruses deal with the potentially inhibitory effect of the large numbers of cellular miRNAs that have at least the potential to target viral RNA genomes and/or mRNAs. Have they simply evolved to inhibit miRNA function and/or minimize the number of functional binding sites for the miRNA species that are expressed in infected cells, or have they instead evolved ways to take advantage of these miRNAs? This issue is one that deserves significant attention, as it may suggest novel methods to selectively perturb the replication of pathogenic viruses in vivo.
Zdroje
1. BartelDP
2004 MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281 297
2. PfefferS
SewerA
Lagos-QuintanaM
SheridanR
SanderC
2005 Identification of microRNAs of the herpesvirus family. Nat Methods 2 269 276
3. CaiX
LiG
LaiminsLA
CullenBR
2006 Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol 80 10890 10893
4. UmbachJL
CullenBR
2009 The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 23 1151 1164
5. NachmaniD
Stern-GinossarN
SaridR
MandelboimO
2009 Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5 376 385
6. ChoyEY
SiuKL
KokKH
LungRW
TsangCM
2008 An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 205 2551 2560
7. UmbachJL
KramerMF
JurakI
KarnowskiHW
CoenDM
2008 MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454 780 783
8. SullivanCS
GrundhoffAT
TevethiaS
PipasJM
GanemD
2005 SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435 682 686
9. SullivanCS
SungCK
PackCD
GrundhoffA
LukacherAE
2009 Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology 387 157 167
10. SeoGJ
FinkLH
O'HaraB
AtwoodWJ
SullivanCS
2008 Evolutionarily conserved function of a viral microRNA. J Virol 82 9823 9828
11. MarshallV
ParksT
BagniR
WangCD
SamolsMA
2007 Conservation of virally encoded microRNAs in Kaposi sarcoma–associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis 195 645 659
12. UmbachJL
WangK
TangS
KrausePR
MontEK
2010 Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J Virol 84 1189 1192
13. WalzN
ChristallaT
TessmerU
GrundhoffA
2010 A global analysis of evolutionary conservation among known and predicted gammaherpesvirus microRNAs. J Virol 84 716 728
14. SchäferA
CaiX
BilelloJP
DesrosiersRC
CullenBR
2007 Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology 364 21 27
15. TribouletR
MariB
LinYL
Chable-BessiaC
BennasserY
2007 Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315 1579 1582
16. CameronJE
FewellC
YinQ
McBrideJ
WangX
2008 Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 382 257 266
17. SkalskyRL
SamolsMA
PlaisanceKB
BossIW
RivaA
2007 Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol 81 12836 12845
18. GottweinE
MukherjeeN
SachseC
FrenzelC
MajorosWH
2007 A viral microRNA functions as an ortholog of cellular miR-155. Nature 450 1096 1099
19. QianS
ZhongX
YuL
DingB
de HaanP
2009 HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc Natl Acad Sci U S A 106 605 610
20. OtsukaM
JingQ
GeorgelP
NewL
ChenJ
2007 Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27 123 134
21. PedersenIM
ChengG
WielandS
VoliniaS
CroceCM
2007 Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449 919 922
22. LecellierCH
DunoyerP
ArarK
Lehmann-CheJ
EyquemS
2005 A cellular microRNA mediates antiviral defense in human cells. Science 308 557 560
23. NathansR
ChuCY
SerquinaAK
LuCC
CaoH
2009 Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34 696 709
24. LuS
CullenBR
2004 Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol 78 12868 12876
25. JoplingCL
SchutzS
SarnowP
2008 Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4 77 85
26. BeckhamCJ
ParkerR
2008 P bodies, stress granules, and viral life cycles. Cell Host Microbe 3 206 212
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek HIV Controller CD4+ T Cells Respond to Minimal Amounts of Gag Antigen Due to High TCR AvidityČlánek Transit through the Flea Vector Induces a Pretransmission Innate Immunity Resistance Phenotype in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 2- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Pathogen Entrapment by Transglutaminase—A Conserved Early Innate Immune Mechanism
- Broadly Protective Monoclonal Antibodies against H3 Influenza Viruses following Sequential Immunization with Different Hemagglutinins
- Neutrophil-Derived CCL3 Is Essential for the Rapid Recruitment of Dendritic Cells to the Site of Inoculation in Resistant Mice
- Differentiation, Distribution and γδ T Cell-Driven Regulation of IL-22-Producing T Cells in Tuberculosis
- IFN-α-Induced Upregulation of CCR5 Leads to Expanded HIV Tropism In Vivo
- An Extensive Circuitry for Cell Wall Regulation in
- TgMORN1 Is a Key Organizer for the Basal Complex of
- Direct Presentation Is Sufficient for an Efficient Anti-Viral CD8 T Cell Response
- Immunoelectron Microscopic Evidence for Tetherin/BST2 as the Physical Bridge between HIV-1 Virions and the Plasma Membrane
- A New Nuclear Function of the Glycolytic Enzyme Enolase: The Metabolic Regulation of Cytosine-5 Methyltransferase 2 (Dnmt2) Activity
- Genome-Wide mRNA Expression Correlates of Viral Control in CD4+ T-Cells from HIV-1-Infected Individuals
- Structural and Biochemical Characterization of SrcA, a Multi-Cargo Type III Secretion Chaperone in Required for Pathogenic Association with a Host
- A Major Role for the ApiAP2 Protein PfSIP2 in Chromosome End Biology
- HIV Controller CD4+ T Cells Respond to Minimal Amounts of Gag Antigen Due to High TCR Avidity
- Fis Is Essential for Capsule Production in and Regulates Expression of Other Important Virulence Factors
- Vaccinia Protein F12 Has Structural Similarity to Kinesin Light Chain and Contains a Motor Binding Motif Required for Virion Export
- A Novel Pseudopodial Component of the Dendritic Cell Anti-Fungal Response: The Fungipod
- Efficacy of the New Neuraminidase Inhibitor CS-8958 against H5N1 Influenza Viruses
- Long-Lived Antibody and B Cell Memory Responses to the Human Malaria Parasites, and
- IPS-1 Is Essential for the Control of West Nile Virus Infection and Immunity
- Transit through the Flea Vector Induces a Pretransmission Innate Immunity Resistance Phenotype in
- Ats-1 Is Imported into Host Cell Mitochondria and Interferes with Apoptosis Induction
- Six RNA Viruses and Forty-One Hosts: Viral Small RNAs and Modulation of Small RNA Repertoires in Vertebrate and Invertebrate Systems
- The Syk Kinase SmTK4 of Is Involved in the Regulation of Spermatogenesis and Oogenesis
- Optineurin Negatively Regulates the Induction of IFNβ in Response to RNA Virus Infection
- On the Diversity of Malaria Parasites in African Apes and the Origin of from Bonobos
- Five Questions about Viruses and MicroRNAs
- A Broad Distribution of the Alternative Oxidase in Microsporidian Parasites
- Caspase-1 Activation via Rho GTPases: A Common Theme in Mucosal Infections?
- Peptides Presented by HLA-E Molecules Are Targets for Human CD8 T-Cells with Cytotoxic as well as Regulatory Activity
- Interaction of Rim101 and Protein Kinase A Regulates Capsule
- Distinct External Signals Trigger Sequential Release of Apical Organelles during Erythrocyte Invasion by Malaria Parasites
- Exacerbated Innate Host Response to SARS-CoV in Aged Non-Human Primates
- Reverse Genetics in Predicts ARF Cycling Is Essential for Drug Resistance and Virulence
- Universal Features of Post-Transcriptional Gene Regulation Are Critical for Zygote Development
- Highly Differentiated, Resting Gn-Specific Memory CD8 T Cells Persist Years after Infection by Andes Hantavirus
- Arterivirus Nsp1 Modulates the Accumulation of Minus-Strand Templates to Control the Relative Abundance of Viral mRNAs
- Lethal Antibody Enhancement of Dengue Disease in Mice Is Prevented by Fc Modification
- Quantitative Comparison of HTLV-1 and HIV-1 Cell-to-Cell Infection with New Replication Dependent Vectors
- The Disulfide Bonds in Glycoprotein E2 of Hepatitis C Virus Reveal the Tertiary Organization of the Molecule
- IL-1β Processing in Host Defense: Beyond the Inflammasomes
- Kaposi's Sarcoma Associated Herpes Virus (KSHV) Induced COX-2: A Key Factor in Latency, Inflammation, Angiogenesis, Cell Survival and Invasion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Caspase-1 Activation via Rho GTPases: A Common Theme in Mucosal Infections?
- Kaposi's Sarcoma Associated Herpes Virus (KSHV) Induced COX-2: A Key Factor in Latency, Inflammation, Angiogenesis, Cell Survival and Invasion
- IL-1β Processing in Host Defense: Beyond the Inflammasomes
- Reverse Genetics in Predicts ARF Cycling Is Essential for Drug Resistance and Virulence
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



