-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Broadly Protective Monoclonal Antibodies against H3 Influenza Viruses following Sequential Immunization with Different Hemagglutinins
As targets of adaptive immunity, influenza viruses are characterized by the fluidity with which they respond to the selective pressure applied by neutralizing antibodies. This mutability of structural determinants of protective immunity is the obstacle in developing universal influenza vaccines. Towards the development of such vaccines and other immune therapies, our studies are designed to identify regions of influenza viruses that are conserved and that mediate virus neutralization. We have specifically focused on viruses of the H3N2 subtype, which have persisted as a principal source of influenza-related morbidity and mortality in humans since the pandemic of 1968. Three monoclonal antibodies have been identified that are broadly-neutralizing against H3 influenza viruses spanning 40 years. The antibodies react with the hemagglutinin glycoprotein and appear to bind in regions that are refractory to the structural variation required for viral escape from neutralization. The antibodies demonstrate therapeutic efficacy in mice against H3N2 virus infection and have potential for use in the treatment of human influenza disease. By mapping the binding region of one antibody, 12D1, we have identified a continuous region of the hemagglutinin that may act as an immunogen to elicit broadly protective immunity to H3 viruses. The anti-H3 monoclonal antibodies were identified after immunization of mice with the hemagglutinin of four different viruses (A/Hong Kong/1/1968, A/Alabama/1/1981, A/Beijing/47/1992, A/Wyoming/3/2003). This immunization schedule was designed to boost B cells specific for conserved regions of the hemagglutinin from distinct antigenic clusters. Importantly, our antibodies are of naturally occurring specificity rather than selected from cloned libraries, demonstrating that broad-spectrum humoral immunity to influenza viruses can be elicited in vivo.
Published in the journal: . PLoS Pathog 6(2): e32767. doi:10.1371/journal.ppat.1000796
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000796Summary
As targets of adaptive immunity, influenza viruses are characterized by the fluidity with which they respond to the selective pressure applied by neutralizing antibodies. This mutability of structural determinants of protective immunity is the obstacle in developing universal influenza vaccines. Towards the development of such vaccines and other immune therapies, our studies are designed to identify regions of influenza viruses that are conserved and that mediate virus neutralization. We have specifically focused on viruses of the H3N2 subtype, which have persisted as a principal source of influenza-related morbidity and mortality in humans since the pandemic of 1968. Three monoclonal antibodies have been identified that are broadly-neutralizing against H3 influenza viruses spanning 40 years. The antibodies react with the hemagglutinin glycoprotein and appear to bind in regions that are refractory to the structural variation required for viral escape from neutralization. The antibodies demonstrate therapeutic efficacy in mice against H3N2 virus infection and have potential for use in the treatment of human influenza disease. By mapping the binding region of one antibody, 12D1, we have identified a continuous region of the hemagglutinin that may act as an immunogen to elicit broadly protective immunity to H3 viruses. The anti-H3 monoclonal antibodies were identified after immunization of mice with the hemagglutinin of four different viruses (A/Hong Kong/1/1968, A/Alabama/1/1981, A/Beijing/47/1992, A/Wyoming/3/2003). This immunization schedule was designed to boost B cells specific for conserved regions of the hemagglutinin from distinct antigenic clusters. Importantly, our antibodies are of naturally occurring specificity rather than selected from cloned libraries, demonstrating that broad-spectrum humoral immunity to influenza viruses can be elicited in vivo.
Introduction
Under non-pandemic conditions, the global mortality attributed to influenza virus infection is considerable, with 200,000–500,000 associated deaths occurring each year [1]. In the setting of the 1918 influenza pandemic, the global mortality reached 50 million people in one year, equivalent to twice the number of people killed by HIV/AIDS since its emergence almost thirty years ago [2]. Notably, in 1918 and in the current swine-origin influenza virus pandemic, the populations normally considered the fittest are observed to be among the most vulnerable [3],[4].
Four kinds of influenza viruses are circulating in the human population at this time: influenza A viruses of the hemagglutinin H3 and H1 subtypes (H1 viruses are further divided into those of human and swine origin) and influenza B viruses. Influenza A viruses are responsible for the bulk of seasonal disease, with H3 viruses dominating eight of the past twelve influenza seasons in the United States [5]. In 1968, an H3 virus caused one of the three major influenza pandemics of the twentieth century and H3 viruses have persisted since that time as a significant agent of human disease. In addition to humans, H3 influenza viruses commonly infect birds, swine, and horses. It is not known whether H3 viruses will persist as human pathogens or how they may evolve to become more or less virulent in humans.
Immunity to influenza viruses is currently achieved by vaccination with strains representing those predicted to circulate in the coming flu season. In a healthy person, the virus acts as a robust immunogen, eliciting neutralizing serum antibody that protects against influenza disease. Both the humoral and cell-mediated arms of the adaptive system are involved in resolution of active influenza infection, with neutralizing antibody titers correlating with protection in vivo [6].
The hemagglutinin glycoprotein is the primary target of antibodies that confer protective immunity to influenza viruses. Antibodies to other influenza proteins likely act in: Fc-receptor mediated uptake of virus particles, antibody-dependent cell cytotoxicity, delay of replication kinetics and, in aggregate, they may contribute to virus neutralization. On a monoclonal level, however, only antibodies specific for the viral hemagglutinin have been shown to block/neutralize infection [7].
Neutralizing monoclonal antibodies (mAbs) act by preventing either of the two functions of the hemagglutinin molecule: virus attachment or virus fusion with the host cell [8]. Antibodies that prevent attachment bind antigenic sites surrounding the receptor binding pocket in the membrane distal HA1 subunit of the hemagglutinin and restrict the association with host cell receptors (sialic acids) [9]. These antibodies drive the outgrowth of antigenic variants, resulting in a continuum of changes in the hemagglutinin structure known as ‘antigenic drift’. Relatively few examples of fusion-inhibiting mAbs are available, but they are most commonly described to interact with the membrane proximal HA2 portion of the hemagglutinin in the region of the fusion peptide [10],[11],[12].
The sixteen subtypes of the influenza hemagglutinin are divided broadly into two phylogenetic groups that correlate with two basic structures taken by the stalk of the molecule [13]. In 1993, mAb C179, an antibody with broad neutralizing activity against viruses in Group 1 (of H1 and H2 subtypes) was described [14]. More recently, several other monoclonal antibodies that neutralize a broad array of Group 1 viruses (including representative H1 and H5 viruses) were identified [11],[12],[15],[16]. These antibodies have consistently been shown to interact with the stalk of the hemagglutinin and neutralize virus by preventing fusion with the host cell. This report constitutes the first description of broadly neutralizing antibodies against viruses in Group 2.
Results
Isolation of broadly-reactive anti-H3 mAbs
In order to enhance the production of cross-reactive antibody specificities, we immunized mice by sequential administration with DNA coding for the hemagglutinin from H3 viruses arising approximately 10 years apart: A/Hong Kong/1/1968, A/Alabama/1/1981, A/Beijing/47/1992. Finally, three days prior to fusion, mice were boosted with the H3 virus A/Wyoming/3/2003. By performing the fusion rapidly after virus boost we ensured that only hemagglutinin-specific B cells were present in the spleen at time of fusion. The hemagglutinins chosen were from viruses that arose over several decades, thus representing multiple H3 antigenic clusters [17]. Post-fusion, hybridoma supernatants were screened for the ability to bind A/Hong Kong/1/1968 by western blot or by ELISA and successive rounds of subcloning were performed on positive supernatants until monoclonal hybridoma populations were isolated.
The immunization schedule we utilized successfully elicited the production of antibodies with broad reactivity against H3 viruses. Approximately 120 clones were isolated that reacted with A/Hong Kong/1/1968; of those, eight mAbs were cross-reactive against all of the H3 hemagglutinins tested. Interestingly, the particular immunization protocol also preferentially elicited the production of antibodies specific for the HA2 subunit of the hemagglutinin. Of the 8 mAbs identified, 5 mAbs react with HA2 and 1 mAb reacts with HA1 by western blot. The remaining 2 mAbs bind conformational epitopes present in the HA trimer as detected by western blot of purified H3 virus proteins separated under non-reducing gel conditions. All mAbs were reactive in a purified H3 virus ELISA. Three of the mAbs, 7A7, 12D1, 39A4, had the highest activity by ELISA and were selected for thorough characterization (Table 1, Figure 1).
Fig. 1. MAbs react with H3 hemagglutinin by western blot. 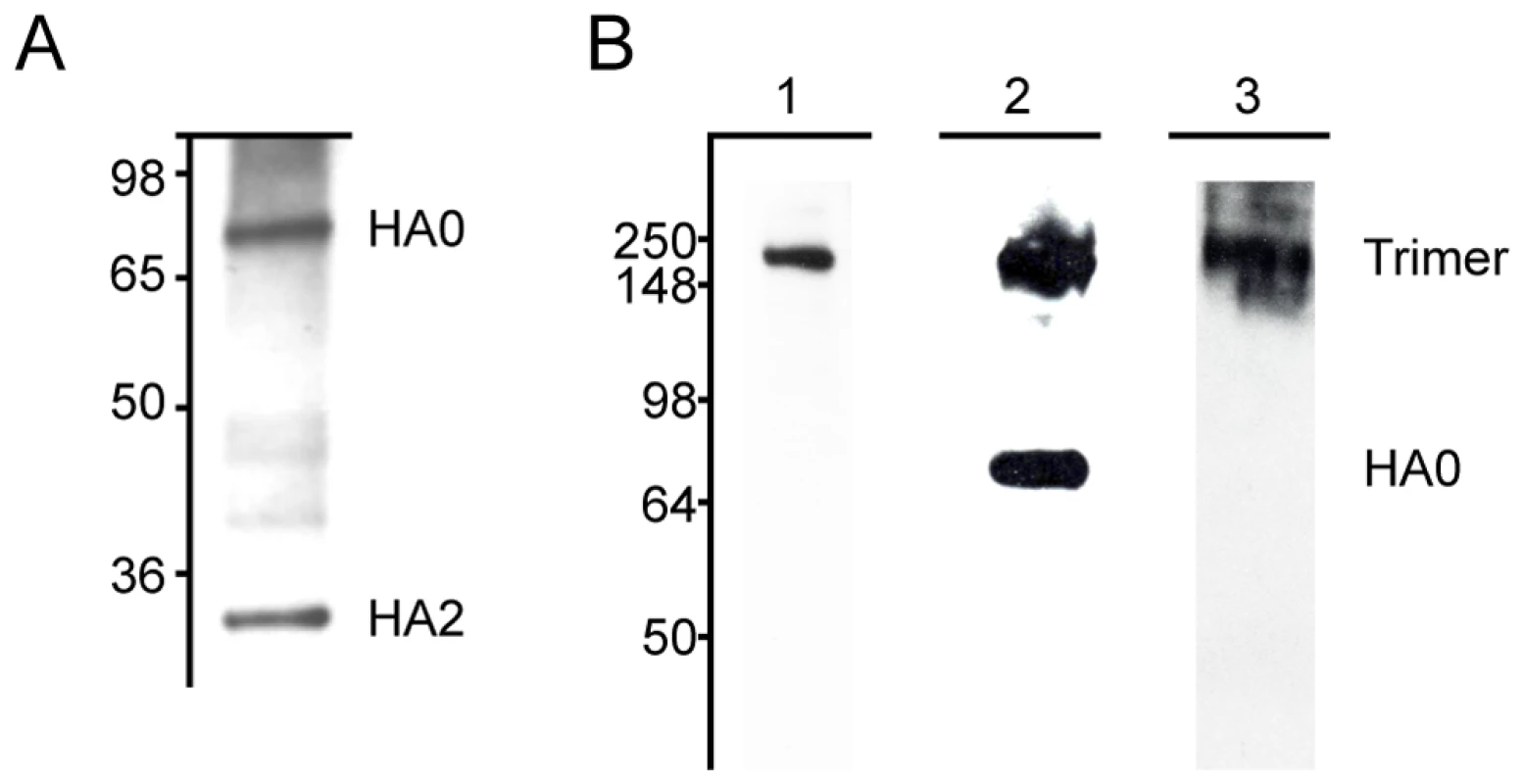
(A) MAb 12D1 binds the A/Pan/2007/1999 hemagglutinin within the HA2 subunit. mAbs 7A7 and 39A4 do not react with hemagglutinin under reducing conditions. (B) MAbs 7A7, 12D1 and 39A4 react with the A/HK/1/1968 hemagglutinin under non-reducing conditions. MAbs 7A7 (lane 1) and 39A4 (lane 3) bind HA trimer complexes. mAb 12D1 (lane 2) binds HA trimer complexes and HA0. Tab. 1. Pattern of reactivity of anti-H3 mAbs. 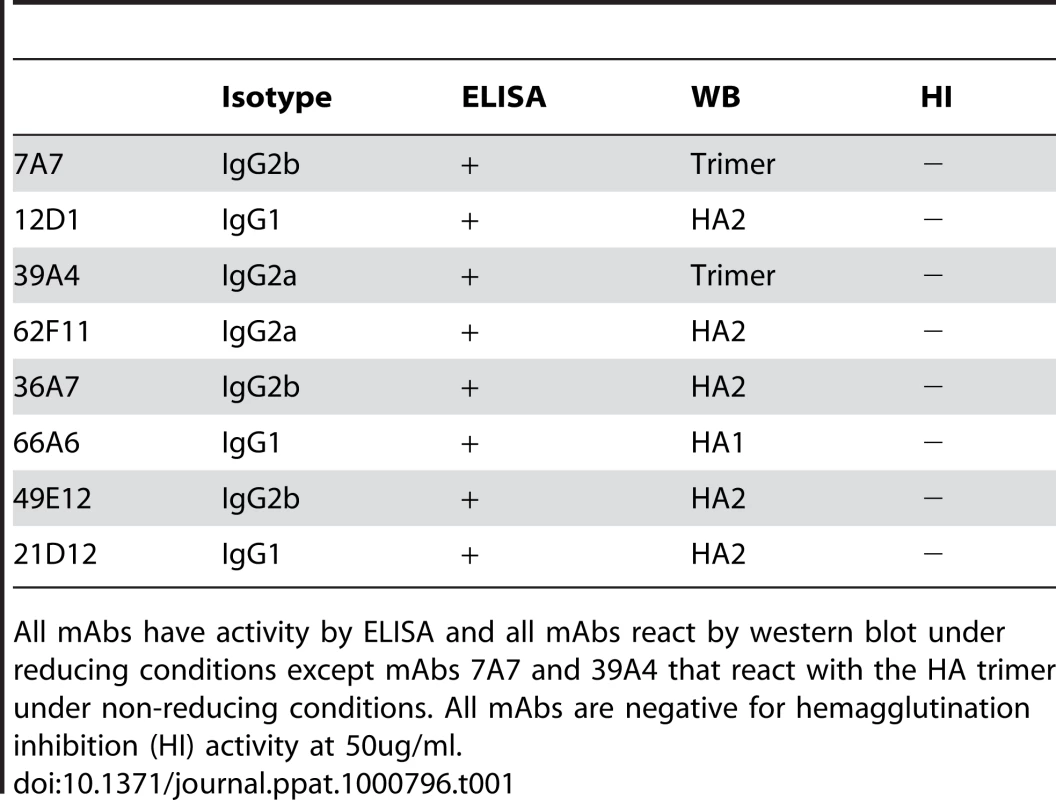
All mAbs have activity by ELISA and all mAbs react by western blot under reducing conditions except mAbs 7A7 and 39A4 that react with the HA trimer under non-reducing conditions. All mAbs are negative for hemagglutination inhibition (HI) activity at 50ug/ml. Antibodies 7A7, 12D1 and 39A4 react by ELISA with purified A/Alabama/1/1981 and purified A/Hong Kong/1/1968 viruses (Figure 2). MAb XY102 is specific for the hemagglutinin of A/Hong Kong/1/1968 virus [18]. MAbs 7A7, 12D1 and 39A4 show broad reactivity by immunofluorescence against cells infected with all H3 viruses spanning 40 drift years. MAbs 7A7 and 39A4 also react by immunofluorescence with other influenza A viruses chosen at random, including representative H1, H2 and equine H3 viruses (Table 2).
Fig. 2. Reactivity of anti-H3 mAbs by ELISA. 
(A) mAbs react with purified A/HK/1968 (H3) virus. (B) mAbs react with purified A/Alabama/1981 (H3) virus. mAb XY102 is specific for the hemagglutinin of A/HK/1968 virus. Tab. 2. Reactivity of mAbs at 5ug/ml by immunofluorescence against MDCK cells infected with a panel of randomly chosen viruses. 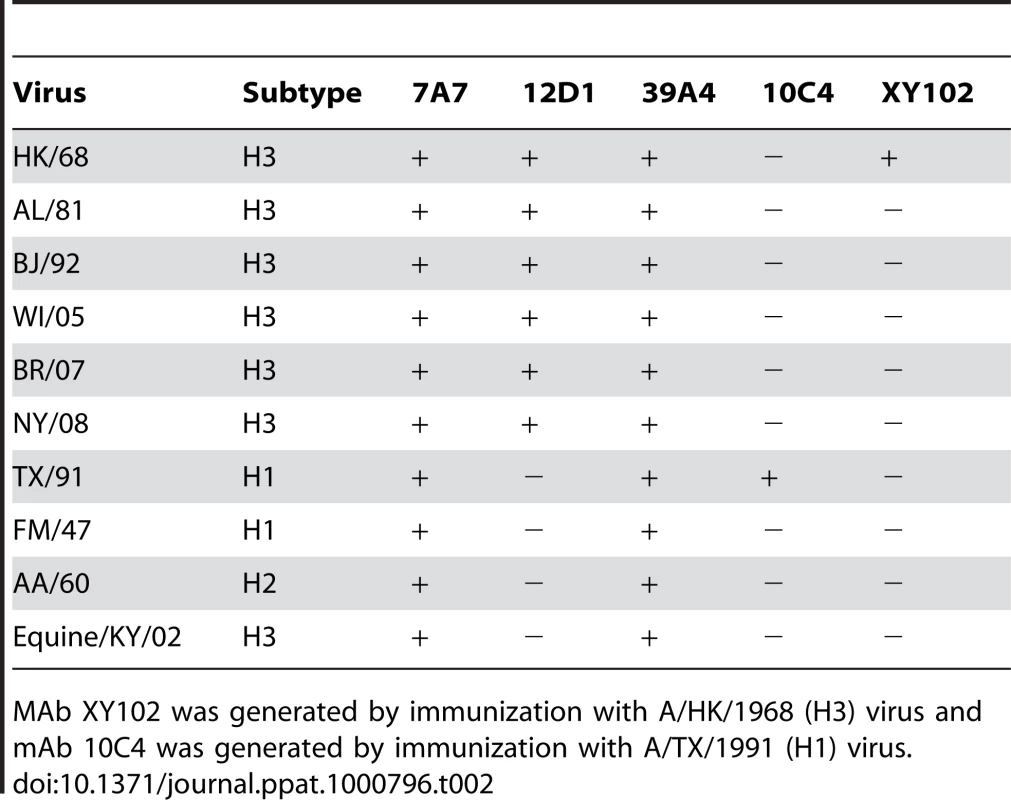
MAb XY102 was generated by immunization with A/HK/1968 (H3) virus and mAb 10C4 was generated by immunization with A/TX/1991 (H1) virus. Anti-H3 mAbs neutralize H3 viruses spanning 40 drift years
The anti-H3 mAbs were first evaluated for their ability to neutralize H3 influenza viruses by microneutralization assay. Viruses used in this assay contain a gene segment coding for firefly luciferase in place of the viral hemagglutinin; a hemagglutinin is present on the viral envelope due to propagation of virus in cells stably expressing a particular H3 hemagglutinin protein (see methods). Luciferase viruses were generated that express the hemagglutinin of A/HK/1968 or A/Panama/99 viruses. Neutralization of viruses by anti-H3 mAbs was determined based on luciferase activity after single-cycle replication; mAbs 7A7, 12D1 and 39A4 were determined to neutralize the hemagglutinin of both A/HK/1968 and A/Pan/99 (Figure 3).
Fig. 3. Anti-H3 mabs in microneutralization assay. 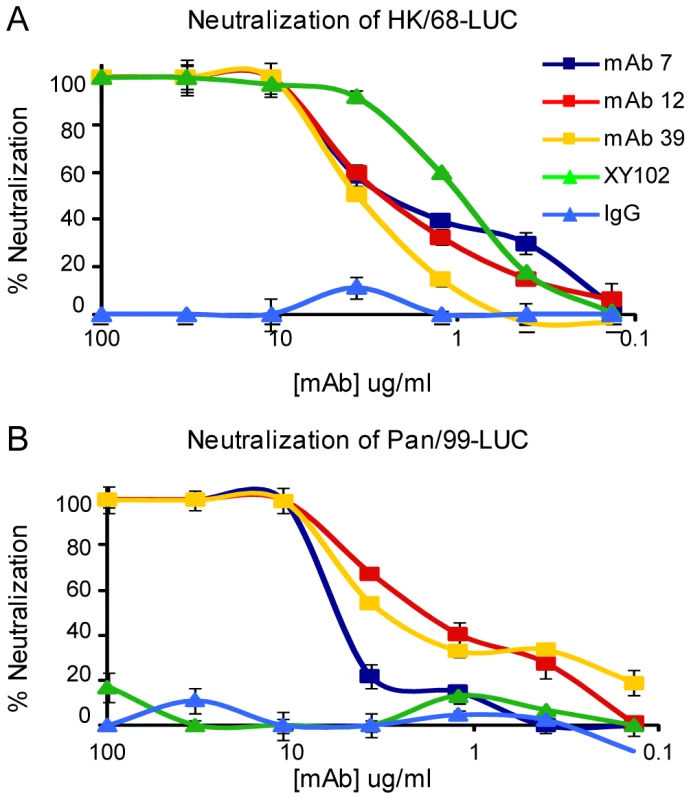
Neutralization of virus expressing the HA from either (A) A/Hong Kong/1/1968 virus or (B) A/Panama/2007/1999 virus. mAb XY102 is specific for A/HK/1968 virus. Purified mouse IgG was used for the negative control. Next, we evaluated neutralization activity by plaque reduction assay. The anti-H3 mAbs were able to prevent infection (not simply reduce plaque size) of Madin Darby canine kidney cells by H3 viruses arising over 40 drift years: A/HK/1968, A/BJ/1992, A/Pan/99, A/Bris/07, A/NY/08 (Figure 4). We tested 7A7, 12D1 and 39A4 against representative H4 and H7 viruses (Group 2) as well as an H1 virus (Group 1) and found that they did not neutralize the non-H3 subtype viruses (Figure 4).
Fig. 4. Activity of anti-H3 mabs in plaque reduction assay on MDCK cells. 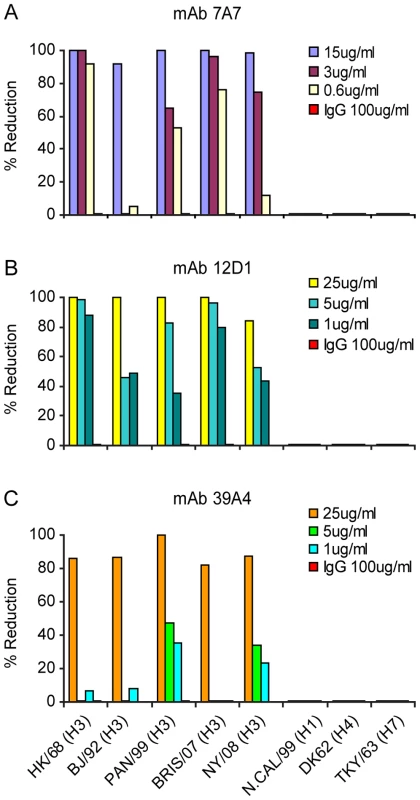
mAb 7A7 (A), 12D1 (B) and 39A4 (C) neutralize all H3 viruses tested by plaque reduction assay but not representative H1, H4 or H7 viruses. Purified mouse IgG was used for the negative control. The plaque reduction assays were performed multiple times and with each new antibody preparation. Data shown are from a single representative experiment. Anti-H3 mAbs in the treatment of influenza in mice
The three mAbs were tested in vivo for use as passive transfer therapies in disease caused by H3 virus infection. Mice were given 30mg/kg mAb intraperitoneally either 1 hour before, 24 hours post or 48 hours post challenge with 10 mouse LD50 reassortant H3 virus (the A/HK/68 reassortant virus contains the six non-hemagglutinin, non-neuraminidase segments from the mouse-adapted A/PR/8 virus). Mice were weighed daily and were sacrificed if they reached 75% of their starting weight. Treatment of mice with mAb 12D1 either prophylactically or therapeutically was 100% protective. mAb 39A4 was evaluated for efficacy by prophylactic treatment and was similarly 100% protective in vivo. Mice treated prophylactically with mAb 7A7 were only 40% protected against the A/HK/68 reassortant virus (Figure 5).
Fig. 5. Anti-H3 mAbs protect against H3 virus in vivo. 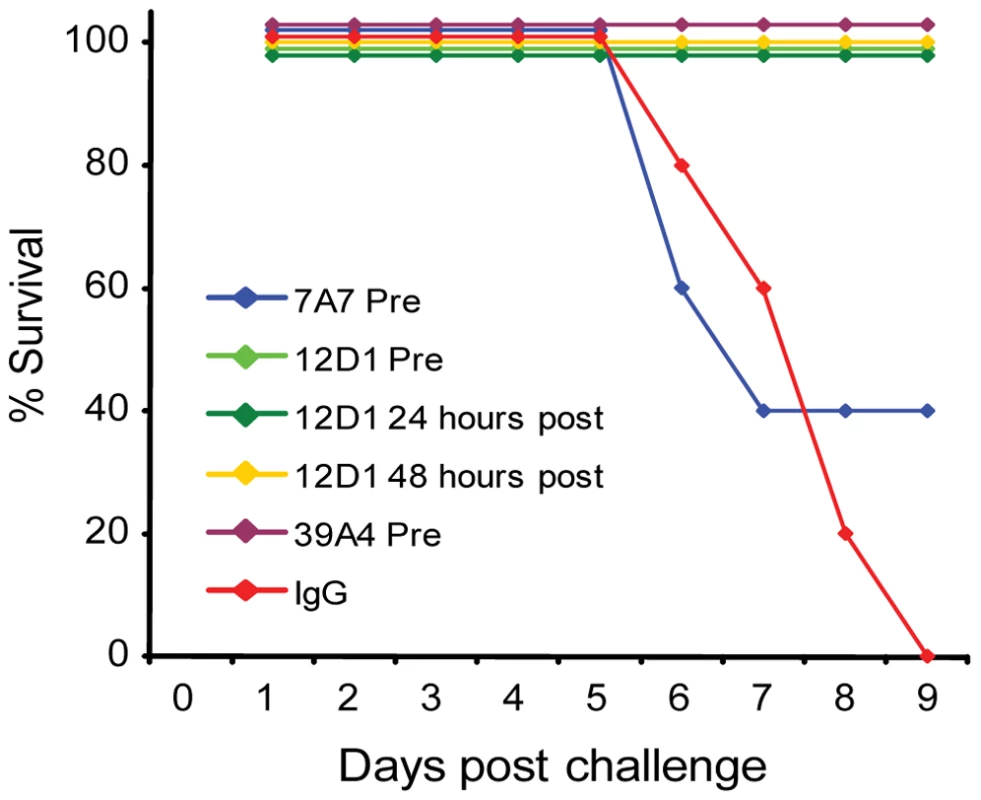
Mice were given 30mg/kg mAb 7A7, 12D1, 39A4 or isotype control by intraperitoneal injection 1 hour prior, 24 hours post (12D1 only) or 48 hours post (12D1 only) challenge with H3N2 reassortant virus (HK68/PR8). N = 5 per group. Next, the effect of prophylactic treatment with mAb 12D1 or 39A4 on lung damage caused by H3 viral pneumonia was assessed by histologic evaluation of tissue taken 4 days post infection with the A/HK/68 reassortant virus. Without treatment, lungs showed degenerative changes with focal hemorrhaging, dense neutrophilic infiltrates and diffuse alveolar damage with edema. Treatment with either anti-H3 mAb significantly diminished pathologic changes (Figure 6).
Fig. 6. Treatment with anti-H3 mAbs diminishes lung damage associated with viral pneumonia caused by HK68/PR8 reassortant virus. 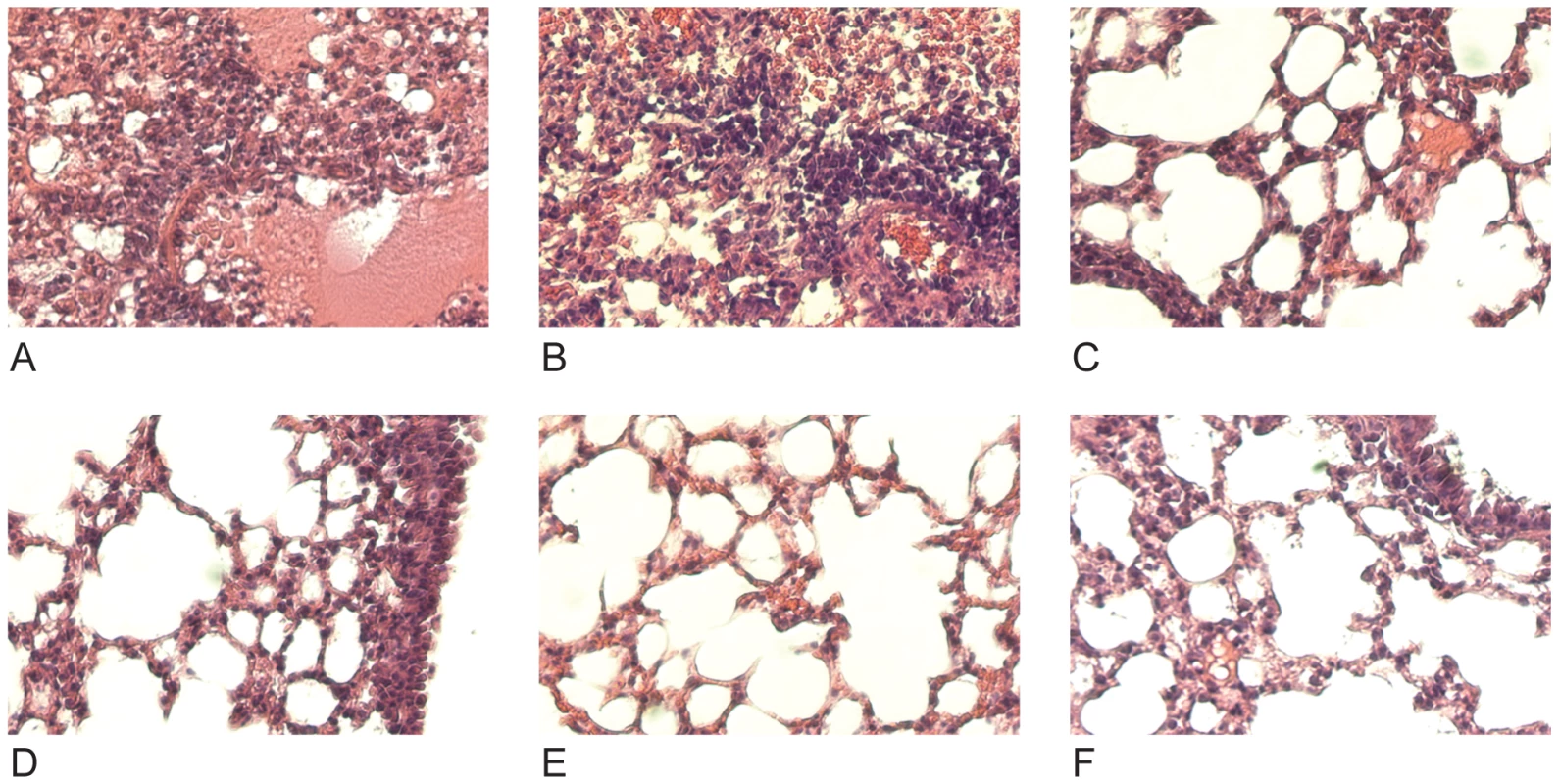
(A,B) Untreated (C,D) mice treated with mAb 39A4 (E,F) mice treated with mAb 12D1. 40× magnification. Having demonstrated protective activity in vivo against the A/HK/68 reassortant virus we sought to evaluate cross-protection mediated by mAbs 12D1 and 39A4 against a second H3 virus, A/Georgia/1981. MAbs 12D1 and 39A4 were administered as described above to BALB/c mice one hour prior to infection. Mice were then infected intranasally with 2700 pfu A/Georgia/1981 and lung titers were evaluated two days post infection. The anti-H3 mAbs were found to reduce lung titers by 97.75% (12D1) or 99.03% (39A4) (Figure 7).
Fig. 7. Anti-H3 mAbs protect against replication of H3 virus in lungs. 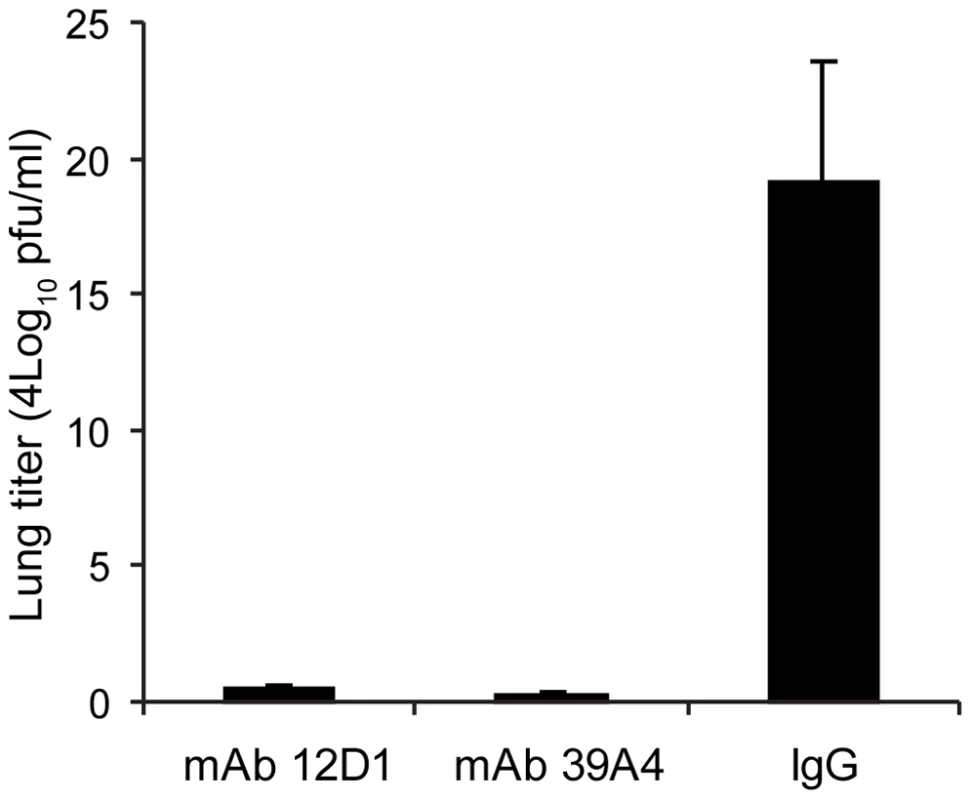
Mice were given 30mg/kg mAb 12D1, 39A4 or isotype control by intraperitoneal injection 1 hour prior to infection with A/Georgia/1981 virus. Data represent lung titers from groups of 5 mice, 2 days post infection. Anti-H3 mAbs act by inhibiting viral fusion
In order to determine the mechanism of virus neutralization by our anti-H3 mAbs, we first looked at the ability of the mAbs to inhibit virus hemagglutination of chicken red blood cells. We found that none of the three mAbs had hemagglutination inhibition activity, suggesting that the mAbs did not act by obstructing the binding of virus to the host-cell.
Next, we tested the effect of the anti-H3 mabs on virus fusion. MAbs 7A7, 12D1 and 39A4 were determined to inhibit the low-pH fusion of A/HK/1968 virus with chicken red blood cells by at least 80% at 10ug/ml (Figure 8).
Fig. 8. Red blood cell fusion assay. 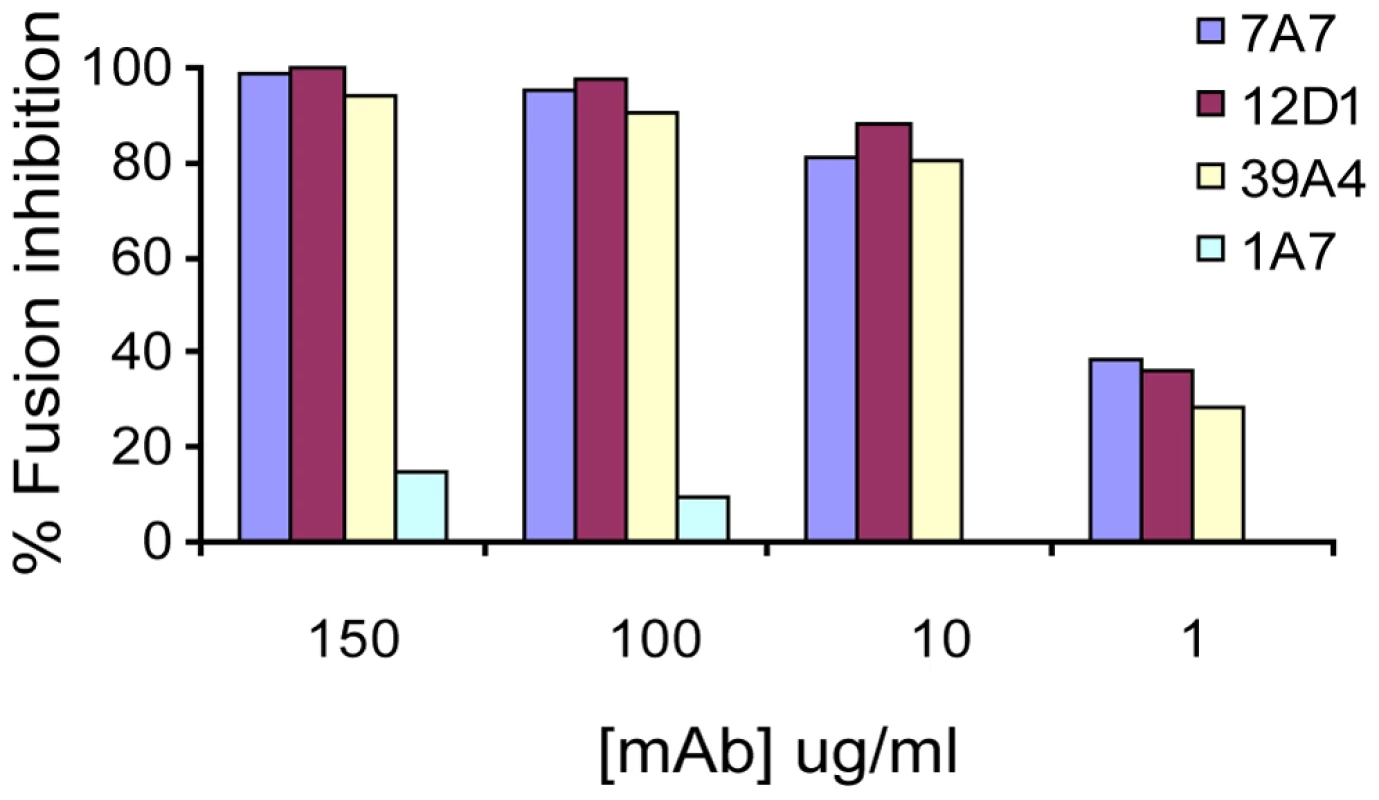
Anti-H3 mabs inhibit low-pH induced fusion of HK/68 hemagglutinin with chicken red blood cells. All mAbs are negative for hemagglutinin-inhibition activity. MAb 1A7 is specific for influenza virus NS1 protein. Binding epitope of mab 12D1
Finally, we aimed to identify the region of the H3 hemagglutinin that might elicit antibodies with fine specificities mirroring those of 12D1 or 39A4. Sixteen passages of A/HK/1968 virus in the presence of the anti-H3 mAbs 12D1 or 39A4 did not yield escape variants that might have assisted in identification of the binding epitopes. Also, the hemagglutinin of six plaques present after incubation of A/HK/1968 virus with 50ug/ml mAb 12D1 or 39A4 in a plaque assay was sequenced and we were surprised to find no changes from the wild-type hemagglutinin.
Because mAb 12D1 mediates protection against influenza disease in vivo and reacts with a continuous epitope of the viral hemagglutinin (no trimeric structure required), as evidenced by reactivity with the denatured hemagglutinin monomer by western blot (Figure 1), we focused on identification of the 12D1 binding epitope. Hemagglutinin truncation mutants consisting of hemagglutinin segments of varying length fused to GFP were generated. GFP expression was utilized to assess expression of the constructs in transfected 293T cells. By analysis of the truncation mutants, it was determined that the 12D1 paratope makes dominant interactions with the HA2 subunit in the region of amino acids 30–106. Diminished 12D1 binding without diminished GFP expression in the 76–184 and 91–184 truncations along with loss of binding with the 106–184 truncation suggested that 12D1 binding is dependent on contacts with amino acids in the HA2 76–106 region (Figure 9). These 30 amino acids fall within the membrane distal half of the long alpha-helix of HA2 (Figure 10). The 12D1 paratope may have additional contacts with amino acids outside of this region (in HA1 or HA2) that are not required for binding by western blot.
Fig. 9. MAb 12D1 reacts by western blot with hemagglutinin truncation mutants. 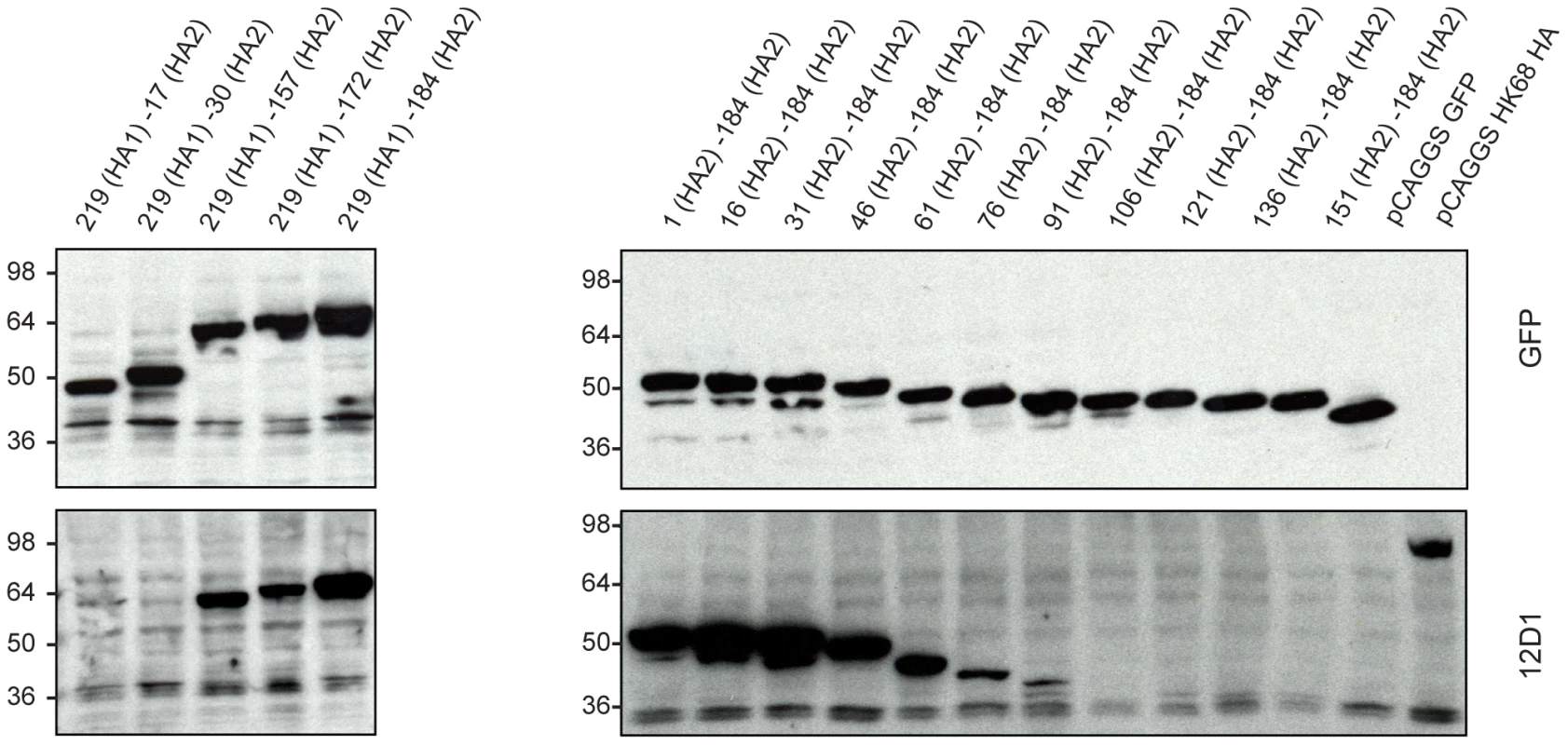
12D1 makes dominant contacts with the HA2 subunit in the region of amino acids 30 to 106 (H3 numbering[21]). Diminished 12D1 binding without diminished GFP expression in the HA2 76–184 and HA2 91–184 truncations along with loss of binding with the HA2 106–184 truncation suggests that the binding epitope lies in the region from amino acids HA2 76–106. These 30 amino acids fall within the membrane distal half of the long alpha-helix of HA2. Fig. 10. Region of dominant contacts between anti-H3 mab 12D1 and HA2. 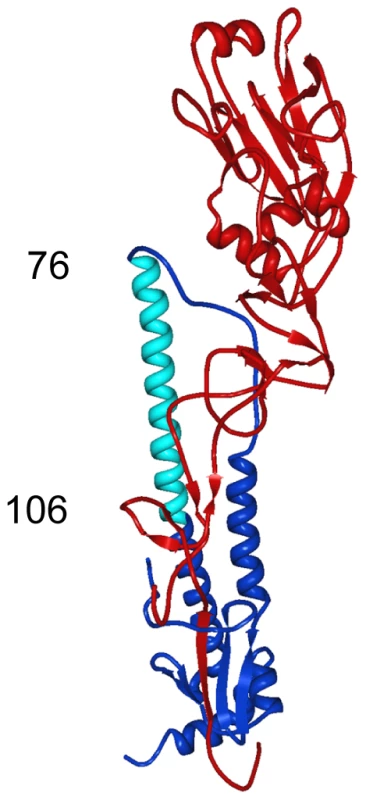
mAb 12D1 makes contact in the HA2 region (blue) of the viral hemagglutinin. Evaluation of binding to hemaggluinin truncation mutants demonstrates binding within the region of amino acids 76–106 of HA2 (light blue). Discussion
For this study, we developed an immunization schedule that elicited broadly-neutralizing antibodies against H3 influenza viruses in vivo. The finding that such antibody specificities can be elicited by vaccination of mice suggests that with the proper immunogen(s) and vaccination protocol, such a response might also be elicited in humans. Several recent studies describe antibodies isolated from human phage display libraries that have cross-neutralizing activity against Group 1 influenza viruses [11],[12],[15],[16]. Mabs isolated from human display libraries have proved extremely useful in the characterization of structural epitopes that mediate heterosubtypic neutralization. Caveats to this methodology exist, however, since the diversity of combinatorial display libraries is typically orders of magnitude greater than the diversity of the true human variable region repertoire [19]. Additionally, phage display libraries are generated by random combination of immunoglobulin VH and VL genes and are therefore not restricted, as the in vivo repertoire is, by mechanisms regulating the production of auto-reactive specificities.
Until now, broadly neutralizing antibodies reactive with H3 viruses have not been described. Interestingly, mAbs 7A7 and 39A4 react by immunofluorescence with the hemagglutinin of multiple subtypes, though neutralizing activity appears to be limited to H3 viruses. Binding by these mAbs to other subtypes may be of relatively low avidity such that they no longer mediate neutralization, or, they may simply bind an epitope of non-H3 hemagglutinins that does not mediate neutralization. The identification of anti-H3 mAbs 12D1 and 39A4 complements recent works describing antibodies F10 and CR6261 that neutralize an array of Group 1 viruses [11],[12],[16]. One might envision a passive transfer therapy consisting of multiple broadly neutralizing mAbs for general use against pandemic and seasonal influenza virus strains. With the increasing resistance of influenza virus isolates to available anti-viral drugs, such an antibody cocktail could be of great value in severe disease.
Mouse monoclonal antibodies such as the anti-H3 mabs described herein are commonly used in the development of therapeutic antibodies for use in humans. Once characterized, rodent antibodies are readily humanized by methods typically involving grafting of non-human complimentary determining regions into appropriate human frameworks followed by cloning of variable region segments into complete human immunoglobulin constructs [20].
The fact that escape mutants were not selected after multiple passages of virus in the presence of anti-H3 mabs 12D1 and 39A4 is intriguing. Sui et al. reported that they were similarly unable to isolate escape mutants using their fusion-inhibiting mAb F10 [11]. MAb F10 makes multiple interactions with the hydrophobic pocket of the hemagglutinin including with the fusion peptide itself and prevents the low-pH triggered conformation change required for fusion. Considering the rigid structural and electrostatic requirements involved in membrane fusion, the hemagglutinin might not readily accommodate mutations at the F10 binding epitope. Anti-H3 mAb 39A4 binds a conformational epitope of the hemagglutinin trimer; the region of binding may bridge two monomers, therefore interacting with two different portions/faces of each monomer. A mutation at one region of contact (that does not affect trimer formation) may not be sufficient to ablate 39A4 binding. Anti-H3 mAb 12D1 likely binds within the long alpha-helix of HA2. This region may not accommodate changes that would affect 12D1 binding due to required secondary helix structure and specific van der Waals interactions that stabilize the hemagglutinin trimer [21]. Generally, mutations in the stalk of the hemagglutinin are more likely to affect the architecture of the entire molecule than are mutations in the classical antigenic sites [9].
The development of HA2-based vaccine constructs is of significant interest given recent reports of anti-HA2 mAbs with broad neutralizing activity against influenza viruses. Original studies of immunogens consisting of virus particles lacking the HA1 subunit demonstrated that design of an effective construct, however, will likely not be straightforward [22]. This is in large part due to the difficulty involved in maintaining the native configuration of the hemagglutinin stalk, which has complex tertiary structure and incorporates a portion of HA1 in addition to the HA2 subunit. Recent reports of mAbs with broad neutralizing activity against influenza viruses that are not active by western blot and that make contacts with amino acids in both HA1 and HA2 underscore the importance of maintaining non-contiguous epitopes in HA2 vaccine contructs [12],[16],[20].
In contrast to these mAbs, anti-H3 mAb 12D1 does not rely on a structural/non-contiguous epitope of the hemagglutinin stalk for binding. The observation that 12D1 makes dominant contacts within a continuous segment of the HA2 subunit suggests the design of an immunogen, perhaps consisting of that HA2 segment coupled to a carrier protein, that would direct an immune response to the region. The identified region, HA2 76–106, is 100% conserved between the H3 viruses used in this study and all other H3 viruses that we have examined. In contrast, the H1 viruses A/New Caledonia/20/99 and A/PR/8/34 share only 56.7% identity with the equivalent region in the H3 hemagglutinin. A vaccine construct incorporating this region, therefore, would likely not provide protection against H1 influenza viruses. This study and other structural studies [11],[12],[14] of the influenza hemagglutinin provide groundwork for the design of novel vaccine constructs aimed at providing broad-spectrum immunity to influenza viruses.
Materials and Methods
Animals
6 week old female BALB/c mice from Jackson Laboratory were used for all experiments. All animal procedures performed in this study are in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines, and have been approved by the IACUC of Mount Sinai School of Medicine.
Viruses and cells
Madin Darby canine kidney cells from ATCC were used for all cell based assays. Cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum, and 100 units/ml of penicillin-100 µg/ml of streptomycin. All viruses were propagated in eggs. Viruses used in various studies: A/Hong Kong/1/1968 (HK/68) (H3), A/Alabama/1/1981 (AL/81) (H3), A/Georgia/1981 (H3), A/Beijing/47/1992 (BJ/92) (H3), A/Wyoming/3/2003 (H3), A/Wisconsin/67/2005 (WI/05) (H3), A/Brisbane/10/2007 (BR/07) (H3), A/New York/2008 (NY08) (H3), A/Texas/36/1991 (TX/91) (H1), A/New Caledonia/20/99 (N.Cal/99) (H1), A/Duck/England/1962 (Dk/62) (H4), A/Turkey/England/1963 (Tky/63) (H7), A/Equine/Kentucky/2002 (e/KY/02) (H3), A/Ann Arbor/6/1960 (AA/60) (H2), A/Fort Monmouth/1/1947 (FM/47) (H1). Purified virus was prepared by high speed centrifugation (43,000 rpm, 1 hour) of allantoic fluid through a 20% sucrose cushion.
Antibody preparations
Hybridoma supernatants were used for screening of mAbs for reactivity by enzyme-linked immunosorbent assay (ELISA) and by western blot. For other assays, purified monoclonal antibody or ascites preparations treated with receptor-destroying enzyme [23] were used. RDE –treated ascites was used for measurement of binding by ELISA, microneutralization, plaque reducion and fusion assays. Antibodies were purified by methods previously described [24]. Because of differences in isotypes, Protein A-agarose (Roche) was used for purification of mAbs 7A7 and 39A4 while protein G-agarose (Roche) was used for purification of mAb 12D1.
Immunization of mice and hybridoma production
6-week old BALB/c mice were immunized with DNA constructs coding for the open-reading frame of influenza virus hemagglutinin in the pCAGGS plasmid [25]. Individual immunizations were given intramuscularly, 3-weeks apart and consisted of 100ug DNA in 100ul PBS. Hemagglutinins utilized in the immunization schedule were cloned from the following parental viruses - primary immunization: A/Hong Kong/1/1968, secondary immunization: A/Alabama/1/1981, tertiary immunization: A/Beijing/47/1992 HA. Three days prior to fusion, mice were boosted with 50ug purified A/Wyoming/3/2003 virus. B cell hybridomas were produced by methods previously described [26],[27].
Screening of hybridoma supernatants
Hybridoma supernatants were screened by blot and by ELISA for reactivity with A/Hong Kong/1/1968 virus. For the ELISA, direct binding to wells coated with 5ug/ml purified virus, 50ul per well was assessed. For the blot assay, 10ug purified virus was adsorbed onto nitrocellulose strips and individual strips were incubated with hybridoma supernatants. For the ELISA and blot assays, binding of antibody to virus was detected using goat anti-mouse γ-chain horse radish peroxidase secondary antibody (SouthernBiotech, Birmingham, Al). All wells that had activity in either assay against A/Hong Kong/1/1968 virus were subcloned repeatedly to ensure the monoclonality of the hybridoma populations.
Western blots
Blots were produced by methods previously described [28]. Samples were boiled for 5 minutes at 100°C in loading buffer containing SDS and 0.6M DTT. SDS migration buffer was used for electrophoresis. For non-reducing gel conditions samples were prepared in loading buffer with SDS but without reducing agent and were not boiled.
Immunofluorescence test
MDCK cells were infected with virus at a multiplicity of infection of 1 and incubated for 6 hours at 37°C. Infected and uninfected cells were incubated with 1ug/ml mAb for 1 hour at room temperature. Goat anti-mouse fluorescein conjugate (SouthernBiotech) was used for detection of mAb binding.
Microneutralization assay
Two stable cell lines were generated that expressed the HA of A/Hong Kong/1/1968 virus or A/Panama/2007/1999 virus. Pseudotyped viruses expressing the HA of either cell line were generated by infection of cells with a virus that carries seven segments from A/WSN/33 virus (all except the HA segment) and one segment encoding Renilla luciferase. Pseudotyped viruses expressing the HA of A/Hong Kong/1/1968 virus or A/Panama/2007/1999 virus were used as the neutralization target. Viruses were incubated with mAb at room temperature for 30 minutes, rocking. Purified polyclonal mouse IgG (Invitrogen) was used for the negative control. The mixture containing virus and mAb was then transferred to wells of a 96-well plate seeded to confluency with MDCK cells and incubated for 12 hours at 37°C. Individual determinants were performed in triplicate. After incubation, luciferase activity in cell-lysates was measured as a read-out of virus infection (Renilla luciferase assay system, Promega).
Plaque reduction assay
Antibody and virus (∼50 pfu/well) were co-incubated at room temperature for 30 minutes, rocking. 6 well plates seeded with MDCK cells were washed once with PBS and 200ul of virus and mAb was added to each well then incubated for 20 minutes, 37°C. Virus with mAb was aspirated from cells and an agar overlay containing antibody was added to each well. Plates were incubated for 3 days, 37°C and plaques were counted by crystal violet staining. Purified mouse IgG (Invitrogen) was used for the negative control.
Passive transfer experiments
Before infection, mice were anesthetized by intraperitoneal administration of a ketamine (75 mg/kg of body weight)/xylazine (15 mg/kg of body weight) mixture. 6 week old BALB/c mice were given 30mg/kg mAb intraperitoneally either one hour before, 24 hours after or 48 hours after challenge with 10 LD50 A/Hong Kong/1/1968, A/PR/8/34 reassortant virus or 2700 pfu A/Georgia/1981 virus (lung titer experiment). Purified mouse IgG (Invitrogen) was used for the negative control. Virus was suspended in PBS and administered intranasally in 50ul (25ul per nostril). Mice were weighed daily and sacrificed if they fell to 75% of starting weight. For the lung titer experiment, mouse lungs were harvested 4 days post infection with A/Georgia/1981 and virus titers in lung homogenates were determined by plaque assay. For histologic evaluation of lung damage, lungs were harvested 4 days post infection with A/Hong Kong/1/1968 - A/PR/8/34 reassortant virus. Tissues were imbedded in paraffin and sections were stained with hematoxylin and eosin.
Hemagglutinin inhibition assay and fusion assay
MAbs were tested in a standard hemagglutination inhibition assay [29] using chicken red blood cells and A/Hong Kong/1/1968 virus. For the red blood cell fusion assay, virus was incubated with chicken red blood cells (2% final red cell concentration) on ice for 10 minutes. Dilutions of antibody were added and samples were incubated on ice for 30 minutes. Sodium citrate buffer, pH 4.6 was then added to bring the final pH to 5.0 and samples were incubated for 30 minutes at room temperature. Samples were centrifuged for 3 minutes at 3000rpm to pellet red blood cells and supernatants were then transferred to an ELISA plate for determination of NADPH content by optical density measurement (340nm). NADPH was present in the supernatant as a function of fusion-induced red blood cell lysis.
Hemagglutinin truncation mutants
DNA constructs were generated in the pCAGGS plasmid that coded for truncations of the A/HK/1/68 virus hemaggluinin fused to green fluorescent protein. All constructs were sequenced and confirmed. 293T cells were then transfected using Lipofectamine 2000 (Invitrogen, Inc.) with the various pCAGGS encoding the HA-GFP fusion gene. Cell lysates were resolved in a 4–20% Tris-HCl SDS-PAGE gel (Bio-Rad Laboratories) and proteins were blotted onto a Protran nitrocellulose membrane (Whatman). GFP and truncated HA fragments were detected using rabbit anti-GFP (Santa Cruz Biotechnology, Inc.) and anti-H3 mAb 12D1 respectively. Secondary antibodies were anti-rabbit IgG HRP (Dako) and anti-mouse Ig HRP(GE Healthcare).
Zdroje
1. WHO April, 2009 Influenza (Seasonal) Fact sheet 211
2. PiotP
BartosM
GhysPD
WalkerN
SchwartlanderB
2001 The global impact of HIV/AIDS. Nature 410 968 973
3. AhmedR
OldstoneMB
PaleseP
2007 Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol 8 1188 1193
4. ChowellG
BertozziSM
ColcheroMA
Lopez-GatellH
Alpuche-ArandaC
2009 Severe Respiratory Disease Concurrent with the Circulation of H1N1 Influenza. N Engl J Med
5. CDC Seasonal flu; United States Surveillance Data
6. CouchRB
KaselJA
1983 Immunity to influenza in man. Annu Rev Microbiol 37 529 549
7. MartinezO
TsibaneT
BaslerCF
2009 Neutralizing anti-influenza virus monoclonal antibodies: therapeutics and tools for discovery. Int Rev Immunol 28 69 92
8. SkehelJJ
WileyDC
2000 Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69 531 569
9. WileyDC
WilsonIA
SkehelJJ
1981 Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289 373 378
10. VareckovaE
MuchaV
WhartonSA
KostolanskyF
2003 Inhibition of fusion activity of influenza A haemagglutinin mediated by HA2-specific monoclonal antibodies. Arch Virol 148 469 486
11. SuiJ
HwangWC
PerezS
WeiG
AirdD
2009 Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16 265 273
12. EkiertDC
BhabhaG
ElsligerMA
FriesenRH
JongeneelenM
2009 Antibody recognition of a highly conserved influenza virus epitope. Science 324 246 251
13. RussellRJ
KerryPS
StevensDJ
SteinhauerDA
MartinSR
2008 Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 105 17736 17741
14. OkunoY
IsegawaY
SasaoF
UedaS
1993 A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67 2552 2558
15. KashyapAK
SteelJ
OnerAF
DillonMA
SwaleRE
2008 Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A 105 5986 5991
16. ThrosbyM
van den BrinkE
JongeneelenM
PoonLL
AlardP
2008 Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3 e3942 doi:10.1371/journal.pone.0003942
17. SmithDJ
LapedesAS
de JongJC
BestebroerTM
RimmelzwaanGF
2004 Mapping the antigenic and genetic evolution of influenza virus. Science 305 371 376
18. MoranTM
MonestierM
LaiAC
NortonG
RealeMA
1987 Characterization of variable-region genes and shared crossreactive idiotypes of antibodies specific for antigens of various influenza viruses. Viral Immunol 1 1 12
19. BowleyDR
JonesTM
BurtonDR
LernerRA
2009 Libraries against libraries for combinatorial selection of replicating antigen-antibody pairs. Proc Natl Acad Sci U S A 106 1380 1385
20. MarascoWA
SuiJ
2007 The growth and potential of human antiviral monoclonal antibody therapeutics. Nat Biotechnol 25 1421 1434
21. WilsonIA
SkehelJJ
WileyDC
1981 Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289 366 373
22. GravesPN
SchulmanJL
YoungJF
PaleseP
1983 Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: unmasking of cross-reactive HA2 determinants. Virology 126 106 116
23. JordanWSJr
OseasohnRO
1954 The use of RDE to improve the sensitivity of the hemagglutination-inhibition test for the serologic diagnosis of influenza. J Immunol 72 229 235
24. HarlowE
LaneD
1988 Antibodies: a laboratory manual Cold Spring Harbor, NY Cold Spring Harbor Laboratory xiii, 726
25. BaslerCF
ReidAH
DybingJK
JanczewskiTA
FanningTG
2001 Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci U S A 98 2746 2751
26. de StGrothSF
ScheideggerD
1980 Production of monoclonal antibodies: strategy and tactics. J Immunol Methods 35 1 21
27. KohlerG
MilsteinC
1975 Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256 495 497
28. TowbinH
StaehelinT
GordonJ
1979 Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76 4350 4354
29. CohenA
NewlandSE
Biddlef
1963 Inhibition of Influenza Virus Haemagglutination: a Difference of Behavior in Sera from a Single Species. Virology 20 518 529
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek HIV Controller CD4+ T Cells Respond to Minimal Amounts of Gag Antigen Due to High TCR AvidityČlánek Transit through the Flea Vector Induces a Pretransmission Innate Immunity Resistance Phenotype in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 2- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Pathogen Entrapment by Transglutaminase—A Conserved Early Innate Immune Mechanism
- Broadly Protective Monoclonal Antibodies against H3 Influenza Viruses following Sequential Immunization with Different Hemagglutinins
- Neutrophil-Derived CCL3 Is Essential for the Rapid Recruitment of Dendritic Cells to the Site of Inoculation in Resistant Mice
- Differentiation, Distribution and γδ T Cell-Driven Regulation of IL-22-Producing T Cells in Tuberculosis
- IFN-α-Induced Upregulation of CCR5 Leads to Expanded HIV Tropism In Vivo
- An Extensive Circuitry for Cell Wall Regulation in
- TgMORN1 Is a Key Organizer for the Basal Complex of
- Direct Presentation Is Sufficient for an Efficient Anti-Viral CD8 T Cell Response
- Immunoelectron Microscopic Evidence for Tetherin/BST2 as the Physical Bridge between HIV-1 Virions and the Plasma Membrane
- A New Nuclear Function of the Glycolytic Enzyme Enolase: The Metabolic Regulation of Cytosine-5 Methyltransferase 2 (Dnmt2) Activity
- Genome-Wide mRNA Expression Correlates of Viral Control in CD4+ T-Cells from HIV-1-Infected Individuals
- Structural and Biochemical Characterization of SrcA, a Multi-Cargo Type III Secretion Chaperone in Required for Pathogenic Association with a Host
- A Major Role for the ApiAP2 Protein PfSIP2 in Chromosome End Biology
- HIV Controller CD4+ T Cells Respond to Minimal Amounts of Gag Antigen Due to High TCR Avidity
- Fis Is Essential for Capsule Production in and Regulates Expression of Other Important Virulence Factors
- Vaccinia Protein F12 Has Structural Similarity to Kinesin Light Chain and Contains a Motor Binding Motif Required for Virion Export
- A Novel Pseudopodial Component of the Dendritic Cell Anti-Fungal Response: The Fungipod
- Efficacy of the New Neuraminidase Inhibitor CS-8958 against H5N1 Influenza Viruses
- Long-Lived Antibody and B Cell Memory Responses to the Human Malaria Parasites, and
- IPS-1 Is Essential for the Control of West Nile Virus Infection and Immunity
- Transit through the Flea Vector Induces a Pretransmission Innate Immunity Resistance Phenotype in
- Ats-1 Is Imported into Host Cell Mitochondria and Interferes with Apoptosis Induction
- Six RNA Viruses and Forty-One Hosts: Viral Small RNAs and Modulation of Small RNA Repertoires in Vertebrate and Invertebrate Systems
- The Syk Kinase SmTK4 of Is Involved in the Regulation of Spermatogenesis and Oogenesis
- Optineurin Negatively Regulates the Induction of IFNβ in Response to RNA Virus Infection
- On the Diversity of Malaria Parasites in African Apes and the Origin of from Bonobos
- Five Questions about Viruses and MicroRNAs
- A Broad Distribution of the Alternative Oxidase in Microsporidian Parasites
- Caspase-1 Activation via Rho GTPases: A Common Theme in Mucosal Infections?
- Peptides Presented by HLA-E Molecules Are Targets for Human CD8 T-Cells with Cytotoxic as well as Regulatory Activity
- Interaction of Rim101 and Protein Kinase A Regulates Capsule
- Distinct External Signals Trigger Sequential Release of Apical Organelles during Erythrocyte Invasion by Malaria Parasites
- Exacerbated Innate Host Response to SARS-CoV in Aged Non-Human Primates
- Reverse Genetics in Predicts ARF Cycling Is Essential for Drug Resistance and Virulence
- Universal Features of Post-Transcriptional Gene Regulation Are Critical for Zygote Development
- Highly Differentiated, Resting Gn-Specific Memory CD8 T Cells Persist Years after Infection by Andes Hantavirus
- Arterivirus Nsp1 Modulates the Accumulation of Minus-Strand Templates to Control the Relative Abundance of Viral mRNAs
- Lethal Antibody Enhancement of Dengue Disease in Mice Is Prevented by Fc Modification
- Quantitative Comparison of HTLV-1 and HIV-1 Cell-to-Cell Infection with New Replication Dependent Vectors
- The Disulfide Bonds in Glycoprotein E2 of Hepatitis C Virus Reveal the Tertiary Organization of the Molecule
- IL-1β Processing in Host Defense: Beyond the Inflammasomes
- Kaposi's Sarcoma Associated Herpes Virus (KSHV) Induced COX-2: A Key Factor in Latency, Inflammation, Angiogenesis, Cell Survival and Invasion
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Caspase-1 Activation via Rho GTPases: A Common Theme in Mucosal Infections?
- Kaposi's Sarcoma Associated Herpes Virus (KSHV) Induced COX-2: A Key Factor in Latency, Inflammation, Angiogenesis, Cell Survival and Invasion
- IL-1β Processing in Host Defense: Beyond the Inflammasomes
- Reverse Genetics in Predicts ARF Cycling Is Essential for Drug Resistance and Virulence
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



