-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
For those studying the effect of genotype on human traits, a collection of genetically diverse renewable cell lines can be an indispensable resource. B-cells immortalized with Epstein-Barr virus, also known as lymphoblastoid cell lines or LCLs, have been particularly favored as such a model because they are easy to generate from donor blood samples and already exist in large panels representing many ethnic and disease populations. However, long-term maintenance of LCL cultures involves practices that reduce the ability of the model to reproduce donor differences in gene expression, potentially compromising the genotype-phenotype relationship. Induced pluripotent stem cells (iPSCs) are increasingly used to study the physiology of primary tissue, and unlike LCLs, have been found to retain a strong donor effect. Recent advances have made it possible to generate iPSCs from LCLs using reprogramming vectors that do not integrate into the genome. Here, we report that reprogramming highly manipulated LCLs to iPSCs can recover donor gene expression signatures that had been lost during long-term LCL maintenance. Our findings suggest that iPSCs generated from LCL panels are well suited for studies of the genetic basis for individual phenotypic variation.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005216
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005216Summary
For those studying the effect of genotype on human traits, a collection of genetically diverse renewable cell lines can be an indispensable resource. B-cells immortalized with Epstein-Barr virus, also known as lymphoblastoid cell lines or LCLs, have been particularly favored as such a model because they are easy to generate from donor blood samples and already exist in large panels representing many ethnic and disease populations. However, long-term maintenance of LCL cultures involves practices that reduce the ability of the model to reproduce donor differences in gene expression, potentially compromising the genotype-phenotype relationship. Induced pluripotent stem cells (iPSCs) are increasingly used to study the physiology of primary tissue, and unlike LCLs, have been found to retain a strong donor effect. Recent advances have made it possible to generate iPSCs from LCLs using reprogramming vectors that do not integrate into the genome. Here, we report that reprogramming highly manipulated LCLs to iPSCs can recover donor gene expression signatures that had been lost during long-term LCL maintenance. Our findings suggest that iPSCs generated from LCL panels are well suited for studies of the genetic basis for individual phenotypic variation.
Introduction
Renewable cell models are widely recognized as valuable platforms for studies of human genotype-phenotype interactions because they are easily manipulated, scalable, and are specific to human physiology (in contrast to lab animal models). Epstein-Barr virus (EBV) transformed lymphoblastoid cell lines (LCLs) are one such commonly-used model. In recent years, LCLs have been used to study genetic influence on disease traits [1], drug response [2–5], and gene regulation [6,7]. In particular, much of what we now know about associations of human genetic variation with differences in gene regulation is based on studies that used data from LCLs. There is little doubt that many fundamental regulatory principles that we have learned by generating and analyzing data from LCLs are generally shared with primary tissues. However, a critical property of any in vitro cellular model is the ability to faithfully recapitulate the specific regulatory properties of the donor’s primary tissue. In that regard, though LCLs have clearly been a convenient and useful model, there is concern that factors related to immortalization and cell line maintenance obscure genetic signal in LCLs [8–10].
A number of studies have characterized differences in gene regulatory phenotypes between LCLs and primary tissues [11–15]. These have shown that a large number of genes are differentially expressed between primary cells and cell lines, and that thousands of CpG sites are differentially methylated between LCLs and primary blood cells. Our group has also demonstrated disruptions in gene regulation in LCLs by studying multiple independent replicates of LCLs from isolated primary B cells of six individuals and repeatedly subjecting the cell lines to cycles of freeze, thaw, and recovery. We found that newly transformed LCLs (within a few passages after the EBV transformation) largely maintained individual differences in gene expression levels. However, LCLs that had been frozen and thawed at least once (we referred to these as mature LCLs) exhibited a substantial loss of inter-individual variation in gene expression levels [14,16].
On the one hand, it is unlikely that the loss of the donor effect on gene expression would lead to false positive findings of genetic influence on gene regulation. Indeed, we reported that genes associated with previously identified eQTLs retain relatively high variation in gene expression levels between individuals even after repeated freeze-thaw culturing cycles. Yet on the other hand, because much of the individual variation observed in primary tissues is not exhibited by LCLs, studies using the LCL model are limited in their ability to detect donor differences.
The induced pluripotent stem cell (iPSC) system is another renewable cell model that is increasingly used to study individual phenotypic variation because it can ultimately provide access to a wide range of tissue types through the use of differentiation protocols. However, the capacity of iPSCs and derived cell types to faithfully recapitulate in vivo physiology is also still largely unknown. Previous studies have noted a significant effect of donor on traits in iPSCs such as hematopoietic [17], neuronal [18] and hepatic [19] differentiation potential. Importantly, the genetic background of iPSCs generated from peripheral blood mononuclear cells and fibroblasts was recently demonstrated to account for more of the variation in gene expression between iPSC lines than any other tested factor such as cell type of origin or reprogramming method [20]. While these findings indicate that reprogramming iPSCs from primary tissues preserves individual variation in gene expression, it is unknown whether reprogramming highly manipulated immortalized cell lines, such as LCLs, to iPSCs can recover the individual gene expression patterns lost during cell line maintenance.
Because LCLs are available in large banks representing disease populations or ethnicities, they are a promising source of starting material for iPSC generation if disruptions in gene regulation do not persist through the reprogramming process. In the present study, we ask whether reprogramming mature LCLs to iPSCs can result in the recovery of individual variation in gene expression that had been lost during the LCL maturation and maintenance process.
Results
To test whether reprogramming LCLs to iPSCs could recover the effect of donor on gene expression profiles, we generated iPSCs from three mature LCLs of each of six Caucasian individuals for a total of 17 pairs of cell lines (one iPSC line failed to reach the requisite ten passages and was excluded from the study; see methods). We have previously collected gene expression data from the LCLs at earlier stages [14,16]. For the current study, we quantified whole genome gene expression microarray data from the 17 mature LCLs immediately prior to reprogramming and from stable and validated iPSCs. See Fig 1 for schematic of the study design and S1 Table for the processed gene expression data from all samples.
Fig. 1. Study design. 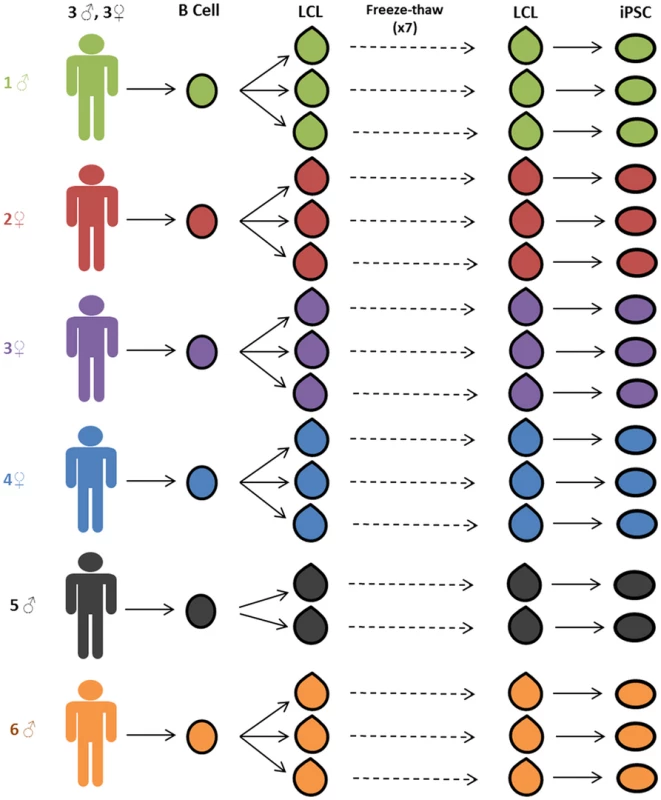
Three independent lymphoblastoid cell lines (LCLs) were generated for each of six unrelated Caucasian individuals. LCLs were frozen and thawed seven times. After the seventh thaw, the LCLs were reprogrammed to iPSCs. Gene expression data was collected from LCLs immediately before reprogramming and from stable iPSC lines. Generation and Validation of the iPSCs
We reprogrammed mature LCLs, which had previously undergone seven freeze-thaw culturing cycles, to iPSCs using an episomal transfection approach [21–23] (see Methods for more details). We reprogrammed the LCLs in four batches; scheduling LCLs derived from the same individual to different reprograming batches to ensure that no artificial correlation structure was introduced between ‘reprograming batch’ and ‘donor individual’ in the process of iPSC generation. All iPSC lines were confirmed to be pluripotent using an embryoid body assay (Fig 2A and S1 Fig), qPCR for pluripotency-associated transcription factors (Fig 2B), genomic PCR to confirm the absence of reprogramming plasmids (S2 and S3 Figs), and PluriTest, a bioinformatic classifier designed to assess pluripotency using gene expression data [24] (S2 Table). Three independently established LCLs were successfully reprogrammed into validated iPSCs for all but one individual, for which only two iPSC lines were obtained.
Fig. 2. iPSC generation and validation. 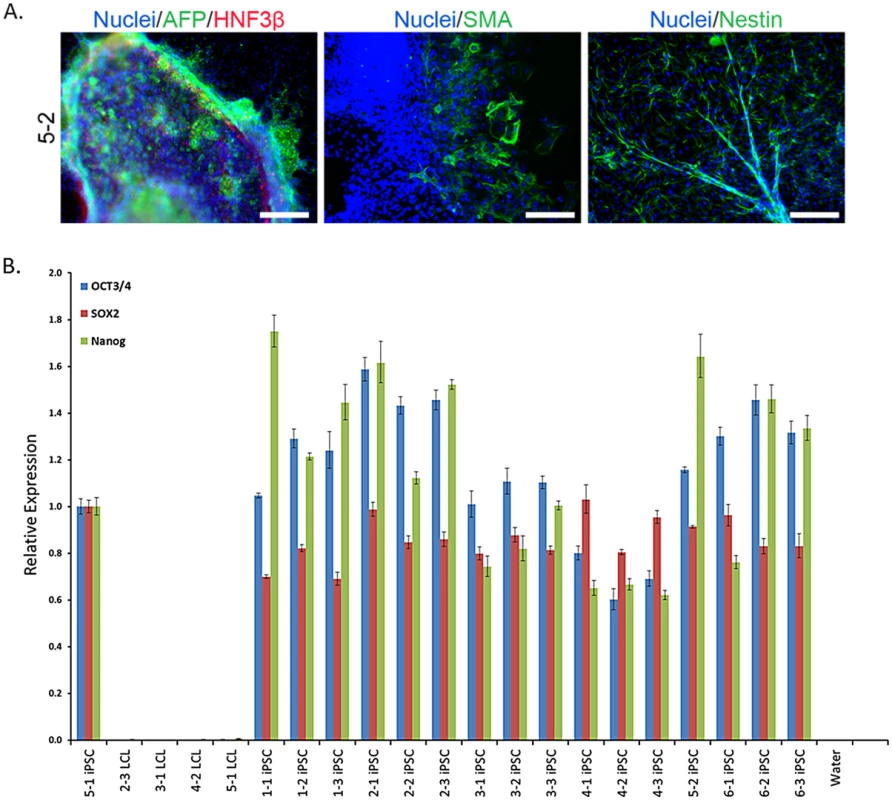
A. Representative embryoid body staining for iPSC line 5–2 demonstrating differentiation potential for endoderm, mesoderm, and ectoderm lineages. See S1 Fig for results from all lines. Scale bars represent 200 μm B. Results from qPCR for three endogenous pluripotency-related transcription factors, normalized to GAPDH. iPSC 5–1 was randomly chosen as a reference sample. Recovery of the Individual Signature of Gene Regulation
We collected high quality RNA (RIN score range: 7.6–9.9; S2 Table) from LCLs immediately prior to reprogramming and from the stable and validated iPSC lines after at least 10 passages (see S2 Table for specific passage information). We quantified gene expression levels for all samples using the Illumina Human HT12v4 microarray platform. As a first step of our analysis, we excluded data from probes whose target transcripts did not map to a unique Ensembl gene ID, those that spanned an exon-exon junction, and those that were not detected as ‘expressed’ in at least two samples from either cell type (we note that our general observations are robust with respect to a wide range of this inclusion criteria). We also excluded from the analysis data from probes with a known SNP with a minor allele frequency > 0.05 in the European population, based on the 1000 Genomes phase I data, to eliminate the possibility of an artificial effect of genotype on the hybridization-based estimates of gene expression levels. We then quantile-normalized the combined data from the remaining probes across all samples. We examined and corrected for array batch using the approach of Johnson et al [25] (see Methods). Finally, we obtained normalized expression levels for 12,243 genes detected as expressed in our samples (S1 Table). Using a linear model-based Empirical Bayes method (implemented in the ‘limma’ R package [26]), we classified 8,185 genes as differentially expressed between iPSCs and LCLs (FDR < 1%; see Methods for more details about modeling and hypothesis testing).
Because the regulation of a large percentage of genes was affected by reprogramming (67% of tested genes), we asked whether gene expression patterns specific to the donor individual were recovered in the process. We addressed this question using two approaches. First, we evaluated the overall degree of similarity across cell lines from the same donor by considering summaries of the gene expression phenotypes using clustering analysis and PCA. The rationale for collapsing our gene-specific expression data and considering overall summaries is that complex phenotypes can often be the result of a large combination of genotype contributions and we are interested to learn whether the overall data from cell lines exhibits a clear signature of the donor. In our second approach, we focused on gene specific patterns by partitioning the variance in expression levels for individual genes and testing for differences between the entire distributions of gene expression levels across cell lines. In this approach we are considering expression patterns of individual genes as independent data points. The rationale for the gene-specific approach is that studies of the genetic basis for regulatory variation (such as eQTL mapping studies) nearly always consider the expression phenotypes of individual genes and we are interested to learn the extent to which the effect of donor genotype on gene expression levels can be studied using a given cell model.
To evaluate overall clustering properties in the expression data from the two cell types, we performed hierarchical clustering analysis and PCA. As we performed these analyses, we consistently observed that data from the second iPSC line of individual 4 (line marked as 4–2 in our figures) accounts for a disproportionate amount of variance (S4B Fig). This individual is a clear outlier and its iPSC is associated with the lowest PluriScore in our study (S2 Table). We have excluded the data from this individual from subsequent analyses. Importantly, we have confirmed (as we show in supplementary figures), that our conclusions our robust with respect to this decision.
Using data from all 12,243 genes detected as expressed, mature LCLs fail to consistently cluster by the individual from whom they were initially derived, in accordance with our previous observations (Fig 3A, S4A and S5A Figs). Data from the corresponding iPSC lines, however, cluster by the individual of origin, indicating a large degree of recovery of donor gene expression patterns (Fig 3B, S4B and S5B Figs). Another method to assess overall clustering properties is through the use of principal components analysis. Taking this approach, we found that clustering of the expression data by individual of origin is substantially more pronounced in the iPSCs than in the LCLs (Fig 3 and S4 Fig). Indeed, the average pairwise Euclidean distances of expression data projections on the first two PCs are significantly smaller within cell lines derived from the same individual than those from different individuals for iPSCs (P < 10-15), but not for LCLs (P = 0.13; S3 Table).
Fig. 3. Improved clustering properties after reprogramming to iPSCs. 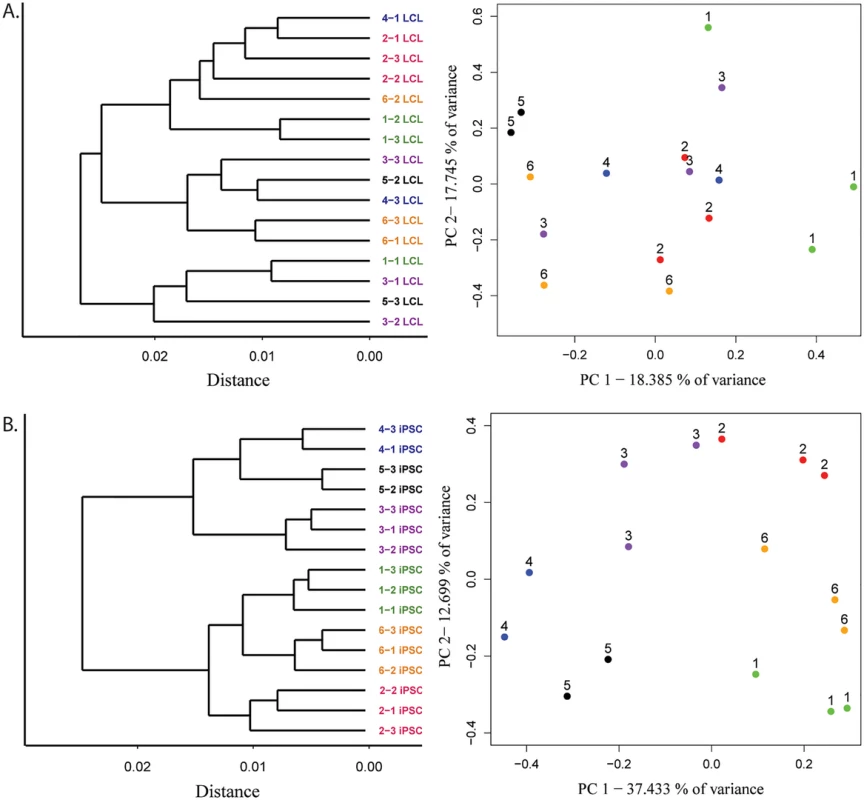
A. Results from hierarchical clustering analysis of microarray gene expression and expression data projections on principal components axes 1 and 2 from cycle 7 LCLs and B. iPSCs. To estimate the magnitude of the donor effect on gene expression patterns in LCLs and iPSCs, we compared the pairwise correlations of expression data from cell lines derived from the same donor to pairwise correlations of data from cell lines derived from different individuals (S6 Fig). On average, both within - and between-donor correlation coefficients are significantly higher in iPSCs than in the LCLs they were initially derived from (p < 10-4 and p < 10-8 for within - and between-donor correlations, respectively). In other words, regardless of the individual of origin, we observed less variation in gene expression between iPSCs than LCLs. Yet, though iPSCs harbor less variation overall, the proportion of variation in gene expression that is explained by donor is significantly higher in the iPSCs compared with the LCLs (P < 10-15). Indeed, using a single factor ANOVA, we estimate that donor explains, on average, 24.5% of the variance in gene expression in iPSCs but only 6.9% in LCLs.
In addition to within-donor correlations, we were specifically interested in identifying genes that were highly variable across donors. We thus proceeded by considering the ratio of between - to within-individual variation in gene expression levels in the two cell types. On average, we found a significantly higher ratio of between-to-within individual variance in gene expression levels in iPSCs compared with data from the LCLs (P < 10-15; recall that in this analysis we consider expression patterns of individual genes as independent data points), despite significantly higher overall variance in LCL gene expression (P < 10-14; Fig 4, and S7 and S8 Figs). We identified 1,620 genes whose expression levels were significantly associated with donor in iPSCs (single factor ANOVA FDR < 0.05; see S9 Fig for histogram of p-values) but only 77 such genes in LCLs.
Fig. 4. Comparison of ability to detect inter-individual gene expression variation. 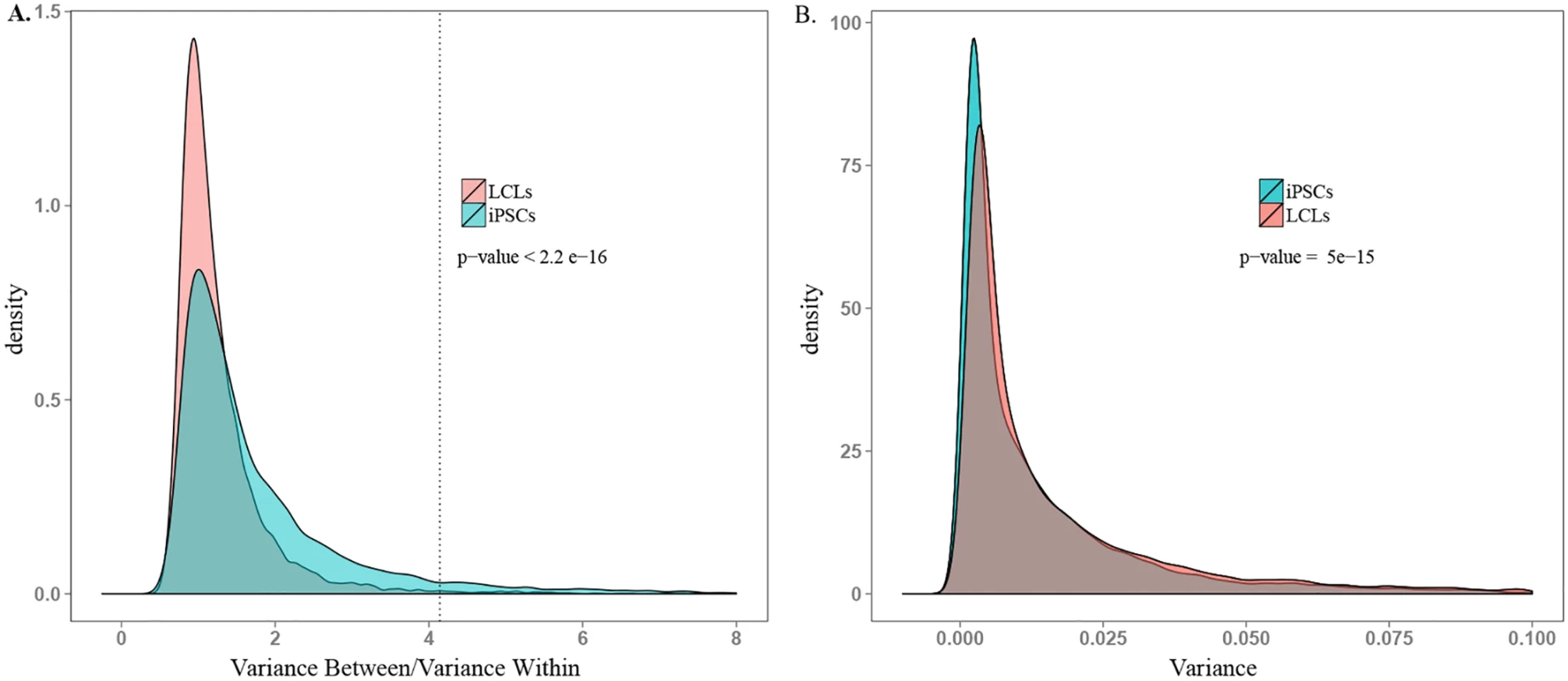
A. Density plot of between donor variance to within donor variance in gene expression for all expressed genes in iPSCs and LCLs. The dotted line indicates the threshold ratio corresponding with significant association between gene expression and donor. X-axis was truncated at 8.0; 0.8% of the data are not plotted here for visualization purposes. B. Density plot of total variance in LCLs and iPSCs. X-axis was truncated at 0.1; 3.7% of the data are not plotted here. See S7 Fig for plots including all the data. Functional Relevance of Highly Variable Genes
We tested for enrichment of functional annotation related to tissue-expression, disease involvement, and biological process (using the online database Lynx [27]) among genes whose expression levels are significantly associated with donor. While these results do not shed much light on the functional importance of these gene sets, we note that genes exhibiting high individual variation in iPSCs are enriched in genes expressed in embryonic tissue while those in LCLs are not significantly enriched in any functional category we tested. The complete set of results for tissue, disease, and biological process enrichment is available in S4 and S5 Tables.
Finally, we considered the relevance of our findings with respect to previously published eQTL studies in LCLs. Our sample of 6 individuals is too small to allow identification of eQTLs. As an alternative, we compared individual variation in expression levels between genes previously associated with an eQTL in LCLs [6], and genes for which an eQTL was not identified. To do so, we randomly selected data from one biological replicate (one LCL and its corresponding iPSC) from each individual.
In both LCLs and iPSCs, the average coefficients of expression variation were significantly higher in genes previously associated with eQTLs than in genes for which eQTLs were not identified (P < 10-10 and P = 0.009, for LCLs and iPSCs, respectively; Fig 5 and S10 Fig). As expected (given that these eQTLs were originally observed in LCLs, and that LCLs have greater overall variation), the coefficients of variation are significantly higher in eQTL-associated genes in LCLs than iPSCs (P < 10-9).
Fig. 5. Genes with eQTLs are highly variable in both cell types. 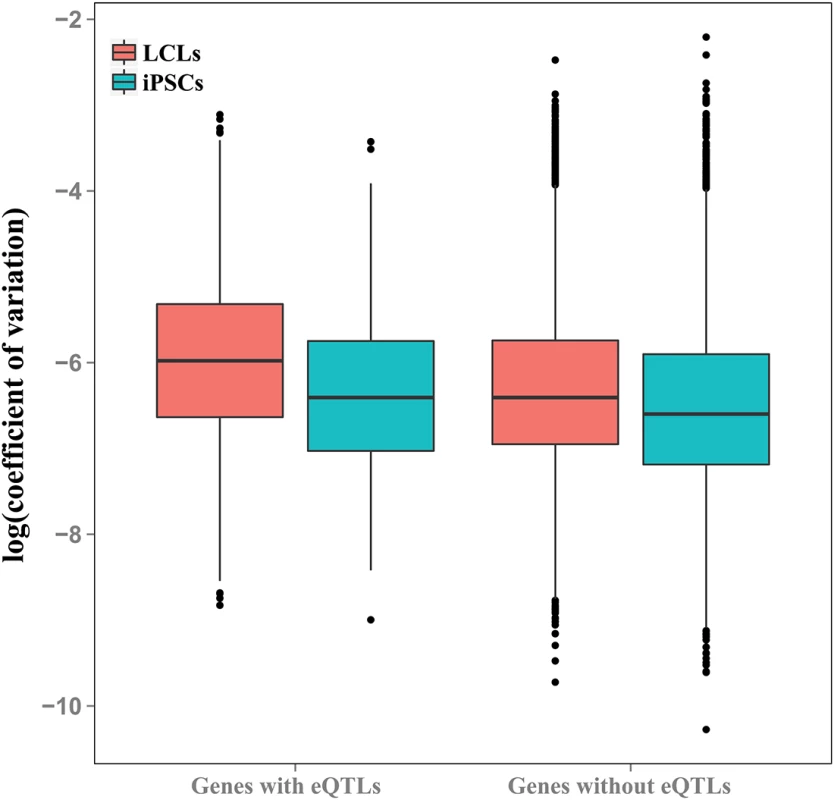
Boxplot of coefficients of variation of gene expression in genes with and without eQTLs previously identified in LCLs [6], plotted for LCLs and iPSCs. Discussion
The utility of renewable cell lines for population genetics studies and as models of complex disease depends on the preservation of the genotype-phenotype relationship in the cell line. Thus, the most useful cell line models would retain a strong influence of individual of origin on their phenotypes, including molecular properties such as gene regulatory patterns. In previous work, we reported that freeze-thaw cycling of LCLs, a standard and required practice in long-term cell line maintenance, reduces the effect of donor on the cell line’s gene expression profile [16]. In fact, we have found that whole-genome gene expression profiles from LCLs that were generated from different individuals are typically as similar to each other as data from independently established replicates of LCLs from the same individual. We suggested that LCLs that have experienced one or more freeze-thaw cycles may be clonally selected for, resulting in a convergent “LCL regulatory phenotype”, which has an advantage growing in culture but masks many of the original gene expression differences between the donor individuals.
Apart from the concern regarding the loss of much of the gene regulatory variation between donors, an intrinsic limitation of the LCL model system is that it theoretically represents the biology of only one primary cell type, B cells. As might be expected, all existing collections of renewable cell lines from human population samples include only easily accessible primary tissues such as blood cells, adipocytes, and skin fibroblasts. Many cell types affected by disease, for example cardiomyocytes, hepatocytes, and neurons, cannot be directly studied using existing human cell line panels. In order to study variation in the most relevant phenotypes and disease processes, we need access to population samples that model additional cellular contexts.
The advent of iPSC technology may have provided the answer. It is now possible to establish renewable iPSC lines from population samples and differentiate them to multiple different cell types for which large collections are currently unavailable. One can establish iPSCs from newly collected fibroblasts or fresh blood samples, but a most attractive possibility is to generate iPSC panels from the already available extensive collections of human LCLs. We thus asked whether individual variation in gene expression levels can be restored by reprograming LCLs into iPSC lines.
Recovery of Individual Variation
We have shown that not only does the iPSC model exhibit a strong effect of donor on overall gene expression, but in fact the process of reprogramming highly manipulated immortalized cell lines to iPSCs recovers the inter-individual variation in gene expression lost during long term cell line maintenance.
The stronger clustering properties of expression data from iPSCs compared to LCLs suggest that iPSCs are better able to capture donor differences in gene regulation than LCLs. We could detect no significant difference between Euclidean distances within - and across - individuals in the projections of gene expression data from LCLs on the first two principal components of variation, indicating that donor is not a significant global source of gene expression variation in the LCL model. This observation is consistent with our previous findings [16]. In contrast, we observed a dramatic increase in the number of genes whose expression was significantly associated with donor in iPSCs, and a higher average variance in expression explained by individual of origin in the iPSCs compared with LCLs. These findings indicate that iPSCs reprogrammed from LCLs are a suitable model for studies of donor differences in gene regulation and genotype-phenotype interactions.
Our work does not provide direct evidence for a mechanism by which iPSCs regain the effect of donor on gene expression. Previously [16] we have hypothesized that the loss of individual variation in gene expression levels in LCLs is due to selection in culture for the fastest growing LCLs. We suggested that this selection results in a convergence to a gene regulatory profile that is common for all mature LCL cultures. In a recent study [28] we have found that DNA methylation profiles in iPSCs reprogrammed from different somatic cell types of the same individuals are practically identical, while we observed hundred of thousands of methylation differences between the precursor somatic cells. Global reprograming of the epigenetic landscape in iPSC lines could potentially be the reason that the LCL gene regulatory signature has been largely replaced by a new regulatory program, which no longer reflects the selection pressures relevant to LCL culturing.
We note that despite their diminished ability to reflect donor differences, attempts to identify instances of genetic regulation of LCL gene expression in mature cell lines have been considered largely successful. Indeed, here we report that genes previously identified as associated with an eQTL in LCLs exhibit higher variance in mature LCLs than those without one. However, our observations suggest that, for future eQTL mapping studies, iPSCs may be a better system than LCLs. While 28.6% of genes with a significant donor effect in LCLs are associated with a previously identified eQTL in LCLs, only 5.9% of genes with a significant donor effect in iPSCs are associated with such an eQTL. Although expressed in LCLs, often at appreciable levels (S11 Fig), the majority (>95%) of genes with a strong donor effect in iPSCs do not show such an effect in LCLs. Put together, these observations support our assertion that iPSC can be a better model than LCLs for detecting eQTLs, and more generally, for studies of inter-individual differences in gene regulation.
Technical Noise Associated with Reprogramming
In any cell model, it is important to consider the magnitude of noise introduced by cell culture relative to biological signal. We note a substantial decrease in within-individual expression correlations for a cell line with a low PluriScore, indicating that we should perhaps reconsider acceptable scores for studies of individual phenotypic variation. However, other technical considerations do not seem to have a marked effect on overall clustering properties. For example, data from the single iPSC line that retained EBV (individual 3, replicate 2; S3 Fig) clustered with the other iPSC lines derived from that individual. Additionally, we reprogrammed iPSCs in four groups and collected expression data at varying passages (between passage 11 and 13, S2 Table) without apparent batch effects. Because it is currently unclear which factors significantly affect our ability to detect donor differences, potential sources of noise need to be more systematically studied and appropriately controlled for.
Much of the excitement surrounding iPSCs is based on their ability to differentiate into terminal cell types, providing a renewable substitute for previously inaccessible tissues. Our study does not provide direct evidence that iPSC-derived differentiated cells will also reflect donor differences, however because the pluripotent state is relatively well-conserved compared to terminal cell types [29,30], we expect that tissues derived from iPSCs will demonstrate an even stronger donor effect on gene expression. That said, we suggest that this expectation needs to be independently confirmed in each differentiated cell type before they are carried into further studies.
Conclusion
Because LCLs are available in large banks that represent panels of ethnic groups and disease populations, they are a popular cell model for genetic research and have been extensively studied. Recent advances in iPSC reprogramming protocols [22,23] have also positioned LCLs as a promising source of starting material for iPSC generation. Here, we have presented the recovery of donor gene expression patterns through the process of reprogramming highly manipulated LCLs to iPSCs, both validating the choice of iPSCs to study donor differences in physiology and the use of LCLs as an appropriate starting material for iPSC generation.
Materials and Methods
Ethics Statement
In this study, blood samples from Research Blood Components were analyzed anonymously. Research Blood Components obtained IRB approval and written informed consent from each donor, giving permission to collect their blood and use or sell it at Research Blood Components's discretion, for research purposes.
Sample Acquisition
Whole blood was collected from six healthy Caucasian donors by Research Blood Components LLC (Brighton, MA) with IRB consent between 2009 and 2010. B-Cell isolation and LCL generation were performed at the University of Chicago as described previously [14]. Between February 2011 and October 2012, each line was thawed, cultured, and re-frozen every three months, for a total of six freeze-thaw cycles prior to use in our study [16]. LCLs were cultured in RPMI with 20% FBS and frozen in Recovery Cell Culture Freezing Media (Life Technologies).
iPSC Generation and Validation
All cell culture was performed at 37°C, 5% CO2, and atmospheric O2. From each individual, three biological replicates of LCLs were reprogrammed to iPSCs using a similar method to that described previously [22,23]. LCLs were transfected in four batches between August 2013 and January 2014 (S2 Table). One million cells were transfected with 2 μg of each episomal plasmid encoding OCT3/4, shP53, Lin28, SOX2, L-MYC, KLF4, and GFP(Addgene plasmids 27077, 27078, 27080, 27082 [21]) using the Amaxa transfection program X-005. For more details see: http://giladlab.uchicago.edu/data/LCL_Reprogramming.pdf. Transfected cells were grown in suspension for a week in hESC media (DMEM/F12 supplemented with 20% KOSR, 0.1mM NEAA, 2mM GlutaMAX, 1% Pen/Strep, 0.1 mM BME, and 12.5 ng/mL human bFGF) supplemented with 0.5mM sodium butyrate between days 2–12 post-nucleofection. After seven days, cells were plated on gelatin-coated plates with CF-1 irradiated mouse embryonic fibroblasts and manually passaged as colonies for at least 10 passages. After day 12, cells were grown in hESC media without sodium butyrate. Media was changed every 48 hours. Cell pellets were collected and stored at -80° C until extraction. One biological replicate from individual five failed to reach passage ten and was excluded from all analyses.
Embryoid body assays were performed following the protocol used by Romero et al [31]. Briefly, embryoid bodies were generated by manual colony detachment and were grown in suspension for seven days on low adherent plates in bFGF-free hESC media. They were then plated on 12 well gelatin-coated plates and grown for another seven days in DMEM-based media. Cells were fixed and stained using antibodies against nestin (1 : 250 SC-71665, Santa Cruz Biotech), α-smooth muscle actin (1 : 1500, CBL171, Millipore), alpha-Fetoprotein (1 : 100, SC-130302, Santa Cruz Biotech), and HNF3β (1 : 100 SC-6554, Santa Cruz Biotech) to detect ectoderm, mesoderm, and endoderm lineages respectively.
DNA was extracted using ZR-Duet DNA/RNA MiniPrep (Zymo) kits according to the manufacturer’s instructions. To assess for the presence of plasmid or EBV genome in iPSCs, PCR was performed using the genomic DNA collected from the iPSCs as template (collected at the same time as expression measurements) with primers designed to amplify the 3’ end of the EBNA-1 gene (present in both the EBV genome and all reprogramming plasmids) and NEBNext High-Fidelity 2X PCR Master Mix. For the sample with detectable EBNA-1, we also performed genomic PCR using primers to amplify a region common to all PXCLE reprogramming plasmids, and primers that amplify the BBRF1/LMP2 gene found only in the EBV genome to determine the source of foreign DNA. Primer sequences are available in S6 Table. Fibroblast DNA containing reprogramming plasmids at 0.02 pg/μL was used as a positive control for the PXCLE and EBNA-1 primer sets. LCL DNA (from YRI lines 18508 and 19238) were used as positive controls for the EBV and EBNA-1 primer sets. Fibroblast DNA was used as a negative control for all primer sets.
RNA was extracted using ZR-Duet DNA/RNA MiniPrep kits according to the manufacturer’s instructions with the addition of a DNAse treatment step prior to RNA extraction. cDNA was then synthesized using Maxima First Strand cDNA Synthesis Kit (Thermo-Scientific.) RT-PCR for endogenous transcripts of three pluripotency-related transcription factors was performed for all iPSC lines using SYBR Select master mix (Life Technologies.) Primers sequences are available in S6 Table. Data were analyzed using Viia7 software (Life Technologies). All expression levels were normalized to GAPDH. Expression was measured relative to a randomly selected iPSC line.
Gene Expression Quantification
Cell pellets were obtained from LCLs immediately before transfection and from stable iPSCs after at least ten passages. RNA concentration and quality was estimated using the Agilent 2100 Bioanalyzer. Donor expression profiles were quantified using Illumina HumanHT-12 v4 Expression BeadChip Microarrays by the Functional Genomics Core at University of Chicago. Samples were hybridized across three array batches. Biological replicates from an individual were assigned to different batches to exclude a relationship between batch and individual. The array data were also used for the PluriTest assay as described previously [24].
Data Processing and Analysis
Raw probe data were filtered for probes whose target transcripts were detected as expressed (P < 0.05) in at least two samples. Probes targeting expressed transcripts were then mapped to the hg19 reference genome. We excluded those with a quality score below 37, those that did not map uniquely to an Ensembl gene ID, that spanned an exon-exon junction, or that contained a SNP with MAF < 0.05 in European populations (calculated using 1000 Genomes phase I integrated call sets). After filtering, probe intensities from all samples were background corrected, quantile-normalized, and log-2-transformed using the R package ‘lumi’ [32]. For genes represented by multiple probes, only the 3’ most probe was included in subsequent analyses to represent the most complete transcript. Finally, array batch was corrected for using an empirical Bayes method implemented in the R package ‘sva’[25,33] This data is available in S1 Table.
Differential expression was estimated using a linear model based empirical bayes method implemented in the R package ‘limma [26]’. Dendrograms were generated for matrices of pairwise Pearson product-moment correlation coefficients. For principal component analysis, expression data was mean-centered by gene across all individuals. The outlier individual 4–2 was omitted prior to hierarchical clustering analysis and PCA. All analyses, figures, and tables presented in the supplement include data from all individuals. Proportion of variance due to donor was estimated as the adjusted R2 value from a linear model including a term for each individual. Genes with FDR-adjusted p-values < 0.05 from a one-way ANOVA across individuals were classified as significantly associated with donor. eQTL data were downloaded from the Pritchard group eQTL browser: http://eqtl.uchicago.edu. Functional group enrichment was assessed using the web-based gene annotation database Lynx: http://lynx.ci.uchicago.edu using all expressed genes subjected to our filtering criteria as background.
Accession Numbers
Gene expression data are available at the GEO database, accession #GSE64263.
Supporting Information
Zdroje
1. Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7 : 118. doi: 10.1186/1471-2164-7-118 16709250; PubMed Central PMCID: PMCPMC1525191.
2. Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Molecular cancer therapeutics. 2008;7(9):3038–46. doi: 10.1158/1535-7163.MCT-08-0248 18765826
3. Wen Y, Gamazon ER, Bleibel WK, Wing C, Mi S, McIlwee BE, et al. An eQTL-based method identifies CTTN and ZMAT3 as pemetrexed susceptibility markers. Hum Mol Genet. 2012;21(7):1470–80. doi: 10.1093/hmg/ddr583 22171072; PubMed Central PMCID: PMCPMC3298275.
4. Ziliak D, O'Donnell PH, Im HK, Gamazon ER, Chen P, Delaney S, et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl Res. 2011;157(5):265–72. doi: 10.1016/j.trsl.2011.01.005 21497773; PubMed Central PMCID: PMCPMC3079878.
5. Moyer AM, Fridley BL, Jenkins GD, Batzler AJ, Pelleymounter LL, Kalari KR, et al. Acetaminophen-NAPQI hepatotoxicity: a cell line model system genome-wide association study. Toxicological sciences: an official journal of the Society of Toxicology. 2011;120(1):33–41. doi: 10.1093/toxsci/kfq375
6. Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–72. Epub 2010/03/12. doi: 10.1038/nature08872 20220758; PubMed Central PMCID: PMC3089435.
7. Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, et al. Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels. PLoS Genet. 2014;10(9):e1004663. doi: 10.1371/journal.pgen.1004663 25233095; PubMed Central PMCID: PMCPMC4169251.
8. Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4(11):e1000287. doi: 10.1371/journal.pgen.1000287 19043577; PubMed Central PMCID: PMCPMC2583954.
9. Plagnol V, Uz E, Wallace C, Stevens H, Clayton D, Ozcelik T, et al. Extreme clonality in lymphoblastoid cell lines with implications for allele specific expression analyses. PLoS One. 2008;3(8):e2966. doi: 10.1371/journal.pone.0002966 18698422; PubMed Central PMCID: PMCPMC2494943.
10. Stark AL, Zhang W, Mi S, Duan S, O'Donnell PH, Huang RS, et al. Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010;10(6):505–12. doi: 10.1038/tpj.2010.3 20142840; PubMed Central PMCID: PMCPMC2975793.
11. Hannula K, Lipsanen-Nyman M, Scherer SW, Holmberg C, Höglund P, Kere J. Maternal and paternal chromosomes 7 show differential methylation of many genes in lymphoblast DNA. Genomics. 2001;73(1):1–9. doi: 10.1006/geno.2001.6502 11352560.
12. Carter KL, Cahir-McFarland E, Kieff E. Epstein-barr virus-induced changes in B-lymphocyte gene expression. J Virol. 2002;76(20):10427–36. 12239319; PubMed Central PMCID: PMCPMC136539.
13. Min JL, Barrett A, Watts T, Pettersson FH, Lockstone HE, Lindgren CM, et al. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics. 2010;11 : 96. doi: 10.1186/1471-2164-11-96 20141636; PubMed Central PMCID: PMCPMC2841682.
14. Caliskan M, Cusanovich DA, Ober C, Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum Mol Genet. 2011;20(8):1643–52. doi: 10.1093/hmg/ddr041 21289059; PubMed Central PMCID: PMCPMC3063990.
15. Powell JE, Henders AK, McRae AF, Wright MJ, Martin NG, Dermitzakis ET, et al. Genetic control of gene expression in whole blood and lymphoblastoid cell lines is largely independent. Genome Res. 2012;22(3):456–66. doi: 10.1101/gr.126540.111 22183966; PubMed Central PMCID: PMCPMC3290781.
16. Calışkan M, Pritchard JK, Ober C, Gilad Y. The effect of freeze-thaw cycles on gene expression levels in lymphoblastoid cell lines. PLoS One. 2014;9(9):e107166. doi: 10.1371/journal.pone.0107166 25192014; PubMed Central PMCID: PMCPMC4156430.
17. Mills JA, Wang K, Paluru P, Ying L, Lu L, Galvão AM, et al. Clonal genetic and hematopoietic heterogeneity among human-induced pluripotent stem cell lines. Blood. 2013;122(12):2047–51. doi: 10.1182/blood-2013-02-484444 23940280; PubMed Central PMCID: PMCPMC3778548.
18. Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(3):279–86. doi: 10.1038/nbt.1783 21293464; PubMed Central PMCID: PMCPMC3229307.
19. Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N. Correction for Kajiwara et al., Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proceedings of the National Academy of Sciences. 2012;109(36):14716-. doi: 10.1073/pnas.1212710109
20. Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10(6):e1004432. doi: 10.1371/journal.pgen.1004432 24901476; PubMed Central PMCID: PMCPMC4046971.
21. Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–12. doi: 10.1038/nmeth.1591 21460823.
22. Choi SM, Liu H, Chaudhari P, Kim Y, Cheng L, Feng J, et al. Reprogramming of EBV-immortalized B-lymphocyte cell lines into induced pluripotent stem cells. Blood. 2011;118(7):1801–5. doi: 10.1182/blood-2011-03-340620 21628406; PubMed Central PMCID: PMCPMC3158714.
23. Rajesh D, Dickerson SJ, Yu J, Brown ME, Thomson JA, Seay NJ. Human lymphoblastoid B-cell lines reprogrammed to EBV-free induced pluripotent stem cells. Blood. 2011;118(7):1797–800. doi: 10.1182/blood-2011-01-332064 21708888.
24. Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8(4):315–7. doi: 10.1038/nmeth.1580 21378979; PubMed Central PMCID: PMCPMC3265323.
25. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037 16632515.
26. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027 16646809.
27. Sulakhe D, Balasubramanian S, Xie B, Feng B, Taylor A, Wang S, et al. Lynx: a database and knowledge extraction engine for integrative medicine. Nucleic Acids Res. 2014;42(Database issue):D1007–12. doi: 10.1093/nar/gkt1166 24270788; PubMed Central PMCID: PMCPMC3965040.
28. Kagan CL, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK, et al. Genetic Variation, Not Cell Type of Origin, Underlies Regulatory Differences in iPSCs. bioRxiv. 2015. doi: 10.1101/013888
29. Garfield DA, Runcie DE, Babbitt CC, Haygood R, Nielsen WJ, Wray GA. The impact of gene expression variation on the robustness and evolvability of a developmental gene regulatory network. PLoS Biol. 2013;11(10):e1001696. doi: 10.1371/journal.pbio.1001696 24204211; PubMed Central PMCID: PMCPMC3812118.
30. Roux J, Robinson-Rechavi M. Developmental constraints on vertebrate genome evolution. PLoS Genet. 2008;4(12):e1000311. doi: 10.1371/journal.pgen.1000311 19096706; PubMed Central PMCID: PMCPMC2600815.
31. Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Banovich NE, Kagan CL, Burnett JE, et al. Generation of a Panel of Induced Pluripotent Stem Cells From Chimpanzees: a Resource for Comparative Functional Genomics. bioRxiv. 2014. doi: 10.1101/008862
32. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–8. doi: 10.1093/bioinformatics/btn224 18467348.
33. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3. doi: 10.1093/bioinformatics/bts034 22257669; PubMed Central PMCID: PMCPMC3307112.
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



