-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
Animal development is regulated by many genes including a class of small RNAs called microRNAs (miRNAs). Nearly half of the miRNAs are located in the protein coding genes but functional importance of this genomic organization is unclear. Here we use Drosophila stem cells in the brain as a model system to investigate the interactions between miR-92a and miR-92b and their host gene jing interacting regulatory protein 1 (jigr1). Our studies reveal that these miRNAs prevent premature differentiation of neural stem cells and they do so in part through directly targeting and suppressing their host gene, jigr1. Our results reveal a novel function of the miR-92 family and identify another negative feedback loop as an essential regulator in neural stem cell development.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005264
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005264Summary
Animal development is regulated by many genes including a class of small RNAs called microRNAs (miRNAs). Nearly half of the miRNAs are located in the protein coding genes but functional importance of this genomic organization is unclear. Here we use Drosophila stem cells in the brain as a model system to investigate the interactions between miR-92a and miR-92b and their host gene jing interacting regulatory protein 1 (jigr1). Our studies reveal that these miRNAs prevent premature differentiation of neural stem cells and they do so in part through directly targeting and suppressing their host gene, jigr1. Our results reveal a novel function of the miR-92 family and identify another negative feedback loop as an essential regulator in neural stem cell development.
Introduction
MicroRNAs (miRNAs) are short (~21–23 nt) noncoding RNAs that regulate gene expression post-transcriptionally in many physiological and pathological processes [1,2]. In the canonical miRNA biogenesis pathway, a long primary transcript (pri-miRNA) is generated by RNA polymerase II and cleaved by a nuclear complex formed by Drosha and DGCR8 [3]. Some pri-miRNAs produce miRNAs only (intergenic miRNAs) while others contain miRNAs in the intronic regions of protein-coding “host” genes (intragenic miRNAs) [4]. Many intronic miRNAs and host gene mRNAs are likely co-expressed [5,6] but others may not be [7,8]. Few cases have been experimentally confirmed, and the functional significance of such a genomic arrangement is largely unknown.
In this study, we used the differentiation of Drosophila neuroblasts as a model system to examine the expression and function of specific miRNAs. Drosophila neuroblasts form during embryonic development and enter a proliferative quiescent state at the end of embryogenesis [9]. In the early larval stage, neuroblasts reenter the cell cycle and undergo a series of proliferative symmetric and self-renewing asymmetric cell divisions to maintain the progenitor pool and generate diverse cell types [10,11]. In each asymmetric cell division, neuroblasts divide to generate a neuroblast cell and a ganglion mother cell, which divides only once to generate two neurons or one neuron and one glial cell. The balance between self-renewal and differentiation is critical for normal development, but the mechanisms are incompletely understood [12].
Here we show that the gene encoding jing-interacting gene regulatory 1 (jigr1), a putative DNA-binding protein containing MADF domain with unknown function [13], hosts miR-92a in the intron and miR-92b in the 3’UTR. We also uncover the functional significance of this intragenic miRNA–host gene interaction through genetic knockout of both miR-92a and miR-92b. During larval development, miR-92 family limits jigr1 expression in neuroblasts and is essential for maintenance of a neuroblast pool. We propose that this genomic arrangement and local feed-back regulatory loop are essential for animal development to ensure the generation of the proper number of neuronal and glial cells.
Results
miR-92a and miR-92b Are Expressed in Neuroblasts of the Drosophila Larval Brain
The miR-92 family is evolutionarily conserved (S1A Fig) but its function in neural development in Drosophila is unknown. In Drosophila, the miR-92 family consists of miR-92a and miR-92b, which have the same seed sequence (miRbase, S1A Fig). We first characterized the expression of miR-92a and miR-92b in Drosophila at different stages of development by northern blot analysis and miRNA Taqman assay. Both miRNAs were expressed at high levels during the embryonic, larval, and pupal stages and at relatively low levels in adult flies; during the third instar larval stage, expression was enriched in the brain (Fig 1A and S1B Fig). miR-92a expression level is mostly higher than that of miR-92b at different developmental stages (S1B Fig).
Fig. 1. Expression profile of miR-92a and miR-92b in third Instar larval brain. 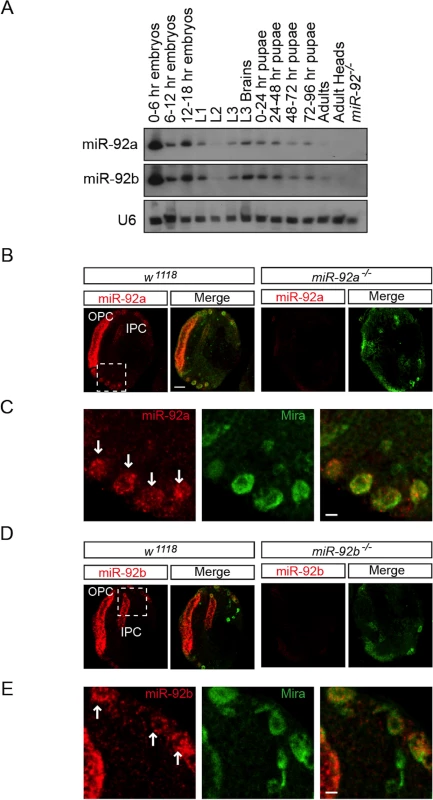
(A) Northern blot analysis of total RNA isolated from staged wild type flies (lanes 1–13) and miR-92–/– mutant third instar larvae (lane 14). RNA was probed for miR-92a (top), miR-92b (middle), and U6 (bottom). (B) In situ analysis of mir-92a in the wild type and miR-92a-/- larval brains sectioned horizontally. Neuroblasts are labeled with Miranda (green). Scale bar: 20 μm. (C) Magnified view of boxed area in B. Arrows indicate neuroblasts. Scale bar: 5 μm. (D) In situ analysis of mir-92b in wild type and miR-92b-/- third instar larval brain horizontal sections. (E) Magnified view of boxed area in B. Arrows indicate neuroblasts. Scale bar: 5 μm. OPC: Outer proliferation center, IPC: Inner proliferation center. To determine which cell types express miR-92a and miR-92b, we analyzed third instar larval brains by RNA in situ hybridization. Both miR-92a and miR-92b were expressed in the optic lobe and central brain in wild type flies; this expression was absent in miR-92a–/– and miR-92b–/– flies (please see detailed generation and characterization of these mutant flies below), respectively (Fig 1B–1E, S2B and S2C Fig). In the optic lobe, miR-92a co-expressed with Discs large (Dlg) in neuroepithelial cells [14] (S2A Fig). In the central brain, miR-92a and miR-92b preferentially co-expressed with the neuroblast markers Miranda (Mira) (Fig 1B–1E) and Deadpan (Dpn) (S2B Fig and Fig 2C). These results raise the possibility that miR-92 family may contribute to neuroblast development.
Fig. 2. Expression profile of Jigr1 in third instar larval brain. 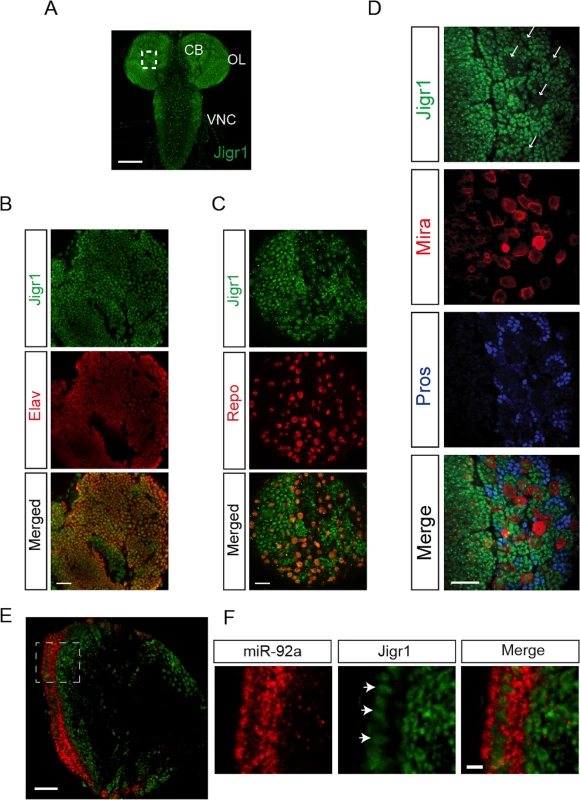
(A) Jigr1 expression (green) in wild type third instar larval brain. Z-projection of optical sections is shown. Scale bar: 100 μm. CB: Central Brain, OL: Optic lobe, VNC: Ventral Nerve Cord. (B) Jigr1 expression (green) in post mitotic neurons labeled with Elav (red). Scale bar: 20 μm (C) Jigr1 expression (green) in glial cells labeled with glial specific transcription factor, Repo, (red) (D) High-magnification images of boxed area in A. Neuroblasts are labeled with Miranda (Mira, red), and ganglion mother cells (GMCs) and neurons are immunostained with Prospero (Pros, blue). Single optical section is shown. Arrows indicate neuroblasts. Scale bar: 20 μm. (E) Horizontal sections of third instar larval brains are labeled with miR-92a (red) and Jigr1 (green). Dashed lines outlines neuroepthelial cells (NE). Scale Bar: 20 μm. (F) Magnified view of dashed box in 2E. Arrows indicate miR-92a positive cells. Scale Bar: 5 μm. Jigr1 and miR-92 Have Complementary Expression Patterns
Based on the FlyBase, miR-92a and miR-92b are located on the right arm of chromosome 3; miR-92a resides in the first intron of jigr1, and miR-92b resides downstream of jigr1 coding region. Since miR-92a is in the intron of jigr1, we asked whether jigr1 is also expressed in neural progenitor cells in larval brain. To assess the expression pattern of jigr1, we generated an antibody specific to first 110 amino acids of Jigr1. Nuclear Jigr1 signal was detected in the third instar larval brain (Fig 2A). High jigr1 expression was present in neurons and glial cells (Fig 2B and 2C). Unexpectedly, we found that jigr1 is expressed at low levels in neuroepithelial cells and neuroblasts, even though it is highly expressed throughout the nervous system (Fig 2D and 2E). Double labeling of miR-92a and Jigr1 in third instar larval brains confirms their complementary expression pattern (Fig 2E and 2F).
Drosophila miR-92a and miR-92b and Their Host Gene jigr1 Are Expressed in the Same Transcriptional Unit in Larval Brain
miR-92a and miR-92b have the same expression profile (Fig 1 and S2 Fig), which suggests that they may be co-transcribed. To investigate this possibility, we generated a series of deletions at the jigr1 locus by FLP-FRT [15] or cre/loxP deletions (Fig 3A and S3A Fig) and analyzed expression of miR-92a and miR-92b by northern blot (Fig 3B). If miR-92a and miR-92b were transcribed together, deleting the promoter region of miR-92a would prevent expression of both miRNAs. Indeed, in Del #38 and Del #4 flies, which lack the sequence upstream of miR-92a, expression of both miR-92a and miR-92b was lost (Fig 3B). Consistent with this, miR-92b expression was also lost in Del #19 flies, which lack miR-92a and the coding region of jigr1 (Fig 3B). Deleting only the coding region of jigr1 in Del #7 also eliminates both miR-92a and miR-92b expression (Fig 3B). Although Del #38 and Del #4 are 12.5 kb and 6.6 kb upstream of miR-92b, respectively, the loss of miR-92b expression resulting from these deletions suggests that a single long transcript contains miR-92a and miR-92b.
Fig. 3. miR-92a and miR-92b are transcribed in the same transcriptional unit. 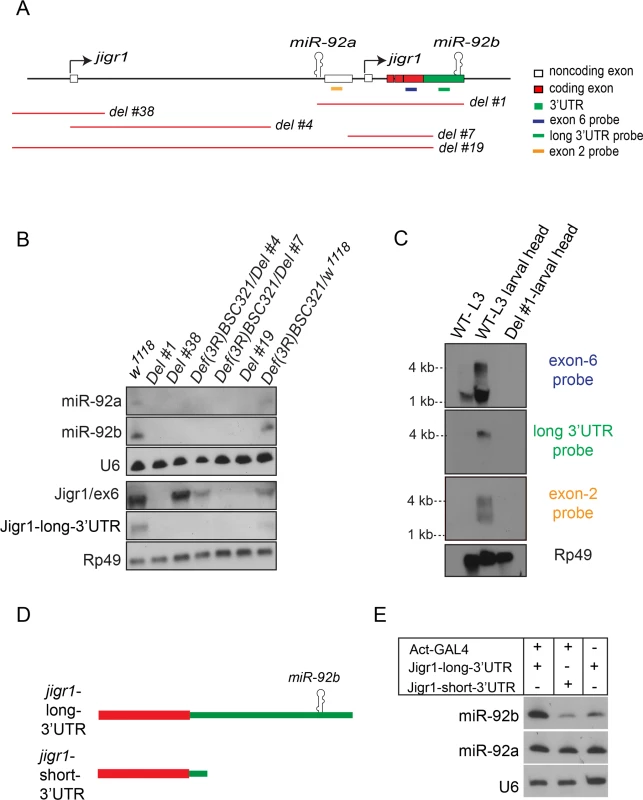
(A) Schematic representation of the genomic arrangement of miR-92a, miR-92b and jigr1 based on results from this study. Red lines indicate deleted regions. (B) Northern blot analysis of total RNA extracted from third instar larval heads of the indicated genotypes. RNA was probed for miR-92a, miR-92b, U6, Jigr1 (exon 6 probe), Jigr1 (long 3’UTR probe) and rp49. Deletion lines, Del #4 and Del #7, are homozygous lethal but in trans to the bigger deficiency covering the locus, Def(3R)BSC321, they are viable and used like this for the analysis. (C) Northern blot analysis of RNA from wild type whole third instar larvae (lane 1), wild type larval heads (lane 2) and Del #1 (lane 3) third instar larval heads. Probes specific for all the isoforms of jigr1 (exon 6 probe, first panel), for the long 3’UTR (second panel), and for alternatively spliced noncoding exon 2 (third panel), and rp49 (bottom panel) were used. (D) Schematic representation of jigr1 constructs. (E) Northern blot analysis of total RNA from HEK 293T cells transfected with UAST-jigr1-long-3’UTR or UAST-jigr1-short-3’UTR plasmids together with the Actin-Gal4 plasmid. Cells transfected with UAST-jigr1-long-3’UTR alone served as negative controls. miR-92b, miR-92a, and control U6 probes were used. The miR-92b probe recognizes endogenous miR-92b in HEK 293T cells. To determine whether miR-92a and miR-92b are co-expressed with jigr1, we analyzed jigr1 expression by northern blot in wild type and deletion lines. In wild type third instar larva brains, a probe specific for jigr1 exon 6, which is found in all the isoforms, identified jigr1 transcripts of 1.5 kb and 4 kb (Fig 3C). Surprisingly, 1.5 kb jigr1 transcript can still be detected in Del #38 and Del #4 flies while the 4 kb transcript was lost (Fig 3B). Since these deletions only remove the first non-coding exon, the short isoforms of jigr1 probably have an alternative promoter downstream of these deletions, as predicted in FlyBase. These results suggest that miR-92a and miR-92b share the same transcriptional unit with a jigr1 isoform that hasn’t been previously annotated. Our results also suggest that miR-92b does not have its own promoter and is not intergenic but is located in the 3’UTR of a longer jigr1 transcript in the Drosophila larval brain.
To confirm this finding, we used 3’ rapid amplification of cDNA ends (3’RACE) to map the 3’UTR of jigr1. We observed two jigr1 3’UTRs of different lengths. One is 203 bp long and the other is 1827 bp long and contains the miR-92b stem loop (S3B and S3C Fig). The presence of the neurally enriched long 3’UTR was also confirmed by RNA-seq analysis as reported in the FlyBase (S3D Fig). Next, we analyzed RNA from whole larvae and larval heads by northern blot. A long 3’UTR-specific probe identified only the 4-kb transcript (Fig 3C). The upper 4 kb transcript was fainter in whole larvae than in larval heads, indicating that it is more abundant in the brain (Fig 3C). We also identified another brain-enriched jigr1 isoform in which intron 2 is retained in the transcript. A probe specific for this noncoding exon 2 recognized transcripts of 2.5 kb and 4 kb in larval brain (Fig 3C). Next, we asked whether the long jigr1 transcript could give rise to miR-92b. In HEK 293T cells, expression of this transcript containing the miR-92b stem loop (Fig 3D) increased the amount of mature miR-92b, confirming that miR-92b can be processed from the 3’UTR of long jigr1 transcript (Fig 3E).
Generation of miR-92a and miR-92b Single - and Double-Knockout Mutant Flies
To analyze the function of the miR-92 family in neurogenesis in Drosophila, we first generated miR-92a and miR-92b single mutants by ends-out homologous recombination in which 140 bp deletions were generated after mini white was removed by Cre/LoxP-mediated recombination (Fig 4A) [16]. Loss of miR-92a or miR-92b was confirmed by PCR (Fig 4B) and northern blot analysis (Fig 4C). In northern blot, miR-92a level seems to be higher in miR-92b-/- mutants and vice versa (Fig 4C). In order to obtain more quantitative data, we performed miRNA Taqman assay. In miR-92a-/- L3 heads, miR-92b expression was increased by 30% and in miR-92b-/- L3 heads, miR-92a expression was increased by 40% (S1B Fig).
Fig. 4. Generation of miR-92a–/– and miR-92b–/– flies by ends-out gene targeting. 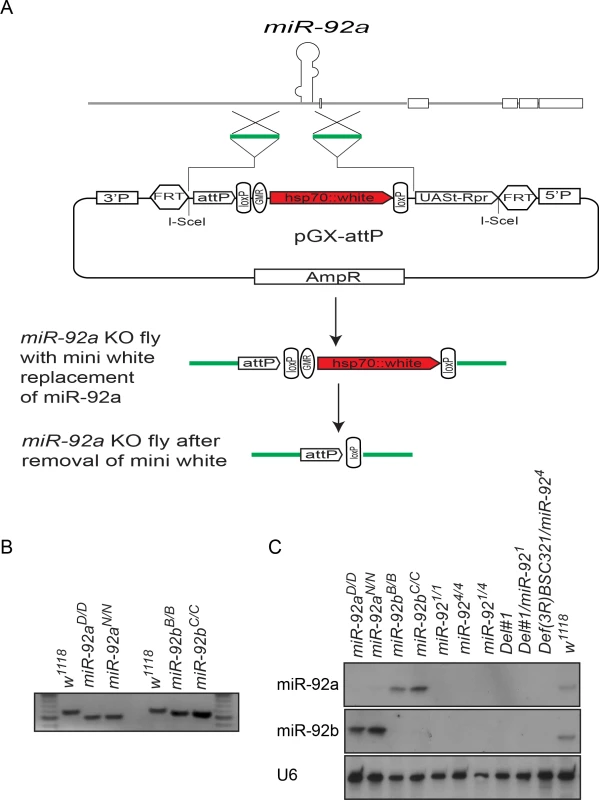
(A) Schematic of ends-out gene targeting strategy (modified from Huang et al., 2009). (B) DNA gel electrophoresis of PCR-amplified miR-92a (left) and miR-92b (right) locus from genomic DNA of wild type and miR-92a–/– and miR-92b–/– single knockout (KO) flies. (C) Northern blot analysis of miR-92a, miR-92b, and U6 expression in wild type, miR-92a–/–, miR-92b–/–, and miR-92–/– mutant third instar larvae. Both miR-92a–/– and miR-92b–/– flies were viable and fertile and had no obvious phenotype, in contrast to a recent report showing miR-92b deletion mutant flies are larval lethal, in which both miR-92b and the jigr1 coding region are deleted [17]. However, our flies homozygous for Del #1, in which both miR-92a/miR-92b and Jigr1 are deleted, are still adult viable. Thus, it is unclear what causes the lethality phenotype in miR-92b mutant flies generated by Chen et al [17]. Since miR-92a and miR-92b have an identical seed sequence and are similarly expressed, they might compensate for each other. So, using the same ends-out gene targeting method [16], we targeted the miR-92b locus on the miR-92a–/– mutant background to generate flies lacking both miR-92a and miR-92b (named as miR-92–/–). Two independent lines, miR-924 and miR-921 were further characterized and used in this study. Unlike the single-knockout flies, which have a normal life span, miR-92–/– flies have a reduced life span; most die within a week (S4 Fig). Expression of miR-92a or miR-92b in the miR-92–/– background with Insc-GAL4 that drives gene expression in neuroblasts largely rescued the life span defect, confirming that this phenotype is indeed due to loss of miR-92a and miR-92b (S4 Fig).
miR-92a and miR-92b Maintain Neuroblast Self-Renewal by Inhibiting Premature Differentiation
Since a high level expression of miR-92a and miR-92b is seen in neuroblasts, we examined larval brain neuroblasts in miR-92–/– flies (S5A Fig). At 96 and 120 hr after larval hatching (ALH), there were about 100 neuroblasts in the central brain in wild type, miR-92a–/–, and miR-92b–/– flies, as shown by staining with the neuroblast marker Dpn (Fig 5A, S5A and S5B Fig). However, miR-92–/– flies had significantly fewer neuroblasts (Fig 5A and S5B Fig). At 120 hr ALH, the reduction was ~23% in miR-921/4 flies (p < 0.005) and ~30% in miR-924/4 flies (p < 0.0001) (Fig 5A). These reductions were fully rescued by expression of either miR-92a or miR-92b with Insc-GAL4, suggesting miR-92a and miR-92b are required cell autonomously in neuroblasts (Fig 5A and S5B Fig).
Fig. 5. miR-92a and miR-92b regulate neuroblast self-renewal by inhibiting premature differentiation. 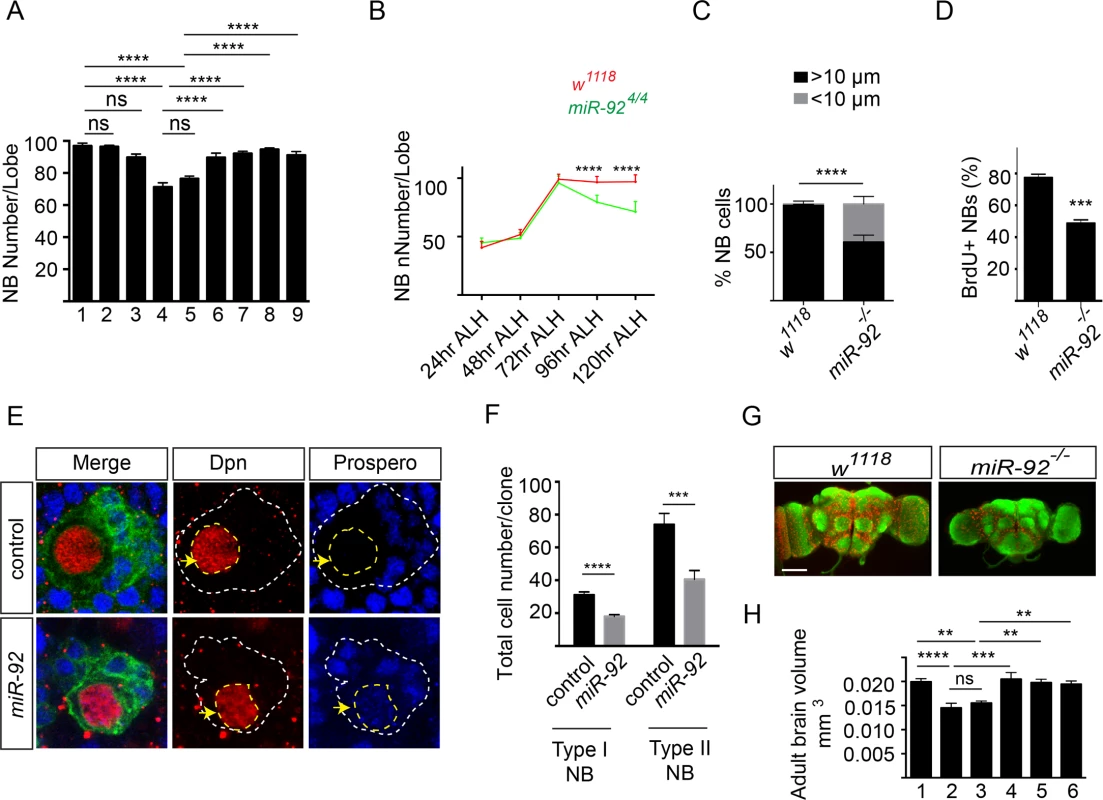
(A) Number of brain neuroblasts at 120 hr ALH. 1: w1118 (n = 17); 2: miR-92a-/- (n = 13); 3: miR-92b-/- (n = 12); 4: miR-924/4 (n = 15); 5: miR-921/4 (n = 20); 6: Insc-GAL4/UAS-miR-92a; miR-924/4 (n = 8); 7: Insc-GAL4/UAS-miR-92b; miR-924/4 (n = 8); 8: Insc-GAL4/UAS-miR-92a; miR-921/4 (n = 16); 9: Insc-GAL4/UAS-miR-92b; miR-921/4 (n = 15). Statistical significance was determined by one-way ANOVA. (B) Number of neuroblasts in wildtype (red) and miR-924/4 (green) mutants from 24 to 120 hr ALH. Statistical significance was determined by Student’s t test. Error bars show S.D. (C) Percentage of neuroblasts with diameter > 10 μm (black bar) and < 10 μm (grey bar) in wild type (n = 60) and miR-92–/– (n = 60) mutant brains at 96 hr ALH. Fisher’s exact test was used for statistical analysis. (D) Percentage of BrdU+ wild type control (n = 15) and miR-92–/– mutant (n = 16) larval neuroblasts. Statistical significance was determined by Student’s t test. (E) Neuroblast clones of wild type control and miR-924 mutants. Clones are marked with CD8::GFP (green). Yellow arrows mark Dpn+ neuroblasts. White dashed lines indicate the position of clones and yellow dashed line encircles the nucleus of neuroblast cell. Single focal planes are shown. (F) Comparison of wild type control (type I; n = 57, type II; n = 27) and miR-92 -/- mutant (type I; n = 67, type II; n = 33) clone sizes. (G) Whole-mount image of wild type and miR-92–/– mutant male adult brains immunostained with the anti-HRP (green) to label axonal membranes and anti-Repo (red) to label glial cell nuclei. Scale bar: 100 μm. (H) Adult brain volume measured using ImageJ. 1: w1118 (n = 10); 2: miR-924/4 (n = 10); 3: miR-921/4 (n = 8); 4: Insc-GAL4/UAS-miR-92a; miR-924/4 (n = 7); 5: Insc-GAL4/UAS-miR-92a; miR-921/4 (n = 9); 6: Insc-GAL4/UAS-miR-92b; miR-921/4 (n = 10). Statistical significance was determined by one-way ANOVA. Values are mean ± s.e.m. in all graphs unless otherwise stated. *: p < 0.05, **: p < 0.005, ***: p < 0.001, ****: p < 0.0001. The decreased number of neuroblasts in miR-92–/– flies could reflect defects in neuroblast formation, cell survival or self-renewal. To distinguish between these possibilities, we first quantified neuroblasts at earlier stages of development. At 24, 48 and 72 hr ALH, wild type and miR-92–/– flies had equal numbers of neuroblasts, suggesting that the phenotype is not due to defect in neuroblast formation or failure of neuroblasts to re-enter the cell cycle (Fig 5B). After 72 hr ALH, the number of neuroblasts in wild type did not change, as they are mainly undergoing asymmetric cell division. However, in miR-92–/– mutants, neuroblasts were lost progressively, resulting in 18% decrease at 96 hr ALH and a further 30% decrease at 120 hr ALH (p < 0.0001) (Fig 5B).
To determine whether neuroblast survival is defective in miR-92–/– flies, we immunostained the cells for activated Caspase-3. No significant increase in caspase activation was detected (S5C Fig), and blocking apoptosis by expressing the anti-apoptotic protein P35 in neuroblasts did not rescue this phenotype (S5D Fig). In larval brain, neuroblasts maintain their population by asymmetric cell division, so a defect in this process might also lead to loss of neuroblasts, if these cells generated two differentiated daughter cells instead of self-renewing. Neither the apical localization of aPKC nor the basal localization of Numb or Miranda was disrupted in miR-92–/– neuroblasts (n = 30) (S6A and S6B Fig). In addition to the differential segregation of cell fate determinants, asymmetric cell division also generates unequal sized daughters and these two mechanisms are independently regulated [18]. In order to determine if size asymmetric division is disrupted in miR-92-/- mutants, we have stained the third instar larval brains with Miranda and phosphor-histone H3. We didn’t observe any size symmetric neuroblast division in mir-92-/- mutant neuroblasts (n = 33) (S6C Fig). Thus, defective asymmetric cell division is unlikely to be the mechanism of neuroblast loss in miR-92–/– larvae.
Another possibility that could result in loss of neuroblasts is premature differentiation of neuroblasts, a process that correlates with a reduction in cell size [19, 20]. Therefore, we measured neuroblast diameter in control and miR-92–/– brains. In controls, almost all neuroblasts were >10 μm in diameter at 96 hr ALH; however, in miR-92–/– mutants, 39% of neuroblasts had a reduced cell diameter (p < 0.0001) (Fig 5C). To determine whether miR-92–/– neuroblasts were still capable of dividing, we performed a BrdU incorporation assay. Almost 80% of wild type neuroblasts incorporated BrdU, versus only 50% of miR-92–/—neuroblasts, suggesting that loss of miR-92a and miR-92b causes premature exit from the cell cycle (Fig 5D).
Next, we analyzed miR-92–/– brains for expression of Prospero, a homeodomain transcription factor that is localized in the cortex in self-renewing larval neuroblasts but transiently moves to the nucleus in quiescent or terminally differentiating neuroblasts [21,22]. In 3–5% neuroblasts in miR-92–/– brains, Prospero was localized in the nucleus and decreased the expression level of the neuroblast nuclear marker Dpn (S5E Fig). Next we analyzed miR-92 phenotype using mosaic analysis of repressible cell marker (MARCM) method [23]. Wild type clones contained single Dpn+, Pros- neuroblast cell surrounded by several Dpn-, Pros+ daughter cells. In miR-92 mutant clones, some Dpn+ neuroblast cells also showed nuclear Prospero expression (Fig 5E and S6D Fig). Moreover, the size of miR-92 mutant clones was significantly reduced (Fig 5F). In BrdU chase experiment in MARCM clones, around 40% of the wild type neurons were BrdU+ while only 23% of miR-92–/– neurons were BrdU+. Thus, control of neuroblast self-renewal by miR-92a and miR-92b is achieved by preventing their early differentiation. Indeed, although no obvious difference in body size was observed for adult flies, the size of the ventral nerve cord of miR-92–/– adult flies is 15% smaller (p < 0.05), the brains were also smaller than those of control flies (Fig 5G and 5H), which is probably due to reduced proliferation (Fig 5D) and premature differentiation (Fig 5E) of neuroblasts that lead to reduced production of neurons and glial cells.
Some jigr1 Isoforms Are Direct Targets of miR-92a and miR-92b In Vivo
miRNAs regulate the expression of their target genes post-transcriptionally, mostly by binding to target sequences in the 3’UTR and mediating mRNA degradation and/or translation inhibition [24]. We were intrigued by the finding that miR-92 family and jigr1 show complementary expression pattern in the larval brain (Fig 2E), which raises the possibility that jigr1 mRNA might be a target of miR-92 family. Indeed, intragenic miRNAs may target their own host genes and function as negative feedback regulators [25–30]. To determine whether jigr1 is a functional target of miR-92 in vivo, we first analyzed jigr1 mRNA levels in control and miR-92–/– third instar larval brains by qRT-PCR. jigr1 transcripts were almost threefold more abundant in miR-92–/– brains (p < 0.0001) (Fig 6A). Moreover, overexpression of miR-92a or miR-92b by Pros-Gal4 in wild type background decreased jigr1 level in third instar larval brain (Fig 6A and S7A Fig). Analysis of Jigr1 expression in third instar larval brains by western blot revealed a single band of ~36 kDa that is absent in Del #1, in which the entire jigr1 coding sequence is deleted, confirming the specificity of the Jigr1 antibody. Consistent with the increase in transcript levels, Jigr1 protein was also substantially more abundant in miR-92–/– larval brains (Fig 6B and 6C).
Fig. 6. Elevated Jigr1 levels in miR-92-/- brains. 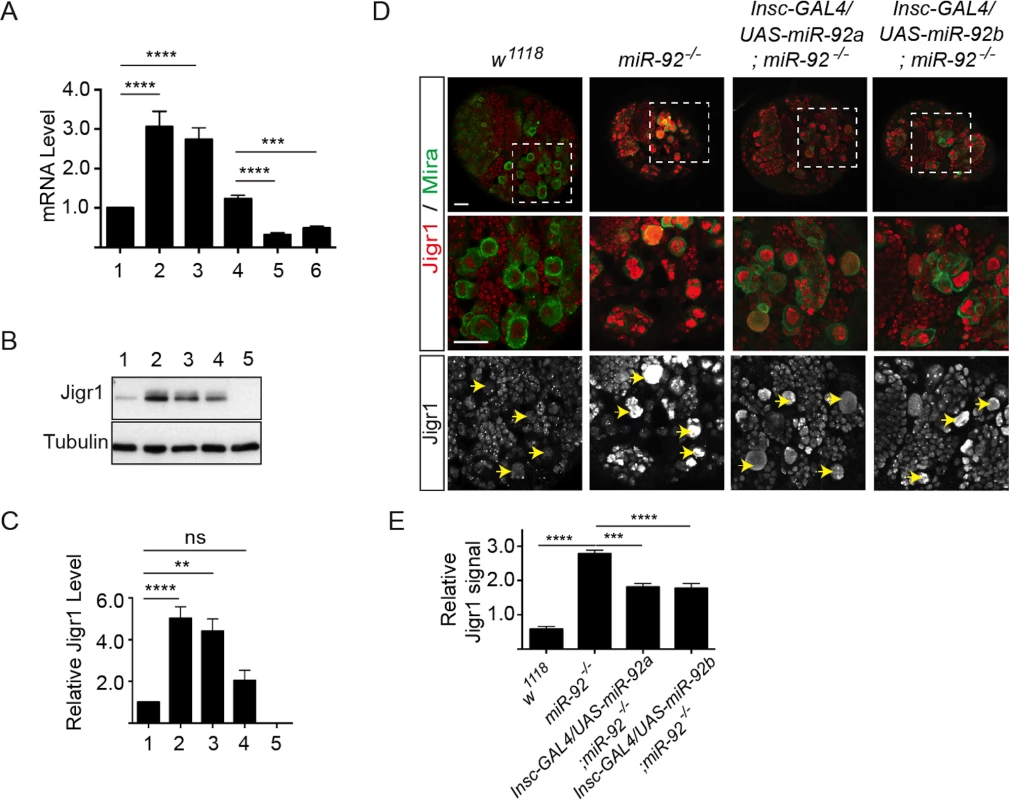
(A) qRT-PCR analysis of relative expression of jigr1 in third instar larval brains. 1: w1118; 2: miR-924/4; 3: miR-921/4. 4: Pros-Gal4/+, 5: Pros-Gal4/UAS-miR-92a; 6: Pros-Gal4/UAS-miR-92b. (B) Western blot analysis of Jigr1 protein in lysates of third instar larval brain. 1: w1118; 2: miR-924/4; 3: miR-921/4; 4: Def(3R)BSC321/miR-924; 5: Del #1. (C) Expression level of Jigr1 normalized to alpha-Tubulin. Western blots are representative of three independent experiments. 1: w1118; 2: miR-924/4; 3: miR-921/4; 4: Def(3R)BSC321/miR-924; 5: Del #1. (D) Single confocal sections of third instar larval brains of the indicated genotypes stained for Jigr1 (red) and Miranda (green). High-magnification images of boxed areas in top panel are shown in the middle panel. Arrowheads indicate neuroblasts. Scale bar: 20 μm. (E) Jigr1 level in neuroblasts relative to that in neighboring ganglion mother cells. Ten larval brains of each genotype were analyzed One-way ANOVA was used for statistical comparisons unless otherwise stated. Values are mean ± s.e.m. in all graphs. *: p < 0.05, **: p < 0.005, ***: p < 0.001, ****: p < 0.0001. Although miR-92a and miR-92b are transcribed together with the long jigr1 transcript, miR-92a and miR-92b are enriched in neuroblasts, which express Jigr1 at a low level. However, in miR-92–/– brains, Jigr1 is expressed at high levels, especially in neuroblasts (Fig 6D). Since all miR-92–/– phenotypes in neuroblasts can be rescued by expression of miR-92a or miR-92b, we wanted to see if the elevated Jigr1 level could be suppressed too. Indeed, expression of miR-92a or miR-92b in miR-92–/– mutants significantly reduced jigr1 expression in neuroblasts (Fig 6D and 6E). These results suggest that miR-92 directly regulates its host gene jigr1.
To further confirm that jigr1 is a direct target of miR-92a and miR-92b, we used a reporter silencing assay. The short 3’UTR of jigr1 has no binding site for miR-92a or miR-92b, while the long 3’UTR has three predicted miR-92 binding sites (Fig 7A). In HEK 293T cells, co-expression of miR-92a and/or miR-92b with a luciferase reporter carrying the jigr1 long 3’UTR in HEK 293T cells significantly suppressed luciferase activity (Fig 7B). Mutating the first two predicted binding sites did not abolish the suppression of luciferase activity by miR-92a and/or miR-92b. However, deletion of site 3 eliminated the suppression, showing that jigr1 mRNA is a direct target of miR-92a and miR-92b (Fig 7B).
Fig. 7. miR-92a and miR-92b target their host gene jigr1. 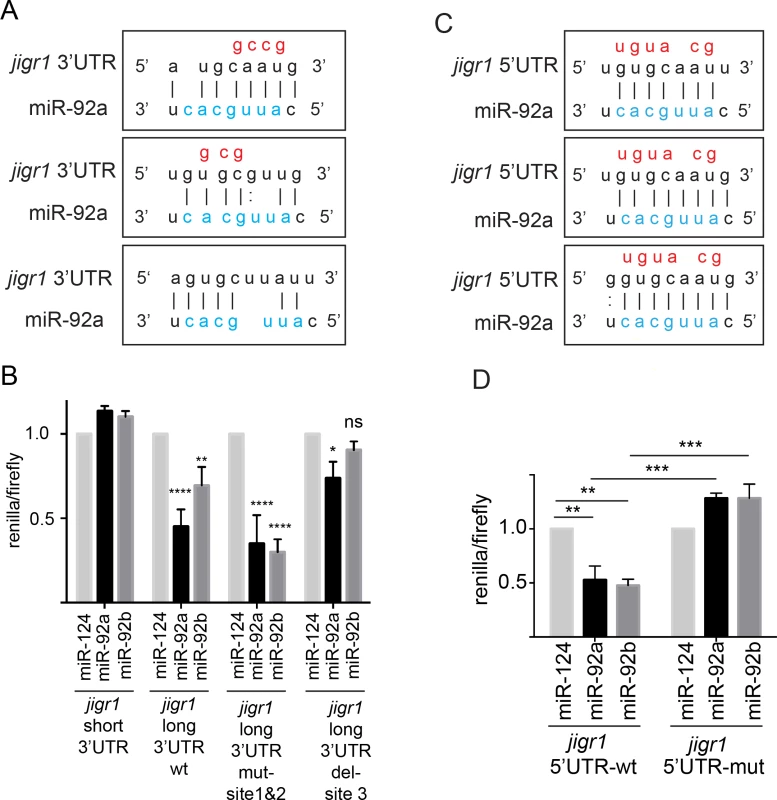
(A) Predicted miR-92a and miR-92b binding sites in the 3’UTR of jigr1. The seed sequences of miR-92a and miR-92b are the same and shown in blue and mutated nucleotides are shown in red. (B) Dual luciferase assay of HEK 293T cell lysates cotransfected with miR-92a, miR-92b or miR-124 expression vectors together with dual luciferase construct containing the wild type or mutant 3’UTR of jigr1 at 3’ of Renilla luciferase. Bar graph shows normalized mean luciferase activity of cells transfected with miR-92 expression plasmid to that of cells transfected with control miRNA from three independent experiments. Statistical significance was determined by two-way ANOVA. (C) Predicted miR-92a and miR-92b binding sites at the 5’UTR of jigr1. Mutated nucleotides are shown in red. (D) Dual luciferase assay of lysates of HEK 293T cells co-transfected with miR-92a, miR-92b, or control miRNA (miR-124) expression vectors together with a firefly luciferase construct containing the wild type or mutant 5’UTR of jigr1. Bar graph shows normalized mean values of the luciferase activity of cells transfected with a miR-92 expression plasmid relative to that of cells transfected with control miRNA from three independent experiments. Statistical significance was determined by two-way ANOVA. Values are mean ± s.e.m. in all graphs. *: p < 0.05, **: p < 0.005, ***: p < 0.001, ****: p < 0.0001. miRNAs can also regulate mRNAs by binding to the 5’UTR [31]. Jigr1 mRNA has three potential miR-92 binding sites in the alternatively spliced noncoding exon 2 (Figs 3A and 7C). We cloned the sequence containing the intact and the mutated binding sites to the 5’UTR of the luciferase gene and co-transfected it together with miR-92a and/or miR-92b and control miRNA. Overexpression of miR-92a or miR-92b suppressed luciferase activity from a reporter with intact binding sites but had no effect on a reporter containing mutated binding sites (Fig 7D). Thus, jigr1 isoforms containing this 5’UTR can be regulated directly by miR-92a and miR-92b through its 5’UTR.
As we showed previously (Fig 5D and S5C Fig), some miR-92-/- mutant neuroblasts have nuclear Prospero expression. Prospero is also a predicted target of miR-92a and miR-92b (Targetscan). In order to investigate whether prospero mRNA can be directly downregulated by miR-92a and miR-92b, we performed luciferase reporter assay. Prospero mRNA has six isoforms and three alternative 3’UTRs with varying lengths (Flybase). Two short 3’UTRs contain one miR-92a/miR-92b binding site and the long 3’UTR contains three. Luciferase activity was measured in HEK 293T cells transfected either with miR-92a, miR-92b or empty vector together with psicheck2-pros-short-UTR or psicheck2-pros-long-UTR plasmids. We couldn’t detect any significant downregulation of luciferase activity by either miR-92a or miR-92b. Thus, prospero mRNA is not a direct target of miR-92a and miR-92b (S7B and S7C Fig).
Jigr1 Upregulation Leads to Premature Neuroblast Differentiation in miR-92–/– Flies
To determine whether upregulation of jigr1contributes to the miR-92 mutant phenotype, we carried out genetic interaction experiments. If an elevated level of Jigr1 causes premature differentiation of neuroblasts in miR-92 mutants, reducing Jigr1 levels in miR-92 mutants could suppress this phenotype. Indeed, limiting the Jigr1 level, either with the deficiency covering the jigr1 locus or by downregulating it in neuroblasts with UAS-jigr1-RNAi, largely rescued reduced neuroblast numbers in miR-92–/– flies (Fig 8A) and brain size (Fig 8B). Moreover, upregulation of jigr1 also contributed significantly to the life span defect in miR-92–/– flies (S8 Fig). Thus, a key function of these miRNAs is to suppress the expression of its host gene in developing neuroblasts. Moreover, in order to assess whether jigr1 upregulation mimics the miR-92–/– phenotype, we overexpressed jigr1 in larval brain neuroblasts (Fig 8C). We observed decreased BrdU uptake and premature localization of Prospero in larval neuroblasts. In addition overexpression of Jigr1 in neuroblasts led to a reduction in adult brain size (Fig 8F) suggesting that Jigr1 overexpression is sufficient to cause premature cell cycle exit and differentiation of neuroblasts (Fig 8D and 8E). Taken together, our results suggest that miR-92 family maintains neuroblast self-renewal in part through limiting Jigr1 expression.
Fig. 8. Jigr1 upregulation is responsible for premature neuroblast differentiation in miR-92–/– flies. 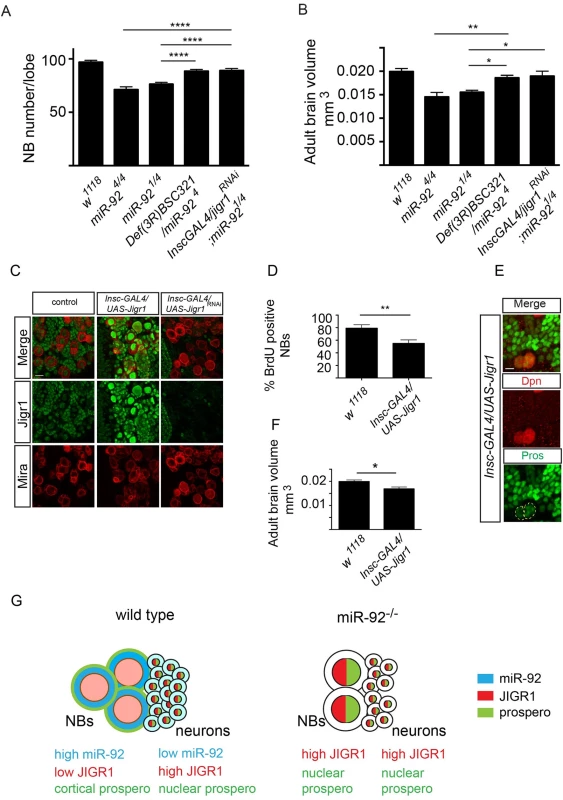
(A) Neuroblast number in third instar larval brains of the indicated genotypes at 120 hr ALH. w1118 (n = 17); miR-924/4 (n = 15); miR-921/4 (n = 20); Def(3R)BSC321/miR-924 (n = 17); Insc-GAL4/UAS-Jigr1RNAi; miR-921/4 (n = 14). (B) The size of Adult brains of indicated genotypes. w1118 (n = 10); miR-924/4 (n = 10); miR-921/4 (n = 8); Def(3R)BSC321/miR-924 (n = 12); Insc-GAL4/UAS-Jigr1RNAi; miR-921/4 (n = 8). (C) Immunostaining of third instar larval brains of wild type (left panel), Insc-GAL4/UAS-jigr1 (middle panel) and Insc-GAL4/UAS-jigr1-RNAi (right panel) flies. Green: Jigr1. Red: Miranda. Scale Bar: 20 m. (D) Percentage of BrdU+ neuroblasts in wild type control (n = 12), Insc-GAL4/UAS-jigr1 (n = 15) and Insc-GAL4/UAS-jigr1-RNAi (n = 9) larval brains. (E) Immunostaining of Insc-GAL4/UAS-jigr1brains with Prospero (green) and Dpn (red) at 120 hr ALH. (F) Adult brain volume of wild type control (n = 10) and Insc-GAL4/UAS-jigr1 (n = 9) flies. (G) Working model summarizing the expression and the role of miR-92 in larval neuroblasts. Statistical significance was determined by One-Way ANOVA. Values are mean ± s.e.m. in all graphs. *: p < 0.05, **: p < 0.005, ***: p < 0.001, ****: p < 0.0001. Discussion
In this study, we report an unusual genomic arrangement in which miR-92a and miR-92b are embedded in the intron and 3’UTR of the host gene jigr1, respectively. In neuroblasts, miR-92a and miR-92b were highly expressed as a single transcriptional unit also containing jigr1 coding region. Genetic analysis in Drosophila showed that downregulation of jigr1 by intragenic miR-92a and miR-92b is required for neuroblast self-renewal, providing an example of the functional significance of miRNA–host gene interactions in animal development.
Nearly half of the miRNAs in mammals and Drosophila are located within protein-coding genes [4, 25]. Most of these intragenic miRNAs are co-expressed with their host genes, but both positive and negative feedback regulation of host gene expression and function by miRNAs remains largely unknown[26–30, 32]. Most intragenic miRNAs are located in the introns of their host genes and are processed by the mirtron pathway, bypassing the microprocessor complex [33]. On the other hand, miRNAs are rarely located in the 3’UTR of a protein-coding gene, and the effect of this organization on host gene expression or miRNA processing is not clear. The exonic miR-198, which is located in the 3’UTR of the gene encoding human folistatin like 1, is processed from a single transcript with its host gene in a mutually exclusive way [34]. In contrast, direct regulation of some jigr1 isoforms by miR-92a and miR-92b largely accounts for the observed complementary expression domains of jigr1 and these miRNAs (Figs 3 and 6), although we cannot completely rule out the possibility other mechanisms may also contribute to jigr1 repression.
Through a genetic knockout of both miR-92a and miR-92b in Drosophila, which has not been done so far in any other model organism, we discovered a novel function for miR-92a and miR-92b in neuroblast self-renewal. Our findings are consistent with results obtained in mammals. However, unlike Drosophila, human and mouse miR-92a and miR-92b genes are not intragenic. Mouse miR-92a genes are located in two clusters: miR-17-92 and miR-106a-303. In developing mouse neocortex, the miR-17-92 cluster promotes neural stem cell expansion and regulates the transition to intermediate progenitors through repression of Pten by miR-19 and Tbr2 by miR-92a [35]. Similarly, acute loss and gain of miR-92b function in mouse cortex showed that miR-92b restricts the generation of intermediate progenitor cells by suppressing Tbr2 [36]. Moreover, miR-92b maintains asymmetric division of neural stem cells by restricting Tis21 expression in mouse neocortex [37]. However, phenotypes caused by loss of both miR-92a and miR-92b in mammals have not been reported yet. Here we found that miR-92a and miR-92b work in concert to restrict Jigr1 expression in Drosophila larval neuroblasts and thereby maintain the neuroblast pool.
Jigr1, a putative MADF-domain-containing transcription factor of unknown function expressed ubiquitously in the larval central nervous system, is expressed at low levels in neural progenitor cells. Our findings suggest that the progressive loss of neuroblasts in miR-92–/– brains is due to premature differentiation of these cells resulted from cell-autonomous effect of loss of miR-92, as shown by ectopic expression of nuclear Prospero, decreased BrdU uptake, and reduced cell size. Upregulation of Jigr1 seems to play a role in nuclear Prospero expression in miR-92–/– neuroblasts, since reducing Jigr1 expression eliminated this phenotype. Moreover, overexpression of Jigr1 in neuroblasts on a wild type background leads to ectopic Prospero expression and a premature differentiation phenotype. In summary, our results reveal a local regulatory loop in which miR-92a and miR-92b are expressed in the jigr1 transcription unit and also work in concert to prevent premature differentiation of neuroblasts by limiting expression of the host gene.
Materials and Methods
Fly Stocks
Fly stocks were maintained at 25°C on standard medium. The following fly stocks were obtained from the Bloomington Drosophila Stock Center: w1118, Elav-GAL4, Def(3R)BSC321, y[1] w[*]/Dp(2;Y)G, P{hs-hid}Y; P{70FLP}11 P{70I-SceI}2B noc[Sco]/CyO, P{hs-hid}4, y[1] w[67c23] P{y[+mDint2] = Crey}1b; D[*]/TM3, Sb[1], hs-flp70; Tub-Gal4 UAS-mCD8:GFP; FRT82B Tub-Gal80, FRT82B. UAS-jigr1-RNAi (#100970) was obtained from the Vienna Drosophila Resource Center. Insc-GAL4 and Pros-GAL4 were kind gift of Chris Doe (University of Oregon).
Generation of Transgenic Flies and Deletion Lines
The UAS-dsRed-miR-92a vector was from the Drosophila RNAi Screening Center, and UAS-miR-92b was generated by amplifying the 500–base pair pri-miR-92b from genomic DNA and cloning it into the pTW vector. Primers for PCR are listed in S1 Table. UAS-miR-92a and UAS-miR-92b transgenic flies were generated by Rainbow Transgenics.
The FRT-bearing transposon lines e04431, e04047, e00089, d03337, e01786, and e03251 (Harvard Exelixis Collection) were used to generate FLP-FRT-based deletions, as described. Deletions were confirmed by PCR and sequencing.
In order to generate Del #1, miR-92-/- lines, which have one loxP site at miR-92a locus and two additional loxP sites at miR-92b locus, were used. Deletions were made as described in [16].
Generation of miR-92a–/–, miR-92b–/–, and miR-92–/– Flies
Ends-out gene targeting was performed as described. Primers used to clone the 5’ and 3’ homology arms of miR-92a and miR-92b into pGX-attp are listed in S1 Table. The gene knockouts in mutant flies were confirmed by PCR, sequencing, and northern blot analysis.
Clonal Analysis
MARCM clones were generated as described before by crossing hs-flp70, Tub-Gal4, UAS-mCD8:GFP; FRT82B, Tub-Gal80 flies with FRT82B flies to generate control clones and with FRT82B, miR-924 flies to generate mutant clones. Clones were induced at 24 ± 6 hr ALH by heat shock in 37°C water bath for an hour and analyzed 72 hr after clone induction. The size of the clones was determined by counting the GFP+ cells in single clones. Type II clones were distinguished by presence of several small GFP+ Dpn+ cells (Intermediate progenitor cells) in addition to single large GFP+ Dpn+ neuroblast.
qRT-PCR
For qRT-PCR, cDNA was synthesized from total RNA with random primers and superscript-II reverse transcriptase (Invitrogen). SYBR Green PCR Master Mix (Applied Biosystems) was used for real-time thermal cycling. The reactions were performed with a StepOne Plus Real-Time PCR System (Applied Biosystems) and StepOne software v2.1. rp49 primers were used for normalization, and relative expression levels were calculated with the comparative CT method. The oligonucleotides used for qRT-PCR are listed in S1 Table.
For quantification of mature miR-92a and miR-92b, miRNA Taqman assay was used (Applied Biosystems assay number 000285 for miR-92a and 000286 for miR-92b). U6 was used as an internal reference.
3’RACE
Total RNA was extracted from larval brains with miRNeasy (Qiagen). 3’RACE was performed as described [38]. Two specific PCR bands of different sizes were gel purified with the QIAquick gel extraction kit (Qiagen), cloned into pGEMT-easy (Promega), and sequenced with vector-specific primers.
Immunoblot Analysis
For western blot, 25 larval brains were dissected and lysed in 200 μl of RIPA buffer. Larval lysate (20 μl) was mixed with 2X Laemmli buffer, boiled, and separated by 12% SDS-PAGE. The blot was probed with rabbit anti-jigr1 (1 : 5000), generated against the N-terminal 110 amino acids of jigr1 protein by SDIX. Mouse anti-tubulin (T6199, Sigma 1 : 5000) was used as a loading control. Horseradish peroxidase–conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories.
Constructs and Mutagenesis
To generate miR-92a and miR-92b expression vectors, Drosophila pri-miR-92a and pri-miR-92b were amplified from genomic DNA and cloned into pSuper-GFP vector. MiR-124 used as a control miRNA. A pri-miR-124 expression plasmid was generated by amplification of 500 bp of primary sequence from mouse genomic DNA and cloning into pSuper-GFP vector.
To generate jigr1 expression constructs, jigr1 coding region containing short 3’UTR and long 3’UTR are amplified from cDNA and cloned into pUASt vector.
To generate a jigr1 3’UTR sensor, jigr1 short and long 3’UTR sequences were amplified from genomic DNA and cloned into psiCHECK-2 downstream of Renilla luciferase. The target sites were mutated by site-directed mutagenesis with QuickChange multi and the QuickChange II site-directed mutagenesis kit (Stratagene). For the Jigr1-5’UTR sensor, oligonucleotides containing wild type and mutated miR-92a and miR-92b binding sites were synthesized, annealed, and cloned at the 5’ of the Renilla luciferase gene using the NheI site in psiCHECK-2 vector. The sequences of the wild type and mutant oligonucleotides are listed in S2 Table.
BrdU Incorporation Assay
Drosophila third instar larvae were fed on food containing 1 mg/mL BrdU for 3 hours. Larval brains were immediately dissected, fixed and stained with the addition of a 2N HCl treatment for 30 min prior to BrdU staining.
For BrdU chase experiments, first clones were induced in hs-flp70, Tub-Gal4, UAS-mCD8:GFP; FRT82B, Tub-Gal80 /FRT82+ and, hs-flp70, Tub-Gal4, UAS-mCD8:GFP; FRT82B, Tub-Gal80/ FRT82B, miR-924 larvae at 24 ± 6 hr ALH. 48 hr later the larvae were first transferred to food containing 1mg/mL BrdU for 4 hours then to normal food for 24hr. Larval brains were dissected, fixed and stained with the addition of a 2N HCl treatment for 30 min prior to BrdU staining.
Northern Blot Analysis
Northern blot for miRNAs was performed as described [39]. The following LNA modified probes from Exiqon were used. miR-92a: 5’-ATAGGCCGGGACAAGTGCAATG-3’; miR-92b: 5’-GCAGGCCGGGACTAGTGCAATT-3’; U6 : 5’-CACGAATTTGCGTGTCATCCTT-3’.
Total RNA was extracted with the miRNAeasy mini kit (Qiagen). RNA (10–20 μg) was loaded onto a 15% polyacrylamide gel (Sequagel, National Diagnostics), transferred to Amersham Hybond-N+ (GE Healthcare), and crosslinked to the membrane with UV crosslinker. Hybridization was done overnight (16 hours) after 1 hour of pre-hybridization in DIG easy Hyb granules (Roche). For detection, DIG wash and block buffer set (Roche) were used according to the manufacturer’s protocol.
For northern blot analysis of jigr1, 10 μg of total RNA was loaded onto an agarose gel (1% agarose, 1x MOPS, and 5% formaldehyde) and transferred to a nylon membrane overnight by capillary transfer. Hybridization and detection were done as described above. To generate probes for jigr1 and rp49, we amplified a ~500-bp region from genomic DNA by PCR. A T7 promoter sequence was added 5’ of the reverse primer. The probes were synthesized by in vitro transcription with T7 RNA polymerase (Roche). The oligonucleotides used to synthesize the jigr1 and rp49 probes are listed in S1 Table.
RNA In Situ Hybridization
Third instar larval brains were dissected and fixed in 4% paraformaldehyde overnight at 4°C. The brains were transferred into 30% sucrose and incubated at 4°C overnight. Then, they were frozen and stored at -80°C until further use. Then 15 um sections were made. 5’ and 3’ DIG-labeled LNA-modified miR-92a and miR-92b probes (Exiqon) were used as recommended by the manufacturer. Anti-DIG-POD primary antibody (Roche; 1 : 500) was used and detected with the TSA Plus Cyanine 3 System (Perkin Elmer).
Immunostaining and Confocal Imaging
Larval brains were dissected and fixed in 4% paraformaldehyde in phosphate-buffered saline for 20 minutes. The primary antibodies used were rabbit anti-Dpn (1 : 500) from Y.-N. Jan (University of California, San Francisco), mouse anti–Dlg 4F3 (1 : 20) from the Developmental Studies Hybridoma Bank (DSHB), rat anti-Miranda (1 : 500) from Chris Doe (University of Oregon), mouse anti-Prospero MR1A (1 : 20) from DSHB, rat anti-BrdU (1 : 500) from Abcam, goat anti-HRP-Cy3 from Jackson ImmunoResearch Laboratories, and mouse anti-nc82 (1 : 5) from DSHB. Secondary antibodies were from Life Technologies. Confocal images were taken with a Nikon D-Eclipse C1 and processed with Image J and Adobe Photoshop.
Luciferase Assay
HEK293T cells were transfected with wild type or mutant miR-92 sensor plasmids or empty psiCHECK-2, pSuper-GFP-pri-miR-92a, or pSuper-GFP-pri-miR-92b expression vectors (described above) or pSuper-GFp-miR-124 using Fugene6 (Promega). 48 hours after transfection, cells were lysed in 1x passive lysis buffer (Promega). The Dual-Glo Luciferase Assay System from Promega was used according to the manufacturer’s protocol. Renilla luciferase activity was normalized to firefly luciferase activity. All the experiments were done in triplicate in three independent experiments.
Supporting Information
Zdroje
1. Ambros V. microRNAs: tiny regulators with great potential. Cell [Internet]. 2001 Dec 28 [cited 2014 Sep 19];107(7):823–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11779458 11779458
2. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell [Internet]. 2012 Apr 27 [cited 2014 Jul 9];149(3):515–24. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3351105&tool=pmcentrez&rendertype=abstract doi: 10.1016/j.cell.2012.04.005 22541426
3. Newman M a, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev [Internet]. 2010 Jun 1 [cited 2014 Jun 5];24(11):1086–92. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2878647&tool=pmcentrez&rendertype=abstract doi: 10.1101/gad.1919710 20516194
4. Kim Y-K, Kim VN. Processing of intronic microRNAs. EMBO J [Internet]. 2007 Feb 7 [cited 2014 Aug 14];26(3):775–83. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1794378&tool=pmcentrez&rendertype=abstract 17255951
5. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res [Internet]. 2004 Oct [cited 2014 Jul 9];14(10A):1902–10. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=524413&tool=pmcentrez&rendertype=abstract 15364901
6. Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA [Internet]. 2005 Mar [cited 2014 Aug 27];11(3):241–7. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1370713&tool=pmcentrez&rendertype=abstract 15701730
7. He C, Li Z, Chen P, Huang H, Hurst LD, Chen J. Young intragenic miRNAs are less coexpressed with host genes than old ones: implications of miRNA-host gene coevolution. Nucleic Acids Res [Internet]. 2012 May [cited 2014 Sep 24];40(9):4002–12. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3351155&tool=pmcentrez&rendertype=abstract doi: 10.1093/nar/gkr1312 22238379
8. Ramalingam P, Palanichamy JK, Singh A, Das P, Bhagat M, Kassab MA, et al. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA [Internet]. 2014 Jan [cited 2014 Jun 2];20(1):76–87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24226766 doi: 10.1261/rna.041814.113 24226766
9. Hartenstein V, Rudloff E, Campos-Ortega JA. The pattern of proliferation of the neuroblasts in the wild-type embryo of Drosophila melanogaster. Roux’s Arch Dev Biol [Internet]. 1987 Dec [cited 2014 Sep 25];196(8):473–85. Available from: http://link.springer.com/10.1007/BF00399871
10. Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol [Internet]. 1988 Jan;125(1):145–57. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3119399 3119399
11. Maurange C, Gould AP. Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci [Internet]. 2005 Jan [cited 2014 Jul 7];28(1):30–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15626494 15626494
12. Jiang Y, Reichert H. Drosophila Neural Stem Cells in Brain Development and Tumor Formation. J Neurogenet [Internet]. 2014 May 12 [cited 2014 Sep 25];1–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24766377
13. Sun X, Morozova T, Sonnenfeld M. Glial and neuronal functions of the Drosophila homolog of the human SWI/SNF gene ATR-X (DATR-X) and the jing zinc-finger gene specify the lateral positioning of longitudinal glia and axons. Genetics [Internet]. 2006 Jul [cited 2014 Jun 11];173(3):1397–415. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1526706&tool=pmcentrez&rendertype=abstract 16648585
14. Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev [Internet]. 2007 Jan [cited 2012 Aug 30];2(January):1. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1779784&tool=pmcentrez&rendertype=abstract
15. Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet [Internet]. 2004 Mar [cited 2014 Jul 14];36(3):288–92. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14981519 14981519
16. Huang J, Zhou W, Dong W, Watson AM, Hong Y. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A [Internet]. 2009 May 19 [cited 2014 Sep 15];106(20):8284–9. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2688891&tool=pmcentrez&rendertype=abstract doi: 10.1073/pnas.0900641106 19429710
17. Chen Z, Liang S, Zhao Y, Han Z. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development [Internet]. 2012 Oct [cited 2013 Aug 12];139(19):3543–52. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22899845 doi: 10.1242/dev.082719 22899845
18. Fuse N, Hisata K, Katzen AL, Matsuzaki F. Heterotrimeric G Proteins Regulate Daughter Cell Size Asymmetry in Drosophila Neuroblast Divisions. Curr Biol. 2003;13(15):947–54.
19. Song Y, Lu B. Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev [Internet]. 2011 Dec 15 [cited 2014 Jun 4];25(24):2644–58. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3248685&tool=pmcentrez&rendertype=abstract doi: 10.1101/gad.171959.111 22190460
20. Xiao Q, Komori H, Lee C-Y. Klumpfuss Distinguishes Stem Cells From Progenitor Cells During Asymmetric Neuroblast Division. Development [Internet]. 2012 Aug [cited 2014 Mar 22];139(15):2670–80. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3392700&tool=pmcentrez&rendertype=abstract doi: 10.1242/dev.081687 22745313
21. Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell [Internet]. 2008 May 30 [cited 2014 Jun 7];133(5):891–902. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18510932 doi: 10.1016/j.cell.2008.03.034 18510932
22. Lai S-L, Doe CQ. Transient nuclear Prospero induces neural progenitor quiescence. Elife [Internet]. 2014 Jan [cited 2015 Jan 31];3 : 1–12. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4212206&tool=pmcentrez&rendertype=abstract doi: 10.7554/eLife.03363 25354199
23. Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron [Internet]. 1999 Mar;22(3):451–61. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10197526 10197526
24. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell [Internet]. 2009 Jan 23 [cited 2014 Jul 9];136(2):215–33. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3794896&tool=pmcentrez&rendertype=abstract doi: 10.1016/j.cell.2009.01.002 19167326
25. Hinske LCG, Galante P a F, Kuo WP, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genomics [Internet]. 2010 Jan;11 : 533. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3091682&tool=pmcentrez&rendertype=abstract doi: 10.1186/1471-2164-11-533 20920310
26. Liu M, Roth A, Yu M, Morris R, Bersani F, Rivera MN, et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev [Internet]. 2013 Dec 1 [cited 2014 Jun 11];27(23):2543–8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3861668&tool=pmcentrez&rendertype=abstract doi: 10.1101/gad.224170.113 24298054
27. Kos A, Olde Loohuis NFM, Wieczorek ML, Glennon JC, Martens GJM, Kolk SM, et al. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS One [Internet]. 2012 Jan [cited 2014 Jun 30];7(2):e31022. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3281898&tool=pmcentrez&rendertype=abstract doi: 10.1371/journal.pone.0031022 22363537
28. Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev [Internet]. 2012 Jan 1 [cited 2014 Jun 14];26(1):25–30. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3258962&tool=pmcentrez&rendertype=abstract doi: 10.1101/gad.177774.111 22215807
29. Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, et al. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun [Internet]. Elsevier Inc.; 2010 Jun 11 [cited 2014 Jun 30];396(4):978–82. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20470754 doi: 10.1016/j.bbrc.2010.05.037 20470754
30. Zhu Y, Lu Y, Zhang Q, Liu J, Li T, Yang J, et al. MicroRNA-26a / b and their host genes cooperate to inhibit the G1 / S transition by activating the pRb protein. Nucleic Acids Res. 2012;40(10):4615–25. doi: 10.1093/nar/gkr1278 22210897
31. Zhou H, Rigoutsos I. MiR-103a-3p targets the 5 ′ UTR of GPRC5A in pancreatic cells. RNA. 2014;20 : 1–9. doi: 10.1261/rna.038182.113 24255166
32. Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36(16):5232–41. doi: 10.1093/nar/gkn513 18684991
33. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell [Internet]. 2007 Jul 13 [cited 2014 May 24];130(1):89–100. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2729315&tool=pmcentrez&rendertype=abstract 17599402
34. Sundaram GM, Common JEA, Gopal FE, Srikanta S, Lakshman K, Lunny DP, et al. “See-saw” expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature [Internet]. 2013 Mar 7 [cited 2014 Jul 12];495(7439):103–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23395958 doi: 10.1038/nature11890 23395958
35. Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep [Internet]. The Authors; 2013 May 30 [cited 2014 Jan 22];3(5):1398–406. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23623502 doi: 10.1016/j.celrep.2013.03.037 23623502
36. Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. 2013;1–6.
37. Fei J-F, Haffner C, Huttner WB. 3’ UTR-dependent, miR-92-mediated restriction of Tis21 expression maintains asymmetric neural stem cell division to ensure proper neocortex size. Cell Rep [Internet]. The Authors; 2014 Apr 24 [cited 2014 Jun 6];7(2):398–411. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24726360 doi: 10.1016/j.celrep.2014.03.033 24726360
38. Scotto-Lavino E, Du G, Frohman M a. 3’ end cDNA amplification using classic RACE. Nat Protoc [Internet]. 2006 Jan [cited 2015 Feb 26];1(6):2742–5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17406530 17406530
39. Li Z, Lu Y, Xu X-L, Gao F-B. The FTD/ALS-associated RNA-binding protein TDP-43 regulates the robustness of neuronal specification through microRNA-9a in Drosophila. Hum Mol Genet [Internet]. 2013 Jan 15 [cited 2014 Sep 25];22(2):218–25. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3526156&tool=pmcentrez&rendertype=abstract doi: 10.1093/hmg/dds420 23042786
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



