-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRegulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
DNA cytosine methylation (5-methylcytosine, 5-meC) plays important roles in genome organization, genomic imprinting, transposon silencing and gene expression. DNA methylation patterns are dynamically controlled by methylation and demethylation reactions during development and reproduction in eukaryotes. In Arabidopsis thaliana, ROS1, a 5-methylcytosine DNA glycosylase, is responsible for active DNA demethylation via a base excision repair pathway. Our previous work showed that a histone acetyltransferase, IDM1, is an important regulator of active DNA demethylation. Here we identified two components, IDL1 and MBD7, as additional novel regulators of active DNA demethylation. Our data suggest that a histone acetyltransferase complex, composed of MBD7, IDL1, IDM2 and IDM1, is required for the recruitment of ROS1 to specific loci for active DNA demethylation. Our findings significantly expand our understanding of the regulatory mechanisms underlying active DNA demethylation in plants.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005210
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005210Summary
DNA cytosine methylation (5-methylcytosine, 5-meC) plays important roles in genome organization, genomic imprinting, transposon silencing and gene expression. DNA methylation patterns are dynamically controlled by methylation and demethylation reactions during development and reproduction in eukaryotes. In Arabidopsis thaliana, ROS1, a 5-methylcytosine DNA glycosylase, is responsible for active DNA demethylation via a base excision repair pathway. Our previous work showed that a histone acetyltransferase, IDM1, is an important regulator of active DNA demethylation. Here we identified two components, IDL1 and MBD7, as additional novel regulators of active DNA demethylation. Our data suggest that a histone acetyltransferase complex, composed of MBD7, IDL1, IDM2 and IDM1, is required for the recruitment of ROS1 to specific loci for active DNA demethylation. Our findings significantly expand our understanding of the regulatory mechanisms underlying active DNA demethylation in plants.
Introduction
DNA methylation is an important epigenetic mark conserved in many eukaryotes. Many studies have demonstrated that DNA methylation plays central roles in genome organization, genomic imprinting, transposon silencing, and gene expression [1,2,3]. DNA methylation levels are coordinately determined by methylation and demethylation reactions during development and reproduction in both plants and animals. In the model plant Arabidopsis thaliana, levels of symmetric CG and CHG methylation are maintained by DNA METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3), respectively, during DNA replication [1]. In contrast, asymmetric CHH methylation needs to be established de novo during each cell cycle [1]. DNA methylation is antagonized by an active DNA demethylation pathway catalyzed by a subfamily of bi-functional DNA glycosylase/lyases represented by REPRESSOROF SILENCING 1 (ROS1) and DEMETER (DME) [4,5,6,7,8].
Although the RNA-directed DNA methylation pathway and its regulation are well-established (Law and Jacobsen, 2010), little is known about the locus-specific targeting of proteins involved in active DNA demethylation in plants. Previous studies suggested that the RNA-binding protein ROS3 is required for the recruitment of ROS1 to some target loci [9]. INCREASED DNA METHYLATION 1(IDM1), a histone acetyltransferase, was recently identified as another important regulator of DNA demethylation that functions upstream of ROS1 [10]. With the assistance of IDM2, an alpha-crystallin domain protein, IDM1 recognizes chromatin regions that are marked by CG methylation and low levels of histone 3 lysine 4 (H3K4) and arginine 2 (H3R2) methylation and subsequently catalyzes H3K18 and H3K23 acetylation, which triggers ROS1-mediated DNA demethylation [10,11]. However, the molecular mechanism(s) by which IDM1 is targeted to these specific genomic loci remains largely unknown.
In plants and animals, a set of proteins containing a methyl-CpG-binding domain (MBD) are capable of specifically recognizing and binding methylated DNA [12,13]. They are believed to function as an interpreter of DNA methylation signals [13]. In mammals, MBD proteins MeCP2, MBD1, MBD2 and MBD4 have methyl-CpG-binding activity and function in targeting of maintenance DNA methylation machinery, H3K9 methylation, transcriptional silencing and X chromosome inactivation [14]. They usually execute their functions by binding to other proteins. For example, MeCP2 was demonstrated to interact with Sin3 and histone deacetylases to transcriptionally silence methylated chromosome, suggesting a direct relationship between histone modification and methylated DNA [15]. In Zebrafish, MBD4, a protein that possesses DNA glycosylase activity, was reported be involved in active DNA demethylation [16].
The Arabidopsis genome contains 13 genes encoding putative MBD proteins orthologous to their mammalian counterparts [17]. Only AtMBD5, AtMBD6 and AtMBD7 bind methylated CpG nucleotide in vitro [17]. AtMBD5, AtMBD6 and AtMBD7 localize in the nuclei, and assemble at highly methylated chromocenters. while, in DNA hypomethylation mutants ddm1 and met1 they dissociate from chromocenters, indicating their methyl-CpG-binding activity in vivo [18]. Different from AtMBD7, AtMBD5 and AtMBD6 mainly localize to chromatin regions covered by ribosomal DNA (rDNA) gene clusters [18,19]. Snf2 family nucleosome remodeler DDM1 is able to bind AtMBD5 and AtMBD6 in vitro and facilitate their heterochromatin localization [18]. AtMBD7, belonging to a unique class of plant MBD proteins that have multiple MBD domains, was hypothesized to bind multiple methylated sites that are not close to each other and promote the formation and/or maintainance of a microenvironment in chromatin [13].
To find out how IDM1 is targeted to specific loci, we identified IDM1 and IDM2 interacting proteins by an approach that combines affinity purification and mass spectrometry. We found that IDM1 and IDM2 copurify with each other and with two novel proteins, IDL1 and MBD7. MBD7 directly binds to the target loci and is required for the H3K18 and H3K23 acetylation in planta. The mbd7 mutants exhibit DNA hypermethylation at thousands of genomic loci and show increased transcriptional silencing of reporter genes and some endogenous genes. Our results reveal novel components involved in DNA demethylation and novel functions for MBD proteins in plants.
Results
Affinity purification and mass-spectrometric identification of IDM1 and IDM2 interacting proteins
To better understand the mechanism of IDM1 targeting in active DNA demethylation, we set out to purify the protein complex associated with IDM1. Transgenic Arabidopsis plants expressing IDM1-3HA-YFP driven by native IDM1 promoter in idm1-3 mutant background were generated. Previous studies showed that IDM1 dysfunction caused the kanamycin sensitive growth phenotype and silencing of NPT II transgene [20]. The IDM1-3HA-YFP was able to complement the kanamycin sensitive growth phenotype (S1A Fig). Real time PCR results showed that the NPT II expression was also restored in the transgenic plants (S1B Fig), suggesting that the IDM1-3HA-YFP fusion protein was functional in vivo. The IDM1-3HA-YFP expression in the transgenic lines was confirmed by Western blot with the HA antibody (S1C Fig). Co-immunoprecipitation (co-IP) of IDM1-3HA-YFP and its interacting proteins was performed using anti-HA antibodies and flower extracts. Tryptic peptides derived from the purified proteins were sequenced by Velos Pro Orbitrap Elite tandem mass spectrometry (MS/MS) (Table 1 and S1 Table). As expected, the MS analysis identified peptides corresponding to a known IDM1 interacting protein IDM2 [11]. Peptides corresponding to the IDM2-related protein IDM2 like 1 (IDL1) and methyl-CpG-binding domain protein 7 (MBD7) were also identified (Table 1), suggesting these two proteins may be new components of active DNA demethylation pathway. Meanwhile, we carried out affinity purification of 6Myc-IDM2 using transgenic plants overexpressing 6Myc-IDM2 in idm2-1 mutants [11] and 6Myc-IDL1 using transgenic plants overexpressing 6Myc-IDL1 in wild type plants. Mass spectrometric analysis revealed that IDM2 and IDL1 copurify with each other. We also identified a large number of peptides corresponding to IDM1 and MBD7 (Table 1). Our data suggest that IDM1, IDM2, IDL1, and MBD7 form a tight complex.
Tab. 1. Mass-spectrometric analysis of IDM1, IDM2 and IDL1 co-purifying proteins. 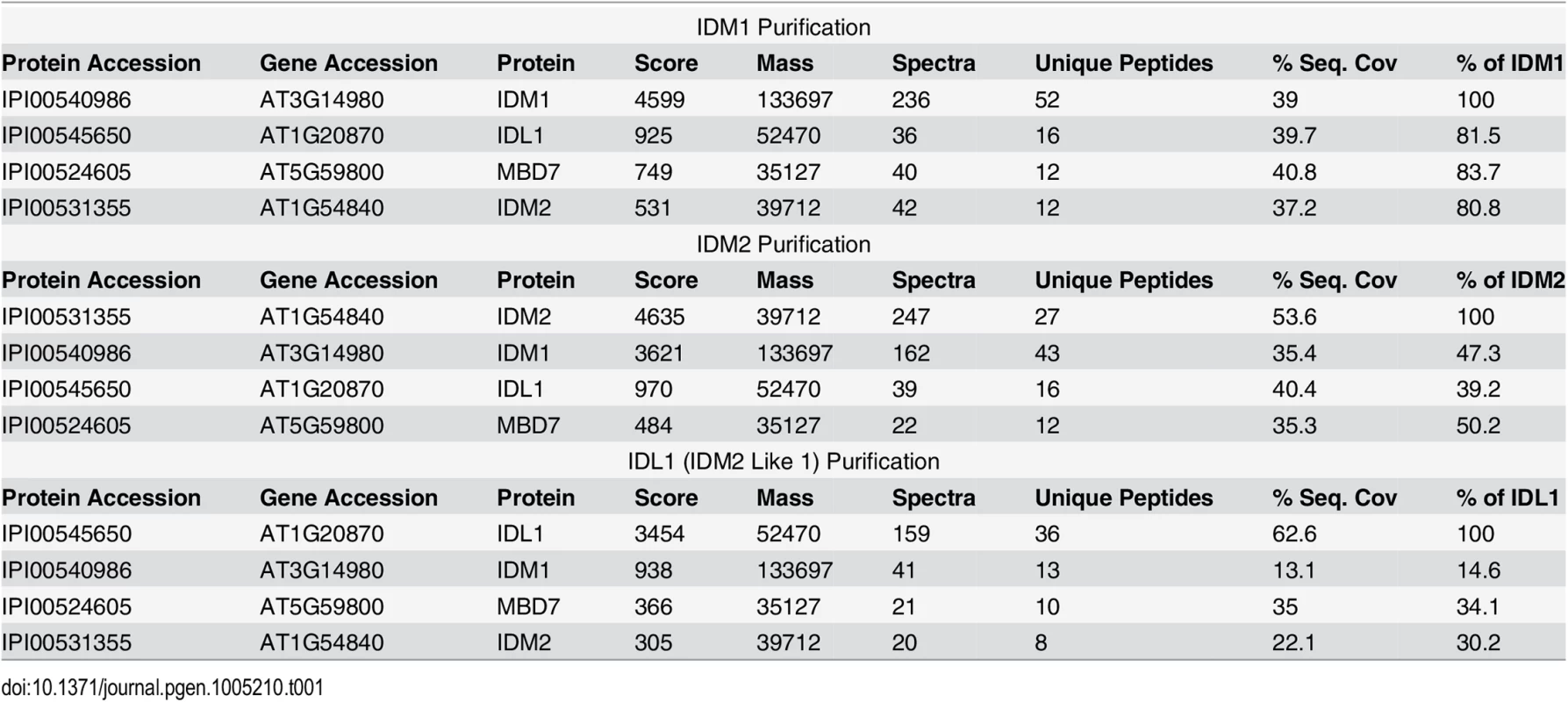
Confirmation of protein-protein interactions in a IDM1 and IDM2 protein complex
The interaction of IDL1 and MBD7 with IDM1 and IDM2 was corroborated using the yeast two-hybrid assay (Fig 1A). Interestingly, IDL1 can interact with the C-terminal of IDM1 while IDM2 can interact with the N-terminal of IDM1 (Fig 1A). MBD7 also interacts with IDM2 and IDL1, but not with IDM1 (Fig 1A). The association between IDM2, IDL1, and MBD7 was also confirmed by a pull down assay (Fig 1B). To confirm these protein-protein interactions in vivo, we performed two assays. First, results from the split-LUC assay demonstrated that IDL1 interacts with IDM2, and that MBD7 interacts with both IDM2 and IDL1 (Fig 1C). Second, experiments using GFP-tagged IDM2 and Flag-tagged MBD7 transiently expressed in Nicotiana benthamiana leaves revealed that MBD7 co-IP’d with IDM2 from IDM2-GFP-expressing leaves, but not from leaves of control plants (Fig 1D). MBD7 also co-IP’d with IDL1 (Fig 1D). The interaction between MBD7 and IDM1 was also detected by co-IP in plants harboring both MBD7-Myc and IDM1-HA-YFP transgenes driven by their native promoters (Fig 1D). Taken together, our results suggest that IDM1, IDM2, IDL1 and MBD7 form a tight multiprotein complex in vivo.
Fig. 1. Protein-protein interactions between IDM1, IDM2, IDL1 and MBD7. 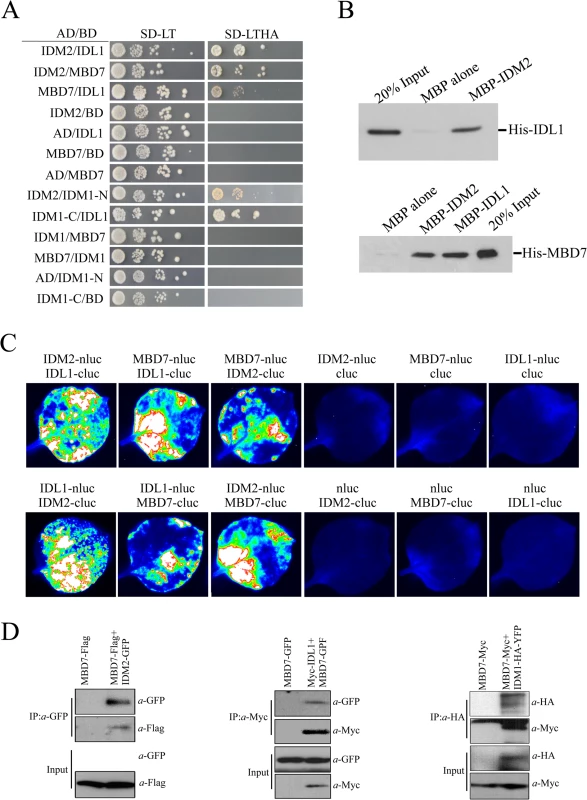
(A) Yeast two-hybrid assays. Yeast cells carrying different fusion protein combinations are listed on the left. Yeast cells expressing the indicated proteins from the pGBK-T7 (BD) and pGAD-T7 (AD) vectors were plated onto medium lacking Leu and Trp (SD-LT) (left) or medium lacking Leu, Trp, Ade and His (SD-LTHA) (right). (B) Pull-down assays showing that IDM2, IDL1 and MBD7 interact with each other. (C) Split-luc assays showing that IDL1 and MBD7 can interact with IDM2 in N. benthamiana leaves. Three biological replicates were performed, and similar results were obtained. (D) Co-immunoprecipitation of MBD7 with IDM2 or IDL1 in tobacco leaves. MYC-tagged IDM2 and GFP-tagged IDM1 were transiently expressed in N. benthamiana leaves. Anti-GFP was used for immunoprecipitation (IP); anti-MYC and anti-GFP were used for immunoblotting; Input, total protein before immunoprecipitation. Transgenic plants expressing MBD7-Myc or IDM1-HA-YFP under their native promoters and their F1 offspring were used for co-IP. The mbd7 mutation leads to DNA hypermethylation at specific loci
IDL1 encodes an α-crystallin domain protein that belongs to the IDM2 protein family. Besides IDM2, the IDM2 protein family includes three IDM2-like proteins including IDL1 (At1g20870), IDL2 (At1g54850), and IDL3 (At1g76640) (S2 Fig). According to sequence alignment and phylogenetic analysis, IDL1 belongs to the same clade as IDM2 and shares maximal sequence similarity with IDM2 [21]. MBD7 encodes a methyl-CpG-binding domain protein, which binds methylated CpG dinucleotides in vitro and localizes to highly CpG-methylated chromocenters in vivo [17,18].
The tight association of IDL1 and MBD7 with IDM1 and IDM2 suggests that IDL1 and MBD7 may also be required for active DNA demethylation. In order to examine the possible role of MBD7 in active DNA demethylation, two T-DNA insertion lines were isolated and RT-PCR was done to confirm that they have a complete loss of mRNA expression (S3 Fig). Whole genome bisulfite sequencing was done using DNA purified from 12-day-old mbd7-1 and wild type seedlings. The overall genome methylation level was slightly increased in mbd7-1mutant plants compared with wild type control (S4 and S5A Figs). Hundreds of differentially methylated regions (DMRs) were apparent in mbd7-1 knockout plants, including 748 loci with DNA hypermethylation and 198 loci with DNA hypomethylation, respectively (S2 and S3 Tables). Six hypermethylated loci in mbd7, idm1, and ros1 plants were selected for validation by PCR-based DNA methylation analysis, and all of them confirmed to be hypermethylated in mbd7 mutants (Fig 2A and 2B and S5B and S5C Fig), as in idm1-1 or ros1-4. However, the chop PCR marker locus DT-77, which was hypermethylated in idm1 and ros1 [10,11], was not hypermethylated in mbd7 mutant plants (Fig 2B), suggesting not all IDM1 target loci were affected by MBD7 loss-of-function.
Fig. 2. Methylome analysis of mbd7-1 mutant. 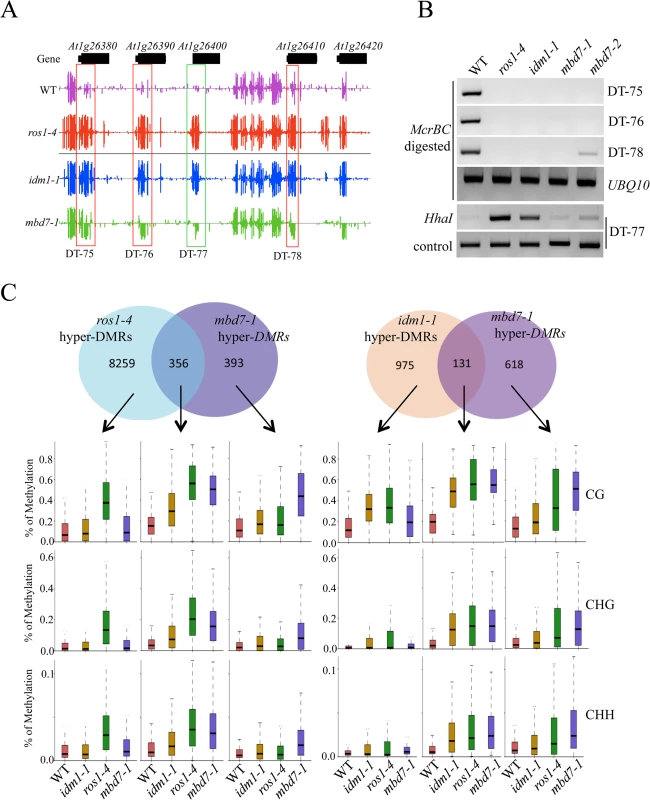
(A) Snapshot in the Integrated Genome Browser showing DNA methylation levels of the FAD-binding berberine gene family in WT, idm1-1, ros1-4 and mbd7-1. Hypermethylated regions in mbd7-1 were highlighted with red boxes. DT-77, which was hypermethylated in idm1-1 or ros1-4 but not in mbd7-1, was highlighted with green box. (B) Confirmation the whole genome bisulfite sequencing results by chop PCR. (C) Overlap of differentially methylated loci between mbd7-1 and ros1-4 and idm1-1. Boxplots represent methylation levels of each class of differentially methylated loci. Same as the results in idm1-1 and ros1-4 mutants, the hypermethylated loci in mbd7-1 mutants spread evenly across the five chromosomes and there was no enrichment at the chromocenters (S5D Fig), although MBD7 are mainly localized at highly methylated chromocenters in nuclei [18]. To determine whether the MBD7 mutation affects DNA demethylation in gene body regions or in TEs, DNA regions were classified into gene body regions, intergenic regions, TEs out of gene regions, and TEs overlapping with gene regions. The ratios of DMRs in each class were calculated. Interestingly, we found the ratios of hypermethylated loci in each class of mbd7-1 were similar to those of ros1-4 (S5E Fig). Both of these two mutants have a small number of hyper DMRs distributed in gene body regions. We also examined the methylation levels of ros1-4 and idm1-1 in those regions that are hypermethylated in mbd7-1 in CG, CHG, and CHH contexts and found most of them are hypermethylated to different degrees (S5F Fig). Approximately 18% and 48% of the 748 hyper-DMRs in mbd7-1 were also hypermethylated in the idm1-1 and ros1-4 mutants, respectively (Fig 2C). We found the hyper-DMRs identified from ros1-4 or idm1-1 that overlap with mbd7-1 mutant showed higher density of CG methylation compared with the non-overlapped loci (S5G Fig). This result suggests MBD7 dysfunction preferentially affects the genomic regions with high CG methylation density. The overlapped loci between mbd7-1 and ros1-4 were more frequently located in TE regions while the overlapped loci between mbd7-1 and idm1-1 were preferentially located in gene body regions (S5H and S5I Fig). The overlapped loci in these mutants are all hypermethylated in all sequence contexts (Fig 2C). The DNA methylation level at the mbd7-specific loci also appeared to be slightly increased in idm1-1 and ros1-4 (Fig 2C). However, the increases in ros1-4 and idm1-1 were not significant to make the loci counted as hyper-DMRs according to the parameters defined in this study, suggesting possible underestimation of the ratio of overlapping (Fig 2C). Taken together, our whole genome bisulfite sequencing results indicate that MBD7, IDM1 and ROS1 may function in the same pathway in active DNA demethylation at some loci.
MBD7 prevents the silencing of a reporter gene and a subset of endogenous genes
Previous studies indicated that IDM1 and IDM2 were required for preventing certain transgenes and endogenous genes from transcriptional silencing [10,11,20,22]. To determine whether MBD7 functions in anti-silencing of transgenes, we introduced the RD29A-LUC and 35S-NPTII into mbd7-1 mutant plants by crossing the mbd7-1 mutant with wild type C24 plants expressing the reporter gene. As shown in Fig 3A, the mbd7-1 mutant exhibited a kanamycin sensitive growth phenotype but normal LUC expression. Real time PCR analysis also showed reduced expression of the NPTII transgene, but the expression level of RD29A-LUC in mbd7-1 was comparable to that of the wild type control (Fig 3B). Thus, the mbd7 mutation preferentially causes the silencing of 35S-NPTII but not RD29A-LUC, consistent with the idm1 or idm2 mutation [20,22].
Fig. 3. MBD7 prevents the silencing of a report gene and some endogenous genes. 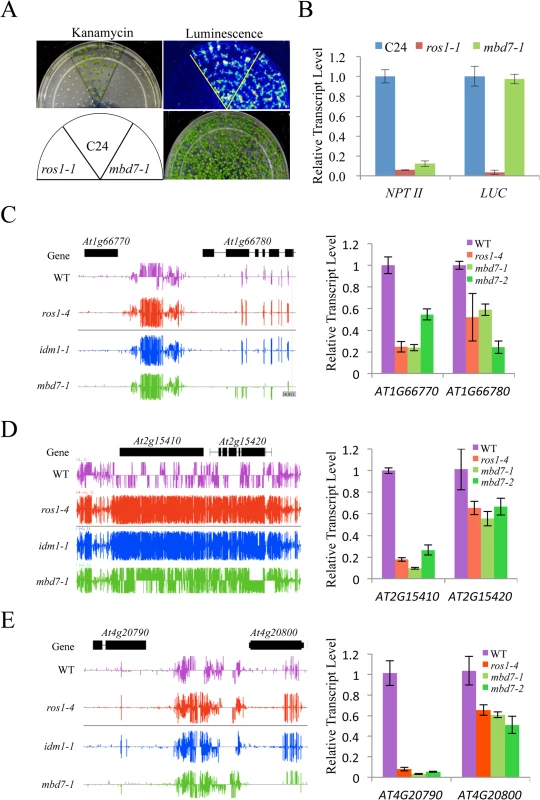
(A) The mbd7-1 mutation causes the silencing of 35S-NPTII but not the RD29A-LUC transgene. Seedlings grown in MS plates were imaged after cold treatment at 4°C for 24 h. For kanamycin resistance test, the seeds were planted on MS medium supplemented with 50 mg/L kanamycin and incubated for 2 weeks before being photographed. The reporter genes were introduced to mbd7-1 mutant plants by crossing. (B) Real-time PCR analysis of the expression level of LUC and NPTII reporter genes in the different genotypes. (C-E) Effect of mbd7 mutation on the expression of endogenous genes. Left column: Snapshot in the IGB showing DNA methylation levels in ros1-4, idm1-1, mbd7-1 mutant plants and wild type control; right column: real-time PCR analysis of the expression level of the hypermethylated genes or nearby genes in different genotypes. TUB8 was used as an internal control. Error bars represent standard error (n = 3). Changes in the DNA methylation level in or near a gene may modulate the expression of that gene. To determine whether DNA hypermethylation affects the expression of adjacent genes in mbd7 mutant plants, we examined the expression levels of 14 genes that have detectable levels of transcript by real time PCR and show increased DNA methylation near or within the gene in mbd7-1, idm1-1 and ros1-4 plants (Fig 3C–3E and S6 Fig). Eleven of the tested genes showed a significant reduction in their transcript levels in these mutants (Fig 3C–3E and S6 Fig). Our results suggest that, like ROS1 and IDM1, MBD7 is critical for preventing the transcriptional silencing of some loci.
MBD7 is important for the in vivo function of IDM1
MBD7 can bind to methylated DNA through the methyl-CpG-binding domain in vitro [17], suggesting that MBD7 may recognize hypermethylated DNA regions and recruit IDM1 complex to the target loci. Previous data showed that IDM1 is critical for H3K18 and H3K23 acetylation in vivo [10]. Chromatin immunoprecipitation (ChIP) assays revealed that the acetylated histone H3K18ac and H3K23ac markers were reduced in idm1-1 and mbd7 mutant plants at hyper-DMRs, although the reduction in mbd7-1 was not as dramatic as in idm1-1 (Fig 4A). ChIP assays also indicated that MBD7 was enriched at all of the hyper-DMRs tested, but not the control regions that contain pretty high CpG DNA methylation in the gene body regions (like At1g01260 and At1g10950) but did not show DNA hypermethylation in mbd7-1 mutant compared with wild type (Fig 4B).
Fig. 4. Histone acetylation marks and MBD7 association with chromatin. 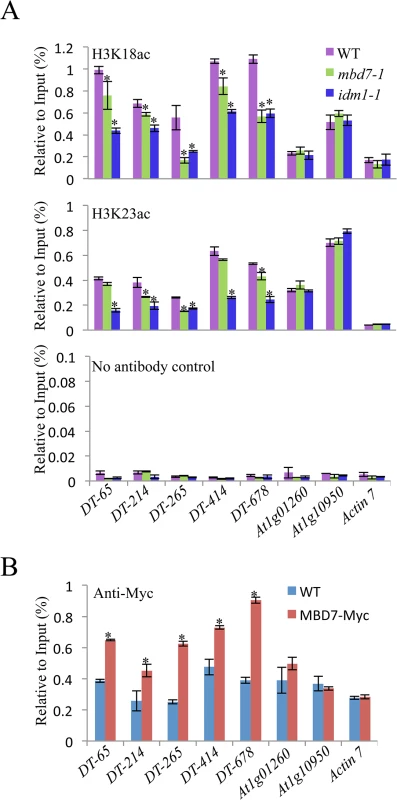
(A) H3K18 acetylation (ac) and H3K23ac levels at the hyper-DMR loci and control regions. ChIP was performed with antibodies against H3K18ac and H3K23ac. The ChIP signal was quantified relative to input DNA. The no-antibody precipitates served as negative control. Two biological replicates were performed, and very similar results were obtained. Standard errors were calculated from three technical repeats, *P < 0.05. (B) Association of MBD7 protein with hyper-DMR loci. ChIP was performed in WT and MBD7-Myc transgenic plants with antibody against Myc. Discussion
ROS1/DME family of 5mC DNA glycosylases initiated DNA demethylation is the major active DNA demethylation pathway in plants [23]. Unlike the well-established DNA methylation pathway, in which enzymes responsible for methylating DNA are guided to specific loci by base-pairing between small RNAs and the scaffold transcripts [1,24], how the DNA demethylation enzymes are recruited to specific genomic loci is poorly understood. Our findings demonstrate that, in addition to IDM1 and IDM2, MBD7 and IDL1 are required for ROS1-mediated active DNA demethylation and suppression of gene silencing at some loci in Arabidopsis.
DNA methylation is a conserved epigenetic marker that is important for TE and gene silencing, whereas DNA demethylation positively regulates gene expression. Some stress responsive genes are silenced by DNA methylation under normal conditions, but are induced by ROS1/DME mediated DNA demethylation under stressed conditions. The dynamic regulation of these genes is important for stress responses in Arabidopsis [25]. For example, the ros1 and rdd (ros1 dml2 dml3) triple mutant shows increased susceptibility to the fungal pathogen Fusarium oxysporum [25]. In light of the important roles of MBD7, IDM1 and IDM2 in DNA demethylation, we speculate that this complex is important for activating stress responsive genes via recruiting ROS1 for active DNA demethylation.
IDM2 encodes an α-crystallin domain (ACD) protein that interact with IDM1 and is required for the full activity of IDM1 in vivo [11]. IDL1 is also an ACD protein and shares the maximal sequence identity with IDM2. Due to the lack of mutants for IDL1 in the public database, we were unable to investigate the effects of IDL1 mutation on DNA methylation level. IDL1 directly interacts with IDM1, IDM2, and MBD7. Our co-IP and MS/MS results suggested that equal amounts of IDM1, IDM2, IDL1, and MBD7 polypeptides are present in the IDM1 complex (Table 1), indicating that IDL1 may be as important as MBD7, IDM1 and IDM2 for active DNA demethylation and gene activation.
Our results also revealed that dysfunction of MBD7 causes DNA hypermethylation at hundreds of genomic loci and silencing of a reporter gene and lots of endogenous genes. Because MBD7 binds to the methylated DNA, the role of MBD7 in DNA demethylation may be facilitating the recruitment of IDM1 to the specific loci. Since the ratio of overlapped hyper-DMRs between mbd7 and idm1 was low, MBD7 may also recruit histone modification enzymes other than IDM1 to create chromatin environment favorable for active DNA demethylation. In addition to MBD7, MBD5 and MBD6 also bind to methylated DNA in vitro [18]. Interestingly, MBD6 was also identified during MS/MS analyses of IDM1 - and IDL1-associated proteins (S7A Fig and S1 Table) and our yeast two-hybrid and split-LUC results confirmed that MBD6 can interact with IDM2 and IDL1 (S7B and S7C Fig). Chop-PCR results shown that the DNA methylation level of two loci were increased in mbd6 and idm1-1 mutants but not in mbd7-1 mutant plants (S7D Fig), suggesting that MBD6 may also involved in recruitment IDM1 to specific DNA regions, promoting active DNA demethylation. This is consistent with the fact only a subset of genomic regions demethylated by ROS1 and other related 5mC DNA glycosylases were influenced in the mbd7 knockout plants.
When this manuscript was under review, Lang et al. and Wang et al. reported that MBD7 and IDM3 were required for active DNA demethylation at some loci in Arabidopsis [26,27]. IDM3 is synonymous with IDL1. The identification of the same two proteins by three independent studies further underlines the importance of MBD7 and IDL1/IDM3 in the active DNA demethylation pathway. More importantly, in our study, we identified MBD7 and IDL1 from affinity purification and mass-spectrometric analysis. We characterized the IDM1-IDM2-IDL1-MBD7 complex and found the ratio of these four proteins should be 1 : 1:1 : 1 in the complex. Moreover, in addition toMBD7, MBD6 was found to be required for the recruitment of IDM1 to some specific loci in our study.
In summary, we propose a model in which MBD7 or MBD6 binds to the DNA hypermethylated region and recruits the histone acetyltransferase complex (IDM1, IDM2 and IDL1) to specific loci. Histone modification (i.e., H3K18 and H3K23 acetylation) creates the favorable chromatin environment for targeting of ROS1 (and other members of the ROS1 subfamily of 5-methycytosine DNA glycosylases) to the loci for active DNA demethylation (Fig 5). In accordance with this model, MBD7 binds the target loci and H3K18ac and H3K23ac markers are reduced in the mbd7 mutant plants, as in idm1 mutants, at the target regions (Fig 4A). Not all the hypermethylated CG sites exhibited MBD7 accumulation (Fig 4B), suggesting that in addition to high CpG DNA methylation, other features such as heterochromatic histone marks may also contribute to MBD7 binding. MBD7 interacting proteins may also be involved in IDM1 targeting. As previous reported, IDM1 also contain a MBD domain at the N-terminal that can bind CG methylated nucleotide in vitro [10]. It seems that single MBD domain in IDM1 is not sufficient to recruit IDM1 to the methylated DNA and three additional MBD domains in MBD7 are required for the recognition of some of the IDM1 target loci. In addition, only some of the ROS1 target loci are affected in idm1, idm2 and mbd7, indicating the existence of IDM1-independent mechanisms for recruiting the ROS1/DME glycosylases.
Fig. 5. Working model for the IDM1-IDM2-IDL1-MBD7 complex functioning in ROS1 mediated active DNA demethylation at some loci in Arabidopsis. 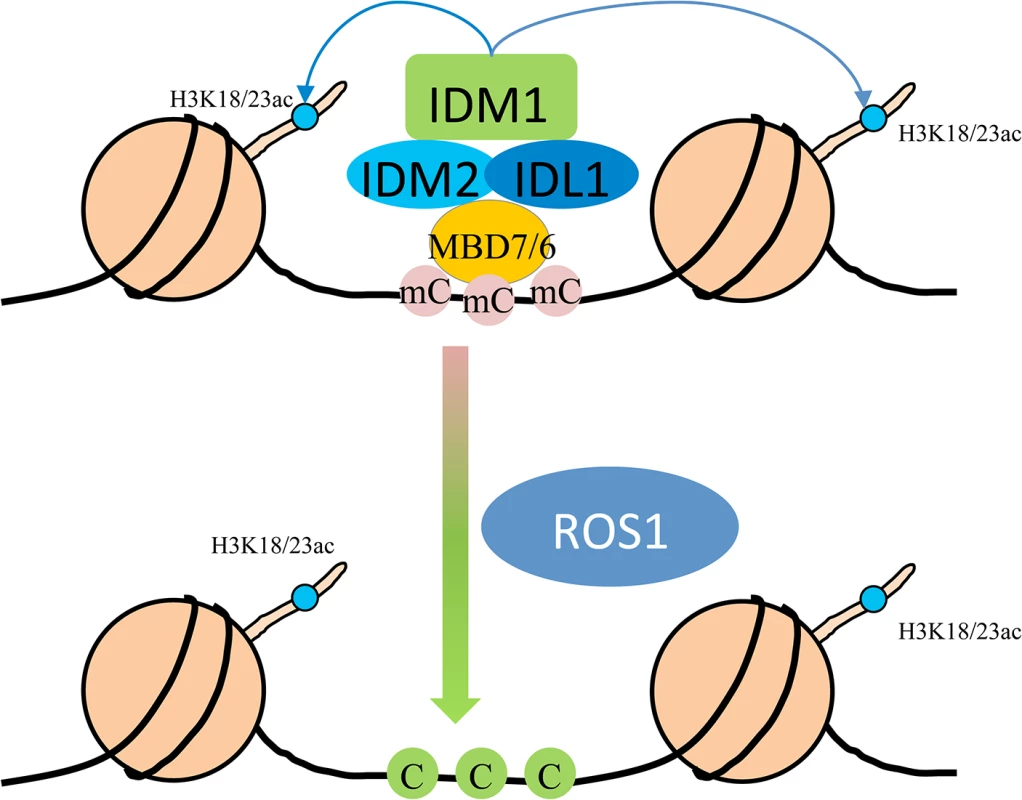
MBD7 forms a complex with IDM1, IDM2 and IDL1, and recognizes methylated DNA through methyl-CpG-binding domains. Then IDM1 is recruited to specific loci and acetylates histone H3 at K18 and K23, facilitating active DNA demethylation by ROS1. Materials and Methods
Plant materials and growth conditions
Two T-DNA insertion mutants of MBD7, mbd7-1 and mbd7-2, were obtained from Arabidopsis Biological Resource Center. The 35S::6Myc-IDM2 transgenic plants were as described [11]. Arabidopsis seedlings were cultivated on Murashige-Skoog (MS) nutrient agar plates at 23°C with 16 h of light and 8 h of darkness for 12 days before DNA or RNA analysis.
Cloning strategy
The IDM1promoter::IDM1-3HA-YFP construct was as described [10]. For the 35S::6Myc-IDL1 construct, full length of IDL1 genomic DNA was amplified from wild type genomic DNA by PCR and sub-cloned into pCAMBIA1307 (6Myc tag). For complementation of mbd7 mutants, a 3.7-kb genomic DNA fragment containing the MBD7 gene was amplified from Col-0 genomic DNA by PCR and cloned into the pCAMBIA1305 vector for plant transformation. Agrobacterium tumefacines strain GV3101 carrying different constructs was used to transform mutant or wild type plants via the standard floral dipping method. Primary transformants were selected on MS plates containing hygromycin.
Affinity purification and mass spectrometry
Approximately 5 g of flower tissue collected from IDM1-HA, IDM2-GFP and 6MYC-IDL1 transgenic plants was ground in liquid N2 with a mortar and pestle. The fine powder was suspended in 25 ml of lysis buffer (50 mM Tris, pH8.0, 230 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, 1 mM PMSF, and 1 protease inhibitor cocktail tablet (Roche, 14696200)) and centrifuged for 15 min at 18,300 g at 4°C. The supernatant was incubated with 50 μl of anti-HA agarose beads (Sigma, A4865), GFP-Trap A beads (ChromoTek), or anti-c-Myc agarose beads (Sigma, A7470) for 5 h at 4°C. The beads were washed twice with 25 ml of lysis buffer and four times with 1 ml of lysis buffer. Proteins bound to HA beads were eluted with HA peptide (Genescript), whereas proteins bound to Myc beads were eluted with 0.1 M ammonium hydroxide (pH 11.5). The GFP-Trap A beads were directly boiled in SDS-sample buffer.
The mass spectrometric identification of purified proteins was done using the method described by Li et al. [28]. Briefly, purified proteins were separated by SDS-PAGE. Then the entire gel lane was cut into 6 equal portions and dehydrated. Proteins were digested in-gel with endoproteinase trypsin (0.5 ng/μL trypsin in 50 mM ammonium bicarbonate, pH 8.5). Extracted peptides were sequenced by LC-MS/MS on the Velos Pro Orbitrap Elite mass spectrometer (Thermo Scientific, USA) equipped with a nano-ESI source. The mass spectrometer was run in data-dependent mode with one MS scan in FT mode at a resolution of 120000. Ten CID (Collision Induced Dissociation) MS/MS scans were applied in the ion trap for each cycle [28]. Mascot server (Matrix Science Ltd., London, UK) and IPI (International Protein Index) Arabidopsis protein database were used as a searching platform.
Yeast two-hybrid assay
The coding sequences of MBD7 and IDL1 were amplified by PCR. After validation of the sequences, the genes were subcloned into pGBK-T7 or pGAD-T7 (Clontech) to generate DNA binding or activation domain fusion constructs, respectively. For protein interaction analysis, two combinatory constructs were transformed simultaneously into the yeast strain AH109 (Clontech) and tested for Leu-, Trp-, and His - auxotrophy according to the manufacturer’s protocols.
Split-LUC assay
The coding sequences of MBD7 and IDL1 were amplified by PCR and subcloned into pCAMBIA1300-NLUC or pCAMBIA1300-CLUC to generate N-terminal or C-terminal luciferase-fusion constructs, respectively. For protein interaction analysis, two combinatory constructs were transformed simultaneously into Nicotiana benthamiana leaves. To prevent the silencing of those genes, a construct encoding virus p19 protein was transformed at the same time [29,30].
Pull down assay
Pull down assays among IDM2, IDL2 and MBD7 were performed as described [31]. Briefly, Maltose binding protein (MBP) either alone or fused to IDM2 (MBP-IDM2) was expressed in E. coli and fixed to an amylose resin. His-tagged IDL1 purified from E.coli (His-IDL1) was incubated with either MBP-IDM2 or MBP bound to the resin. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with antibodies against His6 - tag. To determin the interaction betweem MBD7 and IDM2 or IDL1, MBP alone or MBP-IDM2 or MBP-IDL1 was expressed in E. coli and fixed to an amylose resin. His-tagged MBD7 purified from E.coli (His-MBD7) was incubated with either MBP-IDM2 or MBP-IDL1 or MBP bound to the resin. After washes, the proteins associated to the resin were separated by SDS-PAGE, transferred to a membrane, and immunoblotted with antibodies against His6 - tag.
Agrobacterium tumefaciens-mediated transient co-expression
Over-expression binary constructs were transformed into Agrobaterium tumefaciens GV3101 and cultured overnight. After resuspending in 10 mM MgCl2, 150 μM acetosyringone and 10 mM MES (pH 5.7) at an OD600 of 0.6, and incubating for 3 h at room temperature in dark, the microbodies were syringed into leaves of N. Benthamiana. The infiltrated plants were grown in the greenhouse for 2–3 days.
Co-immunoprecipitation analysis
For co-IP between MBD7 and IDM1, 1 g of flower tissue from MBD7-MYC and IDM1-HA-YFP and MBD7-MYC F1 hybrids were collected and ground in liquid N2. Lysis buffer (4 ml; 50 mM Tris pH8.0, 230 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.2% NP-40, 0.5 mM DTT, 1 mM PMSF, 100 μM MG132 (Gene operation) and 1 protease inhibitor cocktail tablet (Roche, 14696200) was added and mixed carefully. After centrifugation for 15 min at 18,200 g and 4°C, the supernatants were incubated with 8 μL of anti-HA agarose (Sigma, A4865) overnight at 4°C while rotating. After washing the beads 5 times with 1 ml lysis buffer for 5 min each, the beads were resuspended in 60 μL SDS sample buffer and boiled for 5 min. Input and eluted proteins (20 μL and 15 μL, respectively) were resolved by 10% SDS-PAGE and electroblotted onto PVDF membranes for Western blotting. The primary antibodies against HA and MYC were diluted at 1 : 2000 and the second antibody goat anti-mouse IgG (Cwbio) was diluted at 1 : 5000.
For the co-IP between MBD7 and IDM2, MBD7 and IDL1, 0.3 g of co-transformed N. Benthamiana leaves were ground in liquid N2 and incubated in l ml lysis buffer without MG132. After centrifugation, 5 μL GFP-Trap A (ChromoTek) or anti-c-Myc agarose (Sigma, A 7470) were added and incubated for 4 h. The beads were washed and then boiled in 50 μL SDS sample buffer. About 20 μL of input and 10 μl of eluate were analyzed by SDS-PAGE and Western blotting. The primary antibody against GFP (EASYBIO), Flag (Sigma, F1804) and MYC (Cwbio) were diluted at 1 : 2000 and the second antibody goat anti-mouse IgG or goat anti-rabbit IgG (Cwbio) were diluted at 1 : 5000.
Whole-genome bisulfite sequencing and data analysis
DNA was extracted from 2 g of 12-d-old seedlings using the DNeasy Plant Mini Kit (Qiagen) and sent to the Center for High throughput sequencing (Biodynamic Optical Imaging Center, Peking University) for bisulfite treatment, library preparation, and sequencing. For data analysis, adapter, and low-quality sequences (q < 20) were trimmed and clean reads were mapped to Arabidopsis thaliana TAIR 10 (10th release of the Arabidopsis genome sequence from the Arabidopsis Information Resource) genome using Bisulfite Sequence Mapping Program allowing two mismatches. Identification of differentially methylated regions (DMRs) was conducted according to [32]. Heat maps were drawn according to [22].
Chop-PCR
Genomic DNA (100 ng) was cleaved with the methylation-sensitive restriction enzymes HhaI or the methylation-dependent restriction enzyme McrBC, and the products subjected to PCR using the loci-specific primers (S4 Table).
Real-time RT-PCR
Total RNA was extracted from 12-d-old seedlings in plates using the RNeasy plant mini kit (Qiagen), and contaminating DNA removed with RNase-free DNase (Qiagen). About 2 μg mRNA was used for first-strand cDNA synthesis with the Super scriptTM III First-Strand Synthesis System (Invitrogen) for RT-PCR following the manufacturer’s instructions. The cDNA reaction mixture was then diluted five times, and 1 μl used as template in a 20 μl PCR reaction with iQ SYBR Green Supermix (Bio-Rad).
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed according to a published protocol [33]. The following antibodies were used for ChIP assays: anit-H3K18ac (ab1191, Abcam), anti-H3K23ac (07–355, Millipore), and anti-MYC (M4439, Sigma). ChIP products were eluted into 50 μl of TE buffer, and a 2 μl aliquot was used for each qPCR reaction.
Accession numbers
We used whole genome bisulfite sequencing data to analyze the genome-wide methylation status in WT and mbd7-1 mutant plants. The dataset was deposited at NCBI (Col-0:SRX747290, mbd7-1:SRX833671). The ros1-4 and idm1-1 whole genome bisulfite sequencing data were from GEO accession GSE33071 [10].
Supporting Information
Zdroje
1. Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11 : 204–220. doi: 10.1038/nrg2719 20142834
2. Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, et al. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010;6: e1001043. doi: 10.1371/journal.pgen.1001043 20686660
3. Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20 : 244–251. 15145577
4. Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111 : 803–814. 12526807
5. Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124 : 495–506. 16469697
6. Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci U S A. 2006;103 : 11796–11801. 16864782
7. Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol. 2008;67 : 671–681. doi: 10.1007/s11103-008-9346-0 18493721
8. Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci U S A. 2007;104 : 6752–6757. 17409185
9. Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455 : 1259–1262. doi: 10.1038/nature07305 18815596
10. Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336 : 1445–1448. doi: 10.1126/science.1219416 22700931
11. Qian W, Miki D, Lei M, Zhu X, Zhang H, Liu Y, et al. Regulation of active DNA demethylation by an alpha-crystallin domain protein in Arabidopsis. Mol Cell. 2014;55 : 361–371. doi: 10.1016/j.molcel.2014.06.008 25002145
12. Grafi G, Zemach A, Pitto L. Methyl-CpG-binding domain (MBD) proteins in plants. Biochim Biophys Acta. 2007;1769 : 287–294. 17407793
13. Zemach A, Grafi G. Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci. 2007;12 : 80–85. 17208509
14. Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118 : 549–565. doi: 10.1007/s00412-009-0221-9 19506892
15. Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19 : 187–191. 9620779
16. Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135 : 1201–1212. doi: 10.1016/j.cell.2008.11.042 19109892
17. Zemach A, Grafi G. Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 2003;34 : 565–572. 12787239
18. Zemach A, Li Y, Wayburn B, Ben-Meir H, Kiss V, Avivi Y, et al. DDM1 binds Arabidopsis methyl-CpG binding domain proteins and affects their subnuclear localization. Plant cell. 2005;17 : 1549–1558. 15805479
19. Zemach A, Gaspan O, Grafi G. The three methyl-CpG-binding domains of AtMBD7 control its subnuclear localization and mobility. J Biol Chem. 2008;283 : 8406–8411. doi: 10.1074/jbc.M706221200 18211904
20. Li X, Qian W, Zhao Y, Wang C, Shen J, Zhu JK, et al. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109 : 11425–11430. doi: 10.1073/pnas.1208557109 22733760
21. Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones. 2001;6 : 225–237. 11599564
22. Zhao Y, Xie S, Li X, Wang C, Chen Z, Lai J, et al. REPRESSOR OF SILENCING5 Encodes a Member of the Small Heat Shock Protein Family and Is Required for DNA Demethylation in Arabidopsis. Plant cell. 2014;26 : 2660–2675. 24920332
23. Gong Z, Zhu JK. Active DNA demethylation by oxidation and repair. Cell Res. 2011;21 : 1649–51. doi: 10.1038/cr.2011.140 21862972
24. He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21 : 442–465. doi: 10.1038/cr.2011.23 21321601
25. Le TN, Schumann U, Smith NA, Tiwari S, Au P, Zhu QH, et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014;15 : 458. doi: 10.1186/s13059-014-0458-3 25228471
26. Lang Z, Lei M, Wang X, Tang K, Miki D, Zhang H, et al. The Methyl-CpG-Binding Protein MBD7 Facilitates Active DNA Demethylation to Limit DNA Hyper-Methylation and Transcriptional Gene Silencing. Mol Cell. 2015;57 : 971–983. doi: 10.1016/j.molcel.2015.01.009 25684209
27. Wang C, Dong X, Jin D, Zhao Y, Xie S, Li X, et al. Methyl-CpG-Binding Domain Protein MBD7 Is Required for Active DNA Demethylation in Arabidopsis. Plant Physiol. 2015;167 : 905–914. doi: 10.1104/pp.114.252106 25593350
28. Li J, Jia S, Chen PR. Diels-Alder reaction-triggered bioorthogonal protein decaging in living cells. Nat Chem Biol. 2014;10 : 1003–1005. doi: 10.1038/nchembio.1656 25362360
29. Shamloul M, Trusa J, Mett V, Yusibov V. Optimization and utilization of Agrobacterium-mediated transient protein production in Nicotiana. J Vis Exp. 2014 Apr 19;(86). doi: 10.3791/51204
30. Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Näke C, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40 : 428–438. 15469500
31. Martinez-Macias MI, Qian W, Miki D, Pontes O, Liu Y, Tang K, et al. A DNA 3' phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45 : 357–370. doi: 10.1016/j.molcel.2011.11.034 22325353
32. Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci U S A. 2013;110 : 8290–8295. doi: 10.1073/pnas.1300585110 23637343
33. Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008;3 : 1018–1025. doi: 10.1038/nprot.2008.66 18536649
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



