-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
During cell division, chromosomes are divided into two daughter cells by the mitotic spindle, a complex structure made from microtubules. The correct formation of the mitotic spindle is essential, as missegregation of chromosomes can lead to cell death or cancer. Therefore several mechanisms cooperate in nucleating the microtubules needed for the mitotic spindle and focusing them into a bipolar structure. One of these mechanisms, which has only recently been identified, is microtubule nucleation by acentriolar microtubule organizing centers (aMTOCs). These structures have been observed in several cell types, notably also in cancer cells, but is not known how they are formed and which function they might have in mitotic spindle assembly. We identified the pathway of aMTOC formation in Drosophila, which enabled us to perturb their formation in order to study their role during spindle formation. We show that aMTOCs are a source of microtubule nucleation, but their contribution to spindle formation is normally masked by other, more dominant, pathways of microtubule nucleation. Furthermore, we have identified a role for aMTOCs in focusing of mitotic spindle poles by the molecular motor dynein in cells in which centrioles are missing.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005261
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005261Summary
During cell division, chromosomes are divided into two daughter cells by the mitotic spindle, a complex structure made from microtubules. The correct formation of the mitotic spindle is essential, as missegregation of chromosomes can lead to cell death or cancer. Therefore several mechanisms cooperate in nucleating the microtubules needed for the mitotic spindle and focusing them into a bipolar structure. One of these mechanisms, which has only recently been identified, is microtubule nucleation by acentriolar microtubule organizing centers (aMTOCs). These structures have been observed in several cell types, notably also in cancer cells, but is not known how they are formed and which function they might have in mitotic spindle assembly. We identified the pathway of aMTOC formation in Drosophila, which enabled us to perturb their formation in order to study their role during spindle formation. We show that aMTOCs are a source of microtubule nucleation, but their contribution to spindle formation is normally masked by other, more dominant, pathways of microtubule nucleation. Furthermore, we have identified a role for aMTOCs in focusing of mitotic spindle poles by the molecular motor dynein in cells in which centrioles are missing.
Introduction
Centrosomes are the major microtubule organizing centers (MTOCs) in many cells, and they consist of a pair of centrioles surrounded by a cloud of pericentriolar material (PCM), which contains proteins that nucleate and stabilize microtubules (MTs). For many years centrosomes were thought to be the sole drivers of mitotic spindle assembly in animal somatic cells by nucleating MTs, which then randomly search and capture kinetochores in the cytoplasm to form a bipolar spindle [1]. More recently it has become clear that centrosomal MT nucleation cooperates with at least two other pathways of MT nucleation—the chromatin and augmin-mediated pathways—to drive efficient mitotic spindle assembly [2–4]. In current models, MT assembly is induced around the chromatin and the plus ends of these MTs are then captured by the kinetochores and continue to grow from there in a MT bundle [5–8]. This leads to the formation of kinetochore fibers (K fibers) with minus ends that are pushed away from the kinetochores. These K fibers then coalesce into a bipolar spindle together with astral MTs emanating from the centrosomes in a mechanism involving three distinct steps: First, K fibers become crosslinked by the kinesin-14 Ncd/HSET. Second, K fibers are captured by the tips of growing centrosomal MTs, which is also mediated by Ncd/HSET. Third, K fibers are then transported towards centrosomes along astral microtubules by dynein motors, leading to their integration into the bipolar spindle [6,9]. Furthermore, the augmin complex is required for spindle MT amplification by nucleating MTs that branch off the sides of existing MTs, which increases the density of MTs within the mitotic spindle [10–15].
While all three MT nucleation pathways normally make important contributions to spindle assembly, they appear to be partially redundant. Many cell types that usually have centrosomes can assemble bipolar mitotic spindles in their absence, solely relying on chromatin - and augmin-mediated MT nucleation—although spindle assembly without centrosomes takes longer and is usually less accurate [16–20]. Molecular motors are sufficient for focusing MTs into a bipolar spindle without the need for centrosomes to dictate the organization of the two spindle poles (we define spindle poles as the focused collection of MT minus ends of the spindle [21]). In this case the kinesin-14 Ncd/HSET becomes crucial as it crosslinks MTs in a centrosome-independent way; loss of kinesin-14 proteins has been shown to cause severe pole focussing defects in the absence of centrosomes in female meiosis in Drosophila and mouse oocytes, mitosis in Arabidopsis, and in Xenopus egg extracts [22–25]. The dynein complex also plays a crucial role in acentriolar spindle pole focusing in some systems such as acentriolar spindles reconstituted from Xenopus egg extract or from cell free extracts prepared from HeLa cells [25–28]. The exact mechanism by which dynein contributes to acentriolar pole focusing however is unclear, as its normal function in pole focusing relies on the transport of K fibers towards centrosomes [9], which are not present in this case.
While the chromatin-mediated and augmin-dependent MT nucleation pathways are well studied, our knowledge of other acentriolar mechanisms of MT nucleation during mitosis is limited. One such mechanism has been described in centrosome-free mouse oocytes and early mouse embryos where centrosome function is replaced by multiple acentriolar MTOCs (aMTOCs) to which the centrosomal proteins γ-tubulin and Pericentrin localise [29–31]. These aMTOCs form de novo in prophase in the cytoplasm and around the nuclear envelope, and a bipolar spindle is formed in later stages of meiosis through the progressive clustering of multiple aMTOCs into just two poles [30]. In contrast, much less is known about the nature and function of aMTOCs in somatic cells. The presence of γ-tubulin enriched aMTOCs that mediate the de novo formation of MTs has been described in acentriolar Drosophila cultured cells [32,33]. In acentriolar DT40 chicken cells, aMTOCs containing the pericentriolar proteins CDK5RAP2 and γ-tubulin that nucleate MTs have been described [19], while in monkey cells in which the centrosome has been removed by microsurgery aMTOCs containing Pericentrin could be observed integrating into the mitotic spindle [34]. Furthermore, imaging of spindle formation in pig kidney cells showed that even in the presence of centrosomes peripheral, non-centrosomal MT clusters form and are utilized in spindle formation [35]. Interestingly, the ability to form aMTOCs appears to be upregulated in several cancer cell lines that still contain centrosomes; these aMTOCs lead to the formation of multiple spindle poles that need to be clustered into a bipolar spindle [36].
It is unclear, however, how aMTOCs are formed in somatic cells in the absence of centrioles. Moreover, although aMTOCs appear to contribute to spindle assembly in at least some systems [4,35,36] the significance of aMTOC mediated MT generation in spindle formation in somatic cells is still largely uncharacterized. In order to shed light on these open questions, we decided to study aMTOC formation and function in somatic Drosophila cells in vivo. We first set out to elucidate the pathway of aMTOC assembly. We found that aMTOCs consistently form in ~50–60% of mitotic fly somatic brain cells that lack centrioles, and that aMTOC assembly depends on the same proteins that are required to drive mitotic PCM assembly around the centrioles: Asl, Spd-2 and Cnn [37–45]. By identifying the proteins essential for aMTOC formation we then had means to specifically ablate formation of aMTOC formation in the absence of centrioles. Using these tools we performed a detailed genetic analysis to dissect the contribution of aMTOCs to mitotic spindle assembly in somatic Drosophila cells by comparing different acentriolar fly mutants, which either lack centrioles and aMTOCs or lack centrioles but form aMTOCs. Surprisingly, we find that aMTOCs do not detectably contribute to spindle assembly in the absence of centrosomes. In the absence of both centrosomes and MT nucleation from around the chromatin, however, aMTOCs significantly promote the assembly of monopolar spindles. Most importantly, we show for the first time that aMTOCs are essential for dynein-mediated acentriolar spindle pole focusing. On the basis of these observations we propose a revised model for acentriolar spindle organization by the molecular motors dynein and Ncd.
Results
Drosophila cells without centrioles can form aMTOCs in vivo that recruit several PCM components, nucleate microtubules and cluster at mitotic spindle poles
It had previously been shown that γ-tubulin-containing aMTOCs can be found on spindle poles of acentriolar Drosophila cultured cell lines [33] and in Drosophila cultured cells in which centrioles have been depleted by knocking down the levels of the centriole duplication protein Sas-4 by RNAi [32]. In cell lines derived from Sas-4 mutants, however, aMTOCs could not be observed [46]. To test whether PCM proteins were detectable at acentriolar spindle poles in vivo we examined Drosophila Sas-4 mutant larval brain cells. Electron microscopy studies have shown that these mutant cells lack detectable centrioles [16]. Staining for the centriolar protein Ana1 confirmed the lack of centrioles in Sas-4 mutants (Fig 1A). Despite the absence of centrioles, however, we noticed localisation of the PCM protein Cnn at one or both spindle poles in ~56% of Sas-4 mutant brain cells (Fig 1A and 1B). The staining of these Cnn foci was on average ~60% fainter than Cnn staining on centrosomes in WT cells (Fig 1C), and the acentriolar Cnn foci were on average ~60% smaller in diameter than centrosomes (Fig 1D). These observations support the previous conclusion that Sas-4 mutant brain cells lack centrosomes [16], but suggest that about half of these cells have some detectable Cnn at least one spindle pole.
Fig. 1. Drosophila cells without centrioles can form acentriolar microtubule organizing centers (aMTOCs). 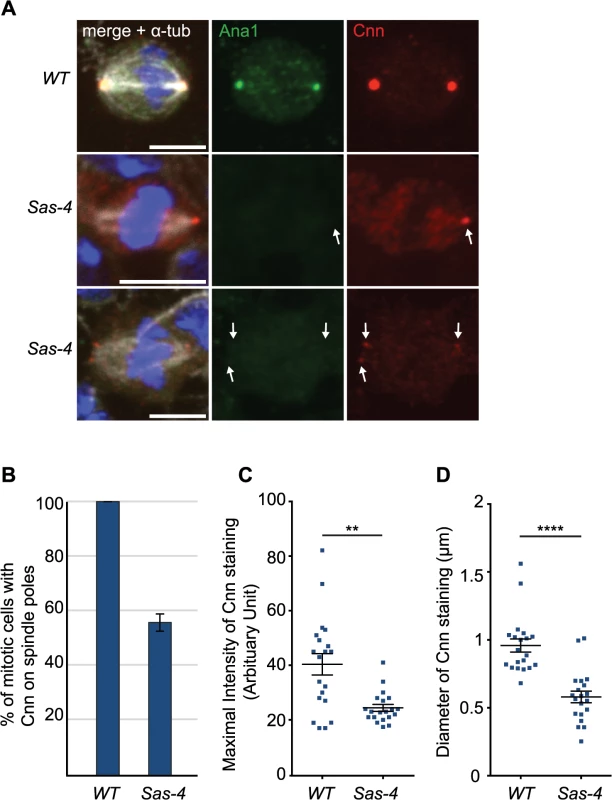
(A) Drosophila larval brain cells were stained with antibodies against Ana1 (green) to mark the centrioles, Cnn (red) to mark the PCM and α-tubulin (white) to mark the spindle. DNA is in blue. Sas-4 mutants do not have centrioles as indicated by the lack of Ana1 on the spindle poles, but often have some Cnn detectable at either one (middle panel) or both (lower panel) of their mitotic spindle poles (arrows). WT and Sas-4 cells were recorded using identical microscope settings, visualising the much fainter staining of Cnn on spindle poles in Sas-4 mutant cells. Scale bars represent 5μm. (B) Quantification of Cnn recruitment to mitotic spindle poles in WT and Sas-4 mutant cells (as judged by Cnn staining on at least one spindle pole) shows that Cnn staining on spindle poles can be detected in 100% ± 0 of WT cells and 55.93% ± 3.16 of Sas-4 cells. A minimum of 120 cells from 4 brains were quantified per genotype. Data are expressed as mean±SEM with average per brain representing one data point. (C, D) Quantification of Cnn intensity (C) and the size of the Cnn foci (D) in WT or Sas-4 mutant Drosophila brain cells stained with anti-Ana1 as centriole marker, anti-α-tubulin to visualise spindles and anti-Cnn. Cnn foci on 20 different spindle poles were analysed for each genotype. Data are expressed as mean±SEM with Cnn staining on each spindle pole representing one data point. P-values were calculated using a Mann-Whitney test. To analyse the mechanism of recruitment of Cnn to acentriolar spindle poles we analysed GFP-Cnn behaviour, together with Jupiter-mCherry as a MT marker, in living Sas-4 mutant cells. As cells prepared to enter mitosis (judged by nuclear envelope breakdown—NEBD), a small number of GFP-Cnn dots began to appear in the cytoplasm of some cells, from which MTs then appeared to emanate (Fig 2A, arrowheads, S1 Movie). As cells entered mitosis, more Cnn dots of variable size and brightness became visible; these often associated with MTs and they tended to become clustered at the spindle poles as mitosis proceeded (Fig 2A, arrows). Importantly, a similar pattern was observed when we followed the behaviour of the centrosomal proteins Spd-2-GFP, γ-tubulin-GFP and Asl-GFP (although the levels of Asl-GFP in these dots was very low and no dots were detectable using anti-Asl antibodies in fixed cells—S1 Fig) (Fig 2B–2D). Based on the existing literature, we hereafter refer to these PCM foci as aMTOCs.
Fig. 2. The formation of aMTOCs in living third instar larval brain cells. 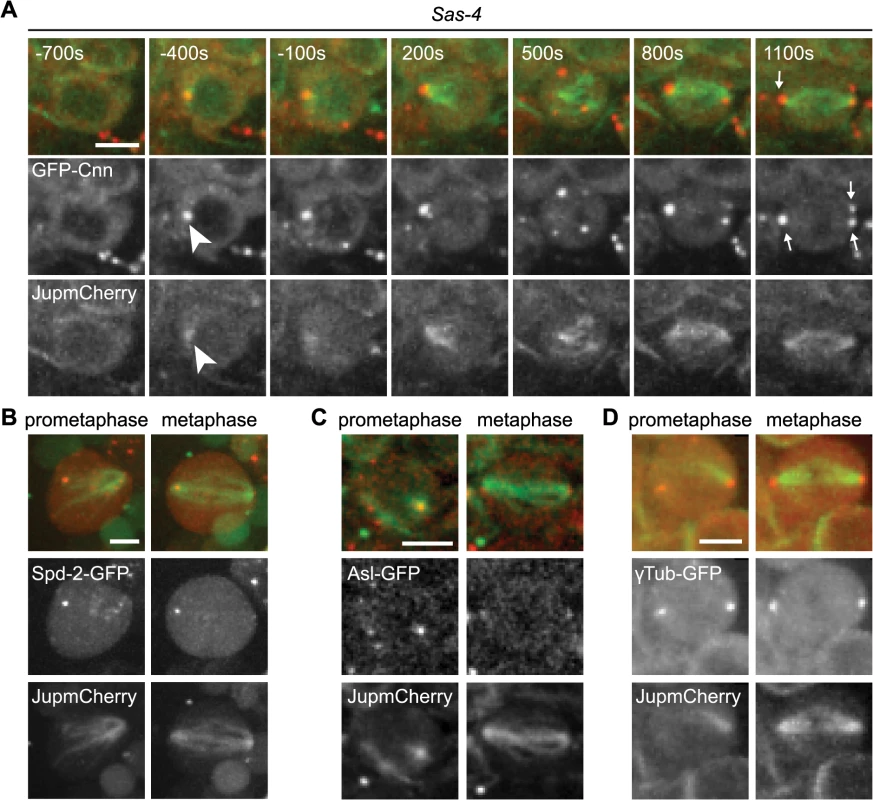
(A) Panels show images from time-lapse movie illustrating spindle formation in Sas-4 mutant larval brain cells expressing GFP-Cnn (red) and Jupiter-mCherry (green). Time (secs) is indicated relative to nuclear envelope breakdown (NEBD). GFP-Cnn foci form in the cytoplasm and nucleate microtubules a few minutes prior to NEBD (arrowheads). These subsequently cluster at the spindle poles by metaphase (arrows). aMTOCs were present at metaphase in 54% of cells filmed (n = 55). aMTOCs are present in prometaphase and metaphase images from Sas-4 mutant brain cells expressing (B) Spd-2-GFP (red) and Jupiter-mCherry (green) (46%, n = 13), (C) Asl-GFP (red) and Jupiter-mCherry (green) (46%, n = 24), (D) γ-Tubulin-GFP (red) and Jupiter-mCherry (green) (54%, n = 59). All scale bars represent 5μm. The PCM proteins PLP, Grip71WD, Msps, Aurora A and TACC all co-localised to some extent with the Cnn-containing aMTOCs in fixed Sas-4 mutant cells, strongly suggesting that these structures contain many of the proteins normally concentrated at centrosomes during mitosis (S1 Fig). We conclude that, as in S2 cells depleted of Sas-4 [32], aMTOCs can form during mitosis in Drosophila cells lacking centrioles in vivo. These aMTOCs appear to contain several known PCM proteins, and are formed prior to mitosis in the cytoplasm where they organize MTs that gradually become localized to the mitotic spindle poles.
aMTOC formation is dependent on the known centrosomal PCM recruitment factors Asl, Spd-2 and Cnn
Next, we wanted to investigate the molecular pathway of aMTOC assembly. It has recently been shown that mitotic PCM recruitment in flies is largely dependent on just three proteins: the centriole duplication protein Asl, which is recruited to the outer region of the mother centriole, and the PCM scaffolding proteins Spd-2 and Cnn [38]. These are recruited to mother centrioles in an Asl-dependent manner, and assemble into a scaffold-structure that spreads outwards from the centrioles and recruits most other mitotic PCM components [37,38,47]. The centriole duplication protein Sas-4 has also been implicated in PCM recruitment [48,49], so we first tested which, if any, of the core centriole duplication proteins Sas-6, Ana2, Sas-4 or Asl are required for aMTOC assembly.
We quantified Cnn staining on mitotic spindle poles in Sas-6, ana2, Sas-4 and asl mutants, which all lack centrioles in third instar larval stages [16,50–52]. Interestingly, robust aMTOCs (identified by Cnn staining on at least one spindle pole) were present in ~50–60% of mitotic cells from Sas-6, ana2 and Sas-4 mutants, but were essentially undetectable in asl mutants (Fig 3A and 3B), and the same was true when we used other PCM proteins as aMTOC markers (S2 Fig). Moreover, Spd-2-GFP, GFP-Cnn and γ-tubulin-GFP did not detectably form aMTOCs in living asl mutant brain cells (Fig 3C). We conclude that Asl is essential for aMTOC formation in these cells.
Fig. 3. The PCM recruitment factor Asl is essential for aMTOC formation. 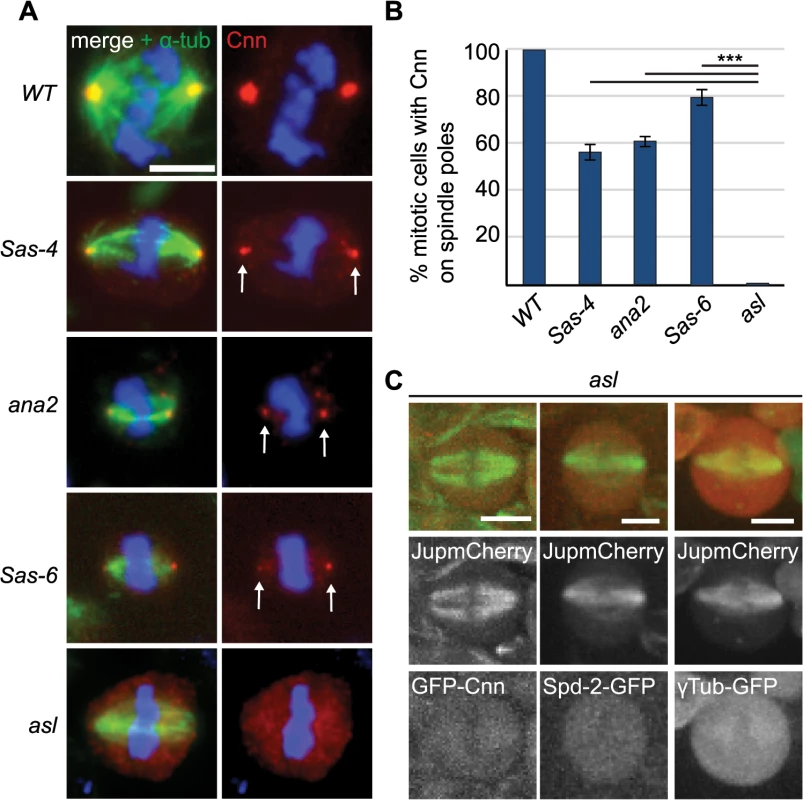
(A) Larval brain cells of acentriolar Drosophila mutants were stained with antibodies against α-tubulin (green) and Cnn (red). DNA is in blue. aMTOCs are indicated by the arrows; note the absence of aMTOCs in asl mutants. (B) Quantification of aMTOC formation in mitotic brain cells (as judged by Cnn staining on at least one spindle pole) in acentriolar mutants shows that Cnn staining on spindle poles can be detected in 100% ± 0 of WT cells, 55.93% ± 3.16 of Sas-4 cells, 60.46% ± 2.42 of ana2 cells, 79.22% ± 3.87 of Sas-6 cells and 0.68% ± 0.44 of asl cells. (A minimum of 120 cells from 4 brains were quantified per genotype). Data are expressed as mean±SEM with average per brain representing one data point. P-values were calculated using a Mann-Whitney test. (C) Spindle formation was followed in live asl mutant brain cells. No aMTOC formation can be observed in asl mutants throughout mitosis in stocks expressing Jupiter-mCherry (green) and either GFP-cnn (red) (n = 21), Spd-2-GFP (red) (n = 12) or γ-Tubulin-GFP (red) (n = 17). All scale bars represent 5μm. We wondered whether Asl, by analogy to the centrosomal mitotic PCM recruitment pathway [38], was able to initiate PCM formation in the cytoplasm even in the absence of centrioles. This might happen by recruitment of Spd-2 and Cnn to cytoplasmic Asl, which could then provide a scaffold for PCM recruitment. We therefore examined PCM localisation on mitotic spindle poles in cnn; Sas-4 and Spd-2 Sas-4 double mutant strains, using the PCM protein γ-tubulin to assess the presence of aMTOCs. While ~51% of mitotic Sas-4 mutant cells had detectable γ-tubulin foci on spindle poles, this was reduced to ~27% in Spd-2 Sas-4 mutant cells and to only ~1% in cnn; Sas-4 mutant cells (Fig 4A and 4B). Thus, Cnn appears to be essential for aMTOC formation, while Spd-2 has an important, but more minor role. Indeed, Spd-2 may function through its effect on Cnn recruitment [38] as Cnn was less efficiently localised at acentriolar spindle poles in the absence of Spd-2 (Fig 4C). Taken together these data suggest that aMTOC formation during mitosis is dependent on the same set of proteins that are essential for mitotic centrosome assembly.
Fig. 4. The PCM scaffolding proteins Spd-2 and Cnn are required for efficient aMTOC formation. 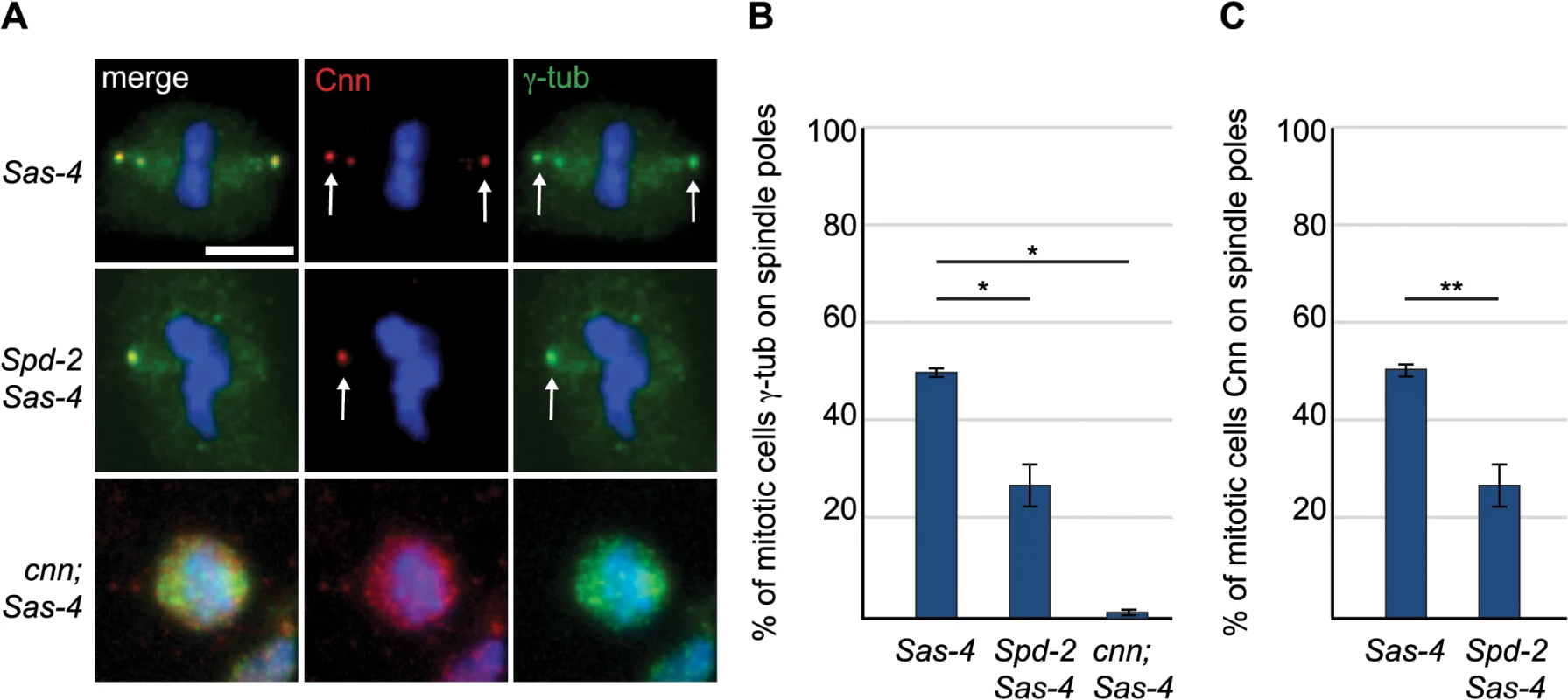
(A) Sas-4 mutant cells and Spd-2 Sas-4 and cnn; Sas-4 double mutant cells were stained with antibodies against Cnn (red) and γ-tubulin (green). DNA is in blue. Scale bar represents 5μm. The aMTOCs present in Sas-4 and Spd-2 Sas-4 mutant cells are indicated by arrows. (B) Quantification of aMTOC formation in mitotic brain cells (as judged by γ-tubulin staining on at least one spindle pole) shows that γ-tubulin staining on spindle poles can be detected in 50.69% ± 0.83 of Sas-4 mutant cells, 27% ± 4.33 of Spd-2 Sas-4 double mutant cells and 1.23% ± 0.67 of cnn; Sas-4 double mutant cells (a minimum of 80 cells from 3 brains were quantified per genotype). (C) Quantification of Cnn recruitment to mitotic spindle poles shows that Cnn staining on spindle poles can be detected in 50.69% ± 0.83 of Sas-4 mutant cells and 27% ± 4.33 of Spd-2 Sas-4 double mutant cells. (A minimum of 137 cells from 4 brains were quantified per genotype). All data are expressed as mean±SEM with average per brain representing one data point. P-values were calculated using a Mann-Whitney test. aMTOCs do not detectably increase the efficiency of spindle assembly in fly larval brain cells that lack centrosomes
These observations provided us with a strategy to genetically investigate the potential function of aMTOCs in acentriolar spindle assembly. Sas-4 mutant cells lack centrioles, but have aMTOCs, while asl mutants lack both structures. Thus, by comparing the behaviour of Sas-4 and asl mutant cells we can infer the contribution of aMTOCs to potentially any acentriolar biological process.
We first assessed the contribution of aMTOCs to acentriolar mitotic spindle assembly using time-lapse microscopy in mutant neuroblasts expressing the MT marker Jupiter-mCherry and the centriole marker GFP-PACT [53]. The latter protein also concentrates in nuclei during interphase, allowing us to precisely determine the time of NEBD [54] (Fig 5A and S2 Movie). As previously observed both Sas-4 and asl mutant cells were able to form acentriolar spindles [16,40]. On average, both asl and Sas-4 mutant neuroblasts were significantly delayed in spindle assembly compared to WT, confirming previous data in Drosophila and DT40 chicken cells lacking centrioles [16,19]. Surprisingly, however, there was no detectable difference in the timing of NEBD to anaphase onset between asl and Sas-4 mutants (Fig 5B), suggesting that aMTOCs do not detectably contribute to the efficiency of spindle assembly in these cells.
Fig. 5. aMTOCs do not detectably contribute to spindle assembly in the absence of centrosomes. 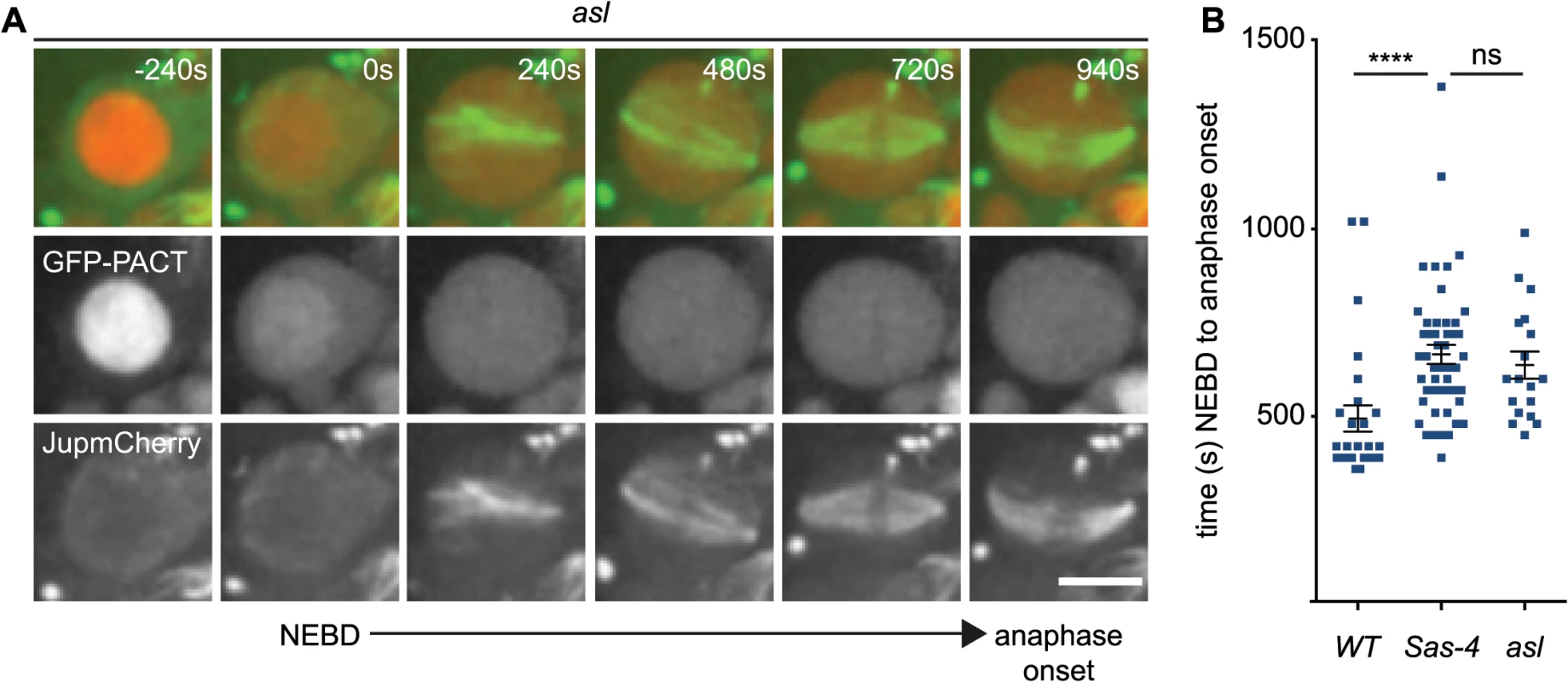
(A) WT, asl and Sas-4 neuroblasts expressing GFP-PACT to mark centrioles and Jupiter-mCherry to visualise spindle formation were filmed to compare spindle assembly dynamics (asl is shown here as an example, time (secs) is relative to NEBD). Note how the GFP-PACT signal present in the nucleus at the first timepoint (-240s) is lost at NEBD. Scale bar represent 5μm. (B) Time between NEBD and anaphase onset was measured for WT (n = 27; 494s ± 35s), Sas-4 (n = 50; 665s ± 25.6s) and asl (n = 18; 637s ± 36s) cells. Spindle assembly takes significantly longer in the mutant cells, but does not vary significantly between Sas-4 and asl mutants indicating that aMTOCs do not increase the efficiency of spindle assembly. Data are expressed as mean±SEM. P-values were calculated using a Mann-Whitney test. aMTOCs increase the efficiency of monopolar spindle formation in acentriolar cells that also lack misato
In the absence of centrosomes, the bulk of spindle MTs are nucleated by the chromatin-mediated pathway [55]. We wondered, therefore, whether the contribution to MT nucleation by aMTOCs could be masked by MT nucleation from around the chromatin. In flies, a mutation in misato (mst) inhibits regrowth of MTs from around the chromatin; misato (mst) mutants are larval lethal, and mutant mitotic cells fail in bipolar spindle assembly and usually only organize monopolar spindles of low MT density [56]. Cells that lack both Mst and centrioles can grow acentriolar MT arrays that appear to be nucleated from cytoplasmic foci, but the nature of these foci is unclear [56].
To test whether aMTOCs are formed in the absence of centrioles and the chromatin-mediated spindle assembly pathway we stained mst; Sas-4 and mst; asl double mutant cells with antibodies against α-tubulin and Cnn. Cnn stained aMTOCs on the poles of monopolar spindles in mst; Sas-4 double mutant cells, but not in mst; asl double mutant cells (Fig 6A). To test whether aMTOCs aid in mitotic array formation in the absence of both centrioles and the chromosome-mediated spindle assembly pathway we quantified mitotic cells with monopolar spindles. We found that approximately 60% of mitotic mst; Sas-4 cells contained monopolar spindles, while only ~25% of mst; asl double mutant cells contained monopolar spindles of MTs (Fig 6B). We conclude that aMTOCs help to establish and/or maintain the monopolar spindles formed in cells lacking both the centrosomal and chromatin pathways of spindle assembly.
Fig. 6. aMTOCs aid formation of monopolar spindles in acentriolar cells that also lack misato. 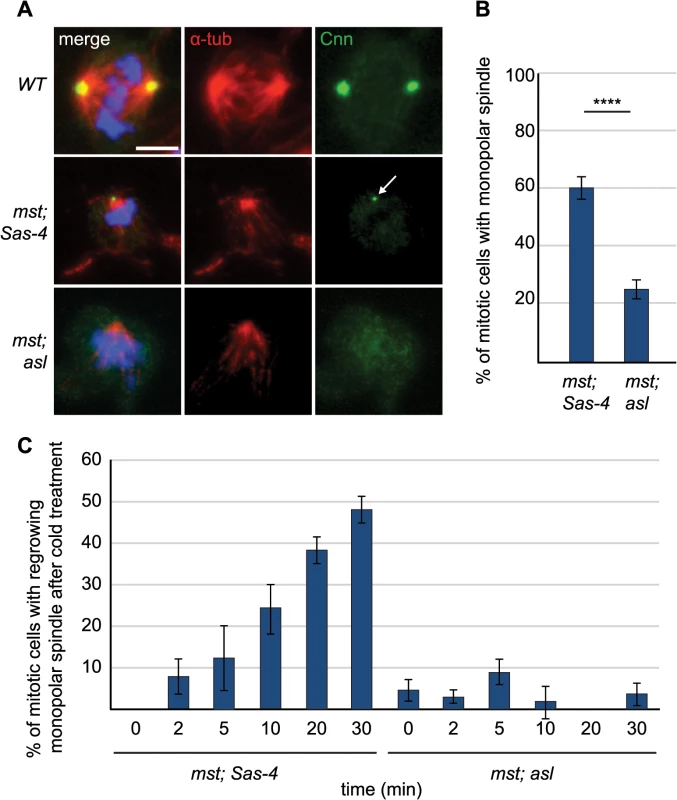
(A) Larval brain cells of WT, mst; Sas-4 and mst; asl double mutants were stained with antibodies against α-tubulin (red) and Cnn (green), DNA is in blue. mst; Sas-4 cells have aMTOCs (arrow) on the poles of monopolar spindles. Scale bar represents 5μm. (B) Quantification of monopolar spindles in mst; Sas-4 and mst; asl cells (a minimum of 263 cells in 8 brains were quantified per genotype). In mst; Sas-4 mutants 59.32% ± 3.81 of cells form monopolar spindles while in mst; asl only 25.15% ± 3.34 of cells form monopolar spindles (the remaining cells do not exhibit any monopolar or bipolar spindles). (C) Quantification of regrowth of monopolar spindles after cold-induced depolymerisation at different time points after release from cold treatment. In mst; Sas-4 brains 0% ± 0 of cells formed monopolar spindles at 0 minutes, 8% ± 4.25 at 2 minutes, 12.5% ± 7.89 at 5 minutes, 24.24% ± 6.1 at 10 minutes, 38.27% ± 3.24 at 20 minutes and 47.96% ± 3.09 at 30 minutes. In mst; asl brains 4.7% ± 2.62 of cells formed monopolar spindles at 0 minutes, 3.03% ± 1.55 at 2 minutes, 8.93% ± 2.9 at 5 minutes, 2% ± 3.93 at 10 minutes, 0% ± 0 at 20 minutes and 3.74% ± 2.74 at 30 minutes (a minimum of 43 cells from 4 brains were quantified for each timepoint). All data are expressed as mean±SEM with average per brain representing one data point. P-values were calculated using a Mann-Whitney test. To test whether aMTOCs nucleate these monopolar MT arrays, or whether they are simply recruited to, and then help to stabilize them, we performed a MT-regrowth assay. In mitotic mst; Sas-4 cells the MTs often grew back from one or several foci in the cytoplasm [56] and we found that these foci were often marked with Cnn (S3A Fig). After ~10 minutes of re-growth, the MTs had all usually coalesced into one acentriolar monopolar array that often had one Cnn-marked aMTOC at the pole (S3A Fig). Quantification of the formation of MT arrays over time showed that in mitotic mst; Sas-4 cells the number of cells with monopolar MT arrays grew steadily over time until, after 30 minutes, ~50% of cells contained monopolar MT arrays again (Fig 6C). Remarkably, in mst; asl cells almost no regrowth of MT arrays was observed, and less than 5% of cells exhibited any MT arrays even after 30 minutes (Figs 6C and S3B). Thus, aMTOCs can nucleate MTs in mitotic cells, and they are an important source of MT generation in mitotic cells lacking centrosomal and chromatin MT nucleation.
aMTOCs and dynein provide a mechanism for acentriolar spindle pole focusing in the absence of Ncd
In addition to providing a site for MT nucleation during mitosis, centrosomes are also involved in spindle pole focusing. In many systems spindle pole focusing in the absence of centrosomes has been shown to rely on the MT crosslinking kinesin Ncd [22–25] and the dynein complex [25–28]. The mechanism by which dynein contributes to acentriolar pole focusing is unclear, as its primary function in pole focusing in unperturbed cells is to transport K fibers towards centrosomes [9].
In Drosophila Ncd is nonessential, but it is crucial for acentriolar meiotic spindle assembly in oocytes [22]. To test if Ncd is essential for acentriolar spindle formation in Drosophila somatic cells we generated Sas-4 ncd double mutants. Surprisingly, these cells were often still capable of forming bipolar mitotic spindles, although many cells exhibited defective spindle phenotypes, ranging from poorly focused and misformed to highly abnormal spindles (Fig 7A, left panels). Quantification of spindle assembly showed that these cells had a reduced ability to form bipolar spindles compared to Sas-4 or ncd mutants alone, but were still able to form a spindle in ~26% of all mitotic cells (Fig 7B). As we often observed aMTOCs at the poles of the Sas-4 ncd cells that formed spindles (Fig 7A, arrow), we wondered whether these structures contributed to spindle formation in the absence of Ncd and centrioles. We therefore generated asl ncd double mutants to assess spindle formation in the absence of Ncd, centrioles and aMTOCs (Fig 7A, right panels). Quantification of the different spindle phenotypes showed that, strikingly, only 4% of mitotic asl ncd cells had formed a bipolar mitotic spindle, suggesting that aMTOCs indeed can ameliorate the severe spindle focusing defects observed in the absence of centrioles and Ncd. To confirm that it was the presence of aMTOCs, and not another potential intrinsic difference between Sas-4 and asl mutants, that enhanced spindle focusing in the absence of Ncd we assessed spindle formation in cnn; Sas-4 ncd triple mutants. In addition to lacking centrioles these cells also cannot form aMTOCs, as Cnn is required for aMTOC formation (Fig 4A and 4B). We could not observe any bipolar mitotic spindles in cnn; Sas-4 ncd triple mutant cells, confirming our hypothesis that aMTOCs likely aid in spindle pole focusing in the absence of centrioles and Ncd (Fig 7B).
Fig. 7. aMTOCs are essential for acentriolar spindle pole focusing by dynein. 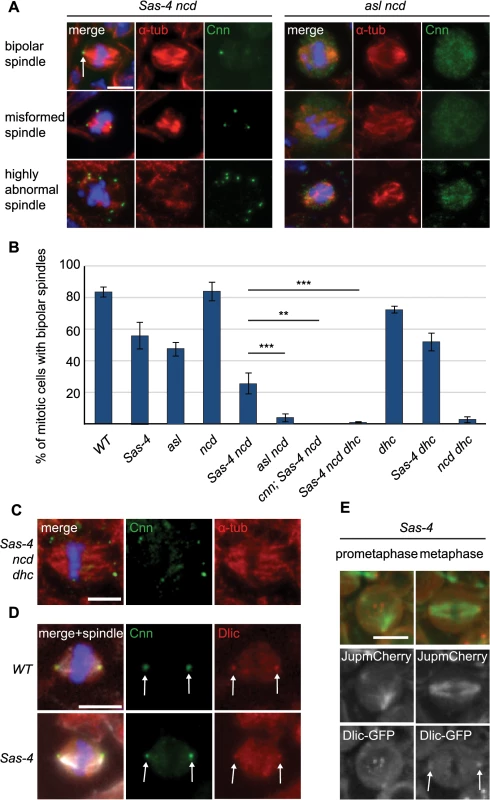
(A) Larval brain cells of Sas-4 ncd and asl ncd were stained with antibodies against Cnn (green) and α-tubulin (red). DNA is in blue. The different spindle phenotypes observed are illustrated; aMTOCs are present in Sas-4 ncd mutant cells (arrow). (B) Quantification of bipolar spindle formation ability in mitotic cells of different mutants (a minimum of 70 cells from 5 brains were quantified per genotype). 82.74% ± 2.94 of WT cells, 55.24% ± 8.11 of Sas-4 cells, 47.08% ± 4.5 of asl cells, 83.65% ± 5.58 of ncd cells, 25.5% ± 6.47 of Sas-4 ncd cells, 4% ± 2.36 of asl ncd cells, 0% ± 0 of cnn; Sas-4 ncd cells, 0.73% ± 0.55 of Sas-4 ncd dhc cells, 71.95% ± 2.39 of dhc cells, 51.71% ± 5.67 of Sas-4 dhc cells and 2.72% ± 1.8 of ncd dhc cells formed bipolar spindles. All data are expressed as mean±SEM with average per brain representing one data point. P-values were calculated using a Mann-Whitney test. aMTOCs cooperating with dynein ameliorate the spindle formation defects observed in the absence of centrioles and Ncd (for a more detailed explanation see main text). Note that Sas-4 and asl have fewer mitotic cells with formed spindles as spindle formation takes longer in the absence of centrioles (Fig 4). (C) Sas-4 ncd dhc triple mutant cells stained with antibodies against Cnn (green) and α-tubulin (red); these cells have a few misformed spindles with splayed spindle poles. (D) WT and Sas-4 mutant cells stained with antibodies against Cnn (green) and Dynein light intermediate chain (red); Dynein localises to centrosomal MTOCs and aMTOCs (arrows). (E) Prometaphase and metaphase images from Sas-4 mutant brain cells expressing Dlic-GFP (red, localising to aMTOCs as marked by the arrows) and Jupiter-mCherry (green) (The punctate signals in the middle region of the cell in prometaphase are likely to be localisation of Dlic-GFP to kinetochores). All scale bars represent 5μm. How do aMTOCs aid spindle formation in the absence of centrioles and Ncd? We hypothesised that dynein might transport K fibers along the MTs nucleated by aMTOCs to help focus spindle poles. If true, then cells which lack centrioles and Ncd but have aMTOCs would not form bipolar spindles if dynein was also lacking. To test this possibility we generated Sas-4 ncd dhc (dynein heavy chain) triple mutants and assessed spindle formation in these cells. Intriguingly, almost none of these cells showed bipolar mitotic spindles (~1%, Fig 7B) and the few spindles we observed invariably exhibited unfocussed poles (Fig 7C). To rule out that this phenotype could originate from another, aMTOC independent, role of dynein we assessed spindle formation in dhc single mutants and Sas-4 dhc double mutants. Both of these had relatively normal spindle morphology, suggesting that dynein in conjunction with aMTOCs is required to ameliorate spindle formation in the absence of Ncd (Fig 7B).
These data suggest that dynein function in spindle focusing might be to transport kinetochore MTs towards MTOCs—either centriolar or acentriolar. If so, cells which have aMTOCs but lack Ncd and dynein should have a very similar phenotype to cells which have centrosomes but lack Ncd and dynein: in both cases K fibers could not be transported towards (either acentriolar or centriolar) MTOCs on the spindle poles. To test this hypothesis, we generated ncd dhc double mutants (which have centrosomes but lack Ncd and dynein). Only ~3% of these mitotic cells formed a bipolar spindle, which was very similar to the situation in Sas-4 ncd dhc triple mutants (which have aMTOCs but lack Ncd and dynein) (Fig 7B). We conclude that dynein is able to focus acentriolar mitotic spindles in an analogous fashion to spindle focusing in centrosomal cells, namely by transporting K fibers towards aMTOCs. In support of this hypothesis, dynein clearly localised to centrosomes (as previously observed [57]) and also aMTOCs on mitotic spindle poles in fixed (Fig 7D) and live larval brain cells (Fig 7E and S3 Movie), but failed to localise in asl mutants (S4 Fig).
Acentriolar spindle pole focusing by dynein and aMTOCs can ameliorate proliferation defects observed in the absence of centrioles and Ncd
Finally, we wanted to assess whether the ability of dynein and aMTOCs to promote spindle formation in acentriolar cells might be sufficiently robust to allow these acentriolar cells to proliferate even in the absence of Ncd. We compared acentriolar Sas-4 ncd tissue (which lacks Ncd, but has aMTOCs) with asl ncd tissue (which lacks both Ncd and aMTOCs). We noticed that while Sas-4 ncd brains were slightly smaller than WT brains they still looked relatively normal and had imaginal discs attached to them (Fig 8A and 8B). In comparison asl ncd brains and imaginal discs were much smaller than normal (Fig 8A and 8B), indicating a much stronger defect in cell proliferation [58]. This suggested that the severe proliferation defect in asl ncd larvae was rescued by aMTOC and dynein-mediated spindle focusing in Sas-4 ncd larvae. In support of this interpretation, Sas-4 ncd dhc larvae (which have aMTOCs but lack dynein) also had very small brains and discs (Fig 8A and 8B). We conclude that aMTOCs and dynein can cooperate to focus acentriolar spindle poles in cells lacking Ncd, and that this pathway substantially increases the efficiency of spindle assembly and thereby the ability of these cells to proliferate.
Fig. 8. Acentriolar spindle pole focusing by dynein can ameliorate proliferation defects observed in the absence of centrioles and Ncd. 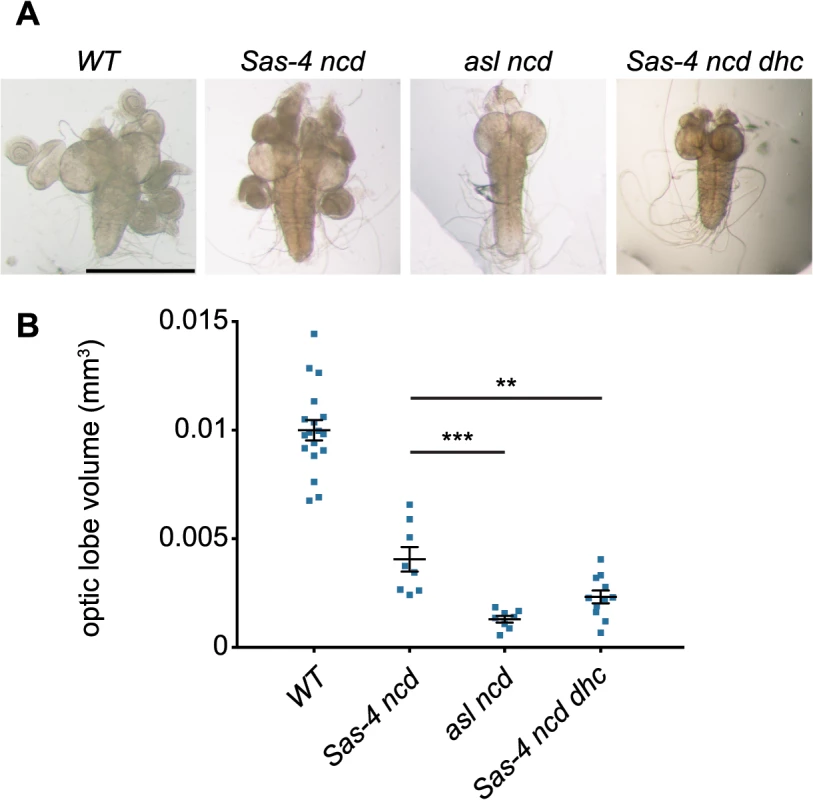
(A) Panels show third instar larval brains from WT and different mutant strains. Note how asl ncd and Sas-4 ncd dhc brain lobes are much smaller than WT or Sas-4 ncd brains and lack fully developed imaginal discs. (B) Quantification of third instar larval brain size in WT, Sas-4 ncd, asl ncd and Sas-4 ncd dhc 3rd instar wandering larvae. Brain size was assessed by measuring brain lobe circumference and calculating brain lobe volume under the assumption that the lobes were spherical. Each data point represents the averaged brain lobe volume of each two lobes of a brain (a minimum of 8 brains were analysed per genotype). A Mann-Whitney test shows that brains that lack aMTOCs and Ncd (asl ncd) or have aMTOCs but lack dynein and Ncd (Sas-4 ncd dhc) brains are both significantly smaller than Sas-4 ncd brains (that lack Ncd but have aMTOCs and dynein) suggesting that acentriolar spindle focusing by aMTOCs and dynein can ameliorate the proliferation defects observed in the absence of Ncd and centrioles. Discussion
Here we confirm the presence of aMTOCs in acentriolar somatic cells and show that the pathway of aMTOC assembly in fly somatic brain cells is genetically very similar to the pathway of mitotic centrosome assembly. At centrosomes, Asl is present exclusively around the mother centriole [59–61], where it helps recruit Spd-2 and Cnn, which then assemble into a scaffold that spreads outwards around the mother centriole and which ultimately recruits the other mitotic PCM components [37–45]. Our data suggest that in ~50–60% of mitotic brain cells that lack centrioles, Asl can still recruit some Spd-2 and Cnn to form cytoplasmic scaffold structures that can then recruit other PCM components to form aMTOCs. Thus, while mother centrioles greatly increase the efficiency of mitotic PCM assembly, these three proteins can self-assemble into a mitotic scaffold structure even in their absence. The number and size of the aMTOCs formed, however, was very variable, suggesting that centrioles not only make mitotic PCM assembly more efficient, but they also serve to regulate the amount of PCM assembled and to ensure that only two MTOCs are normally formed during mitosis.
Structures similar to the aMTOCs described here have also been observed in other Drosophila tissues. In oocytes bundles of MTs were observed in the cytoplasm during acentriolar meiotic spindle formation [22]. No centrosomal markers such as γ-tubulin and CP60 have been observed clustering on meiotic spindle poles [22,62,63] and therefore it is unlikely that these MT bundles represent aMTOCs. In Drosophila embryos, injection of an antibody raised against Spd-2 lead to displacement of PCM from the centrosomes and thereby removed centrosomally nucleated MTs. In these embryos MT asters could be observed to form in the cytoplasm, which were then organised into mitotic spindles [4]. As centrioles are still present in these embryos it is unclear, whether these asters were nucleated from self-assembled aMTOCs as described here, or whether they are fragments of PCM originally nucleated around the mother centriole and then displaced from the centrosomes either through a lack of Cnn or by Spd-2 antibody injection [4]. Finally, a study has described the biochemical purification of cytoplasmic complexes containing Sas-4, Cnn, Asl and Plp (S-CAP complexes; [64]) from Drosophila embryonic extracts. Although S-CAP complexes have a composition similar to the aMTOCs described here, they require Sas-4 for their assembly [64]. The aMTOCs we describe here form in the absence of Sas-4, suggesting that they are unrelated.
Our identification of the factors required for aMTOC assembly in Drosophila allowed us to perform a detailed genetic analysis of the contribution of aMTOCs to different aspects of mitotic spindle assembly in acentriolar fly cells. Surprisingly, we find that aMTOCs do not detectably increase the rate of spindle assembly in somatic brain cells that lack centrioles. This suggests that, in contrast to meiotic spindle assembly in mouse oocytes [30], aMTOCs do not play a major role in generating spindle MTs in the absence of centrosomes, at least in this cell type.
Our data show, however, that aMTOCs can function as a major source of MT regrowth in cells that lack both centrosomal - and chromatin-mediated MT nucleation after the MT cytoskeleton has been depolymerised by cold treatment. Before cold treatment mst; asl mitotic cells (that lack the chromatin-, centrosome - and aMTOC-pathway of MT assembly) still had mitotic MT arrays, but these were significantly fewer than mst; Sas-4 cells (that lack the chromatin - and centrosome-pathways, but still have aMTOCs). We speculate that the MT arrays in mst; asl mitotic cells arise from augmin-mediated MT nucleation from pre-existing MTs, which can no longer happen once all MTs have been removed by cold-treatment.
Interestingly, we find that aMTOCs do play an important part in the mechanism of acentriolar spindle focusing by dynein. Dynein usually transports MTs emanating from kinetochores along astral MTs to the centrosome [6,9]. Our observations suggest that dynein can also transport MTs towards aMTOCs. Based on our results we propose an updated model for acentriolar spindle pole focusing by the minus end directed motors Ncd and dynein (Fig 9). In Drosophila cells with centrosomes, K fibers become crosslinked by Ncd, while dynein transports K fibers along centrosomal MTs towards the centrosome [9] (Fig 9A). When centrosomes and aMTOCs are lost, Ncd becomes essential for focusing acentriolar spindle poles as it can crosslink K fibers independently of centrosomes (Fig 9B). Therefore loss of Ncd leads to severely unfocused poles in acentriolar cells, as dynein, in contrast to Ncd, does not have the ability to statically crosslink MT minus ends (Fig 9C). If aMTOCs are present, however, then dynein can partially compensate for the loss of Ncd by focusing spindle poles through the transport of K fibers towards aMTOCs (Fig 9D).
Fig. 9. A model for acentriolar spindle pole focusing by the molecular motors dynein and Ncd. 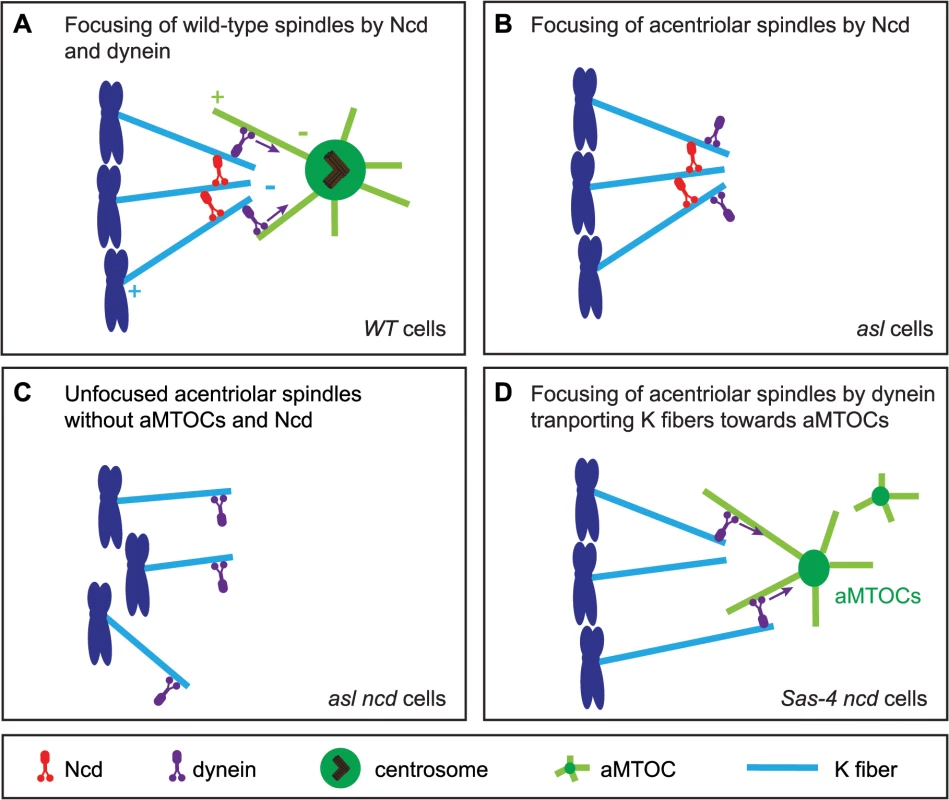
(A) In Drosophila cells with centrosomes K fibers (light blue) are thought to be predominantly crosslinked by the minus end-directed motor Ncd (red), while the minus-end directed motors dynein (purple) transports the K fibers along centrosomal MTs (light green) to the centrosome [9]. (B) In cells lacking centrosomes, Ncd can still focus acentriolar spindle poles by crosslinking the K fibers (as, for example, in an asl mutant cell). (C) Loss of Ncd in acentriolar cells which also lack aMTOCs leads to severe impairment of spindle focusing as dynein cannot crosslink MT minus ends (as, for example, in asl ncd mutant cells). (D) In acentriolar cells with aMTOCs (as, for example, in Sas-4 ncd mutant cells), dynein can transport K fibers towards aMTOCs and so allow some spindle pole focusing even in the absence of Ncd. This last point is important, as previous studies that reported a role for dynein in acentriolar pole focusing did not describe the molecular mechanism by which dynein is able to do this [25–28]. We suspect that structures similar to aMTOCs must have been present in these earlier studies for dynein to fulfil its role. These studies used Xenopus egg or cell extracts for their analyses. Interestingly in Verde et al. 1991 [28] the observation of a MT crosslinking material that accumulated at MT minus ends is described, and electron microscopy showed that this material had electron-dense properties similar to PCM [28]. Our finding that dynein needs to cooperate with aMTOCs to focus acentriolar spindles is an important addition to our understanding of the mechanism of spindle pole coalescence by molecular motors.
Material and Methods
Fly strains
The following mutant alleles were used in this study: Sas-4s2214 [16], Sas-6c02901 [50,51], aslB46 (this study), Spd-2G20143 [43], cnnf04547 [45], cnnHK21 [44,65], ana2169 [66], mstLB20 [56], dhc6-10 [67] and ncd1 [68]. The following transgenic insertion lines were used: Asl::Asl-GFP [69], Ubq-GFP-Cnn [37], Ubq-Spd-2-GFP [43], ncd:: γ-tubulin37C-GFP [70], Ubq-Dlic-GFP (this study—a full length Dlic [dynein light intermediate chain—CG1938] cDNA was cloned into the Ubq-GFPNT Gateway vector [71]; Nina Peel, personal communication) and Jupiter-mCherry [72]. w67 was used as wild type control. An asl null allele was generated by inducing imprecise excision of the P-Element P{EPg}HP37249 located near the 5’UTR about 470bp upstream of the asl start codon. A 2.1kb deletion removing nearly 500bp upstream of the start codon and over half of the coding region was recovered (S4 Fig); the allele was named aslB46. In contrast to previously published alleles of asl [69] this new allele aslB46 produces no detectable N - or C-terminal Asl protein in western blotting or immunofluorescence experiments (S4 Fig). We used this new aslB46 mutant as it represents a true null mutation with no residual part of Asl being expressed, however we observed that aMTOC formation is also ablated in previously published aslmecD allele [69] (Cnn staining on spindle poles can be detected in 0.55% ± 0.55 of aslmecD cells).
Immunofluorescent staining and antibodies and microscopy
Fixation and stainings of third instar larval brains was performed as previously described [53]. The MT regrowth assay in larval brains was performed as described in [56]. The following primary antibodies were used at a 1 : 500 dilution: Rabbit anti-Dlic (this study—raised against amino acids 1–293 of the Drosophila Dlic (CG1938) coding sequence; Nina Peel, personal communication), rat anti-Asl [73], rabbit anti-Asl (N-terminal and C-terminal) [37], guinea pig anti-Cnn [45], rabbit anti-Spd-2 [43], mouse anti - γ-tubulin (GTU88, Sigma), mouse anti-actin (Sigma-Aldrich), rabbit anti-PLP [53], rabbit anti-TACC [74], rabbit anti-Grip71WD [75], rabbit anti-Msps [76], rabbit anti-Aurora A [77], mouse monoclonal anti-α-tubulin (DM1α, Sigma-Aldrich) and rabbit anti-Histone H3 Phospho S10 (Upstate Biotechnology). Alexa488 anti-guinea pig and anti-mouse and Alexa568 anti-rabbit as secondary antibodies were used at a 1 : 1000 dilution (Molecular Probes, Life Technologies). Fixed preparations were examined using either a Zeiss LSM780 confocal microscope using a 63x/1.40 NA objective, or on a Zeiss Axioskop 2 microscope (Carl Zeiss, Ltd) with a CoolSNAP HQ camera (Photometrics), using a 63x/1.25 NA objective (Carl Zeiss, Ltd). Images were processed with Fiji [78] and adjusted to use the full range of pixel intensities.
Quantification of aMTOCs
Larval brains were stained with antibodies against α-tubulin to visualise spindles, Cnn to visualise aMTOCs and anti-Phospho-Histone H3 and were imaged on a Zeiss Axioskop 2 microscope. Only mitotic cells identified by Phospho-Histone H3 staining were scored, and all phenotypes were always quantified blindly, without knowing which genotype was being counted.
Live imaging
Brains were dissected from third instar larvae in PBS and the attached imaginal discs were removed. The brain was transferred to a drop of PBS on a clean coverslip and covered with a slide, which semi-squashed the brain. The coverslip was sealed with a drop of Voltalef 10S oil (VWR) to stop evaporation of the PBS and brains were analysed using a 63x/1.4 NA lens on a Perkin Elmer ERS Spinning Disk confocal system mounted in an inverted microscope (Axiovert 200M; Carl Zeiss Ltd) with a charge-coupled device camera (Orca ER; Hamamatsu) with Ultraview ERS software (Perkin Elmer), or using a 60x/1.4 NA lens on an Andor Revolution XD Spinning Disk confocal system mounted on an inverted Nikon (TE-2000E) with an Electron Multiplying charge-coupled device camera (iXon, Andor) and IQ2 software (Andor).
Analysis of larval brain size
Third instar larval brains were dissected and the brain lobe volume was measured as previously described [79].
Supporting Information
Zdroje
1. Kirschner M, Mitchison TJ (1986) Beyond self-assembly: from microtubules to morphogenesis. Cell: 329–342. 3516413
2. Meunier S, Vernos I (2012) Microtubule assembly during mitosis—from distinct origins to distinct functions? J Cell Sci 125 : 2805–2814. doi: 10.1242/jcs.092429 22736044
3. O'Connell CB, Khodjakov AL (2007) Cooperative mechanisms of mitotic spindle formation. J Cell Sci 120 : 1717–1722. 17502482
4. Hayward D, Metz J, Pellacani C, Wakefield JG (2013) Synergy between Multiple Microtubule-Generating Pathways Confers Robustness to Centrosome-Driven Mitotic Spindle Formation. Dev Cell: 1–13.
5. Khodjakov A (2003) Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol 160 : 671–683. 12604591
6. Maiato H, Rieder CL, Khodjakov A (2004) Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167 : 831–840. 15569709
7. Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F (2008) Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell 19 : 1873–1882. doi: 10.1091/mbc.E07-10-1050 18287525
8. Meunier S, Vernos I (2011) K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol 13 : 1–11. doi: 10.1038/ncb0111-1 21173798
9. Goshima G (2005) Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol 171 : 229–240. 16247025
10. Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD (2008) Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181 : 421–429. doi: 10.1083/jcb.200711053 18443220
11. Kamasaki T, O'Toole E, Kita S, Osumi M, Usukura J, et al. (2013) Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J Cell Biol 202 : 25–33. doi: 10.1083/jcb.201304031 23816620
12. Meireles AM, Fisher KH, Colombie N, Wakefield JG, Ohkura H (2009) Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184 : 777–784. doi: 10.1083/jcb.200811102 19289792
13. Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD (2013) Branching Microtubule Nucleation in Xenopus Egg Extracts Mediated by Augmin and TPX2. Cell 152 : 768–777. doi: 10.1016/j.cell.2012.12.044 23415226
14. Ueharaa R, Nozawab R-S, Tomiokaa A, Petryc S, Ronald D Valec, et al. (2009) The augmin complex plays a critical role in spindlemicrotubule generation for mitotic progressionand cytokinesis in human cells: 1–6.
15. Wainman A, Buster DW, Duncan T, Metz J, Ma A, et al. (2009) A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev 23 : 1876–1881. doi: 10.1101/gad.532209 19684111
16. Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, et al. (2006) Flies without centrioles. Cell 125 : 1375–1386. 16814722
17. Khodjakov A, Cole RW, Oakley BR, Rieder CL (2000) Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 10 : 59–67. 10662665
18. Mahoney NM, Goshima G, Douglass AD, Vale RD (2006) Making microtubules and mitotic spindles in cells without functional centrosomes. Curr Biol 16 : 564–569. 16546079
19. Sir JH, Putz M, Daly O, Morrison CG, Dunning M, et al. (2013) Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol 1 : 7.
20. Bazzi H, Anderson KV (2014) Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci USA 111: E1491–E1500. doi: 10.1073/pnas.1400568111 24706806
21. Compton DA (1998) Focusing on spindle poles. J Cell Sci 111 (Pt 11): 1477–1481.
22. Matthies HJ, McDonald HB, Goldstein LS, Theurkauf WE (1996) Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol 134 : 455–464. 8707829
23. Mountain V, Simerly C, Howard L, Ando A, Schatten G, et al. (1999) The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol 147 : 351–366. 10525540
24. Ambrose JC, Li W, Marcus A, Ma H, Cyr R (2005) A minus-end-directed kinesin with plus-end tracking protein activity is involved in spindle morphogenesis. Mol Biol Cell 16 : 1584–1592. 15659646
25. Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R (1998) A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol 8 : 903–913. 9707401
26. Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, et al. (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382 : 420–425. 8684481
27. Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, et al. (1996) Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol 135 : 399–414. 8896597
28. Verde F, Berrez J-M, Antony C, Karsenti E (1991) Taxol-induced Microtubule Asters in Mitotic Extracts of XenopusEggs: Requirement for Phosphorylated Factors and Cytoplasmic Dynein. J Cell Biol 112 : 1177–1187. 1671864
29. Łuksza M, Queguigner I, Verlhac M-H, Brunet S (2013) Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev Biol 382 : 48–56. doi: 10.1016/j.ydbio.2013.07.029 23954884
30. Schuh M, Ellenberg J (2007) Self-Organization of MTOCs Replaces Centrosome Function during Acentrosomal Spindle Assembly in Live Mouse Oocytes. Cell 130 : 484–498. 17693257
31. Courtois A, Schuh M, Ellenberg J, Hiiragi T (2012) The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol 198 : 357–370. doi: 10.1083/jcb.201202135 22851319
32. Moutinho-Pereira S, Debec A, Maiato H (2009) Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell 20 : 2796–2808. doi: 10.1091/mbc.E09-01-0011 19369414
33. Debec A, Détraves C, Montmory C, Géraud G, Wright M (1995) Polar organization of gamma-tubulin in acentriolar mitotic spindles of Drosophila melanogaster cells. J Cell Sci 108 (Pt 7): 2645–2653. 7593305
34. Hornick JE, Mader CC, Tribble EK, Bagne CC, Vaughan KT, et al. (2011) Amphiastral Mitotic Spindle Assembly in Vertebrate Cells Lacking Centrosomes. Curr Biol 21 : 598–605. doi: 10.1016/j.cub.2011.02.049 21439826
35. Tulu US, Rusan NM, Wadsworth P (2003) Peripheral, Non-Centrosome-Associated Microtubules Contribute to Spindle Formation in Centrosome-Containing Cells. Current Biology 13 : 1894–1899. 14588246
36. Kleylein-Sohn J, Pollinger B, Ohmer M, Hofmann F, Nigg EA, et al. (2012) Acentrosomal spindle organization renders cancer cells dependent on the kinesin HSET. J Cell Sci 125 : 5391–5402. doi: 10.1242/jcs.107474 22946058
37. Conduit PT, Brunk K, Dobbelaere J, Dix CI, Lucas EP, et al. (2010) Centrioles Regulate Centrosome Size by Controlling the Rate of Cnn Incorporation into the PCM. Curr Biol: 1–9.
38. Conduit PT, Richens JH, Wainman A, Holder J, Vicente CC, et al. (2014) A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife 3.
39. Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M (2008) Drosophila SPD-2 Is an Essential Centriole Component Required for PCM Recruitment and Astral-Microtubule Nucleation. Current Biology 18 : 303–309. doi: 10.1016/j.cub.2008.01.058 18291647
40. Bonaccorsi S, Giansanti MG, Gatti M (1998) Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol 142 : 751–761. 9700163
41. Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, et al. (2007) Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol 17 : 1735–1745. 17935995
42. Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, et al. (2010) Asterless is a scaffold for the onset of centriole assembly. Nature 467 : 714–718. doi: 10.1038/nature09445 20852615
43. Dix CI, Raff JW (2007) Drosophila Spd-2 Recruits PCM to the Sperm Centriole, but Is Dispensable for Centriole Duplication. Current Biology 17 : 1759–1764. 17919907
44. Megraw TL, Li K, Kao LR, Kaufman TC (1999) The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126 : 2829–2839. http://dev.biologists.org/content/126/13/2829.long. 10357928
45. Lucas EP, Raff JW (2007) Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol 178 : 725–732. 17709428
46. Lecland N, Debec A, Delmas A, Moutinho-Pereira S, Malmanche N, et al. (2013) Establishment and mitotic characterization of new Drosophila acentriolar cell lines from DSas-4 mutant. Biology Open 2 : 314–323. doi: 10.1242/bio.20133327 23519377
47. Conduit PT, Feng Z, Richens JH, Baumbach J, Wainman A, et al. (2014) The Centrosome-Specific Phosphorylation of Cnn by Polo/Plk1 Drives Cnn Scaffold Assembly and Centrosome Maturation. Dev Cell 28 : 659–669. doi: 10.1016/j.devcel.2014.02.013 24656740
48. Gopalakrishnan J, Chim Y-CF, Ha A, Basiri ML, Lerit DA, et al. (2012) ncb2527. Nat Cell Biol 14 : 865–873. doi: 10.1038/ncb2527 22729084
49. Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, et al. (2011) Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nature Communications 2 : 359–11. doi: 10.1038/ncomms1367 21694707
50. Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, et al. (2007) DSAS-6 Organizes a Tube-like Centriole Precursor, and Its Absence Suggests Modularity in Centriole Assembly. Current Biology 17 : 1465–1472. 17689959
51. Peel N, Stevens NR, Basto R, Raff JW (2007) Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol 17 : 834–843. 17475495
52. Cottee MA, Muschalik N, Wong YL, Johnson CM, Johnson S, et al. (2013) Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife 2: e01071–e01071. doi: 10.7554/eLife.01071 24052813
53. Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW (2004) The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J Cell Biol 165 : 673–683. 15184400
54. Conduit PT, Raff JW (2010) Cnn Dynamics Drive Centrosome Size Asymmetry to Ensure Daughter Centriole Retention in Drosophila Neuroblasts. Current Biology 20 : 2187–2192. doi: 10.1016/j.cub.2010.11.055 21145745
55. Hinchcliffe EH (2014) Centrosomes and the Art of Mitotic Spindle Maintenance. International Review of Cell and Molecular Biology 313 : 180–206.
56. Mottier-Pavie V, Cenci G, Verni F, Gatti M, Bonaccorsi S (2011) Phenotypic analysis of misato function reveals roles of noncentrosomal microtubules in Drosophila spindle formation. J Cell Sci 124 : 706–717. doi: 10.1242/jcs.072348 21285248
57. Quintyne NJ (2002) Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol 159 : 245–254. 12391026
58. Gatti M, Baker BS (1989) Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev: 438–453.
59. Novak ZA, Conduit PT, Wainman A, Raff JW (2014) Asterless Licenses Daughter Centrioles to Duplicate for the First Time in Drosophila Embryos. Curr Biol 24 : 1276–1282. doi: 10.1016/j.cub.2014.04.023 24835456
60. Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, et al. (2012) Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 14 : 1159–1168. doi: 10.1038/ncb2597 23086239
61. Fu J, Glover DM (2012) Structured illumination of the interface between centriole and peri-centriolar material. Open Biology 2 : 120104–120104. doi: 10.1098/rsob.120104 22977736
62. Tavosanis G, Llamazares S, Goulielmos G, Gonzalez C (1997) Essential role for gamma-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J 16 : 1809–1819. 9155007
63. Riparbelli MG (2005) The meiotic spindle of the Drosophila oocyte: the role of Centrosomin and the central aster. J Cell Sci 118 : 2827–2836. 15976443
64. Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, et al. (2011) Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nature Communications 2 : 359–11. doi: 10.1038/ncomms1367 21694707
65. Vaizel-Ohayon D, Schejter ED (1999) Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol 9 : 889–898. 10469591
66. Wang C, Li S, Januschke J, Rossi F, Izumi Y, et al. (2011) An Ana2/Ctp/Mud Complex Regulates Spindle Orientation in Drosophila Neuroblasts. Dev Cell 21 : 520–533. doi: 10.1016/j.devcel.2011.08.002 21920316
67. Gepner J, Li M, Ludmann S, Kortas C, Boylan K, et al. (1996) Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics 142 : 865–878. 8849893
68. Endow SA, Chandra R, Komma DJ, Yamamoto AH, Salmon ED (1994) Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J Cell Sci 107 (Pt 4): 859–867.
69. Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, et al. (2008) Drosophila asterless and Vertebrate Cep152 Are Orthologs Essential for Centriole Duplication. Genetics 180 : 2081–2094. doi: 10.1534/genetics.108.095141 18854586
70. Hallen MA, Ho J, Yankel CD, Endow SA (2008) Fluorescence Recovery Kinetic Analysis of γ-Tubulin Binding to the Mitotic Spindle. Biophys J 95 : 3048–3058. doi: 10.1529/biophysj.108.134593 18567627
71. Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, et al. (2008) Centrosome amplification can initiate tumorigenesis in flies. Cell 133 : 1032–1042. doi: 10.1016/j.cell.2008.05.039 18555779
72. Cabernard C, Doe CQ (2009) Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell 17 : 134–141. doi: 10.1016/j.devcel.2009.06.009 19619498
73. Franz A, Roque H, Saurya S, Dobbelaere J, Raff JW (2013) CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J Cell Biol 203 : 785–799. doi: 10.1083/jcb.201305109 24297749
74. Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW (2000) D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J 19 : 241–252. 10637228
75. Vérollet C, Colombié N, Daubon T, Bourbon H - M, Wright M, et al. (2006) Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol 172 : 517–528. 16476773
76. Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW (2001) Msps/XMAP215 interacts with thecentrosomal protein D-TACC toregulate microtubule behaviour: 1–8.
77. Barros TP, Kinoshita K, Hyman AA, Raff JW (2005) Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol 170 : 1039–1046. 16186253
78. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nature Publishing Group 9 : 676–682.
79. Baumbach J, Levesque MP, Raff JW (2012) Centrosome loss or amplification does not dramatically perturb global gene expression in Drosophila. Biology Open 1 : 983–993. doi: 10.1242/bio.20122238 23213376
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



