-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAutoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
The rather wide-spread and extremely A/T rich yeast virus like elements (VLEs, also termed linear plasmids) which encode toxic anticodon nucleases (ACNases) ensure autoselection in the cytoplasm by preventing functional nuclear capture of the cognate immunity genes, but how? When expressed in the nucleus, the mRNA of the VLE immunity genes is split into fragments to which poly(A) tails are added. Consistently, lowering the A/T content by gene synthesis prevented transcript cleavage and permitted functional nuclear expression providing full immunity against the respective ACNase toxin. Thus, internal poly(A) cleavage is likely to prevent functional nuclear immunity gene expression.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005005
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005005Summary
The rather wide-spread and extremely A/T rich yeast virus like elements (VLEs, also termed linear plasmids) which encode toxic anticodon nucleases (ACNases) ensure autoselection in the cytoplasm by preventing functional nuclear capture of the cognate immunity genes, but how? When expressed in the nucleus, the mRNA of the VLE immunity genes is split into fragments to which poly(A) tails are added. Consistently, lowering the A/T content by gene synthesis prevented transcript cleavage and permitted functional nuclear expression providing full immunity against the respective ACNase toxin. Thus, internal poly(A) cleavage is likely to prevent functional nuclear immunity gene expression.
Introduction
Pichia acaciae and Kluyveromyces lactis each contain two cytoplasmic virus-like elements (VLEs, also known as linear plasmids); i.e. pPac1-1 (12.6 kb), pPac1-2 (6.8 kb) and pGKL2 (13.5 kb), pGKL1 (8.9 kb) respectively [1,2]. The respective larger elements display substantial similarities to each other in terms of organization and gene content. They can exist without the smaller ones as they encode all proteins required for nucleus-independent cytoplasmic replication and maintenance [3]. The smaller VLEs pPac1-2 and pGKL1, respectively, which depend on the larger ones in terms of cytoplasmic transcription and/or replication, encode for the production of killer toxin complexes, zymocin (pGKL1) and PaT (pPac1-2) [reviewed in 4]. One subunit in either zymocin or PaT is highly conserved; it carries chitin binding and chitinase domains that recognize cell wall associated chitin of target cells as primary toxin receptor for subsequent import and/or activation [5,6,7]. In both zymocin and PaT, a rather hydrophobic stretch or subunit appears to manage membrane transfer of the cytotoxic subunits, PaOrf2 (encoded by pPac1-2 ORF2) and γ-toxin (encoded by pGKL1 ORF4). Although they hardly show any sequence similarity, they both act as anticodon nucleases (ACNases). The recently solved crystal structure of PaOrf2 revealed a unique fold, which shows no similarity to any known ribonuclease [8]. PaOrf2 specifically attacks tRNAGln in vivo and additionally cleaves in vitro tRNAGlu and tRNALys or synthetic stem-loop RNA derived from the tRNAGln sequence [8,9]. γ-toxin cleaves the same tRNAs in vitro, but in vivo its preferred target is tRNAGlu [10,11]. While γ-toxin cleaves its target tRNA once at the 3`side of the wobble uridine, PaOrf2 apparently cleaves at the same position and additionally two nucleotides upstream, as judged from the appearance of two alternative cleavage products with full length tRNA from S. cerevisiae [9,10]. Since PaOrf2 but not γ-toxin evades a possible repair of the tRNA halves by cellular tRNA ligases, it was speculated that the presence of two cleavage sites might allow the excision of a di-nucleotide, rendering the target tRNA non-repairable [12,13,14,15].
VLE cured strains of P. acaciae and K. lactis are sensitive to their own respective toxins, proving that not only the killer phenotype but also the cognate immunity are encoded by the elements [1,2]. Indeed, PaT immunity is conferred by the only protein encoded by pPac1-2 (ORF4) that lacks a signal peptide for secretion [16] and immunity against zymocin had been postulated to be encoded by KlORF3 of pGKL1 [17]. There is hardly any homology among PaORF4 and KlORF3, and consistent with it, no cross-protection has been observed against zymocin or PaT [16]. PaT and zymocin are the most thoroughly studied VLE encoded ACNase killer toxins, but there are other systems in yeast [reviewed in 18], such as PiT from Pichia inositovora, a ribonuclease inducing specific fragmentation of 25S and 18S rRNAs [19], or DrT from Debaryomyces robertsiae, an ACNase resembling PaT and cleaving tRNAGln [20]. From an evolutionary point of view, toxin and immunity functions implemented in VLEs have to be considered as players of an autoselection system rather than providing advantages for the respective host [16,21], although the latter, clearly benefits from the conferred killer phenotype.
Intriguingly, PaORF4 encoding PaT immunity could be heterologously functionally expressed solely from VLEs in the cytoplasm [16], i.e. when the gene was integrated into the pGKL-system of Kluyveromyces lactis. All efforts to express the immunity phenotype with PaORF4 on nuclear episomal and centromeric vectors failed to establish self-protection against the ACNase toxin. Here, we show that PaORF4 as well as the immunity genes from other VLEs (KlORF3 and DrORF5) are nevertheless transcribed when the genes are governed by a yeast nuclear promoter in episomal vector backbones, but the mRNA becomes immediately fragmented thereby preventing translation that otherwise would yield a functional immunity protein. As exemplified for PaORF4 and KlORF3, changing the primary structure from a rather high A/T bias to a much lower degree allowed for functional nuclear immunity expression, proving that the gene’s primary sequence information is sufficient to provide ACNase self-protection and that the native ORF context ensures autoselection of the VLE.
Results
VLE encoded immunity factors cannot be functionally expressed in the nucleus
For the three known VLE encoded ACNase toxin complexes PaT, zymocin and DrT, immunity functions were proposed to be encoded by PaORF4, KlORF3 and DrORF5, respectively [16,17,20]. Subsequently, PaORF4 and DrORF5 were functionally expressed from their native promoters in the cytoplasm after integration of the genes into a VLE system (the pGKL1/2 system transferred to S. cerevisiae) [16,20,22]. Attempts to express both immunity genes in the nucleus after fusion of the ORFs to the constitutive ADH1 promoter (ADH1pr), however, did not establish toxin immunity. This is in contrast to the putative zymocin immunity gene (KlORF3) which was previously identified on the basis of functional expression from the nucleus after fusion of the KlORF3 to the PGK promoter [17]. Upon expression of the PGKpr-KlORF3 construct, partial zymocin protection was observed in S. cerevisiae cells. However, the phenotype required the presence of the autonomous VLE pGKL2 while, nuclear expression in a standard S. cerevisiae strain devoid of any VLE did not confer detectable zymocin immunity [17]. To reconfirm this latter notion, we fused KlORF3 to the alternative strong constitutive promoter ADH1pr and analyzed the zymocin response of the sensitive S. cerevisiae strain BY4741 containing the ADH1pr-KlORF3 fusion in comparison to the wild type by the microdilution method. As shown in Fig 1, zymocin sensitivity indeed remains unaltered in the presence of the ADH1pr-KlORF3 construct, supporting the conclusion that the VLE encoded immunity factors cannot be functionally expressed in the nucleus in a standard S. cerevisiae strain. In contrast, both DrORF5 and PaORF4 provide resistance to their cognate ACNase toxins (DrT and PaT) when expressed in the cytoplasm of a sensitive S. cerevisiae strain and in all assays conducted, complete, rather than partial immunity was observed [16,20].
Fig. 1. The zymocin immunity gene (KlORF3) does not confer immunity when expressed in the nucleus. 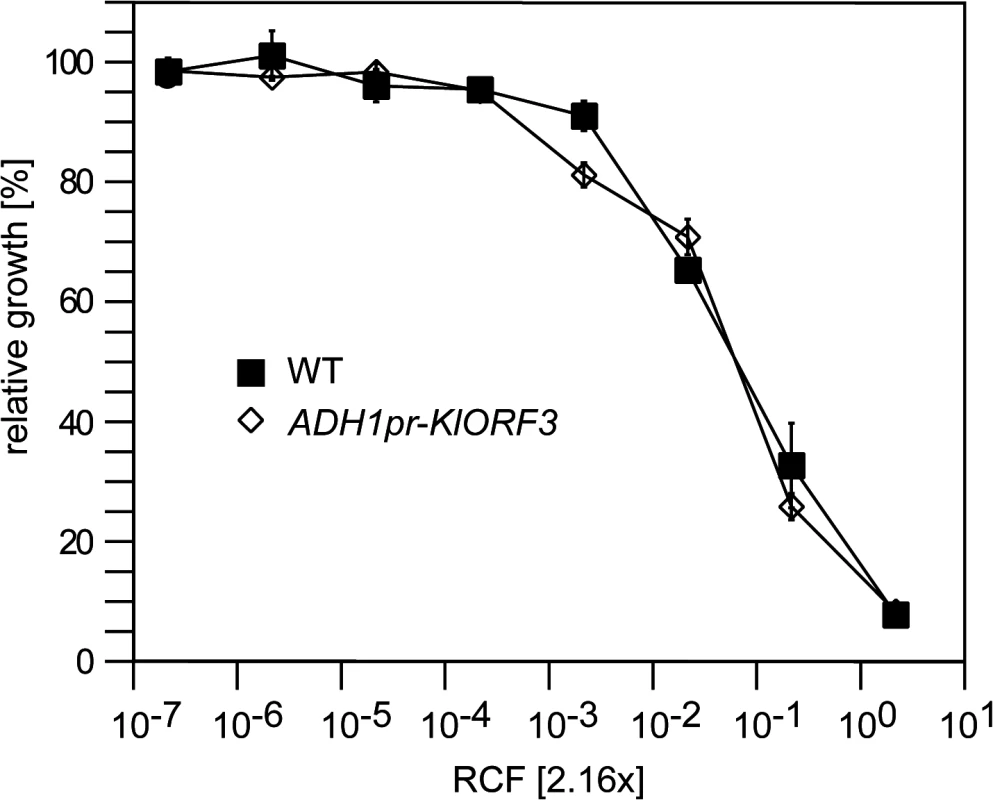
S. cerevisiae strains without any vector (WT) or with YEKlO3 (ADHpr-KlORF3) were analyzed for zymocin resistance using the microtiter plate assay method. Relative growth was determined (OD600nm) after 24 hours at 30°C; values refer to strains grown in toxin-free medium. A relative concentration factor (RCF) of 1 relates to the toxin concentration in non-diluted supernatants. Each value represents a mean of triplicates. Northern analysis of immRNA reveals specific fragmentation in the nucleus
To analyze whether the observed failure of immunity expression from the nuclear vectors was due to a barrier in transcription or due to transcript instability, we analyzed the levels of mRNAs encoding immunity (immRNA) and their stability. ImmRNA from S. cerevisiae strains carrying nuclear fusions of the immunity factor encoding ORF (immORF) and the ADH1pr was compared with immRNA from natural expression hosts, where immORFs are expressed from the cytoplasm. In all cases, cytoplasmic expression yielded stable immRNA that exceeded the size of the corresponding full-length immORF (Fig 2). In contrast, nuclear expression of immORFs produced one (KlORF3), two (DrORF5) or four (PaORF4) distinct signal bands in Northern blots, which were significantly smaller in size than their corresponding full-length immORF (Fig 2). Thus, while immRNAs are stable, when expressed from their cognate VLEs in the cytoplasm, they are prone to fragmentation and get particularly instable when expressed in the nucleus. The lack of full length immRNA in the latter case is in line with the observed general lack of functional ACNase immunity when immORFs are expressed in the nucleus.
Fig. 2. Northern blot analysis of immRNA from different genetic backgrounds. 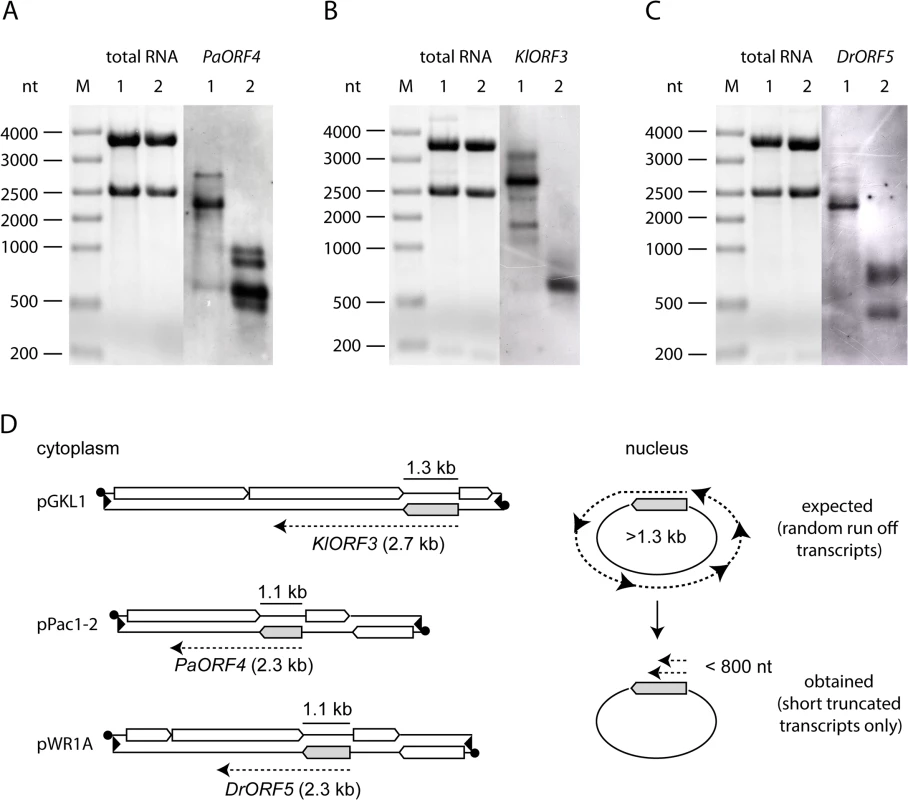
Total RNA isolated from strains expressing the respective immORFs from VLEs or nuclear vectors was probed using the respective immRNA specific DIG-labelled probes. A: PaORF4 (1092 bp) expressed in Pichia acaciae from pPac1-1, pPac1-2 (1) or in S. cerevisiae 301 YEPaO4 (2). B: KlORF3 (1287 bp) expressed in K. lactis from pGKL1, pGKL2 (1) or in S. cerevisiae CEN.PK2-1c YEKlO3 (2). C: DrORF5 (1062 bp) expressed in D. robertsiae from pWR1A, pWR1B (1) or in S. cerevisiae CEN.PK2-1c YEDrO5 (2). D: Scheme of immORF transcription and mRNA sizes transcribed from cytoplasmic VLEs (pGKL1, pPac1-2 and pWR1B, left side) or from episomal vectors, which are located in the nucleus (right side). ImmORFs are grey shaded. Poly(A)-site processing leads to fragmentation of immRNA expressed in the nucleus
Since the poly(A) site processing machinery recognizes UA rich elements that appear to be quite diverse in S. cerevisiae [reviewed in 23], we explored the possibility that immRNA fragmentation of the highly A/T biased transcripts could be associated with recognition of random, internal poly(A) sites, leading to immORF fragmentation with the addition of poly(A) tails at the cleavage sites. We isolated total RNA from S. cerevisiae strains expressing ADH1pr-KlORF3, ADH1pr-PaORF4 and ADH1pr-DrORF5 and primed cDNA synthesis with a poly(A)-specific oligonucleotide. Such cDNAs were analyzed by PCR using immORF specific oligonucleotides, binding at the 5’ end together with an oligonucleotide complementary to the poly(A)-specific anchor. As a control, the ERG3 mRNA was amplified from the same cDNA preparations. For all immORFs, several 3’ RACE products were obtained, all of which were smaller than the minimum expected gene size (Fig 3), suggesting the presence of poly(A) stretches at the 3´ ends of the immRNA fragments. Such result agrees with the specific fragmentation of the nuclearly expressed immRNAs followed by the addition of poly(A) tails. Fragments of each PCR reaction were extracted from the gel and cloned; sequencing identified the fragments to perfectly match the 5`terminal regions of the respective immRNAs, which are truncated at their 3´ends and extended by attachment of several (16 to 66) adenines (Fig 4).
Fig. 3. Amplification of immRNA fragments by 3´-RACE. 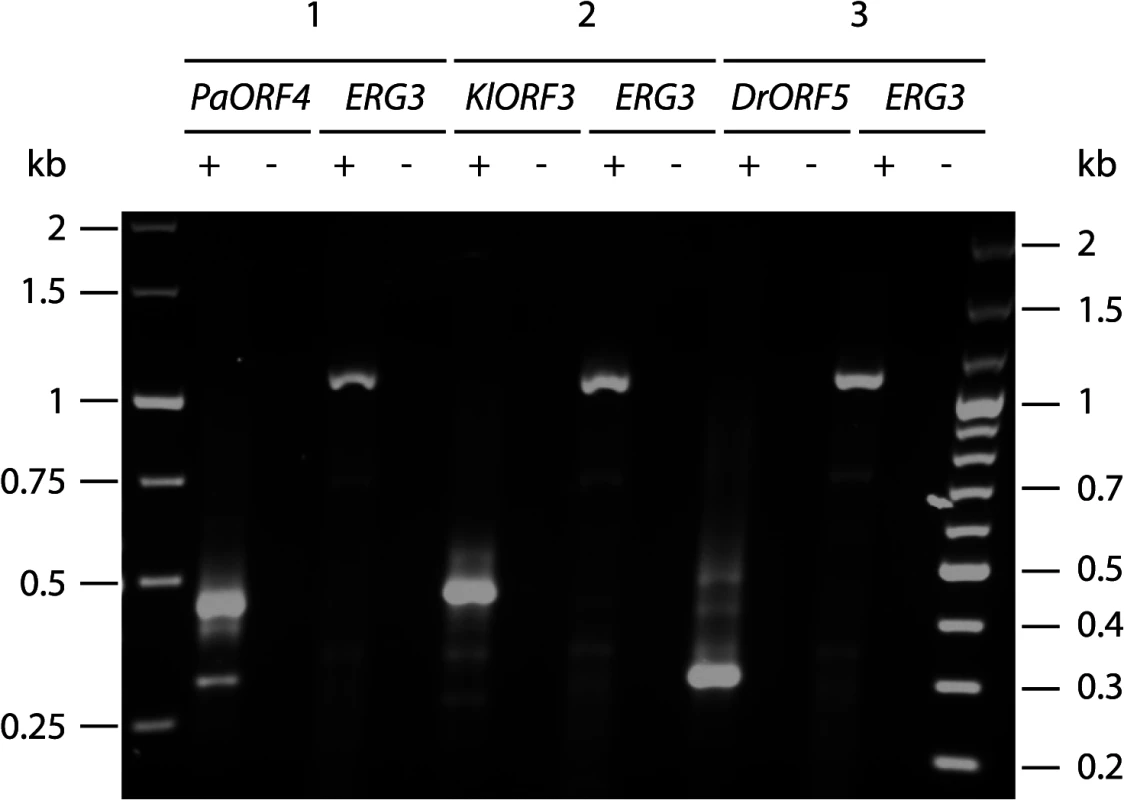
Total RNA from S. cerevisiae CEN.PK2-1c strains carrying vectors YEPaO4 (1), YEKlO3 (2) or YEDrO5 (3) was transcribed to cDNA using a poly(A)-specific primer (+). Absence of DNA was checked by control reactions lacking the reverse transcriptase (-). ImmRNA was then amplified with one primer specific for the 5´ end of the immORF and a second primer complimentary to the poly(A)-specific anchor. ERG3 cDNA was amplified using ERG3 specific primers. Fig. 4. Schematic representation of the identified mRNA fragments of the immORFs PaORF4 (A), KlORF3 (B) and DrORF5 (C). 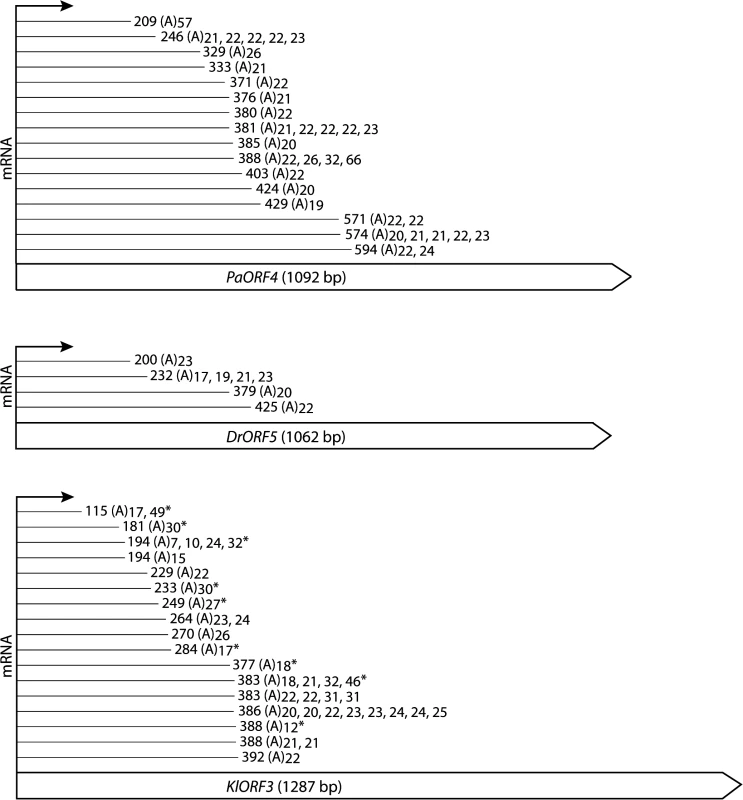
The cDNA fragments reached the indicated indicated nucleotide positions and were extended by a stretch of 7–66 adenyl (A) nucleotides, the length of which is indicated for each particular molecule that was obtained. Asterisks (*) mark fragments which were identified by the linker ligation method; all of the other fragments were identified by 3´-RACE. For complete gene sequences including the identified mRNA truncation sites we refer to S1 Fig To ensure that the data obtained from 3’RACE experiments did not result from unspecific internal priming within A-rich mRNA regions, a linker ligation method was applied to exemplarily identify the immRNA ends of KlORF3. The linked KlORF3 fragments were amplified using a gene specific and a linker specific primer and after cloning analyzed by sequencing. All identified fragments contained a poly(A) tail consisting of 7–49 adenyl nucleotides. The results confirm the previous observations. Both methods identified that cleavage and polyadenylation of each of the immRNAs happens not only at one definite, but at multiple positions. For example, among the 33 sequenced PaORF4 mRNA fragments, 15 different polyadenylation sites in a region between positions 209 nt and 594 nt were mapped (Fig 4 and S1 Fig). The identified positions in the mRNA truncation products and their frequency of occurrence for each immRNA are summarized in Fig 4.
Lowering immORF A/T content enables functional nuclear expression
Since the above results suggested the possibility that the high A/T content of immRNAs limits their nuclear expression due to ORF-internal poly(A) site processing, we analysed whether a reduction of the A/T content of PaORF4 and KlORF3 improves their expression from the nucleus. Synthetic variants of both genes were generated, where most of the A/U rich codons were replaced by synonymous more G/C rich codons via gene synthesis (S2 Fig). As a result, the G/C content for PaORF4 increased from 21% to 45% in the synthetic variant PaORF4ms and KlORF3 increased in G/C content from 22% to 54% in the synthetic variant KlORF3ms without altering the amino acid sequence. Both variants were cloned into the same vector backbone previously used to study the native, unchanged genes, resulting in a set of multicopy plasmids carrying immORF fusions to the ADH1 promoter, where immORFs remain either unchanged in codon usage (78–79% A/T) or exhibit a significantly reduced A/T content (46–55%). All constructs were expressed in a PaT or zymocin sensitive VLE-free S. cerevisiae strain and the presence of full length immRNA was comparatively analysed by RT-PCR (Fig 5). As a control, the ERG3 mRNA was detected in all strains in parallel. Full length immRNA was generally absent in the strains expressing the A/T-rich native (non-modified) versions of the immORFs (Fig 5A), which is in agreement with the results obtained by Northern analysis (Fig 2). In striking contrast, however, full length immRNA becomes detectable when low A/T content codon usage variants PaORF4ms and KlORF3ms are expressed from the same nuclear constructs (Fig 5B). Thus, lowering the A/T content clearly improves immORF expression in the nucleus compared to the natural gene variants being expressible only in the cytoplasm.
Fig. 5. RT-PCR analysis of immRNA. 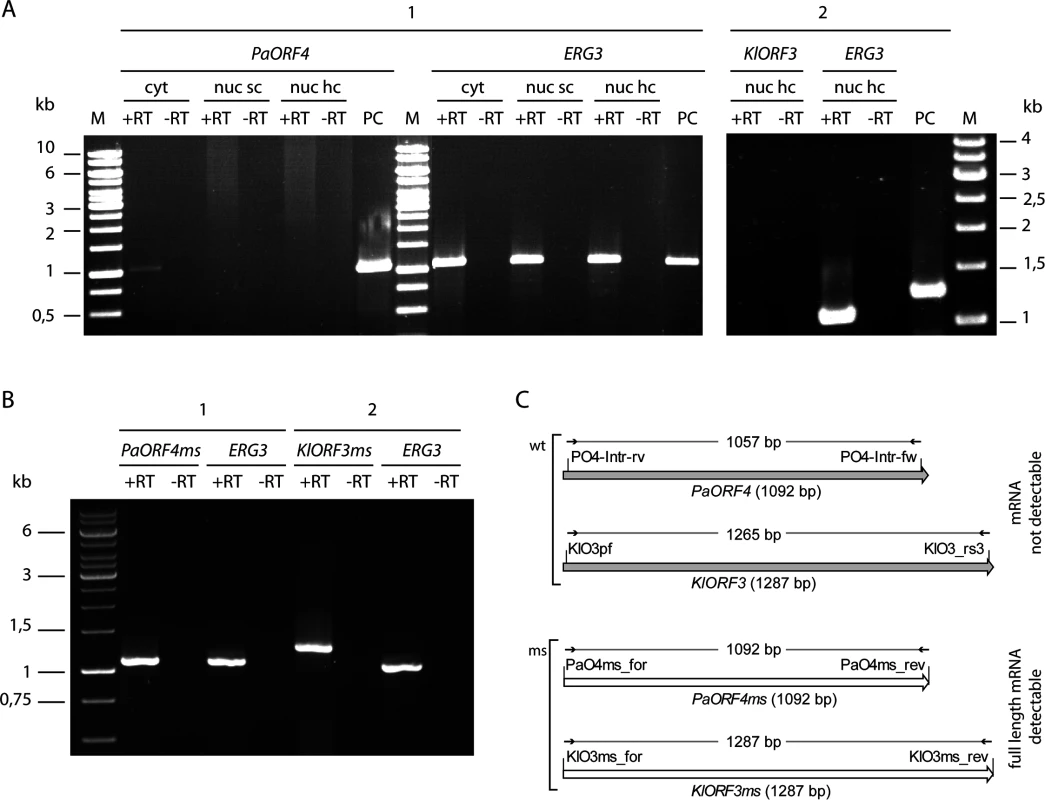
mRNA of PaORF4/PaORF4ms and KlORF3/KlORF3ms or the control gene ERG3 was analysed by reverse transcription (+RT) using total RNA as the template. The absence of contaminating DNA was checked by control reactions lacking the reverse transcriptase (-RT). A: mRNA of the original A/T-rich immORFs PaORF4 and KlORF3 was analysed using total RNA isolated from S. cerevisiae CEN.PK2-1c strains expressing ADH1pr-PaORF4 (1) or ADH1pr-KlORF3 (2) from nuclear single copy vectors (nuc sc) or nuclear high copy vectors (nuc hc) or from S. cerevisiae F102.2 MS1607 expressing PaORF4 from a cytoplasmic VLE (cyt). PC: as a positive control for checking primer binding vector DNA carrying the immunity genes PaORF4 or KlORF3 was used. B: Reverse transcription of mRNA of the codon adjusted, A/T-decreased immunity genes PaORF4ms and KlORF3ms using as the template total RNA isolated from strains expressing ADH1pr-PaORF4ms (1) or ADH1pr-KlORF3ms (2) from high copy vectors. C: Schematic representation of RT-PCR analyses for the detection of immRNAs. Primers, depicted as small arrows, are denoted as such (for sequences see S3 Table). wt denotes the original immunity gene sequences (grey shaded large arrows), ms denotes the modified (G/C rich) sequences (white large arrows). The expected sizes of RT-PCR products refer to the size of the structural genes and are given in base pairs (bp). To check whether such improvement also enables functional immORF expression, ACNase immunity of strains carrying the different immORF expression constructs was scored by the eclipse plate and microdilution assays (Fig 6). Consistent with previous results, expression of A/T-rich versions of PaORF4 or KlORF3 did not confer a detectable immunity phenotype to the PaT or zymocin producers, respectively. When the low A/T-content variants PaORF4ms or KlORF3ms were expressed, however, sensitivity to the cognate ACNase toxin producer was entirely lost. At the same time, the PaORF4ms expressing strain remained sensitive to the zymocin producer (Fig 6A and Fig 6B) and the KlORF3ms expressing strain remained sensitive to the PaT producer (Fig 6A). These results indicate that although lowering the A/T contents in two functionally distinct immORFs suffices to overcome the observed nuclear expression barrier, the immunity factors, once being expressed, do not confer ACNase cross-protection to non-self yeast strains.
Fig. 6. The codon adapted immORFs PaORF4ms and KlORF3ms confer resistance to the respective toxins PaT and zymocin. 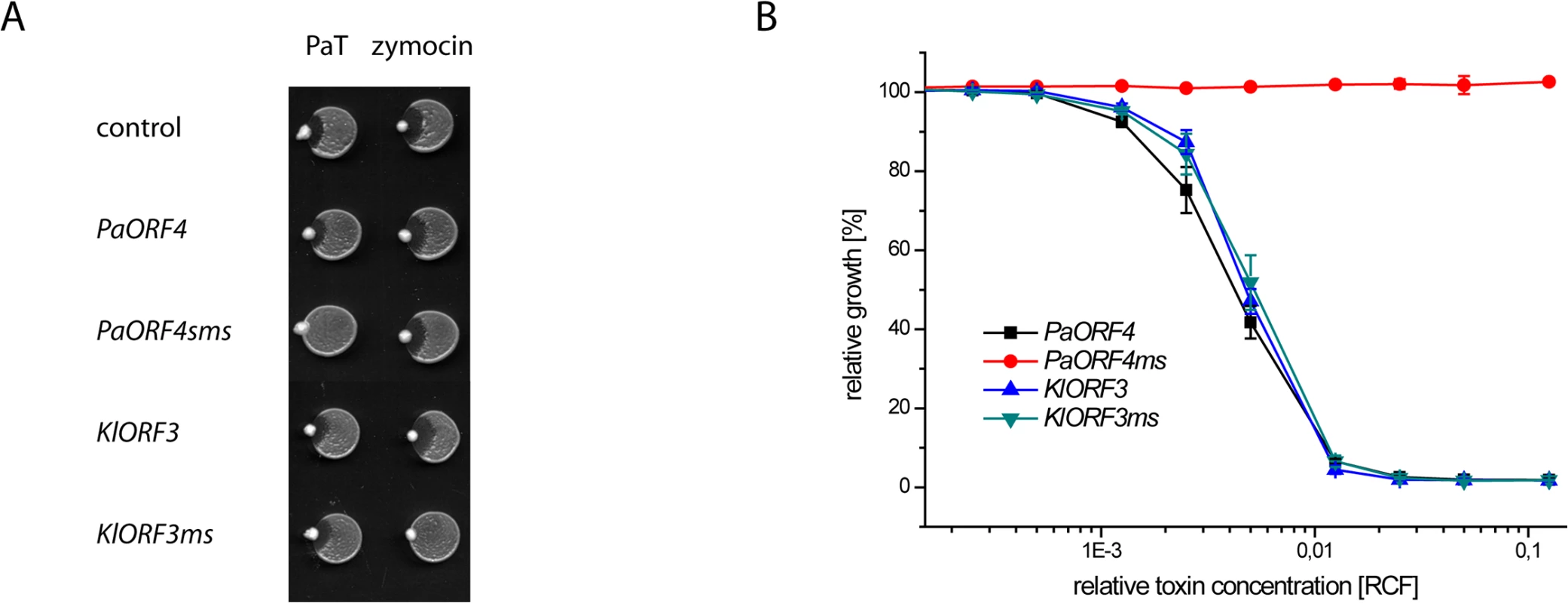
A: S. cerevisiae CEN.PK2-1c transformed with yeast episomal vectors carrying ADH1pr fusions of either the A/T-rich (PaORF4 and KlORF3) or the A/T-decreased (PaORF4ms and KlORF3ms) immunity genes or the empty vector (control) were dropped on YPD agar, inoculated with P. acaciae (PaT) or K. lactis (zymocin) at the rim and incubated over night at 30°C. B: Microtiter plate assays. Supernatants from a P. acaciae culture were added in different concentrations (RCF) to micro cultures of the yeast strains described in A. Relative growth was determined (OD600nm) after 24 hours at 30°C; values refer to strains grown in toxin-free medium. A relative concentration factor (RCF) of 1 relates to the toxin concentration in non diluted supernatants. Each value represents a mean of triplicates. KlORF3ms confers full, pGKL2 independent immunity to intracellular γ-toxin
Our KlORF3ms expression studies (Fig 6A) confirm the previous proposal by Tokunaga et al., [17] that pGKL1 encoded Orf3 protein confers zymocin immunity. As KlORF3ms yields protection to the zymocin producer K. lactis in the background of S. cerevisiae strain BY4741, there is evidently no general requirement for the presence of the pGKL2 VLE to establish immunity. In previous experiments, the immunity phenotype associated with the PGKpr-KlORF3 construct and the pGKL2 VLE was only partial, since exogenous purified zymocin induced detectable growth inhibition [17]. To check whether the pGKL2 independent immunity conferred by nuclear expression of ADH1pr-KlORF3ms also is partial, we analyzed the zymocin response using the microdilution assay. We compared KlORF3ms-induced zymocin protection in WT cells with elp3 cells not carrying any immORF. The latter condition prevents zymocin induced tRNA cleavage due to absence of the crucial wobble uridine mcm5s2-modification and confers full toxin resistance [10,11]. We observed no difference between the zymocin response of elp3 cells not expressing any immORF and ELP3 cells expressing KlORF3ms; only ELP3 wild type cells without KlORF3ms showed sensitivity to zymocin (Fig 7A). To further check the dominant nature of KlORF3ms induced immunity and to analyze whether the KlOrf3 protein is capable of intracellular inactivation of γ-toxin, as suggested in earlier work [17], we constructed strains co-expressing KlORF3/KlORF3ms and GAL1pr-driven, multi copy KlORF4 devoid of its signal peptide encoding region, leading to intracellular accumulation of the ACNase subunit γ-toxin (KlOrf4). As a control, both immORF constructs were introduced into the elp3 strain, where the need for an immORF is overcome by preventing tRNA cleavage in the first place. Galactose induced expression of the γ-subunit proved inhibitory to the strain expressing the A/T-rich variant KlORF3 only; as expected, the elp3 mutation prevented toxic effects of γ-toxin but also KlORF3ms entirely prevented growth inhibition by intracellular γ-toxin and the additional removal of ELP3 did not improve growth of the strain under inducing conditions (Fig 7B). Thus, zymocin immunity acts, similar to the previously studied PaT and DrT immunity functions at the intracellular stage and provides true immunity rather than partial resistance, independent of any pGKL2 encoded functions. Since we detected no cross resistance of KlORF3ms expressing strains to the non-cognate ACNase toxin PaT (Fig 6A and Fig 6B) and PaORF4ms did not protect detectably from zymocin (Fig 6A), the two immunity proteins PaOrf4 and KlOrf3 are highly specific for each of their cognate ACNase subunits.
Fig. 7. The zymocin immORF confers full immunity to exo zymocin and intracellularly expressed ACNase subunit γ-toxin. 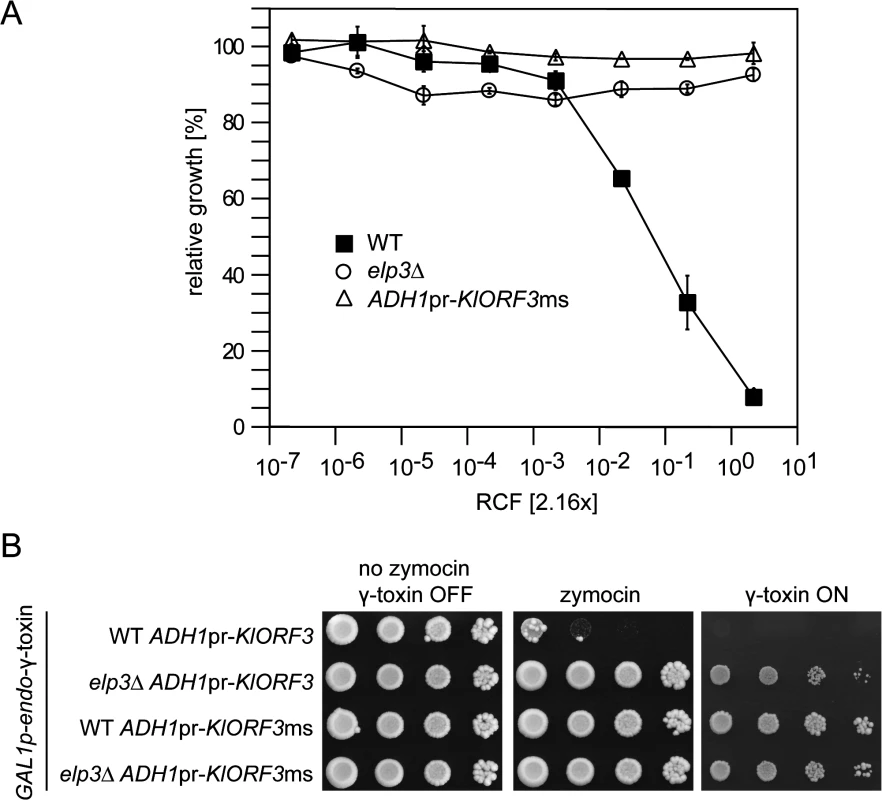
(A) Exo-zymocin resistance of isogenic WT, elp3∆, and WT strains carrying YEKlO3ms (ADHpr-KlORF3ms) analyzed by microtiter plate assay as described in Fig 1. (B) Intracellular coexpression of zymocin immORF and γ-toxin. Tenfold serial dilutions of cultures were spotted on YPD media (no zymocin, γ-toxin OFF and zymocin), YPD media supplemented with zymocin (zymocin) or YNB media with galactose as the sole carbon source (γ-toxin ON). All strains analyzed in (B) carry pRK57 for GAL driven, intracellular expression of γ-toxin and additionally carry YEKlO3 (ADH1pr-KlORF3) or YEKlO3ms (ADH1pr-KlORF3ms). WT and elp3∆ indicate the additional presence or absence of a deletion in ELP3 (elp3∆). Discussion
PaT, DrT and zymocin are the three known examples of eukaryotic protein toxins with ACNase activity. All three are encoded by non-autonomous VLEs (pPac1-2; pWR1A and pGKL1) persisting in the cytoplasm of different yeast species. Crucial functions for cytoplasmic transcription and DNA replication, processes normally occurring in the nucleus, are supplied in each case by a larger VLE (pPac1-1; pWR1B and pGKL2). Among these are a uniquely structured RNA polymerase [24] as well as a virus like mRNA capping enzyme [25,26] to generate capped, cytoplasmic mRNAs from unique cytoplasmic promoters [27,28,29]. Generally, VLE genes cannot be expressed in the nucleus due to non-recognition of the cytoplasmic promoters by the nuclear (host encoded) RNA polymerases. The presence of genes on pPac1-2 and pWR1A mediating immunity against PaT and DrT was previously shown by integration into the pGKL1/2 system transferred to S. cerevisiae. The parental pGKL1/2 carrying S. cerevisiae strain produces zymocin and its cognate immunity factor but was sensitive to DrT and PaT. This sensitivity was entirely lost upon integration of DrORF5 and PaORF4, respectively [16,20]. Since both, DrOrf5/PaOrf4 and DrOrf3/PaOrf2 display detectable sequence homology and there is significant DrT/PaT cross protection mediated by PaOrf4, a direct recognition of the matching (PaOrf2) or nearly matching (DrOrf3) ACNase by the immunity factor was suggested. In support, PaOrf4 can disable toxic in vivo effects of intracellular PaOrf2 and both proteins were shown to form a complex in vitro that inhibits the ACNase activity of PaOrf2, resembling the mode of action of tRNase colicin immunity factors, which tightly bind and occlude the tRNase active site [8,30]. Similarly, in vivo studies with DrOrf5 showed that it protects against the in vivo tRNase activity of the intracellular DrOrf3 subunit [20], hinting at a similar immunity principle as for PaOrf4. Co-crystal structures of VLE-encoded tRNases with their cognate immunity proteins will be required to determine whether immunity factors against toxic tRNases as evolutionary diverse as prokarytotic colicins and VLE encoded killer toxins indeed share a similar mechanistic strategy to bind and occlude the tRNase active site.
For zymocin, understanding of the immunity factor function had been less advanced; published data [17] indicated an as yet undefined requirement for the VLE pGKL2 to establish KlOrf3 mediated immunity and in contrast to PaT and DrT immunity functions, the zymocin immunity factor appeared to provide partial protection only. Additionally, it was suggested that the zymocin immunity factor protects from intracellular γ-toxin based on the observation that pGKL1/2 carrying cells are resistant to galactose-induced expression of a signalpeptide-less KlORF4 gene [17], but no similar protection has been shown for the isolated KlORF3 gene. Since heterologous expression of KlORF3 precluded the use of the cytoplasmic pGKL1/2 based expression system, we encountered the general problem that even the established immunity genes PaORF4 and DrORF5 could not be expressed in the nucleus after replacement of the cytoplasmic promoter by well characterized nuclear promoters. As the same outcome was observed for the zymocin immunity gene, a general principle inhibitory to nuclear expression of these immunity genes became obvious. Since AT rich immunity genes are efficiently translated by the host’s translational machinery when the corresponding mRNA is generated in the cytoplasm by a VLE encoded transcriptional machinery but not when the same mRNA is generated in the nucleus, a nuclear transcriptional rather than a cytoplasmic translational barrier appeared to exist. Northern, RACE and linker ligation analysis now shows that nuclear expression generally ends up in fragmentation of the immRNAs which goes along with the addition of poly(A) tails.
Poly(A) site recognition is thought to basically involve the presence of the AAUAAA poly(A) signal (PAS), together with a GU-rich sequence as a downstream element [reviewed in 23]. However, unlike higher eukaryotes, yeast apparently tolerates a high degree of variation in individual poly(A) site recognition elements, as was derived from the analysis of expressed sequence tags generated by oligo(dT) primed cDNA synthesis [31]. For example, the PAS element in yeast is simply characterized by being A-rich. Thus, extremely AU-rich transcripts, such as VLE derived genes exhibit a high probability of ORF internal poly(A) site recognition and processing when moved to the nucleus. In support of this, we show that lowering the A/T content by defined gene synthesis is sufficient to prevent immRNA fragmentation in the nucleus allowing for functional expression of PaT and zymocin immunity phenotypes.
Our analysis with the synthetic KlORF3ms construct shows that the zymocin immunity protein indeed acts intracellularly and provides true self-protection rather than partial resistance. Thus, all eukaryotic ACNase immunity proteins may, like PaOrf4, recognize and inhibit their cognate ACNase. In support of a specific recognition, cross immunity against DrT but not zymocin can be provided by PaOrf4, whereas KlOrf3 provides immunity solely and specifically to zymocin. Since ACNase subunits of DrT and PaT are detectably similar but no similarity exists between DrT/PaT and zymocin, such similarity/non-similarity is apparently recognized by the immunity proteins. The initial detection of partial zymocin resistance in a strain carrying PGKpr-KlORF3 in the nucleus as well as pGKL2 in the cytoplasm [17] may be correlated to the fact that the fusion of KlORF3 to the PGKpr did not eliminate the upstream conserved sequence (UCS) element of KlORF3. Since UCS sequences have been shown to be sufficient for mediating cytoplasmic transcription [28] and toxin resistance was only seen when pGKL2, i.e. the UCS-recognizing RNA polymerase was present, it appears possible, that partial zymocin immunity was due to cytoplasmic transcription of the PGKpr-KlORF3 construct, which may have been located transiently in the cytoplasm after transformation. Such transient cytoplasmic availability may constitute the basis for the observed partial zymocin resistance as opposed to full immunity in this study.
Only rather recently nuclear sequences of plasmid and viral origin (NUPAVs) were detected which—eponymously—result from evolutionary capture of plasmid and VLE-genes by the yeast nucleus [32]. Indeed, cytoplasmic VLE based genes can frequently and repeatedly be trapped by the nucleus, as explicitly shown for pDH1A from Debaryomyces hansenii, which represents the most recent ancestor of NUPAVs so far known [33]. Taken also into consideration that some ORFs of VLEs, such as the toxin genes have been cloned and successfully expressed from nuclear vectors [19,34,35] the risk is immediately imposed on a VLE system that upon nuclear immunity gene capture autoselection is disabled, which is yet mandatory for VLE long term propagation. While chromosomally encoded yeast killer toxins are traditionally considered as factors beneficial to the producer cell due to the ability to eliminate competitors, VLE encoded toxins additionally or even predominantly serve to counterselect for spontaneous plasmid free segregants, clearly resembling the autoselective properties of bacterial toxin/antitoxin systems [16,21,36]. In Pichia acaciae, Kluyveromyces lactis and Debaryomyces robersiae toxin encoding cytoplasmic VLEs can be easily eliminated under laboratory conditions, which in all cases generates toxin sensitive segregants. Such situation differs from the vast majority of chromosomally encoded toxins, which are routinely not active against the producing species which supports that VLE encoded killer toxins function to kill spontaneous VLE-free segregants. However, such function does not exclude additional benefits to producer cells that are provided by the ability to kill other yeasts in a given environment. Since VLE-derived NUPAVs in different stages of degeneration can be detected in various yeast genomes [32,33], the high A/T content of VLEs in general may serve to minimize the potential for domestication of VLE based genes by the host, which might be particularly relevant for the immunity function that, if domesticated and separated from a toxin encoding VLE, would eliminate the positive selective pressure incurred by the toxin on the maintenance of the VLE system. In other words, spontaneous VLE-free segregants would no longer be eliminated in an environment of VLE containing, toxin secreting sister cells. Interestingly, among the VLE derived NUPAVs described by Frank and Wolfe, 2009, immunity genes constitute the largest group. We identified an additional immunity derived NUPAV in the yeast Pichia sorbitophila [37], which appears to be almost intact and closely related to the DrORF5/PaORF4 genes (S3 Fig). The gene (Piso0_001880) located close to the end of chromosome F spans 729 bp and contains at its 5’end an extended region of similarity to the immORFs where, however, the ATG and the VLE promoter appear to be lost, leading to the annotation of an internal ATG as the gene’s startcodon. Importantly, Piso0_001880 has an A/T content of 75.5% which resembles the typical VLE characteristics and differs significantly from P. sorbitophila genome average (58.6%). We assume that Pios0_001880 represents a rather recent VLE derived domestication in an early stage of degeneration that may be related to the nuclear expression barrier caused by extreme A/T content. In support, no entirely intact immunity-NUPAV has been identified so far, suggesting a general incompatibility of A/T rich VLE genes with the nuclear transcript processing machinery.
Materials and Methods
Strains, media and general methods
The cloning host Escherichia coli DH5αF’ was grown in Luria-Bertani (LB) medium (0.5% yeast extract, 1% peptone, 0.5% NaCl) supplemented with ampicillin (100 μg ml-1) at 37°C. Yeast strains used in this study are listed in S1 Table. They were grown either in YEPD medium (1% yeast extract, 2% peptone, 2% glucose) or in yeast nitrogen base (Difco, Detroit, MI, USA) at 30°C. Transformation of S. cerevisiae was performed according to the PEG/lithium acetate method [38].
Construction of plasmids
Plasmids used in this study are listed in S2 Table. The immORFs were amplified by PCR using total DNA of the killer yeasts P. acaciae, D. robertsiae, K. lactis as template and the primers listed in S3 Table (PaORF4: PO4-NdeI-rv and PO4-fw, KlORF3: KlO3rev_NdeI and KlO3for, DrORF5: DrO5rev_NdeI and DrO5for). The PCR products were blunt-end cloned into EcoRV restricted pSK - plasmid to yield pSKPaO4, pSKKlO3 and pSKDrO5. The pSKDrO5 plasmid was modified by site-directed mutagenesis to remove a DrORF5-internal NdeI restriction site using the primers mut_NdeI_for and mut_NdeI_rev. The A/T decreased gene versions PaORF4ms and KlORF3ms were synthesized by GeneArt (Regensburg, Germany) and delivered in a vector containing NdeI and HindIII restriction sites upstream and downstream of the respective ORF. All ORFs were released from their vectors via NdeI and HindIII and cloned into a likewise restricted pSKpADH1. The ADH1pr-immORF fusions were then ligated into the 2μ vector YEplac195 using the restriction sites KpnI and SacI (YEPaO4, YCPaO4 YEKlO3, YEDrO5, YEPaO4ms) or SmaI and HindIII (YEKlO3ms). For coexpression of KlORF3ms and γ-toxin, the EcoRI-BglII insert of pABY1643 (GAL1pr-γ-toxin-GST; [10]) was subcloned into EcoRI-BamHI digested YEplac181.
Killer toxin assays
For the microtiter plate assay the partially purified toxins PaT and zymocin were obtained from culture supernatants of P. acaciae NRRL Y-18665 and K. lactis AWJ137 by ultrafiltration as described previously [20]. Different toxin concentrations were applied in microtiter plates as described in Klassen et al., [39]. Cell growth was monitored photometrically in a Multiscan FC Microplate Photometer (Thermo Fisher Scientific, Waltham, MA, USA) at 620 nm.
To check the sensitivity of a strain in the eclipse assay, a drop of 7 μl of the cell suspension was spotted on a YEPD agar plate. The killer strain was then placed at the rim of the drop and the plate was incubated at 30°C overnight.
Effects of intracellularly expressed, galactose-inducible toxin subunits on the growth of certain strains were checked with the drop dilution assay. Cultures were serially diluted and 5 μl aliquots were spotted on YNB medium containing glucose (repressing condition) or galactose (inducing condition) and incubated for several days at 30°C. Zymocin containing YPD plates were prepared by spreading 300 μl of filter sterilized, concentrated supernatant (RCF 5) of K. lactis AWJ137.
Northern analysis
Total RNA of different immunity ORF expressing yeast strains was isolated after over night cultivation in YEPD medium. 1.5 μg of each RNA sample were separated by a denaturating 1.5% agarose gel electrophoresis (20 mM MOPS, 8 mM sodium acetate, 1 mM EDTA, 0.74% formaldehyde, pH 7.0) and blotted onto a positively charged nylon membrane (Roche Diagnostic GmbH, Mannheim, Germany). The blotting success and RNA integrity were controlled by methylene blue staining (0.02% methylene blue, 0.3 M sodium acetate, pH 5.2). Hybridization was performed at 57–61°C overnight in hybridization buffer containing 50% formamid and a DIG-labelled RNA probe specific for the mRNAs of PaORF4, KlORF3 and DrORF5, respectively. Probes were prepared by amplifying the gene sequences using the primers (PaORF4_probe_for/PaORF4_probe_revT7, KlO3proberevT7/KlO3res1, DrO5_probe_rev_T7/DrO5_rs1, S3 Table) and labelled with the DIG RNA Labeling Kit (SP6/T7) (Roche Diagnostic GmbH, Mannheim, Germany) according to the manufacturer’s instructions. For detection a phosphatase-conjuncted anti-DIG antibody and a chemilumiscent alkaline phosphatase substrate CDPstar (Roche Diagnostic GmbH, Mannheim, Germany) were applied, and signals were visualized by exposure to X-Ray films.
cDNA synthesis
For cDNA synthesis the RevertAid H minus first strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany) was applied according to the manufacturer’s instructions. All primers used for cDNA synthesis are listed in S3 Table.
Isolation of total RNA
Total RNA was isolated as previously described (Klassen et al., 2008) or, alternatively, by making use of the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
mRNA detection by RT-PCR analysis
cDNA was synthesized using random hexamer primers and total RNA as template. 1 μl of cDNA was used as the template for PCR reactions applying the Phusion High fidelity DNA polymerase (Thermo Fisher Scientific) and primer combinations PaO4intr-fw/PaO4intr-rv (PaORF4), PaO4ms_for/PaO4ms_rev (PaORF4ms), KlO3_rs3/KlO3pf (KlORF3) and KlO3ms_rev/KlO3ms_for (KlORF3ms) (for primer binding positions see Fig 5C).
Identification of immRNA 3´ ends (3´ RACE and linker ligation)
Primer design for 3´ RACE experiments was based on the protocol of Scotto-Lavino et al., [40]. Polyadenylated RNA was transcribed to cDNA using the poly(A) complimentary primer QT22. The cDNA was used as template for PCR with primer Q0 (binds to a QT22 anchor) and an immORF specific primer at an annealing temperature of 58°C. PCR products were separated on an 1% agarose gel and extracted fragments were cloned into vector pSKpADH1 via NdeI and HindIII restriction sites. Isolated plasmids were analyzed by sequencing.
For linker ligation 2 μg of total RNA were mixed with 2 μg of a phosphorylated DNA oligo nucleotide (Oligo_5P_3ddC) and ligated using T4 RNA ligase (NEB, Frankfurt, Germany) for 1.5 h at 37°C. Column purified reactions were used for cDNA synthesis with Oligo_rev as primer. cDNA of the immunity gene was amplified using the primers Oligo_rev and KlO3rev_NdeI as and then cloned into pSKpADH1 via NdeI and EcoRV restriction sites. Cloned fragments were analyzed by sequencing.
Supporting Information
Zdroje
1. Worsham PL, Bolen PL (1990) Killer toxin production in Pichia acaciae is associated with linear DNA plasmids. Curr Genet 18 : 77–80. 2245477
2. Gunge N, Tamaru A, Ozawa F, Sakaguchi K (1981) Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J Bacteriol 145 : 382–390. 6257636
3. Jeske S, Meinhardt F (2006) Autonomous cytoplasmic linear plasmid pPac1-1 of Pichia acaciae: molecular structure and expression studies. Yeast 23 : 479–486. 16652393
4. Schaffrath R, Meinhardt F, editors (2005) Kluyveromyces lactis zymocin and other plasmid-encoded yeast killer toxins. Berlin: Springer. 337–352 p.
5. Jablonowski D, Fichtner L, Martin VJ, Klassen R, Meinhardt F, et al. (2001) Saccharomyces cerevisiae cell wall chitin, the Kluyveromyces lactis zymocin receptor. Yeast 18 : 1285–1299. 11571753
6. Mehlgarten C, Schaffrath R (2004) After chitin docking, toxicity of Kluyveromyces lactis zymocin requires Saccharomyces cerevisiae plasma membrane H+-ATPase. Cell Microbiol 6 : 569–580. 15104597
7. Zink S, Mehlgarten C, Kitamoto HK, Nagase J, Jablonowski D, et al. (2005) Mannosyl-diinositolphospho-ceramide, the major yeast plasma membrane sphingolipid, governs toxicity of Kluyveromyces lactis zymocin. Eukaryot Cell 4 : 879–889. 15879522
8. Chakravarty AK, Smith P, Jalan R, Shuman S (2014) Structure, mechanism, and specificity of a eukaryal tRNA restriction enzyme involved in self-nonself discrimination. Cell Rep 7 : 339–347. doi: 10.1016/j.celrep.2014.03.034 24726365
9. Klassen R, Paluszynski JP, Wemhoff S, Pfeiffer A, Fricke J, et al. (2008) The primary target of the killer toxin from Pichia acaciae is tRNA(Gln). Mol Microbiol 69 : 681–697. doi: 10.1111/j.1365-2958.2008.06319.x 18532979
10. Lu J, Huang B, Esberg A, Johansson MJ, Byström AS (2005) The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. Rna 11 : 1648–1654. 16244131
11. Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R (2006) tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 59 : 677–688. 16390459
12. Nandakumar J, Schwer B, Schaffrath R, Shuman S (2008) RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell 31 : 278–286. doi: 10.1016/j.molcel.2008.05.019 18657509
13. Meineke B, Schwer B, Schaffrath R, Shuman S (2011) Determinants of eukaryal cell killing by the bacterial ribotoxin PrrC. Nucleic Acids Res 39 : 687–700. doi: 10.1093/nar/gkq831 20855293
14. Meineke B, Kast A, Schwer B, Meinhardt F, Shuman S, et al. (2012) A fungal anticodon nuclease ribotoxin exploits a secondary cleavage site to evade tRNA repair. RNA 18 : 1716–1724. doi: 10.1261/rna.034132.112 22836353
15. Klassen R, Meineke B, Kast A, Wünsche G, Krampe L, et al. (2013) A secondary cleavage site in tRNA prevents anticodon nuclease toxin resistance via RNA repair. Yeast 30 : 230–230.
16. Paluszynski JP, Klassen R, Meinhardt F (2007) Pichia acaciae killer system: genetic analysis of toxin immunity. Appl Environ Microbiol 73 : 4373–4378. 17483256
17. Tokunaga M, Wada N, Hishinuma F (1987) Expression and identification of immunity determinants on linear DNA killer plasmids pGKL1 and pGKL2 in Kluyveromyces lactis. Nucleic Acids Res 15 : 1031–1046. 3029695
18. Satwika D, Klassen R, Meinhardt F (2012) Anticodon nuclease encoding virus-like elements in yeast. Appl Microbiol Biotechnol 96 : 345–356. doi: 10.1007/s00253-012-4349-9 22899498
19. Kast A, Klassen R, Meinhardt F (2014) rRNA fragmentation induced by a yeast killer toxin. Mol Microbiol 91 : 606–617. doi: 10.1111/mmi.12481 24308908
20. Klassen R, Kast A, Wünsche G, Paluszynski JP, Wemhoff S, et al. (2014) Immunity factors for two related tRNAGln targeting killer toxins distinguish cognate and non-cognate toxic subunits. Curr Genet 60 : 213–222. doi: 10.1007/s00294-014-0426-1 24719080
21. Klassen R, Meinhardt F (2007) Linear protein-promed replicating plasmids in eukaryotic microbes. In: Meinhardt F, Klassen R, editors. Microbiology monographs Microbial linear plasmids. Berlin: Springer. pp. 187–226.
22. Gunge N, Sakaguchi K (1981) Intergeneric transfer of deoxyribonucleic acid killer plasmids, pGKl1 and pGKl2, from Kluyveromyces lactis into Saccharomyces cerevisiae by cell fusion. J Bacteriol 147 : 155–160. 7016841
23. Proudfoot NJ (2011) Ending the message: poly(A) signals then and now. Genes Dev 25 : 1770–1782. doi: 10.1101/gad.17268411 21896654
24. Wilson DW, Meacock PA (1988) Extranuclear gene expression in yeast: evidence for a plasmid-encoded RNA polymerase of unique structure. Nucleic Acids Res 16 : 8097–8112. 3138657
25. Larsen M, Gunge N, Meinhardt F (1998) Kluyveromyces lactis killer plasmid pGKL2: evidence for a viral-like capping enzyme encoded by ORF3. Plasmid 40 : 243–246. 9806862
26. Tiggemann M, Jeske S, Larsen M, Meinhardt F (2001) Kluyveromyces lactis cytoplasmic plasmid pGKL2: heterologous expression of Orf3p and proof of guanylyltransferase and mRNA-triphosphatase activities. Yeast 18 : 815–825. 11427964
27. Meinhardt F, Wodara C, Larsen M, Schickel J (1994) A novel approach to express a heterologous gene on Kluyveromyces lactis linear killer plasmids: expression of the bacterial aph gene from a cytoplasmic promoter fragment without in-phase fusion to the plasmid open reading frame. Plasmid 32 : 318–327. 7899517
28. Schickel J, Helmig C, Meinhardt F (1996) Kluyveromyces lactis killer system: analysis of cytoplasmic promoters of the linear plasmids. Nucleic Acids Res 24 : 1879–1886. 8657569
29. Schaffrath R, Meinhardt F, Meacock PA (1996) Yeast killer plasmid pGKL2: molecular analysis of UCS5, a cytoplasmic promoter element essential for ORF5 gene function. Mol Gen Genet 250 : 286–294. 8602143
30. Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, et al. (2007) Colicin biology. Microbiol Mol Biol Rev 71 : 158–229. 17347522
31. Graber JH, Cantor CR, Mohr SC, Smith TF (1999) In silico detection of control signals: mRNA 3'-end-processing sequences in diverse species. Proc Natl Acad Sci U S A 96 : 14055–14060. 10570197
32. Frank AC, Wolfe KH (2009) Evolutionary capture of viral and plasmid DNA by yeast nuclear chromosomes. Eukaryot Cell 8 : 1521–1531. doi: 10.1128/EC.00110-09 19666779
33. Satwika D, Klassen R, Meinhardt F (2012) Repeated capture of a cytoplasmic linear plasmid by the host nucleus in Debaryomyces hansenii. Yeast 29 : 145–154. doi: 10.1002/yea.2893 22434608
34. Butler AR, Porter M, Stark MJ (1991) Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast 7 : 617–625. 1767590
35. Klassen R, Teichert S, Meinhardt F (2004) Novel yeast killer toxins provoke S-phase arrest and DNA damage checkpoint activation. Mol Microbiol 53 : 263–273. 15225320
36. Guglielmini J, Van Melderen L (2011) Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mob Genet Elements 1 : 283–290. 22545240
37. Louis VL, Despons L, Friedrich A, Martin T, Durrens P, et al. (2012) Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3 (Bethesda) 2 : 299–311. doi: 10.1534/g3.111.000745 22384408
38. Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11 : 355–360. 7785336
39. Klassen R, Krampe S, Meinhardt F (2007) Homologous recombination and the yKu70/80 complex exert opposite roles in resistance against the killer toxin from Pichia acaciae. DNA Repair (Amst) 6 : 1864–1875. 17765020
40. Scotto-Lavino E, Du G, Frohman MA (2006) 3' end cDNA amplification using classic RACE. Nat Protoc 1 : 2742–2745. 17406530
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



