-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
Histone variants, including histone H2A.Z, are incorporated into specific genomic sites and participate in transcription regulation. The role of H2A.Z at these sites remains poorly characterized. Our study investigates changes in the chromatin environment at the Cyclin D1 gene (CCND1) during transcriptional initiation in response to estradiol in estrogen receptor positive mammary tumour cells. We show that H2A.Z is present at the transcription start-site and downstream enhancer sequences of CCND1 when the gene is poorly transcribed. Stimulation of CCND1 expression required release of H2A.Z concomitantly from both these DNA elements. The AAA+ family members TIP48/reptin and the histone variant H2A.Z are required to remodel the chromatin environment at CCND1 as a prerequisite for binding of the estrogen receptor (ERα) in the presence of hormone. TIP48 promotes acetylation and exchange of H2A.Z, which triggers a dissociation of the CCND1 3′ enhancer from the promoter, thereby releasing a repressive intragenic loop. This release then enables the estrogen receptor to bind to the CCND1 promoter. Our findings provide new insight into the priming of chromatin required for transcription factor access to their target sequence. Dynamic release of gene loops could be a rapid means to remodel chromatin and to stimulate transcription in response to hormones.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003387
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003387Summary
Histone variants, including histone H2A.Z, are incorporated into specific genomic sites and participate in transcription regulation. The role of H2A.Z at these sites remains poorly characterized. Our study investigates changes in the chromatin environment at the Cyclin D1 gene (CCND1) during transcriptional initiation in response to estradiol in estrogen receptor positive mammary tumour cells. We show that H2A.Z is present at the transcription start-site and downstream enhancer sequences of CCND1 when the gene is poorly transcribed. Stimulation of CCND1 expression required release of H2A.Z concomitantly from both these DNA elements. The AAA+ family members TIP48/reptin and the histone variant H2A.Z are required to remodel the chromatin environment at CCND1 as a prerequisite for binding of the estrogen receptor (ERα) in the presence of hormone. TIP48 promotes acetylation and exchange of H2A.Z, which triggers a dissociation of the CCND1 3′ enhancer from the promoter, thereby releasing a repressive intragenic loop. This release then enables the estrogen receptor to bind to the CCND1 promoter. Our findings provide new insight into the priming of chromatin required for transcription factor access to their target sequence. Dynamic release of gene loops could be a rapid means to remodel chromatin and to stimulate transcription in response to hormones.
Introduction
Transcription activation relies on a choreography of local chromatin remodeling events that include posttranslational histone modifications and replacement of canonical histones by variants [1]–[4]. Chromatin immunoprecipitation (ChIP) studies have provided extensive information on the recruitment of these complexes by the hormone bound estrogen receptor in ERα-positive breast cancer cells [5]. The first complex to occupy promoter sequences is the ATP-dependent SWI/SNF chromatin remodeling complex and its catalytic subunit Brg1. Its activity enables subsequent binding of a plethora of histone and protein modifying assemblies which lead to transcription initiation by polymerase II [6]–[10]. In contrast, it is less clear how local chromatin structure prepares for rapid and massive recruitment of the estrogen receptor itself in the presence of estrogen.
Incorporation of histone variants constitutes a means to alter nucleosome properties and positioning at specific genomic loci. The histone H2A variant H2A.Z is frequently found within nucleosomes at regulatory sequences [11]–[14]. In particular, H2A.Z occupancy characterizes inducible and constitutive DNAseI hypersensitive sites to which nuclear receptors bind [15]. This variant is believed to induce a chromatin conformation that poises genes for transcription in human cells [16]. In yeast, H2A.Z exchange is mediated by the SWR1 complex [17], [18]. However, in mammalian cells, the mechanisms of H2A.Z deposition are still poorly characterized and may require several distinct protein complexes depending on the cellular context (for a review see [19]). Among ATP-dependent chromatin remodeling complexes the TIP48/TIP49 containing SWR1/SRCAP [20] or TIP60/p400 [21], [22] complexes have been shown to play a role in H2A.Z deposition [17], [18], [23]. p400 was also reported to be required for H2A.Z incorporation into the TFF1/pS2 gene concomitant to estrogen receptor binding [24]. In an in vitro study Choi et al. demonstrated that the AAA+ family (ATPases Associated with various cellular Activities) members TIP48/TIP49 participate in the replacement of H2A by H2A.Z [25]. This H2A.Z exchange was facilitated by TIP60-mediated H2A acetylation. TIP48/TIP49 proteins (also known as TIP49b and TIP49a, Rvb2 and Rvb1, reptin and pontin) are important for assembly and activity of the histone TIP60 acetyltransferase complex [26].
To gain a better understanding of the early steps required in estrogen receptor mediated transcription activation and the coordination between remodeling complexes and chromatin structure, we analyzed transcription of the cyclin D1 gene (CCND1) in ERα-positive MCF-7 breast cancer cells. This oncogene is frequently overexpressed in human breast tumors [27]. Its down-regulation increases migratory capacity and is linked to unfavorable prognosis [28], [29]. Cyclin D1 is a mitogenic sensor that modulates cell cycle progression. CCND1 transcription is stimulated by 17β-estradiol (E2), inhibited by antiestrogens and cell cycle regulated in ERα-positive breast cancer cells [30], [31].
Here we show that TIP48 and H2A.Z associate with CCND1 promoter and enhancer sequences. TIP48 is required for chromatin reorganization which is initiated by release of H2A.Z and opening of a repressive promoter-enhancer gene loop enabling TIP60 and the E2 bound estrogen receptor to be loaded to stimulate CCND1 transcription.
Results
TIP48 and H2A.Z association with CCND1 regulatory sites is required for activation by estradiol
In ERα-positive MCF-7 cells grown in steroid stripped media only basal transcription levels of the Cyclin D1 gene (CCND1) were measured. Addition of E2 lead to a 2.5-fold increase in CCND1 mRNA levels in cells treated 6 h with 100 nM E2 (Figure 1A). H2A.Z has been reported to act in concert with ER to regulate the TFF1 gene [24] prompting us to examine H2A.Z association with CCND1 regulatory elements. Analysis of ChIP-on-chip data revealed that H2A.Z was highly enriched at sequences 5′ and 3′ flanking the CCND1 gene, and largely absent from the open reading frame (Figure 1B) [32]. By conventional ChIP, we found that the amount of H2A.Z present at the CCND1 promoter was reduced by 50% at the TSS (Figure 1C). Eeckhoute et al. identified an enhancer (enh2) at the 3′ end of the CCND1 gene which acts as the primary site for ERα and cofactor binding during CCND1 transcriptional regulation [7]. We thus analyzed the chromatin organization of enh2. Similar to the promoter, H2A.Z present at enh2 was removed during transcription activation (Figure 1C). Reduced binding was not due to a decrease in H2AFZ expression in the presence of E2 (Figure 1A). Chromatin modifications at the CCND1 promoter and enhancer appear to be coordinated. Removal of H2A.Z from promoter sequences upon transcription activation correlates with observations in yeast and several mammalian cells and points to a mechanism of regulation distinct from the one of the ERα target gene TFF1 [33], [24], [34]. Replacement of nucleosomal H2A with H2A.Z has been shown to be catalyzed by the TIP48/49 complex in vitro [25]. The TIP48/49 complex was thus a good candidate for regulating H2A.Z dynamics at CCND1 regulatory sequences. TIP48 and TIP49 are ubiquitously expressed and are often part of the same complex. In most cell types, and in particular in epithelial cancer cells such as MCF-7 cells, silencing one of the partners by interference RNA lead to degradation of the other partner [33]. Better antibody specificity and efficiency prompted us to investigate TIP48. TIP48 was associated with the promoter and enh2 of CCND1 in non-induced ERα-positive MCF-7 cells (Figure 1D). Binding of TIP48 to the CCND1 TSS and enh2 decreased rapidly following addition of 100 nM estradiol (E2). Expression of the gene coding for TIP48 was insensitive to E2 (Figure S1A). To examine the relationship between TIP48 and H2A.Z, we selectively depleted TIP48 by siRNA (Figure S1A). In siTIP48 transfected cells treated or not with E2, H2A.Z binding to CCND1 was reduced compared to control, non–specific siRNA transfected cells. Levels of H2A.Z binding in the absence of TIP48 were roughly equivalent to levels in E2 treated control cells (Figure 2A). Moreover, nucleosome density assessed by immunoprecipitating histone H3 was unchanged near the TSS (Figure 2B). Thus, eviction of H2A.Z upon initiation of E2 stimulated transcription was not due to general chromatin decondensation around the CCND1 gene and its promoter region in particular. TIP48 appears to be necessary for recruiting H2A.Z to the CCND1 gene in MCF-7 mammary tumor cells.
Fig. 1. TIP48 and H2A.Z are required for CCND1 activation by estradiol. 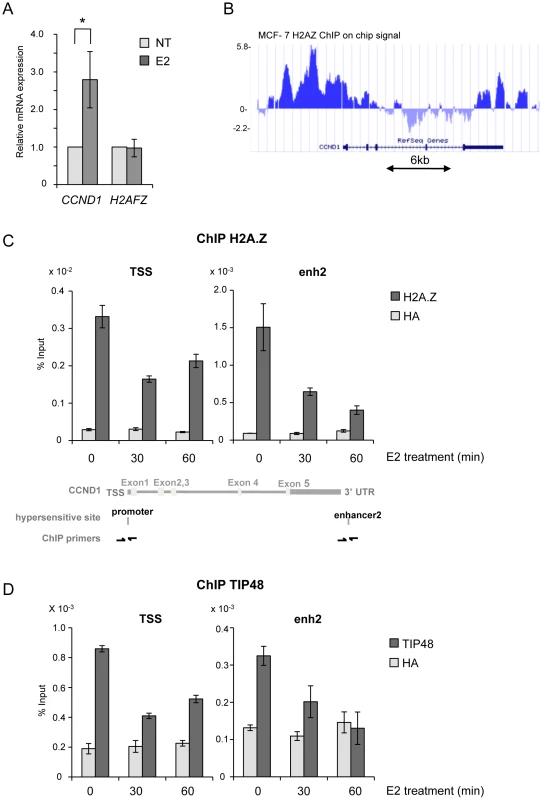
A) MCF-7 cells were cultivated 3 days in steroid stripped (white) medium and then induced by E2 10−7 M for 6 h. CCND1, H2AFZ mRNA was quantified by real-time RT-PCR. The mean and SD from three independent experiments are shown. (*) indicates a p value<0.05 (Student t-test). B) MCF-7 H2A.Z ChIP on chip signal detected around the CCND1 gene on human chromosome 11 loaded from [32] on UCSC. C, D) H2A.Z (C) and TIP48 (D) occupancy at CCND1 TSS and enh2 before and after 30 min and 60 min of E2 10−7 M treatment analysed by ChIP. The values of ChIP efficiencies are given in % of input with s.e.m indicated (n = 3). Positions of DNAseI hypersensitive sites and primers used for ChIP are indicated in C). Fig. 2. TIP48 regulates estrogen-activated transcription via H2A.Z release. 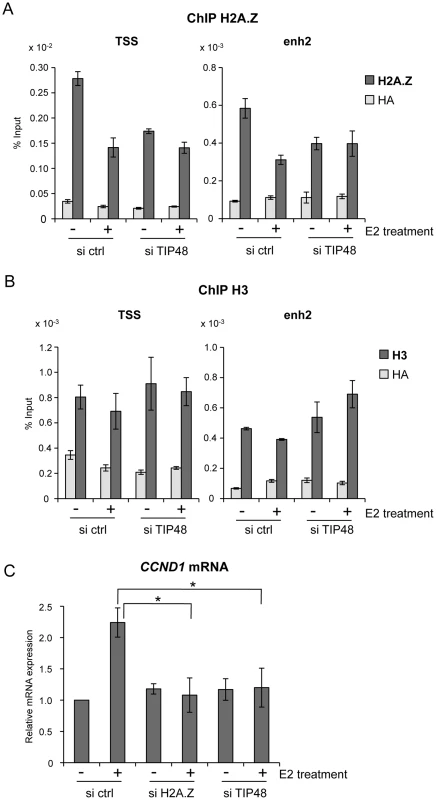
A, B) MCF-7 cells were cultivated 3 days in steroid free medium, transfected with, scramble (ctrl) or TIP48 siRNA for 72 h and induced by E2 10−7 M for 30 min. Association of H2A.Z (A) and histone H3 (B) with TSS and enh2 sites was analyzed by ChIP and shown as % input (n = 2). C) MCF-7 cells were cultivated 3 days in steroid free medium and transfected at day 0 with a Scramble (Ctrl), a H2A.Z siRNA or a TIP48 siRNA for 72 h and then induced by E2 10−7 M for 6 h. CCND1 mRNA levels were determined by qRT-PCR. Control (Ctrl) without E2 treatment was set to 1. The mean and SD from three independent experiments are shown. (*) indicates a p value<0.05. We thus asked whether binding of H2A.Z or its release were important for regulating CCND1 transcription. H2A.Z mRNA expression levels were reduced ∼5-fold 48 h post transfection with a smartpool siRNA directed against H2A.Z compared to control cells (Figure S1B). Reduced levels of H2A.Z did not alter basal CCND1 expression levels, but impeded activation by E2 (Figure 2C). Similarly, in the absence of TIP48, basal transcription levels were conserved, while activation of CCND1 by E2 was compromised (Figure 2C). H2AFZ mRNA levels were not affected by selective knockdown of TIP48 (Figure S2A). Thus, Stimulation of CCND1 expression required release of H2A.Z concomitantly from both these DNA elements.
TIP48 promotes ERα binding during E2 activated transcription
Absence of activation was likely due to failure of ERα fixation to the CCND1 promoter. Under standard conditions, E2 stimulated ERα binding to both the promoter and enh2 of CCND1 (Figure 3A) [7]. Selective knock down of TIP48 hindered ERα binding to these sites (Figure 3A). Reduced binding could not be attributed to altered or decreased ESR1 expression patterns in cells transfected with control or TIP48 siRNAs (Figure 3B). Therefore, TIP48 appears to be necessary to remodel CCND1 chromatin structure for productive ERα binding in the presence of hormone.
Fig. 3. TIP48 is required for ERα binding during E2 activated transcription. 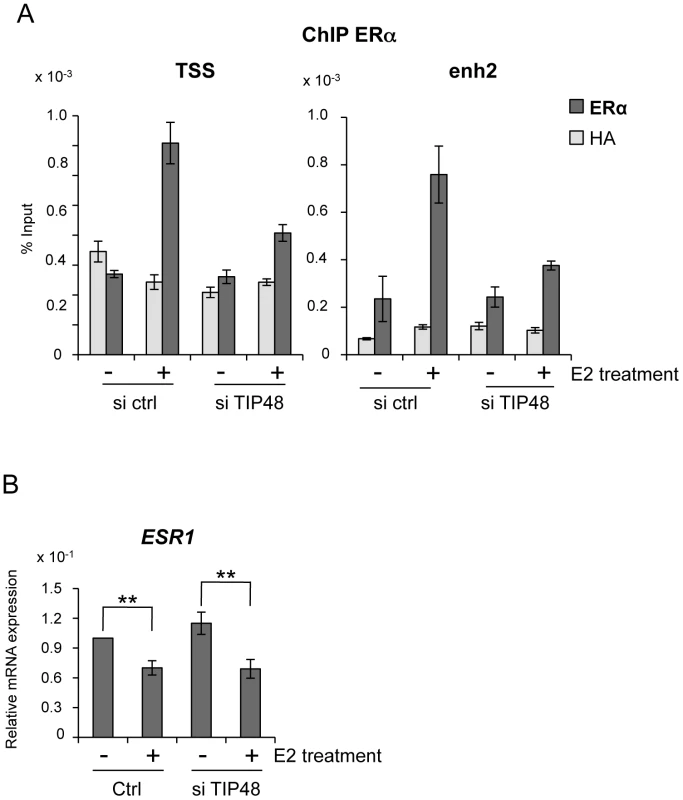
MCF-7 cells were cultivated 3 days in steroid free medium, transfected with scramble (ctrl) or TIP48 siRNA for 72 h and induced by E2 10−7 M for 30 min. A) ChIP analysis of ERα occupancy at the CCND1 TSS and enh2 sites. Results are shown as % input (n = 2). B) qRT-PCR quantification of ESR1 mRNA expression relative to RPLO. The mean and SD from three independent experiments are shown. (**) indicates a p value<0.01. TIP48 promotes recruitment of TIP60 to CCND1
TIP48 and TIP60 have been found as part of the same complex [20], [25]. TIP60 also cooperates with ERα and other chromatin-remodeling enzymes during estrogen-induced transcription [34], [35]. We tested whether TIP48 and TIP60 binding to the CCND1 promoter was coordinated. In the presence of E2, TIP60 was recruited to the CCND1 promoter (Figure 4A). Upon depletion of TIP48, TIP60 no longer associated with the CCND1 TSS (Figure 4A). Cooperation between TIP48, ERα and TIP60 binding was likely to be necessary for transcription activation. To unravel a functional link, we first over-expressed TIP60 in MCF-7 cells (Figure S2B). TIP60 overexpression stimulated E2 activated CCND1 transcription nearly 5-fold compared to control untreated cells, without affecting neither basal, non-induced mRNA levels (Figure 4B) nor the expression pattern of the H2AFZ gene (Figure S2C). In siTIP48 transfected cells, overexpression of TIP60 was no longer able to stimulate CCND1 transcription upon E2 stimulation (Figure 4C), suggesting that TIP48 is required for TIP60 function.
Fig. 4. TIP48 promotes TIP60 binding and CCND1 transcription activation. 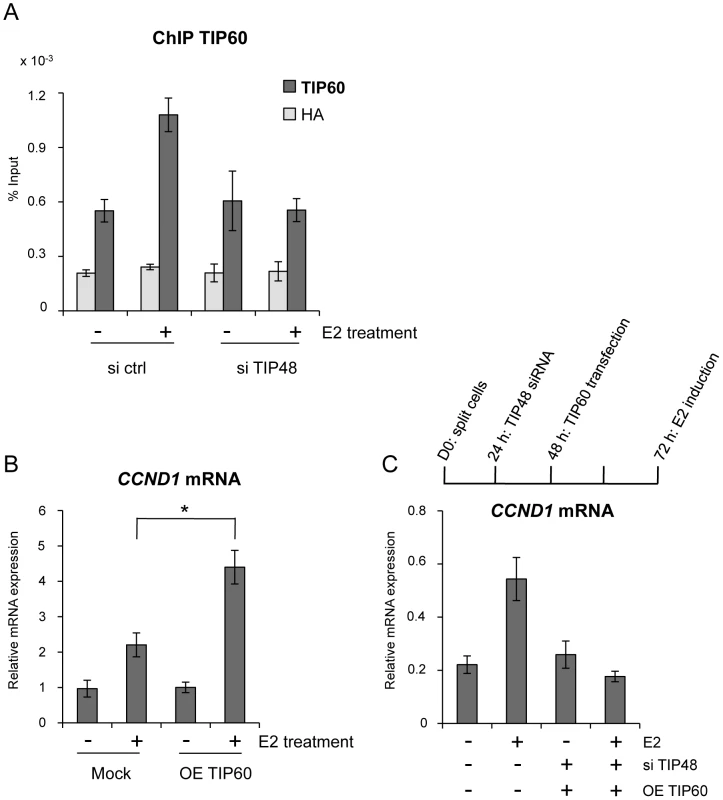
MCF-7 cells were cultivated 3 days in steroid free medium and induced by E2 10−7 M for 6 h. A) Cells were transfected with scramble (-) or TIP48 siRNA for 72 h and induced by E2 10−7 M for 30 min. TIP60 binding to the CCND1 promoter was analyzed by ChIP and shown as percent of input (n = 2). B) After 24 h in steroid free medium, MCF-7 cells were transfected with a mock vector or a vector expressing TIP60 for 48 h. CCND1 mRNA expression was analyzed by qRT-PCR. The mean and SD from three independent experiments are shown. (*) indicates a p value<0.05. C) MCF-7 cells were first transfected with scramble (-) or TIP48 siRNA as in (A). After 24 h, same cells were transfected with a mock vector or a vector expressing TIP60 for 48 h as in (B). CCND1 gene expression was analyzed by qRT-PCR after 30 min of E2 10−7 M induction. TIP60 is found in protein complexes able to acetylate histones, with a preference for lysine 5 of H2A [36]. Core histones are generally acetylated in the promoter region of transcribed genes. Acetylation of the histone variant H2A.Z was shown to characterize active genes in yeast and recently also in prostate cancer cells [13], [37]. Using an antibody that specifically recognizes H2A.Z acetylated at 3 N-terminal lysines, we determined that a large fraction of H2A.Z bound to the CCND1 promoter and to the 3′ enh2, was highly acetylated (Figure 5A). Acetylation levels of H2A.Z did not vary following E2 induced CCND1 gene activation in control samples (Figure 5A). However, because H2A.Z was released during transcription activation, the ratio of acetylated H2A.Z/total H2A.Z increased nearly 2-fold at these sites (Figure 5B). In siTIP48 transfected cells, we observed a decrease in acetylated H2A.Z present at the TSS and the enh2 (Figure 5A). The increased ratio of acetylated H2A.Z associated with the CCND1 gene following E2 was abolished in cells transfected with siTIP48 (Figure 5B). The reduced ratio of H2A.Z acetylation thus correlated with impeded transcription activation in siTIP48 transfected MCF-7 cells (Figure 5B and Figure 2C).
Fig. 5. TIP48 promotes acetylation of H2A.Z. 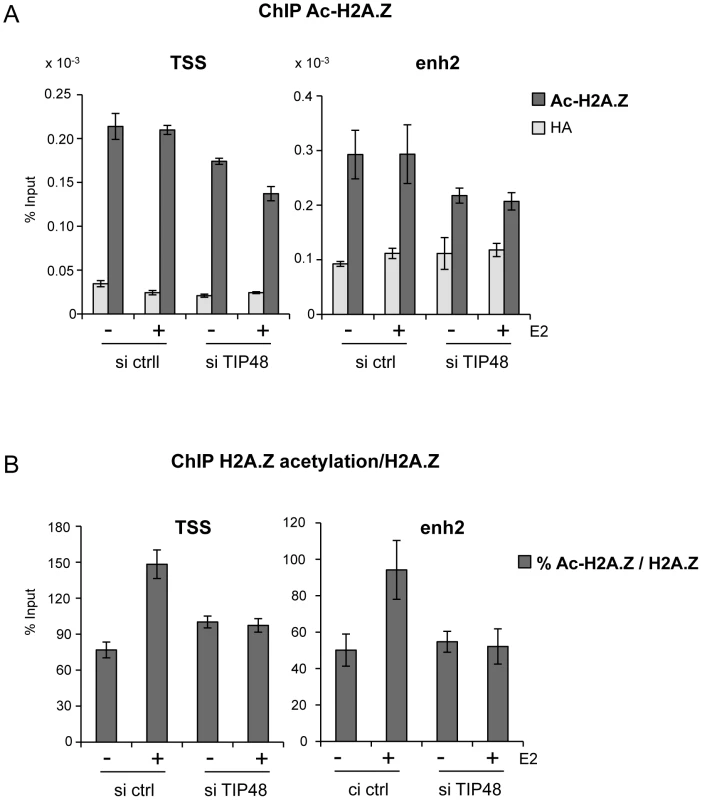
MCF-7 cells were cultivated and transfected as in Figure 2. A) Association of acetyl H2A.Z with CCND1 TSS and enh2 sequences analyzed by ChIP and shown as % input (n = 2). B) Ratio of acetylated versus total H2A.Z in %. In conclusion, failure of TIP60 to associate with CCND1 in the absence of TIP48 correlated with reduced binding of ERα (Figure 3), reduced levels of H2A.Z acetylation at the CCND1 gene (Figure 5) and the inability to activate this gene by estrogen (Figure 2C).
TIP48 modulates CCND1 chromatin structure by controlling gene looping
Long-range chromatin interactions between ERa recognition sequences and enhancers have been proposed to regulate ERa-target genes in breast cancer cells [38], [39]. The main enhancer regulating CCND1 is located at the 3′ end of the gene, 14 kb distant from the promoter [7]. Gene looping via promoter-enhancer crosstalk is associated with repressed, low CCND1 expression in ERa-negative, MDA-MB231 cells [40]. Thus we asked whether this loop also existed in MCF-7 cells and more importantly, whether looping was sensitive to hormone.
We used a chromatin conformation capture (3C) assay. The 3C method detects physical proximity between distal DNA sites by ligation of cross-linked restricted DNA fragments [41], [42]. Ligation products between enh2 and promoter, and between enh2 and a control fragment inside the CCND1 ORF were amplified and normalized to an amplified enh2 PCR product (see Materials and Methods) (Figure 6A). We measured significant interaction frequencies between enh2 and promoter sequences in MCF-7 cells grown in hormone-stripped media (-E2). Interaction frequencies were reduced ∼10-fold 45 min after addition of E2 to the cell culture (Figure 6B). No significant amplification of ligation products between enh2 and the internal control fragment was detectable. Hence, an extragenic loop mediated by specific promoter enhancer interactions was present when CCND1 expression is low (Figure 6B and Figure 2D). Upon transcription activation, gene looping is markedly reduced.
Fig. 6. Release of CCND1 gene looping requires TIP48. 
A) Schematic representation of the CCND1 locus on h.s. chromosome 11 (adapted from the UCSS genome browser http://genome.ucsc.edu/cgi-bin/hgGateway). DNAseI hypersensitive sites [7], Csp6I restriction sites and fragments assayed by 3C are represented. B) Quantitative PCR amplification of ligation events in a 3C assay in MCF-7 cells treated or not with E2 10−7 M for 45 min and transfected 72 h before with scramble (ctrl) or TIP48 siRNA. Crosslinked DNA was digested with the Csp6I restriction enzyme and ligated. qRT-PCR was performed with primers designed for the 4 possible ligation events (only one is shown). It was tempting to speculate that TIP48 plays a role in regulating looping. We assessed the relative frequencies of interaction between enh2/promoter and enh2/internal control fragments in MCF-7 cells transfected or not by siTIP48. Depletion of TIP48 had no impact on enh2/promoter contacts in the absence of E2 (Figure 6B). This observation correlated with identical basal expression levels of CCND1 in control and siTIP48 transfected cells (Figure 2C).
45 min after addition of E2 to the cells, the frequency of enh2/promoter interaction was 5-fold greater in siTIP48 transfected cells compared to control cells (Figure 6B). Conservation of significant repressive gene looping could thus account for impeded E2 bound ERa binding to the CCND1 promoter and compromised transcription activation. We propose a model (Figure 7) in which TIP48 is required at early steps during transcription activation which is initiated by release of H2A.Z and subsequent dissociation of the enhancer from the promoter. E2 bound estrogen receptor can then recognize the promoter and stimulate transcription of CCND1.
Fig. 7. Model for the role of TIP48, H2A.Z, and TIP60 in initiation of transcription activation of <i>CCND1</i> via promoter enhancer crosstalk. 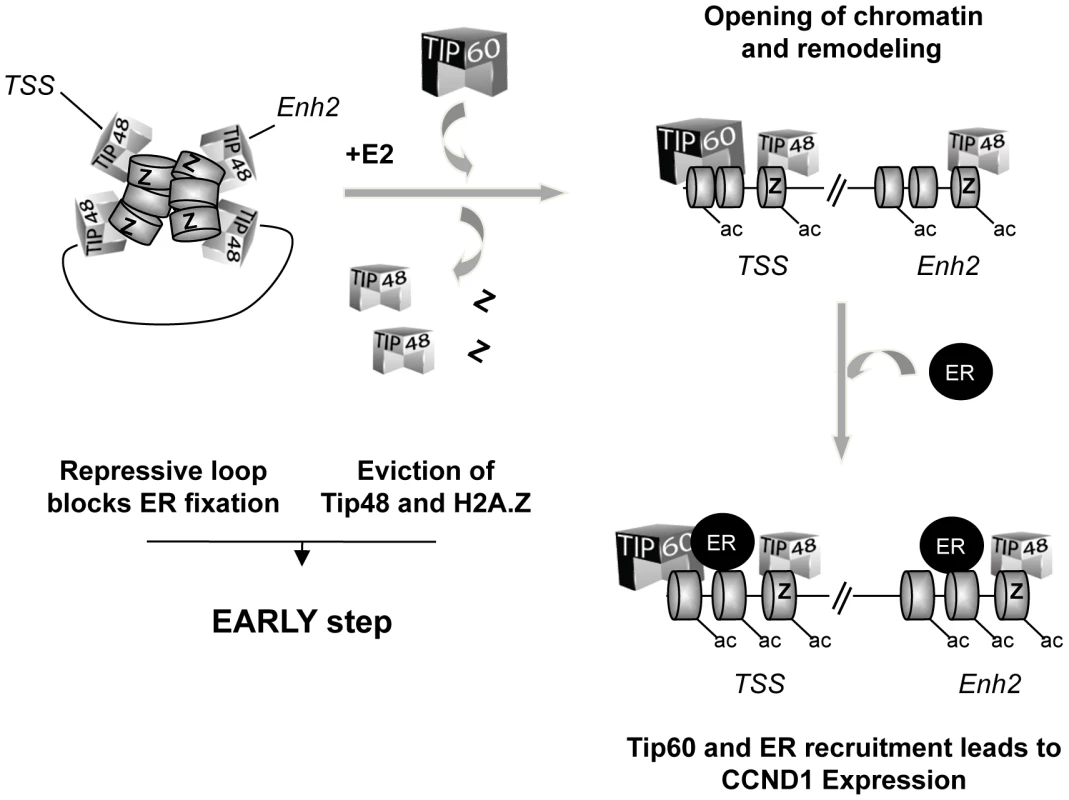
Discussion
We unraveled a role for TIP48 in initiating transcription activation of the CCND1 oncogene. Recruitment of the histone acetyltransferase TIP60 is dependent on TIP48 and H2A.Z binding to the promoter and 3′ enhancer of the CCND1 gene. We propose that low levels of CCND1 expression are regulated because the associated gene loop is transcription-dependent. This regulation is brought about by the activity of TIP48 containing complexes which locally act upon chromatin structure to release a disabling loop. Such a mechanism allows fine-tuning transcription regulation of genes pivotal for the cellular equilibrium in rapidly changing environments.
Our work describes early events implicated in E2 induction of CCND1. These events include dynamic exchange of a series of cofactors, namely the TIP48 complex and histone variant H2A.Z, recruitment of TIP60 and acetylation of H2A.Z enabling the main transcription factor, the estrogen receptor, to associate with its target sequences. TIP60 can directly interact with ERa and its acetyltransferase activity is important during transcription initiation once ERa is bound to target gene promoters [34].
TIP60 is a versatile enzyme that functions with a variety of partners in a gene and cell specific manner [34], [43]. Selective knock-down of TIP60 by siRNA compromises activation of some, but not all ERa target genes in MCF-7 cells, as well as nuclear receptor independent genes in several cell lines (unpublished observations). CCND1 was one of the genes found to be insensitive to siTIP60 [34]. This observation denotes that TIP60 can be replaced by other histone acetyltransferases in CCND1 transcription activation. Thus, dependency of early chromatin remodeling steps on TIP48 and H2A.Z may be more generally applicable to allow cofactor recruitment for productive ERa binding in stimulated transcription.
We found that H2A.Z was removed from CCND1 regulatory elements while this variant had previously been shown to be recruited to the promoter of the TFF1 gene upon E2 treatment of MCF-7 cells [24]. Differences in promoter structure are a plausible explanation for divergent remodeling mechanisms. It is also likely that post-translational modifications of H2A.Z are important as shown in a recent genome wide study by Valdes-Mora et al. who found that H2A.Z acetylation at the TSS correlates with active transcription in prostate cancer cells [13]. Indeed, the level of acetylation of H2A.Z near the TSS of CCND1 was equivalent at non-activated and E2 stimulated cells. We propose that, in ERα-positive breast cancer cells, the ratio of acetylated H2A.Z/H2A.Z rather than the total amount of H2A.Z bound to the CCND1 promoter correlates with transcriptional activity.
Chromatin remodeling events are crucial for hormone stimulated activation of estrogen receptor target genes. However, so far, all data available describe the recruitment of remodeling complexes and cofactors once the estrogen receptor is bound. The Brg1 subunit of the SWI/SNF complex is one of the first proteins to associate with ERa and, although transcription is no longer activated in its absence, ERa remains bound in siBrg1 transfected cells [34]. Here we demonstrate that chromatin remodeling events prior to ERa binding are essential for initiating transcription. These events depend on TIP48 and H2A.Z specific nucleosome conformation. Chromatin structure impedes ERa loading via intragenic looping. Notably, interaction between promoter and enhancer sequences forms a repressive complex. Reduced distances between 5′ and 3′ ends of gene loci have been attributed to greater chromatin density. In this case, looping does not require changes in chromatin compaction. Dynamic release of gene loops is consistent with rapid chromatin remodeling and transcription activation by hormone.
Finally, addition of hormone triggers large scale chromatin remodeling. In breast cancer cells gene response to progestin is mediated by nucleosomes [44] and estradiol treatment leads to expansion of chromosome territories within minutes [45]. This latter phenomenon was also observed in ERα-negative cells (unpublished) suggesting that chromatin decondensation is independent of the receptor and may prepare its binding in ERα-positive cells. It is thus tempting to speculate that the signaling mechanism by which hormone addition primes chromatin triggers histone exchange and remodeling prior to ERα binding.
Materials and Methods
Cell lines, transfection, and Western blotting
MCF-7 cells were purchased from ATCC and were maintained in DMEM/F12 without phenol red with Glutamax containing 50 mg/ml gentamicin, 1 mM sodium pyruvate and 10% heat-inactivated and steroid free fetal calf serum (FCS) (Invitrogen). MCF-7 cells were treated with 10−7 M estrogen E2 (Sigma) for the indicated times. 5×106 MCF-7 cells were transfected with 20 nM of H2A.Z siRNA ON-TARGET plus SMARTpool, TIP48 siRNA ON-TARGET plus SMARTpool or scrambled (scr) siRNA (Dharmacon Thermo Scientific) using Interferine (Ozyme). Cells were mock-transfected (pcDNA3.1) or transfected with 1 µg of pcDNA3.1/TIP60 (gift from Dr. Didier Trouche) using the Amaxa Cell line Nucleofactor Kit V program P-020 according to the manufacturer's protocol. TIP60 siRNA [43] was purchased from Eurogentec, and transfected using Interferine (Ozyme). 5×105 MCF-7 cells were seeded in 6 well plates. 72 h following siRNA transfection, total cell extracts were isolated and protein levels of H2A.Z, TIP48 and TIP60 analyzed by immunoblotting on gel SDS-page 15% using antibodies against H2A.Z (ABCAM, ab4174), TIP48 (gift of Dr. Mikhaïl Grigoriev) TIP60 [43] or GAPDH (Millipore, mab374).
RNA analysis
Total RNA was extracted using an RNeasy mini-kit (Qiagen) and eluted with 35 µl of RNAase-free water. First strand cDNA was generated using 2 µg of total RNA in a reaction containing random oligonucleotides as primers with the ThermoScript RT-PCR system (Invitrogen). Real-time PCR was performed on a Mastercycler ep realplex 4 (Eppendorf) using the platinum SYBR Green q-PCR SuperMix (Invitrogen) according to the manufacturer's instructions. Amplification conditions: 1 min at 50°C, 3 min at 95°C followed by 40 cycles (20 s at 95°C, 20 s at 60°C, 20 s at 72°C). mRNA expression were normalized against expression levels of the RPLP0 ribosomal gene used as an internal control. qRT-PCR primers: H2AFZ: 5′-CCTTTTCTCTGCCTTGCTTG-3′ and 5′-CGGTGAGGTACTCCAGGATG-3′, CCND1: 5′-GCGTCCATGCGGAAGATC-3′ and 5′-ATGGCCAGCGGGAAGAC-3′, RPLP0: 5′-TGGCAGCATCTACAACCCTGAA-3′ and 5′ - CACTGGCAACATTG CGGACA-3′, TIP48 : 5′-TGAAGAGCACTACGAAGACGC-3′ and 5′-CCTTACTACCCAGCTC CTGA - 3′.
ChIP assays
ChIP analyses were performed as described previously [46]. Samples were sonicated to generate DNA fragments <500 bp. Chromatin fragments were immunoprecipitated using antibodies against H2A.Z (ab4174, ABCAM), acetyl H2A.Z (ab18262, ABCAM), TIP48 (gift of Dr. Mikhaïl Grigoriev), ERα (sc-543, Santa Cruz), H3 (ab1791, ABCAM), TIP60 [47] or an irrelevant HA antibody (H6908, Sigma). The precipitated DNA was amplified by real-time PCR, with primer sets designed to amplify the promoter (TSS) and enh2 enhancer regions of the CCND1 gene (Figure 1B). qRT-PCR primers: CCND1 (TSS): 5′-CGGGCTTTGATCTTTGCTTA-3′ and 5′-ACTCTGCTGCTCGCTGCTAC-3′, distal CCND1 enhancer (enh2): 5′-CAGTTTGTCTTCCCGGGTTA-3′ and 5′ - CATCCAGAGCAAACAGCAG-3′. All ChIP data are shown as percent input.
3C assays
3C assays were performed essentially as described [48], [49], with minor modifications. MCF-7 cells were treated with E2 10−7 M for 45 mn or transfected with a scrambled control siRNA, with TIP48 SMARTpool siRNA (Dharmacon Thermo Scientific), and cultured in phenol red-free DMEM containing 10% FBS-T for 72 h before cross-linking. The culture medium was removed, and cells were fixed with 1.5% formaldehyde for 10 min at room temperature. Cells were then washed twice with cold phosphate-buffered saline solution, and resuspended in ice-cold lysis buffer (10 mm Tris-HCl, pH 8.0, 10 mm NaCl, 0.2% Nonidet P-40, and protease inhibitor mixture). Nuclei were resuspended in 1 ml of Buffer B 1.2× buffer (MBI Fermentas) supplemented with SDS 0,3%. Triton X-100 1,8% was added to sequester the SDS and incubated for 1 h at 37°C. The cross-linked DNA was digested overnight with 400 units of restriction enzyme Csp6I (MBI Fermentas). The restriction enzyme was inactivated by incubation at 65°C for 20 min. The reactions were diluted with ligase buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 10 mm dithiothreitol, 1 mm ATP, and 25 µg/ml bovine serum albumin), supplemented with Triton X-100 (1% final concentration). The DNA was ligated using T4 DNA ligase (New England Biolabs, Ipswich, MA) overnight at 16°C and an additional 100 units for 2 h at 37°C. RNase was added for 30 min at 37°C, and samples were incubated with SDS overnight at 70°C to reverse the crosslink. The following day, samples were incubated for 2 h at 45°C with proteinase K, and the DNA was purified by phenol-chloroform extractions and ethanol precipitation. Interaction between chromatin domains was assessed by real-time-PCR amplification for each predicted ligation event [48], [50]. Primers have been designed on the digested BAC fragments, directly around the putative site of ligation for the four possibilities. BAC clones RP11-300ID (BACPAC Resources Center at Childrens Hospital Oakland Research Institute, Oakland, CA) containing the CCND1 gene and downstream 160-kb region were used. 40 ug of BAC was digested by Csp6I overnight and ligated. This product was purified by phenol chloroform and precipitated in order to generate 3C control templates. PCR primer efficiency was measured by amplifying 0.01 to 50 ng of digested BAC product and also tested on a fixed amount (50 ng) of digested genomic DNA. All primers have an annealing temperature between 65 to 70°C and a product size around 150–300 bp. All primer combinations showed PCR efficiency between 90 and 100%. 3C assay results are presented as the average from three independent preparations of 3C DNA, followed by qPCR analysis in triplicate. qPCR for enh2 (PCR primers design inside the Csp6I restriction fragment enh2) was used as an internal control to verify ligation events. Non-digested sample and ligation between a control fragment and enh2 were also performed (data not shown). Primers used for one of the four ligation event tested: Enh2/Prom: 5′-CTGGGAGAGATGGAGCTGAG-3′ and 5′-GGTTTTGTTGGGGGTGTAGA-3′, Enh2/ctrl: 5′-AAGCTCTCCCACAACCCATT-3′ and 5′-GTCAGCCCCACTGTTGACTC-3′. Other primers available upon request.
Supporting Information
Zdroje
1. BhaumikSR, SmithE, ShilatifardA (2007) Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14 : 1008–1016.
2. DalvaiM, BystrickyK (2010) The role of histone modifications and variants in regulating gene expression in breast cancer. J Mammary Gland Biol Neoplasia 15 : 19–33.
3. HenikoffS, AhmadK (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21 : 133–153.
4. StrahlBD, AllisCD (2000) The language of covalent histone modifications. Nature 403 : 41–45.
5. VinckeviciusA, ChakravartiD (2012) Chromatin immunoprecipitation: advancing analysis of nuclear hormone signaling. J Mol Endocrinol 49: R113–123.
6. McKennaNJ, LanzRB, O'MalleyBW (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20 : 321–344.
7. EeckhouteJ, CarrollJS, GeistlingerTR, Torres-ArzayusMI, BrownM (2006) A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 20 : 2513–2526.
8. MetivierR, PenotG, HubnerMR, ReidG, BrandH, et al. (2003) Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115 : 751–763.
9. MetivierR, StarkA, FlouriotG, HubnerMR, BrandH, et al. (2002) A dynamic structural model for estrogen receptor-alpha activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol Cell 10 : 1019–1032.
10. ShangY, HuX, DiRenzoJ, LazarMA, BrownM (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103 : 843–852.
11. GuillemetteB, BatailleAR, GevryN, AdamM, BlanchetteM, et al. (2005) Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3: e384.
12. LiB, PattendenSG, LeeD, GutierrezJ, ChenJ, et al. (2005) Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A 102 : 18385–18390.
13. Valdes-MoraF, SongJZ, StathamAL, StrbenacD, RobinsonMD, et al. (2011) Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res 22 : 307–321.
14. ZhangH, RobertsDN, CairnsBR (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123 : 219–231.
15. JohnS, SaboPJ, JohnsonTA, SungMH, BiddieSC, et al. (2008) Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29 : 611–624.
16. FarrisSD, RubioED, MoonJJ, GombertWM, NelsonBH, et al. (2005) Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J Biol Chem 280 : 25298–25303.
17. MorrisonAJ, ShenX (2009) Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10 : 373–384.
18. ZhangH, RichardsonDO, RobertsDN, UtleyR, Erdjument-BromageH, et al. (2004) The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol 24 : 9424–9436.
19. BillonP, CoteJ (2012) Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta 1819 : 290–302.
20. CaiY, JinJ, FlorensL, SwansonSK, KuschT, et al. (2005) The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem 280 : 13665–13670.
21. IkuraT, OgryzkoVV, GrigorievM, GroismanR, WangJ, et al. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102 : 463–473.
22. KuschT, FlorensL, MacdonaldWH, SwansonSK, GlaserRL, et al. (2004) Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306 : 2084–2087.
23. GevryN, ChanHM, LaflammeL, LivingstonDM, GaudreauL (2007) p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev 21 : 1869–1881.
24. GevryN, HardyS, JacquesPE, LaflammeL, SvotelisA, et al. (2009) Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev
25. ChoiJ, HeoK, AnW (2009) Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res 37 : 5993–6007.
26. JhaS, ShibataE, DuttaA (2008) Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol 28 : 2690–2700.
27. ArnoldA, PapanikolaouA (2005) Cyclin D1 in breast cancer pathogenesis. J Clin Oncol 23 : 4215–4224.
28. LehnS, TobinNP, BerglundP, NilssonK, SimsAH, et al. (2010) Down-regulation of the oncogene cyclin D1 increases migratory capacity in breast cancer and is linked to unfavorable prognostic features. Am J Pathol 177 : 2886–2897.
29. TobinNP, SimsAH, LundgrenKL, LehnS, LandbergG (2011) Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer 11 : 417.
30. DalvaiM, BystrickyK (2010) Cell cycle and anti-estrogen effects synergize to regulate cell proliferation and ER target gene expression. PLoS One 5: e11011.
31. SutherlandRL, PrallOW, WattsCK, MusgroveEA (1998) Estrogen and progestin regulation of cell cycle progression. J Mammary Gland Biol Neoplasia 3 : 63–72.
32. SerandourAA, AvnerS, PercevaultF, DemayF, BizotM, et al. (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21 : 555–565.
33. GrigolettoA, LestienneP, RosenbaumJ (2011) The multifaceted proteins Reptin and Pontin as major players in cancer. Biochim Biophys Acta 1815 : 147–157.
34. JeongKW, KimK, SituAJ, UlmerTS, AnW, et al. (2011) Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol 18 : 1358–1365.
35. SvotelisA, GevryN, GaudreauL (2009) Regulation of gene expression and cellular proliferation by histone H2A.Z. Biochem Cell Biol 87 : 179–188.
36. KimuraA, HorikoshiM (1998) Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells 3 : 789–800.
37. MillarCB, XuF, ZhangK, GrunsteinM (2006) Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev 20 : 711–722.
38. FullwoodMJ, LiuMH, PanYF, LiuJ, XuH, et al. (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462 : 58–64.
39. Tan-WongSM, FrenchJD, ProudfootNJ, BrownMA (2008) Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A 105 : 5160–5165.
40. DalvaiM, BellucciL, FleuryL, LavigneAC, MoutahirF, et al. (2012) H2A.Z-dependent crosstalk between enhancer and promoter regulates Cyclin D1 expression. Oncogene
41. DekkerJ (2006) The three 'C' s of chromosome conformation capture: controls, controls, controls. Nat Methods 3 : 17–21.
42. HagegeH, KlousP, BraemC, SplinterE, DekkerJ, et al. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2 : 1722–1733.
43. MatteraL, EscaffitF, PillaireMJ, SelvesJ, TytecaS, et al. (2009) The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene 28 : 1506–1517.
44. BallareC, CastellanoG, GavegliaL, AlthammerS, Gonzalez-VallinasJ, et al. (2013) Nucleosome-driven transcription factor binding and gene regulation. Mol Cell 49 : 67–79.
45. KocanovaS, KerrEA, RafiqueS, BoyleS, KatzE, et al. (2010) Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet 6: e1000922.
46. IacovoniJS, CaronP, LassadiI, NicolasE, MassipL, et al. (2010) High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29 : 1446–1457.
47. LegubeG, LinaresLK, TytecaS, CaronC, ScheffnerM, et al. (2004) Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem 279 : 44825–44833.
48. LiG, ThomasAM, HartSN, ZhongX, WuD, et al. (2010) Farnesoid X receptor activation mediates head-to-tail chromatin looping in the Nr0b2 gene encoding small heterodimer partner. Mol Endocrinol 24 : 1404–1412.
49. SplinterE, GrosveldF, de LaatW (2004) 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol 375 : 493–507.
50. DeschenesJ, BourdeauV, WhiteJH, MaderS (2007) Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem 282 : 17335–17339.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



