-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
Alkylating agents comprise a major class of front-line cancer chemotherapeutic compounds, and while these agents effectively kill tumor cells, they also damage healthy tissues. Although base excision repair (BER) is essential in repairing DNA alkylation damage, under certain conditions, initiation of BER can be detrimental. Here we illustrate that the alkyladenine DNA glycosylase (AAG) mediates alkylation-induced tissue damage and whole-animal lethality following exposure to alkylating agents. Aag-dependent tissue damage, as observed in cerebellar granule cells, splenocytes, thymocytes, bone marrow cells, pancreatic β-cells, and retinal photoreceptor cells, was detected in wild-type mice, exacerbated in Aag transgenic mice, and completely suppressed in Aag−/− mice. Additional genetic experiments dissected the effects of modulating both BER and Parp1 on alkylation sensitivity in mice and determined that Aag acts upstream of Parp1 in alkylation-induced tissue damage; in fact, cytotoxicity in WT and Aag transgenic mice was abrogated in the absence of Parp1. These results provide in vivo evidence that Aag-initiated BER may play a critical role in determining the side-effects of alkylating agent chemotherapies and that Parp1 plays a crucial role in Aag-mediated tissue damage.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003413
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003413Summary
Alkylating agents comprise a major class of front-line cancer chemotherapeutic compounds, and while these agents effectively kill tumor cells, they also damage healthy tissues. Although base excision repair (BER) is essential in repairing DNA alkylation damage, under certain conditions, initiation of BER can be detrimental. Here we illustrate that the alkyladenine DNA glycosylase (AAG) mediates alkylation-induced tissue damage and whole-animal lethality following exposure to alkylating agents. Aag-dependent tissue damage, as observed in cerebellar granule cells, splenocytes, thymocytes, bone marrow cells, pancreatic β-cells, and retinal photoreceptor cells, was detected in wild-type mice, exacerbated in Aag transgenic mice, and completely suppressed in Aag−/− mice. Additional genetic experiments dissected the effects of modulating both BER and Parp1 on alkylation sensitivity in mice and determined that Aag acts upstream of Parp1 in alkylation-induced tissue damage; in fact, cytotoxicity in WT and Aag transgenic mice was abrogated in the absence of Parp1. These results provide in vivo evidence that Aag-initiated BER may play a critical role in determining the side-effects of alkylating agent chemotherapies and that Parp1 plays a crucial role in Aag-mediated tissue damage.
Introduction
DNA damage continually arises from environmental agents and reactive byproducts of normal cellular function. Moreover, DNA damage is deliberately induced during the course of cancer chemotherapy. Such damage can result in cell death, mutagenesis, and genetic instability thus promoting tissue degeneration, aging, cancer, and sometimes death. DNA repair pathways have evolved to cope with recurring DNA damage, providing protection against carcinogenesis, neurodegeneration, and premature aging [1]–[5]. Understandably, loss of function mutations have been extensively studied, whereas genetic variants that result in increased DNA repair activity have not received the same attention, primarily because decreased DNA repair is thought to be more relevant for increased cancer risk. While this concept is accurate for many DNA repair proteins [1]–[3], a growing body of evidence suggests that increased levels of certain DNA repair enzymes can result in loss of coordination between the enzymatic steps within a particular DNA repair pathway; such loss of coordination can negatively impact cellular homeostasis [6]–[9].
The base excision repair (BER) pathway acts on a wide range of DNA base lesions including alkylated, oxidized, and deaminated bases, as well as abasic (AP) sites and DNA single-strand breaks (SSBs) (reviewed in [6], [10]). In its most simplified form, BER is coordinated into 4 main steps (Figure 1A). DNA glycosylases recognize and excise specific base lesions by cleaving the N-glycosyl bond, forming an AP site. AP endonuclease (APE1) then hydrolyzes the phosphodiester backbone, generating a single-stranded DNA break (SSB) with 3′OH and 5′deoxyribose-5-phosphate (5′dRP) termini. DNA polymerase β (Pol β) contains a lyase domain that removes the 5′dRP terminus and a polymerase domain that replaces the missing nucleotide. Finally, BER is completed upon ligation of the nick by DNA Ligase I or the Xrcc1/Ligase IIIα complex (Figure 1A).
Fig. 1. Cellular processing and repair of DNA base lesions in DNA. 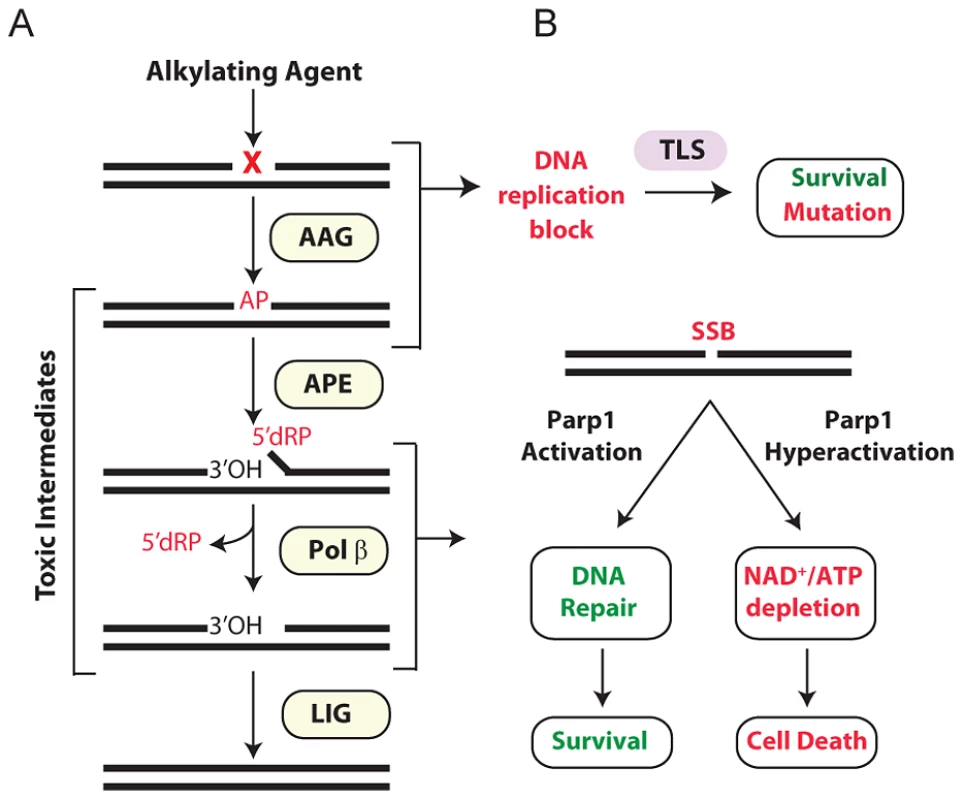
(A) DNA base lesions induced by SN1 or SN2 alkylating agent are recognized and excised by the DNA glycosylase, Aag, to generate an AP site. The base excision repair (BER) pathway continues when an AP endonuclease cleaves the DNA backbone to generate 3′ hydroxyl and 5′ deoxyribose phosphate (5′dRP) termini. Polymerase β removes the 5′dRP species, and inserts the missing DNA bases; DNA ligase completes BER by sealing the nicked DNA. (B) Error-prone translesion (TLS) polymerases can assist in the tolerance or bypass of base lesions and AP sites. Parp1 has an important role in regulating the response to DNA damage. During times of moderate DNA damage, Parp1 activation facilitates BER. Upon high levels of DNA damage, Parp1 undergoes hyperactivation; cells consequently suffer NAD+/ATP depletion, triggering cell death. Importantly, numerous BER intermediates (AP sites, 5′dRP termini, and SSBs) are toxic if allowed to accumulate rather than being efficiently shuttled through the downstream BER steps (Figure 1A). Both SSBs and AP sites exert their toxicity as a function of blocking transcription and replication [11]. Further, large numbers of SSBs can indirectly induce toxicity through the hyperactivation of poly(ADP-ribose) polymerase 1 (Parp1) [12] (Figure 1B). AP sites can also be mutagenic; although translesion DNA polymerases can prevent toxicity by bypassing AP sites, such bypass can generate point mutations [13]–[17]. The 5′dRP intermediate is particularly toxic in mouse embryonic fibroblasts (MEFs) and the alkylation sensitivity of Polβ deficient MEFs is almost completely suppressed upon expression of the Pol β 5′dRP lyase domain [18]. The toxic nature of BER intermediates underscores why this pathway must be tightly regulated and why alterations in any step of the pathway, without compensatory changes in upstream/downstream steps, can result in the accumulation of toxic intermediates. A clear example of this was illustrated by the fact that hypersensitivity to the alkylating agent methyl methanesulfonate (MMS) in Polβ−/− MEFs is completely suppressed if BER is not initiated by the alkyladenine DNA glycosylase (AAG, also known as MPG, ANPG) [19]. Therefore, although BER is essential for the repair of many different types of DNA damage, it must be carefully regulated to avoid the accumulation of toxic BER intermediates.
Aag has a wide substrate specificity, excising numerous structurally-diverse lesions, some of which are innocuous (e.g. 7-methylguanine), while others can be replication-blocking and cytotoxic (e.g. 3-methyladenine) [20]–[27]. The absence of Aag should therefore result in unrepaired alkylated DNA bases that are replication-blocking lesions, thus increasing cytotoxicity; strikingly, the converse is seen in certain Aag deficient tissues. Aag−/− bone marrow cells are MMS resistant in ex vivo survival assays [28], and Aag−/− retinal photoreceptor cells are remarkably refractory to MMS-induced death [29]. Thus, when BER is not initiated, MMS-induced cytotoxicity is avoided, presumably by preventing the accumulation of toxic intermediates, and by translesion DNA synthesis (TLS) bypassing lesions in replicating cells (Figure 1B).
The multi-functional protein, Parp1, mediates several cellular processes including stress responses, transcriptional regulation, and DNA SSB repair and BER [30]–[32]. Parp1's role as a molecular sensor of SSBs is well established; upon binding DNA breaks, Parp1 adds poly(ADP-ribose) (PAR) polymers to numerous nuclear proteins including itself, DNA polymerases, DNA ligases, transcription factors, and histones [31], [33]. Parp1 automodification facilitates BER by recruiting the scaffold protein XRCC1 that in turn facilitates the formation of a BER repair complex comprising APE1, DNA Pol β, and DNA ligase III [34]–[36]. Further, PARylation of histones, Parp1, and chromatin remodeling enzymes relaxes chromatin allowing DNA repair proteins access to damaged DNA [37]–[39]. Importantly, Parp1 is also a cell death mediator [12]; upon excessive levels of DNA damage, Parp1 hyperactivation vastly increases NAD+ consumption resulting in depletion of both NAD+ and ATP, such that cells succumb to bioenergetic failure (Figure 1B). Independent of NAD+/ATP depletion, the PAR polymer can also stimulate cell death by facilitating translocation of apoptosis inducing factor (AIF) from mitochondria to the nucleus, resulting in chromatin condensation, caspase-independent DNA degradation, and ultimately cell death [12], [40], [41]. While the various roles of Parp1 in programmed necrosis are still being elucidated, it is quite clear that Parp1 is a central player.
Imbalanced BER can arise either by increased DNA glycosylase activity, or by a decrease in any downstream BER step (reviewed in [6]). For example, decreased Pol β activity, as observed in the PolβY265C/Y265C knock-in mice, results in an accumulation of BER intermediates, causing severe physiological consequences [42]. Interestingly, recent studies generated imbalanced BER by both increasing Aag activity and eliminating Pol β activity; such cells displayed enhanced alkylation sensitivity [43], [44]. Although BER imbalance increases alkylation sensitivity in cultured cells, the effects of BER imbalance on in vivo alkylation sensitivity have not yet been extensively studied. Using transgenic mice exhibiting modestly increased Aag activity, we investigated the effects of imbalanced BER in many tissues. We show that AagTg mice exhibit dramatic alkylation sensitivity, at both the tissue and the whole-body level, consistent with imbalanced BER leading to the accumulation of toxic intermediates. Moreover, we show that Parp1 deficiency prevents alkylation-induced damage in numerous tissues, indicating that the Aag-dependent alkylation sensitivity observed in vivo occurs in a Parp1-dependent manner.
Results
Generation of AagTg mice to model BER imbalance in vivo
BER modulation has recently attracted attention as a way to potentiate alkylation sensitivity [6], [30], [44]–[48]. To investigate the consequences of imbalanced BER in vivo, we generated Aag transgenic (AagTg) mice; Table S1 displays the Aag activity levels in three transgenic founder (Fo) lines. Fo line 243 exhibits increased Aag activity in all tissues examined, with a ∼2–9-fold increase compared to WT levels (Table S1 and Figure S1A). Fo line 8756 displays negligible increases in Aag activity in every tissue except the cerebellum. Finally, Aag activity in Fo line 943 tissues falls in-between, with 1.5–4-fold increases compared to WT levels. To add context to the range of Aag activities in our AagTg mice, we examined human AAG activity in peripheral blood mononuclear cells (PBMC) of healthy individuals. We observe >10-fold variation in AAG activity in this healthy population, as measured by excision of 1′N6 ethenoadenine (εA) bases from DNA (Figure 2); εA represents one of AAG's many substrates [23], [26]. This wide range of AAG activity among healthy individuals is similar to that recently reported [7].
Fig. 2. Human peripheral blood mononuclear cells (PBMCs) exhibit a wide range in AAG activity. 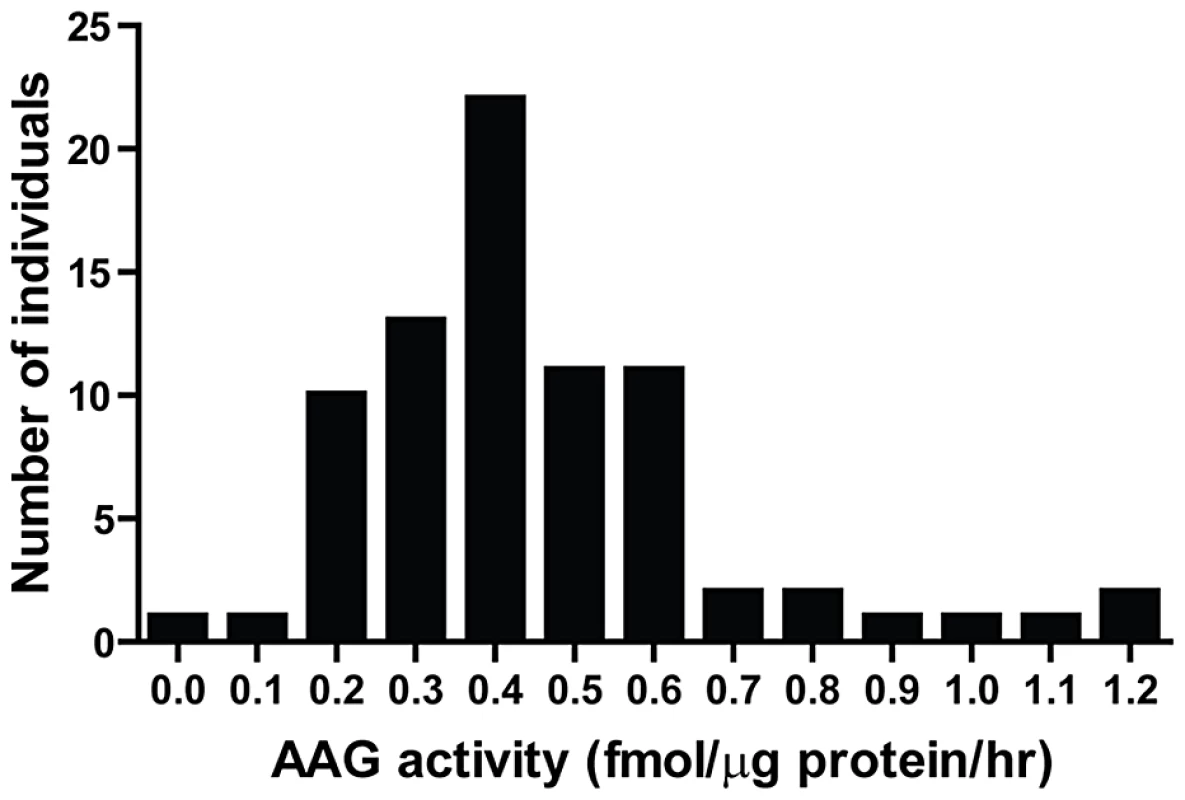
An in vitro glycosylase assay determined AAG activity in PBMCs isolated from 80 healthy individuals. Imbalanced BER increases whole-body sensitivity to alkylating agents
AagTg mice are viable and fertile, and an aging study revealed no apparent differences in lifespan or tumor incidence between WT, Aag−/− and AagTg Fo line 243 (Figure S1B and data not shown). Although increased Aag activity in vivo does not significantly alter longevity or spontaneous tumor incidence, it does profoundly affect how mice respond to DNA damage. Similar to our previous published findings, WT and Aag−/− mice display the same approximate LD50 for MMS (150 mg/kg) (Table 1) [28]. However, AagTg Fo 243, with a 2–9-fold increase in Aag activity, exhibits a dramatic increase in MMS sensitivity (Table 1). Mice with intermediate Aag levels (AagTg Fo 943) have an intermediate LD50, and AagTg Fo 8756, with negligible Aag activity in most tissues, exhibits the same MMS LD50 as WT and Aag−/− mice (Table 1). Therefore, increased Aag activity sensitizes animals to MMS-induced whole-body lethality.
Tab. 1. Approximate MMS LD50 for Aag transgenic mice. 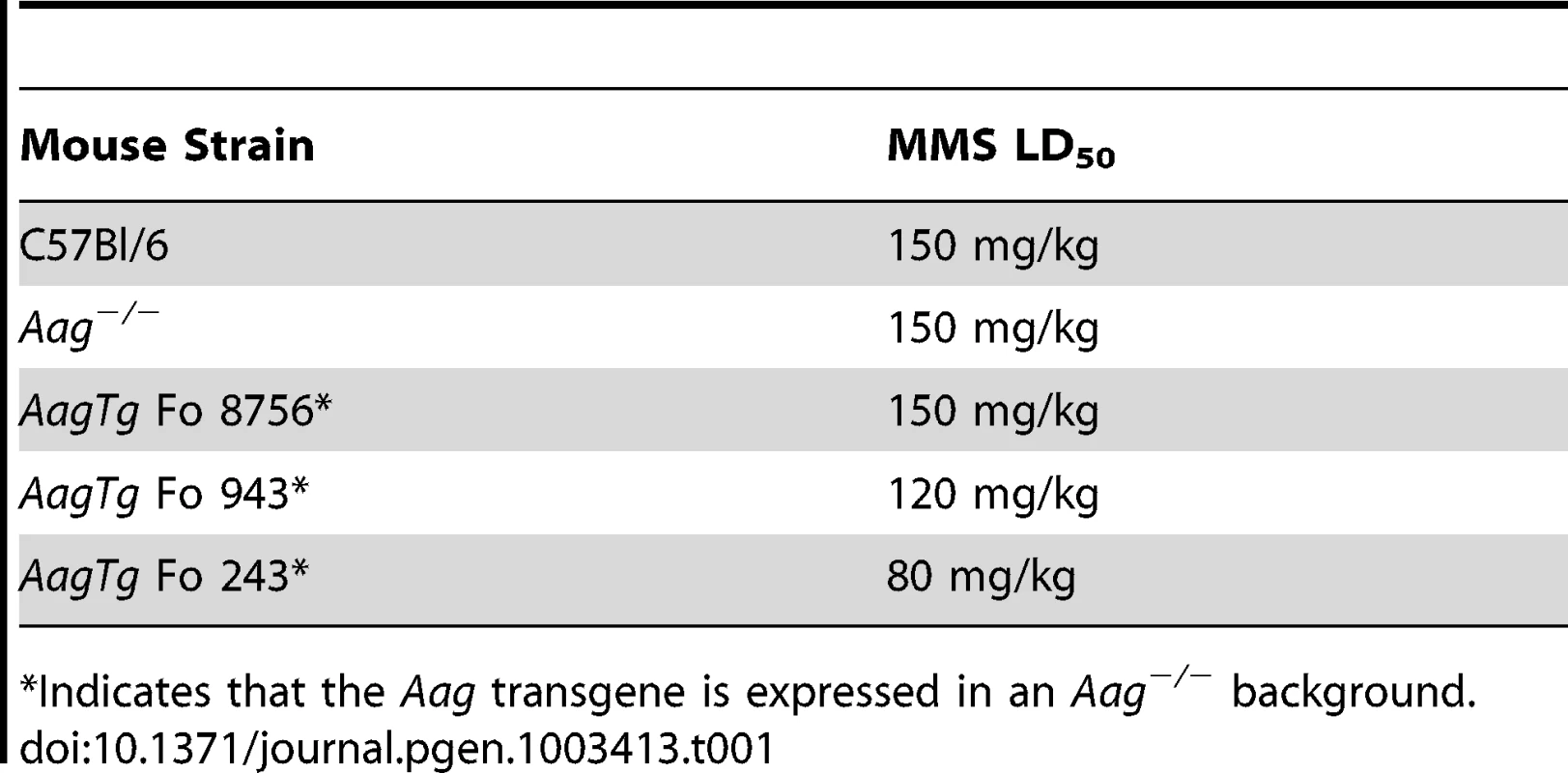
Indicates that the Aag transgene is expressed in an Aag−/− background. We next determined whether Aag activity affects the approximate LD50 for other genotoxic agents: N-methyl-N-nitrosourea (MNU), azoxymethane (AOM), mitomycin C (MMC), and chloroacetaldehyde (CAA). Table 2 illustrates that while the AagTg Fo 243 mice show dramatically increased whole-body sensitivity to three different methylating agents (MMS, MNU and AOM), they were not sensitized to non-methylating genotoxic agents (MMC and CAA). Thus, Aag activity dictates sensitivity to both SN1 (MNU and AOM) and SN2 methylating agents (MMS), but not to the other genotoxic agents examined. Since the increased Aag activity in AagTg Fo 243 mice falls within the range observed in PBMCs of a healthy human population, this founder line was chosen to further examine the consequences of BER imbalance, and are henceforth referred to as AagTg mice.
Tab. 2. Approximate LD50 of Aag−/− and Aag transgenic mice to various genotoxic agents. 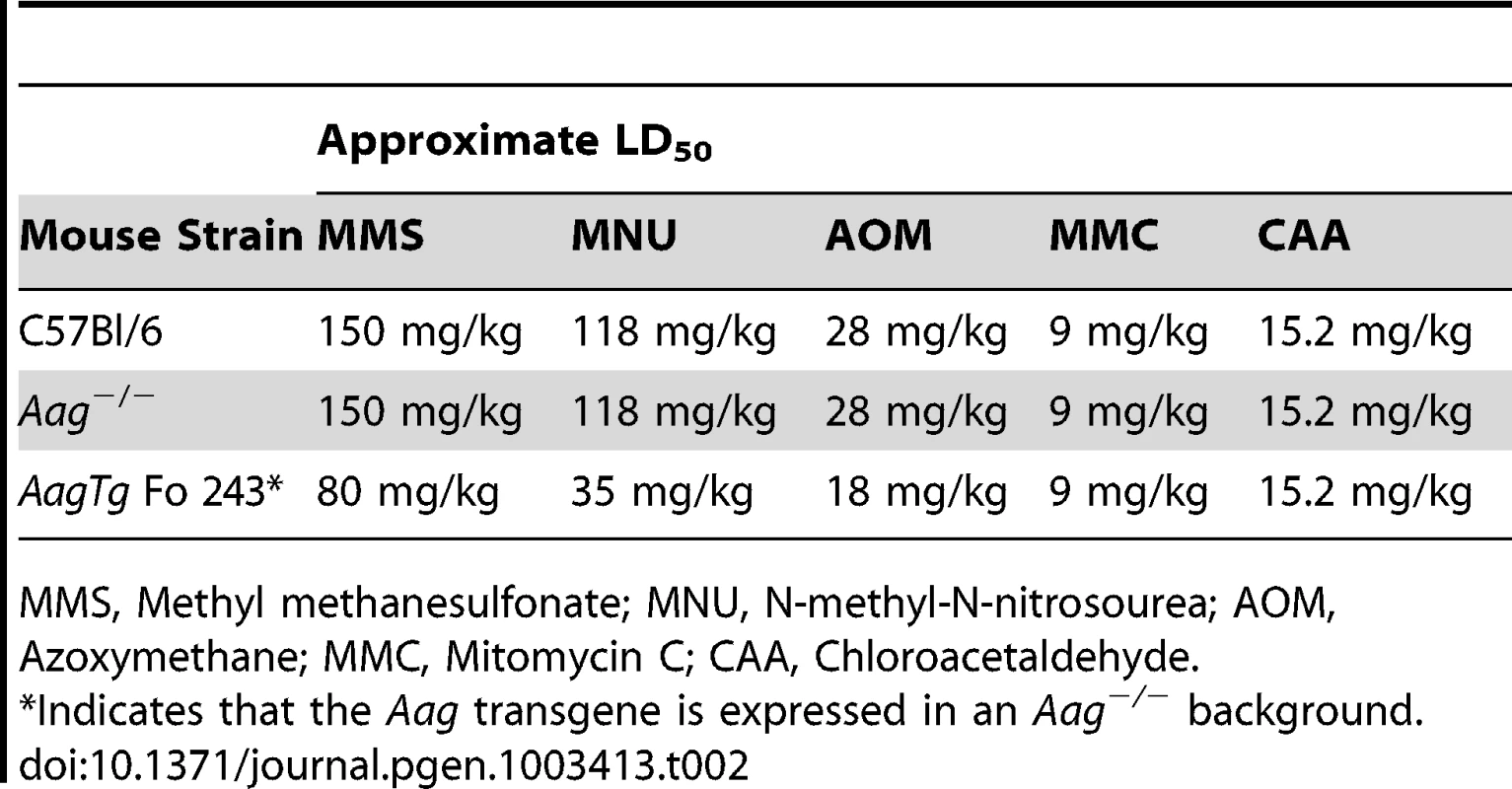
MMS, Methyl methanesulfonate; MNU, N-methyl-N-nitrosourea; AOM, Azoxymethane; MMC, Mitomycin C; CAA, Chloroacetaldehyde. AagTg mice exhibit increased MMS cytotoxicity in numerous, but not all, tissues
Histopathological analysis was performed on tissues harvested from WT, Aag−/−, and AagTg mice 24 h following MMS treatment (150 mg/kg). Because massive cell death was observed in rapidly-proliferating tissues including the spleen, thymus, and bone marrow (BM) for all genotypes, we reduced the MMS dose to 75 mg/kg to better discern any differences in sensitivity in these tissues. Even with this reduced MMS dose, AagTg mice displayed evidence of whole-body toxicity whereas WT and Aag−/− mice did not. Remarkably, as early as 24 h following MMS treatment, AagTg mice exhibit greater reductions in body weight than WT or Aag−/− mice (Figure 3A), losing >10% of their BW by 24 h; the decreased body weight remains significantly greater than that for WT and Aag−/− mice for over 3 weeks (Figure 3B). Moreover, 24 h following MMS treatment (75 mg/kg), we observe gross tissue atrophy in the thymus and spleen in AagTg mice; AagTg mice exhibit 46% and 53% decreases in thymus and spleen weight, respectively, compared to untreated tissues (Figure 3C). WT mice also exhibit a slight (26%) but significant decrease in spleen weight following MMS treatment but no evidence of thymic atrophy (compared to untreated WT mice). Strikingly, Aag−/− mice are completely protected from the MMS-induced atrophy in both the thymus and spleen (Figure 3C). Further, ex vivo clonogenic survival assays illustrate that AagTg bone marrow (BM) cells display dramatically increased MMS sensitivity, compared to WT and Aag−/− mice; as previously published, Aag−/− BM cells are less sensitive than WT BM cells to MMS (Figure 3D) [28]. However, not all tissues that exhibit increased Aag activity reveal evidence of gross tissue atrophy. Figure S2 illustrates that tissue weights of the heart, kidney, brain, gonadal fat pad, skeletal muscle and liver remain unchanged following MMS treatment (75 mg/kg). Finally, Figure S3 illustrates that AagTg mice exhibit severe cell death within the pancreatic β-islets following MMS treatment (150 mg/kg), which is not observed in WT or Aag−/− mice. The β-cells exhibit nuclear fragmentation and pyknosis, a state of increased chromatin condensation. Taken together, these results reveal that the ∼2–9 fold increase in Aag activity in the thymus, spleen, BM, and pancreas relative to WT (Table S1) renders these tissues dramatically more sensitive to the toxic effects of MMS. Further, we observe increased MMS toxicity in only a subset of tissues expressing the Aag transgene, underscoring the importance of cellular context in determining MMS sensitivity in AagTg mice.
Fig. 3. AagTg mice are more susceptible to MMS-induced toxicity. 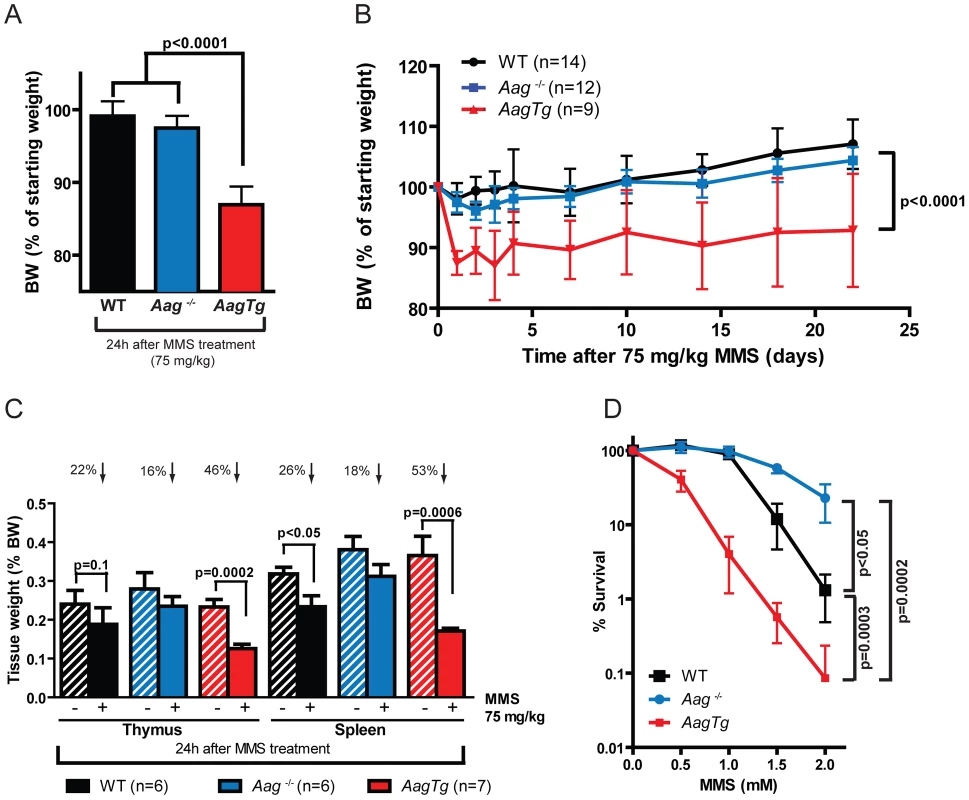
(A) Body weight (BW) of WT (n = 14), Aag−/− (n = 12) and AagTg (n = 12) mice 24 h following MMS treatment (75 mg/kg). Representative data (mean ± standard deviation) from 3 independent experiments are shown. (B) BW is illustrated for WT (n = 14), Aag−/− (n = 12) and AagTg (n = 9) mice following MMS treatment (75 mg/kg). Data represent mean ± standard deviation. (C) Tissue weights of spleen and thymus are illustrated for n>6 per genotype. Striped bars represent untreated tissue weights and solid bars represent tissue weights 24 h following MMS treatment (75 mg/kg). Percent decrease in tissue weight observed following MMS treatment is shown above bars. Data represent mean ± SEM. (D) Ex vivo bone marrow (BM) clonogenic survival assays were performed using BM isolated from WT (n = 3), Aag−/− (n = 3) and AagTg (n = 3). Data represent mean ± SEM. All the mice used in this figure are males on a pure C57BL/6 background. Unexpectedly, 24 h following a high MMS dose (150 mg/kg), we observe cell death in the cerebellar granule cells, the cell type which comprises 99% of the granular layer of the cerebellum. Following MMS treatment, there is a striking change in cerebellar morphology in AagTg mice. We observe severe cerebellar lesions containing numerous pyknotic nuclei surrounded by white spaces, indicative of edema (Figure 4A). Pyknotic nuclei, but not edema, are also observed, albeit at a lower frequency, in the cerebella of treated WT mice, whereas the cerebella of treated Aag−/− mice are indistinguishable from untreated mice (Figure 4A). The regions of edema were quantitated using image analysis software; examples of colorized lesions are shown in the lowest panel of Figure 4A. In untreated mice, no edema is observed (Figure 4B). However, 24 h following MMS (150 mg/kg), there is an obvious increase in edema in AagTg mice compared to either WT or Aag−/− mice, and a trend towards an increase in WT compared to Aag−/− mice (p = 0.308), suggesting that Aag−/− mice are protected against MMS-mediated cerebellar toxicity. Here we illustrate that MMS treatment results in severe cerebellar damage that is Aag-dependent. Although cerebellar damage has been described following treatment of early postnatal mice (PND3) with the alkylating agents methylazoxymethanol and mechlorethamine, to our knowledge, it has not previously been demonstrated following treatment of adult mice [49], [50].
Fig. 4. MMS induces severe cerebellar lesions AagTg mice. 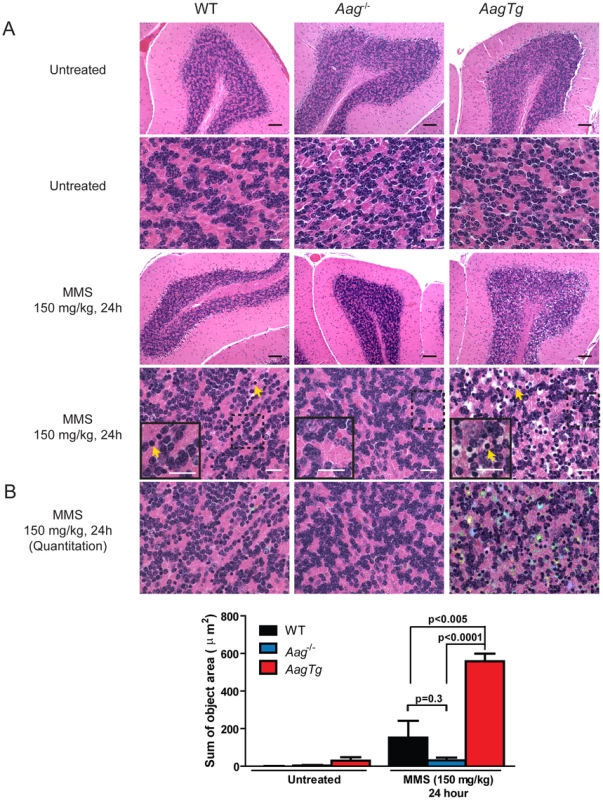
(A) H&E stained image of cerebellar granule cells from WT, Aag−/− and AagTg mice either in untreated conditions or 24 h following MMS treatment (150 mg/kg). Representative images are shown of n>6 experiments. Yellow arrows indicate pyknotic nuclei. Scale bar is 100 µm on low magnification images (black bar) and 15 µm on high-magnification images (white bar). Insets contain magnified images of area in dashed boxes. (B) Quantitation of cerebellar phenotype was performed on images from WT (n = 4), Aag−/− (n = 3) and AagTg mice (n = 4). Representative images with identified objects (edema) colorized for visualization. Greater than 3 images/cerebella were quantitated per mouse, and the average sum of object area per image is presented. Data represent mean ± SEM. All the mice in this figure are on a pure C57BL/6 background. Aag-dependent, MMS-induced cerebellar degeneration results in impaired motor function
Given the importance of the cerebellum in coordinating motor function, we investigated whether MMS-induced cerebellar lesions result in diminished motor control. Decreased mobility was observed in all genotypes following a high MMS dose (150 mg/kg) (data not shown), so we reduced the MMS dose (90 mg/kg) for gait comparisons between WT, Aag−/−, and AagTg mice. The gait of all genotypes was indistinguishable under untreated conditions (Figure S4). However, three hours following MMS treatment, the gait of WT and Aag−/− mice is unchanged whereas AagTg mice exhibit severe gait abnormalities including immobility, circling, and walking backwards (Figure 5A). We quantitated motor defects by performing an accelerating speed rotarod test. To ensure all genotypes were capable of performing for >30 seconds on the rotarod test, we further reduced the MMS dose (60 mg/kg). Without MMS exposure, all genotypes performed comparably (Figure 5B). However, three hours following MMS treatment, we observed a dramatic decrease in rotarod performance for AagTg mice compared to WT and Aag−/− mice (Figure 5B). Although the AagTg mice slightly improved their performance by 4 hours, it remained significantly decreased compared to WT and Aag−/− mice (Figure 5B). Treated WT and Aag−/− mice performed similarly to untreated mice, indicating that MMS at this dose (60 mg/kg) caused no motor dysfunction in WT or Aag−/− mice.
Fig. 5. MMS induces an Aag-dependent decrease in motor function. 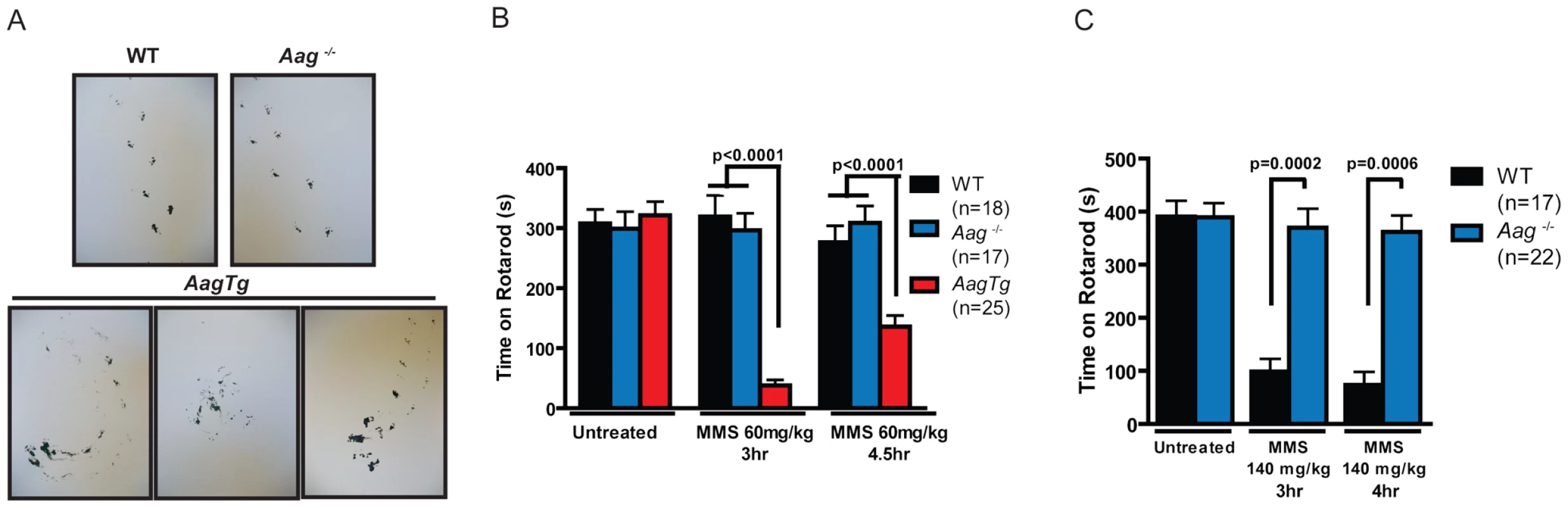
(A) Representations of gait are shown for WT (n = 3), Aag−/− (n = 3) and AagTg (n = 3) mice three hours following MMS treatment (90 mg/kg). (B) Rotarod performance is shown for WT (n = 18), Aag−/− (n = 17) and AagTg (n = 25) mice under untreated conditions and following MMS treatment (60 mg/kg). Data represent mean ± SEM. (C) Performance for the rotarod challenge is shown for WT (n = 17) and Aag−/− (n = 22), 3 and 4 h following MMS treatment (140 mg/kg). Data represent mean ± SEM. All the mice in this figure are on a pure C57BL/6 background. We next increased the MMS dose to one that resulted in obviously impaired motor function in WT mice (140 mg/kg), and again examined motor function using the rotarod test (Figure 5C); the AagTg mice could not be included in this experiment due to their extreme sensitivity at this MMS dose. Strikingly, even at this high dose, Aag−/− mice remain completely protected against MMS-induced motor dysfunction; they not only exhibit significantly better performance than WT mice, but their performance following MMS remains the same as in untreated conditions (Figure 5C). This observation is consistent with the absence of histological cerebellar lesions in Aag−/− mice following MMS (150 mg/kg) treatment (Figure 4A). Together, these data underscore the importance of BER coordination in neuronal homeostasis; MMS induces cerebellar degeneration that is exacerbated by imbalanced BER in AagTg mice. Perhaps most importantly, the elimination of BER initiation in Aag−/− mice completely suppresses MMS-induced cerebellar toxicity.
The absence of Parp1 suppresses Aag-dependent MMS-induced toxicity in several tissues
Given the role for Parp1 in mediating alkylation toxicity, we next set out to determine whether eliminating Parp1 could modulate the MMS-induced cytotoxicity observed in WT and AagTg mice; to explore this possibility, we utilized Parp1−/− mice [51]. We first investigated MMS-induced retinal degeneration (RD). As previously illustrated, MMS induces the selective degeneration of the photoreceptors in the retinal outer nuclear layer (ONL) in WT mice, whereas juxtaposed retinal layers are unaffected [29]. While Aag−/− mice are completely protected from such RD, AagTg mice are hypersensitive compared to WT and Aag−/− mice, as observed by decreased number of cells found within the ONL (Figure 6 and [29]). Strikingly, we observe that Parp1−/− mice are also completely protected against MMS-induced RD (Figure 6). Moreover, MMS-induced RD is completely abrogated in AagTg/Parp1−/− mice, indicating that Parp1 deficiency completely suppresses the Aag-dependent MMS-hypersensitivity in photoreceptors of AagTg mice (Figure 6). To confirm that Parp1 enzymatic activity is stimulated during MMS-induced Aag-dependent RD, we evaluated Parp1 activation by immunodetection of the PAR polymer. Figure S5 shows that increased PAR polymer staining is observed 24 h following MMS treatment, in an Aag-dependent manner, confirming that MMS-induced RD is preceded by Parp1 activation. Similarly, we observe that Parp1 deficiency is able to suppress the pancreatic β-cell death observed in AagTg mice following an acute MMS treatment (Figure S6). Together, these data indicate that the BER intermediates exert their toxicity through the hyperactivation of Parp1. Further, we find that deletion of Parp1 prevents MMS-induced toxicity and that both Aag and Parp1 are required for MMS-induced cell death.
Fig. 6. Parp1 deficiency protects against Aag-dependent, MMS-induced toxicity in retina photoreceptors. 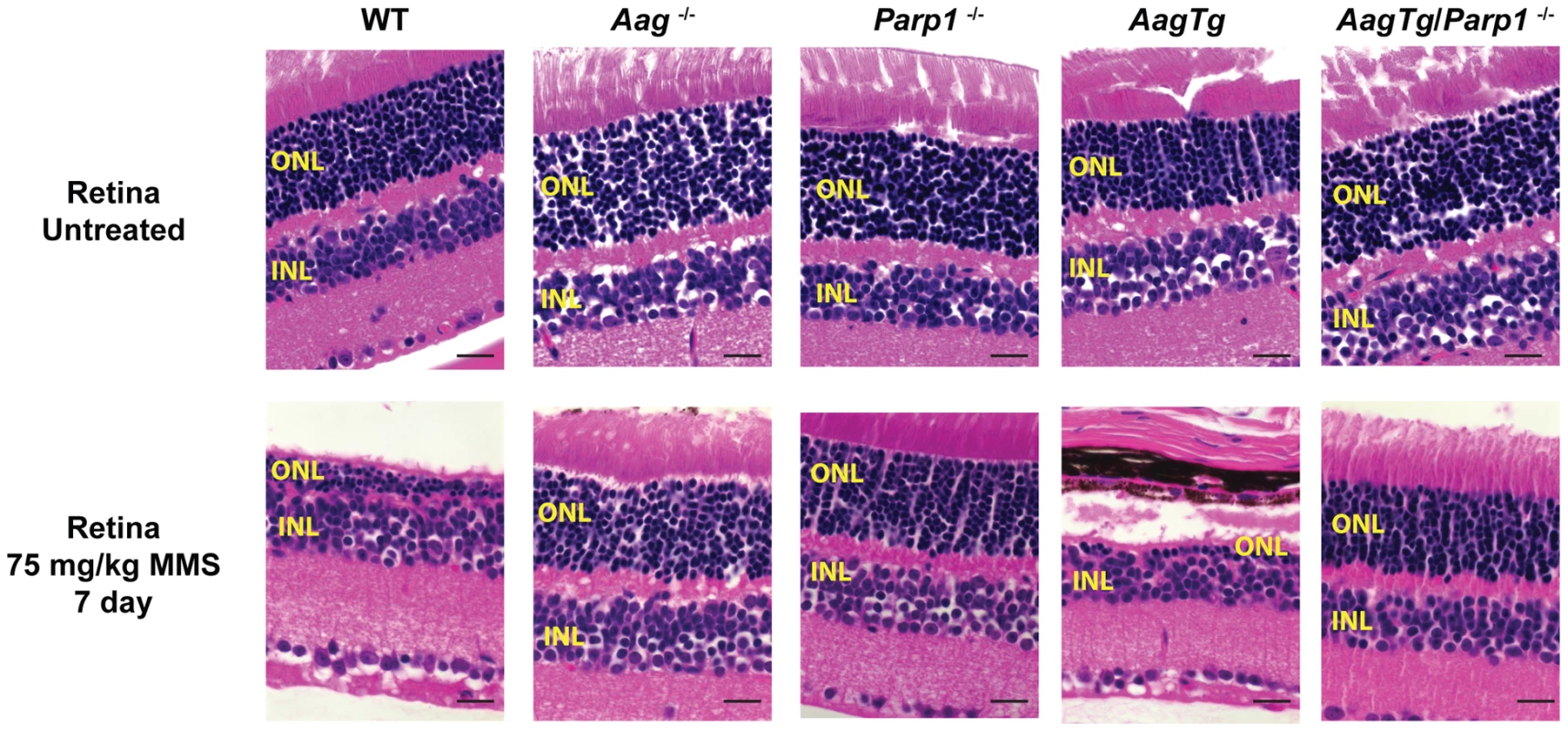
H&E stained retinal sections for WT, Aag−/−, Parp1−/−, AagTg, and AagTg/Parp1−/− under untreated conditions or 7 d following MMS treatment (75 mg/kg). Scale bar is 15 µm. Representative images for n = 5 mice/genotype are shown. All the mice used in this figure are mixed C57BL/6:129S background. ONL, Outer nuclear layer; INL, inner nuclear layer. We next investigated the requirement for Parp1 in MMS-induced cerebellar degeneration. While MMS (150 mg/kg) induces severe cerebellar lesions in AagTg mice (Figure 4A), Parp1 deficiency completely suppresses this Aag-dependent, alkylation-induced cerebellar toxicity (Figure 7A). Image analysis confirms that the drastic increase in the edema in MMS-treated AagTg mice was completely abrogated in AagTg/Parp1−/− mice (Figure 7B). Consistent with rescue of cerebellar lesions in AagTg/Parp1−/− mice (Figure 7B), we illustrate using gait analysis that Parp1 deficiency prevents the motor dysfunction observed following MMS treatment in AagTg mice (Figure S7). Additionally, rotarod assays were performed in WT, AagTg and AagTg/Parp−/− mice to quantitate motor function. Following MMS (60 mg/kg), the motor function in WT mice remain unaffected, whereas AagTg mice exhibit significantly diminished performance (Figure 7C). Importantly, following MMS, AagTg/Parp1−/− mice perform just as well as WT mice, indicating that the Aag-dependent MMS-hypersensitivity in the cerebella of AagTg mice is completely dependent on Parp1 (Figure 7C). Further, the Parp1 deficiency was sufficient to prevent the motor dysfunction observed at high MMS doses. As described above, at high MMS (140 mg/kg), WT mice exhibit severe motor dysfunction (Figure 5C and Figure 7D); however we find that like Aag−/− mice, Parp1−/− mice are protected against MMS-induced motor dysfunction and exhibit enhanced rotarod performance compared to MMS-treated WT mice (Figure 7D). The mice in this experiment are on a mixed C57Bl/6 : 129S6 background, and the slight decrease in rotarod performance observed here, compared to Figure 5, can be attributed to differences in genetic background, as previously shown [52], [53]. Together, these data (Figure 6, Figure 7, and Figure S6) indicate that both Aag and Parp1 are required for the severe MMS-mediated cytotoxicity observed in retinal photoreceptors, pancreatic β-cells, and cerebellar granule cells.
Fig. 7. Parp1 deficiency protects against Aag-dependent, MMS-induced motor dysfunction. 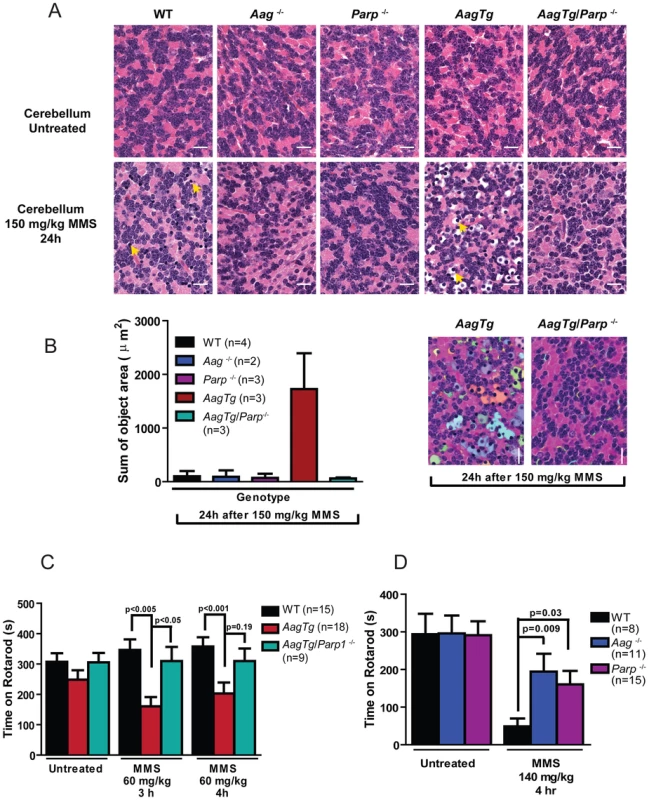
(A) H&E stained cerebellar sections are shown from WT, Aag−/−, Parp1−/−, AagTg, and AagTg/Parp1−/− under untreated conditions or 24 h following MMS treatment (150 mg/kg). Scale bar is 15 µm. Representative images for n = 5 mice/genotype are shown. (B) Quantitation of cerebellar phenotype is shown. Three or more images/cerebella were quantitated per mouse, and 3 mice per genotype analyzed for quantitation; the average sum of object area (edema) per image is presented. (C) Performance during the rotarod challenge in WT (n = 15), AagTg (n = 18), and AagTg/Parp1−/− (n = 9) is illustrated under untreated conditions and following MMS treatment (60 mg/kg). (D) Performance for the rotarod challenge is shown for WT (n = 8), Aag−/− (n = 11), and Parp1−/− (n = 15) mice four hours following MMS treatment (140 mg/kg). All the mice used in this figure are mixed C57BL/6:129S background. All data represent mean ± SEM. Discussion
Using AagTg mice, we have investigated the in vivo consequences of imbalanced BER. Although increased human AAG activity was recently linked to elevated lung cancer risk [7], [9] and decreased survival of glioma patients [54], [55], increased Aag activity in mice does not affect spontaneous tumorigenesis or overall longevity. However, dramatic in vivo consequences of imbalanced BER were revealed upon treating AagTg mice with alkylating agents. AagTg mice exhibited increased whole-animal lethality to both SN1 and SN2 methylating agents, but not to other genotoxic agents. Our data suggest that under basal conditions, the level of BER intermediates produced during the repair of spontaneous DNA damage is readily accommodated by the downstream BER enzymes. However, in the presence of higher levels of DNA base damage generated by alkylating agents, Aag initiates BER at a rate such that downstream BER enzymes may be unable to efficiently process the toxic BER intermediates, resulting in cell death and tissue damage, and potentially in the death of the animal (Figure 1). Therefore, it may be predicted that patients exhibiting increased AAG activity may exhibit increased sensitivity or more detrimental side-effects to alkylating chemotherapeutic agents. Indeed, AAG expression does predict temozolomide sensitivity in glioblastoma and ovarian cancer cell lines [48], [56], and AAG expression inversely correlates with survival of glioma patients following treatment [54]. Together, the data presented here as well as published findings provide justification for an epidemiological study examining alkylation sensitivity in correlation with AAG activity levels.
Imbalanced BER in AagTg mice confers increased MMS sensitivity to cells in the thymus, spleen, bone marrow, retina, pancreas, and cerebellum. Further, Aag activity predicts MMS toxicity in vivo, such that relative sensitivities are as follows: AagTg>WT>Aag−/−. However, Aag activity is not the sole determinant of MMS-mediated tissue cytotoxicity; numerous tissues in the AagTg mice exhibit ∼5–8 fold increases in Aag activity (e.g. heart, kidney, liver) but show no evidence of increased MMS sensitivity (Figure S2 and data not shown), underscoring the importance of cellular context in determining Aag-mediated alkylation sensitivity. It remains to be determined why only subsets of cell types are susceptible to Aag-mediated alkylation toxicity. In the highly-proliferative thymus, spleen, and bone marrow, unrepaired BER intermediates presumably result in replication fork collapse and DSBs, thus triggering cell death. However, pancreatic β-cells are not highly-proliferative and both the adult cerebellar granule cells and retinal photoreceptor cells are non-replicating, post-mitotic tissues [57], indicating cell death must be replication-independent. It is important to note that even within tissues exhibiting MMS sensitivity, toxicity is not uniform across all cell types in the tissue. For example, only the ONL of the retina undergoes MMS-induced degeneration whereas the juxtaposed retinal layers remain intact. Similarly, following MMS treatment, cerebellar purkinje cells remain unaffected while neighboring cerebellar granule cells are ablated. Why are some cells so sensitive, whereas others are resistant? The activity of downstream BER proteins in the sensitive cells may simply be insufficient to process accumulating BER intermediates; this and other possibilities are currently under investigation. Together, our data emphasize the concept that BER imbalance and the resulting intermediates can profoundly affect cellular and tissue homeostasis, and reveal that in certain contexts, the absence of DNA repair can actually be beneficial to an organism.
Although it has been known for 30 years that Parp inhibition potentiates alkylation-induced toxicity [58], the recent discovery of synthetic lethality in BRCA1/2 homologous recombination-deficient tumors upon Parp inhibition has renewed intense interest in using PARP inhibitors for cancer chemotherapy [59], [60]. However, in stark contrast to this well-documented potentiation of alkylation toxicity by Parp inhibitors [30], [61], we observe complete suppression of alkylation toxicity by genetic deletion of Parp1. This is likely due to multiple inherent differences between Parp1 deficiency and Parp inhibition [62]. The main difference is the proposed trapping of Parp1 on DNA substrates by Parp inhibitors, which thereby prevents BER and interferes with replication [63], [64]. Although this ‘DNA trapping’ phenomenon has been demonstrated for many Parp inhibitors, it may not be the case for all Parp inhibitors [64]–[67]. In fact, uncovering the relationship between the inhibition of the catalytic activity of Parp, the potency of DNA trapping, and overall toxicity by Parp inhibitors has recently garnered interest [66]. Another difference between Parp1 deficiency and Parp inhibition resides in the fact that Parp inhibitors are not specific for the inhibition of Parp1, but can potentially inhibit the catalytic activity of 17 other members of the Parp superfamily [68]. Regardless of the differences between Parp1 deletion and Parp inhibition, suppression of alkylation toxicity upon treatment of cultured cells with Parp inhibitors is not unprecedented [43], [69]–[71].
Although Parp1 deficiency mechanistically differs from Parp inhibition, it was surprising to observe that Parp1 deficiency was capable of completely suppressing MMS hypersensitivity in numerous tissues under conditions of imbalanced BER (AagTg mice). This in vivo data reveals that Parp1 acts downstream of Aag to govern alkylation sensitivity, presumably through Parp1's alternate function in mediating programmed necrosis (Figure 1B) [12], [71]. Interestingly, interrupting Parp1's function has been shown to be protective in several other models of neuronal damage including ischemia/reperfusion and glutamate excitotoxity [72]–[74], and retinal degeneration induced by ischemia/reperfusion or by treatment with PDE6 inhibitor, which mimics the rd1 mutation [75], [76]. Whether Aag also plays a role upstream from Parp1 in these modes of tissue damage remains to be determined.
Using genetic experiments, we show here that modestly increased Aag activity results in dramatic increases in tissue and whole-animal sensitivity to alkylating agents. Given that human AAG activity varies greatly among healthy individuals (Figure 2), and also that BER protein levels are known to be altered in numerous human cancers [77]–[79], the resulting imbalanced BER may have dramatic consequences in patients undergoing chemotherapy involving alkylating agents [6]. Moreover, PARP1 expression and activity varies greatly in human tumors [80], and SNPs in PARP1 have been associated with numerous cancers [81]–[83]. We illustrate that Parp1 deficiency protects against alkylation sensitivity at both the tissue and whole-animal level. Therefore, decreased PARP1 activity may result in a decreased response during a chemotherapeutic regimen. Indeed, leukemic patients expressing decreased PARP1 levels exhibit resistance to standard chemotherapy therapy [84], [85]. Taken together, our findings illustrate that monitoring for both BER imbalance and PARP1 expression is warranted prior to selecting a chemotherapeutic regimen that includes alkylating agents.
Materials and Methods
Ethics statement
The MIT Committee on the Use of Humans as Experimental Subjects reviewed and approved the research involving human subjects. Written informed consent was obtained from all participants. All animal procedures were approved by the MIT Committee on Animal Care.
Animals
Aag−/− and Aag transgenic (AagTg) mice were described previously [21], [29]. Parp −/− mice (Jackson Laboratories) were described previously [51]. Mice were fed a standard diet ad libitum, housed in an AAALAC-accredited facility, and euthanized by CO2 asphyxiation. Additional details about the mice utilized are included in the Methods S1.
Reagents
Methyl methanesulfonate (MMS), chloroacetaldehyde (CAA), N-methyl-N-nitrosurea (MNU), and Mitomycin C (MMC) were obtained from Sigma (St. Louis, MO). Azoxymethane (AOM) was obtained from the Midwest Research Institute, NCI Chemical Repository.
Treatments
Approximate LD50 (whole-animal sensitivity to genotoxic agents) was determined as in Deichmann and LeBlanc [86]. Acute MMS treatments were performed by i.p. injecting mice with varying doses of MMS (60–150 mg/kg).
Tissue processing and histopathology
Tissues were processed at the David H. Koch Institute for Integrative Cancer Research, Histology Core Facility; they were paraffin-embedded, sectioned at 5 µm, and stained with hematoxylin and eosin (H&E). All H&E stained slides were blindly analyzed by a pathologist (R.T.B) for the cause of death as well as for the identification of any tumors/lesions.
Image quantitation
Volocity (Perkin Elmer) image analysis software was used to quantitate edema, as observed by white spaces in cerebellar histological sections. Thresholding was performed using the Red/Green/Blue quantitation tool and objects smaller than 5 microns were excluded. The sum of all object areas/image was calculated and greater than 3 representative images were analyzed for each of 3 animals.
Ex vivo bone marrow clonogenic assays
Bone marrow cells were harvested from the femurs of 6–12 weeks old WT, Aag−/− and AagTg mice. Cells were treated with varying dose of MMS, washed, resuspended in complete media mixed with methylcellulose (Stem Cell Technologies), plated in duplicate and the percent survival calculated as in [28]. Experiments were performed in triplicate.
Evaluation of Aag activity in mouse tissues
Cell extracts were made from tissues harvested from WT, AagTg, and Aag−/− mice. Tissues were sonicated in Aag glycosylase assay buffer (20 mM Tris-Cl pH 7.6, 100 mM KCl, 5 mM EDTA, 1 mM EGTA and 5 mM β-mercaptoethanol) with protease inhibitors. Protein concentration was measured using micro BCA Kit (Pierce). Glycosylase assays were performed as previously published [21]. A [32P]γ-labeled double-stranded 25mer oligonucleotide containing a single centrally located hypoxanthine residue [5′-GCAATCTAGCTTTTT(Hx)CGATGTATGC-3′] was incubated with an amount of extract determined to be in the linear range for activity at 37°C for 1 h. The resulting AP sites were cleaved by incubation with 0.1 N NaOH at 70 °C for 20 min. A phosphorimager was used to visualize and quantitate Aag DNA glycosylase activity.
Evaluation of AAG activity in PBMC
Peripheral blood samples were obtained from healthy individual at the MIT Catalyst Clinical Research Center, and PBMC were isolated using standard Ficoll-Paque (Sigma) density gradient centrifugations. The glycosylase assay was performed as above with the following exceptions; the [32P]γ-labeled lesion-containing oligonucleotide used was 5′-GCAATCTAGCCA(εA)GTCGATGTATGC-3′, and the glycosylase reaction was incubated for 37°C for 2 h.
Evaluation of motor function
To capture gait abnormalities, the mouse hind paws were dipped into non-toxic paint; mice were placed in a closed container on a sheet of white paper and allowed to walk freely. The Ugo Basile 7650 Accelerating Rotarod was used to evaluate fore-limb and hind-limb motor coordination. The rotarod testing protocol was two weeks long; the first week examined motor function under untreated conditions and the second week following MMS-treatment. An accelerating speed rotarod protocol was used, which linearly accelerates the rotating rod from 4 to 40 RPM in 10 minutes. During each week, day 1 to 4 consisted of training the mice for two 5-minute runs to familiarize the mice with the apparatus. On day 5, the mice underwent two tests, with an hour rest between the tests; the time spent on the rotatod is recorded as a measurement of performance. The 2nd week follows the same schedule, 4 days training and test on the fifth day. However, on the 5th day of the second week, the mice were treated with either 60 or 140 mg/kg MMS by i.p. injection and the test performed 3 and 4–4.5 hours post-treatment.
Immunofluorescence
5 µm unstained sections were deparaffinized and rehydrated in a graded ethanol series, incubated in citrate buffer and thermally processed for epitope retrieval. Sections were permeabilized three times (5 minutes) with PBS-T (1× PBS+0.1% Triton X-100). Non-specific antibody binding sites were blocked by incubating sections with 1× PBS-T+heat-inactivated goat serum (HIGS; 10%) for 30 minutes. Sections were then incubated with primary anti-PAR antibody (1∶250; BD Pharmingen) for 2 hours at RT. After 5 washes in PBS-T, sections were incubated for 30 min with secondary antibody DyLight 488 (1∶1000; Vector Labs). Nuclear counterstaining was done using TOPRO-3 (Invitrogen). All staining was performed in humidified chambers. A Zeiss Axiovert LSM 510 META confocal microscope (Germany) with a 63× oil objective was used to image the retinal sections. Images were viewed and analyzed using LSM Image Browser.
Statistics
Statistical analyses were performed using GraphPad Prism software. Data are presented as mean ± SEM or mean ± SD (as stated in figure legends). Statistical significance was determined using unpaired t-test or two-way ANOVA. Kaplan-Meier survival curves were generated and survival differences determined using the Log-Rank test. A p-value is considered significant if less than 0.05.
Supporting Information
Zdroje
1. HoeijmakersJ (2009) DNA damage, aging, and cancer. N Engl J Med 361 : 1475–1485.
2. HewishM, LordCJ, MartinSA, CunninghamD, AshworthA (2010) Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol 7 : 197–208.
3. RoyR, ChunJ, PowellSN (2012) BRCA1 and BRCA2: important differences with common interests. Nat Rev Cancer 12 : 372.
4. McKinnonPJ (2009) DNA repair deficiency and neurological disease. Nature reviews Neuroscience 10 : 100–112.
5. KatyalS, McKinnonPJ (2008) DNA strand breaks, neurodegeneration and aging in the brain. Mech Ageing Dev 129 : 483–491.
6. FuD, CalvoJA, SamsonLD (2012) Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12 : 104–120.
7. CrosbiePA, WatsonAJ, AgiusR, BarberPV, MargisonGP, et al. (2012) Elevated N3-methylpurine-DNA glycosylase DNA repair activity is associated with lung cancer. Mutat Res 732 : 43–46.
8. CerdaSR, TurkPW, ThorAD, WeitzmanSA (1998) Altered expression of the DNA repair protein, N-methylpurine-DNA glycosylase (MPG) in breast cancer. FEBS Lett 431 : 12–18.
9. Leitner-DaganY, SevilyaZ, PinchevM, KramerR, ElingerD, et al. (2012) N-Methylpurine DNA Glycosylase and OGG1 DNA Repair Activities: Opposite Associations With Lung Cancer Risk. J Natl Cancer Inst 104(22):1765–1769 doi:10.1093/jnci/djs445.
10. RobertsonAB, KlunglandA, RognesT, LeirosI (2009) DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci 66 : 981–993.
11. BoiteuxS, GuilletM (2004) Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 3 : 1–12.
12. AndrabiSA, DawsonTM, DawsonVL (2008) Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann N Y Acad Sci 1147 : 233–241.
13. SchaaperRM, KunkelTA, LoebLA (1983) Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A 80 : 487–491.
14. PagèsV, JohnsonRE, PrakashL, PrakashS (2008) Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc Natl Acad Sci U S A 105 : 1170–1175.
15. AvkinS, AdarS, BlanderG, LivnehZ (2002) Quantitative measurement of translesion replication in human cells: Evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci U S A 99 : 3764–3769.
16. GoodmanMF, CreightonS, BloomLB, PetruskaJ, KunkelTA (1993) Biochemical Basis of DNA Replication Fidelity. Critical Reviews in Biochemistry and Molecular Biology 28 : 83–126.
17. StraussBS (1991) The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays 13 : 79–84.
18. SobolRW, PrasadR, EvenskiA, BakerA, YangX-P, et al. (2000) The lyase activity of the DNA repair protein [beta]-polymerase protects from DNA-damage-induced cytotoxicity. Nature 405 : 807–810.
19. SobolRW, KartalouM, AlmeidaKH, JoyceDF, EngelwardBP, et al. (2003) Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J Biol Chem 278 : 39951–39959.
20. SaparbaevM, LavalJ (1994) Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A 91 : 5873–5877.
21. EngelwardBP, WeedaG, WyattMD, BroekhofJL, de WitJ, et al. (1997) Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci U S A 94 : 13087–13092.
22. GallagherPE, BrentTP (1984) Further purification and characterization of human 3-methyladenine-DNA glycosylase. Evidence for broad specificity. Biochim Biophys Acta 782 : 394–401.
23. HangB, SingerB, MargisonGP, ElderRH (1997) Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc Natl Acad Sci U S A 94 : 12869–12874.
24. MiaoF, BouzianeM, O'ConnorTR (1998) Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res 26 : 4034–4041.
25. O'ConnorTR (1993) Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Research 21 : 5561–5569.
26. LeeCY, DelaneyJC, KartalouM, LingarajuGM, Maor-ShoshaniA, et al. (2009) Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG). Biochemistry 48 : 1850–1861.
27. FuD, SamsonLD (2012) Direct repair of 3,N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair (Amst) 11 : 46–52.
28. RothRB, SamsonLD (2002) 3-Methyladenine DNA glycosylase-deficient Aag null mice display unexpected bone marrow alkylation resistance. Cancer Res 62 : 656–660.
29. MeiraLB, Moroski-ErkulCA, GreenSL, CalvoJA, BronsonRT, et al. (2009) Aag-initiated base excision repair drives alkylation-induced retinal degeneration in mice. Proc Natl Acad Sci U S A 106 : 888–893.
30. RouleauM, PatelA, HendzelMJ, KaufmannSH, PoirierGG (2010) PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10 : 293–301.
31. SchreiberV, DantzerF, AmeJC, de MurciaG (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7 : 517–528.
32. KrishnakumarR, GambleMJ, FrizzellKM, BerrocalJG, KininisM, et al. (2008) Reciprocal Binding of PARP-1 and Histone H1 at Promoters Specifies Transcriptional Outcomes. Science 319 : 819–821.
33. KrausWL, LisJT (2003) PARP Goes Transcription. Cell 113 : 677–683.
34. El-KhamisySF, MasutaniM, SuzukiH, CaldecottKW (2003) A requirement for PARP1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Research 31 : 5526–5533.
35. MassonM, NiedergangC, SchreiberV, MullerS, Menissier-de MurciaJ, et al. (1998) XRCC1 Is Specifically Associated with Poly(ADP-Ribose) Polymerase and Negatively Regulates Its Activity following DNA Damage. Mol Cell Biol 18 : 3563–3571.
36. VidalAE, BoiteuxS, HicksonID, RadicellaJP (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J 20 : 6530–6539.
37. AhelD, HořejšíZ, WiechensN, PoloSE, Garcia-WilsonE, et al. (2009) Poly(ADP-ribose)–Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science 325 : 1240–1243.
38. TiminszkyG, TillS, HassaPO, HothornM, KustatscherG, et al. (2009) A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16 : 923–929.
39. GottschalkAJ, TiminszkyG, KongSE, JinJ, CaiY, et al. (2009) Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA 106 : 13770–13774.
40. HeeresJT, HergenrotherPJ (2007) Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol 11 : 644–653.
41. JozaN, PospisilikJA, HangenE, HanadaT, ModjtahediN, et al. (2009) AIF: not just an apoptosis-inducing factor. Ann N Y Acad Sci 1171 : 2–11.
42. SenejaniAG, DalalS, LiuY, NottoliTP, McGrathJM, et al. (2012) Y265C DNA polymerase beta knockin mice survive past birth and accumulate base excision repair intermediate substrates. Proc Natl Acad Sci U S A 109 : 6632–6637.
43. TangJ-b, SvilarD, TrivediRN, WangX-h, GoellnerEM, et al. (2011) N-methylpurine DNA glycosylase and DNA polymerase β modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro-Oncology 13 : 471–486.
44. TangJ-b, GoellnerEM, WangX-h, TrivediRN, St CroixCM, et al. (2010) Bioenergetic Metabolites Regulate Base Excision Repair–Dependent Cell Death in Response to DNA Damage. Molecular Cancer Research 8 : 67–79.
45. LiuL, GersonSL (2004) Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr Opin Investig Drugs 5 : 623–627.
46. WilsonD, SimeonovA (2010) Small molecule inhibitors of DNA repair nuclease activities of APE1. Cellular and Molecular Life Sciences 67 : 3621–3631.
47. JelezcovaE, TrivediRN, WangX-h, TangJ-b, BrownAR, et al. (2010) Parp1 activation in mouse embryonic fibroblasts promotes Pol [beta]-dependent cellular hypersensitivity to alkylation damage. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 686 : 57–67.
48. TrivediRN, WangX-h, JelezcovaE, GoellnerEM, TangJ-b, et al. (2008) Human Methyl Purine DNA Glycosylase and DNA Polymerase β Expression Collectively Predict Sensitivity to Temozolomide. Molecular Pharmacology 74 : 505–516.
49. KisbyGE, LesselrothH, OlivasA, SamsonL, GoldB, et al. (2004) Role of nucleotide - and base-excision repair in genotoxin-induced neuronal cell death. DNA Repair (Amst) 3 : 617–627.
50. KisbyGE, OlivasA, ParkT, ChurchwellM, DoergeD, et al. (2009) DNA repair modulates the vulnerability of the developing brain to alkylating agents. DNA Repair (Amst) 8 : 400–412.
51. WangZQ, AuerB, StinglL, BerghammerH, HaidacherD, et al. (1995) Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes & Development 9 : 509–520.
52. TarantinoLM, GouldTJ, DruhanJP, BucanM (2000) Behavior and mutagenesis screens: the importance of baseline analysis of inbred strains. Mammalian Genome 11 : 555–564.
53. BrooksSP, PaskT, JonesL, DunnettSB (2004) Behavioural profiles of inbred mouse strains used as transgenic backgrounds. I: motor tests. Genes Brain Behav 3 : 206–215.
54. LiuC, TuY, YuanJ, MaoX, HeS, et al. (2012) Aberrant Expression of N-Methylpurine-DNA Glycosylase Influences Patient Survival in Malignant Gliomas. J Biomed Biotechnol 2012 : 760679 doi: 10.1155/2012/760679.
55. AgnihotriS, GajadharAS, TernamianC, GorliaT, DiefesKL, et al. (2012) Alkylpurine–DNA–N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. The Journal of Clinical Investigation 122 : 253–266.
56. FishelML, HeY, SmithML, KelleyMR (2007) Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res 13 : 260–267.
57. GouldE (2007) How widespread is adult neurogenesis in mammals? Nat Rev Neurosci 8 : 481–488.
58. DurkaczB, OmidijiO, GrayD, ShallS (1980) (ADP-ribose)n participates in DNA excision repair. Nature 283 : 593–593.
59. BryantHE, SchultzN, ThomasHD, ParkerKM, FlowerD, et al. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434 : 913–917.
60. FarmerH, McCabeN, LordCJ, TuttANJ, JohnsonDA, et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434 : 917–921.
61. Mégnin-ChanetF, BolletM, HallJ (2010) Targeting poly(ADP-ribose) polymerase activity for cancer therapy. Cellular and Molecular Life Sciences 67 : 3649–3662.
62. HainceJ-F, RouleauM, HendzelMJ, MassonJ-Y, PoirierGG (2005) Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends in Molecular Medicine 11 : 456–463.
63. StromCE, JohanssonF, UhlenM, SzigyartoCA, ErixonK, et al. (2011) Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res 39 : 3166–3175.
64. KedarPS, StefanickDF, HortonJK, WilsonSH (2012) Increased PARP-1 Association with DNA in Alkylation Damaged, PARP-Inhibited Mouse Fibroblasts. Molecular Cancer Research 10 : 360–368.
65. MaW, HalwegCJ, MenendezD, ResnickMA (2012) Differential effects of poly(ADP-ribose) polymerase inhibition on DNA break repair in human cells are revealed with Epstein–Barr virus. Proceedings of the National Academy of Sciences 109 : 6590–6595.
66. MuraiJ, HuangS-yN, DasBB, RenaudA, ZhangY, et al. (2012) Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Research 72 : 5588–5599.
67. StromCE, JohanssonF, UhlenM, Al-Khalili SzigyartoC, ErixonK, et al. (2010) Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res 39(8): 3166–75 doi: 10.1093/nar/gkq1241.
68. HottigerMO, HassaPO, LüscherB, SchülerH, Koch-NolteF (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends in Biochemical Sciences 35 : 208–219.
69. ZongWX, DitsworthD, BauerDE, WangZQ, ThompsonCB (2004) Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev 18 : 1272–1282.
70. XuY, HuangS, LiuZG, HanJ (2006) Poly(ADP-ribose) polymerase-1 signaling to mitochondria in necrotic cell death requires RIP1/TRAF2-mediated JNK1 activation. J Biol Chem 281 : 8788–8795.
71. HaHC, SnyderSH (1999) Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci USA 96 : 13978–13982.
72. EliassonMJ, SampeiK, MandirAS, HurnPD, et al. (1997) Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med 10 : 1089–1095.
73. MandirAS, PoitrasMF, BerlinerAR, HerringWJ, GuastellaDB, et al. (2000) NMDA but not non-NMDA excitotoxicity is mediated by Poly(ADP-ribose) polymerase. J Neurosci 20 : 8005–8011.
74. YangZ, ZingarelliB, SzaboC (2000) Effect of genetic disruption of poly (ADP-ribose) synthetase on delayed production of inflammatory mediators and delayed necrosis during myocardial ischemia-reperfusion injury. Shock 13 : 60–66.
75. SahabogluA, TanimotoN, KaurJ, Sancho-PelluzJ, HuberG, et al. (2010) PARP1 Gene Knock-Out Increases Resistance to Retinal Degeneration without Affecting Retinal Function. PLoS ONE 5: e15495 doi:10.1371/journal.pone.0015495.
76. LiC, WangL, KernTS, ZhengL (2012) Inhibition of poly(ADP-ribose) polymerase inhibits ischemia/reperfusion induced neurodegeneration in retina via suppression of endoplasmic reticulum stress. Biochemical and Biophysical Research Communications 423 : 276–281.
77. Al-AttarA, GossageL, FareedKR, ShehataM, MohammedM, et al. (2010) Human apurinic/apyrimidinic endonuclease (APE1) is a prognostic factor in ovarian, gastro-oesophageal and pancreatico-biliary cancers. Br J Cancer 102 : 704–709.
78. SweasyJB, LangT, DiMaioD (2006) Is Base Excision Repair a Tumor Suppressor Mechanism? Cell Cycle 5 : 250–259.
79. StarcevicD, DalalS, SweasyJB (2004) Is There a Link Between DNA Polymerase Beta and Cancer? Cell Cycle 3 : 996–999.
80. ZarembaT, KetzerP, ColeM, CoulthardS, PlummerER, et al. (2009) Poly(ADP-ribose) polymerase-1 polymorphisms, expression and activity in selected human tumour cell lines. Br J Cancer 101 : 256–262.
81. HaoB, WangH, ZhouK, LiY, ChenX, et al. (2004) Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res 64 : 4378–4384.
82. LockettKL, HallMC, XuJ, ZhengSL, BerwickM, et al. (2004) The ADPRT V762A genetic variant contributes to prostate cancer susceptibility and deficient enzyme function. Cancer Res 64 : 6344–6348.
83. ZhangX, MiaoX, LiangG, HaoB, WangY, et al. (2005) Polymorphisms in DNA base excision repair genes ADPRT and XRCC1 and risk of lung cancer. Cancer Res 65 : 722–726.
84. BacaliniMG, TavolaroS, PeragineN, MarinelliM, SantangeloS, et al. (2012) A subset of chronic lymphocytic leukemia patients display reduced levels of PARP1 expression coupled with a defective irradiation-induced apoptosis. Experimental Hematology 40 : 197–206.
85. HollemanA, BoerMLd, KazemierKM, BeverlooHB, von BerghARM, et al. (2005) Decreased PARP and procaspase-2 protein levels are associated with cellular drug resistance in childhood acute lymphoblastic leukemia. Blood 106 : 1817–1823.
86. DeichmannWB, LeBlancTJ (1943) Determination of the approximate lethal dose with about six animals. Journal of Industrial Hygiene and Toxicology 25 : 415–417.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



